Abstract
Several natural isolates of Escherichia coli O157:H7 have previously been shown to exhibit stationary-phase-dependent variation in their resistance to inactivation by high hydrostatic pressure. In this report we demonstrate that loss of the stationary-phase-inducible sigma factor RpoS resulted in decreased resistance to pressure in E. coli O157:H7 and in a commensal strain. Furthermore, variation in the RpoS activity of the natural isolates of O157:H7 correlated with the pressure resistance of those strains. Heterogeneity was noted in the rpoS alleles of the natural isolates that may explain the differences in RpoS activity. These results are consistent with a role for rpoS in mediating resistance to high hydrostatic pressure in E. coli O157:H7.
In recent years there has been growing interest in the use of high hydrostatic pressure (HHP) as a means of food preservation. The attraction of HHP lies in the production of microbiologically safe foodstuffs with minimal use of chemical additives and without adversely affecting the organoleptic qualities of the food (11). HHP processing could replace traditional thermal pasteurization or be used in conjunction with existing techniques (3, 10, 14, 22). However, exploitation of this potential requires a better understanding of the effects of HHP on microorganisms. HHP resistance varies among genera and species and is dependent on the physiological state of the organisms at the time of pressurization (6, 18). It is of concern that certain E. coli O157:H7 strains are among the most pressure-resistant vegetative bacteria known (2, 18). It is therefore critically important to characterize the innate HHP resistance in these strains.
We demonstrated recently that the pressure resistance of certain natural isolates of E. coli O157:H7 varied greatly (2, 17). Strains C9490 and 30-2C4 were the most pressure resistant and were able to withstand 500 MPa for 5 min with little viability loss; strains NCTC 12079 and W2-2 were of intermediate pressure resistance (ca. 3 to 4 log units decrease under the same conditions), whereas H1071 and an O124 strain, NCTC 8003, were the least pressure resistant (5 to 6 log decrease). However, this variation in pressure resistance was stationary-phase dependent, with the strains exhibiting similar pressure resistance in exponential phase (2). This led us to speculate that the differences in pressure resistance among the isolates were related to differences in RpoS. This sigma factor changes the specificity of RNA polymerase, allowing it to activate more than 30 genes, some of which are involved in stationary-phase stress survival (8). Stationary-phase bacteria are generally more resistant to other stresses, such as oxidative and osmotic stress (21). It is thus possible that resistance to inactivation by HHP is also controlled by mechanisms used to survive stationary-phase stress. These studies attempted to identify the genetic basis, and specifically the role of RpoS, in the wide variation observed in HHP resistance of natural isolates of E. coli O157:H7 strains. Details of the E. coli strains used in these experiments are shown in Table 1.
TABLE 1.
E. coli strains used
| Serotype | Strain | Description | Source (reference) |
|---|---|---|---|
| O157:H7 | C9490 | Clinical isolate associated with hamburger patty | M. P. Doyle |
| O157:H7 | 30-2C4 | Clinical isolate associated with dry-cured salami | M. P. Doyle |
| O157:H7 | W2-2 | Poultry isolate | M. P. Doyle |
| O157:H7 | H1071 | Clinical isolate | M. F. Patterson |
| O157:H7 | NCTC 12079 | Clinical isolate | M. F. Patterson |
| O157:H7 | ATCC 43895 | Clinical isolate associated with raw hamburger meat | M. T. Rowe (4) |
| O157:H7 | FRIK 816-3 | ATCC 43895 rpoS::pRR10 | M. T. Rowe (4) |
| O124 | NCTC 8003 | Clinical isolate | M. F. Patterson |
| K−:H− | BJ4 | Streptomycin-resistant rat gut isolate | K. McClean (15) |
| K−:H− | BJ4L1 | rpoS null mutant | K. McClean (15) |
| K−:H− | BJ4L1C | BJ4L1(pCR2.1), encoding rpoS from BJ4 on a 1.6-kb PCR fragment | This work |
Pressure resistance of rpoS mutants.
To investigate the effect of loss of RpoS on the pressure resistance of E. coli O157:H7, we used the characterized rpoS insertion mutant FRIK 816-3 (4). Analysis of HHP resistance was performed as previously described (2, 17). Briefly, bacteria grown to stationary phase in tryptone-soy-yeast extract (TSYE) broth were harvested by centrifugation and resuspended in phosphate-buffered saline. HHP was applied to 2-ml bacterial suspensions in sealed plastic pouches using a 300-ml pressure vessel (model S-FL-850-9-W; Stanstead Fluid Power, Stanstead, United Kingdom). The pressure-transmitting fluid used was ethanol-castor oil (80:20 [vol/vol]). Viability was assessed by plating pressure-treated bacteria onto TSYE agar supplemented with 0.1% pyruvate, which was incubated for 24 to 48 h at 37°C until CFU were evident.
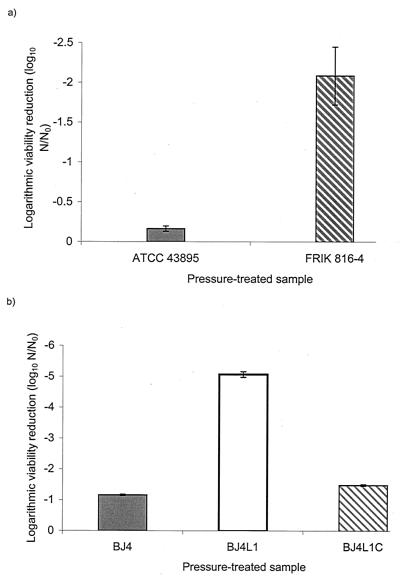
Following treatment at 500 MPa for 15 min, viability of FRIK 816-3 was significantly decreased compared with the parent, ATCC 43895 (Fig. 1a), suggesting a role for rpoS in HHP resistance in E. coli O157:H7. To confirm that rpoS did indeed play a general role in pressure resistance, the effect of an rpoS null mutation on the pressure resistance of a commensal strain of E. coli was also examined. As with the O157:H7 strain, the rpoS null mutant, E. coli strain BJ4L1, exhibited decreased pressure resistance at 400MPa compared with its wild-type parent, BJ4. trans-Complementation of BJ4L1 with the rpoS allele from BJ4 cloned into plasmid pCR2.1 (as described below) resulted in restoration of the pressure-resistant phenotype to the levels observed with the wild type (Fig. 1b). Taken together, these results suggest a role for rpoS in pressure resistance of both E. coli O157:H7 and a commensal strain of E. coli.
FIG. 1.
Effect of loss of rpoS on pressure resistance of E. coli strains. (a) Stationary-phase O157:H7 strains ATCC 43895 (parent) and FRIK 816-4 (rpoS) were pressure treated at 400 MPa for 15 min; (b) commensal strain BJ4, BJ4L1 (rpoS), and BJ4L1C (complemented) were pressure treated at 400 MPa for 8 min. The logarithmic viability reduction was calculated as log N/N0, where N0 is the CFU per milliliter at time zero and N is the CFU per milliliter of the pressurized sample. Data are the averages of triplicate experiments, and standard deviations between experiments are shown. Significant differences in HHP resistance were noted between ATCC 43895 and FRIK 816-4 (P < 0.005, Student's t test). Significant differences were also noted between the commensal wild type and rpoS mutant (P < 0.005, Student's t test) and the wild type and the complemented strain, BJ4L1C (P < 0.005, Student's t test).
RpoS activity of O157:H7 natural isolates.
To investigate the stationary-phase-dependent pressure resistance of these strains, an spvR/spvA′::luxCDABE reporter plasmid, pSB367, was used to measure RpoS activity in the strains (23). This plasmid contains the spvR gene and the spvA promoter transcriptionally fused to the luxCDABE genes. RpoS activates spvR, and in turn the spvA promoter is activated by SpvR and RpoS. As activation of the spvA promoter leads to bioluminescence via expression of the luxCDABE genes, this system is an indirect reporter of RpoS activity. Bioluminescence reflects both the total amounts of RpoS and the ability of RpoS to bind to the spvA promoter. Plasmids were introduced into bacteria by electroporation by standard methods (20). Cultures were prepared by inoculating Luria-Bertani broth supplemented with kanamycin with 1/103 of an overnight culture and incubating at 30°C with shaking. At various times, the optical density at 600 nm (OD600)was measured, and bioluminescence was assessed using a Biocounter Lumac luminometer (Lumac, Landgraat, The Netherlands; model M1500).
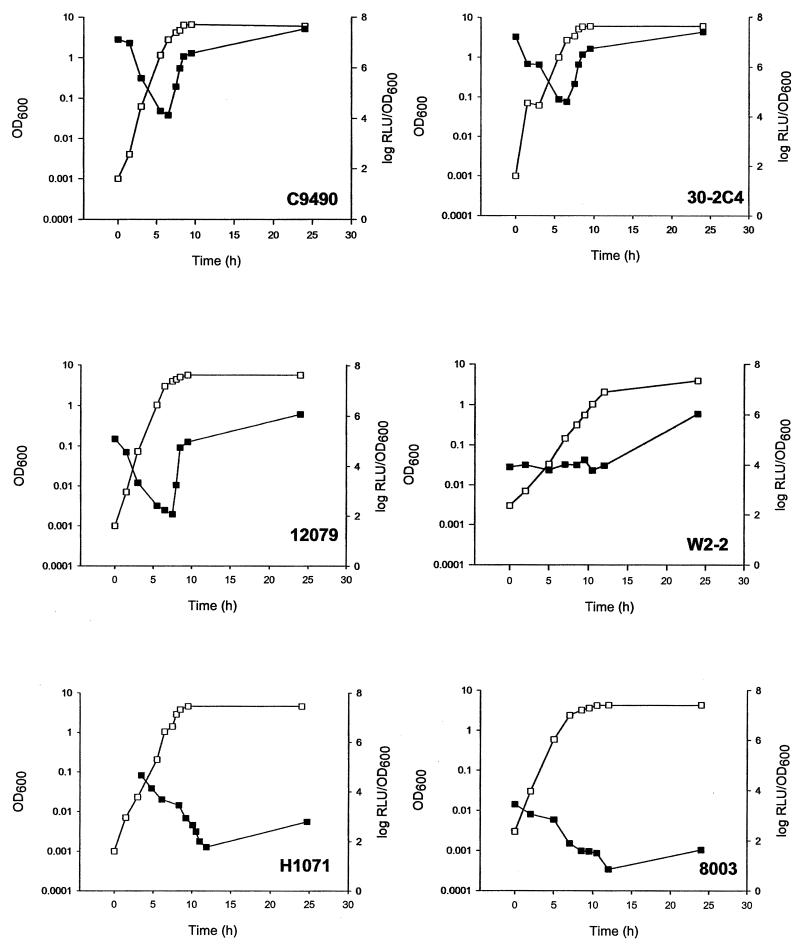
The two most pressure-resistant strains, C9490 and 30-2C4, had very similar RpoS activity kinetics (Fig. 2). At 0 h, a high level of bioluminescence was observed that declined upon entry into exponential phase. This was most likely due to carryover of preformed luciferase from the stationary-phase inocula used for culture preparation because lux expression would previously have been induced by RpoS in these cells. As the cells entered exponential phase, the levels of bioluminescence declined, and this was most likely due to the dilution of the carried-over luciferase during exponential growth in the absence of RpoS-mediated induction of the spvA promoter. Both C9490 and 30-2C4 exhibited sharp increases in RpoS activity as bacteria entered stationary phase, attaining levels similar to that seen at 0 h.
FIG. 2.
Changes in RpoS activity during growth of E. coli O157 natural isolates C9490, 30-2C4, NCTC 12079, W2-2, H1071, and NCTC 8003 (non-O157). All strains contained the spvR/spvA′::luxCDABE fusion plasmid. Growth was followed by OD measurements at 600 nm (open symbols). RpoS activity was expressed as log relative light units (RLU)/OD600 (solid symbols). Experiments were repeated twice, and representative results are shown.
The moderately pressure-resistant strain NCTC 12079 had a profile of RpoS activity similar to that of C9490 and 30-2C4, but had 2 log units lower total levels of RpoS activity (Fig. 2). The other moderately pressure-resistant strain, W2-2, had total levels of RpoS activity similar to those in NCTC 12079 at 25 h (Fig. 2). However, strain W2-2 did not appear to induce RpoS activity in the same rapid growth phase-dependent manner as C9490, 30-2C4, and 12079. A difference of 3 to 4 log units between the minimum and maximum RpoS activity was observed in C9490, 30-2C4, and 12079, whereas a 2 log difference was noted in strain W2-2.
The strains which exhibited the lowest pressure resistance of the natural isolates, H1071 and NCTC 8003, showed little induction of RpoS activity as the cells entered stationary phase (Fig. 2) and after 24 h of growth exhibited the lowest levels of RpoS activity. The level of RpoS activity was lower at 24 h than at 0 h in strains H1071 and NCTC 8003. This may indicate that induction of RpoS activity in these strains did not coincide with the onset of stationary phase at 10 h, but may occur later in stationary phase.
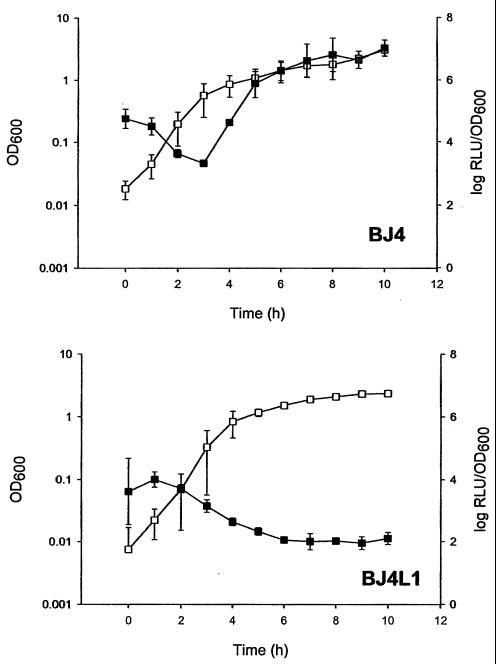
While it is conceivable that luciferase activity may be affected by factors other than the level and activity of RpoS, this is not likely. When the RpoS reporter plasmid pSB367 was introduced into BJ4 (parent strain) and BJ4L1 (containing a defined rpoS mutation but otherwise isogenic), the pattern of luminescence seen in BJ4 (Fig. 3a) showed a clear growth phase-dependent induction of luminescence which resembled that seen in C9490, 30-2C4, and 12079. In contrast, the pattern of luminescence in BJ4L1 (Fig. 3b) was similar to that seen in H1071 and NCTC 8003. Given that the presence or absence of RpoS has no effect on luminescence when luciferase is expressed from constitutive promoters, with luminescence remaining constant throughout growth (13), the difference in light emission from BJ4 and BJ4L1 can only reflect levels of luciferase expression from the reporter system and thus levels of RpoS. Given this, it is highly likely that the differences in luminescence described here are due solely to variations in the level or activity of RpoS between strains. Consequently, the levels of RpoS activity of the natural isolates, as measured by the bioluminescence reporter assay, appear to correlate very closely with their relative pressure resistance.
FIG. 3.
Changes in RpoS activity in E. coli commensal strain BJ4 (parent) and BJ4L1 (rpoS). Both strains contained the spvR/spvA′::luxCDABE fusion plasmid. Growth was followed by OD measurements at 600 nm (open symbols). RpoS activity was expressed as log RLU/OD600 (solid symbols). The experiment was performed twice with similar results, and standard deviations between experiments are given. Significant differences in RpoS activity were noted between BJ4 and BJ4L1 after 9 h of growth (P < 0.001).
Cloning and sequencing of rpoS gene from E. coli strains.
As the natural isolates showed different RpoS activities, we speculated that there might be differences in their rpoS alleles. To investigate this hypothesis, we cloned and sequenced the rpoS genes from the natural isolates. A 1.6-kb PCR fragment specific to rpoS and the upstream promoter region was generated using primers, RPOS-1 (5′-GGA ACA GCG CTT CGA TAT TCA G-3′) (24) and RPOS-2 (5′-GCA GAG CAA GGA GTT GTG AT-3′) (5) and was then cloned into the pCR2.1-TOPO T vector according to the manufacturer's recommendations (Invitrogen, NV Leek, The Netherlands). Sequencing was performed on at least two independent clones using an ABI Prism Rhodamine terminator cycle sequencing reaction kit in an ABI Prism 377 automated sequencer (Perkin Elmer, Cambridge, United Kingdom). The following primers were used for sequencing: RPOS-1, RPOS-2, RPOS-3 (5′-TGA TTA CCT GAG TGC CTA CG-3′), RPOS-4 (5′-TTG GTG AGA TTG GTT ATT CA-3′), and RPOS-5 (5′-TAC CAC CAG ACG CAA GTT AC-3′) (24). Primers were generated by Eurogentec (Abingdon, United Kingdom). Sequences were analyzed using the DNAStar biocomputing software (Lasergene) and compared with the recently published O157:H7 genome database from strain EDL933, a hamburger isolate (GenBank accession no. AE005174) (19). Comparisons were also made with O157:H7 strains SEA 13B28, a salami isolate (GenBank accession no. AF002207), SEA 6318, a hamburger isolate (GenBank accession no. AF002208), and E. coli K-12 strain DH1 (GenBank accession no. D13548) (5). The main promoter region lies 601 bases upstream of rpoS in the nlpD gene, encoding a lipoprotein putatively involved in cell wall formation (16, 24). The rpoS sequences of the natural isolates have been deposited in the GenBank database under accession numbers AF182102 (C9490), AF182103 (W2-2), AF182104 (H1071), AF182105 (NCTC 12079), AF182106 (NCTC 8003), and AF182107 (30-2C4).
The two strains with the greatest pressure resistance and RpoS activity, C9490 and 30-2C4, had rpoS sequences that were identical to each other and to those of SEA 13B28 and SEA 6318 (Table 2, Fig. 2). However, the EDL933 rpoS sequence contained two differences at nucleotide (nt) 57 and nt 61 resulting in an aspartic acid instead of a glutamic acid and an arginine instead of a glycine as in the rpoS sequences of C9490, 30-2C4, SEA 13B28, and SEA 6318 (Table 2). The DNA sequences of the promoter region of rpoS in C9490 and 30-2C4 were identical to each other and to the corresponding sequence in EDL933 (Table 2). Nine base changes were noted in the coding and promoter regions when comparing C9490 and 30-2C4 with K-12 DH1, four of which were reported previously by Ferreira et al. (5) (Table 2). The remaining five changes did not change the amino acid sequence (Table 2). It therefore appears that the rpoS allele contained in C9490 and 30-2C4 produces a high level of RpoS activity during stationary phase.
TABLE 2.
Variation in rpoS sequences of E. coli O157:H7, O124, and K-12 strains
| Strain | Serotype | Base (amino acid) at position:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −306 | −294 | −183 | −162 | −94/−87 | −96/−98a | 55–57 | 61–63 | 214–216 | 382–384 | 387a | 423 | 442–444 | 543a | 606 | 714 | 819a | 910–91 | ||
| DH1 | —(K-12) | C | T | T | G | CTG (Leu) | GAG (Glu) | GGA (Gly) | ACG (Thr) | ATC (IIe) | C | C | TGG (Trp) | C | G | G | G | CAG (Glu) | |
| EDL933 | O157:H7 | T | C | G | A | GAG (Glu) | GAC (Asp) | CGA (Arg) | ACG (Thr) | ATC (IIe) | C | C | TGG (Trp) | A | G | G | A | CAG (Glu) | |
| C9490 | O157:H7 | T | C | G | A | GAG (Glu) | GAG (Glu) | GGA (Gly) | ACG (Thr) | ATC (IIe) | T | C | TGG (Trp) | A | G | G | A | CAG (Glu) | |
| 30-2C4 | O157:H7 | T | C | G | A | GAG (Glu) | GAG (Glu) | GGA (Gly) | ACG (Thr) | ATC (IIe) | T | C | TGG (Trp) | A | G | G | A | CAG (Glu) | |
| 12079 | O157:H7 | T | C | G | A | GAG (Glu) | GAG (Glu) | GGA (Gly) | ACG (Thr) | ACC (Thr) | T | C | TGG (Trp) | A | G | G | A | CAG (Glu) | |
| W2-2b | O157:H7 | T | C | G | A | (TAAACCCG) | GAG (Glu) | GAG (Glu) | GGA (Gly) | CCG (Pro) | ATC (IIe) | T | C | TGG (Trp) | A | G | G | A | CAG (Glu) |
| H1071 | O157:H7 | T | C | G | A | GAG (Glu) | GAG (Glu) | GGA (Gly) | ACG (Thr) | ATC (IIe) | T | C | TAG | C | G | G | A | CAG (Glu) | |
| 8003 | O124 | C | C | G | A | GAG (Glu) | GAG (Glu) | GGA (Gly) | ACG (Thr) | ATC (IIe) | C | T | TGG (Trp) | C | A | A | A | TAG | |
Mutations already noted by and in agreement with Ferreira et al. (5).
The transcriptional start site of rpoS in W2-2 differs from that of the other strains by 8 nt resulting from the insertion of a duplicated region at nt −94 to −87. Therefore, in strain W2-2 the positions of the bases before the start site are displaced compared with the other strains, with nt −306, −294, −183 and −162 in the other strains being equivalent to nt −314, −302, −191, and −170, respectively, in strain W2-2.
Polymorphisms were observed in the rpoS sequences of the other natural isolates compared with the rpoS alleles from C9490 and 30-2C4. These differences correlated well with the HHP resistance and RpoS activity of those strains. For example, strain NCTC 12079, which exhibited moderate pressure resistance and RpoS activity, had an rpoS allele identical to that of C9490 and 30-2C4 except for substitution of a threonine for the usual isoleucine observed in the other O157:H7 strains and K-12 DH1 (Table 2). This single polymorphism appears to be sufficient to generate a strain that has lower total RpoS activity than C9490 and 30-2C4 but is still able to induce RpoS activity in stationary phase. That this strain shows moderate RpoS activity correlates well with its intermediate HHP resistance.
Strain W2-2 had pressure resistance and level of total RpoS activity similar to those in NCTC 12079 but did not appear to exhibit the same rapid induction of RpoS activity on entering stationary phase (Table 2, Fig. 2). There were two sequence alterations in W2-2 compared with C9490 and 30-2C4; a change in the amino acid sequence from a threonine to a proline within the coding region and the insertion of a duplicated region resulting in a stop codon in the upstream nlpD gene. The stationary-phase-inducible promoter for rpoS lies within the nlpD gene (16, 24), and thus sequence changes in this region may drastically affect rpoS expression. As the rpoS coding region is unaffected by this insertion, W2-2 may still be capable of producing a fully functional RpoS. This may explain why W2-2 has a total level of RpoS activity similar to that of NCTC 12079 at 24 h but has a different pattern of induction during growth.
Strains H1071 and NCTC 8003 both had base changes resulting in early stop codons (TAG) within the rpoS coding region (Table 2). These stop codons may result in a truncated RpoS, which could be less active than full-length RpoS. As these strains are largely genetically uncharacterized, the presence of amber suppressor mutations is unknown, but the possibility of limited expression of rpoS due to translational readthrough cannot be excluded. A truncated RpoS could have reduced ability to interact with RNA polymerase, thus affecting its role as a transcriptional activator. It could be for this reason that H1071 and NCTC 8003 exhibit the lowest RpoS activities of any of the natural isolates and did not induce RpoS activity upon entry into stationary phase.
Common laboratory E. coli strains have also been shown to harbor and even accumulate mutant rpoS alleles (12, 25, 26, 28, 29). Some alleles that do not result in a null rpoS phenotype nevertheless cause distinct phenotypic changes in stress resistance and in other properties (1, 26, 27). Recent evidence suggests that O157:H7 strains also show polymorphisms in the downstream mutS-rpoS region compared to K-12 serotypes (9). This indicates that despite its obvious importance to the cell, rpoS is a highly mutable gene, and it is feasible that various rpoS alleles could be found within natural O157:H7 isolates. Furthermore, evidence suggests that subpopulations, which often dominate in stationary phase, have an rpoS mutation (28). Maintenance of a less active RpoS at first may appear somewhat paradoxical, as this is likely to affect stationary-phase survival; however, such mutations may confer a selective advantage under certain conditions. For example, for subpopulations able to utilize a novel substrate in spent medium, delaying the activation of RpoS-dependent survival determinants may allow the population to dominate the culture (28).
Concluding remarks.
Available evidence suggests that HHP resistance is multifactorial in nature. For example, calcium and membrane stability have been shown to be important in pressure resistance (2, 7). Factors controlling bacterial pressure resistance are likely to be numerous, and our results suggest that rpoS is an important contributing factor. How does RpoS affect pressure resistance? Because RpoS controls a large number of genes involved in protection against stress, resistance could be the cumulative effect of many genes switched on by RpoS in stationary phase. Alternatively, it may be that only a specific subset of genes are critically important in the increased pressure resistance seen in the O157:H7 strains. Evidence that specific functions may be involved in pressure resistance was provided by Hauben et al. (6), who generated several mutants by pressure cycling and found that two of the pressure-resistant mutants were also more heat resistant but other mutants were unaltered in their heat resistance phenotype. Further studies are required to discover which particular gene(s) involved in stationary phase survival is responsible for the pressure-resistant phenotype of E. coli. The wide differences in pressure resistance among strains of E. coli O157:H7 make it imperative that care be taken in choosing and maintaining strains used for testing the efficacy of pressure processes. To be confident that a pressure treatment (or other mild process) can eliminate E. coli O157:H7 from food, it will be necessary to use strains at least as resistant as those described here and to ensure that resistance properties are regularly monitored.
Acknowledgments
We are grateful to the Food Standards Agency/Ministry of Agriculture Fisheries and Food London, United Kingdom, for financial support of this work.
We thank the following for providing strains: M. Doyle (University of Georgia), M. Patterson (Queen's University, Belfast), M. Rowe (Queen's University, Belfast), and K. McClean (University of Nottingham, United Kingdom). We also thank S. Swift (University of Nottingham, United Kingdom) for providing the lux reporter plasmid pSB367.
REFERENCES
- 1.Allen-Vercoe E, Collighan R, Woodward M J. The variant rpoS allele of S. enteritidis strain 27655R does not affect virulence in a chick model nor constitutive curliation but does generate a cold-sensitive phenotype. FEMS Microbiol Lett. 1998;167:245–253. doi: 10.1111/j.1574-6968.1998.tb13235.x. [DOI] [PubMed] [Google Scholar]
- 2.Benito A, Ventoura G, Casadei M, Robinson T, Mackey B. Variation in resistance of natural isolates of Escherichia coli O157 to high hydrostatic pressure, mild heat, and other stresses. Appl Environ Microbiol. 1999;65:1564–1569. doi: 10.1128/aem.65.4.1564-1569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheftel J C. Review: high pressure, microbial inactivation and food preservation. Food Sci Technol Int. 1995;1:75–90. [Google Scholar]
- 4.Cheville A M, Arnold K W, Buchrieser C, Cheng C M, Kaspar C W. rpoS regulation of acid, heat, and salt tolerance in Escherichia coli O157:H7. Appl Environ Microbiol. 1996;62:1822–1824. doi: 10.1128/aem.62.5.1822-1824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferreira A, Rendano L, Wiedmann M, Boor K J. Characterization of rpoS alleles in Escherichia coli O157:H7 and in other E. coli serotypes. J Appl Microbiol. 1999;86:295–301. doi: 10.1046/j.1365-2672.1999.00664.x. [DOI] [PubMed] [Google Scholar]
- 6.Hauben K J, Bartlett D H, Soontjens C C, Cornelis K, Wuytack E Y, Michiels C W. Escherichia coli mutants resistant to inactivation by high hydrostatic pressure. Appl Environ Microbiol. 1997;63:945–950. doi: 10.1128/aem.63.3.945-950.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hauben K J, Bernaerts K, Michiels C W. Protective effect of calcium on inactivation of Escherichia coli by high hydrostatic pressure. J Appl Microbiol. 1998;85:678–684. doi: 10.1111/j.1365-2672.1998.00577.x. [DOI] [PubMed] [Google Scholar]
- 8.Hengge-Aronis R. Regulation of gene expression during entry into stationary phase. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasnik B, Reznikoff W S, Riley M, Schaecter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1996. [Google Scholar]
- 9.Herbelin C J, Chirillo S C, Melnick K A, Whittam T S. Gene conservation and loss in the mutS-rpoS genomic region of pathogenic Escherichia coli. J Bacteriol. 2000;182:5381–5390. doi: 10.1128/jb.182.19.5381-5390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoover D. Minimally processed fruits and vegetables: reducing microbial load by nonthermal physical treatments. Food Technol. 1997;51:66–71. [Google Scholar]
- 11.Hoover D G, Metrick C, Papineau A M, Farkas D, Knorr D. Biological effects of high hydrostatic pressure on food microorganisms. Food Technol. 1989;43:99–107. [Google Scholar]
- 12.Ivanova A, Renshaw M, Guntaka R V, Eisenstark A. DNA base sequence variability in katF (putative sigma factor) gene of Escherichia coli. Nucleic Acids Res. 1992;20:5479–5480. doi: 10.1093/nar/20.20.5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jorgensen F, Leach S, Wilde S J, Davies A, Stewart G S A B, Humphrey T J. Invasiveness in chickens, stress resistance and RpoS status of wild-type Salmonella enterica subsp enterica serovar Typhimurium definitive type 104 and serovar Enteritidis phage type 4 strains. Microbiol-UK. 2000;146:3227–3235. doi: 10.1099/00221287-146-12-3227. [DOI] [PubMed] [Google Scholar]
- 14.Knorr D. Effects of high hydrostatic pressure processes on food safety and quality. Food Technol. 1993;47:156–161. [Google Scholar]
- 15.Krogfelt K A, Hjulgaard M, Sorensen K, Cohen P S, Givskov M. rpoS gene function is a disadvantage for Escherichia coli BJ4 during competitive colonization of the mouse large intestine. Infect Immun. 2000;68:2518–2524. doi: 10.1128/iai.68.5.2518-2524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lange R, Fischer D, Hengge-Aronis R. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the sigma S subunit of RNA polymerase in Escherichia coli. J Bacteriol. 1995;177:4676–4680. doi: 10.1128/jb.177.16.4676-4680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pagan R, Mackey B. Relationship between membrane damage and cell death in pressure-treated Escherichia coli cells: differences between exponential- and stationary-phase cells and variation among strains. Appl Environ Microbiol. 2000;66:2829–2834. doi: 10.1128/aem.66.7.2829-2834.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patterson M F, Quinn M, Simpson R, Gilmour A. Sensitivity of vegetative pathogens to high hydrostatic pressure treatment in phosphate-buffered saline and foods. J Food Prot. 1995;58:524–529. doi: 10.4315/0362-028X-58.5.524. [DOI] [PubMed] [Google Scholar]
- 19.Perna N T, Plunkett G, Burland I I I, V, Mau B, Glasner J D, Rose D J, Mayhew G F, Evans P S, Gregor J, Kirkpatrick H A, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck E J, Wayne Davis N, Lim A, Dimalantal E T, Potamousis K D, Apodaca J, Anantharaman T S, Lin J, Yen G, Schwartz D C, Welch R A, Blattner F R. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Schellhorn H E, Audia J P, Wei L I, Chang L. Identification of conserved, RpoS-dependent stationary-phase genes of Escherichia coli. J Bacteriol. 1998;180:6283–6291. doi: 10.1128/jb.180.23.6283-6291.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smelt J P P M. Recent advances in the microbiology of high pressure processing. Trends Food Sci Technol. 1998;9:152–158. [Google Scholar]
- 23.Swift S, Stewart G S A B. Luminescence as a signal of spvA expression. In: Campbell A K, Kricka L J, Stanley P E, editors. Bioluminescence and chemiluminescence: fundamentals and applied aspects. Chichester, England: John Wiley and Sons; 1994. pp. 93–96. [Google Scholar]
- 24.Takayanagi Y, Tanaka K, Takahashi H. Structure of the 5′ upstream region and the regulation of the rpoS gene of Escherichia coli. Mol Gen Genet. 1994;243:525–531. doi: 10.1007/BF00284200. [DOI] [PubMed] [Google Scholar]
- 25.Visick J E, Clarke S. RpoS- and OxyR-independent induction of HPI catalase at stationary phase in Escherichia coli and identification of rpoS mutations in common laboratory strains. J Bacteriol. 1997;179:4158–4163. doi: 10.1128/jb.179.13.4158-4163.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waterman S R, Small P L. Characterization of the acid resistance phenotype and rpoS alleles of shiga-like toxin-producing Escherichia coli. Infect Immun. 1996;64:2808–2811. doi: 10.1128/iai.64.7.2808-2811.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilmes-Riesenberg M R, Foster J W, Curtiss R., 3rd An altered rpoS allele contributes to the avirulence of Salmonella typhimurium LT2. Infect Immun. 1997;65:203–210. doi: 10.1128/iai.65.1.203-210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zambrano M M, Siegele D A, Almiron M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 29.Zinser E R, Kolter R. Prolonged stationary-phase incubation selects for lrp mutations in Escherichia coli K-12. J Bacteriol. 2000;182:4361–4365. doi: 10.1128/jb.182.15.4361-4365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]