Summary
Current crop yield of the best ideotypes is stagnating and threatened by climate change. In this scenario, understanding wild plant adaptations in extreme ecosystems offers an opportunity to learn about new mechanisms for resilience. Previous studies have shown species specificity for metabolites involved in plant adaptation to harsh environments.
Here, we combined multispecies ecological metabolomics and machine learning‐based generalized linear model predictions to link the metabolome to the plant environment in a set of 24 species belonging to 14 families growing along an altitudinal gradient in the Atacama Desert.
Thirty‐nine common compounds predicted the plant environment with 79% accuracy, thus establishing the plant metabolome as an excellent integrative predictor of environmental fluctuations. These metabolites were independent of the species and validated both statistically and biologically using an independent dataset from a different sampling year. Thereafter, using multiblock predictive regressions, metabolites were linked to climatic and edaphic stressors such as freezing temperature, water deficit and high solar irradiance.
These findings indicate that plants from different evolutionary trajectories use a generic metabolic toolkit to face extreme environments. These core metabolites, also present in agronomic species, provide a unique metabolic goldmine for improving crop performances under abiotic pressure.
Keywords: adaptation, extreme environments, multiple species, plant metabolism, predictive metabolomics
Introduction
Humans domesticated plants 10 000 yr ago in the hostile environments of the Fertile Crescent (Dai et al., 2012; Riehl et al., 2012). Over the years, selected crops have been improved by a variety of methods. However, current yields of domesticated therophytes are stagnating and threatened by climate change despite significant efforts to develop abiotic stress tolerance for the best ideotypes (Long et al., 2015). Wild plants naturally evolved mechanisms to meet abiotic constraints in natural habitats from which they cannot escape (Fatima et al., 2020; Signori‐Müller et al., 2021). In this scenario, returning to wild plant species that live and thrive in some of the harshest environments on Earth offers an opportunity to find new strategies for crop improvement (Castañeda‐Álvarez et al., 2016). Recent studies have pinpointed relevant metabolic clusters in adaptation to extreme environments in plants harvested in high mountains, deserts and salt lands (Dussarrat et al., 2021). These adaptive mechanisms involved the accumulation of amino acids (Lugan et al., 2010) as precursors of secondary metabolites, and carotenoids (Cui et al., 2019) and polyphenols (Hashim et al., 2020) as processors of reactive oxygen species (ROS). In addition, most of these studies were carried out on a unique or limited number of species (Dussarrat et al., 2021), which, combined with high biochemical diversity, led to highly specific metabolic markers involved in adaptive mechanisms exclusive to the species or environment (Peters et al., 2018; Dussarrat et al., 2021).
The metabolome is an excellent integrative system to predict plant environment because it carries imprints of omic inferences and environmental influences (Kosmacz et al., 2020; Lewis & Kemp, 2021). Ecological metabolomics aims to study the environmental impact on metabolic responses, acclimation and adaptation processes in natural ecosystems. Applying untargeted ecological metabolomics on multiple species could unravel universal plant adaptive strategies to abiotic factors in their natural environment (Poorter et al., 2012; Umair et al., 2019; Sardans et al., 2020; Wong et al., 2020). However, this approach has focused primarily on analysis of phytochemical diversity. Plant metabolomes were recently used to predict phenotypic traits such as yield and stress resistance (Zhu et al., 2018; Luna et al., 2020; Szymański et al., 2020) within specific species. By exploiting multiple species, previous studies reported strong relationships between growth rate and biomass composition (Roch et al., 2020) and between phytochemical diversity and environmental conditions of plants growing in alpine regions (Defossez et al., 2021). Interestingly, several phenotypic traits predicted from plant metabolism were further used to predict complex output such as plant fitness but remain tedious to collect (Laughlin et al., 2012; Laughlin & Messier, 2015). Thus, ecological metabolomics could be used to uncover readily measurable soft traits that can predict complex outputs such as plant fitness. Adaptation to extreme environments is thought to rely on specialized secondary metabolic pathways often considered to be species‐specific (Moghe & Last, 2015; Scossa & Fernie, 2020). However, generic mechanisms may also exist. To test this hypothesis, large‐scale metabolomics in multiple wild species are needed to unveil general metabolic interactions with environmental factors and propose adaptive roles for specific metabolites (Wong et al., 2020).
The Atacama Desert is the driest nonpolar desert on Earth. In addition to extreme aridity, the Atacama is characterized by high solar radiation, extreme daily temperature oscillations, high soil salinity and low nitrogen content (Eshel et al., 2021). Although multiple abiotic factors are intense enough to severely limit plant life, this desert hosts tens of plant species (Jordan & Kirk‐Lawlor, 2014; Díaz et al., 2016, 2019), thus bestowing a unique opportunity to analyse adaptive metabolic plant responses to abiotic stress in an entire ecosystem. The present study aimed to characterize the metabolic profiles of 24 dominant plant species in 19 different sites along an altitudinal transect in the Atacama Desert. Biological and environmental diversity was used to question the extent to which adaptation to extreme environments relies on generic metabolic mechanisms.
To meet these ambitious objectives, multiplatform metabolomics covering primary compounds including carbohydrates, amino and organic acids, fatty acids and secondary metabolites revealed metabolic features that participate in environmental adaptation. Subsequent machine learning modelling of this comprehensive dataset via a generalized multilinear‐based statistical approach established that the metabolome of these 24 extremophile plants was an excellent integrative predictor of plant environments. Moreover, our analysis uncovered a common set of metabolites associated with extreme climate resilience.
Materials and Methods
Plant materials and sampling
The aerial parts of 24 plant species belonging to 14 plant families (Supporting Information Table S1) were collected in their natural conditions from the Chilean Atacama Desert (Talabre‐Leja transect (Díaz et al., 2016), 22–24°S). For each species, a minimum of three biological replicates composed of multiple plants was collected. Each species was collected at one to six distinct elevation levels (Fig. 1) depending on biological availability, directly snap frozen in liquid nitrogen, brought back to the laboratory on dry ice and stored at −80°C until freeze‐drying. Sampling was performed during two consecutive days (6–7 April 2019) between 09:30 and 17:30 h. The time variation between samplings in different environments did not impact starch content, suggesting stable central metabolism during the sampling period (Fig. S1). Additionally, crops and ornamental plant species including Capsicum annuum, Phaseolus vulgaris, Spinacia oleracea, Vicia faba, Pisum sativum, Beta vulgaris, Portulaca oleracea, Helianthus annuus, Zea mays, Nicotiana tabacum and Solanum lycopersicum were grown in multiple natural conditions in France. The aerial parts of those plants were harvested, snap‐frozen in liquid nitrogen and stored at −80°C until freeze‐drying. All freeze‐dried material from extremophiles and common plants was kept at −80°C until further analysis.
Fig. 1.
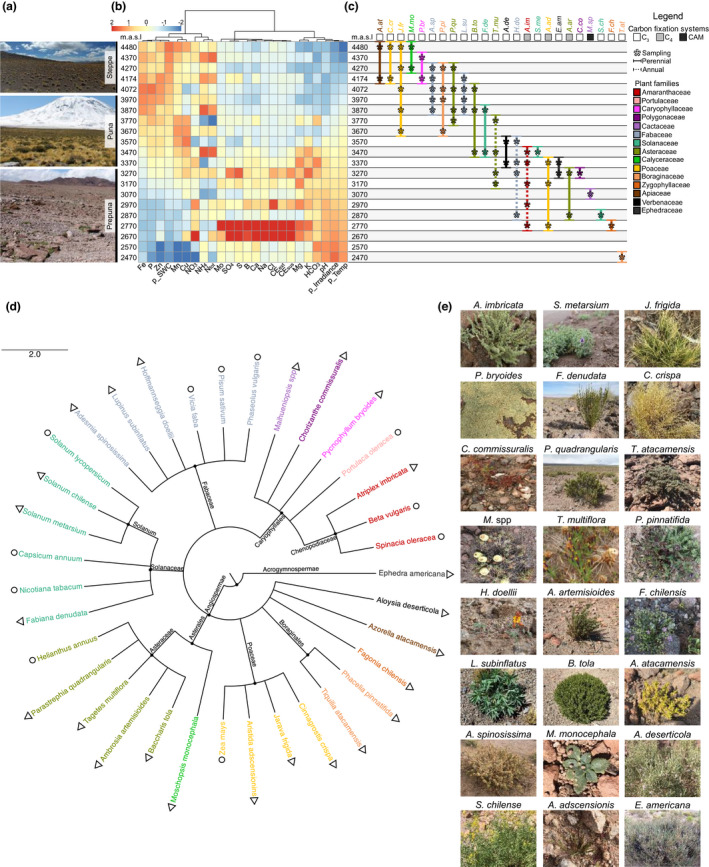
Depiction of Atacama plant diversity despite extreme conditions. (a) Picture of the three vegetation belts. (b) Description of the environmental conditions observed along the elevation gradient (Pearson correlation, P < 0.05). CE, electrical conductivity; Ntot, total nitrogen; SWC, soil water content; Temp, temperature; p_ represents a partially predicted parameter. (c) Description of the sampling site ranges and main characteristics (carbon fixation systems or lifespan) of the collected plant species. (d) Analysis of the taxonomic relationships between Atacama species and between Atacama and agronomic or ornamental plant species. Triangles represent the Atacama plants while circles represent the agronomic and ornamental species. (e) Pictures of the Atacama plant species collected.
Environmental data
Climatic conditions were characterized using two meteorological stations (at 3060 and 4090 m above sea level (m asl)), which measured temperature, humidity and solar irradiance as well as precipitation or soil moisture levels every hour throughout 2018–2019 (Eshel et al., 2021). In addition, soil chemical properties including pH and contents of nitrate, ammonium, Olsen phosphorous, zinc, potassium, manganese, copper, iron, boron, molybdenum, sulphur, calcium, manganese, sodium, chlorine, bicarbonate salt and silt were measured and described for over 3 yr (Eshel et al., 2021).
Metabolite extraction
Using 20 mg of lyophilized plant material (from crops, ornamental and Atacama plants), robotized extractions of metabolites were performed according to an ethanol fractionation protocol (Luna et al., 2020), which targets a wide range of semipolar plant biochemicals including primary compounds (soluble sugars and starch, organic and amino acids, total proteins) and specialized metabolites (terpenes, phenolics, alkaloids). In parallel, 10 mg of lyophilized plant samples were used to extract fatty acyls from total lipids as described previously (Domergue et al., 2010).
Metabolomics
Ethanol extracts were screened for multiple compounds (Chl, glucose, fructose, sucrose, malate, free amino acids, nitrate, total proteins and starch) based on coupled enzyme assays (Luna et al., 2020). The same extracts were also subjected to untargeted metabolic profiling by UHPLC‐LTQ‐Orbitrap MS (liquid chromatography‐mass spectrometry (LCMS)) using an Ultimate 3000 ultra‐high‐pressure liquid chromatography (UHPLC) system coupled to an LTQ‐Orbitrap Elite mass spectrometer interfaced with an electrospray ionization (ESI) source (ThermoScientific, Bremen, Germany) operating in both negative and positive ion modes as described previously (Luna et al., 2020). Separation was performed using a C18 column (C18‐Gemini, 2.0 × 150 mm, 3 µm, 110 Å; Phenomenex, Torrance, CA, USA). Full scan high‐resolution MS spectra were acquired at 240k resolution power at m/z = 200 Da. In addition, LCMS/MS acquisitions were acquired in higher‐energy collisional dissociation (HCD) mode at a normalized collision energy of 60% and 35% (ESI− and + respectively). Fatty acyls were analysed using gas chromatography coupled to a flame ionization detector (GCFID) or a mass spectrometric detector (GCMS) as detailed previously (Domergue et al., 2010). Biochemical phenotyping and LCMS experiments were performed for all plants (i.e. Atacama plants from 2014 and 2019, crops and ornamental plants), while GCFID and GCMS experiments were performed for 2019 Atacama plants and crops and ornamental plants exclusively.
Processing of metabolomic data
Raw LCMS data were processed via Xcms (v.4.2) in R (v.3.6.1) (Smith et al., 2006) using in‐house optimized parameters(Luna et al., 2020) yielding 8750 detected RT–m/z pairs for 5130 ESI− and for 3620 ESI+ modes. Subsequent data cleaning (blank check, ΔRT < 60 s, Δ m/z < 0.025 Da, coefficient of variation in quality controls < 30%) generated 4540 metabolic variables (2564 ESI− and 1976 ESI+) that were retained for chemometric analyses. Both untargeted and targeted metabolomics data were first normalized by median normalization, cube‐root transformation and Pareto scaling using MetaboAnalyst v.3 (Xia et al., 2015) before applying multivariate and univariate statistical analyses. The nonnormalized dataset obtained after preprocessing is available in Table S2 and deposited online (see ‘Data availability’ section).
Generalized multilinear models
Generalised linear modelling (GLM) was performed to determine the quantitative correlation between metabolism and elevation levels used as a proxy of the plant environment. All metabolic variables that could not be measured based on detection limitations were inputted as 0 in the data matrix. The linear models were generated using the glmnet package (Friedman et al., 2010) in the R software (R Core Team, 2020) (v.3.6.1). Three model types were constructed (lasso, elastic net and ridge) by varying the penalty value of the elastic net as a proxy to modulate the number of variables used by the models. Thousand values ranging from 0 to 1 were tested. Internal cross‐validation was performed for construction of the models to mitigate the overfitting. The best model was chosen based on the mean square error (MSE), and the most parsimonious model within one SE of the minimal MSE was selected to perform predictions. The datasets were divided into three parts: 70% of the plants in a ‘training’ set, 20% in a ‘test’ set and 10% in a ‘validation’ set to perform real predictions using the best model developed with both training and testing sets. Stratified sampling was used to perform a uniform sampling of the individuals based on the measured elevation levels. Due to this random partitioning, 500 different simulations were performed to sample the solution space of possible predictions. In addition, 500 sets of randomly assigned elevation levels were created to test the likelihood of spurious predictions. Student tests were performed to compare the 500 results from models performed with permutated and real elevation levels. The occurrences of the metabolic variables among the 500 simulations were analysed to extract the best predictors (Table 1).
Table 1.
Annotation of the 39 best metabolic markers.
| Variable ID | Occurrence in the model | Correlation | Observation | Detected m/z | Detected RT | P value FDR | Ion type | Predicted m/z | Δ m/z (ppm) | Putative formula | MSI level | Putative compound |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Starch | 98.6 | Negative | – | – | – | < 1.64E‐15 | – | – | – | – | ‐ | – |
| n_2561 | 90.0 | Negative | Mg(C2H2O4) n + HCOO− | 386.93908 | 0.9 | < 1.64E‐15 | – | – | – | Mg(C2H2O4) n + HCOO− | – | Mg(C2H2O4) n + HCOO− |
| p_1777 | 87.4 | Negative | Na(HCO2) n + HCOO− | 274.87218 | 1.1 | < 1.64E‐15 | – | – | – | Na(HCO2) n + HCOO− | – | Na(HCO2) n + HCOO− |
| n_0601 | 86.6 | Negative | – | 233.10296 | 3.9 | < 1.64E‐15 | [M‐H]− | 234.11024 | 0 | C10H18O6 | MSI3 | 3,6‐Dihydroxy‐2,7‐dimethyloctanedioic acid |
| p_2029 | 85.7 | Negative | – | 298.09560 | 3.6 | 1.03E‐12 | [M+H]+ | 297.08780 | 4.8 | C11H15N5O3S | MSI2 | 5′‐Methylthioadenosine |
| n_0615 | 84.8 | Negative | – | 236.05628 | 5.6 | < 1.64E‐15 | [M‐H]− | 237.06370 | 0.4 | C11H11NO5 | MSI3 | N‐Benzoylaspartic acid |
| p_2329 | 83.7 | Positive | – | 323.07473 | 7.8 | 8.91E‐08 | [M+H]+ | 322.06890 | 4.9 | C15H14O8 | MSI3 | Leucodelphinidin |
| p_0179 | 82.7 | Negative | Fragment of 251.16 | 116.05708 | 11.5 | 1.64E‐15 | [M+H]+ | 250.15640 | 4.2 | C15H22O3 | MSI3 | Xanthoxin |
| Qt027 | 82.5 | Positive | – | – | – | 1.01E‐10 | – | – | – | – | – | ND |
| p_3344 | 82.0 | Negative | – | 584.27182 | 9.1 | < 1.64E‐15 | [M+H]+ | 583.26820 | 5.9 | C34H37N3O6 | MSI2 | N1,N5,N10‐Tricoumaroyl spermidine |
| n_3183 | 81.4 | Negative | – | 436.22628 | 5.4 | < 1.64E‐15 | [M‐H]− | 437.02315 | 0.8 | C25H31O4N3 | MSI2 | N1,N10‐Dicoumaroylspermidine |
| n_2074 | 79.3 | Negative | – | 349.15000 | 7.3 | < 1.64E‐15 | [M‐H]− | 350.15727 | 0.4 | C15H26O9 | MSI3 | Azelaic acid glycoside |
| n_4005 | 78.2 | Positive | – | 503.16151 | 1.4 | 1.10E‐05 | [M‐H]− | 504.16900 | 0.4 | C18H32O16 | MSI2 | Raffinose |
| p_0421 | 79.2 | Negative | – | 144.10132 | 1.3 | < 1.64E‐15 | [M+H]+ | 143.09460 | 3.7 | C7H13O2N | MSI2 | Proline betaine |
| n_2571 | 78.3 | Negative | – | 387.16572 | 5.2 | 1.64E‐15 | [M‐H]− | 388.17330 | 1.3 | C18H28O9 | MSI2 | 7‐Epi‐12‐hydroxyjasmonic acid glucoside |
| n_4749 | 77.2 | Negative | M: 625.14020, ESI relative: p_3426 | 627.14616 | 6.0 | < 1.64E‐15 | [M‐H]− | 626.14830 | 1.3 | C27H30O17 | MSI3 | 3,3′,4′,5,7,8‐Hexahydroxyflavone; 7‐O‐α‐l‐rhamnopyranoside, 8‐O‐β‐d‐glucopyranoside |
| n_1843 | 77.1 | Negative | M: 331.04562 | 332.04902 | 8.9 | < 1.64E‐15 | [M‐H]− | 332.05320 | 0.9 | C16H12O8 | MSI3 | Quercetagetin methyl ether |
| p_0586 | 73.6 | Negative | – | 161.09152 | 1.1 | < 1.64E‐15 | [M+H]+ | 161.09207 | 3.9 | C3H7NO2 | MSI2 | d‐Alanyl‐d‐alanine |
| p_0184 | 73.0 | Negative | M: 116.06969 | 117.07322 | 1.3 | 6.79E‐05 | [M+H]+ | 115.06330 | 0.5 | C5H9O2N | MSI2 | Proline |
| p_2208 | 71.1 | Positive | – | 315.04863 | 11.1 | 0.000000151 | [M+H]+ | 314.04270 | 3.8 | C16H10O7 | MSI3 | Wedelolactone |
| n_2574 | 70.2 | Negative | – | 387.20213 | 5.2 | 1.96E‐14 | [M‐H]− | 388.20970 | 0.9 | C19H32O8 | MSI3 | 9,13‐Dihydroxy‐4‐megastigmen‐3‐one 9‐glucoside |
| n_5122 | 69.4 | Negative | – | 925.47889 | 10.0 | < 1.64E‐15 | [M‐H]− | 926.48750 | 1.1 | C47H74O18 | MSI3 | Araloside A |
| n_4791 | 69.0 | Negative | – | 639.13506 | 8.1 | 1.09E‐08 | [M‐H]− | 640.14280 | 0 | C31H28O15 | MSI3 | Quercetin 3‐(6′‐ferulylglucoside) |
| n_0657 | 68.3 | Positive | – | 241.07186 | 5.0 | 8.10E‐06 | ND | ND | ND | ND | ND | ND |
| n_2605 | 67.6 | Negative | – | 389.21773 | 8.4 | < 1.64E‐15 | [M‐H]− | 390.22540 | 0.4 | C19H34O8 | MSI3 | (3S,5R,6S,7E,9x)‐7‐Megastigmene‐3,6,9‐triol 9‐glucoside |
| n_2530 | 66.9 | Negative | – | 383.13785 | 5.6 | < 1.64E‐15 | [M‐H]− | 383.13562 | 2.1 | C19H28O4S2 | MSI3 | Unkwown |
| n_0378 | 65.0 | Negative | – | 197.04552 | 3.9 | < 1.64E‐15 | [M‐H]− | 198.05280 | 0.3 | C9H10O5 | MSI3 | 3‐(3,4‐Dihydroxyphenyl)lactic acid |
| p_1078 | 64.4 | Positive | – | 209.15278 | 5.0 | 1.20E‐05 | [M+H]+ | 208.14630 | 4.1 | C13H20O2 | MSI3 | 4‐Hydroxy β‐ionone |
| p_2652 | 64.0 | Negative | – | 365.10471 | 1.3 | < 1.64E‐15 | [M+Na]+ | 342.11622 | 1.6 | C12H22O11 | MSI2 | Trehalose |
| p_3452 | 63.6 | Negative | M1: 641.16797 | 642.17144 | 6.4 | < 1.64E‐15 | [M+H]+ | 640.16390 | 5.0 | C28H32O17 | MSI3 | Quercetagetin 7‐methyl ether 3‐neohesperidoside |
| n_1201 | 62.5 | Negative | – | 288.06021 | 1.2 | 1.14E‐11 | [M‐H]− | 289.06740 | 1.2 | C17H11N3S | MSI4 | 2‐[5‐(Pyrimidin‐4‐yl)thiophen‐2‐yl]quinoline |
| n_0421 | 62.5 | Negative | – | 204.08774 | 1.3 | < 1.64E‐15 | [M‐H]− | 205.09500 | 0.2 | C8H15NO5 | MSI3 | N‐Acetyl‐d‐fucosamine |
| n_4973 | 62.5 | Negative | M: 719.16089 | 720.16461 | 7.6 | < 1.64E‐15 | [M‐H]− | 720.16900 | 1.2 | C36H32O16 | MSI2 | Sagerinic acid |
| n_4127 | 62.2 | Negative | – | 515.21332 | 7.7 | 2.34E‐03 | [M‐H]− | 516.21340 | 0.5 | C24H36O12 | MSI3 | CucurbitosideF |
| p_1908 | 61.4 | Negative | Fragment of 449.10553 | 287.05382 | 4.0 | 1.35E‐12 | [M+H]+ | 449.10501 | 6.2 | C21H20O11 | MSI2 | Luteolin‐4′‐O‐glucoside |
| n_4969 | 61.3 | Negative | – | 717.14574 | 7.7 | < 1.64E‐15 | [M‐H]− | 718.15340 | 0.2 | C36H30O16 | MSI3 | Salvianolic acid L |
| p_2660 | 60.8 | Negative | Fragment of 394.20529 | 366.17393 | 7.2 | < 1.64E‐15 | [M+H]+ | 393.19400 | 7.3 | C24H27NO4 | MSI3 | Tylophorine |
| p_0576 | 60.5 | Negative | – | 160.09622 | 1.3 | < 1.64E‐15 | [M+H]+ | 159.08950 | 6.2 | C7H13O3N | MSI3 | trans‐4‐Hydroxy‐l‐proline betaine |
| p_1252 | 60.2 | Negative | – | 227.12687 | 5.1 | 3.95E‐13 | [M+H]+ | 226.12050 | 4.6 | C12H18O4 | MSI3 | 12‐Hydroxyjasmonic acid |
ND, not determined.
Finally, statistical validation was complemented with biological confirmation to validate the predictive capacity of the metabolic markers using an independent sample set harvested in 2014 (Eshel et al., 2021) as for 2019. Using the equation calculated based on the entire 2019 dataset, a validation model predicted the elevation for each plant from the 2014 dataset. The quality of the prediction was evaluated by both the coefficient of determination and the P‐value observed when comparing real and predicted altitudes (Fig. 2). The same permutation protocol was used to test the likelihood of spurious prediction of 2014 validation models.
Fig. 2.
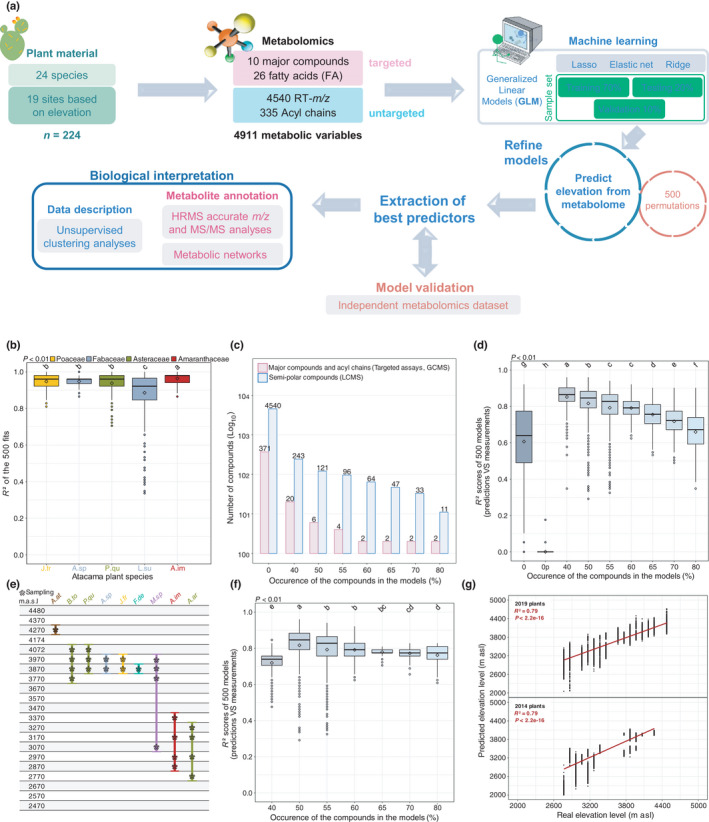
Predictive metabolomics of Atacama plants. (a) A simplified scheme of the predictive metabolomics approach used in this study. (b) Species‐specific level: R 2 scores of the fit between calculated and real elevation levels with letters indicating statistical significance (Tukey's test, P < 0.01). Theoretical elevations were calculated from the plant metabolome. (c) Global level: threshold of the variable occurrence defined by 500 models performed on all variables for all species. The 13 variables used in 80% represent the most relevant compounds for predicting elevation. (d) R 2 scores depending on the variable occurrence threshold (Tukey's test, P < 0.01). (e) Biological validation using an independent sample set from 2014. (f) R 2 scores obtained by predicting the elevation level from plants from 2014 using the multilinear equation calculated based on plants from 2019 depending on the variable occurrence threshold (Tukey's test, P < 0.01). (g) Predicted elevations from 2019 and 2014 plants using the best 66 markers (Pearson correlation). Compounds in (c, d, f) refer to metabolic variables stricto sensu before annotation.
Multivariate statistical analyses
The normalized dataset was processed through multivariate analysis such as principal components analysis (PCA) via the factominer package (Lê et al., 2008) in R (v.3.6.1) and two‐way orthogonal partial least square (O2PLS) via simca 16.0.1 (Umetrics, Umeå, Sweden). Tukey's tests were performed to compare the expression of the metabolic markers between species or between environments using the agricolae package (Mendiburu, 2020) with a threshold of significance established at P < 0.01. Finally, box plots, scatter plots, correlation plots and heatmaps were realized using the ggplot2, ggpubr, hmisc and pheatmap packages (Wickham, 2016; Kolde, 2019; Harrell Jr, 2020; Kassambara, 2020) (Pearson correlation, Ward algorithm) in R, respectively.
Annotation
The best metabolic predictors were annotated using two different methods. First, MS spectra were used to analyse the isotopic patterns (13C, 18O, 15N and 34S) and speculate on the ion composition. In addition, all putative chemical formulas were calculated by the FreeStyle function within the Xcalibur 4.2 software with the following minimal and maximal constraints on chemical elements: 14N: 0–60, 16O: 0–6, 12C: 0–100, 1H: 0–200, 32S: 0–60, 35Cl: 0–60 and 31P: 0–60, and a mass tolerance at 10 ppm. Thus, the best candidates were chosen based on the MS analysis and screened on chemical databases (Chebi (de Matos et al., 2010), Metlin (Xue et al., 2020), DNP (http://dnp.chemnetbase.com) and Knapsack (Shinbo et al., 2006)). In parallel, accurate m/z values of the most discriminant monoisotopic ions were screened using the Metlin database (Smith et al., 2005) for putative annotation. The resulting outputs from the two methods were compared to select the best putative annotation for each ion (Table 1). In addition, MS/MS spectra of all samples were used to improve the annotation level by comparing experimental fragments with experimental MS/MS spectra available in multiple libraries such as MassBank (Horai et al., 2010), MzCloud (https://www.mzcloud.org) and Metlin (Xue et al., 2020). The annotation level of each predictor was therefore attributed following the metabolomics standards initiative confidence level (MSI levels) (Sumner et al., 2014).
Pathway analysis and metabolic networks
The 39 predictors were screened through chemical databases and integrated into a metabolic network to better interpret their role in the plant's response to extreme conditions. The KEGG identifiers were determined using the KEGG database (Kanehisa et al., 2014) on the 39 molecules if available or on chemically related compounds (Table S3). Thereafter, a pathway enrichment analysis was realized using the MetaboAnalyst (Xia et al., 2015) and PlantReactome (Naithani et al., 2019) databases, and combined with the best markers integration into a preexisting Arabidopsis thaliana metabolic network via MetExplore (Cottret et al., 2010).
Results
Plant diversity in the extreme conditions of the Atacama Desert
The Atacama Desert represents one of the harshest environments for plant life (Jordan & Kirk‐Lawlor, 2014; Díaz et al., 2016), where plants must endure the major abiotic stresses currently threatening agriculture. The Talabre–Lejía Transect (TLT) spans an elevation gradient covering three different plant communities: the poorly vegetated Prepuna (2400–3300 m asl), the Puna shrubland (3300–4000 m asl) and the high Andean Steppe (4000–4500 m asl) (Fig. 1a) (Díaz et al., 2016; Eshel et al., 2021). Water availability increases and temperature decreases with altitude, while high solar irradiance and very low nitrogen levels are critical constraints throughout the TLT (Eshel et al., 2021). Rainfall ranges from 20 mm yr−1 in the prepuna to 160 mm yr−1 in the steppe, illustrating the extreme aridity as compared to other plant‐sheltering deserts (Báez & Collins, 2008; Li et al., 2015; Díaz et al., 2016; Ziaco et al., 2018). The daily average solar irradiance of 600 W m−2 d−1 along this transect is three times higher than many deserts and high mountain ecosystems (Bo et al., 2009; Zhang et al., 2010; Arancibia‐Bulnes et al., 2014). In addition, low total nitrogen (average 9 mg kg−1) throughout the transect, low phosphorus levels (6–20 mg kg−1) and high salinity in Prepuna sites add to the harsh conditions plants must endure. Nonetheless, plant life in this ecosystem of the Atacama can be traced back to 45 000 yr ago (Latorre et al., 2002; Díaz et al., 2019) and has thrived in such extreme conditions since probably 12 million years ago (Jordan & Kirk‐Lawlor, 2014). Hence, this ecosystem represents a unique resource of adaptive mechanisms potentially relevant to engineer crop resilience. Interestingly, deep‐sequencing of 32 dominant species representing the major clades highlighted common and specific strategies relevant for plant survival (Eshel et al., 2021). In this context, we collected 21 of these 32 plant species based on their coverage in their natural ecosystem. We complemented this set with one Cactaceae, one Solanaceae and one Boraginaceae to finally represent relevant biodiversity covering annual and perennial plants, different carbon fixation systems (i.e. C3, C4 and CAM) and different lifespans such as shrubs and herbs (Fig. 1c; Table S1). Clear distinctions regarding the distribution of life‐form and carbon fixation types have been highlighted where all annuals and C4 plants were observed at an elevation of < 3870 m asl. Additionally, while some species were relatively specific to a single environment (e.g. Moschopsis monocephala), others had a wide distribution along the transect area that we divided into 19 sites (each 100 m asl) (Fig. 1b). We also selected 11 agronomic and ornamental species based on their plant family to analyse and compare using the same experimental procedures (as explained in the ‘Materials and Methods’ section). A taxonomic analysis performed on the Atacama and agronomic plant species via the NCBI taxonomy browser revealed the relationships between the 14 Atacama plant families (Fig. 1d). Interestingly, this sample set of 23 angiosperms and one gymnosperm included well‐known resilient plant families such as Cactaceae and Boraginaceae (Ma et al., 2010) together with species of economic interest such as those in Poaceae, Asteraceae, Fabaceae and Solanaceae. In addition, the 11 agronomic and ornamental plant species covered five of the 14 Atacama plant families (including the most widespread ones in Poaceae, Fabaceae, Asteraceae and Solanaceae).
Predictive metabolomics reveals a core metabolic set in multiple resilient species
To gain insight into the mechanisms by which these extremophile plants adapt to the extreme conditions of the Atacama Desert and respond to environmental variations, we performed multiplatform metabolomics to screen both primary and secondary metabolisms from the aerial tissues of Atacama plants and agronomic species (Fig. 2a). Quantitative evaluation of 10 major compounds by biochemical phenotyping (used as key physiological indicators) and 26 fatty acids by GCFID highlighted a significant reduction in Chl, nitrate and protein content in Atacama species when compared to 11 known crops and ornamental plants (Fig. S2). In addition, the unknown biochemical diversity of these extreme Atacama plants was analysed through untargeted metabolomics using GCMS and LCMS, which resulted in 335 acyl chains and 4540 semipolar features after preprocessing (Fig. S2). Given that the phytochemical diversity fluctuated with environmental conditions along the elevation gradient (400–2000 m asl) (Defossez et al., 2021), GLM was used to test whether the metabolome (4911 variables) could predict environmental conditions (Fig. 2a). Elevation represents the integration of abiotic factors (Carpenter, 2005), among which climatic and edaphic factors have been previously described (Eshel et al., 2021) (Fig. 1b). Thus, the elevation level of the 19 sampling spots was used as a proxy of the 19 environmental conditions analysed. First, the possibility of calculating the elevation levels from five different plant species selected based on both their biomass and coverage along the elevation gradient was evaluated. For each species, 80% of the sample set (i.e. training sets) was used for the regression analysis. The equation was then used to calculate elevation for the 20% of the sample set remaining (i.e. testing set). Interestingly, the resulting average R 2 from 500 models (i.e. fits between calculated and measured elevation) ranged between 0.88 and 0.96 depending on the species (Fig. 2b). These results indicate the plant metabolome integrates environmental variations. Environmental conditions elicited characteristic metabolic patterns in which compounds correlate with elevation, allowing us to infer the altitude from which the sample was collected.
Moreover, estimating the altitude (i.e. resulting as an environment proxy) from metabolic data alone for species from several plant families raised the question of whether generic mechanisms serve as a basis for adaptation to extreme environments such as the Atacama Desert. To address this question, we used GLM on the entire dataset divided into a training set (70%), a testing set (20%) and a validation set (10%) since the total size of the dataset was sufficient (n = 224). We thus predicted the plant environment (i.e. elevation level) and highlighted the shared metabolic predictors (Fig. 2a). A first modelling step determined the predictive capacity of the 4911 metabolic variables, represented by their percentage of use in the models. Consequently, a threshold of 40% (i.e. variables used in more than 40% of the 500 models) included 263 features while 80% involved the best 13 metabolic predictors (Fig. 2c). Subsequently, each threshold was processed to exclude the nonpredictive features and tightly select the best ratio between the predictive capacity and the number of metabolic variables. The plant environment was considerably predictable at 66% and 79% using 13 or 66 markers, respectively (Fig. 2d). Lower thresholds (e.g. 40%) allowed better predictions but yielded less robust predictors (i.e. higher SD). Importantly, 500 permutation sets involving randomly assigned elevation levels were developed to test the likelihood of spurious predictions, which led to a mean R 2 of 0% and thus statistically validated the GLM‐based modelling approach. Hence, we demonstrated that common features could greatly predict plant environments (79%), independently of the species and family.
To further test the robustness of such predictions, we biologically confirmed the predictive capacity of the metabolic features using an independent dataset composed of nine Atacama plant species harvested in 2014 and covering 12 environments (2770–4270 m asl) (Fig. 2e). The linear equation developed using the 2019 samples was then applied to the 2014 dataset to estimate elevation levels, thereby resulting in similar predictive patterns between 2019 and 2014 (Fig. 2f). Altogether, both mean R 2 prediction and SD results pointed towards an ideal threshold of 60% (66 variables), which allowed a prediction at 79% for both years (P < 2.2e−16) (Fig. 2g). These results hence confirm that plants harbour a core set of metabolites to adapt to the environmental constraints.
A not‐so‐specialized set of secondary metabolites also detected in agronomic and ornamental plant species
Next, we annotated the best 66 predictors using both accurate m/z values and MS/MS analysis. This annotation process allowed us to exclude the fragments observed among the 66 features (Table S4), finally retaining 39 metabolic predictors without remarkable impact on the average R 2 (Fig. S2a). The MSI annotation level for each predictor is presented in Table 1. Notably, the best predictor was starch, while 37 metabolites were referred to semipolar compounds (Table 1). Only six markers were positively correlated to elevation, while the intensity of the remaining compounds decreased with elevation (Fig. S3b). Remarkably, predictors in Atacama plant species were also found in several agronomic and ornamental plants (Tables S5, S6), demonstrating the ubiquitous nature of these metabolites. These 39 compounds were queried in biochemical databases (e.g. Kegg, PlantReactome) to perform a pathway analysis (Table S3) and placed into a preexisting A. thaliana metabolic network available on MetExplore (Kanehisa et al., 2014) (Fig. S4). More than half of the markers were involved in secondary metabolism (56%), while primary metabolism and regulators (e.g. jasmonates) covered 31% in total (Fig. 3). The remaining 13% included three unknown compounds and two salt artefacts that combined sodium and magnesium to formic acid, suggesting salt hyperaccumulation processes (Figs 3, S3a). Notably, starch, trehalose and amino acid‐related pathways were involved in crosstalk with the biosynthesis of secondary metabolites, while the central place of raffinose was highlighted in galactose metabolism involving other oligosaccharides known for their role in abiotic stress tolerance (Vinson et al., 2020) (Fig. S4a). In addition, phenolics represented the major enrichment observed in Atacama plant species with 14 of the 39 markers. While alkaloids and N‐containing compounds (e.g. proline betaine, or polyamines combined with flavonoids) were included in the best markers, flavonoid, phenylpropanoid and terpenoid pathways were clearly overrepresented (Figs 3, S4). Last but not least, despite their classification into primary or secondary metabolisms, a relevant proportion of these 39 markers also referred to redox homeostasis based on their chemical nature or interactions with ascorbate or glutathione pathways (Fig. S4), suggesting the importance of redox homeostasis in the adaptation to hostile environments. Overall, predictive metabolomics reveals that plant metabolism greatly reflects environmental fluctuations in extreme ecosystems, as also pinpointed by a core set of metabolites (involved in secondary, primary and redox pathways) capable of predicting at 79% the plant environment independently of the plant species. These findings thus confirm a central place for generic metabolic pathways underpinning plant adaptation to environmental constraints.
Fig. 3.
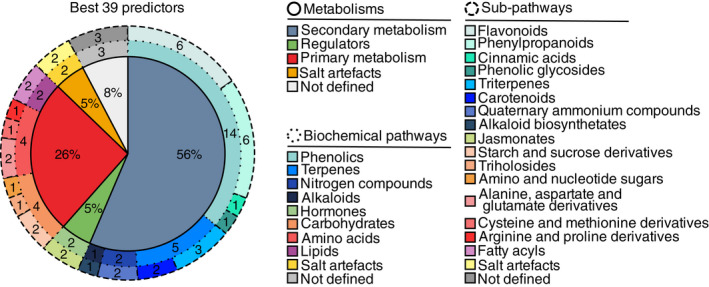
Pathway analysis of the 39 markers. Metabolism, biochemical pathways and subpathways were elucidated by screening the KEGG identifiers through the MetaboAnalyst, PlantReactome and MetExplore databases.
The plant metabolome is tailored to environmental constraints
Elevation integrates a wide range of abiotic factors, among which edaphic variables were measured in each of the 19 sampling spots. Climatic variables such as temperature, soil water content (SWC, representing the interaction between precipitation and soil properties), precipitation and solar irradiance were measured via two stations (at 3060 and 4090 m asl). Theoretical values of these factors along the elevation gradient for the 19 environments were predicted considering a linear distribution that was confirmed by field measurements (Fig. S5). Elevation correlated closely with most environmental parameters in the Atacama Desert (Fig. S6). Thus, an analysis combining PCA and O2PLS was performed to unravel the elevation factor and highlight the relationship between the 39 best predictors and environmental factors.
First, PCA was used to reveal the influence of elevation on the climatic and edaphic conditions. The first two components of the PCA model explained 82.4% of the total variance of the dataset (Fig. 4a) and showed clear discrimination of the plant communities (i.e. Prepuna, Puna and Steppe) along a multivariate vector that represented the elevation gradient. Also, a second plane defined by several minerals divided the different environments belonging to the Prepuna ecosystem, which did not occur with Steppe spots. Hence, the previously predicted elevation factor was here depicted by a multivariate vector represented mostly by edaphic variables (e.g. pH, P, K), temperature and solar irradiance.
Fig. 4.
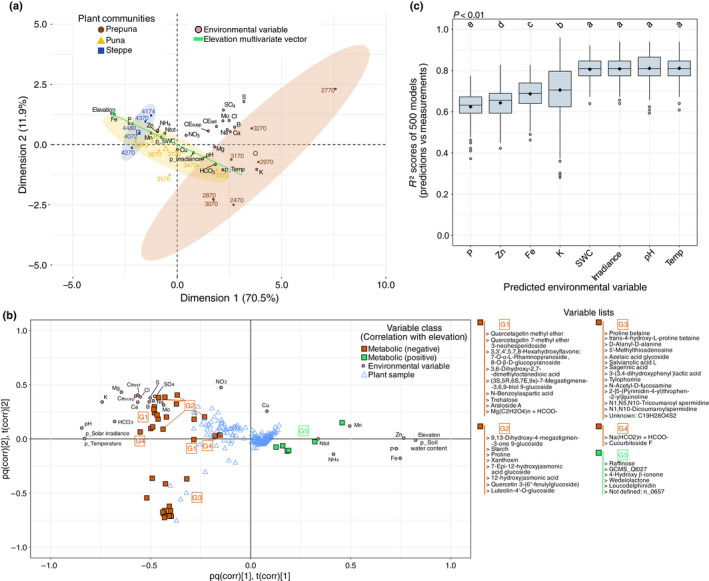
Decomposition of the elevation factor and environment–metabolome covariation. (a) Principal component analysis biplot. Discrimination of the sampling spots by the environmental data. SWC represents the soil water content while p_ represents a partially predicted parameter. (b) Two‐way orthogonal partial least squares analysis describing the covariation between environmental and metabolic data. Hierarchical clustering analysis was realized with Pearson correlation and Ward algorithm. (c) Boxplot showing the average R 2 scores (500 models) performed on the best discriminant environmental variables using the 66 best metabolic markers. Letters indicate statistical significance (Tukey's test, P < 0.01). SWC, soil water content; Temp, temperature.
Second, we predicted the covariation between environmental factors and the best 39 markers using an O2PLS analysis (Fig. 4b). Remarkably, 95% of the variation observed in the environmental dataset was covered by the metabolic features (Table S7). Congruently with the correlation matrix and PCA (Fig. 4), the best predictors were distributed primarily along the first component representing elevation and, to a lesser extent, linked to several edaphic factors such as sulphur. The O2PLS biplot further highlighted a remarkable separation between metabolic compounds positively or negatively correlated with elevation along the first plane. In particular, five phytochemicals determined by Pearson clustering and including raffinose were plotted with regard to temperature. These results indicate that the discriminant capacity of elevation resulted primarily from temperature, solar irradiance, SWC and several edaphic factors. These PLS predictions were also confirmed by GLM, where the best predictions using the 39 best markers on independent environmental parameters were obtained for SWC, solar irradiance, pH and temperature (Fig. 4c).
Overall, we provide a valuable approach combining metabolomics, GLM and multivariate statistical analyses. Exploiting multiple species in a natural environment can successively reveal generic mechanisms of interest and disentangle complex systems into specific environmental parameters (i.e. climatic and edaphic).
Discussion
Predictive metabolomics demonstrates a generic metabolic toolbox for plant adaptation to extreme habitats
Ecological metabolomics, which allowed study of the interaction between plant metabolism and environment, has attracted scientific curiosity for over 50 yr (Sardans et al., 2020). While studies on single plant species led to limited results when transferred to crops, a metaanalysis of individual studies highlighted metabolic convergences in plant species inhabiting extreme environments (Dussarrat et al., 2021), encouraging plant researchers to move towards a more holistic approach. Strikingly, our approach combining ecological metabolomics with GLM‐based modelling was able to predict the plant environment with an R 2 as high as 0.96 within given species (Fig. 2b) and 0.79 between species (Fig. 2). Such values are far above correlation coefficients usually obtained with phenotypic traits (Laughlin et al., 2012; Poorter et al., 2019), which are also more difficult to score (Laughlin & Messier, 2015), making metabolic markers ideal soft traits.
All Atacama plant species harboured low Chl levels as compared to agronomic species, which could result from an adaptive response to high solar irradiance or the meagre availability of other resources such as water or nitrogen (Hikosaka et al., 2003), as further illustrated by the very low nitrate and protein contents (Fig. S2). Our results suggest that these 24 species, belonging to 14 families, also use a common metabolic toolbox, underpinned by the 39 metabolic markers revealed by our modelling approach, to cope with their environment (Fig. 2g). This toolbox is certainly generic as the same metabolic traits were found in several agronomic and ornamental families, validating the ubiquitous nature of this core set. Main differences were observed for flavonoid‐ and terpenoid‐related predictors, which accumulated to high levels in Atacama plants (Fig. S7). In addition, levels of raffinose and 4‐hydroxy β‐ionone (a compound related to carotenoid degradation) were higher in Steppe species than most temperate species, while Prepuna species accumulated proline derivative compounds. Several markers such as quercetin glucoside and coumaroyl‐spermidine relatives were only detectable using a lower threshold in agronomic families (Table S6). By contrast, several hormone and primary metabolism‐related predictors (e.g. jasmonates, trehalose) did not present major changes between extreme and nonadapted species, except for Chls and proteins, which were lower in Atacama plants. Overall, these observations question the potential adaptive capacity of these agronomic and ornamental species, which naturally develop in mainly temperate regions. The possibility that the genome of agronomic plants would already permit the synthesis of metabolites relevant for plant survival in harsh lands is supported by the presence of Solanum chilense (closely related to the cultivated S. lycopersicum) in the 24 Atacama species studied. From an evolutionary point of view, this further suggests that it is easier to modify the regulation of existing metabolic pathways than to create new ones. Also, the high R 2 score prompts the question of how species‐specific metabolic adaptations could provide a selective advantage. Hence, the relationship between these metabolic markers and the genetic adaptations discovered in Atacama plants (Eshel et al., 2021) deserves further investigation. In particular, several species colonized a wide elevation gradient, which suggests high plasticity (Fig. 1). Consequently, metabolic adjustments enabling the acclimation or adaptation to extreme conditions are not necessarily the result of a long evolutionary process.
Involvement of the best metabolic predictors in extreme environment adaptation
The influence of the elevation gradient on metabolic patterns is mainly reflected in a multivariate vector involving temperature, SWC, irradiance and pH (Fig. 4a). However, extremophiles also face other environmental pressures such as the mineral imbalance observed in the Atacama Desert that deserves closer examination (Fig. 1b). Thus, the success of thriving in the Atacama Desert elevation gradient would result from the ability to cope with daily subzero temperatures at the top of the transect and hyperaridity and high salinity at the bottom, or to manage the balance between carbon input and access to other critical resources such as water.
Starch was the best predictor (Table 1), while trehalose and raffinose were among the top predictors, confirming a suitable place for carbohydrates in the resilience mechanism (Fig. S4). In the Atacama Desert, solar irradiance is not a limiting factor for carbon entry, and even threatens plant survival (Eshel et al., 2021). The shallow protein level observed in all Atacama plant species (Fig. S2) suggests that plant growth is very low (Elser et al., 2008). Therefore, carbon that does not fuel plant growth and protein turnover could be transiently stored as starch or allocated to protective systems against oxidative stress induced by other environmental factors such as water availability, temperature and salinity. Starch, metabolism of which is known to play a major role against abiotic stress (Thalmann & Santelia, 2017), can be used as a carbon source for the synthesis of protective compounds when environmental conditions become harsher, while its accumulation could be linked to sodium scavenging in halophytes (Thalmann & Santelia, 2017), for instance. The strong negative correlation of starch with elevation would result from a trade‐off with the production of osmolytes and other protective compounds required at the highest elevations, where daily subzero temperatures occur. Alternatively, the lower efficiency of transitory starch remobilization under low temperature could explain low starch contents at high levels. Conversely, raffinose negatively correlated with temperature, validating the central place of raffinose family oligosaccharides in cold tolerance (Vinson et al., 2020). However, several predictors were fatty acyls within the primary metabolism and jasmonates, which supported a role of lipid remodelling in extreme environments (Cao et al., 2016; Dussarrat et al., 2021).
More than half of the 39 best markers were involved in secondary metabolism (Fig. 3). Remarkably, phenolics (14/39 compounds) were increased at lower elevations, which would help plants cope with both the very low water availability and high salinity of these lands. This protective process has already been described in multiple extremophile plant species (Dussarrat et al., 2021). Phenolic antioxidant properties, mainly for cinnamic acid and quercetin derivatives (extensively represented in the best predictors), enhance photoprotection and resilience to abiotic stresses (Agati & Tattini, 2010). Regarding terpenoids, the presence of xanthoxin and 4‐hydroxy β‐ionone (a carotenoid degradation product) within the 39 best markers supported the role of carotenoids per se as well as their degradation in extreme climate resilience (Table 1). Despite their well‐described antioxidant role, Havaux (2014) discussed the link between the catabolism of carotenoids and plant defence, as their cleavage leads to hormonal compounds (e.g. abscisic acid) or redox signalling. Also, the accumulation of N‐related compounds could be attributed to their role in osmoregulation (e.g. proline betaine). The contribution of phenolics that conjugate polyamines and other molecules (e.g. tricoumaroyl spermidine) is more complex, despite a growing body of evidence for their implication in stress mitigation (Pál et al., 2018). Most importantly, our study linked plant survival under harsh conditions to redox metabolism since the majority of metabolic markers directly or indirectly involve redox homeostasis. This was exemplified by primary compounds of the glutathione and ascorbate pathways, metabolites for ROS processing including carotenoids, as well as potential links between the biosynthesis of several compounds and NAD metabolism (Fig. S4), all participating in oxidative stress signalling (Decros et al., 2019; Dussarrat et al., 2021). Among the best predictors is proline, accumulation of which in response to osmotic stress has been widely documented and recently attributed to redox homeostasis (Szabados & Savouré, 2010). Alternatively, accumulated levels of amino acids could serve as metabolic intermediates for the synthesis of more complex secondary metabolites with stress‐responsive functions.
Overall, uncovering the metabolic characteristics of Atacama species highlighted the linear encapsulation of environmental fluctuations by the plant metabolome (involving primary, secondary and redox pathways), and the use of generic metabolic mechanisms to adapt to extreme growth conditions. Such an approach (multispecies harvested in extreme environments) offers promising perspectives in both ecological chemistry and stress physiology worldwide. A fascinating perspective will be to research the genetic and molecular mechanisms that control the levels of these metabolic markers.
Author contributions
TD, YG, DR, RG and PP designed and planned the project. TD, CL, FD and RG performed the fieldwork. TD, SP, PP, SB, AF, CC, PVD, KV and JJ conducted metabolic or bioinformatic experiments and analyses. DJ uploaded all data online. TD, YG, DR, RG and PP integrated and analysed the results of all the experiments. TD, RG and PP wrote the paper with feedback form all the coauthors.
Supporting information
Fig. S1 Correlation between starch content and sampling time. Absence of correlation between the time of sampling and starch content in plant samples.
Fig. S2 Changes in major compounds in Atacama plants. (a) Depiction of the biochemical diversity observed in Atacama plants and 11 agronomic or ornamental species.
Fig. S3 Best metabolomics predictors in Atacama plants. (a) Predictive capacity and clustering of the best metabolic markers.
Fig. S4 Metabolic networks. Best markers were mapped into a preexisting A. thaliana metabolic network using MetExplore.
Fig. S5 Validation of the environmental prediction. Linearity of environmental parameters with elevation.
Fig. S6 Decomposition of the elevation parameter. Correlation plot of the environmental data.
Fig. S7 Best metabolic predictors in agronomic and ornamental plant species. Clustering analysis of the best metabolic predictors between Atacama and agronomic and ornamental plant species.
Table S1 (a) Sample data from Atacama plants. (b) Sample data from agronomic and ornamental plants.
Table S2 Metabolic dataset.
Table S3 Pathway analysis.
Table S4 Annotation table of markers.
Table S5 Markers in control plant. Intensity of the best metabolic markers in agronomic and ornamental plants. The detection limits were fixed to 25 000 or 10 000.
Table S6 Markers threshold 100. Intensity of the metabolic markers that could not be observed with a 10k noise threshold. Those markers were manually checked on one sample of each nonextreme species and one Atacama species with a threshold of 100.
Table S7 O2PLS model.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We acknowledge the GenOuest bioinformatics core facility (https://www.genouest.org) and the Genotoul bioinformatics platform Toulouse Occitanie (Bioinfo Genotoul, https://doi.org/10.15454/1.5572369328961167E12) for providing computing infrastructures and resources. We also acknowledge the Comunidad de Talabre. We are grateful to Christiane Gallet, Guillaume Tcherkez and Fabien Jourdan for their analytical advice, and MetaboHUB (ANR‐11‐INBS‐0010) and PHENOME (ANR‐11‐INBS‐0012) for financial support to INRAE. We acknowledge the support from the European Commission's Horizon 2020 Research and Innovation Program via the GLOMICAVE project under grant agreement no. 952908. We thank Fondo de Desarrollo de Áreas Prioritarias (FONDAP) Center for Genome Regulation (15200002), Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT 1180759), DOI EVONET Proyect and ANID – Millennium Science Initiative Program – iBio ICN17_022 for financial support to RAG.
Contributor Information
Rodrigo A. Gutiérrez, Email: rgutierrez@bio.puc.cl.
Pierre Pétriacq, Email: pierre.petriacq@inrae.fr.
Data availability
The metabolic dataset and all metadata were deposited online using Dataverse INRAE (https://dx.doi.org/10.15454/UUBXIF) and following the Findable, Accessible, Interoperable, Reusable (FAIR) principles (Jacob et al., 2020).
References
- Agati G, Tattini M. 2010. Multiple functional roles of flavonoids in photoprotection: letters. New Phytologist 186: 786–793. [DOI] [PubMed] [Google Scholar]
- Arancibia‐Bulnes CA, Peón‐Anaya R, Riveros‐Rosas D, Quiñones JJ, Cabanillas RE, Estrada CA. 2014. Beam solar irradiation assessment for Sonora, Mexico. Energy Procedia 49: 2290–2296. [Google Scholar]
- Báez S, Collins SL. 2008. Shrub invasion decreases diversity and alters community stability in Northern Chihuahuan desert plant communities. PLoS ONE 3: e2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bo H, Yue‐Si W, Guang‐Ren L. 2009. Properties of solar radiation over Chinese Arid and Semi‐Arid areas. Atmospheric and Oceanic Science Letters 2: 183–187. [Google Scholar]
- Cao J, Li M, Chen J, Liu P, Li Z. 2016. Effects of MeJA on Arabidopsis metabolome under endogenous JA deficiency. Scientific Reports 6: 37674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter C. 2005. The environmental control of plant species density on a Himalayan elevation gradient: plant species density on a Himalayan elevation gradient. Journal of Biogeography 32: 999–1018. [Google Scholar]
- Castañeda‐Álvarez NP, Khoury CK, Achicanoy HA, Bernau V, Dempewolf H, Eastwood RJ, Guarino L, Harker RH, Jarvis A, Maxted N et al. 2016. Global conservation priorities for crop wild relatives. Nature Plants 2: 16022. [DOI] [PubMed] [Google Scholar]
- Cottret L, Wildridge D, Vinson F, Barrett MP, Charles H, Sagot M‐F, Jourdan F. 2010. MetExplore: a web server to link metabolomic experiments and genome‐scale metabolic networks. Nucleic Acids Research 38: W132–W137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui G, Ji G, Liu S, Li B, Lian L, He W, Zhang P. 2019. Physiological adaptations of Elymus dahuricus to high altitude on the Qinghai‐Tibetan plateau. Acta Physiologiae Plantarum 41: 115. [Google Scholar]
- Dai F, Nevo E, Wu D, Comadran J, Zhou M, Qiu L, Chen Z, Beiles A, Chen G, Zhang G. 2012. Tibet is one of the centers of domestication of cultivated barley. Proceedings of the National Academy of Sciences, USA 109: 16969–16973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decros G, Beauvoit B, Colombié S, Cabasson C, Bernillon S, Arrivault S, Guenther M, Belouah I, Prigent S, Baldet P et al. 2019. Regulation of pyridine nucleotide metabolism during tomato fruit development through transcript and protein profiling. Frontiers in Plant Science 10: 1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defossez E, Pitteloud C, Descombes P, Glauser G, Allard P‐M, Walker TWN, Fernandez‐Conradi P, Wolfender J‐L, Pellissier L, Rasmann S. 2021. Spatial and evolutionary predictability of phytochemical diversity. Proceedings of the National Academy of Sciences, USA 118: e2013344118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz FP, Frugone M, Gutiérrez RA, Latorre C. 2016. Nitrogen cycling in an extreme hyperarid environment inferred from δ15N analyses of plants, soils and herbivore diet. Scientific Reports 6: 22226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz FP, Latorre C, Carrasco‐Puga G, Wood JR, Wilmshurst JM, Soto DC, Cole TL, Gutiérrez RA. 2019. Multiscale climate change impacts on plant diversity in the Atacama Desert. Global Change Biology 25: 1733–1745. [DOI] [PubMed] [Google Scholar]
- Domergue F, Vishwanath SJ, Joubès J, Ono J, Lee JA, Bourdon M, Alhattab R, Lowe C, Pascal S, Lessire R et al. 2010. Three Arabidopsis fatty acyl‐coenzyme a reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiology 153: 1539–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussarrat T, Decros G, Díaz FP, Gibon Y, Latorre C, Rolin D, Gutiérrez RA, Pétriacq P. 2021. Another tale from the harsh world: how plants adapt to extreme environments. Annual Plant Reviews Online 4: 551–603. [Google Scholar]
- Elser JJ, Sterner RW, Gorokhova E, Fagan WF, Markow TA, Cotner JB, Harrison JF, Hobbie SE, Odell GM, Weider LW. 2008. Biological stoichiometry from genes to ecosystems. Ecology Letters 3: 540–550. [Google Scholar]
- Eshel G, Araus V, Undurraga S, Soto DC, Moraga C, Montecinos A, Moyano T, Maldonado J, Díaz FP, Varala K et al. 2021. Plant ecological genomics at the limits of life in the Atacama Desert. Proceedings of the National Academy of Sciences, USA 118: e2101177118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima Z, Ahmed M, Hussain M, Abbas G, Ul‐Allah S, Ahmad S, Ahmed N, Ali MA, Sarwar G, Haque EU et al. 2020. The fingerprints of climate warming on cereal crops phenology and adaptation options. Scientific Reports 10: 18013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. 2010. Regularization paths for generalized linear models via coordinate descent. Journal of Statistical Software 33: 1–22. [PMC free article] [PubMed] [Google Scholar]
- Harrell FE Jr. 2020. With contributions from C. In: Dupont and many others many. Hmisc: Harrell miscellaneous. R package v.4.2‐0. [Google Scholar]
- Hashim AM, Alharbi BM, Abdulmajeed AM, Elkelish A, Hozzein WN, Hassan HM. 2020. Oxidative stress responses of some endemic plants to high altitudes by intensifying antioxidants and secondary metabolites content. Plants 9: 869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M. 2014. Carotenoid oxidation products as stress signals in plants. The Plant Journal 79: 597–606. [DOI] [PubMed] [Google Scholar]
- Hikosaka K, Yamano T, Nagashima H, Hirose T. 2003. Light‐acquisition and use of individuals as influenced by elevated CO2 in even‐aged monospecific stands of Chenopodium album: effects of elevated CO2 on light‐acquisition and use. Functional Ecology 17: 786–795. [Google Scholar]
- Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K et al. 2010. MassBank: a public repository for sharing mass spectral data for life sciences. Journal of Mass Spectrometry 45: 703–714. [DOI] [PubMed] [Google Scholar]
- Jacob D, David R, Aubin S, Gibon Y. 2020. Making experimental data tables in the life sciences more FAIR: a pragmatic approach. GigaScience 9: giaa144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan TE, Kirk‐Lawlor NE, Blanco NP, Rech JA, Cosentino NJ. 2014. Landscape modification in response to repeated onset of hyperarid paleoclimate states since 14 Ma, Atacama Desert, Chile. Geological Society of America Bulletin 126: 1016–1046. [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Research 42: D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassambara A. 2020. ggpubr: ‘ggplot2’ based publication ready plots, vol. 438. R package version 0.4.0. [Google Scholar]
- Kolde R. 2019. pheatmap: pretty heatmaps. R package version 1.0. 12. R Package version 1.0, 8. [Google Scholar]
- Kosmacz M, Sokołowska EM, Bouzaa S, Skirycz A. 2020. Towards a functional understanding of the plant metabolome. Current Opinion in Plant Biology 55: 47–51. [DOI] [PubMed] [Google Scholar]
- Latorre C, Betancourt JL, Rylander KA, Quade J. 2002. Vegetation invasions into absolute desert: a 45 000 yr rodent midden record from the Calama‐Salar de Atacama basins, northern Chile (lat 22°–24°S). GSA Bulletin 114: 349–366. [Google Scholar]
- Laughlin DC, Joshi C, van Bodegom PM, Bastow ZA, Fulé PZ. 2012. A predictive model of community assembly that incorporates intraspecific trait variation. Ecology Letters 15: 1291–1299. [DOI] [PubMed] [Google Scholar]
- Laughlin DC, Messier J. 2015. Fitness of multidimensional phenotypes in dynamic adaptive landscapes. Trends in Ecology & Evolution 30: 487–496. [DOI] [PubMed] [Google Scholar]
- Lê S, Josse J, Husson F. 2008. FactoMineR: an R package for multivariate analysis. Journal of Statistical Software 25: 1–18. [Google Scholar]
- Lewis JE, Kemp ML. 2021. Integration of machine learning and genome‐scale metabolic modeling identifies multi‐omics biomarkers for radiation resistance. Nature Communications 12: 2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wada H, Matsuzaki H. 2015. Radial growth rate through successive cambia in Haloxylon ammodendron (Chenopodiaceae) from the Gurbantünggüt Desert, Northwestern China, determined by a series of radiocarbon dating. Geochemical Journal 49: 39–51. [Google Scholar]
- Long SP, Marshall‐Colon A, Zhu X‐G. 2015. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161: 56–66. [DOI] [PubMed] [Google Scholar]
- Lugan R, Niogret M‐F, Leport L, Guégan J‐P, Larher FR, Savouré A, Kopka J, Bouchereau A. 2010. Metabolome and water homeostasis analysis of Thellungiella salsuginea suggests that dehydration tolerance is a key response to osmotic stress in this halophyte. The Plant Journal 64: 215–229. [DOI] [PubMed] [Google Scholar]
- Luna E, Flandin A, Cassan C, Prigent S, Chevanne C, Kadiri CF, Gibon Y, Pétriacq P. 2020. Metabolomics to exploit the primed immune system of tomato fruit. Metabolites 10: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma WB, Zhao XJ, Tan DY, Baskin CC, Baskin JM, Xue JH. 2010. Nutlet dimorphism in individual flowers of two cold desert annual Lappula species (Boraginaceae): implications for escape by offspring in time and space. Plant Ecology 209: 361–374. [Google Scholar]
- de Matos P, Alcántara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. 2010. Chemical entities of biological interest: an update. Nucleic Acids Research 38: D249–D254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mendiburu F. 2020. agricolae: statistical procedures for agricultural research. R package version 1.3‐2. [Google Scholar]
- Moghe G, Last RL. 2015. Something old, something new: conserved enzymes and the evolution of novelty in plant specialized metabolism. Plant Physiology 169, 1512–1523. pp.00994.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naithani S, Gupta P, Preece J, D’Eustachio P, Elser JL, Garg P, Dikeman DA, Kiff J, Cook J, Olson A et al. 2019. Plant reactome: a knowledgebase and resource for comparative pathway analysis. Nucleic Acids Research 48: 1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál M, Tajti J, Szalai G, Peeva V, Végh B, Janda T. 2018. Interaction of polyamines, abscisic acid and proline under osmotic stress in the leaves of wheat plants. Scientific Reports 8: 12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters K, Worrich A, Weinhold A, Alka O, Balcke G, Birkemeyer C, Bruelheide H, Calf O, Dietz S, Dührkop K et al. 2018. Current challenges in plant eco‐metabolomics. International Journal of Molecular Sciences 19: 1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poorter H, Niinemets Ü, Ntagkas N, Siebenkäs A, Mäenpää M, Matsubara S, ThijsL P. 2019. A meta‐analysis of plant responses to light intensity for 70 traits ranging from molecules to whole plant performance. New Phytologist 223: 1073–1105. [DOI] [PubMed] [Google Scholar]
- Poorter H, Niklas KJ, Reich PB, Oleksyn J, Poot P, Mommer L. 2012. Biomass allocation to leaves, stems and roots: meta‐analyses of interspecific variation and environmental control: tansley review. New Phytologist 193: 30–50. [DOI] [PubMed] [Google Scholar]
- R Core Team . 2020. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Riehl S, Benz M, Conard NJ, Darabi H, Deckers K, Nashli HF, Zeidi‐Kulehparcheh M. 2012. Plant use in three pre‐pottery neolithic sites of the northern and eastern fertile Crescent: a preliminary report. Vegetation History and Archaeobotany 21: 95–106. [Google Scholar]
- Roch L, Prigent S, Klose H, Cakpo C‐B, Beauvoit B, Deborde C, Fouillen L, van Delft P, Jacob D, Usadel B et al. 2020. Biomass composition explains fruit relative growth rate and discriminates climacteric from non‐climacteric species. Journal of Experimental Botany 71: 5823–5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardans J, Gargallo‐Garriga A, Urban O, Klem K, Walker TWN, Holub P, Janssens IA, Peñuelas J. 2020. Ecometabolomics for a better understanding of plant responses and acclimation to abiotic factors linked to global change. Metabolites 10: 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scossa F, Fernie AR. 2020. The evolution of metabolism: how to test evolutionary hypotheses at the genomic level. Computational and Structural Biotechnology Journal 18: 482–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinbo Y, Nakamura Y, Altaf‐Ul‐Amin M, Asahi H, Kurokawa K, Arita M, Saito K, Ohta D, Shibata D, Kanaya S. 2006. II.6 KNApSAcK: a comprehensive species‐metabolite relationship database. Plant Metabolomics 17: 165–181. [Google Scholar]
- Signori‐Müller C, Oliveira RS, Barros FV, Tavares JV, Gilpin M, Diniz FC, Zevallos MJM, Yupayccana CAS, Acosta M, Bacca J et al. 2021. Non‐structural carbohydrates mediate seasonal water stress across Amazon forests. Nature Communications 12: 2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Maille GO, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. 2005. mETLIN: a metabolite mass spectral database. Therapeutic Drug Monitoring 27: 747–751. [DOI] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. 2006. Xcms: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical Chemistry 78: 779–787. [DOI] [PubMed] [Google Scholar]
- Sumner LW, Lei Z, Nikolau BJ, Saito K, Roessner U, Trengove R. 2014. Proposed quantitative and alphanumeric metabolite identification metrics. Metabolomics 10: 1047–1049. [Google Scholar]
- Szabados L, Savouré A. 2010. Proline: a multifunctional amino acid. Trends in Plant Science 15: 89–97. [DOI] [PubMed] [Google Scholar]
- Szymański J, Bocobza S, Panda S, Sonawane P, Cárdenas PD, Lashbrooke J, Kamble A, Shahaf N, Meir S, Bovy A et al. 2020. Analysis of wild tomato introgression lines elucidates the genetic basis of transcriptome and metabolome variation underlying fruit traits and pathogen response. Nature Genetics 52: 1111–1121. [DOI] [PubMed] [Google Scholar]
- Thalmann M, Santelia D. 2017. Starch as a determinant of plant fitness under abiotic stress. New Phytologist 214: 943–951. [DOI] [PubMed] [Google Scholar]
- Umair M, Sun N, Du H, Yuan J, Abbasi AM, Wen J, Yu W, Zhou J, Liu C. 2019. Differential metabolic responses of shrubs and grasses to water additions in arid karst region, southwestern China. Scientific Reports 9: 9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson CC, Mota APZ, Porto BN, Oliveira TN, Sampaio I, Lacerda AL, Danchin EGJ, Guimaraes PM, Williams TCR, Brasileiro ACM. 2020. Characterization of raffinose metabolism genes uncovers a wild Arachis galactinol synthase conferring tolerance to abiotic stresses. Scientific Reports 10: 15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. New York, NY, USA: Springer‐Verlag. [Google Scholar]
- Wong C, Ling YS, Wee JLS, Mujahid A, Müller M. 2020. A comparative UHPLC‐Q/TOF–MS‐based eco‐metabolomics approach reveals temperature adaptation of four Nepenthes species. Scientific Reports 10: 21861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Sinelnikov IV, Han B, Wishart DS. 2015. MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Research 43: W251–W257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Guijas C, Benton HP, Warth B, Siuzdak G. 2020. METLIN MS2 molecular standards database: a broad chemical and biological resource. Nature Methods 17: 953–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gu S, Zhao X, Cui X, Zhao L, Xu S, Du M, Jiang S, Gao Y, Ma C et al. 2010. Radiation partitioning and its relation to environmental factors above a meadow ecosystem on the Qinghai‐Tibetan Plateau. Journal of Geophysical Research 115: D10106. [Google Scholar]
- Zhu G, Wang S, Huang Z, Zhang S, Liao Q, Zhang C, Lin T, Qin M, Peng M, Yang C et al. 2018. Rewiring of the fruit metabolome in tomato breeding. Cell 172: 249–261.e12. [DOI] [PubMed] [Google Scholar]
- Ziaco E, Truettner C, Biondi F, Bullock S. 2018. Moisture‐driven xylogenesis in Pinus ponderosa from a Mojave Desert mountain reveals high phenological plasticity: moisture‐driven xylogenesis in Pinus ponderosa . Plant, Cell & Environment 41: 823–836. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Correlation between starch content and sampling time. Absence of correlation between the time of sampling and starch content in plant samples.
Fig. S2 Changes in major compounds in Atacama plants. (a) Depiction of the biochemical diversity observed in Atacama plants and 11 agronomic or ornamental species.
Fig. S3 Best metabolomics predictors in Atacama plants. (a) Predictive capacity and clustering of the best metabolic markers.
Fig. S4 Metabolic networks. Best markers were mapped into a preexisting A. thaliana metabolic network using MetExplore.
Fig. S5 Validation of the environmental prediction. Linearity of environmental parameters with elevation.
Fig. S6 Decomposition of the elevation parameter. Correlation plot of the environmental data.
Fig. S7 Best metabolic predictors in agronomic and ornamental plant species. Clustering analysis of the best metabolic predictors between Atacama and agronomic and ornamental plant species.
Table S1 (a) Sample data from Atacama plants. (b) Sample data from agronomic and ornamental plants.
Table S2 Metabolic dataset.
Table S3 Pathway analysis.
Table S4 Annotation table of markers.
Table S5 Markers in control plant. Intensity of the best metabolic markers in agronomic and ornamental plants. The detection limits were fixed to 25 000 or 10 000.
Table S6 Markers threshold 100. Intensity of the metabolic markers that could not be observed with a 10k noise threshold. Those markers were manually checked on one sample of each nonextreme species and one Atacama species with a threshold of 100.
Table S7 O2PLS model.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
The metabolic dataset and all metadata were deposited online using Dataverse INRAE (https://dx.doi.org/10.15454/UUBXIF) and following the Findable, Accessible, Interoperable, Reusable (FAIR) principles (Jacob et al., 2020).


