Abstract
The cell density-dependent acid sensitivity phenotypes of Escherichia coli strains K-12 and O157:H7 were examined with reference to three possible mechanisms of acid resistance. There was no evidence of any diffusible substance released from dead cells which could influence the cell density-dependent acid survival phenotype. Instead, cell density-dependent acid survival phenotype was associated with induction of glutamate- and arginine-decarboxylase acid survival pathways and concomitant availability of glutamate and arginine during acid challenge.
Because of their pathogenic and commensal lifestyle, an acidic environment is a common stress encountered by enteric bacteria such as Salmonella enterica and Escherichia coli. In order to survive such potentially lethal acid conditions, bacteria have evolved common as well as different strategies (10, 14). Several studies have described how enteric microorganisms cope with this form of environmental stress and referred to the acid survival systems as the acid tolerance response, acid resistance, and acid habituation (14, 17, 18, 20). Direct comparison of acid survival results among various groups (and microorganisms) has been difficult due to the use of complex versus minimal medium, log-phase versus stationary-phase cells, and acid challenge at various pHs (11, 14, 24). To add further complexity to the analysis, there appear to be substances secreted by cells that apparently influence acid sensitivity (22, 23). Among the various extracellular components which have been reported to influence acid tolerance, synthesis of some of the diffusible components from enterohemorrhagic E. coli strains appeared to depend on the synthesis of an alternative sigma transcription factor, rpoS (7). All the enterohemorrhagic E. coli strains and Shigella spp. examined reportedly synthesized the diffusible substances irrespective of their serotype or their ability to synthesize Shiga-like toxins. These substances were postulated to regulate cell density-dependent acid survival responses in a manner similar to N-acyl-l-homoserine lactones (7, 8, 16).
Few studies have directly examined the effect of cell density, acid pH, and growth conditions on survival of pathogenic E. coli strains. Earlier studies examined this relationship without considering the possibility that exposure to different growth conditions might influence the ultimate outcome of acid challenge (1, 2, 13). Recent analysis of the molecular aspects of acid tolerance pathways in E. coli has opened up new strategies capable of dissecting cell density-dependent acid sensitivity phenotypes (6, 12). Our aim was to identify putative diffusible substances involved in cell density-dependent acid sensitivity in E. coli, with the knowledge that multiple acid resistance systems may be involved.
Three pathways have been identified which enable E. coli to survive acid challenge. One is a glucose-repressible oxidative pathway regulated by the alternative sigma transcription factor rpoS, which is induced in cells grown on complex media as they enter the stationary growth phase. Once the oxidative system is active, how it protects cells during acid challenge remains a mystery. The rpoS-mediated system is not operative in fermentatively metabolizing cells (grown in complex medium containing glucose). Two other acid resistance systems are activated in cells under this growth condition, which attempt to alkalinize cytoplasmic pH and require the presence of amino acids during acid challenge (4). These two systems are known as the glutamate decarboxylase system (gadABC operon) (15) and the arginine decarboxylase system (adiA) (17). The glutamate decarboxylase pathway is also induced at somewhat reduced levels during aerobic growth as cultures enter the stationary growth phase.
In this study we have addressed the issue of diffusible substances released from E. coli strains during acid challenge (7, 23). Our data confirmed cell density-dependent acid survival in E. coli. However, no direct evidence for the occurrence of diffusible substances which could induce cell density-dependent acid sensitivity was obtained. On the contrary, the data indicated that the absence of certain amino acids or their limited availability during acid challenge gives a phenotype of cell density-dependent acid sensitivity.
Bacterial strains and culture conditions.
The E. coli and S. enterica serovar Typhimurium strains used in this study are listed in Table 1. Cultures were streaked on Luria-Bertani (LB) agar plates from freezer stocks, and a single colony was inoculated in LB broth. Cultures were inoculated in 10 ml of LB broth in a 125-ml flask, which was incubated on a shaker incubator at 37°C and 150 rpm for 18 to 20 h (oxidative growth). For fermentative growth, cultures were started from a single colony in LB broth containing 0.4% glucose (adjusted to pH 5.0) as described by Lin et al. (17). Briefly, 3 ml of broth was placed in a sterile tube (100 by 11 mm), which was placed at a 45° angle in a shaker incubator at 37°C at150 rpm. After incubation for 18 to 20 h, cultures had a pH of 4.5 to 4.7.
TABLE 1.
E. coli and S. enterica strains used in this study
| Strain | Serotype or genotypea | Reference |
|---|---|---|
| E. coli | ||
| MG1655 | K-12 wild type | 21 |
| EK274 | O157:H7 wild type (ATCC 43895), Nar Rfr | 19 |
| EK275 | EK274 rpoS::pRR10 Apr | 19 |
| EK484 | EK274 gadC::pRR10 Apr | S. B. Price and J. W. Foster, unpublished data |
| EK489 | EK274 adiA::pRR10 Apr | S. B. Price and J. W. Foster, unpublished data |
| S. enterica serovar Typhimurium | ATCC 14028s (wild type) | 9 |
Abbreviations for antibiotics: Na, nalidixic acid; Rf, rifampin; Ap, ampicillin.
Acid challenge assays.
Cultures grown for 18 to 20 h were centrifuged, washed once in sterile saline, and resuspended in saline at various cell densities. The washed cell suspensions were subjected to acid challenge in either acidified LB broth (pH 2.5, acidified with HCl) or synthetic gastric juice (2, 5) for 2 h at 37°C. Acid challenge was performed in the presence of glutamate or arginine at different concentrations where indicated. Cells were diluted in sterile phosphate-buffered saline before viable cells were counted.
To obtain used synthetic gastric juice, cell suspensions after acid challenge in fresh gastric juice (using ≥2 × 109 cells ml−1) were centrifuged at 10,000 × g, and the supernatant was passed through a 0.2-μm nylon filter. The filtered sterilized synthetic gastric juice was referred to as used gastric juice and either used immediately for acid challenge assay or stored at −20°C until use.
Acid challenge at various cell densities.
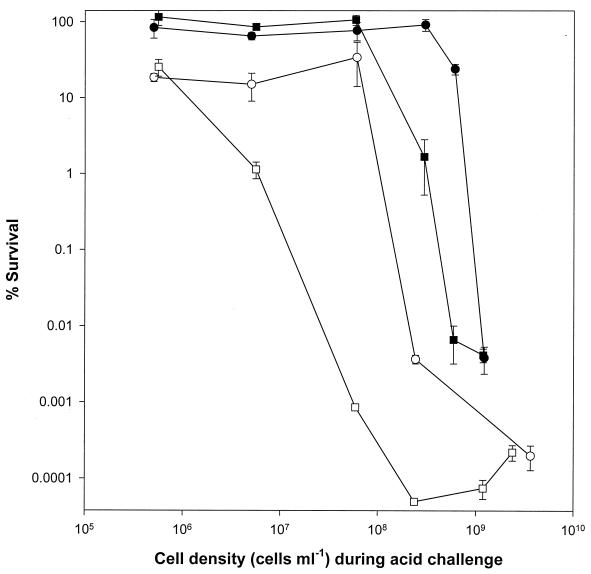
We designed experiments to examine the potential presence of a factor(s) associated with cell-to-cell communication, especially those which may be associated with cell density-dependent acid sensitivity. High (>2 × 109 cells ml−1) and low (<5 × 106 cells ml−1) cell density suspensions of E. coli O157:H7 and E. coli K-12 MG1665 were subjected to acid challenge in an acidified LB medium (pH 2.5, 37°C, 2 h). In order to facilitate subsequent purification of diffusible substances which may be released from dead cells (7, 23), experiments were also repeated in synthetic gastric juice under identical conditions. Both strains survived poorly under the test conditions of high cell density during acid challenge. However, the same cell preparations were resistant to an identical acid challenge at lower cell densities (Fig. 1). Next, we examined the properties of the acidic LB medium and synthetic gastric juice in which the acid challenge assays were performed. Cells challenged at both low and high cell densities survived poorly in the used synthetic gastric juice (Table 2). The failure of the low-cell-density population to survive in the used synthetic gastric juice was further examined to determine whether it was due to substances released from the dying high-cell-density population during acid challenge.
FIG. 1.
Effect of cell density during acid challenge on survival of E. coli strains. Strains MG1665 (○, ●) and EK274 (□, ▪) were acid challenged in fresh synthetic gastric juice (open symbols) or in acidified LB broth (solid symbols). Acid challenge was performed at pH 2.5 and 37°C for 2 h. Error bars represent the standard deviation (not shown if smaller than the symbol).
TABLE 2.
Survival of high- and low-density cell suspensions during acid challenge
| Strain | Cell density during acid challenge (cells ml−1) | Mean % survival ± SD in:
|
|
|---|---|---|---|
| Fresh medium (pH 2.5, 37°C, 2 h) | Used medium (pH 2.5, 37°C, 2 h) | ||
| E. coli K-12 | 2 × 109 | 0.0037 ± 0.0002 | 0.0051 ± 0.00004 |
| MG1655 (wild type) | 2 × 106 | 15.00 ± 6.01 | 0.0026 ± 0.0002 |
| E. coli O157:H7 EK274 | 2 × 109 | 0.0014 ± 0.00002 | ≤0.00001 |
| 2 × 106 | 25.5 ± 6.37 | ≤0.00001 | |
Acid challenge in the presence of substances released from dead cells.
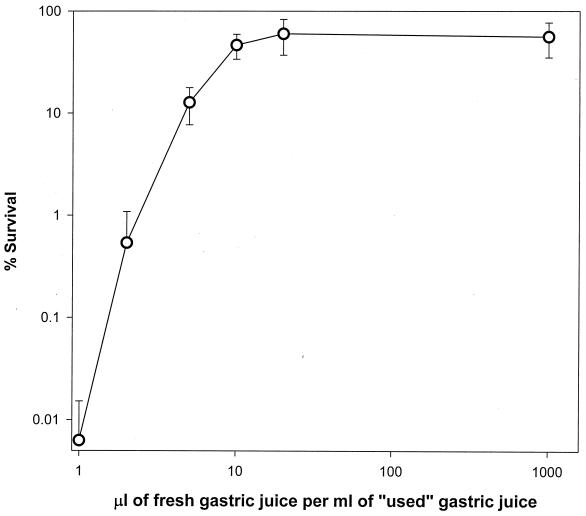
In order to obtain large quantities of the putative factor(s) at higher concentration, cell suspensions of very high cell density (ca. 109 to 1010 cells ml−1) were subjected to acid challenge in synthetic gastric juice. It was reasoned that killing large cell populations in small volumes would provide large quantities of crude diffusible substances that might be responsible for cell density-dependent acid sensitivity. We continued to observe poor survival rates (≤0.004% survival) at cell densities as high as 1010 cells of synthetic gastric juice per ml (data not shown). After filter sterilization, various dilutions of used synthetic gastric juice made in fresh synthetic gastric juice were tested for potency in acid challenge assays using low-cell-density suspensions (Fig. 2). It was observed that irrespective of the initial cell concentration used to obtain the used synthetic gastric juice (i.e., 108 cells ml−1 or 1010 cells ml−1), only undiluted preparations were effective in providing acid-mediated killing of cells at low cell density. In contrast, addition of small quantities of fresh synthetic gastric juice was sufficient to restore acid survival (Fig. 2), indicating that it may be the absence of a factor(s) in the spent synthetic gastric juice that is responsible for the low survival rate during acid challenge.
FIG. 2.
Survival of E. coli O157:H7 in used synthetic gastric juice supplemented with various quantities of fresh synthetic gastric juice. Acid challenge was performed at low cell density (2 × 106 cells ml−1). Error bars represent the standard deviation.
Role of individual acid survival pathways.
Of the three pathways by which E. coli cells survive acid challenge (6), i.e., oxidative pathway (rpoS mediated) and two amino acid decarboxylase pathways (mediated by glutamate decarboxylase [gadABC operon] and arginine decarboxylase [adiA]), the glutamate decarboxylase pathway is expressed under both aerobic and fermentative growth conditions, while arginine decarboxylase is induced strictly under fermentative growth. Using strains carrying mutations in each of the three acid survival pathways, it was determined whether cell density-dependent acid sensitivity is due to limited availability of an amino acid(s) during acid challenge.
Effect of RpoS on cell density-dependent acid sensitivity.
The oxidative acid resistance pathway is fully expressed during the stationary growth phase of aerobically grown cells and requires the rpoS gene product, an alternative ς-factor for transcription. E. coli strains defective in rpoS were shown previously to be independent of cell density-dependent killing during acid challenge (7). We tested an rpoS mutant of E. coli O157:H7, EK275, grown under oxidative and fermentative growth conditions. The rpoS mutant grown aerobically was extremely sensitive to acid, and acid sensitivity was independent of cell density during acid challenge (Table 3). The likely reason for this is that the rpoS mutant, under aerobic growth conditions, does not synthesize glutamate decarboxylase (6) or use the glutamic acid-dependent acid survival pathway. As a consequence, the strain has no acid resistance mechanisms that are operative during aerobic growth. This is reflected in the strain's extremely acid-sensitive phenotype.
TABLE 3.
Effect of oxidative and fermentative growth on utilization of three acid tolerance pathways in high- and low-density cultures of E. coli O157:H7
| Strain | Growth conditions | Mean % survival ± SD in fresh acid challenge medium at starting cell density (cells ml−1):
|
|
|---|---|---|---|
| 2 × 106 | 2 × 109 | ||
| EK274 (wild type) | Oxidative | 62.0 ± 3.6 | 0.00084 ± 0.0001 |
| Fermentative | 91.0 ± 14.7 | 0.46 ± 0.07 | |
| EK275 (rpoS::pRR10) | Oxidative | ≤0.0001 | ≤0.0001 |
| Fermentative | 15.0 ± 2.6 | 0.25 ± 0.04 | |
| EK484 (gadC::pRR10) | Oxidative | 0.13 ± 0.003 | 0.09 ± 0.024 |
| Fermentative | 100.0 ± 9.3 | 0.51 ± 0.13 | |
| EK489 (adiA::pRR10) | Oxidative | 94.0 ± 9.1 | 0.0009 ± 0.0002 |
| Fermentative | 66.0 ± 21.3 | 0.14 ± 0.06 | |
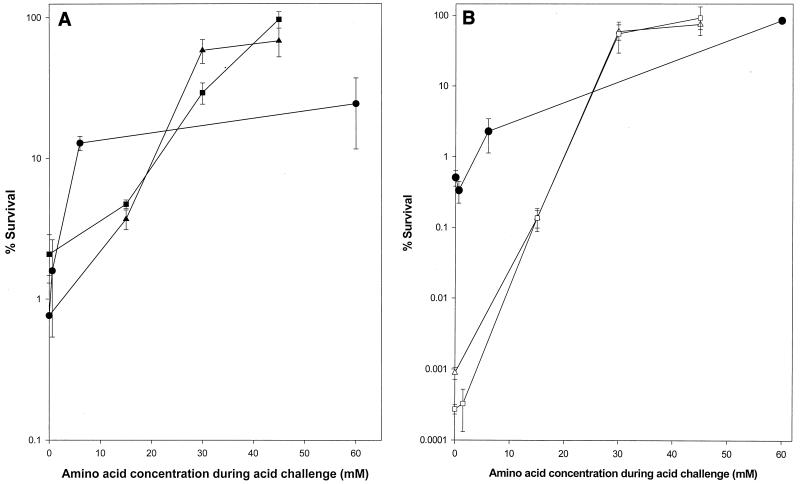
Activation of the glutamate decarboxylase acid tolerance pathway is complex. It requires functional RpoS as cells enter the stationary growth phase under aerobic conditions, but its activation is RpoS independent under fermentative growth conditions in the presence of glucose (6). We took advantage of the dual regulatory aspects of the glutamate decarboxylase pathway by examining cell density-dependent acid sensitivity of the rpoS mutant grown fermentatively. Wild-type cells survived in a cell density-dependent manner irrespective of aerobic or fermentative growth conditions. The rpoS mutant strain grown on LB-glucose did exhibit the cell density-dependent acid-sensitive phenotype (Table 3). This phenotype was most likely due to limited quantities of available glutamate and arginine in the acidified LB medium. Based on the information found at the Organotechnie web site (www.organotechnie.com), the estimated concentrations of free glutamate and arginine in the LB medium (at 10 g of tryptone and 5 g of yeast extract liter−1) are 2.58 mM and 0.55 mM, respectively. The free glutamate levels in the synthetic gastric juice (at 8.3 g of peptone liter−1) are much lower (0.28 mM), while free arginine is estimated to be 1.29 mM. Addition of glutamate and arginine rescued the rpoS mutant during acid challenge (Fig. 3A). Acid survival in these experiments was dependent on cell density and the concentration of glutamate and arginine during acid challenge. No synergistic protection was observed using a combination of arginine and glutamate (data not shown).
FIG. 3.
Effect of rpoS, gadC, and adiA mutations and amino acid availability during acid challenge on cell survival. Cells were grown aerobically (open symbols) or fermentatively (solid symbols) and subjected to acid challenge at high cell density (2 × 109 cells ml−1). Glutamate (▪, □, ▴, ▵) or arginine (●) was added at the indicated concentration to the acid challenge assay. (A) E. coli O157:H7 strains EK275 (●, ▴) and EK274 (▪). (B) E. coli O157:H7 strains EK 274 (□), EK484 (●), and EK489 (▵). Error bars represent standard deviation (not shown if smaller than the symbol).
Role of glutamate- and arginine-dependent acid survival pathways in cell density-dependent acid sensitivity.
We further confirmed that limited availability of glutamate (and arginine) causes the cell density-dependent phenotype by using gadC and adiA mutant strains. The cell density-dependent acid sensitivity in a gadC mutant (strain EK484, defective in glutamate:γ-aminobutyric acid antiporter) was examined. Strain EK484 was grown under two different culture conditions, aerobic growth in LB medium and fermentative growth in LB-glucose medium. The cells were subjected to acid challenge in the acidified LB medium at high and low cell densities (Table 3). The survival of strain EK484 grown aerobically was independent of cell density, while cells obtained after fermentative growth on LB-glucose broth did exhibit cell density-dependent acid sensitivity. The acid sensitivity of strain EK489, defective in arginine decarboxylase, was cell density dependent irrespective of aerobic or fermentative growth. In this strain, the glutamate decarboxylase pathway is expected to be functional under aerobic as well as fermentative growth conditions. It was examined whether aerobically grown cells of strain EK489 could be rescued during acid challenge by addition of glutamate (Fig. 3). The arginine decarboxylase pathway is expected to be induced in strain EK484 when it is grown on LB-glucose. Thus, we determined if arginine could be the limiting factor during acid challenge for strain EK484 cells that were grown on LB-glucose (Fig. 3B). Availability of arginine and glutamate helped cells of strains EK484 and EK489 to overcome acid challenge, and there was a clear dose-response relationship between available arginine or glutamate, cell density, and cell survival (Fig. 3A and B). This indicated that arginine and glutamate are probably the limiting components in the acidified LB medium responsible for cell density-dependent acid sensitivity.
In addition to the defect in the glutamate decarboxylase pathway, aerobically grown gadC mutant cells do not utilize the arginine decarboxylase pathway (17). Thus, the aerobically grown gadC mutant has only an rpoS-mediated oxidative acid survival pathway that is functional when challenged in acidified LB medium. Since the strain did not exhibit cell density-dependent acid sensitivity, the rpoS-mediated acid survival pathway appears to be independent of glutamate and arginine availability during acid challenge. S. enterica serovar Typhimurium cells do not possess glutamate or arginine decarboxylase-mediated acid resistance mechanisms (3), and we confirmed the absence of glutamate- or arginine-dependent acid sensitivity in this organism (data not shown). Contrary to the previous report (7), the wild-type E. coli strain K-12 MG1665, which possessed all three acid resistance pathways, like several other O157:H7 strains, showed cell density-dependent acid sensitivity.
In summary, the data revealed that when acid challenge assays are performed at high cell density, the limited availability of glutamate and/or arginine creates the illusion of an involvement of cell-to-cell signaling or quorum sensing-type phenomena due to the observed cell density-dependent acid survival.
Acknowledgments
We thank J. F. Foster for sharing unpublished strains and D. W. Bauer, K. C. Gross, J. McEvoy, and M. Wachtel for critical reading of the manuscript.
REFERENCES
- 1.Abdul-Raouf U M, Beuchat L R, Ammar M S. Survival and growth of Escherichia coli O157:H7 in ground, roasted beef as affected by pH, acidulants, and temperature. Appl Environ Microbiol. 1993;59:2364–2368. doi: 10.1128/aem.59.8.2364-2368.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold K W, Kaspar C W. Starvation- and stationary-phase-induced acid tolerance in Escherichia coli O157:H7. Appl Environ Microbiol. 1995;61:2037–2039. doi: 10.1128/aem.61.5.2037-2039.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bang I S, Kim B H, Foster J W, Park Y K. OmpR regulates the stationary-phase acid tolerance response of Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182:2245–2252. doi: 10.1128/jb.182.8.2245-2252.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bearson S, Bearson B, Foster J W. Acid stress responses in enterobacteria. FEMS Microbiol Lett. 1997;147:173–180. doi: 10.1111/j.1574-6968.1997.tb10238.x. [DOI] [PubMed] [Google Scholar]
- 5.Beumer R R, de Vries J, Rombouts F M. Campylobacter jejuni non-culturable coccoid cells. Int J Food Microbiol. 1992;15:153–163. doi: 10.1016/0168-1605(92)90144-r. [DOI] [PubMed] [Google Scholar]
- 6.Castanie-Cornet M P, Penfound T A, Smith D, Elliott J F, Foster J W. Control of acid resistance in Escherichia coli. J Bacteriol. 1999;181:3525–3535. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datta A R, Benjamin M M. Cell density dependent acid sensitivity in stationary phase cultures of enterohemorrhagic Escherichia coli O157:H7. FEMS Microbiol Lett. 1999;181:289–295. doi: 10.1111/j.1574-6968.1999.tb08857.x. [DOI] [PubMed] [Google Scholar]
- 8.de Kievit T R, Iglewski B H. Bacterial quorum sensing in pathogenic relationships. Infect Immun. 2000;68:4839–4849. doi: 10.1128/iai.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang F C, Libby S J, Buchmeier N A, Loewen P C, Switala J, Harwood J, Guiney D G. The alternative sigma factor KatF (RpoS) regulates Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster J W. When protons attack: microbial strategies of acid adaptation. Curr Opin Microbiol. 1999;2:170–174. doi: 10.1016/S1369-5274(99)80030-7. [DOI] [PubMed] [Google Scholar]
- 11.Foster J W, Hall H K. Inducible pH homeostasis and the acid tolerance response of Salmonella typhimurium. J Bacteriol. 1991;173:5129–5135. doi: 10.1128/jb.173.16.5129-5135.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster J W, Moreno M. Inducible acid tolerance mechanisms in enteric bacteria. Novartis Found Symp. 1999;221:55–69. doi: 10.1002/9780470515631.ch5. [DOI] [PubMed] [Google Scholar]
- 13.Glass K A, Loeffelholz J M, Ford J P, Doyle M P. Fate of Escherichia coli O157:H7 as affected by pH or sodium chloride and in fermented, dry sausage. Appl Environ Microbiol. 1992;58:2513–2516. doi: 10.1128/aem.58.8.2513-2516.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorden J, Small P L. Acid resistance in enteric bacteria. Infect Immun. 1993;61:364–367. doi: 10.1128/iai.61.1.364-367.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hersh B M, Farooq F T, Barstad D N, Blankenhorn D L, Slonczewski J L. A glutamate-dependent acid resistance gene in Escherichia coli. J Bacteriol. 1996;178:3978–3981. doi: 10.1128/jb.178.13.3978-3981.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hussain N H, Goodson M, Rowbury R J. Recent advances in biology: intercellular communication and quorum sensing in microorganisms. Sci Prog. 1998;81:69–80. [PubMed] [Google Scholar]
- 17.Lin J, Lee I S, Frey J, Slonczewski J L, Foster J W. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol. 1995;177:4097–4104. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Hara G W, Glenn A R. The adaptive acid tolerance response in root nodule bacteria and Escherichia coli. Arch Microbiol. 1994;161:286–292. doi: 10.1007/BF00303582. [DOI] [PubMed] [Google Scholar]
- 19.Price S B, Cheng C M, Kaspar C W, Wright J C, DeGraves F J, Penfound T A, Castanie-Cornet M P, Foster J W. Role of RpoS in acid resistance and fecal shedding of Escherichia coli O157:H7. Appl Environ Microbiol. 2000;66:632–637. doi: 10.1128/aem.66.2.632-637.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raja N, Goodson M, Chui W C, Smith D G, Rowbury R J. Habituation to acid in Escherichia coli: conditions for habituation and its effects on plasmid transfer. J Appl Bacteriol. 1991;70:59–65. doi: 10.1111/j.1365-2672.1991.tb03787.x. [DOI] [PubMed] [Google Scholar]
- 21.Rao N N, Liu S, Kornberg A. Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J Bacteriol. 1998;180:2186–2193. doi: 10.1128/jb.180.8.2186-2193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rowbury R J. Acid tolerance induced by metabolites and secreted proteins, and how tolerance can be counteracted. Novartis Found Symp. 1999;221:93–106. doi: 10.1002/9780470515631.ch7. [DOI] [PubMed] [Google Scholar]
- 23.Rowbury R J. Killed cultures of Escherichia coli can protect living organisms from acid stress. Microbiology. 2000;146:1759–1760. doi: 10.1099/00221287-146-8-1759. [DOI] [PubMed] [Google Scholar]
- 24.Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski J L. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]