Abstract
Genomes of cyanobacteria feature a variety of cryptic biosynthetic pathways for complex natural products, but the peculiarities limiting the discovery and exploitation of the metabolic dark matter are not well understood. Here we describe the discovery of two cell density‐dependent chemical mediators, nostoclide and nostovalerolactone, in the symbiotic model strain Nostoc punctiforme, and demonstrate their pronounced impact on the regulation of specialized metabolism. Through transcriptional, bioinformatic and labeling studies we assigned two adjacent biosynthetic gene clusters to the biosynthesis of the two polyketide mediators. Our findings provide insight into the orchestration of specialized metabolite production and give lessons for the genomic mining and high‐titer production of cyanobacterial bioactive compounds.
Keywords: Biosynthesis, Cyanobacteria, Genomic Mining, Quorum Sensing, Specialized Metabolism
Nostovalerolactones are a novel class of cyanobacterial natural products that are derived from a tetronate‐like biosynthesis pathway and team up with nostoclides to orchestrate the specialized metabolism in a symbiotic cyanobacterium. The data show how an understanding of regulation can be used to achieve high‐titer production of bioactive specialized metabolites in cyanobacteria.
Introduction
Cyanobacteria are oxygenic phototrophic bacteria that flourish in a wide range of aquatic and terrestrial habitats. These phototrophic prokaryotes are of great significance for global carbon and nitrogen fixation, but they are also notorious for the production of a range of potent toxins, thereby posing a serious risk to humans and animals. [1] On the other hand, cyanobacteria produce a wealth of bioactive molecules with potential for pharmaceutical development featuring intricate structures as well as unprecedented biosynthetic enzymes. [2] Yet, the full biosynthetic potential revealed by genome sequencing programs of cyanobacteria is still largely untapped. [3] Although there are diverse examples for the genome‐based discovery of novel families of natural products from cyanobacteria and their heterologous production, concepts for the exploitation of specialized metabolites from cyanobacteria are in their infancy.
A major reason for the lack of fundamental studies is the fact that the majority of cyanobacteria harboring a multitude of orphan secondary metabolite biosynthetic gene clusters (BGCs) are non‐model organisms. Genomic surveys have revealed that a high content in BGCs encoding nonribosomal peptide synthetases (NRPS), polyketide synthases (PKS) and ribosomally produced and posttranslationally modified peptides (RiPPs) is primarily found in late‐branching lineages of cyanobacteria that frequently show either a transient or permanent multicellularity. [3] Facultative symbiotic cyanobacteria of the genus Nostoc are among these versatile genera and show a multitude of chemical interactions with plant hosts and other microorganisms. N. punctiforme PCC 73102 is the only strain that currently combines an accessibility to genetic manipulation with a large number of unexplored BGCs. While the antiSMASH [4] platform (version 5.2) identifies 21 BGC regions, only five of the pathways could be assigned to a product, namely geosmin, heterocyst glycolipids, anabaenopeptins, nostopeptolides and microviridins.[ 5 , 6 , 7 ]
A role of cell density‐dependent factors in the control of BGC expression in N. punctiforme was suggested by two recent studies. Time‐series analyses of N. punctiforme grown on plates using RNA sequencing revealed a pronounced upregulation of several BGCs at high cell densities. [5] High‐density cultivation of N. punctiforme in liquid culture also led to an enhanced transcription of more than 50 % of BGCs and facilitated the discovery of novel anabaenopeptin and microviridin variants.[ 5 , 7 ] A systematic dissection of abiotic and biotic factors contributing to the increased expression using a transcriptional reporter library pointed to interconnected effects of high light conditions, high CO2 partial pressure and extracellular signaling factors on the control of the specialized metabolism in N. punctiforme. [5] Yet, the nature of the signaling molecules triggering the cell density effects in N. punctiforme is currently unknown. In heterotrophic microorganisms, cell density effects are commonly conveyed by quorum sensing (QS) mediators. Different bacterial phyla have evolved a diverse chemical language for intra‐ and interspecies communication, including acylhomoserine lactones (AHLs), dialkylresorcinols (DARs) and γ‐butyrolactones (GBLs). The diffusible signals show an autoinduction and thereby control a QS regulon that frequently includes BGCs as major target genes. The specialized metabolism in the prolific model strain Streptomyces coelicolor, for example, is largely controlled by signals of the γ‐butyrolactone family (GBLs).[ 8 , 9 ] Understanding the QS regulons can inspire the design of genetic switches and contribute to the engineering of strains and populations. [10] Although the production of N‐octanoyl‐homoserine lactone and (+)‐α(S)‐butyramido‐γ‐butyrolactone have been reported for the cyanobacterial strains Gloeothece sp. PCC 6909 and Lyngbya majuscula, respectively, [11] QS mediators in cyanobacteria are largely unknown. No study has so far elucidated a QS network in cyanobacteria or connected this type of signal to the specialized metabolism.
Here, we have combined a reporter‐based and bioactivity‐guided approach with the overexpression of BGC regulator genes to elucidate cell density‐dependent signaling factors of N. punctiforme PCC 73102. Our study not only reveals for the first time the influence of cell density‐dependent mediators for the specialized metabolism in cyanobacteria but highlights the discovery of the first tetronate biosynthesis pathway in cyanobacteria. Further, we demonstrate a method that can be used for the pathway‐specific high‐titer production of target metabolites in native cyanobacterial hosts. Our study provides lessons for the development of cyanobacteria into sustainable production systems.
Results and Discussion
We started our study with two hypotheses: 1) a putative QS mediator or the underlying biosynthetic genes show cell density‐dependent expression, and 2) a putative QS mediator is the product of one of the 16 orphan BGCs in N. punctiforme. Hence, we took a closer look at cell density‐dependent BGCs in N. punctiforme. Amongst several candidates, the polyketide synthase gene cluster pks1 that is silent under conventional growth conditions but highly transcribed under high‐density conditions was the most promising candidate. In contrast to other BGCs, the cell density‐dependent induction of pks1 transcription was observed independent of light and CO2 conditions and was also evident in time‐course experiments on plates (Figure 1A). [5] Further, transcription of pks1 could be induced by the addition of high‐density culture supernatant under conventional growth conditions. [5]
Figure 1.

Cell‐density dependent expression of the pks1 BGC, and establishment of an AraC_PKS1 overexpression mutant. A) Expression values of a representative biosynthetic core gene of the BGC pks1 (RS10485) from a 9 d time‐course experiment performed on filters grown on diazotrophic agar plates. [5] The baseline level estimated for constitutively expressed BGCs is indicated by the dashed line. TPM: transcripts per million. B) Schematic representation of the pks1 BGC. C) Schematic overview of the overexpression plasmid containing the potential pathway‐specific regulatory gene RS10525 from the pks1 BGC, a strong promoter region from N. punctiforme (5′ region of RS16340) and a streptomycin resistance cassette for selection. D) Transcription level of the potential pathway‐specific regulator AraC_PKS1 (RS10525) in the AraC_PKS1 mutant strain compared to the transcription level in the WT strain. E) HPLC profiles of cell pellets and culture supernatants from AraC_PKS1 mutant strain and WT under conventional and HL/HC cultivation conditions. HPLC profiles are shown at a wavelength of 199 nm. Known metabolite peaks are labeled (Mvd: microviridin, Nos1052: nostopeptolide 1052, NosA: nostopeptolide A, Apt: nostamide A).
In addition to putative biosynthetic enzymes, the pks1 BGC encodes a transcription factor (RS10525) of the AraC‐type (Figure 1B). A bioinformatic survey for related AraC‐type transcription factors revealed the presence of close homologues in a number of cyanobacterial genomes, but not outside the cyanobacterial phylum. The majority of these AraC homologues are encoded in the proximity of BGCs including RiPP as well as NRPS gene clusters (Figure S1).
As AraC‐type regulators commonly act as positive regulators, [12] we designed an N. punctiforme overexpression mutant with the aim to constitutively upregulate the pks1 BGC and to identify the pks1 products. N. punctiforme was therefore transformed with an autonomously replicating plasmid carrying the AraC_PKS1 encoding gene fused to a strong N. punctiforme promoter region identified in a previous study (5′ region of RS16340 [5] ) (Figure 1C). Transcriptional analysis of the resulting AraC_PKS1 mutant showed that the respective regulatory gene is strongly overexpressed when compared to the transcription of the wild‐type (WT) strain under conventional growth conditions (Figure 1D).
The comparison of metabolite profiles of the WT and the AraC_PKS1 overexpression mutant under conventional growth conditions and under high light and high CO2 conditions (hereafter HL/HC conditions) revealed remarkable differences. Both strains show a pronounced stimulation of metabolite production under HL/HC conditions as reported earlier. [7] The HPLC profiles of both the cell pellet and the supernatant of the AraC_PKS1 strain, however, showed a considerable number of additional peaks (Figure 1E). These findings indicate that AraC_PKS1 overexpression leads to further stimulation of the specialized metabolism in N. punctiforme. To test this assumption, BGC transcription of both strains was compared by RT‐qPCR. As expected, the AraC_PKS1 overexpression strongly induces the transcription of the pks1 BGC (Figure S2). Moreover, a considerable number of BGCs are upregulated under conventional growth conditions in the AraC_PKS1 overexpression strain. The most pronounced upregulation was observed for the cryptic lanthipeptide BGCs ripp3 and ripp4 (Figure S2). These data support the hypothesis that the pks1 product might act as a global chemical mediator and that additional BGCs might represent putative target genes within a global signaling network.
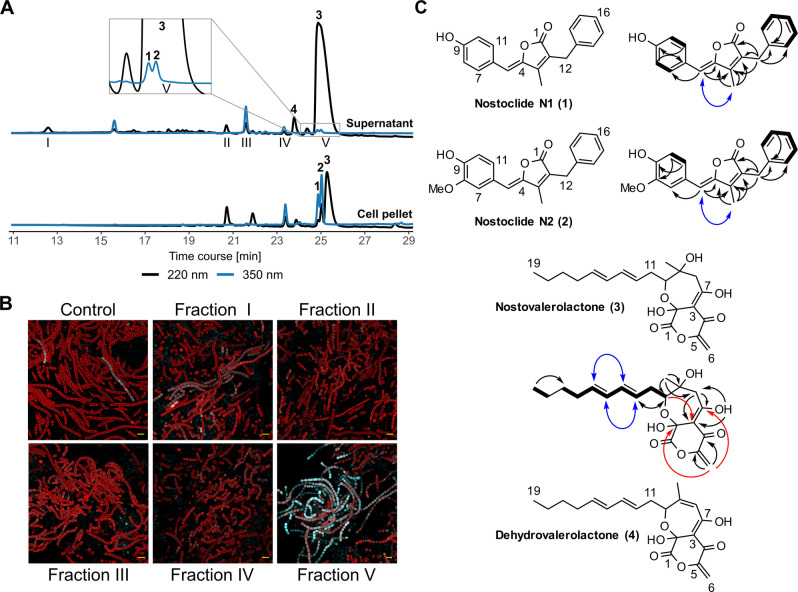
As we anticipated that the cryptic ripp4 lanthipeptide BGC might respond to a pks1‐derived diffusible signaling compound, we designed a bioactivity‐guided search for the pks1 product utilizing the ripp4 transcriptional reporter strain. Therefore, major metabolites accumulating in the supernatant of the AraC_PKS1 strain were purified and added to a freshly inoculated culture of the reporter strain. We found that only the hydrophobic fraction V (Figure 2A) is able to strongly induce ripp4 expression after 24 h of cultivation (Figure 2B).
Figure 2.
Identification of bioactive metabolites from AraC_PKS1 strain. A) HPLC profiles of the extracts of culture supernatant and cells from AraC_PKS1 grown under HL/HC cultivation condition for 20 d. Several yet uncharacterized metabolite peaks are visible in the mutant strain. Main supernatant peaks were fractionated and labelled with I–V as indicated below the chromatogram. HPLC profiles are shown at the wavelengths of 220 nm (black) and 350 nm (light blue). B) Fluorescence micrographs of ripp4 reporter strain treated with fractions I–V and of an untreated ripp4 reporter control culture. All cultures were grown conventionally for 24 h. The reporter strain shows a strongly increased expression upon treatment with fraction V. CFP fluorescence signal (blue) indicating promoter activity of the ripp4 BGC. Chlorophyll‐a autofluorescence (red) indicating living vegetative cells. Scale bar (yellow)=10 μm. C) Obtained structures of new compounds, nostoclide N1 (1), N2 (2), and nostovalerolactone (3) from fraction V, and a minor congener 9‐dehydronostovalerolactone (4). These structures were elucidated by a combination of MS and NMR analyses. Selected 1H‐1H COSY (black bold line), 2,3 J‐HMBC (black arrow), 4,5 J‐HMBC (red arrow), and NOESY (blue arrow) correlations are shown.
These findings indicated that one or more hydrophobic metabolites, likely the products of the pks1 pathway, act as inducer(s) for the ripp4 BGC. Mass spectrometric analysis of the active fraction showed the presence of three potential pks1 metabolites (1–3, Figure 2C), which were isolated from an upscaled fermentation broth and purified by preparative HPLC. As compounds 1 and 2 predominantly accumulate in the cell pellet (Figure 2A), both the supernatants and the cell pellets were included in the upscaling process. Their structures were elucidated by a combination of HR‐MS and one‐ and two‐dimensional NMR analysis (Figure S3–S51, Tables S1–S6).
To get further insight into the impact of the AraC_PKS1 overexpression on the transcription of the pks1 BGC and the global specialized metabolism we performed comparative RNA sequencing studies for the WT and the AraC_PKS1 strain under conventional and HL/HC conditions. While in the WT strain the pks1 BGC is only actively transcribed under HL/HC conditions, its transcription is strongly enhanced in the AraC_PKS1 strain under both conventional growth conditions and HL/HC conditions (Figure 3). Concomitantly, the accumulation of nostovalerolactone and nostoclide was compared for both strains using HPLC. In agreement with the transcriptional data, the production level of both substances is greatly increased in the AraC_PKS1 strain, especially under HL/HC conditions (Figure 3). Yet, the production level of both compounds is low under conventional conditions even after constitutive upregulation of the AraC_PKS1 regulator (Table S8). These data suggest that the upregulation of transcription alone is not sufficient for the high‐titer production of the two novel compounds but likely depends on a better supply of precursors under HL/HC conditions. Further inspection of the expression of secondary metabolite BGCs revealed a pronounced upregulation of a number of additional BGCs in the AraC_PKS1 strain under HL/HC conditions including the characterized BGCs for nostopeptolide, anabaenopeptin, microviridin and orphan BGCs such as the polyketide BGCs pks2 and pks3 and the lanthipeptide BGCs ripp3 and ripp4 (Figure S52 and Dataset S1). Together with the pks1 BGCs thus eight of the secondary metabolite BGCs predicted by AntiSMASH are upregulated in the AraC_PKS1 strain.
Figure 3.

Production of nostoclides and nostovalerolactone in N. punctiforme WT and the AraC_PKS1 mutant strain. HPLC profiles are shown at a wavelength of 350 nm. Culture supernatant (3) and cell pellets (1 and 2) were used for HPLC analysis. The heatmaps visualize the TPMs (transcripts per million) for each gene of the pks1 BGC.
The strict dependency of nostovalerolactone and nostoclide production on the expression of the regulator gene AraC_PKS1 suggests an involvement of the pks1 BGC in the biosynthesis of both types of compounds. This is matched by the fact that the pks1 cluster can be split into two subclusters (Figure 4A) by comparison with other characterized or predicted BGCs. Notably, many gene products of the first subcluster (nvl) share homology to enzymes from well‐characterized tetronate biosynthetic pathways (Table S7). [13] Given that the nostovalerolactones and linear tetronate antibiotics like agglomerin A have various structural features in common, like a linear hydrocarbon tail, a terminal lactone ring, and an exocyclic methylene group, an involvement of the first subcluster in nostovalerolactone biosynthesis seems plausible. Based on the homology on the structural and genetic levels a model for the biosynthesis of nostovalerolactones is proposed (Figure 4B). Accordingly, the biosynthesis would start with the activation of octanoic acid by the fatty acyl‐AMP ligase (FAAL) domain of NvlI. Next, the fatty acid moiety would be oxidized by an adjacent acyl‐CoA dehydrogenase (ACAD) domain and then elongated by four extension modules of the type I polyketide synthases NvlJ, K, L and M. Interestingly, double bond formation seems to occur in β,γ‐position, which is rare in polyketide biosynthesis. [14] After fusion of an ACP‐bound glyceryl moiety with the linear PKS intermediate, the bicyclic ring system would be formed with help of the FabH‐like 3‐oxoacyl‐ACP synthase III protein NvlD. The glyceryl residue in tetronate biosynthetic pathways is commonly derived from 1,3‐bisphosphoglycerate, which is derived from glycolysis and transferred to an ACP by a glyceryl‐S‐ACP synthase. [13] However, addition of 13C‐labeled glucose or 13C‐glycerol to the cultivation medium did not yield 13C‐labeled nostovalerolactone (Figure S47–50, Table S4). In addition, the production of nostovalerolactone was strongly reduced after glucose feeding (Figure S46). In contrast to that, nostovalerolactone production was highly stimulated by HL/HC conditions (Figure 3). Thus, 1,3‐bisphosphoglycerate or a related C3 precursor, which is transferred to the ACP NvlF by the glyceryl‐S‐ACP synthase NvlE may be derived from the Calvin‐Benson cycle, which is highly active under HL/HC conditions but downregulated under mixotrophic growth conditions in cyanobacteria. [15] NvlD, E, and F are homologs of RkD, E, and F, which facilitate tetronate ring formation in the biosynthesis of the protein phosphatase inhibitor RK‐682. [16]
Figure 4.
Assignment of nostovalerolactones and nostoclides to the pks1 BGC. A) The pks1 BGC can be split into the nostovalerolactone BGC (nvl) and the nostoclide subcluster (ncl) based on sequence homology to characterized BGCs of structurally related tetronate antibiotics, [13] as well as the BGC of cyanobacterin. [20] B) Model for the biosynthesis of 9‐dehydronostovalerolactone (4) and nostovalerolactone (3) considering the results of labeling studies with 1‐13C acetate and 1,2‐13C2 acetate (Figure S46–S51, Table S4). The proposed biosynthesis shares many parallels with the biosynthesis of tetronate antibiotics, [13] but also comprises a non‐canonical cyclization sequence together with an oxidative rearrangement reaction to yield nostovalerolactone's unprecedented bicyclic ring system as evidenced by the unconventional labeling pattern of carbon atoms C7 and C3 of 3 and the increased number of oxygen atoms in the first proposed cyclization product (O7) compared to its linear precursors (O3+O3). FAAL: fatty acyl‐AMP ligase, ACAD: acyl‐CoA dehydrogenase, ACP: acyl carrier protein, KS: ketoacylsynthase, AT: acyl transferase, KR: ketoreductase, DH: dehydratase, MT: C‐methyltransferase.
Notably, cyclization in nostovalerolactone biosynthesis does not yield a tetronate ring but a highly unusual δ‐valerolactone ring instead, which is fused to an oxepane‐like unit. To gain first insights into this unusual cyclization sequence we performed labeling studies with 1‐13C‐ and 1,2‐13C2‐labeled acetate (Figure S46–S51, Table S4). Intriguingly, the unconventional labeling pattern of carbon atoms C7 and C3 of nostovalerolactone indicates a rearrangement reaction in the course of cyclization (Figure 4B). In addition, ring formation seems to be accompanied by an oxygenation. This is evident from the increased number of oxygen atoms in the first proposed cyclization product (O7) compared to its linear precursors (O3+O3) (Figure 4B). Possible candidates for an involvement in the oxidative rearrangement would be the putative Baeyer–Villiger monooxygenase (BVMO) NvlQ, [17] which shares similarity to a group of oxygenases catalyzing Baeyer–Villiger oxidations in various trans‐AT PKS pathways as well as NvlH, a homolog of the BVMO MtmOIV, which catalyzes oxidative ring cleavage in the biosynthesis of mithramycin. [18] However, the mechanistic details and order of the rearrangement and cyclization reactions, as well as putative enzymes involved remain to be investigated.
After the formation of the bicyclic ring system, dehydration to yield the exocyclic double bond is proposed to be facilitated in two steps: First, O‐acetylation is catalyzed by NvlG followed by elimination of acetic acid catalyzed by NvlN to yield compound 4. This enzyme couple is homologous to Agg4 and Agg5, which catalyze the analogous acetylation–elimination reaction in the biosynthesis of the acyltetronate antibiotic agglomerin. [19] Finally, hydration of 4 yields nostovalerolactone.
The second subcluster adjacent to the proposed nostovalerolactone BGC (ncl) has pronounced similarity to the cyanobacterin BGC that was recently elucidated using both heterologous expression in E. coli and in vitro reconstitution. [20] Further support for the involvement of this subcluster in nostoclide biosynthesis comes from the biochemical and structural characterization of the gene product of RS10365 (NclD in our nomenclature; Figure 4A). This enzyme has been shown to be a phenylalanine ammonia lyase (PAL) that converts phenylalanine to cinnamic acid, [21] which is an essential building block in the biosynthesis of related furanolides. [20]
As nostoclide and nostovalerolactone are demonstrably able to induce the promoter activity of the ripp4 BGC (Figure 2), we were considering the possibility that the two metabolites might also directly stimulate the transcription of other BGCs which are upregulated in the AraC_PKS1 strain. To test this possibility, we performed an additional series of RNA sequencing studies comparing the transcriptome of the WT strain grown under conventional conditions and treated with either nostoclide N1 and N2, nostovalerolactone or a combination of both types of compounds for 24 h (Figure 5). Seven of the secondary metabolite BGCs are directly responding to the treatment with either of the compounds (nostopeptolide BGC, anabaenopeptin BGC, microviridin BGC, ripp3 and ripp4 lanthipeptide BGCs) or to the combined treatment with both compounds (pks3 and pks4 polyketide BGCs) (Figure 5). Strikingly, neither nostoclide nor nostovalerolactone or a combination thereof could induce the transcription of the pks1 subclusters themselves (Dataset S2). These findings suggest that nostoclide and nostovalerolactone act as global chemical mediators and are significantly responsible for the cell‐density‐dependent effects previously described for the specialized metabolism in N. punctiforme, [5] however, the compounds do not exhibit an autoinduction. The induction of pks1 itself presumably involves another signal acting upstream of the nostoclide/nostovalerolactone dependent regulon (Figure 6). The existence of a chemical mediator that activates the AraC_PKS1 gene itself could indeed be verified by bioactivity‐guided fractionation (Figure S53); structural elucidation of the yet unknown compound will however require further upscaling.
Figure 5.
Effect of nostoclide and nostovalerolactone addition on BGC expression. The heatmaps visualize the log2 fold change between treated and untreated WT. A schematic representation of the corresponding BGC is given above the heatmap.
Figure 6.

Potential signaling pathway of the pks1‐derived products N1, N2 and nostovalerolactone are involved in. The factor required to activate pks1 BGC expression is not yet identified. *The PKS BGCs pks3 and pks4 are upregulated only in presence of nostovalerolactone and both nostoclides.
Nostoclide and nostovalerolactone thus fulfil only part of the characteristics of QS mediators. The observed signaling activity of the nostoclides and nostovalerolactone is nevertheless in line with the structural features that these compounds share with many other bacterial QS mediators. The nostoclides belong to the class of butenolides, which share a 2‐furanone moiety.[ 22 , 23 ] Butenolides are well‐known signaling molecules in Gram‐positive bacteria but recently also gained attention as potential signaling compounds in Pseudomonas. [23] They are structurally related to tetronates and γ‐butyrolactones. Many linear tetronates are inhibitors of phosphatase enzymes and interfere with signaling pathways that depend on phosphorylation, [13] whereas γ‐butyrolactones, like the A‐factor, are prototype QS signals. [8] For nostovalerolactone structural resemblance with bacterial pheromones is less obvious. However, besides its already mentioned remote similarity with linear tetronates (Figure 4) the δ‐valerolactone substructure is reminiscent of an α‐pyrone ring. A set of α‐pyrones from Photorhabdus luminescens has recently been shown to take part in a novel type of cell‐cell communication circuit. [24] Although the complete pks1 BGC is not well conserved in cyanobacterial genomes, biosynthetic components of the pathway are widely distributed. The pks1 BGC was recently included in a phylogenetic and literature survey on putative alkylresorcinol BGCs and scaffolds in cyanobacteria indicating a common phylogenetic and biosynthetic origin. [25] Interestingly, also dialkylresorcinols have been shown to act as QS‐like mediators. [26] We believe that a mosaic‐like evolution has contributed to the diversification of the compounds that may perform related functions in diverse cyanobacterial strains.
As nostoclides are structurally resembling cyanobacterin, an inhibitor of photosynthetic electron transport targeting photosystem II, [27] it has been suggested that nostoclides may also act as allelopathic agents against most cyanobacteria and several eukaryotic algae. [28] We thus tested purified nostoclides N1 and N2 in growth and allelopathy assays against the two cyanobacterial strains N. punctiforme PCC 73102 and Anabaena sp. PCC 7120 and the green alga Chlamydomonas reinhardtii. Both nostoclides did not induce any visible allelopathic effects in the three organisms tested (Figure S54). To evaluate possible effects on photosystem II (PSII), the maximum PSII quantum yield (Fv/Fm) and the effective PSII quantum yield (Fv′/Fm′) were determined using PAM fluorometry (Figure S54C). The two compounds did not cause any changes in the variable chlorophyll fluorescence measurements in the eukaryotic or the prokaryotic algae, suggesting that in contrast to cyanobacterin, nostoclide N1 and N2 likely do not act as algicidal mediators in the strains tested (Figure S54).
Overall, our data provide evidence that nostoclides and nostovalerolactone jointly control the expression of a significant part of secondary metabolite BGCs in N. punctiforme. With nostopeptolide and anabaenopeptin the regulatory network includes two characterized compounds that were shown to be involved in the negotiation with the symbiotic host Blasia pusilla and the intraspecific competition with other Nostoc strains in the habitat.[ 7 , 29 ] The two novel chemical mediators thus enforce multilateral interactions of N. punctiforme that include inter‐ as well as intraspecific interactions. Further research is needed to understand the physiological role of the extracellular signaling.
Conclusion
In the present study, we demonstrate that the combination of transcription factor engineering and transcriptional reporter‐guided purification of compounds is an effective method to mine cryptic metabolites in cyanobacteria. Although the remarkable stimulation of the specialized metabolism through high‐density cultivation was principally known, assignment of products of individual BGCs is complicated because of the complexity of the extracts and the simultaneous upregulation of a large number of BGCs and extracellular proteins.
Transcriptional and bioinformatic analyses as well as labeling studies enabled us to assign two adjacent polyketide pathways to the biosynthesis of the novel metabolites and to establish a first model for the biosynthesis of the unprecedented δ‐valerolactones.
Thus, our study not only extends the amazing diversity of products of tetronate‐like biosynthetic pathways, but also assigns this type of biosynthesis to a cyanobacterium for the first time. Analysis of the transcriptional response towards the cell density‐dependent mediators under low cell density allows to dissect the individual impact of high light and high CO2 conditions and chemical mediators, respectively, for the reprogramming of the specialized metabolism in N. punctiforme. Understanding the role of cell density‐dependent mediators can not only guide the elucidation of the full genetic circuit underlying the regulation of the specialized metabolism in N. punctiforme but also be used for the design of genetic tools to awaken the specialized metabolism in other prolific cyanobacterial strains. The study can thereby contribute to the development of cyanobacteria into the sustainable production systems for bioactive natural products and can inspire customized screening programs of cyanobacteria.
Conflict of interest
The authors declare no conflict of interest.
1.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgements
We thank A. Perner and F. Trottmann for LC HR‐MS measurements, H. Heinecke and F. Trottmann for NMR measurements, C. Weigel for performing antimicrobial agar diffusion assays and Dr. T. Jakob for help with the photosynthesis measurements. This study was supported by Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Project‐ID 239748522‐ SFB 1127 to E.D., C.H. and S.S. and by the German Research Foundation (DFG, GRK 2473 “Bioactive Peptides”—project number 392923329) to E.D. Open Access funding enabled and organized by Projekt DEAL.
J. Krumbholz, K. Ishida, M. Baunach, J. E. Teikari, M. M. Rose, S. Sasso, C. Hertweck, E. Dittmann, Angew. Chem. Int. Ed. 2022, 61, e202204545; Angew. Chem. 2022, 134, e202204545.
Data Availability Statement
The data that support the findings of this study are available in the Supporting Information of this article.
References
- 1.
- 1a. Dittmann E., Fewer D. P., Neilan B. A., FEMS Microbiol. Rev. 2013, 37, 23–43; [DOI] [PubMed] [Google Scholar]
- 1b. Breinlinger S., Phillips T. J., Haram B. N., Mareš J., Martínez Yerena J. A., Hrouzek P., Sobotka R., Henderson W. M., Schmieder P., Williams S. M., Lauderdale J. D., Wilde H. D., Gerrin W., Kust A., Washington J. W., Wagner C., Geier B., Liebeke M., Enke H., Niedermeyer T. H. J., Wilde S. B., Science 2021, 371, eaax9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.
- 2a. Jones A. C., Monroe E. A., Eisman E. B., Gerwick L., Sherman D. H., Gerwick W. H., Nat. Prod. Rep. 2010, 27, 1048–1065; [DOI] [PubMed] [Google Scholar]
- 2b. Demay J., Bernard C., Reinhardt A., Marie B., Mar. Drugs 2019, 17, 320; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2c. Kleigrewe K., Gerwick L., Sherman D. H., Gerwick W. H., Nat. Prod. Rep. 2016, 33, 348–364. [DOI] [PubMed] [Google Scholar]
- 3. Calteau A., Fewer D. P., Latifi A., Coursin T., Laurent T., Jokela J., Kerfeld C. A., Sivonen K., Piel J., Gugger M., BMC Genomics 2014, 15, 977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blin K., Shaw S., Kautsar S. A., Medema M. H., Weber T., Nucleic Acids Res. 2021, 49, D639–D643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dehm D., Krumbholz J., Baunach M., Wiebach V., Hinrichs K., Guljamow A., Tabuchi T., Jenke-Kodama H., Süssmuth R. D., Dittmann E., ACS Chem. Biol. 2019, 14, 1271–1279. [DOI] [PubMed] [Google Scholar]
- 6.
- 6a. Giglio S., Jiang J., Saint C. P., Cane D. E., Monis P. T., Environ. Sci. Technol. 2008, 42, 8027–8032; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6b. Campbell E. L., Cohen M. F., Meeks J. C., Arch. Microbiol. 1997, 167, 251–258; [DOI] [PubMed] [Google Scholar]
- 6c. Rouhiainen L., Jokela J., Fewer D. P., Urmann M., Sivonen K., Chem. Biol. 2010, 17, 265–273. [DOI] [PubMed] [Google Scholar]
- 7. Guljamow A., Kreische M., Ishida K., Liaimer A., Altermark B., Bähr L., Hertweck C., Ehwald R., Dittmann E., Appl. Environ. Microbiol. 2017, 83, e01510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Horinouchi S., Beppu T., Mol. Microbiol. 1994, 12, 859–864. [DOI] [PubMed] [Google Scholar]
- 9. Palonen E. K., Neffling M. R., Raina S., Brandt A., Keshavarz T., Meriluoto J., Soini J., Microorganisms 2014, 2, 111–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.
- 10a. McNerney M. P., Styczynski M. P., Wiley Interdiscip. Rev. Syst. Biol. Med. 2018, 10, 10.1002/wsbm.1405; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10b. Schuster M., Sexton D. J., Diggle S. P., Greenberg E. P., Annu. Rev. Microbiol. 2013, 67, 43–63. [DOI] [PubMed] [Google Scholar]
- 11.
- 11a. Sharif D. I., Gallon J., Smith C. J., Dudley E., ISME J. 2008, 2, 1171–1182; [DOI] [PubMed] [Google Scholar]
- 11b. Marner F.-J., Moore R. E., Phytochemistry 1978, 17, 553–554. [Google Scholar]
- 12. Gallegos M., Schleif R., Bairoch A., Hofmann K., Ramos J., Microbiol. Mol. Biol. Rev. 1997, 61, 393–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vieweg L., Reichau S., Schobert R., Leadlay P. F., Süssmuth R. D., Nat. Prod. Rep. 2014, 31, 1554–1584. [DOI] [PubMed] [Google Scholar]
- 14.
- 14a. Kusebauch B., Busch B., Scherlach K., Roth M., Hertweck C., Angew. Chem. Int. Ed. 2010, 49, 1460–1464; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2010, 122, 1502–1506; [Google Scholar]
- 14b. Brandt P., García-Altares M., Nett M., Hertweck C., Hoffmeister D., Angew. Chem. Int. Ed. 2017, 56, 5937–5941; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 6031–6035. [Google Scholar]
- 15. Takahashi H., Uchimiya H., Hihara Y., J. Exp. Bot. 2008, 59, 3009–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun Y., Hahn F., Demydchuk Y., Chettle J., Tosin M., Osada H., Leadlay P. F., Nat. Chem. Biol. 2010, 6, 99–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meoded R. A., Ueoka R., Helfrich E. J. N., Jensen K., Magnus N., Piechulla B., Piel J., Angew. Chem. Int. Ed. 2018, 57, 11644–11648; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2018, 130, 11818–11822. [Google Scholar]
- 18. Beam M. P., Bosserman M. A., Noinaj N., Wehenkel M., Rohr J., Biochemistry 2009, 48, 4476–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kanchanabanca C., Tao W., Hong H., Liu Y., Hahn F., Samborskyy M., Deng Z., Sun Y., Leadlay P. F., Angew. Chem. Int. Ed. 2013, 52, 5785–5788; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2013, 125, 5897–5900. [Google Scholar]
- 20.P. M. D'Agostino, C. J. Seel, T. Gulder, T. Gulder, ChemRxiv 2021 https://chemrxiv.org/engage/chemrxiv/article-details/60c75901469df4117bf457b7, accessed 16 March 2022.
- 21. Moffitt M. C., Louie G. V., Bowman M. E., Pence J., Noel J. P., Moore B. S., Biochemistry 2007, 46, 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.
- 22a. Niehs S. P., Kumpfmüller J., Dose B., Little R. F., Ishida K., Flórez L. V., Kaltenpoth M., Hertweck C., Angew. Chem. Int. Ed. 2020, 59, 23122–23126; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 23322–23326; [Google Scholar]
- 22b. Nakou I. T., Jenner M., Dashti Y., Romero-Canelón I., Masschelein J., Mahenthiralingam E., Challis G. L., Angew. Chem. Int. Ed. 2020, 59, 23145–23153; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 23345–23353. [Google Scholar]
- 23. Klapper M., Schlabach K., Paschold A., Zhang S., Chowdhury S., Menzel K.-D., Rosenbaum M. A., Stallforth P., Angew. Chem. Int. Ed. 2020, 59, 5607–5610; [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 5656–5659. [Google Scholar]
- 24. Brachmann A. O., Brameyer S., Kresovic D., Hitkova I., Kopp Y., Manske C., Schubert K., Bode H. B., Heermann R., Nat. Chem. Biol. 2013, 9, 573–578. [DOI] [PubMed] [Google Scholar]
- 25. Martins T., Rouger C., Glasser N., Freitas S., de Fraissinette N., Balskus E., Tasdemir D., Leão P., Nat. Prod. Rep. 2019, 36, 1437–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brameyer S., Kresovic D., Bode H. B., Heermann R., Proc. Natl. Acad. Sci. USA 2015, 112, 572–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mallipudi L. R., Gleason F. K., Plant Sci. 1989, 60, 149–154. [Google Scholar]
- 28. Teixeira R. R., Barbosa L. C. A., Forlani G., Piló-Veloso D., Walkimar de Mesquita Carneiro J., J. Agric. Food Chem. 2008, 56, 2321–2329. [DOI] [PubMed] [Google Scholar]
- 29. Liaimer A., Helfrich E. J. N., Hinrichs K., Guljamow A., Ishida K., Hertweck C., Dittmann E., Proc. Natl. Acad. Sci. USA 2015, 112, 1862–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Data Availability Statement
The data that support the findings of this study are available in the Supporting Information of this article.