Abstract
Breast cancer is the most commonly diagnosed cancer in women worldwide. Major advances have been made towards breast cancer prevention and treatment. Unfortunately, the incidence of breast cancer is still increasing globally. Metabolomics is the field of science which studies all the metabolites in a cell, tissue, system, or organism. Metabolomics can provide information on dynamic changes occurring during cancer development and progression. The metabolites identified using cutting-edge metabolomics techniques will result in the identification of biomarkers for the early detection, diagnosis, and treatment of cancers. This review briefly introduces the metabolic changes in cancer with particular focus on breast cancer.
Keywords: breast cancer, metabolomics, metabolites, glycolysis
1. Introduction
Breast cancer is the most common cancer in women and it is one of the leading causes of cancer-related death among women. There is already strong evidence that multiple risk factors make women more susceptible to breast cancer. Even so, the incidence of breast cancer is increasing in women globally [1,2]. Reproductive factors such as early menarche, late menopause, and age at first full-term pregnancy influence the risk of breast cancer [3]. Beside these, other factors such as obesity or first-degree family history of breast cancer also alter the risk of breast cancer [4]. Family history of breast cancer increases the chances of getting breast cancer by 2.5-fold or higher [5,6]. Having the BRCA1 and BRCA2 mutation also puts women at an even higher risk [7]. Some other known risk factors include body fat, alcohol consumption, and high child birth weight [3,8]. There is a 40% increase in the risk of breast cancer in obese postmenopausal women [9]. There are even non-genetic, modifiable risk factors which affect the risk of breast cancer, such as income, education, and insurance status [10]. Ethnicity is another significant risk factor, with breast cancer predominately affecting Non-Hispanic white females, followed by Non-Hispanic blackfemales [11]. Although the risk of breast cancer is lower in Hispanics, according to the American Cancer Society Cancer Facts and Figures for Hispanics/Latinos 2018–2020, it is still the most frequently diagnosed cancer and is the leading cause of cancer-related death, as well in this group of women. Interestingly, it is also well-known that majority of women who are newly diagnosed with breast cancer do not have any known risk factors [12].
The World Health Organization estimates that breast cancer accounted for 12% of all new cancers diagnosed worldwide in 2021 [2]. By 2030, it is estimated that the number of breast cancer cases worldwide will increase to almost 3.2 million new cases per year [13]. It is also estimated that, in the US, 287,850 new cases of invasive breast cancer and 51,700 new cases of non-invasive breast cancers will be diagnosed in 2022. Approximately 43,250 women are expected to die due to breast cancer in 2022 in the US. The five-year survival rate of breast cancer varies depending on stage and subtype of breast cancer.
2. Breast Cancer Subtypes
Breast cancer is a highly complex and heterogeneous disease with very distinctive characteristics. It also has an array of clinical presentations and responses to therapy [14]. They are mainly classified based on histological and molecular subtypes [15,16,17]. Histological subtypes include mainly ductal and lobular carcinomas. Besides ductal and lobular breast carcinomas, other histological types include: mucinous, tubular, comedo, inflammatory, medullary and papillary carcinomas, which account for about 10% of the cases [18]. The molecular classification of breast cancer was made using microarray-based gene expression analysis and unbiased hierarchical clustering. Based on these findings, breast cancers are classified as luminal subtype A (estrogen receptor (ER)-positive and/or progesterone receptor (PR)-positive, human epidermal growth factor receptor 2 (HER2)-negative and low Ki67), luminal subtype B (ER-positive, PR-positive or negative, HER2-positive and has high Ki-67), HER2-positive (HER2 overexpressing), basal-like/triple-negative (ER-negative, PR-negative and HER2-negative), normal breast-like and claudin-low [16,19]. Over 60% of all breast cancers are ER+, 15–20% are HER2+ and 10–20% accounts for triple-negative breast cancer (TNBC) [20,21,22]. Generally, hormone-receptor-positive breast cancers are less aggressive than hormone-receptor-negative breast cancers. Even though breast cancer-related death has decreased by 1.0% from 2013 to 2018 [23], the incidence rate for invasive breast cancer has increased approximately by 0.5% annually in the US [2,24]. There are several reasons that have been attributed to the increased incidence of breast cancer worldwide, which include adoption of a Western lifestyle, delayed childbearing, combined hormone replacement therapy, etc. [25]. Another reason for this increase in breast cancer incidence is that early diagnosis and treatment are still challenging.
3. Metabolome and Metabolomics
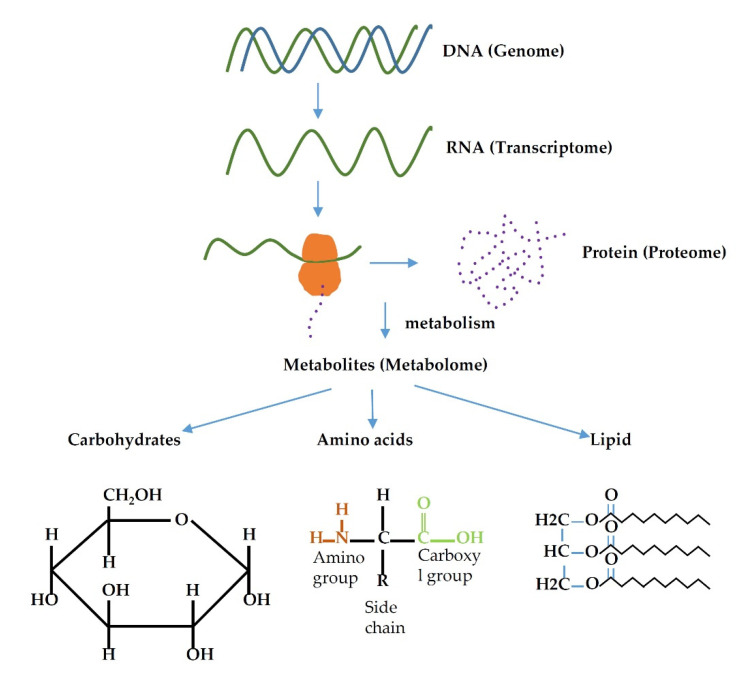
The term metabolome/metabolomics was coined in 1998 [26]. The metabolome is the total number of metabolites present within the cell, tissue, organ, or organism and have a wide range of functions. Metabolomics is a relatively new field focusing on the analysis of metabolites in the metabolome. The message in the DNA (genome) of an organism are transcribed into RNA (transcriptome), translated into protein (proteome) and finally results in the formation of small molecules (result of metabolism) known as metabolites (metabolome) (Figure 1). Thus, any change in the gene, whether that be mutation, over-expression, or under-expression, alters the metabolomics profile of an organism. Many diseases, including cancer, are the result of alteration in gene expression profile, e.g., BRAC1/BRAC2 genes are the most mutated genes in hereditary breast cancer [27]. Therefore, alterations in genes could cause changes in the metabolic profile, and these changes eventually could facilitate cancer development.
Figure 1.
Relation flow of different omics family.
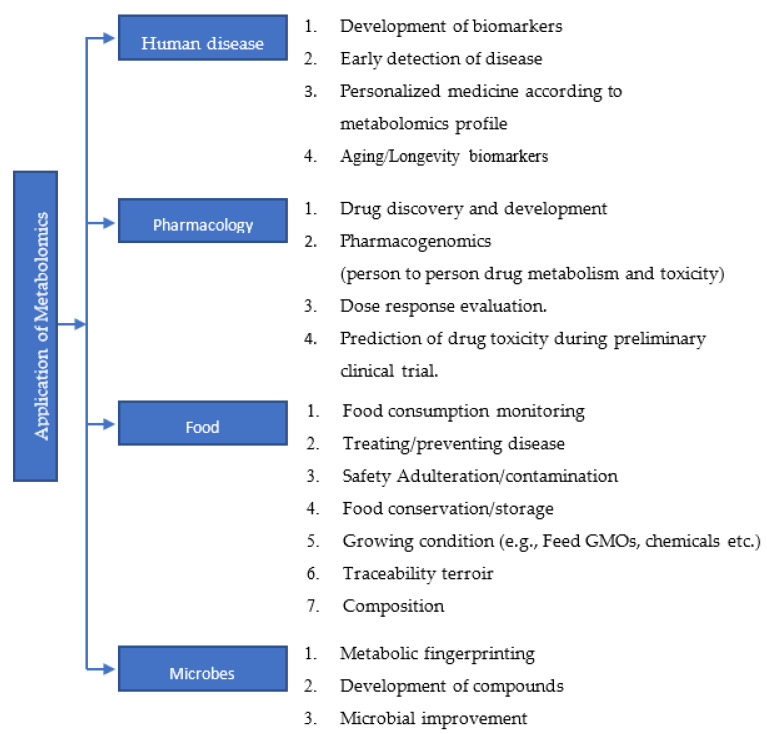
The metabolome includes all the products of catabolism and anabolism. Like genomics, transcriptomics, and proteomics, metabolomics study has also gained its own importance. Metabolites are closely linked to the phenotype of an organism; a metabolome is largely defined by its genome. Metabolomics or metabolic profiling is the measurement in biological systems of low-molecular-weight metabolites and intermediates that reflect the dynamic response to genetic modification. It is considered as a very powerful and reliable tool that has high reproducibility, and which can have significant impact on the health of humans [28]. A comprehensive metabolomics study using 928 cell lines from over 20 different cancer types resulted in the identification of 225 metabolites specific to cancer metabolism [29]. Changes in the levels of metabolites can be used as diagnostic markers [30,31], prognostic markers [32] and also as therapeutic targets [33,34,35]. Many biological processes associated with age, gender, obesity, medication, cancers, cardiovascular disease, diabetes, etc., can alter the metabolomics profile of an individual [36,37,38]. The study carried out by Mansell et al. highlighted how the metabolome profile can be used as a marker to predict the future risk of cardiovascular disease in a new born child of a mother with gestational diabetes [39]. Further, epidemiological and experimental data demonstrate that critical changes in metabolites are observed during aging, age-related diseases and geriatric syndromes [40]. It is well-known that various types of cancers are age-related diseases. Endocrine and mitochondrial functions, signaling pathways and calorie restrictions in aging have been associated with amino acid metabolism, lipid metabolism, redox homeostasis and nutrient sensing [41,42,43,44]. Since changes in metabolomics profiles can be associated with various pathological conditions, studying the metabolome will be of significant help towards the development of personalized medicine and also in the early detection and diagnosis of diseases. Application of metabolomics study in various field is summarized in flow chart (Figure 2).
Figure 2.
A brief list of applications of metabolomics study in various fields.
4. Analysis of the Metabolome
Metabolites can be measured by 3 major techniques: gas chromatography-mass spectrometry (GC-MS), liquid chromatography (LC-MS), and nuclear magnetic resonance spectroscopy (NMR) [45]. There are two approaches in mass spectrometry: untargeted and targeted. An untargeted approach focuses on a broad range of metabolites; these metabolites are then identified and characterized. A targeted approach focuses on finding the pre-defined and characterized metabolites. In general, the targeted approach has higher sensitivity and selectivity. NMR is a reliable and sensitive technique, which can detect relatively small changes in metabolite concentration among biological specimens [46]. Metabolomics profiling can also be performed and visually observed by using Scanning Electron Microscopy (SEM), Matrix-Assisted Laser Desorption Ionization (MALDI), and Nanostructure-Imaging Mass Spectrometry (NIMS) [47]. The data collected are processed for statistical analysis using various platforms such as XCMS Online, DeviumWEb and MetaboAnalyst [47]. Once metabolites are identified, they are correlated to a particular phenotype, physiological state, or aberration [47]. Metabolites can also be identified by comparing the result obtained from a mass spectral and RI index with databases such as GOLM, NIST05, METLIN, Mass Bank, or the most widely used, Human Metabolome Database (HMDB) [47,48]. As promising as metabolomics sounds, there are a few disadvantages. The latest release by the HMDB contained 247 inborn error metabolites (IEMs) for humans, including both endogenous and exogenous metabolites [49]. There are also the challenges in metabolites extraction, purification, fractionation, and identification. Lastly, there is the possibility of false-positive metabolites identification [50]. Metabolomics is still an emerging field and has a lot of promise to help identify new disease conditions, biomarkers, and therapeutic targets.
5. Metabolomics Profile of Breast Cancers
Metabolomics is promising in the area of personalized medicine because it reflects a patient’s phenotype most closely [51]. The cancer metabolome is constituted by different factors. These include metabolites that are the product of the oncologic process, as well as the systemic response of the body to tumors [51]. Studying metabolites and their associated pathways in human diseases such as cancer will provide a better understanding of how the dysregulated metabolism could lead to the initiation and progression of cancer [52]. There are studies that have demonstrated that many types of cancers, including breast cancer, alter the metabolic system in profound ways. In order to understand the development and progression of cancer, it is clearly necessary to understand the cellular metabolome and its metabolic changes [28]. Metabolomics can thus provide a measurement of these phenotypic changes that reflect genetic alterations in breast cancer [52]. An example of this is tumorigenesis, which is a consequence of oncogenic mutations and is dependent on the reprogramming of cellular metabolism. Moreover, since it is well know that cancer cells need to sustain abnormal growth and proliferation rates, which require supplements of metabolic precursors, it makes intuitive sense that it would also lead to an altered metabolism [48]. These secreted metabolites then enter circulation and are transferred to target tissues. These in turn exert biological effects that modulate cells [12]. Metabolic changes play important pathological roles by inducing proliferation, angiogenesis and epithelial-to-mesenchymal transition (EMT). Metabolites can also have a profound effect on mitochondrial metabolism. An example of this is with estrogen, which has been shown to induce carcinogenesis by causing changes in the expression of mitochondrial genes [12]. Metabolome alterations can also be specific to breast cancer subtypes. Metabolomics data has been able to distinguish ER and HER2 molecular subtypes by using glutamate-to-glutamine ratio and aerobic glycolysis as biomarkers [45]. Metabolites can be used as an potential indicator which also describes the aggressiveness of the breast cancer [53]. Similarly, metabolites of the energy-generating metabolic pathways such as glycolysis, TCA cycle, beta-oxidation are higher in hormone-receptor-negative breast cancer and TNBC compared to hormone-receptor-positive breast cancer, which is positively correlated to the aggressiveness of breast cancer [54]. Hence, metabolomics study is important in categorizing breast cancer into different types and stages, as it uses accurate methods and requires less time for analysis. Metabolites from secondary bile acid metabolism, amino acid degradation, short-chain fatty acid production, and deconjugated hormones can be measured to predict tamoxifen resistance, hormone-induced apoptosis, cancer aggressiveness, and histone deacetylase (HDAC) inhibition [55,56,57].
Morphological changes of the cancer cell are also regulated by metabolites such as collagen, cytokines, or by adipocytes in the tumor microenvironment. Currently, more than 30 endogenous metabolites in breast tissue have been identified including elevated choline, low glycerophosphocholine, and low glucose in breast cancer tissue compared to healthy tissue or benign tumors [51]. There are also several pathways that influence the levels of glutamine, lipids, serine, protein translation and cholesterol metabolism, which have been shown to be upregulated in breast cancer [12]. These metabolic changes, and their impact on the progression or inhibition of breast cancer, can serve as a biomarker for early diagnosis and potential target for treatment.
5.1. Carbohydrate Metabolism
A hallmark of cancer is the altered utilization of energy by cancer cells relative to normal cells due to increased rates of proliferation [58]. Glycolysis in breast cancer cells is characterized by decreased level of glucose [59,60,61,62] and increased levels of lactate [63,64,65]. Earlier, it had been reported that glucose metabolism was the sole source of energy for cancer cells [66], but more complex processes are required to accomplish this [67]. Cancer cells rely on aerobic glycolysis for energy, popularly known as Warburg effect [68]. This preference for aerobic glycolysis even in the presence of oxygen [68] is an effective way for attaining sufficient energy and evading immune suppression [69]. Furthermore, this could also facilitate complete oxidation of carbon in cancer cells compared to normal cells [59].
The metabolites that are involved in pathways associated with energy metabolism, such as glycolysis, glycogenolysis, tricarboxylic acid cycle (TCA cycle) proliferation and redox pathways are found to be significantly altered in breast cancer [54]. These metabolic processes also produce many metabolites, which are the precursors for many macromolecules required for cancer cell growth and proliferation. Glucose-6-phosphate undergoes the pentose phosphate pathway to yield ribose-5-phosphate precursor for nucleic acid biosynthesis; NADPH and acetyl CoA are used for lipid synthesis; intermediate 3-phosphoglycerate serves as the precursor for synthesis of amino acid such as glycine and cysteine. Targeting the glycolytic enzyme can be beneficial for the inhibiting cancer growth and progression. GLUT-1, a glucose transporter, is present in high levels in TNBC and could serve as potential therapeutic target [70]. Recently, GLUT-1 inhibitor BAY-876 has been shown to selectively inhibit the growth of TNBC cell lines [70]. Aggressive forms of breast cancers tend to have increased rates of glycolysis, TCA cycle, proliferation, and redox pathways [12,71]. It has been well established that metabolites such as ATP, acetyl-coA, and NAD regulate post-translational modifications that affect protein activity adversely, and these co-substrates counteract normal biological pathways. Cancer cells can adapt well to an increase in lactate production and survive in acidic microenvironments [47]. In addition, lower 5-year survival rates have been associated with higher levels of lactate [72]. These findings demonstrate that the metabolites of carbohydrate metabolism play a major role in breast cancer growth and progression. Further understanding the roles and mechanisms by which these metabolites influence breast cancer is of importance.
5.2. Lipid Metabolism
Lipid metabolism is significantly altered in cancers. Increased cell growth and tumor formation needs increased synthesis and uptake of lipids. Lipids are key molecules that form the structural basis of biological membranes. They also function as signaling molecules and as an energy source. Several cancers have enhanced fatty acid synthesis, and lipogenesis is vital for tumor growth [73]. Fatty acids are the source of energy as well as the precursor for lipid molecules. The cancer cells undergo two processes: First, de novo synthesis of fatty acid, which utilizes citrate from the TCA cycle and synthesizes many lipid molecules required for cancer cell growth. Second, beta oxidation of exogenous fatty acids, whereby fatty acids are transported into the cell by fatty acid transporters and undergo beta oxidation, yielding huge amounts of energy required for cancer cells. Earlier it was demonstrated that lipid uptake can also be achieved through the receptor-mediated endocytosis of low-density lipoprotein (LDL) particles, CD36 fatty acid translocase, fatty acid transport proteins (FATP) and fatty acid binding proteins (FABP) [74,75,76]. Upregulation of the metabolites that are involved in fatty acid transportation and fatty acid synthesis molecules have been observed in breast cancer [77] and these molecules can be used as biomarkers of breast cancer diagnosis and treatment.
Cancer cells and adipocytes interact to promote tumorigenesis, including breast cancers [78,79]. Coculture of cancer cells with adipocytes increased fatty acid oxidation by activating AMPK activation [80,81,82]. Leptin secreted by adipocytes enhanced fatty acid oxidation by activating JAK/STAT3 signaling pathway in breast cancer stem cells [83]. Acetylcarnitine plays an important role in the production of energy. It transports fatty acid to mitochondria for beta-oxidation and is positively correlated with the risk of breast cancer [84]. Higher levels of acetylcarnitine result in breast cancer development via insulin resistance [84,85]. CD36 is a fatty acid translocase which interacts with FABP4 for transporting fatty acid into the cell. CD36 expression promotes breast cancer by increasing STAT3 signaling and beta oxidation, providing energy for growing breast cancer cell [86]. The inhibition of CD36 and FABP4 induces apoptosis in breast cancer cell lines. Thus CD36 can be used as a biomarker for diagnosis and can also be used as a target for treatment of breast cancer [86]. Phospholipids lysophosphatidlycholine (LPC) correlates with lower risk of breast cancer, while higher level of phosphotidylcholine (PC) correlates with increased risk of breast cancer [87]. Lysophosphatidylcholine acyltransferase 1 (LPCAT1) converts LPC to PC and it is overexpressed in breast cancer with poor prognosis [88], which demonstrates that a higher level of PC is associated with a high risk of breast cancer. Hence, cellular levels of LPC and PC could predict the risk of breast cancer, and targeting LPCAT1 can add a new therapeutic approach for breast cancer. Overall, increased fatty acid synthesis alters cellular lipid composition in cancer cells, and could serve as potential diagnostics and therapeutic targets. The increase in de novo FA synthesis in cancer cells alters cellular lipid composition and can be used for diagnostics [89].
5.3. Amino Acid Metabolism
Amino acid metabolism plays a critical role in cancer cell proliferation. Non-essential amino acids and semi-essential amino acids are also needed in addition to essential amino acids to sustain breast cancer cell proliferation [90,91]. It is accomplished by establishing pools of amino acids for the production of non-essential amino acids to be used in protein synthesis, conversion of glucose and lipids, and the activation of key signaling pathways. Amino acids facilitate epigenetic modification, such as methylation, produce α-ketoglutarate, which is oxidized by the tricarboxylic acid (TCA) cycle, oxidative phosphorylation for ATP production, and also maintain intracellular redox status [90]. Amino acid consumption and utilization helps to sustain the growth of cancer cells [92,93,94,95,96].
Glutamine metabolism plays an important role in serving the energy demand of the cancer cell. Multiple transporters, including the Na+-dependent transporters, system ASC, which preferentially transports alanine/serine/cysteine and the Na+-coupled neutral amino acid transporters (SNATs) import glutamine into cancer cells [97,98]. Tumors have high expression of glutamine transporters ASCT2, SNAT1, SNAT2, and SNAT5 [99]. Inhibition of ASCT2 has been shown to decrease the growth of various cancers including triple-negative breast cancer [100]. Reductive glutamine metabolism for lipid biosynthesis supports tumor growth under hypoxia or mitochondrial dysfunction [101]. In addition, citrate, fumarate and malate derived from glutamine are increased in glucose-deprived cancers, demonstrating that glutamine can support the growth of tumors in a nutrient-poor microenvironment [102]. The glutaminase I (GLS-I) enzyme converts glutamine via glutaminolysis into glutamate, which in turn is converted to alpha ketoglutarate by glutamate dehydrogenase. This alpha ketoglutarate enters the TCA cycle. The glutaminolysis pathway generates a high amount of energy (via TCA cycle) and also produces macromolecules required for cancer cell growth and proliferation. Therefore, the glutamate to glutamine ratio (GGR) can be used as a biomarker for breast cancer diagnosis [103]. A higher GGR has been observed in ER-ve breast cancer and TNBC. Thus GLS-I enzyme can be a target for the treatment of breast cancer. Molecule 7, a derivative of withangulatin A, is an inhibitor of GLS-I enzyme, which inhibited the growth of TNBC cell line by decreasing the amount of glutamate [104].
Serine is a neutral amino acid and is imported into the cells via ASCT1. Breast cancers have high expression of ASCT1 [105]. Serine acts as a carbon source for nucleotide synthesis and as a source for DNA methylation. It has been shown to play a vital role in cancer growth and progression. Increased rates of proliferation in tumor cells relies on the availability of extracellular serine. The reduction in serine and glycine inhibits tumor growth and increases survival time of mice with tumors [106,107]. Some cancers prefer serine as a source of nutrition for rapid cell proliferation while others prefer glycine [93,108,109,110,111,112].
LAT1 is a transporter of branched-chain amino acids such as leucine, isoleucine, and valine. It is highly expressed in many cancers including breast cancers [99,113,114]. Cysteine is another metabolite that is indicative of cancer development. Elevated levels of cysteine are associated with oxidative damage and overproduction of free radicals that lead to gene mutation [115]. Dramatic metabolic shifts in choline and proline levels are known to be characteristic of metastatic breast cancer [28]. Altered levels of arginine and asparagine are also correlated with breast cancer. It has been shown that breast cancers are high L-arginine-dependent [113]. Further, it has been demonstrated that L-arginine supplementation enhances innate and adaptive immune responses and inhibits the growth of breast cancer [114]. A study in premenopausal women found an increase in plasma arginine levels to have resulted in decreased plasma levels of insulin such as growth factor 1 and estradiol [116]. Lowering the bioavailability of asparagine either by reducing dietary asparagine, or by knockdown of asparagine synthetase, reduced breast cancer metastasis [117]. These findings demonstrate that amino acid metabolism could play a vital role in cancer. Understanding how this metabolism can be utilized to prevent or treat breast cancer will be of significance.
Some of the changes in metabolomics profile of breast cancer is presented in Table 1.
Table 1.
Changes in metabolomics profile of breast cancer.
| Study Subject | Comparison Within | Metabolomics Technique Used | Change in Metabolites (Pathways) | References |
|---|---|---|---|---|
| Breast cancer tissue specimen from African-American Women |
Metabolites change in ER + ve versus TNBC tissue specimen | GC-MS LC-MS |
Glycolysis, glycogenolysis, TCA cylcle, proliferation and redox pathways metabolites, e.g., NAD+ synthesis pathway; increase in intermediates of transmethylation were increased in TNBC compared to ER + ve. | [54] |
| 267 Human Breast Tissue | Lipid metabolite was compared between breast cancer and normal breast tissue. | UPLC-MS/MS | Membrane phospholipids (phosphatidylcholine, phosphatidylethanolamine, and sphingomyelins ceramides) were increased in breast cancer tissue sample (more in ER-ve samples) than normal breast tissue. | [89] |
| Breast Cancer tissue from DUKE University Medical center | ER+ve versus ER-ve tumor |
GC-MS LC-MS |
Glycolytic and glycogenolytic intermediates; glutathione pathway intermediates; onco-metabolites 2-hydroxyglutrate; tryptophan metabolite Kynurenine were elevated in ER-ve tumor compared to ER +ve. | [118] |
| Serum sample from breast cancer patient | Change in metabolites between obese versus non-obese breast cancer patients | LC-MS | Lipid, carbohydrate, amino acid metabolism metabolites; oxidative phosphorylation, uric acid, ammonia recycling vitamin metabolism (all having role in ATP generation) are increased significantly in obese compared to non-obese breast cancer serum sample. Neurotransmitter metabolites such as serotonin, histamine; acetylcholine is also increased in obese compared to non-obese breast cancer patient serum | [119] |
| Plasma sample from healthy and breast cancer patient | Breast cancer patient verus healthy control | LC-MS | Increase in antioxidative metabolites (taurine and uric acid); increase in metabolites for bioenergetics (fatty acids capric acid, myristic acid); increase in three branched-chain amino acid which provides carbon for gluconeogenesis (2-hydroxy-3-methylbutiric acid, 2-hydroxy-3-methylpentanoic acid, and 3-methylglutaric acid); increase in nucleic acid biosynthesis substrate (cystidine and inosine diphosphate) in breast cancer patients plasma compared to healthy controls. | [120] |
| Blood (plasma) sample from healthy and breast cancer patient after overnight fasting | Plasma Metabolomics comparison carried out between breast cancer versus healthy individual | LC-MS | Arginine proline metabolism pathway metabolites and tryptophan metabolism pathway metabolites decreased and fatty acid biosynthesis pathway metabolites increases in plasma of breast cancer when compared to normal healthy individual. | [121] |
| Breast cancer patient tissue specimen | Comparison was made between metabolites in different sub-group of luminal A (A1, A2 and A3) | HR MAS MRS (High resolution magic angle spinning magnetic resonance spectroscopy) | Glucose signal was lower in A2 compared to A1 and A3. α-hydrogen amino acid signal was lower in A1 higher in A3 compared to A2. Al anine signal was higher in A2 compared to A3. Myo-inositol signal was lower in A1 than A2 and A3. | [122] |
| Breast cancer cell line MCF-7S (Adriamycin-sensitive) and MCF-7Adr (Adriamycin-resistant) | Effect of Adriamycin in metabolic profile of MCF-7S and MCF-7Adr Cell lines | GC-MS | Adriamycin significantly increases the metabolite as glucose, glutamine; amino acids such as valine isoleucine serine threonine, etc., while adriamycin slightly changed metabolites such as serine isoleucine glutamic acid after long-term exposure. | [123] |
| Serum sample from breast cancer patient | Comparing the metabolites in full response/pCR (pathological complete response), partial response (PR) and no response/SD (stable disease) to neoadjuvant chemotherapy | NMR LC-MS |
Four metabolites were detected with threonine and glutamine decreased in pCR group compared to SD group. Isoleucine increased in pCR group compared to SD and PR and linolenic acid was decreased in pCR group and increased in both PR and SD group. | [124] |
| Fasting blood (serum and plasma) sample from healthy and breast cancer patients | LC-TOF-MS (Liquid chromatography time of flight mass spectrometry) GC-TOFMS (Gas chromatography time of flight mass spectrometry) | Taurine pathway metabolite (hypotaurine, pyruvate); pyruvate the metabolite for glycine, serine threonine metabolism is increased in breast cancer than in normal healthy individual. While amino acid like succinate, choline, serine, glycine and alanine and glycerol 3 phosphate, metabolite in phospholipid biosynthesis are decreased in both plasma and serum sample of breast cancer patient when compared to normal healthy individual. | [125] |
6. Conclusions
Metabolic reprograming is a hallmark of cancer. The field of metabolomics provides various opportunities to understand cancer initiation, promotion, progression, and metastasis. Using the metabolomics profile could result in mechanism-based prevention and treatment options for various cancers including breast cancer. Metabolic profiling is expected to differentiate between aggressive and non-aggressive breast cancers. Further, it can also predict the outcome and response to treatments. Metabolomics provides us with the knowledge of dynamic changes that occur in cancer cells, which could lead to a better understanding of the process of tumorigenesis. This could result in a major advance in elucidating whether a premalignant lesion has the potential to grow further and metastasize. This will result in reducing over-diagnosis and over-treatment. Finally, metabolomics is a highly promising field of science, which is expected to advance our knowledge of human health and reduce the burden of cancer.
Acknowledgments
We would like to thank Texas Tech Health Sciences Center of El Paso for their support.
Author Contributions
Supervision, review and editing, R.L.; original draft preparation, writing and editing, R.S. and S.P.; literature survey, data collection A.E. and K.D.S. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Konat-Bąska K., Matkowski R., Błaszczyk J., Błaszczyk D., Staszek-Szewczyk U., Piłat-Norkowska N., Maciejczyk A. Does Breast Cancer Increasingly Affect Younger Women? Int. J. Environ. Res. Public Health. 2020;17:4884. doi: 10.3390/ijerph17134884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 3.Ataollahi M., Sharifi J., Paknahad M., Paknahad A. Breast cancer and associated factors: A review. J. Med. Life. 2015;8:6. [PMC free article] [PubMed] [Google Scholar]
- 4.Colditz G.A., Rosner B. Cumulative risk of breast cancer to age 70 years according to risk factor status: Data from the Nurses’ Health Study. Am. J. Epidemiol. 2000;152:950–964. doi: 10.1093/aje/152.10.950. [DOI] [PubMed] [Google Scholar]
- 5.Sun Y.-S., Zhao Z., Yang Z.-N., Xu F., Lu H.-J., Zhu Z.-Y., Shi W., Jiang J., Yao P.-P., Zhu H.-P. Risk factors and preventions of breast cancer. Int. J. Biol. Sci. 2017;13:1387. doi: 10.7150/ijbs.21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colditz G.A., Willett W.C., Hunter D.J., Stampfer M.J., Manson J.E., Hennekens C.H., Rosner B.A., Speizer F.E. Family history, age, and risk of breast cancer: Prospective data from the Nurses’ Health Study. JAMA. 1993;270:338–343. doi: 10.1001/jama.1993.03510030062035. [DOI] [PubMed] [Google Scholar]
- 7.Kuchenbaecker K.B., Hopper J.L., Barnes D.R., Phillips K.-A., Mooij T.M., Roos-Blom M.-J., Jervis S., Van Leeuwen F.E., Milne R.L., Andrieu N. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA. 2017;317:2402–2416. doi: 10.1001/jama.2017.7112. [DOI] [PubMed] [Google Scholar]
- 8.Bukowski R., Chlebowski R.T., Thune I., Furberg A.-S., Hankins G.D., Malone F.D., D’Alton M.E. Birth weight, breast cancer and the potential mediating hormonal environment. PLoS ONE. 2012;7:e40199. doi: 10.1371/journal.pone.0040199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qureshi R., Picon-Ruiz M., Aurrekoetxea-Rodriguez I., de Paiva V.N., D’Amico M., Yoon H., Radhakrishnan R., Morata-Tarifa C., Ince T., Lippman M.E. The major pre-and postmenopausal estrogens play opposing roles in obesity-driven mammary inflammation and breast cancer development. Cell Metab. 2020;31:1154–1172.e1159. doi: 10.1016/j.cmet.2020.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Dean L.T., Gehlert S., Neuhouser M.L., Oh A., Zanetti K., Goodman M., Thompson B., Visvanathan K., Schmitz K.H. Social factors matter in cancer risk and survivorship. Cancer Causes Control. 2018;29:611–618. doi: 10.1007/s10552-018-1043-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Power E.J., Chin M.L., Haq M.M. Breast cancer incidence and risk reduction in the Hispanic population. Cureus. 2018;10:e2235. doi: 10.7759/cureus.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mikó E., Kovács T., Sebő É., Tóth J., Csonka T., Ujlaki G., Sipos A., Szabó J., Méhes G., Bai P. Microbiome—microbial metabolome—cancer cell interactions in breast cancer—familiar, but unexplored. Cells. 2019;8:293. doi: 10.3390/cells8040293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginsburg O., Bray F., Coleman M.P., Vanderpuye V., Eniu A., Kotha S.R., Sarker M., Huong T.T., Allemani C., Dvaladze A. The global burden of women’s cancers: A grand challenge in global health. Lancet. 2017;389:847–860. doi: 10.1016/S0140-6736(16)31392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weigelt B., Geyer F.C., Reis-Filho J.S. Histological types of breast cancer: How special are they? Mol. Oncol. 2010;4:192–208. doi: 10.1016/j.molonc.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perou C.M., Sørlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A., et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 16.Malhotra G.K., Zhao X., Band H., Band V. Histological, molecular and functional subtypes of breast cancers. Cancer Biol. 2010;10:955–960. doi: 10.4161/cbt.10.10.13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eliyatkın N., Yalçın E., Zengel B., Aktaş S., Vardar E. Molecular Classification of Breast Carcinoma: From Traditional, Old-Fashioned Way to a New Age, and A New Way. J. Breast Health. 2015;11:59–66. doi: 10.5152/tjbh.2015.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C., Uribe D., Daling J. Clinical characteristics of different histologic types of breast cancer. Br. J. Cancer. 2005;93:1046–1052. doi: 10.1038/sj.bjc.6602787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho N. Molecular subtypes and imaging phenotypes of breast cancer. Ultrasonography. 2016;35:281. doi: 10.14366/usg.16030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boyle P. Triple-negative breast cancer: Epidemiological considerations and recommendations. Ann. Oncol. 2012;23:vi7–vi12. doi: 10.1093/annonc/mds187. [DOI] [PubMed] [Google Scholar]
- 21.Howlader N., Altekruse S.F., Li C.I., Chen V.W., Clarke C.A., Ries L.A., Cronin K.A. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J. Natl. Cancer Inst. 2014;106:dju055. doi: 10.1093/jnci/dju055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg P.S., Barker K.A., Anderson W.F. Estrogen Receptor Status and the Future Burden of Invasive and In Situ Breast Cancers in the United States. J. Natl. Cancer Inst. 2015;107:djv159. doi: 10.1093/jnci/djv159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 24.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA Cancer J. Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 25.Schneider A.P., 2nd, Zainer C.M., Kubat C.K., Mullen N.K., Windisch A.K. The breast cancer epidemic: 10 facts. Linacre Q. 2014;81:244–277. doi: 10.1179/2050854914Y.0000000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver S.G., Winson M.K., Kell D.B., Baganz F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998;16:373–378. doi: 10.1016/S0167-7799(98)01214-1. [DOI] [PubMed] [Google Scholar]
- 27.Hedenfalk I., Duggan D., Chen Y., Radmacher M., Bittner M., Simon R., Meltzer P., Gusterson B., Esteller M., Raffeld M. Gene-expression profiles in hereditary breast cancer. N. Engl. J. Med. 2001;344:539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- 28.Nittoli A.C., Costantini S., Sorice A., Capone F., Ciarcia R., Marzocco S., Budillon A., Severino L. Effects of α-zearalenol on the metabolome of two breast cancer cell lines by 1H-NMR approach. Metabolomics. 2018;14:33. doi: 10.1007/s11306-018-1330-3. [DOI] [PubMed] [Google Scholar]
- 29.Li H., Ning S., Ghandi M., Kryukov G.V., Gopal S., Deik A., Souza A., Pierce K., Keskula P., Hernandez D., et al. The landscape of cancer cell line metabolism. Nat. Med. 2019;25:850–860. doi: 10.1038/s41591-019-0404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan L., Liu Q., Liang D., Guo Y., Liu G., Ren J., He Y., Shan B. Circulating Tumor Cell and Metabolites as Novel Biomarkers for Early-Stage Lung Cancer Diagnosis. Front. Oncol. 2021;11:1465. doi: 10.3389/fonc.2021.630672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao C., Wang M., Li L., Tang J.H. Circulating metabolites serve as diagnostic biomarkers for HER2-positive breast cancer and have predictive value for trastuzumab therapy outcomes. J. Clin. Lab. Anal. 2022;36:e24212. doi: 10.1002/jcla.24212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joshi S., Garlapati C., Bhattarai S., Su Y., Rios-Colon L., Deep G., Torres M.A., Aneja R. Exosomal Metabolic Signatures Are Associated with Differential Response to Neoadjuvant Chemotherapy in Patients with Breast Cancer. Int. J. Mol. Sci. 2022;23:5324. doi: 10.3390/ijms23105324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossi C., Cicalini I., Cufaro M.C., Consalvo A., Upadhyaya P., Sala G., Antonucci I., Del Boccio P., Stuppia L., De Laurenzi V. Breast cancer in the era of integrating "Omics" approaches. Oncogenesis. 2022;11:17. doi: 10.1038/s41389-022-00393-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nees J., Schafferer S., Yuan B., Tang Q., Scheffler M., Hartkopf A., Golatta M., Schneeweiß A., Burwinkel B., Wallwiener M. How previous treatment changes the metabolomic profile in patients with metastatic breast cancer. Arch. Gynecol. Obs. 2022 doi: 10.1007/s00404-022-06558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiggs A., Molina S., Sumner S.J., Rushing B.R. A Review of Metabolic Targets of Anticancer Nutrients and Nutraceuticals in Pre-Clinical Models of Triple-Negative Breast Cancer. Nutrients. 2022;14:1990. doi: 10.3390/nu14101990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito K., Maekawa K., Kinchen J.M., Tanaka R., Kumagai Y., Saito Y. Gender-and age-associated differences in serum metabolite profiles among Japanese populations. Biol. Pharm. Bull. 2016;39:1179–1186. doi: 10.1248/bpb.b16-00226. [DOI] [PubMed] [Google Scholar]
- 37.Cheng S., Shah S.H., Corwin E.J., Fiehn O., Fitzgerald R.L., Gerszten R.E., Illig T., Rhee E.P., Srinivas P.R., Wang T.J. Potential impact and study considerations of metabolomics in cardiovascular health and disease: A scientific statement from the American Heart Association. Circ. Cardiovasc. Genet. 2017;10:e000032. doi: 10.1161/HCG.0000000000000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chashmniam S., Madani N.H., Ghoochani B.F.N.M., Safari-Alighiarloo N., Khamseh M.E. The metabolome profiling of obese and non-obese individuals: Metabolically healthy obese and unhealthy non-obese paradox. Iran. J. Basic Med. Sci. 2020;23:186. doi: 10.22038/IJBMS.2019.37885.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mansell T., Vlahos A., Collier F., Ponsonby A.-L., Vuillermin P., Ellul S., Tang M.L., Burgner D., Saffery R. The newborn metabolome: Associations with gestational diabetes, sex, gestation, birth mode, and birth weight. Pediatric Res. 2021;91:1864–1873. doi: 10.1038/s41390-021-01672-7. [DOI] [PubMed] [Google Scholar]
- 40.Franceschi C., Garagnani P., Morsiani C., Conte M., Santoro A., Grignolio A., Monti D., Capri M., Salvioli S. The Continuum of Aging and Age-Related Diseases: Common Mechanisms but Different Rates. Front. Med. 2018;5:61. doi: 10.3389/fmed.2018.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balasubramanian P., Howell P.R., Anderson R.M. Aging and Caloric Restriction Research: A Biological Perspective With Translational Potential. EBioMedicine. 2017;21:37–44. doi: 10.1016/j.ebiom.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barbosa M.C., Grosso R.A., Fader C.M. Hallmarks of Aging: An Autophagic Perspective. Front. Endocrinol. 2018;9:790. doi: 10.3389/fendo.2018.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pignatti C., D’Adamo S., Stefanelli C., Flamigni F., Cetrullo S. Nutrients and Pathways that Regulate Health Span and Life Span. Geriatrics. 2020;5:95. doi: 10.3390/geriatrics5040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chung K.W. Advances in Understanding of the Role of Lipid Metabolism in Aging. Cells. 2021;10:880. doi: 10.3390/cells10040880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alakwaa F.M., Chaudhary K., Garmire L.X. Deep learning accurately predicts estrogen receptor status in breast cancer metabolomics data. J. Proteome Res. 2018;17:337–347. doi: 10.1021/acs.jproteome.7b00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Günther U.L. Metabolomics biomarkers for breast cancer. Pathobiology. 2015;82:153–165. doi: 10.1159/000430844. [DOI] [PubMed] [Google Scholar]
- 47.Johnson C.H., Ivanisevic J., Siuzdak G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016;17:451–459. doi: 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kapoore R.V., Coyle R., Staton C.A., Brown N.J., Vaidyanathan S. Influence of washing and quenching in profiling the metabolome of adherent mammalian cells: A case study with the metastatic breast cancer cell line MDA-MB-231. Analyst. 2017;142:2038–2049. doi: 10.1039/C7AN00207F. [DOI] [PubMed] [Google Scholar]
- 49.Mandal R., Chamot D., Wishart D.S. The role of the Human Metabolome Database in inborn errors of metabolism. J. Inherit. Metab. Dis. 2018;41:329–336. doi: 10.1007/s10545-018-0137-8. [DOI] [PubMed] [Google Scholar]
- 50.Schrimpe-Rutledge A.C., Codreanu S.G., Sherrod S.D., McLean J.A. Untargeted metabolomics strategies—challenges and emerging directions. J. Am. Soc. Mass Spectrom. 2016;27:1897–1905. doi: 10.1007/s13361-016-1469-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aboud O.A., Weiss R.H. New opportunities from the cancer metabolome. Clin. Chem. 2013;59:138–146. doi: 10.1373/clinchem.2012.184598. [DOI] [PubMed] [Google Scholar]
- 52.Wu J., Yang R., Zhang L., Li Y., Liu B., Kang H., Fan Z., Tian Y., Liu S., Li T. Metabolomics research on potential role for 9-cis-retinoic acid in breast cancer progression. Cancer Sci. 2018;109:2315–2326. doi: 10.1111/cas.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dougan M.M., Li Y., Chu L.W., Haile R.W., Whittemore A.S., Han S.S., Moore S.C., Sampson J.N., Andrulis I.L., John E.M., et al. Metabolomic profiles in breast cancer:a pilot case-control study in the breast cancer family registry. BMC Cancer. 2018;18:532. doi: 10.1186/s12885-018-4437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanaan Y.M., Sampey B.P., Beyene D., Esnakula A.K., Naab T.J., Ricks-Santi L.J., Dasi S., Day A., Blackman K.W., Frederick W., et al. Metabolic Profile of Triple-negative Breast Cancer in African-American Women Reveals Potential Biomarkers of Aggressive Disease. Cancer Genom. —Proteom. 2014;11:279–294. [PubMed] [Google Scholar]
- 55.Kisanga E.R., Mellgren G., Lien E.A. Excretion of hydroxylated metabolites of tamoxifen in human bile and urine. Anticancer Res. 2005;25:4487–4492. [PubMed] [Google Scholar]
- 56.Visekruna A., Luu M. The Role of Short-Chain Fatty Acids and Bile Acids in Intestinal and Liver Function, Inflammation, and Carcinogenesis. Front. Cell Dev. Biol. 2021;9:703218. doi: 10.3389/fcell.2021.703218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arnone A.A., Cline J.M., Soto-Pantoja D.R., Cook K.L. Investigating the role of endogenous estrogens, hormone replacement therapy, and blockade of estrogen receptor-α activity on breast metabolic signaling. Breast Cancer Res. Treat. 2021;190:53–67. doi: 10.1007/s10549-021-06354-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hanahan D., Weinberg R.A. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 59.Meadows A.L., Kong B., Berdichevsky M., Roy S., Rosiva R., Blanch H.W., Clark D.S. Metabolic and morphological differences between rapidly proliferating cancerous and normal breast epithelial cells. Biotechnol. Prog. 2008;24:334–341. doi: 10.1021/bp070301d. [DOI] [PubMed] [Google Scholar]
- 60.Bathen T.F., Geurts B., Sitter B., Fjøsne H.E., Lundgren S., Buydens L.M., Gribbestad I.S., Postma G., Giskeødegård G.F. Feasibility of MR metabolomics for immediate analysis of resection margins during breast cancer surgery. PLoS ONE. 2013;8:e61578. doi: 10.1371/journal.pone.0061578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mamtimin B., Hizbulla M., Kurbantay N., You L., Yan X., Upur H. An magnetic resonance-based plasma metabonomic investigation on abnormal Savda in different complicated diseases. J. Tradit. Chin. Med. 2014;34:166–172. doi: 10.1016/S0254-6272(14)60073-X. [DOI] [PubMed] [Google Scholar]
- 62.Maria R.M., Altei W.F., Andricopulo A.D., Becceneri A.B., Cominetti M.R., Venâncio T., Colnago L.A. Characterization of metabolic profile of intact non-tumor and tumor breast cells by high-resolution magic angle spinning nuclear magnetic resonance spectroscopy. Anal. Biochem. 2015;488:14–18. doi: 10.1016/j.ab.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 63.Cala M.P., Aldana J., Medina J., Sánchez J., Guio J., Wist J., Meesters R.J.W. Multiplatform plasma metabolic and lipid fingerprinting of breast cancer: A pilot control-case study in Colombian Hispanic women. PLoS ONE. 2018;13:e0190958. doi: 10.1371/journal.pone.0190958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suman S., Sharma R.K., Kumar V., Sinha N., Shukla Y. Metabolic fingerprinting in breast cancer stages through (1)H NMR spectroscopy-based metabolomic analysis of plasma. J. Pharm. Biomed. Anal. 2018;160:38–45. doi: 10.1016/j.jpba.2018.07.024. [DOI] [PubMed] [Google Scholar]
- 65.Murata T., Yanagisawa T., Kurihara T., Kaneko M., Ota S., Enomoto A., Tomita M., Sugimoto M., Sunamura M., Hayashida T., et al. Salivary metabolomics with alternative decision tree-based machine learning methods for breast cancer discrimination. Breast Cancer Res. Treat. 2019;177:591–601. doi: 10.1007/s10549-019-05330-9. [DOI] [PubMed] [Google Scholar]
- 66.Lin X., Xiao Z., Chen T., Liang S.H., Guo H. Glucose Metabolism on Tumor Plasticity, Diagnosis, and Treatment. Front. Oncol. 2020;10:317. doi: 10.3389/fonc.2020.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mukha A., Dubrovska A. Metabolic Targeting of Cancer Stem Cells. Front. Oncol. 2020;10:537930. doi: 10.3389/fonc.2020.537930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garber K. Energy boost: The Warburg effect returns in a new theory of cancer. J. Natl. Cancer Inst. 2004;96:1805–1806. doi: 10.1093/jnci/96.24.1805. [DOI] [PubMed] [Google Scholar]
- 69.Choi S.Y., Collins C.C., Gout P.W., Wang Y. Cancer-generated lactic acid: A regulatory, immunosuppressive metabolite? J. Pathol. 2013;230:350–355. doi: 10.1002/path.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu Q., Deblois G., Cruickshank J., Duan S., Lima-Fernandes E., Haight J., Tonekaboni S.A.M., Fortier A.-M., Kuasne H., McKee T.D. GLUT1 inhibition blocks growth of RB1-positive triple negative breast cancer. Nat. Commun. 2020;11:4205. doi: 10.1038/s41467-020-18020-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tayyari F., Gowda G.N., Olopade O.F., Berg R., Yang H.H., Lee M.P., Ngwa W.F., Mittal S.K., Raftery D., Mohammed S.I. Metabolic profiles of triple-negative and luminal A breast cancer subtypes in African-American identify key metabolic differences. Oncotarget. 2018;9:11677. doi: 10.18632/oncotarget.24433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Giskeødegård G.F., Lundgren S., Sitter B., Fjøsne H.E., Postma G., Buydens L.M., Gribbestad I.S., Bathen T.F. Lactate and glycine-potential MR biomarkers of prognosis in estrogen receptor-positive breast cancers. NMR Biomed. 2012;25:1271–1279. doi: 10.1002/nbm.2798. [DOI] [PubMed] [Google Scholar]
- 73.Röhrig F., Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer. 2016;16:732–749. doi: 10.1038/nrc.2016.89. [DOI] [PubMed] [Google Scholar]
- 74.Brown M.S., Goldstein J.L. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 75.Furuhashi M., Hotamisligil G.S. Fatty acid-binding proteins: Role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008;7:489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kazantzis M., Stahl A. Fatty acid transport proteins, implications in physiology and disease. Biochim. Biophys. Acta. 2012;1821:852–857. doi: 10.1016/j.bbalip.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koundouros N., Poulogiannis G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer. 2020;122:4–22. doi: 10.1038/s41416-019-0650-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang B., Rong X., Palladino E.N.D., Wang J., Fogelman A.M., Martín M.G., Alrefai W.A., Ford D.A., Tontonoz P. Phospholipid Remodeling and Cholesterol Availability Regulate Intestinal Stemness and Tumorigenesis. Cell Stem Cell. 2018;22:206–220.e204. doi: 10.1016/j.stem.2017.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Snaebjornsson M.T., Janaki-Raman S., Schulze A. Greasing the Wheels of the Cancer Machine: The Role of Lipid Metabolism in Cancer. Cell Metab. 2020;31:62–76. doi: 10.1016/j.cmet.2019.11.010. [DOI] [PubMed] [Google Scholar]
- 80.Nieman K.M., Kenny H.A., Penicka C.V., Ladanyi A., Buell-Gutbrod R., Zillhardt M.R., Romero I.L., Carey M.S., Mills G.B., Hotamisligil G.S., et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 2011;17:1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wen Y.A., Xing X., Harris J.W., Zaytseva Y.Y., Mitov M.I., Napier D.L., Weiss H.L., Mark Evers B., Gao T. Adipocytes activate mitochondrial fatty acid oxidation and autophagy to promote tumor growth in colon cancer. Cell Death Dis. 2017;8:e2593. doi: 10.1038/cddis.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y.Y., Attané C., Milhas D., Dirat B., Dauvillier S., Guerard A., Gilhodes J., Lazar I., Alet N., Laurent V., et al. Mammary adipocytes stimulate breast cancer invasion through metabolic remodeling of tumor cells. JCI Insight. 2017;2:e87489. doi: 10.1172/jci.insight.87489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang T., Fahrmann J.F., Lee H., Li Y.J., Tripathi S.C., Yue C., Zhang C., Lifshitz V., Song J., Yuan Y., et al. JAK/STAT3-Regulated Fatty Acid β-Oxidation Is Critical for Breast Cancer Stem Cell Self-Renewal and Chemoresistance. Cell Metab. 2018;27:136–150.e135. doi: 10.1016/j.cmet.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.His M., Viallon V., Dossus L., Gicquiau A., Achaintre D., Scalbert A., Ferrari P., Romieu I., Onland-Moret N.C., Weiderpass E. Prospective analysis of circulating metabolites and breast cancer in EPIC. BMC Med. 2019;17:178. doi: 10.1186/s12916-019-1408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Longo N., Frigeni M., Pasquali M. Carnitine transport and fatty acid oxidation. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2016;1863:2422–2435. doi: 10.1016/j.bbamcr.2016.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gyamfi J., Yeo J.H., Kwon D., Min B.S., Cha Y.J., Koo J.S., Jeong J., Lee J., Choi J. Interaction between CD36 and FABP4 modulates adipocyte-induced fatty acid import and metabolism in breast cancer. NPJ Breast Cancer. 2021;7:129. doi: 10.1038/s41523-021-00324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kühn T., Floegel A., Sookthai D., Johnson T., Rolle-Kampczyk U., Otto W., von Bergen M., Boeing H., Kaaks R. Higher plasma levels of lysophosphatidylcholine 18: 0 are related to a lower risk of common cancers in a prospective metabolomics study. BMC Med. 2016;14:13. doi: 10.1186/s12916-016-0552-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Abdelzaher E., Mostafa M.F. Lysophosphatidylcholine acyltransferase 1 (LPCAT1) upregulation in breast carcinoma contributes to tumor progression and predicts early tumor recurrence. Tumor Biol. 2015;36:5473–5483. doi: 10.1007/s13277-015-3214-8. [DOI] [PubMed] [Google Scholar]
- 89.Hilvo M., Denkert C., Lehtinen L., Müller B., Brockmöller S., Seppänen-Laakso T., Budczies J., Bucher E., Yetukuri L., Castillo S., et al. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 2011;71:3236–3245. doi: 10.1158/0008-5472.CAN-10-3894. [DOI] [PubMed] [Google Scholar]
- 90.Wei Z., Liu X., Cheng C., Yu W., Yi P. Metabolism of Amino Acids in Cancer. Front. Cell Dev. Biol. 2020;8:603837. doi: 10.3389/fcell.2020.603837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Geck R.C., Toker A. Nonessential amino acid metabolism in breast cancer. Adv. Biol. Regul. 2016;62:11–17. doi: 10.1016/j.jbior.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 92.Sugimoto M., Wong D.T., Hirayama A., Soga T., Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010;6:78–95. doi: 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jain M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., Souza A.L., Kafri R., Kirschner M.W., Clish C.B., Mootha V.K. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Willmann L., Schlimpert M., Halbach S., Erbes T., Stickeler E., Kammerer B. Metabolic profiling of breast cancer: Differences in central metabolism between subtypes of breast cancer cell lines. J. Chromatogr B Anal. Technol. Biomed. Life Sci. 2015;1000:95–104. doi: 10.1016/j.jchromb.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 95.Wang Q., Sun T., Cao Y., Gao P., Dong J., Fang Y., Fang Z., Sun X., Zhu Z. A dried blood spot mass spectrometry metabolomic approach for rapid breast cancer detection. Onco. Targets Ther. 2016;9:1389–1398. doi: 10.2147/OTT.S95862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Du S., Wang Y., Alatrash N., Weatherly C.A., Roy D., MacDonnell F.M., Armstrong D.W. Altered profiles and metabolism of l- and d-amino acids in cultured human breast cancer cells vs. non-tumorigenic human breast epithelial cells. J. Pharm. Biomed. Anal. 2019;164:421–429. doi: 10.1016/j.jpba.2018.10.047. [DOI] [PubMed] [Google Scholar]
- 97.Cha Y.J., Kim E.S., Koo J.S. Amino Acid Transporters and Glutamine Metabolism in Breast Cancer. Int. J. Mol. Sci. 2018;19:907. doi: 10.3390/ijms19030907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kandasamy P., Gyimesi G., Kanai Y., Hediger M.A. Amino acid transporters revisited: New views in health and disease. Trends Biochem. Sci. 2018;43:752–789. doi: 10.1016/j.tibs.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 99.Bhutia Y.D., Babu E., Ramachandran S., Ganapathy V. Amino Acid transporters in cancer and their relevance to “glutamine addiction”: Novel targets for the design of a new class of anticancer drugs. Cancer Res. 2015;75:1782–1788. doi: 10.1158/0008-5472.CAN-14-3745. [DOI] [PubMed] [Google Scholar]
- 100.van Geldermalsen M., Wang Q., Nagarajah R., Marshall A.D., Thoeng A., Gao D., Ritchie W., Feng Y., Bailey C.G., Deng N., et al. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene. 2016;35:3201–3208. doi: 10.1038/onc.2015.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Metallo C.M., Gameiro P.A., Bell E.L., Mattaini K.R., Yang J., Hiller K., Jewell C.M., Johnson Z.R., Irvine D.J., Guarente L., et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Le A., Lane A.N., Hamaker M., Bose S., Gouw A., Barbi J., Tsukamoto T., Rojas C.J., Slusher B.S., Zhang H., et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Budczies J., Pfitzner B.M., Györffy B., Winzer K.-J., Radke C., Dietel M., Fiehn O., Denkert C. Glutamate enrichment as new diagnostic opportunity in breast cancer. Int. J. Cancer. 2015;136:1619–1628. doi: 10.1002/ijc.29152. [DOI] [PubMed] [Google Scholar]
- 104.Zhou W.-X., Chen C., Liu X.-Q., Li Y., Lin Y.-L., Wu X.-T., Kong L.-Y., Luo J.-G. Discovery and optimization of withangulatin A derivatives as novel glutaminase 1 inhibitors for the treatment of triple-negative breast cancer. Eur. J. Med. Chem. 2021;210:112980. doi: 10.1016/j.ejmech.2020.112980. [DOI] [PubMed] [Google Scholar]
- 105.Pollari S., Käkönen S.M., Edgren H., Wolf M., Kohonen P., Sara H., Guise T., Nees M., Kallioniemi O. Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res. Treat. 2011;125:421–430. doi: 10.1007/s10549-010-0848-5. [DOI] [PubMed] [Google Scholar]
- 106.Maddocks O.D., Labuschagne C.F., Adams P.D., Vousden K.H. Serine Metabolism Supports the Methionine Cycle and DNA/RNA Methylation through De Novo ATP Synthesis in Cancer Cells. Mol. Cell. 2016;61:210–221. doi: 10.1016/j.molcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Maddocks O.D.K., Athineos D., Cheung E.C., Lee P., Zhang T., van den Broek N.J.F., Mackay G.M., Labuschagne C.F., Gay D., Kruiswijk F., et al. Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature. 2017;544:372–376. doi: 10.1038/nature22056. [DOI] [PubMed] [Google Scholar]
- 108.Labuschagne C.F., van den Broek N.J., Mackay G.M., Vousden K.H., Maddocks O.D. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7:1248–1258. doi: 10.1016/j.celrep.2014.04.045. [DOI] [PubMed] [Google Scholar]
- 109.Altman B.J., Stine Z.E., Dang C.V. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat. Rev. Cancer. 2016;16:619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bernfeld E., Foster D.A. Glutamine as an Essential Amino Acid for KRas-Driven Cancer Cells. Trends Endocrinol. Metab. 2019;30:357–368. doi: 10.1016/j.tem.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 111.Maggi M., Scotti C. Enzymes in Metabolic Anticancer Therapy. Adv. Exp. Med. Biol. 2019;1148:173–199. doi: 10.1007/978-981-13-7709-9_9. [DOI] [PubMed] [Google Scholar]
- 112.Vettore L., Westbrook R.L., Tennant D.A. New aspects of amino acid metabolism in cancer. Br. J. Cancer. 2020;122:150–156. doi: 10.1038/s41416-019-0620-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Abdelmagid S.A., Rickard J.A., McDonald W.J., Thomas L.N., Too C.K. CAT-1-mediated arginine uptake and regulation of nitric oxide synthases for the survival of human breast cancer cell lines. J. Cell Biochem. 2011;112:1084–1092. doi: 10.1002/jcb.23022. [DOI] [PubMed] [Google Scholar]
- 114.Cao Y., Feng Y., Zhang Y., Zhu X., Jin F. L-Arginine supplementation inhibits the growth of breast cancer by enhancing innate and adaptive immune responses mediated by suppression of MDSCs in vivo. BMC Cancer. 2016;16:343. doi: 10.1186/s12885-016-2376-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lin J., Lee I.M., Song Y., Cook N.R., Selhub J., Manson J.E., Buring J.E., Zhang S.M. Plasma homocysteine and cysteine and risk of breast cancer in women. Cancer Res. 2010;70:2397–2405. doi: 10.1158/0008-5472.CAN-09-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nagata C., Wada K., Tsuji M., Hayashi M., Takeda N., Yasuda K. Plasma amino acid profiles are associated with biomarkers of breast cancer risk in premenopausal Japanese women. Cancer Causes Control. 2014;25:143–149. doi: 10.1007/s10552-013-0316-8. [DOI] [PubMed] [Google Scholar]
- 117.Knott S.R.V., Wagenblast E., Khan S., Kim S.Y., Soto M., Wagner M., Turgeon M.-O., Fish L., Erard N., Gable A.L., et al. Asparagine bioavailability governs metastasis in a model of breast cancer. Nature. 2018;554:378–381. doi: 10.1038/nature25465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tang X., Lin C.C., Spasojevic I., Iversen E.S., Chi J.T., Marks J.R. A joint analysis of metabolomics and genetics of breast cancer. Breast Cancer Res. 2014;16:415. doi: 10.1186/s13058-014-0415-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hassan M.A., Al-Sakkaf K., Shait Mohammed M.R., Dallol A., Al-Maghrabi J., Aldahlawi A., Ashoor S., Maamra M., Ragoussis J., Wu W., et al. Integration of Transcriptome and Metabolome Provides Unique Insights to Pathways Associated With Obese Breast Cancer Patients. Front. Oncol. 2020;10:804. doi: 10.3389/fonc.2020.00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jové M., Collado R., Quiles J.L., Ramírez-Tortosa M.C., Sol J., Ruiz-Sanjuan M., Fernandez M., de la Torre Cabrera C., Ramírez-Tortosa C., Granados-Principal S., et al. A plasma metabolomic signature discloses human breast cancer. Oncotarget. 2017;8:19522–19533. doi: 10.18632/oncotarget.14521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jasbi P., Wang D., Cheng S.L., Fei Q., Cui J.Y., Liu L., Wei Y., Raftery D., Gu H. Breast cancer detection using targeted plasma metabolomics. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019;1105:26–37. doi: 10.1016/j.jchromb.2018.11.029. [DOI] [PubMed] [Google Scholar]
- 122.Borgan E., Sitter B., Lingjærde O.C., Johnsen H., Lundgren S., Bathen T.F., Sørlie T., Børresen-Dale A.L., Gribbestad I.S. Merging transcriptomics and metabolomics--advances in breast cancer profiling. BMC Cancer. 2010;10:628. doi: 10.1186/1471-2407-10-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cao B., Li M., Zha W., Zhao Q., Gu R., Liu L., Shi J., Zhou J., Zhou F., Wu X., et al. Metabolomic approach to evaluating adriamycin pharmacodynamics and resistance in breast cancer cells. Metabolomics. 2013;9:960–973. doi: 10.1007/s11306-013-0517-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wei S., Liu L., Zhang J., Bowers J., Gowda G.A., Seeger H., Fehm T., Neubauer H.J., Vogel U., Clare S.E., et al. Metabolomics approach for predicting response to neoadjuvant chemotherapy for breast cancer. Mol. Oncol. 2013;7:297–307. doi: 10.1016/j.molonc.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huang S., Chong N., Lewis N.E., Jia W., Xie G., Garmire L.X. Novel personalized pathway-based metabolomics models reveal key metabolic pathways for breast cancer diagnosis. Genome Med. 2016;8:34. doi: 10.1186/s13073-016-0289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]