Abstract
Mycorrhizal fungi can colonize multiple trees of a single or multiple taxa, facilitating bidirectional exchange of carbon between trees. Mycorrhiza‐induced carbon transfer was shown in the forest, but it is unknown whether carbon is shared symmetrically among tree species, and if not, which tree species are better donors and which are better recipients. Here, we test this question by investigating carbon transfer dynamics among five Mediterranean tree species in a microcosm system, including both ectomycorrhizal (EM) and arbuscular (AM) plants. Trees were planted together in “community boxes” using natural soil from a mixed forest plot that serves as a habitat for all five tree species and their native mycorrhizal fungi. In each box, only the trees of a single species were pulse‐labelled with 13CO2. We found that carbon transfer was asymmetric, with oak being a better donor, and pistacia and cypress better recipients. Shared mycorrhizal species may have facilitated carbon transfer, but their diversity did not affect the amount, nor timing, of the transfer. Overall, our findings in a microcosm system expose rich, but hidden, belowground interactions in a diverse population of trees and mycorrhizal fungi. The asymmetric carbon exchange among cohabiting tree species could potentially contribute to forest resilience in an uncertain future.
Keywords: 13CO2 labelling, carbon transfer, fungal community, mycorrhizal networks, resource sharing, rhizosphere
1. INTRODUCTION
Roots of vascular plants interact with mycorrhizal fungi along with other components of the soil microbiome including nonmycorrhizal fungi, archaea, and bacteria (Högberg et al., 2008). The symbiotic interaction between the fungus and the root relies on transmission of soil‐derived nutrients from the fungus to the host tree (Collins Johnson et al., 2010), increasing root absorption of water by the fungal hyphae and mediating the interaction of the root with other microbes in the soil (Aroca et al., 2007; Hestrin et al., 2019). The heterotrophic fungus benefits from the interaction by receiving carbon from the autotrophic host tree (Högberg et al., 2008). There are two main functional groups of mycorrhizae: ectomycorrhiza (EM) which do not penetrate the root cortex of the host and interact mainly with trees that are located in seasonally cold and dry climates, and arbuscular mycorrhiza (AM) whose hyphae penetrate the root cortex and interact mainly with plants that are located in seasonally warm and wet climates (Steidinger et al., 2019). Each individual tree may interact with tens to hundreds of different mycorrhizal taxa at the same time (Bahram et al., 2011). It is generally thought that plants associate exclusive with a single mycorrhizal type, but it has also been shown that some plants can be colonized by fungi of several mycorrhizal types, including both arbuscular mycorrhizal fungi (AMF) and ectomycorrhizal fungi (EMF) (Teste et al., 2020). The fungal affinity to their hosts is also complex since fungi have a wide range of specificity with some species that are specialist to their host trees and others that are generalist, having multiple partners (Massicotte et al., 1994; Heijden et al., 2015). The formation of common mycelial networks among trees depends on both the neighbouring trees specificity to compatible fungal species and on the fungal specificity to the host trees.
Trees in the forest compete over limited resources such as light and nutrients (Lindenmayer & Laurance, 2017). However, some studies demonstrated carbon exchange among trees which might serve as a mutualistic interaction between them. Studies showing asymmetrical carbon transfer to young seedlings and supporting their establishment (Van Der Heijden & Horton, 2009) or studies presenting increased carbon transfer to trees under starvation (Simard, et al., 1997) certainly point that way. Several studies showed different benefits from CMNs among tree species. In a microcosm with the EM trees Pinus and Larix, Pinus got more carbon when having Suillus bovinus fungi and Larix got more carbon when ectomycorrhizal fungus was Suillus grevillei or Boletinus cavipes (Finlay, 1989). In AM plants it was shown that AMF favoured legume over grasses (Scheublin et al., 2007). Carbon transfer has been shown between legumes of the same species and between trees of different, usually related, species in the laboratory and later in the forest (Fitter et al.,1998; Francis & Read, 1984; Högberg et al., 2008; Simard, et al., 1997). Recently it has been shown that carbon is bidirectionally transferred among trees of different taxa in a mature forest (Klein et al., 2016). There are several possible mechanisms underlying the transfer of carbon between trees: Roots of different trees form fusions (natural root grafts) and can exchange carbon, water, or nutrients among them (Fraser et al., 2006; Graham & Bormann, 1966; Nara, 2006). However, root grafts occur mostly between trees of the same species (Fraser et al., 2006; Graham & Bormann, 1966; Woods & Brock, 1964) and therefore it does not provide a good explanation for carbon transfer between trees of different taxa. Another possible explanation is that carbon of one tree is secreted to the soil and is being absorbed by a root of a different tree (Pérez‐Pazos et al., 2021). A third possibility is that trees exchange carbon through mycorrhizal networks (Fitter et al., 1998; Francis & Read, 1984; Högberg et al., 2008; Simard, et al., 1997). Some evidence have been found to support this theory in mature trees (Klein et al., 2016). First, fruit bodies of mycorrhizal fungi that interacted with carbon‐labelled trees contained labelled carbon in contrast to identical fungi located far from the carbon‐labelled trees and to fungi that did not interact with the labelled trees (Klein et al., 2016). In addition, trees that shared more mycorrhizal fungi tended to exchange more carbon (Rog et al., 2020).
The temporal and quantitative dynamics of carbon transfer between trees is still unclear. Once carbon is assimilated in the leaf of a tree it can reach three major sinks: respiration, biomass, and exudation from the root (Epron et al., 2012; Klein & Hoch, 2015). Carbon reaching the phloem moves in both directions and reaches the roots according to the source‐sink gradient from the leaf to the root (Liesche et al., 2015). Studies have shown that carbon flux in the phloem is faster in angiosperms compared to gymnosperms with the rate of 0.2–6 m/h and 0.1–0.2 m/h, respectively (Epron et al., 2012). This is probably due to the anatomical differences in their transport system (Liesche et al., 2015). A pulse labelling experiment on individual Mediterranean saplings showed carbon allocation to roots three days post labelling in saplings of Pinus halepensis, Cupressus sempervirens, Quercus calliprinos, Ceratonia siliqua and Pistacia lentiscus (Rog, Jakoby, et al., 2021 ). The partitioning of carbon to belowground compartments was 28%–38% in gymnosperms (Pinus halepensis and Cupressus sempervirens), and 5%–10% in angiosperms (Quercus calliprinos, Ceratonia siliqua and Pistacia lentiscus) (Rog, Jakoby, et al., 2021). These quantities refer to allocation of carbon from the leaf to the root of the same tree. After reaching the roots, carbon can transfer to the rhizosphere, which is composed of the soil and the rhizosphere microbiota, including mycorrhizal fungi. Carbon is expected to further decrease while transferring to mycorrhizal fungi and to the roots of a neighbouring tree 2021. Indeed, in an experiment studying carbon transfer between paper birch and Douglas fir saplings it was shown that small amounts of 4.7% of the carbon fixed by paper birch were transferred to Douglas fir (Simard, et al., 1997). While most common mycorrhizal networks (CMNs) were studied in temperate ecosystems with rich organic soil and wet climates, less in known on CMNs in water limited environments such as the Mediterranean forest. In such environments the trees must deal with frequent droughts and often limit their activity to specific seasons or times of the day. This can affect the dependency of trees on the mycorrhizal network for resource uptake. It can also possibly change the specificity level of mycorrhizal fungi, connecting to additional hosts to reduce risk and have an advantage in a harsher environment. Considering that drought periods are becoming longer and harsher also in temperate forests (Klein et al., 2022), ecosystems such as the Mediterranean forest can help us predict how mycorrhizal networks will look like in the future.
Here, saplings of five Mediterranean tree species, EM: Pinus halepensis and Quercus calliprinos (Torres & Honrubia, 1994; Trocha et al., 2012), and AM: Cupressus sempervirens (Zarik et al., 2016), Ceratonia siliqua (Essahibi et al., 2018; Lahcen et al., 2012) and Pistacia lentiscus (Caravaca et al., 2002; Green et al., 2005) were labelled with 13CO2. We measured the amount and direction of belowground carbon transfer to other saplings of the same species and of different species. We aimed to identify which tree species serve as carbon donors and which serve as carbon recipients in this microforest community, and to map the mycorrhizal networks connecting them. To do so, we planted trees in microcosm system and used 13CO2 pulse labelling to track carbon allocation within and between trees. Then we used high throughput DNA sequencing to identify the mycorrhizal fungi colonizing each tree. We hypothesized that carbon would transfer: (i) in a bidirectional way in an asymmetrical manner, (ii) among tree species saplings that share mycorrhizal species, and (iii) among EMF hosts and among AMF hosts but not between these two guilds of plants.
2. MATERIALS AND METHODS
2.1. Experimental design
Two‐year old saplings were obtained from KKL forest nursery (Jewish National Fund, Eshtaol, Israel), where they were grown on clean peat soil. Saplings of four tree species were planted together in four boxes at the size of 240 L, width: 80 cm, height: 50 cm, depth: 60 cm (October 2017): Pinus halepensis, Cupressus sempervirens, Quercus calliprinos, and Ceratonia siliqua (n = 9 for each species). For height, diameter, and dry weight of final biomass for each sapling, see Table S1. In each “community box” we planted six saplings of one of the four species and one sapling from each of the other species. In each box we also planted three saplings of Pistacia lentiscus, the major understory tree in the mixed Mediterranean forest, to get a total of 12 saplings per box (schematic diagram of the saplings arrangement in the boxes in Figure 1). While Pistacia was not tested as a donor in our layout, it served as a verification of C transfer, since it was not exposed to any labelling. In addition, we had three pots with an individual tree for each of the species. The five tree species studied in the experiment are dominant in the Mediterranean forest (Lapidot et al., 2019; Rog, Tague, et al., 2021) and share territory in many other mixed evergreen forests. Pinus halepensis and Cupressus sempervirens are gymnosperms, Quercus calliprinos, Ceratonia siliqua, and Pistacia lentiscus are evergreen angiosperms. For brevity, we hereby refer to each of the five species by its genus name, that is, Pinus, Cupressus, Quercus, Ceratonia and Pistacia. To maintain the natural environment of the root and its associations with microorganisms in the soil, we grew the saplings in sand mixed with soil from the Yishi forest (ratio of 1:4), which serves as habitat for these five species (Lapidot et al., 2019; Rog, Jakoby, et al., 2021). The soil is composed mainly with terra rossa and characterized by neutral acidity (pH 6.9–7.2) and the same mixture of soil was used to grow all trees.
FIGURE 1.
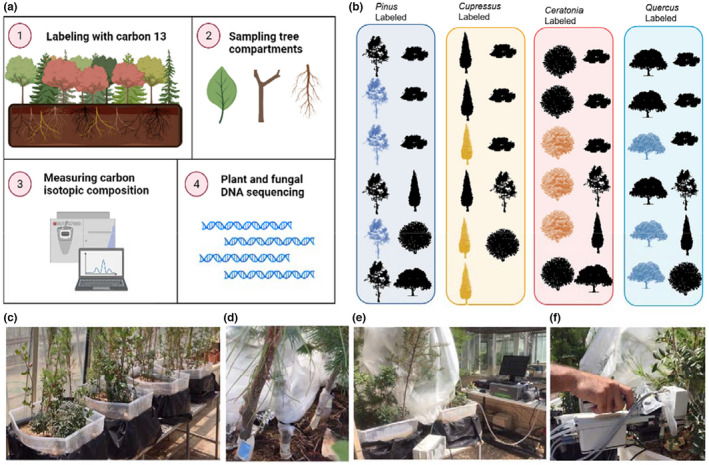
Experimental design. (a) Schematic representation of the experimental procedures. (b) The arrangement of the saplings in the four boxes. In each box,12 saplings were planted. Three saplings of one species were covered with plastic bags and enriched with 13CO2. (c) Four community boxes containing 12 saplings. (d) Sealed plastic bag covering the labelled plants. (e) Continued carbon detection by cavity ring down spectrometer (CRDS). (f) Concentration and flux measurements using infrared gas analyser (IRGA)
The saplings were grown in the Weizmann institute glasshouse in Rehovot, Israel. The glasshouse had light extinction of 25% and temperature was kept at ~25°C with no humidity control. The trees were irrigated well. For minimizing labelled C contamination, the trees were transferred to ambient environment in the Weizmann institute garden before and post labelling. Climate conditions in Rehovot from the time of labelling to the end of the experiment (26 September 2019 to 2 May 2020) were as follows: overall minimum temperature of 3.1°C and maximum temperature of 39.3°C, average temperature of 17.4°C, average midday global light intensity of 598.9 W m−2 and average midday relative humidity of 70.8%. For more details see Figure S1. Environmental conditions around Rehovot during the day of labelling (October 2019) were as follows: minimum temperature of 19.2°C and maximum temperature of 30.5°C, average temperature at night and day were 24.5°C and 27.9°C respectively, midday global light intensity of 799 W * m−2, midday relative humidity of 53.5% (Beit Dagan meteorological station, 10 km North of the Weizmann institute).
2.2. 13CO2 labelling
Three trees of a single species from each box were labelled with 13CO2 for a 4‐h pulse starting at noon in October 2019 (11:05 AM–15:00 PM). The canopy of the labelled trees was wrapped and sealed by an isolating plastic bag (Figure 1) and the surface of the soil was covered with plastic to prevent contamination of the soil. The bag was flushed with zero air containing filtered ambient air with no CO2 to keep low levels of 12CO2. The bag was then enriched with 13CO2 by adding 50% hydrochloric acid to labelled sodium bicarbonate (NaH13CO3, 13C, 99%, Cambridge Isotope Laboratories), producing salt, water and 13CO2. To validate there were no leakages of labelled carbon out of the bag, the concentration of 13CO2 outside of the bag was monitored during the labelling by Cavity Ring Down spectrometer (CRDS) G2121‐i isotopic CO2 (Picarro G2131i, Picarro).
2.3. Leaf and root gas exchange
Gas exchange of the leaves and roots was measured before and post labelling, and at the evening of each sampling day (0, 1, 2, 4, 6, 10, 15, 28, 42, 56, 110 and 240 days post labelling). Root samples were disconnected and immediately measured. Mature leaf samples were taken from similar positions on the plant. Both respiration and net assimilation rate were measured by a portable photosynthesis infrared gas analyser (IRGA; GFS −3000, Walz), with the following conditions: Standard leaf chamber (Walz 3010 – S); ambient CO2 concentration of 400 ppm; flow rate of 750 µmol s−1; and impeller speed of 7. Net assimilation rate in leaves was measured to estimate the amount of labelled carbon assimilated in the trees. To determine the net assimilation rate, dark respiration rate measurements were subtracted from the assimilation rate. The respiration of leaves and roots was measured during the evenings together with carbon isotopic ratio to quantify the amount of labelled carbon in the respiration of each tree. The exhaust ventilation of the IRGA module was directly interfaced to a CO2 analyser (Picarro G2131i) which measured the ratio between 13CO2 and 12 CO2 in the respired CO2 from each sapling (more details on the setup in Rog, Jakoby, et al.,2021).
2.4. Biomass sampling
For labelled carbon measurements, lateral roots, branches and leaves of all saplings in the community boxes were sampled before labelling and through 8 months post labelling at 12 time points (0, 1, 2, 4, 6, 10, 15, 28, 42, 56, 110 and 240 days post labelling). For root samples we collected one replicate of living lateral fine root from each tree in each time point. The roots were followed by tracing the stem gently, using tweezers and screws, in order to minimize misidentification of roots. Several leaves (9–12) and branches were taken from three directions of the tree so it would represent the whole tree. During the sampling four saplings died: Cupressus 4, Quercus 8 and Ceratonia 8 from box 2 and Ceratonia 1 from box 3. All samples were oven dried at 60°C for 48 h (Thermo Fisher Scientific). Then, the samples were cut and ground to fine powder with a bead beater (Restch GmbH, Haan, Germany). Three replicates of soil samples were collected from the centre of each box at four time points: 0.5, 2, 113 and 230 days post labelling and were kept in –80°C. All samples were oven dried at 60°C for 48 h. Then, 10 mg of sample was weighed in tin capsules and measured using the combustion module and the 13C analyser (Picarro G2131i), similarly to the plant biomass measurements.
After 8 months, the community boxes were taken apart, saplings were sacrificed and two replicates of roots with the biomass of ~0.7 g were taken from each tree and kept in –80°C for fungal DNA extraction. Then the saplings were divided into three compartments: leaves, aboveground woody tissues, and roots (images of root systems are shown in Figure S2). Each of the compartments was weighed separately. To calculate the ratio of mass and surface area, three subsamples were taken from each compartment (Table S2). Each subsample was weighed and scanned by a tabletop scanner (MFP ‐M477fdh, HP). The scanning was analysed by ImageJ software (Schneider et al., 2012) using a threshold tool and area calculation. Then, the ratio of the tree subsamples was averaged.
2.5. Root tip sampling for mycorrhizal species identification
For root tip collection, another set of root samples was collected after the termination of the experiment. Two replicates for lateral root branchlets of ~500 mg and ~0.3 mm diameter were collected. The lateral roots were washed with tap water on a 1 mm sieve to remove attached soil and were directly stored in an ice box. The samples were kept in −80°°C until root tips collection. Each root tip found on each lateral root replicate was picked using a dissecting scope and tweezers that were flame sterilized and washed with 70% EtOH between each sample. In addition, 16 root tips were collected from each of the individual trees. The root tips were lyophilized for 48 h and ground using a bead beater for DNA extraction and for carbon measurements.
2.6. Labelled carbon measurement
One mg of each grounded root was weighed (Sartorius Stedim Biotech) and packed in a tin capsule that was inserted and burned in PICARRO combustion module (ECS 4010, Costech Analytical). δ13C was analysed by G2121‐i Isotopic CO2 (CRDS). The C isotope ratio was calculated as:
| (1) |
is represented in ‰ units where R = 13 C/12 C. The sample ratio is relative to the ViennaPeeDee Belemnite (V‐PDB) standard. An international standard (IAEA‐CH‐3, Cellulose, International Atomic Energy Agency, Vienna, Austria) was used every 11 samples and values of each sample were calibrated according to it.
| (2) |
| (3) |
The molar fractional abundance (F) was calculated as:
| (4) |
The mass based fractional abundance (MF) was then calculated as:
| (5) |
The background MF, the natural abundance in each species before labelling (for Pistacia stac, n = 12. For other species n = 9), was subtracted from MF to get the change in MF.
| (6) |
Samples of acetanilide C8H9NO and atropine C17H23NO3 were used for calculating carbon percentage in each sample. Together with the sample mass and change in MF, 13 C excess was computed:
| (7) |
13 C excess was converted to 12 C equivalent according to their atomic masses:
| (8) |
In order to assess how much of the carbon mass in fine roots of a recipient originated from donor trees, we applied a simple mixing model:
| (9) |
2.7. DNA extraction, PCR amplification, and Sanger sequencing of plants
Plant DNA was extracted from 3–4 mg of dried ground root samples by adding 500 µl 0.5 M NaOH with 1.5% PVP to each sample, incubating it in room temperature for 10 min, and centrifuging for 2.5 min at 16,000 g. Then, 5 μl of the supernatant was added to 150 µl 100 mM TRIS, followed by vortexing and a spin down. We amplified the internal transcribed spacer (ITS) region of plant samples. Plant DNA was amplified using ITS2‐S2L (5′‐ATGCGATACTTGGTGTGAAT) and ITS2‐S3R (5′‐GACGCTTCTCCAGACTACAAT) Primers (Yao et al., 2010). For the PCR reaction, we used 15 µl PCRBIO HS Taq Mix Red (PCR Biosystems Ltd), 2 µl of DNA, 1.5 µl of each primer and 10 µl DDW. PCR reactions were performed as follows: initial 5 min at 95°C followed by 35 cycles of 30 s 95°C, 40 s 56°C, 1 min 72°C and final cycle with 10 min 72°C. Then, 10 µl of the PCR product was run on 1.5% agarose gel. PCR reaction samples with one clear band were purified by Exo1 and SAP enzymes. Then, the DNA samples were Sanger sequenced by the Biological Services Department, Weizmann Institute of Science, Rehovot, Israel.
2.8. DNA extraction, PCR amplification, Sanger, and Illumina sequencing of fungi
To estimate the fungal colonization of root tips, eight root tips were collected from one root of each tree species that were picked at the termination of the experiment. Fungal DNA from these eight root tips and from root tips of individual trees, was extracted using the Extract‐N‐Amp Tissue PCR kit (SIGMA XNAT2‐1KT). We added 16 µl extraction solution and 2 µl tissue preparation solution for each well. The samples were incubated at room temperature for 10 min and then were heated at 95°C for 4 min. After that, 15 µl neutralizing inhibitor solution was added to the samples. ITS2 region was amplified using ITS1(5′‐CTTGGTCATTTAGAGGAAGTAA) and ITS4 (5′‐ TCCTCCGCTTATTGATATGC) primers (Gardes & Bruns, 1993; White et al., 1989). The PCR reaction, DNA purification and sequencing were performed as described in the previous section. To find the fungal community of each root, we extracted DNA from lyophilized root tips (Livne‐Luzon et al., 2017). DNA was extracted using DNeasy 96 blood and tissue kit (Qiagen) according to its protocol with slight modifications in the lysis step to adjust it to plant and fungal material. Ground root tips were suspended with 1 ml of freshly prepared 2× CTAB, and heated to 65°C. To separate phases, 600 µl of chloroform was added, followed by 10 min centrifugation. The supernatant containing the DNA was transferred to a new tube with 1,200 ul EtOH 96%. Next, it was transferred to Qiagen filters and the protocol of Qiagen blood and tissue kit was followed (Qiagen) (Glassman et al., 2015).
DNA was diluted ×50 and two steps protocol for library preparation was performed according to Straussman laboratory (Nejman et al., 2020). The forward primer we used for the first PCR reaction is 5.8S‐Fun (5′‐ AACTTTYRRCAAYGGATCWCT) (Taylor et al., 2016). The reverse primer we used is RD2‐ITS4Fun, consisted of ITS4Fun (Taylor et al., 2016), a linker adapter RD2 and RD2‐ITS4Fun (5′AGACGTGTGCTCTTCCGATCT‐AGCCTCCGCTTATTGATATGCTTAART). We used 25 μl of KAPA HiFi Hot Start Ready‐mix DNA polymerase (Hoffmann‐La Roch), 2 µl of diluted DNA, 1ul of each primer and DNase free water to a final volume of 50 µl. PCR reactions were performed as follows: initial 2 min at 98°C followed by 35 cycles of 10 s 98°C, 15 s 51°C, and 35 s 72°C final cycle with 5 min 72°C. The primers for the second PCR were.
P5‐rd1‐5.8S‐Fun (5′‐ AATGATACGGCGACCACCGAGATCT‐ACACTCTTTCCCTACACGACGCTCTTCCGATCT‐AACTTTYRRCAAYGGATCWCT) and RD2‐Barcode (5′ AGACGTGTGCTCTTCCGATCT‐BARCODE). Second PCR reactions were performed as the first PCR with six cycles instead of 35. We measured the purity and concentration of the DNA using qubit 2.0 fluorometer (Invitrogen). The size of the libraries was selected by AMPure magnetic beads (Beckman Coulter Inc.) and was validated using the Agilent 2100 Bioanalyser (Agilent Technologies). The libraries were sequenced in the Grand Israel National Center for Personalized Medicine (Weizmann Institute of Science, Rehovot, Israel) using Illumina MiSeq with 300 bp paired end reads (PE300_V3).
2.9. Bioinformatic analysis
Raw sequences were demultiplexed and both adapters and barcodes were removed for 94 samples. The sequences were analysed using the amplicon sequencing dada2 package v. 1.7.9 in R (Callahan et al., 2016). Sequences were qualified, filtered and trimmed (maxN = 0, maxEE = c(2,5) minLen = 50 truncQ = 2). We removed sequences with a length of less than 50 bases from the analysis, and sequences with more than two consecutive errors on average. We used the MergePairs function (dada2) to merge paired end sequences. To dereplicate each site we performed the derepFastq function. Finally, we combined all the files into a single Fastq file containing amplicon sequence variant (ASVs) in each sample. Using the removeBimeraDenovo function, sequences that appeared only once and chimera sequences were removed. Then, sequences were clustered (id = 0.97), fungal identities were assigned by referencing the sequences to the UNITE database (Nilsson et al., 2019). We further used the FUNguild tool (Nguyen et al., 2016) to identify the fungal guild of each fungal species. ASVs that were assigned as nonfungal were removed from the analysis. To normalize the data, it was rarefied according to the sample with the lowest number of sequences. Samples below the tenth percentile were discarded. We had 10 negative control samples: three with ultrapure water, four with all PCR reagent and two having only the second PCR reagents. The negative control samples had low numbers of 50 to 190 reads. To avoid contamination, we summed all ASVs that were found on them and removed these from each of the other samples. We had two replicates of roots for each tree, and after validating their tree species by Sanger sequencing, their ASVs outputs were merged. Two roots with wrong identification were removed from the analysis. To find the most abundant ASVs, we set the read count threshold to 10x. Further analyses were performed using the Phyloseq tool implemented in (McMurdie & Holmes, 2013). Specifically, nonmetric multidimensional scaling (NMDS) was used to visualize community composition differences between trees. NMDS plots were plotted using the ggplot2 package and ellipses showing the multivariate normal distribution with 95% confidence level using the stat_ellipse function in r. Tree Quercus 5 had suspiciously low reads, within the negative control range, and was hence termed an outlier (Figure S3) so we removed it from the final figure.
2.10. Statistical analysis
Statistical analysis was done using R Core Team, 2018a version 4.0.3 and the interface R Studio R Core Team, 2018b. δ13C level in leaves of labelled trees before and post labelling, and baseline difference of δ13C among tree species were computed using one‐way analysis of variance (ANOVA) followed by Tukey's post‐hoc test. To detect significant differences in roots δ13C values across days of measurement, among tree species (5 species, three replicates), and the interactions between them post labelling, we used repeated‐measures ANOVA within days between species (Stats, version 3.6.2). The main factors discriminating between fungal communities of individual trees were identified by computing a Bray–Curtis dissimilarity matrix followed by a permutational MANOVA (Vegan, version 2.5–7, functions vegdist and adonis, Oksanen et al., 2012). To test for correlations between shared ASV’s and δ13C in the recipient trees, we used Spearman's rank test. Prior to any ANOVA test, ANOVAs assumption of variance homogeneity, independence and normal distribution were checked using a Levene's test for homogeneity of the variances and Shapiro‐Wilk test for normality of the residuals.
2.11. Bootstrap resampling procedure
The constructed data set from the 4 microcosms was unbalanced (with 24, 11, 16, 17 and 15 root samples for Pistacia, Cupressus, Quercus, Pinus and Ceratonia, respectively). We used a bootstrap resampling procedure (Dixon & Hillis, 1993) to produce a balanced ASV table with the minimal number of samples per species (i.e., 11, Cupressus). We repeatedly (1,000 times) selected 11 random samples per species and calculated the basic statistics on each of the balanced ASV tables and on the averaged balanced ASV table. To illustrate the main axes discriminating between saplings’ species, a permutational MANOVA (PERMANOVA) was performed based on a Bray‐Curtis dissimilarity matrix, using the distance and adonis functions embedded in the Phyloseq package (version 1.28) (Figure S4). No significant differences were found between the balanced and the original data set.
3. RESULTS
3.1. Labelling of saplings
The baseline δ13C measured in the roots of the saplings prior to labelling varied among tree species (ANOVA, F(4,24) = [11.34], p <.001) with higher values in Pinus than Ceratonia and Pistacia (p = .02, 95% CI: [0.22, 3.42]; p <.001, 95% CI: [−5.81, –2.24] respectively) and higher values in Cupressus than Pistacia (p = .002, 95% CI: [–4.32, –0.75]). Still, all samples collected prior to labelling had δ13C below –20‰ as expected (Figures S5, S6). In each of the community boxes, roots of labelled trees had higher δ13C than nonlabelled trees, 4–240 days post labelling (Figure 2, Figures S5, S6 ANOVA, F(1,99) = [9.882], F(1,96) = [108.7], F(1,108) = [60.49], F(1,77) = [34.51], p < .001 for all boxes). We found high δ13C values in the leaves of labelled saplings that were sampled one day post labelling, but not in the leaves of nonlabelled saplings, indicating no contamination through the leaves during the labelling. Assimilation rates can affect the amount of transferred carbon, making it important to know how much carbon is assimilated by the tree, and how much is getting to its root (Rog et al., 2020). Here, assimilation rates were measured right before labelling in the different trees, with Ceratonia having the highest assimilation rates (Table 1). Correspondingly, among species of labelled saplings, Ceratonia had the highest leaves δ13C values one day post labelling (Figure S7A, ANOVA, F(3,6) = [16.82], p = .003), with higher levels in Ceratonia than Cupressus, Pinus and Quercus (p = .004, 95% CI: [–2548.5, –1623.8]; p = .004, 95% CI: [–2491.66, –1566.9]; p = .004, 95% CI: [–2767.0, –1754.04] respectively). The level of roots δ13C in labelled saplings over 1–240 days post labelling was also different among species (ANOVA, F(3,119) = [4.322], p = .006, Figure S7B), with higher values in Ceratonia compared to Pinus and Quercus (p = .02, 95% CI: [–37.4, –2.319] and p = .009, 95% CI: [–5.76, –2.287] respectively). To compare carbon allocation rates in the saplings, δ13C was measured in biomass in leaves and roots of the labelled trees over time. Tissues of labelled sapling roots showed increase in δ13C relative to prelabelling baseline values in different times: as soon as two days post labelling in Cupressus, four days in Ceratonia and Quercus and six days in Pinus (Figure S6B). δ13C of leaf respiration increased 1–3 days post labelling and δ13C of root respiration increased 2–9 days post labelling and then decreased and had another peak 54 days post labelling (Figure S8). The level of soil δ13C in the boxes changed throughout the experiment. It had an increase as soon as after labelling, then it decreased and had another small peak 230 days post labelling (Figure S9, ANOVA, F(4,48) = [6.228], p < .001).
FIGURE 2.
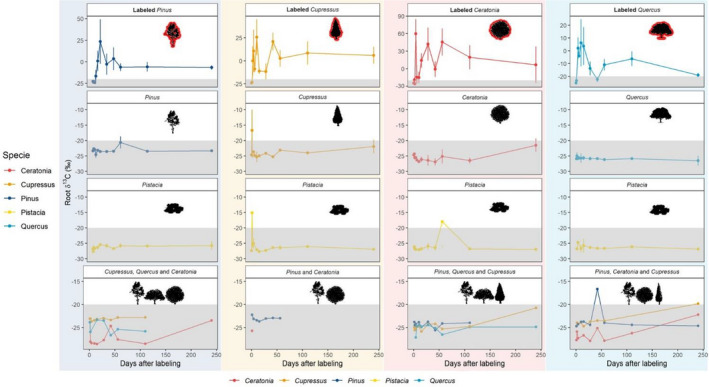
Dynamics of δ13C in roots of labelled and nonlabelled saplings of different Mediterranean tree species. δ13C values through time since labelling in saplings are shown by average and standard error in plots with different background colour for each box (box 1 – green, box 2 – yellow, box 3 – red, box 4 – blue). Saplings of the following species: Pinus halepensis; Cupressus sempervirens; Quercus calliprinos; Pistacia lentiscus and Ceratonia siliqua, in each box are represented by four facets from top to bottom: (1) Three labelled saplings of one species, (2) three nonlabelled saplings of same species, (3) three nonlabelled saplings of Pistacia, and (4) three nonlabelled saplings of the remaining species. Grey area represents δ13C values below natural values of roots of C3 plants = (–20 to –34‰) (Kohn, 2010)
TABLE 1.
Net assimilation rate in the labelled saplings as measured right before labelling
| A (µmol CO2 m− 2 s−1) | |
|---|---|
| Cupressus | 5.026 (4.5) |
| Ceratonia | 7.67 (4.85) |
| Pinus | 2.72 (1.01) |
| Quercus | 5.245 (1.16) |
Standard errors of the mean are indicated by brackets.
3.2. Carbon transfer between saplings
To characterize carbon trade dynamics in the community of trees, root δ13C was measured in all nonlabelled trees of each box after 13C labelling. The species identity of the roots was validated by sequencing their rRNA ITS2 region. We identified carbon donors and recipients in the community based on high δ13C values (δ13C >–20‰) in their roots (Figure 2). In most cases when labelled carbon was found in a nonlabelled tree, it was from the same mycorrhizal group as the labelled tree sharing its box. Among the AM trees, Pistacia seedlings received carbon from Cupressus and from Ceratonia (δ13C = –6.15‰, –1.82‰; carbon from the donor trees 62.9 and 29%, respectively). One seedling of Pistacia received carbon from Quercus (δ13C = –21.35‰ in respect to –27‰ as baseline, carbon from the donor tree 27.1%). Among the EM tree species, Quercus transferred carbon to Pinus (δ13C = –16.65‰; carbon from the donor tree 6.51%). In addition, Cupressus, Ceratonia and Pinus transferred carbon to nonlabelled saplings of their own species, although our genetic identification method is not to the individual resolution. (Cupressus δ13C = 35.2‰, –10.01‰, –19.74‰; Ceratonia δ13C = –19.42‰; Pinus δ13C = –16.76‰). Interestingly, Quercus saplings also transferred carbon to the AM tree Cupressus (δ13C = –19.8‰; carbon from the donor tree 15.1%). They also did not get labelled carbon from any other tree in any of the boxes. The temporal dynamics were broad, with transfer occurrences documented 1, 2, 15, 42, and 240 days post labelling. The faster dynamics were characterized with higher δ13C levels. We found significant interaction between time and tree species affecting root δ13C. There was an increase of δ13C in nonlabelled saplings of Pistacia in box 2 (donor = Cupressus) 1 day post labelling and in nonlabelled saplings of Pinus in box 1 (donor = Pinus) 56 days post labelling. However, following Bonferroni adjustment of the p‐value for multiple comparisons the result was not significant (Figure 2, Figure S6).
3.3. Shared fungal species among tree species growing in a community and in individual trees
Prior to coplanting in boxes, individual trees were associated with a low number of mycorrhizal fungal species; four in Pinus and Cupressus and two in Quercus and Pistacia. They had only few shared fungi among them, Pinus and Quercus shared Tomentella sublilacina and Sphaerosoporella brunnea, and Pistacia and Cupressus shared Plectosphaerella cucumerina (Figure S10 ). Among the eight major fungal species identified in our community boxes (Figure 3), only Sphaerosoporella brunnea and Tomentella ellisii existed in the individual saplings (on Pinus and Quercus), of which only T. ellisii dominated in the boxes. After the seedlings were coplanted in microcosms, mycorrhizal fungi colonization of the plants was examined by sequencing several root tips from each species. In Pinus and Quercus we found two colonized root tips with mycorrhiza out of eight that were collected (2 tuber species in Pinus, Tommentella and Scleroderma in Quercus). In Ceratonia and Cupressus the sequencing didn't work and in Pistacia we found two species of Tuber (Figure S11). Root tips of Pinus and Quercus were scanned by SEM to show fungal colonization (Figure S12). To examine whether mycorrhizal networks could explain the carbon transfer between saplings, we identified the mycorrhizal community of all the root tips of the saplings post identifying the roots species by DNA. We have computed the abundance of shared amplicon sequence variants (ASVs) for each donor‐recipient species couple and found shared ASVs in each case where carbon transferred between trees (Figures S13 and S14). However, there was no significant correlation between the δ13C level in the recipients and the abundance of shared mycorrhiza fungal ASVs with the donor tree (ρ = 0.309, p = .198). We found 30 abundant ASVs, 14 of which were shared among all five host tree species, and only seven were unique to a specific tree species. Among the most abundant ASVs, six species were ectomycorrhizal fungi and two were arbuscular mycorrhizal fungi; the rest were identified as saprotroph, pathogen, or endophyte fungi (Figure S15). The three most abundant species were ectomycorrhizal fungi: Suillus collintus, Tomentella ellisi and Terezia pini. They were found on roots of all tree species involved in this experiment (Figure 3). Overall, we found abundant ASVs of both AMF and EMF, with several generalist species colonizing multiple tree species.
FIGURE 3.
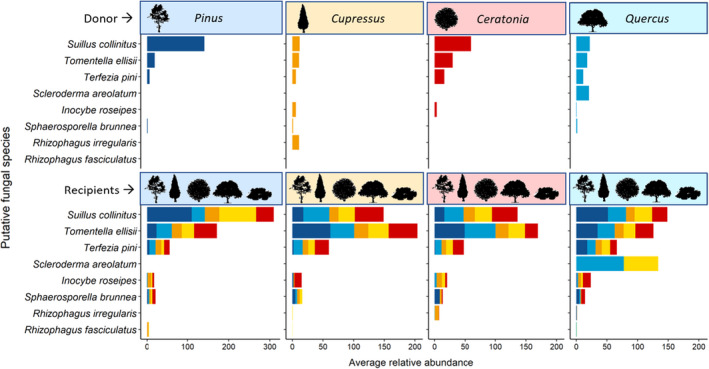
The distribution of abundant Mycorrhizal fungi on different tree species. The most abundant mycorrhizal fungi are represented here. the average abundance of each fungus is represented on the x‐axis. Different colours stand for different tree species: Pinus halepensis, Cupressus sempervirens, Quercus calliprinos, Pistacia lentiscus and Ceratonia siliqua. silhouettes represent the community composition of trees in each box
3.4. Mycorrhizal networks in a tree community
To evaluate community composition differences among roots, a Bray‐Curtis dissimilarity matrix was calculated. We found that both tree species (PERMANOVA, F(4,81) = [4.8192], p = .001) and container identity (box) significantly affected (PERMANOVA, F(3,81) = [3.3338], p = .002) the fungal community composition (i.e., trees from the same box or from the same host‐species have a more similar fungal community composition). Nonmetric multidimensional scaling (NMDS) with a stress level of 0.18 indicated that the fungal community of different boxes and species indeed differed (Figure 4). In agreement with the PERMANOVA results, a better visual separation based on NMDS is achieved when comparing the different species versus the box (r 2 = 18%, r 2 = 10%, respectively). To detect the dominant EMF that led to these observed differences, we created a heatmap showing the abundance of the most prevalent EMF on all trees over boxes and tree species. Focusing on the different boxes, we could see that the trees in box 4 were distinguished by high abundance of Scleroderma compared to trees in other boxes. Box 1 was distinguished by high abundance of Suillus. Focusing on the clustering of different species, Pinus and Quercus trees were well clustered while Pistacia were the most scattered (Figure 4).
FIGURE 4.
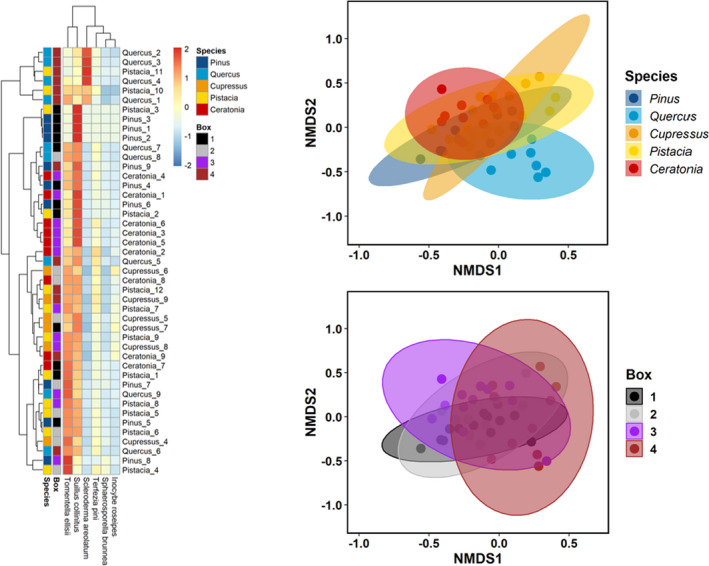
Fungal community composition in different tree species and different boxes. (left) Heatmap representing the most abundant EMF in all saplings across boxes and species: Pinus halepensis, Cupressus sempervirens, Quercus calliprinos, Pistacia lentiscus and Ceratonia siliqua. Saplings identities are indicated in the y‐axis by species and individual number. NMDS plot representing the distance of each tree's fungal communities among abundant mycorrhiza across (top right) species (r 2 = 18%) and across (bottom right) boxes (r 2 = 10%). Ellipses show the multivariate normal distribution with 95% confidence level. NMDS stress level is 0.18
4. DISCUSSION
Here, we studied belowground carbon allocation in a microcosm system for the mixed Mediterranean forest. We identified carbon donor and recipient trees in this community and found overlapping mycorrhizal fungi species among them. We hypothesized that carbon would transfer in a bidirectional and asymmetrical way. Indeed, most species had the positions of both carbon recipients and carbon donors (Figure 2, Figures S13 and S14). This aligns with the results of previous studies and with our perception of the mycorrhizal network as a pathway that transports materials in both directions (Simard, et al., 1997). Surprisingly, Quercus trees demonstrated a pattern of unidirectional carbon transfer. They transferred carbon to Pistacia, Cupressus and Pinus, while not recicarbon from any other tree, including other Quercus trees (Figure 2, Figure S14). Previous studies have suggested that carbon transfers among trees according to a carbon gradient (Francis & Read, 1984; Simard, et al., 1997). Indeed, we found high carbon percentage in Quercus roots, making the Quercus trees a potential physiological source relative to their neighbours (Figure S16). This result could also be explained by the small sample size in this study together with low quantities of transferred carbon, making it hard to detect. In other experiments, Quercus was shown to receive carbon from both other Quercus trees and pines, implying bidirectional transfer could happen also in Quercus trees (Cahanovitc et al., 2022). There were some differences between the studies that can explain why we did not see carbon transfer to Quercus in the current study. First, the mycorrhizal community differed between the two experiments, with the previous study dominated by Tomentella fungi and the current study containing mainly Scleroderma fungi. Second, in this current study, we had many trees of several species in each box creating a highly competitive environment.
We found that carbon transfer was asymmetrical as expected, in line with previous studies (Finlay, 1989; Scheublin et al., 2007; Simard, et al., 1997). A mixing model indicated that 4–29% of fine root carbon in recipients originated from donor trees (Table 2). However, the utility of this approach is limited in the case of pulse labelling with restricted root sampling. For example, in the box with labelled Cupressus we collected a Cupressus root that according to the calculation was composed entirely by carbon from its neighbour, and a Pistacia root with ~63% carbon from its neighbour (Table 2). To get these values we used the peak in the donor tree, however, there were large variations in the roots of labelled trees and perhaps the root that served as the source for the labelled carbon in the Pistacia had much higher levels, and thus transferred lower quantities of labelled carbon than estimated. The movement of carbon can be either symmetrical or asymmetrical, forming a platform for competition when specific species exploit the network to their advantage or for collaboration when stronger species can support the weaker ones. It also fits the results of previous studies showing different benefits from CMNs among tree species. In a microcosm with the EM trees Pinus and Larix, Pinus got more carbon when interacting with Suillus bovinus fungi and Larix got more carbon when associating with the ectomycorrhizal fungus Suillus grevillei or Boletinus cavipes (Finlay, 1989). In AM plants it was shown that AMF favoured legumes over grasses (Scheublin et al., 2007).
TABLE 2.
Carbon transfer between donor and recipient trees. Carbon excess was calculated using Equations (2), (3), (4), (5), (6), (7), (8), and the % of donor carbon in recipients was calculated by the mixing model in Equation 9
| Donor | Recipient | Mycorrhizal group | Days post labelling | Shared mycorrhizal ASVs | δ13C in recipient (‰) | Carbon excess (mg) | Donor carbon (%) |
|---|---|---|---|---|---|---|---|
| Quercus | Pinus | EM | 42 | 137 | –16.66 | 0.22 | 6.51 |
| Quercus | Cupressus | EM +AM | 240 | 57 | –19.836 | 0.19 | 15.09 |
| Quercus | Pistacia | EM +AM | 15 | 103 | –21.35 | 0.07 | 27.12 |
| Ceratonia | Cupressus | AM | 240 | 73 | –20.75 | 0.28 | 4.41 |
| Ceratonia | Ceratonia | AM | 240 | 130 | –19.419 | 0.01 | 6.35 |
| Ceratonia | Pistacia | AM | 56 | 45 | –1.818 | 0.21 | 29.01 |
| Cupressus | Cupressus | AM | 2 | 52 | 35.195 | 0.37 | 118.88 |
| Cupressus | Pistacia | AM | 1 | 40 | 6.147 | 1.28 | 62.87 |
| Pinus | Pinus | EM | 56 | 300 | 16.762 | 0.04 | 13.38 |
Abbreviations: AM, arbuscular mycorrhizal; EM, ectomycorrhizal.
We further hypothesized that carbon will be shared among tree species that host the same mycorrhizal species. Here, various fungal species were colonizing roots of all the five dominant tree species in the Mediterranean forest. These mycorrhizal fungi could form belowground networks among trees and serve as a platform for the carbon transfer among trees. Our last hypothesis was that carbon will move among EMF hosts and among AMF hosts but not between these two guilds of plants. Here, most cases of carbon transfer indeed occurred within the same mycorrhizal groups. In the fourth box having Quercus trees as donors, trees of different guilds served as recipients: Pinus (EM), Pistacia and Cupressus (AM). This could be explained by carbon movement in the soil and not by direct CMNs connecting the trees. Several studies have shown that Quercus trees can have both EM and AM fungi, allowing it to move carbon to both groups of trees (Egerton‐Warburton & Allen, 2001; Rothwell et al., 1983). Here, we have not verified and identified the mycorrhizal association between trees and fungi and therefore this explanation is merely speculation. Carbon had transferred among saplings that share the same phylogenetic groups (gymnosperms vs. angiosperms) but also among trees from other groups (e.g., Cupressus to Pistacia). These results suggest that the similarity of the fungal community (guild) of the trees is more relevant to carbon transfer than the phylogenetic proximity among the trees.
4.1. Different dynamics of carbon transfer suggest multiple mechanisms
We found several cases in which carbon was found in roots of nonlabelled trees. This carbon was found over a broad time range, between 1 day and 240 days post labelling. We have focused on the dynamics among several species and did not have many replicates of each donor‐recipient couple. Together with the complex dynamics of carbon within the compartments of each tree it reduced our ability to point out significant differences of carbon transfer timing across species. However, since natural levels of root δ13C are well studied, we can say with confidence that carbon transfer had occurred in the cases mentioned. In our experiment we did not include a separation between carbon exchange by roots, soil, or CMNs. However, trees were taken apart after the termination of the experiment, and no root grafting that could explain our results were found (Figure S2). A plausible mechanism underlying carbon transfer belowground is diffusion in soil. In another experiment using the same soil, diffusion rate was calculated at 0.2–0.3 cm day−1 (Cahanovitc et al., 2022). Saplings were planted 10 cm apart from each other (Figure 1), and their root systems were ~5 cm apart (Figure S2), and hence C transfer through soil was possible after ~17 days. Here, levels of δ13C in the soil increased 2 days post labelling then decreased and had another peak after 230 days (Figure S9). Several cases of carbon transfer demonstrated in our results certainly fit these dynamics, for example carbon transfer between Quercus and Cupressus 240 days post labelling. In other cases, such as the transition between Quercus and Pinus this rate did not correspond to the level of labelled carbon in the soil, making the CMN more plausible than simple diffusion.
4.2. Generalist mycorrhiza as a possible mechanism underlying carbon exchange
Most studied forests are dominated by either EMF or AMF and accordingly present each tree as either EM host or AM host. In the Mediterranean mixed forest, however, both AMF and EMF naturally cohabit, allowing trees to host both types simultaneously. There is prior evidence for trees hosting AMF and EMF within the same root system in seedlings of Quercus in different ages (Egerton‐Warburton & Allen, 2001) and next to different host plants (Dickie et al., 2001). Also, in a temperate forest in Japan, where AM and EM trees co‐occur, it was shown that the fungal composition of the AMF host Chamaecyparis obtuse contained EMF in addition to AMF (Toju & Sato, 2018). Here, we have found multiple overlapping mycorrhizal fungi among our trees. These fungi could potentially form networks connecting tree roots and allowing carbon transfer. While many of the fungi were host specific, most of the highly abundant fungi had multiple hosts and were found on several species (Figure 3). In this experiment we had Quercus and Pinus, which are considered as EMF hosts (Torres & Honrubia, 1994; Trocha et al., 2012), alongside the AMF hosts Pistacia (Caravaca et al., 2002; Green et al., 2005), Cupressus (Zarik et al., 2016) and Ceratonia (Essahibi et al., 2018; Lahcen et al., 2012). Most cases of transferred carbon occurred within the same guild, suggesting that the mycorrhizal fungi were active only in their traditional hosts. There was no correlation between the number of shared mycorrhizal fungi and the amount of shared carbon among trees, in contrast to other studies (Rog et al., 2020), raising the possibility that the identity of the shared fungus is more relevant than its abundance. Here, Quercus and Pinus shared carbon and were colonized with Tomentella eliisi and Suillus collintus, that were shown to mediate carbon transition between these two tree species (Cahanovitc et al., 2022). In addition, we identified two abundant AMFs‐ Rhizophagus irregularis and Rhizophagus fasciculatus, mostly on the AMF hosts Cupressus and Pistacia. This finding increases the likelihood that they have mycorrhizal association with the trees, especially due to the high and fast carbon exchange that was documented between Cupressus and Pistacia saplings. However, in the fourth box, we did document these two AMF species on the EM tree species Pinus and Quercus (Figure 4). Unexpectedly, we also found seven abundant EMF that colonized all tree species, including the AM tree species Cupressus and Pistacia. The fact that we detected EMF species on non‐EMF hosts does not necessarily mean that these fungi functioned as mycorrhizal symbionts. Since we did not verify the mycorrhizal colonization status using microscopy approaches, we cannot rule out the possibility that these are simply hyphae growing on the surface of roots. However, we can neither rule out that tree species in our experiment were dual‐mycorrhizal (Teste et al., 2020). It can stem from the conditions of our experiment, having trees of several species growing densely by each other (Figure S2), as they grow in the Mediterranean mixed forest.
4.3. Effects on mycorrhizal community composition in roots
The young saplings were planted in microcosm boxes along with their primary fungal community. Prior studies have shown that initial fungal colonization of roots can affect the fungal community in a system through priority effect (Hausmann & Hawkes, 2010). Here, the mycorrhizal fungi in the individual trees were identified before the start of the experiment and contained a small and specific community of mycorrhizal fungi on each tree species (Figure S10). The separation between the functional groups of AMF and EMF hosts was clear in the individual trees, however trees grown in mixed microcosm presented a more diverse and mixed community of mycorrhizal fungi. Only two mycorrhizal fungi were overlapping among fungi in individual trees and in microcosms. These are the Tomentella ellisi that was found only in individual Pinus trees, and Sphaerosporella brunnea found in Pinus and Quercus trees. In the microcosm on the other hand, these two fungi were observed on all the tree species that participated in the experiment.
One of the most abundant EMF taxa we found was Suillus, known to be restricted to members of the pinoid clade of Pinaceae (Kretzer et al., 1996). Yet, there are new reports of various Suillus species sporocarps from forests in which their Pinaceae hosts are absent. In bioassays, Suillus was found to form mycorrhiza with Abies and Tsuga that belong to the abietoid clade of Pinaceae, indicating that Suillus host specificity is more flexible than previously thought (Pérez‐Pazos et al., 2021). Here, Suillus collinitus was indeed most abundant on Pinus saplings, yet we found it on roots of all species. Accordingly, in other Mediterranean forest species, Suillus collinitus was found on the classic Pinus host but also on Quercus roots (Cahanovitc et al., 2022). Root tips with Scleroderma were found only on Quercus saplings and on Pistacia saplings that were planted in the fourth box (with many Quercus trees), but not in other boxes even though sharing the same soil. Both results can be explained by a host neighbour effect. It was previously shown that the presence and composition of neighbouring plants can affect the ability of some mycorrhizal fungi to develop mycorrhizae with hosts (Hausmann & Hawkes, 2010; Kohout & Sýkorová, 2011; Massicotte et al., 1994; Molina et al., 1997). Specifically, there are prior examples of specialist mycorrhizal fungi that formed symbiosis with other trees when growing in a mixed community near to their main host (Pringle, 2009). For example, the Larix specialist Suillus larcinus was detected on a Betula sapling when growing next to a Larix sapling (Nara, 2006). In another study it was shown that Suillus subaureus can germinate and associate with both Pinus and Quercus hosts, both in the laboratory and in the forest. Two other fungi, Suillus americanus and Suillus clintonianus, germinated by spores only in the presence of their primary Pinus hosts but could also form mycorrhizal association with Quercus and Larix trees when colonizing via mycelial networks (Lofgren et al., 2018).
4.4. Similarity of fungal composition in the community boxes and in the Mediterranean forest
The current experimental setup serves as a microcosm for the water limited Mediterranean forest. It is important to understand CMNs composition and function in such environments due to the effects of drought on the fungal community abundance and diversity in soil (Hawkes et al., 2011; Yang et al., 2010). In addition, tree species vary in their spatial root distribution. In our system, Quercus is more deep rooted than Pinus (Rog, Tauge, et al., 2021). This variation can also affect resource sharing by CMNs. We compared the fungal communities in our microcosms with fungal communities of roots from the Mediterranean mixed forest tree species (Cahanovitc et al., 2022). Among the most abundant mycorrhizal fungi, four species were found in both communities: Suillus collinitus, Tomentella ellisii, Terefezia pini, Inocybe (roseipes vs. rhodiola). In other studies on mycorrhizal colonization of Pinus halepensis saplings in natural Mediterranean soil and in the forest itself, Suillus collintus and Inocybe were highly abundant, as was shown here (Livne‐luzon et al., 2017; Querejeta et al., 1998). The composition similarity between fungal communities in the natural forest and in the community boxes in this experiment reinforces our results. Yet, the ability of plants and fungi to create mycorrhizal association and trade resources may be different from what was demonstrated by experimental syntheses. Therefore, it is important to study the dynamics of carbon exchange and quantify its extent under different conditions in the mixed forest itself.
5. CONCLUSION
Carbon transfer between trees of different species has so far been demonstrated mainly in temperate and boreal forests (Fitter et al., 1998; Francis & Read, 1984; Högberg et al., 2008; Klein et al., 2016; Simard, Perry, et al., 1997). Here, it was shown that carbon was moving between tree species of the Mediterranean forest whether by CMNs or by diffusion through the soil. Considering that our microcosm experiment simulated a productive mixed Mediterranean forest, it is possible that asymmetric resource distribution is part of a healthy forest ecosystem. Carbon transferred among the trees from a donor species to a recipient species, and sometimes vice versa. Overall, among our five species, Quercus served only as a carbon donor; Cupressus, Ceratonia and Pinus had both donor and recipient functions, and Pistacia was the most dominant carbon recipient (we did not test it as a donor). The function of carbon trade induced by mycorrhizal networks is still unknown. This mechanism can have large effects on trees dynamics in the forest, whether it underlies a competitive symbiosis in which highly connected trees have an advantage over not well‐connected trees or a mutual symbiosis allowing trees to support each other in times of need. Further research needs to be done to examine the effect of carbon trading on various conditions such as establishment of young seedlings, deficiency in nutrients such as sugars, N and P, and drought.
CONFLICT OF INTEREST
The authors declare no conflict of interests.
AUTHOR CONTRIBUTIONS
Tamir Klein conceived the research idea, further developed by Ido Rog. Ido Rog performed the experiment under the supervision of Tamir Klein. Shifra Avital analysed the samples, with help from Ido Rog and Rotem Cahanovitc. Stav Livne‐Luzon worked with Shifra Avital and Ido Rog on the interpretation of the results. Shifra Avital wrote the manuscript under the guidance of Tamir Klein and with the assistance of all authors.
Supporting information
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Ravid Sraussman and Lian Narunsky Haziza (WIS) for the NGS library preparation; Shacham Megidish and Gilad Jakoby (WIS) for laboratory assistance.
Avital, S. , Rog, I. , Livne‐Luzon, S. , Cahanovitc, R. , & Klein, T. (2022). Asymmetric belowground carbon transfer in a diverse tree community. Molecular Ecology, 31, 3481–3495. 10.1111/mec.16477
Handling Editor: Mitchell Cruzan
Shifra Avital and Ido Rog authors contributed equally.
Funding information
The project was funded by the European Research Council project RHIZOCARBON (TK). IR is supported by the Sustainability and Energy Research Initiative PhD Fellowship.
DATA AVAILABILITY STATEMENT
Sequences were submitted to the National Centre for Biotechnology Information Sequence Read Archive with the accession codes: Bioproject: PRJNA824025. Isotope data, plant physiological measurements, and R analysis script, are accessible through Zenodo: doi 0.5281/zenodo.6411835.
REFERENCES
- Aroca, R. , Porcel, R. , & Ruiz‐Lozano, J. M. (2007). How does arbuscular mycorrhizal symbiosis regulate root hydraulic properties and plasma membrane aquaporins in Phaseolus vulgaris under drought, cold or salinity stresses? New Phytologist, 173(4), 808–816. [DOI] [PubMed] [Google Scholar]
- Bahram, M. , Põlme, S. , Kõljalg, U. , & Tedersoo, L. (2011). A single European aspen (Populus tremula) tree individual may potentially harbour dozens of Cenococcum geophilum ITS genotypes and hundreds of species of ectomycorrhizal fungi. FEMS Microbiology Ecology, 75(2), 313–320. 10.1111/j.1574-6941.2010.01000.x [DOI] [PubMed] [Google Scholar]
- Cahanovitc, R. , Livne‐Luzon, S. , Angel, R. , & Klein, T. (2022). Ectomycorrhizal fungi mediate belowground carbon transfer between pines and oaks. ISME Journal, 1305(94), 1–10. 10.1038/s41396-022-01193-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan, B. J. , Mcmurdie, P. J. , Rosen, M. J. , Han, A. W. , Johnson, A. J. A. , & Holmes, S. P. (2016). DADA2: High‐resolution sample inference from Illumina amplicon data. Nature Methods, 13(7), 581–583. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caravaca, F. , Barea, J. M. , & Rolda, A. (2002). Synergistic influence of an arbuscular mycorrhizal fungus and organic amendment on Pistacia lentiscus L. seedlings afforested in a degraded semiarid soil. Soil Biology & Biochemistry, 34, 1139–1145. [Google Scholar]
- Collins Johnson, N. , Wilson, G. W. T. , Bowker, M. A. , Wilson, J. A. , & Miller, R. M. (2010). Resource limitation is a driver of local adaptation in mycorrhizal symbioses. PNAS, 107(5), 2093–2098. 10.1073/pnas.0906710107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickie, I. A. , Koide, R. T. , Fayish, A. C. , & Koide, R. T. (2001). Vesicular – arbuscular mycorrhizal infection of Quercus rubra seedlings. New Phytologist, 151, 257–264. [DOI] [PubMed] [Google Scholar]
- Dixon, M. T. , & Hillis, D. M. (1993). Ribosomal RNA secondary structure: Compensatory mutations and implications for phylogenetic analysis. Molecular Biology and Evolution, 10(1), 256–267. [DOI] [PubMed] [Google Scholar]
- Egerton‐Warburton, L. , & Allen, M. F. (2001). Endo‐ and ectomycorrhizas in Quercus agrifolia Nee. (Fagaceae): patterns of root colonization and effects on seedling growth. Mycorrhiza, 11, 283–290. 10.1007/s005720100134 [DOI] [PubMed] [Google Scholar]
- Epron, D. , Bahn, M. , Derrien, D. , Lattanzi, F. A. , Pumpanen, J. , Gessler, A. , Hogberg, P. , Maillard, P. , Dannoura, M. , Gerant, D. , & Buchmann, N. (2012). Pulse‐labelling trees to study carbon allocation dynamics: A review of methods, current knowledge and future prospects. Tree Physiology, 32(6), 776–798. 10.1093/treephys/tps057 [DOI] [PubMed] [Google Scholar]
- Essahibi, A. , Benhiba, L. , Babram, M. A. , Ghoulam, C. , & Qaddoury, A. (2018). Influence of arbuscular mycorrhizal fungi on the functional mechanisms associated with drought tolerance in carob (Ceratonia siliqua L.). Trees, 32(1), 87–97. 10.1007/s00468-017-1613-8 [DOI] [Google Scholar]
- Finlay, R. D. (1989). Functional aspects of phosphorus uptake and carbon translocation in incompatible ectomycorrhizal associations between Pinus sylvestris and Suillus grevillei and Boletinus cauipes . New Phytologist, 112(2), 185–192. 10.1111/j.1469-8137.1989.tb02373.x [DOI] [Google Scholar]
- Fitter, A. H. , Graves, J. D. , Watkins, N. K. , Robinson, D. , & Scrimgeour, C. (1998). Carbon transfer between plants and its control in networks of arbuscular mycorrhizas. Functional Ecology, 1998(12), 406–412. 10.1046/j.1365-2435.1998.00206.x [DOI] [Google Scholar]
- Francis, R. , & Read, D. J. (1984). Direct transfer of carbon between plants connected by vesiculararbuscular mycorrhizal mycelium. Nature, 307, 53–56. 10.1038/307053a0 [DOI] [Google Scholar]
- Fraser, E. C. , Lieffers, V. J. , & Landhausser, S. M. (2006). Carbohydrate transfer through root grafts to support shaded trees. Tree Physiology, 26, 1019–1023. 10.1093/treephys/26.8.1019 [DOI] [PubMed] [Google Scholar]
- Gardes, M. , & Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Molecular Ecology, 2(2), 113–118. 10.1111/j.1365-294X.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- Glassman, S. I. , Peay, K. G. , Talbot, J. M. , Smith, D. P. , Chung, J. A. , Taylor, J. W. , Vilgalys, R. , & Bruns, T. D. (2015). A continental view of pine‐associated ectomycorrhizal fungal spore banks: A quiescent functional guild with a strong biogeographic pattern. New Phytologist, 205(4), 1619–1631. 10.1111/nph.13240 [DOI] [PubMed] [Google Scholar]
- Graham, B. F. , & Bormann, F. H. (1966). Natural root grafts. The Botanical Review, 32(3), 255–292. 10.1007/BF02858662 [DOI] [Google Scholar]
- Green, J. J. , Baddeley, J. A. , Cortina, J. , & Watson, C. A. (2005). Root development in the Mediterranean shrub Pistacia lentiscus as affected by nursery treatments. Journal of Arid Environments, 61(1), 1–12. 10.1016/j.jaridenv.2004.09.001 [DOI] [Google Scholar]
- Hausmann, N. T. , & Hawkes, C. V. (2010). Order of plant host establishment alters the composition of arbuscular mycorrhizal communities. Ecology, 91(8), 2333–2343. 10.1890/09-0924.1 [DOI] [PubMed] [Google Scholar]
- Hawkes, C. V. , Kivlin, S. N. , Rocca, J. D. , Huguet, V. , Thomsen, M. A. , & Suttle, K. B. (2011). Fungal community responses to precipitation. Global Change Biology, 17(4), 1637–1645. 10.1111/j.1365-2486.2010.02327.x [DOI] [Google Scholar]
- Hestrin, R. , Hammer, E. C. , Mueller, C. W. , & Lehmann, J. (2019). Synergies between mycorrhizal fungi and soil microbial communities increase plant nitrogen acquisition. Communications Biology, 2(233), 1–9. 10.1038/s42003-019-0481-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Högberg, P. , Högberg, M. N. , Göttlicher, S. G. , Betson, N. R. , Keel, S. G. , Metcalfe, D. B. , Campbell, C. , Schindlbacher, A. , Hurry, V. , Lundmark, T. , Linder, S. , & Näsholm, T. (2008). High temporal resolution tracing of photosynthate carbon from the tree canopy to forest soil microorganisms. New Phytologist, 177(1), 220–228. 10.1111/j.1469-8137.2007.02238.x [DOI] [PubMed] [Google Scholar]
- Klein, T. , & Hoch, G. (2015). Tree carbon allocation dynamics determined using a carbon mass balance approach. New Phytologist, 205(1), 147–159. 10.1111/nph.12993 [DOI] [PubMed] [Google Scholar]
- Klein, T. , Siegwolf, R. T. W. , & Korner, C. (2016). Belowground carbon trade among tall trees in a temperate forest. Science, 352(6283), 342–344. 10.1126/science.aad6188 [DOI] [PubMed] [Google Scholar]
- Klein, T. , Torres‐Ruiz, J. M. , & Albers, J. J. (2022). Conifer desiccation in the 2021 NW heatwave confirms the role of hydraulic damage. Tree Physiology, 42(4), 722–726. 10.1093/treephys/tpac007 [DOI] [PubMed] [Google Scholar]
- Kohn, M. J. (2010). Carbon isotope compositions of terrestrial C3 plants as indicators of (paleo) ecology and (paleo) climate. PNAS, 107(46), 19691–19695. 10.1073/pnas.1004933107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohout, P. , Sýkorová, Z. , Bahram, M. , Hadincová, V. , Albrechtová, J. , Tedersoo, L. , & Vohník, M. (2011). Ericaceous dwarf shrubs affect ectomycorrhizal fungal community of the invasive Pinus strobus and native Pinus sylvestris in a pot experiment. Mycorrhiza, 21, 403–412. 10.1007/s00572-010-0350-2 [DOI] [PubMed] [Google Scholar]
- Kretzer, A. , Li, Y. , Szaro, T. , & Bruns, T. D. (1996). Internal transcribed spacer sequences from 38 recognized species of Suillus sensu lato: Phylogenetic and taxonomic implications. Mycologia, 88(5), 776–785. 10.1080/00275514.1996.12026715 [DOI] [Google Scholar]
- Lahcen, O. , Ibrahima, N. , Abdessadek, M. , & Abderrahim, F. (2012). Inoculation of Ceratonia siliqua L. with native arbuscular mycorrhizal fungi mixture improves seedling establishment under greenhouse conditions. African Journal of Biotechnology, 11(98), 16422–16426. 10.5897/AJB12.1163 [DOI] [Google Scholar]
- Lapidot, O. , Ignat, T. , Rud, R. , Rog, I. , Alchanatis, V. , & Klein, T. (2019). Agricultural and Forest Meteorology Use of thermal imaging to detect evaporative cooling in coniferous and broadleaved tree species of the Mediterranean maquis. Agricultural and Forest Meteorology, 271, 285–294. 10.1016/j.agrformet.2019.02.014 [DOI] [Google Scholar]
- Liesche, J. , Windt, C. , Bohr, T. , Schulz, A. , & Jensen, K. H. (2015). Slower phloem transport in gymnosperm trees can be attributed to higher sieve element resistance. Tree Physiology, 35(4), 376–386. 10.1093/treephys/tpv020 [DOI] [PubMed] [Google Scholar]
- Lindenmayer, D. B. , & Laurance, W. F. (2017). The ecology, distribution, conservation and management of large old trees. Biological Reviews, 92, 1434–1458. 10.1111/brv.12290 [DOI] [PubMed] [Google Scholar]
- Livne‐luzon, S. , Avidan, Y. , Weber, G. , Migael, H. , & Bruns, T. (2017). Wild boars as spore dispersal agents of ectomycorrhizal fungi : consequences for community composition at different habitat types. Mycorrhiza, 27, 165–174. 10.1007/s00572-016-0737-9 [DOI] [PubMed] [Google Scholar]
- Lofgren, L. , Nguyen, N. H. , & Kennedy, P. G. (2018). Ectomycorrhizal host specificity in a changing world: can legacy effects explain anomalous current associations? New Phytologist, 220(4), 1273–1284. 10.1111/nph.15008 [DOI] [PubMed] [Google Scholar]
- Massicotte, H. B. , Molina, R. , Luoma, D. , & Smith, J. E. (1994). Biology of the ectomycorrhizal genus. Rhizopogon. New Phytologist, 126(4), 677–690. 10.1111/j.1469-8137.1994.tb02962.x [DOI] [Google Scholar]
- McMurdie, P. J. , & Holmes, S. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census Data. PLoS One, 8(4), e61217. 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina, R. , Smith, J. E. , McKay, D. , & Melville, L. H. (1997). Biology of the ectomycorrhizal genus, Rhizopogon: III. Influence of co‐cultured conifer species on mycorrhizal specificity with the arbutoid hosts Arctostaphylos uva‐ursi and Arbutus menziesii. The New Phytologist, 137(3), 519–528. 10.1046/j.1469-8137.1997.00836.x [DOI] [PubMed] [Google Scholar]
- Nara, K. (2006). Pioneer dwarf willow may facilitate tree succession by providing late colonizers with compatible ectomycorrhizal fungi in a primary successional volcanic desert. New Phytologist, 171, 187–198. 10.1111/j.1469-8137.2006.01744.x [DOI] [PubMed] [Google Scholar]
- Nejman, D. , Livyatan, I. , Fuks, G. , Gavert, N. , Zwang, Y. , Geller, L. T. , Rotter‐Maskowitz, A. , Weiser, R. , Mallel, G. , Gigi, E. , Meltser, A. , Douglas, G. M. , Kamer, I. , Gopalakrishnan, V. , Dadosh, T. , Levin‐Zaidman, S. , Avnet, S. , Atlan, T. , Cooper, Z. A. , … Straussman, R. (2020). The human tumor microbiome is composed of tumor type–specific intracellular bacteria. Science, 368, 973–980. 10.1126/science.aay9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, N. H. , Song, Z. , Bates, S. T. , Branco, S. , & Tedersoo, L. (2016). FUNGuild : An open annotation tool for parsing fungal community datasets by ecological guild FUNGuild : An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecology. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006 [DOI] [Google Scholar]
- Nilsson, R. H. , Larsson, K. , Taylor, A. F. S. , Bengtsson‐palme, J. , Jeppesen, T. S. , Schigel, D. , Urmas, K. (2019). The UNITE database for molecular identification of fungi : Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Research, 47, 259–264. doi: 10.1093/nar/gky1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen, J. , Blanchet, F. G. , Kindt, R. , Legendre, P. , Minchin, P. , O’Hara, R. B. , & Wagner, H. (2012). Vegan: Community ecology package. [Google Scholar]
- Pérez‐Pazos, E. , Certano, A. , Gagne, J. , Lebeuf, R. , Nguyen, N. , Kennedy, P. G. , & Siegel, N. (2021). The slippery nature of ectomycorrhizal host specificity : Suillus fungi associated with novel pinoid (Picea) and abietoid (Abies) hosts with novel pinoid (Picea) and abietoid (Abies) hosts. Mycologia, 113(5), 891–901. 10.1080/00275514.2021.1921525 [DOI] [PubMed] [Google Scholar]
- Pringle, A. (2009). Mycorrhizal networks. Current Biology, 19(18), R838–R839. 10.1016/j.cub.2009.07.003 [DOI] [PubMed] [Google Scholar]
- Querejeta, J. I. , Roldn, A. , Albaladejo, J. , & Castillo, V. (1998). The role of mycorrhizae, site preparation, and organic amendment in the afforestation of a semi‐arid mediterranean site with pinus halepensis. Forest Science, 44(2), 203–211. [Google Scholar]
- R Core Team (2018a). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Retrieved from https://www.R‐project.org. [Google Scholar]
- R Core Team (2018b). RStudio: Integrated development environment for R (Version 1.2.1335). Boston, MA: RStudio, Inc. Retrieved from http://www.rstudio.com/100 [Google Scholar]
- Rog, I. , Jakoby, G. , & Klein, T. (2021). Forest Ecology and Management Carbon allocation dynamics in conifers and broadleaved tree species revealed by pulse labeling and mass balance. Forest Ecology and Management, 493(119258), 1–12. 10.1016/j.foreco.2021.119258 [DOI] [Google Scholar]
- Rog, I. , Rosenstock, N. P. , Körner, C. , & Klein, T. (2020). Share the wealth: Trees with greater ectomycorrhizal species overlap share more carbon. Molecular Ecology, 29(13), 2321–2333. 10.1111/mec.15351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog, I. , Tague, C. , Jakoby, G. , Megidish, S. , Yaakobi, A. , Wagner, Y. & Klein, T. (2021). Interspecific soil water partitioning as a driver of increased productivity in a diverse mixed Mediterranean forest. Journal of Geophysical Research: Biogeosciences, 126(9), e2021JG006382. 10.1029/2021JG006382 [DOI] [Google Scholar]
- Rothwell, F. M. , Hacskaylo, E. , & Fisher, D. (1983). Ecto‐ and endomycorrhizal fungus associations with Quercus. Plant and Soil, 71, 309–312. [Google Scholar]
- Scheublin, T. R. , Van Logtestijn, R. S. P. , & Van Der Heijden, M. G. A. (2007). Presence and identity of arbuscular mycorrhizal fungi influence competitive interactions between plant species. Journal of Ecology, 95, 631–638. 10.1111/j.1365-2745.2007.01244.x [DOI] [Google Scholar]
- Schneider, C. A. , Rasband, W. S. , Eliceiri, K. W. , & Instrumentation, C. (2012). NIH image to ImageJ : 25 years of image analysis. Nature Methods, 9(7), 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard, S. W. , Perry, D. A. , Jones, M. D. , Myrold, D. D. , Durall, D. M. , & Molina, R. (1997). Net transfer of carbon between ectomycorrhizal tree species in the field. Nature, 388(6642), 579–582. 10.1038/41557 [DOI] [Google Scholar]
- Steidinger, B. S. , Crowther, T. W. , Liang, J. , Van Nuland, M. E. , Werner, G. D. A. , Reich, P. B. , Nabuurs, G. J. , de‐Miguel, S. , Zhou, M. , Picard, N. , Herault, B. , Zhao, X. , Zhang, C. , Routh, D. , & Peay, K. G. (2019). Climatic controls of decomposition drive the global biogeography of forest‐tree symbioses. Nature, 569, 404–413. 10.1038/s41586-019-1128-0 [DOI] [PubMed] [Google Scholar]
- Taylor, D. L. , Walters, W. A. , Lennon, N. J. , Bochicchio, J. , Krohn, A. , Caporaso, J. G. , & Pennanen, T. (2016). Accurate estimation of fungal diversity and abundance through improved lineage‐specific primers optimized for Illumina amplicon sequencing. Applied and Environmental Microbiology, 82(24), 7217–7226. 10.1128/AEM.02576-16.Editor [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teste, F. P. , Jones, M. D. , & Dickie, I. (2020). Tansley review Dual‐mycorrhizal plants: Their ecology and relevance. New Phytologist, 225, 1835–1851. 10.1111/nph.16190 [DOI] [PubMed] [Google Scholar]
- Toju, H. , & Sato, H. (2018). Root‐associated fungi shared between arbuscular mycorrhizal and ectomycorrhizal conifers in a temperate forest. Frontiers in Microbiology, 9(433), 1–11. 10.3389/fmicb.2018.00433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, P. , & Honrubia, M. (1994). Ectomycorrhizal assocations proven for Pinus Halepensis. Israel Journal of Plant Sciences, 42(1), 51–58. 10.1080/07929978.1994.10676557 [DOI] [Google Scholar]
- Trocha, L. K. , Kałucka, I. , Stasińska, M. , Nowak, W. , Dabert, M. , Leski, T. , Rudawska, M. , & Oleksyn, J. (2012). Ectomycorrhizal fungal communities of native and non‐native Pinus and Quercus species in a common garden of 35‐year‐old trees. Mycorrhiza, 22, 121–134. 10.1007/s00572-011-0387-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Heijden, M. G. A. , & Horton, T. R. (2009). Socialism in soil ? The importance of mycorrhizal fungal networks for facilitation in natural ecosystems. Journal of Ecology, 2009(97), 1139–1150. 10.1111/j.1365-2745.2009.01570.x [DOI] [Google Scholar]
- Van Der Heijden, M. G. A. , Martin, F. M. , Selosse, M.‐A. , & Sanders, I. R. (2015). Tansley review Mycorrhizal ecology and evolution : the past, the present, and the future. New Phytologist, 205, 1406–1423. [DOI] [PubMed] [Google Scholar]
- White, T. J. , Bruns, T. , Lee, S. , & Taylor, J. (1989). Amplification and direct sequencing of fungal ribosomal rna genes for phylogenetics. (1). [Google Scholar]
- Woods, F. W. , & Brock, K. (1964). Interspecific transfer of Ca‐45 and P‐32 by root systems. Ecology, 45(4), 886–889. 10.2307/1934943 [DOI] [Google Scholar]
- Yang, C. , Hamel, C. , Schellenberg, M. P. , Perez, J. C. , & Berbara, R. L. (2010). Diversity and functionality of arbuscular mycorrhizal fungi in three plant communities in semiarid Grasslands National Park, Canada. Microbial Ecology, 59(4), 724–733. 10.1007/s00248-009-9629-2 [DOI] [PubMed] [Google Scholar]
- Yao, H. , Song, J. , Liu, C. , Luo, K. , Han, J. , Li, Y. , Pang, X. , Xu, H. , Zhu, Y. , Xiao, P. , & Chen, S. (2010). Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS One, 5(10), e13102. 10.1371/journal.pone.0013102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarik, L. , Meddich, A. , Hijri, M. , Hafidi, M. , Ouhammou, A. , Ouahmane, L. , Duponnois, R. , & Boumezzough, A. (2016). Use of arbuscular mycorrhizal fungi to improve the drought tolerance of Cupressus atlantica G. Comptes Rendus – Biologies, 339(5–6), 185–196. 10.1016/j.crvi.2016.04.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
Sequences were submitted to the National Centre for Biotechnology Information Sequence Read Archive with the accession codes: Bioproject: PRJNA824025. Isotope data, plant physiological measurements, and R analysis script, are accessible through Zenodo: doi 0.5281/zenodo.6411835.


