Abstract
The microbial communities on the skin of dogs include several species of bacteria, which contribute to skin health and disease. Staphylococcus pseudintermedius, cultured at high frequency from the skin of dogs, is an opportunistic pathogen causing superficial pyoderma. Effective treatment against S. pseudintermedius infections is an important issue in veterinary medicine. However, multiple antibiotic-resistant mechanisms gradually developed by bacteria make treatment more challenging nowadays. Drug-resistant genes may have the chance to be transferred from infected dogs to other staphylococci in humans. The objective of this survey is to investigate the bacterial species that cause canine superficial pyoderma and characterize the antibiotic-resistant profiles and drug-resistant genes of isolated S. pseudintermedius. In addition, the possible risk factors causing S. pseudintermedius colonizing owners were also evaluated by a questionnaire survey. Sixty-five bacteria were isolated from dogs with superficial pyoderma, which included 47 S. pseudintermedius (72.3%), 12 other staphylococci (18.5%), 4 other Gram-positive bacteria (6.2%) and 2 Gram-negative bacteria (3.1%). Strains containing mecA and blaZ genes showed multiple-drug resistance characteristics. Dogs that received antimicrobial treatment within a recent month were at significantly higher risk of MRSP infections. Only five S. pseudintermedius strains (8.33%) were isolated from 60 samples of owners. Risk factor analysis indicated there was no significant association between S. pseudintermedius isolated from dogs and owners, but the “Keeping three or more dogs” and “Dogs can lick the owner’s face” have high odds ratios of 3.503 and 5.712, respectively. MRSP isolates belonged to three different dru types, including dt11y (29.41%), dt11a (47.06%) and dt10cp (23.53%). In conclusion, the major pathogen of canine superficial pyoderma is found to be S. pseudintermedius in Taiwan, and isolates which are mecA- or blaZ-positive are generally more resistant to commonly used antibiotics. Although S. pseudintermedius isolated from the owners might be transferred from their dogs, definite risk factors should be examined in the future study.
Keywords: superficial pyoderma, Staphylococcus pseudintermedius, MRSP, dru type, risk factors, owners
1. Introduction
In clinical cases in dogs, diseases related to the skin are very important. In general, the normal skin of dogs has a variety of defense mechanisms against foreign pathogens at the outermost periphery of the entire body. In addition to physical and chemical barriers, the regular microbial flora on the skin surface is also an important protective layer for the maintenance of healthy skin. The microbial flora with a wide diversity can maintain the balance of the entire microenvironment. They can actively secrete certain substances with antibiotic properties to inhibit the proliferation of foreign microorganisms or perform a scavenging effect [1,2]. However, even with multiple layers of protection, if the dog has a primary or secondary bacterial infection caused by local wounds, skin parasitic infection, or sebum leakage, it will cause a variety of skin lesions. In bacterial infections of the skin, superficial pyoderma is the most common disease in dogs [3].
Superficial pyoderma in dogs refers to bacterial infections involving the dog’s epidermis and epithelium at the hair follicles and can be further subdivided according to the site of bacterial infection. For example, bacterial infections that occur in the hair follicles are called superficial bacterial folliculitis (SBF), while bacterial infections that occur around the mouth or on the lips are called mucocutaneous pyoderma [3]. In dogs, SBF is more common than in other mammalian species and is usually caused by S. pseudintermedius [4]. Routine treatment with systemic antimicrobial agents has increased the multi-resistant bacteria, particularly methicillin-resistant S. pseudintermedius (MRSP). In staphylococci, resistance to beta-lactam antibiotics, such as methicillin, occurs due to the acquisition of a mobile gene segment called the Staphylococcal cassette chromosome mec (SCCmec) [5]. In SCCmec, the mecA gene enables bacteria to produce a “penicillin-binding protein 2a” (PBP2a) that is different from the normal penicillin-binding protein, making bacteria resistant to methicillin. The increasing frequency of multidrug resistance of MRSP complicates the selection of antimicrobial therapy in veterinary medicine.
In addition to the skin of dogs, S. pseudintermedius can also be isolated from other organs or systems. However, it can hardly be isolated from healthy humans. S. pseudintermedius isolates from humans have been found to be associated with frequent contact between their own dogs, so this species has been regarded as an important zoonotic pathogen in recent years [6,7,8]. In addition to the direct transmission of S. pseudintermedius to dog owners, the drug-resistant gene fragments carried by the strain also have the opportunity to exchange with other staphylococci in humans [7,8]. The risk of horizontal gene transfer from the dog to owner strain may induce more multi-drug-resistant bacteria in the future, eventually making it difficult for humans to treat bacterial infections.
As mentioned above, there were fewer studies focusing on risk factors analysis of Staphylococcus species isolated from dogs with superficial pyoderma and their owners. Therefore, this study collected samples from dogs with superficial pyoderma and their owners at the Veterinary Medical Teaching Hospital, Department of Veterinary Medicine, National Chung-Hsing University from 2017 to 2018. In addition to the isolation of canine pathogens, the drug resistance profiles of S. pseudintermedius isolates, the detection of drug resistance genes mecA and blaZ, and the risk factors for MRSP infection in dogs were also investigated. The detection rate of S. pseudintermedius will be confirmed in the owner’s samples, and the questionnaire results will be combined to explore the possible risk factors for their owners. Finally, dru gene type of the isolated MRSP will also be classified in order to understand the type of strains in Taiwan currently and compare the differences with foreign countries.
2. Materials and Methods
2.1. Clinical Cases Collection
From 2017 to 2018, dogs presenting superficial pyoderma were identified by a veterinarian at the Veterinary Medical Teaching Hospital, Department of Veterinary Medicine, National Chung-Hsing University. Sixty cases, including skin swabs from dogs and nasal swabs of owners, were submitted to the laboratory for microbiological analysis.
2.2. Questionnaire Survey
A survey was designed as a 12-question questionnaire for the owners to fill out. The content of the questions is related to the basic information of the owners and mainly focuses on the interaction between the dogs and the owners. The questionnaire data were further applied for statistical analysis together with the bacterial survey results in the follow-up.
2.3. Bacterial Isolation and Identification
The skin and nasal swabs were submitted to the microbiology laboratory within 12 h for bacterial isolation and identification. Swabs were cultured on Columbia agar with 5% sheep blood (BD, Heidelberg, Germany) at 37 °C for 24 to 48 h. Suspected colonies were picked and stained with Gram stain. Gram-positive coccus was subcultured for catalase and coagulase biochemical testing. Genomic DNA was extracted from bacterial samples using a commercial kit (GenoMaker, Blossom Biotech, Inc., Taiwan). 16S rDNA sequencing was used for bacterial identification. 16S rDNA was amplified by 27F and 1492R primer pair and sequences were blasted with the NCBI BLAST database (https://blast.ncbi.nlm.nih.gov/Blast.cgi; accessed on 11 May 2018) according to the references [9,10]. Staphylococcus intermedius group (SIG) was further distinguished as S. intermedius, S. pseudintermedius, and S. delphini groups A and B by targeting the nuc gene locus using multiplex PCR [11].
2.4. Antimicrobial Susceptibility Testing
According to the CLSI (Clinical and Laboratory Standards Institute) standard method [12], the disk diffusion method was selected for antimicrobial susceptibility testing. Ten antimicrobial agents, including AMC30 (amoxicillin 20 μg + clavulanic acid 10 μg), AMP10 (ampicillin 10 μg), KZ30 (cephazolin 30 μg), CL30 (cephalexin 30 μg), DA2 (clindamycin 2 μg), DO30 (doxycycline 30 μg), ENR5 (enrofloxacin 5 μg), CN10 (gentamycin 10 μg), OX1 (oxacillin 1 μg), P10 (penicillin 10 units), were used in this study. MRSP or MSSP was identified according to the CLSI oxacillin standard [12].
2.5. PCR Detection of blaZ and mecA Gene from S. pseudintermeidus
The condition of blaz and mecA gene detection was described as follows: The PCR reaction mixtures contained 100 ng chromosomal DNA, oligonucleotide primers (10 pmols), and 2X Taq DNA Polymerase Mastermix-Red® (Ampliqon, Denmark) at a final volume of 20 μL. The PCR condition for the blaZ gene was designed as an initial denaturation step (94 °C, 2 min), 30 cycles of denaturation (94 °C, 1 min), annealing (52 °C, 1 min), and extension (72 °C, 1 min) step, and a final extension step (72 °C, 5 min). The primer pairs used for PCR experiments include the forward primer blaZ F (5′-AAGAGATTTGCCTATGCTTC-3′) the reverse primer blaZ R (5′-GCTTGACCACTTTTATCAGC-3′); the product size was 512 base pairs [13]. The PCR condition for mecA gene was designed as an initial denaturation step (94 °C, 4 min), 35 cycles of denaturation (94 °C, 60 s), annealing (55 °C, 60 s), and extension (72 °C, 60 s) step, and a final extension step (72 °C, 10 min). The primer pairs used for mecA detection included the forward primer mecA F (5′-GTAGAAATGACTGAACGTCCGATAA-3′), the reverse primer mecA R (5′-CCAATTCCACATTGTTTCGGTCTAA-3′); the final product size was 310 base pairs [14].
2.6. Dru Gene Typing
The condition of dru gene typing was described as follows: The PCR reaction mixtures contained 100 ng chromosomal DNA, oligonucleotide primers (10 pmols), 2X Taq DNA Polymerase Mastermix-Red® (Ampliqon, Denmark) at a final volume of 20 μL. The PCR condition for the dru gene was designed as an initial denaturation step (95 °C, 5 min), 30 cycles of denaturation (94 °C, 45 s), annealing (52 °C, 45 s), and extension (72 °C, 60 s) step, and a final extension step (72 °C, 5 min). The primer pairs used for PCR experiments included the forward primer dru GF (5′-GTTAGCATATTACCTCTCCTTGC-3′), the reverse primer dru GR (5′-GCCGATTGTGCTTGATGAG-3′), and the product size was about 900 base pairs. The PCR product was further sequenced and the sequence was compared with the data bank (http://dru-typing.org; accessed on 11 May 2018) for dru gene typing.
2.7. Statistical Analysis
Statistical analysis and charting of data were performed using SAS 9.4 (SAS Institute Inc., Cary, NC, USA) and Excel 2010 (Microsoft, Washington, WA, USA). The Chi-squared test was used to compare whether the presence or absence of drug resistance genes (mecA or blaZ) and the dru typing were related to the resistance of the isolated strains to antibiotics. The questionnaire data were analyzed with the bacterial results using the Chi-squared test and odds ratio (OR). If the expected value in the Chi-square test is less than 5, use Fisher’s Exact Test for statistical analysis. The statistical result was expressed as p-values and was considered statistically significant when p < 0.05.
3. Results
3.1. Bacterial Identification in Dogs with Superficial Pyoderma and Their Owners
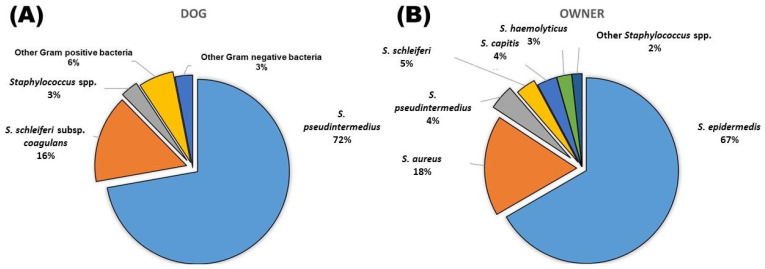
In this study, swabbed samples from the lesion areas of 60 dogs with superficial pyoderma were collected from the Veterinary Teaching Hospital of National Chung-Hsing University, in addition to nasal swabbed samples from the owners of these 60 dogs. A total of 65 strains were isolated from dogs, including 47 strains of S. pseudintermedius (72.3%), 10 strains of S. schleiferi subsp. coagulans (15.4%), 2 strains of other staphylococci (3.1%), 4 strains of other Gram-positive bacteria (6.2%) and 2 strains of Gram-negative bacteria (3.1%) (Figure 1). Two strains were identified as S. epidermedis and S. hominis in the other staphylococci group. The remaining four Gram-positive bacteria included Enterococcus gallinarum, Enterococcus faecalis, Streptococcus halichoeri and Streptococcus sanguinis, respectively. In addition, Gram-negative bacteria were identified as Sphingonas mucosissima and Acinetobacter schindleri, respectively.
Figure 1.
Bacterial isolation and identification from dogs with superficial pyoderma (A) and their owner (B).
In terms of owners, a total of 114 Staphylococcus strains were isolated, including 76 strains of S. epidermedis (66.7%), 20 strains of S. aureus (17.5%), 5 strains of S. pseudintermedius (4.4%), 4 strains of S. schleiferi (3.5%), 4 strains of S. capitis (3.5%), 3 strains of S. haemolyticus (2.6%), and 1 strain of each S. gallinarum (0.9%) and S. pasteuri (0.9%) (Figure 1).
3.2. Antimicrobial Susceptibility Testing
3.2.1. Antimicrobial Susceptibility Testing of Staphylocuccus Isolated from Dogs
Among the 47 strains of S. pseudintermedius, 76.6% of the strains were found to be resistant to ampicillin, 72.34% to penicillin G, 55.32% to doxycycline, 48.94% to gentamicin, 40.43% to clindamycin, 31.91% to enrofloxacin, 25.53% to cephalexin and cephazolin and 21.28% to augmentin. Strains of S. pseudintermedius isolated in this study were highly resistant to ampicillin but relatively sensitive to augmentin. In the non-S. pseudintermedius Staphylococcus group, 83.33% of the strains were resistant to ampicillin and penicillin G, 50% to clindamycin, 33.33% to cephalexin, 16.67% to doxycycline, gentamicin, cephazolin, augmentin and 8.33% to enrofloxacin. The Chi-square test indicated that S. pseudintermedius had significant resistance to gentamicin and doxycycline compared to other Staphylococcus strains isolated from the skin of dogs (Table 1, p < 0.05).
Table 1.
Antibiotics resistant profiles comparing between S. pseudintermedius and other Staphylococcus spp. from dogs.
| Antibiotics | S. pseudointermedius (n = 47) | Other Staphylococcus spp. (n = 12) | p Value |
|---|---|---|---|
| Gentamicin | 48.94% (23) | 16.67% (2) | 0.044 |
| Clindamycin | 40.43% (19) | 50.00% (6) | 0.549 |
| Ampicillin | 76.60% (36) | 83.30% (10) | 1.000 |
| Doxycycline | 55.32% (26) | 16.67% (2) | 0.017 |
| Augmentin * | 21.28% (10) | 16.67% (2) | 1.000 |
| Cephalexin | 25.53% (12) | 33.33% (4) | 0.718 |
| Penicillin G | 72.34% (34) | 83.33% (10) | 0.712 |
| Cephazolin | 25.53% (12) | 16.67% (2) | 0.712 |
| Enrofloxacin | 31.91% (15) | 8.33% (1) | 0.151 |
| Multi-drug resistant | 70.21% (33) | 83.33% (10) | 0.581 |
* Augmentin: amoxicillin + clavulanic acid.
In multi-drug resistant analysis, 70.21% of the S. pseudintermedius strains and 83.33% of the other staphylococcus strains were multi-resistant, respectively. There was no statistically significant difference between these two groups (p = 0.581).
3.2.2. Correlation between Antibiotics Resistant Profiles and mecA Gene of S. pseudintermedius from Dogs
All S. pseudintermedius isolates were submitted for mecA gene detection. Sixteen strains were found to be mecA positive, and the remaining 31 strains were mecA negative. All mecA-positive S. pseudintermedius strains were found to be resistant to ampicillin and penicillin G, 93.75% to clindamycin, 87.5% to doxycycline and enrofloxacin, 81.25% to gentamicin and oxacillin, and 75% to cephalexin and cephazolin and 62.5% to augmentin. In mecA-negative S. pseudintermedius group, 64.52% of the strains were found to be resistant to ampicillin, 38.71% for doxycycline, 32.26% for gentamicin, 18% for penicillin G, 12.9% for clindamycin and oxacillin, and 3.23% for enrofloxacin. It was found that mecA-negative S. pseudintermedius strains were sensitive to augmentin, cephalexin and cephazolin, respectively. In addition, all of the mecA-positive strains were MRSP. The Chi-square test indicated that the presence or absence of mecA gene significantly influences the profile of antibiotic resistance in S. pseudintermedius (Table 2. p < 0.05).
Table 2.
Antibiotics resistant profiles comparing between mecA positive and negative of S. pseudintermedius from dogs.
| Antibiotics | mecA Positive (n = 16) | mecA Negative (n = 31) | p-Value |
|---|---|---|---|
| Gentamicin | 81.25% (13) | 32.26% (10) | 0.002 |
| Clindamycin | 93.75% (15) | 12.90% (4) | <0.0001 |
| Ampicillin | 100.00% (16) | 64.52% (20) | 0.009 |
| Doxycycline | 87.50% (14) | 38.71% (12) | 0.001 |
| Augmentin * | 62.50% (10) | 0.00% (0) | <0.0001 |
| Cephalexin | 75.00% (12) | 0.00% (0) | <0.0001 |
| Penicillin G | 100.00% (16) | 58.06% (18) | 0.002 |
| Cephazolin | 75.00% (12) | 0.00% (0) | <0.0001 |
| Enrofloxacin | 87.50% (14) | 3.23% (1) | <0.0001 |
| Oxacillin | 81.25% (13) | 12.90% (4) | <0.0001 |
| Multi-drug resistant | 100.00% (16) | 54.84% (17) | 0.004 |
* Augmentin: amoxicillin + clavulanic acid.
3.2.3. Correlation between Antibiotics Resistant Profiles and blaZ Gene of S. pseudintermedius from Dogs
All S. pseudintermedius isolates were submitted for blaZ gene detection; 38 strains were found to be blaZ positive, and the remaining nine strains were blaZ negative; 94.74% of blaZ -positive S. pseudintermedius strains were found to be resistant to ampicillin, 89.47% to penicillin G, 68.42% to doxycycline, 60.53% to clindamycin, enrofloxacin, and gentamicin, and 31.58% to cephalexin and cephazolin and 26.32% to augmentin. It was found that blaZ-negative S. pseudintermedius strains were sensitive to all tested antibiotics. The Chi-square test indicated that the presence or absence of blaZ gene significantly influences the gentamicin, clindamycin, ampicillin, doxycycline, penicillin G and enrofloxacin resistance in S. pseudintermedius (Table 3. p < 0.05).
Table 3.
Antibiotics resistant profiles comparing between blaZ positive and negative of S. pseudointermedius from dogs.
| Antibiotics | blaZ Positive (n = 38) | blaZ Negative (n = 9) | p Value |
|---|---|---|---|
| Gentamicin | 60.53% (23) | 0.00% (0) | 0.002 |
| Clindamycin | 50.00% (19) | 0.00% (0) | 0.007 |
| Ampicillin | 94.74% (36) | 0.00% (0) | <0.0001 |
| Doxycycline | 68.42% (26) | 0.00% (0) | 0.0002 |
| Augmentin * | 26.32% (10) | 0.00% (0) | 0.172 |
| Cephalexin | 31.58% (12) | 0.00% (0) | 0.087 |
| Penicillin G | 89.47% (34) | 0.00% (0) | <0.0001 |
| Cephazolin | 31.58% (12) | 0.00% (0) | 0.087 |
| Enrofloxacin | 39.47% (15) | 0.00% (0) | 0.041 |
| Multi-drug resistant | 86.84% (33) | 0.00% (0) | <0.0001 |
* Augmentin: amoxicillin + clavulanic acid.
In addition, most of the blaZ-positive S. pseudintermedius strains (86.84%) were multi-drug resistant. The chi-square test indicated that the presence or absence of blaZ gene significantly influences the multi-drug resistance in S. pseudintermedius (Table 3. p < 0.0001).
3.2.4. Antibiotics Resistant Profiles and dru Gene Typing of S. pseudintermedius from Dogs
A total of 17 MRSP strains were isolated in this study, of which 16 strains were from dogs and only one strain was from the owner. The dru genes of the 17 MRSP strains were amplified, sequenced and compared with an online database to determine the dru gene types. There were three types of dru genes found in 17 MRSP strains, including eight strains of dt11a, five strains of dt11y and four strains of dt10cp. In the statistical analysis, the results showed that there was no significant correlation between antibiotic resistance and dru gene typing (Table 4).
Table 4.
Antibiotics resistant profiles and dru gene typing of S. pseudointermedius from dogs.
| Antibiotics | dt11a (n = 8) | dt11y (n = 5) | dt10cp (n = 4) | p Value |
|---|---|---|---|---|
| Gentamicin | 87.5% (7) | 60.0% (3) | 100.0% (4) | 0.394 |
| Clindamycin | 87.5% (7) | 100.0% (5) | 100.0% (4) | 1.000 |
| Ampicillin | 100.0% (8) | 100.0% (5) | 100.0% (4) | - |
| Doxycycline | 87.5% (7) | 60.0% (3) | 100.0% (4) | 0.394 |
| Augmentin * | 50.0% (4) | 60.0% (3) | 100.0% (4) | 0.344 |
| Cephalexin | 62.5% (5) | 80.0% (4) | 100.0% (4) | 0.630 |
| Penicillin G | 100.0% (8) | 100.0% (5) | 100.0% (4) | - |
| Cephazolin | 62.5% (5) | 80.0% (4) | 100.0% (4) | 0.630 |
| Enrofloxacin | 100.0% (8) | 60.0% (3) | 100.0% (4) | 0.118 |
* Augmentin: amoxicillin + clavulanic acid.
3.3. Risk Factors Analysis
3.3.1. Risk Factors Analysis of MRSP and MSSP from Dogs
In the questionnaire survey, two questions were asked about “whether the owner works in a medical institution” and “whether the dog has received any form of antibiotic treatment within a month”. Of the 45 dogs with S. pseudintermedius infection, 16 strains were MRSP and 29 were MSSP. The Chi-square test indicated that the association of owners’ workplace with MRSP or MSSP isolated from dogs in the medical facility had no significant association (p = 0.608). However, dogs who received antibiotic treatment within one month had a statistical correlation (p = 0.0004) between the isolation of MRSP and MSSP from dogs, with an odds ratio of 11.5 (Table 5). Results indicated that dogs who had been treated with antibiotics within one month had a significantly higher isolation rate of MRSP.
Table 5.
Risk factors analysis of MRSP and MSSP from dogs.
| Risk Factors | MRSP from Dogs (n = 16) | MSSP from Dogs (n = 29) | OR | 95% Confidence Interval | p Value |
|---|---|---|---|---|---|
| Owner works in medical institution | 12.5% (2) | 6.9% (2) | 1.93 | 0.245–15.185 | 0.608 |
| Dogs treated with antibiotics within a month | 75.0% (12) | 20.7% (6) | 11.5 | 2.711–48.777 | 0.0004 |
3.3.2. Risk Factors Analysis for S. pseudintermedius Isolated from Owners
In this study, only five S. pseudintermedius strains were isolated from the nasal cavity of sixty dog owners. The results showed that the dogs raised by the owners who had S. pseudintermedius isolated also had S. pseudintermedius isolated in the skin lesion, and the detection results of the mecA and blaZ genes of these five pairs of S. pseudintermedius strains were consistent, respectively. However, the results of antibiotic susceptibility tests were slightly different among the five groups of strains (Table 6). Results showed S. pseudintermedius strains isolated from dogs generated more antibiotic resistant profiles than the strains isolated from humans, especially for resistance to doxycycline (C9 versus H9) and gentamycin (C41 vs. H41; C45 vs. H45). In addition, strain H35 did not resist any antibiotics tested in this study.
Table 6.
Comparison of S. pseudintermedius isolated from dogs and their owners.
| Isolated Strains * | mecA | blaZ | Antibiotics Resistant Profiles ** |
|---|---|---|---|
| C9 | + | + | CN, DA, AMP, DO, AMC, CL, OX, P, KZ, ENR |
| H9 | + | + | CN, DA, AMP, AMC, CL, OX, P, KZ, ENR |
| C35 | – | + | CN, AMP, DO, P |
| H35 | – | + | – |
| C41 | – | + | CN, AMP, P |
| H41 | – | + | AMP, P |
| C45 | – | + | CN, AMP, P |
| H45 | – | + | AMP, P |
| C48 | – | + | AMP, P |
| H48 | – | + | AMP, P |
* The name of isolated strains: C indicates isolated from canine and H is from human. The same strain number means the dog is kept by the same owner. ** AMC: amoxicillin + clavulanic acid, AMP: ampicillin, KZ: cephazolin, CL: cephalexin, DA: clindamycin, DO: doxycycline, ENR: enrofloxacin, CN: gentamycin, OX: oxacillin, P: penicillin.
There are 10 questions in the questionnaire about the relationship between the owner and the dog, including “whether there are more than 3 dogs at home”, “whether the dogs are kept indoors”, “whether the dogs can rest on the sofa or seat in the living room”, “whether the dogs can enter the owner’s bedroom, “whether the dogs can rest or be active in the owner’s bedroom for a long time”, “whether the dogs can move on the owner’s bed”, “Contact with dogs more than three times a day”, “whether the dogs can lick owner’s hands”, “whether the dogs can lick owner’s face”, and “whether the dogs can bathe in owner’s bathroom”. The correlation is analyzed by a Chi-square test and the odds ratio at the same time for the above 10 questions and whether S. pseudintermedius was isolated from the owner. Results indicated that there was no statistical relationship between the S. pseudintermedius isolated from the owner and any item in the questionnaire, but the “Keeping three or more dogs” and “Dogs can lick the owner’s face” have high odds ratios of 3.503 and 5.712, respectively (Table 7).
Table 7.
Possible risk factors analysis for S. pseudintermedius isolated from owners.
| Risk Factors | Positive (n = 5) | Negative (n = 55) | OR | 95% Confidence Interval | p Value |
|---|---|---|---|---|---|
| Keeping three or more dogs | 20.0% (1) | 10.9% (6) | 3.053 | 0.246–37.892 | 0.475 |
| Dogs are kept indoors | 100.0% (5) | 96.3% (53) | - | - | 1.000 |
| Dogs can rest on the sofa seat in the living room | 100.0% (5) | 72.7% (40) | - | - | 0.318 |
| Dogs can enter the owner’s bedroom | 80.0% (4) | 76.3% (42) | 1.089 | 0.108–10.980 | 1.000 |
| Dogs can rest in the owner’s bedroom for a long time | 80.0% (4) | 70.9% (39) | 1.641 | 0.170–15.841 | 1.000 |
| Dogs can move on the owner’s bed | 40.0% (2) | 47.2% (26) | 0.744 | 0.115–4.805 | 1.000 |
| Contact with dogs more than three times a day | 100.0% (5) | 92.7% (51) | - | - | 1.000 |
| Dogs can lick owner’s hands | 80.0% (4) | 70.9% (39) | 1.641 | 0.170–15.841 | 1.000 |
| Dogs can lick owner’s face | 80.0% (4) | 38.1% (21) | 5.712 | 0.732–44.556 | 0.150 |
| Dogs can take a bathe in owner’s bathroom | 60.0% (3) | 67.2% (37) | 0.548 | 0.071–4.216 | 1.000 |
4. Discussion
Superficial pyoderma is a superficial infection by bacteria, such as S. pseudintermedius, S. schleiferi, Escherichia coli, and species of the genera Pseudomonas and Proteus. S. pseudintermedius is a species that has only been identified in the past ten years, and it was often identified as another species of Staphylococcus due to insufficient development of classification technology [15,16]. S. pseudintermedius is a member of the S. intermedius group, which also includes S. intermedius and S. delphini. The species in this group have similar biochemical properties and high 16s rDNA sequence similarity [17]. According to the evolution of molecular biology, there are various tools that can assist in the identification of S. pseudintermedius species and even further classify them according to their genotypes [18,19]. The results of genotyping can be used in epidemiological surveillance and improve the medical care of dogs and their owners.
Previous studies showed that Staphylococcus was the main species isolated from dog skins and most of them can be identified as S. pseudintermedius [20,21]. A one-year study in Australia found that 70.8% of the bacteria isolated from companion animals were S. pseudintermedius [21]. Another study in South America from 2007 to 2012 found that staphylococci were isolated in 26.5% of samples from various body parts of dogs, and 71.7% were identified as S. pseudintermedius [20]. According to the results of this study, S. pseudintermedius had the highest rate (72.3%) in dogs with superficial pyoderma, indicating that S. pseudintermedius plays an important role in the skin infection of dogs.
Among staphylococci, the most frequently isolated species from human nasal mucosa have been reported to be S. epidermidis and S. aureus, but the composition ratio varies from person to person [22]. In this study, only 4.4% of the isolates in the owner’s survey were identified as S. pseudintermedius. Compared with previous reports, the rate of S. pseudintermedius isolated from the nasal cavity of ordinary humans is much higher, probably because the humans in this survey are all dog owners. Many studies have also shown that dog owners have a higher chance of severing SIG or S. pseudintermedius, indicating the risk factors of pathogens transfer from pets to owners [23,24].
In this study, S. pseudintermedius was significantly more resistant to doxycycline and gentamicin than the rest of the isolated staphylococci among the tested antibiotics. These two antibiotics, doxycycline and gentamicin, are not the first-line treatment for dogs with superficial pyoderma in most veterinary hospitals in Taiwan. Our results may reflect two facts: First, relative to other staphylococci, in facing the treatment of S. pseudintermedius, there is a more frequent treatment with non-first-line drugs. Second, S. pseudintermedius is a pathogen that is often isolated from canine infections (whether skin or other organs and systems) [25]. Therefore, S. pseudintermedius exposure to more diverse antibiotics is predictable. The bacteria have more opportunities to screen out individuals with resistance to multiple antibiotics and cause clinical treatment challenges finally.
According to the results, a total of 47 strains of S. pseudintermedius were isolated from the skin lesions of dogs, among which 16 strains carried the mecA gene. The proportion of mecA-positive S. pseudintermedius resistant to the 10 antibiotics tested in this survey was significantly higher than that of mecA-negative strains. Interestingly, the 16 mecA-positive strains were all multi-drug resistant strains, indicating the presence of mecA was significantly correlated with multi-resistant characteristics of S. pseudintermedius. S. pseudintermedius with the mecA gene can be identified as MRSP because of its resistance to β-lactam antibiotics, such as methicillin. Although different SCCmec species have been reported to have different degrees of multi-drug resistance, most research studies agree that MRSP is related to strains with multi-drug resistance, which is consistent with the findings of this study [26,27,28].
In the results of the blaZ gene analysis, most strains of S. pseudintermedius were blaZ positive and resistant to gentamicin, clindamycin, ampicillin, doxycycline, penicillin G and enrofloxacin. Interestingly, the blaZ-negative strains were all susceptible to the antibiotics tested in this study. The possible reason for this phenomenon is blaZ-negative S. pseudintermedius is more sensitive to commonly used first-line penicillin antibiotics than blaZ-positive strains, so it has less chance to encounter other antibiotics and the chance of developing resistance to multiple antibiotics is also reduced.
Questionnaires were also conducted for two items of interest in this survey, namely “Owner works in medical institution” and “Dogs treated with antibiotics within a month”. Originally, speculation had been raised that the owner’s job attributes were prone to encountering severe drug-resistant staphylococci, the interaction between the owner and the dog may lead to the spread of strains, and even further exchange of gene fragments that make bacteria resistant to drugs. However, the results indicated that there was no statistical support for such speculation. A previous study on risk factors for S. aureus isolates in dogs and owners showed that colonization of dogs was not associated with close human contact but was strongly associated with health-care occupations [29]. Under the condition that the transmission of strains does exist, the drug resistance gene of Staphylococcus in the owner is likely to be transmitted to the Staphylococcus in dogs. Although this survey has not obtained the predicted results, it may be possible to increase the number of samples and exclude possible interference factors in the future. Moreover, a number of studies indicated that in the dogs who received antibiotic treatment shortly before sampling, the isolation rate of MRSP was significantly increased [30,31,32]. These findings are consistent in this survey and may represent that MRSP has a better survival advantage than MSSP when S. pseudintermedius in dogs is under pressure caused by antibiotics, so the possibility of being isolated in dogs is also relatively increased.
Although the probability of S. pseudintermedius in dogs being transmitted to humans and causing a direct infection is not high, S. pseudintermedius is likely to exchange drug-resistant gene segments with other staphylococci after transmission to humans [23,33,34]. This survey combined questionnaires to clarify whether certain risk factors were associated with the isolation of S. pseudintermedius in owners. From the statistical results, it can be seen that “Keeping three or more dogs” and “Dogs can lick the owner’s face” have high odds ratios of 3.503 and 5.712, respectively. A similar study on the statistical analysis of “Keeping three or more dogs” found that it was significantly related to the isolation of S. pseudintermedius from the owner [24]. In addition, the high odds ratio of the factor “dogs could lick the owner’s face” suggested that the act of licking the owner’s face may increase the chance of severing S. pseudintermedius in the owner. Several studies have shown that MRSP can also be isolated from other healthy dogs and cats living with MRSP-infected dogs and cats [35,36]. Therefore, if the number of dogs raised is greater, the sources of S. pseudintermedius transmission may increase for owners, and a higher probability of isolation of the strain from owners can be expected.
In this study, dru typing was used to confirm the genetic diversity of the isolated MRSPs in Taiwan. It was found that three types of MRSPs were included in our isolated MRSPs, namely dt11a (47.06%), dt11y (29.41%) and dt10cp (23.53%). All strains of the three types were all multi-drug resistant, and there was no significant correlation between the type and the tested antibiotic resistance. Compared to other countries, it was found that the MRSP isolated in Canada was dominated by four types of dt11a, dt10h, dt9a and dt11af [37]. A comprehensive survey of MRSP isolates in Canada and the United States found that dt11a and dt9a were predominant [38]. Another report also found that dt11a and dt9a were dominant in MRSP collected in Europe and North America [39]. In Asia, the MRSPs isolated from Korea are dt11a and dt11y, while in Thailand they are dt11a and dt11cj, respectively [40,41]. According to the above literature records and our results, dt11a is widely distributed and is the predominant type in the northern hemisphere. Although dt11y also has sporadic been isolated in Europe and the United States, it was identified in South Korea and Thailand in Asia with significantly higher rates of 28.5% and 10.26%. It is considered to be a more prevalent type in Asia. The dru type dt10cp, which accounted for about a quarter of our results, was relatively undocumented. This type was first recorded in 2016 and only 7.69% of MRSPs were identified as dt10cp from Thailand [40,41].
This research involves several aspects, from the isolation and identification of skin pathogens from dogs to the transmission of these zoonotic bacteria to owners, and scientific statistical analysis has also been carried out. Although S. pseudintermedius isolated from the owners might be transferred from their dogs, definite risk factors should be examined in the future study. Therefore, future research on the S. pseudintermedius isolated in Taiwan can use this paper as a stepping stone for more in-depth analysis.
5. Conclusions
This research investigates the pathogenic bacteria of superficial pyoderma in dogs, the antibiotics resistant profiles and drug resistance genes of S. pseudintermedius, and dru gene typing of MRSP in Taiwan. The detection rate of S. pseudintermedius isolated from owners and possible risk factors were also statistically analyzed in this survey to provide a possible direction for the prevention of zoonotic transmission of S. pseudintermedius. Results showed that strains that contained the mecA and blaZ gene generated multiple-drug resistance characteristics. Recently antimicrobial treated dogs were at significantly higher risk of MRSP infections. Dru gene typing indicated MRSP isolates belonged to dt11a, dt11y and dt10cp. The dt11y and dt11a were the most commonly detected type of MRSP in Asia. Further studies on definite risk factors should be examined in the future.
Acknowledgments
The authors gratefully acknowledge the Veterinary Medical Teaching Hospital, Department of Veterinary Medicine, National Chung-Hsing University, Taiwan for supporting the process of obtaining samples of skin swabs from superficial pyoderma dogs.
Abbreviations
| MLST | multi-locus sequence typing |
| MRSA | methicillin-resistant Staphylococcus aureus |
| MRSP | methicillin-resistant S. pseudintermedius |
| MSSP | methicillin-sensitive S. pseudintermedius |
| PFGE | pulsed-field gel electrophoresis |
| SCCmec | Staphylococcal cassette chromosome mec |
| SIG | Staphylococcus intermedius group |
Author Contributions
Conceptualization, C.-H.L. and C.-M.W.; methodology, Y.-C.M., W.-Y.S. and Y.-L.H.; software, Y.-C.M.; validation, C.-H.L. and C.-M.W.; formal analysis, Y.-C.M. and W.-Y.S.; investigation, Y.-C.M., W.-Y.S. and Y.-L.H.; resources, C.-H.L. and Y.-C.M.; data curation, Y.-C.M., W.-Y.S. and Y.-L.H.; writing—original draft preparation, C.-H.L. and C.-M.W.; writing—review and editing, C.-H.L. and C.-M.W.; visualization, Y.-C.M. and C.-M.W.; supervision, C.-H.L. and C.-M.W.; project administration, C.-H.L. and C.-M.W.; funding acquisition, C.-M.W. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of National Cheng Kung University Hospital (protocol code B-ER-106-114; 2 June 2017) for studies involving humans.
Informed Consent Statement
Informed Consent Statement was obtained from the owners of Sixty dogs presenting with superficial pyoderma.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to institutional privacy policy.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was financially supported, in part, by research grants from the Ministry of Science and Technology (MOST 110-2313-B-415-001-MY2) in Taiwan awarded to C.-M.W.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Cusco A., Sanchez A., Altet L., Ferrer L., Francino O. Individual Signatures Define Canine Skin Microbiota Composition and Variability. Front. Vet. Sci. 2017;4:6. doi: 10.3389/fvets.2017.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drake D.R., Brogden K.A., Dawson D.V., Wertz P.W. Thematic review series: Skin lipids. Antimicrobial lipids at the skin surface. J. Lipid Res. 2008;49:4–11. doi: 10.1194/jlr.R700016-JLR200. [DOI] [PubMed] [Google Scholar]
- 3.Bloom P. Canine superficial bacterial folliculitis: Current understanding of its etiology, diagnosis and treatment. Vet. J. 2014;199:217–222. doi: 10.1016/j.tvjl.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 4.Hillier A., Lloyd D.H., Weese J.S., Blondeau J.M., Boothe D., Breitschwerdt E., Guardabassi L., Papich M.G., Rankin S., Turnidge J.D., et al. Guidelines for the diagnosis and antimicrobial therapy of canine superficial bacterial folliculitis (Antimicrobial Guidelines Working Group of the International Society for Companion Animal Infectious Diseases) Vet. Dermatol. 2014;25:163-e43. doi: 10.1111/vde.12118. [DOI] [PubMed] [Google Scholar]
- 5.Katayama Y., Ito T., Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2000;44:1549–1555. doi: 10.1128/AAC.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robb A.R., Wright E.D., Foster A.M.E., Walker R., Malone C. Skin infection caused by a novel strain of Staphylococcus pseudintermedius in a Siberian husky dog owner. JMM Case Rep. 2017;4:jmmcr005087. doi: 10.1099/jmmcr.0.005087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ventrella G., Moodley A., Grandolfo E., Parisi A., Corrente M., Buonavoglia D., Guardabassi L. Frequency, antimicrobial susceptibility and clonal distribution of methicillin-resistant Staphylococcus pseudintermedius in canine clinical samples submitted to a veterinary diagnostic laboratory in Italy: A 3-year retrospective investigation. Vet. Microbiol. 2017;211:103–106. doi: 10.1016/j.vetmic.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 8.Soedarmanto I., Kanbar T., Ulbegi-Mohyla H., Hijazin M., Alber J., Lammler C., Akineden O., Weiss R., Moritz A., Zschock M. Genetic relatedness of methicillin-resistant Staphylococcus pseudintermedius (MRSP) isolated from a dog and the dog owner. Res. Vet. Sci. 2011;91:e25–e27. doi: 10.1016/j.rvsc.2011.01.027. [DOI] [PubMed] [Google Scholar]
- 9.Wang C.M., Shia W.Y., Jhou Y.J., Shyu C.L. Occurrence and molecular characterization of reptilian Campylobacter fetus strains isolated in Taiwan. Vet. Microbiol. 2013;164:67–76. doi: 10.1016/j.vetmic.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Wang C.M., Li T.C., Jhan Y.L., Weng J.H., Chou C.H. The impact of microbial biotransformation of catechin in enhancing the allelopathic effects of Rhododendron formosanum. PLoS ONE. 2013;8:e85162. doi: 10.1371/journal.pone.0085162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki T., Tsubakishita S., Tanaka Y., Sakusabe A., Ohtsuka M., Hirotaki S., Kawakami T., Fukata T., Hiramatsu K. Multiplex-PCR method for species identification of coagulase-positive staphylococci. J. Clin. Microbiol. 2010;48:765–769. doi: 10.1128/JCM.01232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CLSI . Performance Standards for Antimicrobial Susceptibility Tests. 27th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2017. [Google Scholar]
- 13.Ferreira A.M., Martins K.B., Silva V.R., Mondelli A.L., Cunha M.L. Correlation of phenotypic tests with the presence of the blaZ gene for detection of beta-lactamase. Braz. J. Microbiol. 2017;48:159–166. doi: 10.1016/j.bjm.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bannoehr J., Ben Zakour N.L., Waller A.S., Guardabassi L., Thoday K.L., van den Broek A.H., Fitzgerald J.R. Population genetic structure of the Staphylococcus intermedius group: Insights into agr diversification and the emergence of methicillin-resistant strains. J. Bacteriol. 2007;189:8685–8692. doi: 10.1128/JB.01150-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hájek V. Staphylococcus intermedius, a New Species Isolated from Animals. Int. J. Syst. Evol. Microbiol. 1976;26:401–408. doi: 10.1099/00207713-26-4-401. [DOI] [Google Scholar]
- 16.Devriese L.A., Vancanneyt M., Baele M., Vaneechoutte M., De Graef E., Snauwaert C., Cleenwerck I., Dawyndt P., Swings J., Decostere A., et al. Staphylococcus pseudintermedius sp. nov. a coagulase-positive species from animals. Int. J. Syst. Evol. Microbiol. 2005;55:1569–1573. doi: 10.1099/ijs.0.63413-0. [DOI] [PubMed] [Google Scholar]
- 17.Sasaki T., Kikuchi K., Tanaka Y., Takahashi N., Kamata S., Hiramatsu K. Reclassification of phenotypically identified staphylococcus intermedius strains. J. Clin. Microbiol. 2007;45:2770–2778. doi: 10.1128/JCM.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malisova L., Safrankova R., Keklakova J., Petras P., Zemlickova H., Jakubu V. Correct species identification (reclassification in CNCTC) of strains of Staphylococcus intermedius-group can improve an insight into their evolutionary history. Folia Microbiol. 2019;64:231–236. doi: 10.1007/s12223-018-0647-7. [DOI] [PubMed] [Google Scholar]
- 19.Bannoehr J., Franco A., Iurescia M., Battisti A., Fitzgerald J.R. Molecular diagnostic identification of Staphylococcus pseudintermedius. J. Clin. Microbiol. 2009;47:469–471. doi: 10.1128/JCM.01915-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qekwana D.N., Oguttu J.W., Sithole F., Odoi A. Burden and predictors of Staphylococcus aureus and S. pseudintermedius infections among dogs presented at an academic veterinary hospital in South Africa (2007–2012) PeerJ. 2017;5:e3198. doi: 10.7717/peerj.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saputra S., Jordan D., Worthing K.A., Norris J.M., Wong H.S., Abraham R., Trott D.J., Abraham S. Antimicrobial resistance in coagulase-positive staphylococci isolated from companion animals in Australia: A one year study. PLoS ONE. 2017;12:e0176379. doi: 10.1371/journal.pone.0176379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C.M., Price L.B., Hungate B.A., Abraham A.G., Larsen L.A., Christensen K., Stegger M., Skov R., Andersen P.S. Staphylococcus aureus and the ecology of the nasal microbiome. Sci. Adv. 2015;1:e1400216. doi: 10.1126/sciadv.1400216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanselman B.A., Kruth S.A., Rousseau J., Weese J.S. Coagulase positive staphylococcal colonization of humans and their household pets. Can. Vet. J. 2009;50:954–958. [PMC free article] [PubMed] [Google Scholar]
- 24.Walther B., Hermes J., Cuny C., Wieler L.H., Vincze S., Abou Elnaga Y., Stamm I., Kopp P.A., Kohn B., Witte W., et al. Sharing more than friendship—Nasal colonization with coagulase-positive staphylococci (CPS) and co-habitation aspects of dogs and their owners. PLoS ONE. 2012;7:e35197. doi: 10.1371/journal.pone.0035197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch S.A., Helbig K.J. The Complex Diseases of Staphylococcus pseudintermedius in Canines: Where to Next? Vet. Sci. 2021;8:11. doi: 10.3390/vetsci8010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasaki T., Kikuchi K., Tanaka Y., Takahashi N., Kamata S., Hiramatsu K. Methicillin-resistant Staphylococcus pseudintermedius in a veterinary teaching hospital. J. Clin. Microbiol. 2007;45:1118–1125. doi: 10.1128/JCM.02193-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perreten V., Kadlec K., Schwarz S., Gronlund Andersson U., Finn M., Greko C., Moodley A., Kania S.A., Frank L.A., Bemis D.A., et al. Clonal spread of methicillin-resistant Staphylococcus pseudintermedius in Europe and North America: An international multicentre study. J. Antimicrob. Chemother. 2010;65:1145–1154. doi: 10.1093/jac/dkq078. [DOI] [PubMed] [Google Scholar]
- 28.Maluping R.P., Paul N.C., Moodley A. Antimicrobial susceptibility of methicillin-resistant Staphylococcus pseudintermedius isolated from veterinary clinical cases in the UK. Br. J. Biomed. Sci. 2014;71:55–57. doi: 10.1080/09674845.2014.11669965. [DOI] [PubMed] [Google Scholar]
- 29.Boost M.V., O’Donoghue M.M., James A. Prevalence of Staphylococcus aureus carriage among dogs and their owners. Epidemiol. Infect. 2008;136:953–964. doi: 10.1017/S0950268807009326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nienhoff U., Kadlec K., Chaberny I.F., Verspohl J., Gerlach G.-F., Kreienbrock L., Schwarz S., Simon D., Nolte I. Methicillin-resistant Staphylococcus pseudintermedius among dogs admitted to a small animal hospital. Vet. Microbiol. 2011;150:191–197. doi: 10.1016/j.vetmic.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 31.Lehner G., Linek M., Bond R., Lloyd D.H., Prenger-Berninghoff E., Thom N., Straube I., Verheyen K., Loeffler A. Case–control risk factor study of methicillin-resistant Staphylococcus pseudintermedius (MRSP) infection in dogs and cats in Germany. Vet. Microbiol. 2014;168:154–160. doi: 10.1016/j.vetmic.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 32.Gronthal T., Eklund M., Thomson K., Piiparinen H., Sironen T., Rantala M. Antimicrobial resistance in Staphylococcus pseudintermedius and the molecular epidemiology of methicillin-resistant S. pseudintermedius in small animals in Finland. J. Antimicrob. Chemother. 2017;72:1021–1030. doi: 10.1093/jac/dkx086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joosten P., Van Cleven A., Sarrazin S., Paepe D., De Sutter A., Dewulf J. Dogs and Their Owners Have Frequent and Intensive Contact. Int. J. Environ. Res. Public Health. 2020;17:4300. doi: 10.3390/ijerph17124300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pomba C., Rantala M., Greko C., Baptiste K.E., Catry B., van Duijkeren E., Mateus A., Moreno M.A., Pyorala S., Ruzauskas M., et al. Public health risk of antimicrobial resistance transfer from companion animals. J. Antimicrob. Chemother. 2017;72:957–968. doi: 10.1093/jac/dkw481. [DOI] [PubMed] [Google Scholar]
- 35.Laarhoven L.M., de Heus P., van Luijn J., Duim B., Wagenaar J.A., van Duijkeren E. Longitudinal study on methicillin-resistant Staphylococcus pseudintermedius in households. PLoS ONE. 2011;6:e27788. doi: 10.1371/journal.pone.0027788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Windahl U., Gren J., Holst B.S., Borjesson S. Colonization with methicillin-resistant Staphylococcus pseudintermedius in multi-dog households: A longitudinal study using whole genome sequencing. Vet. Microbiol. 2016;189:8–14. doi: 10.1016/j.vetmic.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 37.Saab M.E., Weese J.S., McClure J.T. Direct repeat unit (dru) typing and antimicrobial resistance of methicillin-resistant Staphylococcus pseudintermedius isolated from dogs in Atlantic Canada. Can. J. Vet. Res. 2017;81:192–198. [PMC free article] [PubMed] [Google Scholar]
- 38.Weese J.S., Sweetman K., Edson H., Rousseau J. Evaluation of minocycline susceptibility of methicillin-resistant Staphylococcus pseudintermedius. Vet. Microbiol. 2013;162:968–971. doi: 10.1016/j.vetmic.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Kadlec K., Schwarz S., Goering R.V., Weese J.S. Direct Repeat Unit (dru) Typing of Methicillin-Resistant Staphylococcus pseudintermedius from Dogs and Cats. J. Clin. Microbiol. 2015;53:3760–3765. doi: 10.1128/JCM.01850-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kadlec K., Weiss S., Wendlandt S., Schwarz S., Tonpitak W. Characterization of canine and feline methicillin-resistant Staphylococcus pseudintermedius (MRSP) from Thailand. Vet. Microbiol. 2016;194:93–97. doi: 10.1016/j.vetmic.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 41.Kang J.H., Chung T.H., Hwang C.Y. Clonal distribution of methicillin-resistant Staphylococcus pseudintermedius isolates from skin infection of dogs in Korea. Vet. Microbiol. 2017;210:32–37. doi: 10.1016/j.vetmic.2017.08.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to institutional privacy policy.