Abstract
In the present study, we surveyed the ascomycetes from bamboo of Phyllostachys across Sichuan Province, China. A biphasic approach based on morphological characteristics and multigene phylogeny confirmed seven species, including one new genus, two new species, and five new host record species. A novel genus Paralloneottiosporina is introduced to accommodate Pa. sichuanensis that was collected from leaves of Phyllostachys violascens. Moreover, the newly introduced species Bifusisporella sichuanensis was isolated from leaves of P. edulis, and five species were newly recorded on bamboos, four species belonging to Apiospora, viz. Ap. yunnana, Ap. neosubglobosa, Ap. jiangxiensis, and Ap. hydei, and the last species, Seriascoma yunnanense, isolated from dead culms of P. heterocycla. Morphologically similar and phylogenetically related taxa were compared. Comprehensive descriptions, color photo plates of micromorphology are provided.
Keywords: bambusicolous fungi, molecular phylogeny, one new genus, systematics, two new species
1. Introduction
Bamboo is currently classified in the subfamily Bambusoideae of the extensive grass family Poaceae, and distributed worldwide. It comprises circa 1000 to 1500 species in up to 90 genera [1] and more than 70 species in Phyllostachys (Bambusoideae, Poaceae) [2,3]. Most bamboos are distributed in Southeast Asia, with China as the distribution center [4]. There are about 21 species of Phyllostachys in Sichuan, including Phyllostachys edulis (Carriere) J. Houzea, P. heteroclada Oliver, and P. violascens ‘Prevernalis’ S.Y. Chen et C.Y. Yao. Bamboos of Phyllostachys play an important role in native economy and ecology. They are used in furniture, and construction (e.g., fishing rods, flutes, flooring materials, chairs.) [5,6]. Bamboo shoots are used as food for humans and animals such as pandas [7,8]. In addition, it is an important ornamental plant for the landscape in China because of its evergreen and graceful appearance [9].
A review of the literature on bamboo-associated fungi reveals that nearly 1500 species have been described or recorded worldwide [10], including economically important pathogenic fungi, and a large number of saprobic and endophytic fungi [1,11,12,13]. Most bambusicolous fungi have been reported from Asia, especially Japan and Thailand, a few known from India and South America [1,12,14,15,16,17,18]. However, few studies have investigated the diversity and phylogeny on bamboo in China. The taxonomic studies on bambusicolous fungi are of great significance [19,20,21]. According to the literature review, about 85 species associated with Phyllostachys have been recorded. Teng [22] first reported the fungus Oedocephalum glomerulosum (Bull.) Sacc. on Phyllostachys in 1932. Tai listed 36 species of Phyllostachys from bamboo based on the reports on Chinese fungal resource until 1973 [23]. Chen investigated the phytogeography of forest fungi in China, North America, and Siberia, from which 33 species were found associated with Phyllostachys [24]. However, most of those identifications were conducted lacking molecular data and detailed micromorphology, and as most bamboos are unidentified, the relationship of bambusicolous fungi with bamboo species is not clear.
Due to the high fungal diversity on Phyllostachys, an ongoing investigation was conducted in several main producing or planting areas of bamboo Phyllostachys in Sichuan Province, China, including Ya’an City, Qionglai City, Chengdu City, and Yibin City. In this study, we provide detailed taxonomic features combining morphology and phylogeny on the fungi associated with Phyllostachys from Sichuan Province, China, which is a fundamental task for the bioresource collection on bambusicolous fungi.
2. Materials and Methods
2.1. Specimen Collection and Morphological Study
From 2020 to 2021, the specimens were collected from leaves, branches, and culms. The samples were kept in plastic bags and taken back to the laboratory after being photographed with a Sony DSC-HX3 digital camera. The fungi were isolated into pure culture based on single spore isolation [25]. Glass slide specimens were prepared by free-hand slicing with double-sided blades for morphologic observation. Morphological characteristics of ascomata and sporodochia were observed using a dissecting microscope, the NVT-GG (Shanghai Advanced Photoelectric Technology Co. Ltd., Shanghai, China), and photographed with a VS-800C micro-digital camera (Shenzhen Weishen Times Technology Co. Ltd., Shenzhen, China). An Olympus BX43 compound microscope with an Olympus DP22 digital camera was used to observe and photograph the microstructure of asci, ascospores, conidiophores, and conidia. Measurements were performed using Tarosoft® Image Frame Work v.0.9.7 (Tarosoft (R), Nontha Buri, Thailand). Specimens were deposited at the Herbarium of Sichuan Agricultural University, Chengdu, China (SICAU), and pure cultures were deposited at the Culture Collection in Sichuan Agricultural University (SICAUCC).
2.2. DNA Extraction, PCR Amplification, and Nucleotide Sequencing
Genomic DNA was extracted from fresh mycelia which was cultured on PDA at 25 °C for 15–30 days, using a TreliefTM Plant Genomic DNA Kit. Primers ITS5/ITS4 [26], NS1/NS4 [26], LR0R/LR5 [27], T1/Bt2b [28,29], RPB1-Ac/RPB1-Cr [30,31], and fRPB2-5F/fRPB2-7cR [32] were used for the amplification of internal transcribed spacers (ITS), the partial small subunit nuclear rDNA (SSU), the partial large subunit nuclear rDNA (LSU), the β-tubulin gene (tub2), the large subunit of RNA polymerase I (rpb1), and RNA polymerase II second largest subunit (rpb2) genes, respectively. Primers EF1-983F/EF1-2218R [33] and EF1-728F/EF2 [34,35] were employed for translation elongation factor 1-alpha (tef1-α) genes.
Amplification reactions were performed in 25 µL of total reaction that contained 22 µL Master Mix (Beijing TsingKe Biotech Co., Ltd., Beijing, China), 1 µL each of forward and reverse (10 µM) primers and 1 µL of DNA template. The amplification reactions were performed as described by Dai et al. [16] and Wang et al. [36]. PCR products were purified and sequenced at TsingKe Biological Technology Co., Ltd. (Chengdu, China). The resulting sequences were submitted to GenBank.
2.3. Sequence Alignment and Phylogenetic Analyses
Based on blast searches in GenBank, using ITS, LSU, SSU, tef1-α, tub2, rpb1, or rpb2 sequence data, separate phylogenetic analyses were carried out to determine the placements of each fungal group (Table 1). Sequences for phylogenetic analyses were selected mainly from recently published literature and phylogenetic related sequences based on BLAST searches in GenBank (Table A1). Datasets were aligned using MAFFT v.7.407 [37], and ambiguous regions were excluded with BioEdit version 7.0.5.3 [38]. Maximum likelihood (ML) and Bayesian inference (BI) were constructed as described in Xu et al. [39]. The phylogram was visualized with FigureTree v. 1.4.3 and edited using Adobe Illustrator CS6 (Adobe Systems Inc., San Jose, CA, USA).
Table 1.
Selected genes for polymerase chain reaction of each genus.
| Genera | Sequences Dataset |
|---|---|
| Apiospora | ITS, LSU, tub2, tef1-α |
| Bifusisporella | ITS, LSU, tef1-α, rpb1 |
| Paralloneottiosporina | ITS, LSU, SSU, tef1-α |
| Seriascom | ITS, LSU, SSU, tef1-α, rpb2 |
3. Results
3.1. Phylogenetic Analyses
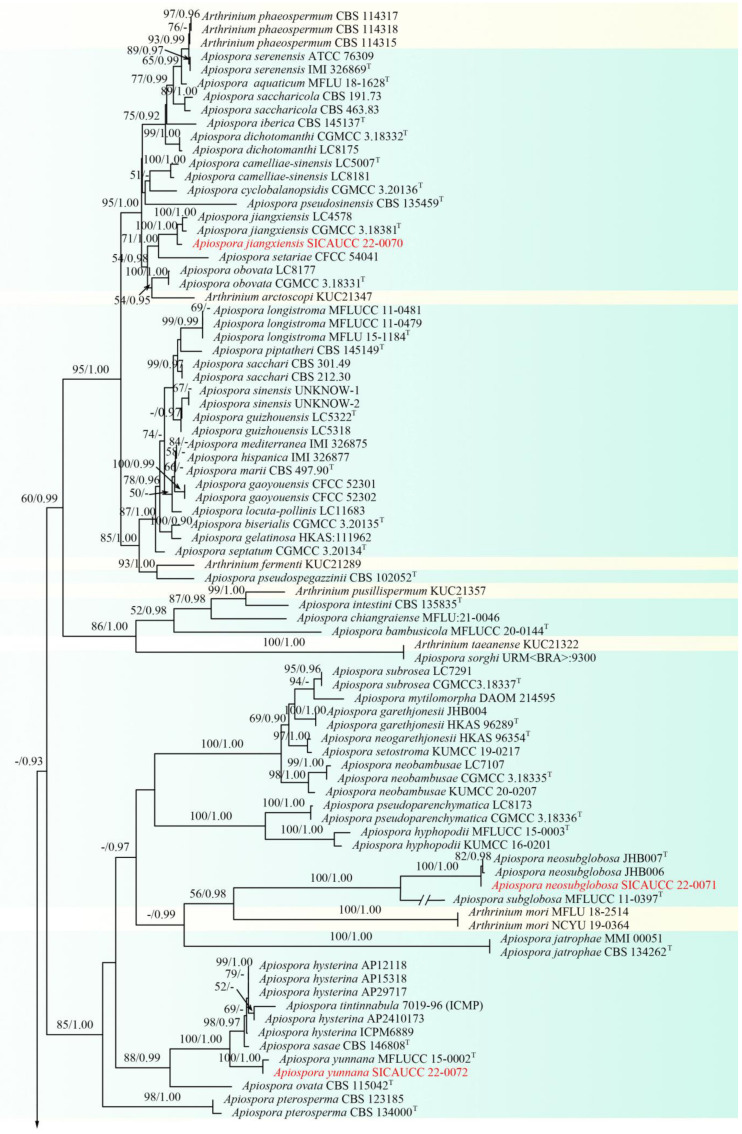
A combined dataset (ITS, LSU, tef1-α, tub2) comprising 138 taxa within Apiosporaceae, which is rooted with Pestalotiopsis chamaeropis (CBS 237.38) and Pe. colombiensis (CBS 118553) (Pestalotiopsidaceae, Amphisphaeriales), was used for the phylogenetic analyses. The alignment contained 5875 characters (ITS = 999, LSU = 1382, tef1-α = 1651, tub2 = 1844), including gaps. The best scoring RAxML tree with a final likelihood value of −36198.939448 is presented. The matrix had 2337 distinct alignment patterns, with 64.85% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.237208, C = 0.257370, G = 0.253511, T = 0.251911, with substitution rates AC = 1.104968, AG = 2.746651, AT = 1.143208, CG = 0.910079, CT = 4.335389, GT = 1.000000. The gamma distribution shape parameter α = 0.269105, and the tree length = 3.509694. In the phylogenetic trees generated from ML and BI analyses, the strain SICAUCC 22-0032 clustered with the known species Apiospora hydei (KUMCC 16-0204, CBS 114990) in a clade with 97% ML and 0.99 BYPP support value, strain SICAUCC 22-0070 clustered with Ap. jiangxiensis (CGMCC 3.18381, LC4578) with high support values (100% ML and 1.00 BYPP), strain SICAUCC 22-0071 clustered with Ap. neosubglobosa (JHB006, JHB007) in a clade with 100% ML and 1.00 BYPP support value, and strain SICAUCC 22-0072 clustered with the Ap. yunnana (MFLUCC 15-0002) in a clade with 100% ML and 1.00 BYPP support values (Figure 1).
Figure 1.
Phylogram generated from RAxML analysis based on combined ITS, LSU, tub2, and tef1-α sequence data of Apiosporaceae. Bootstrap support values for maximum likelihood (ML, left) higher than 50% and Bayesian posterior probabilities (BYPP, right) equal to or greater than 0.90 are indicated at the nodes, respectively. The sequences from ex-type strains are marked by a superscript symbol T. The newly generated sequences are written in red. Arthrinium species with yellow background were temporarily not combined to Apiospora.
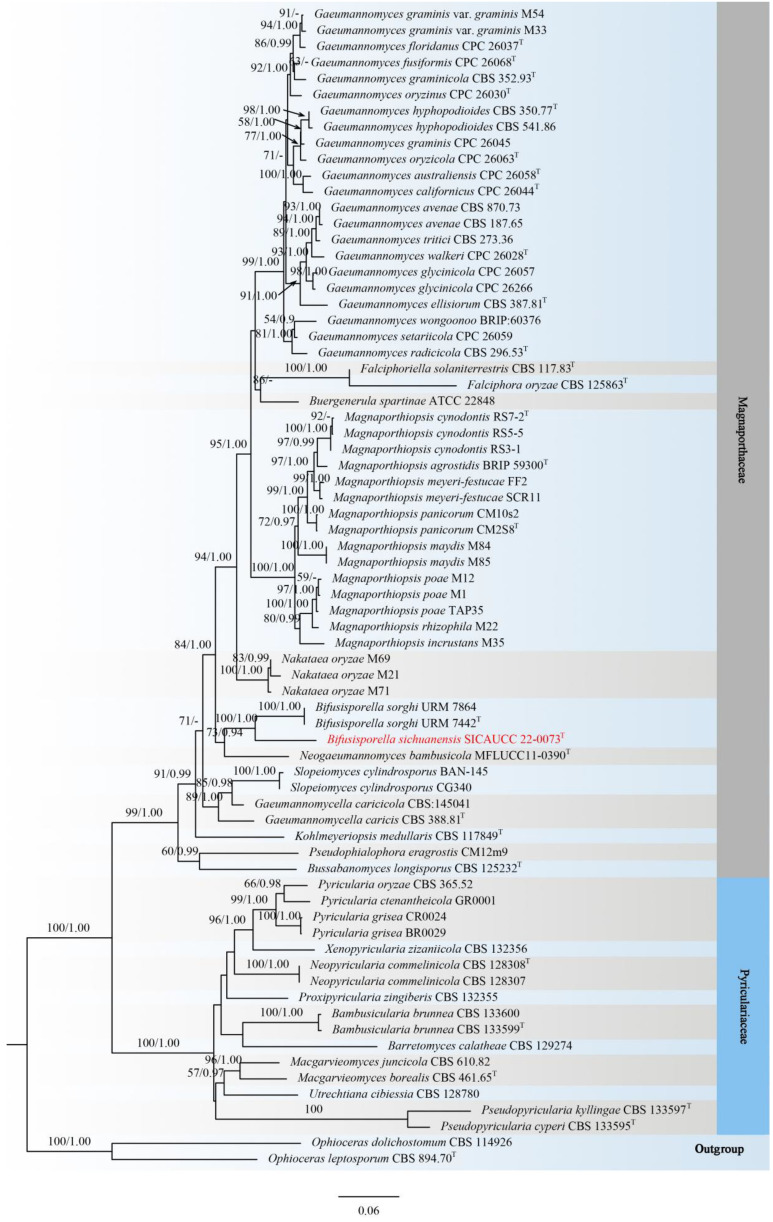
Phylogenetic analyses of a concatenated aligned dataset (ITS, LSU, rpb1, tef1-α), including 70 taxa within Magnaporthaceae and Pyriculariaceae, were conducted and rooted with Ophioceras dolichostomum (CBS 114926) and O. leptosporum (CBS 894.70) (Ophioceraceae, Magnaporthales). The alignment contained 4094 characters (ITS = 899, LSU = 1105, rpb1 = 1047, tef1-α = 1043), including gaps. The best scoring RAxML tree with a final likelihood value of −31022.648763 is presented. The matrix had 1923 distinct alignment patterns, with 36.77% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.243596, C = 0.275654, G = 0.281915, T = 0.198836, with substitution rates AC = 1.103727, AG = 2.292134, AT = 1.431191, CG = 0.918700, CT = 5.773674, GT = 1.000000. The gamma distribution shape parameter α = 0.319184, and the tree length = 3.313974. In the phylogenetic tree (Figure 2), the novel species Bifusisporella sichuanensis constitutes a highly supported independent lineage (ML = 100%, BYPP = 1.00) with B. sorghi (URM 7864, URM 7442).
Figure 2.
Phylogram generated from RAxML analysis based on combined ITS, LSU, rpb1, and tef1-α sequence data of Magnaporthaceae and Pyriculariaceae. Bootstrap support values for maximum likelihood (ML, left) higher than 50% and Bayesian posterior probabilities (BYPP, right) equal to or greater than 0.90 are indicated at the nodes, respectively. The sequences from ex-type strains are marked by a superscript symbol T. The newly generated sequence is written in red.
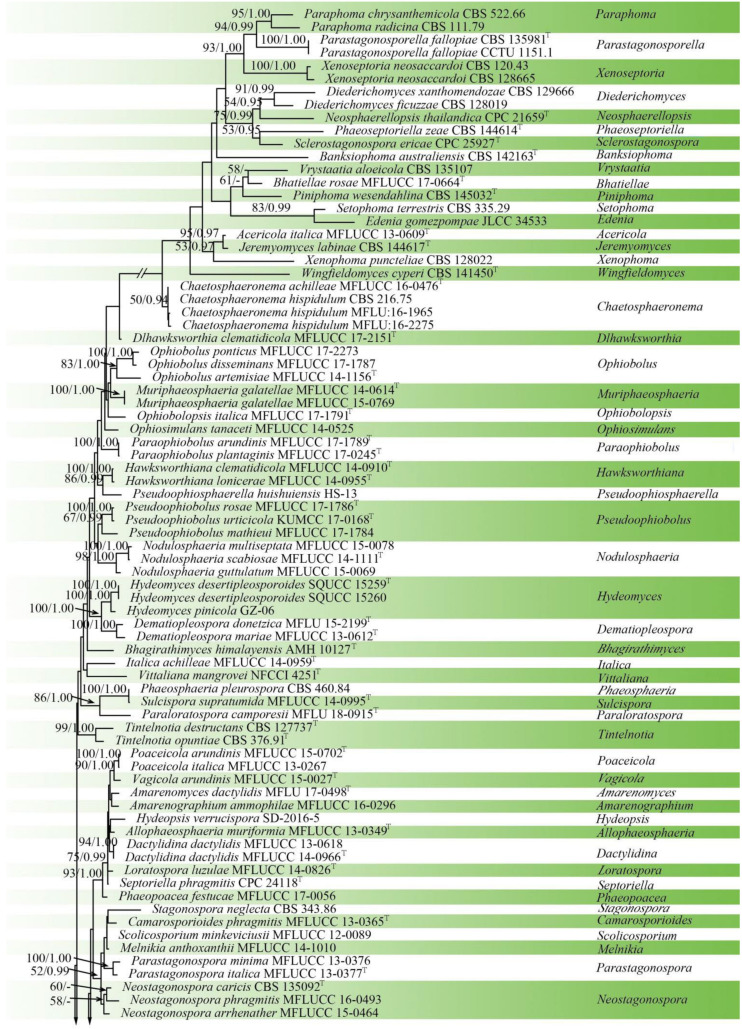
The concatenated aligned dataset of ITS, LSU, SSU, tef1-α sequences, including 124 ingroup taxa within Phaeosphaeriaceae and two outgroup taxa in Leptosphaeriaceae, were used for the phylogenetic analyses of Paralloneottiosporina. The alignment contained 5851 characters (ITS = 1469, LSU = 1433, SSU = 1548, tef1-α = 1401), including gaps. The best scoring RAxML tree with a final likelihood value of −46908.078740 is presented. The matrix had 2382 distinct alignment patterns, with 55.68% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.246158, C = 0.236637, G = 0.264322, T = 0.252883, with substitution rates AC = 1.087661, AG = 2.657942, AT = 2.045792, CG = 0.863381, CT = 6.106747, GT = 1.000000. The gamma distribution shape parameter α = 0.263651, and the tree length = 7.503091. In the phylogenetic tree generated from ML and BI analyses, the novel species Paralloneottiosporina sichuanensis (SICAUCC 22-0074, SICAUCC 22-0075) constitutes a moderately supported independent lineage (63% ML/0.99 BYPP statistical support) with the species Alloneottiosporina thailandica (MFLUCC 15-0576) (Figure 3).
Figure 3.
Phylogram generated from RAxML analysis based on combined ITS, LSU, SSU, and tef1-α sequence data of Phaeosphaeriaceae. Bootstrap support values for maximum likelihood (ML, left) higher than 50% and Bayesian posterior probabilities (BYPP, right) equal to or greater than 0.90 are indicated at the nodes, respectively. The sequences from ex-type strains are marked by a superscript symbol T. The newly generated sequences are written in red.
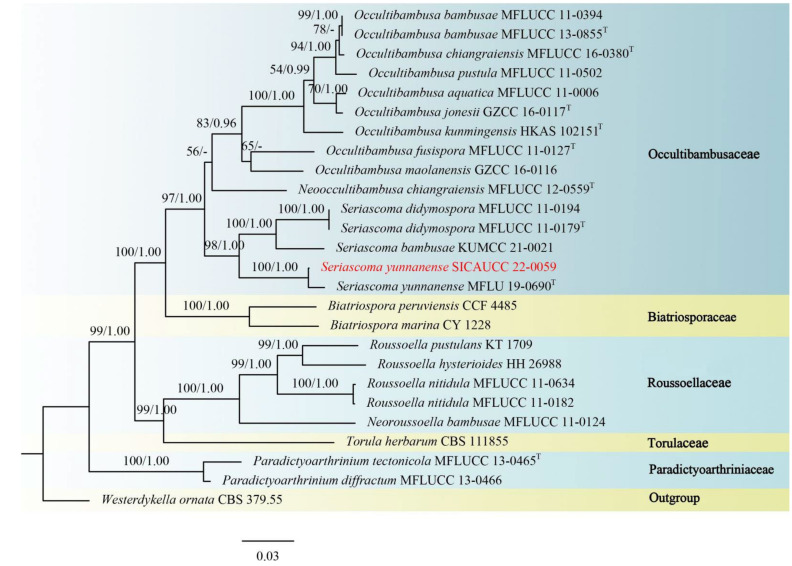
A combined dataset (ITS, LSU, SSU, tef1-α, rpb2) comprising 25 taxa within Bambusicolaceae, Biatriosporaceae, Roussoellaceae, Torulaceae, and Paradictyoarthriniaceae was used for phylogenetic analyses of Seriascoma, and the Westerdykella ornata (CBS 379.55) (Sporormiaceae) was used as outgroup taxon. The alignment contained 6569 characters (LSU = 1383, SSU = 1741, tef1-α = 1346, rpb2 = 2099), including gaps. The best scoring RAxML tree with a final likelihood value of −22606.776997 is presented. The matrix had 1406 distinct alignment patterns, with 48.40% of undetermined characters or gaps. Estimated base frequencies were as follows: A = 0.250203, C = 0.247742, G = 0.269455, T = 0.232600, with substitution rates AC = 1.348170, AG = 4.119625, AT = 1.278817, CG = 1.296090, CT = 9.080955, GT = 1.000000. The gamma distribution shape parameter α = 0.146142, and the tree length = 1.192279. According to the phylogenetic tree (Figure 4), the strain (SICAUCC 22-0059) clustered with Seriascoma yunnanense (MFLU 19-0690) in a clade with 100% ML and 1.00 BYPP statistical support.
Figure 4.
Phylogram generated from RAxML analysis based on combined ITS, LSU, rpb2, and tef1-α sequence data of isolates within Bambusicolaceae and other representative species in Biatriosporaceae, Roussoellaceae, Torulaceae, and Paradictyoarthriniaceae. Bootstrap support values for maximum likelihood (ML, left) higher than 50% and Bayesian posterior probabilities (BYPP, right) equal to or greater than 0.90 are indicated at the nodes, respectively. The sequences from ex-type strains are marked by a superscript symbol T. The newly generated sequence is written in red.
3.2. Taxonomy
Apiosporaceae K.D. Hyde, J. Fröhl., Joanne E. Taylor & M.E. Barr, Sydowia. 50 (1): 23 (1998).
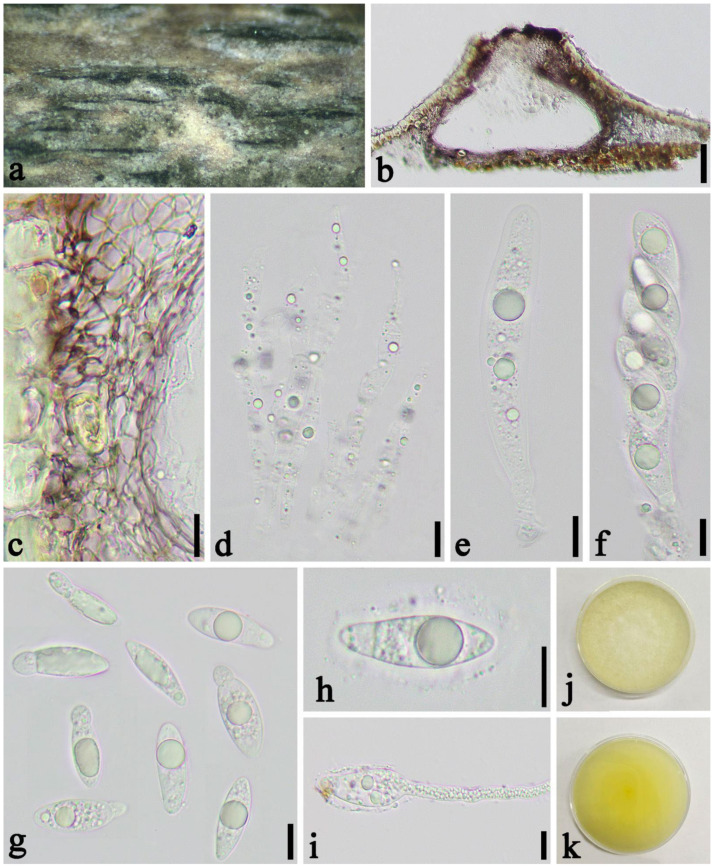
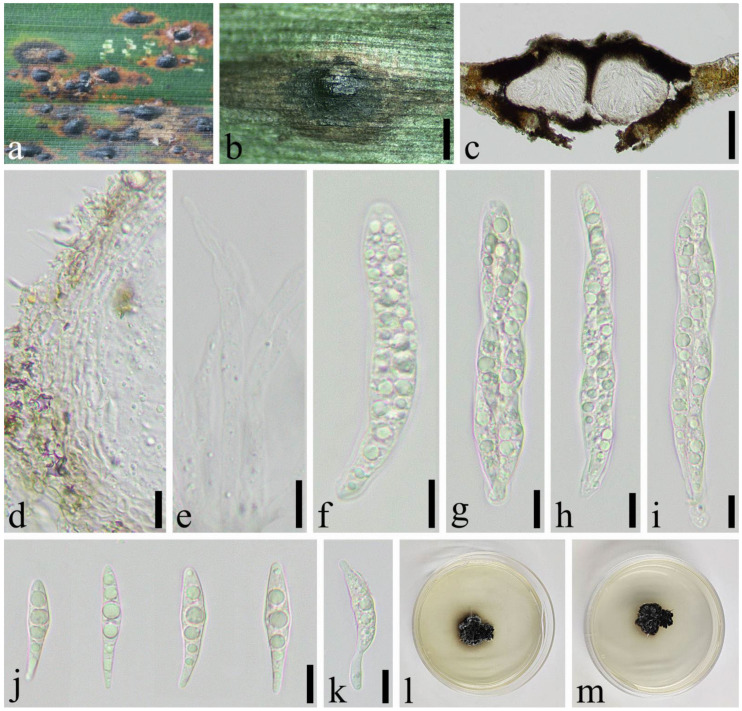
Apiospora hydei (Crous) Pintos & P. Alvarado, Fungal Systematics and Evolution. 7: 206 (2021) (Figure 5).
Figure 5.
Apiospora hydei (SICAU 22-0032). (a) Ascostromata developing on bamboo branches. (b) Vertical sections of ascostromata. (c) Peridium. (d) Paraphyses. (e,f) Asci. (g,h) Ascospores. (i) Germinating ascospore. (j,k) Cultures on PDA. Scale bars: (b) = 50 μm, (c–i) = 10 μm.
≡ Arthrinium hydei Crous, IMA Fungus 4(1): 142 (2013).
Saprobic on dead culms of Phyllostachys nigra (Lodd. ex Lindl.) Munro. Sexual morph: Ascostromata 421–1343 × 174–387 × 176–245 μm ( = 705 × 267 × 198 μm, n = 30), solitary to gregarious, immersed, fusiform to ellipsoid, dark brown to black, multi-loculate, with long axis. Peridium 17–46 μm wide, composed of 8–15 layers of brown to hyaline cells of textura angularis to prismatica. Hamathecium 2–6.5 μm wide, composed of dense, long, septate, and unbranched paraphyses. Asci 81–123 × 16–23 μm, ( = 116 × 180 μm, n = 50), 8–spored, unitunicate, broadly cylindrical, slightly curved, with a short pedicel, apically rounded. Ascospores 24–30 × 7–11 μm, ( = 26 × 10 μm, n = 50), 2-seriate, elliptical, 1–septate, with a large, curved upper cell and small lower cell, with narrowly rounded ends, hyaline, guttules, smooth-walled, surrounded by gelatinous sheath. Asexual morph: see Crous et al. [40].
Material examined: China, Sichuan Province, Chengdu City, Wenjiang District (19°30′42.22″ N, 103°51′19″ E, Alt. 528 m), on dead culms of Phyllostachys nigra, 14 March 2021, Yi-cong Lv, LYC202103003 (SICAU 22-0032), living culture SICAUCC 22-0032.
Culture characters: Ascospores germinate within 24 h. Colonies grow fast on PDA, reaching 6 cm after one week at 25 °C, under 12 h light/12 h dark, and are cottony, circular, and white from above and light yellow below, with irregular edge.
Notes: Apiospora hydei was introduced based on the asexual morph characters and phylogeny analyses by Crous et al. [40]. Morphological comparisons were impossible due to the lack of sexual morph between our isolates and the ex-type strain (CBS 114990), but it is similar to A. hydei in sexual descriptions provided by Dai et al. [41]. Nucleotide comparisons of ITS, LSU, tef1-α and tub2 (SICAUCC 22-0033) showed high homology with the sequences of A. hydei (CBS 114990), similarities are 100% (528/528, 0 gaps), 99.77% (896/898, 0 gaps), 99.71% (355/356, 0 gaps), and 98.82% (754/763, 0 gaps), respectively.
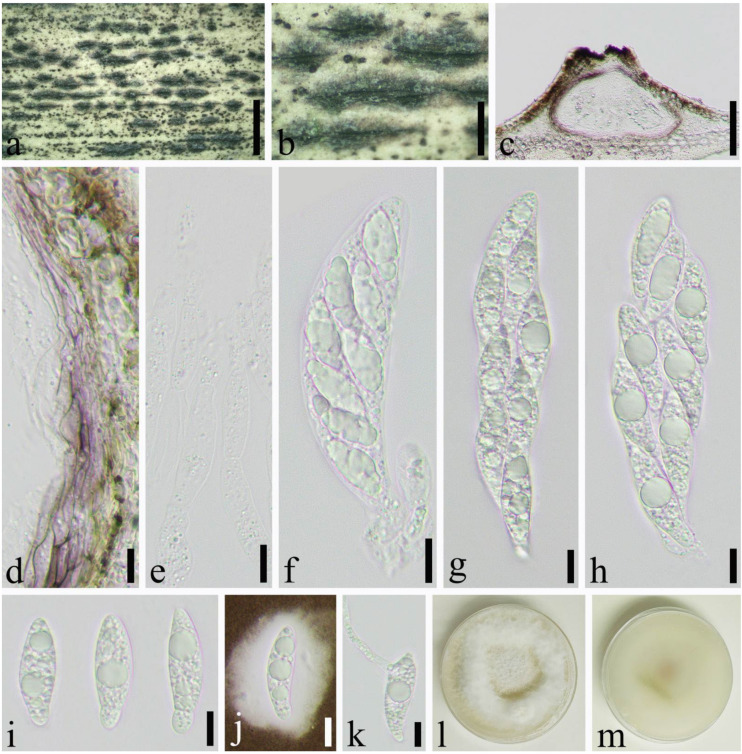
Apiospora jiangxiensis (M. Wang & L. Cai) Pintos & P. Alvarado, Fungal Systematics and Evolution 7: 206 (2021) (Figure 6).
Figure 6.
Apiosporajiangxiensis (SICAU 22-0070). (a,b) Ascostromata developing on bamboo culm. (c) Vertical sections of ascostromata. (d) Peridium. (e) Paraphyses. (f–h) Asci. (i,j) Ascospores. (k) Germinating ascospore. (l,m) Cultures on PDA. Scale bars: (a) = 2 mm, (b) = 500 μm, (c) = 100 μm, (d–k) = 10 μm.
≡ Arthrinium jiangxiense M. Wang & L. Cai, in Wang, Tan, Liu & Cai, MycoKeys 34(1): 14 (2018).
Saprobic on dead culms of Phyllostachys heteroclada Oliver. Sexual morph: Ascostromata 575–1334 × 274–444 × 134–157 μm ( = 876 × 355 × 143 μm, n = 30), solitary to gregarious, multi-loculate, immersed, fusiform to ellipsoid, black, with long axis broken at the top. Peridium 9.0–44 μm wide ( = 21 μm, n = 25), composed of several layers of brown to hyaline cells of textura angularis to prismatica. Hamathecium 4.0–11 μm wide, composed of dense, long, septate, unbranched, paraphyses. Asci 83–114 × 18–28 μm ( = 104 × 23 μm, n = 50), 8–spored, unitunicate, broadly cylindrical to long clavate, with a short pedicel, slightly curved, apically rounded. Ascospores 32–37 × 9.6–11 μm ( = 34 × 10 μm, n = 50), 2–seriate, 1–septate, elliptical, with a large, curved, upper cell and small lower cell, with narrowly rounded ends, hyaline, smooth-walled, with many guttules, surrounded by gelatinous sheath attached. Asexual morph: see Wang et al. [36].
Material examined: China, Sichuan Province, Luzhou City, Xuyong District (27°53′28″ N, 105°16′36″ E, Alt. 1350 m), on dead culm of Phyllostachys heteroclada, 26 July 2021, Qian Zeng, ZQ202107133 (SICAU 22-0070), living culture SICAUCC 22-0070.
Culture characters: Ascospores germinate on PDA within 24 h. Colonies grow fast on PDA, reaching 6 cm after 1 week at 25 °C, under 12 h light/12 h dark, and are cottony, white, circular, with irregular edge.
Notes: Specimen in our study shared similar morphology with the original description of Apiospora jiangxiensis by Wang et al. [36]. Nucleotide comparisons of ITS, LSU, and tub2 (SICAUCC 22-0070) showed high homology with the sequences of Ap. jiangxiensis (CGMCC 3.18381), similarities are 100% (541/541, 0 gaps), 99.09% (436/440, 0 gaps), and 98.22% (717/730, 0 gaps), respectively. However, the latter lack tef1-α sequences for further comparisons.
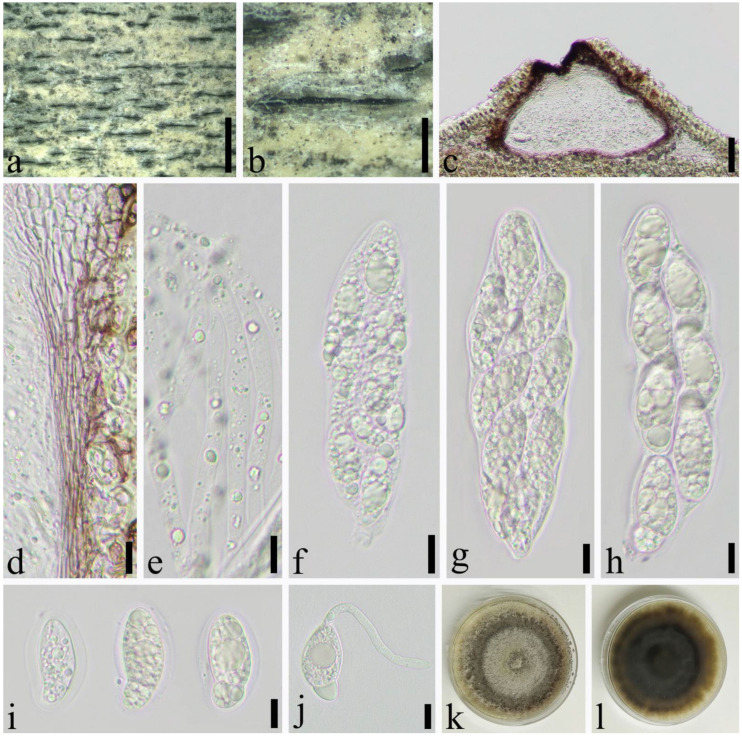
Apiospora neosubglobosa (D.Q. Dai & H.B. Jiang) Pintos & P. Alvarado, Fungal Systematics and Evolution 7: 206 (2021) (Figure 7).
Figure 7.
Apiosporaneosubglobosa (SICAU 22-0071). (a,b) Ascostromata developing on bamboo culm. (c) Vertical sections of ascostromata. (d) Peridium. (e) Paraphyses. (f–h) Asci. (i) Ascospores. (j) Germinating ascospore. (k,l) Cultures on PDA. Scale bars: (a) = 2 mm, (b) = 500 μm, (c) = 50 μm, (d–j) = 10 μm.
≡ Arthrinium neosubglobosum D.Q. Dai & H.B. Jiang, Mycosphere 7(9): 1337 (2017).
Saprobic on dead culms of Phyllostachys bissetii McClure. Sexual morph: Ascostromata 330–1092 × 198–354 × 134–224 μm ( = 632 × 250 × 174 μm, n = 30), gregarious, immersed, multi-loculate, fusiform to ellipsoid, dark brown to black, with long axis broken at the top. Peridium 17.0–46 μm wide ( = 19 μm, n = 25), composed of several layers of brown to hyaline, cells of textura angularis to prismatica. Hamathecium 3.5–6.0 μm wide, composed of dense, long, septate, unbranched, paraphyses. Asci 94–137 × 23–40 μm ( = 125 × 31 μm, n = 50), 8-spored, unitunicate, broadly cylindrical to long clavate, with a short pedicel, slightly curved, apically rounded. Ascospores 28–36 × 13–15 μm ( = 32 × 14 μm, n = 50), 2–seriate, 1–septate, elliptical, with a large, curved, upper cell and small lower cell, with narrowly rounded ends, hyaline, smooth-walled, with many guttules, surrounded by gelatinous sheath attached. Asexual morph: see Dai et al. [16].
Material examined: CHINA, Sichuan Province, Luzhou City, Xuyong District (27°52′5″ N, 105°16′23″ E, Alt. 1470 m), on dead culm of Phyllostachys bissetii, 26 July 2021, Qian Zeng, ZQ202107128 (SICAU 22-0071), living culture SICAUCC 22-0071.
Cultural characters: Ascospores germinate on PDA within 24 h. Colonies grow fast on PDA, reaching 4 cm after 1 week at 25 °C, under 12 h light/12 h dark, and are cottony, circular, initially white, then brown, with regular edge.
Notes: Apiospora neosubglobosa was described by Dai et al. based on the morphological characteristics and molecular phylogeny [16]. Strain SICAUCC 22-0071 clustered with ex–type strain (JHB007) with high bootstrap support (100% ML and 1.00 BYPP). Nucleotide comparisons of ITS and LSU (SICAUCC 22-0071) showed high homology with the sequences of Ap. neosubglobosa (JHB007), similarities are 99.84% (649/650, 0 gaps), 100% (1173/1173, 0 gaps), respectively.
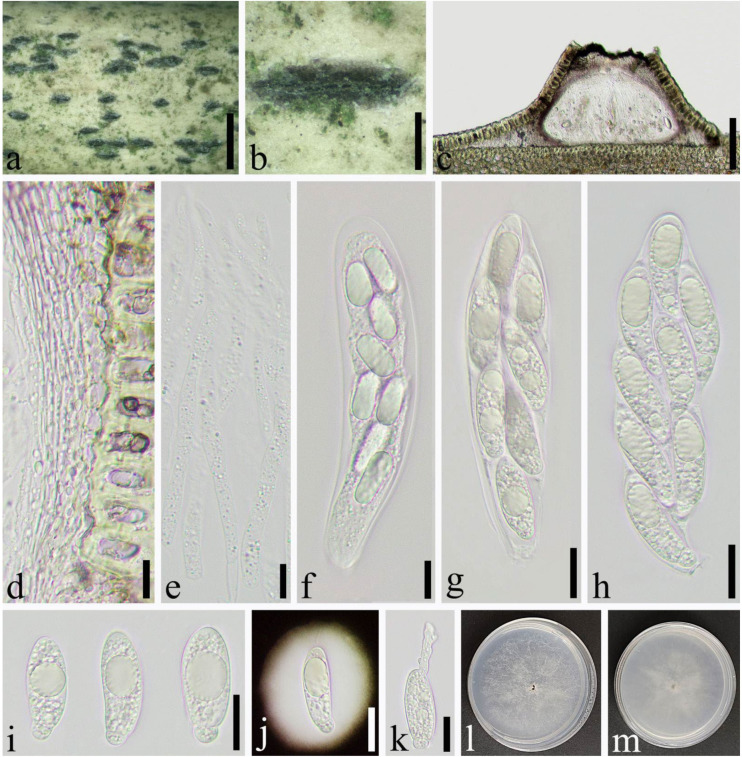
Apiospora yunnana (D.Q. Dai & K.D. Hyde) Pintos & P. Alvarado, Fungal Systematics and Evolution 7: 207 (2021) (Figure 8).
Figure 8.
Apiosporayunnana (SICAU 22-0072). (a,b) Ascostromata developing on bamboo culm. (c) Vertical sections of ascostromata. (d) Peridium. (e) Paraphyses. (f–h) Asci. (i,j) Ascospores. (k) Germinating ascospore. (l,m) Cultures on PDA. Scale bars: (a) = 2 mm, (b) = 500 μm, (c) = 100 μm, (d–f) = 10 μm, (g–k) = 20 μm.
≡ Arthrinium yunnanum D.Q. Dai & K.D. Hyde, Fungal Diversity 82: 69 (2016).
Saprobic on culms of Phyllostachys aurea Carr. ex A. et C. Riv. Sexual morph: Ascostromata 624–1307 × 253–510 × 165–211 μm ( = 892 × 359 × 188 μm, n = 30), gregarious, multi-loculate, immersed, fusiform to ellipsoid, black, with long axis broken at the top. Peridium 8.5–43 μm wide ( = 17 μm, n = 25), composed of several layers of brown to hyaline cells of textura angularis to prismatica. Hamathecium 3.5–8.0 μm wide, composed of dense, long, septate, unbranched paraphyses. Asci 89–144 × 18–40 μm ( = 120 × 32 μm, n = 50), 8–spored, unitunicate, broadly cylindrical to long clavate, no pedicel, slightly curved, apically rounded. Ascospores 30–42 × 10–13 μm ( = 36 × 12 μm, n = 50), 2–seriate, 1–septate, elliptical, with a large, curved, upper cell and a small lower cell, with narrowly rounded ends, hyaline, smooth-walled, with many guttules, surrounded by gelatinous sheath attached. Asexual morph: see Dai et al. [16].
Material examined: China, Sichuan Province, Yibin City, Changning District (28°28′8″ N, 105°0′16″ E, Alt. 890 m), on dead culm of Phyllostachys aurea, 23 July 2021, Qian Zeng, ZQ202107027 (SICAU 22-0072), living culture, SICAUCC 22-0072.
Culture characters: Ascospores germinate on PDA within 24 h and germ tubes produced from sides. Colonies grow fast on PDA, reaching 6 cm after 1 week at 25 °C, under 12 h light/12 h dark, and are cottony, circular, and white with irregular edge.
Notes: The sexual and asexual morph of Apiospora yunnana was reported by Dai et al. [16]. Morphologically, our observations were identical to the sexual descriptions provided by Daiet et al. [16]. Nucleotide comparisons of ITS and LSU (SICAUCC 22-0072) showed high homology with the sequences of Ap. yunnana (MFLUCC 15-0002), similarities are 99.85% (667/668, 0 gaps), 100% (847/847, 0 gaps), respectively. However, the latter lack tef1-α and tub2 sequences for further comparisons.
Magnaporthales Thongkantha, Vijaykrishna & K.D. Hyde. Fungal Diversity. 34: 157–173 (2009).
Magnaporthaceae P.F. Cannon, Systema Ascomycetum 13: 26 (1994).
Bifusisporella R.M.F. Silva, R.J.V. Oliveira, J.D.P. Bezerra, J.L. Bezerra, C.M. Souza-Motta & G.A. Silva, Mycological Progress 18(6): 852 (2019).
Type species: Bifusisporella sorghi R.M.F. Silva, R.J.V. Oliveira, J.D.P. Bezerra, J.L. Bezerra, C.M. Souza-Motta & G.A. Silva.
Description: Endophytic and parasitic fungi on Poaceae. Sexual morph: Ascomata separate or gregarious, subglobose, black, coriaceous, semi-immersed, unilocular or multilocular. Peridium with hyaline to brown cells of textura angularis. Hamathecium hyaline, with distinct septa, wider at the base, tapering towards the apex. Asci 8–spored, cylindrical, with a J-, apical ring, developing from the base and periphery of the ascomata, with a short pedicel. Ascospores biseriate, hyaline, fusiform, with distinct septa, with narrowly rounded ends, without appendages. Asexual morph: Found in Bifusisporella sorghi cultures by Silva et al. [42].
Notes: Bifusisporella was introduced as a new genus to accommodate B. sorghi based on morphology and phylogeny. At present, Bifusisporella comprises only the ex-type species B. sorghi, and no records on its sexual morph. The new species B. sichuanensis is well-supported within Bifusisporella, which suggests that there is a need to amend the morphological circumscriptions of the genus.
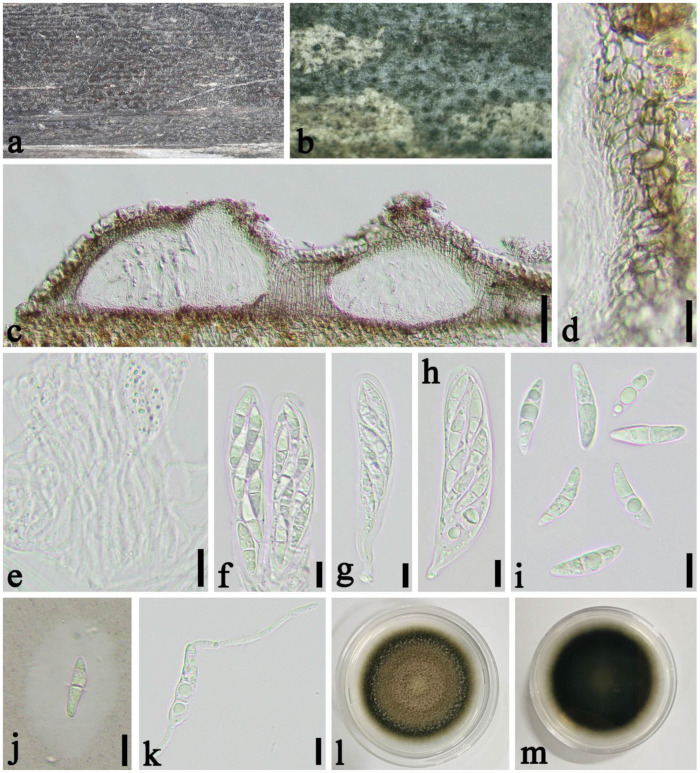
Bifusisporella sichuanensis Q. Zeng, Y.C. Lv & C.L. Yang, sp. nov. (Figure 9).
Figure 9.
Bifusisporella sichuanensis (SICAU 22-0073). (a,b) Ascostromata developing on the host. (c) Vertical sections of ascostromata. (d) Peridium. (e) Pseudoparaphyses. (f–i) Asci. (j) Ascospores. (k) Germinating ascospore. (l,m) Cultures on PDA. Scale bars: (b) = 500 µm, (c) = 100 µm, (d–k) = 10 µm.
Index Fungorum: IF559625
Etymology: Refers to the region from where the fungus was collected.
Holotype: SICAU 22-0073
Parasitic on living leaves of Phyllostachys edulis (Carriere) J. Houzeau. Sexual morph: Ascostromata 536–1672 × 332–849 × 125–245 μm ( = 1103 × 591 × 193 μm, n = 30), separate or gregarious, subglobose, black, coriaceous, semi-immersed, unilocular or multilocular, glabrous. Peridium 14–34 μm wide ( = 20 μm, n = 30), composed of 3–9 layers, with hyaline to brown cells of textura angularis. Hamathecium, hyaline, cellular, with distinct septa. Asci 79–126 × 9.5–13 μm ( = 99 × 11 μm, n = 30), 8–spored, bitunicate, cylindrical, with an apical chamber and a short pedicel. Ascospores 22–35 × 5.0–6.5 μm ( = 29 × 5.5 μm, n = 50), overlapping, biseriate, hyaline, fusiform, 3–septate, rarely constricted at septate, with narrowly rounded ends, smooth-walled, guttules, without gelatinous sheath. Asexual morph: Undetermined.
Material examined: China, Sichuan Province, Yibin City, Xingwen District (28°15′22″ N, 105°6′29″ E, Alt. 850 m), on living to nearly dead leaves of Phyllostachys edulis, 25 July 2021, Qian Zeng, ZQ202107111 (SICAU 22-0073 holotype), ex-type living culture, SICAUCC 22-0073.
Culture characters: Ascospores germinate in sterilized water within 12 h at 25 °C. Colonies grow slow on PDA, reaching approximately 2 cm in 30 days at 25 °C, under 12 h light/12 h dark, and are irregular, black, frilly with white margin, and black on the back of colonies.
Notes: Bifusisporella sichuanensis is phylogenetically close (100% ML and 1.00 BYPP) to B. sorghi (URM 7442) introduced by Silva et al. [42], which is described with asexual morph. However, striking base-pair differences are noted, viz. 11.43% (55/481, 0 gaps), 3.36% (27/803, 0 gaps), 5.11% (24/469, 0 gaps), 9.04% (64/708, 0 gaps) in the ITS, LSU, tef1-α and rpb1, respectively. Hence, our collection is proposed as a new species.
Pleosporales Luttr. ex M.E. Barr, Prodromus to class Loculoascomycetes: 67 (1987).
Phaeosphaeriaceae M.E. Barr, Mycologia 71: 948 (1979).
Paralloneottiosporina Q. Zeng, Y.C. Lv & C.L. Yang, gen. nov.
Index Fungorum: IF559626.
Type species: Paralloneottiosporina sichuanensis Q. Zeng, Y.C. Lv & C.L. Yang.
Etymology: Name reflects the morphological similarity to the genus Alloneottiosporina.
Parasitic on living to nearly dead leaves of Phyllostachys violascens ‘Prevernalis’ S.Y. Chen et C.Y. Yao. Sexual morph: Ascomata visible as raised to superficial on host, gregarious, globose to subglobose or dome shape, dark brown to black, unilocular, glabrous. Ostiole single, circular, centrally located. Peridium multi-layered, brown to dark brown cells of textura angularis. Hamathecium hyaline, numerous, septate, often constricted at septa. Asci 8-spored, bitunicate, rounded at apex, cylindrical, curved, with a short pedicel. Ascospores hyaline, fusiform, 1–2 septate, constricted at the septum, guttules, smooth-walled, with narrowly rounded ends. Asexual morph: Conidiomata brown to dark brown, globose to long ellipsoid, coriaceous, semi-immersed, unilocular, gregarious, glabrous. Conidiomatal wall comprising multi-layered, dark brown to black cells of textura angularis. Conidia ellipsoid to ovoid, 1–septate, slightly constricted at the septum, smooth-walled, hyaline, with a rounded apex and a truncated base, guttules.
Notes: Paralloneottiosporina resembles Alloneottiosporina in asexual status having semi-immersed, unilocular, gregarious, glabrous conidiomata, but Paralloneottiosporina differs in absent of microconidia, conidia without mucoid appendages, bigger conidia, fewer layers of conidiomatal wall. The macroconidia of Alloneottiosporina species are usually accompanied with mucoid appendages at both ends, and microconidia are produced near the ostiolar channel. Moreover, colonies are whitish to bright orange-pink on PDA in Paralloneottiosporina, but olivaceous-black in Alloneottiosporina [43]. Based on morphological characteristics and molecular phylogeny, the new genus is introduced in Phaeosphaeriaceae.
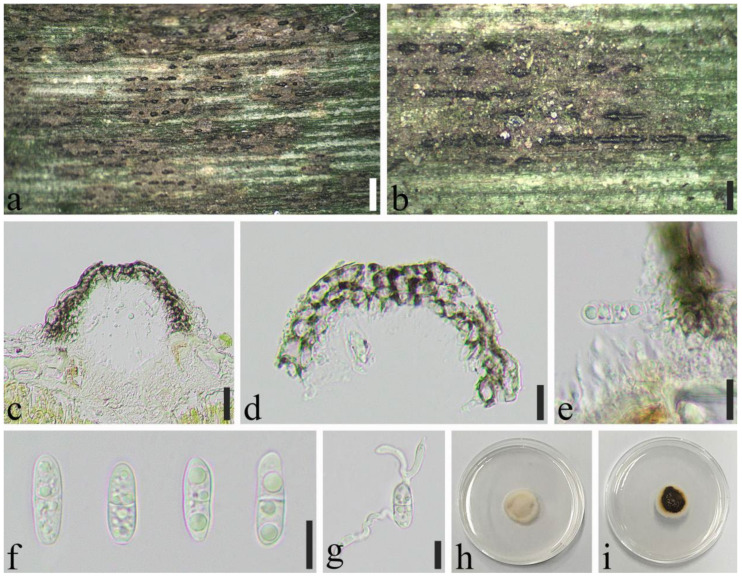
Paralloneottiosporina sichuanensis Q. Zeng, Y.C. Lv & C.L. Yang, sp. nov. (Figure 10 and Figure 11).
Figure 10.
Paralloneottiosporina sichuanensis (SICAU 22-0074, holotype). (a,b) Ascostromata developing on the host. (c) Vertical sections of ascostromata. (d) Peridium. (e–g) Asci. (h) Ascospores. (i) Germinating ascospore. (j,k) Cultures on PDA. Scale bars: (a) = 1 mm, (b) = 500 µm, (c,d) = 20 µm, (e–i) = 10 µm.
Figure 11.
Paralloneottiosporina sichuanensis (SICAU 22-0075, paratype). (a,b) Conidiomata on the host. (c) Vertical sections of conidiomata. (d) Peridium. (e) Conidiogenous cells and developing conidia. (f) Conidia. (g) Germinating conidium. (h,i) Cultures on PDA. Scale bars: (a) = 500 µm, (b) = 200 µm, (c) = 20 µm, (d–g) = 10 µm.
Index Fungorum: IF559627.
Etymology: In reference to Sichuan Province where the specimens were collected.
Holotype: SICAU 22-0074.
Associated with leaf blight on living to nearly dead leaves of Phyllostachys violascens (Poaceae). Sexual morph: Ascomata 106–343 × 39–196 × 55–112 μm ( = 168 × 111 × 89 μm, n = 30), separate, gregarious to confluent, globose to subglobose, dark brown to black, superficial, unilocular, glabrous. Ostiole single, circular, centrally located. Peridium 17–38 μm wide ( = 29 μm, n = 30), composed of 7–12 layers, with brown cells of textura angularis. Hamathecium hyaline, dense, cellular, with distinct septa. Asci 49–97 × 8.5–19 μm ( = 71 × 13 μm, n = 30), 8-spored, bitunicate, cylindrical, curved, with a short pedicel. Ascospores 15–21 × 5.0–7.5 μm ( = 18 × 6.0 μm, n = 50), overlapping biseriate, straight, hyaline, fusiform, 1–2 septate, constricted at the septum, smooth-walled, with narrowly rounded ends. Asexual morph: Conidiomata 90–191 × 61–132 × 81–123 μm ( = 132 × 102 × 105 μm, n = 30), globose to long ellipsoid, coriaceous, semi-immersed, black, unilocular, gregarious, glabrous. Conidiomatal wall 7.5–21 μm wide ( = 13 μm), comprising 3–6 layers, brown cells of textura angularis. Conidiophores reduced to conidiogenous cells. Conidiogenous cell 3.0–6.5 × 2.5–5.0 μm ( = 5.0 × 3.5 μm, n = 20), hyaline, ampulliform to subcylindrical, smooth. Conidia 11–20 × 4.0–6.5 μm ( = 17 ×5.0 μm, n = 50), ellipsoid to ovoid, 1–septate, slightly constricted at the septum, smooth-walled, hyaline, with a rounded apex and a truncated base.
Material examined: China, Sichuan Province, Ya’an City, Yucheng District (29°56′49.54″ N, 102°56′46.03″ E, Alt. 807 m), on living to nearly dead leaves of Phyllostachys violascens, 13 May 2020, Qian Zeng, ZQ202005002 (SICAU 22-0074, holotype), ex-type living culture, SICAUCC 22-0074; CHINA, Sichuan Province, Qionglai City, Linjiang Town (30°19′4.42″ N, 103°17′23.06″ E, Alt. 518 m), on living leaves of Ph. violascens, 8 November 2020, Qian Zeng, ZQ202011012 (SICAU 22-0075, paratype), living culture, SICAUCC 22-0075.
Culture characteristics: Ascospores germinate in sterilized water within 24 h at 25 °C. Colonies grow slow on PDA, reaching approximately 2.5 cm in 30 days at 25 °C, circular, white aerial mycelium, whitish to bright orange-pink on the surface, and brown on the back.
Pleosporales Luttr. ex M.E. Barr, Prodromus to class Loculoascomycetes: 67 (1987).
Bambusicolaceae D.Q. Dai & K.D. Hyde, Fungal Diversity. 63 (1): 49 (2013).
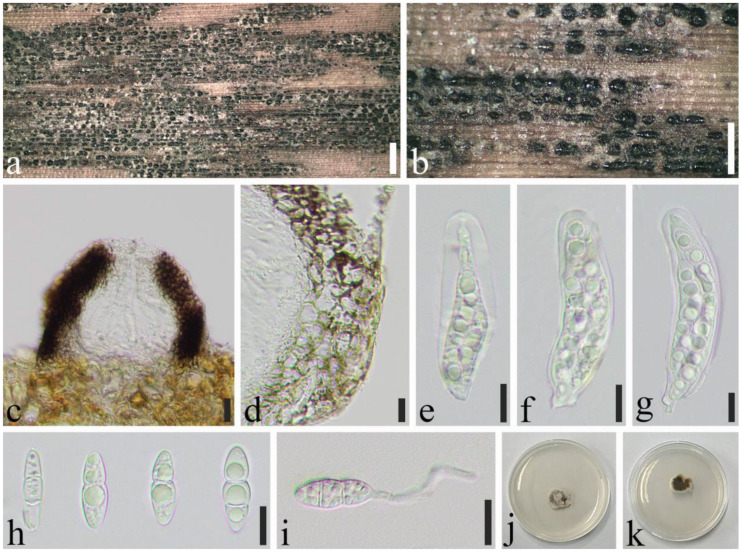
Seriascoma yunnanense Rathnayaka & K.D. Hyde, Asian Journal of Mycology 2(1): 250 (2019) (Figure 12).
Figure 12.
Seriascoma yunnanense (SICAU 22-0059). (a,b) Ascostromata developing on the host. (c) Vertical sections of ascostromata. (d) Peridium. (e) Pseudoparaphyses. (f–h) Asci. (i,j) Ascospores. (k) Germinating ascospore. (l,m) Cultures on PDA. Scale bars: (c) = 50 µm, (d–k) = 10 µm.
Saprobic on dead culm of Phyllostachys edulis (Carriere) J. Houzeau. Sexual morph: Ascostromata 110–200 × 120–150 × 120–140 μm ( = 160 × 140 × 130 μm, n = 20), solitary to gregarious, immersed, globose to subglobose, coriaceous, dark brown to black. Peridium 12–26 μm wide ( = 4.0 μm, n = 20), composed of 4–9 layers of brown to hyaline cells of textura angularis. Hamathecium 1.5–2.0 μm wide, composed of dense, branched, long, septate. Asci 52–80 × 12–16 μm, ( = 60 × 14 μm, n = 50), 8-spored, bitunicate, broadly cylindrical, with a short pedicel, straight or slightly curved, with an apical chamber. Ascospores 20–30 × 6.0–7.5μm ( = 23 × 7.0 μm, n = 50), 2–seriate, 1–septate, slightly constricted at the septum, fusiform, narrowly acute at both ends, straight to curved, hyaline, smooth-walled, surrounded by a gelatinous sheath. Asexual morph: Undetermined.
Material examined: China, Sichuan Province, Chengdu City, Jin’niu District (30°45′57″ N, 104°7′34″ E, Alt. 539 m), on dead culm of Phyllostachys edulis, 8 April 2021, Yicong Lv, LYC202104043 (SICAU 22-0059), living culture SICAUCC 22-0059.
Culture characteristics: Ascospores germinate in sterile water within 12 h at 25 °C. Colonies grow slowly on PDA, and reach 6 cm after 30 days at 25 °C, circular, brown to dark brown.
Notes: On the morphology, our observations were identical to the descriptions of Seriascoma yunnanense provided by Rathnayaka et al. [44]. Nucleotide comparisons of SSU, LSU, tef1-α and rpb2 (SICAUCC 22-0059) showed high homology with the sequences of S. yunnanense (MFLU 19-0690), similarities are 98.37% (847/861, 0 gaps), 100% (841/841, 0 gaps), 96.59% (396/410, 0 gaps), 99.65% (855/858, 0 gaps), respectively. We report our collection as S. yunnanense.
4. Discussion
In this study, we confirmed seven species of saprophyte or parasitism from leaves and culms of Phyllostachys, corresponding to four genera. Microfungi are abundant on culms and leaves of bamboo as pointed out by Dai et al. [45]. Ascomycetes are the most abundant species on bamboo, with about 1150 taxa having been recorded [45]. Furthermore, the number of saprophytic fungi is more than that of pathogenic fungi [16,36].
The genus Apiospora Sacc. was recognized and described by Saccardo considering Ap. montagnei designated as the type species [46]. Apiospora has been widely accepted as a synonym for Arthrinium after Ellis [47]. Crous and Groenewald combined Apiospora species to be sexual morphs of Arthrinium species and synonymized under Arthrinium [40]. However, Pintos and Alvarado found that the morphological and ecological differences between Apiospora and Arthrinium are sufficient to support the taxonomic separation of the two genera. As a result, fifty-five species of Arthrinium were combined to Apiospora [48]. In this study, given the phylogenetic analysis with species of Apiospora and Arthrinium, in which 10 species of Arthrinium (Ar. agari, Ar. arctoscopi, Ar. fermenti, Ar. koreanum, Ar. mori, Ar. phaeospermum, Ar. pusillispermum, Ar. sargassi, Ar. taeanense, Ar. marinum) are clustered in a well-supported clade within Apiospora, future studies are needed to better understand the combination of previous Arthrinium species with Apiospora. Apiospora species have a worldwide distribution and can be found on various hosts. Most species occurred on the plants in Poaceae, although some were known from Amaranthaceae, Juncaceae, Euphorbiaceae, Cyperaceae, Restionaceae, Fagaeaeand, even seaweeds [48,49]. To date, more than 25 species have been found on bamboo, most species were saprobic on dead bamboo culms, and a few species have been reported as pathogens. For example, Ap. arundinis causes brown culm streak of Phyllostachys praecox, and Ap. kogelbergensis causes blight disease of Bambusa intermedia [16,41,50,51]. Apiospora. hydei, Ap. neosubglobosa, and Ap. jiangxiensis were saprophytic on unidentified bamboo culms and leaves [41,52]. Apiospora yunnansis has been reported on bamboo culms of Phyllostachys nigra and P. heteroclada, which can cause bamboo blight disease of P. heteroclada [53,54]. In this study, four known species, Apiospora hydei, Ap. neosubglobosa, Ap. jiangxiensis, and Ap. yunnansis, were newly recorded on Phyllostachys nigra, P. heteroclada, P. bissetii, and P. aurea respectively.
At present, Bifusisporella only comprises the ex-type species B. sorghi. In this study, we provide taxonomic details for another new species, B. sichuanensis, that was collected from living leaves of Phyllostachys edulis. B. sorghi was isolated as an endophyte from healthy sorghum leaves in Brazil by Silva et al. [42]. However, B. sichuanensis is pathogenic, causing tar spot on bamboo leaves. In addition, the sexual stage in this genus is supplemented.
Phaeosphaeriaceae is one of the most important and species-rich families in Pleosporales with diverse lifestyles [55,56], and may be found on herbaceous stems or monocotyledonous culms, branches, leaves, flowers, and woody substrates [57,58]. Currently, more than 70 genera are accommodated in Phaeosphaeriaceae [59]. Most genera in this family were introduced as monotypic genera, such as Acericola, Banksiophoma, Bhagirathimyces, Bhatiellae, Brunneomurispora, Camarosporioides, Elongaticollum, Equiseticola, Hydeopsis, Jeremyomyces, Mauginiella, Melnikia, Neoophiobolus, Neosphaerellopsis, Neostagonosporella, Ophiobolopsis, and Parastagonosporella, among others. Due to these genera being represented by a single species, resulting in few samples that could be used for taxon, the phylogenetic relationships with the related genera are sometimes not well-resolved. Based on morphological characteristics and multigene phylogeny, a novel genus, Paralloneottiosporina, is introduced to accommodate Pa. sichuanensis sp. nov. According to the field investigation, Pa. sichuanensis can cause leaf blight that eventually leads to leaf necrosis and plant decline in severe cases. Besides Ph. violascens, leaf blight caused by Pa. sichuanensis has also been observed on P. heterocycla and P. tianmuensis. This indicates that Pa. sichuanensis may be a common parasitic fungus on bamboos.
As only three species are accommodated within Seriascoma, more research is also needed for better understanding this genus [60]. Seriascoma is presently known as saprobic on decaying wood and dead bamboo in the terrestrial or freshwater habitats distributed in China and Thailand [16,44,61,62]. Seriascoma. yunnanense is found on dead branches of bamboo in Yunnan. In this study, S. yunnanense was saprophytic on Phyllostachys edulis.
The previous studies have revealed a high fungal diversity associated with bamboo Phyllostachys. In recent years, 10 species belonging to seven genera have been described from bamboo of Phyllostachys, including two new genera, Neostagonosporella and Parakarstenia, established by Yang et al. on P. heteroclada in Sichuan Province [54,58,63,64,65,66,67,68,69]. However, the knowledge about bambusicolous fungi is incomplete and mainly remains at cataloguing stage [14]. The previous studies of identification were mostly based on morphological characteristics, and lacked molecular data. Moreover, their hosts were poorly documented or unknown [70], and specimens were absent for further re-examination. Therefore, these species need to be recollected, epitypified, and sequenced [10], and new species need to be discovered and described.
Appendix A. Molecular Data Used in This Study and GenBank Accession Numbers
Table A1.
Isolates and GenBank accession numbers of sequences used in this study.
| Species | Strains | GenBank Accession Numbers | |||||||
|---|---|---|---|---|---|---|---|---|---|
| ITS | LSU | tub2 | tef1-a | rpb1 | SSU | rpb2 | References | ||
| Apiospora acutiapica | KUMCC 20-0209 | MT946342 | MT946338 | – | – | – | – | – | [71] |
| Apiospora acutiapica | KUMCC 20-0210 T | MT946343 | MT946339 | – | – | – | – | [71] | |
| Apiospora aquaticum | MFLU 18-1628 T | MK828608 | MK835806 | – | – | – | – | – | [55] |
| Apiospora arundinis | CBS 114316 | KF144884 | KF144928 | KF144974 | KF145016 | – | – | – | [40] |
| Apiospora arundinis | CBS 450.92 | AB220259 | – | AB220306 | – | – | – | – | [71] |
| Apiospora arundinis | AP11118A | MK014868 | MK014835 | MK017974 | MK017945 | – | – | – | [72] |
| Apiospora aureum | CBS 244.83 T | AB220251 | KF144935 | KF144981 | KF145023 | – | – | – | NCBI |
| Apiospora balearica | CBS 145129 T | MK014869 | MK014836 | MK017975 | MK017946 | – | – | – | [72] |
| Apiospora bambusicola | MFLUCC 20-0144 T | MW173030 | MW173087 | – | MW183262 | – | – | – | [73] |
| Apiospora biserialis | CGMCC 3.20135 T | MW481708 | – | MW522955 | MW522938 | – | – | – | [52] |
| Apiospora camelliae-sinensis | LC5007 T | KY494704 | KY494780 | KY705173 | KY705103 | – | – | – | [36] |
| Apiospora camelliae-sinensis | LC8181 | KY494761 | KY494837 | KY705229 | KY705157 | – | – | – | [36] |
| Apiospora chiangraiense | MFLU:21-0046 | MZ542520 | MZ542524 | MZ546409 | – | – | – | – | [49] |
| Apiospora chromolaenae | MFLUCC 17-1505 T | MT214342 | MT214436 | – | – | – | – | – | [74] |
| Apiospora cyclobalanopsidis | CGMCC 3.20136 T | MW481713 | – | MW522962 | MW522945 | – | – | – | [52] |
| Apiospora descalsii | CBS 145130 T | MK014870 | MK014837 | MK017976 | MK017947 | – | – | – | [72] |
| Apiospora dichotomanthi | CGMCC 3.18332 T | KY494697 | KY494832 | KY705167 | KY705096 | – | – | – | [36] |
| Apiospora dichotomanthi | LC8175 | KY494755 | KY494831 | KY705223 | KY705151 | – | – | – | [36] |
| Apiospora esporlensis | CBS 145136 T | MK014878 | MK014845 | MK017983 | MK017954 | – | – | – | [72] |
| Apiospora euphorbiae | IMI 285638b | AB220241 | – | AB220288 | – | – | – | – | [71] |
| Apiospora gaoyouensis | CFCC 52301 | MH197124 | – | MH236789 | MH236793 | – | – | – | [53] |
| Apiospora gaoyouensis | CFCC 52302 | MH197125 | – | MH236790 | MH236794 | – | – | – | [53] |
| Apiospora garethjonesii | JHB004 | KY356086 | KY356091 | – | – | – | – | – | [41] |
| Apiospora garethjonesii | HKAS 96289 T | NR_154736 | NG_057131 | – | – | – | – | – | [41] |
| Apiospora gelatinosa | HKAS:111962 | – | – | MW5229 | MW522941 | – | – | – | [52] |
| Apiospora guizhouensis | LC5318 | KY494708 | KY494784 | KY705177 | KY705107 | – | – | – | [36] |
| Apiospora guizhouensis | CGMCC 3.18334 T = LC5322 | KY494709 | KY494785 | KY705178 | KY705108 | – | – | – | [36] |
| Apiospora hispanica | IMI 326877 | AB220242 | AB220336 | AB220289 | – | – | – | – | [71] |
| Apiospora hydei | CBS 114990 T | KF144890 | KF144936 | KF144982 | KF145024 | – | – | – | [40] |
| Apiospora hydei | KUMCC 16-0204 | KY356087 | KY356092 | – | – | – | – | – | [41] |
| Apiospora hydei | SICAUCC 22-0032 | ON183998 | ON185553 | ON221313 | ON221312 | – | – | – | This study |
| Apiospora hyphopodii | MFLUCC 15-003 T | KR069110 | KR069111 | – | – | – | – | – | [75] |
| Apiospora hyphopodii | KUMCC 16-0201 | KY356088 | KY356093 | – | – | – | – | – | [41] |
| Apiospora hysterina | AP15318 | MK014873 | MK014840 | MK017979 | MK017950 | – | – | – | [72] |
| Apiospora hysterina | ICPM6889 | MK014874 | MK014841 | MK017980 | MK017951 | – | – | – | [72] |
| Apiospora hysterina | AP29717 | MK014875 | MK014842 | MK017981 | MK017952 | – | – | – | [72] |
| Apiospora hysterina | AP2410173 | MK014876 | MK014843 | – | – | – | – | – | [72] |
| Apiospora hysterina | AP12118 | MK014877 | MK014844 | MK017982 | MK017953 | – | – | – | [72] |
| Apiospora iberica | CBS 145137 T | MK014879 | MK014846 | MK017984 | MK017955 | – | – | – | [72] |
| Apiospora intestini | CBS 135835 T | KR011352 | MH877577 | KR011350 | KR011351 | – | – | – | [76] |
| Apiospora italica | CBS 145138 T | MK014880 | MK014847 | MK017985 | MK017956 | – | – | – | [72] |
| Apiospora italica | AP221017 T | MK014881 | MK014848 | MK017986 | MK017957 | – | – | – | [72] |
| Apiospora jatrophae | CBS 134262 T | NR_154675 | – | – | – | – | – | – | [77] |
| Apiospora jatrophae | MMI 00051 = CBS:134262 | JQ246355 | – | – | – | – | – | – | [77] |
| Apiospora jiangxiensis | CGMCC 3.18381 T | KY494693 | – | KY705163 | KY705092 | – | – | – | [36] |
| Apiospora jiangxiensis | LC4578 | KY494694 | KY494770 | KY705164 | KY705093 | – | – | – | [36] |
| Apiospora jiangxiensis | SICAUCC 22-0070 | ON227094 | ON227098 | ON244432 | ON244431 | – | – | – | This study |
| Apiospora kogelbergensis | CBS 113332 | KF144891 | KF144937 | KF144983 | KF145025 | – | – | – | [40] |
| Apiospora kogelbergensis | CBS 113333 T | KF144892 | KF144938 | KF144984 | KF145026 | – | – | – | [40] |
| Apiospora kogelbergensis | CBS 113335 | KF144893 | KF144939 | KF144985 | KF145027 | – | – | – | [40] |
| Apiospora kogelbergensis | CBS 117206 | KF144895 | KF144941 | KF144987 | KF145029 | – | – | – | [40] |
| Apiospora locuta-pollinis | LC11683 | MF939595 | – | MF939622 | MF939616 | – | – | – | [78] |
| Apiospora longistroma | MFLUCC 11-0479 | KU940142 | KU863130 | – | – | – | – | – | [16] |
| Apiospora longistroma | MFLUCC 11-0481 | KU940141 | KU863129 | – | – | – | – | – | [16] |
| Apiospora longistroma | MFLU 15-1184 T | NR_154716 | – | – | – | – | – | – | [16] |
| Apiospora malaysiana | CBS 102053 T | KF144896 | KF144942 | KF144988 | KF145030 | – | – | – | [40] |
| Apiospora marii | CBS 497.90 T | AB220252 | KF144947 | KF144993 | KF145035 | – | – | – | [40] |
| Apiospora mediterranea | IMI 326875 | AB220243 | – | AB220290 | – | – | – | – | [71] |
| Apiospora minutispora | 17E-042 | LC517882 | – | LC518888 | LC518889 | – | – | – | [79] |
| Apiospora montagnei | LSU0093 | MT000394 | MT000490 | – | – | – | – | – | [80] |
| Apiospora mytilomorpha | DAOM 214595 | KY494685 | – | – | – | – | – | – | [36] |
| Apiospora neobambusae | CGMCC 3.18335 T | KY494718 | KY494794 | KY705186 | KY806204 | – | – | – | [36] |
| Apiospora neobambusae | KUMCC 20-0207 | MT946346 | MT946340 | – | – | – | – | – | [71] |
| Apiospora neobambusae | LC7107 | KY494719 | KY494795 | KY705187 | KY705117 | – | – | – | [36] |
| Apiospora neochinensis | CFCC 53037 | MK819292 | – | MK818548 | MK818546 | – | – | – | [81] |
| Apiospora neochinensis | CFCC 53036 T | MK819291 | – | MK818547 | MK818545 | – | – | – | [81] |
| Apiospora neogarethjonesii | HKAS 96354 T | MK070897 | MK070898 | – | – | – | – | – | [82] |
| Apiospora neosubglobosa | JHB006 | KY356089 | KY356094 | – | – | – | – | – | [41] |
| Apiospora neosubglobosa | JHB007 T | KY356090 | KY356095 | – | – | – | – | – | [41] |
| Apiospora neosubglobosa | SICAUCC 22-0071 | ON227095 | ON227099 | ON244430 | ON244429 | – | – | – | This study |
| Apiospora obovata | CGMCC 3.18331 T | KY494696 | KY494834 | KY705166 | KY705095 | – | – | – | [41] |
| Apiospora obovata | LC8177 | KY494757 | KY494833 | KY705225 | KY705153 | – | – | – | [41] |
| Apiospora ovata | CBS 115042 T | KF144903 | KF144950 | KF144995 | KF145037 | – | – | – | [40] |
| Arthrinium paraphaeospermum | NCYU 19-0341 | MW114315 | MW293936 | – | MW288020 | – | – | – | NCBI |
| Apiospora paraphaeosperma | MFLUCC 13-0644 T | KX822128 | KX822124 | – | – | – | – | – | [71] |
| Apiospora phragmitis | CPC 18900 T | KF144909 | – | KF145001 | KF145043 | – | – | – | [40] |
| Apiospora phragmitis | AP3218 | MK014891 | MK014858 | MK017996 | MK017967 | – | – | – | [72] |
| Apiospora phragmitis | AP2410172A | MK014890 | MK014857 | MK017995 | MK017966 | – | – | – | [72] |
| Apiospora phyllostachydis | MFLUCC 18-1101 | – | – | MK291949 | – | – | – | – | [65] |
| Apiospora piptatheri | CBS 145149 T | MK014893 | MK014860 | – | MK017969 | – | – | – | [72] |
| Apiospora pseudoparenchymatica | CGMCC 3.18336 T | KY494743 | KY494819 | KY705211 | KY705139 | – | – | – | [36] |
| Apiospora pseudoparenchymatica | LC8173 | KY494753 | KY494829 | KY705221 | KY705149 | – | – | – | [36] |
| Apiospora pseudorasikravindrae | KUMCC 20-0208 T | MT946344 | – | – | – | – | – | – | [71] |
| Apiospora pseudorasikravindrae | KUMCC 20-0211 | MT946345 | – | – | – | – | – | – | [71] |
| Apiospora pseudosinensis | CBS 135459 T | KF144910 | KF144957 | – | KF145044 | – | – | – | [40] |
| Apiospora pseudospegazzinii | CBS 102052 T | KF144911 | KF144958 | KF145002 | KF145045 | – | – | – | [40] |
| Apiospora pterosperma | CBS 123185 | KF144912 | KF144959 | KF145003 | – | – | – | [40] | |
| Apiospora pterosperma | CBS 134000 T | KF144913 | KF144960 | KF145004 | KF145046 | – | – | – | [40] |
| Apiospora qinlingensis | CFCC 52303 T | MH197120 | – | MH236791 | MH236795 | – | – | – | [53] |
| Apiospora qinlingensis | CFCC 52304 | MH197121 | – | MH236792 | MH236796 | – | – | – | [53] |
| Apiospora rasikravindrae | NFCCI 2144 T | KF144914 | – | – | – | – | – | – | [83] |
| Apiospora rasikravindrae | MFLUCC 11-0616 | KU940144 | KU863132 | – | – | – | – | – | [16] |
| Apiospora rasikravindrae | LC5449 | KY494713 | KY494789 | KY705182 | KY705112 | – | – | – | [36] |
| Apiospora rasikravindrae | LC7115 | KY494721 | KY494797 | KY705189 | KY705118 | – | – | – | [36] |
| Apiospora rasikravindrae | KUC21351 | MH498540 | MH498498 | MN868932 | – | – | – | [84] | |
| Apiospora rasikravindrae | KUC21327 | MH498541 | MH498499 | MH544670 | – | – | – | [84] | |
| Apiospora sacchari | CBS 212.30 | KF144916 | KF144962 | KF145005 | KF145047 | – | – | – | [40] |
| Apiospora sacchari | CBS 301.49 | KF144917 | KF144963 | KF145006 | KF145048 | – | – | – | [40] |
| Apiospora saccharicola | CBS 191.73 | KF144920 | KF144966 | KF145009 | KF145051 | – | – | – | [40] |
| Apiospora saccharicola | CBS 463.83 | KF144921 | KF144968 | KF145010 | KF145052 | – | – | – | [40] |
| Apiospora sasae | CBS 146808 T | MW883402 | MW883797 | MW890120 | MW890104 | – | – | – | [85] |
| Apiospora septatum | CGMCC 3.20134 T | MW481711 | – | MW522960 | MW522943 | – | – | – | [52] |
| Apiospora serenensis | IMI 326869 T | AB220250 | – | AB220297 | – | – | – | – | [71] |
| Apiospora serenensis | ATCC 76309 | AB220240 | – | AB220287 | – | – | – | – | [71] |
| Apiospora setariae | CFCC 54041 | MT492004 | – | MT497466 | MW118456 | – | – | – | [86] |
| Apiospora setostroma | KUMCC 19-0217 | MN528012 | MN528011 | – | MN527357 | – | – | – | [87] |
| Apiospora sinensis | UNKNOW-1 = HKUCC 3143 | – | AY083831 | – | – | – | – | – | NCBI |
| Apiospora sinensis | UNKNOW-2 | – | DQ810215 | – | – | – | – | – | NCBI |
| Apiospora sorghi | URM<BRA>:9300 | MK371706 | – | – | – | – | – | – | NCBI |
| Apiospora stipae | CBS 146804 | MW883403 | MW883798 | MW890121 | MW890105 | – | – | – | [85] |
| Apiospora subglobosa | MFLUCC 11-0397 T | KR069112 | KR069113 | – | – | – | – | – | [75] |
| Apiospora subrosea | LC7291 | KY494751 | KY494827 | KY705219 | KY705147 | – | – | – | [36] |
| Apiospora subrosea | CGMCC3.18337 T | KY494752 | KY494828 | KY705220 | KY705148 | – | – | – | [36] |
| Apiospora thailandica | MFLUCC 15-0199 | KU940146 | KU863134 | – | – | – | – | – | [16] |
| Apiospora thailandica | MFLUCC 15-0202 T | KU940145 | KU863133 | – | – | – | – | – | [16] |
| Apiospora thailandica | LC5630 | KY494714 | KY494790 | KY806200 | KY705113 | – | – | – | [36] |
| Apiospora tintinnabula | 7019-96 (ICMP) | – | DQ810216 | – | – | – | – | – | [71] |
| Apiospora vietnamensis | IMI 99670 | KX986096 | KX986111 | KY019466 | – | – | – | – | [88] |
| Apiospora xenocordella | CBS 478.86 T | KF144925 | KY494763 | – | – | – | – | – | [40] |
| Apiospora xenocordella | CBS 595.66 | KF144926 | KF144971 | KF145013 | KF145055 | – | – | – | [40] |
| Apiospora yunnana | MFLUCC 15-0002 T | KU940147 | KU863135 | – | – | – | – | – | [16] |
| Apiospora yunnana | SICAUCC 22-0072 | ON227096 | ON227100 | ON244426 | ON244425 | – | – | – | This study |
| Arthrinium agari | KUC21364 | MH498516 | – | MH498474 | MN868917 | – | – | – | [84] |
| Arthrinium arctoscopi | KUC21347 | MH498525 | – | MH498483 | MN868922 | – | – | – | [84] |
| Arthrinium fermenti | KUC21289 | MF615226 | – | MF615231 | MH544667 | – | – | – | [84] |
| Arthrinium koreanum | KUC21350 | MH498521 | – | MH498479 | MN868929 | – | – | – | [84] |
| Arthrinium marinum | KUC21328 | MH498538 | MH498496 | MH544669 | [84] | ||||
| Arthrinium marinum | KUC21356 | MH498534 | – | MH498492 | MN868926 | – | – | – | [84] |
| Arthrinium marinum | KUC21355 | MH498535 | MH498493 | MN868925 | [84] | ||||
| Arthrinium marinum | KUC21354 | MH498536 | MH498494 | MN868924 | [84] | ||||
| Arthrinium mori | MFLU 18-2514 | MW114313 | MW114393 | – | – | – | – | – | [89] |
| Arthrinium mori | NCYU 19-0364 | MW114314 | MW114394 | – | – | – | – | – | [89] |
| Arthrinium phaeospermum | CBS 114315 | KF144905 | KF144952 | KF144997 | KF145039 | – | – | – | [40] |
| Arthrinium phaeospermum | CBS 114317 | KF144906 | KF144953 | KF144998 | KF145040 | – | – | – | [40] |
| Arthrinium phaeospermum | CBS 114318 | KF144907 | KF144954 | KF144999 | KF145041 | – | – | – | [40] |
| Arthrinium pusillispermum | KUC21357 | MH498532 | – | MH498490 | MN868931 | – | – | – | [84] |
| Arthrinium sargassi | KUC21232 | KT207750 | – | KT207648 | MH544676 | – | – | – | [84] |
| Arthrinium taeanense | KUC21322 | MH498515 | – | MH498473 | MH544662 | – | – | – | [84] |
| Pestalotiopsis chamaeropis | CBS 237.38 | MH855954 | MH867450 | KM199392 | KM199474 | – | – | – | [76] |
| Pestalotiopsis colombiensis | CBS 118553 T | KM199307 | KM116222 | KM199421 | KM199488 | – | – | – | [90] |
| Bambusicularia brunnea | CBS 133599 T | KM484830 | KM484948 | – | – | KM485043 | – | – | [91] |
| Bambusicularia brunnea | CBS 133600 | AB274436 | KM484949 | – | – | KM485044 | – | – | [91,92] |
| Barretomyces calatheae | CBS 129274 = CPC 18464 | KM484831 | KM484950 | – | – | KM485045 | – | – | [76] |
| Bifusisporella sichuanensis | SICAUCC 22-0073 T | ON227097 | ON227101 | – | ON244427 | ON244428 | – | – | This study |
| Bifusisporella sorghi | URM 7442 T | MK060155 | MK060153 | – | MK060157 | MK060159 | – | – | [42] |
| Bifusisporella sorghi | URM 7864 | MK060156 | MK060154 | – | MK060158 | MK060160 | – | – | [42] |
| Buergenerula spartinae | ATCC 22848 | JX134666 | DQ341492 | – | JX134692 | JX134720 | – | – | [93] |
| Bussabanomyces longisporus | CBS 125232 T | KM484832 | KM484951 | – | KM009202 | KM485046 | – | – | [94] |
| Falciphora oryzae | CBS 125863 T | EU636699 | KJ026705 | – | JN857963 | KJ026706 | – | – | [95] |
| Falciphoriella solaniterrestris | CBS 117.83 T | KM484842 | KM484959 | – | – | KM485058 | – | – | [91] |
| Gaeumannomycella caricicola | CBS:145041 | MK442584 | MK442526 | – | – | – | – | – | [96] |
| Gaeumannomycella caricis | CBS 388.81 T | KM484843 | KM484960 | – | KX306674 | – | – | – | [91] |
| Gaeumannomyces australiensis | CPC 26058 T | KX306480 | KX306550 | – | KX306683 | KX306619 | – | – | [97] |
| Gaeumannomyces avenae | CBS 187.65 | JX134668 | JX134680 | – | – | JX134722 | – | – | [93] |
| Gaeumannomyces avenae | CBS 870.73 = DAR 20999 | KM484833 | DQ341495 | – | – | KM485048 | – | – | [91] |
| Gaeumannomyces californicus | CPC 26044 T | KX306490 | KX306560 | – | KX306691 | KX306625 | – | – | [97] |
| Gaeumannomyces ellisiorum | CBS 387.81 T | KM484835 | KM484952 | – | KX306692 | KM485051 | – | – | [91] |
| Gaeumannomyces floridanus | CPC 26037 T | KX306491 | KX306561 | – | KX306693 | KX306626 | – | – | [97] |
| Gaeumannomyces fusiformis | CPC 26068 T | KX306492 | KX306562 | – | KX306694 | KX306627 | – | – | [97] |
| Gaeumannomyces glycinicola | CPC 26266 | KX306494 | KX306564 | – | KX306696 | KX306629 | – | – | [97] |
| Gaeumannomyces glycinicola | CPC 26057 | KX306493 | KX306563 | – | KX306695 | KX306628 | – | – | [97] |
| Gaeumannomyces graminicola | CBS 352.93 T | KM484834 | DQ341496 | – | KX306697 | KM485050 | – | – | [91] |
| Gaeumannomyces graminis | CPC 26045 | KX306505 | KX306575 | – | KX306708 | KX306640 | – | – | [97] |
| Gaeumannomyces graminis var. graminis | M33 | JF710374 | JF414896 | – | JF710411 | JF710442 | – | – | [98] |
| Gaeumannomyces graminis var. graminis | M54 | JF414848 | JF414898 | – | JF710419 | JF710444 | – | – | [98] |
| Gaeumannomyces hyphopodioides | CBS 350.77 T | KX306506 | KX306576 | – | – | – | – | – | [97] |
| Gaeumannomyces hyphopodioides | CBS 541.86 | KX306507 | KX306577 | – | KX306709 | – | – | – | [97] |
| Gaeumannomyces oryzicola | CPC 26063 T | KX306516 | KX306586 | – | KX306717 | KX306646 | – | – | [97] |
| Gaeumannomyces oryzinus | CPC 26030 T | KX306517 | KX306587 | – | KX306718 | KX306647 | – | – | [97] |
| Gaeumannomyces radicicola | CBS 296.53 T | KM009170 | KM009158 | – | KM009206 | KM009194 | – | – | [94] |
| Gaeumannomyces setariicola | CPC 26059 | KX306524 | KX306594 | – | KX306725 | KX306654 | – | – | [97] |
| Gaeumannomyces tritici | CBS 273.36 | KX306525 | KX306595 | – | KX306729 | KX306655 | – | – | [97] |
| Gaeumannomyces walkeri | CPC 26028 T | KX306543 | KX306613 | – | KX306746 | KX306670 | – | – | [97] |
| Gaeumannomyces wongoonoo | BRIP:60376 | KP162137 | KP162146 | – | – | – | – | – | [99] |
| Kohlmeyeriopsis medullaris | CBS 117849 T = JK5528S | KM484852 | KM484968 | – | – | KM485068 | – | – | [91] |
| Macgarvieomyces borealis | CBS 461.65 T | MH858669 | DQ341511 | – | KM009198 | KM485070 | – | – | [94] |
| Macgarvieomyces juncicola | CBS 610.82 | KM484855 | KM484970 | – | KM009201 | KM485071 | – | – | [91] |
| Magnaporthiopsis agrostidis | BRIP 59300 T | KT364753 | KT364754 | – | KT364756 | KT364755 | – | – | [100] |
| Magnaporthiopsis cynodontis | RS7-2 = CBS 141700 T | KJ855508 | KM401648 | – | KP282714 | KP268930 | – | – | [101] |
| Magnaporthiopsis cynodontis | RS5-5 | KJ855506 | KM401646 | – | KP282712 | KP268928 | – | – | [101] |
| Magnaporthiopsis cynodontis | RS3-1 | KJ855505 | KM401645 | – | KP282711 | KP268927 | – | – | [101] |
| Magnaporthiopsis incrustans | M35 | JF414843 | JF414892 | – | JF710412 | JF710437 | – | – | [98] |
| Magnaporthiopsis maydis | M84 | KM009160 | KM009148 | – | KM009196 | KM009184 | – | – | [94] |
| Magnaporthiopsis maydis | M85 | KM009161 | KM009149 | – | KM009197 | KM009185 | – | – | [94] |
| Magnaporthiopsis meyeri-festucae | FF2 | MF178146 | MF178151 | – | MF178167 | MF178162 | – | – | [102] |
| Magnaporthiopsis meyeri-festucae | SCR11 | MF178150 | MF178155 | – | MF178171 | MF178166 | – | – | [102] |
| Magnaporthiopsis panicorum | CM2S8 T | KF689643 | KF689633 | – | KF689623 | KF689613 | – | – | [103] |
| Magnaporthiopsis panicorum | CM10s2 | KF689644 | KF689634 | – | KF689624 | KF689614 | – | – | [103] |
| Magnaporthiopsis poae | TAP35 | KJ855511 | KM401651 | – | KP282717 | KP268933 | – | – | [104] |
| Magnaporthiopsis poae | M1 | JF414827 | JF414876 | – | JF710400 | JF710425 | – | – | [98] |
| Magnaporthiopsis poae | M12 | JF414828 | JF414877 | – | JF710401 | JF710426 | – | – | [98] |
| Magnaporthiopsis rhizophila | M22 | JF414833 | JF414882 | – | JF710407 | JF710431 | – | – | [98] |
| Nakataea oryzae | M21 | JF414838 | JF414887 | – | JF710406 | JF710441 | – | – | [98] |
| Nakataea oryzae | M69 | JX134672 | JX134685 | – | JX134698 | JX134726 | – | – | [93] |
| Nakataea oryzae | M71 | JX134673 | JX134686 | – | JX134699 | JX134727 | – | – | [93] |
| Neogaeumannomyces bambusicola | MFLUCC11-0390 T | KP744449 | KP744492 | – | – | – | – | – | [105] |
| Neopyricularia commelinicola | CBS 128307 = KACC 44083 | FJ850125 | KM484984 | – | KM009199 | KM485086 | – | – | [91,106] |
| Neopyricularia commelinicola | CBS 128308 T | FJ850122 | KM484985 | – | – | KM485087 | – | – | [91,106] |
| Ophioceras dolichostomum | CBS 114926 = HKUCC 3936 = KM 8 | JX134677 | JX134689 | – | JX134703 | JX134731 | – | – | [93] |
| Ophioceras leptosporum | CBS 894.70 T = ATCC 24161 = HME 2955 | JX134678 | JX134690 | – | JX134704 | JX134732 | – | – | [83] |
| Proxipyricularia zingiberis | CBS 132355 = MAFF 240221 | AB274433 | KM484987 | – | – | KM485090 | – | – | [91] |
| Pseudophialophora eragrostis | CM12m9 | KF689648 | KF689638 | – | KF689628 | KF689618 | – | – | [103] |
| Pseudopyricularia cyperi | CBS 133595 T = MAFF 240229 | KM484872 | KM484990 | – | – | AB818013 | – | – | [91] |
| Pseudopyricularia kyllingae | CBS 133597 T = MAFF 240227 | KM484876 | KM484992 | – | KT950880 | KM485096 | – | – | [91] |
| Pyricularia ctenantheicola | GR0001 = Ct-4 = ATCC 200218 | KM484878 | KM484994 | – | – | KM485098 | – | – | [91] |
| Pyricularia grisea | BR0029 | KM484880 | KM484995 | – | – | KM485100 | – | – | [91] |
| Pyricularia grisea | CR0024 | KM484882 | KM484997 | – | – | KM485102 | – | – | [91] |
| Pyricularia oryzae | CBS 365.52 = MUCL 9451 | KM484890 | KM485000 | – | – | KM485110 | – | – | [76] |
| Slopeiomyces cylindrosporus | BAN-145 | JF508361 | – | – | – | – | – | – | [107] |
| Slopeiomyces cylindrosporus | CG340 | AY428776 | – | – | – | – | – | – | [108] |
| Utrechtiana cibiessia | CBS 128780 = CPC 18916 | JF951153 | JF951176 | – | – | KM485047 | – | – | [76] |
| Xenopyricularia zizaniicola | CBS 132356 | KM484946 | KM485042 | – | KM009203 | KM485160 | – | – | [91] |
| Acericola italica | MFLUCC 13-0609 T | MF167428 | MF167429 | – | – | – | MF167430 | – | [109] |
| Alloneottiosporina thailandica | MFLUCC 15-0576 T | MT177913 | MT177940 | – | MT454002 | – | MT177968 | – | [43] |
| Allophaeosphaeria muriformia | MFLUCC 13-0349 T | KP765680 | KP765681 | – | – | – | KP765682 | – | [105] |
| Amarenographium ammophilae | MFLUCC 16-0296 | KU848196 | KU848197 | – | MG520894 | – | KU848198 | – | [109] |
| Amarenomyces dactylidis | MFLU 17-0498 T | KY775577 | KY775575 | – | – | – | – | – | [110] |
| Ampelomyces quisqualis | CBS 129.79 T | – | EU754128 | – | – | – | EU754029 | – | [111] |
| Banksiophoma australiensis | CBS 142163 T | KY979739 | KY979794 | – | KY979889 | – | – | – | [112] |
| Bhagirathimyces himalayensis | AMH 10127 T = NFCCI 4580 | MK836021 | MK836020 | – | – | – | MN121697 | – | [113] |
| Bhatiellae rosae | MFLUCC 17-0664 T | MG828873 | MG828989 | – | – | – | MG829101 | – | [114] |
| Brunneomurispora lonicerae | KUMCC 18-0157 T | MK356373 | MK356346 | – | MK359065 | – | MK356360 | – | [59] |
| Camarosporioides phragmitis | MFLUCC 13-0365 T | KX572340 | KX572345 | – | KX572354 | – | KX572350 | – | [115] |
| Chaetosphaeronema achilleae | MFLUCC 16-0476 T | KX765265 | KX765266 | – | – | – | – | [115] | |
| Chaetosphaeronema hispidulum | MFLU:16-1965 | MT177915 | MT177942 | – | – | – | MT177970 | – | [43] |
| Chaetosphaeronema hispidulum | MFLU:16-2275 | MT177914 | MT177941 | – | MT454003 | – | MT177969 | – | [43] |
| Chaetosphaeronema hispidulum | CBS 216.75 | KF251148 | KF251652 | – | KF253108 | – | – | [116] | |
| Dactylidina dactylidis | MFLUCC 13-0618 | KP744432 | KP744473 | – | – | – | KP753946 | – | [105] |
| Dactylidina dactylidis | MFLUCC 14-0966 T | MG828886 | MG829002 | – | MG829199 | – | MG829113 | – | [114] |
| Dematiopleospora donetzica | MFLU 15-2199 T | – | MG829005 | – | – | – | MG829116 | – | [114] |
| Dematiopleospora mariae | MFLUCC 13-0612 T | KJ749654 | KJ749653 | – | KJ749655 | – | KJ749652 | – | [117] |
| Diederichomyces ficuzzae | CBS 128019 | KP170647 | – | – | KP170673 | – | – | – | [118] |
| Diederichomyces xanthomendozae | CBS 129666 | KP170651 | – | – | KP170677 | – | – | – | [118] |
| Dlhawksworthia clematidicola | MFLUCC 17-2151 T | MT310619 | MT214574 | – | MT394633 | – | MT226687 | – | [119] |
| Edenia gomezpompae | JLCC 34533 | KC193601 | – | – | – | – | – | – | [120] |
| Elongaticollum hedychii | MFLUCC 18-1638 T | MT321796 | MT321810 | – | MT328753 | – | MT321803 | – | [115] |
| Elongaticollum hedychii | NCYUCC 19-0286 | MT321797 | MT321811 | – | MT328754 | – | MT321804 | – | [115] |
| Embarria clematidis | MFLUCC 14-0652 | KT306949 | KT306953 | – | – | – | KT306956 | – | [121] |
| Embarria clematidis | MFLUCC 14-0976 | MG828871 | MG828987 | – | MG829194 | – | MG829099 | – | [114] |
| Equiseticola fusispora | MFLUCC 14-0522 T | KU987668 | KU987669 | – | MG520895 | – | KU987670 | – | [122] |
| Galliicola pseudophaeosphaeria | MFLUCC 14-0524 | – | – | – | MG520896 | – | – | – | [109] |
| Hawksworthiana clematidicola | MFLUCC 14-0910 T | MG828901 | MG829011 | – | MG829202 | – | MG829120 | – | [114] |
| Hawksworthiana lonicerae | MFLUCC 14-0955 T | MG828902 | MG829012 | – | MG829203 | – | MG829121 | – | [114] |
| Hydeomyces desertipleosporoides | SQUCC 15260 | MK290842 | MK290840 | – | MK290849 | – | MK290844 | – | [123] |
| Hydeomyces desertipleosporoides | SQUCC 15259 T | MK290841 | MK290839 | – | MK290848 | – | MK290843 | – | [123] |
| Hydeomyces pinicola | GZ-06 | MK522506 | MK522496 | – | MK523386 | – | MK522502 | – | [124] |
| Hydeopsis verrucispora | SD-2016-5 | MK522508 | MK522498 | – | MK523388 | – | MK522504 | – | [124] |
| Italica achilleae | MFLUCC 14-0959 T | MG828903 | MG829013 | – | MG829204 | – | MG829122 | – | [114] |
| Jeremyomyces labinae | CBS 144617 T | MK442589 | – | – | MK442695 | – | – | – | [96] |
| Juncaceicola italica | MFLUCC 13-0750 | KX500110 | KX500107 | – | MG520897 | – | KX500108 | – | [109] |
| Juncaceicola luzulae | MFLUCC 13-0780 | KX449529 | KX449530 | – | MG520898 | – | KX449531 | – | [125] |
| Kwanghwana miscanthi | FU31017 | MK503817 | MK503823 | – | MT009126 | – | MK503829 | – | [126] |
| Leptosphaeria doliolum | CBS 505.75 T | JF740205 | GU301827 | – | GU349069 | – | GU296159 | – | [127,128] |
| Leptospora galii | KUMCC 15-0521 T | KX599547 | KX599548 | – | MG520899 | – | KX599549 | – | [109] |
| Leptospora rubella | CPC 11006 | DQ195780 | DQ195792 | – | – | – | DQ195803 | – | [129] |
| Leptospora thailandica | MFLUCC 16-0385 T | KX655559 | KX655549 | – | KX655564 | – | KX655554 | – | [130] |
| Loratospora luzulae | MFLUCC 14-0826 T | KT328497 | KT328495 | – | – | – | KT328496 | – | [121] |
| Mauginiella scaettae | CBS 239.58 | MH857770 | MH869303 | – | – | – | – | – | [76] |
| Melnikia anthoxanthii | MFLUCC 14-1010 | – | KU848204 | – | – | – | KU848205 | – | [131] |
| Murichromolaenicola chiangraiensis | MFLUCC 17-1488 T | MN994582 | MN994559 | – | MN998163 | – | MN994605 | – | [74] |
| Muriphaeosphaeria galatellae | MFLUCC 15-0769 | – | KT438330 | – | – | – | KT438332 | – | [132] |
| Muriphaeosphaeria galatellae | MFLUCC 14-0614 T | KT438333 | KT438329 | – | MG520900 | – | KT438331 | – | [132] |
| Neoophiobolus chromolaenae | MFLUCC 17-1467 T | MN994583 | MN994562 | – | MN998164 | – | MN994606 | – | [74] |
| Neosetophoma garethjonesii | MFLUCC 14-0528 | – | – | – | KY514402 | – | KY501126 | – | [133] |
| Neosetophoma rosigena | MFLUCC 17-0768 T | MG828928 | MG829037 | – | – | – | MG829143 | – | [114] |
| Neosphaerellopsis thailandica | CPC 21659 T | KP170652 | KP170721 | – | KP170678 | – | – | – | [118] |
| Neostagonospora arrhenather | MFLUCC 15-0464 | KX926417 | KX910091 | – | MG520901 | – | KX950402 | – | [134] |
| Neostagonospora caricis | CBS 135092 T | KF251163 | KF251667 | – | – | – | – | – | [76] |
| Neostagonospora phragmitis | MFLUCC 16-0493 | KX926416 | KX910090 | – | MG520902 | – | KX950401 | – | [134] |
| Neostagonosporella sichuanensis | MFLUCC 18-1223 | MH394690 | MH394687 | – | MK313854 | – | MK296469 | – | [58] |
| Neostagonosporella sichuanensis | MFLUCC 18-1228 T | MH368073 | MH368079 | – | MK313851 | – | MH368088 | – | [58] |
| Neosulcatispora strelitziae | CPC 25657 | KX228253 | KX228305 | – | – | – | – | – | [112] |
| Nodulosphaeria guttulatum | MFLUCC 15-0069 | – | – | – | KY514394 | – | KY501115 | – | [133] |
| Nodulosphaeria multiseptata | MFLUCC 15-0078 | KY496748 | KY496728 | – | – | – | – | – | [133] |
| Nodulosphaeria scabiosae | MFLUCC 14-1111 T | KU708850 | KU708846 | – | KU708854 | – | KU708842 | – | [135] |
| Ophiobolopsis italica | MFLUCC 17-1791 T | MG520939 | MG520959 | – | MG520903 | – | MG520977 | – | [109] |
| Ophiobolus artemisiae | MFLUCC 14-1156 T | KT315508 | KT315509 | – | MG520905 | – | MG520979 | – | [109] |
| Ophiobolus disseminans | MFLUCC 17-1787 | MG520941 | MG520961 | – | MG520906 | – | MG520980 | – | [109] |
| Ophiobolus ponticus | MFLUCC 17-2273 | MG520943 | MG520963 | – | MG520908 | – | MG520982 | – | [109] |
| Ophiosimulans tanaceti | MFLUCC 14-0525 | KU738890 | KU738891 | – | MG520910 | – | KU738892 | – | [109] |
| Ophiosphaerella herpotricha | KY423 | KP690989 | – | – | KP691011 | – | – | – | [136] |
| Ophiosphaerella korrae | ATCC 56289 | KC848509 | – | – | KC848515 | – | – | – | [136] |
| Ophiosphaerella narmari | ATCC 64688 | KC848510 | – | – | KC848516 | – | – | – | [136] |
| Paraleptosphaeria dryadis | CBS 643.86 | JF740213 | GU301828 | – | GU349009 | – | KC584632 | – | [127,128] |
| Paraleptospora chromolaenae | MFLUCC 17-1481 T | MN994587 | MN994563 | – | MN998167 | – | MN994609 | – | [74] |
| Paralloneottiosporina sichuanensis | SICAUCC 22-0074 T | ON226746 | ON227102 | – | ON244423 | – | ON227129 | – | This study |
| Paralloneottiosporina sichuanensis | SICAUCC 22-0075 | ON226747 | ON227103 | – | ON244424 | – | ON227130 | – | This study |
| Paraloratospora camporesii | MFLU 18-0915 T | MN756639 | MN756637 | – | – | – | MN756635 | – | [113] |
| Paraophiobolus arundinis | MFLUCC 17-1789 T | MG520945 | MG520965 | – | MG520912 | – | MG520984 | – | [109] |
| Paraophiobolus plantaginis | MFLUCC 17-0245 T | KY797641 | KY815010 | – | – | – | KY815012 | – | [109] |
| Paraphoma chrysanthemicola | CBS 522.66 | KF251166 | KF251670 | – | KF253124 | – | – | – | [116] |
| Paraphoma radicina | CBS 111.79 | KF251172 | KF251676 | – | KF253130 | – | – | – | [116] |
| Parastagonospora italica | MFLUCC 13-0377 T | KU058714 | KU058724 | – | MG520915 | – | MG520985 | – | [109,137] |
| Parastagonospora minima | MFLUCC 13-0376 | KU058713 | KU058723 | – | MG520916 | – | MG520986 | – | [109,137] |
| Parastagonosporella fallopiae | CCTU 1151.1 | MH460544 | MH460546 | – | MH460550 | – | – | – | [138] |
| Parastagonosporella fallopiae | CBS 135981 T | MH460543 | MH460545 | – | MH460549 | – | – | – | [138] |
| Phaeopoacea festucae | MFLUCC 17-0056 | KY824766 | KY824767 | – | – | – | KY824769 | – | [134] |
| Phaeoseptoriella zeae | CBS 144614 T | MK442611 | MK442547 | – | MK442702 | – | – | – | [96] |
| Phaeosphaeria chiangraina | MFLUCC 13-0231 T | KM434270 | KM434280 | – | KM434298 | – | KM434289 | – | [57] |
| Phaeosphaeria oryzae | CBS 110110 T | KF251186 | KF251689 | – | – | – | GQ387530 | – | [139] |
| Phaeosphaeria pleurospora | CBS 460.84 | AF439498 | – | – | – | – | – | – | [140] |
| Phaeosphaeriopsis glaucopunctata | MFLUCC 13-0265 | KJ522473 | KJ522477 | – | MG520918 | – | KJ522481 | – | [109,141] |
| Phaeosphaeriopsis triseptata | MFLUCC 13-0271 | KJ522475 | KJ522479 | – | MG520919 | – | KJ522484 | – | [109,141] |
| Phaeosphaeriopsis yuccae | MFLUCC 16-0558 | KY554482 | KY554481 | – | MG520920 | – | KY554480 | – | [109] |
| Piniphoma wesendahlina | CBS 145032 T | MK442615 | MK442551 | – | MK442706 | – | – | – | [96] |
| Poaceicola arundinis | MFLUCC 15-0702 T | KU058716 | KU058726 | – | MG520921 | – | MG520988 | – | [109] |
| Poaceicola italica | MFLUCC 13-0267 | KX926421 | KX910094 | – | MG520924 | – | KX950409 | – | [109,134] |
| Populocrescentia ammophilae | MFLUCC 17-0665 T | MG828949 | MG829059 | – | MG829231 | – | MG829164 | – | [114] |
| Populocrescentia forlicesenensis | MFLUCC 14-0651 T | KT306948 | KT306952 | – | MG520925 | – | KT306955 | – | [121] |
| Populocrescentia rosae | TASM 6125 T | – | MG829060 | – | MG829232 | – | MG829165 | – | [114] |
| Pseudoophiobolus mathieui | MFLUCC 17-1784 | MG520949 | MG520969 | – | MG520928 | – | MG520991 | – | [109] |
| Pseudoophiobolus rosae | MFLUCC 17-1786 T | MG520952 | MG520972 | – | MG520930 | – | MG520993 | – | [109] |
| Pseudoophiobolus urticicola | KUMCC 17-0168 T | MG520955 | MG520975 | – | MG520933 | – | MG520996 | – | [109] |
| Pseudoophiosphaerella huishuiensis | HS-13 | MK522509 | MK522499 | – | MK523389 | – | MK522505 | – | [124] |
| Pseudophaeosphaeria rubi | MFLUCC 14-0259 T | KX765298 | KX765299 | – | MG520934 | – | KX765300 | – | [130] |
| Sclerostagonospora ericae | CPC 25927 T | KX228268 | KX228319 | – | KX228375 | – | – | – | [112] |
| Scolicosporium minkeviciusii | MFLUCC 12-0089 | – | KF366382 | – | – | – | KF366383 | – | [142] |
| Septoriella phragmitis | CPC 24118 T | KR873251 | KR873279 | – | – | – | – | – | [143] |
| Setomelanomma holmii | CBS 110217 | KT389542 | GU301871 | – | GU349028 | – | GU296196 | – | [127,144] |
| Setophoma terrestris | CBS 335.29 | KF251246 | KF251749 | – | KF253196 | – | – | – | [116] |
| Stagonospora neglecta | CBS 343.86 | AJ496630 | – | – | – | – | – | – | [145] |
| Sulcispora supratumida | MFLUCC 14-0995 T | KP271443 | KP271444 | – | MH665366 | – | KP271445 | – | [146] |
| Tintelnotia destructans | CBS 127737 T | KY090652 | KY090664 | – | – | – | KY090698 | – | [147] |
| Tintelnotia opuntiae | CBS 376.91 T | KY090651 | GU238123 | – | – | – | GU238226 | – | [147,148] |
| Vagicola arundinis | MFLUCC 15-0027 T | KY706139 | KY706129 | – | MG520936 | – | KY706134 | – | [109] |
| Vittaliana mangrovei | NFCCI 4251 T | MG767311 | MG767312 | – | MG767314 | – | MG767313 | – | [149] |
| Vrystaatia aloeicola | CBS 135107 | KF251278 | KF251781 | – | – | – | – | – | [116] |
| Wingfieldomyces cyperi | CBS 141450 T | KX228286 | KX228337 | – | MK540163 | – | – | – | [150] |
| Wojnowicia italica | MFLUCC 13-0447 T | KX342923 | KX430001 | – | KX430003 | – | KX430002 | – | [130] |
| Wojnowicia rosicola | MFLUCC 15-0128 T | MG828979 | MG829091 | – | – | – | MG829191 | – | [114] |
| Wojnowiciella eucalypti | CBS 139904 T | KR476741 | KR476774 | – | – | – | – | – | [76] |
| Xenophoma puncteliae | CBS 128022 | – | JQ238619 | – | KP170686 | – | – | – | [118,151] |
| Xenoseptoria neosaccardoi | CBS 120.43 | KF251280 | KF251783 | – | KF253227 | – | – | – | [116] |
| Xenoseptoria neosaccardoi | CBS 128665 | KF251281 | KF251784 | – | KF253228 | – | – | – | [116] |
| Yunnanensis phragmitis | MFLUCC 17-1361 T | MF684869 | MF684865 | – | – | – | MF684864 | – | [152] |
| Yunnanensis phragmitis | MFLUCC 17-0315 | MF684862 | MF684863 | – | MF683624 | – | MF684867 | – | [152] |
| Biatriospora marina | CY 1228 | – | GQ925848 | – | GU479848 | – | GQ925835 | GU479823 | [153] |
| Biatriospora peruviensis | CCF 4485 | – | LN626683 | – | LN626671 | – | LN626677 | LN626665 | [154] |
| Neooccultibambusa chiangraiensis | MFLUCC 12-0559 T | – | KU764699 | – | – | – | KU712458 | – | [155] |
| Neoroussoella bambusae | MFLUCC 11-0124 | – | KJ474839 | – | KJ474848 | – | – | KJ474856 | [156] |
| Occultibambusa aquatica | MFLUCC 11-0006 | – | KX698110 | – | – | – | KX698112 | – | [130] |
| Occultibambusa bambusae | MFLUCC 11-0394 | – | KU863113 | – | KU940194 | – | KU872117 | KU940171 | [16] |
| Occultibambusa bambusae | MFLUCC 13-0855 T | – | KU863112 | – | KU940193 | – | KU872116 | KU940170 | [16] |
| Occultibambusa chiangraiensis | MFLUCC 16-0380 T | – | KX655546 | – | – | – | KX655551 | KX655566 | [130] |
| Occultibambusa fusispora | MFLUCC 11-0127 T | – | KU863114 | – | KU940195 | – | – | KU940172 | [16] |
| Occultibambusa jonesii | GZCC 16-0117 T | – | KY628322 | – | KY814756 | – | KY628324 | KY814758 | [157] |
| Occultibambusa kunmingensis | HKAS 102151 T | – | MN913733 | – | MT954407 | – | MT864342 | MT878453 | [61] |
| Occultibambusa maolanensis | GZCC 16-0116 | – | KY628323 | – | KY814757 | – | KY628325 | KY814759 | [157] |
| Occultibambusa pustula | MFLUCC 11-0502 | – | KU863115 | – | – | – | KU872118 | – | [16] |
| Paradictyoarthrinium diffractum | MFLUCC 13-0466 | – | KP744498 | – | – | – | KP753960 | KX437764 | [105,158] |
| Paradictyoarthrinium tectonicola | MFLUCC 13-0465 T | – | KP744500 | – | – | – | KP753961 | KX437763 | [105,158] |
| Roussoella hysterioides | HH 26988 | – | AB524622 | – | AB539115 | – | AB524481 | AB539102 | [127] |
| Roussoella nitidula | MFLUCC 11-0182 | – | KJ474843 | – | KJ474852 | – | – | KJ474859 | [156] |
| Roussoella nitidula | MFLUCC 11-0634 | – | KJ474842 | – | KJ474851 | – | – | KJ474858 | [156] |
| Roussoella pustulans | KT 1709 | – | AB524623 | – | AB539116 | – | AB524482 | AB539103 | [1,127] |
| Seriascoma bambusae | KUMCC 21-0021 | – | MZ329035 | – | MZ325468 | – | MZ329031 | MZ325470 | [159] |
| Seriascoma didymospora | MFLUCC 11-0179 T | – | KU863116 | – | KU940196 | – | – | KU940173 | [16] |
| Seriascoma didymospora | MFLUCC 11-0194 | – | KU863117 | – | KU940197 | – | – | KU940174 | [16] |
| Seriascoma yunnanense | MFLU 19-0690 T | – | NG_068303 | – | MN381858 | – | MN174694 | MN210324 | [44] |
| Seriascoma yunnanense | SICAUCC 22-0059 | – | ON226771 | – | ON567182 | – | ON227356 | ON567183 | This study |
| Torula herbarum | CBS 111855 | – | KF443386 | – | KF443403 | – | KF443391 | KF443396 | [160] |
| Westerdykella ornata | CBS 379.55 | – | GU301880 | – | GU349021 | – | GU296208 | GU371803 | [127] |
Notes: superscript T represents ex-type or ex-epitype isolates. “–” means that the sequence is missing, unavailable or unused. New sequences are listed in bold. Abbreviation: AP: Culture Collection of A. Pintos; ATCC: American Type Culture Collection, U.S.A.; BRIP: Queensland Plant Pathology Herbarium, Brisbane, Australia; CBS: Culture Collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CFCC: China Forestry Culture Collection Center, Beijing, China; CGMCC: China General Microbiological Culture Collection Center, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China; CPC: Culture Collection of P.W. Crous; DAOM: Plant Research Institute, Department of Agriculture (Mycology), Ottawa, Canada; GZCC: Guizhou Academy of Agricultural Sciences Culture Collection, Guizhou, China; IMI: Culture Collection of CABI Europe UK Centre, Egham, UK; JHB: Culture Collection of H.B. Jiang; KUMCC: Kunming Institute of Botany Culture Collection, Yunnan, China; LC: Working collection of Lei Cai, housed at the Institute of Microbiology, Chinese Academy of Sciences, Beijing, China; MFLU: Herbarium of Mae Fah Luang University, Chiang Rai, Thailand; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; NCYUCC: National Chiayi University Culture Collection, Chiayi, Taiwan; SICAUCC: Sichuan Agricultural University Culture Collection, Sichuan, China; URM: Culture Collection of the Universidade Federal de Pernambuco, Brazil.
Author Contributions
Q.Z. and C.-L.Y.: conceptualization. Q.Z.: data curation. Q.Z. and Y.-C.L.: formal analysis, methodology, and writing—original draft. Q.Z., Y.-C.L., Y.D. and F.-H.W.: investigation. C.-L.Y. and Y.-G.L.: project administration. C.-L.Y. and X.-L.X.: supervision. C.-L.Y., X.-L.X., S.-Y.L. and L.-J.L.: writing—review and editing. All authors contributed to the article and approved the submitted version. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
The datasets presented in this study can be found in the NCBI GenBank (https://www.ncbi.nlm.nih.gov/), Index Fungorum (http://www.indexfungorum.org/Names/Names.asp) (all accessed on 8 May 2022).
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tanaka K., Hirayama K., Yonezawa H., Hatakeyama S., Harada Y., Sano T., Shirouzu T., Hosoya T. Molecular taxonomy of bambusicolous fungi: Tetraplosphaeriaceae, a new pleosporalean family with Tetraploa-like anamorphs. Stud. Mycol. 2009;64:175–209. doi: 10.3114/sim.2009.64.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shi J.Y., Zhou D.Q., Ma L.S., Yao J., Zhang D.M. Diversity of bamboo species in China. World Bamboo Ratt. 2020;18:55–65. doi: 10.12168/sjzttx.2020.04.011. [DOI] [Google Scholar]
- 3.Du S.S. Classification and arrangement of Phyllostachys (Poaceae: Bambusoideae), China. J. Fujian Forestry Sci. Technol. 2020;47:120–123. doi: 10.13428/j.cnki.fjlk.2020.03.022. [DOI] [Google Scholar]
- 4.Li Y.G., Xue L., Fan L.L., Ye L.T., Cheng L.Y., He T.Y., Zheng Y.S. Research progress in germplasm resources and applications of Phyllostachy. J. Sic. For. Sci. Technol. 2019;40:117–122. doi: 10.16779/j.cnki.1003-5508.2019.04.024. [DOI] [Google Scholar]
- 5.Bystriakova N., Kapos V., Lysenko I., Stapleton C.M.A. Distribution and conservation status of forest bamboo biodiversity in the Asia-Pacific Region. Biodivers. Conserv. 2003;12:1833–1841. doi: 10.1023/A:1024139813651. [DOI] [Google Scholar]
- 6.Scurlock J.M.O., Dayton D.C., Hames B. Bamboo: An overlooked biomass resource? Biomass. Bioener. 2000;19:229–244. doi: 10.1016/S0961-9534(00)00038-6. [DOI] [Google Scholar]
- 7.Idris M.A., Mohamad A. Bamboo shoot utilization in peninsular Malaysia: A case study in Pahang. J. Bamboo Rattan. 2002;1:141–155. doi: 10.1163/156915902760181612. [DOI] [Google Scholar]
- 8.Shi J.Y., Chen Q.B., Huang J.Y., Zhou D.Q., Ma L.S., Yao J. Biodiversity of the staple food bamboos of giant panda and its important value. World Bamboo Rattan. 2020;18:10–19. doi: 10.12168/sjzttx.2020.05.002. [DOI] [Google Scholar]
- 9.Wang X.J., Wang T., Chi M., Li L.B. Research progress of ornamental bamboos in China. J. Bamboo Res. 2019;38:3–9. doi: 10.19560/j.cnki.issn1000-6567.2019.04.002. [DOI] [Google Scholar]
- 10.Hyde K.D., Zhou D.Q., Mckenzie E.H.C., Ho W.H., Dalisay T. Vertical distribution of saprobic fungi on bamboo culms. Fungal Divers. 2002;11:109–118. [Google Scholar]
- 11.Tanaka E., Shimizu K., Imanishi Y., Yasuda F., Tanaka C. Isolation of basidiomycetous anamorphic yeast-like fungus Meira argovae found on Japanese bamboo. Mycoscience. 2008;49:329–333. doi: 10.1007/S10267-008-0429-1. [DOI] [Google Scholar]
- 12.Dai D.Q., Bhat D.J., Liu J.K., Chukeatirote E., Zhao R.L., Hyde K.D. Bambusicola, a new genus from bamboo with asexual and sexual morphs. Cryptogamie Mycol. 2012;33:363–379. doi: 10.7872/crym.v33.iss3.2012.363. [DOI] [Google Scholar]
- 13.Doungporn M., Hiroko K., Tatsuji S. Molecular diversity of bamboo–associated fungi isolated from Japan. FEMS Microbiol. Lett. 2007;266:10–19. doi: 10.1111/j.1574-6968.2006.00489.x. [DOI] [PubMed] [Google Scholar]
- 14.Hyde K.D., Zhou D., Dalisay T. Bambusicolous fungi: A review. Fungal Divers. 2002;9:1–14. [Google Scholar]
- 15.Tanaka K., Harada Y. Bambusicolous fungi in Japan (1): Four Phaeosphaeria species. Mycoscience. 2004;45:377–382. doi: 10.1007/S10267-004-0197-5. [DOI] [Google Scholar]
- 16.Dai D.Q., Phookamsak R., Wijayawardene N.N., Li W.J., Bhat D.J., Xu J.C., Taylor J.E., Hyde K.D., Chukeatirote E. Bambusicolous fungi. Fungal Divers. 2017;82:1–105. doi: 10.1007/s13225-016-0367-8. [DOI] [Google Scholar]
- 17.Hatakeyama S., Tanaka K., Harada Y. Bambusicolous fungi in Japan (7): A new coelomycetous genus, Versicolorisporium. Mycoscience. 2008;49:211–214. doi: 10.1007/S10267-008-0409-5. [DOI] [Google Scholar]
- 18.Hatakeyama S., Tanaka K., Harada Y. Bambusicolous fungi in Japan (5): Three species of Tetraploa. Mycoscience. 2005;46:196–200. doi: 10.1007/S10267-005-0233-0. [DOI] [Google Scholar]
- 19.Zhang Z.Y., Zhang X. Potentials of bamboo in traditioanl Chinese medicine and development of heath products. World Sci. Technol. 2000;3:54–56. doi: 10.3969/j.issn.1674-3849.2000.03.018. [DOI] [Google Scholar]
- 20.Zhou B.Z., Fu M.Y., Xie J.Z., Yang X.S., Li Z.C. Ecological functions of bamboo forest: Research and application. J. Forestry Res. 2005;16:143–147. doi: 10.1007/BF02857909. [DOI] [Google Scholar]
- 21.Singhal P., Bal L.M., Satya S., Sudhakar P., Naik S.N. Bamboo shoots: A novel source of nutrition and medicine. Crit. Rev. Food Sci. Nutr. 2013;53:517–534. doi: 10.1080/10408398.2010.531488. [DOI] [PubMed] [Google Scholar]
- 22.Teng S.C. Fungi of China. Mycotaxon, Ltd.; New York, NY, USA: 1996. pp. 1–728. [Google Scholar]
- 23.Tai F.L. Sylloge Fungorum Sinicorum. Science Press, Academica Sinica; Beijing, China: 1979. pp. 1–1527. [Google Scholar]
- 24.Chen M.M. Forest Fungi Phytogeography: Forest Fungi Phytogeography of China, North America, and Siberia and International Quarantine of Tree Pathogens. Pacific Mushroom Research and Education Center; Sacramento, CA, USA: 2002. pp. 1–469. [Google Scholar]
- 25.Chomnunti P., Hongsanan S., Hudson B.A., Tian Q., Peršoh D., Dhami M.K., Alias A.S., Xu J.C., Liu X.Z., Stadler M., et al. The sooty moulds. Fungal Divers. 2014;66:1–36. doi: 10.1007/s13225-014-0278-5. [DOI] [Google Scholar]
- 26.White T.J., Bruns T., Lee S., Taylor J. Amplifcation and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfaud D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; San Diego, CA, USA: 1990. pp. 315–322. [Google Scholar]
- 27.Vilgalys R., Hester M. Rapid genetic identifification and mapping of enzymatically amplifified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Donnell K., Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- 29.Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microb. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matheny P.B., Liu Y.J., Ammirati J.F., Hall B.D. Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales) Am. J. Bot. 2002;89:688–698. doi: 10.3732/ajb.89.4.688. [DOI] [PubMed] [Google Scholar]
- 31.Castlebury L., Rossman A., Sung G., Hyten A., Spatafora J. Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycol. Res. 2004;108:864–872. doi: 10.1017/S0953756204000607. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 33.Rehner S.A., Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98. doi: 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- 34.O’Donnell K., Kistler H.C., Cigelnik E., Ploetz R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carbone I., Kohn L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. doi: 10.1080/00275514.1999.12061051. [DOI] [Google Scholar]
- 36.Wang M., Tan X.M., Liu F., Cai L. Eight new Arthrinium species from China. MycoKeys. 2018;34:1–24. doi: 10.3897/mycokeys.34.24221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 39.Xu X.L., Yang C.L., Jeewon R., Wanasinghe D.N., Xiao Q.G. Morpho-molecular diversity of Linocarpaceae (Chaetosphaeriales): Claviformispora gen. nov. from decaying branches of Phyllostachys heteroclada. MycoKeys. 2020;69:113–129. doi: 10.3897/mycokeys.69.54231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crous P.W., Groenewald J.Z. A phylogenetic re-evaluation of Arthrinium. IMA Fungus. 2013;4:133–154. doi: 10.5598/imafungus.2013.04.01.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai D.Q., Jiang H.B., Tang L.Z., Bhat D.J. Two new species of Arthrinium (Apiosporaceae, Xylariales) associated with bamboo from Yunnan, China. Mycosphere. 2016;7:1332–1345. doi: 10.5943/mycosphere/7/9/7. [DOI] [Google Scholar]
- 42.Silva R.M., Oliveira R.J., Bezerra J.D., Bezerra J.L., Souza–Motta C.M., Silva G.A. Bifusisporella sorghi gen. et sp. nov. (Magnaporthaceae) to accommodate an endophytic fungus from Brazil. Mycol. Prog. 2019;18:847–854. doi: 10.1007/s11557-019-01494-2. [DOI] [Google Scholar]
- 43.Li W.J., McKenzie E.H.C., Liu J.K., Bhat D.J., Dai D.Q., Camporesi E., Tian Q., Maharachchikumbura S.S.N., Luo Z.L., Shang Q.J., et al. Taxonomy and phylogeny of hyaline-spored coelomycetes. Fungal Divers. 2020;100:279–801. doi: 10.1007/s13225-020-00440-y. [DOI] [Google Scholar]
- 44.Rathnayaka A.R., Dayarathne M.C., Maharachchikumbura S.S.N., Liu J.K., Tennakoon D.S., Hyde K.D. Introducing Seriascoma yunnanense sp. nov. (Occultibambusaceae, Pleosporales) based on evidence from morphology and phylogeny. Asian J. Mycol. 2019;2:245–253. doi: 10.5943/ajom/2/1/15. [DOI] [Google Scholar]
- 45.Dai D.Q., Tang L.Z., Wang H.B. A Review of Bambusicolous Ascomycetes. Bamboo Curr. Future Prospect. 2018:165–183. doi: 10.5772/intechopen.76463. [DOI] [Google Scholar]
- 46.Saccardo P. Conspectus generum pyrenomycetum italicorum additis speciebus fungorum Venetorum novis vel criticis, systemate carpologico dispositorum. Atti Soc. Veneziana-Trent. Istriana Sci. Nat. 1875;4:77–100. [Google Scholar]
- 47.Ellis M.B. Dematiaceous hyphomycetes VI. Commonw. Mycol. Inst. 1965;103:1–46. [Google Scholar]
- 48.Pintos Á., Alvarado P. Phylogenetic delimitation of Apiospora and Arthrinium. Fungal Syst. Evol. 2021;7:197–221. doi: 10.3114/fuse.2021.07.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian X.G., Karunarathna S.C., Mapook A., Promputtha I., Xu J.C., Bao D.F., Tibpromma S. One new species and two new host records of Apiospora from bamboo and maize in Northern Thailand with thirteen new combinations. Life. 2021;11:1071. doi: 10.3390/life11101071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen K., Wu X.Q., Huang M.X., Han Y.Y. First report of brown culm ctreak of Phyllostachys praecox caused by Arthrinium arundinis in Nanjing, China. Plant Dis. 2014;98:1274. doi: 10.1094/PDIS-02-14-0165-PDN. [DOI] [PubMed] [Google Scholar]
- 51.Yin C.W., Luo F.Y., Zhang H., Fang X.M., Zhu T.H., Li S.J. First report of Arthrinium kogelbergense causing blight disease of Bambusa intermedia in Sichuan Province, China. Plant Dis. 2021;105:214. doi: 10.1094/PDIS-06-20-1159-PDN. [DOI] [PubMed] [Google Scholar]
- 52.Feng Y., Liu J.K., Lin C.G., Chen Y.Y., Xiang M.M., Liu Z.Y. Additions to the genus Arthrinium (Apiosporaceae) from bamboos in China. Front. Microbiol. 2021;12:661281. doi: 10.3389/fmicb.2021.661281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang N., Li J., Tian C.M. Arthrinium species associated with bamboo and reed plants in China. Fungal Syst. Evol. 2018;2:1–9. doi: 10.3114/fuse.2018.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang C.L., Xu X.L., Liu Y.G., Xu X.L. First report of bamboo blight disease caused by Arthrinium yunnanum on Phyllostachys heteroclada in Sichuan, China. Plant Dis. 2018;103:1026. doi: 10.1094/PDIS-10-18-1740-PDN. [DOI] [Google Scholar]
- 55.Luo Z.L., Hyde K.D., Liu J.K., Maharachchikumbura S.S.N., Jeewon R., Bao D.F., Bhat D.J., Lin C.G., Li W.L., Yang J., et al. Freshwater Sordariomycetes. Fungal Divers. 2019;99:451–660. doi: 10.1007/s13225-019-00438-1. [DOI] [Google Scholar]
- 56.Tennakoon D.S., Jeewon R., Gentekaki E., Kuo C.H., Hyde K.D. Multi-gene phylogeny and morphotaxonomy of Phaeosphaeria ampeli sp. nov. from Ficus ampelas and a new record of P. musae from Roystonea regia. Phytotaxa. 2019;406:111–128. doi: 10.11646/phytotaxa.406.2.3. [DOI] [Google Scholar]
- 57.Phookamsak R., Liu J.K., McKenzie E.H.C., Manamgoda D.S., Ariyawansa H., Thambugala K.M., Dai D.Q., Camporesi E., Chukeatirote E., Wijayawardene N.N., et al. Revision of Phaeosphaeriaceae. Fungal Divers. 2014;68:159–238. doi: 10.1007/s13225-014-0308-3. [DOI] [Google Scholar]
- 58.Yang C.L., Xu X.L., Wanasinghe D.N., Jeewon R., Phookamsak R., Liu Y.G., Liu L.J., Hyde K.D. Neostagonosporellasichuanensis gen. et sp. nov. (Phaeosphaeriaceae, Pleosporales) on Phyllostachys heteroclada (Poaceae) from Sichuan Province, China. MycoKeys. 2019;46:119–150. doi: 10.3897/mycokeys.46.32458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Phookamsak R., Hyde K.D., Jeewon R., Bhat D.J., Jones E.B.J., Maharachchikumbura S.S.N., Raspé O., Karunarathna S.C., Wanasinghe D.N., Hongsanan S., et al. Fungal diversity notes 929–1035: Taxonomic and phylogenetic contributions on genera and species of fungal. Fungal Divers. 2019;95:1–273. doi: 10.1007/s13225-019-00421-w. [DOI] [Google Scholar]
- 60.Hyde K.D., Jeewon R., Chen Y.J., Bhunjun C.S., Calabon M.S., Jiang H.B., Lin C.G., Norphanphoun C., Sysouphanthong P., Pem D., et al. The numbers of fungi: Is the descriptive curve flattening? Fungal Divers. 2020;103:219–271. doi: 10.1007/s13225-020-00458-2. [DOI] [Google Scholar]
- 61.Dong W., Wang B., Hyde K.D., McKenzie E.H.C., Raja H.A., Tanaka K., Abdel-Wahab M.A., Abdel-Aziz F.A., Doilom M., Phookamsak R., et al. Freshwater Dothideomycetes. Fungal Divers. 2020;105:319–575. doi: 10.1007/s13225-020-00463-5. [DOI] [Google Scholar]
- 62.Boonmee S., Wanasinghe D.N., Calabon M.S., Huanraluek N., Chandrasiri S.K.U., Jones G.E.B., Rossi W., Leonardi M., Singh S.K., Rana S., et al. Fungal diversity notes 1387–1511: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2021;111:1–135. doi: 10.1007/s13225-021-00489-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang C.L., Xu X.L., Liu Y.G. Two new species of Bambusicola (Bambusicolaceae, Pleosporales) on Phyllostachys heteroclada from Sichuan, China. Nova Hedwigia. 2019;108:527–545. doi: 10.1127/nova_hedwigia/2019/0526. [DOI] [Google Scholar]
- 64.Yang C.L., Xu X.L., Liu Y.G., Hyde K.D., Mckenzie E.H.C. A new species of Phyllachora (Phyllachoraceae, Phyllachorales) on Phyllostachys heteroclada from Sichuan, China. Phytotaxa. 2019;392:186–196. doi: 10.11646/phytotaxa.392.3.2. [DOI] [Google Scholar]
- 65.Yang C.L., Xu X.L., Dong W., Wanasinghe D.N., Liu Y.G., Hyde K.D. Introducing Arthrinium phyllostachium sp. nov. (Apiosporaceae, Xylariales) on Phyllostachys heteroclada from Sichuan Province, China. Phytotaxa. 2019;406:91–110. doi: 10.11646/phytotaxa.406.2.2. [DOI] [Google Scholar]
- 66.Yang C.L., Xu X.L., Liu Y.G. Podonectria sichuanensis, a potentially mycopathogenic fungus from Sichuan Province in China. Phytotaxa. 2019;402:219–231. doi: 10.11646/phytotaxa.402.5.1. [DOI] [Google Scholar]
- 67.Yang C.L., Xu X.L., Jeewon R., Boonmee S., Liu Y.G., Hyde K.D. Acremonium arthrinii sp. Nov., a mycopathogenic fungus on Arthrinium yunnanum. Phytotaxa. 2019;420:283–299. doi: 10.11646/phytotaxa.420.4.4. [DOI] [Google Scholar]
- 68.Yang C.L., Baral H.O., Xu X.L., Liu Y.G. Parakarstenia phyllostachydis, a new genus and species of non-lichenized Odontotremataceae (Ostropales, Ascomycota) Mycol. Prog. 2019;18:833–845. doi: 10.1007/s11557-019-01492-4. [DOI] [Google Scholar]
- 69.Yan H., Jiang N., Liang L.Y., Yang Q., Tian C.M. Arthriniumtrachycarpum sp. nov. from Trachycarpus fortunei in China. Phytotaxa. 2019;400:203–210. doi: 10.11646/phytotaxa.400.3.7. [DOI] [Google Scholar]
- 70.Alves-Silva G., Drechsler-Santos E.R., da Silveira R.M.B. Bambusicolous Fomitiporia revisited: Multilocus phylogeny reveals a clade of host-exclusive species. Mycologia. 2020;112:633–648. doi: 10.1080/00275514.2020.1741316. [DOI] [PubMed] [Google Scholar]
- 71.Senanayake I.C., Bhat J.D., Cheewangkoon R., Xie N. Bambusicolous Arthrinium species in Guangdong Province, China. Front. Microbiol. 2020;11:2981. doi: 10.3389/fmicb.2020.602773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pintos A., Alvarado P., Planas J., Jarling R. Six new species of Arthrinium from Europe and notes about A.caricicola and other species found in Carex spp. hosts. MycoKeys. 2019;49:15–48. doi: 10.3897/mycokeys.49.32115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang X., Goonasekara I.D., Jayawardena R.S., Jiang H.B., Li J.F., Hyde K.D., Kang J.C. Arthrinium bambusicola (Fungi, Sordariomycetes), a new species from Schizostachyum brachycladum in northern Thailand. Biodivers Data J. 2020;8:e58755. doi: 10.3897/BDJ.8.e58755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mapook A., Hyde K.D., McKenzie E.H.C., Jones E.B.G., Bhat D.J., Jeewon R., Stadler M., Samarakoon M.C., Malaithong M., Tanunchai B., et al. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed) Fungal Divers. 2020;101:1–175. doi: 10.1007/s13225-020-00444-8. [DOI] [Google Scholar]
- 75.Senanayake I.C., Maharachchikumbura S.S.N., Hyde. K.D., Bhat J.D., Jones E.B.G., McKenzie E.H.C., Dai D.Q., Daranagama D.A., Dayarathne M.C., Goonasekara I.D., et al. Towards unraveling relationships in Xylariomycetidae (Sordariomycetes) Fungal Divers. 2015;73:73–144. doi: 10.1007/s13225-015-0340-y. [DOI] [Google Scholar]
- 76.Vu D., Groenewald M., de Vries M., Gehrmann T., Stielow B., Eberhardt U., Al-Hatmi A., Groenewald J.Z., Cardinali G., Houbraken J., et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2018;91:23–36. doi: 10.1016/j.simyco.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sharma R., Kulkarni G., Sonawane M.S., Shouche Y.S. A new endophytic species of Arthrinium (Apiosporaceae) from Jatropha podagrica. Mycoscience. 2014;55:118–123. doi: 10.1016/j.myc.2013.06.004. [DOI] [Google Scholar]
- 78.Zhao Y.Z., Zhang Z.F., Cai L., Peng W.J., Liu F. Four new filamentous fungal species from newly-collected and hive-stored bee pollen. Mycosphere. 2018;9:1089–1116. doi: 10.5943/mycosphere/9/6/3. [DOI] [Google Scholar]
- 79.Das K., Lee S.Y., Choi H.W., Eom A.H., Choe Y.J., Jung H.Y. Taxonomy of Arthrinium minutisporum sp. nov., Pezicula neosporulosa, and Acrocalymma pterocarpi: New records from soil in Korea. Mycobiology. 2020;48:450–463. doi: 10.1080/12298093.2020.1830741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Allen W.J., de Vries A.E., Bologna N.J., Bickford W.A., Kowalski K.P., Meyerson L.A., Cronin J.T. Intraspecific and biogeographical variation in foliar fungal communities and pathogen damage of native and invasive Phragmites australis. Glob. Ecol. Biogeogr. 2020;29:1199–1211. doi: 10.1111/geb.13097. [DOI] [Google Scholar]
- 81.Jiang N., Liang Y.M., Tian C.M. A novel bambusicolous fungus from China, Arthrinium chinense (Xylariales) Sydowia. 2020;72:77–83. doi: 10.12905/0380.sydowia72-2020-0077. [DOI] [Google Scholar]
- 82.Hyde K.D., Norphanphoun C., Maharachchikumbura S., Bhat D.J., Jones E.B.G., Bundhun D., Chen Y.J., Bao D.F., Boonmee S., Calabon M., et al. Refined families of Sordariomycetes. Mycosphere. 2020;11:305–1059. doi: 10.5943/mycosphere/11/1/7. [DOI] [Google Scholar]
- 83.Schoch C.L., Robbertse B., Robert V., Vu D., Cardinali G., Irinyi L., Meyer W., Nilsson R.H., Hughes K., Miller A.N., et al. Finding needles in haystacks: Linking scientific names, reference specimens and molecular data for fungi. Database. 2014:336–341. doi: 10.1093/database/bau061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kwon S.L., Park M.S., Jang S., Lee Y.M., Heo Y.M., Hong J.H., Lee H., Jang Y., Park J.H., Kim C., et al. The genus Arthrinium (Ascomycota, Sordariomycetes, Apiosporaceae) from marine habitats from Korea, with eight new species. IMA Fungus. 2021;12:13. doi: 10.1186/s43008-021-00065-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Crous P., Hernandez-Restrepo M., Schumacher R.K., Cowan D.A., Maggs-Koelling G., Marais E., Wingfield M.J., Yilmaz N., Adan O.C.G., Akulov A., et al. New and interesting fungi. 4. Fungal Syst. Evol. 2021;7:255–343. doi: 10.3114/fuse.2021.07.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jiang N., Tian C.M. The holomorph of Arthrinium setariae sp. nov. (Apiosporaceae, Xylariales) from China. Phytotaxa. 2021;483:149–159. doi: 10.11646/phytotaxa.483.2.7. [DOI] [Google Scholar]
- 87.Jiang H.B., Hyde K.D., Doilom M., Karunarathna S.C., Xu J.C., Phookamsak R. Arthrinium setostromum (Apiosporaceae, Xylariales), a novel species associated with dead bamboo from Yunnan, China. Asian J. Mycol. 2019;2:254–268. doi: 10.5943/ajom/2/1/16. [DOI] [Google Scholar]
- 88.Wang M., Liu F., Crous P.W., Cai L. Phylogenetic reassessment of Nigrospora: Ubiquitous endophytes, plant and human pathogens. Persoonia. 2017;39:118–142. doi: 10.3767/persoonia.2017.39.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tennakoon D.S., Kuo C.H., Maharachchikumbura S.S.N., Thambugala K.M., Gentekaki E., Phillips A.J.L., Bhat D.J., Wanasinghe D.N., de Silva N.I., Promputtha I., et al. Taxonomic and phylogenetic contributions to Celtis formosana, Ficus ampelas, F. septica, Macaranga tanarius and Morus australis leaf litter inhabiting microfungi. Fungal Divers. 2021;108:1–215. doi: 10.1007/s13225-021-00474-w. [DOI] [Google Scholar]
- 90.Maharachchikumbura S.S.N., Hyde K.D., Groenewald J.Z., Xu J., Crous P.W. Pestalotiopsis revisited. Stud. Mycol. 2014;79:121–186. doi: 10.1016/j.simyco.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Klaubauf S., Tharreau D., Fournier E., Groenewald J.Z., Crous P.W., de Vries R.P., Lebrun M.H. Resolving the polyphyletic nature of Pyricularia (Pyriculariaceae) Stud. Mycol. 2014;79:85–120. doi: 10.1016/j.simyco.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hirata K., Kusaba M., Chuma I., Osue J., Nakayashiki H., Mayama S., Tosa Y. Speciation in Pyricularia inferred from multilocus phylogenetic analysis. Mycol. Res. 2007;111:799–808. doi: 10.1016/j.mycres.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 93.Luo J., Zhang N. Magnaporthiopsis, a new genus in Magnaporthaceae (Ascomycota) Mycologia. 2013;105:1019–1029. doi: 10.3852/12-359. [DOI] [PubMed] [Google Scholar]
- 94.Luo J., Walsh E., Zhang N. Toward monophyletic generic concepts in Magnaporthales: Species with Harpophora asexual states. Mycologia. 2015;107:641–646. doi: 10.3852/14-302. [DOI] [PubMed] [Google Scholar]
- 95.Yuan Z.L., Lin F.C., Zhang C.L., Kubicek C.P. A new species of Harpophora (Magnaporthaceae) recovered from healthy wild rice (Oryza granulata) roots, representing a novel member of a beneficial dark septate endophyte. FEMS Microbiol. Lett. 2010;307:94–101. doi: 10.1111/j.1574-6968.2010.01963.x. [DOI] [PubMed] [Google Scholar]
- 96.Crous P.W., Schumacher R.K., Akulov A., Thangavel R., Hernandez-Restrepo M., Carnegie A.J., Cheewangkoon R., Wingfield M.J., Summerell B.A., Quaedvlieg W., et al. New and interesting fungi. 2. Fungal Syst. Evol. 2019;3:57–134. doi: 10.3114/fuse.2019.03.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hernández-Restrepo M., Groenewald J.Z., Elliott M.L., Canning G., Mcmillan V.E., Crous P.W. Take-all or nothing. Stud. Mycol. 2016;83:19–48. doi: 10.1016/j.simyco.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang N., Zhao S., Shen Q. A six-gene phylogeny reveals the evolution of mode of infection in the rice blast fungus and allied species. Mycologia. 2011;103:1267–1276. doi: 10.3852/11-022. [DOI] [PubMed] [Google Scholar]
- 99.Wong P.T.W. Gaeumannomyces wongoonoo sp. nov., the cause of a patch disease of buffalo grass (St Augustine grass) Mycol. Res. 2002;106:857–862. doi: 10.1017/S0953756202006147. [DOI] [Google Scholar]
- 100.Khemmuk W., Geering A.D., Shivas R.G. Wongia gen. nov. (Papulosaceae, Sordariomycetes), a new generic name for two root-infecting fungi from Australia. IMA Fungus. 2016;7:247–252. doi: 10.5598/imafungus.2016.07.02.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vines P.L., Hoffmann F.G., Meyer F., Allen T.W., Luo J., Zhang N., Tomaso-Peterson M. Magnaporthiopsis cynodontis, a novel turfgrass pathogen with widespread distribution in the United States. Mycologia. 2020;112:52–63. doi: 10.1080/00275514.2019.1676614. [DOI] [PubMed] [Google Scholar]
- 102.Luo J., Vines P.L., Grimshaw A., Hoffman L., Walsh E., Bonos S.A., Clarke B.B., Murphy J.A., Meyer W.A., Zhang N. Magnaporthiopsis meyeri-festucae, sp. nov., associated with a summer patch-like disease of fine fescue turfgrasses. Mycologia. 2017;109:780–789. doi: 10.1080/00275514.2017.1400306. [DOI] [PubMed] [Google Scholar]
- 103.Luo J., Walsh E., Zhang N. Four new species in Magnaporthaceae from grass roots in New Jersey Pine Barrens. Mycologia. 2014;106:580–588. doi: 10.3852/13-306. [DOI] [PubMed] [Google Scholar]
- 104.Vines P.L. Master Thesis. Mississippi State University; Starkville, MI, USA: 2015. Evaluation of Ultradwarf Bermudagrass Cultural Management Practices and Identification, characterization, and Pathogenicity of Ectotrophic Rootinfecting Fungi Associated with Summer Decline of Ultradwarf Bermudagrass Putting Greens. [Google Scholar]
- 105.Liu J.K., Hyde K.D., Jones E.B.G., Ariyawansa H.A., Bhat D.J., Boonmee S., Maharachchikumbura S.S.N., McKenzie E.H.C., Phookamsak R., Phukhamsakda C., et al. Fungal diversity notes 1–110: Taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 2015;72:1–197. doi: 10.1007/s13225-015-0324-y. [DOI] [Google Scholar]
- 106.Park M.J., Shin H.D. A new species of Pyricularia on Commelina communis. Mycotaxon. 2009;108:449–456. doi: 10.5248/108.449. [DOI] [Google Scholar]
- 107.Ban Y., Tang M., Chen H., Xu Z., Zhang H., Yang Y. The response of dark septate endophytes (DSE) to heavy metals in pure culture. PLoS ONE. 2018. 7:e47968. doi: 10.1371/journal.pone.0047968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Saleh A.A., Leslie J.F. Cephalosporium maydis is a distinct species in the Gaeumannomyces-Harpophora species complex. Mycologia. 2004;96:1294–1305. doi: 10.1080/15572536.2005.11832879. [DOI] [PubMed] [Google Scholar]
- 109.Phookamsak R., Wanasinghe D.N., Hongsanan S., Phukhamsakda C., Huang S.K., Tennakoon D.S., Norphanphoun C., Camporesi E., Bulgakov T.S., Promputtha I., et al. Towards a natural classification of Ophiobolus and ophiobolus-like taxa; introducing three novel genera Ophiobolopsis, Paraophiobolus and Pseudoophiobolus in Phaeosphaeriaceae (Pleosporales) Fungal Divers. 2017;87:299–339. doi: 10.1007/s13225-017-0393-1. [DOI] [Google Scholar]
- 110.Hyde K.D., Norphanphoun C., Abreu V.P., Bazzicalupo A., Chethana K.W.T., Clericuzio M., Dayarathne M.C., Dissanayake A.J., Ekanayaka A.H., He M.Q., et al. Fungal diversity notes 603–708: Taxonomic and phylogenetic notes on genera and species. Fungal Divers. 2017;87:1–235. doi: 10.1007/s13225-017-0391-3. [DOI] [Google Scholar]
- 111.De Gruyter J., Aveskamp M.M., Woudenberg J.H., Verkley G.J., Groenewald J.Z., Crous P.W. Molecular phylogeny of Phoma and allied anamorph genera: Towards a reclassification of the Phoma complex. Mycol. Res. 2009;113:508–519. doi: 10.1016/j.mycres.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 112.Crous P.W., Wingfield M.J., Guarro J., Cheewangkoon R., van der Bank M., Swart W.J., Stchigel A.M., Cano-Lira J.F., Roux J., Madrid H., et al. Fungal planet description sheets: 154–213. Persoonia. 2013;31:188–296. doi: 10.3767/003158513X675925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Manawasinghe I.S., Pem D., Bundhun D., Karunarathna A., Ekanayaka A.H., Bao D.F., Li J.F., Samarakoon M.C., Chaiwan N., Lin C.G., et al. Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020;100:5–277. doi: 10.1007/s13225-020-00439-5. [DOI] [Google Scholar]
- 114.Wanasinghe D.N., Phukhamsakda C., Hyde K.D., Jeewon R., Lee H.B., Jones G.E.B., Tibpromma S., Tennakoon D.S., Dissanayake A.J., Jayasiri S.C., et al. Fungal diversity notes 709–839: Taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018;89:1–236. doi: 10.1007/s13225-018-0395-7. [DOI] [Google Scholar]
- 115.Tennakoon D.S., Thambugala K.M., Wanasinghe D.N., Gentekaki E., Promputtha I., Kuo C.H., Hyde K.D. Additions to Phaeosphaeriaceae (Pleosporales): Elongaticollum gen. nov., Ophiosphaerella taiwanensis sp. nov., Phaeosphaeriopsis beaucarneae sp. nov. and a new host record of Neosetophoma poaceicola from Musaceae. MycoKeys. 2020;70:59–88. doi: 10.3897/mycokeys.70.53674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Quaedvlieg W., Verkley G.J., Shin H.D., Barreto R.W., Alfenas A.C., Swart W.J., Groenewald J.Z., Crous P.W. Sizing up Septoria. Stud. Mycol. 2013;75:307–390. doi: 10.3114/sim0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wanasinghe D.N., Jones E.B.G., Camporesi E., Boonmee S., Karunarathna S.C., Thines M., Mortimer P.E., Xu J., Hyde K.D. Dematiopleospora mariae gen. sp. nov., from Ononis Spinosa in Italy. Cryptogamie. Mycol. 2014;35:105–117. doi: 10.7872/crym.v35.iss2.2014.105. [DOI] [Google Scholar]
- 118.Trakunyingcharoen T., Lombard L., Groenewald J.Z., Cheewangkoon R., To-Anun C., Alfenas A.C., Crous P.W. Mycoparasitic species of Sphaerellopsis, and allied lichenicolous and other genera. IMA Fungus. 2014;5:391–414. doi: 10.5598/imafungus.2014.05.02.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Phukhamsakda C., McKenzie E.H.C., Phillips A.J.L., Jones E.B.G., Bhat D.J., Stadler M., Bhunjun C.S., Wanasinghe D.N., Thongbai B., Camporesi E., et al. Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 2020;102:1–203. doi: 10.1007/s13225-020-00448-4. [DOI] [Google Scholar]
- 120.Cui Y., Jia H., He D., Yu H., Gao S., Yokoyama K., Li J., Wang L. Characterization of Edenia gomezpompae isolated from a patient with keratitis. Mycopathologia. 2013;176:75–81. doi: 10.1007/s11046-013-9667-7. [DOI] [PubMed] [Google Scholar]
- 121.Ariyawansa H.A., Hyde K.D., Jayasiri S.C., Buyck B., Chethana K.W.T., Dai D.Q., Dai Y.C., Daranagama D.A., Jayawardena R.S., Lücking R., et al. Fungal diversity notes 111–252—taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015;75:27–274. doi: 10.1007/s13225-015-0346-5. [DOI] [Google Scholar]
- 122.Abd-Elsalam K.A., Tibpromma S., Wanasinghe D.N., Camporesi E., Hyde K.D. Equiseticola gen. nov. (Phaeosphaeriaceae), from Equisetum sp. in Italy. Phytotaxa. 2016;284:169–180. doi: 10.11646/phytotaxa.284.3.2. [DOI] [Google Scholar]
- 123.Maharachchikumbura S.S.N., Ariyawansa H.A., Wanasinghe D.N., Dayarathne M.C., Al-Saady N.A., Al-Sadi A.M. Phylogenetic classification and generic delineation of Hydeomyces desertipleosporoides gen. et sp. nov., (Phaeosphaeriaceae) from Jebel Akhdar Mountain in Oman. Phytotaxa. 2019;391:28–38. doi: 10.11646/phytotaxa.391.1.2. [DOI] [Google Scholar]
- 124.Zhang J.F., Liu J.K., Jeewon R., Wanasinghe D.N., Liu Z.Y. Fungi from Asian Karst formations III. Molecular and morphological characterization reveal new taxa in Phaeosphaeriaceae. Mycosphere. 2019;10:202–220. doi: 10.5943/mycosphere/10/1/3. [DOI] [Google Scholar]
- 125.Tennakoon D.S., Hyde K.D., Phookamsak R., Wanasinghe D.N., Camporesi E., Promputtha I. Taxonomy and phylogeny of Juncaceicola gen. nov. (Phaeosphaeriaceae, Pleosporinae, Pleosporales) Cryptogamie Mycol. 2016;37:135–156. doi: 10.7872/crym/v37.iss2.2016.135. [DOI] [Google Scholar]
- 126.Karunarathna A., Phookamsak R., Jayawardena R.S., Hyde K.D., Kuo C.H. Kwanghwana miscanthi Karun., C.H. Kuo & K.D. Hyde, gen. et sp. nov. (Phaeosphaeriaceae, Pleosporales) on Miscanthus floridulus (Labill.) Warb. ex K. Schum. & Lauterb. (Poaceae) Cryptogamie. Mycol. 2020;41:119–132. doi: 10.5252/cryptogamiemycologie2020v41a6. [DOI] [Google Scholar]
- 127.Schoch C.L., Crous P.W., Groenewald J.Z., Boehm E.W., Burgess T.I., de Gruyter J., de Hoog G.S., Dixon L.J., Grube M., Gueidan C., et al. A class-wide phylogenetic assessment of Dothideomycetes. Stud. Mycol. 2009;64:1–15. doi: 10.3114/sim.2009.64.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.De Gruyter J., Woudenberg J.H., Aveskamp M.M., Verkley G.J., Groenewald J.Z., Crous P.W. Redisposition of phoma-like anamorphs in Pleosporales. Stud. Mycol. 2013;75:1–36. doi: 10.3114/sim0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Crous P.W., Verkley G.J., Groenewald J.Z. Eucalyptus microfungi known from culture. 1. Cladoriella and Fulvoflamma genera nova, with notes on some other poorly known taxa. Stud. Mycol. 2006;55:53–63. doi: 10.3114/sim.55.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hyde K.D., Hongsanan S., Jeewon R., Bhat D.J., McKenzie E.H.C., Jones E.B.G., Phookamsak R., Ariyawansa H.A., Boonmee S., Zhao Q., et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;80:1–270. doi: 10.1007/s13225-016-0373-x. [DOI] [Google Scholar]
- 131.Wijayawardene N.N., Hyde K.D., Wanasinghe D.N., Papizadeh M., Goonasekara I.D., Camporesi E., Bhat D.J., McKenzie E.H.C., Phillips A.J.L., Diederich P., et al. Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Divers. 2016;77:1–316. doi: 10.1007/s13225-016-0360-2. [DOI] [Google Scholar]
- 132.Phukhamsakda C., Ariyawansa H.A., Phookamsak R., Chomnunti P., Bulgakov T.S., Yang J.B., Bhat D.J., Bahkali A.H., Hyde K.D. Muriphaeosphaeria galatellae gen. et sp. nov. in Phaeosphaeriaceae (Pleosporales) Phytotaxa. 2015;227:55–65. doi: 10.11646/phytotaxa.227.1.6. [DOI] [Google Scholar]
- 133.Tibpromma S., Hyde K.D., Jeewon R., Maharachchikumbura S.S.N., Liu J.K., Bhat D.J., Jones E.B.G., McKenzie E.H.C., Camporesi E., Bulgakov T.S., et al. Fungal diversity notes 491–602: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2017;83:1–261. doi: 10.1007/s13225-017-0378-0. [DOI] [Google Scholar]
- 134.Thambugala K.M., Wanasinghe D.N., Phillips A.J.L., Camporesi E., Bulgakov T.S., Phukhamsakda C., Ariyawansa H.A., Goonasekara I.D., Phookamsak R., Dissanayake A., et al. Mycosphere notes 1–50: Grass (Poaceae) inhabiting Dothideomycetes. Mycosphere. 2017;8:697–796. doi: 10.5943/mycosphere/8/4/13. [DOI] [Google Scholar]
- 135.Mapook A., Boonmee S., Ariyawansa H.A., Tibpromma S., Campesori E., Jones E.B.G., Bahkali A.H., Hyde K.D. Taxonomic and phylogenetic placement of Nodulosphaeria. Mycol. Progs. 2016;15:1–15. doi: 10.1007/s11557-016-1176-x. [DOI] [Google Scholar]
- 136.Flores F.J., Marek S.M., Orquera G., Walker N.R. Molecular identification and multilocus phylogeny of Ophiosphaerella species associated with spring dead spot of bermudagrass. Crop. Sci. 2017;57:249. doi: 10.2135/cropsci2016.05.0437. [DOI] [Google Scholar]
- 137.Li W.J., Bhat D.J., Camporesi E., Tian Q., Wijayawardene N.N., Dai D.Q., Phookamsak R., Chomnunti P., Bahkali A.H., Hyde K.D. New asexual morph taxa in Phaeosphaeriaceae. Mycosphere. 2015;6:681–708. doi: 10.5943/mycosphere/6/6/5. [DOI] [Google Scholar]
- 138.Bakhshi M., Arzanlou M., Groenewald J.Z., Quaedvlieg W., Crous P.W. Parastagonosporella fallopiae gen. et sp. nov. (Phaeosphaeriaceae) on Fallopia convolvulus from Iran. Mycol. Prog. 2019;18:203–214. doi: 10.1007/s11557-018-1428-z. [DOI] [Google Scholar]
- 139.De Gruyter J., Woudenberg J.H.C., Aveskamp M.M., Verkley G.J.M., Groenewald J.Z., Crous P.W. Systematic reappraisal of species in Phoma section Paraphoma, Pyrenochaeta and Pleurophoma. Mycologia. 2010;102:1066–1081. doi: 10.3852/09-240. [DOI] [PubMed] [Google Scholar]
- 140.Camara M.P.S., Palm M.E., van Berkum P., O’Neill N.R. Molecular phylogeny of Leptosphaeria and Phaeosphaeria. Mycologia. 2002;94:630–640. doi: 10.1080/15572536.2003.11833191. [DOI] [PubMed] [Google Scholar]
- 141.Thambugala K.M., Camporesi E., Ariyawansa H.A., Phookamsak R., Liu Z.Y., Hyde K.D. Phylogeny and morphology of Phaeosphaeriopsis triseptata sp. nov., and Phaeosphaeriopsis glaucopunctata. Phytotaxa. 2014;176:238–250. doi: 10.11646/phytotaxa.176.1.23. [DOI] [Google Scholar]
- 142.Wijayawardene N.N., Camporesi E., Song Y., Dai D.Q., Hyde K.D. Multi-gene analyses reveal taxonomic placement of Scolicosporium minkeviciusii in Phaeosphaeriaceae (Pleosporales) Cryptogamie Mycol. 2013;34:357–366. doi: 10.7872/crym.v34.iss2.2013.357. [DOI] [Google Scholar]
- 143.Crous P.W., Carris L.M., Giraldo A., Groenewald J.Z., Hawksworth D.L., Hernandez-Restrepo M., Jaklitsch W.M., Lebrun M.H., Schumacher R.K., Stielow J.B., et al. The genera of fungi—Fixing the application of the type species of generic names—G 2: Allantophomopsis, Latorua, Macrodiplodiopsis, Macrohilum, Milospium, Protostegia, Pyricularia, Robillarda, Rotula, Septoriella, Torula, and Wojnowicia. IMA Fungus. 2015;6:163–198. doi: 10.5598/imafungus.2015.06.01.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chen Q., Jiang J.R., Zhang G.Z., Cai L., Crous P.W. Resolving the Phoma enigma. Stud. Mycol. 2015;82:137–217. doi: 10.1016/j.simyco.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ernst M., Mendgen K.W., Wirsel S.G. Endophytic fungal mutualists: Seed-borne Stagonospora spp. enhance reed biomass production in axenic microcosms. Mol. Plant-Microb. MPMI. 2003;16:580–587. doi: 10.1094/MPMI.2003.16.7.580. [DOI] [PubMed] [Google Scholar]
- 146.Senanayake I.C., Jeewon R., Camporesi E., Hyde K.D., Zeng Y.J., Tian S.L., Xie N. Sulcisporasupratumida sp. nov. (Phaeosphaeriaceae, Pleosporales) on Anthoxanthumodoratum from Italy. MycoKeys. 2018;38:35–46. doi: 10.3897/mycokeys.38.27729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ahmed S.A., Hofmuller W., Seibold M., de Hoog G.S., Harak H., Tammer I., van Diepeningen A.D., Behrens-Baumann W. Tintelnotia, a new genus in Phaeosphaeriaceae harbouring agents of cornea and nail infections in humans. Mycoses. 2016;60:244–253. doi: 10.1111/myc.12588. [DOI] [PubMed] [Google Scholar]
- 148.Aveskamp M.M., de Gruyter J., Woudenberg J.H., Verkley G.J., Crous P.W. Highlights of the Didymellaceae: A polyphasic approach to characterise Phoma and related pleosporalean genera. Stud Mycol. 2010;65:1–60. doi: 10.3114/sim.2010.65.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Devadatha B., Mehta N., Wanasinghe D.N., Baghela A., Venkateswara V., Vittaliana S. Vittaliana mangrovei Devadatha, Nikita, A. Baghela & V.V. Sarma, gen. nov, sp. nov. (Phaeosphaeriaceae), from mangroves near Pondicherry (India), based on morphology and multigene phylogeny. Cryptogamie Mycol. 2019;40:117–132. doi: 10.5252/cryptogamiemycologie2019v40a7. [DOI] [Google Scholar]
- 150.Marin-Felix Y., Hernández-Restrepo M., Iturrieta-González I., García D., Gené J., Groenewald J.Z., Cai L., Chen Q., Quaedvlieg W., Schumacher R.K., et al. Genera of phytopathogenic fungi: GOPHY 3. Stud. Mycol. 2019;94:1–124. doi: 10.1016/j.simyco.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lawrey J.D., Diederich P., Nelsen M.P., Freebury C., Van den Broeck D., Sikaroodi M., Ertz D. Phylogenetic placement of lichenicolous Phoma species in the Phaeosphaeriaceae (Pleosporales, Dothideomycetes) Fungal Divers. 2012;55:195–213. doi: 10.1007/s13225-012-0166-9. [DOI] [Google Scholar]
- 152.Karunarathna A., Papizadeh M., Senanayake I.C., Jeewon R., Phookamsak R., Goonasekara I.D., Wanasinghe D.N., Wijayawardene N.N., Amoozegar M.A., Shahzadeh-Fazeli S.A., et al. Novel fungal species of Phaeosphaeriaceae with an asexual/sexual morph connection. Mycosphere. 2017;8:1818–1834. doi: 10.5943/mycosphere/8/10/8. [DOI] [Google Scholar]
- 153.Suetrong S., Schoch C.L., Spatafora J.W., Kohlmeyer J., Volkmann-Kohlmeyer B., Sakayaroj J., Phongpaichit S., Tanaka K., Hirayama K., Jones E.B.G. Molecular systematics of the marine Dothideomycetes. Stud. Mycol. 2009;64:155–173. doi: 10.3114/sim.2009.64.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kolarik M., Spakowicz D., Gazis R., Shaw J., Novakova A., Chudickova M., Forcina G.C., Kang K.W., Kelnarova I., Skaltsas D., et al. Biatriospora (Ascomycota: Pleosporales) is an ecologically diverse genus including facultative marine fungi and endophytes with biotechnological potential. Plant Syst. Evol. 2017;303:35–50. doi: 10.1007/s00606-016-1350-2. [DOI] [Google Scholar]
- 155.Doilom M., Dissanayake A.J., Wanasinghe D.N., Boonmee S., Liu J.K., Bhat D.J., Taylor J.E., Bahkali A.H., McKenzie E.H.C., Hyde K.D. Microfungi on Tectona grandis (teak) in Northern Thailand. Fungal Divers. 2017;82:107–182. doi: 10.1007/s13225-016-0368-7. [DOI] [Google Scholar]
- 156.Liu J.K., Phookamsak R., Dai D.Q., Tanaka K., Jones E.B.G., Xu J.C., Chukeatirote E., Hyde K.D. Roussoellaceae, a new pleosporalean family to accommodate the genera Neoroussoella gen. nov., Roussoella and Roussoellopsis. Phytotaxa. 2014;181:1–33. doi: 10.11646/phytotaxa.181.1.1. [DOI] [Google Scholar]
- 157.Zhang J.F., Liu J.K., Hyde K.D., Yang W., Liu Z.Y. Fungi from Asian Karst formations II. Two new species of Occultibambusa (Occultibambusaceae, Dothideomycetes) from karst landforms of China. Mycosphere. 2017;8:550–559. doi: 10.5943/mycosphere/8/4/4. [DOI] [Google Scholar]
- 158.Li J., Bhat D.J., Phookamsak R., Mapook A., Lumyong S., Hyde K.D. Sporidesmioides thailandica gen. et sp nov (Dothideomycetes) from northern Thailand. Mycol. Prog. 2016;15:1169–1178. doi: 10.1007/s11557-016-1238-0. [DOI] [Google Scholar]
- 159.Jiang H.B., Phookamsak R., Hyde K.D., Mortimer P.E., Xu J.C., Kakumyan P., Karunarathna S.C., Kumla J. A taxonomic appraisal of bambusicolous fungi in Occultibambusaceae (Pleosporales, Dothideomycetes) with new collections from Yunnan Province, China. Life. 2021;11:932. doi: 10.3390/life11090932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ahmed S.A., van de Sande W.W., Stevens D.A., Fahal A., van Diepeningen A.D., Menken S.B., de Hoog G.S. Revision of agents of black-grain eumycetoma in the order Pleosporales. Persoonia. 2014;33:141–154. doi: 10.3767/003158514X684744. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in the NCBI GenBank (https://www.ncbi.nlm.nih.gov/), Index Fungorum (http://www.indexfungorum.org/Names/Names.asp) (all accessed on 8 May 2022).