Abstract
Background and Objective
Periodontitis is a multifactorial chronic inflammatory disease that can lead to the irreversible destruction of dental support tissues. As an epigenetic factor, the expression of circRNA is tissue‐dependent and disease‐dependent. This study aimed to identify novel periodontitis‐associated circRNAs and predict relevant circRNA‐periodontitis regulatory network by using recently developed bioinformatic tools and integrating sequencing profiling with clinical information for getting a better and more thorough image of periodontitis pathogenesis, from gene to clinic.
Material and Methods
High‐throughput sequencing and RT‐qPCR were conducted to identify differentially expressed circRNAs in gingival tissues from periodontitis patients. The relationship between upregulated circRNAs expression and probing depth (PD) was performed using Spearman's correlation analysis. Bioinformatic analyses including GO analysis, circRNA‐disease association prediction, and circRNA‐miRNA‐mRNA network prediction were performed to clarify potential regulatory functions of identified circRNAs in periodontitis. A receiver‐operating characteristic (ROC) curve was established to assess the diagnostic significance of identified circRNAs.
Results
High‐throughput sequencing identified 70 differentially expressed circRNAs (68 upregulated and 2 downregulated circRNAs) in human periodontitis (fold change >2.0 and p < .05). The top five upregulated circRNAs were validated by RT‐qPCR that had strong associations with multiple human diseases, including periodontitis. The upregulation of circRNAs were positively correlated with PD (R = .40–.69, p < .05, moderate). A circRNA‐miRNA‐mRNA network with the top five upregulated circRNAs, differentially expressed mRNAs, and overlapped predicted miRNAs indicated potential roles of circRNAs in immune response, cell apoptosis, migration, adhesion, and reaction to oxidative stress. The ROC curve showed that circRNAs had potential value in periodontitis diagnosis (AUC = 0.7321–0.8667, p < .05).
Conclusion
CircRNA‐disease associations were predicted by online bioinformatic tools. Positive correlation between upregulated circRNAs, circPTP4A2, chr22:23101560‐23135351+, circARHGEF28, circBARD1 and circRASA2, and PD suggested function of circRNAs in periodontitis. Network prediction further focused on downstream targets regulated by circRNAs during periodontitis pathogenesis.
Keywords: bioinformatics analysis, circRNA, circRNA‐disease association, high‐throughput sequencing, periodontitis
1. INTRODUCTION
Periodontitis, characterized by irreversible pathologic loss of periodontal ligament and alveolar bone, is a chronic complex infectious disease associated with a destructive host immune response to microbial dysbiosis. 1 , 2 In addition to an initial factor of periodontal biofilms, genetic and epigenetic factors were identified to affect periodontitis progression. 3 , 4 Apart from well‐revealed typical expression profiles of genes in diseased periodontal tissues, epigenetic factors, especially noncoding RNAs (ncRNAs) have been recently described as important players involved in periodontitis pathogenesis. 4 , 5 , 6
Among the noncoding RNAs, circular RNAs (circRNAs) are formed from the covalent linkage of the end of a single RNA molecule due to a back‐splicing variant of expression. 7 Through the formation of the covalently closed‐loop structure, the stability of circRNA could be substantially improved. 8 Most well‐known function of circRNAs is its action as “miRNA sponges,” in which circRNAs can competitively bind to miRNAs and thereby regulate target gene transcription. 9 Studies have indicated that circRNA expression is tissue‐dependent and disease‐dependent. 8 , 10 For periodontal tissue and disease, studies have looked at the difference in circRNA expression during human periodontal ligament stem cells (hPDLSCs) osteogenic differentiation. 11 , 12 , 13 Profiling of circRNA in the cell line provided evidences for functional validation of circRNAs regulation in hPDLSCs osteogenic differentiation in several more studies. 14 , 15 One recent study profiled circRNA expression in exosomes that were derived from hPDLSCs, whose results indicated that extracellular vesicles could act as a source of circRNA to regulate hPDLSCs osteogenic differentiation. 16 In addition to the regulation of osteogenic differentiation, other functional validation studies suggested that circRNAs are also involved in the regulation of hPDLSCs cell viability, apoptosis, proliferation, migration, response to inflammation and oxidative stress, and the maintenance of stemness. 17 , 18 , 19 , 20 , 21 Some recent studies have used human gingival tissue samples to profile circRNA expression, including studies that profiled circRNA expression in human gingival tissues from patients with periodontitis and loss‐of‐function studies that presented circRNA as a promising therapeutical target for periodontitis using human gingival fibroblasts derived from human gingival tissues. 22 , 23 , 24 , 25
While studies on the regulation of circRNA in periodontitis are progressing, emerging roles of circRNAs, such as RNA‐binding protein sponges, regulators of splicing and transcription, translation, and biomarkers, could act as a “game changer.” 7 , 8 , 10 , 26 , 27 , 28 Fortunately, multiple bioinformatic approaches have been developed to help us cope with challenges for circRNA detection and functional prediction. 26 , 29 , 30 , 31 , 32 , 33 , 34 Bioinformatic tools used in circRNA research can be classified into three main categories, including circRNA identification tools, circRNA annotation databases, and other tools for circRNA‐disease association prediction, and network analysis of competing endogenous RNAs. 26 These tools might allow us to have access to higher accuracy of target selection and improve the sensitivity of the prediction on circRNA regulations. At the same time, integrating sequencing profiling with clinical information could help broaden the knowledge for a better understanding of periodontitis pathogenesis. 35 , 36 , 37 , 38
The purpose of this study was to assess the perturbation in circular RNA expression profiles by high‐throughput sequencing and interrogate the association of aberrantly expressed circRNAs and the pathology of periodontitis. RT‐qPCR and correlation between circRNA expression levels and clinical parameter probing depth (PD) were conducted to strengthen the reliability of prediction. The circRNA‐miRNA‐mRNA network was predicted to suggest roles circRNAs played in periodontitis.
2. MATERIALS AND METHODS
2.1. Ethics statement
The study was approved by the Institutional Review Board (Ethics number: [2018]226) and complies with the Helsinki Declaration. Written informed consent was obtained from all participants involved in the study.
2.2. Subjects and sample collection
In total, 32 periodontal healthy patients who had extractions of impacted teeth and 36 patients with periodontitis who underwent periodontal flap surgery were included in this study. Patients with generalized periodontitis on Stage II to IV were classified as the group of Periodontitis. Among them, six paired samples of gingival tissues were used for transcriptome high‐throughput sequencing, and another 26 healthy and 30 inflamed gingival samples for further RT‐qPCR validation (Figure 1A). The inclusion criteria of periodontitis subjects were as follows: interdental clinical attachment loss (CAL) at the site of greatest loss ≥3 mm, radiographic bone loss >15%, tooth loss due to periodontitis ≤4 teeth, and more than 30% of teeth involved according to Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri‐Implant Diseases and Conditions. 2 The inclusion criteria of periodontal healthy subjects were as follows: individuals with PD ≤3 mm, CAL <1 mm, bleeding on probing (BOP) sites ≤10%, no tooth loss due to periodontitis, and no history of periodontal diseases. The exclusion criteria were as follows: (1) smoking history, (2) taking special medication that could affect periodontal status over the past 3 months, (3) pregnant, lactating, or menopausal status, (4) suffering from any systemic disease affecting the progression of periodontal diseases, such as obesity, diabetes, chronic renal disease, rheumatoid arthritis, and cardiovascular diseases. Samples of gingival tissues comprising the oral epithelium and connective tissue were obtained from the above‐mentioned individuals. After sample collection, the gingival tissue samples were washed with 0.9% normal saline and immediately stored at −80°C for further analysis.
FIGURE 1.
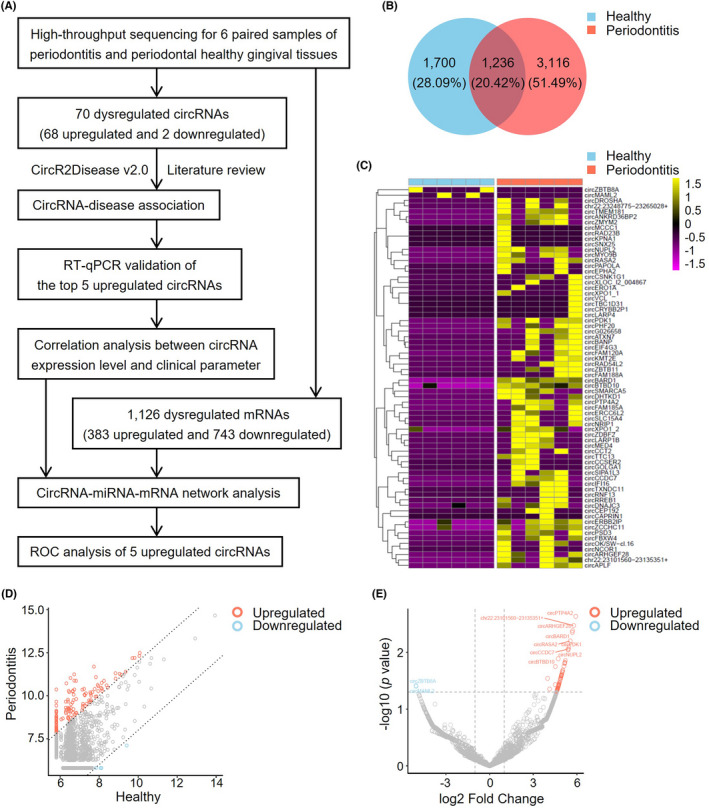
Analysis of differentially expressed circRNAs in human gingival tissues from patients with or without periodontitis. (A) Workflow of circRNA expression profiling and analysis in human periodontitis. (B) The number of the shared circRNAs and differentially expressed circRNAs. There are 1236 circRNA shared, 1700 circRNA expressed in healthy gingival tissues, and 3116 circRNA expressed in gingival tissues of periodontitis. (C) Heatmap of circRNA expression for gingival tissues from periodontitis patients when compared to periodontal healthy ones. Hierarchical clustering indicates differences in circRNA expression profiling between the two groups. The golden color indicates the upregulated expression and purple color for the downregulated expression. (D) Scatter plot displaying the changes in circRNA expression. The red scatters indicate the upregulated circRNAs, and the blue scatters indicate the downregulated circRNAs with more than a 2.0‐fold change between the two compared groups. The gray scatters indicate otherwise. (E) Volcano plot showing the expression profiling between the two compared groups. The vertical gray lines refer to a 2.0‐fold upregulation and downregulation, respectively. The horizontal gray line corresponds to a p value of .05 (−log10 scaled). The red points in the plot represent the significantly upregulated circRNAs, and the blue points represent the significantly downregulated circRNAs. The most significantly dysregulated circRNAs were highlighted in the figure
2.3. RNA‐sequencing analysis of circRNAs and mRNAs
Twelve samples of gingival tissues in the group of periodontal healthy (n = 6) and the group of periodontitis (n = 6) were collected for high‐throughput sequencing for circRNAs and mRNAs. For transcriptome high‐throughput sequencing, the total RNA was isolated from the gingival tissue samples and the RNA concentration was determined by NanoDrop ND‐1000 analysis (Thermo Fisher Scientific). Denatured agarose gel electrophoresis was used to measure RNA integrity and gDNA (genomic DNA) contamination. Ribo‐Zero rRNA Removal Kits (Illumina) were used to remove rRNAs from the total RNA. Libraries were prepared with rRNA‐depleted RNAs using TruSeq Stranded Total RNA Library Prep Kit (Illumina). RNA libraries were controlled for quality and further quantified using the BioAnalyzer 2100 system (Agilent Technologies). Libraries (10 pM) were denatured into single‐stranded DNA molecules, then captured on Illumina flow cells, and amplified in situ as clusters. It was followed by 150‐cycle sequence runs on an Illumina HiSeq™ 4000 sequencer (Illumina). Paired‐end reads were harvested and quality controlled by Q30. After 3′ adaptor‐trimming and low‐quality reads removal, the high‐quality reads were used for the analysis of circRNAs and mRNAs.
For circRNAs, the high‐quality trimmed reads were aligned to the reference genome/transcriptome with STAR software (v2.5.1b). DCC software (v0.4.4) was used to detect and identify circRNAs. edgeR software (v3.16.5) was used for normalization and to perform differentially expressed circRNA analysis. Significant differentially expressed transcripts were screened by fold change >2.0 and p value <.05. The location of parental genes of each circRNAs on chromosomes was reported. For mRNAs, were aligned with the human reference genome (UCSC hg19) using hisat2 software (v2.0.4). Under the guidance of the Ensembl gtf gene annotation file, cuffdiff software (v2.2.1, part of cufflinks) was used to get the FPKM as mRNA expression profile. Fold change and p value were calculated based on FPKM. Significant differential expressed transcripts were screened by fold change ≥1.5 and p value <.05.
2.4. Gene ontology analysis
The Gene Ontology project provides a controlled vocabulary to describe gene and gene product attributes in any organism (http://www.geneontology.org). The Fisher's exact test is used to find if there is more overlap between the gene list and the GO annotation list than would be expected by chance. The p value denotes the significance of GO terms enrichment in the host genes of differentially expressed circRNAs. The biological items exhibiting p < .05 were regarded as indicative of statistical significance in GO analysis, including molecular functions (MF), biological processes (BP), and cellular components (CC).
2.5. Predicting circRNA‐disease associations using CircDis model based on heterogeneous networks
The investigation of circRNA‐disease associations was conducted using the CircR2Disease v2.0 database. 39 CircR2Disease v2.0 collects experimentally validated circRNA‐disease associations across 3077 circRNAs and 312 diseases. The study examined the circRNA‐disease association score provided by the CircDis model, a computational method that combines graph convolutional networks and gradient boosting decision trees to predict the potential circRNA‐disease associations. CircDis outputs a predicted score for each circRNA‐disease association, ranging from 0 to 1 (significant with a score >0.5; insignificant otherwise).
At the same time, we confirmed disease‐circRNA associations by searching the latest published literature (Figure 1A). Search strategy was as follows: Sources of studies included the PubMed and Web of Science online databases for the circRNAs research published in English until December 3, 2021. The key words we used in the research were (“circPTP4A2” or “hsa_circ_0007364” or “circARHGEF28” or “hsa_circ_0005777” or “circBARD1” or “hsa_circ_0002999” or “circRASA2” or “hsa_circ_0067582” or “chr22:23101560–23135351+”) and (“circRNA” or “circular RNA”). Two researchers searched the titles, abstracts, and full‐text articles. Other researchers were involved in data extraction with the two previous researchers. Any disagreement was solved by a third researcher. Data were extracted from the articles as entries in the appropriate table. Articles in which circRNAs were reported with dysregulation in expression profile or analyzed circRNA‐disease association were included. Articles not in English, reviews, conference summaries, editorials, commentaries, or articles with incomplete data were excluded.
2.6. RT‐qPCR validation for circRNAs expression levels and clinical significance
The sample size of the periodontal healthy (n = 26) and the periodontitis (n = 30) were expanded for the RT‐qPCR validation. To confirm the high‐throughput sequencing data, RT‐qPCR was performed on the top five dysregulated circRNAs (fold change >2.0, p < .05) in high‐throughput sequencing. Total RNA was isolated from the gingival tissue samples with TRIzol reagent (Invitrogen Life Technologies). The RNA was reverse transcribed into cDNA using PrimeScript™ RT Master Mix (Takara Bio), which contains a random primer. RT‐qPCR was performed by FastStart Universal SYBR Green Master Mix (Roche) on the QuantStudio 7 Flex Real‐Time PCR System (Applied Biosystems). The divergent primers were used, which were designed and synthesized by Sangon Biotech according to the circRNAs’ back‐splice sites. The primer sequences are listed in Table S1. After normalization to GAPDH, the relative RNA expression was presented via the delta C t method. All experiments were repeated in triplicate. Correlation between the expression level of the top 5 upregulated circRNAs and clinical parameter PD was conducted by Spearman's correlation analysis. Significance was expressed as: *p < .05, **p < .01, ***p < .001, and ****p < .0001.
2.7. Prediction of circRNA‐miRNA‐mRNA networks
Pathway analysis was performed on the mRNA‐seq data using gene set enrichment analysis (GSEA) by function GSEA in R package clusterProfiler (v 4.0.5), with hallmark gene sets obtained from Molecular Signatures Database (v7.5.1, https://www.gsea‐msigdb.org/gsea/msigdb/index.jsp). Normalized Enrichment Score (NES) >0.5 and adjusted p value <.05 were set as the cut‐off criteria.
A circRNA‐miRNA‐mRNA network was constructed to reveal the interactions in circRNA, miRNA, and mRNA. The top 5 upregulated circRNAs were selected for prediction of circRNA‐miRNA interaction based on miRanda and Targetscan databases, as previously reported. 13 The binding potential of miRNAs to upregulated mRNAs was predicted by the prediction software TargetScan and starBase 2.0. The overlap of predicted miRNAs from upregulated circRNAs and upregulated mRNAs was investigated to construct a circRNA‐miRNA‐mRNA network. Cytoscape software (v 3.9.0) was employed to construct networks to visualize interactions between these molecules. Gene Ontology terms were used to indicate the potential functions of enrolled mRNAs.
2.8. Statistical analysis
All data were statistically analyzed using R v4.0.3 (Foundation for Statistical Computing) and GraphPad Prism 8 (GraphPad Software), and the data were presented as mean ± standard deviation (SD). The Chi‐square test, Student's t‐test, and Mann‐Whitney test were used to compare two groups of independent samples as appropriate. The study participants were divided based on their gender, and two‐way ANOVA was used among subgroups. Correlation between the expression level of the top 5 upregulated circRNAs and mRNAs was conducted based on RNA‐sequencing by Spearman's correlation analysis. The potential diagnostic values of significantly differentially expressed circRNAs were evaluated through receiver‐operating characteristic (ROC) analysis. Significance was expressed as: *p < .05, **p < .01, ***p < .001, and ****p < .0001.
3. RESULTS
3.1. High‐throughput sequencing identified upregulated circRNAs that have strong associations with multiple human diseases, including periodontitis
Six paired gingival samples of healthy and periodontitis were used for high‐throughput sequencing analysis of the expression profile of circular RNAs and bioinformatic analysis (Figure 1A, Table 1). In total, 6052 circRNA transcripts were detected in the human gingival tissue samples, located in genes across all chromosomes. Of these transcripts, there were 1236 shared circRNAs and 4816 differentially expressed transcripts in periodontitis gingival tissues compared with healthy ones (Figure 1B). A total of 70 circRNAs were significantly differentially regulated (Table S2), with 68 upregulated and 2 downregulated circRNAs in gingival tissues of periodontitis compared with healthy ones (fold change >2.0 and p < .05). The circRNA expression levels for two groups were quite different (Figure 1C–E). RNA integrity and gDNA contamination test by Denaturing Agarose Gel Electrophoresis were presented in Figure S1.
TABLE 1.
Demographic and clinical characteristics of the group of periodontal healthy and the group of periodontitis for high‐throughput sequencing
| Characteristics | Healthy (n = 6) | Periodontitis (n = 6) |
|---|---|---|
| Age range (years) | 20–29 | 25–37 |
| Age (years) | 24.83 ± 3.31 | 31.33 ± 5.13* |
| Gender (male/female) | 2/4 | 3/3 |
| BMI (kg/m2) | 19.84 ± 1.11 | 20.39 ± 2.71 |
| Tooth loss due to periodontitis | 0.00 ± 0.00 | 1.67 ± 0.82** |
| Tooth number | 28.00±0.00 | 26.33 ±0.82** |
| PD (mm) | 1.94 ± 0.38 | 3.67 ± 0.90** |
| CAL (mm) | 0.00 ± 0.00 | 3.08 ± 0.87** |
| BOP % | 7.94 ± 1.29 | 92.24 ± 5.10** |
Data are expressed as mean ± SD. Statistical significance is indicated as *p < .05, **p < .01, ***p < .001, and ****p < .0001.
Abbreviations: BMI, body mass index; BOP, bleeding on probing; CAL, clinical attachment loss; PD, probing depth.
Among the 68 upregulated circRNAs, 58 were included in the circBase and 10 circRNAs were identified as novel. These circRNAs belong to several catalogs, with 57 being exonic, 2 intergenic, 5 sense overlapping, 2 intronic and 2 antisense. The details of the top 10 upregulated and 2 downregulated circRNAs were presented (Table 2). GO analysis presented a total number of 309 terms about the potential functions of the differentially expressed circRNAs, including cellular component (CC), biological process (BP) and molecular function (MF) (Figure 2A–C). The largest proportion of target genes was in the nuclear part. The primary BP identified associated with the upregulated circRNAs was about the catabolic and metabolic process. The MF predicted among the upregulated circRNAs were binding, protein transporter activity, and Ras GTPase activator activity. The parental genes of 70 differentially expressed circRNAs were located in almost all chromosomes (Figure 2D).
TABLE 2.
Top 10 significantly upregulated and two downregulated circRNAs ranked by fold change in high‐throughput sequencing
| CircRNA | Catalog | Chromosomal location | CircBase ID | Log FC | p value |
|---|---|---|---|---|---|
| circPTP4A2 | Exonic | chr1:32381496‐32385259‐ | hsa_circ_0007364 | 5.9157 | .0023 |
| chr22:23101560‐23135351+ | Intergenic | chr22:23101560‐23135351+ | novel | 5.7801 | .0034 |
| circARHGEF28 | Exonic | chr5:73136305‐73136585+ | hsa_circ_0005777 | 5.7015 | .0042 |
| circBARD1 | Exonic | chr2:215617171‐215661841‐ | hsa_circ_0002999 | 5.6773 | .0044 |
| circRASA2 | Exonic | chr3:141231005‐141259451+ | hsa_circ_0067582 | 5.5417 | .0063 |
| circPDK1 | Exonic | chr2:173423436‐173460751+ | novel | 5.4459 | .0080 |
| circCCDC7 | Sense overlapping | chr10:32854486‐32873232+ | hsa_circ_0008679 | 5.4090 | .0087 |
| circNUPL2 | Exonic | chr7:23224689‐23226765+ | hsa_circ_0001683 | 5.3956 | .0090 |
| circANKRD36BP2 | Exonic | chr2:89082251‐89092011+ | novel | 5.2080 | .0139 |
| circMYO9B | Exonic | chr19:17212470‐17213367+ | hsa_circ_0000907 | 5.1745 | .0150 |
| circZBTB8A | Exonic | chr1:33058532‐33059355+ | hsa_circ_0011422 | −5.0565 | .0385 |
| circMAML2 | Exonic | chr11:95825056‐95826681‐ | hsa_circ_0024085 | −5.0384 | .0395 |
FIGURE 2.
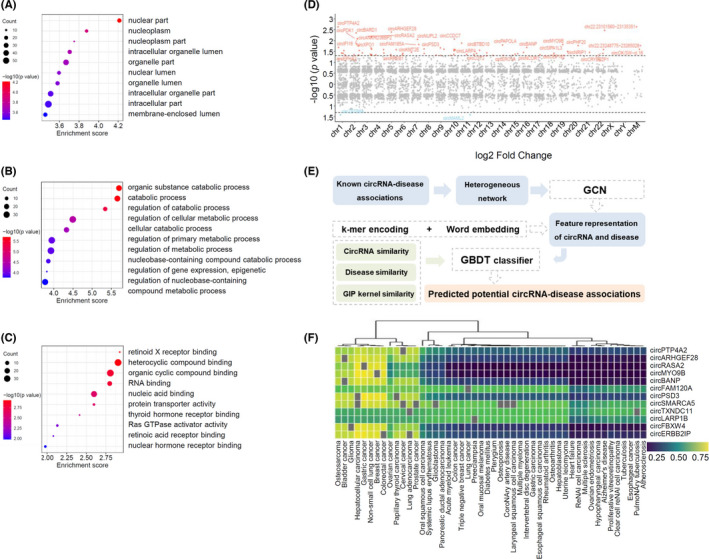
Functional prediction of differentially expressed circRNAs by GO analysis, chromosome location, and online database. (A–C) Gene ontology analysis of selected circRNAs, including cellular component (A), biological process (B), and molecular function (C). Term is the functional description information of GO and count is the number of genes associated with the listed terms. Dot plot shows the enrichment score values of the top most significant enrichment terms. The p value denotes the significance of GO terms enrichment in the genes. The enrichment score was calculated as ‐log10 (p value). (D) Distribution of significantly differentially expressed circRNAs in human chromosomes. (E) Schematic of circRNA‐disease associations prediction by CircDis trained on CircR2Disease v2.0. GCN is used to extract circRNA features and disease features from known circRNA‐disease associations. k‐mer encoding and word embedding help workout feature representation of circRNAs and diseases. Then, circRNA functional similarity, disease similarity, and GIP kernel for circRNAs and diseases are calculated respectively. And GBDT classifier is applied to predict the potential circRNA‐disease associations. GBDT, gradient boosting decision tree; GCN, graph convolutional network; GIP, Gaussian interaction profile. (F) CircRNA‐disease associations predicted by CircR2Disease v2.0. Twelve upregulated circRNAs are predicted to associate with 48 diseases based on CircDis model, with circRNA‐disease association score ranging from 0 to 1 (significant with score >.5; insignificant otherwise)
The investigation of circRNA‐disease associations was based on CircR2Disease v2.0 database (Figure 2E). Among the CircDis‐predicted association for 202 diseases, 48 diseases were predicted to associate with at least one of the 12 upregulated circRNAs reported in CircDis (Figure 2F). In particular, CircDis predicted that the top five upregulated circRNAs in this study are associated with many cancer‐related diseases, such as circPTP4A2‐prostate cancer (0.758), circPTP4A2‐hepatocellular carcinoma (0.762), circARHGEF28‐prostate cancer (0.747), circARHGEF28‐hepatocellular carcinoma (0.778), and circRASA2‐hepatocellular carcinoma (0.790). Upregulated circRNAs were reported to associate with diseases, including circPTP4A2, circARHGEF28, and circRASA2 (Table 3). CircPTP4A2 was dysregulated in glioma and diabetic kidney disease and could promote cervical cancer. 40 , 41 , 42 CircARHGEF28 was associated with cancers, such as bladder cancer, gastric cancer, and non‐small‐cell lung cancer. 43 , 44 , 45 CircRASA2 was downregulated in gastric cancer and associated with tumor diameter and stages. 46 , 47 Some of the putative circRNA‐disease associations were overlapped on experimentally validated CircR2Disease v2.0 database. Of note, circARHGEF28, one of the upregulated circRNAs in this study, was reported to have a higher expression level than normal controls in periodontitis. 22
TABLE 3.
Function of identified circRNAs in other tissues and disease models
| CircRNA | Disease | Function | Cell and tissue type | Study method | PMID |
|---|---|---|---|---|---|
| hsa_circ_0007364 (circPTP4A2) | Cervical cancer |
Upregulated in cervical cancer Promote cervical cancer progression via miR‐101‐5p/MAT2A axis Promote the proliferation and invasion of cervical cancer cell lines Promote cervical cancer growth in vivo |
Cervical cancer and adjacent nontumor tissues Cervical cancer cell lines (HeLa, CaSki, SiHa, C‐33A, C‐4I, SW756) Normal human cervical epithelial cell line (End1/E6E7) |
GEO microarray datasets (GSE113696, GSE102686), RT‐qPCR, Luciferase reporter assay, RIP, cell proliferation assay, cell invasion assay, tumor xenograft in vivo | 33138667 a |
| hsa_circ_0007364 (circPTP4A2) | Glioma | Downregulated in glioma | Glioma and normal brain tissues | GEO microarray dataset (GSE86202) | 31895689 |
| hsa_circ_0007364 (circPTP4A2) | Diabetic kidney disease | Upregulated in human renal proximal tubular epithelial cell line in response to glucose stress | Immortalized human renal proximal tubular epithelial cell line (HK‐2) | Microarray analysis, RT‐qPCR | 32115515 |
| hsa_circ_0005777 (circARHGEF28) | Bladder cancer |
Downregulated in bladder cancer Promote bladder cancer progression through miR‐1305/Tgf‐β2/smad3 pathway targeting EMT Accelerate bladder cancer cells’ progression, including cell viability, proliferation, invasion, migration, and wound healing potential of bladder cancer cell lines |
Bladder cancer and adjacent nontumor tissues Bladder cancer cell lines (5637, UM‐UC‐3) |
RNA‐sequencing, RT‐qPCR, FISH, Dual‐luciferase reporter assay, RNA pulldown assay, cell viability assay, cell proliferation assay, cell migration and matrigel invasion assay, wound healing assay, Western blot, tumor subcutaneous mice model | 32019579 a |
| hsa_circ_0005777 (circARHGEF28) | Gastric cancer | Downregulated in gastric cancer | Gastric cancer and adjacent nontumor tissues | GEO microarray datasets (GSE78092, GSE83521, GSE93541, and GSE100170) | 31346318 |
| hsa_circ_0005777 (circARHGEF28) | Non‐small cell lung cancer |
Upregulated in non‐small‐cell lung cancer Promote non‐small‐cell lung cancer progression via miR‐671‐5p/FOXM1 axis CircRNA expression associated with tumor size, nodule metastasis, and cancer staging |
Non‐small‐cell lung cancer and adjacent nontumor tissues Immortalized lung epithelial cell line (16HBE) Non‐small‐cell lung cancer cell lines (A549, H460, HCC827) |
RT‐qPCR, Western blot, RIP, FISH, RNA pulldown assay, Luciferase reporter assay, cell proliferation assay, cell migration assay | 34291441 |
| hsa_circ_0005777 (circARHGEF28) | Periodontitis | Upregulated in periodontitis | Human gingival tissues | RNA‐sequencing | 31021476 |
| hsa_circ_0067582 (circRASA2) | Gastric cancer |
Downregulated in gastric cancer Associated with gastric cancer patients’ tissue CEA level positively Biomarker for gastric cancer diagnosis |
Gastric cancer and adjacent non‐tumor tissues Healthy gastric mucosa, gastritis mucosa, and gastric intestinal metaplasia tissues |
RT‐qPCR | 31328820 a |
| hsa_circ_0067582 (circRASA2) | Gastric cancer |
Downregulated in gastric cancer Associated with tumor diameter positively and CA19‐9 level negatively Biomarker for gastric cancer diagnosis and prognosis evaluation |
Gastric cancer and adjacent nontumor tissues | RT‐qPCR | 31721300 a |
Abbreviations: CA19‐9, carbohydrate antigen 19‐9; CEA, carcinoembryonic antigen; EMT, epithelial‐mesenchymal transition; FISH, florescent in situ hybridization; GEO, Gene Expression Omnibus; RIP, RNA immunoprecipitation.
CircRNA‐disease associations predicted by online CircR2Disease v2.0 database.
3.2. Validation of top five upregulated circRNAs identified with clinical profiling
Five circRNAs with largest fold change and p < .05 in high‐throughput sequencing, including circPTP4A2, chr22:23101560‐23135351+, circARHGEF28, circBARD1, and circRASA2 were validated in RT‐qPCR (Table 4). The relative expression levels of selected circRNAs on RNA‐sequencing expressed as log2 (Fold Change) (Periodontitis vs. Healthy), ranged from 5.54 to 5.92 (Figure 3A). All five circRNAs were significantly upregulated in periodontitis than in healthy tissues with statistical significance (Figure 3B–F).
TABLE 4.
Demographic and clinical characteristics of the group of periodontal healthy and the group of periodontitis for RT‐qPCR
| Characteristics | Healthy (n = 26) | Periodontitis (n = 30) |
|---|---|---|
| Age range (years) | 24–50 | 26–51 |
| Age (years) | 33.46 ± 6.25 | 36.67 ± 7.84 |
| Gender (male/female) | 14/12 | 15/15 |
| BMI (kg/m2) | 20.74 ± 1.04 | 20.46 ± 1.49 |
| Tooth loss due to periodontitis | 0.00 ± 0.00 | 0.40 ± 0.81* |
| Tooth number | 28.00 ± 0.00 | 27.60 ± 0.81* |
| PD (mm) | 2.19 ± 0.57 | 6.53 ± 1.76**** |
| CAL (mm) | 0.00 ± 0.00 | 6.97 ± 2.44**** |
| BOP % | 6.87 ± 1.97 | 76.60 ± 13.34**** |
Data are expressed as mean ± SD. Statistical significance is indicated as *p < .05, **p < .01, ***p < .001, and ****p < .0001.
Abbreviations: BMI, body mass index; BOP, bleeding on probing; CAL, clinical attachment loss; PD, probing depth.
FIGURE 3.
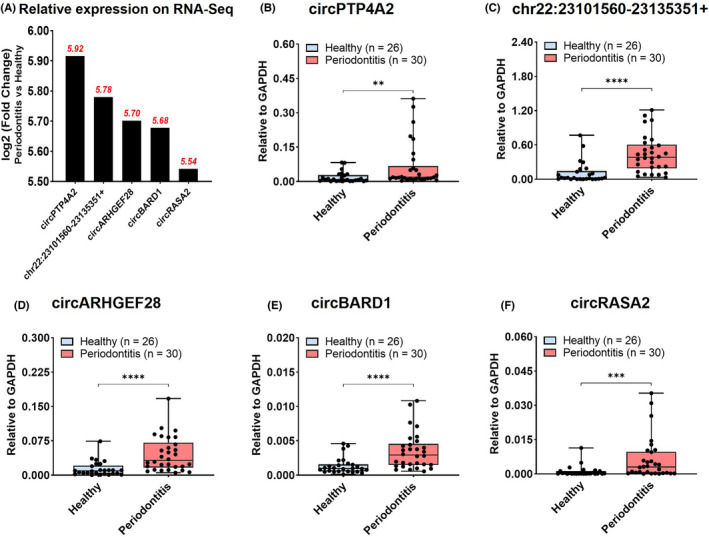
Identification and validation of the top five upregulated circRNAs expression. (A) Relative expression of five selected circRNAs on high‐throughput sequencing results. (B‐F) Relative expression of circRNAs in gingival tissues from patients with periodontitis (n = 30) and periodontal healthy donors (n = 26) by RT‐qPCR analysis, including circPTP4A2 (B), chr22:23101560‐23135351+ (C), circARHGEF28 (D), circBARD1 (E) and circRASA2 (F). Box and whiskers from Min to Max, showing all points. Significance was expressed as: *p < .05, **p < .01, ***p < .001 and ****p < .0001
The relationships between upregulated circRNAs, including circPTP4A2, chr22:23101560‐23135351+, circARHGEF28, circBARD1 and circRASA2, and clinical parameter PD were investigated. As shown in Figure 4A, there were significant and positive correlations between the top five upregulated circRNAs in PD level (R = .40–.69, p < .05, moderate). We then further grouped study participants based on their gender. While significant differences still existed between healthy and periodontitis in the expression levels of top five circRNAs, no significant differences were identified between males and females (Figure 4B). Significant and positive correlations between the five upregulated circRNAs and PD levels were observed in males and females, respectively (R = .40–.69, p < .05, moderate) (Figure 4C).
FIGURE 4.
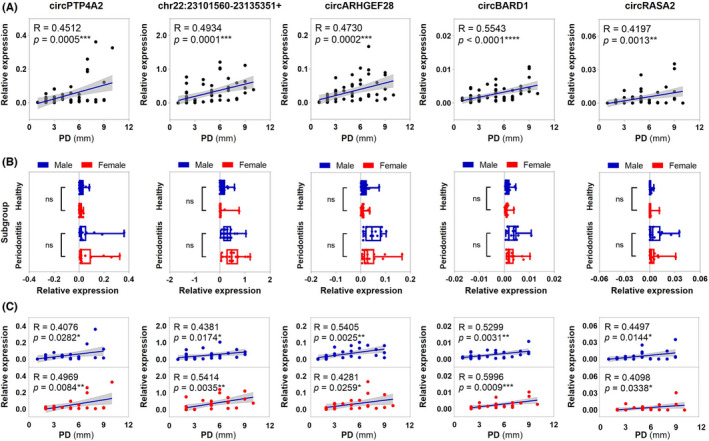
Correlation between relative expression levels of the top five upregulated circRNAs and probing depth. (A) Spearman's correlation analysis was used to assess correlations between circRNA relative expression and probing depth. p < .05 was considered statistically significant. (B) Participants were further divided based on their gender. Four subgroups included male healthy, female healthy, male periodontitis, and female periodontitis. Two‐way ANOVA was used to compare relative expression levels among gender subgroups. (C) Correlations between circRNA relative expression and probing depth were analyzed using Spearman's correlation analysis in male and female participants respectively. Significance was expressed as: *p < .05, **p < .01, ***p < .001, and ****p < .0001
3.3. Construction of the circRNA‐miRNA‐mRNA network
High‐throughput sequencing analysis of the expression profile of mRNAs detected 20 308 transcripts in the six paired human gingival tissue samples. Of these transcripts, there were 1126 differentially expressed mRNAs, with 383 upregulated and 743 downregulated mRNAs in gingival tissues of periodontitis compared with healthy ones (fold change ≥1.5 and p < .05). The expression profiles showed the mRNA expression levels were quite distinguished and different between two groups (Figure 5A–C). To identify the biological functional mechanism in periodontitis, the expression profile of mRNAs was used for GSEA. GSEA revealed that the top significantly enriched hallmark gene sets in periodontitis included TNF‐α signaling via NF‐κB, IL2‐STAT5 signaling, KRAS signaling, inflammatory response, allograft rejection, and myogenesis, inferring that differentially expressed mRNAs were closely associated with the inflammatory characteristics of periodontitis (Figure 5D).
FIGURE 5.
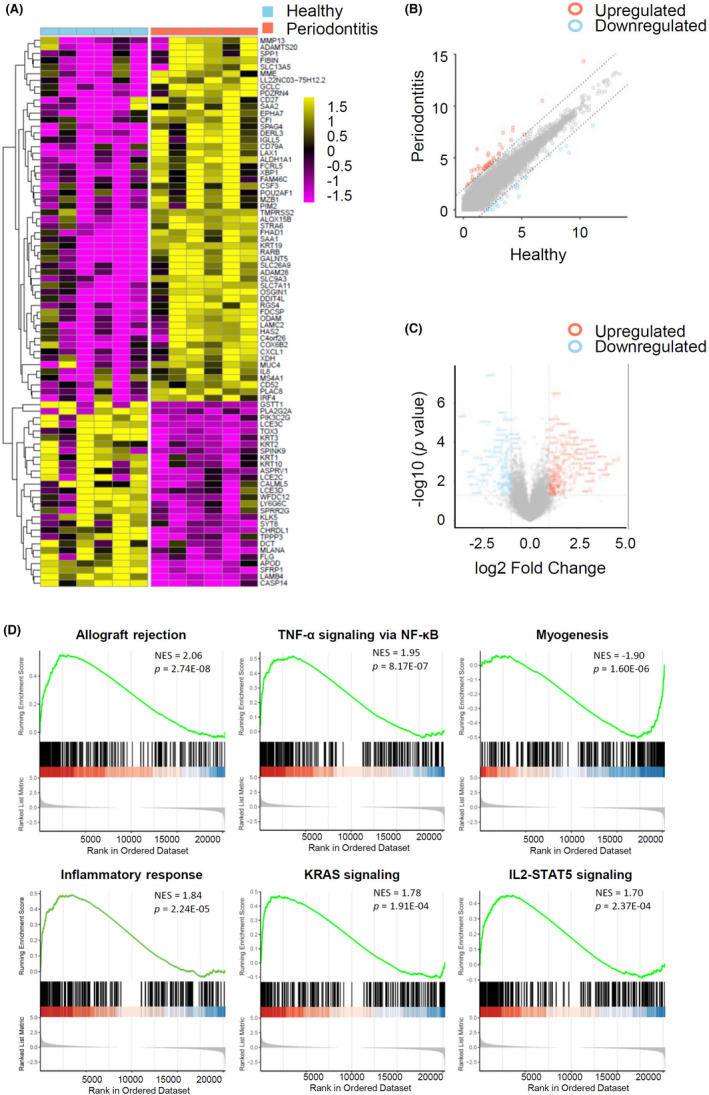
Expression profiling and GSEA of mRNA. (A) Heatmap displaying differentially expressed mRNAs across gingival tissues from periodontitis patients and periodontal healthy individuals. Gene expression was z‐scored transformed by row. The golden color indicates the upregulated expression and purple color for the downregulated expression (log2 FC >1.8, p < .05). (B) Scatter plot displaying the changes in mRNA expression. The red scatters indicate the upregulated mRNAs, and the blue scatters indicate the downregulated mRNAs with log2 FC >2.0 between the two compared groups. The gray scatters indicate otherwise. (C) Volcano plot showing the expression profiling between the two compared groups. The vertical gray lines refer to a 2.0‐fold change upregulation and downregulation, respectively. The horizontal gray line corresponds to a p value of .05 (−log10 scaled). The red points in the plot represent the significantly upregulated mRNAs, and the blue points represent the significantly downregulated mRNAs. (D) The GSEA of DEGs datasets. The top portion of plots shows the enrichment scores for each gene, and the bottom portion shows the ranked genes. y‐axis: ranking metric, x‐axis: individual ranks for all genes
Based on the upregulated mRNAs, a circRNA‐miRNA‐mRNA network was constructed regarding the top five upregulated circRNAs identified. Using miRanda, Targetscan and starBase 2.0 database, the predicted miRNAs for circRNA‐miRNA‐mRNA network have conserved binding sites to both circRNAs and mRNAs. There were 5 circRNAs, 30 miRNAs, and 67 mRNAs shown in Figure 6A. Predicted mRNAs regulated by the identified upregulated circRNAs were further categorized into nine functional clusters, namely positive regulation of cell adhesion, positive regulation of cytokine production, regulation of B cell activation, regulation of T cell activation, leukocyte aggregation, regulation of innate immune response, response to oxidative stress, regulation of apoptotic signaling pathway and myeloid leukocyte migration (Figure 6A).
FIGURE 6.
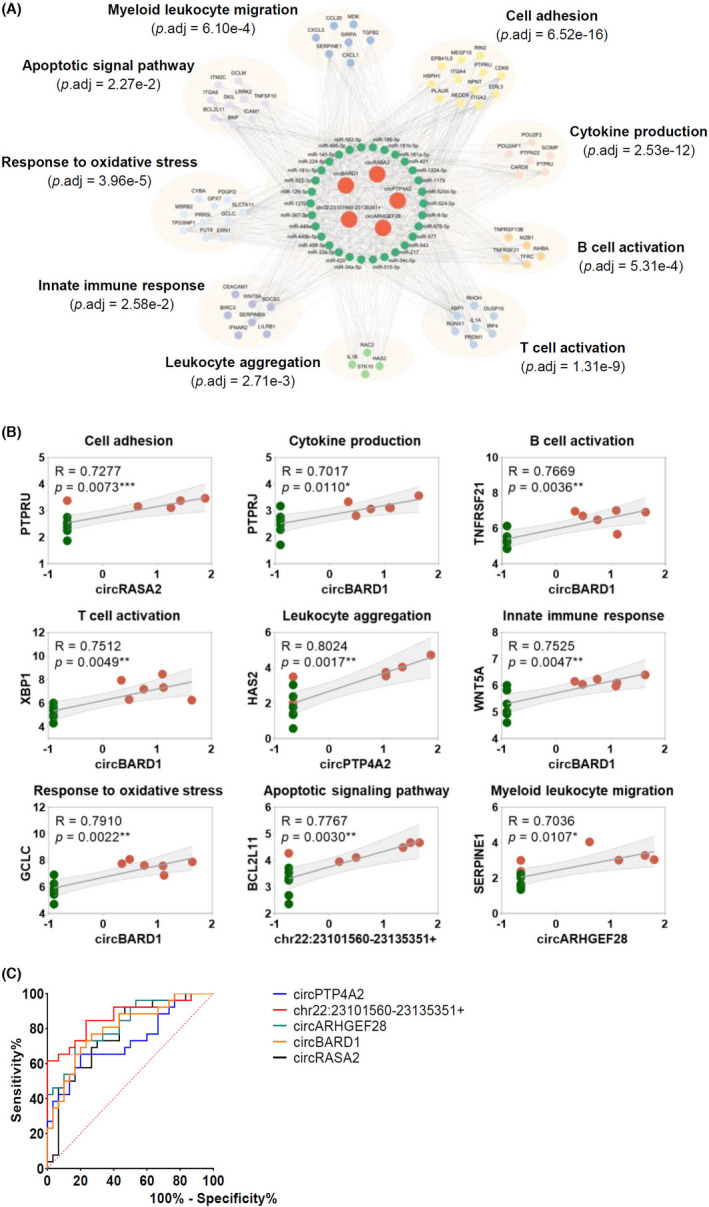
Construction of circRNA‐miRNA‐mRNA network and ROC analysis of identified upregulated circRNAs. (A) CircRNA‐miRNA‐mRNA network analysis for the five upregulated circRNAs and differentially expressed mRNAs. (B) Correlations between the five upregulated circRNAs expression and 9 mRNAs expression in high‐throughput sequencing were analyzed by Spearman's correlation analysis. The 9 mRNAs were selected as representatives for nine clusters of circRNA network. p < .05 was considered statistically significant. (C) ROC curves of identified circRNAs in gingival tissues from patients with periodontitis. The largest AUC was demonstrated for chr22:23101560‐23135351+ (0.8667), followed by circARHGEF28 (0.8244), circBARD1 (0.8013), circRASA2 (0.7731), and circPTP4A2 (0.7321). y‐axis indicates the true positive rate of the risk prediction. x‐axis indicates the false‐positive rate of the risk
The correlation between the expression levels of top 5 upregulated circRNAs and mRNAs was also conducted. In each mRNA cluster, the circRNA encompassing the highest correlation coefficient was reported and shown in Figure 6B, ranging between .70 and .89 (p < .05), indicating a strong correlation between circRNA and mRNA expression levels. Details of circRNA‐miRNA‐mRNA networks were presented in Table S3.
3.4. Diagnostic values of circRNAs in periodontitis
ROC curve analysis was performed on circPTP4A2, chr22:23101560‐23135351+, circARHGEF28, circBARD1, and circRASA2 in periodontitis diagnosis (Figure 6C). The largest area under the curve (AUC) was demonstrated for chr22:23101560‐23135351+ (0.8667), followed by circARHGEF28 (0.8244), circBARD1 (0.8013), circRASA2 (0.7731), and circPTP4A2 (0.7321). Details of sensitivity, specificity, and cut‐off were presented in Table 5 as well. The ROC analysis indicated the potential diagnostic values of circRNAs in periodontitis.
TABLE 5.
ROC analysis of identified circRNAs in gingival tissues from patients with periodontitis
| CircRNA | AUC | SEM | 95% CI | p value | Sensitivity | Specificity | Cut‐off |
|---|---|---|---|---|---|---|---|
| circPTP4A2 | 0.7321 | 0.0684 | 0.5981, 0.8660 | .0029 | 0.6538 | 0.8000 | 0.01235 |
| chr22:23101560‐23135351+ | 0.8667 | 0.0505 | 0.7677, 0.9656 | <.0001 | 0.6154 | 1.000 | 0.02933 |
| circARHGEF28 | 0.8244 | 0.0547 | 0.7171, 0.9316 | <.0001 | 0.7308 | 0.8333 | 0.01120 |
| circBARD1 | 0.8013 | 0.0591 | 0.6855, 0.9170 | .0001 | 0.7692 | 0.7333 | 0.001554 |
| circRASA2 | 0.7731 | 0.0631 | 0.6493, 0.8968 | .0005 | 0.9231 | 0.5333 | 0.002835 |
Abbreviations: AUC, area under the curve; ROC, receiver‐operating characteristic.
4. DISCUSSION
In this study, Illumina high‐throughput sequencing was used to determine the expression profile of circRNAs and mRNAs in gingival tissues of human periodontitis. Bioinformatics analysis using CircR2Disease v2.0 database suggested that the top five upregulated circRNAs identified, including circPTP4A2, chr22:23101560‐23135351+, circARHGEF28, circBARD1, and circRASA2, are likely associated with multiple human diseases, including periodontitis. The expression levels of the top five upregulated circRNAs identified were significantly correlated with the PD of subjects. To the best of our knowledge, this correlation was reported for the first time. A circRNA‐miRNA‐mRNA network was predicted combined with high‐throughput sequencing, bioinformatics analysis, and online database prediction, suggesting regulatory roles of circRNAs in cell adhesion, immune response, oxidative stress, apoptosis and production of cytokines. ROC analysis also indicated that the top five upregulated circRNAs may serve as biomarkers for the diagnosis of human periodontitis.
4.1. Potential roles of the top five upregulated circRNAs
The upregulated circRNAs were reported to be dysregulated and associated with diseases (Table 3). CircPTP4A2 was upregulated and correlated with PD in periodontitis gingival tissues in this study. Apart from periodontitis, circPTP4A2 was dysregulated in diabetic kidney disease and glioma and reported to promote cervical cancer. 40 , 41 , 42 This kind of circRNA correlated with clinical features and promote cell proliferation and invasion in cervical cancer by sponging miR‐101‐5p. 42 In addition, circPTP4A2 expression also correlated with advanced clinical features in cervical cancer patients, including International Federation of Gynecology and Obstetrics (FIGO) stage and lymph node metastasis. 42
CircARHGEF28, also named circRIP2, was reported to have a higher expression level in periodontitis and upregulated during PDLSC osteogenic differentiation. 12 , 22 In bladder cancer and gastric cancer, circARHGEF28 was downregulated, while in non‐small‐cell lung cancer (NSCLC), it was upregulated. 43 , 44 , 45 CircARHGEF28 could promote proliferation and metastasis of bladder cancer cells and negatively associated with the grade, metastasis, stage, and outcome of bladder cancer. 43 For circARHGEF28 in NSCLC, it promoted cancer progression and indicated relatively poorer overall survival and disease‐free survival. 45 CircARHGEF28 exerted functions through sponging miR‐1305 and inducing EMT via Tgf‐β2/smad3 pathway in bladder cancer, and sponging miR‐671‐5p to regulate downstream FOXM1 expression in NSCLC. 43 , 45
CircRASA2 was downregulated in gastric cancer in several studies. 47 , 48 It was considered as a potential indicator of gastric cancer and was closely related to gastric carcinogenesis and clinicopathological factors, such as tumor diameter, stages, tissue carcinoembryonic antigen (CEA) level, and carbohydrate antigen 19‐9 (CA19‐9) level. 47 , 48 The clinical significance of circRASA2 has been emphasized more than a functional understanding, so further identification of the roles of circRNAs could enhance the comprehension of the mechanisms of such circRNAs in diseases.
The other two circRNAs, circBARD1 and chr22:23101560‐23135351+ were also upregulated in periodontitis. CircBARD1 was downregulated during PDLSC osteogenic differentiation and suggested to be a potential regulator in the intracellular signaling associated with neuronal excitation. 12 , 49 As an intergenic circRNA, chr22:23101560‐23135351+ was first identified upregulated in periodontitis with 2,342 predicted binding sites to miRNAs. Sequence analyses indicated that intergenic or intronic circRNAs generally showed weak conservation when compared to exons, and the function of intergenic circRNAs remains unknown at this time. 7 Studies around intergenic‐type circRNAs are still limited, therefore more studies are needed for this special kind of circRNA.
4.2. Function of circRNAs identified and their potential targets in periodontal diseases
While there are scant studies concerning the mechanism underlying periodontitis, research about the functions of circRNAs in periodontitis and periodontal cells may shed light on its development. 50 Each circRNAs may play a distinct role in periodontitis pathogenesis. (Table 6).
TABLE 6.
Function of circRNAs in periodontal tissues and cells
| CircRNA | miRNA | Target | Function | Cell/tissue Type | Study method |
|---|---|---|---|---|---|
| hsa_circ_0081572 (circSERPINE1) | miR‐378h | RORA |
Downregulated in periodontitis Enhance viability, suppress apoptosis, inflammatory response, and oxidative stress to inhibit the progression of periodontitis via miR‐378h/RORA axis |
Human gingival tissues hPDLSCs |
RT‐qPCR, CCK‐8 assay, flow cytometry, Western blot; Caspase 3 activity assay; ELISA; ROS level detection; dual‐luciferase reporter assay, RIP, RNA pulldown assay |
| hsa_circ_0085289 (circCTHRC1) | let‐7f‐5p | SOCS6 |
Downregulated in periodontitis Promote cell viability Suppress proinflammatory cytokines production and apoptosis Alleviate injury of hPDLSCs via let‐7f‐5p/SOCS6 axis in response to LPS |
Human PDL tissues hPDLSCs |
RT‐qPCR, ELISA, cell viability assay, Caspase 3 activity assay; flow cytometry, Western blot; dual‐luciferase reporter assay, RNA pulldown assay, RIP |
| hsa_circ_002284 (circMAP3K11) | miR‐511‐3p | TLR4 |
Upregulated in periodontitis Promote the cell viability, proliferation, migration and the osteogenic potential Reduce cell apoptosis Promote the proliferation and inhibit the apoptosis of hPDLSCs via miR‐511‐3p/TLR4 axis in vivo |
Human PDL tissues hPDLSCs Periodontitis murine model |
Immunohistochemistry assay, target prediction, cell viability assay, cell proliferation assay, cell migration assay, cell apoptosis assay, RT‐qPCR, Western blot, dual‐luciferase reporter assay, TUNEL assay, and Ki‐67 assay in vivo |
| hsa_circ_0003948 (circKDELR2) | miR‐144‐3p | NR2F2 |
Downregulated in periodontitis Promote cell proliferation and inhibit apoptosis in response to LPS treatment via miR‐144‐3p/NR2F2/PTEN axis |
Human gingival tissues hPDLSCs |
RNA‐sequencing, RT‐qPCR, cell proliferation assay, bioinformatic analyses, cell apoptosis assay, dual‐luciferase reporter assay |
| circ‐Amotl1 | miR‐17‐5p | STAT3, Dnmt3a, Fibronectin |
Promote skin wound healing in vivo Promote Stat3 expression and facilitate Stat3 nuclear translocation Promote cell adhesion, migration, proliferation, survival, and wound repair of fibroblasts |
Human gingival fibroblast cell line (CRL‐2014) NIH 3T3 fibroblast cell line Full‐thickness excisional wound murine model |
Wound healing experiment in vivo, cell adhesion and migration assays, cell proliferation and survival assays, ChIP, Western blot, RT‐qPCR, RNA pulldown assay, FISH, dual‐luciferase reporter assay |
| circ_0138959 | miR‐527 | CASP5 |
Upregulated in periodontitis Suppress cell viability and increase pyroptosis in response to LPS via miR‐527/CASP5 axis Promote the secretion of LDH, IL‐1β, and IL‐18, and increase the protein levels of pyroptosis‐related proteins, including caspase‐1, caspase‐4, and GSDMD‐N |
Human PDL tissues Human gingival fibroblasts (HGFs) |
RT‐qPCR, CCK‐8 assay, LDH measurement, ELISA, flow cytometry, Western blot, dual‐luciferase reporter assay, RNA pulldown assay |
| hsa_circ_0003489 (circCDK8) | / | mTOR signaling |
Upregulated in periodontitis Inhibit the osteogenic differentiation of hPDLSCs by triggering autophagy activation and apoptosis through mTOR signaling in a hypoxic microenvironment |
Human PDL tissues hPDLSCs |
RT‐qPCR; Western blot; transmission electron microscopy; immunofluorescence analysis; cell apoptosis assay; cell proliferation assay |
| CDR1as | miR‐7 | GDF5/SMAD and p38 MAPK signaling pathway |
Upregulated during osteogenic differentiation Promote osteoblastic differentiation of hPDLSCs via miR‐7/GDF5/SMAD and p38 MAPK signaling pathway in vitro and in vivo |
hPDLSCs Critical‐sized calvarial defect murine model |
RNA oligoribonucleotides, ALP staining and activity assay, ARS staining and quantification, RT‐qPCR, Western blot, dual‐luciferase reporter assay, bone formation assay in vivo, micro‐CT, H&E staining, immunofluorescence staining analysis |
| CDR1as | miR‐7 | ERK signal pathway |
Downregulated in periodontitis Regulate the proliferation of hPDLSCs under an LPS‐induced inflammatory condition via miR‐7/ERK signal pathway |
Human PDL tissues hPDLSCs |
RT‐qPCR, cell proliferation assay, Western blot |
| CDR1as | miR‐7 | KLF4 | Upregulated in response to hnRNPM Maintain stemness of hPDLSCs, including expression levels of stemness‐associated genes (SOX2, OCT4, and Nanog), osteogenic differentiation, adipogenic differentiation and migration via miR‐7/KLF4 axis | hPDLSCs | Osteogenic induction, ALP, and alizarin red staining, Adipogenic induction, Oil Red O staining, RT‐qPCR, cell proliferation assay, cell migration assay, RNA pulldown assay, dual‐luciferase reporter assay |
Abbreviations: CCK‐8, Cell counting kit‐8; ChIP, chromatin immunoprecipitation; ELISA, enzyme‐linked immunosorbent assay; FISH, florescent in situ hybridization; H&E staining, hematoxylin and eosin staining; LDH, lactate dehydrogenase; LPS, lipopolysaccharide; PDL, periodontal ligament; RIP, RNA immunoprecipitation.
In this study, the circRNA‐miRNA‐mRNA network predicted potential roles for the top five upregulated circRNAs in cell adhesion, cytokine production, B cell activation, T cell activation, leukocyte aggregation, innate immune response, response to oxidative stress, apoptotic signal pathway, and myeloid leukocyte migration. These predicted functions of circRNAs in periodontitis were closely related to identified functions in previous studies and related to immune response. 51
Several circRNAs have been suggested that might exert regulations in hPDLSCs cell viability, proliferation, migration, apoptosis, osteogenic differentiation, inflammatory response, oxidative stress, cytokine production, as well as stemness maintenance, including circSERPINE1, circCTHRC1, circMAP3K11, circKDELR2, circCDK8, and CDR1as. 14 , 15 , 17 , 18 , 19 , 20 , 21 , 23 Among these circRNAs, further validation studies suggested that circSERPINE1 has a negative impact on inflammatory response and oxidative stress by regulating the miR‐378h /RORA axis, while circCTHRC1 suppressed cytokine production via sponging let‐7f‐5p to regulate SOCS6. 19 , 21 CircMAP3K11 promoted the proliferation and inhibited the apoptosis of hPDLSCs via the miR‐511‐3p/TLR4 axis, and circKDELR2 promoted cell proliferation and inhibited apoptosis in response to LPS via the miR‐144‐3p/NR2F2/PTEN axis. 18 , 23
For human gingival fibroblasts, circ‐Amotl1 could promote cell adhesion, migration, proliferation, and wound healing through suppressing miR‐17‐5p and increasing the expression of fibronectin, Dnmt3a, and Stat3. 24 On the other hand, pyroptosis of gingival fibroblasts could be induced by circ_0138959, which was accompanied by increased levels of pro‐inflammatory cytokines and pyroptosis‐related proteins. 25
For the regulation of osteogenic differentiation, circRNAs expression profiling of hPDLSCs cell lines in vitro provided clues for functional validation of dysregulated circRNAs identified, such as circCDK8 and CDR1as. CircCDK8 was upregulated in periodontitis and repressed the osteogenic differentiation of hPDLSCs by triggering autophagy activation and apoptosis through mTOR signaling in a hypoxic microenvironment. 15 CircRNA CDR1as was downregulated in periodontitis as the sponge of miR‐7. The function of CDR1as was validated to promote osteogenic differentiation of hPDLSCs, promote cell proliferation and maintain stemness, including expression levels of stemness‐associated genes (SOX2, OCT4, and Nanog), osteogenic differentiation, adipogenic differentiation, and migration capability. 14 , 17 , 20
There are several studies focusing on the role of circRNA in the regulation of osteoblast differentiation and cementoblast differentiation, which is important in the context of periodontal regeneration and tissue engineering. 50 For osteoblast, circ_0008500 remained upregulated during osteoblast differentiation and promoted osteoblast mineralization via sponging miR‐1301‐3p. 52 One recent study showed that circLrp6 promoted cementoblast differentiation by regulating Zeb2. 53
4.3. Bridging the gap between bioinformatics and clinical needs: potential use of circRNA as a biomarker for periodontitis
Numerous algorithms have been developed to detect genome‐wide circRNA expression profiling from RNA‐sequencing, but there is hardly overlap in their predictions and no gold standard method to assess the accuracy of the algorithms. 32 Experimental challenges, bioinformatic challenges, as well as biases from statistical analysis complicate the accurate discovery of circRNAs. To solidify the accuracy of disease‐associated, especially periodontitis‐associated, circRNA prediction, the following factors must be considered, as detailed in Table 7.
TABLE 7.
Toolbox: how to validate the functions of circRNAs
| Sample preparation | Bioinformatics tools | Clinical indicators | Experimental validation |
|---|---|---|---|
|
Sample collection and preservation Collect clinical features Apply liquid biopsy technology Decrease sampling time RNA extraction and selection Assess RNA integrity Correct for sample‐to‐sample variations Deplete ribosomal RNA Deplete linear RNAs with RNase R treatment Deplete Poly(A)+ RNA Retain linear and circular RNAs without RNase R treatment Library preparation Estimate the false discovery rate Select appropriate thresholds for high‐confidence circRNA detection Test for biochemical artefacts Conduct normalization procedures Standardize tissue sample banks |
CircRNA identification tools DCC* ACFS find_circ CircRNA annotation database CircBase* CircBank deepBase2.0 CircFunBase CircNet circAtlas Tools for circRNA‐disease associations CircR2Disease v2.0* circRNADisease Circ2Traits Tools for circRNA network prediction starBase v2.0* miRanda* TargetScan* miRBase CircInteractome Visualization Cytoscape* Bioinformatics |
Probing depth (PD) Clinical attachment loss (CAL) Plaque index (PI) Bleeding index (BI) Gingival index (GI) Bleeding on probing (BOP) |
Detection of expression profiling Next‐generation sequencing Microarray Single cell sequencing Experimental validation in vitro Expression level detection RT‐qPCR Sanger sequencing Northern blot Denaturing agarose gel electrophoresis Functional experiments RIP FISH ChIP Western blot Dual‐luciferase reporter RNA pull‐down Loss and gain of function model Experimental validation in vivo Animal model, e.g., murine model |
4.3.1. Sample preparation
In this study, inflamed gingival tissues samples were obtained from patients with periodontitis who underwent periodontal flap surgery, and healthy gingival samples were collected from donors who had tooth extraction with no periodontitis. Samples from each patient were clearly marked regarding clinical features, including clinical attachment loss, probing depth, bleeding, and so on, for further analysis. 54 The tissue samples included epithelium and connective tissue. To enhance RNA quality and consistency, periodontal samples were stringently collected and stored freshly frozen at −80°C until RNA extraction and analysis. Liquid biopsy may be used to study diseases with high tissue heterogeneity. 55 For future studies, decreasing sampling time is highly recommended. 56 The assessment of RNA integrity and correction for sample‐to‐sample variations in efficiency and errors in sample quantification might help obtain more meaningful data. 55 , 56 , 57 Various approaches are available to enrich circRNAs from the total RNA pool, including rRNA depletion, poly(A)+ RNA depletion, and RNase R treatment to deplete linear RNAs before sequencing. 32 , 58 The RNase R treatment is specially designed for circRNA detection to increase sensitivity and to achieve isolation of highly pure circRNA populations. 59 , 60 However, RNase R degradation is variable, and there are rare cases of RNase R resistant linear RNAs and RNase R sensitive circRNAs. 8 , 10 , 26 , 59 , 61 This treatment also requires a high RNA input, which could be limiting for some kinds of tissues. 61 Other circRNA studies choose to sequence either total, non‐polyadenylated (polyA‐), or ribosomal‐depleted (ribo‐) RNA, where both linear and circular RNAs can be identified without RNase R treatment. 8 This approach reduces the RNA amount needed for the sequencing and profiles the expression of other types of RNAs from the same dataset. 59 True circRNAs can be detected from the whole transcriptome sequencing with good sequencing depth, quality, careful data analysis, and a later circularity confirmation for circRNAs. 59 , 61 In this study, the expression profiles of circRNAs and mRNAs were detected from the same sequencing, in which library preparation with rRNA removal was conducted without RNase R treatment to obtain a comprehensive regulatory network. Similar approach has also been reported in several other studies. 62 , 63 , 64 Future circRNA studies to use RNase R treatment for the removal of linear RNA contamination in modified conditions can help to improve the sensitivity of circRNA detection. To deal with the experimental challenges in RNA‐seq library preparation, methods to estimate the false discovery rate, selection of appropriate thresholds for high‐confidence circRNA detection, and tests for biochemical artifacts are needed. 32 Approaches such as normalization addressing enrichment will help reduce false positives. 65 , 66 Standard tissue sample banks would provide a tremendous opportunity for future studies. 67
4.3.2. Bioinformatics tools
Bioinformatics tools support circRNA detection and functional prediction. 7 , 30 , 31 , 32 , 33 , 34 , 39 , 84 One circRNA identification tool used in this study was DCC software, which systematically detects back‐splice junctions in RNA‐sequencing data with output from the STAR read mapper. 32 , 68 Other tools such as ACFS and find_circ were available for circRNA identification. 7 , 69 The nomenclature of circRNAs used was circGeneName (e.g., circPTP4A2), representing the circRNA generated from parental gene PTP4A2. The circBase ID and chromosome location were also referred to in the circRNA annotation. 70 Apart from circBase, there are several circRNA annotation databases, such as CircBank, deepBase2.0, CircFunBase, CircNet, and circAtlas. 71 , 72 , 73 , 74 , 75 There being no gold standard to normalize the nomenclature of circRNAs, novel circRNAs are difficult to annotate properly. The investigation of circRNA‐disease associations was conducted on CircR2Disease v2.0 database, with the largest number of associations and features among the current comparison. 39 Online databases like miRanda (http://www.microrna.org/), Targetscan (www.targetscan.org), and starBase 2.0 are popular tools for network prediction. 78 GSE54710 dataset reported miRNA and mRNA expression in gingival tissues, while dataset for circRNA expression in periodontitis was still limited. 79 Online tools Cytoscape and Bioinformatics (http://www.bioinformatics.com.cn/) were helpful for visualization. 81
4.3.3. Clinical indicators
The probing depth (PD), clinical attachment loss (CAL), plaque index (PI), bleeding index (BI), and bleeding on probing (BOP) were important indicators for the diagnosis of periodontitis, which can represent the severity and development of periodontitis. Studies indicated that microRNA expression levels were significantly associated with the clinical parameters of periodontitis, such as miR‐1226, miR‐155, and miR‐146a and possess diagnostic value in this disease. 35 , 36 There is little research on the correlation between circRNA expression levels and clinical indicators of periodontitis; we reported a positive correlation between the expression levels of the top five upregulated circRNAs identified and PD. Further studies are needed to test and validate the usefulness of these biomarkers in clinical practice.
4.3.4. Experimental validation
Apart from “miRNA‐sponge,” other functions of circRNAs, such as miRNA “reservoirs” and protein‐coding, are not fully understood in diseases. 8 , 26 To validate novel functions of circRNAs, experiments in vitro and in vivo help confirm their possible application in diagnosis, prognosis, and therapy. 85 Considering the high false‐positive rates of the accessible computational methods, experimental approaches are in need to validate results of computational prediction and select high‐confidence circRNAs for further study. The expression profiling could be detected by next‐generation sequencing (NGS) or microarray. 48 , 86 , 87 Regarding cell‐type‐specific gene regulation, the single‐cell RNA‐sequencing technique is powerful for profiling “cell‐to‐cell” variability at the genomic level and equipped NGS with a much better cell‐specificity. 88 , 89 , 90 Targeted validation of the accuracy of a circRNA prediction is accomplished by RT‐qPCR using outward‐facing primers, Sanger sequencing, Northern blot, and RNase R treatment. In addition, RNA immunoprecipitation (RIP), fluorescent in situ hybridization (FISH), chromatin immunoprecipitation (ChIP), Western blot, Dual‐luciferase reporter assay, RNA pull‐down, and validation in vivo were all reliable experimental tools for validation. 8 , 91
4.4. Limitations and future directions
This study implied that circRNAs may serve as regulators in periodontitis pathogenesis using bioinformatic tools, and the correlation between upregulated circRNAs expression levels and the severity of periodontitis strengthened the reliability of circRNA‐disease associations. Our integrated analysis provides foundational information that may inspire further experimental investigation into the behavior of circRNAs and to further identify their regulatory mechanisms on target genes in human diseases, especially periodontitis. RNase R treatment designed to deplete all linear RNAs is able to increase sensitivity and to achieve isolation of highly pure circRNA populations, accompanied with rRNA depletion or poly(A)+ RNA depletion, although such approach requires a high RNA input and extra attention is necessary as RNase R degradation is highly variable depending on the tissue type used. There are several other shortcomings in this study, including but not limited to the fact that bulk sequencing is not cell‐specific, the role of circRNA in adipogenic, neurogenic, myogenic, and ligament differentiation in periodontal tissues still requires future investigation, and the molecular regulation of candidate circRNAs has not been unveiled. More recently, several novel functions of circRNAs have been reported, and as such, future studies will need to heed these advances.
5. CONCLUSIONS
CircRNAs and mRNAs had different expression levels between periodontitis and periodontal healthy gingival tissues. Upregulated circRNAs identified in periodontitis through high‐throughput sequencing have strong associations with multiple human diseases via bioinformatics analysis, including periodontitis. Positive correlation between upregulated circRNAs, circPTP4A2, chr22:23101560‐23135351+, circARHGEF28, circBARD1, and circRASA2, and PD suggested a correlation between circRNA expression levels and severity of periodontitis. The upregulated circRNAs were predicted to influence leukocyte aggregation, myeloid leukocyte migration, regulation of B cell activation, regulation of T cell activation, regulation of innate immune response, regulation of apoptotic signaling pathway, response to oxidative stress, positive regulation of cytokine production, and positive regulation of cell adhesion. The upregulated circRNAs might work as potential biomarkers in periodontitis diagnosis. To deal with experimental challenges, bioinformatic challenges and biases, aspects concerning sample preparation, bioinformatics tools, clinical indicators, and experimental validation need to be standardized to discover and validate circRNA.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Fig S1
Table S1
Table S2
Table S3
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Bin Jiang for advice on sample collection. The authors would like to thank Dr. Fengzhi Xin for assistance in paper revision and thank CloudSeq Biotech Inc. (Shanghai, China) for high‐throughput sequencing technical support. This study was supported by the National Natural Science Foundation of China (81900565, 81570948), the Program of Science and Technology Commission of Shanghai Municipality (19ZR1430900, 201409006300), the Key Program of Biomedical Engineering Cross Research Foundation of Shanghai Jiao Tong University (YG2017ZD06), and the Incubating Program for Clinical Research and Innovation of Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University (PYII‐17‐014).
Yu W, Gu Q, Wu D, et al. Identification of potentially functional circRNAs and prediction of circRNA‐miRNA‐mRNA regulatory network in periodontitis: Bridging the gap between bioinformatics and clinical needs. J Periodont Res. 2022;57:594–614. doi: 10.1111/jre.12989
Funding information
National Natural Science Foundation of China, Grant/Award Number: 81900565 and 81570948; the Program of Science and Technology Commission of Shanghai Municipality, Grant/Award Number: 19ZR1430900 and 201409006300; the Key Program of Biomedical Engineering Cross Research Foundation of Shanghai Jiao Tong University, Grant/Award Number: YG2017ZD06; the Incubating Program for Clinical Research and Innovation of Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University, Grant/Award Number: PYII‐17‐014
Contributor Information
Michael Z. Miao, Email: zhemiao@unc.edu.
Yuhua Gong, Email: rjgongyuhua@163.com.
Yifei Zhao, Email: rjzhaoyifei@163.com.
Eryi Lu, Email: lueryi222@outlook.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Slots J. Periodontitis: facts, fallacies and the future. Periodontology. 2000;2017(75):7‐23. [DOI] [PubMed] [Google Scholar]
- 2. Papapanou PN, Sanz M, Buduneli N, et al. Periodontitis: consensus report of workgroup 2 of the 2017 world workshop on the classification of periodontal and peri‐implant diseases and conditions. J Periodontol. 2018;89(Suppl 1):s173‐s182. [DOI] [PubMed] [Google Scholar]
- 3. Masumoto R, Kitagaki J, Fujihara C, et al. Identification of genetic risk factors of aggressive periodontitis using genomewide association studies in association with those of chronic periodontitis. J Periodontal Res. 2019;54:199‐206. [DOI] [PubMed] [Google Scholar]
- 4. Loos BG, Van Dyke TE. The role of inflammation and genetics in periodontal disease. Periodontology. 2000;2020(83):26‐39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lin L, Yu W, Zhang W, et al. Expression profile of lipoxygenases in gingival tissues of human periodontitis. Oral Dis. 2021;27:567‐576. [DOI] [PubMed] [Google Scholar]
- 6. Liu Y, Liu Q, Li Z, et al. Long non‐coding RNA and mRNA expression profiles in peri‐implantitis vs periodontitis. J Periodontal Res. 2020;55:342‐353. [DOI] [PubMed] [Google Scholar]
- 7. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333‐338. [DOI] [PubMed] [Google Scholar]
- 8. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675‐691. [DOI] [PubMed] [Google Scholar]
- 9. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384‐388. [DOI] [PubMed] [Google Scholar]
- 10. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428‐442. [DOI] [PubMed] [Google Scholar]
- 11. Zheng Y, Li X, Huang Y, Jia L, Li W. The circular RNA landscape of periodontal ligament stem cells during osteogenesis. J Periodontol. 2017;88:906‐914. [DOI] [PubMed] [Google Scholar]
- 12. Gu X, Li M, Jin Y, Liu D, Wei F. Identification and integrated analysis of differentially expressed lncRNAs and circRNAs reveal the potential ceRNA networks during PDLSC osteogenic differentiation. BMC Genet. 2017;18:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang H, Feng C, Jin Y, Tan W, Wei F. Identification and characterization of circular RNAs involved in mechanical force‐induced periodontal ligament stem cells. J Cell Physiol. 2019;234:10166‐10177. [DOI] [PubMed] [Google Scholar]
- 14. Li X, Zheng Y, Zheng Y, et al. Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR‐7/GDF5/SMAD and p38 MAPK signaling pathway. Stem Cell Res Ther. 2018;9:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zheng J, Zhu X, He Y, et al. CircCDK8 regulates osteogenic differentiation and apoptosis of PDLSCs by inducing ER stress/autophagy during hypoxia. Ann N Y Acad Sci. 2021;1485:56‐70. [DOI] [PubMed] [Google Scholar]
- 16. Xie L, Chen J, Ren X, et al. Alteration of circRNA and lncRNA expression profile in exosomes derived from periodontal ligament stem cells undergoing osteogenic differentiation. Arch Oral Biol. 2021;121:104984. [DOI] [PubMed] [Google Scholar]
- 17. Wang F, Chen X, Han Y, Xi S, Wu G. circRNA CDR1as regulated the proliferation of human periodontal ligament stem cells under a lipopolysaccharide‐induced inflammatory condition. Mediators Inflamm. 2019;2019:1625381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu B, Hu J, Li Q, Wang F. CircMAP3K11 contributes to proliferation, apoptosis and migration of human periodontal ligament stem cells in inflammatory microenvironment by regulating TLR4 via miR‐511 sponging. Front Pharmacol. 2021;12:633353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang J, Du C, Xu L. Circ_0081572 inhibits the progression of periodontitis through regulating the miR‐378h/RORA axis. Arch Oral Biol. 2021;124:105053. [DOI] [PubMed] [Google Scholar]
- 20. Gu X, Li X, Jin Y, et al. CDR1as regulated by hnRNPM maintains stemness of periodontal ligament stem cells via miR‐7/KLF4. J Cell Mol Med. 2021;25:4501‐4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Du W, Wang L, Liao Z, Wang J. Circ_0085289 alleviates the progression of periodontitis by regulating let‐7f‐5p/SOCS6 pathway. Inflammation. 2021;44:1607‐1619. [DOI] [PubMed] [Google Scholar]
- 22. Li J, Xie R. Circular RNA expression profile in gingival tissues identifies circ_0062491 and circ_0095812 as potential treatment targets. J Cell Biochem. 2019;120:14867‐14874. [DOI] [PubMed] [Google Scholar]
- 23. Li W, Zhang Z, Li Y, Wang Z. Abnormal hsa_circ_0003948 expression affects chronic periodontitis development by regulating miR‐144‐3p/NR2F2/PTEN signaling. J Periodontal Res. 2021;57:316‐323. [DOI] [PubMed] [Google Scholar]
- 24. Yang ZG, Awan FM, Du WW, et al. The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR‐17 function. Mol Ther. 2017;25:2062‐2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pan J, Zhao L, Liu J, Wang G. Inhibition of circular RNA circ_0138959 alleviates pyroptosis of human gingival fibroblasts via the microRNA‐527/caspase‐5 axis. Bioengineered. 2022;13:1908‐1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen L, Wang C, Sun H, et al. The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform. 2021;22:1706‐1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lu Y, Li Z, Lin C, Zhang J, Shen Z. Translation role of circRNAs in cancers. J Clin Lab Anal. 2021;35:e23866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132‐136. [DOI] [PubMed] [Google Scholar]
- 29. Aghaee‐Bakhtiari SH. Online databases and circular RNAs. Adv Exp Med Biol. 2018;1087:35‐38. [DOI] [PubMed] [Google Scholar]
- 30. Wang L, You ZH, Li YM, Zheng K, Huang YA. GCNCDA: a new method for predicting circRNA‐disease associations based on Graph Convolutional Network Algorithm. PLoS Comput Biol. 2020;16:e1007568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Westholm JO, Miura P, Olson S, et al. Genome‐wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age‐dependent neural accumulation. Cell Rep. 2014;9:1966‐1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17:679‐692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zeng X, Lin W, Guo M, Zou Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput Biol. 2017;13:e1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li G, Luo J, Wang D, et al. Potential circRNA‐disease association prediction using DeepWalk and network consistency projection. J Biomed Inform. 2020;112:103624. [DOI] [PubMed] [Google Scholar]
- 35. Du Y, Qi YS, Chen H, Shen G. The expression and clinical significance of miR‐1226 in patients with periodontitis. BMC Oral Health. 2021;21:487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu P, Feng J, Wang W. Expression of miR‐155 and miR‐146a in the saliva of patients with periodontitis and its clinical value. Am J Transl Res. 2021;13:6670‐6677. [PMC free article] [PubMed] [Google Scholar]
- 37. Rovas A, Puriene A, Snipaitiene K, et al. Gingival crevicular fluid microRNA associations with periodontitis. J Oral Sci. 2022;64:11‐16. [DOI] [PubMed] [Google Scholar]
- 38. Nik Mohamed Kamal NNS, Awang RAR, Mohamad S, Shahidan WNS. Plasma‐ and saliva exosome profile reveals a distinct microRNA signature in chronic periodontitis. Front Physiol. 2020;11:587381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fan C, Lei X, Tie J, Zhang Y, Wu F, Pan Y. CircR2Disease v2.0: an updated web server for experimentally validated circRNA‐disease associations and its application. Genomics Proteomics Bioinformatics. 2021. 10.1016/j.gpb.2021.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xiong Z, Zhou C, Wang L, et al. Circular RNA SMO sponges miR‐338‐3p to promote the growth of glioma by enhancing the expression of SMO. Aging. 2019;11:12345‐12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wen S, Li S, Li L, Fan Q. circACTR2: a novel mechanism regulating high glucose‐induced fibrosis in renal tubular cells via pyroptosis. Biol Pharm Bull. 2020;43:558‐564. [DOI] [PubMed] [Google Scholar]
- 42. Chen H, Gu B, Zhao X, et al. Circular RNA hsa_circ_0007364 increases cervical cancer progression through activating methionine adenosyltransferase II alpha (MAT2A) expression by restraining microRNA‐101‐5p. Bioengineered. 2020;11:1269‐1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Su Y, Feng W, Shi J, Chen L, Huang J, Lin T. circRIP2 accelerates bladder cancer progression via miR‐1305/Tgf‐β2/smad3 pathway. Mol Cancer. 2020;19:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guan YJ, Ma JY, Song W. Identification of circRNA‐miRNA‐mRNA regulatory network in gastric cancer by analysis of microarray data. Cancer Cell Int. 2019;19:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu Y, Feng X, Kang S, Lv F, Ni Y, Wu H. CircRIP2 promotes NSCLC progression by sponging for miR‐671‐5p to regulate FOXM1 expression. Histol Histopathol. 2021;37:18360. [DOI] [PubMed] [Google Scholar]
- 46. Lu R, Shao Y, Tao X, Ye G, Xiao B, Guo J. Clinical significances of hsa_circ_0067582 and hsa_circ_0005758 in gastric cancer tissues. J Clin Lab Anal. 2019;33:e22984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yu X, Ding H, Yang L, et al. Reduced expression of circRNA hsa_circ_0067582 in human gastric cancer and its potential diagnostic values. J Clin Lab Anal. 2020;34:e23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li S, Teng S, Xu J, et al. Microarray is an efficient tool for circRNA profiling. Brief Bioinform. 2019;20:1420‐1433. [DOI] [PubMed] [Google Scholar]
- 49. Mahmoudi E, Kiltschewskij D, Fitzsimmons C, Cairns MJ. Depolarization‐associated CircRNA regulate neural gene expression and in some cases may function as templates for translation. Cells. 2019;9:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jiao K, Walsh LJ, Ivanovski S, Han P. The emerging regulatory role of circular RNAs in periodontal tissues and cells. Int J Mol Sci. 2021;22:4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen X, Yang T, Wang W, et al. Circular RNAs in immune responses and immune diseases. Theranostics. 2019;9:588‐607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhai Q, Zhao Y, Wang L, et al. CircRNA hsa_circ_0008500 acts as a miR‐1301‐3p sponge to promote osteoblast mineralization by upregulating PADI4. Front Cell Dev Biol. 2020;8:602731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li M, Du M, Wang Y, et al. CircRNA Lrp6 promotes cementoblast differentiation via miR‐145a‐5p/Zeb2 axis. J Periodontal Res. 2021;56:1200‐1212. [DOI] [PubMed] [Google Scholar]
- 54. Jeon YS, Cha JK, Choi SH, Lee JH, Lee JS. Transcriptomic profiles and their correlations in saliva and gingival tissue biopsy samples from periodontitis and healthy patients. J Periodontal Implant Sci. 2020;50:313‐326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wen G, Zhou T, Gu W. The potential of using blood circular RNA as liquid biopsy biomarker for human diseases. Protein Cell. 2021;12:911‐946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fleige S, Pfaffl MW. RNA integrity and the effect on the real‐time qRT‐PCR performance. Mol Aspects Med. 2006;27:126‐139. [DOI] [PubMed] [Google Scholar]
- 57. Miao MZ, Wang B, Wu D, et al. Temporomandibular joint positional change accompanies post‐surgical mandibular relapse‐A long‐term retrospective study among patients who underwent mandibular advancement. Orthod Craniofac Res. 2018;21:33‐40. [DOI] [PubMed] [Google Scholar]
- 58. Pandey PR, Rout PK, Das A, Gorospe M, Panda AC. RPAD (RNase R treatment, polyadenylation, and poly(A)+ RNA depletion) method to isolate highly pure circular RNA. Methods. 2019;155:41‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang J, Chen S, Yang J, Zhao F. Accurate quantification of circular RNAs identifies extensive circular isoform switching events. Nat Commun. 2020;11:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xiao MS, Wilusz JE. An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G‐quadruplexes or structured 3’ ends. Nucleic Acids Res. 2019;47:8755‐8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Iparraguirre L, Prada‐Luengo I, Regenberg B, Otaegui D. To be or not to be: circular RNAs or mRNAs from circular DNAs? Front Genet. 2019;10:940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xu H, Wang C, Song H, Xu Y, Ji G. RNA‐Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers. Mol Cancer. 2019;18:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ma C, Wang X, Yang F, et al. Circular RNA hsa_circ_0004872 inhibits gastric cancer progression via the miR‐224/Smad4/ADAR1 successive regulatory circuit. Mol Cancer. 2020;19:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hu K, He C, Sun X, et al. Integrated study of circRNA, lncRNA, miRNA, and mRNA networks in mediating the effects of testicular heat exposure. Cell Tissue Res. 2021;386:127‐143. [DOI] [PubMed] [Google Scholar]
- 65. Hansen TB. Improved circRNA identification by combining prediction algorithms. Front Cell Dev Biol. 2018;6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Miao Z, Wang XD, Mao LX, et al. Influence of temporomandibular joint disc displacement on mandibular advancement in patients without pre‐treatment condylar resorption. Int J Oral Maxillofac Surg. 2017;46:328‐336. [DOI] [PubMed] [Google Scholar]
- 67. Khaseb S, Orooji M, Pour MG, Safavi SM, Eghbal MJ, Rezai RM. Dental stem cell banking: techniques and protocols. Cell Biol Int. 2021;45:1851‐1865. [DOI] [PubMed] [Google Scholar]
- 68. Cheng J, Metge F, Dieterich C. Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics. 2016;32:1094‐1096. [DOI] [PubMed] [Google Scholar]
- 69. You X, Conrad TO. Acfs: accurate circRNA identification and quantification from RNA‐Seq data. Sci Rep. 2016;6:38820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666‐1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu M, Wang Q, Shen J, Yang BB, Ding X. Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol. 2019;16:899‐905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zheng LL, Li JH, Wu J, et al. deepBase v2.0: identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep‐sequencing data. Nucleic Acids Res. 2016;44:D196‐202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Meng X, Hu D, Zhang P, Chen Q, Chen M. CircFunBase: a database for functional circular RNAs. Database. 2019;2019:baz003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Liu YC, Li JR, Sun CH, et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44:D209‐215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wu W, Ji P, Zhao F. CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol. 2020;21:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhao Z, Wang K, Wu F, et al. circRNA disease: a manually curated database of experimentally supported circRNA‐disease associations. Cell Death Dis. 2018;9:475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA‐ceRNA, miRNA‐ncRNA and protein‐RNA interaction networks from large‐scale CLIP‐Seq data. Nucleic Acids Res. 2014;42:D92‐97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Stoecklin‐Wasmer C, Guarnieri P, Celenti R, Demmer RT, Kebschull M, Papapanou PN. MicroRNAs and their target genes in gingival tissues. J Dent Res. 2012;91:934‐940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ji P, Wu W, Chen S, et al. Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep. 2019;26:3444‐3460.e3445. [DOI] [PubMed] [Google Scholar]
- 81. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498‐2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kozomara A, Birgaoanu M, Griffiths‐Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155‐d162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chandran V, Bermúdez ML, Koka M, et al. Large‐scale genomic study reveals robust activation of the immune system following advanced Inner Engineering meditation retreat. Proc Natl Acad Sci USA. 2021;118:e2110455118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34‐42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Cui H, Shan H, Miao MZ, et al. Identification of the key genes and pathways involved in the tumorigenesis and prognosis of kidney renal clear cell carcinoma. Sci Rep. 2020;10:4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wong SW, Han D, Zhang H, et al. Nine novel PAX9 mutations and a distinct tooth agenesis genotype‐phenotype. J Dent Res. 2018;97:155‐162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Fu Q, Liu CJ, Zhai ZS, Zhang X, Qin T, Zhang HW. Single‐cell non‐coding RNA in embryonic development. Adv Exp Med Biol. 2018;1068:19‐32. [DOI] [PubMed] [Google Scholar]
- 89. Jacox LA, Tang N, Li Y, et al. Orthodontic loading activates cell‐specific autophagy in a force‐dependent manner. Am J Orthod Dentofacial Orthop. 2022;161:423‐436.e421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Li X, Wang L, Zhou L, et al. The imbalance between regulatory and memory B cells accompanied by an increased number of circulating T‐follicular helper cells in MOG‐antibody‐associated demyelination. Mult Scler Relat Disord. 2019;36:101397. [DOI] [PubMed] [Google Scholar]
- 91. Marchesan J, Girnary MS, Jing L, et al. An experimental murine model to study periodontitis. Nat Protoc. 2018;13:2247‐2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Table S1
Table S2
Table S3
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


