Abstract
Canola varieties exhibit variation in drought avoidance and drought escape traits, reflecting adaptation to water‐deficit environments. Our understanding of underlying genes and their interaction across environments in improving crop productivity is limited. A doubled haploid population was analysed to identify quantitative trait loci (QTL) associated with water‐use efficiency (WUE) related traits. High WUE in the vegetative phase was associated with low seed yield. Based on the resequenced parental genome data, we developed sequence‐capture‐based markers and validated their linkage with carbon isotope discrimination (Δ13C) in an F2 population. RNA sequencing was performed to determine the expression of candidate genes underlying Δ13C QTL. QTL contributing to main and QTL × environment interaction effects for Δ13C and yield were identified. One multiple‐trait QTL for Δ13C, days to flower, plant height, and seed yield was identified on chromosome A09. Interestingly, this QTL region overlapped with a homoeologous exchange (HE) event, suggesting its association with the multiple traits. Transcriptome analysis revealed 121 significantly differentially expressed genes underlying Δ13C QTL on A09 and C09, including in HE regions. Sorting out the negative relationship between vegetative WUE and seed yield is a priority. Genetic and genomic resources and knowledge so developed could improve canola WUE and yield.
Keywords: carbon isotope discrimination, drought avoidance, gene expression, genetic analysis, physiology, water use
Summary Statement
Growing concerns about reduced water availability and climate change are putting pressure on sustainable food production worldwide. Identification of varieties, suitable for cultivation under reduced water inputs is critical to developing resilience to drought conditions. We identified 29 genetic and environmental determinants underlying plasticity in water use efficiency (WUE) and productivity traits. We show that one of the quantitative trait loci underlying variation in Δ13C, flowering time, plant height, and seed yield is subjected to homoeologous exchange and causes dosage‐dependent gene expression changes in WUE. Our results provide insights into the complexity of WUE genes and provide a strategy for improving canola adaptation under water‐deficit conditions.
1. INTRODUCTION
Drought is the major abiotic stress that reduces the yield potential of various crops including canola (Brassica napus L.), especially in arid and semiarid regions, of which 89% of regions are prevalent in Oceania (Koohafkan & Stewart, 2008). No doubt the impact of drought stress on crop productivity can be alleviated through irrigation at the “critical” stages of plant development. However, in recent years, freshwater, suitable for irrigation, is becoming scarce for crop production, required to meet the demand of a burgeoning human population (Gleick, 2000). Predicted climatic patterns such as debilitating drought and heat‐wave episodes and their possible increased frequency further pose a significant threat to crop production (Mills et al., 2018; S. Smith et al., 2012). The proportion of arable land per capita is also decreasing at a significant rate due to population growth and land degradation (http://www.fao.org/sustainability/). Therefore, improving crop varieties that have high yield potential and utilize water more effectively or require less water could provide a part of the solution to reduce the negative impacts of drought stress and increase productivity and food security (Bertolino et al., 2019; Blum, 2009; Kijne et al., 2003; Leakey et al., 2019; Passioura, 1977).
Canola is an allotetraploid (AACC, 2n = 38) member of the family Brassicaceae derived from interspecific hybridization between the diploid progenitors, Brassica rapa (AA, 2n = 20) and Brassica oleracea (CC, 2n = 18). Genomes of these diploid and allotetraploid species have undergone extensive evolutionary events, including homoeologous exchange (HE; Bayer et al., 2021; Chalhoub et al., 2014; Cheung et al., 2009; Hurgobin et al., 2018; Liu et al., 2014; Lysak et al., 2005; Parkin et al., 2005; Schranz et al., 2006; Town et al., 2006; Wang et al., 2011). Currently, canola is the second most important oilseed crop, grown worldwide with a global production of 75 million tons and used to produce 15% of vegetable oil (FAO STAT, http://www.fao.org/). Water stress reduces canola seed yield by up to 60% (Champolivier & Merrien, 1996; Gan et al., 2004; Qian et al., 2018). However, its effect varies depending upon the stage of plant development, and the severity and frequency of water stress (Hall et al., 1994). Despite its economic significance to the oilseed industry as well as being an essential rotational crop in agricultural production systems, little research has been conducted on traits contributing to drought avoidance and drought tolerance in canola (Fletcher et al., 2016, 2015; Hossain et al., 2020; Knight et al., 1994; Matus et al., 1995; McVetty et al., 1989; Pater et al., 2017; Raman, Raman, Mathews, et al., 2020; Raman, Raman, McVittie, et al., 2020).
In nature, to cope with water‐deficit conditions, plants have evolved three strategies known as drought escape, drought avoidance, and drought tolerance (Levitt, 1980; Ludlow, 1989; Rodrigues et al., 2019; Zhu et al., 2016). The drought escape strategy enables plants to complete their life cycle before the onset of drought. Drought avoidance involves maintaining internal water status under drought conditions by minimizing water loss/or maximizing water uptake (water‐use efficiency, WUE) while the drought tolerance strategy enables the plant to survive under internal water deficits (Ludlow, 1989; McKay et al., 2003). Through tiny microscopic pores on the surface of leaves called stomata, plants assimilate CO2 for photosynthesis by trading‐off water, required for transpiration and other biological processes. This close intimacy between productivity and water use contributes to the adaptation of plants to their growing environments. Therefore, genetic variation in WUE and transpiration efficiency (TE, biomass production/transpirational water loss) that occurs as a result of intentional (via breeding/selection) and unintentional selection in nature provides an opportunity to identify and assemble useful alleles for improving the productivity of various crops.
Genetic variation in WUE has been described variously at the single leaf level as intrinsic WUE (iWUE), defined as the ratio of the photosynthetic CO2 assimilation rate (A) over stomatal conductance, g sw, as TE, defined as the ratio of A over transpirational water loss, as whole‐plant vegetative WUE, as the ratio of total dry matter production to total water transpired (dmWUE) or as an integrated whole‐plant WUE, as the ratio of above‐ground shoot biomass (sbWUE) or seed yield (syWUE) to evapotranspiration (Farquhar et al., 1982; Farquhar & Richards, 1984; Leakey et al., 2019; Raman et al., 2019; Zhengbin et al., 2011). However, iWUE assessments using the gas‐exchange method are very challenging to be accurately performed, particularly in large breeding populations, as WUE is regulated by a myriad of plant development, physiological, biochemical, and molecular networks (Moore et al., 2009; Takahashi et al., 2018). Farquhar and Richards (1984) proposed Δ13C (the discrimination against 13C which is negatively related to δ13C) as a time‐integrated surrogate trait for assessing WUE both at the single leaf level (mmol C/mol H2O) and the whole plant level (dmWUE, dry matter/kg H2O), as C3 plants discriminate less against 13C during photosynthesis with increased water‐deficit stress. The negative relationship between TE and Δ13C (mostly in the vegetative growth periods) has been verified in Arabidopsis thaliana (Campitelli et al., 2016; Masle et al., 2005) and some agricultural crop plants, including canola (Des Marais et al., 2014; Ehleringer, 1993; Farquhar & Richards, 1984; Fischer et al., 1998; Hall et al., 1994; Hossain et al., 2020; Rebetzke et al., 2008), with some exceptions where nil or weak positive relationships were observed between syWUE and Δ13C (Devi et al., 2011; Hammer et al., 1997; Mekonnen et al., 2020; Monneveux et al., 2007; Raman, Raman, McVittie, et al., 2020).
Earlier genetic analysis studies suggest that drought tolerance is a complex trait, controlled by multiple quantitative trait loci (QTL) in canola. So far, QTL linked to drought avoidance traits; Δ13C, lateral root number, root mass, root diameter, taproot length, and root pulling force have been identified in canola by traditional simple‐, interval‐, and composite QTL mapping approaches (Fletcher et al., 2016, 2015; Mekonnen et al., 2020; Raman, Raman, Mathews, et al., 2020; Raman, Raman, McVittie, et al., 2020). Some of these QTL associated with drought avoidance traits also map in the vicinity or colocalize with QTL for flowering time, shoot biomass, plant height, and seed yield in canola, and with QTL for iWUE, photosynthetic capacity, leaf thickness, nitrogen content, and stomatal density in B. oleracea (Fletcher et al., 2015; Hall et al., 2005; Mekonnen et al., 2020). It is yet to be established which canola genotypes and loci show association with phenotypic plasticity and genotype (G) × environment (E) interaction in Δ13C and other integrated WUE traits in response to different environmental conditions. Thus, a comprehensive understanding of the genetical and physiological bases, and identification of stable loci, and also subjected to G × E interaction across environments underlying WUE is central to developing strategies for resilience to water‐deficit conditions. Recently, it was shown that two Australian canola inbred lines, BC1329, and BC9102, differ by ~2‰ in their Δ13C signatures with corresponding opposite differences in TE (Hossain et al., 2020). However, the genetic and environmental determinants underlying variation in Δ13C and other integrated WUE traits, such as plant biomass, flowering time, and seed yield were not deciphered. To uncover environmentally stable and environment‐specific loci associated with WUE traits, we carried out multienvironment QTL analyses based on extensive phenotypic and physiological measurements and genomic and transcriptomic studies. This multienvironment QTL approach was not used to uncover the genetic basis of drought avoidance and drought escape traits in earlier studies (Fletcher et al., 2016, 2015; Mekonnen et al., 2020; Raman, Raman, Mathews, et al., 2020; Raman, Raman, McVittie, et al., 2020). We demonstrate that multiple genetic and environmental determinants underlie phenotypic variation in drought avoidance traits such as Δ13C, early vigour, and plant height and drought escape traits such as flowering time in canola. We show that one QTL which is associated with multiple traits; Δ13C, days to flower, plant height, and seed yield on chromosome A09 was subjected to homoeologous recombination. Our research also describes candidate genes and differentially expressed genes (DEGs) underlying two stable QTL for Δ13C on chromosomes A09 and C09 that account for the majority of genotypic variance in the canola doubled haploid (DH) mapping population.
2. MATERIALS AND METHODS
2.1. Plant materials
A DH population of 223 lines (06‐5101DH) derived from the F1 cross between advanced breeding lines “BC1329” (maternal parent) and “BC9102” (paternal parent) were evaluated in different genetic analysis experiments. In addition, an F2 population comprising 733 lines derived from a single F1 plant from BC1329/BC9102 was employed for validation of QTL associated with Δ13C.
2.2. Experimental trials
Six experiments were conducted to (i) determine the genetic basis of variation in WUE‐related traits (Experiments 1–4), (ii) determine the relationship between physiological (Δ13C, iWUE) and integrated WUE‐related traits under wet and dry conditions (Experiment 5), and (iii) identify DEGs/candidate genes underlying QTL for Δ13C variation (Experiment 6) under field, glasshouse, and rain‐out shelter conditions. Details of the experiments and their designs are presented (Supporting Information: Method S1). Monthly weather statistics for average atmospheric temperatures and rainfall are also presented (Supporting Information: Figure S1).
2.3. Phenotypic trait measurements
Several plant development, agronomic, gas exchange, and other physiological traits were measured for genetic analysis. A summary of the experiments in terms of their aim, genetic material evaluated, and traits measured are presented (Table 1, Supporting Information: Figure S2). Details of trait measurement protocols are given in our recent study (Raman, Raman, McVittie, et al., 2020) and summarized in Methods S1. A brief description of the traits measured is given below.
Table 1.
Description of phenotyping experiments conducted to uncover the genetic basis of carbon isotope discrimination (Δ13C) and water‐use efficiency‐related traits in canola (B. napus L). The plant development stage at which traits were measured is detailed (Method S1)
| Experiment | Research question | Material | Phenotyping environment | Traits measured |
|---|---|---|---|---|
| 1 | Genetic basis of Δ13C variation | 223 DH lines from BC1329/BC9102, plus parental lines | Field (2017) |
Δ13C Days to flower (DTF) Plant height (PH) Seed yield (SY) |
| 2 | 223 DH lines from BC1329/BC9102, plus parental lines and commercial cultivars | Field (2018) |
Δ13C Normalised difference in the vegetative index (NDVI) DTF PH SY |
|
| 3 | 217 Doubled haploid lines from BC1329/BC9102, plus parental lines | Pots under rain‐out shelter (2017) |
Δ13C DTF PH |
|
| 4 | 733 F2 lines derived from BC1329/BC9102, plus parental lines | Pots in glasshouse (2020) under wet (field capacity) conditions |
Δ13C DTF Specific leaf weight (SLW) Leaf‐water content (LWC) |
|
| 5 | Relationship between physiological (intrinsic water‐use efficiency, Δ13C) and agronomic water‐use efficiency (canola productivity) related traits | Selected 70 DH lines representing extreme (low and high Δ13C values) plus parental lines under wet and dry conditions | Rain‐out shelter (2019) |
Δ13C DTF PH SY Photosynthesis (A) Stomatal conductance (g sw) Intrinsic water‐use efficiency (iWUE) SLW LWC |
| 6 | Identify differentially expressed genes underlying QTL for Δ13C variation | Parental lines of DH population (BC1329 and BC9102) | Pots in glasshouse (2020) under wet and dry conditions | None |
Note: The plant development stage at which traits were measured is detailed in Supporting Information: Method S1.
Abbreviations: DH, doubled haploid; QTL, quantitative trait loci.
2.4. Plant development and agronomic traits
Δ13C, days to flower, plant height, and seed yield were measured for Experiments 1–3 and 5, and normalized difference in the vegetative index (NDVI) was measured only for Experiment 2. The δ13C composition (13C/12C) was determined at laboratory phase experiments for dried leaf samples collected from field/pot phase experiments with appropriate multiphase experimental designs (Supporting Information: Method S1) to account for the variations attributed to field/pot and laboratory conditions (Smith et al., 2006), as described previously (Raman, Raman, McVittie, et al., 2020). Δ13C was determined from the leaf δ13C composition and that of the source CO2 in the air (taken as −7.8‰) as proposed by Farquhar and Richards (1984). For Experiment 4, Δ13C, days to flower, specific leaf weight, and leaf‐water content (LWC) were measured.
2.5. Physiological traits
The gas exchange measurements were taken at the single leaf level for the plots under wet conditions in Experiment 5, as the relationship between Δ13C and WUE varies under different water‐deficit levels. We determined iWUE by measuring light‐saturated assimilation rate (A) and stomatal conductance to the diffusion of water vapour (g sw). The fifth fully expanded leaf of each of the 70 lines of the DH population (representing extreme phenotypes), including parental lines, was tagged and utilized for gas exchange measurements. Details of parameters used for gas exchange measurements are given in our earlier study (Raman, Raman, McVittie, et al., 2020).
2.6. Light microscopy
A leaf disc (9.08 cm2 size) was taken from each of the two replicate plots in the wet block from Experiment 5, fixed, and stored in 70% ethanol as detailed (Supporting Information: Method S1). Leaf sections were stained using a method modified from Rae et al. (2020) and were imaged using 488 nm excitation and 500–560 nm emission on a Leica SP8 confocal microscope.
2.7. Genotyping and linkage map construction
Genotyping of DH lines was carried out using the PstI‐MseI restriction enzymes based genotyping‐by‐sequencing DArTseq approach (Raman et al., 2014). Sequence polymorphisms were used for linkage map construction, following the DArT P/L's mapping software OCDmap® detailed in Raman et al. (2014). Recombination frequencies were converted to centiMorgan (cM) map distances using the Kosambi function. The markers that showed cosegregation were “binned” into a unique locus and the resulting “bin” map was used to identify trait–marker associations. To obtain the physical position of markers, DArTseq sequences were aligned with the Darmor‐bzh reference assembly version 4.1 using the default parameter settings with the Bowtie programme.
2.8. Statistical methods
Commensurate with the aims of the experiments and the structure of the datasets, for Experiments 1–3 whole genome, single‐step QTL analyses were performed on each trait using an extension of the approach developed by Verbyla and Cullis (2012) within a multienvironment trial (MET) analysis framework using factor analytic linear mixed models (FA‐LMM) (Smith et al., 2015). A genome scan within a linear mixed model (LMM) framework was performed for Δ13C measured at Experiment 4 to validate the QTL identified from Experiments 1 to 3. Each trait measured for Experiment 5 was analysed individually using appropriate LMM. A detailed description of the methods is presented (Supporting Information: Method S1).
All analyses were performed in ASReml‐R (Butler et al., 2018), which provides residual maximum likelihood (REML) estimates of variance parameters, empirical best linear unbiased predictions (EBLUPs) of random effects, and empirical best linear unbiased estimates of fixed effects. The extent of genetic control of traits in each environment was investigated by calculating line mean H 2 (broad‐sense heritability) as the mean of the squared accuracy of the predicted line effects as described previously (Cullis et al., 2006) and found to be dependent on the environment. The across environment summary measure of overall performance (OP) proposed by Smith and Cullis (2018) was used to identify lines of interest. OP is one of the so‐called factor analytic selection tools to summarize the predictions from the fit of an FA‐LMM for a MET dataset in a concise yet informative manner. OP is defined for a line using the EBLUP of the first‐factor score in the FA‐LMM representing the generalized genotype main effect. We examined the genetic correlations between pairs of traits, especially Δ13C with agronomic traits (seed yield, days to flowering, plant height, and NDVI) using bivariate analyses where data for each pair of traits were combined and analysed together within the LMM framework. Three sets of bivariate analyses were performed: (1) Experiments 1–3, (2) Experiment 4, and (3) Experiment 5. Correlation between traits was modelled using an unstructured variance model and the inference on the genetic correlation between traits was obtained (see Supporting Information: Method S1).
2.9. Resequencing and structural variation analysis of parental lines
Libraries from high‐quality genomic DNA from both parental lines, BC1329, and BC9102, were constructed using the Illumina TruSeq DNA Preparation Kit, following the manufacturer's instructions (Illumina). Whole‐genome resequencing (2 × 150 bp) was performed at the Novogene facility (Novogene Co., Ltd.) using the Illumina HiSeq 2000 sequencing platform. The coverage of the parental lines ranged from 77.6 (BC1329, 102.6 Gb) to 83.8× (BC 9102, 112.4 Gb). Read mapping to the “Darmor‐bzh” reference assembly (version 4.1, http://www.genoscope.cns.fr/brassicanapus/data/), single nucleotide polymorphisms (SNP) and InDel (<50 bp) calling, structural variation (SV, ≥50 bp) detection, and identification of HE event (≥10‐kb windows) were performed as described in Raman et al. (2021). Details of HE analysis are described in Supporting Information: Method S1.
2.10. Development of sequence‐capture‐based DArTag markers
We processed resequence data for target Δ13C QTL regions on A09 and C09 chromosomes and selected 148 SNPs for DArTag oligo synthesis. Oligos were synthesized by IDT (Ultramer DNA Oligos, http://idtdna.com) at a 200 pmol scale, pooled in the equimolar amount into a single assay, and used for processing eight plates (96 well/plate) of DNA with the F2 population and a control canola sample using a proprietary DArTag assay (Targeted Genotyping—Diversity Arrays Technology) using 384 plate format. For each plate, a sample of the pooled product was also run on agarose gel and compared against positive control before proceeding with the sequencing process. The libraries were sequenced on Illumina Hiseq 2500 with an average volume of sequencing per sample at 43 225 sequencing reads (median at 46 389) and average read depth per assay at 280. Marker data were extracted using DArT PL's proprietary algorithm deployed as a plugin in the KDCompute application framework (https://www.kddart.org/kdcompute.html).
2.11. RNA sequencing and differential gene expression analysis
Parental lines, BC1329 and BC9102, of the DH population were grown with three replicates under both wet (100% field capacity) and dry (50% field capacity) conditions in a glasshouse (Experiment 6). Details of the layout of the experiment, sampling, RNA isolation, and sequence analysis are presented (Supporting Information: Method S1). A generalized linear model approach was then used to quantify the differential expression between the groups. The DEGs were obtained using a false discovery rate (<0.05). The p‐value was adjusted for multiple hypothesis testing using the Benjamini–Hochberg adjustment (p < 0.05). Heatmaps showing the expression pattern of genes in A09 and C09 QTL regions were produced using the ComplexHeatmap R package (Gu et al., 2016).
2.12. Identification of candidate genes for WUE
Arabidopsis thaliana genes which had been annotated with various WUE‐related terms were retrieved from the TAIR 10 database (https://www.arabidopsis.org/). These genes were then used to identify putative homologs in canola. We implemented an improved syntenic approach to exclude genes as causal candidates in syntenic QTL regions combining genomic and transcriptomics data (Zhang et al., 2019).
3. RESULTS
3.1. Substantial genetic variation in Δ13C and other productivity traits
We observed high levels of genetic variation in Δ13C and other WUE‐related traits in the DH population. The significant source of genetic variation was from the additive component (genetic markers), where the mean values ranged from 21.5% for NDVI to 79.1% for days to flower (Supporting Information: Table S1, Additive M1, %). Broad sense heritability estimates for Δ13C and other integrated WUE‐related traits (plant height, NDVI, flowering time, and seed yield) were variable, ranging from low (56%) to high (98%), depending on the nature of the trait and growing environment (Supporting Information: Table S2). Across traits, pot experiments had lower H 2 values compared to field experiments. The frequency distributions of the traits from DH lines evaluated across environments are presented (Supporting Information: Figure S3). Estimated additive and total (additive plus non‐additive) genetic correlations between environments revealed that there are strong correlations between environments for both additive and total genetic variance with values greater than 0.89 and 0.83, respectively, for all traits (Supporting Information: Table S3). Total (additive plus non‐additive) OP estimates for Δ13C ranged from 18.73‰ to 21.25‰ and displayed transgressive segregation among DH lines across environments (Figure 1a, Supporting Information: Table S4). Up to 2.52‰ variation in Δ13C was observed among DH lines which equates to a fivefold increase compared with the parental lines.
Figure 1.
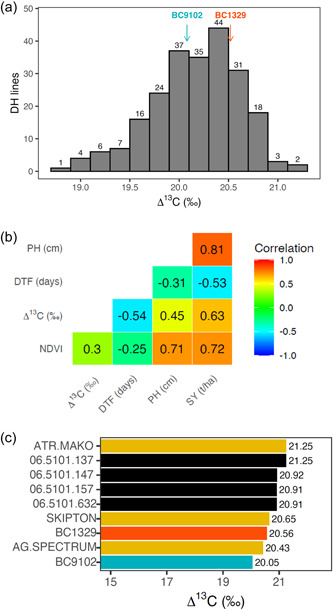
Genetic variation in WUE traits and their relationships among doubled haploid lines derived from the cross, BC1329/BC9102 (Experiments 1–3). (a) Frequency distribution of the OP for Δ13C. Estimates for the parental lines are shown with arrows; (b) genetic correlations between Δ13C (‰) and other WUE‐related traits; and (c) the top four DH lines that showed the highest Δ13C based on OP across environments in relation to control commercial varieties of canola and the parental lines are shown. DH, doubled haploid; DTF, days to flower; NDVI, normalised difference in the vegetative index; OP, overall performance; PH, plant height; SY, seed yield; WUE, water use efficiency.
3.2. Genetic correlations between WUE and productivity traits at plot level
To determine the relationships between Δ13C and other WUE‐related traits, between trait total (additive plus non‐additive) genetic correlations were obtained using bivariate analyses (Figure 1b). The Δ13C showed a negative correlation with days to flower (r = −0.54), while positive correlations were observed with NDVI, a proxy for plant vigour (r = 0.30), plant height (r = 0.45), and seed yield (r = 0.63). Flowering time showed a negative correlation with seed yield (r = −0.53). The promising DH lines that had high seed yield (high Δ13C) for use in canola breeding programmes based on the OP estimates are presented in Figure 1c. DH line 06‐5101‐137 had the maximum Δ13C (21.25‰) among the DH progenies.
3.3. Genetic correlations between physiological WUE (single leaf level) and integrated WUE (whole plant level)
Significant genetic variation for both A and g sw was observed, although the H 2 estimate of iWUE was low (Supporting Information: Table S5). Genotype EBLUPs for A and g sw ranged from 4.97 to 17.15, and 0.11 to 0.38, respectively (Supporting Information: Table S5). The estimated between trait genetic (non‐additive) correlations revealed that both A and g sw are dependent on each other with a correlation of 0.61 (Figure 2a). We observed a negative correlation between Δ13C and iWUE (r = −0.38), indicating that DH lines with low Δ13C have higher iWUE, consistent with the findings made earlier (Adiredjo et al., 2014; Farquhar & Richards, 1984). There was a more negative correlation between iWUE and g sw (r = −0.68) in comparison to A (r = −0.33), suggesting that g sw is the predominant driver for variation in iWUE parameters.
Figure 2.
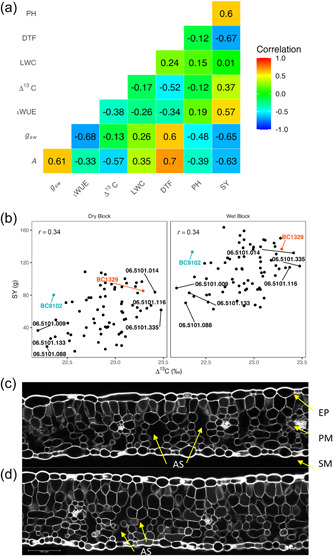
Relationships between Δ13C, gas exchange measurements (CO2 assimilation (A), stomatal conductance (g sw), intrinsic water‐use efficiency (iWUE), and plant developmental and agronomic traits of selected 70 DH lines of the BC1329/BC9102 population, representing extremes (high and low values) in Δ13C and their parents (Experiment 5). (a) Genetic correlations from the bivariate analyses are shown. (b) Relationships between Δ13C and seed yield (SY) for wet and dry blocks. Genotype EBLUPs for Δ13C and SY are plotted. Parental lines and the DH lines with high and low Δ13C are labelled. Leaf sections showing differences in air spaces (AS, marked with arrow) between parental lines BC1329 (c) and BC9102 (d). DH, doubled haploid; DTF, days to flower; EBLUP, empirical best linear unbiased predictions; EP, epidermis; LWC, leaf‐water content; PH, plant height; PM, palisade mesophyll (comparatively regular elongated cells); SM, spongy mesophyll (irregular cells).
This study showed that Δ13C correlates negatively with iWUE but it (Δ13C) correlates positively with seed yield (Figure 2a). Under well‐watered conditions, there were negative correlations between Δ13C and days to flower, A, and iWUE. We further investigated relationships between LWC at a single leaf level and WUE traits at the whole plant level and found that LWC shows a negative relationship with Δ13C, but it did not show any relationship with seed yield (Figure 2a). Furthermore, the estimated genetic correlations between wet and dry blocks for seed yield (Figure 2b) and plant height, the only two traits measured after imposing water stress at the first flowering stage, were very high (0.93 for both traits). This suggests that genotype by irrigation block interaction is small. High Δ13C lines revealed higher yield across irrigation blocks compared to low Δ13C lines (Figure 2b). Our anatomical analysis of leaf discs revealed both parental lines BC1329 and BC9102 differ in thickness and arrangement of palisade and spongy mesophyll cells: BC1329 (192 μm) had high porosity with large air spaces compared to BC9102 (184 μm, Figure 2c,d), which may facilitate gas exchange, thus leading to efficient water use.
3.4. Genetic basis underlying Δ13C and WUE‐related traits
We constructed a linkage map that includes 8985 DArTseq markers onto 24 linkage groups, representing all the 19 chromosomes of B. napus (Supporting Information: Table S6a). To reduce computation time for genetic analysis, we produce a “bin” map of 1793 markers that spanned a total of 1965.29 cM, with an average interval of 1.10 cM between adjacent loci (Supporting Information: Table S6c). Multienvironment QTL analyses identified a total of 29 QTL (15 QTL for main effects); corresponding to genotypic (G) effects, and 14 for QTL (Q) × environment (E) interactions; corresponding to G × E effects for variation in leaf Δ13C and other WUE‐related traits (Supporting Information: Table S7). For Δ13C, three QTL main effects that showed statistically significant (logarithm of the odds [LOD] ≥ 3) associations were identified on chromosomes A08, A09, and C09, while one “suggestive” QTL (LOD > 2.5 but less than 3) was located on chromosome A07 (Supporting Information: Table S7, Figure 3a, Supporting Information: Figure S5). We identified QTL for phenotypic variation in different traits between three growing environments (Q × E interaction) on A02, A05, A08, A09, A10, C02, C03, C06, C07, and C09 chromosomes (Supporting Information: Table S7). For Δ13C, two QTL for Q × E interactions were identified on chromosomes A02 and C06 (Figure 3a). Collectively, QTL explained 38% of genotypic variation in Δ13C (Supporting Information: Table S1, VAFm) of which the QTL delimited with DArTseq marker 3153720 accounted for the most R 2 (Supporting Information: Table S7). Environment‐specific QTL showed a range of allelic effects, varying from consistent (in the same direction with same/similar magnitude) to opposite (in opposite direction with similar/variable magnitude, Supporting Information: Table S7). The strongest Q × E interactions with opposite allelic effects (different parental alleles) were observed for Δ13C (A02, FT18), days to flower (A05, FT18; C02‐FT17), plant height (C07 and C09; rainout shelter17, FT18), with variable allelic effects; depending on the environment (Supporting Information: Table S7). This implies that G × E interactions contribute to observed trait variation. Reaction norm of Δ13C estimates from DH lines evaluated under two rainfed (field‐2017, 2018, Experiments 1 and 2) and two irrigated (pot‐2017, wet block of rainout shelter‐2019, Experiments 3 and 5) environments showed a change in scale (non‐crossover interaction) and as well as ranks (crossover interaction), suggesting that variation in Δ13C is also due to G × E interactions (Figure 4).
Figure 3.
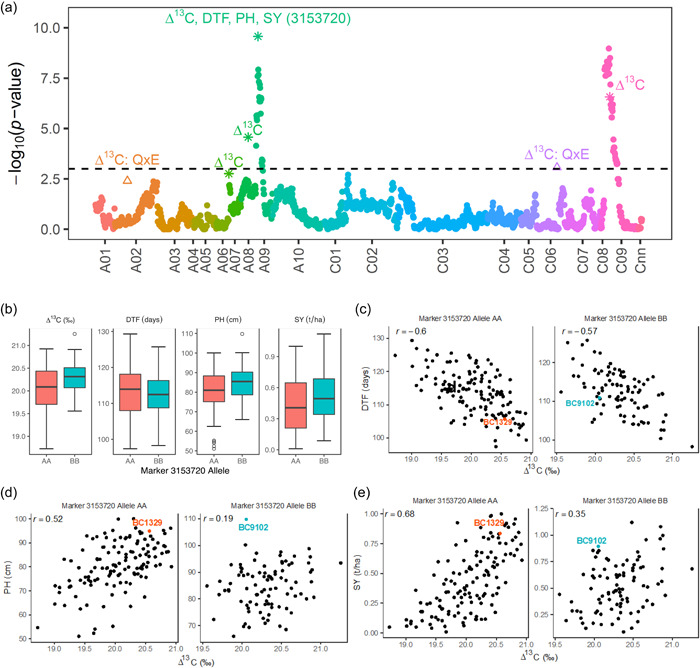
Distribution and relationships between Δ13C, days to flower (DTF), plant height (PH) and seed yield (SY), and DArTseq marker alleles for the QTL (3153720) that colocalized in the same genomic region on chromosome A09. (a) Manhattan plot showing LOD scores for associations between DArTseq markers and Δ13C. QTL main effects (depicted by*) are labelled with the respective trait (for DTF, PH, and SY only the 3153720 QTL is shown) and QTL × environment (E) interactions (depicted by Δ) are labelled with the trait followed by “Q × E” (only shown for Δ13C). LOD scores presented in the Manhattan plot are from the genome scan for the QTL main effects where the LOD scores of the significant QTL are replaced with the ones from the final model. The black dash line indicates the threshold value for significant SNPs at LOD ≥ 3. Manhattan plot for each trait, are presented in Supporting Information: Figure S4. (b) Box plots showing the distribution of the OP estimates for Δ13C, DTF, PH, and SY partitioned into allele combinations, “AA (BC1329)” and “BB (BC9102),” for the SNP marker 3153720. Pair‐wise correlations of OP estimates between Δ13C versus DTF (c), Δ13C versus PH (d), and Δ13C versus SY (e) are partitioned into different allelic combinations. LOD, logarithm of the odds; OP, overall performance; QTL, quantitative trait loci; SNP, single‐nucleotide polymorphisms.
Figure 4.
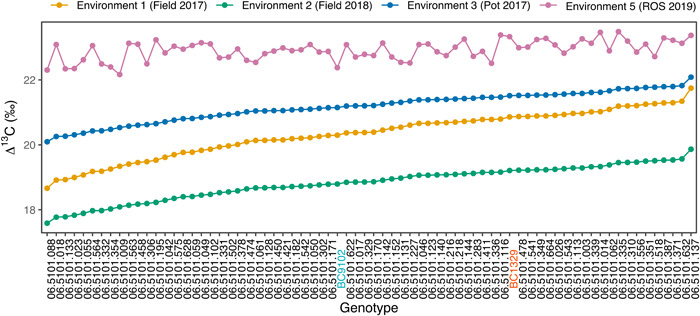
Reaction norm and phenotypic variation in Δ13C (‰) among a subset of 70 DH lines and the parental lines evaluated under different environments (Experiments 1–3, 5). CGE EBLUPs for Experiments 1–3 and genotype EBLUPs for Experiment 5 are plotted to show the change in scale and ranks. Genotypes are ordered based on the ranks for Environment 1. DH, doubled haploid; EBLUP, empirical best linear unbiased predictions; CGE, common genotype by environment.
3.5. Comparative localization of QTL
Three QTL for multiple traits on chromosomes A01, A08, and A09 were colocalized to the same genomic regions (Supporting Information: Table S7, Supporting Information: Figure S5). One QTL delimited with marker 3153720 for variation in Δ13C was colocated with QTL for days to flower, plant height, and seed yield on chromosome A09 (Supporting Information: Table S7, Figure 3a). We further sought a correlation between allelic effects of markers and variation in Δ13C, days to flower, plant height, and seed yield (Figure 3b–e). Up to 68% of allelic effects were explained by the same marker allele (Figure 3e), suggesting pleiotropic relationships between these traits and/or tight genetic linkage between them.
3.6. Verification of Δ13C QTL in an F2 population
We validated the genetic control, the linkage between 148 DArTseq markers and Δ13C (in DH population), and focused on the identification of candidate gene(s) underlying most of the genetic variation in Δ13C at QTL regions on chromosomes A09 and C09 (Supporting Information: Table S7). The Δ13C values showed normal segregation among F2 lines (Figure 5a). Consistent with DH lines, Δ13C exhibited a negative correlation with flowering time and SLW, and a positive correlation with LWC (Figure 5b). Genetic analysis revealed that the majority of polymorphic DArTag markers (83.3%, 65/78) show significant segregation distortion (deviating from the normal segregation consistent with a 1:2:1 ratio for codominance, or 3:1 ratio for dominance) on chromosomes A09 and C09 (Supporting Information: Table S8) suggesting that the corresponding multiple trait QTL regions could be subjected to SV. Genome scan within LMM framework revealed that DArTag markers positioned at 29 499 141 bp on chromosome A09, and 46 318 271 bp on C09 of the Darmor‐bzh genome exhibit statistically significant association with Δ13C in an F2 population (Supporting Information: Figure S4). Physical mapping verified that the genomic regions for Δ13C variation are localized to the same genetic intervals in both DH and F2 populations (Figure 5c–f).
Figure 5.
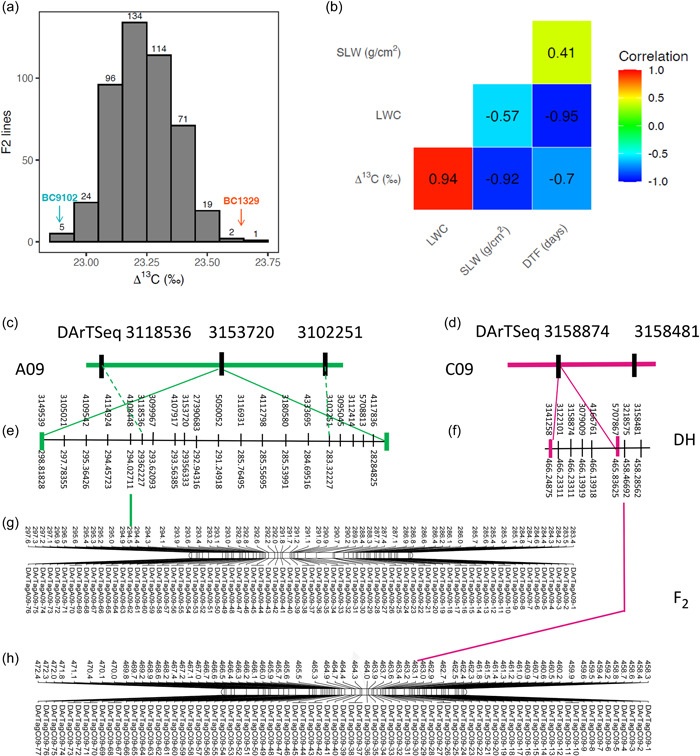
Validation of the genetic linkage between Δ13C and molecular markers in an F2 validation population derived from the BC1329/BC9102, grown under non‐stress conditions (Experiment 4). The frequency distribution of Δ13C (‰) among 733 F2 lines (a). Genetic correlations between Δ13C, DTF, LWC, and SLW from bivariate analyses are shown (b). The genetic position of flanking markers for Δ13C QTL on A09 (c) and C09 (d) and their corresponding physical positions (e, f) in the DH population and physical map positions of DArTseq markers on A09 and C09 chromosomes in an F2 population. (g, h) Comparative physical locations of markers are shown with lines (in colour). For clarity, physical positions are given in 1/10th fraction of Mb. DH, doubled haploid; DTF, days to flower; LWC, leaf‐water content; QTL, quantitative trait loci; SLW, specific leaf weight; Δ13C, carbon isotope discrimination.
3.7. Δ13C QTL region on chromosome A09 is subjected to HE
To investigate whether significant segregation distortion among marker alleles on chromosomes A09 and C09 (Supporting Information: Table S8) is due to SV, we resequenced the genomes of parental lines of mapping/validation populations (NCBI, Accession No. PRJNA743730). Sequence mapping revealed 26 genomic regions undergone HE events, varying from 90 to 870 kb, including the A09 multiple trait QTL regions (29.1 to 29.3 Mb) from the C08 chromosome in BC9102, as a result of homoeologous recombination (Figure 6a, Supporting Information: Table S9). However, in the maternal line BC1329, no such events were identified (Figure 6b).
Figure 6.
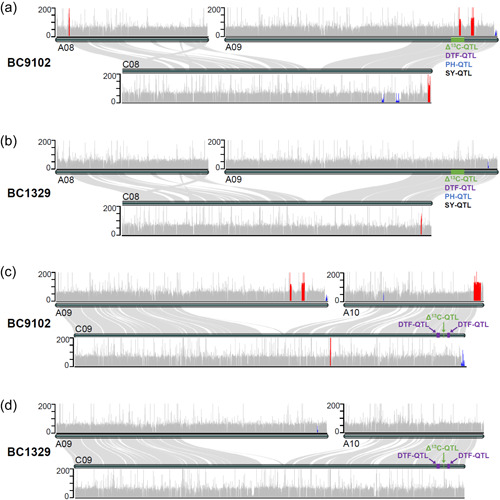
Homoeologous exchange (HE) events were detected between parental lines of the doubled haploid population derived from the BC1329/BC9102 Coverage depth (vertical axis) obtained for each chromosomes was calculated by mapping the Illumina resequencing reads to the reference genome of B. napus Darmor‐bzh. The height of each line (a 10‐kb window) from the horizontal axis represents the coverage depth. Genomic sequences (regions) that have undergone HE is shown in Supporting Information: Table S9. Substituted and “translocated” reads are highlighted in blue and red colour, respectively. DTF, days to flower; PH, plant height; QTL, quantitative trait loci; SY, seed yield.
3.8. Candidate genes associated with WUE near Δ13C QTL
We first interrogated genomic regions underlying the significant marker associations in both the mapping (DH) and validation (F2) populations (Figure 3c–h). DArTseq “bin” marker 3153720 on A09 revealed the complete linkage with another 18 markers in the DH population, which were localized within the 1.54 Mb region, spanning 28.28 to 29.82 Mb (Supporting Information: Tables S6 and S9). To identify potential candidate genes involved in the Δ13C variation, we first excluded the syntenic genes in QTL regions and their corresponding homologs without any SNP and gene expression (logFC ≥ 1.5) variations between parental lines (Supporting Information: Table S10). In addition, we also excluded the genes in QTL regions with synonymous SNPs and InDels. We then focused on the remaining genes especially non‐syntenic genes in QTL regions with different expression levels between parental lines. Finally, we identified a total of eight genes as candidates for Δ13C variation at three QTL regions of chromosomes A08, A09, and C09 (Supporting Information: Table S10). Annotation of genomic intervals revealed that haloacid dehalogenase‐like hydrolase superfamily protein (BnaA08g17740D) on chromosome A08/C03, myelin transcription factor‐like protein (BnaA09g40350D), C3HC4‐type RING finger protein (BnaA09g40860D), serine carboxypeptidase‐like 23 (BnaA09g41720D), poly(A) binding protein 4 (BnaA09g42030D), and cation‐transporting P‐type ATPase (BnaA09g42040D) and cytochrome P450 (BnaA09g42330D) on A09 and its homoeologous chromosome C08, and serine carboxypeptidase‐like 19 (BnaC09g47000D) on C09/A10 chromosomes are likely candidates to be involved in Δ13C variation (Supporting Information: Table S10, Supporting Information: Figure S4). The leaf tissue‐specific transcriptome analyses revealed a total of 60 genes on A09 and 51 genes on C09 underlying Δ13C QTL regions that were significantly differentially expressed between the two parental lines: BC1329 and BC9102 (Supporting Information: Table S11, Figure 7). Several of the DEGs, such as myelin transcription factor‐like protein, casein kinase 2 α4, cation‐transporting P‐type ATPase, C3HC4‐type RING finger protein, and serine carboxypeptidase, are related to water use, WUE, and response to water stress (https://www.arabidopsis.org/). We also found that the expression levels of genes in BC9102 (with HE event) such as BnaA09g41850D, BnaA09g41970D (wall‐associated receptor kinase‐like 14), BnaA09g41990D (cyclin‐dependent kinase inhibitor), BnaA09g42000D (nicotinate phosphoribosyltransferase 2), BnaA09g42030D (RNA recognition motif domain), and BnaA09g42040D were significantly higher (at least twofold) than those of BC1329 (without HE event) (Figure 8, Supporting Information: Table S11), suggesting that HE may be responsible for expression variation at the Δ13C‐QTL region on A09.
Figure 7.
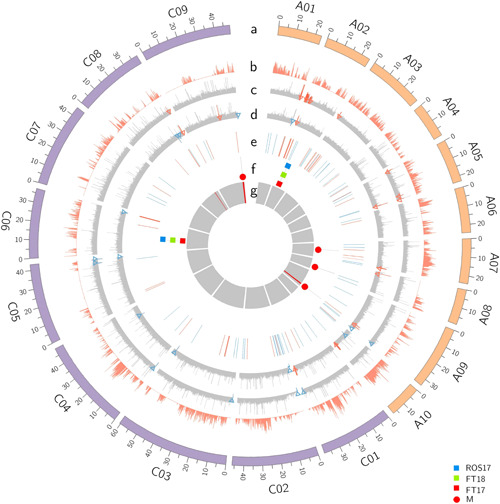
Physical map positions of QTL controlling Δ13C variation in a doubled haploid population derived from the BC1329/BC9102. a: the outermost circle represents the physical length of the chromosomes (A genome in orange, C genome in purple), b: polymorphic SNPs between BC1329 and BC9102, c: homoeologous exchanges identified in BC1329, d: homoeologous exchanges identified in BC9102, e: DEGs identified between BC1329 and BC9102 (under dry and wet conditions), f: QTL associated with Δ13C variation; blue, green and red, g: DEGs located within QTL regions. DEG, differentially expressed gene; QTL, quantitative trait loci; SNP, single nucleotide polymorphism.
Figure 8.
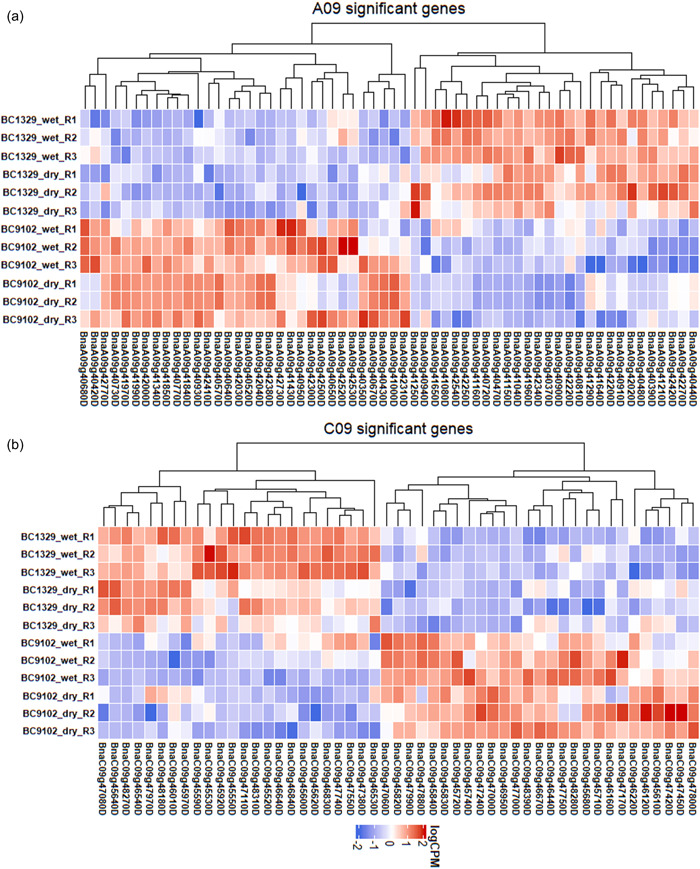
Expression profiles of differentially expressed genes (DEGs) in A09 (a) and C09 (b) QTL regions under water‐deficit and water non‐deficit conditions (Experiment 6) of the parental lines of the doubled haploid population derived from the BC1329/BC9102. The normalized read counts were plotted as a heatmap and genes were clustered according to the basis of their expression pattern. Detailed information on DEGs of parental lines (in triplicate: R1, R2, and R3) is presented in Supporting Information: Table S11. QTL, quantitative trait loci.
4. DISCUSSION
Delineating the genetic and environmental determinants underlying variation in WUE traits is essential for the optimized breeding of high‐yielding varieties in water‐limited environments. In this study, we explored the genetic basis of variation in Δ13C and related WUE traits in the biparental population derived from a cross between BC1902 and BC9102 lines that differ in Δ13C and TE signatures (Hossain et al., 2020). We employed multienvironment‐based QTL analysis which is considered a more powerful approach to dissecting complex traits, controlled by G, E, and G × E interactions than the traditional QTL approaches (Zhang et al., 2010). We show that the growing environment plays an important role in the phenotypic expression of trait variation; thus, in the identification of environmental stable and environment‐specific QTL effects. To reveal candidate genes underlying stable QTL for Δ13C, our work also identifies SV (HE), and DEGs between the parental lines of the mapping population, suggesting that homoeologous recombination may contribute to phenotypic variation for multiple traits at the chromosome A09 QTL.
4.1. Canola reveals considerable variation for Δ13C
We found substantial genotypic variation in Δ13C, from 18.78‰ to 21.23‰ among DH, and 22.89‰ to 23.67‰ among F2 lines. An earlier study has shown that an increase of 0.5‰ in δ13C can lead to 25% more TE in Arabidopsis (Juenger et al., 2005). Extrapolating this relationship, which is positive between Δ13C and syWUE, and negative between Δ13C and iWUE (Figure 2A), canola lines with 2.52‰ higher Δ13C values than parental lines should reduce WUE theoretically by 126%. It reflects the dependence of the sensitivity on the general level of Δ13C. For example, Masle et al. (2005) found that at the level they saw in Arabidopsis, an increase in Δ13C of 1‰ was associated with a 15% decrease in TE.
4.2. Integrated WUE is partly driven by fitness traits
This study showed that DH lines that discriminate less between 12C and 13C as a carbon source for photosynthesis (low Δ13C) show higher iWUE at the single leaf level (Figure 2a). However, low Δ13 lines did not produce high seed yield) reiterating that Δ13C estimates TE at the leaf level which is not syWUE. Our results also suggest that selection for high iWUE at a single leaf level is useful for improving seed yield (r = 0.57, Figure 2a), rather than using low Δ13C as a surrogate trait for predicting high seed yield in canola, consistent with our earlier findings made in the Skipton/Ag‐spectrum DH population as well as in diverse canola accessions (Raman, Raman, McVittie, et al., 2020). The positive relationship observed between Δ13C and seed yield could be due to genotypic variation in WUE being driven by variation in soil water capture and water use rather than by variation in assimilation per unit of water applied (Blum, 2005; Kobata et al., 1996; Sinclair, 2018). Δ13C estimates TE at the leaf level which is not syWUE. In this study, we did not establish the relationship between Δ13 and total dry matter production. Canola plants often senesce their leaves before the crop reaches maturity, and, therefore, it was not possible to estimate WUE accurately without measuring leaf mass. WUE, being a multidimensional trait can also be driven by other “fitness” traits that reduce evapotranspiration rate and crop water use. For example, high Δ13C lines with faster growth (NDVI, a proxy for plant vigour and plant height) could provide quicker canopy cover, which enables plants to reduce water loss from soil evaporation, thus increasing seed yield (r = 0.63 to 0.81, Figure 1b). This is partly supported by this study showing a high correlation between plant fitness and seed yield and tight linkage of corresponding QTL (Figures 1, 2, 3). In addition, Δ13C exhibited negative correlations with flowering time (r = −0.53; DH population), and a positive correlation with NDVI, plant height, and seed yield (Figure 1b) suggesting that high Δ13C lines tend to “escape” via accelerating growth and flowering—an evolutionary trait for adaptation to terminal drought stress. Our results showed that genotypes with low Δ13C had less canopy cover, late flowering, and lower seed yield; these characteristics are typical for plants with a drought avoidance strategy. However, under terminal water‐deficit situations, low Δ13C lines could yield poorly due to the shorter seed filling period, accompanied by high temperatures. It remains to establish how low Δ13C lines which require a longer season for seed filling, perform in climates that are not prone to environmental constraints (non‐water deficit/heat stress). Research findings suggest that canola breeding programmes should focus on adaptative traits such as early flowering, and drought avoidance traits such as early vigour for effective water use for achieving high yield under water‐limited conditions.
4.3. Genetic and environmental determinants affect phenotypic trait expression
Our comprehensive multienvironment QTL analysis showed that by using well‐designed multiphase experiments across three environments (Experiments 1–3, Table 1) and accounting for sources of variation in field/pot and laboratory conditions (Supporting Information: Method S1), both genetic and environmental determinants underpinning phenotypic variation can be deciphered for traits of interest (Supporting Information: Table S7). For example, we substantiated phenotypic plasticity for Δ13C/WUE‐related traits (Figure 4) and identified QTL for the main effects and Q × E interaction effects underlying variation in Δ13C, days to flower, seed yield, and plant height under field conditions (Supporting Information: Table S7). In addition, the magnitude and inconsistent direction of allelic effects also hint that observed variation in Δ13C, days to flower, seed yield, and plant height across environments is due to Q × E interaction. A substantial proportion of QTL (51.7%, 15/29) that exhibited Q × E interaction, suggests their contribution to phenotypic trait variation across different environments (Supporting Information: Table S7). Similar estimates for Q × E interaction effects are reported in other studies (Borevitz et al., 2002; El‐Soda et al., 2014; Ungerer et al., 2003).
Consistent detection of Δ13C‐QTL (QTL main effects on A07, A08, A09, and C09) across three environments suggests that these loci contribute to the adaptive capacity of DH lines from BC9102/BC1902) to water‐deficit stress conditions and thus translate to economic seed yield (~1 t/ha). Across field environments, DH lines were subjected to water‐deficit conditions, right from stem elongation to seed maturity (rainfall ranged from 225 to 235 mm over 7 months of the growing season, Supporting Information: Figure S1). Colocation of QTL for seed yield, Δ13C, days to flower and plant height at the same genomic regions, and stable allele (BC9102), contributing to trait variation suggest multiple trait QTL on chromosome A09 is associated with effective water use. Early flowering showed a negative relationship with seed yield (Figure 1b), reiterating the crosstalk between drought stress signalling and flowering time pathways (Des Marais et al., 2012).
It was interesting that none of the Δ13C QTL that we identified for the main effect and Q × E interactions (Supporting Information: Table S7) was detected in the Skipton/Ag‐spectrum population (Raman, Raman, McVittie, et al., 2020). In an independent study, Mekonnen et al. (2020) identified three QTL for δ 13C on chromosomes A02, A09, and C08 in the North American B. napus mapping population. However, none of the QTL was consistently detected across environments. It is yet to establish whether the genomic region on chromosome A09 or its homoeologous counterpart C08 (QTL for root pulling force, plant height, and δ 13C) is the same as found in our study, as the authors did not report the physical positions of QTL marker intervals. In addition, there was a poor marker coverage on chromosome C08 in our genetic mapping population due to low recombination frequency (13 markers, Supporting Information: Table S6c), which may have led to QTL (if any) being undetected in the unmapped regions, especially in the HE region. Nevertheless, these studies suggest that several genomic regions on A02, A03, A07, A09, C03, C06, C08, and C09 control variation in Δ13C; thus, the genetic architecture of Δ13C is rather complex.
4.4. A priori genes regulating WUE and efficient water use underlie QTL for Δ13C
Genetic and genomic approaches utilized herein facilitated the identification (in a DH population from BC1902/BC9102) and validation (in an F2 population from BC1902/BC9102) of genomic regions for Δ13 variation and delimited candidate genes in canola, which are implicated in leaf‐level WUE (Cutler et al., 2010; Hersen et al., 2008; Menéndez et al., 2019; Tao et al., 2018; Youn et al., 2016). For example, this study identified a QTL that influences multiple traits; Δ13C, days to flower, plant height, and seed yield on chromosome A09 that map within 92 kb of the ERECTA gene (see Masle et al., 2005) (Supporting Information: Table S12). We also localized several stress‐responsive genes, including DEGs that may contribute to drought avoidance strategies via signal transduction pathways, encoding functional proteins (LEA18, RD20, glycine metabolism, and CAT) and regulatory proteins, including transcription factors (bHLH, MYB, TINY2, ATHB6), protein kinases (tyrosine‐protein kinase, wall‐associated receptor kinase‐like 14, MAPK, SNF1‐related protein kinase), and receptors (ABA receptor PYL12), phosphatases (PP2C), and calmodulins (CPK17) (Des Marais et al., 2014; Jagodzik et al., 2018; Jonak et al., 2002; Yong et al., 2019) within QTL intervals associated with Δ13C variation (Table 1, Supporting Information: Tables S11 and S12). Plant expressing PYL12, SRK2C, and AtTPPF genes are shown to improve the water use and drought tolerance (Lin et al., 2019; Yang et al., 2016) whereas ABC transporter (ABCG22) and ABA‐responsive kinase gene, MPK12 reduced the WUE (Des Marais et al., 2014). Our data hint that genes affecting stomatal characteristics (RD20, ERECTA), leaf thickness, and water‐deficit responsive genes described above likely underlie WUE and drought avoidance traits, while Q × E interactions are likely driven by environmental cues such as water stress and temperature (phytochrome C was mapped with 6.2 kb from Δ13C‐QTL on C06, Supporting Information: Table S12). Our results suggest that a QTL region underlying Δ13C, flowering time, plant height, and seed yield on chromosome A09 may be subjected to HE. Homoeologous recombination is associated with presence‐absence variation (Hurgobin et al., 2018; Nicolas et al., 2007). Recently, a major QTL for homoeologous recombination, BnaPh1 was mapped on A09 (Higgins et al., 2021) and this was located within 5 Mbp of the QTL region that is associated with multiple traits. The same genomic region may be involved in regulating WUE in diverse canola accessions and require further research.
The most pressing need for research is to clarify the bases of the observation of a positive relation between seed yield and leaf Δ13C, despite the negative relation between shoot growth and leaf Δ13C. Condon et al. (1987) reported the same phenomenon in wheat and discussed some possible biophysical explanations, including the idea that the major source of variation might be stomatal conductance rather than photosynthetic capacity. The negative association between time to flowering and Δ13C may also have contributed, but the most revealing observation (Condon & Richards, 1993) was that wheat crop yields were positively correlated with Δ13C in wet environments, but negatively correlated in dry environments, with no association under intermediate levels of water limitation.
In summary, this current study demonstrates that measures of iWUE, Δ13C, and integrated WUE (dmWUE and syWUE) are complex and modulated by genetic and environmental determinants, including that subject to HE. Our findings identify useful variation in Δ13C (up to 2.52‰) and its underlying genetic basis of variation in WUE traits, including their plasticity across the natural field and rainout shelter environments. Identification of favourable alleles for increasing WUE would provide potential resources for developing new drought‐tolerant varieties for drier environments to continue making genetic gains in the breeding programmes via genomic‐assisted selection strategies. Resources developed in this study could also enable us to understand molecular mechanisms underlying phenotypic plasticity and G × E interactions.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Simon Diffey (Apex Biometry) for the multiphase experimental designs for Δ13C and Dr. Alison Smith (UOW) for constructive discussions on the statistical methods. They are grateful to Mr. Warren Bartlett and Mr. Dean McCullum, for their assistance in sowing and management of field experiments; Hannah Roe and Wayne Pitt for grinding leaf samples for Δ13C analysis; and Advanta for providing F1 cross. This study was supported by the Australian Grains Research and Development Corporation and NSW Department of Primary Industries and partners (projects: DAN00117 and DAN00208. Open access publishing facilitated by New South Wales Department of Planning Industry and Environment, as part of the Wiley‐New South Wales Department of Planning Industry and Environment agreement via the Council of Australian University Librarians.
Raman, H. , Raman, R. , Pirathiban, R. , McVittie, B. , Sharma, N. , Liu, S. et al. (2022) Multienvironment QTL analysis delineates a major locus associated with homoeologous exchanges for water‐use efficiency and seed yield in canola. Plant, Cell & Environment, 45, 2019–2036. 10.1111/pce.14337
Contributor Information
Harsh Raman, Email: harsh.raman@dpi.nsw.gov.au.
Yuanyuan Zhang, Email: zhangyy@caas.cn.
DATA AVAILABILITY STATEMENT
The raw sequence data reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive (Accession No. PRJNA743730 for RNA‐sequencing data, PRJNA743989 for whole‐genome resequencing data).
REFERENCES
- Adiredjo, A.L. , Navaud, O. , Lamaze, T. & Grieu, P. (2014) Leaf carbon isotope discrimination as an accurate indicator of water‐use efficiency in sunflower genotypes subjected to five stable soil water contents. Journal of Agronomy and Crop Science, 200(6), 416–424. [Google Scholar]
- Bayer, P.E. , Scheben, A. , Golicz, A.A. , Yuan, Y. , Faure, S. , Lee, H. et al. (2021) Modelling of gene loss propensity in the pangenomes of three Brassica species suggests different mechanisms between polyploids and diploids. Plant Biotechnology Journal, 19, 2488–2500. 10.1111/pbi.13674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino, L.T. , Caine, R.S. & Gray, J.E. (2019) Impact of stomatal density and morphology on water‐use efficiency in a changing world. Frontiers in Plant Science, 10, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, A. (2005) Drought resistance, water‐use efficiency, and yield potential—are they compatible, dissonant or mutually exclusive? Australian Journal of Agricultural Research, 56, 1159–1168. [Google Scholar]
- Blum, A. (2009) Effective use of water (EUW) and not water‐use efficiency (WUE) is the target of crop yield improvement under drought stress. Field Crops Research, 112, 119–123. [Google Scholar]
- Borevitz, J.O. , Maloof, J.N. , Lutes, J. , Dabi, T. , Redfern, J.L. , Trainer, G.T. et al. (2002) Quantitative trait loci controlling light and hormone response in two accessions of Arabidopsis thaliana . Genetics, 160(2), 683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, D.G. , Cullis, B.R. , Gilmour, A.R. , Gogel, B.J. & Thompson, R. (2018) ASReml‐R reference manual version 4. VSN International Ltd. Technical Report.
- Campitelli, B.E. , Des Marais, D.L. & Juenger, T.E. (2016) Ecological interactions and the fitness effect of water‐use efficiency: competition and drought alter the impact of natural MPK12 alleles in Arabidopsis . Ecology Letters, 19(4), 424–434. [DOI] [PubMed] [Google Scholar]
- Chalhoub, B. , Denoeud, F. , Liu, S. , Parkin, I.A. , Tang, H. , Wang, X. et al. (2014) Early allopolyploid evolution in the post‐Neolithic Brassica napus oilseed genome. Science, 345(6199), 950–953. [DOI] [PubMed] [Google Scholar]
- Champolivier, I. & Merrien, A. (1996) Effects of water stress applied at different growth stages to Brassica napus L. var. Oleifera on yield, yield components and seed quality. Journal of Agronomy, 93(3), 53–58. [Google Scholar]
- Cheung, F. , Trick, M. , Drou, N. , Lim, Y.P. , Park, J.Y. , Kwon, S.‐J. et al. (2009) Comparative analysis between homoeologous genome segments of Brassica napus and its progenitor species reveals extensive sequence‐level divergence. The Plant Cell, 21, 1912–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon, A. & Richards, R. (1993) Exploiting genetic variation in transpiration efficiency in wheat: an agronomic view. In: Ehleringer, J.R. , Hall, A.E. & Farquhar, G.D. (Eds.) Stable isotopes and plant carbon‐water relations. San Diego, CA: Academic Press, pp. 435–450. [Google Scholar]
- Condon, A.G. , Richards, R.A. & Farquhar, G.D. (1987) Carbon isotope discrimination is positively correlated with grain yield and dry matter production in field‐grown wheat. Crop Science, 27, 96–1001. [Google Scholar]
- Cullis, B.R. , Smith, A.B. & Coombes, N.E. (2006) On the design of early generation variety trials with correlated data. Journal of Agricultural, Biological, and Environmental Statistics, 11, 381–393. 10.1198/108571106X154443 [DOI] [Google Scholar]
- Cutler, S.R. , Rodriguez, P.L. , Finkelstein, R.R. & Abrams, S.R. (2010) Abscisic acid: emergence of a core signaling network. Annual Review of Plant Biology, 61(1), 651–679. [DOI] [PubMed] [Google Scholar]
- Des Marais, D.L. , Auchincloss, L.C. , Sukamtoh, E. , McKay, J.K. , Logan, T. , Richards, J.H. et al. (2014) Variation in MPK12 affects water use efficiency in Arabidopsis and reveals a pleiotropic link between guard cell size and ABA response. Proceedings of the National Academy of Sciences of the United States of America, 111(7), 2836–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais, D.L. , McKay, J.K. , Richards, J.H. , Sen, S. , Wayne, T. & Juenger, T.E. (2012) Physiological genomics of response to soil drying in diverse Arabidopsis accessions. Plant Cell, 24(3), 893–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi, M.J. , Bhatnagar‐Mathur, P. , Sharma, K.K. , Serraj, R. , Anwar, S.Y. & Vadez, V. (2011) Relationships between transpiration efficiency and its surrogate traits in the rd29A:DREB1A transgenic lines of groundnut. Journal of Agronomy and Crop Science, 197(4), 272–283. [Google Scholar]
- Ehleringer, J.R. (1993) Gas‐exchange implications of isotopic variation in arid‐land plants. In: Griffiths, H. & Smith, J. (Eds.) Plant responses to water deficit. London: BIOS Scientific Publishers, pp. 265–284. [Google Scholar]
- El‐Soda, M. , Malosetti, M. , Zwaan, B.J. , Koornneef, M. & Aarts, M.G.M. (2014) Genotype × environment interaction QTL mapping in plants: lessons from Arabidopsis . Trends in Plant Science, 19(6), 390–398. [DOI] [PubMed] [Google Scholar]
- Farquhar, G.D. , O'Leary, M.H. & Berry, J.A. (1982) On the relationship between carbon isotope discrimination and intercellular carbon dioxide concentration in leaves. Australian Journal of Plant Physiology, 9, 121–137. [Google Scholar]
- Farquhar, G.D. & Richards, R.A. (1984) Isotopic composition of plant carbon correlates with water‐use efficiency of wheat genotypes. Australian Journal of Plant Physiology, 11, 539–552. [Google Scholar]
- Fischer, R.A. , Rees, D. , Sayre, K.D. , Lu, Z.M. , Condon, A.G. & Larque, S.A. (1998) Wheat yield progress associated with higher stomatal conductance and photosynthetic rate, and cooler canopies. Crop Science, 38, 1467–1475. [Google Scholar]
- Fletcher, R.S. , Herrmann, D. , Mullen, J.L. , Li, Q. , Schrider, D.R. , Price, N. et al. (2016) Identification of polymorphisms associated with drought adaptation QTL in Brassica napus by resequencing. G3: Genes|Genomes|Genetics, 6(4), 793–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher, R.S. , Mullen, J.L. , Heiliger, A. & McKay, J.K. (2015) QTL analysis of root morphology, flowering time, and yield reveals trade‐offs in response to drought in Brassica napus . Journal of Experimental Botany, 66(1), 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan, Y. , Angadi, S.V. , Cutforth, H. , Potts, D. , Angadi, V.V. & McDonald, C.L. (2004) Canola and mustard response to short periods of temperature and water stress at different developmental stages. Canadian Journal of Plant Science, 84, 697–704. [Google Scholar]
- Gleick, P.H. (Ed.) (2000) The world water 2000‐2001: the biennial report on freshwater resources. Washington, DC: Island Press, p. 53. [Google Scholar]
- Gu, Z. , Eils, R. & Schlesner, M. (2016) Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics, 32(18), 2847–2849. [DOI] [PubMed] [Google Scholar]
- Hall, N.M. , Griffiths, H. , Corlett, J.A. , Jones, H.G. , Lynn, J. & King, G.J. (2005) Relationships between water‐use traits and photosynthesis in Brassica oleracea resolved by quantitative genetic analysis. Plant Breeding, 124, 557–564. [Google Scholar]
- Hall, A.E. , Richards, R.A. , Condon, A.G. , Wright, G.C. & Farquhar, G.D. (1994) Carbon isotope discrimination and plant breeding. Plant Breeding Reviews, 12, 81–113. [Google Scholar]
- Hammer, G.L. , Farquhar, G.D. & Broad, I.J. (1997) On the extent of genetic variation for transpiration efficiency in sorghum. Australian Journal of Agricultural Research, 48(5), 649–656. [Google Scholar]
- Hersen, P. , McClean, M.N. , Mahadevan, L. & Ramanathan, S. (2008) Signal processing by the HOG MAP kinase pathway. Proceedings of the National Academy of Sciences of the United States of America, 105(20), 7165–7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, E.E. , Howell, E.C. , Armstrong, S.J. & Parkin, I.A.P. (2021) A major quantitative trait locus on chromosome A9, BnaPh1, controls homoeologous recombination in Brassica napus . New Phytologist, 229(6), 3281–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain, S.M. , Masle, J. , Easton, A. , Hunter, M.N. , Godwin, I.D. , Farquhar, G.D. et al. (2020) Genetic variation for leaf carbon isotope discrimination and its association with transpiration efficiency in canola (Brassica napus). Functional Plant Biology, 47(4), 355–367. [DOI] [PubMed] [Google Scholar]
- Hurgobin, B. , Golicz, A.A. , Bayer, P.E. , Chan, C.K. , Tirnaz, S. , Dolatabadian, A. et al. (2018) Homoeologous exchange is a major cause of gene presence/absence variation in the amphidiploid Brassica napus . Plant Biotechnology Journal, 16(7), 1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagodzik, P. , Tajdel‐Zielinska, M. , Ciesla, A. , Marczak, M. & Ludwikow, A. (2018) Mitogen‐activated protein kinase cascades in plant hormone signaling. Frontiers in Plant Science, 9, 1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak, C. , Okrész, L. , Bögre, L. & Hirt, H. (2002) Complexity, cross talk and integration of plant MAP kinase signalling. Current Opinion in Plant Biology, 5(5), 415–424. [DOI] [PubMed] [Google Scholar]
- Juenger, T.E. , McKay, J.K. , Hausmann, N. , Keurentjes, J.J.B. , Sen, S. , Stowe, K.A. et al. (2005) Identification and characterization of QTL underlying whole‐plant physiology in Arabidopsis thaliana: δ13C, stomatal conductance and transpiration efficiency. Plant, Cell & Environment, 28(6), 697–708. [Google Scholar]
- Kijne, J. W. , Barker, R. & Molden, D. (Eds.) (2003) Water productivity in agriculture: limits and opportunities for improvement, 19. Wallingford: CABI, p. 332. (Comprehensive Assessment of Water Management in Agriculture Series 1). [Google Scholar]
- Knight, J.D. , Livingston, N.J. & Van Kessel, C. (1994) Carbon isotope discrimination and water‐use efficiency of six crops grown under wet and dryland conditions. Plant, Cell & Environment, 17(2), 173–179. [Google Scholar]
- Kobata, T. , Okuno, T. & Yamamoto, T. (1996) Contributions of capacity for soil water extraction and water use efficiency to maintenance of dry matter production in rice subjected to drought. Japanese Journal of Crop Science, 65(4), 652–662. 10.1626/jcs.65.652 [DOI] [Google Scholar]
- Koohafkan, P. & Stewart, B.A. (2008) Water and cereals in dryland. London: The Food and Agriculture Organisation of the United Nations and Earthscan. [Google Scholar]
- Leakey, A.D.B. , Ferguson, J.N. , Pignon, C.P. , Wu, A. , Jin, Z. , Hammer, G.L. et al. (2019) Water use efficiency as a constraint and target for improving the resilience and productivity of c3 and c4 crops. Annual Review of Plant Biology, 70(1), 781–808. [DOI] [PubMed] [Google Scholar]
- Levitt, J. (1980) Responses of plants to environmental stresses, Water, radiation, salt, and other stresses, II. New York, NY: Academic Press. [Google Scholar]
- Lin, Q. , Yang, J. , Wang, Q. , Zhu, H. , Chen, Z. , Dao, Y. et al. (2019) Overexpression of the trehalose‐6‐phosphate phosphatase family gene AtTPPF improves the drought tolerance of Arabidopsis thaliana . BMC Plant Biology, 19(1), 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Liu, Y. , Yang, X. , Tong, C. , Edwards, D. , Parkin, I.A.P. et al. (2014) The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nature Communications, 5, 3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow, M.M. (1989) Strategies in response to water stress. In: Kreeb, H.K. , Richter, H. & Hinkley, T.M. (Eds.) Structural and functional response to environmental stresses: water shortage. Amsterdam: SPB Academic Press, pp. 269–281. [Google Scholar]
- Lysak, M.A. , Koch, M.A. , Pecinka, A. & Schubert, I. (2005) Chromosome triplication found across the tribe Brassiceae. Genome Research, 15(4), 516–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masle, J. , Gilmore, S.R. & Farquhar, G.D. (2005) The ERECTA gene regulates plant transpiration efficiency in Arabidopsis . Nature, 436(7052), 866–870. [DOI] [PubMed] [Google Scholar]
- Matus, A. , Slinkard, A. & van Kessel, C. (1995) Carbon isotope discrimination: Potential for indirect selection for seed yield in canola. Crop Science, 35(5), 1267–1271. [Google Scholar]
- McKay, J.K. , Richards, J.H. & Mitchell‐Olds, T. (2003) Genetics of drought adaptation in Arabidopsis thaliana: I. Pleiotropy contributes to genetic correlations among ecological traits. Molecular Ecology, 12, 1137–1151. 10.1046/j.1365-294X.2003.01833.x [DOI] [PubMed] [Google Scholar]
- McVetty, P.B.E. , Austin, R.B. & Morgan, C.L. (1989) A comparison of the growth, photosynthesis, stomatal conductance and water use efficiency of Moricandia and Brassica species. Annals of Botany, 64, 87–94. [Google Scholar]
- Mekonnen, M.D. , Mullen, J.L. , Arathi, H.S. , Assefa, Y. , McKay, J.K. & Byrne, P.F. (2020) Quantitative trait locus mapping for carbon isotope ratio and root pulling force in canola. Agrosystems . Geosciences & Environment, 3(1), e20095. [Google Scholar]
- Menéndez, A.B. , Calzadilla, P.I. , Sansberro, P.A. , Espasandin, F.D. , Gazquez, A. & Bordenave, C.D. et al. (2019) Polyamines and legumes: joint stories of stress, nitrogen fixation and environment. Frontiers in Plant Science, 10, 1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills, G. , Sharps, K. , Simpson, D. , Pleijel, H. , Frei, M. , Burkey, K. et al. (2018) Closing the global ozone yield gap: Quantification and cobenefits for multistress tolerance. Global Change Biology, 24(10), 4869–4893. [DOI] [PubMed] [Google Scholar]
- Monneveux, P. , Sheshshayee, M.S. , Akhter, J. & Ribaut, J.‐M. (2007) Using carbon isotope discrimination to select maize (Zea mays L.) inbred lines and hybrids for drought tolerance. Plant Science, 173(4), 390–396. [Google Scholar]
- Moore, J.P. , Le, N.T. , Brandt, W.F. , Driouich, A. & Farrant, J.M. (2009) Towards a systems‐based understanding of plant desiccation tolerance. Trends in Plant Science, 14(2), 110–117. [DOI] [PubMed] [Google Scholar]
- Nicolas, S.D. , Le Mignon, G. , Eber, F. , Coriton, O. , Monod, H. , Clouet, V. et al. (2007) Homeologous recombination plays a major role in chromosome rearrangements that occur during meiosis of Brassica napus haploids. Genetics, 175(2), 487–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin, I.A.P. , Gulden, S.M. , Sharpe, A.G. , Lukens, L. , Trick, M. , Osborn, T.C. et al. (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana . Genetics, 171(2), 765–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passioura, J.B. (1977) Grain yield, harvest index and water use of wheat. The Journal of the Australian Institute of Agricultural Science, 43, 117–121. [Google Scholar]
- Pater, D. , Mullen, J.L. , McKay, J.K. & Schroeder, J.I. (2017) Screening for natural variation in water use efficiency traits in a diversity set of Brassica napus l. identifies candidate variants in photosynthetic assimilation. Plant and Cell Physiology, 58(10), 1700–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, B. , Jing, Q. , Bélanger, G. , Shang, J. , Huffman, T. , Liu, J. et al. (2018) Simulated canola yield responses to climate change and adaptation in Canada. Agronomy Journal, 110(1), 133–146. [Google Scholar]
- Rae, A.E. , Wei, X. , Flores‐Rodriguez, N. , McCurdy, D.W. & Collings, D.A. (2020) Super‐resolution fluorescence imaging of Arabidopsis thaliana transfer cell wall ingrowths using pseudo‐Schiff labelling adapted for the use of different dyes. Plant and Cell Physiology, 61, 1775–1787. [DOI] [PubMed] [Google Scholar]
- Raman, H. , Raman, R. , Kilian, A. , Detering, F. , Carling, J. , Coombes, N. et al. (2014) Genome‐wide delineation of natural variation for pod shatter resistance in Brassica napus . PLoS One, 9(7), e101673. 10.1371/journal.pone.0101673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman, H. , Raman, R. , Mathews, K. , Diffey, S. & Salisbury, P. (2020) QTL mapping reveals genomic regions for yield based on an incremental tolerance index to drought stress and related agronomic traits in canola. Crop and Pasture Science, 71(6), 562–577. 10.1071/CP20046 [DOI] [Google Scholar]
- Raman, H. , Raman, R. , McVittie, B. , Borg, L. , Diffey, S. & Yadav, A.S. et al. (2020) The genetic and physiological basis for phenotypic variation in effective water use in Brassica napus . Food and Energy Security, 9:e237. 10.1002/fes3.237 [DOI] [Google Scholar]
- Raman, H. , Raman, R. , Qiu, Y. , Zhang, Y. , Batley, J. & Liu, S. (2021) The Rlm13 gene, a new player of Brassica napus‐Leptosphaeria maculans interaction maps on chromosome C03 in canola. Frontiers in Plant Science, 12, 654604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman, H. , Uppal, R.K. & Raman, R. (2019) Genetic solutions to improve resilience of canola to climate change. In: Kole, C. Genomic designing of climate‐smart oilseed crops. Cham: Springer International Publishing, pp. 75–131. [Google Scholar]
- Rebetzke, G.J. , Condon, A.G. , Farquhar, G.D. , Appels, R. & Richards, R.A. (2008) Quantitative trait loci for carbon isotope discrimination are repeatable across environments and wheat mapping populations. Theoretical and Applied Genetics, 118(1), 123–137. [DOI] [PubMed] [Google Scholar]
- Rodrigues, J. , Inzé, D. , Nelissen, H. & Saibo, N.J.M. (2019) Source–sink regulation in crops under water deficit. Trends in Plant Science, 24(7), 652–663. [DOI] [PubMed] [Google Scholar]
- Schranz, M. , Lysak, M. & Mitchell‐Olds, T. (2006) The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends in Plant Science, 11, 535–542. [DOI] [PubMed] [Google Scholar]
- Sinclair, T.R. (2018) Effective water use required for improving crop growth rather than transpiration efficiency. Frontiers in Plant Science, 9, 1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A.B. & Cullis, B.R. (2018) Plant breeding selection tools built on factor analytic mixed models for multi‐environment trial data. Euphytica, 214(8), 143. 10.1007/s10681-018-2220-5 [DOI] [Google Scholar]
- Smith, S. & De Smet, I. (2012) Root system architecture: insights from Arabidopsis and cereal crops. Philosophical Transactions of the Royal Society B: Biological Sciences, 367(1595), 1441–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A. , Ganesalingam, A. , Kuchel, H. & Cullis, B. (2015) Factor analytic mixed models for the provision of grower information from national crop variety testing programmes. Theoretical and Applied Genetics, 128, 55–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A. , Lim, P. & Cullis, B. (2006) The design and analysis of multi‐phase plant breeding experiments. Journal of Agricultural Science 144, 5, 393–409. [Google Scholar]
- Takahashi, F. , Kuromori, T. , Sato, H. & Shinozaki, K. (2018) Regulatory gene networks in drought stress responses and resistance in plants. Advances in Experimental Medicine and Biology, 1081, 189–214. [DOI] [PubMed] [Google Scholar]
- Tao, Y. , Chen, M. , Shu, Y. , Zhu, Y. , Wang, S. , Huang, L. et al. (2018) Identification and functional characterization of a novel BEL1‐LIKE homeobox transcription factor GmBLH4 in soybean. Plant Cell, Tissue and Organ Culture, 134(2), 331–344. [Google Scholar]
- Town, C.D. , Cheung, F. , Maiti, R. , Crabtree, J. , Haas, B.J. , Wortman, J.R. et al. (2006) Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. The Plant Cell, 18, 1348–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer, M.C. , Halldorsdottir, S.S. , Purugganan, M.D. & Mackay, T.F. (2003) Genotype‐environment interactions at quantitative trait loci affecting inflorescence development in Arabidopsis thaliana . Genetics, 165(1), 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbyla, A.P. & Cullis, B.R. (2012) Multivariate whole genome average interval mapping: QTL analysis for multiple traits and/or environments. Theoretical and Applied Genetics, 125, 933–953. 10.1007/s00122-012-1884-9 [DOI] [PubMed] [Google Scholar]
- Wang, X. , Wang, H. , Wang, J. , Sun, R. , Wu, J. , Liu, S. et al. (2011) The genome of the mesopolyploid crop species Brassica rapa . Nature Genetics, 43, 1035–1039. [DOI] [PubMed] [Google Scholar]
- Yang, Z. , Liu, J. , Tischer, S.V. , Christmann, A. , Windisch, W. & Schnyder, H. et al. (2016) Leveraging abscisic acid receptors for efficient water use in Arabidopsis . Proceedings of the National Academy of Sciences of the United States of America, 113(24), 6791–6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong, Y. , Zhang, Y. & Lyu, Y. (2019) A MYB‐related transcription factor from Lilium lancifolium L. (LlMYB3) is involved in anthocyanin biosynthesis pathway and enhances multiple abiotic stress tolerance in Arabidopsis thaliana . International Journal of Molecular Sciences, 20(13), 3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn, H.‐S. , Kim, T.G. , Kim, M.‐K. , Kang, G.B. , Kang, J.Y. , Lee, J.‐G. et al. (2016) Structural insights into the quaternary catalytic mechanism of hexameric human quinolinate phosphoribosyltransferase, a key enzyme in de novo NAD biosynthesis. Scientific Reports, 6(1), 19681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, F. , Huang, J. , Tang, M. , Cheng, X. , Liu, Y. , Tong, C. et al. (2019) Syntenic quantitative trait loci and genomic divergence for Sclerotinia resistance and flowering time in Brassica napus . Journal of Integrative Plant Biology, 61(1), 75–88. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Li, Y. , Wang, Y. , Liu, Z. , Liu, C. , Peng, B. et al. (2010) Stability of QTL across environments and QTL‐by‐environment interactions for plant and ear height in maize. Agricultural Sciences in China, 9(10), 1400–1412. [Google Scholar]
- Zhengbin, Z. , Ping, X. , Hongbo, S. , Mengjun, L. , Zhenyan, F. & Liye, C. (2011) Advances and prospects: biotechnologically improving crop water use efficiency. Critical Reviews in Biotechnology, 31(3), 281–293. [DOI] [PubMed] [Google Scholar]
- Zhu, M. , Monroe, J.G. , Suhail, Y. , Villiers, F. , Mullen, J. , Pater, D. et al. (2016) Molecular and systems approaches towards drought‐tolerant canola crops. The New Phytologist, 210(4), 1169–1189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Supplementary information.
Data Availability Statement
The raw sequence data reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive (Accession No. PRJNA743730 for RNA‐sequencing data, PRJNA743989 for whole‐genome resequencing data).


