Abstract
Objectives
We conducted a systematic review and meta‐analysis to determine the diagnostic yield of exome sequencing (ES) for prenatal diagnosis of fetal structural anomalies, where karyotype/chromosomal microarray (CMA) is normal.
Methods
Following electronic searches of four databases, we included studies with ≥10 structurally abnormal fetuses undergoing ES or whole genome sequencing. The incremental diagnostic yield of ES over CMA/karyotype was calculated and pooled in a meta‐analysis. Sub‐group analyses investigated effects of case selection and fetal phenotype on diagnostic yield.
Results
We identified 72 reports from 66 studies, representing 4350 fetuses. The pooled incremental yield of ES was 31% (95% confidence interval (CI) 26%–36%, p < 0.0001). Diagnostic yield was significantly higher for cases pre‐selected for likelihood of monogenic aetiology compared to unselected cases (42% vs. 15%, p < 0.0001). Diagnostic yield differed significantly between phenotypic sub‐groups, ranging from 53% (95% CI 42%–63%, p < 0.0001) for isolated skeletal abnormalities, to 2% (95% CI 0%–5%, p = 0.04) for isolated increased nuchal translucency.
Conclusion
Prenatal ES provides a diagnosis in an additional 31% of structurally abnormal fetuses when CMA/karyotype is non‐diagnostic. The expected diagnostic yield depends on the body system(s) affected and can be optimised by pre‐selection of cases following multi‐disciplinary review to determine that a monogenic cause is likely.
Key points
What's already known about this topic?
Prenatal exome sequencing (ES) increases genetic diagnoses in fetuses with structural abnormalities and a normal karyotype and chromosomal microarray.
Published diagnostic yields from ES are varied and may be influenced by study size, case selection and fetal phenotype.
What does this study add?
This study provides a comprehensive systematic review of the literature to date and investigates the diagnostic yield of ES for a range of isolated system anomalies, to support clinical decision‐making on how to offer prenatal ES.
1. INTRODUCTION
Ultrasound scanning in pregnancy can reveal fetal structural abnormalities from the first trimester onwards, 1 ranging from isolated minor anomalies to severe, multisystem disorders. These abnormalities may indicate an underlying genetic condition but making a definitive diagnosis can be difficult because many conditions have overlapping fetal presentations. A genetic diagnosis can clarify the prognosis for the fetus, informing parental decision making and clinical management during pregnancy and the perinatal period.
Conventional genetic testing for fetal anomalies comprises quantitative fluorescence PCR (QF‐PCR) for rapid aneuploidy exclusion, and karyotype or chromosomal microarray (CMA) for detection of chromosomal and sub‐chromosomal abnormalities. Together, these can provide a diagnosis in up to 40% of cases 2 , 3 , 4 , but leave the majority undiagnosed. Targeted sequencing of individual genes can be offered in cases where clinical features or family history suggest a specific monogenic disorder but genome‐wide next generation sequencing (NGS) is now being used more frequently to allow interrogation of multiple genes in a single test. Exome sequencing (ES) examines the 1%–2% of the genome that encodes proteins and harbours around 85% of known disease‐causing genetic variants, and is generally less costly and easier to interpret than whole genome sequencing (WGS). ES can be applied in practice as whole ES (sequencing or analysing all exons), medical/clinical ES (sequencing or analysing only the exons of known Mendelian disease genes), or targeted ES (applying ‘virtual’ gene panels to ES data to limit analysis to genes of interest for a specific phenotype).
In the postnatal setting ES has proven a powerful tool for the diagnosis of children with neurodevelopmental delay but its application in prenatal diagnosis initially lagged behind. This is largely because long turnaround times precluded the return of results within the timeframe of a pregnancy. Additionally, incomplete or unrecognised fetal phenotypes complicate the interpretation of genetic variants from prenatal ES. Over recent years, rapid analytical pipelines have made it possible to return ES results during an ongoing pregnancy and it is now considered for prenatal diagnosis of fetal structural anomalies in the guidance of several professional bodies. 9 , 10 , 11 Two large prospective studies demonstrated the diagnostic utility of ES for prenatal diagnosis of unselected fetal structural abnormalities, reporting diagnostic rates of 8.5%–10%, 12 , 13 while numerous smaller studies report diagnostic rates above 80% in highly‐selected case series. 7 , 14 , 15 This body of literature has been previously reviewed 4 , 16 , 17 but has rapidly expanded over the last 2 years.
As prenatal ES is increasingly adopted in clinical settings there remain important questions around best practice for implementation. It will be of interest to health professionals and policy makers to define expected diagnostic yields from prenatal ES for different fetal presentations. This information may help to manage parental expectations during pre‐test counselling and will inform development of further guidelines on when and how to offer prenatal ES. In this systematic review and meta‐analysis we aim to address the question ‘what is the diagnostic yield of ES for prenatal diagnosis in pregnancies complicated by fetal structural anomalies where fetal karyotype/microarray is normal?’ We also explore the diagnostic yield in different phenotypic sub‐groups.
2. METHODS
2.1. Protocol and registration
We devised a systematic review protocol in line with PRISMA guidance 18 , 19 and registered it prospectively on the PROSPERO international register of systematic reviews (reference CRD42020200600). Study authors agreed the protocol prior to conducting the searches. Any required small amendments were made with the consensus of all authors.
2.2. Eligibility criteria
Studies were included in this review if they met the following criteria: (i) Retrospective or prospective cohorts of 10 or more pregnancies undergoing ES (whole, clinical or targeted) or WGS for diagnosis of fetal structural anomalies, (ii) CMA/karyotype was negative or non‐diagnostic, (iii) Testing was initiated based on the prenatal phenotype, (iv) Full text report was available in English language.
The focus of this review is prenatal diagnosis of structural anomalies, therefore studies with mixed pre‐ and postnatal presentations were only included if it was possible to extract the prenatal cases. Similarly, for studies with mixed indications for testing (e.g. some cases initiated because of fetal phenotype and some because of family history) we only included studies where fetal anomaly cases could be extracted. Studies where ES was completed postnatally for some or all cases but had been initiated based on the prenatal phenotype alone were included. Only variants classified as pathogenic/likely pathogenic according to diagnostic‐standard guidelines and determined to be causative of the fetal phenotype were considered in diagnostic yield, therefore studies using ES purely for novel candidate gene discovery were not included. We also excluded case reports, series of fewer than 10 cases, reviews, editorials and commentaries, and studies using gene panel testing rather than ES.
2.3. Information sources and search strategy
Electronic searches of four databases (MEDLINE, Embase, Cochrane library, and Web of Science) were conducted for records published between January 2010 and October 2021. This date range was chosen because prenatal ES was not used prior to 2010. The search was initially conducted on 27/08/2020 and repeated on 15/10/2021. Additional potentially relevant studies were identified by manually searching reference lists of relevant studies and published reviews, and conference proceedings of prenatal and genetics conferences of relevant major societies in last 3 years.
Search terms were variations on the keywords ‘prenatal diagnosis’ and ‘exome sequencing’. Alternative terms for ‘prenatal diagnosis’ included ‘fetal’, ‘fetus’, ‘prenatal’, and ‘antenatal’. Alternative terms for ‘exome sequencing’ included ‘whole exome sequencing’, ‘exome’, ‘whole genome sequencing’, ‘genome, human’ and ‘sequence analysis, DNA’. The full search strategy is detailed in supporting information S1. We chose relatively broad search terms in an effort to capture as many relevant studies as possible; for the same reason no search limits were applied apart from date range.
2.4. Study selection
After the removal of duplicates, two reviewers (RM and KO) independently screened titles and abstracts. For abstracts identified as potentially relevant, full text articles were retrieved and reviewed against the inclusion and exclusion criteria. Any disagreement between reviewers was resolved by discussion. Where the same data were presented more than once, the most recent study was selected.
2.5. Data extraction
The following data, where available, were extracted by two reviewers (RM and ES) into a datasheet: study setting, sample size, study inclusion criteria, ES approach, phenotypes used for interpretation (prenatal only or prenatal plus postnatal/post‐mortem), number of fetuses with diagnostic variants, variants of uncertain significance (VUS), incidental findings, gestation at testing, test turnaround time, pregnancy outcomes, and impact on management. For studies that performed CMA in parallel with sequencing, the cases with negative CMA were extracted, in order to be comparable with other studies where chromosomal abnormalities were ruled out prior to ES.
2.6. Quality assessment of included studies
The quality of the studies was assessed using a modified Standards for Reporting of Diagnostic Accuracy (STARD) checklist, 20 comprising 22 items. The items for reporting considered most important for these studies (as they may influence the reported diagnostic yield) were: (i) clearly described selection criteria, (ii) whether diagnostic variants were confirmed by Sanger sequencing, (iii) whether American College of Medical Genetics and Genomics (ACMG) or equivalent standardised classification was used for variant interpretation, (iv) approach to analysing and reporting VUS and incidental findings addressed. For eight studies the assessment was done independently by two reviewers (RM and MH) with discrepancies resolved by discussion. Once consensus was reached on how to apply the criteria and reviewers' scores were concordant on new study assessments, the remaining studies were assessed by RM only.
2.7. Data synthesis
The primary outcome of interest was the incremental diagnostic yield of ES (i.e. the proportion of cases in which a diagnostic genetic variant is detected by ES) over CMA/karyotype, expressed as a risk difference. This was calculated for each study with 95% confidence intervals (CI) and pooled for all studies in a meta‐analysis, using a random effects model with inverse variance weighting. A subgroup analysis was performed to investigate the effect on diagnostic yield of pre‐test case selection for higher likelihood of monogenic disease, where this information was available. Where studies reported data on isolated system anomalies classified by different phenotypic groups, a further subgroup analysis was performed to estimate pooled incremental yields by phenotypic group. Only phenotype groups reported in at least three studies were included in this analysis. Findings were displayed using forest plots. Between‐study heterogeneity was assessed visually and using an I2 statistic with a Q‐test. Publication bias was assessed graphically using a funnel plot with Egger's test. All statistical analyses were performed in R studio 21 v1.3.1093 using the ‘metafor’ package. 22
3. RESULTS
Database searches identified 8123 unique records in total. After screening by title and abstract, and retrieving full text reports, 148 articles were assessed, of which 72 reports from 66 studies were deemed eligible and included in the final review (Figure 1, Table 1). Six reports examined specific phenotypic sub‐groups from previously published larger cohorts, 23 , 24 , 25 , 26 , 27 , 28 hence were not separate studies. These were excluded from the overall meta‐analysis of incremental yield and the subgroup analysis by case selection, to avoid double‐counting of cases. However, these reports provide updated information in their respective phenotypic groups so were included in the subgroup analysis by phenotypic group, in place of the previously reported sub‐groups from their parent studies. Across 66 studies, this review represents 4350 probands undergoing prenatal ES.
FIGURE 1.
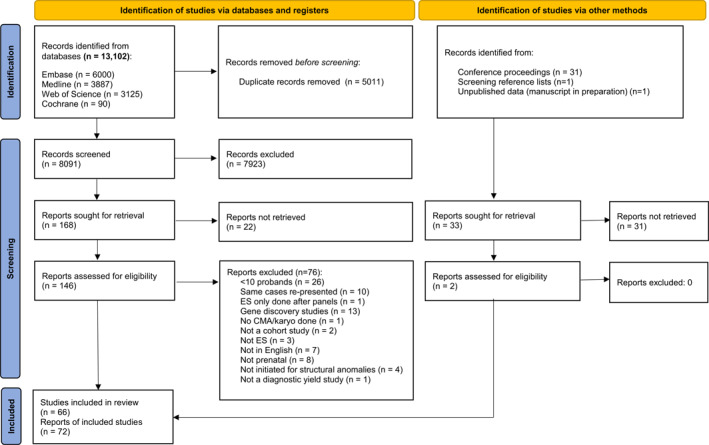
PRISMA flow diagram showing study screening and selection
TABLE 1.
Characteristics and results of 72 prenatal ES study reports included in the systematic review
| Study | No. of cases with normal CMA | Inclusion criteria | Sequencing and analysis approach | Diagnostic variants (%) | VUS or candidate genes (%) | Incidental findings (%) | Clinical impact (%) | Average TAT in days (range, if given) |
|---|---|---|---|---|---|---|---|---|
| Carss et al, 2014 a | 28 | Fetuses and neonates with prenatal structural anomalies | Trio WES | 3/28 (11%) | 5/28 (18%) | ND | ND | ND |
| Drury et al, 2015 a | 24 | Fetuses with structural abnormalities, and/or increased NT (>3.5 mm) | Solo or Trio CES | 5/24 (21%) | 1/24 (4%) | 2/24 (8%) | ND | ND |
| Pangalos et al, 2016 | 14 | Fetuses with (mild or severe) structural anomalies | Solo TES (758 prenatal disorders gene panel) | 6/14 (43%) | 1/14 (7%) | 0/14 | ND | <10 |
| Yates et al, 2017 | 84 | Fetuses with US anomalies resulting in TOP or IUFD | Solo or Trio WES | 17/84 (20%) | 45/84 (54%) | 2/84 (2%) | ND | ND |
| Aarabi et al, 2018 a | 20 | ≥1 major structural defect detected prenatally | Trio WES or CES | 1/20 (5%) | 2/20 (10%) | ND | ND | ND |
| Armes et al, 2018 a | 16 | Fetal or early neonatal death, having undergone full autopsy, genetic aetiology suspected (PM) | Trio WGS with panel analysis | 2/16 (13%) | 6/16 (38%) | ND | ND | ND |
| Boissel et al, 2018 a , b | 101 | Fetuses or stillborns with severe anomalies: | Solo or Trio WES | 19/101 (19%) | 7/101 (7%) | ND | ND | ND |
| Chandler et al, 2018 a , b | 16 | Fetuses with suspected skeletal dysplasia on prenatal US | Trio TES (240 skeletal gene panel) | 13/16 (81%) | 2/16 (13%) | ND | NQ | 16.5 (11–41) |
| Fu et al, 2018 a , b | 196 | Fetuses with ultrasound detected structural malformations, excluding isolated increased NT ≥ 3.5 mm/cystic hygroma, and isolated soft markers | Solo or Trio CES | 47/196 (24%) | 25/196 (13%) | 12/196 (6%) | ND | ∼21 |
| Leung et al, 2018 a | 33 | Fetuses with structural anomalies detected on prenatal US | Trio WES | 3/33 (9%) | 6/33 (18%) | ND | ND | ND |
| Shamseldin et al, 2018 a | 44 | Severe fetal malformations, lethal NIHF or unexplained IUFD | Solo WES | 22/44 (50%) | 15/44 (34%) | ND | ND | ND |
| Zhou, X et al, 2018 a , b | 12 | Fetuses with suspected skeletal dysplasia on US | Solo TES (363 skeletal dysplasia gene panel) | 10/12 (83%) | ND | ND | ND | 42 |
| Choy et al, 2019 a , b | 42 | Fetuses with increased NT ≥ 3.5 mm +/− structural anomalies | Solo WGS | 7/42 (17%) | 18/42 (43%) | ND | None ‐ results not returned to families | 10 |
| Daum et al, 2019 a , b | 77 | Fetuses with US‐detected malformations OR family history suggestive of a genetic diagnosis | Solo or Trio WES | 16/77 (21%) | ND | ND | NQ | 26 (19–33) |
| De Koning et al, 2019 a | 20 | Fetuses with one or more structural anomalies, genetic aetiology suspected | Trio WES | 8/20 (40%) | 1/20 (5%) | 3/20 (15%) | 9/12 (75%) | 10.5 (7–19) |
| Greenbaum et al, 2019 | 45 | Fetuses with abnormal US findings and/or relevant family history | Solo or Trio WES | 13/45 (29%) | ND | ND | ND | Ongoing pregnancies: 15–20 |
| Post‐mortem: 15–60 | ||||||||
| Liu et al, 2019 a , b | 28 | Fetuses with skeletal abnormalities on US | Solo TES (363 skeletal dysplasia gene panel) | 16/28 (57%) | 5/28 (18%) | ND | NQ | ND |
| Lord et al, 2019 a , b | 610 | Fetuses with any structural anomaly on US, including isolated increased NT ≥ 4 mm | Trio TES (1628 developmental disorder gene panel) | 52/610 (9%) | 24/610 (4%) | ND | NQ | ND |
| Meier et al, 2019 a | 19 | Severe fetal anomalies identified by US scan: (i) Two or more anomalies associated with a high risk for fetal or perinatal lethality and suspected genetic aetiology, or (ii) familial recurrence of the fetal anomaly phenotype | Trio WES | 6/19 (32%) | 6/19 (32%) | ND | ND | ND |
| Normand et al, 2019 a | 146 | Fetuses with at least one structural anomaly detected by prenatal imaging or autopsy | Solo or Trio WES | 46/146 (32%) | ND | 3/62 (5%) | 15/19 (79%) | Rapid: 14 (7–38) |
| Trio: 43 (13–78) | ||||||||
| Solo: 88 (18–141) | ||||||||
| Petrovski et al, 2019 a , b | 234 | Any ultrasound identified fetal structural anomalies, including isolated NT ≥3·5 mm | Trio WES | 24/234 (10%) | 46/234 (20%) | 4/234 (2%) | NQ | 28–56 |
| Quinlan‐Jones et al, 2019 a | 27 | Fetuses with a significant structural anomaly resulting in TOP, IUFD, or NND | Trio TES (1628 developmental disorder gene panel) | 10/27 (37%) | 6/27 (22%) | ND | NQ | ND |
| Westphal et al, 2019 a , b | 30 | Fetuses and neonates with prenatally diagnosed CHDs (focus on severe and syndromic) | Trio WES | 6/30 (20%) | 4/30 (13%) | 3/30 (10%) | ND | ND |
| Aggarwal et al, 2020 a | 32 | Fetuses with structural abnormalities or unexplained IUFD, genetic aetiology suspected (PM) | Solo and Trio, WES or CES | 18/32 (56%) | 4/32 (13%) | ND | ND | ND |
| Becher et al, 2020 a | 35 | One or more fetal malformations or severe fetal hydrops, genetic aetiology suspected | Trio WES | 9/35 (26%) | 7/35 (20%) | 1/35 (3%) | NQ | Urgent: 12 (9–16) |
| Non‐urgent: 120–180 | ||||||||
| Chen et al, 2020 a , b | 105 | Fetuses with structural anomalies, including isolated increased NT >3.5mm | Trio CES | 20/105 (19%) | 12/105 (11%) | ND | ND | 25 |
| Corsten‐Janssen et al, 2020 a | 23 | Fetuses with major structural anomalies | Trio TES (∼3850 OMIM disease gene panel with late onset diseases excluded) | 8/23 (35%) | ND | 11/23 (48%) | 6/23 (26%) | 14 (8–20) |
| Deden et al, 2020 a , b | 54 | Fetuses with multiple congenital anomalies. Isolated major anomaly or multiple soft markers only if high suspicion of genetic aetiology | Trio WES | 18/54 (33%) | 2/54 (4%) both later re‐classified as pathogenic | ND | 24/37 (68%) | 10 (4–28) |
| Deng et al, 2020 a , b | 21 | Fetuses with NIHF +/‐ other structural anomalies | Trio CES | 3/21 (14%) | 2/21 (10%) | ND | NQ | ND |
| Guo et al, 2020 a | 40 | Recurrent fetal congenital abnormalities or dysmorphic features | Trio WES | 12/40 (30%) | 20/40 (50%) | ND | NQ | ND |
| Han et al, 2020 a , b | 26 | Fetal anomalies suggestive of a skeletal dysplasia | Trio CES | 23/26 (89%) | 1/26 (4%) | 1/26 (4%) | NQ | <14 |
| Heide et al, 2020 a , b | 62 | Fetuses with abnormal corpus callosum (complete or partial agenesis, or short CC) detected on US +/‐ other anomalies | Trio TES (425 brain anomaly gene panel) | Or 12/62 (19%) | 6/65 (9%) | ND | NQ | 21.5 (9–53) |
| Lefebvre et al, 2020 a | 95 | Fetuses with multiple congenital anomalies | Solo WES | 24/95 (25%) | 14/95 (15%) | ND | ND | ND |
| Lei et al, 2020 a , b | 163 | Fetuses with CAKUT +/‐ other structural anomalies | Trio CES | 20/163 (12%) | 2/163 (1%) | 9/163 (6%) | NQ | 14–84 |
| Li, L et al, 2020 a , b | 19 | Fetuses with cerebellar vermis defects +/‐ other structural anomalies | Solo or Trio CES | 8/19 (42%) | 2/19 (11%) | 1/19 (5%) | ND | 28–56 |
| Li, R et al, 2020 a , b | 260 | Fetuses with congenital heart defects +/‐ other structural anomalies | Trio CES | 26/260 (10%) | 16/260 (6%) | 7/260 (3%) | ND | 21–56 |
| Mone et al, 2020 b , c | 197 | Fetuses with prenatally detected congenital heart disease (excluding small muscular VSDs), extracted from the PAGE study extended cohort (Lord et al, 2019) | Trio TES (1628 developmental disorder gene panel) | 25/197 (13%) | 10/197 (5%) | ND | ND | ND |
| Pooh et al, 2020 a | 16 | Fetuses with anomalies detected on US at 11‐13 weeks, genetic disease strongly suspected | Trio CES | 7/16 (44%) | ND | ND | ND | 7–10 |
| Reischer et al, 2020 a , b | 11 | Fetuses with decreased/absent fetal movement plus arthrogryposis of limbs | Solo WES | 6/11 (55%) | 1/11 (9%) | ND | ND | ND |
| Sparks et al, 2020 a , b | 127 | Fetuses with NIHF, fetal ascites, pleural or pericardial effusions, skin oedema, cystic hygroma, increased NT, or a combination of these conditions | Trio WES | 37/127 (29%) | 12/127 (9%) | 4/115 | NQ | Ongoing pregnancies: 14–28 |
| Post‐mortem: 56–84 | ||||||||
| Sun et al, 2020 a , b | 66 | Fetuses with cardiac left‐sided lesions | Solo or Trio WES | 13/66 (20%) | 11/66 (17%) | 1/66 (2%) | ND | ND |
| Vora et al, 2020 a | 102 | Fetuses with isolared or multiple congenital anomalies | Trio WES (n = 99) or WGS (n = 3) | 21/102 (21%) | 10/102 (10%) 2 re‐classified as benign after 5 yrs | 9/204 (4%) in consenting parents | NQ | 182–365 |
| Xue et al, 2020 a , b | 24 | Fetuses with increased NT and no detectable structural malformations on US | Trio WES | 3/24 (13%) | 2/24 (8%) | ND | ND | ND |
| Yang et al, 2020 a , b | 73 | Fetuses with isolated first‐trimester increased NT ≥3.5 mm | Trio CES | 4/73 (6%) | 7/73 (10%) | ND | ND | ND |
| Zhou, X et al, 2020 a , b | 41 | Fetuses with isolated renal abnormalities on US | Solo or Trio WES | 3/41 (7%) | ND | 1/41 (2%) | NQ | ND |
| Qi et al, 2020 a , b | 80 | Fetuses with at least one ultrasonographic structural anomaly, including isolated increased NT | Trio CES (5000 OMIM genes) +/‐ WES (where sufficient DNA) | 27/80 (33.8%) | 5/80 (6.3%) | ND | ND | 14–28 |
| Rinaldi et al, 2020 a | 30 | Fetuses with structural malformation or ‘severe condition’ on US (e.g. growth restriction, absence of spontaneous movements) | Trio CES (5817 OMIM genes) | 19/30 (63%) | 0/30 | ND | None in affected pregnancy ‐ sequenced after end of pregnancy | ND |
| Al‐Kouatly et al, 2021 a | 22 | Fetuses with confirmed NIHF after first trimester +/‐ other structural anomalies (strict phenotypic description for NIHF: at least two abnormal fluid collections) | Trio CES | 11/22 (50%) | 5/22 (22.7%) | ND | NQ | 14–21 |
| Aoi et al, 2021 a | 17 | Fetuses with ultrasonographic structural anomalies | Solo or Trio WES | 5/17 (29.4%) | 1/17 (5.9%) | ND | ND | ND |
| Cao et al, 2021 a , b | 11 | Fetuses with abnormalities suggestive of fetal akinesia | Trio CES (4200 OMIM genes) | 5/11 (45.5%) | 3/11 (27.3%) | ND | None in affected pregnancy ‐ sequenced after end of pregnancy | ND |
| Correa et al, 2021 a , b | 19 | Fetuses with confirmed NIHF after first trimester +/‐ other structural anomalies (strict phenotypic description for NIHF: at least two abnormal fluid collections) | Solo WES | 7/19 (36.8%) | 1/19 (5.3%) | ND | ND | ND |
| De Koning et al, 2021 b , c | 18 | Fetuses with CNS malformations detected by prenatal US; isolated or with other structural anomalies. (Some cases previously published in De Koning et al, 2019) | Trio WES | 10/18 (56%) | 2/18 (11.1%) | 2/18 (11.1%) | 12/18 (67%) | ND |
| Dempsey et al, 2021 a | 52 | Ongoing pregnancies with US‐detected fetal structural abnormalities where MDT judged a high likelihood of phenotype being explained by a single gene disorder, and where the result may influence management of the pregnancy, labour or early neonatal care | Trio WES with phenotype‐specific panel analysis | 19/52 (37%) | 13/52 (25%) | ND | 17/43 (39.5%) in pregnancies ongoing at time of result | 17 |
| He et al, 2021 b | 94 | Fetuses with structural anomalies detected by prenatal US in the second trimester | Solo or Trio WES | 37/94 (39.4%) | 14/94 (14.9%) | 4/94 (4.3%) | ND | 42–56 |
| Kucińska‐Chahwan et al, 2021 a , b | 26 | Fetuses with skeletal abnormalities, excluding isolated talipes | Solo WES | 18/26 (69.2%) | ND | ND | ND | 84 |
| Lei et al, 2021 a , b | 85 | Fetuses with US‐detected structural anomalies, other than skeletal abnormalities | Solo WES | 16/85 (18.8%) | 16/85 (18.8%) | ND | ND | 14–21 |
| Liao et al, 2021 b | 24 | Fetuses with ultrasonographic diagnosis of abnormal sylvian fissure | Solo, Duo or Trio WGS | 12/24 (50%) | ND | ND | ND | ND |
| Mellis et al, 2021 b , c | 213 | Fetuses with increased NT ≥3.5 mm at 11–14 weeks of gestation +/‐ other anomalies, extracted from the PAGE and Columbia studies' extended cohorts (Lord et al, 2019; Petrovski et al, 2019) | Trio TES (1628 developmental disorder gene panel) or WES | 28/213 (13.1%) | 8/213 (3.8%) | ND | ND | ND |
| Mone et al, 2021 b , c | 28 | Fetuses with NIHF +/‐ other anomalies, extracted from the PAGE study extended cohort (Lord et al, 2019) | Trio TES (1628 developmental disorder gene panel) | 7/28 (25%) | 2/28 (7.1%) | ND | ND | ND |
| Peng et al, 2021 a , b | 38 | Fetuses with suspected skeletal dysplasia on prenatal US | Solo or Trio WES | 25/38 (65.8%) | 4/38 (10.5%) | 12/38 (31.6%) | ND | ND |
| Qiao et al, 2021 a , b | 300 | Fetuses with CHD detected by echocardiography | Solo or Trio WES | 24/300 (8%) | 32/300 (10.7%) | incidental: 37/300 (12.3%) | ND | ND |
| secondary “looked for”: 11/300 (3.7%) | ||||||||
| Sun et al, 2021 a , b | 19 | Fetuses with non‐compaction cardiomyopathy on fetal echocardiogram | Trio WES | 8/19 (42.1%) | 8/19 (42.1%) | ND | ND | ND |
| Tang et al, 2021 a , b | 15 | Fetuses with suspected skeletal dysplasias on US scanning | Trio WES | 10/15 (66.7%) | 1/15 (6.7%) | ND | ND | ND |
| Tolusso et al, 2021 a | 20 | Fetuses with congenital anomalies judged likely to be caused by an underlying monogenic condition | Trio WES | 8/20 (40%) | 7/20 (35%) | 1/20 (5%) | 9/20 (45%) | Ongoing pregnancies: 19.3 |
| Terminated pregnancies: 56.8 | ||||||||
| Wagner et al, 2021 b , c | 15 | Fetuses with NIHF detected on US. (Some cases previously published in Daum et al, 2019) | Trio WES | 7/15 (47%) | ND | ND | ND | ND |
| Zhang, F et al, 2021 b | 12 | Fetuses with isolated absent or hypoplastic nasal bone | Solo CES (2742 OMIM genes) | 2/12 (16.7%) | ND | ND | ND | ND |
| Zhang, L et al, 2021 a , b | 55 | Fetuses with suspected skeletal dysplasias based on US features | Solo or Trio WES | 35/55 (64%) | 4/55 (7.3%) | ND | ND | 21–70 |
| Zhang, X et al, 2021 a , b | 27 | Fetuses with suspected skeletal dysplasias undergoing ToP before the third trimester | Trio WES (n = 17) or TES (n = 10 (505 skeletal dysplasia gene panel)) | 21/27 (77.8%) | ND | ND | ND | ∼28 |
| Zhen et al, 2021 b | 13 | Fetuses with apparently isolated micrognathia identified on US at 11–14 weeks | Trio CES (4200 OMIM disease genes) | 8/13 (61.5%) | ND | ND | ND | ND |
| Zhou, J et al, 2021 a , b | 102 | Fetuses with structural or growth anomalies identified by US scanning | Trios WES and WGS in parallel | 13/102 (12.7%) | ND | ND | NQ | 21 (15–27) |
| Zhou, X et al, 2021 a , b | 28 | Fetuses with recurrent NIHF +/− additional structural anomalies, where immune, infectious causes or thalassaemia ruled out | Trio WES | 10/28 (36%) | 5/28 (18%) | ND | ND | 28–42 |
| Baptiste et al, 2022 b , c | 268 | Fetuses with CNS abnormalities +/− other anomalies, extracted from the PAGE and Columbia studies' extended cohorts (Lord et al, 2019; Petrovski et al, 2019) | Trio TES (1628 developmental disorder gene panel) or WES | 36/268 (13.4%) | 11/268 (4.1%) | ND | ND | ND |
Abbreviations: CES, clinical exome sequencing; CHD, congenital heart defect, IUFD, intrauterine fetal demise, MDT, multi‐disciplinary team; ND, not described; NIHF, non‐immune hydrops fetalis; NND, neonatal death; NQ, not quantified; NT, nuchal translucency; OMIM, Online Mendelian Inheritance in Man; PM, post‐mortem; TAT, turnaround time; TES, targeted exome sequencing; TOP, termination of pregnancy; US, ultrasound; VUS, variants of unknown significance; WES, whole exome sequencing.
studies included in the case selection subgroup analysis (those which it was possible to determine whether cases were selected based on higher likelihood of monogenic aetiology, and where all cases in the cohort were subject to these selection criteria).
studies included in the phenotype subgroup analysis (those which recruited a single phenotypic group or reported breakdown of cases by phenotypic group/affected body system and distinguished single system anomalies from multisystem).
reports of phenotypic sub‐groups from previously published larger cohorts, where cases overlap with previous publication.
3.1. Study characteristics
The characteristics of the included studies are summarised in Table 1. Of the 72 included reports, 48 (67%) were published in 2020‐21. Three studies used WGS rather than ES, with or without applying phenotype‐specific panels for analysis. 29 , 30 , 31 Eighteen studies used a clinical exome sequencing (CES) approach in some or all cases, which here we define as targeting either the sequencing or the analysis to Online Mendelian Inheritance in Man disease genes. 14 , 32 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 Eight studies targeted ES analysis more tightly using phenotype‐specific gene panels, 7 , 12 , 15 , 49 , 50 , 51 , 52 , 53 while the remainder took a whole ES approach. 8 , 13 , 62 , 63 , 64 , 65 , 66 , 67 , 68 , 69 , 70 , 71 , 54 , 72 , 73 , 74 , 75 , 76 , 77 , 78 , 79 , 80 , 81 , 55 , 82 , 83 , 84 , 85 , 86 , 87 , 88 , 56 , 57 , 58 , 59 , 60 , 61 Where stated (15 studies), 7 , 8 , 74 , 75 , 78 , 82 , 84 , 15 , 29 , 36 , 52 , 53 , 55 , 62 , 73 the median turnaround time for ES was 20 days (range 4–141). Studies included fetuses with a range of structural abnormality phenotypes, with some studies recruiting cases with fetal anomalies in a specific body system (e.g. renal anomalies or cardiac defects) and others including any structural anomaly. In total, 42/66 studies recruited cases with anomalies in a specific body system or broke down fetal abnormalities within the cohort into distinct phenotypic groups (Table 1). The quality of included reports, assessed using modified STARD criteria, was generally high (Figure 2).
FIGURE 2.
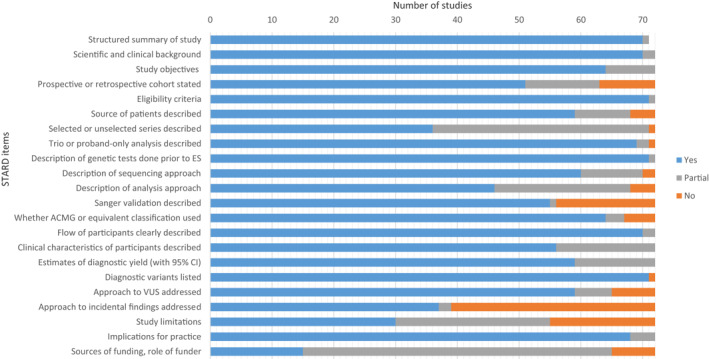
Quality assessment of included study reports using modified Standards for Reporting of Diagnostic Accuracy (STARD) criteria. (n = 72, except or the item “structured summary of study”, where n = 71 because one report was a Letter to the Editor, which would not be expected to contain a structured abstract)
3.2. Incremental yield of diagnostic variants from ES
The diagnostic yield of ES for fetal structural anomalies among the included studies ranged from 5% (95% CI −8 to 18%) to 89% (95% CI 72%–99%). The pooled incremental yield of ES over CMA/karyotype from all studies was 31% (95% CI 26%–36%, p < 0.0001), illustrated in the forest plot (Figure 3). Between‐study heterogeneity was high, indicated by an I2 statistic of 94% (Q = 682, p < 0.001). Testing for publication bias showed significant funnel plot asymmetry (z = 5.6, p < 0.0001 by Egger's test; supporting information S1), indicating possible publication bias.
FIGURE 3.
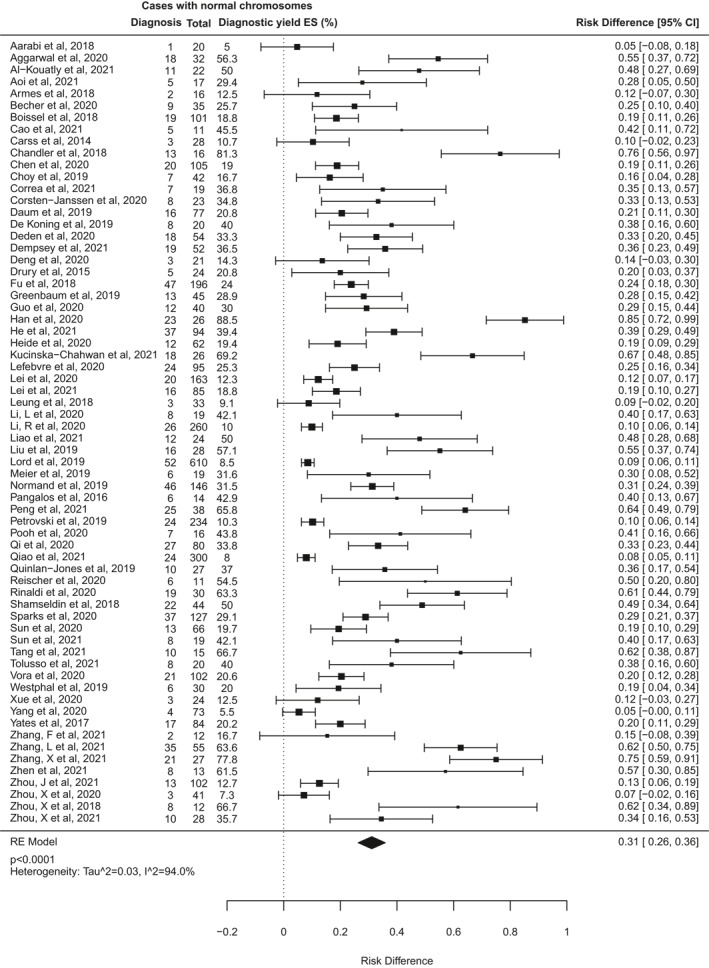
Forest plot showing individual and pooled incremental yield of prenatal exome sequencing (ES) over karyotype/chromosomal microarray from all 66 studies included in this review
3.3. Effect of case selection on diagnostic yield
Some of the heterogeneity between included studies may be due to differing approaches to case selection. Some studies included fetuses with structural anomalies without any prior review of the clinical features to ascertain risk of a monogenic aetiology, whereas others selected only pregnancies where the pattern of fetal anomalies was highly suggestive for a monogenic disorder (as determined by multidisciplinary teams [MDTs] usually including clinical genetics specialists). Seven studies were excluded from this analysis because it was not possible to determine how the cases were selected, or because mixed selection criteria were used. The other 59 studies were classified as selected or unselected cohorts (Table 2) for a subgroup analysis (Figure 4). This demonstrated that the pooled incremental diagnostic yield of ES for selected cohorts was 42% (95% CI 35%–48%), which is significantly higher than that in unselected cohorts (15%, 95% CI 11%–18%; p < 0.0001).
TABLE 2.
Effect of case selection on incremental diagnostic yield of ES over CMA
| Study | Number of probands with normal CMA | Diagnostic yield (%) |
|---|---|---|
| Unselected fetuses: NO prior review/selection for features indicating higher likelihood of monogenic disease | ||
| Quinlan‐Jones et al, 2019 | 27 | 10/27 (37%) |
| Qi et al, 2020 | 80 | 27/80 (33.8%) |
| Lefebvre et al, 2020 | 95 | 24/95 (25.3%) |
| Fu et al, 2018 | 196 | 47/196 (24%) |
| Drury et al, 2015 | 24 | 5/24 (20.8%) |
| Sun et al, 2020 | 66 | 13/66 (19.7%) |
| Heide et al, 2020 | 62 | 12/62 (19.4%) |
| Chen et al, 2020 | 105 | 20/105 (19%) |
| Boissel et al, 2018 | 101 | 19/101 (18.8%) |
| Lei et al, 2021 | 85 | 16/85 (18.8%) |
| Choy et al, 2019 | 42 | 7/42 (16.7%) |
| Zhou, J et al, 2021 | 102 | 13/102 (12.7%) |
| Xue et al, 2020 | 24 | 3/24 (12.5%) |
| Lei et al, 2020 | 163 | 20/163 (12.3%) |
| Carss et al, 2014 | 28 | 3/28 (10.7%) |
| Petrovski et al, 2019 | 234 | 24/234 (10.3%) |
| Li, R et al, 2020 | 260 | 26/260 (10%) |
| Leung et al, 2018 | 33 | 3/33 (9.1%) |
| Lord et al, 2019 | 610 | 52/610 (8.5%) |
| Qiao et al, 2021 | 300 | 24/300 (8%) |
| Zhou, X et al, 2020 | 41 | 3/41 (7.3%) |
| Yang et al, 2020 | 73 | 4/73 (5.5%) |
| Aarabi et al, 2018 | 20 | 1/20 (5%) |
| Pooled estimated diagnostic yield for unselected subgroup | 2771 | 15% [95% CI = 11%–18%] p < 0.0001 |
| Selected fetuses: Prior expert review/selection for clinical features indicating higher likelihood of monogenic disorder or ruling out non‐genetic conditions | ||
| Han et al, 2020 | 26 | 23/26 (88.5%) |
| Chandler et al, 2018 | 16 | 13/16 (81.3%) |
| Zhang, X et al, 2021 | 27 | 21/27 (77.8%) |
| Kucińska‐Chahwan et al, 2021 | 26 | 18/26 (69.2%) |
| Tang et al, 2021 | 15 | 10/15 (66.7%) |
| Zhou, X et al, 2018 | 12 | 8/12 (66.7%) |
| Peng et al, 2021 | 38 | 25/38 (65.8%) |
| Zhang, L et al, 2021 | 55 | 35/55 (63.6%) |
| Rinaldi et al, 2020 | 30 | 19/30 (63.3%) |
| Liu et al, 2019 | 28 | 16/28 (57.1%) |
| Aggarwal et al, 2020 | 32 | 18/32 (56.3%) |
| Reischer et al, 2020 | 11 | 6/11 (54.5%) |
| Al‐Kouatly et al, 2021 | 22 | 11/22 (50%) |
| Shamseldin et al, 2018 | 44 | 22/44 (50%) |
| Cao et al, 2021 | 11 | 5/11 (45.5%) |
| Pooh et al, 2020 | 16 | 7/16 (43.8%) |
| Li, L et al, 2020 | 19 | 8/19 (42.1%) |
| Sun et al, 2021 | 19 | 8/19 (42.1%) |
| De Koning et al, 2019 | 20 | 8/20 (40%) |
| Tolusso et al, 2021 | 20 | 8/20 (40%) |
| Correa et al, 2021 | 19 | 7/19 (36.8%) |
| Dempsey et al, 2021 | 52 | 19/52 (36.5%) |
| Zhou, X et al, 2021 | 28 | 10/28 (35.7%) |
| Corsten‐Janssen et al, 2020 | 23 | 8/23 (34.8%) |
| Deden et al, 2020 | 54 | 18/54 (33.3%) |
| Meier et al, 2019 | 19 | 6/19 (31.6%) |
| Normand et al, 2019 | 146 | 46/146 (31.5%) |
| Guo et al, 2020 | 40 | 12/40 (30%) |
| Aoi et al, 2021 | 17 | 5/17 (29.4%) |
| Sparks et al, 2020 | 127 | 37/127 (29.1%) |
| Becher et al, 2020 | 35 | 9/35 (25.7%) |
| Daum et al, 2019 | 77 | 16/77 (20.8%) |
| Vora et al, 2020 | 102 | 21/102 (20.6%) |
| Westphal et al, 2019 | 30 | 6/30 (20%) |
| Deng et al, 2020 | 21 | 3/21 (14.3%) |
| Armes et al, 2018 | 16 | 2/16 (12.5%) |
| Pooled estimated diagnostic yield for selected subgroup | 1293 | 42% [95% CI = 35%–48%] p < 0.0001 |
Note: The bold and underlined formatting was just for emphasis in defining the two sub‐groups.
Abbreviations: CMA, chromosomal microarray; ES, exome sequencing.
FIGURE 4.
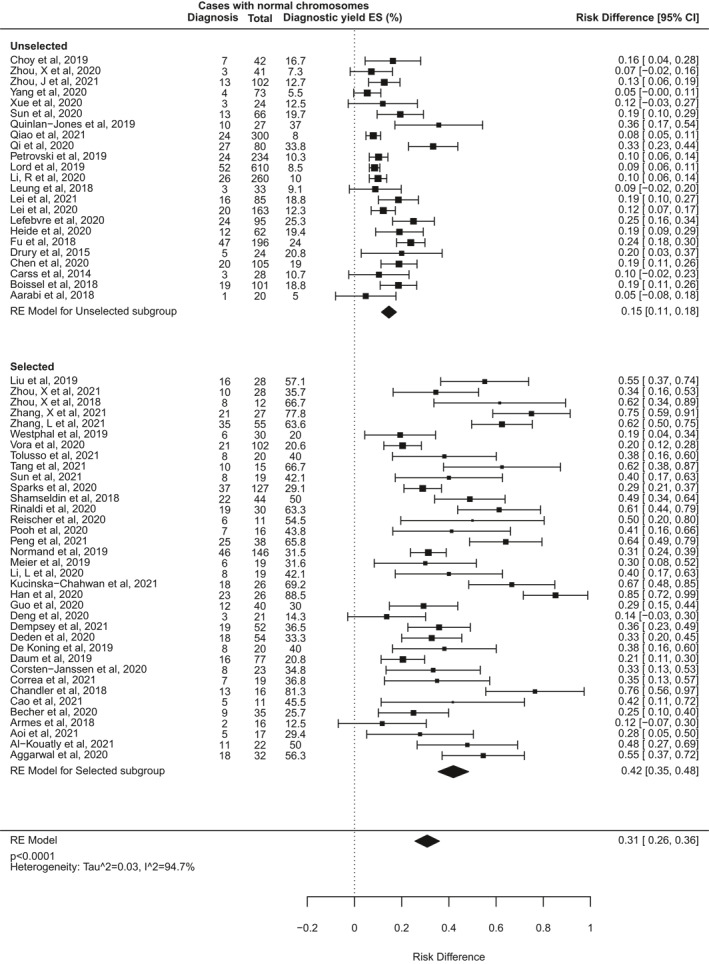
Forest plot showing individual and pooled incremental yield of prenatal exome sequencing (ES) over karyotype/chromosomal microarray analysed by pre‐test case selection criteria: “unselected” cohorts underwent no prior review to determine clinical features indicating higher likelihood of monogenic aetiology; “selected” cohorts underwent pre‐test expert review to select fetuses with clinical features indicating higher likelihood of monogenic disorder or to rule out non‐genetic conditions
3.4. Effect of fetal phenotypic group on diagnostic yield
Further heterogeneity may arise from the varied types of fetal structural anomaly included in different studies. To investigate this, a second subgroup analysis was performed for 42 studies that reported cases disaggregated by phenotypic group, and where it was possible to distinguish isolated system anomalies from multisystem anomalies (Figure 5). Data from at least 3 studies were available for fetuses in each of 13 phenotypic subgroups based on affected body system (Table 3). The number of fetuses analysed in each subgroup varied widely, from seven cases with isolated abdominal wall abnormalities, to 773 cases with isolated cardiac abnormalities. The subgroups for multisystem anomalies, isolated skeletal anomalies, and isolated brain or central nervous system (CNS) anomalies were the next largest, with over 400 cases each. The subgroup analysis found that the incremental diagnostic yield of ES differed significantly between different phenotypic groups (I 2 96.4%, Q = 177, p < 0.0001). Table 3 details the pooled estimated incremental diagnostic yield for each subgroup, from highest to lowest. The highest diagnostic yields occurred in fetuses with skeletal abnormalities (53% [95% CI 42%–63%], p < 0.0001), neuromuscular abnormalities, usually presenting as fetal akinesia deformation sequence (FADS), (37% [20%–54%], p < 0.0001), and abnormalities in multiple systems (29% [22%–35%], p < 0.0001). By contrast, the lowest rates of ES diagnoses occurred in fetuses with isolated increased nuchal translucency (NT; 2% [0%–5%], p = 0.04), and those with gastrointestinal abnormalities (2% [−4 to 8%], p = 0.5). There were no diagnoses from ES in 7 fetuses with abdominal wall defects and 38 fetuses with chest abnormalities.
FIGURE 5.
Forest plot showing individual and pooled incremental yield of prenatal exome sequencing (ES) over karyotype/chromosomal microarray analysed by phenotypic sub‐group in studies where fetuses with anomalies confined to a single body system were distinguishable from those with multi‐system anomalies
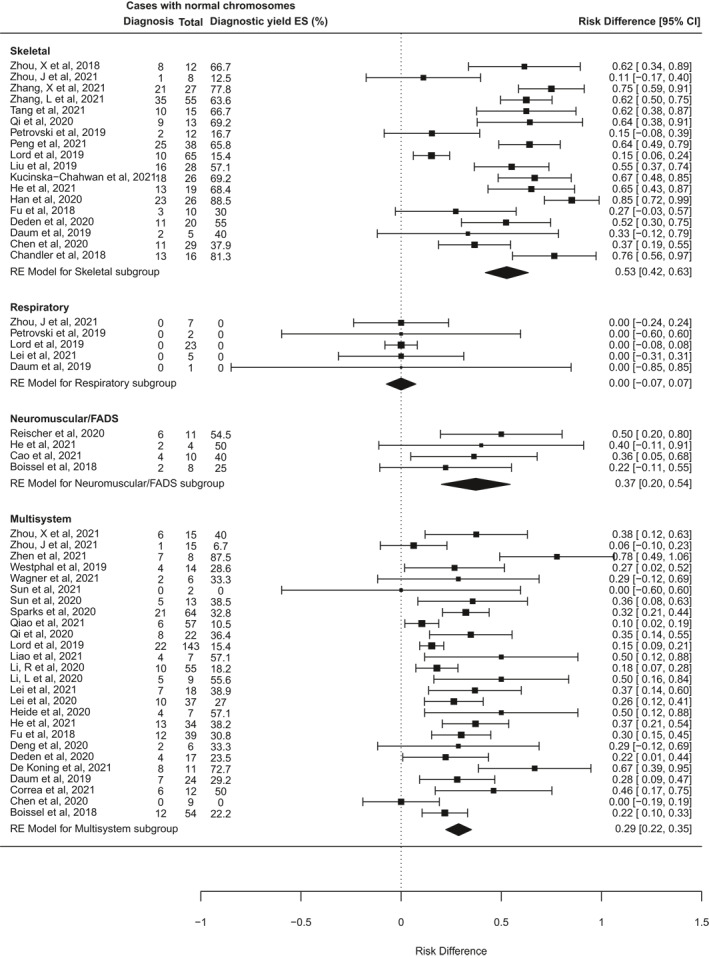
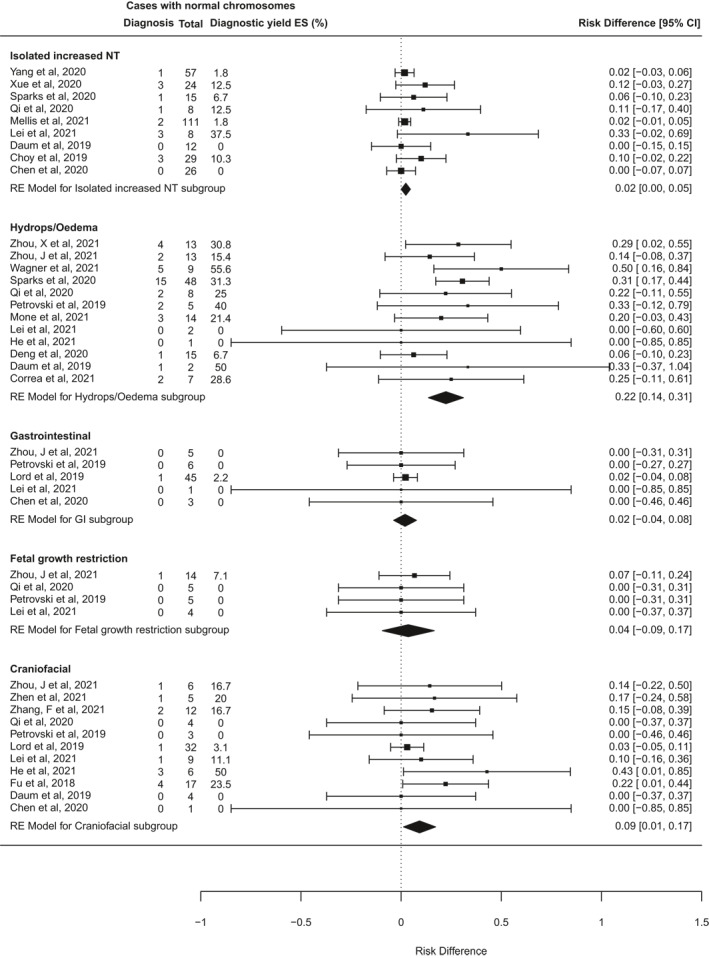
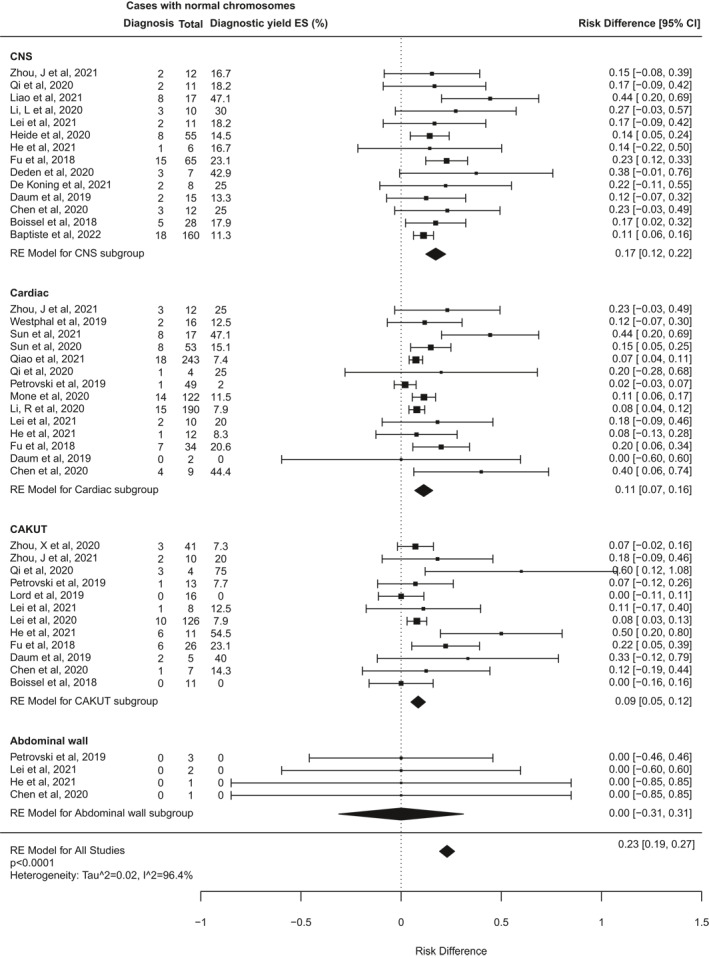
TABLE 3.
Pooled effect size for incremental diagnostic yield of ES over CMA in different phenotypic groups
| Phenotypic group | Cases analysed | Pooled estimated diagnostic yield [95% CI], p‐value |
|---|---|---|
| Skeletal | 424 | 53% [42%–63%], p < 0.0001 |
| Neuromuscular/Fetal akinesia deformation sequence (FADS) | 33 | 37% [20%–54%], p < 0.0001 |
| Multisystem | 698 | 29% [22%–35%], p < 0.0001 |
| Hydrops/Oedema | 137 | 22% [14%–31%], p < 0.0001 |
| Central nervous system (CNS) | 417 | 17% [12%–22%], p < 0.0001 |
| Cardiac | 773 | 11% [7%–16%], p < 0.0001 |
| Craniofacial | 99 | 9% [1%–17%], p = 0.02 |
| Congenital anomalies of kidneys and urinary tract (CAKUT) | 278 | 9% [5%–12%], p < 0.0001 |
| Fetal growth restriction | 28 | 4% [−9 to 17%], p = 0.59 |
| Isolated increased nuchal translucency (NT) | 290 | 2% [0%–5%], p = 0.04 |
| Gastrointestinal | 60 | 2% [−4 to 8%], p = 0.5 |
| Respiratory/Chest | 38 | 0 [−7 to 7%], p = 1 |
| Abdominal wall | 7 | 0 [−31% to 31%], p = 1 |
Note: Phenotypic groups refer to fetuses with one or more anomalies in a single body system. Fetuses with anomalies in more than one system are classified as ‘Multisystem’. The ‘isolated increased NT’ group contains a mixture of (i) cases with isolated increased NT at presentation where it was unspecified whether additional anomalies developed later in pregnancy, and (ii) cases where isolated increased NT remained isolated throughout pregnancy. The bold and underlined formatting was just for emphasis in defining the two sub‐groups.
Abbreviations: CMA, chromosomal microarray; ES, exome sequencing.
3.5. Impact of prenatal ES on decision‐making and clinical management
Only seven reports from six studies explicitly assessed the impact of prenatal ES on clinical management. These studies reported that ES results influenced at least one of (i) decisions about termination of pregnancy, (ii) pre‐ or perinatal management, (iii) future reproductive planning, or (iv) counselling on recurrence risk in 26%–79% of cases. 8 , 26 , 52 , 62 , 73 , 74 , 82
4. DISCUSSION
The findings of our review underscore the value of applying ES in the prenatal context. Meta‐analysis results show an incremental diagnostic yield of 31% from ES over CMA/karyotype alone for prenatal diagnosis of fetal structural anomalies. This diagnostic yield could be considered a conservative estimate because we included only definite diagnoses (variants classified as pathogenic/likely pathogenic and deemed to be causing the fetal phenotype) in our definition of diagnostic yield. Variants reported as ‘possibly diagnostic’, ‘probably diagnostic’, or ‘potentially relevant’ were not included in our analysis but may have been re‐classified as diagnostic after further study. For example, one study of 19 fetuses with severe multiple anomalies of suspected genetic aetiology found a diagnostic yield of 31.6% when strictly using ACMG guidelines, but this rose to 63% when including variants not reportable by ACMG criteria but which were plausible based on animal models, in silico prediction tools or functional studies. 68 Other studies have also shown potentially increased diagnostic yields when probable diagnoses are included. 30 , 71
In this way, diagnostic yields can increase over time, as has been previously documented in large postnatal ES studies such as the UK Deciphering Developmental Disorders (DDD) study, where iterative reanalysis was performed after several years. 89 This results from improved bioinformatics methodologies, newly discovered gene‐disease associations and, in the case of prenatal ES, evolving knowledge of fetal presentations of known disorders. For example, in one study from our review, Petrovski et al. reported four variants which were re‐classified (from bioinformatic signatures) to diagnostic variants over the course of the study period in light of new published evidence or additional phenotypes emerging postnatally. 13
Our second key finding is that the diagnostic yield of ES for fetal anomalies is higher when cases are pre‐selected for likelihood of a monogenic aetiology by MDTs with expertise in genetics. This is intuitive but, as prenatal ES is increasingly implemented in clinical diagnostic settings, it highlights the value of pre‐test review and MDT collaboration to direct testing where it is most likely to yield a diagnosis. Furthermore, our findings demonstrate that diagnostic yield varies significantly between phenotypic groups: the highest yield observed was for skeletal dysplasias (53%) and the lowest yields were for fetuses with isolated increased NT (2%) and isolated gastrointestinal anomalies (2%).
This understanding of likely diagnostic yield for different fetal presentations may inform clinical guidelines on offering prenatal ES, however our findings should be interpreted with care. Classification of cases by phenotype and comparison across studies is challenging because of different approaches to the classification (e.g. whether ‘soft markers’ are classed as separate abnormalities, and whether cases were categorised strictly based on isolated system anomalies or based on the system that best describes the predominant phenotype in the fetus). Moreover, fetal phenotypes evolve as organ development progresses, so classification of the same case may differ depending on the gestation at assessment. 23 Further research is needed to clarify the diagnostic utility of prenatal ES for isolated anomalies in systems that have been less thoroughly investigated to date. For example, in this review, for fetuses with chest (respiratory) abnormalities, fetal growth restriction, gastrointestinal anomalies and abdominal wall defects, the sample sizes were too small to reach statistical significance in estimates of pooled diagnostic yield.
Studies examining other aspects of clinical utility such as impact on clinical management and parental decision‐making will be an important complement to this, 90 however we found only six to include in our review. 8 , 26 , 52 , 62 , 73 , 74 , 82 Three of these studies found that negative as well as diagnostic results impacted upon parental and clinical decision‐making, 73 , 74 , 82 leading Deden et al. to conclude that ‘the efficacy of prenatal [ES] should be evaluated by more variables than diagnostic yield alone.’ 73 This is an area where the need for further research is pressing, including more studies exploring the perspectives of parents undergoing prenatal ES to investigate what personal utility they derive from it.
4.1. Strengths and limitations
This systematic review is the most detailed and comprehensive in this area to date. It included 72 reports of 66 studies, many of which were published in the last 12 months. The reporting of research in the included studies was generally of high quality with a standardised approach to reporting which made it easier to compare study characteristics, and we were able to perform meta‐analyses for a range of phenotypic groups as well as overall. There are however signs of possible publication bias, as indicated by funnel plot asymmetry showing a paucity of published studies with small sample size and low diagnostic yield (supporting information S1). This could reflect publication bias or indeed may represent a true relationship between small sample size and higher diagnostic yield: among the studies included in this review, smaller cohorts were often highly selected (usually after review by clinical genetics specialists) for cases where the clinical presentation suggested a monogenic aetiology. The higher a priori likelihood of a monogenic disorder among such cases translates to higher diagnostic yields, whereas the largest studies to date have been unselected cohorts, including fetuses with isolated and minor abnormalities and anomalies associated with multifactorial aetiology, as well as those more likely to have a monogenic disorder.
The main limitation of our review was the high degree of heterogeneity between included studies, limiting the precision of comparisons. On performing subgroup analyses to investigate the effect on ES diagnostic yield of case selection criteria or fetal phenotypic group, there remained substantial heterogeneity within many of the subgroups as well as between them, which suggests that neither of these factors alone fully explain the observed heterogeneity amongst the studies. Studies also varied in sample size, sequencing and analysis approach (gene agnostic or targeted), approach to variant interpretation, and whether or not interpretation considered postnatal/post‐mortem phenotype information in addition to prenatal phenotypes. All of these factors are likely to influence the diagnostic yield. For example, even when adhering to standardised ACMG variant interpretation guidelines, there may be some subjectivity in how the guidelines are applied and how it is judged whether a variant explains the fetal phenotype. Studies done in a purely research setting without reporting results back to families may have interpreted variants with candidate causality more generously, leading to higher reported diagnostic yields than in studies that used more conservative reporting standards. In addition, for the phenotype subgroup analysis we only included studies that disaggregated cases by phenotypic group and distinguished cases with a single affected body system from those with multiple system anomalies. This resulted in the exclusion of 24/66 studies from this subgroup analysis. Manual inspection of the raw phenotype data from these studies could enable extraction and classification of more cases in their phenotypic groups and allow a more granular analysis of the incremental yield of ES in different phenotypic groups, as has been reported recently for congenital heart disease, fetal hydrops, and isolated increased NT respectively. 24 , 25 , 91
5. CONCLUSION
Prenatal ES is a powerful tool to improve the diagnosis of monogenic conditions in fetuses with structural abnormalities. However, it also presents challenges for interpreting variants in the face of incomplete and non‐specific fetal phenotypes while also aiming to minimise identification of VUS and incidental findings. In ongoing pregnancies, results must be returned quickly if they are to inform decisions on termination of pregnancy or prenatal management. Prenatal ES is rapidly being adopted in many clinical settings and its successful implementation will require robust laboratory infrastructure and bioinformatics pipelines, expert parental counselling, multidisciplinary collaboration between fetal medicine and clinical and laboratory genetics specialists to offer and interpret tests appropriately, and clear guidelines to ensure equity of access. The need to sequence fetus‐parent trios in parallel to minimise turn‐around times means that ES remains relatively costly, so in many health systems it may be necessary to triage cases to allocate finite resources. Further research is needed on the clinical impact of prenatal ES, to better understand which pregnancies benefit most and how to prioritise potential cases in a way that maximises benefit to families, and is efficient for healthcare systems.
CONFLICT OF INTEREST
None of the authors have any conflict of interests to declare.
ETHICS STATEMENT
This work is a systematic review of literature and no ethical committee approval was required.
Supporting information
Supplementary Material 1
ACKNOWLEDGEMENTS
With thanks to Dr Michelle Peter for her advice and assistance with the use of R software. This work is funded by the NIHR Biomedical Research Centre at Great Ormond Street Hospital. RM is fully funded and MH and LSC are partially funded by the NIHR Biomedical Research Centre at Great Ormond Street Hospital. All research at Great Ormond Street Hospital NHS Foundation Trust and UCL Great Ormond Street Institute of Child Health is made possible by the NIHR Great Ormond Street Hospital Biomedical Research Centre. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the UK Department of Health.
Mellis R, Oprych K, Scotchman E, Hill M, Chitty LS. Diagnostic yield of exome sequencing for prenatal diagnosis of fetal structural anomalies: a systematic review and meta‐analysis. Prenat Diagn. 2022;42(6):662‐685. 10.1002/pd.6115
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analysed in this study. The data that support the findings of this study are derived from the cited publicly available published studies included in the review.
REFERENCES
- 1. Syngelaki A, Hammami A, Bower S, Zidere V, Akolekar R, Nicolaides KH. Diagnosis of fetal non‐chromosomal abnormalities on routine ultrasound examination at 11–13 weeks’ gestation. Ultrasound Obstet Gynecol. 2019;54(4):468‐476. [DOI] [PubMed] [Google Scholar]
- 2. Wapner RJ, Martin CL, Levy B, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med. 2012;367(23):2175‐2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Callaway JLA, Shaffer LG, Chitty LS, Rosenfeld JA, Crolla JA. The clinical utility of microarray technologies applied to prenatal cytogenetics in the presence of a normal conventional karyotype: a review of the literature. Prenat Diagn. 2013;33(12):1119‐1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Best S, Wou K, Vora N, Van der Veyver IB, Wapner R, Chitty LS. Promises, pitfalls and practicalities of prenatal whole exome sequencing. Prenat Diagn. 2017;38(1):10‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Srivastava S, Love‐Nichols JA, Dies KA, et al. Meta‐analysis and multidisciplinary consensus statement: exome sequencing is a first‐tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet Med. 2019;21(11):2413‐2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clark MM, Stark Z, Farnaes L, et al. Meta‐analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genom Med. 2018;3(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chandler N, Best S, Hayward J, et al. Rapid prenatal diagnosis using targeted exome sequencing: a cohort study to assess feasibility and potential impact on prenatal counseling and pregnancy management. Genet Med. 2018;20(11):1430‐1437. [DOI] [PubMed] [Google Scholar]
- 8. Normand EA, Braxton A, Nassef S, et al. Clinical exome sequencing for fetuses with ultrasound abnormalities and a suspected Mendelian disorder. Genome Med. 2018;10(1):74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mone F, McMullan D, Williams D, et al. Evidence to support the clinical utility of prenatal exome sequencing in evaluation of the fetus with congenital anomalies. BJOG An Int J Obstet Gynaecol. 2021;128(9). [DOI] [PubMed] [Google Scholar]
- 10. ISPD, SMFM, PQF . Joint Position Statement from the International Society of Prenatal Diagnosis (ISPD), the Society of Maternal Fetal Medicine (SMFM) and the Perinatal Quality Foundation (PQF) on the use of genome‐wide sequencing for fetal diagnosis. Prenat Diagn. 2018;38:6‐9. [DOI] [PubMed] [Google Scholar]
- 11. Monaghan KG, Leach NT, Pekarek D, Prasad P, Rose NC. The use of fetal exome sequencing in prenatal diagnosis: a points to consider document of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2020;22(4):675‐680. [DOI] [PubMed] [Google Scholar]
- 12. Lord J, McMullan DJ, Eberhardt RY, et al. Prenatal exome sequencing analysis in fetal structural anomalies detected by ultrasonography (PAGE): a cohort study. Lancet. 2019;393(10173):747‐757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Petrovski S, Aggarwal V, Giordano JL, et al. Whole‐exome sequencing in the evaluation of fetal structural anomalies: a prospective cohort study. Lancet. 2019;393(10173):758‐767. [DOI] [PubMed] [Google Scholar]
- 14. Han J, Yang Y‐D, He Y, et al. Rapid prenatal diagnosis of skeletal dysplasia using medical trio exome sequencing: benefit for prenatal counseling and pregnancy management. Prenat Diagn. 2020;40(5):577‐584. [DOI] [PubMed] [Google Scholar]
- 15. Zhou X, Chandler N, Deng L, Zhou J, Yuan M, Sun L. Prenatal diagnosis of skeletal dysplasias using a targeted skeletal gene panel. Prenat Diagn. 2018;38(9):692‐699. [DOI] [PubMed] [Google Scholar]
- 16. Pratt M, Garritty C, Thuku M, et al. Application of exome sequencing for prenatal diagnosis: a rapid scoping review. Genet Med. 2020;22(12):1925‐1934. [DOI] [PubMed] [Google Scholar]
- 17. Guadagnolo D, Mastromoro G, Di Palma F, Pizzuti A, Marchionni E. Prenatal exome sequencing: background, current practice and future perspectives—a systematic review. Diagnostics. 2021;11(2):224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta‐analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372:n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bossuyt PM, Reitsma JB, Bruns DE, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ. 2015;351:h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2020. [Google Scholar]
- 22. Viechtbauer W. Conducting meta‐analyses in R with the metafor package. J Stat Softw. 2010;36(3):1‐48. [Google Scholar]
- 23. Mellis R, Eberhardt R, Hamilton S, et al. Fetal exome sequencing for isolated increased nuchal translucency: should we be doing it? BJOG An Int J Obstet Gynaecol. 2022;129(1):52‐61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mone F, Eberhardt RY, Morris RK, et al. COngenital heart disease and the Diagnostic yield with Exome sequencing (CODE) study: prospective cohort study and systematic review. Ultrasound Obstet Gynecol. 2021;57(1):43‐51. [DOI] [PubMed] [Google Scholar]
- 25. Mone F, Eberhardt RY, Hurles ME, et al. Fetal hydrops and the Incremental yield of Next‐generation sequencing over standard prenatal Diagnostic testing (FIND study): prospective cohort study and meta‐analysis. Ultrasound Obstet Gynecol. 2021;58(4):509‐518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Koning MA, Hoffer MJV, Nibbeling EAR, et al. Prenatal exome sequencing: a useful tool for the fetal neurologist. Clin Genet. 2022;101(1):65‐77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wagner T, Fahham D, Frumkin A, et al. The many etiologies of nonimmune hydrops fetalis diagnosed by exome sequencing. Prenat Diagn. 2021;5977. [DOI] [PubMed] [Google Scholar]
- 28. Baptiste C, Mellis R, Aggarwal V, et al. Fetal central nervous system anomalies: When should we offer exome sequencing? Prenat Diagn. 2022:1‐8. [DOI] [PubMed] [Google Scholar]
- 29. Choy KW, Wang H, Shi M, et al. Prenatal diagnosis of fetuses with increased nuchal translucency by genome sequencing analysis. Front Genet. 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Armes JE, Williams M, Price G, et al. Application of whole genome sequencing technology in the investigation of genetic causes of fetal, perinatal, and early infant death. Pediatr Dev Pathol. 2018;21(1):54‐67. [DOI] [PubMed] [Google Scholar]
- 31. Liao Y, Yang Y, Wen H, et al. Abnormal Sylvian fissure at 20–30 weeks as an indicator of malformations of cortical development: role for prenatal whole‐genome sequencing. Ultrasound Obstet Gynecol. 2021. [DOI] [PubMed] [Google Scholar]
- 32. Drury S, Williams H, Trump N, et al. Exome sequencing for prenatal diagnosis of fetuses with sonographic abnormalities. Prenat Diagn. 2015;35(10):1010‐1017. [DOI] [PubMed] [Google Scholar]
- 33. Aarabi M, Sniezek O, Jiang H, et al. Importance of complete phenotyping in prenatal whole exome sequencing. Hum Genet. 2018;137(2):175‐181. [DOI] [PubMed] [Google Scholar]
- 34. Fu F, Li R, Li Y, et al. Whole exome sequencing as a diagnostic adjunct to clinical testing in fetuses with structural abnormalities. Ultrasound Obstet Gynecol. 2018;51(4):493‐502. [DOI] [PubMed] [Google Scholar]
- 35. Aggarwal S, Vineeth VS, Das Bhowmik A, et al. Exome sequencing for perinatal phenotypes: the significance of deep phenotyping. Prenat Diagn. 2020;40(2):260‐273. [DOI] [PubMed] [Google Scholar]
- 36. Chen M, Chen J, Wang C, et al. Clinical application of medical exome sequencing for prenatal diagnosis of fetal structural anomalies. Eur J Obstet Gynecol Reprod Biol. 2020;251:119‐124. [DOI] [PubMed] [Google Scholar]
- 37. Deng Q, Fu F, Yu Q, et al. Nonimmune hydrops fetalis: genetic analysis and clinical outcome. Prenat Diagn. 2020;40(7):803‐812. [DOI] [PubMed] [Google Scholar]
- 38. Lei T‐Y, Fu F, Li R, et al. Whole‐exome sequencing in the evaluation of fetal congenital anomalies of the kidney and urinary tract detected by ultrasonography. Prenat Diagn. 2020. [DOI] [PubMed] [Google Scholar]
- 39. Li L, Fu F, Li R, et al. Genetic tests aid in counseling of fetuses with cerebellar vermis defects. Prenat Diagn. 2020. [DOI] [PubMed] [Google Scholar]
- 40. Li R, Fu F, Yu Q, et al. Prenatal exome sequencing in fetuses with congenital heart defects. Clin Genet. 2020. [DOI] [PubMed] [Google Scholar]
- 41. Pooh RK, Machida M, Nakamura T, Matsuzawa N, Chiyo H. Early sonographic findings for suspecting de novo single‐gene mutation. Donald Sch J Ultrasound Obstet Gynecol. 2020;14(2):125‐130. [Google Scholar]
- 42. Yang X, Huang L, Pan M, et al. Exome sequencing improves genetic diagnosis of fetal increased nuchal translucency. Prenat Diagn. 2020;5789. [DOI] [PubMed] [Google Scholar]
- 43. Qi Q, Jiang Y, Zhou X, et al. Simultaneous detection of CNVs and SNVs improves the diagnostic yield of fetuses with ultrasound anomalies and normal karyotypes. Genes (Basel). 2020;11(12):1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rinaldi B, Race V, Corveleyn A, et al. Next‐generation sequencing in prenatal setting: some examples of unexpected variant association. Eur J Med Genet. 2020;63(5):103875. [DOI] [PubMed] [Google Scholar]
- 45. Al‐Kouatly HB, Makhamreh MM, Rice SM, et al. High diagnosis rate for nonimmune hydrops fetalis with prenatal clinical exome from the Hydrops‐Yielding Diagnostic Results of Prenatal Sequencing (HYDROPS) Study. Genet Med. 2021;23(7):1325‐1333. [DOI] [PubMed] [Google Scholar]
- 46. Cao Q, Yang Y, Pan M, Han J, Yang X, Li D‐Z. Fetal akinesia: the application of clinical exome sequencing in cases with decreased fetal movement. Eur J Obstet Gynecol Reprod Biol. 2021;260:59‐63. [DOI] [PubMed] [Google Scholar]
- 47. Zhang F, Long W, Zhou Q, et al. Is prenatal diagnosis necessary for fetal isolated nasal bone absence or hypoplasia? Int J Gen Med. 2021;14:4435‐4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhen L, Yang Y‐D, Xu L‐L, Cao Q, Li D‐Z. Fetal micrognathia in the first trimester: an ominous finding even after a normal array. Eur J Obstet Gynecol Reprod Biol. 2021;263:176‐180. [DOI] [PubMed] [Google Scholar]
- 49. Pangalos C, Hagnefelt B, Lilakos K, Konialis C. First applications of a targeted exome sequencing approach in fetuses with ultrasound abnormalities reveals an important fraction of cases with associated gene defects. PeerJ. 2016;4:e1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu Y, Wang L, Yang Y‐K, et al. Prenatal diagnosis of fetal skeletal dysplasia using targeted next‐generation sequencing: an analysis of 30 cases. Diagn Pathol. 2019;14(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Quinlan‐Jones E, Lord J, Williams D, et al. Molecular autopsy by trio exome sequencing (ES) and postmortem examination in fetuses and neonates with prenatally identified structural anomalies. Genet Med. 2019;21(5):1065‐1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Corsten‐Janssen N, Bouman K, Diphoorn JCD, et al. A prospective study on rapid exome sequencing as a diagnostic test for multiple congenital anomalies on fetal ultrasound. Prenat Diagn. 2020;40(10):1300‐1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Heide S, Spentchian M, Valence S, et al. Prenatal exome sequencing in 65 fetuses with abnormality of the corpus callosum: contribution to further diagnostic delineation. Genet Med. 2020;22(11):1887‐1891. [DOI] [PubMed] [Google Scholar]
- 54. Zhou X, Zhou J, Wei X, et al. Value of exome sequencing in diagnosis and management of recurrent non‐immune hydrops fetalis: a retrospective analysis. Front Genet. 2021;12(616392). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhou J, Yang Z, Sun J, et al. Whole genome sequencing in the evaluation of fetal structural anomalies: a parallel test with chromosomal microarray plus whole exome sequencing. Genes (Basel). 2021;12(3):1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang X, Ren Y, Song R, et al. Combined exome sequencing and deep phenotyping in highly selected fetuses with skeletal dysplasia during the first and second trimesters improves diagnostic yield. Prenat Diagn. 2021;41(11):1401‐1413. [DOI] [PubMed] [Google Scholar]
- 57. Zhang L, Pan L, Teng Y, Liang D, Li Z, Wu L. Molecular diagnosis for 55 fetuses with skeletal dysplasias by whole‐exome sequencing: a retrospective cohort study. Clin Genet. 2021;100(2):219‐226. [DOI] [PubMed] [Google Scholar]
- 58. Yates CL, Monaghan KG, Copenheaver D, et al. Whole‐exome sequencing on deceased fetuses with ultrasound anomalies: expanding our knowledge of genetic disease during fetal development. Genet Med. 2017;19(10):1171‐1178. [DOI] [PubMed] [Google Scholar]
- 59. Xue S, Yan H, Chen J, et al. Genetic examination for fetuses with increased fetal nuchal translucency by genomic technology. Cytogenet Genome Res. 2020;160(2):57‐62. [DOI] [PubMed] [Google Scholar]
- 60. Westphal DS, Leszinski GS, Rieger‐Fackeldey E, et al. Lessons from exome sequencing in prenatally diagnosed heart defects: a basis for prenatal testing. Clin Genet. 2019;95(5):582‐589. [DOI] [PubMed] [Google Scholar]
- 61. Vora NL, Gilmore K, Brandt A, et al. An approach to integrating exome sequencing for fetal structural anomalies into clinical practice. Genet Med. 2020;22(8):1426. [DOI] [PubMed] [Google Scholar]
- 62. Tolusso LK, Hazelton P, Wong B, Swarr DT. Beyond diagnostic yield: prenatal exome sequencing results in maternal, neonatal, and familial clinical management changes. Genet Med. 2021;23(5):909‐917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tang H, Zhang Q, Xiang J, et al. Whole exome sequencing aids the diagnosis of fetal skeletal dysplasia. Front Genet. 2021;12(599863). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sun H, Yi T, Hao X, et al. Contribution of single‐gene defects to congenital cardiac left‐sided lesions in the prenatal setting. Ultrasound Obstet Gynecol. 2020;56(2):225‐232. [DOI] [PubMed] [Google Scholar]
- 65. Sparks TN, Lianoglou BR, Adami RR, et al. Exome sequencing for prenatal diagnosis in nonimmune hydrops fetalis. N Engl J Med. 2020;383(18):1746‐1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shamseldin HE, Kurdi W, Almusafri F, et al. Molecular autopsy in maternal–fetal medicine. Genet Med. 2018;20(4):420‐427. [DOI] [PubMed] [Google Scholar]
- 67. Reischer T, Liebmann‐Reindl S, Bettelheim D, Balendran‐Braun S, Streubel B. Genetic diagnosis and clinical evaluation of severe fetal akinesia syndrome. Prenat Diagn. 2020;40(12):1532‐1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Meier N, Bruder E, Lapaire O, et al. Exome sequencing of fetal anomaly syndromes: novel phenotype‐genotype discoveries. Eur J Hum Genet. 2019;27(5):730‐737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Leung GKC, Mak CCY, Fung JLF, et al. Identifying the genetic causes for prenatally diagnosed structural congenital anomalies (SCAs) by whole‐exome sequencing (WES). BMC Med Genomics. 2018;11(1):93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lefebvre M, Bruel A‐L, Tisserant E, et al. Genotype‐first in a cohort of 95 fetuses with multiple congenital abnormalities: when exome sequencing reveals unexpected fetal phenotype‐genotype correlations. J Med Genet. 2021;58(6):400‐413. [DOI] [PubMed] [Google Scholar]
- 71. Guo W, Lai Y, Yan Z, et al. Trio‐whole‐exome sequencing and preimplantation genetic diagnosis for unexplained recurrent fetal malformations. Hum Mutat. 2020;41(2):432‐448. [DOI] [PubMed] [Google Scholar]
- 72. Greenbaum L, Pode‐Shakked B, Eisenberg‐Barzilai S, et al. Evaluation of diagnostic yield in fetal whole‐exome sequencing: a report on 45 consecutive families. Front Genet. 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Deden C, Neveling K, Zafeiropopoulou D, et al. Rapid whole exome sequencing in pregnancies to identify the underlying genetic cause in fetuses with congenital anomalies detected by ultrasound imaging. Prenat Diagn. 2020;40(8):972‐983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. de Koning MA, Haak MC, Adama van Scheltema PN, et al. From diagnostic yield to clinical impact: a pilot study on the implementation of prenatal exome sequencing in routine care. Genet Med. 2019;21(10):2303‐2310. [DOI] [PubMed] [Google Scholar]
- 75. Daum H, Meiner V, Elpeleg O, et al. Fetal exome sequencing: yield and limitations in a tertiary referral center. Ultrasound Obstet Gynecol. 2019;53(1):80‐86. [DOI] [PubMed] [Google Scholar]
- 76. Carss KJ, Hillman SC, Parthiban V, et al. Exome sequencing improves genetic diagnosis of structural fetal abnormalities revealed by ultrasound. Hum Mol Genet. 2014;23(12):3269‐3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Boissel S, Fallet‐Bianco C, Chitayat D, et al. Genomic study of severe fetal anomalies and discovery of GREB1L mutations in renal agenesis. Genet Med. 2018;20(7):745‐753. [DOI] [PubMed] [Google Scholar]
- 78. Becher N, Andreasen L, Sandager P, et al. Implementation of exome sequencing in fetal diagnostics‐Data and experiences from a tertiary center in Denmark. Acta Obstet Gynecol Scand. 2020;99(6):783‐790. [DOI] [PubMed] [Google Scholar]
- 79. Aoi H, Mizuguchi T, Suzuki T, et al. Whole exome sequencing of fetal structural anomalies detected by ultrasonography. J Hum Genet. 2021;66(5):499‐507. [DOI] [PubMed] [Google Scholar]
- 80. Zhou X, Wang Y, Shao B, et al. Molecular diagnostic in fetuses with isolated congenital anomalies of the kidney and urinary tract by whole‐exome sequencing. J Clin Lab Anal. 2020;e23480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Correa ARE, Naini K, Mishra P, et al. Utility of fetal whole exome sequencing in the etiological evaluation and outcome of nonimmune hydrops fetalis. Prenat Diagn. 2021;41(11):1414‐1424. [DOI] [PubMed] [Google Scholar]
- 82. Dempsey E, Haworth A, Ive L, et al. A report on the impact of rapid prenatal exome sequencing on the clinical management of 52 ongoing pregnancies: a retrospective review. BJOG An Int J Obstet Gynaecol. 2021;128(6):1012‐1019. [DOI] [PubMed] [Google Scholar]
- 83. He M, Du L, Xie H, et al. The added value of whole‐exome sequencing for anomalous fetuses with detailed prenatal ultrasound and postnatal phenotype. Front Genet. 2021;12:627204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kucińska‐Chahwan A, Roszkowski T, Nowakowska B, et al. Genetic causes of the skeletal system abnormalities diagnosed by prenatal sonography with the use of exome sequencing: single institution experience. Ultrasound Obstet Gynecol. 2021;01:uog.23722. [Google Scholar]
- 85. Lei L, Zhou L, Xiong JJ. Whole‐exome sequencing increases the diagnostic rate for prenatal fetal structural anomalies. Eur J Med Genet. 2021;64(9):104288. [DOI] [PubMed] [Google Scholar]
- 86. Peng Y, Yang S, Huang X, et al. Whole exome sequencing analysis in fetal skeletal dysplasia detected by ultrasonography: an analysis of 38 cases. Front Genet. 2021;12(728544). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Qiao F, Wang Y, Zhang C, et al. Comprehensive evaluation of genetic variants using chromosomal microarray analysis and exome sequencing in fetuses with congenital heart defect. Ultrasound Obstet Gynecol. 2021;58(3):377‐387. [DOI] [PubMed] [Google Scholar]
- 88. Sun HR, Hao XY, Wang X, et al. Genetics and clinical features of noncompaction cardiomyopathy in the fetal population. Front Cardiovasc Med. 2021;7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wright CF, McRae JF, Clayton S, et al. Making new genetic diagnoses with old data: iterative reanalysis and reporting from genome‐wide data in 1,133 families with developmental disorders. Genet Med. 2018;20(10):1216‐1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Diderich K, Joosten M, Govaerts L, et al. Is it feasible to select fetuses for prenatal WES based on the prenatal phenotype? Prenat Diagn. 2019;39(11):1039‐1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Pauta M, Martinez‐Portilla R, Borrell A. Diagnostic yield of exome sequencing in fetuses with an isolated increased nuchal translucency: systematic review and meta‐analysis. Ultrasound Obstet Gynecol. 2021; uog.23746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed in this study. The data that support the findings of this study are derived from the cited publicly available published studies included in the review.


