Highlights
-
•
The UPR is an adaptive stress response network that is tightly linked to the ability of Aspergillus fumigatus, and other pathogenic fungi, to sustain viability in the presence of adverse environmental conditions, including the stress of infection. In this review, we summarize the evidence that supports the concept of targeting the A. fumigatus UPR as a strategy to reduce the ability of the fungus to withstand stress.
Keywords: Aspergillus fumigatus, UPR, HacA, IreA, ER stress, cell death
Abstract
One of the most potent opportunistic fungal pathogens of humans is Aspergillus fumigatus, an environmental mold that causes a life-threatening pneumonia with a high rate of morbidity and mortality. Despite advances in therapy, issues of drug toxicity and antifungal resistance remain an obstacle to effective therapy. This underscores the need for more information on fungal pathways that could be pharmacologically manipulated to either reduce the viability of the fungus during infection, or to unleash the fungicidal potential of current antifungal drugs. In this review, we summarize the emerging evidence that the ability of A. fumigatus to sustain viability during stress relies heavily on an adaptive signaling pathway known as the unfolded protein response (UPR), thereby exposing a vulnerability in this fungus that has strong potential for future therapeutic intervention.
Graphical abstract
Introduction
Aspergillus fumigatus is an environmental mold and opportunistic pathogen of humans that causes invasive aspergillosis (IA), a life-threatening pulmonary infection that affects patients with immune defects that disrupt the barrier functions of the lung (Cadena et al., 2021). The infection is acquired by the inhalation of airborne conidia, which are widespread in nature and hence practically impossible to avoid. Individuals with mild immunosuppression, or those with pre-existing pulmonary structural defects, are unable to efficiently clear the inhaled conidia, which allows them to germinate in the lung. Upon germination, the spores begin to release hydrolytic enzymes, creating damage to the surrounding tissue and triggering a chronic inflammatory process that can evolve over months to years (Barac et al., 2019). In the context of more severe immunosuppression, most frequently in patients undergoing hematopoietic stem cell or solid organ transplantation, the germinating spores can develop into hyphae. This filamentous form of the organism is highly invasive, capable of breaching the epithelial barrier and spreading through the circulation. The outcome of the resulting IA is very poor, even when treated, with mortality rates that can exceed 50%. IA may also arise in patients with acute respiratory distress syndrome (ARDS), including those infected with SARS-CoV-2 (Batah and Fabro, 2021). This emerging COVID-19-associated pulmonary aspergillosis (CAPA) syndrome is associated with increased mortality, the incidence of which may be underestimated due to difficulties with the accurate diagnosis of A. fumigatus infection in this patient population (Arastehfar et al., 2020; Borman et al., 2020; Marr et al., 2021). The precise reason for the susceptibility of COVID-19 patients to CAPA is likely to be multifactorial, involving impaired mucociliary clearance, damage to the lung epithelial barrier, and the use of immunomodulatory treatment regimens to dampen the hyperinflammatory signaling that contributes to COVID-19 pathology (Marr et al., 2021). However, recent evidence has also implicated COVID-19 in the suppression of host functional adaptive and innate immunity, which may account for some reports of CAPA arising even in the absence of well-defined immune compromising disease (Remy et al., 2020).
The ability of A. fumigatus to cause disease in a human host involves diverse phenotypic characteristics that initially evolved to benefit the fungus in its environmental niche, but are similarly advantageous to its ability to cause infection (Abad et al., 2010; Casadevall et al., 2019). One of the most important of these traits involves nutrient acquisition. A. fumigatus obtains nutrients from environmental debris, or the tissues of an infected human host, by secreting hydrolytic enzymes that degrade extracellular biopolymers. These enzymes are synthesized on endoplasmic reticulum (ER)-bound ribosomes and inserted into the ER lumen as linear polymers, which then begin a process of self-assembly that is facilitated by transient interactions with ER-resident molecular chaperones. Accurate protein folding is also assisted by the activity of glycosylating enzymes, as well as ER-lumenal folding enzymes that provide protein structural support in the form of disulfide bonds (Braakman and Bulleid, 2011). However, if an increase in the demand for secretion distorts the homeostatic balance of protein folding, the consequent accumulation of unfolded proteins increases the risk for illegitimate protein interactions that can trigger toxic protein aggregation and cell death. To counter this unfolded protein stress, eukaryotic cells possess an adaptive stress response pathway known as the unfolded protein response (UPR), which allows them to adjust multiple aspects of ER function in order to optimize protein folding equilibrium (Hetz et al., 2020).
The extensive armamentarium of pathogenicity factors that is available to A. fumigatus is a major challenge to the rational design of anti-virulence strategies that seek to neutralize one or more of the most relevant virulence traits. This raises the possibility that adaptive cellular networks that indirectly control the expression of a broad spectrum of homeostatic processes during infection could have merit as therapeutic targets. The UPR represents one such adaptive network that is tightly linked to the ability of fungal species to maintain viability in the presence of stress. In this review, we summarize the evidence that supports the concept of targeting the A. fumigatus UPR as a strategy to enhance fungicidal activity.
The UPR pathway in fungi and mammals
The UPR is a signaling pathway that continually monitors the efficiency of protein folding in the ER lumen and communicates that information to the nucleus.
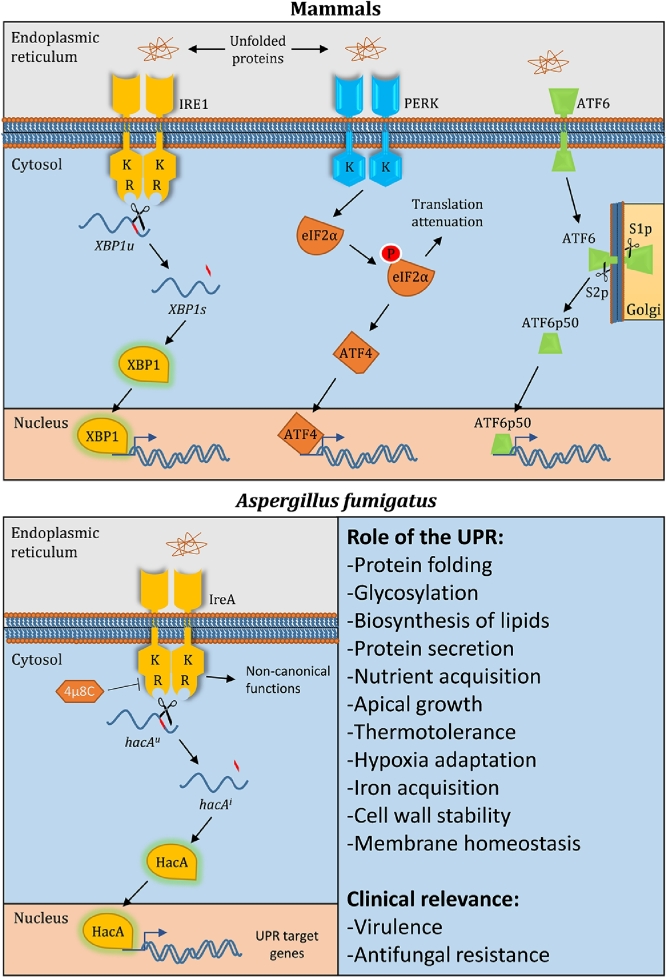
This triggers a reprogramming of the transcriptome to increase protein folding capacity, in addition to augmenting other aspects of cell function that contribute to ER homeostasis (Fig. 1). In humans, three ER-transmembrane sensors initiate the signal transducing process: IRE1 (inositol requiring enzyme 1), ATF6 (activating transcription factor 6), and PERK (PKR-like endoplasmic reticulum kinase) (FIG 1) (Hetz et al., 2020). All three sensors are activated by the accumulation of unfolded or misfolded proteins, which arises whenever the demand for secretion exceeds ER protein folding capacity. IRE1 and ATF6 counter this stress by directing the synthesis of separate transcription factors, XBP1 (X-box binding protein 1) and ATF6p50 respectively, each of which controls the expression of UPR target genes involved in ER homeostasis. Human IRE1 accomplishes this by regulating the translation of a cytoplasmic mRNA that encodes the XBP1 transcription factor, whereas the ATF6 transcription factor is a component of the ATF6 sensor itself and is released from the sensor by proteolytic cleavage in the Golgi apparatus (FIG 1). PERK is unique among the three sensors in that it employs both transcriptional and translational outputs. Upon activation, PERK phosphorylates the alpha subunit of the eukaryotic initiation factor 2 (eIF2α), resulting in a broad attenuation of translational activity that relieves ER stress by temporarily reducing the protein load entering the ER. The transcriptional activity of PERK involves the mRNA encoding ATF4 (activating transcription factor 4), which is able to bypass the translation block and produce the ATF4 transcription factor that contributes to the overall transcriptional rewiring that brings stressed cells back to the homeostatic state. IRE1 directs the most ancient branch of the UPR, which is the only known pathway in the fungal kingdom and discussed further below.
Fig. 1.
UPR signaling in mammals and fungi. Top: The mammalian UPR comprises three ER-transmembrane sensors that are activated by the accumulation of unfolded proteins in the ER lumen: IRE1, PERK and ATF6. The IRE1 protein has a cytosolic region containing both kinase (K) and RNase (R) domains. Upon activation, the RNase cleaves an intron (shown in red) from the XBP1u (unspliced) mRNA. The resulting XBP1s (spliced) mRNA contains a translational frame-shift that directs the synthesis of the XBP1 transcription factor required for UPR target gene induction in the nucleus. The PERK sensor contains a cytosolic kinase domain which, when activated, phosphorylates eIF2α (eukaryotic initiation factor 2 alpha), resulting in a global attenuation of protein synthesis that serves to reduce further influx of client proteins into the ER. However, the mRNA encoding the ATF4 transcription factor is able to bypass this translational block, allowing it to contribute to the transcriptional rewiring necessary to restore ER homeostasis. The ATF6 sensor has a cytosolic transcription factor domain (ATF6p50). Upon activation, the protein moves to the Golgi apparatus, where the ATF6p50 transcription factor is released from the sensor by the activity of site-1 protease (S1p) and site-2 protease (S2p). Bottom: The UPR pathway in A. fumigatus. Unfolded proteins activate the IreA RNase to cleave an intron from the hacAu mRNA, creating a frame-shift in the resulting hacAi mRNA that translates the HacA transcription factor (the canonical pathway). Both canonical and non-canonical functions for IreA contribute to the expression of virulence-related traits, which jointly support both pathogenicity during infection and resistance to antifungal drugs.
In fungi, the canonical IRE1-driven UPR was first defined in Saccharomyces cerevisiae and shown to comprise the Ire1 sensor and its downstream bZIP transcription factor Hac1 (FIG 1) (Mori, 2009). In yeast, and in most other fungal species, the mRNA encoding Hac1 is unusual in that it is first synthesized as a cytoplasmic precursor mRNA (HAC1u for uninduced). The HAC1u mRNA is unable to translate the UPR transcription factor due to the presence of an atypical intron that is not excised by the conventional splicing machinery in the nucleus. However, upon transport to the cytoplasm the unconventional intron becomes a target for regulated splicing by the Ire1 sensor. Ire1 is a type I ER-membrane protein with three functional domains: an ER-luminal unfolded protein sensing domain, a cytosolic kinase domain, and a cytosolic C-terminal endoribonuclease (RNase) domain. When unfolded or misfolded proteins accrue in the ER lumen, Ire1 oligomerizes in the ER membrane, resulting in trans-autophosphorylation and activation of its RNase domain. The activated RNase then cleaves the unconventional intron from the HAC1u mRNA, creating an induced form of the mRNA, HAC1i (for induced). The open reading frame in the spliced HAC1i mRNA is shifted relative to that of HAC1u, allowing for the translation of the Hac1 transcription factor. After migrating to the nucleus, the Hac1 protein reorganizes the transcriptome towards a program that elevates the capacity of the ER to handle the influx of proteins in the secretory pathway. Any ER proteins that ultimately fail to achieve their native confirmation are destroyed by ER-associated degradation (ERAD), a UPR-linked quality control step that returns abnormal proteins to the cytosol and targets them for proteasomal degradation (Hwang and Qi, 2018; Travers et al., 2000).
In A. fumigatus, the canonical Ire1 pathway follows the same paradigm established in S. cerevisiae and humans but with modified nomenclature: the IreA RNase converts the hacAu mRNA into hacAi, resulting in the translation of the HacA transcription factor (FIG 1). As in yeast, ER stress triggers the HacA transcription factor to increase the expression of chaperones and folding enzymes that are well-established UPR target genes in other species. In addition, the A. fumigatus UPR coordinates chaperone upregulation with increased transcription of genes encoding P-type ATPases that transport different cargoes across ER and Golgi membranes. For example, the P2-type Ca2+ ATPases SrcA and PmrA provide a mechanism to ensure that sufficient quantities of Ca2+ are available to support Ca2+-dependent chaperones in the secretory pathway. By contrast P5-type ATPase SpfA supports fungal ER homeostasis through more pleiotropic effects on lipid homeostasis, Ca2+ and Mn+2 transport, as well as selective extraction of transmembrane proteins that are mistargeted to the ER (Cohen et al., 2013; Cronin et al., 2002; Guirao-Abad et al., 2021; McKenna et al., 2020; Sørensen et al., 2019; Weichert et al., 2020). However, in contrast to S. cerevisiae, where processing of HAC1u mRNA into HAC1i occurs mostly under conditions of an acute ER stress stimulus, a substantial amount of hacAu mRNA processing into hacAi is always detected in A. fumigatus mycelia grown in the absence of any exogenous stress (Richie et al., 2009). This suggests that the demands of polarized filamentous growth may require greater support from the UPR in A. fumigatus relative to budding yeast.
Therapeutic prospects for targeting fungal PCD-like mechanisms
If the adaptive responses of the metazoan UPR fail to restore homeostasis in the presence of chronic ER stress, the UPR switches to an alternative signaling mechanism that triggers apoptotic death (Hetz et al., 2020). Apoptosis and related programmed cell death (PCD) pathways have been well characterized in metazoan species, which has provided a rational foundation for the development of novel anticancer compounds that elicit the demise of the cancer cell (Fulda, 2015). Regulated forms of cell death have also been described in fungi (Gonçalves et al., 2017; Hamann et al., 2007; Hutchison et al., 2009), including a recent report that immune cells can initiate an apoptosis-like program in A. fumigatus (Shlezinger et al., 2017). This raises the possibility that pharmacologic manipulation of fungal cell death mechanisms could be a useful adjunct to current therapies. Although fungal genomes encode some homologs of established mammalian apoptosis death regulators, their functions in bona fide PCD pathways are less clear relative to their mammalian counterparts (Fedorova et al., 2005). For example, fungi possess metacaspases, which are related to the metazoan caspases that participate in the execution of apoptosis (Tsiatsiani et al., 2011). In contrast to caspases, no evidence was found that A. fumigatus metacaspases actively promote cell death in since a mutant lacking both of the only two metacaspases in the A. fumigatus genome showed no reduction in stress response viability or virulence (Richie et al., 2007). However, this mutant was growth impaired under conditions of ER stress (Richie et al., 2007), suggesting that metacaspase activity in A. fumigatus provides a function that is more adaptive than pro-apoptotic. It is interesting to note that subsequent studies have demonstrated a role for fungal metacaspases in the removal of insoluble protein aggregates (Fernandez et al., 2021; Lee et al., 2010; Shrestha et al., 2019), raising the possibility that these proteolytic enzymes work in collaboration with the UPR and ERAD pathways to prevent the accumulation of damaged proteins that would be deleterious to cell viability. This implies that any therapeutic strategy to manipulate metacaspase function in A. fumigatus should be designed to inhibit metacaspase function rather than triggering their activity.
Therapeutic prospects for cell death induction by targeting the UPR: inspiration from cancer biology
The central role occupied by the ER in numerous aspects of cell function suggests that intracellular pathways that sustain ER homeostasis could represent a point of vulnerability that may be amenable to pharmacologic induction of cell death. For example, human tumor cells are predisposed to ER stress because of their elevated metabolic activity, their creation of nutrient-limited or hypoxic microenvironments when they expand without sufficient vascular support, or their exposure to chemotherapy and radiotherapy (Oakes, 2020). This is particularly true for neoplasms that are derived from professional secretory cells such as myeloma-related disorders or pancreatic tumors, placing them at high risk for ER stress (Oakes, 2020). Indeed, activation of all three branches of the UPR has been reported in several human cancers, which allows the tumor cells to thrive in the adverse tumor microenvironment and to resist anticancer therapies (Cubillos-Ruiz et al., 2017; Logue et al., 2018; Zhao et al., 2018). This is driving the search for novel strategies to control UPR outputs in cancer cells, with the goal of activating tumor cell death pathways or sensitizing the tumor to cell death induction by existing methods of chemotherapy or radiotherapy (Doultsinos et al., 2021; Logue et al., 2018; Sun et al., 2016).
The presence of dual kinase and RNase domains in IRE1, both of which are necessary to induce the UPR, makes this protein a strong candidate for therapeutic intervention. The rationale for this approach is based on the notion that blocking the IRE1 branch of the UPR in a cell type that relies heavily on IRE1-derived signals for sustained ER homeostasis would create a situation of unresolved ER stress that either kills the cell directly, or renders the cell inviable in the presence of other therapeutic compounds. A theoretical concern about this strategy to treat human cancer is the potential for collateral damage to normal cells that also rely on IRE1 for normal homeostasis. However, several small molecule inhibitors of human IRE1 have already been developed for this purpose and have shown efficacy in vitro and in vivo, either as monotherapy or as chemosensitizing agents in combination with other cancer treatments (Korbelik et al., 2020; Le Reste et al., 2020; Logue et al., 2018; Maly and Papa, 2014; McCarthy et al., 2020; Shao et al., 2020; Zhao et al., 2018). The successful application of these IRE1-targeting agents with minimal side effects is likely to be due to functional redundancy among the three UPR sensors.
Prospects for cell death induction by targeting the A. fumigatus UPR
Cancer cells encounter ER stress in the host environment or when exposed to anticancer therapy (Oakes, 2020). The following lines of evidence argue that A. fumigatus also encounters ER stress in the host, and that its reliance on UPR intervention to maintain homeostasis exposes a vulnerability in the fungus that could be harnessed to manipulate cell death.
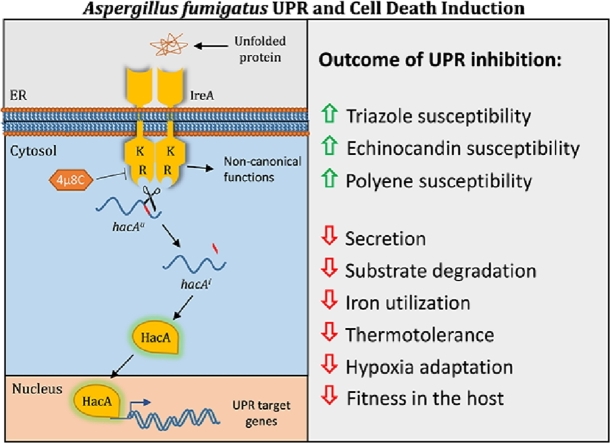
1. The canonical HacA pathway supports virulence and drug susceptibility
As a typical filamentous fungus, A. fumigatus is highly specialized for protein secretion (Conesa et al., 2001; Vivek-Ananth et al., 2018; Wang et al., 2020), suggesting that it would be particularly vulnerable to pharmacologic disruption of ER homeostasis. To test this, we and others have taken genetic approaches to determine how fungal viability is affected by loss of UPR signaling. The data obtained from the deletion of the hacA gene encoding the UPR transcription factor in A. fumigatus (Richie et al., 2009) could prove useful in predicting the effects of a drug that would prevent the IreA sensor pathway from activating the canonical UPR. Gene expression analysis demonstrated that the ΔhacA mutant was unable to upregulate the expression of known UPR target genes when treated with an acute ER stress agent, confirming that the absence of hacA effectively blocks the canonical pathway. In addition, although the ΔhacA deletion mutant grew similarly to wild type on standard medium, it rapidly lost viability in the presence of chemical agents that disrupt ER protein folding, indicating that the UPR is dispensable in the absence of stress but becomes critical for viability in situations that require UPR intervention. Importantly, despite the ability of ΔhacA mutant to grow at a similar rate to wild type in vitro, it was attenuated for virulence in an immunosuppressed mouse model of IA. These data, provide evidence that one of the situations that requires UPR involvement is the challenge of growing in the host environment.
A second environment where the UPR was required for viability involved growth at elevated temperatures. A. fumigatus is notoriously thermotolerant, capable of growing rapidly at 37°C and surviving extended periods above 50°C, a characteristic that allows this environmental fungus to thrive in composting plant debris where intense microbial activity raises the temperature (Bhabhra and Askew, 2005). By contrast, the ΔhacA mutant rapidly lost viability at 45°C, which is only a few degrees above the optimum temperature range of 37-42°C for this species (Araujo and Rodrigues, 2004). The inability of this mutant to survive at 45°C correlated with extensive tip lysis, suggesting that the absence of hacA creates a defect in cell wall integrity that is incompatible with growth at high temperature. This was confirmed through a biochemical analysis of its wall, revealing a deficiency in β-(1,3)- and α-(1,3)-glucan composition. Interestingly, the corresponding mutant in S. cerevisiae did not exhibit a temperature-sensitive phenotype, demonstrating a reliance of A. fumigatus on the UPR for thermotolerant hyphal growth that is absent in this yeast.
The abnormal cell wall in the ΔhacA mutant suggested that a third situation where loss of UPR function would be lethal is in the presence of antifungal drugs. The current armamentarium of antifungal drugs against A. fumigatus includes agents that impair cell wall synthesis (the echinocandin class) or disrupt membrane homeostasis (the triazole and polyene classes). The ΔhacA mutant showed a striking increase in susceptibility to all three of these drug classes, providing proof-of-principle that pharmacologic inhibition of the canonical UPR could be used to enhance the efficacy of existing antifungal agents. Of particular note was caspofungin, a member of the echinocandin class that is normally fungistatic towards A. fumigatus. In the absence of a functional UPR, caspofungin was fungicidal for A. fumigatus, indicating that UPR intervention is a major reason why the echinocandins are unable to induce the death of this fungus. These findings provide a rational foundation for the future design of combination therapies that unite IreA inhibition with current antifungal therapy as a way to enhance fungicidal activity against A. fumigatus. The notion of enhancing triazole potency through UPR blockade is particularly attractive in light of emerging environmental and clinical isolates of A. fumigatus that are resistant to this crucial class of antifungal drugs (Bastos et al., 2021; Nywening et al., 2020). However, the extent to which UPR inhibition could mitigate the effects of different mutations that confer triazole resistance remains to be tested.
2. The canonical HacA pathway works in conjunction with ERAD
Cytosolic proteasomal degradation is the final step involved in the disposal of aberrant ER proteins by ERAD (Hwang and Qi, 2018; Travers et al., 2000). In humans, proteasome inhibitors have been developed to exploit cellular differences in the dependence on proteasomal degradation, and their clinical application can dramatically improve the outcome for patients with some forms of cancer, particularly lymphoma and myeloma-related disorders (Manasanch and Orlowski, 2017; Park et al., 2018). Since ERAD and UPR pathways jointly support protein quality control (Travers et al., 2000), the impact of ERAD dysfunction in A. fumigatus was examined by deleting the derA gene, encoding a major component of the ER-membrane complex involved in ERAD (Krishnan et al., 2013; Richie et al., 2011). Although the ΔderA mutant grew normally and showed wild-type virulence, it exhibited constitutive upregulation of UPR activity suggesting that the UPR compensates for a deficiency in ERAD. Consistent with this, a double deletion mutant lacking both derA and hacA genes was avirulent, demonstrating that cooperation between ERAD and the UPR is necessary for the fungus to thrive in the host environment. The implication of this finding is that combination therapies that target both the UPR and ERAD arms of ER protein quality control could be an innovative approach to generate critical levels of ER stress and induce fungal death.
3. The IreA sensor of A. fumigatus has functions that are independent of HacA.
The attenuated virulence of the ΔhacA mutant suggests that the canonical UPR pathway is necessary to support the fitness of A. fumigatus in the host environment. In contrast to the ΔhacA mutant, which retained partial virulence (Richie et al., 2009), a ΔireA mutant was avirulent in mouse infection models. This indicates that IreA contributes to pathogenesis not only through the canonical IreA-HacA pathway but also with its own HacA-independent functions (Feng et al., 2011). Indeed, the ΔireA mutant exhibited phenotypic defects that were much more severe than those displayed by ΔhacA, including virulence-related defects in hypoxia adaptation, growth on complex substrates, iron acquisition, thermotolerance, and antifungal drug susceptibility. Gene expression profiling confirmed that A. fumigatus IreA has both HacA-dependent and HacA-independent functions that contribute to fitness in the host environment. Together, these findings demonstrated that IreA function is not limited to the processing of hacAu into hacAi mRNA. In fact, by virtue of its role as an ER stress sensor, IreA indirectly serves as a regulatory hub for the expression of multiple phenotypic characteristics that are essential during infection. This implies that pharmacologic interruption of both the canonical IreA-HacA UPR and the independent functions of IreA would sensitize the fungus to the adverse environmental conditions encountered in the host environment, thereby limiting growth and/or inducing cell death.
4. Pharmacological inhibition of the canonical UPR is feasible in A. fumigatus
Despite the genetic evidence implicating the UPR in A. fumigatus pathogenesis and antifungal drug susceptibility, surprisingly little is known about pharmacologic inhibition of the fungal pathway. We recently reported that a salicylaldehyde-based inhibitor of the human IRE1 RNase domain, known as 4μ8C, can block an acute ER stress stimulus from triggering the accumulation of A. fumigatus hacAi mRNA and, consequently, prevent downstream UPR target gene induction (Guirao-Abad et al., 2020). This provided proof-of-principle that 4μ8C can cross the A. fumigatus cell wall and effectively block the IreA RNase from splicing the hacAu mRNA. The 4μ8C compound was only slightly toxic to the fungus at the concentrations used to inhibit the UPR, similar to the mild phenotype of the UPR-deficient mutant ΔhacA grown in the absence of stress (Richie et al., 2009). However, 4μ8C treatment weakened the ability of the wild-type fungus to grow on a collagen substrate, mirroring a similar poor growth phenotype of the ΔhacA mutant on polymeric substrates (Richie et al., 2009). Interestingly, A. fumigatus rapidly lost viability when 4μ8C treatment was combined with the plant-derived compound carvacrol (Guirao-Abad et al., 2020), a monoterpene with antifungal activity that has been shown to disrupt ER morphology and trigger the UPR in C. albicans (Chaillot et al., 2015; Wang et al., 2019). This suggests that the ER stress prompted by carvacrol is more toxic to fungal viability when UPR activation is inhibited by 4μ8C. Despite the dramatic effects of hacA deletion on antifungal drug susceptibility and stress response, only a subset of the phenotypes displayed by the ΔhacA mutant were recapitulated by pharmacologic treatment with 4μ8C. For example, unlike genetic deletion of the hacA gene, treatment with 4μ8C did not increase the azole sensitivity of A. fumigatus. However, these 4μ8C-induced phenotypes were strikingly similar to those displayed by an IreA RNase domain-deficient mutant of A. fumigatus that is unable to process hacAu mRNA into hacAi (Guirao-Abad et al., 2020). Thus, although pharmacological inhibition of the canonical UPR pathway in A. fumigatus is achievable, the evidence suggests that additional branch points exist in the fungal UPR that may be able to support the growth of A. fumigatus and potentially other fungi when this IreA/Ire1-dependent pathway is blocked. Future identification and molecular dissection of these non-canonical pathway(s) of the UPR are needed to expand our understanding of fungal stress adaptation and uncover points of vulnerability that trigger fungal cell death.
Summary and future prospects
Accumulating evidence from A. fumigatus and other fungal pathogens has demonstrated that UPR function regulates many aspects of virulence. To date, these studies have employed genetic approaches to determine the impact of UPR inhibition in diverse fungal species. These include human-pathogenic fungi (Candida spp., A. fumigatus, Cryptococcus spp., Trichoderma rubrum) and plant-pathogenic fungi (Ustilago maydis, Verticillium dahliae, Alternaria brassicicola), encompassing yeasts, polymorphic species and molds (Bitencourt et al., 2020; Cheon et al., 2011; Feng et al., 2011; Heimel et al., 2013; Joubert et al., 2011; Jung et al., 2018; Miyazaki et al., 2013; Pinter et al., 2019; Richie et al., 2009; Sircaik et al., 2021; Starke et al., 2021). Although the UPR underpins ER homeostasis in all of these organisms, the transcriptional circuitry activated by the UPR has been uniquely wired in each species to achieve different outputs. This is likely to reflect the enormous range of habitats in which each fungus resides, such as organic debris in the environment, living plants, human skin, and mucosal surfaces. Despite fundamental differences in UPR output between fungal pathogens, there are two common threads. The first is virulence; all pathogenic species in which the UPR has been studied thus far have been shown to exploit this pathway for the expression of virulence-related traits that support fitness in the host environment. Depending on the fungus, these features include hypoxia adaptation, iron acquisition, hydrolase secretion to digest host tissues, effector secretion for modification of host physiology, capsule formation to evade host defenses, biofilm formation, and developmental transitions linked to virulence such as hyphal switching or appressoria formation. The second is susceptibility to antifungal agents; all UPR-deficient mutants of human-pathogenic fungi display increased death in the presence of one or more classes of the major antifungal drug classes. Also, in the case of plant pathogens, loss of UPR function has been shown to increase fungal susceptibility to plant-derived defenses molecules (Joubert et al., 2011). As usual, however, nature knows more than we do. If we are to effectively harness the UPR as a future drug target, more research is needed to understand the branch points and networks that allow this pathway to represent such an important aspect of intracellular homeostasis and cell integrity.
Credit author statement
D.S.A.: Conceptualization; Formal analysis; Funding acquisition; Writing-original draft preparation.
J.P.G.A.: Formal analysis; Investigation; Writing - review & editing.
M.W.: Formal analysis; Investigation; Writing - review & editing.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: David Askew reports financial support was provided by National Institute of Health.
Acknowledgements
This work was supported by National Institutes of Health (NIH) grant R01 AI123158-01A1 to DSA and a University of Cincinnati Department of Pathology & Laboratory Medicine pilot research grant to JPGA.
References
- Abad A., Fernández-Molina J.V., Bikandi J., Ramírez A., Margareto J., Sendino J., Hernando F.L., Pontón J., Garaizar J., Rementeria A. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev. Iberoam. Micol. 2010;27:155–182. doi: 10.1016/j.riam.2010.10.003. 10.1016/j.riam.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Arastehfar A., Carvalho A., van de Veerdonk F.L., Jenks J.D., Koehler P., Krause R., Cornely O.A., S., Perlin D., Lass-Flörl C., Hoenigl M. COVID-19 Associated Pulmonary Aspergillosis (CAPA)—From Immunology to Treatment. J. Fungi. 2020;6:91. doi: 10.3390/jof6020091. 10.3390/jof6020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo R., Rodrigues A.G. Variability of germinative potential among pathogenic species of Aspergillus. J. Clin. Microbiol. 2004;42:4335–4337. doi: 10.1128/JCM.42.9.4335-4337.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barac A., Kosmidis C., Alastruey-Izquierdo A., Salzer H.J.F., CPAnet Chronic pulmonary aspergillosis update: A year in review. Med. Mycol. 2019;57:S104–S109. doi: 10.1093/mmy/myy070. 10.1093/mmy/myy070. [DOI] [PubMed] [Google Scholar]
- Bastos R.W., Rossato L., Goldman G.H., Santos D.A. Fungicide effects on human fungal pathogens: Cross-resistance to medical drugs and beyond. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1010073. 10.1371/journal.ppat.1010073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batah S.S., Fabro A.T. Pulmonary pathology of ARDS in COVID-19: A pathological review for clinicians. Respir. Med. 2021;176 doi: 10.1016/j.rmed.2020.106239. 10.1016/j.rmed.2020.106239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhabhra R., Askew D.S. Thermotolerance and virulence of Aspergillus fumigatus: role of the fungal nucleolus. Med. Mycol. 2005;43(Suppl 1):S87–S93. doi: 10.1080/13693780400029486. 10.1080/13693780400029486. [DOI] [PubMed] [Google Scholar]
- Bitencourt T.A., Lang E.A.S., Sanches P.R., Peres N.T.A., Oliveira V.M., Fachin A.L., Rossi A., Martinez-Rossi N.M. HacA Governs Virulence Traits and Adaptive Stress Responses in Trichophyton rubrum. Front. Microbiol. 2020;11:193. doi: 10.3389/fmicb.2020.00193. 10.3389/fmicb.2020.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borman A.M., Palmer M.D., Fraser M., Patterson Z., Mann C., Oliver D., Linton C.J., Gough M., Brown P., Dzietczyk A., Hedley M., McLachlan S., King J., Johnson E.M. COVID-19-Associated Invasive Aspergillosis: Data from the UK National Mycology Reference Laboratory. J. Clin. Microbiol. 2020:59. doi: 10.1128/JCM.02136-20. 10.1128/JCM.02136-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I., Bulleid N.J. Protein folding and modification in the mammalian endoplasmic reticulum. Annu. Rev. Biochem. 2011;80:71–99. doi: 10.1146/annurev-biochem-062209-093836. [DOI] [PubMed] [Google Scholar]
- Cadena J., Thompson G.R., Patterson T.F. Aspergillosis: Epidemiology, Diagnosis, and Treatment. Infect. Dis. Clin. North Am. 2021;35:415–434. doi: 10.1016/j.idc.2021.03.008. [DOI] [PubMed] [Google Scholar]
- Casadevall A., Fu M.S., Guimaraes A.J., Albuquerque P. The “Amoeboid Predator-Fungal Animal Virulence” Hypothesis. J. fungi (Basel, Switzerland) 2019:5. doi: 10.3390/jof5010010. 10.3390/jof5010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaillot J., Tebbji F., Remmal A., Boone C., Brown G.W., Bellaoui M., Sellam A. The Monoterpene Carvacrol Generates Endoplasmic Reticulum Stress in the Pathogenic Fungus Candida albicans. Antimicrob. Agents Chemother. 2015;59:4584–4592. doi: 10.1128/AAC.00551-15. 10.1128/AAC.00551-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheon S.A., Jung K.-W., Chen Y.-L., Heitman J., Bahn Y.-S., Kang H.A. Unique evolution of the UPR pathway with a novel bZIP transcription factor, Hxl1, for controlling pathogenicity of Cryptococcus neoformans. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002177. 10.1371/journal.ppat.1002177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y., Megyeri M., Chen O.C.W., Condomitti G., Riezman I., Loizides-Mangold U., Abdul-Sada A., Rimon N., Riezman H., Platt F.M., Futerman A.H., Schuldiner M. The yeast p5 type ATPase, spf1, regulates manganese transport into the endoplasmic reticulum. PLoS One. 2013;8:e85519. doi: 10.1371/journal.pone.0085519. 10.1371/journal.pone.0085519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A., Punt P.J., van Luijk N., van den Hondel C.A. The secretion pathway in filamentous fungi: a biotechnological view. Fungal Genet. Biol. 2001;33:155–171. doi: 10.1006/fgbi.2001.1276. 10.1006/fgbi.2001.1276 [DOI] [PubMed] [Google Scholar]
- Cronin S.R., Rao R., Hampton R.Y. Cod1p/Spf1p is a P-type ATPase involved in ER function and Ca2+ homeostasis. J. Cell Biol. 2002;157:1017–1028. doi: 10.1083/jcb.200203052. 10.1083/jcb.200203052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos-Ruiz J.R., Bettigole S.E., Glimcher L.H. Tumorigenic and Immunosuppressive Effects of Endoplasmic Reticulum Stress in Cancer. Cell. 2017;168:692–706. doi: 10.1016/j.cell.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doultsinos D., Carlesso A., Chintha C., Paton J.C., Paton A.W., Samali A., Chevet E., Eriksson L.A. Peptidomimetic-based identification of FDA-approved compounds inhibiting IRE1 activity. FEBS J. 2021;288:945–960. doi: 10.1111/febs.15372. 10.1111/febs.15372. [DOI] [PubMed] [Google Scholar]
- Fedorova N.D., Badger J.H., Robson G.D., Wortman J.R., Nierman W.C. Comparative analysis of programmed cell death pathways in filamentous fungi. BMC Genomics. 2005;6:177. doi: 10.1186/1471-2164-6-177. 10.1186/1471-2164-6-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X., Krishnan K., Richie D.L., Aimanianda V., Hartl L., Grahl N., Powers-Fletcher M.V., Zhang M., Fuller K.K., Nierman W.C., Lu L.J., Latgé J.-P., Woollett L., Newman S.L., Cramer R.A., Rhodes J.C., Askew D.S. HacA-independent functions of the ER stress sensor IreA synergize with the canonical UPR to influence virulence traits in Aspergillus fumigatus. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002330. 10.1371/journal.ppat.1002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J., Lopez V., Kinch L., Pfeifer M.A., Gray H., Garcia N., Grishin N.V, Khang C.-H., Orth K. Role of Two Metacaspases in Development and Pathogenicity of the Rice Blast Fungus Magnaporthe oryzae. MBio. 2021;12:1–15. doi: 10.1128/mBio.03471-20. 10.1128/mBio.03471-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda S. Targeting extrinsic apoptosis in cancer: Challenges and opportunities. Semin. Cell Dev. Biol. 2015;39:20–25. doi: 10.1016/j.semcdb.2015.01.006. [DOI] [PubMed] [Google Scholar]
- Gonçalves A.P., Heller J., Daskalov A., Videira A., Glass N.L. Regulated Forms of Cell Death in Fungi. Front. Microbiol. 2017;8:1837. doi: 10.3389/fmicb.2017.01837. 10.3389/fmicb.2017.01837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirao-Abad J.P., Weichert M., Albee A., Deck K., Askew D.S. A Human IRE1 Inhibitor Blocks the Unfolded Protein Response in the Pathogenic Fungus Aspergillus fumigatus and Suggests Noncanonical Functions within the Pathway. mSphere. 2020:5. doi: 10.1128/mSphere.00879-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirao-Abad J.P., Weichert M., Luengo-Gil G., Sze Wah Wong S., Aimanianda V., Grisham C., Malev N., Reddy S., Woollett L., Askew D.S. Pleiotropic Effects of the P5-Type ATPase SpfA on Stress Response Networks Contribute to Virulence in the Pathogenic Mold Aspergillus fumigatus. MBio. 2021;12 doi: 10.1128/mBio.02735-21. 10.1128/mBio.02735-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann A., Brust D., Osiewacz H.D. Deletion of putative apoptosis factors leads to lifespan extension in the fungal ageing model Podospora anserina. Mol. Microbiol. 2007;65:948–958. doi: 10.1111/j.1365-2958.2007.05839.x. 10.1111/j.1365-2958.2007.05839.x. [DOI] [PubMed] [Google Scholar]
- Heimel K., Freitag J., Hampel M., Ast J., Bölker M., Kämper J. Crosstalk between the unfolded protein response and pathways that regulate pathogenic development in Ustilago maydis. Plant Cell. 2013;25:4262–4277. doi: 10.1105/tpc.113.115899. 10.1105/tpc.113.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C., Zhang K., Kaufman R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020;21:421–438. doi: 10.1038/s41580-020-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison E., Brown S., Tian C., Glass N.L. Transcriptional profiling and functional analysis of heterokaryon incompatibility in Neurospora crassa reveals that reactive oxygen species, but not metacaspases, are associated with programmed cell death. Microbiology. 2009;155:3957–3970. doi: 10.1099/mic.0.032284-0. 10.1099/mic.0.032284-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J., Qi L. Quality Control in the Endoplasmic Reticulum: Crosstalk between ERAD and UPR pathways. Trends Biochem. Sci. 2018;43:593–605. doi: 10.1016/j.tibs.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert A., Simoneau P., Campion C., Bataillé-Simoneau N., Iacomi-Vasilescu B., Poupard P., François J.M., Georgeault S., Sellier E., Guillemette T. Impact of the unfolded protein response on the pathogenicity of the necrotrophic fungus Alternaria brassicicola. Mol. Microbiol. 2011;79:1305–1324. doi: 10.1111/j.1365-2958.2010.07522.x. 10.1111/j.1365-2958.2010.07522.x. [DOI] [PubMed] [Google Scholar]
- Jung K.-W., Lee K.-T., Averette A.F., Hoy M.J., Everitt J., Heitman J., Bahn Y.-S. Evolutionarily Conserved and Divergent Roles of Unfolded Protein Response (UPR) in the Pathogenic Cryptococcus Species Complex. Sci. Rep. 2018;8:8132. doi: 10.1038/s41598-018-26405-5. 10.1038/s41598-018-26405-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbelik M., Zhao J., Zeng H., Bielawska A., Szulc Z.M. Mechanistic insights into ceramidase inhibitor LCL521-enhanced tumor cell killing by photodynamic and thermal ablation therapies. Photochem. Photobiol. Sci. 2020;19:1145–1151. doi: 10.1039/d0pp00116c. [DOI] [PubMed] [Google Scholar]
- Krishnan K., Feng X., Powers-Fletcher M.V, Bick G., Richie D.L., Woollett L.A., Askew D.S. Effects of a defective endoplasmic reticulum-associated degradation pathway on the stress response, virulence, and antifungal drug susceptibility of the mold pathogen Aspergillus fumigatus. Eukaryot. Cell. 2013;12:512–519. doi: 10.1128/EC.00319-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Reste P.J., Pineau R., Voutetakis K., Samal J., Jégou G., Lhomond S., Gorman A.M., Samali A., Patterson J.B., Zeng Q., Pandit A., Aubry M., Soriano N., Etcheverry A., Chatziioannou A., Mosser J., Avril T., Chevet E. Local intracerebral inhibition of IRE1 by MKC8866 sensitizes glioblastoma to irradiation/chemotherapy in vivo. Cancer Lett. 2020;494:73–83. doi: 10.1016/j.canlet.2020.08.028. 10.1016/j.canlet.2020.08.028. [DOI] [PubMed] [Google Scholar]
- Lee R.E.C., Brunette S., Puente L.G., Megeney L.A. Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proc. Natl. Acad. Sci. U. S. A. 2010;107:13348–13353. doi: 10.1073/pnas.1006610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue S.E., McGrath E.P., Cleary P., Greene S., Mnich K., Almanza A., Chevet E., Dwyer R.M., Oommen A., Legembre P., Godey F., Madden E.C., Leuzzi B., Obacz J., Zeng Q., Patterson J.B., Jäger R., Gorman A.M., Samali A. Inhibition of IRE1 RNase activity modulates the tumor cell secretome and enhances response to chemotherapy. Nat. Commun. 2018;9:3267. doi: 10.1038/s41467-018-05763-8. 10.1038/s41467-018-05763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maly D.J., Papa F.R. Druggable sensors of the unfolded protein response. Nat. Chem. Biol. 2014;10:892–901. doi: 10.1038/nchembio.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manasanch E.E., Orlowski R.Z. Proteasome inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 2017;14:417–433. doi: 10.1038/nrclinonc.2016.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr K.A., Platt A., Tornheim J.A., Zhang S.X., Datta K., Cardozo C., Garcia-Vidal C. Aspergillosis Complicating Severe Coronavirus Disease. Emerg. Infect. Dis. 2021;27:18–25. doi: 10.3201/eid2701.202896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy N., Dolgikh N., Logue S., Patterson J.B., Zeng Q., Gorman A.M., Samali A., Fulda S. The IRE1 and PERK arms of the unfolded protein response promote survival of rhabdomyosarcoma cells. Cancer Lett. 2020;490:76–88. doi: 10.1016/j.canlet.2020.07.009. 10.1016/j.canlet.2020.07.009. [DOI] [PubMed] [Google Scholar]
- McKenna M.J., Sim S.I., Ordureau A., Wei L., Harper J.W., Shao S., Park E. The endoplasmic reticulum P5A-ATPase is a transmembrane helix dislocase. Science. 2020;369 doi: 10.1126/science.abc5809. eabc5809. 10.1126/science.abc5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T., Nakayama H., Nagayoshi Y., Kakeya H., Kohno S. Dissection of Ire1 functions reveals stress response mechanisms uniquely evolved in Candida glabrata. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003160. 10.1371/journal.ppat.1003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K. Signalling pathways in the unfolded protein response: development from yeast to mammals. J. Biochem. 2009;146:743–750. doi: 10.1093/jb/mvp166. 10.1093/jb/mvp166. [DOI] [PubMed] [Google Scholar]
- Nywening A.V., Rybak J.M., Rogers P.D., Fortwendel J.R. Mechanisms of triazole resistance in Aspergillus fumigatus. Environ. Microbiol. 2020;22:4934–4952. doi: 10.1111/1462-2920.15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes S.A. Endoplasmic Reticulum Stress Signaling in Cancer Cells. Am. J. Pathol. 2020;190:934–946. doi: 10.1016/j.ajpath.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.E., Miller Z., Jun Y., Lee W., Kim K.B. Next-generation proteasome inhibitors for cancer therapy. Transl. Res. 2018;198:1–16. doi: 10.1016/j.trsl.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter N., Hach C.A., Hampel M., Rekhter D., Zienkiewicz K., Feussner I., Poehlein A., Daniel R., Finkernagel F., Heimel K. Signal peptide peptidase activity connects the unfolded protein response to plant defense suppression by Ustilago maydis. PLoS Pathog. 2019;15 doi: 10.1371/journal.ppat.1007734. 10.1371/journal.ppat.1007734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy K.E., Mazer M., Striker D.A., Ellebedy A.H., Walton A.H., Unsinger J., Blood T.M., Mudd P.A., Yi D.J., Mannion D.A., Osborne D.F., Martin R.S., Anand N.J., Bosanquet J.P., Blood J., Drewry A.M., Caldwell C.C., Turnbull I.R., Brakenridge S.C., Moldwawer L.L., Hotchkiss R.S. Severe immunosuppression and not a cytokine storm characterizes COVID-19 infections. JCI insight. 2020:5. doi: 10.1172/jci.insight.140329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richie D.L., Feng X., Hartl L., Aimanianda V., Krishnan K., Powers-Fletcher M.V., Watson D.S., Galande A.K., White S.M., Willett T., Latgé J.P., Rhodes J.C., Askew D.S. The virulence of the opportunistic fungal pathogen Aspergillus fumigatus requires cooperation between the endoplasmic reticulum-associated degradation pathway (ERAD) and the unfolded protein response (UPR) Virulence. 2011;2:12–21. doi: 10.4161/viru.2.1.13345. 10.4161/viru.2.1.13345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richie D.L., Hartl L., Aimanianda V., Winters M.S., Fuller K.K., Miley M.D., White S., McCarthy J.W., Latgé J.-P., Feldmesser M., Rhodes J.C., Askew D.S. A role for the unfolded protein response (UPR) in virulence and antifungal susceptibility in Aspergillus fumigatus. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000258. 10.1371/journal.ppat.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richie D.L., Miley M.D., Bhabhra R., Robson G.D., Rhodes J.C., Askew D.S. The Aspergillus fumigatus metacaspases CasA and CasB facilitate growth under conditions of endoplasmic reticulum stress. Mol. Microbiol. 2007;63:591–604. doi: 10.1111/j.1365-2958.2006.05534.x. 10.1111/j.1365-2958.2006.05534.x. [DOI] [PubMed] [Google Scholar]
- Shao A., Xu Q., Spalek W.T., Cain C.F., Kang C.W., Tang C.-H.A., Del Valle J.R., Hu C.-C.A. Development of Tumor-Targeting IRE-1 Inhibitors for B-cell Cancer Therapy. Mol. Cancer Ther. 2020;19:2432–2444. doi: 10.1158/1535-7163.MCT-20-0127. 10.1158/1535-7163.MCT-20-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlezinger N., Irmer H., Dhingra S., Beattie S.R., Cramer R.A., Braus G.H., Sharon A., Hohl T.M. Sterilizing immunity in the lung relies on targeting fungal apoptosis-like programmed cell death. Science. 2017;357:1037–1041. doi: 10.1126/science.aan0365. 10.1126/science.aan0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha A., Brunette S., Stanford W.L., Megeney L.A. The metacaspase Yca1 maintains proteostasis through multiple interactions with the ubiquitin system. Cell Discov. 2019;5:6. doi: 10.1038/s41421-018-0071-9. 10.1038/s41421-018-0071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sircaik S., Román E., Bapat P., Lee K.K., Andes D.R., Gow N.A.R., Nobile C.J., Pla J., Panwar S.L. The protein kinase Ire1 impacts pathogenicity of Candida albicans by regulating homeostatic adaptation to endoplasmic reticulum stress. Cell. Microbiol. 2021;23:e13307. doi: 10.1111/cmi.13307. 10.1111/cmi.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen D.M., Holen H.W., Pedersen J.T., Martens H.J., Silvestro D., Stanchev L.D., Costa S.R., Günther Pomorski T., López-Marqués R.L., Palmgren M. The P5A ATPase Spf1p is stimulated by phosphatidylinositol 4-phosphate and influences cellular sterol homeostasis. Mol. Biol. Cell. 2019;30:1069–1084. doi: 10.1091/mbc.E18-06-0365. 10.1091/mbc.E18-06-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starke J., Harting R., Maurus I., Leonard M., Bremenkamp R., Heimel K., Kronstad J.W., Braus G.H. Unfolded Protein Response and Scaffold Independent Pheromone MAP Kinase Signaling Control Verticillium dahliae Growth, Development, and Plant Pathogenesis. J. fungi (Basel, Switzerland) 2021:7. doi: 10.3390/jof7040305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Lin D.-C., Guo X., Kharabi Masouleh B., Gery S., Cao Q., Alkan S., Ikezoe T., Akiba C., Paquette R., Chien W., Müller-Tidow C., Jing Y., Agelopoulos K., Müschen M., Koeffler H.P. Inhibition of IRE1α-driven pro-survival pathways is a promising therapeutic application in acute myeloid leukemia. Oncotarget. 2016;7:18736–18749. doi: 10.18632/oncotarget.7702. 10.18632/oncotarget.7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers K.J., Patil C.K., Wodicka L., Lockhart D.J., Weissman J.S., Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Tsiatsiani L., Van Breusegem F., Gallois P., Zavialov A., Lam E., Bozhkov P.V. Metacaspases. Cell Death Differ. 2011;18:1279–1288. doi: 10.1038/cdd.2011.66. 10.1038/cdd.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivek-Ananth R.P., Mohanraj K., Vandanashree M., Jhingran A., Craig J.P., Samal A. Comparative systems analysis of the secretome of the opportunistic pathogen Aspergillus fumigatus and other Aspergillus species. Sci. Rep. 2018;8:6617. doi: 10.1038/s41598-018-25016-4. 10.1038/s41598-018-25016-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Jiang S., Yang Y., Fan L., Su F., Ye M. Synthesis and antifungal activity of carvacrol and thymol esters with heteroaromatic carboxylic acids. Nat. Prod. Res. 2019;33:1924–1930. doi: 10.1080/14786419.2018.1480618. 10.1080/14786419.2018.1480618. [DOI] [PubMed] [Google Scholar]
- Wang Q., Zhong C., Xiao H. Genetic Engineering of Filamentous Fungi for Efficient Protein Expression and Secretion. Front. Bioeng. Biotechnol. 2020;8:293. doi: 10.3389/fbioe.2020.00293. 10.3389/fbioe.2020.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert M., Guirao-Abad J.P., Aimanianda V., Krishnan K., Grisham C., Snyder P., Sheehan A., Abbu R.R., Liu H., Filler S.G., Gruenstein E.I., Latgé J.-P., Askew D.S. Functional Coupling between the Unfolded Protein Response and Endoplasmic Reticulum/Golgi Ca2+-ATPases Promotes Stress Tolerance, Cell Wall Biosynthesis, and Virulence of Aspergillus fumigatus. MBio. 2020;11 doi: 10.1128/mBio.01060-20. e01060-20. 10.1128/mBio.01060-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N., Cao J., Xu L., Tang Q., Dobrolecki L.E., Lv X., Talukdar M., Lu Y., Wang X., Hu D.Z., Shi Q., Xiang Y., Wang Y., Liu X., Bu W., Jiang Y., Li M., Gong Y., Sun Z., Ying H., Yuan B., Lin X., Feng X.-H., Hartig S.M., Li F., Shen H., Chen Y., Han L., Zeng Q., Patterson J.B., Kaipparettu B.A., Putluri N., Sicheri F., Rosen J.M., Lewis M.T., Chen X. Pharmacological targeting of MYC-regulated IRE1/XBP1 pathway suppresses MYC-driven breast cancer. J. Clin. Invest. 2018;128:1283–1299. doi: 10.1172/JCI95873. [DOI] [PMC free article] [PubMed] [Google Scholar]