Highlights
-
•
The inaccessible extreme environments harbor a large majority of anaerobic microbes which remain unknown.
-
•
Anaerobic microbes are used in a variety of industrial applications.
-
•
In the future, metagenomic-assisted techniques can be used to identify novel anaerobic microbes from the unexplored extreme environments.
-
•
Genetic engineering can be used to enhance the efficiency of anaerobic microbes for various processes.
Keywords: Anaerobe, Anaerobic, Extreme environments, Metagenomics, Microbes
Abstract
The genome of an organism is directly or indirectly correlated with its environment. Consequently, different microbes have evolved to survive and sustain themselves in a variety of environments, including unusual anaerobic environments. It is believed that their genetic material could have played an important role in the early evolution of their existence in the past. Presently, out of the uncountable number of microbes found in different ecosystems we have been able to discover only one percent of the total communities. A large majority of the microbial populations exists in the most unusual and extreme environments. For instance, many anaerobic bacteria are found in the gastrointestinal tract of humans, soil, and hydrothermal vents. The recent advancements in Metagenomics and Next Generation Sequencing technologies have improved the understanding of their roles in these environments. Presently, anaerobic bacteria are used in various industries associated with biofuels, fermentation, production of enzymes, vaccines, vitamins, and dairy products. This broad applicability brings focus to the significant contribution of their genomes in these functions. Although the anaerobic microbes have become an irreplaceable component of our lives, a major and important section of such anaerobic microbes still remain unexplored. Therefore, it can be said that unlocking the role of the microbial genomes of the anaerobes can be a noteworthy discovery not just for mankind but for the entire biosystem as well.
Graphical abstract
1. Introduction
Microbes - the small tiny forms of life, have become an indispensable part of our lives, however, their omnipresent existence and their contribution to the environment are underappreciated. Microbes are classified into two broad categories, i.e., aerobic and anaerobic, based on their oxygen requirement for growth and metabolism (Singh et al., 2017). There is a third category of microbes that are “facultative” in nature that lies between aerobic and anaerobic microorganisms. An obligate aerobe is defined as a microorganism that requires oxygen for its growth and survival (Scheld, 2012). On the other hand, obligate anaerobes are microorganisms that do not require oxygen for growth, and the presence of oxygen can be toxic to them (Lu and Imlay, 2021). Facultative microorganisms can grow in the presence or absence of oxygen and are further classified as facultative aerobes or facultative anaerobes, respectively (André et al., 2021).
A wide range of unicellular and multicellular microorganisms, including several bacteria, fungi, and protozoa, fall under the category of facultative or obligate anaerobes (Pitt and Barer, 2012). These anaerobic microorganisms can survive in the absence of oxygen with the help of the special enzymes encoded by their genes (Lu and Imlay, 2021). Furthermore, owing to their genomes and the very nature of these microbes to survive in oxygen-less environments, these anaerobic microbes are of immense value to a variety of commercial and non-commercial sectors (Nguyen et al., 2019; Andrade et al., 2020). The anaerobic microbes have great versatility in terms of the metabolic products they can synthesize. Therefore, understanding the genetic makeup and its roles is very useful as this can help us discover the possible applications of these anaerobic microorganisms.
In this review, we will be discussing the applications of the microbial genomes in the anaerobic environments. Also, the review presents detailed information on what possible roles the microbial genomes could have played in the past and the present scenarios. Lastly, the future scope of these microbial genomes in the anaerobic conditions is discussed.
2. The past
During the early evolution of life on earth, the conditions were not very stable and it is believed that the environment back then lacked oxygen. This could have possibly supported the growth of anaerobic heterotrophs. The Last Common Ancestor (Luca) is believed to be an anaerobic prokaryote (Sousa et al., 2016). Later, the first aerobic bacteria evolved around 2.4 billion years ago as the conditions became favourable for their growth. Methanogens and Clostridia are the most ancient lineages not just amongst anaerobes but also prokaryotes in general (Decker and Jungermann, 1970).
The first evidence of anaerobic microbial life was proposed by Antonie van Leeuwenhoek, who observed that some "animalcules" were capable of living in the absence of oxygen (Finegold, 1993). Louis Pasteur discovered the first pathogenic anaerobic bacterium, the septic vibrio (later termed Clostridium septicum) (Sebald and Finegold, 1995). In 1863, Pasteur coined the terms aerobes and anaerobes based on their requirement of oxygen for growth. In 1877, for the first time, Louis Pasteur and Jules Francois Joubert successfully cultured an anaerobic human pathogen, Clostridium septicum (earlier known as Vibrion septique) (Sebald and Hauser, 1995).
Since the culturing of anaerobes is difficult, there was not much information available on them and most of the time they were poorly or even wrongly classified. Several attempts were made to culture them, but most of them suffered from significant drawbacks. However, in 1916, a milestone discovery was seen when McIntosh and Fildes introduced the anaerobic jars to isolate and cultivate anaerobic microbes (Mcintosh et al., 1916). In 1977, Carl Woese and George Fox used the 16S rRNA gene as a genetic marker for taxonomic classification and phylogenetic analyses (Woese and Fox, 1977). After almost two decades, Craig Venter and his colleagues published the first complete genome of the free-living bacteria, Haemophilus influenzae, a facultative anaerobe (Fleischmann et al., 1995). Later in 1988, the 18S rRNA gene was used for the first time as a genetic marker to study the evolutionary relationships among Metazoa, a group of multicellular eukaryotic animals (Field et al., 1988). Fungi are also usually studied using the 18S rRNA gene marker. Another important genetic marker that is commonly used for fungi is Internal Transcribed Spacer (ITS) sequences. An ITS is a non-coding DNA region situated between the rRNA genes and these are useful universal genetic markers for taxonomic classification and phylogenetic analysis of fungi.
Schoch and colleagues had successfully evaluated six different ITS regions as DNA barcode markers for phylogenetic analysis and classification of fungi at different taxonomic levels (Schoch et al., 2012). To assess the barcoding performance, Schoch et al. carried out a comparative analysis of four different DNA biomarkers for fungi, namely ITS, LSU (28S rRNA gene), SSU (18S rRNA gene), and RPB1 (RNA polymerase II subunit) across 742 strains or specimens and two additional protein-coding markers (MCM7 and RPB) across a subset of about 200 fungi. DNA was isolated from cultures, purified, and subjected to PCR amplification and sequencing. PCR experiments showed that ribosomal RNA genes are more reliable compared to the protein-coding genes. However, the PCR success rate varied across taxonomic group. Nonetheless, maximum success was observed for ITS, that is, 65% to 100% depending on the taxonomic group. This study implies that ITS is generally superior to LSU in species discrimination, have a more clearly defined barcode gap, and exhibit good overall probability of correct species identification. ITS combines the highest resolving power for discriminating closely related species with a high PCR and sequencing success rate across a broad range of Fungi. Furthermore, based on the data, the authors also suggest the use of two-maker (ITS and LSU) based system for the taxonomic classification of fungi (Schoch et al., 2012).
Albeit all the challenges faced in handling the anaerobic microorganisms, they have been extensively used in the past for several important commercial applications, which are discussed in the sections below.
2.1. Anaerobes in the fermentation and brewing processes
Alcoholic fermentation under anaerobic conditions has been one of the oldest applications of anaerobes. However, the involvement of microbes remained unknown, partly due to the lack of suitable microscopic facilities (Alba-Lois and Segal-Kischinevzky, 2010). Only after the development of a microscope, the role of Saccharomyces cerevisiae could be sufficiently understood in this process (Barnett, 1998). Moreover, the natural fermentation is a slow and time-consuming process and a lack of suitable facilities hindered the large-scale commercialization using microbes.
2.2. Anaerobes in the baking industry
The earliest records of the role of Saccharomyces cerevisiae in the baking process come from ancient Egypt and China (Samuel, 1996). The commercialization of yeast began around the 1700s, however, it did not involve any modern pure culture methods (Frey, 1931). As a result, the identification of individual yeast strains was difficult. Further, failure to maintain hygiene, suitable culturing conditions, and lack of understanding of the genetics adversely affected the commercialization. Nonetheless, the baking industry continued to evolve alongwith the expansion of modern microbiology (Linko et al., 1997).
2.3. Anaerobes in the dairy industry
Anaerobes have been used in the dairy industries for years as markers to check the quality of milk (Martin et al., 2016). For example, spore-forming Bacillus spp. spores have been used in the past to indicate the presence of contamination in milk (Doyle et al., 2015). In addition, Clostridia spp. have also been found as pathogenic contaminants in milk (Doyle et al., 2015).
3. The present
Presently, innumerable anaerobic microbes, including fungi, bacteria, and protozoan, are widely used in various industrial and non-industrial applications. Their wide applicability comes from the fact that their genomes can code for proteins that are not only useful to the microbe itself but are involved in the production of a variety of other products, which are of industrial relevance. Despite their wide applicability, we have not yet been able to make efficient use of their presence. One of the major challenges towards this is the uncultivable nature of some species, specifically bacteria (Steen et al., 2019). Many Microbiologists believe that around ninety-nine percent of the microbes are unculturable (Locey and Lennon, 2016). Furthermore, the anaerobic microorganisms are even more difficult to culture, given the unusual environmental condition they require for their growth.
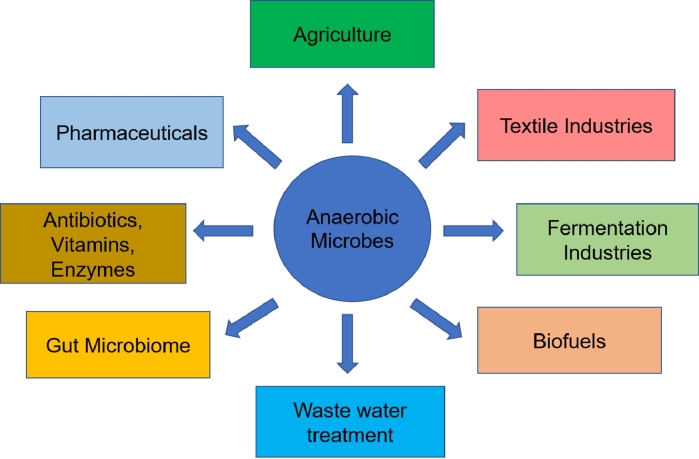
The taxonomic exploration of natural environments has been performed since quite long ago with the help of the traditional 16S rRNA based genetic marker (Johnson et al., 2019). However, significant advancements in this direction have been possible in the recent past with the emergence of the field of metagenomics. The major applications of “Metagenomics” in this direction have been benefited by the advancements and developments of the next generation sequencing (NGS) techniques (Metzker, 2010). These developments together have made it possible to identify and classify the microorganisms from the extreme anaerobic habitats by recovering their complete genomic materials for further exploration from their natural environmental habitats. Hence, metagenomics has brought a paradigm shift in modern microbiology, especially in the microbiology of uncultivable anaerobes. Nevertheless, at least one percent of the entire anaerobic microorganisms are known and are extensively used at present for various applications. They have wide applicability and are presently used in various fields, some of which have been listed below. Fig. 1 provides a comprehensive overview of the different commercial and non-commercial applications of the anaerobes.
Fig. 1.
Applications of anaerobic microbes in various industrial and non-industrial fields.
3.1. Agriculture
Soil naturally comprises a large number of microbial communities belonging to a variety of taxonomic groups. Anaerobes are helpful in the process of biological soil disinfestation (BSD) (Ueki et al., 2018). It is a process that involves the suppression of plant pathogens, soil pathogens, parasites, and plant weeds by stimulating the microbial activity of the indigenous microbiome of the soil using organic materials (Rokunuzzaman et al., 2016). It is an efficient, environment-friendly, and cost-effective process as compared to the other chemical methods (Wen et al., 2016; Ueki et al., 2018). Several anaerobes carry out this process via the production and release of organic acids such as acetate and butyrate (Zhou et al., 2021) and also metal ions, such as Fe+2 and Mn+2 (Momma et al., 2011). Moreover, it has been reported that the Clostridium spp. produce antifungal enzymes to eliminate soil-borne fungal pathogens (Ueki et al., 2018). These microbes are also known to synthesize enzyme Chitosanases, which degrade chitosan or chitin, present in the cell wall of pathogenic fungi such as Fusarium (Mahawar et al., 2019).
3.2. Fermentation industry
With the growing demand for fermented products, the use of anaerobic microbes in this industry has increased exponentially. For instance, various benefits of probiotics, increased consumption of fibers, and an increased health-consciousness amongst people has led to a significant boost in probiotics consumption (Sanders et al., 2019). Likewise, an increasing energy demand, a decline in petrol and other fossil fuels, and environmental concerns have led to an increased demand for eco-friendly compounds. Ethanol, for example, is the most commonly produced fermentation product currently (Tse et al., 2021). And, it is rapidly emerging as an efficient, environment-friendly alternative. It is produced by the fermentation of different agro-based biomasses. It has been studied that Saccharomyces cerevisiae is capable of utilizing starch-rich microalgae such as Chlorella, Chlamydomonas, Spirulina, and others, in the anaerobic fermentation process of Bio-ethanol production (Tsolcha et al., 2021).
The process of Bio-ethanol production requires enzymes such as cellulase. This cellulose enzyme can be obtained from Clostridium, Cellulomonas, Bacteroides, Ruminococcus, and Erwinia (Gupta and Verma, 2015). The production of cellulases is not only limited to the bacterial kingdom, but it is also produced by anaerobic fungi, such as Piromyces and Neocallimastix (Lee et al., 2015). In addition, the anaerobic microorganisms produce other fermentation products. For example, vitamin B2 (riboflavin) (Zhao et al., 2016), acetone (Tondro et al., 2020), and butanol (Al-Shorgani et al., 2019) are produced using Clostridium acetobutylicum. Likewise, ethanol is produced using Saccharomyces cerevisiae (Ahmad et al., 2021).
3.3. Textile industry
With the rapid increase in the demand for synthetic colorants and dyes in the textile industries (Pavithra et al., 2019), the environment is facing a wrath of harmful waste generated by these toxic dyes. The waste released from the textile industries can pollute water bodies (Bhatia and Devraj, 2017). Therefore, it is important to degrade and remove these toxic compounds from the environment. The most commonly used dyes are the azo dyes (Gičević et al., 2020), which have a complex molecular structure that makes it challenging to degrade them. As a result, these dyes remain in the environment as recalcitrant for a longer time (Sarkar et al., 2017).
However, anaerobic microbes can be used to overcome this problem. Anaerobes produce different types of enzymes that have efficient degradation capacity (Ajaz et al., 2020). Hence, they are used for the biodegradation of azo dyes that are present in the industrial waste. Anaerobic bacteria belonging to the Proteobacteria phylum are prominent microbes involved in dye biodegradation. Besides, some of the anaerobic sulfate-reducing bacteria are also responsible for this process (Dai et al., 2020). In addition, a few anaerobes found in the human gut are capable of dye degradation, including Eubacterium hadrum, Clostridium paraputrificum, Eubacterium sp., Clostridium clostridiiforme, Bacteroides sp., Clostridium nexile, and Butyrivibrio sp. (Chung, 2016). These anaerobes produce enzymes, including oxidases and peroxidases, to degrade the azo dyes. Furthermore, Saccharomyces cerevisiae, a facultative anaerobic yeast, is known to produce an enzyme known as “ferric reductase”, which can reduce azo dyes (Chen, 2006). In addition, other anaerobic fungi, for example Penicillium spp., can degrade a wide range of polycyclic aromatic hydrocarbons, xenobiotics, dyes, and phenol derivatives under anaerobic production conditions (Leitão, 2009).
3.4. Sewage and wastewater treatment
Anaerobic microorganisms play a significant role in the wastewater treatment process. Even though aerobic microbes predominate waste and sewage water, some anaerobic microbes are also present in these environments (Cyprowski et al., 2018). The anaerobic bacteria, which are responsible for methane fermentation in sewage include Methanosarcina (Hardegen et al., 2018), Methanosaeta (Vítěz et al., 2020), Clostridium, and Bifidobacterium (Cyprowski et al., 2018). Moreover, it has been found that Clostridium is predominantly found in the sludge and can be used as a microbial indicator of water pollution (Saxena et al., 2015; Li et al., 2021). The presence of anaerobes improves the efficiency of wastewater treatment in many ways. These primarily include low energy input of the system as no energy is required for oxygenation, lower production of excess sludge, lower nutrient requirement due to lower biological synthesis, and production of biogas, a valuable energy source, from the degradation of waste organic material (Muralikrishna and Manickam, 2017).
3.5. Enzyme source
Anaerobic microbes are found in extreme environments. They have evolved with the capacity to efficiently utilize a variety of compounds, including the recalcitrant (Hatti-Kaul and Mattiasson, 2016). They produce highly stable enzymes, which can be used for the hydrolysis of polysaccharides, lipids, biopolymers, and others (Blair et al., 2021). Interestingly, the cellulolytic Clostridium thermocellum has been reported to contain a multienzyme complex termed as Cellulosome, which is more efficient than a free enzyme as it comprises multiple enzymes (including hemicellulases) capable of acting on several different substrates (Barth et al., 2018).
These enzymes are also useful in food industries. For example, cellulase is used in the juice industry in combination with other enzymes to extract and clarify juices (Santana et al., 2021). The highly thermostable enzyme “α-amylase” is used for starch saccharification, brewing, and baking and is obtained from Bacillus amyloliquefaciens (Devaraj et al., 2019), Bacillus licheniformis (Rakaz et al., 2021), or Bacillus stearothermophilus (Ravindran et al., 2019).
Catalase enzyme is used to remove excess Hydrogen Peroxide from fabric and food processing industries (Raveendran et al., 2018). Although catalase enzyme is mainly found in aerobic organisms, few anaerobic bacteria have also been reported to exhibit catalase activity. For example, the bacteria Bacteroides fragilis shows increased catalase levels in the presence of haem in the culture media (Paunkov et al., 2021). Bacillus maroccanus, a facultative anaerobe, can produce catalase enzyme (Kauldhar and Sooch, 2016).
3.6. Gut microbiome
Many anaerobic microbes have been found to coexist in the gut or digestive tract of herbivores such as cattle (Alipour et al., 2018). It has been reported that anaerobes are present in the gut of herbivores in a symbiotic relationship (Moraïs and Mizrahi, 2019). Anaerobic fungi belonging to the Neocallimastigomycota phylum play an important role in the digestion of fiber in the host gut (Hartinger and Zebeli, 2021). Anaerobic fungi produce cellulolytic enzymes for fiber digestion and exhibit better tissue penetration capacity than bacteria (Hess et al., 2020). Further, these anaerobic fungi can coexist with the other aerobic and anaerobic bacteria to carry out the process of fiber digestion more efficiently (Azad et al., 2020). A recent review has highlighted the role of anaerobic fungi in the gut of herbivores (Hartinger and Zebeli, 2021).
3.7. Probiotics
Anaerobic microorganisms are under investigation for their role as probiotics. However, their applicability as probiotics has not been explored well enough. Nevertheless, some anaerobes are being used as probiotics, such as Lactobacillus spp. and Bifidobacterium spp. (Masoumi et al., 2021). These are associated with various benefits such as immunological activity, anti-cancer activity, and enhancement of gastro-intestinal digestion (Silva et al., 2020).
Recent advancements in molecular and genomic technologies have played a significant role in building our knowledge of the genomic construct of the anaerobes. More valuable information is on its way as we continue unlocking the role of microbial genomes of these anaerobes and a plethora of anaerobic microorganisms is awaiting discovery. The genomes of these microorganisms comprise of genes that encode enzymes which are useful in various sectors such as biodegradation, dairy industry, textile industry, food industry, etc. Many microbial genes and the metabolic pathways associated with their gene products are of immense value to several industries. Table 1 shows a list of some potential industrially important anaerobic microorganisms and their genes along with the products produced by them.
Table 1.
List of potential anaerobic microorganisms and their genes involved in the industrial applications.
| References | (Ravi et al., 2021; Qi et al., 2011; Gruninger et al., 2016) | (Luo et al., 2014) | (Luo et al., 2014) | (Invernici et al., 2018) | (Luo et al., 2014) | (Luo et al., 2014) | (Luo et al., 2014) | (Pidot et al., 2014) | (Dunbar et al., 2018; Li et al., 2019) | (Lueders and von Netzer, 2014) | (Mamo, 2016; Trmčić et al., 2011) | (Town et al., 2014) | (Rabah et al., 2018; Falentin et al., 2010) | (Wang et al., 2020; Piwowarek et al., 2018) | (Luo et al., 2014) | (Ahmad et al., 2021; Walker and Stewart, 2016; Chatsurachai et al., 2020) | (Rischer et al., 2018) | (Franchi et al., 2018) | (Lueders and von Netzer, 2014) | (Ma'As et al., 2020) |
| Potential industrial application | Lignocellulose biomass degradation | Benzoate degradation | Benzoate degradation | Probiotics | Benzene degradation | Benzene degradation | Benzene degradation | Antibiotic | Antibiotic | Hydrocarbon degradation | Antimicrobial peptide | Anaerobic wastewater treatment | Swiss Cheese | Vitamin B12 | Benzoate degradation | Bioethanol, alcoholic beverages | Pharmacological compound | Anaerobic digestion of Phenolics in sludge | Nitrate reduction during hydrocarbon degradation | Biofuel |
| Product/Metabolic Pathway | Esterase | Benzoate CoA ligase | Benzoyl-CoA reductase | – | Benzene | Putative iron-sulfur binding protein | Putative anaerobic benzene carboxylases | Clostrubin A pigment | Closthioamide | Benzylsuccinate synthase | Nisin | – | Carbon dioxide, Propionate/Glycolysis, Wood-Werkman cycle | Tetrapyrrolic derivatives synthesis pathway | 6-Hydroxycyclohex-1-ene-1-carboxyl-CoA dehydrogenase | Glycolysis, anaerobic fermentation (pyruvate to ethanol) pathway | Barnesin A | Hydrolase/Anaerobic degradation pathway (4-hydroxybenzoate to benzoyl-CoA pathway) | Benzylsuccinate synthase (fumarate-adding enzymes) | Bioethanol/Entner-Doudoroff pathway, PPP pathway |
| Gene | AmCE1/Fae1A | bzdA | bzdQ | – | bamD | bamE | abcA, abcD | PKS gene cluster | cta | bssA | nisABTCIPRKFEG operon | – | lac, gal, tnp, murQ, mutA, mutB, cat, and others | hemB, hemC, hemD, cbi, cob | had | ADH, HXT, TPI1, SUC2 | brn | bamA | bssA | xylA/B operon |
| Anaerobic microbe | Anaeromyces mucronatus | Azoarcus | Azoarcus | Bifidobacterium spp. | Clostridiales | Clostridiales | Clostridiales | Clostridium beijerinckii | Clostridium cellulolyticum | Georgfuchsia toluolica | Lactococcus lactis | Methanosarcina barkeri | Propionibacterium freudenreichii | Propionibacterium freudenreichii | Proteobacteria | Saccharomyces cerevisiae | Sulfurospirillum barnesii | Syntrophorhabdus | Thauera aromatica | Zymomonas mobilis |
4. The future
Given the important roles that have been known for various anaerobic microbes, the future holds a variety of opportunities for their wider applications. The major bottlenecks in the wider applications of anaerobic microbes include their extreme habitats, which are still unexplored, their unculturable nature, and the lack of knowledge of their genomic and metabolic capabilities. However, the advancements in sequencing techniques, metagenomics approaches, and functional metagenomics screening-based methods have opened new avenues for exploring the wide applications of microbes residing in the extreme anaerobic environments. The use of metagenomic approaches to decipher the taxonomic diversity of the yet unexplored environments will lead to the identification of novel anaerobic microbial strains with previously unknown metabolic capabilities (Alalawy et al., 2021; Fischer et al., 2021). This can be followed with the application of the functional metagenomic screening based strategies to explore the novel metabolic capabilities of anaerobic microbes living in an environment (Macdonald et al., 2019). Additionally, the efficiency of the potential microbes can be improved further by modifying their genomes by following the genetic engineering principles, for various anaerobic applications (Croux et al., 2016).
In this direction, the gut microbiome of patients (Armour et al., 2019), Himalayan region (Sahay et al., 2017), Spacecraft clean-room facilities (Wood et al., 2021), and others can provide valuable sources of novel anaerobes. These microbial genomes are valuable targets for several new industrial and non-industrial applications. For example, the gut of herbivores is an important source of microbial diversity. Anaerobic fungi, which are found in the gut of ruminant host help in digestion of plant material including fibres. Despite the unknown mechanism, boosting the indigenous anaerobic fungi population is of great interest. The underlying mechanism through which the indigenous anaerobic fungi improve the digestibility can be identified by understanding the role of their genomes. With the decline in natural resources, such as farmable land area, it will be essential to improve the utilization of low-quality animal feed. Hence, exploring the enzyme repertoire by decoding their genomes will immensely contribute to the improved utilization of animal feed as well as animal derived products (Hess et al., 2020). In the subsequent sections, we have discussed the role of metagenomics and metagenomics-assisted techniques to explore the anaerobic diversity in the extreme environments.
4.1. Metagenomics - a field of future
Metagenomics has opened several avenues for microbial analysis of the most unexplored and otherwise inaccessible sites, including the hydrothermal vents, hot springs, gut, and others, which are major habitats for anaerobic microbes. One of the most unexplored sites in India includes the Himalayan region and only limited information about the microbial diversity of this region is available currently. The extremely ‘cold’ regions and some extremely high temperature ‘hot-spring’ regions (Manikaran, Ringigad, and Soldhar) (Narsing Rao et al., 2018) make it a highly diversified and valuable site for comprehensive metagenomic studies (Sahay et al., 2017). The anaerobic thermophilic microbes isolated from such sites can produce highly thermostable biocatalysts, which can be used in commercial industries.
Another clinically important target for metagenomic studies to analyze the novel gene pools includes the anaerobic human gut environment to perform studies related to gut associated diseases and their potential treatment methods (Mobeen et al., 2020). Overcoming the challenges in the field of “Probiotics” can be another important area of research in this direction. Highly specific strains for personalized treatment, identification of the underlying mechanism used by them, identification of other health benefits, and modification of the gut microbiome complexity are some of the important prospects that could be investigated (Jain, 2020).
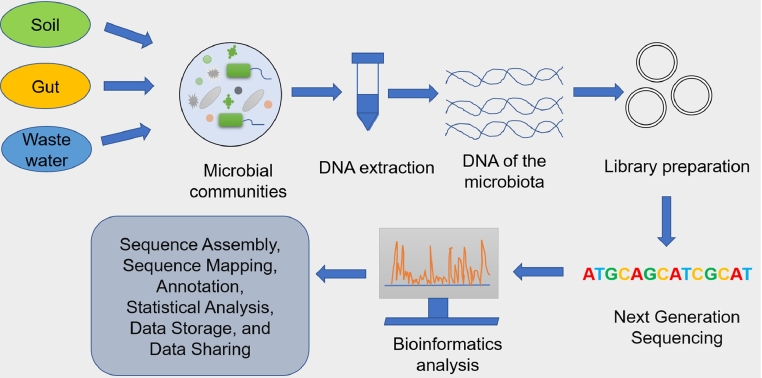
The uncultivable anaerobes can also be identified, characterized, and analyzed based on their genomes in a culture-independent manner (Forbes et al., 2017). This will further contribute to our better understanding of their taxonomic classification, which is presently vague and undefined. Presently, metagenomics, is used for the analysis of these anaerobic bacteria (Zhang et al., 2019). It is a culture-independent study of microbial diversity in a given environment (Escobar-Zepeda et al., 2015). This powerful tool can be extensively used to study and analyze more and more anaerobic genomes. Fig. 2 represents a general scheme used in the metagenomic analysis of samples from different habitats.
Fig. 2.
A general schematic for metagenomic analysis.
However, there are certain limitations of metagenomics that need to be addressed in future in order to improve the understanding of microbial diversity. Marker-based (16S rRNA or 18S rRNA) and whole-genome shotgun are the two commonly used metagenomic techniques. Both differ in their approach, but both suffer from some limitations. Marker-based technique employs the use of 16S rRNA or 18S rRNA genes to identify and classify the microbial diversity. However, it involves PCR-based amplification step, often leading to inevitable PCR bias. Other limitations are its inapplicability to viruses, failure to identify strain level differences, and the lack of functional analysis (Gupta et al., 2019). In contrast, the shotgun metagenomic sequencing enables study of viruses and functional analysis. However, it is very costly, involves greater risk of contamination, generates large number of reads leading to difficulty in assembly, and involves use of highly complex bioinformatics analysis (Petersen et al., 2019). More efforts are required to address these bottlenecks in order to expand the use of metagenomics in exploring the anaerobic microbial diversity.
As metagenomics is dependent on the sequencing techniques, the limitations of a sequencing platform can also adversely affect the metagenomic study. Each sequencing platform itself suffers from some shortcomings. For instance, Illumina, though being the most commonly used sequencing technique, generates shorter reads which hinder the downstream analysis (Wang et al., 2022). Furthermore, PacBio, a third-generation sequencing technique, overcomes the limitation of Illumina, but results in high error rate (Wang et al., 2022). The read length and error frequencies are very critical aspects while exploring the novel microbial diversity. Hence, it is important to overcome these problems of sequencing platforms in order to expand the use of metagenomics in exploring the novel microbial diversity.
Functional metagenomic screening is a metagenomic assisted approach involving the isolation of metagenomic DNA from the microbial communities residing in the extreme environments to study the functional roles of the microbial proteins (Ngara and Zhang, 2018). Therefore, functional metagenomic screening enables culture-independent study of microbial genomes and their metabolic pathways. In a recent study by Macdonald and colleagues, functional metagenomic screening was used to screen microbial communities with Glycoside phosphorylases (GPs) activity (Macdonald et al., 2019). The methods for the identification of GP activity containing microbes involves collection of metagenomic DNA from the cellulose-rich environments enriched with cellulose-degrading microbial communities, followed by generation of Fosmid metagenomic libraries and screening of these libraries. GP has valuable biocatalytic application in conversion of high-value carbohydrates and hence identification of microbial strains with GP activity can be of valuable application to various industries. Further, the study also suggest that the metabolic efficiency advantage of GPs is more impactful in anaerobes than in aerobic microorganisms and therefore the emphasis is on the importance of constructing GP screening libraries sourced from the anaerobic environments (Macdonald et al., 2019). Thus, such functional metagenomic studies can be very useful in the identification of microbial communities with valuable metabolic potential, particularly uncultivable anaerobic microbial communities.
Further, combining metagenomics with metatranscripomics, proteomics, and metabolomics will enable the study of species, functions, gene expression profiles, and metabolic pathways in an anaerobic microbial community. Therefore, more efforts are needed in understanding the microbial diversity of anaerobic environments using metagenomics techniques (Zhang et al., 2021).
4.2. Genetic engineering
As we are reminded of the fact that we cannot cultivate around ninety-nine percent of the microbes, can we modify their genomes without culturing them in the lab? With a combined approach involving functional metagenomics and genetic engineering, this has become possible recently. Genetic engineering can be used to modify the genomes of anaerobes to produce products of our interest or improve their efficiency in various present-day applications.
Multiple Clostridium spp. are used in biogas degradation and industrial-scale production of enzymes, solvents, and organic acids. Their genomes can be genetically modified to increase the yield and efficiency of the production process using multiple methods, including strain improvement, vector-based method, gene deletion, genetic recombination, and others (Croux et al., 2016; Wang et al., 2016; Herman et al., 2017). CRISPR (clustered regularly interspaced short palindromic repeats)/Cas (CRISPR-associated proteins) is emerging as a powerful and revolutionary tool for genome editing. Several unsuccessful attempts have been made to use CRISPR/Cas tool for Clostridium acetobutylicum (Wang et al., 2016). However, using a tightly controlled CRISPR/Cas9 system can provide promising results for Clostridium acetobutylicum genome editing (Wilding-Steele et al., 2021).
In a recent study, the utilization of recombinant DNA application to the genome of anaerobic fungi enabled cloning and expression of xylAr10 gene from Anaeromyces robustus fungus in Pichia pastoris to produce the recombinant xylanase. This enzyme has application in various industries including bread making, paper and others (Wen et al., 2021). Such genetic modifications can help in production of industrially important gene products from anaerobic microbes.
Currently, gene sequences from uncultured microbes are introduced into the expression vectors to obtain proteins of our interest (Kumar et al., 2015). Also, the gene sequences can be synthesized in vitro and can be introduced into the vectors (Veneziano et al., 2018). This eliminates the need for culturing in case of uncultivable anaerobic microbes and reduces the tedious process of culturing and cloning for cultivable ones. Although metagenomics and genetic engineering are already being used, their integration can lead to the development of metagenomic assisted genetic engineering techniques which can be used to produce the desired product in culture independent manner from anaerobic microbes (Macdonald et al., 2019).
The already existing applications of anaerobic microbial genomes can be improved, and the scope can be further extended. For example, with the reduced availability of land area, it has become essential to look at alternatives that can be used as feedstock for animal consumption. The indigenous anaerobic fungi present in the herbivore gut can be stimulated and their accessibility for plant fiber degradation can be improved (Hess et al., 2020). The future approaches will be focused on boosting the indigenous population of anaerobic fungi in the gut of the host organism. The same is applicable for anaerobic bacteria found in the gut of these herbivore animals. The anaerobic microbial community has a lot to offer to nature and mankind. In the future, as we see advancements in various genomic technologies, there is a good possibility of knowing the unknown.
5. Conclusion remarks and future perspective
We are surrounded by microorganisms in our environment and these small invisible forms of life are of immense importance to our ecosystem. They are playing an important role in maintaining the very balance required in the ecosystem. Hence, it can be said that any form of existence is incomplete without microbes. Anaerobic microbes grow and survive in extreme environments and they can do so with the help of their genomes. They tend to adapt to such harsh conditions because of their unique genomic composition. For instance, anaerobes have certain enzymes that scavenge oxygen and protect them from oxygen. It is noteworthy that most of these coping mechanisms are enabled due to the genes and their corresponding gene products.
Anaerobes have been used in the past in various fields. However, due to the lack of a suitable cultivation method, their application was limited, and most of them remained unidentified. Nevertheless, today they are extensively used across fields. In the past two decades, we have seen enormous development in molecular biology and genomics. Moreover, the use of 16S rRNA genetic marker in metagenomics has helped us to unlock the roles of microbial genomes of some important anaerobes. These have further opened new avenues for the microbiology of anaerobes. In the future, linking of different OMICs will lead us to study genes to proteins and structures to functions of anaerobic microbes. As more advancements are expected in science and research, we will be able to achieve more accessibility to these anaerobic microorganisms which will build our knowledge of these tiny yet giant life forms found in nature.
The COVID-19 pandemic has brought light to a crucial fact that we should be well aware of the microbial communities found in nature. These microbes, including the anaerobes, can be of immense benefits, but at the same time, they can have an adverse impact on our health. Some of them are known to cause severe infections and diseases in animals and humans (Tang et al., 2020). Hence, it becomes even more critical to identify them and understand their biology and also their epidemiology.
CRediT authorship contribution statement
Pratyusha Patidar: Conceptualization, Writing – review & editing. Tulika Prakash: Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
References
- Ahmad, A., Naqvi, S.A., Muhammad, Jaskani, J., Waseem, Muhammad, Ali, E., Khan, I.A., Manzoor, F., Siddeeg, A., Aadil, R.M., 2021. Efficient utilization of date palm waste for the bioethanol production through Saccharomyces cerevisiae strain. https://doi.org/10.1002/fsn3.2175. [DOI] [PMC free article] [PubMed]
- Ajaz M., Shakeel S., Rehman A. Microbial use for azo dye degradation—A strategy for dye bioremediation. Int. Microbiol. 2020;23:149–159. doi: 10.1007/s10123-019-00103-2. [DOI] [PubMed] [Google Scholar]
- Alalawy A.I., Guo Z., Almutairi F.M., El Rabey H.A., Al-Duais Mohammed A., Mohammed Ghena M., Almasoudi F.M., Alotaibi M.A., Salama E.-.S., Abomohra Abd, El-Fatah Sakran M.I. Explication of structural variations in the bacterial and archaeal community of anaerobic digestion sludges: an insight through metagenomics. J. Environ. Chem. Eng. 2021;9 doi: 10.1016/j.jece.2021.105910. [DOI] [Google Scholar]
- Alba-Lois L., Segal-Kischinevzky C. Yeast fermentation and the making of beer and wine. Nat. Educ. 2010;3:1–4. [Google Scholar]
- Alipour M.J., Jalanka J., Pessa-Morikawa T., Kokkonen T., Satokari R., Hynönen U., Iivanainen A., Niku M. The composition of the perinatal intestinal microbiota in cattle. Sci. Rep. 2018;8:10437. doi: 10.1038/s41598-018-28733-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shorgani N.K.N., Al-Tabib A.I., Kadier A., Zanil M.F., Lee K.M., Kalil M.S. Continuous butanol fermentation of dilute acid-pretreated de-oiled rice bran by clostridium acetobutylicum YM1. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-40840-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade J.C., Almeida D., Domingos M., Seabra C.L., Machado D., Freitas A.C., Gomes A.M. Commensal obligate anaerobic bacteria and health: production, storage, and delivery strategies. Front. Bioeng. Biotechnol. 2020;8:550. doi: 10.3389/fbioe.2020.00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- André A.C., Debande L., Marteyn B.S. The selective advantage of facultative anaerobes relies on their unique ability to cope with changing oxygen levels during infection. Cell. Microbiol. 2021;23 doi: 10.1111/cmi.13338. [DOI] [PubMed] [Google Scholar]
- Armour, C.R., Nayfach, S., Pollard, K.S., Sharpton, T.J., 2019. A metagenomic meta-analysis reveals functional signatures of health and disease in the human gut microbiome. mSystems 4. https://doi.org/10.1128/msystems.00332-18. [DOI] [PMC free article] [PubMed]
- Azad E., Fehr K.B., Derakhshani H., Forster R., Acharya S., Khafipour E., McGeough E., McAllister T.A. Interrelationships of fiber-associated anaerobic fungi and bacterial communities in the rumen of bloated cattle grazing alfalfa. Microorganisms. 2020;8:1–17. doi: 10.3390/microorganisms8101543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett J.A. A history of research on yeasts 1: work by chemists and biologists 1789-1850. Yeast. 1998;14:1439–1451. doi: 10.1002/(SICI)1097-0061(199812)14:16<1439::AID−YEA339>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Barth A., Hendrix J., Fried D., Barak Y., Bayer E.A., Lamb D.C. Dynamic interactions of type I cohesin modules fine-tune the structure of the cellulosome of Clostridium thermocellum. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E11274–E11283. doi: 10.1073/pnas.1809283115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S.C., Devraj S. 1st ed. Taylor & Francis; 2017. Pollution Control in Textile Industry. [DOI] [Google Scholar]
- Blair E.M., Dickson K.L., O'Malley M.A. Microbial communities and their enzymes facilitate degradation of recalcitrant polymers in anaerobic digestion. Curr. Opin. Microbiol. 2021;64:100–108. doi: 10.1016/j.mib.2021.09.008. [DOI] [PubMed] [Google Scholar]
- Chatsurachai S., Watanarojanaporn N., Phaengthai S., Sakulsombat M., Sriroth K. Genetic variation in genes involved in ethanol production among saccharomyces cerevisiae strains. Sugar Tech. 2020;22:250–258. doi: 10.1007/s12355-019-00771-4. [DOI] [Google Scholar]
- Chen H. Recent advances in Azo dye degrading enzyme research. Curr. Protein Pept. Sci. 2006;7:101–111. doi: 10.2174/138920306776359786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K.-.T. Azo dyes and human health: a review. J. Environ. Sci. Health. C. Environ. Carcinog. Ecotoxicol. Rev. 2016;34:233–261. doi: 10.1080/10590501.2016.1236602. [DOI] [PubMed] [Google Scholar]
- Croux C., Nguyen N.P.T., Lee J., Raynaud C., Saint-Prix F., Gonzalez-Pajuelo M., Meynial-Salles I., Soucaille P. Construction of a restriction-less, marker-less mutant useful for functional genomic and metabolic engineering of the biofuel producer Clostridium acetobutylicum. Biotechnol. Biofuels. 2016;9:1–13. doi: 10.1186/s13068-016-0432-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyprowski M., Stobnicka-Kupiec A., Ławniczek-Wałczyk A., Bakal-Kijek A., Gołofit-Szymczak M., Górny R.L. Anaerobic bacteria in wastewater treatment plant. Int. Arch. Occup. Environ. Health. 2018;91:571–579. doi: 10.1007/s00420-018-1307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q., Zhang S., Liu H., Huang J., Li L. Sulfide-mediated azo dye degradation and microbial community analysis in a single-chamber air cathode microbial fuel cell. Bioelectrochemistry. 2020;131 doi: 10.1016/j.bioelechem.2019.107349. [DOI] [PubMed] [Google Scholar]
- Decker, B.K., Jungermann, K., 1970. Energy Production i n Anaerobic Organisms[**’ 9, 138–158. [DOI] [PubMed]
- Devaraj K., Aathika S., Periyasamy K., Manickam Periyaraman P., Palaniyandi S., Subramanian S. Production of thermostable multiple enzymes from Bacillus amyloliquefaciens KUB29. Nat. Prod. Res. 2019;33:1674–1677. doi: 10.1080/14786419.2018.1425857. [DOI] [PubMed] [Google Scholar]
- Doyle C.J., Gleeson D., Jordan K., Beresford T.P., Ross R.P., Fitzgerald G.F., Cotter P.D. Anaerobic sporeformers and their significance with respect to milk and dairy products. Int. J. Food Microbiol. 2015;197:77–87. doi: 10.1016/j.ijfoodmicro.2014.12.022. [DOI] [PubMed] [Google Scholar]
- Dunbar K.L., Büttner H., Molloy E.M., Dell M., Kumpfmüller J., Hertweck C. Genome editing reveals novel thiotemplated assembly of polythioamide antibiotics in anaerobic bacteria. Angew. Chemie. 2018;130:14276–14280. doi: 10.1002/ange.201807970. [DOI] [PubMed] [Google Scholar]
- Escobar-Zepeda A., Vera-Ponce de Leon A., Sanchez-Flores A. The road to metagenomics: from microbiology to DNA sequencing technologies and bioinformatics. Front. Genet. 2015;6:348. doi: 10.3389/fgene.2015.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falentin H., Deutsch S.M., Jan G., Loux V., Thierry A., Parayre S., Maillard M.B., Dherbecourt J., Cousin F.J., Jardin J., Siguier P. The complete genome of Propionibacterium freudenreichii CIRM-BIA1T, a hardy Actinobacterium with food and probiotic applications. PLoS ONE. 2010;5(7) doi: 10.1371/journal.pone.0011748. p. e11748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field K.G., Olsen G.J., Lane D.J., Giovannoni S.J., Ghiselin M.T., Raff E.C., Pace N.R., Raff R.A. Molecular phylogeny of the animal kingdom. Science. 1988;239:748–753. doi: 10.1126/science.3277277s. (80-.) [DOI] [PubMed] [Google Scholar]
- Finegold S.M. A century of anaerobes: a look backward and a call to arms. Clin. Infect. Dis. 1993;16:S453–S457. doi: 10.1093/clinids/16.Supplement_4.S453. [DOI] [PubMed] [Google Scholar]
- Fischer P.Q., Sánchez-Andrea I., Stams A.J.M., Villanueva L., Sousa D.Z. Anaerobic microbial methanol conversion in marine sediments. Environ. Microbiol. 2021;23:1348–1362. doi: 10.1111/1462-2920.15434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann R.D., Adams M.D., White O., Clayton R.A., Kirkness E.F., Kerlavage A.R., Bult C.J., Tomb J.F., Dougherty B.A., Merrick J.M., McKenney K., Sutton G., FitzHugh W., Fields C., Gocayne J.D., Scott J., Shirley R., Liu L.I., Glodek A., Kelley J.M., Weidman J.F., Phillips C.A., Spriggs T., Hedblom E., Cotton M.D., Utterback T.R., Hanna M.C., Nguyen D.T., Saudek D.M., Brandon R.C., Fine L.D., Fritchman J.L., Fuhrmann J.L., Geoghagen N.S.M., Gnehm C.L., McDonald L.A., Small K.V., Fraser C.M., Smith H.O., Venter J.C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science (80-.). 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Forbes J.D., Knox N.C., Ronholm J., Pagotto F., Reimer A. Metagenomics: the next culture-independent game changer. Front. Microbiol. 2017;8:1–21. doi: 10.3389/fmicb.2017.01069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi O., Rosenkranz F., Chamy R. Key microbial populations involved in anaerobic degradation of phenol and p-cresol using different inocula. Electron. J. Biotechnol. 2018;35:33–38. doi: 10.1016/j.ejbt.2018.08.002. [DOI] [Google Scholar]
- Frey C.N. History and development of the modern yeast industry. Ind. Eng. Chem. 1931;23:340. doi: 10.1021/ie50255a036. [DOI] [Google Scholar]
- Gičević A., Hindija L., Karačić A. IFMBE Proceedings. Springer; Cham: 2020. Toxicity of Azo dyes in pharmaceutical industry; pp. 581–587. [DOI] [Google Scholar]
- Gruninger R.J., Cote C., McAllister T.A., Abbott D.W. Contributions of a unique β-clamp to substrate recognition illuminates the molecular basis of exolysis in ferulic acid esterases. Biochem. J. 2016;473:839–849. doi: 10.1042/BJ20151153. [DOI] [PubMed] [Google Scholar]
- Gupta A., Verma J.P. Sustainable bio-ethanol production from agro-residues: a review. Renew. Sustain. Energy Rev. 2015;41:550–567. doi: 10.1016/j.rser.2014.08.032. [DOI] [Google Scholar]
- Gupta S., Mortensen M.S., Schjørring S., Trivedi U., Vestergaard G., Stokholm J., Bisgaard H., Krogfelt K.A., Sørensen S.J. Amplicon sequencing provides more accurate microbiome information in healthy children compared to culturing. Commun. Biol. 2019;2:1–7. doi: 10.1038/s42003-019-0540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardegen J., Latorre-Pérez A., Vilanova C., Günther T., Porcar M., Luschnig O., Simeonov C., Abendroth C. Methanogenic community shifts during the transition from sewage mono-digestion to co-digestion of grass biomass. Bioresour. Technol. 2018;265:275–281. doi: 10.1016/j.biortech.2018.06.005. [DOI] [PubMed] [Google Scholar]
- Hartinger T., Zebeli Q. The present role and new potentials of anaerobic fungi in ruminant nutrition. J. Fungi. 2021;7:200. doi: 10.3390/jof7030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatti-Kaul R., Mattiasson B. Anaerobes in industrial- and environmental biotechnology. Adv. Biochem. Eng. Biotechnol. 2016;156:1–33. doi: 10.1007/10_2016_10. [DOI] [PubMed] [Google Scholar]
- Herman N.A., Kim S.J., Li J.S., Cai W., Koshino H., Zhang W. The industrial anaerobe Clostridium acetobutylicum uses polyketides to regulate cellular differentiation. Nat. Commun. 2017;8 doi: 10.1038/s41467-017-01809-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess M., Paul S.S., Puniya A.K., van der Giezen M., Shaw C., Edwards J.E., Fliegerová K. Anaerobic fungi: past, present, and future. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.584893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Invernici M.M., Salvador S.L., Silva P.H.F., Soares M.S.M., Casarin R., Palioto D.B., Souza S.L.S., Taba M., Novaes A.B., Furlaneto F.A.C., Messora M.R. Effects of Bifidobacterium probiotic on the treatment of chronic periodontitis: a randomized clinical trial. J. Clin. Periodontol. 2018;45:1198–1210. doi: 10.1111/jcpe.12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N. The need for personalized approaches to microbiome modulation. Front. Public Heal. 2020;8 doi: 10.3389/fpubh.2020.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James McIntosh, Fildes Paul, Bulloch W. A new apparatus for the isolation and cultivation of anaerobic micro-organisms. Lancet. 1916;187:768–770. [Google Scholar]
- Johnson J.S., Spakowicz D.J., Hong B.-.Y., Petersen L.M., Demkowicz P., Chen L., Leopold S.R., Hanson B.M., Agresta H.O., Gerstein M., Sodergren E., Weinstock G.M. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019;10:1–11. doi: 10.1038/s41467-019-13036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauldhar B.S., Sooch B.S. Tailoring nutritional and process variables for hyperproduction of catalase from a novel isolated bacterium Geobacillus sp. BSS-7. Microb. Cell Fact. 2016;15 doi: 10.1186/s12934-016-0410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Krishnani K.K., Bhushan B., Brahmane M.P. Metagenomics: retrospect and prospects in high throughput age. Biotechnol. Res. Int. 2015:1–13. doi: 10.1155/2015/121735. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.M., Guan L.L., Eun J.-.S., Kim C.-.H., Lee S.J., Kim E.T., Lee S.S. The effect of anaerobic fungal inoculation on the fermentation characteristics of rice straw silages. J. Appl. Microbiol. 2015;118:565–573. doi: 10.1111/jam.12724. [DOI] [PubMed] [Google Scholar]
- Leitão A.L. Potential of penicillium species in the bioremediation field. Int. J. Environ. Res. Public Health. 2009;6:1393–1417. doi: 10.3390/ijerph6041393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E., Saleem F., Edge T.A., Schellhorn H.E. Biological indicators for fecal pollution detection and source tracking: a review. Processes. 2021;9:2058. doi: 10.3390/pr9112058. [DOI] [Google Scholar]
- Li J.S., Barber C.C., Zhang W. Natural products from anaerobes. J. Ind. Microbiol. Biotechnol. 2019;46:375–383. doi: 10.1007/s10295-018-2086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linko Y.Y., Javanainen P., Linko S. Biotechnology of bread baking. Trends Food Sci. Technol. 1997;8:339–344. doi: 10.1016/S0924-2244(97)01066-2. [DOI] [Google Scholar]
- Locey K.J., Lennon J.T. Scaling laws predict global microbial diversity. Proc. Natl. Acad. Sci. U. S. A. 2016;113:5970–5975. doi: 10.1073/pnas.1521291113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Imlay J.A. When anaerobes encounter oxygen: mechanisms of oxygen toxicity, tolerance and defence. Nat. Rev. Microbiol. 2021;19:774–785. doi: 10.1038/s41579-021-00583-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lueders T., von Netzer F. Hydrocarbon and Lipid Microbiology Protocols. Springer; Berlin Heidelberg: 2014. Primers: functional genes for anaerobic hydrocarbon degrading microbes; pp. 39–55. [DOI] [Google Scholar]
- Luo F., Gitiafroz R., Devine C.E., Gong Y., Hug L.A., Raskin L., Edwards E.A. Metatranscriptome of an anaerobic benzene-degrading, nitrate-reducing enrichment culture reveals involvement of carboxylation in benzene ring activation. Appl. Environ. Microbiol. 2014;80:4095–4107. doi: 10.1128/AEM.00717-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma'As M.F., Ghazali H.M., Chieng S. Bioethanol production from Brewer's rice by Saccharomyces cerevisiae and Zymomonas mobilis: evaluation of process kinetics and performance. energy sources, Part A recover. Util. Environ. Eff. 2020;00:1–14. doi: 10.1080/15567036.2020.1815901. [DOI] [Google Scholar]
- Macdonald S.S., Armstrong Z., Morgan-Lang C., Osowiecka M., Robinson K., Hallam S.J., Withers S.G. Development and application of a high-throughput functional metagenomic screen for glycoside phosphorylases. Cell Chem. Biol. 2019;26:1001–1012. doi: 10.1016/j.chembiol.2019.03.017. e5. [DOI] [PubMed] [Google Scholar]
- Mahawar H., Prasanna R., Gogoi R. Elucidating the disease alleviating potential of cyanobacteria, copper nanoparticles and their interactions in Fusarium solani challenged tomato plants. Plant Physiol. Reports. 2019;24:533–540. doi: 10.1007/s40502-019-00490-8. [DOI] [Google Scholar]
- Mamo G. Anaerobes as sources of bioactive compounds and health promoting tools. Adv. Biochem. Eng. Biotechnol. 2016;156:433–464. doi: 10.1007/10_2016_6. [DOI] [PubMed] [Google Scholar]
- Martin N.H., Trmčić A., Hsieh T.H., Boor K.J., Wiedmann M. The evolving role of coliforms as indicators of unhygienic processing conditions in dairy foods. Front. Microbiol. 2016;7:1549. doi: 10.3389/fmicb.2016.01549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoumi S.J., Mehrabani D., Saberifiroozi M., Fattahi M.R., Moradi F., Najafi M. The effect of yogurt fortified with Lactobacillus acidophilus and Bifidobacterium sp . probiotic in patients with lactose intolerance. Food Sci. Nutr. 2021;9:1704–1711. doi: 10.1002/fsn3.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker M.L. Sequencing technologies - the next generation. Nat. Rev. Genetics. 2010;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- Mobeen F., Sharma V., Prakash T. Comparative gut microbiome analysis of the Prakriti and Sasang systems reveals functional level similarities in constitutionally similar classes. 3 Biotech. 2020;10:1–15. doi: 10.1007/s13205-020-02376-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momma N., Kobara Y., Momma M. Fe2+ and Mn2+, potential agents to induce suppression of Fusarium oxysporum for biological soil disinfestation. J. Gen. Plant Pathol. 2011;77:331–335. doi: 10.1007/s10327-011-0336-8. [DOI] [Google Scholar]
- Moraïs S., Mizrahi I. Islands in the stream: from individual to communal fiber degradation in the rumen ecosystem. FEMS Microbiol. Rev. 2019;43:362–369. doi: 10.1093/femsre/fuz007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralikrishna I.V., Manickam V. Elsevier; 2017. Wastewater Treatment Technologies, in: Environmental Management; pp. 249–293. [DOI] [Google Scholar]
- Ngara T.R., Zhang H. Recent advances in function-based metagenomic screening. Genomics. Proteomics Bioinf. 2018;16:405–415. doi: 10.1016/j.gpb.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.N., Nguyen A.Q., Johir M.A.H., Guo W., Ngo H.H., Chaves A.V., Nghiem L.D. Application of rumen and anaerobic sludge microbes for bio harvesting from lignocellulosic biomass. Chemosphere. 2019;228:702–708. doi: 10.1016/j.chemosphere.2019.04.159. [DOI] [PubMed] [Google Scholar]
- Paunkov A., Gutenbrunner K., Sóki J., Leitsch D. Haemin deprivation renders Bacteroides fragilis hypersusceptible to metronidazole and cancels high-level metronidazole resistance. J. Antimicrob. Chemother. 2021 doi: 10.1093/jac/dkab485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavithra K.G., P S.K., V J., P S.R. Removal of colorants from wastewater: a review on sources and treatment strategies. J. Ind. Eng. Chem. 2019;75:1–19. doi: 10.1016/j.jiec.2019.02.011. [DOI] [Google Scholar]
- Petersen L.M., Martin I.W., Moschetti W.E., Kershaw C.M., Tsongalis G.J. Third-generation sequencing in the clinical laboratory: exploring the advantages and challenges of nanopore sequencing. J. Clin. Microbiol. 2019;58:e01315–e01319. doi: 10.1128/JCM.01315-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidot S., Ishida K., Cyrulies M., Hertweck C. Discovery of clostrubin, an exceptional polyphenolic polyketide antibiotic from a strictly anaerobic bacterium. Angew. Chemie. 2014;126:7990–7993. doi: 10.1002/ange.201402632. [DOI] [PubMed] [Google Scholar]
- Pitt T.L., Barer M.R. Elsevier; 2012. Classification, Identification and Typing of micro-organisms, in: Medical Microbiology; pp. 24–38. [DOI] [Google Scholar]
- Piwowarek K., Lipińska E., Hać-Szymańczuk E., Kieliszek M., Ścibisz I. Propionibacterium spp.—Source of propionic acid, vitamin B12, and other metabolites important for the industry. Appl. Microbiol. Biotechnol. 2018;102:515–538. doi: 10.1007/s00253-017-8616-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M., Wang P., Selinger L.B., Yanke L.J., Forster R.J., Mcallister T.A. Isolation and characterization of a ferulic acid esterase (Fae1A) from the rumen fungus Anaeromyces mucronatus. J. Appl. Microbiol. 2011;110:1341–1350. doi: 10.1111/J.1365-2672.2011.04990.X. [DOI] [PubMed] [Google Scholar]
- Rabah H., Ferret-Bernard S., Huang S., Le Normand L., Cousin F.J., Gaucher F., Jeantet R., Boudry G., Jan G. The cheese matrix modulates the immunomodulatory properties of propionibacterium freudenreichii CIRM-BIA 129 in healthy piglets. Front. Microbiol. 2018;9:1–15. doi: 10.3389/fmicb.2018.02584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakaz M.A., Hussien M.O., Ibrahim H.M. Isolation, extraction, purification, and molecular characterization for thermostable α-amylase from locally isolated bacillus species in Sudan. Biochem. Res. Int. 2021:1–8. doi: 10.1155/2021/6670380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveendran S., Parameswaran B., Ummalyma S.B., Abraham A., Mathew A.K., Madhavan A., Rebello S., Pandey A. Applications of microbial enzymes in food industry. Food Technol. Biotechnol. 2018;56:16–30. doi: 10.17113/ftb.56.01.18.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi B., Nkongndem Nkemka V., Hao X., Yanke J., McAllister T.A., Lee H., Veluchamy C., Gilroyed B.H. Effect of Bioaugmentation with anaerobic fungi isolated from ruminants on the hydrolysis of corn silage and phragmites Australis. Appl. Sci. 2021;11:9123. doi: 10.3390/app11199123. [DOI] [Google Scholar]
- Ravindran R., Williams G.A., Jaiswal A.K. Evaluation of brewer's spent grain hydrolysate as a substrate for production of thermostable α-amylase by Bacillus stearothermophilus. Bioresour. Technol. Reports. 2019;5:141–149. doi: 10.1016/j.biteb.2019.01.004. [DOI] [Google Scholar]
- Rischer M., Raguž L., Guo H., Keiff F., Diekert G., Goris T., Beemelmanns C. Biosynthesis, synthesis, and activities of barnesin A, a NRPS-PKS hybrid produced by an anaerobic epsilonproteobacterium. ACS Chem. Biol. 2018;13:1990–1995. doi: 10.1021/acschembio.8b00445. [DOI] [PubMed] [Google Scholar]
- Rokunuzzaman M., Hayakawa A., Yamane S., Tanaka S., Ohnishi K. Effect of soil disinfection with chemical and biological methods on bacterial communities. Egypt. J. Basic Appl. Sci. 2016;3:141–148. doi: 10.1016/j.ejbas.2016.01.003. [DOI] [Google Scholar]
- Sahay H., Yadav A.N., Singh A.K., Singh S., Kaushik R., Saxena A.K. Hot springs of Indian Himalayas: potential sources of microbial diversity and thermostable hydrolytic enzymes. 3 Biotech. 2017;7:1–11. doi: 10.1007/s13205-017-0762-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel D. Investigation of ancient Egyptian baking and brewing methods. Science. 1996;273:488–490. doi: 10.1126/science.273.5274.488. [DOI] [PubMed] [Google Scholar]
- Sanders M.E., Merenstein D.J., Reid G., Gibson G.R., Rastall R.A. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 2019;16:605–616. doi: 10.1038/s41575-019-0173-3. [DOI] [PubMed] [Google Scholar]
- Santana M.L., Bispo J.A.C., de Sena A.R., Teshima E., de Brito A.R., Costa F.S., Franco M., de Assis S.A. Clarification of tangerine juice using cellulases from Pseudoyma sp. J. Food Sci. Technol. 2021;58:44–51. doi: 10.1007/s13197-020-04511-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Banerjee A., Halder U., Biswas R., Bandopadhyay R. Degradation of synthetic Azo dyes of textile industry: a sustainable approach using microbial enzymes. Water Conserv. Sci. Eng. 2017;2:121–131. doi: 10.1007/s41101-017-0031-5. [DOI] [Google Scholar]
- Saxena G., Bharagava R.N., Kaithwas G., Raj A. Microbial indicators, pathogens and methods for their monitoring in water environment. J. Water Health. 2015;13:319–339. doi: 10.2166/wh.2014.275. [DOI] [PubMed] [Google Scholar]
- Scheld W.M. Introduction to microbial disease: host-pathogen interactions. Goldman's cecil med. Twenty Fourth Ed. 2012;2:1761–1762. doi: 10.1016/B978-1-4377-1604-7.00286-4. [DOI] [Google Scholar]
- Schoch C.L., Seifert K.A., Huhndorf S., Robert V., Spouge J.L., Levesque C.A., Chen W., Bolchacova E., Voigt K., Crous P.W., Miller A.N., Wingfield M.J., Aime M.C., An K.-.D., Bai F.-.Y., Barreto R.W., Begerow D., Bergeron M.-.J., Blackwell M., Boekhout T., Bogale M., Boonyuen N., Burgaz A.R., Buyck B., Cai L., Cai Q., Cardinali G., Chaverri P., Coppins B.J., Crespo A., Cubas P., Cummings C., Damm U., de Beer Z.W., de Hoog G.S., Del-Prado R., Dentinger B., Diéguez-Uribeondo J., Divakar P.K., Douglas B., Dueñas M., Duong T.A., Eberhardt U., Edwards J.E., Elshahed M.S., Fliegerova K., Furtado M., García M.A., Ge Z.-.W., Griffith G.W., Griffiths K., Groenewald J.Z., Groenewald M., Grube M., Gryzenhout M., Guo L.-.D., Hagen F., Hambleton S., Hamelin R.C., Hansen K., Harrold P., Heller G., Herrera C., Hirayama K., Hirooka Y., Ho H.-.M., Hoffmann K., Hofstetter V., Högnabba F., Hollingsworth P.M., Hong S.-.B., Hosaka K., Houbraken J., Hughes K., Huhtinen S., Hyde K.D., James T., Johnson E.M., Johnson J.E., Johnston P.R., Jones E.B.G., Kelly L.J., Kirk P.M., Knapp D.G., Kõljalg U., Kovács G.M., Kurtzman C.P., Landvik S., Leavitt S.D., Liggenstoffer A.S., Liimatainen K., Lombard L., Luangsa-ard J.J., Lumbsch H.T., Maganti H., Maharachchikumbura S.S.N., Martin M.P., May T.W., McTaggart A.R., Methven A.S., Meyer W., Moncalvo J.-.M., Mongkolsamrit S., Nagy L.G., Nilsson R.H., Niskanen T., Nyilasi I., Okada G., Okane I., Olariaga I., Otte J., Papp T., Park D., Petkovits T., Pino-Bodas R., Quaedvlieg W., Raja H.A., Redecker D., Rintoul T.L., Ruibal C., Sarmiento-Ramírez J.M., Schmitt I., Schüßler A., Shearer C., Sotome K., Stefani F.O.P., Stenroos S., Stielow B., Stockinger H., Suetrong S., Suh S.-.O., Sung G.-.H., Suzuki M., Tanaka K., Tedersoo L., Telleria M.T., Tretter E., Untereiner W.A., Urbina H., Vágvölgyi C., Vialle A., Vu T.D., Walther G., Wang Q.-.M., Wang Y., Weir B.S., Weiß M., White M.M., Xu J., Yahr R., Yang Z.L., Yurkov A., Zamora J.-.C., Zhang N., Zhuang W.-.Y., Schindel D. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebald M., Finegold S.M. Pasteur: the anaerobic life and the anaerobes. Clin. Infect. Dis. 1995;20 doi: 10.1093/clinids/20.Supplement_2.S111. S111. [DOI] [Google Scholar]
- Sebald M., Hauser D. Pasteur, oxygen and the anaerobes revisited. Anaerobe. 1995;1:11–16. doi: 10.1016/s1075-9964(95)80353-x. [DOI] [PubMed] [Google Scholar]
- Silva D.R., Sardi J.de C.O., Pitangui N.de S., Roque S.M., Silva A.C.B.da, Rosalen P.L. Probiotics as an alternative antimicrobial therapy: current reality and future directions. J. Funct. Foods. 2020;73 doi: 10.1016/j.jff.2020.104080. [DOI] [Google Scholar]
- Singh S.P., Kaur S., Singh D. Elsevier Inc; 2017. Toxicological Profile of Indian foods-Ensuring Food Safety in India, Food Safety in the 21st Century: Public Health Perspective. [DOI] [Google Scholar]
- Sousa F.L., Nelson-Sathi S., Martin W.F. One step beyond a ribosome: the ancient anaerobic core. Biochim. Biophys. Acta - Bioenerg. 2016;1857:1027–1038. doi: 10.1016/j.bbabio.2016.04.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen A.D., Crits-Christoph A., Carini P., DeAngelis K.M., Fierer N., Lloyd K.G., Cameron Thrash J. High proportions of bacteria and archaea across most biomes remain uncultured. ISME J. 2019;13:3126–3130. doi: 10.1038/s41396-019-0484-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Prem A., Tjokrosurjo J., Sary M., Van Bel M.A., Rodrigues-Hoffmann A., Krumbeck J.A. The canine skin and ear microbiome: a comprehensive survey of pathogens implicated in canine skin and ear infections using a novel next-generation-sequencing-based assay. Vet. Microbiol. 2020;247 doi: 10.1016/j.vetmic.2020.108764. [DOI] [PubMed] [Google Scholar]
- Tondro H., Musivand S., Zilouei H., Bazarganipour M., Zargoosh K. Biological production of hydrogen and acetone- butanol-ethanol from sugarcane bagasse and rice straw using co-culture of Enterobacter aerogenes and Clostridium acetobutylicum. Biomass and Bioenergy. 2020;142 doi: 10.1016/j.biombioe.2020.105818. [DOI] [Google Scholar]
- Town J.R., Links M.G., Fonstad T.A., Dumonceaux T.J. Molecular characterization of anaerobic digester microbial communities identifies microorganisms that correlate to reactor performance. Bioresour. Technol. 2014;151:249–257. doi: 10.1016/j.biortech.2013.10.070. [DOI] [PubMed] [Google Scholar]
- Trmčić A., Monnet C., Rogelj I., Bogovič Matijašić B. Expression of nisin genes in cheese—a quantitative real-time polymerase chain reaction approach. J. Dairy Sci. 2011;94:77–85. doi: 10.3168/jds.2010-3677. [DOI] [PubMed] [Google Scholar]
- Tse T.J., Wiens D.J., Reaney M.J.T. Production of bioethanol—a review of factors affecting ethanol yield. Fermentation. 2021;7:268. doi: 10.3390/fermentation7040268. [DOI] [Google Scholar]
- Tsolcha O.N., Patrinou V., Economou C.N., Dourou M., Aggelis G., Tekerlekopoulou A.G. Utilization of biomass derived from cyanobacteria-based agro-industrial wastewater treatment and raisin residue extract for bioethanol production. Water (Switzerland) 2021;13 doi: 10.3390/w13040486. [DOI] [Google Scholar]
- Ueki A., Kaku N., Ueki K. Role of anaerobic bacteria in biological soil disinfestation for elimination of soil-borne plant pathogens in agriculture. Appl. Microbiol. Biotechnol. 2018;102:6309–6318. doi: 10.1007/s00253-018-9119-x. [DOI] [PubMed] [Google Scholar]
- Veneziano R., Shepherd T.R., Ratanalert S., Bellou L., Tao C., Bathe M. In vitro synthesis of gene-length single-stranded DNA. Sci. Rep. 2018;8:1–7. doi: 10.1038/s41598-018-24677-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsing Rao, MP, Liu, L, Jiao, J-Y, Xiao, M, Li, W. J, 2018. Hot springs of India: occurrence and microbial diversity 29–55. https://doi.org/10.1007/978-981-13-0329-6_2.
- Vítěz, T., Novák, D., Lochman, J., Vítězová, M., 2020. Methanogens diversity during anaerobic sewage sludge stabilization and the effect of temperature. Processes 8, 822. https://doi.org/10.3390/pr8070822.
- Walker G.M., Stewart G.G. Saccharomyces cerevisiae in the production of fermented beverages. Beverages. 2016;2:1–12. doi: 10.3390/beverages2040030. [DOI] [Google Scholar]
- Wang L., Zhu P., Mo Q., Luo W., Du Z., Jiang J., Yang S., Zhao L., Gong Q., Wang Y. Comprehensive analysis of full-length transcriptomes of Schizothorax prenanti by single-molecule long-read sequencing. Genomics. 2022;114:456–464. doi: 10.1016/j.ygeno.2021.01.009. [DOI] [PubMed] [Google Scholar]
- Wang P., Shen C., Li L., Guo J., Cong Q., Lu J. Simultaneous production of propionic acid and vitamin B12 from corn stalk hydrolysates by Propionibacterium freudenreichii in an expanded bed adsorption bioreactor. Prep. Biochem. Biotechnol. 2020;50:763–767. doi: 10.1080/10826068.2020.1734942. [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang Z.T., Seo S.O., Lynn P., Lu T., Jin Y.S., Blaschek H.P. Bacterial genome editing with CRISPR-Cas9: deletion, integration, single nucleotide modification, and desirable “clean” mutant selection in clostridium beijerinckii as an example. ACS Synth. Biol. 2016;5:721–732. doi: 10.1021/acssynbio.6b00060. [DOI] [PubMed] [Google Scholar]
- Wen S., Wu G., Wu H. Biochemical characterization of a GH10 xylanase from the anaerobic rumen fungus Anaeromyces robustus and application in bread making. 3 Biotech. 2021;11:406. doi: 10.1007/s13205-021-02956-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen T., Huang X., Zhang J., Cai Z. Effects of biological soil disinfestation and water regime on suppressing Artemisia selengensis root rot pathogens. J. Soils Sediments. 2016;16:215–225. doi: 10.1007/s11368-015-1172-9. [DOI] [Google Scholar]
- Wilding-Steele T., Ramette Q., Jacottin P., Soucaille P. Improved crispr/cas9 tools for the rapid metabolic engineering of clostridium acetobutylicum. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22073704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C.R., Fox G.E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. U. S. A. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J.M., Singh N.K., Guan L., Seuylemezian A., Benardini J.N., Venkateswaran K. Performance of multiple metagenomics pipelines in understanding microbial diversity of a low-biomass spacecraft assembly facility. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.685254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen F., Zeng Z., Xu M., Sun F., Yang L., Bi X., Lin Y., Gao Y., Hao H., Yi W., Li M., Xie Y. Advances in Metagenomics and Its Application in Environmental Microorganisms. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.766364. 766364–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Loh K.C., Lim J.W., Zhang J. Bioinformatics analysis of metagenomics data of biogas-producing microbial communities in anaerobic digesters: a review. Renew. Sustain. Energy Rev. 2019;100:110–126. doi: 10.1016/j.watres.2018.12.041. [DOI] [Google Scholar]
- Zhao X., Condruz S., Chen J., Jolicoeur M. A quantitative metabolomics study of high sodium response in Clostridium acetobutylicum ATCC 824 acetone-butanol-ethanol (ABE) fermentation. Sci. Rep. 2016;6:1–13. doi: 10.1038/srep28307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Lu Y., Huang L., Zhang Q., Wang X., Zhu J. Effect of pH on volatile fatty acid production and the microbial community during anaerobic digestion of Chinese cabbage waste. Bioresour. Technol. 2021;336 doi: 10.1016/j.biortech.2021.125338. [DOI] [PubMed] [Google Scholar]