Highlights
-
•
Negative correlation of RV prevalence in suppressing influenza viruses is independent of meteorological effects.
-
•
A low odds of RV/IV co-detection infers a unique interference effect.
-
•
Sequential RV infection model demonstrates the inhibitory effect of RV on the replication of influenza A and B viruses.
Keywords: Viral interference, Rhinovirus, Influenza, Co-infection, Viral epidemiology
Abstract
Background
Influenza virus (IV) and the rhinovirus (RV) are the two most common circulating respiratory viruses circulating. Natural viral interference has been suggested between them. The effect of such at the population level has been described in temperate region, while its effect at the individual and cellular levels warrants further validation. In this study, we described the respiratory virus epidemiology and the co-infection landscape in the hospitalized population and investigated the distinct molecular pathways involved in the inhibition of virus replication.
Methods
Nasopharyngeal aspirates (NPAs) collected from patients during 2015 to 2019 were examined for the presence of respiratory viruses. The correlation of the monthly prevalence between all the tested respiratory viruses, the co-infection rate and the temporal interference of RV and IV were tested. The viral interference was validated in vitro by conducting sequential RV and IV infections in the well-differentiated primary human airway epithelial cells. The contributing molecular pathways were determined by transcriptome analysis.
Findings
A total of 112,926 NPAs were evaluated, and the Enterovirus/RV was the most prevalent respiratory virus detected. The negative correlation between EV/RV and IVs prevalence was independent of age and meteorological factors. Compare with other viruses, EV/RV had a significantly lower incidence of co-infection with IVs. Prior exposure to RV inhibited the replication of IV species A, B and oseltamivir-resistance stain in vitro. RV uniquely downregulated genes related to processing of viral mRNA, ribosomal proteins, translation and influenza infection.
Interpretation
Epidemiological surveillance and the sequential infection in vitro suggested viral interference between EV/RV and IV operates at the population, individual and cellular levels.
Funding
This study was supported by the General Research Fund (Ref: 24107017 and 14103119 to RWYC) and the Chinese University Direct Grant for Research (Ref: 2019·073 to RWYC).
Graphical abstract

1. Introduction
Influenza virus (IV) confers substantial morbidity and mortality worldwide every year. The existence of natural reservoirs of IV makes it impossible to be eradicated in humans. IV vaccine is a classic preventive measure to attenuate disease severity. However, the viral antigenic drift may limit the duration of vaccine effectiveness (Cao et al., 2021), while the antigenic shift may result in the emergence of new strains and a vaccine mismatch (Treanor, 2004). Antiviral drug is another prophylactic and therapeutic option, however the effective time frame is limited to the first few days after infection (Burch et al., 2009). Moreover, the detection of antiviral resistant IV strains have been reported sporadically, with an increasing number of oseltamivir-resistant influenza A (R-IAV) being found (Lackenby et al., 2011). Whether or not there is natural viral interference in counteracting the influence of IV at the population or at the individual levels is an interesting area to explore. Identifying novel interfering factors that provide temporal immunity against pan-IV virus and other virulent viral infections would provide novel options for disease prevention and treatment.
Rhinovirus (RV), on the other hand, is the most frequent respiratory pathogen being detected throughout the year. However, it catches less attention in the public health aspect as it generally causes mild and self-limiting symptoms, and sometimes affected individuals would remain asymptomatic (Self et al., 2016). Nevertheless, infants could have up to six to eight RV infections per year, while adults could have around two to four episodes annually (Chen et al., 2015). Having RV infections repeatedly seems to be unavoidable as it comprises more than 160 distinct genotypes. Among genotypes, they do not confer substantial immunity against each other (Glanville and Johnston, 2015). Intriguingly, though the prevalence of RV is high, the molecular epidemiology study on RV is uncommon due to its high genotypic diversity. The enterovirus/rhinovirus (EV/RV) test has not been included as a standard test in clinical labs until recent years. In Hong Kong, it has become available in public hospitals since September 2015.
With the advancement for the co-detection of viruses through multiplex PCR, multiple population-wide surveillance programs suggested that the seasonality of respiratory viral infection is not only contributed by meteorological factors but also the biological interactions among different viruses. The concept of viral interference, a phenomenon in which a primary virus infection could transiently prevent or inhibit the secondary superinfecting virus (Henle, 1950), may also play an important role in influencing the pattern of virus outbreaks. Multiple epidemiological analyses, including studies performed in the United Kingdom and the United States, identified a negative interaction between the prevalence of IVs and RVs (Nickbakhsh et al., 2019; Wu et al., 2020), and viral interactions operate at multiple levels (Opatowski et al., 2018). In the epidemiological part of this study, we investigated the correlation of virus prevalence and the temporal interference between RV and IV at the population level and the chance of co-infection at the individual level. We validated this concept further by conducting a sequential infection study in the differentiated primary human nasopharyngeal epithelial cells (HNEC) and human bronchial epithelial cells (HBEC) to elucidate the inhibitory effect in vitro. RNA sequencing of HBEC was performed to pinpoint the responsible biological pathways in the gene expression level.
2. Method
2.1. Epidemiological analysis
All patients who were hospitalized during 2015 to 2019 with their NPA collected for respiratory pathogen detection were included in the analysis. Parameters including the odds of viral infection, co-infection rate, demographics of the patients and meteorological factors at the moment of collection were analyzed by epidemiological approaches. Please refer to online supplemental file for more detailed description of the analysis.
2.2. Sequential RV-A16 and IV virus infection in vitro
Primary airway epithelial cells were isolated and maintained in controlled Air-Liquid Interface (ALI). Differentiated cells were challenged with either RV-A16 only, IVs only, IVs with prior RV exposure for 48 h, or sham control. Supernatants were collected for the determination of viral load by qPCR and viral titration assay and expressed in viral gene copy per milliliter and 50% tissue culture infectious dose (TCID50) per milliliter, respectively. Cell lysate were harvested for total mRNA collection and subjected to RNA sequencing (RNA-seq). Differentially expressed genes (DEGs) analysis and Gene Set Enrichment Analysis (GSEA) were performed to identify pathways that were enriched upon RV and Influenza A virus (IAV) infection. Detailed protocol on the virus preparation, ALI culture, in vitro infection, RNA extraction and transcriptomic analysis were provided in the online supplemental file. Gene expression profiles using the HBECs were available at GEO with accession number GSE193164.
2.3. Statistical analysis
The association of the monthly prevalence between all pairs of virus infections was tested using Pearson correlation coefficients. To assess whether IV prevalence was statistically associated with the change of the future EV/RV prevalence, the Granger causality test was conducted and the significant lagged week of IV prevalence was determined. To examine the association between EV/RV and IV prevalence independent of meteorological effects at different lagged times, a quasi-Poisson generalized additive model was used to control the total number of weekly collected samples (i.e. model offset), long-term trend, and seasonal trend. The technical detail is provided in the online supplemental file. The effect of IV was quantified using adjusted relative risk (ARR) along with its corresponding 95% confidence interval (CI). The reference value was set as its median value.
The likelihood of viral co-infection was computed by Fisher's exact test and logistic regression after adjustment to age and gender. The expected number of co-infections in the absence of interference was estimated as the product of the incidence of the virus pair multiplied by the total sample size. Age group stratification with toddlers (age <2), preschool (age 2–5), school-age (age 6–17), adult (age 18–64) and elderly (age ≥65) were segregated for regression analysis. Differences in IV titers and viral gene expression were compared at respective time points with or without prior EV exposure using two-way repeated-measures ANOVA followed by Bonferroni post-test for multiple comparisons. All statistical tests were performed using Graphpad version 9·2·0 and IBM SPSS Statistics. Differences were considered statistically significant at p < 0·05.
3. Results
3.1. Opposing seasonality of EV/RV and IVs in the hospitalized population
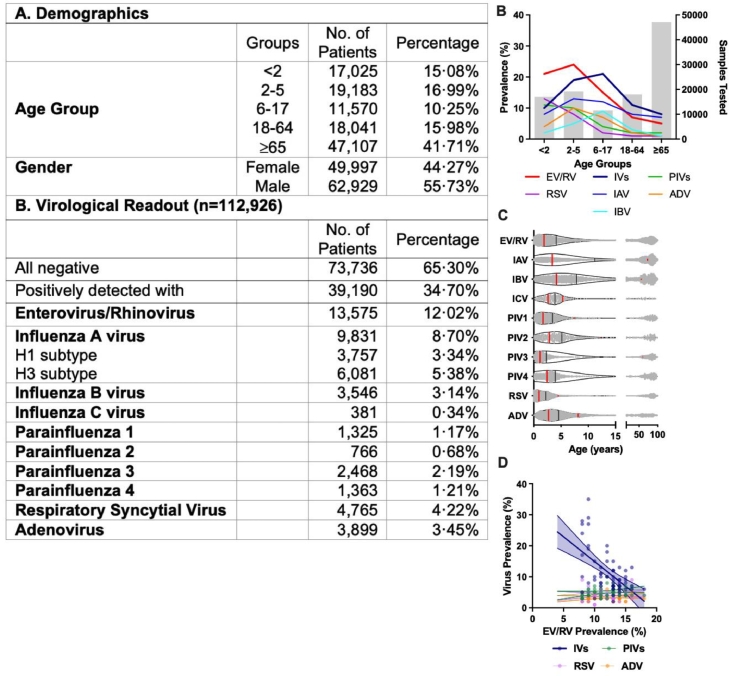
A total of 112,926 NPAs were included in this study. EV/RV was the most prevalent viral infection. It contributed a monthly positive rate of at least 8% throughout the study period (Fig. 1, red line). IVs were the second most dominant viral group being detected (Fig. 1, navy line). Combining influenza A, B and C viruses (IAV, IBV, and ICV), the positive rate of IVs reached up to 35% during flu season but remained below 10% low for the rest of the year. A strong seasonal pattern was observed in both EV/RV and IVs, with robust biannual peaks of EV/RV occurred, and one to two peaks of IV occurred in summers and winters of Hong Kong, yet the onset, magnitude, duration and dominating subtypes of the peaks varied extensively (Fig.1). A staggered pattern between EV/RV and IVs could be observed.
Fig. 1.
Temporal dynamics of respiratory virus prevalence in the inpatient cohort from September 2015 to December 2019. Monthly prevalence of individual virus infections (left y-axis) with the respective number of tested samples showed in gray bars (right y-axis) throughout the study period. EV/RV = enterovirus/rhinovirus; IVs = influenza viruses; PIVs = parainfluenza viruses; RSV = respiratory syncytial viruses; ADV = adenovirus. There are typically two influenza seasons in Hong Kong, winter peak from January to April and summer peak from July to August. The number indicates the ten influenza seasons within the study period.
3.2. Time series causality
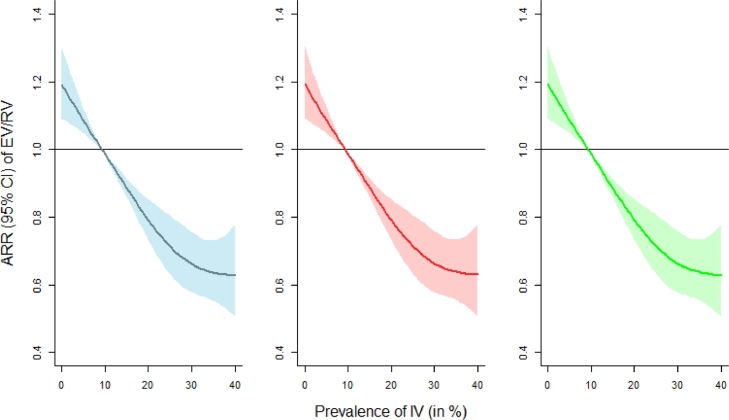
IV prevalence was significantly associated with the evolution of EV/RV (p < 0·001) and the effect of IV was highly significant at lag zero (p < 0·001), indicating a non-lagged interference between IV and EV/RV as assessed by the Granger causality test. The disease-disease association at lag zero was further examined via quasi-Poisson generalized additive model analysis and a significant negative association between IV and EV/RV was shown (Fig. 2). The ARR of EV/RV was 0·652 (95% CI: 0·571 to 0·745) when the prevalence of IV increased to 31·3% (i.e. 95th percentile of IV), whereas the ARR of EV/RV was 1·159 (95% CI: 1·079 to 1·244) when the prevalence of IV decreased to 1·5% (i.e. 5th percentile of IV), with a median reference value (9·3%).
Fig. 2.
Adjusted relative risks (ARRs) with 95% confidence interval on EV/RV against IV prevalence. The estimated ARRs without controlling meteorological effects, with ambient temperature plus relative humidity controlled, and with actual vapor pressure adjusted are expressed as blue, red, and green colors (from left to right), respectively. Median IV prevalence was used as the reference value for comparison. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Moreover, the negative relationship between EV/RV and IV was independent of meteorological variations from both ambient temperature & relative humidity and absolute humidity. After controlling the effect of temperature and relative humidity, the ARR of EV/RV was 0·654 (95% CI: 0·572 to 0·748) when the prevalence of IV was at its 95th percentile. An increase of lagged time of IV demonstrated a sinusoidal change in the ARR of EV/RV, highlighting a counteracting oscillation between the two viruses (Supplementary Figure 1).
3.3. Population level: a negative correlation between the prevalence of EV/RV with IVs
Apart from meteorological factors, interactions between viruses may be confounded by other factors such as the age of the subjects. Overall, 12·02% of the respiratory virus positive NPAs were EV/RV positive (Fig. 3A). When we analyzed the virological data by age stratification, EV/RV was the most prevalent in those aged between 2-to-5 (Fig. 3B), while the median age of IAV and IBV positive cases was significantly higher (11·19 and 7·84 years old, respectively) than that of EV/RV (4·01 years old) (Fig. 3C). Moreover, linear regression analysis revealed a significant negative correlation between the monthly prevalence of EV/RV against IVs (−1.60, p < 0·0001, Fig. 3D, navy) but not with other virus pairs, such as PIVs, RSV, and ADV. Pearson bivariate analysis showed a similar result in which a significant negative correlation was identified between EV/RV against total IVs (p = −0·477) or IAV (p = −0·421) (Table 1). Significant negative correlations between the monthly IVs prevalence and PIV2 (p = −0·428) and PIV4 (p = −0·109) were documented though their intensities were not as strong as those between IVs and EV/RV.
Fig. 3.
(A) Demographics of the patients included in the inpatient study from September 2015 to December 2019. Virological readouts were obtained from all 112,926 NPA samples for the detection of virus infection by multiplex PCR. Subtyping of H1 and H3 were performed after positive IAV detection. Due to the variation of the dominant IV species and subtypes each year, IAV-H1, IAV-H3, IBV, and ICV were combined as IVs for regression analyses. (B) Prevalence of individual virus infections of different age groups is represented by the color lines (left y-axis). The gray bar represents the number of samples tested in each group (right y-axis). (C) The age distribution of virus infections from test-positive 39,190 NPA samples presents individual data as gray dots. The black line shows the median age, and the red lines show the interquartile range. (D) Logistic regression between the prevalence of EV/RV with other viruses with 95% CI marked in dotted lines. Significant negative correlation (−1·60 ± 0·31, p < 0·0001*** with R2 = 0·3123) was identified only between EV/RV and IVs. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Correlation of viral prevalence in hospitalized subjects. Bivariate Spearman's cross-correlation coefficients (rs) between respiratory viruses using the monthly prevalence are shown. Asterisks indicate significance at p < 0·05*, p < 0·01** and p < 0·001***. IVs in the last column indicates the sum of IAV, IBV and ICV, the cross-correlation within the IVs and IAV, IBV and ICV are therefore excluded from the analysis. Red and blue values indicate a negative and positive correlation, respectively.
| p | IAV | IBV | ICV | PIV1 | PIV2 | PIV3 | PIV4 | RSV | ADV | IVs |
|---|---|---|---|---|---|---|---|---|---|---|
| EV/RV | −0·421** | 0·096 | −0·223 | 0·117 | 0·114 | 0·240 | 0·147 | 0·052 | 0·138 | −0·477*** |
| IAV | −0·217* | −0·025 | 0·126 | −0·350** | −0·184 | 0·042 | 0·078 | −0·120 | – | |
| IBV | 0·081 | −0·186 | −0·289* | 0·224 | −0·232 | −0·022 | −0·100 | – | ||
| ICV | −0·147 | 0·277* | 0·138 | 0·025 | −0·426** | −0·125 | – | |||
| PIV1 | 0·038 | 0·275 | 0·415* | −0·172** | −0·051 | 0·047 | ||||
| PIV2 | 0·098 | −0·053 | −0·178 | −0·090 | −0·428** | |||||
| PIV3 | 0·084 | −0·396* | −0·011 | −0·061 | ||||||
| PIV4 | −0·047 | 0·059 | −0·109** | |||||||
| RSV | −0·032 | −0·044 | ||||||||
| ADV | −0·199 |
3.4. Individual level: a reduced likelihood of EV/RV co-infected case with IVs at individual level
Co-infection of respiratory viruses was common in hospitalized patients. Overall, 6·58% (n = 2582) of the virus positive NPA samples were co-detected with two or more respiratory viruses, and 59·8% of these co-infection cases were contributed by EV/RV (n = 1545) (Fig. 4A). In our dataset, co-infection in children contributed over 80% of cases, except those with IAV (Fig. 4B). We then investigated this co-infected subset for the likelihood of virus co-detection. Interestingly, the odds to have both EV/RV-IVs detected in the same specimen was the lowest (OR = 0·15). It was also significantly lower when compared with 0·75 for EV/RV-PIVs, 0·54 for EV/RV-RSV and 0·94 for EV/RV-ADV co-detection using Fisher's Exact Test (Fig. 4C). A further reduction in odds to 0·09 (0·08 – 0·10) was observed between EV/RV and IVs after the adjustment of the confounding effect due to age and gender by binary logistic regression (Fig. 4D).
Fig. 4.
Co-infection statistics. (A) Rate of co-detecting more than one respiratory virus in different respiratory virus infection. The number of NPA samples detected with more than one pathogen detected was divided by the total number of the sample test-positive with the agent listed in each row. (B) Breakdown of co-infection cases according to age group. (C) The odd ratio of EV/RV and IVs co-infection with other respiratory pathogens using Fisher's exact test with null hypothesis assuming the likelihood of individual infection events were not interrelated. (D) Logistic regression analysis of EV/RV and IV infection after adjustment to gender and age group.
3.5. At cellular level: a suppression of IAV replication by a prior exposure to RV-A16 in HBECs and HNECs
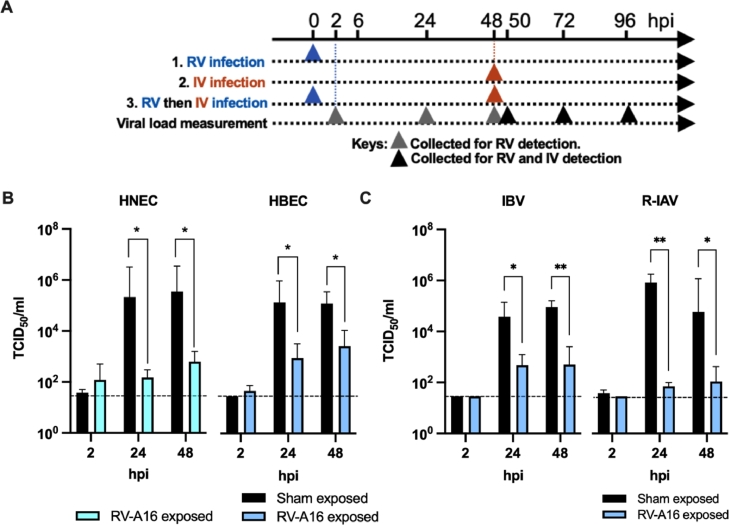
We Showed the productive replication of RV-A16 and seasonal IAV, IBV and R-IAV achieved a 2-log10 increase in viral titer at 48 hpi in our well-differentiated primary human respiratory epitheial cultures (Supplementary Fig. 2A and 2B). To address if viral interference between IV and RV occured at the cellular level, we performed sequential infection RV and IV as shown in Fig. 5A.
Fig. 5.
RVs suppress subsequent IVs infection in vitro. (A) Experimental plan of the model. Fully differentiated ALI cultures were first infected (or sham-exposed) with RV-A16 for 2 days, followed by secondary infection of IVs. Supernatant and cell lysates were collected at 2, 24 and 48 h post infection (hpi) for determination of viral load and gene expression. (B) Suppression of IAV titers with prior RV-A16 infection in HBEC (n = 7) and HNEC (n = 4). Y-axis represents the difference in titer in RV exposed to the sham-exposed cells. (C) Suppression of IBV and oseltamivir-resistance stain of IAV (R-IAV) in HBEC with prior RV-A16 exposure. Error bars showing the SEM of means and asterisks indicating significance of p < 0·05*, and p < 0·01** compared with sham treatment as examined by two way ANOVA followed by Bonferroni's multiple comparisons test.
With a prior exposure to RV-A16, the IV replication had a reduced viral titer of −3·16 log10 and −2·75 log10 in HNECs, and −2·18 log10 and −1·69 log10 in HBECs at 24 h and 48 h post IV virus infection, respectively, compared with those exposed to sham treatment (Fig. 5B). Similarly, significant reductions in the normalized matrix gene copies in the total mRNA collected from the cell lysate and in the viral matrix gene copies in the culture supernatant were observed (Supplementary Fig. 2C and 2D). Furthermore, RV exposure protected HBEC from IAV induced cell death. Extensive CPE developed in IAV-infected HBECs at 48 hpi, while no observable CPE was observed for at least 7 days if the cells were pre-exposed to RV-A16 before IAV infection (Supplementary Fig. 2E to 2G). Lactate dehydrogenase (LDH) cytotoxicity assay demonstrated the prolonged protective effect exerted by RV to the IV-infected cell (Supplementary Fig. 2H).
3.6. Suppression of IBV and R-IAV with prior exposure to RV-A16 in HBECs
The suppression by the prior exposure of RV-A16 was not limited to IAV. A significant reduction in the replication of IBV (1·91 log10, and 2·25 log10 at 24 hpi and 48 hpi, p < 0·05) and oseltamivir-resistant IAV strain (4·06 log10 and 2·73 log10 at 24 and 48hpi respectively, p < 0·01, Fig. 5C) was observed in RV-A16 exposed than sham-exposed HBECs.
3.7. Regulation of key pathways by RV to inhibit IAV replication
We examined the differential expressed genes (DEGs) induced in HBECs by RV-A16 and IAV compared with the sham-treated cells at 48 hpi. The RV and IAV commonly dysregulated 411 genes of which 380 were upregulated and 31 were downregulated when compared to the control sham HBECs. The top upregulated genes were found to be chemokines and cytokines (CXCL10, CCL5, CXCL9), interferons (IFNL1, IFNL2, IFNL3), interferon-induced proteins (IFIT1, IFIT2, IFIT3), nucleotidyltransferase (OAS1, OASL) and RNA processing factor (APOBEC3A) (Supplementary Fig. 3A). The enriched Reactome pathways including IFN and antiviral responses, innate sensing pathways, innate immunity and cell death (Supplementary Fig. 3B). In contrast, the downregulated genes (BBS9, DYNC2H1, UNC119B) were related to cilium assembly (R-HSA-56,178) (Supplementary Fig. 3C).
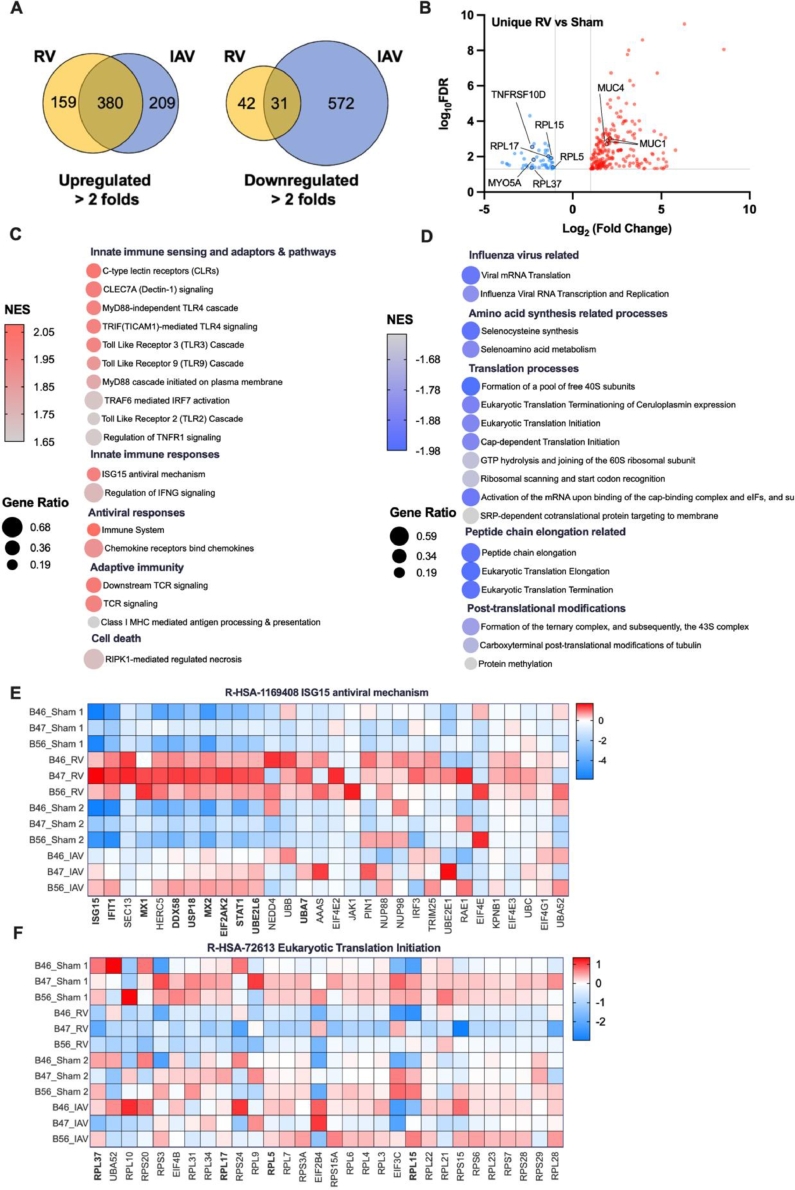
The DEGs induced by RV reflected the cellular environments that the IAV would encounter. The unique profile contributed by RV-A16 was selected for in-depth investigation. To characterize if specific classes of genes were dysregulated by RV infection and distinct from IAV infected cells, Venn analysis was carried out (Fig. 6A). RV-A16 uniquely upregulated mucin genes MUC1 and MUC4 (Fig. 6B) which encodes membrane-tethered mucins on the airways and provides anti-inflammatory function (Li et al., 2021) and are inhibitory to influenza virus infection (Kesimer et al., 2009). The enriched Reactome pathways identified by ranked gene list of the RV-A16 infected HBECs included innate immune sensing and adaptors and pathways, innate immune response, adaptive immunity (Fig. 6C) and antiviral responses (Fig. 6E). Moreover, Reactome pathways related to viral mRNA translation, amino acid synthesis, translation processes, peptide chain elongation and post-translational modifications were negatively enriched (Fig. 6D). Of particular interest, a group of ribosomal proteins (RPL5, RPL15, RPL17, RPL37), which generally promotes eukaryotic translation initiation (R-HSA-72613) (Fig. 6F) was suppressed by RV but not IAV.
Fig. 6.
RNA-seq analysis revealed signature gene sets enriched in HBECs upon RV infection. (A) Venn analysis showing the number of DEGs that are specifically or commonly enriched in RV or IAV infected cells when compared to respective sham controls. (B) Volcano plot displaying transcripts which were differentially expressed upon RV but not by IAV infection. Only transcripts with fold change >2 and FDR <0.05 were displayed. GSEA analysis showing top pathways that were (C) positively or (D) negatively enriched in RV-infected cells in contrast to IAV infected counterparts. Gene expression in selected Reactome pathways that were uniquely enriched upon RV infection. (E) Positive enrichment in ISG15 antiviral mechanism pathway. (F) Negative enrichment in eukaryotic translation initiation pathway.
4. Discussion
IV exerts a great burden on the health system each year in terms of frequent medical visits, hospitalization and IV-related death. We demonstrated the viral interference between RV and IVs using epidemiological data and biological experiments, suggesting a broad protective role of EV/RV in inhibiting subsequent IVs. We evaluated this interaction using the epidemiological data collected from hospitalized patients from September 2015, when the EV/RV test was first introduced as a routine test in the clinical settings, to December 2019, the last normal month before the SARS-CoV-2 pandemic began. In this study, a total of 112,926 NPAs obtained from all ages were examined. During these ten IV seasons, EV/RV prevalence oscillated in a counteracting manner. The negative association between EV/RV and IVs prevalence was independent of subject age and meteorological factors. Consistent with studies performed in different climatic parameters (Nickbakhsh et al., 2019; Wu et al., 2020), an interference effect at the population-level is suggested. We also demonstrated the competitive effect between RV and IV also operates at the individual level. Compared with other viruses, EV/RV had an exceptionally lower likelihood to be co-detected with IVs.
Referral bias may be a potential confounding factor in this study as the positive rate of respiratory viruses was higher in hospitalized patients than generalized public. Large scale study on virus prevalence and co-infection in asymptotic or healthy community is challenging considered the low detection rate of viruses except EV/RV. We extended our finding on the virus interference by comparing prevalence of our inpatient data with the figures from community surveillance program conducted by Center of Health Protection (CHP), the government of Hong Kong SAR. Sixty-four general out-patient clinics and fifty general practitioners in private practice were enrolled to this scheme as a better representation of the community. The seasonality pattern of virus prevalence was highly similar between inpatient and out-patient, and strong linearity was observed between the two for both EV/RV (0·79, p < 0·0001) and IVs (0·63, p < 0·0001). Similar suppression of IVs was observed as revealed using out-patient (−1·30, p < 0·001) or inpatient datasets (−1·57, p < 0·0001) (Supplementary Fig. 4). This suggest that the RV interference effect was also observable in community setting, though further investigation such as multivariate analysis nor co-infection statistics were not possible due to the lack of demographics of the dataset.
Our in vitro data validated the observation of viral interference at the population. RV-A16 replication significantly inhibited the influenza viral matrix gene expression and the productive replication. We attempted to investigate if any of the RV altered host genes would inhibit the replication of IAV. In our transcriptomic analysis, RV and IAV infection commonly upregulated genes related to innate sensing pathways, innate immune cytokine and chemokine signaling pathways, IFNs and antiviral signaling pathways, which agree with previous reports (Bui et al., 2019; Murphy et al., 2020; Chen et al., 2020). Using a similar dual infection model, Manel Essaidi-Laziosi et al. and Fage et al. studied the interference between RV, RSV, influenza and SARS-CoV-2 by simultaneous or sequential infection in the human ALI culture system (Essaidi-Laziosi et al., 2022; Fage et al., 2022). While both studies indicated the effect of influenza in suppressing the subsequent SARS-CoV-2 replication and both of them suggested that the viral interference was dependent on interferon pathways but not related to blockage of viral entry, the effect of SARS-CoV-2 in inhibiting the subsequent infection of influenza virus diverged in these two studies. Similarly, Essaidi-Laziosi et al. showed a conflicting result to our finding that RV-A16 was not able to interfere with influenza H1N1 infection Essaidi-Laziosi et al. (2020). This might be due to the different virus strains, virus preparation method and the calculation of the multiplicity of infection. Nevertheless, in vivo animal study by Gonzalez et al. (2018) and Van Leuven et al. (2021) provided the reproducible result that RV exposure can inhibition the infection of IAV (PR8) and attenuate the disease outcome through the type I IFN signaling pathway.
Moreover, the Reactome pathway involving cilium assembly (R-HSA-56178) was negatively enriched after RV and IAV infection. The impairment of the respiratory cilia would result in the alternation of cilia density, length, beating frequency and synchronization (Fu et al., 2018). Nevertheless, this result may also be a reflection of the ciliated cell detachment from the infected HBEC cultures after infection, as ciliated cells are the target cells of RV and IAV (Bui et al., 2019). The decreased proportion of ciliated cells on the RV-infected epithelium could be another possible reason for the attenuated IV infection in this in vitro system.
Furthermore, we explored if any of the DEGs in RV infected HBECs contributed to the suppression of the IAV life cycle. Though the initial mRNA synthesis is transcribed by viral ribosomal nucleoprotein associated polymerases together with cap snatching (Plotch et al., 1981), the import and export of viral ribosomal nucleoproteins (vRNPs), viral protein synthesis and virions assembly processes are entirely dependent on the cellular machinery of the host. Therefore, ribosomal proteins (RPs) and ribosomal RNAs play an essential role in the complete virus replication cycle. In our transcriptome result, RV specifically downregulated ribosomal proteins (RPL5, RPL15, RPL17, RPL37) which took part in the cap-dependent translation initiation (R-HSA-72737), influenza viral RNA transcription and replication (R-HSA-168273), formation of a pool of free 40S subunits (R-HSA-72689) and the GTP hydrolysis and joining of the 60S ribosomal subunit (R-HSA-72706). Of note, RPL5 is not only a chaperone of 5S rRNA but also a cellular interaction partner of influenza vRNPs (Mayer et al., 2007). The downregulation of such might negatively interfere its direct interaction with nucleophosmin and therefore hindering the maturation of 60S ribosomal subunits and the associated nuclear export (Yu et al., 2006). Besides, the downregulated MYH10 and TNFRF10D are found to be involved in the viral ribosomal nucleoprotein (vRNP) incorporation into virions (Watanabe et al., 2014) while the downregulated WNT9A gene can inhibit influenza A/WSN/33 and A/Hamburg/04/2009 viruses in the A549 lung epithelial cells (Karlas et al., 2010). Thus, the suppressed influenza virus replication by RV-A16 exposure were interfered at multiple stages in our HBECs system. An RNAi-based assay of the DEGs downregulated by RV-A16 would be a systematic next step to validate these hypotheses.
How viral interference is occurring at the population level would be our next exploration using dynamic models. Since RV infection occurs predominantly in younger children, at least in the hospital setting, how would this drive the interference to hamper influenza infection among other age groups warrants further investigation. In addition, precise molecules inherited from the EV/RV effect that is responsible for the viral suppression effect may serve as a novel antiviral defense on the respiratory mucosa against emerging outbreaks and shed light on the design of novel prophylactics design against a broad range of viral infections.
5. Limitations
In the population level study, data was from patients hospitalized in public hospital only, the pattern in the outpatient clinic and in the community was not assessed comprehensively. Moreoever, we have exclude data during the period of SARS-CoV-2 pandemic, as the implementation of social distancing measures and enhanced personal hygiene has a great effect in suppressing the transmissibility of enveloped viruses, e.g. Influenza virus (Chen et al., 2021). In the individual level study on viral co-infection, the data was solely from the multiplex PCR, whether the co-detected viral agents are actively infecting the patients, or a bystander were not defined. In addition, the clinical specimens analyzed in the epidemiological study were collected from the upper tract only and the pathogens contributing the disease in the lower tract only might be missed. Moreover, with the retrospective nature of this large inpatient dataset virological readout, the detection of EV/RV was expressed as one entity in the multiplex assay, and we were not able to perform further resolution for every specimen tested. In the cellular level study, we demonstrated that the inhibitory effect was significant for at least 48 h post RV exposure. The maximal viral interference window is yet to define. Moreover, the action of prior infection of IV to subsequent RV infection or the competition between RV and IV by adding the viruses at the same time point were not evaluated. The former could not be performed in our human primary respiratory epithelial cell culture, as the IV would cause a significant CPE even at a low MOI. Lastly, the current transcriptome analysis was done in the form of bulk sequencing, the contribution of individual epithelial cell types cannot be assessed. Their possible interaction with immune cells should be validated in the future study.
6. Conclusion
The cumulative evidence suggests the occurrence of viral interference at the population, individual and cellular levels. The understudied role of rhinovirus in providing the baseline immunity to influenza virus replication warrants further attention.
CRediT authorship contribution statement
Kin P. Tao: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Visualization, Writing – original draft, Writing – review & editing. Marc K.C. Chong: Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. Kathy Y.Y. Chan: Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. Jason C.S. Pun: Data curation, Investigation, Methodology. Joseph G.S. Tsun: Data curation, Formal analysis, Investigation, Methodology, Project administration. Samuel M.W. Chow: Investigation, Methodology, Writing – review & editing. Calvin S.H. Ng: Investigation, Methodology, Writing – review & editing. Maggie H.T. Wang: Investigation, Methodology, Writing – review & editing. Paul K.S. Chan: Investigation, Methodology, Resources, Writing – review & editing. Albert M. Li: Investigation, Methodology, Writing – review & editing. Renee W.Y. Chan: Conceptualization, Formal analysis, Funding acquisition, Supervision, Investigation, Methodology, Project administration, Resources, Visualization, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Renee WY Chan reports financial support was provided by University Grants Committee Research Grants Council. Renee WY Chan reports financial support was provided by Hong Kong Food and Health Bureau. Renee WY Chan reports financial support was provided by The Chinese University of Hong Kong Faculty of Medicine.
Acknowledgements
We would like to acknowledge Professor Gillian M Air of the University of Oklahoma Health Sciences Center who provides IAV strain A/Oklahoma/447/2008 H1N1 for the study; Drs Yan Wang, Steffi Long and Louisa LY Chan for culturing the human primary epithelial cells the study; Ms Waii WY Yu for her assistance in the molecular biology experiments.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.crmicr.2022.100147.
Appendix. Supplementary materials
References
- Bui C.H.T., Chan R.W.Y., Ng M.M.T., et al. Tropism of influenza B viruses in human respiratory tract explants and airway organoids. Eur. Respir. J. 2019;54(2) doi: 10.1183/13993003.00008-2019. [DOI] [PubMed] [Google Scholar]
- Burch J., Corbett M., Stock C., et al. Prescription of anti-influenza drugs for healthy adults: a systematic review and meta-analysis. Lancet Infect. Dis. 2009;9(9):537–545. doi: 10.1016/S1473-3099(09)70199-9. [DOI] [PubMed] [Google Scholar]
- Cao L., Lou J., Zhao S., et al. In silico prediction of influenza vaccine effectiveness by sequence analysis. Vaccine. 2021;39(7):1030–1034. doi: 10.1016/j.vaccine.2021.01.006. [DOI] [PubMed] [Google Scholar]
- Chen A.P., Chu I.Y., Yeh M.L., et al. Differentiating impacts of non-pharmaceutical interventions on non-coronavirus disease-2019 respiratory viral infections: hospital-based retrospective observational study in Taiwan. Influenza Other Respir. Viruses 2021. [DOI] [PMC free article] [PubMed]
- Chen Q., Tan K.S., Liu J., et al. Host antiviral response suppresses ciliogenesis and motile ciliary functions in the nasal epithelium. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.581340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.J., Arnold J.C., Fairchok M.P., et al. Epidemiologic, clinical, and virologic characteristics of human rhinovirus infection among otherwise healthy children and adults: rhinovirus among adults and children. J. Clin. Virol. 2015;64:74–82. doi: 10.1016/j.jcv.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essaidi-Laziosi M., Alvarez C., Puhach O., et al. Sequential infections with rhinovirus and influenza modulate the replicative capacity of SARS-CoV-2 in the upper respiratory tract. Emerg. Microbes. Infect. 2022;11(1):412–423. doi: 10.1080/22221751.2021.2021806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essaidi-Laziosi M., Geiser J., Huang S., Constant S., Kaiser L., Tapparel C. Interferon-dependent and respiratory virus-specific interference in dual infections of airway epithelia. Sci. Rep. 2020;10(1):10246. doi: 10.1038/s41598-020-66748-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fage C., Hénaut M., Carbonneau J., Piret J., Boivin G. Influenza A(H1N1)pdm09 virus but not respiratory syncytial virus interferes with SARS-CoV-2 replication during sequential infections in human nasal epithelial cells. Viruses. 2022;14(2):395. doi: 10.3390/v14020395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Tong J., Meng F., et al. Ciliostasis of airway epithelial cells facilitates influenza A virus infection. Vet. Res. 2018;49(1):65. doi: 10.1186/s13567-018-0568-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanville N., Johnston S.L. Challenges in developing a cross-serotype rhinovirus vaccine. Curr. Opin. Virol. 2015;11:83–88. doi: 10.1016/j.coviro.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Gonzalez A.J., Ijezie E.C., Balemba O.B., Miura T.A. Attenuation of influenza a virus disease severity by viral coinfection in a mouse model. J. Virol. 2018;92(23):e00881–e008818. doi: 10.1128/JVI.00881-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle W. Interference phenomena between animal viruses; a review. J. Immunol. 1950;64(3):203–236. [PubMed] [Google Scholar]
- Karlas A., Machuy N., Shin Y., et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463(7282):818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- Kesimer M., Scull M., Brighton B., et al. Characterization of exosome-like vesicles released from human tracheobronchial ciliated epithelium: a possible role in innate defense. FASEB J. 2009;23(6):1858–1868. doi: 10.1096/fj.08-119131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackenby A., Moran Gilad J., Pebody R., et al. Continued emergence and changing epidemiology of oseltamivir-resistant influenza A(H1N1)2009 virus, United Kingdom, winter 2010/11. Euro. Surveill. 2011;16(5) [PubMed] [Google Scholar]
- Li K., Cao P., McCaw J.M. Modelling the effect of MUC1 on influenza virus infection kinetics and macrophage dynamics. Viruses. 2021;13(5) doi: 10.3390/v13050850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer D., Molawi K., Martinez-Sobrido L., et al. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J. Proteome Res. 2007;6(2):672–682. doi: 10.1021/pr060432u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R.C., Lai Y., Barrow K.A., et al. Effects of asthma and human rhinovirus A16 on the expression of SARS-CoV-2 entry factors in human airway epithelium. Am. J. Respir. Cell Mol. Biol. 2020;63(6):859–863. doi: 10.1165/rcmb.2020-0394LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickbakhsh S., Mair C., Matthews L., et al. Virus-virus interactions impact the population dynamics of influenza and the common cold. Proc. Natl. Acad. Sci. U. S. A. 2019 doi: 10.1073/pnas.1911083116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opatowski L., Baguelin M., Eggo R.M. Influenza interaction with cocirculating pathogens and its impact on surveillance, pathogenesis, and epidemic profile: a key role for mathematical modelling. PLoS Pathog. 2018;14(2) doi: 10.1371/journal.ppat.1006770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotch S.J., Bouloy M., Ulmanen I., Krug R.M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23(3):847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- Self W.H., Williams D.J., Zhu Y., et al. Respiratory viral detection in children and adults: comparing asymptomatic controls and patients with community-acquired pneumonia. J. Infect. Dis. 2016;213(4):584–591. doi: 10.1093/infdis/jiv323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treanor J. Influenza vaccine–outmaneuvering antigenic shift and drift. N. Engl. J. Med. 2004;350(3):218–220. doi: 10.1056/NEJMp038238. [DOI] [PubMed] [Google Scholar]
- Van Leuven J.T., Gonzalez A.J., Ijezie E.C., et al. Rhinovirus reduces the severity of subsequent respiratory viral infections by interferon-dependent and -independent mechanisms. mSphere. 2021;6(3) doi: 10.1128/mSphere.00479-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Kawakami E., Shoemaker J.E., et al. Influenza virus-host interactome screen as a platform for antiviral drug development. Cell Host Microbe. 2014;16(6):795–805. doi: 10.1016/j.chom.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Mihaylova V.T., Landry M.L., Foxman E.F. Interference between rhinovirus and influenza A virus: a clinical data analysis and experimental infection study. Lancet Microbe. 2020;1(6):e254–e262. doi: 10.1016/s2666-5247(20)30114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Maggi Jr L.B., Brady S.N., et al. Nucleophosmin is essential for ribosomal protein L5 nuclear export. Mol. Cell. Biol. 2006;26(10):3798–3809. doi: 10.1128/MCB.26.10.3798-3809.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.