Abstract
Recent studies suggest that bacterial abundance and species diversity in the ocean's water column are variable at the millimeter scale, apparently in response to the small-scale heterogeneity in the distribution of organic matter. We hypothesized that bacterium-bacterium antagonistic interactions may contribute to variations in community structure at the microscale. We examined each of the 86 isolates for their inhibition of growth of the remaining 85 isolates by the Burkholder agar diffusion assay. More than one-half of the isolates expressed antagonistic activity, and this trait was more common with particle-associated bacteria than with free-living bacteria. This was exemplified by members of the α subclass of the class Proteobacteria (α-proteobacteria), in which production of antagonistic molecules was dominated by attached bacteria. We found that γ-proteobacteria (members of the orders Alteromonadales and Vibrionales) are the most prolific producers of inhibitory materials and also the most resilient to them, while members of the Bacteriodetes were the organisms that were least productive and most sensitive to antagonistic interactions. Widespread interspecies growth inhibition is consistent with the role of this phenomenon in structuring bacterial communities at the microscale. Furthermore, our results suggest that bacteria from pelagic marine particles may be an underutilized source of novel antibiotics.
The pelagic ocean is replete with physical and chemical gradients at microscales that may be important in creating microniches that maintain high species richness and diversity (discussed in detail in reference 6; R. A. Long and F. Azam, submitted for publication). The detection of high levels of colloids, submicrometer particles, and transparent organic particles (2, 29, 32, 52) and the dynamic nature of polymers and colloids that form a gel matrix (13) have changed our perception of the aqueous environment that is the ocean's water column. This matrix forms the physical context for bacterium-bacterium interactions.
The perception that microbes are homogeneously distributed in seawater is changing to a perception that microbes are distributed heterogeneously (6). Bacterial abundance is now known to vary at the millimeter scale (42). Bacterial species richness is also variable at the millimeter scale (Long and Azam, submitted), and the variability increases in response to increases in the concentration of particulate organic matter in seawater. In light of the heterogeneous distribution and potential for spatial structuring of bacterial populations, we considered antagonistic interactions involving growth inhibition as one mechanism that may cause and maintain millimeter scale variations in the patterns of bacterial species composition.
Bacterium-bacterium antagonistic interactions involving antibiotics are well documented in soils. In situ production of antibiotics in soil has been detected only in association with organically rich microenvironments (e.g., seeds, rhizospheres, and straw fragments in soil) (9, 47, 55, 56). Organically rich microenvironments in the marine pelagial environment include phytoplankton and zooplankton and their detritus, as well as marine snow particles that range in size from millimeters to centimeters and are nutrient-rich microscale hot spots. Nair and Simidu (36) found that isolates derived from marine particles were more effective than free-living bacteria in inhibiting the growth of two human-pathogenic bacteria. Studies examining the frequency of antagonistic interactions of marine bacterial isolates have found that 5 to 8% of the isolates express some level of activity (30, 36, 37). While these studies focused on marine isolates as potential producers of antibiotics, nonmarine bacteria, often human pathogens, were employed as the target species. Similarly, our knowledge of the sensitivity of environmental isolates to antibiotics is more limited than our knowledge of production of antibiotics. Classical antibiotics (e.g., penicillin and kanamycin) have different levels of inhibition with pigmented and nonpigmented marine bacteria (35). Plasmids carrying genes for resistance to antibiotics have been found in marine Vibrio species and are thought to be laterally exchanged (31). However, previous studies relied on antibiotics that were not derived from indigenous species, so we lack information concerning interactions between marine bacteria and their potential antagonistic products. As a result, the conclusions drawn from such studies (e.g., that only 5 to 8% of marine isolates produce antagonistic substances) cannot a priori be extrapolated to address our question concerning the interactions in an ecological context (see below).
In mesotrophic and eutrophic waters or during phytoplankton blooms, heterotrophic bacteria on particles can account for large fractions of the bacterial activity (e.g., ectoenzymatic hydrolysis of organic particles and polymers and utilization of organic matter for respiration and growth) (40, 43, 44). The cell-specific levels of activity of particle-attached bacteria are often 2 to 3 orders of magnitude greater than those of cooccurring, free-living bacteria (44). Attached bacterial hydrolytic enzymatic activity significantly influences the quantity and quality of biogenic matter that sinks from the upper water column into the ocean's depths (43). Since different bacteria express different arrays of hydrolytic enzymatic activities (33), changes in the bacterial species composition, such as those potentially caused by microscale antagonism, could alter the hydrolytic activity exerted by bacteria on organic particles. Furthermore, the species richness and diversity on particles could be influenced by bacterium-bacterium antagonisms, and in turn, this could affect the nature and rates of biogeochemical transformation of the particles. Thus, bacterium-bacterium antagonisms could be important variables in the ecology of pelagic bacteria and in bacterium-mediated carbon cycling in the ocean.
Recent studies in which small-subunit ribosomal gene sequence comparison was used as the basis for phylogenetic analyses demonstrated that there are distinct differences in species composition between free-living and attached bacteria. DeLong et al. (15) found that the marine snow population consisted predominately of members of the phylum Bacteroidetes (20a) that were distinct from the bacteria in the surrounding water. In the Chesapeake Bay, the dominant particle-attached bacteria were found to be different from the dominant free-living bacteria (8).
In this study, we examined whether bacterial antagonism is a common phenomenon in the pelagic ocean. We also tried to determine whether particle-attached bacteria are more likely than free-living bacteria to use chemical antagonism as a potential mechanism to maintain distinct microscale populations and/or to cause succession of species on particles. Furthermore, we characterized isolates by 16S rRNA phylogenetic analysis to determine if the ability to use chemical antagonism is concentrated in a specific group(s) of cultivable pelagic bacteria that may be specialized for antagonistic interactions.
MATERIALS AND METHODS
Isolation and culture of bacteria.
Most bacterial isolates were collected from southern California coastal waters (Table 1). The natural assemblage was sampled off the pier at Scripps Institution of Oceanography, La Jolla, Calif., and approximately 1 km due west of the pier. Seawater was also collected from these sites and from the ocean off the University of California, Santa Barbara, and used for mesocosm enrichment with phytoplankton nutrients (F/2 medium) (1, 24) to stimulate diatom blooms. Bacterial strains were isolated after the peak of the blooms, when aggregates were readily visible. We operationally defined attached or particle-associated bacteria as bacteria that were isolated from visible particles in our samples; thus, this situation met the definition of marine snow (3). The particles were individually picked and rinsed with filtered (pore size, 0.22 μm) seawater several times prior to plating of the samples (see below). Free-living bacteria were defined as either bacteria that were gravity filterable through a 1.0-μm-pore-size filter or bacteria that were isolated in bulk or unfiltered seawater that contained no visible particles. As stated above, the distinction between attached bacteria and free-living bacteria was operational; it is possible that the free-living isolates included some attached isolates that were dislodged from particles during sampling or were progeny of attached bacteria released into seawater. It is less likely that the reverse occurred since particles were individually picked and thoroughly rinsed to remove free-living contaminants.
Pelagic bacteria were isolated on ZoBell 2216E plates. Each liter of seawater was filtered with GF/F glass fiber filters (nominal pore size, 0.7 μm; Whatman, Maidstone, England) and contained 5 g of peptone, 1 g of yeast extract, and 15 g of of Bacto Agar; FeCl3 was not included in the medium. Isolates were initially grown at the ambient sea surface temperature at the time of isolation, which ranged from 12.5 to 25°C. Colonies were picked after 1 to 3 days of incubation. Bacterial isolates were streaked a minimum of three times until a monospecific culture was obtained, based on uniform colony morphology and color. Isolates were maintained as frozen stock preparations. The bacterial isolates used for the Burkholder assay (see below) were taken from the frozen stock preparations and streaked on ZoBell 2216E plates. The isolates were then grown in ZoBell 2216E broth (containing [per liter of GF/F-filtered seawater] 5 g of peptone and 1 g of yeast extract) without FeCl3 at room temperature (20 ± 2°C) on a rotary shaker at 100 rpm. Isolate suspensions were adjusted to an optical density at 600 nm (OD600) of 1 with GF/F-filtered autoclaved seawater.
Screening of isolates for inhibitory interactions.
We used the Burkholder agar diffusion assay (10) to screen for inhibitory interactions. A lawn of a target isolate was prepared by mixing 2.5 ml of molten (44°C) 0.6% ZoBell agar with 30 μl of a suspension of the isolate (OD600, ∼1), immediately vortexing the preparation for ∼1 s, and pouring the agar onto a ZoBell plate. In a 3 × 3 matrix, 10-μl portions (OD600, ∼0.1) of nine potential producers were then spotted on the lawn. The plates were incubated face up for 6 days at room temperature (20 to 24°C) and examined daily for zones of inhibition (areas where the target isolate failed to grow). Potential producers were considered positive when the diameter of the zone of inhibition was at least 4 mm greater than the diameter of the colony formed by the potential producer. Isolates that were positive were tested again to confirm the initial observation. If ambiguous results were observed in the first two assays, a third set of assays was performed. In all such cases the third assay failed to detect inhibition; therefore, the isolates in these cases were not considered producers.
Sequencing of the 16S rRNA gene of bacterial isolates.
One microliter of a bacterial suspension was transferred to a sterile thin-wall PCR tube. Five microliters of Lyse-N-Go (Pierce, Rockford, Ill.) was added to the PCR tube, and the thermocycling protocol for lysis was carried out according to the manufacturer's instructions (65°C for 30 s, 8°C for 30 s, 65°C for 90 s, 97°C for 180 s, 8°C for 60 s, 65°C for 180 s, 97°C for 60 s, and 65°C for 60 s, followed by incubation at 80°C until the PCR reagents were added). Upon completion of the lysis cycle, 44 μl of a PCR master mixture (2 U of Qiagen Taq DNA polymerase, each primer at a concentration of 1 μM, each deoxynucleoside triphosphate at a final concentration of 0.8 mM) was added to the tube, and a touchdown PCR was performed with universal primer 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and eubacterium-specific primer 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) to amplify the majority of the 16S rRNA gene (51).
An approximately 1,460-bp segment of the 16S rRNA gene was amplified by using a modification of the touchdown PCR protocol (16). An initial denaturing step consisting of 94°C for 5 min was followed by 30 cycles of amplification (3 min of denaturation at 94°C; 1 min of annealing with a temperature of 65°C for the first cycle which was then reduced 0.5°C per cycle to 50°C; 3 min of extension at 72°C) and then by five additional cycles of amplification (3 min at 94°C, 1 min at 50°C, 3 min at 72°C) and a final extension step consisting of 10 min at 72°C. The PCR products were separated by electrophoresis on a 0.8% agarose gel and stained with ethidium bromide to confirm that the approximately 1,460-bp product was present. Sequencing was performed by using ABI BigDye chemistry with an automated ABI DNA sequencer and 27F primer. This resulted in data for approximately 500 bp. The sequences obtained were aligned with those in the Ribosome Database Project by using BLAST (4). A phylogenetic tree was generated by using the neighbor-joining method with NJPlot, based on alignments from CLUSTAL W (48). The bootstrap values obtained were from 100 iterations.
Statistics.
To test for heterogeneous production of inhibitory molecules by isolates, we used the Fisher exact test (20) for analysis of a 2 × 2 contingency table:
 |
where f is the observed characteristics, R1 = f11 + f12, C1 = f11 + f21, and n = R1 + R1 = C1 + C2. P is the probability that the table is random, as calculated by:
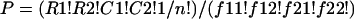 |
The two-sided t test with unequal variance was used to compare means for production and sensitivity of the various classes.
Nucleotide sequence accession numbers.
The nucleotide sequences of the isolates sequenced in this study have been deposited in the GenBank database under the following accession numbers: N67, AF365990; BB2AT3, AF365991; BBAT1, AF365992; BBAT2, AF365993; BBAT3, AF365994; BBAT4, AF365995; BBFL1, AF365996; BB2AT1, AF365997; SB11, AF365998; N43, AF365999; S3, AF366000; SB15, AF366001; SB12, AF366002; SB9, AF366003; SB17, AF366004; BBFL7, AF366005; LHAT9, AF366006; SWFL2, AF366007; AF1, AF366008; DC, AF366009; JSL61, AF366010; LHAT1A, AF366011; LHAT4, AF366012; LHAT5, AF366013; LHFL4, AF366014; SB10, AF366015; SB13, AF366016; SB16, AF366017; SB3, AF366018; SB5, AF366019; SB7, AF366020; SWAT2, AF366021; SWAT3, AF366022; SWAT8, AF366023; SWAT4, AF366024; SWFL1, AF366025; LHAT7, AF366026; LHAT2, AF366027; JSL101, AF366028; LHFL1, AF366029; JSL91, AF366030; BB2FL1, AF366031; BBFL4, AF366032; JSL121, AF366033; JSL122, AF366034; S5, AF366035; SB18, AF366036; SB2, AF366037; SWAT1, AF366038; SWAT7, AF366039; SWAT9, AF366040; LHAT3, AF366041; BB2AT2, AF366042; BB2FL3, AF366043; BB2FL2, AF366044; SB6, AF366045; SB8, AF366046; SB14, AF366047; BBFL3, AF366048; SB4, AF366049; and SWAT5, AF366050.
Nucleotide sequences of the following organisms were obtained from other studies (22; L. B. Fandino and F. Azam, unpublished data): GAI5 (accession number AF007256), GAI21 (AF007257), GAI16 (AF007258), EE36 (AF007254), SP18 (AJ276036), SP25 (D88520), EL1 (AB010981), EL2 (AF025556), 5.3.10 (AF367849), AA (AF025554), AC (AB008046), AG (AF367853), B12 (AF125327), BA (AF367854), BD (AY030102), BE (AY030102), Cytophaga lytica (M62796), D8 (AF125323), F12 (AF125325), F27 (AY030100), and G20 (AF125324).
RESULTS
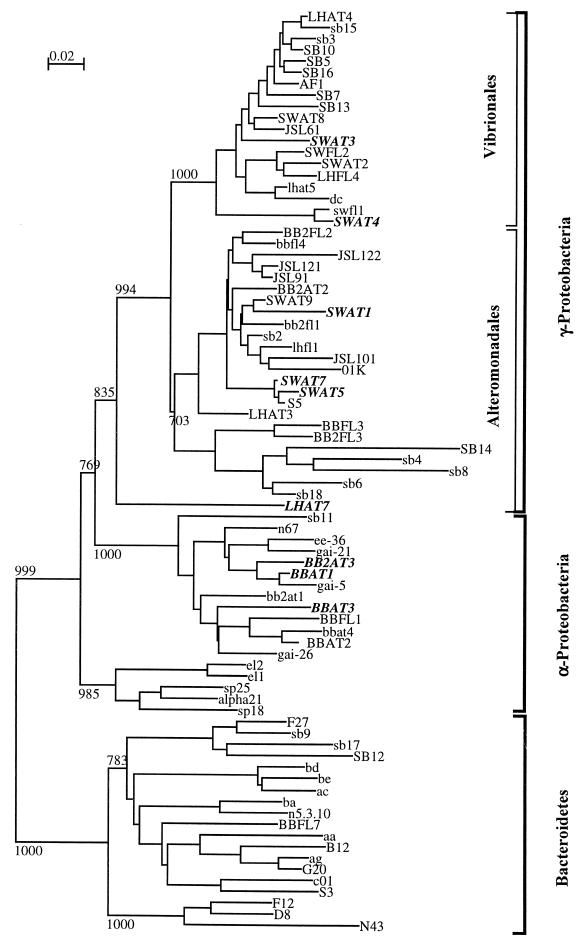
Eighty-six unique bacterial isolates representing four classes of cultivable marine bacteria were examined to determine their production of antibiotics and their sensitivity to antibiotics produced by one another. Thus, we used an 86 × 86 array of tests (7,396 tests) to study inhibitory interactions. The isolates were grouped in the α subclass of the class Proteobacteria (α-Proteobacteria), the Bacteroidetes (primarily the class Sphingobacterium), and the orders Alteromonadales and Vibrionales (the latter two groups are orders in the γ-proteobacteria) based on 16S rRNA gene sequence analysis data (Fig. 1). These groups represent the predominant cultivable marine pelagic bacteria (21). The bacteria isolated were either free-living or attached. Comparisons were made on the basis of source of isolation (i.e., free-living versus particle) and on the basis of phylogenetic relationships.
FIG. 1.
Phylogenetic tree of isolates. Phylogenetic relationships of isolates were inferred from a comparison of an approximately 500-bp region of the 16S rRNA gene sequence by using CLUSTAL W for multiple alignments and NJPlot for construction of the tree. The bootstrap values at the nodes are based on 1,000 iterations. Boldface type indicates producers that inhibited 10 or more isolates, lightface uppercase type indicates nonproducers that inhibited one to nine isolates; and lightface lowercase type indicates nonproducers.
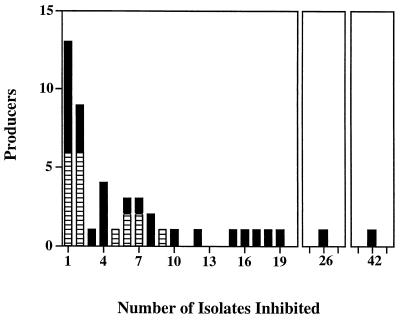
Production of inhibitory compounds was found in 53.5% of the isolates. The producers inhibited a mean of 6.4 isolates per producer. While the majority of producers inhibited only one or two isolates, there were nine isolates that inhibited 10 or more isolates (Fig. 2). The zones of inhibition varied and had diameters that ranged from 4 to 30 mm.
FIG. 2.
Range of inhibitor producers. Eighty-six marine bacteria were examined to determine their inhibitory interactions with one another by using the Burkholder assay on ZoBell 2216E plates. Inhibition was considered to be positive if the diameter of the zone of inhibition was at least 4 mm greater than the diameter of the colony of the producer. Inhibition had to be observed at least twice to be considered positive. Solid columns, particle-attached bacteria; cross-hatched columns, free-living bacteria.
Production of inhibitory compounds by attached and free-living bacteria.
A significantly greater percentage of attached bacteria than of free-living bacteria produced inhibitory compounds (66.7 and 40.9%; Fisher exact test, P = 0.01). Not only did more attached bacteria cause inhibition, but these bacteria tended to inhibit more isolates. Attached bacteria accounted for all of the isolates that inhibited 10 or more isolates (Fig. 2). The mean number of isolates inhibited by attached bacteria was 8.5, compared to 3.2 for free-living bacteria.
When we examined individual isolate-isolate interactions between attached bacteria (n = 42) and free-living bacteria (n = 44), we found that the attached bacteria inhibited the free-living bacteria in 161 of 1,848 cases (8.7%) (Table 2). However, many fewer cases of free-living bacteria that inhibited attached bacteria were observed (7 of 1,848 cases [0.4%]) (Table 2). Attached isolates were derived from several different systems or sources. All seawater attached (SWAT) isolates from particles found in the mesotrophic water column had some capacity to inhibit the growth of other isolates. The SWAT isolates inhibited an average of 16.8 isolates, and these isolates accounted for five of the nine isolates that inhibited 10 or more isolates. One isolate, SWAT3, inhibited nearly one-half of the isolates examined. All of the SWAT isolates were found to be γ-proteobacteria (Fig. 1).
The attached isolates from mesocosms that had been enriched with F/2 medium (22) to generate diatom blooms (BBAT and BB2AT isolates) were predominately α-proteobacteria. Five of seven of these isolates inhibited other isolates and inhibited a mean of 10.2 targets. Three of the isolates inhibited 10 or more isolates (Fig. 2). The bacteria isolated from abandoned larvacean houses (LHAT isolates) included a similar percentage of producers; however, the producers were not as potent, inhibiting a mean of 5.8 targets. One LHAT isolate inhibited 10 or more targets.
The last group of attached isolates (SB isolates) came from a 1,200-liter mesocosm (1). Only 8 of 18 isolates had inhibitory properties, and they inhibited a mean of 2.8 targets. These isolates were distributed throughout the phylogenetic groups examined.
Production of inhibitory compounds: phylogenetic comparison.
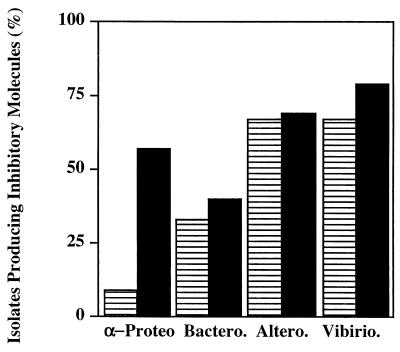
Bacterial isolates were placed into phylogenetic groups based on the 16S rRNA gene sequence analysis. Additional phylogenetically identified isolates from our culture collection of pelagic bacteria were added to the test pool in order to balance the distribution of isolates in the four phylogenetic groups. Inhibitory activity was not equally spread throughout the four groups. Members of the Alteromonadales and Vibrionales were the dominant producers, while members of the Bacteroidetes were not as prolific (Fig. 3).
FIG. 3.
Production of inhibitory molecules by members of different phylogenetic groups. Eighty-six marine bacterial isolates belonging to the α-proteobacterial (α-Proteo), Bacteroidetes (Bactero.), Alteromonadales (Altero.), and Vibrionales (Vibirio.) groups were examined for production of inhibitory molecules that were active against at least one other isolate. Cross-hatched columns, free-living bacteria; solid columns, particle-associated bacteria.
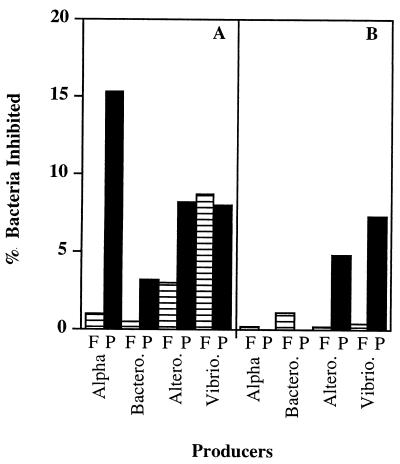
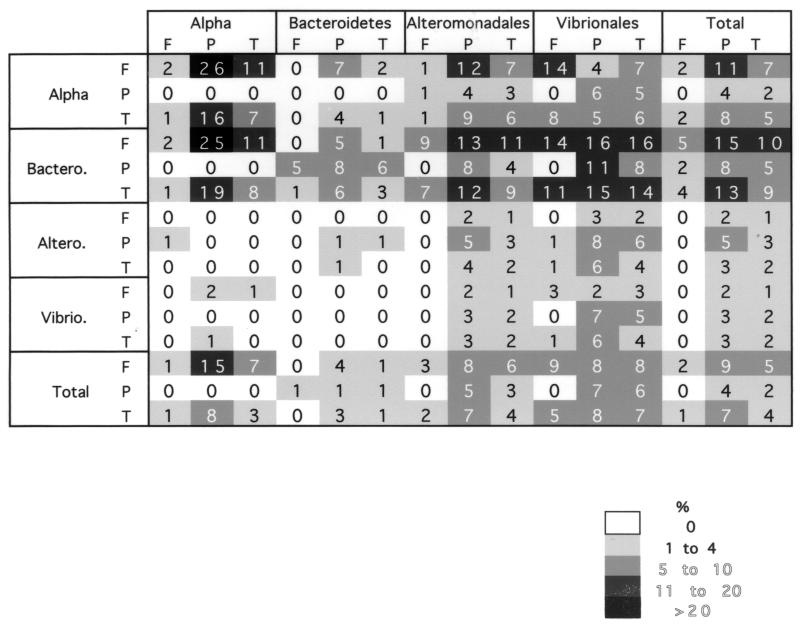
Isolates belonging to the Alteromonadales and Vibrionales expressed inhibitory activity against members of all four groups examined (Table 2). When the data were expressed as percentages of producers, there was no difference between the free-living and attached isolates belonging to these groups (Fisher exact test; P = 0.31 and P = 0.35 for the Alteromonadales and Vibrionales, respectively). However, the attached bacteria belonging to these groups inhibited more targets than the free-living bacteria (Fig. 4). Inhibition by attached members of the Alteromonadales was nearly four times more common than inhibition by free-living members of the Alteromonadales when isolate-isolate interactions were examined. For the Vibrionales the relationship was similar, and the ratio approached 2:1 (Table 2). Members of the Vibrionales accounted for 67% of the broad-range inhibitors (Fig. 2).
FIG. 4.
Inhibition of free-living and particle-attached bacteria according to phylogenetic groups. Eighty-six marine bacterial isolates belonging to the α-proteobacterial (Alpha), Bacteroidetes (Bactero.), Alteromonadales (Altero.), and Vibrionales (Vibrio.) groups were examined for sensitivity to inhibitory molecules produced by at least one other isolate. (A) Free-living targets; (B) particle-associated targets. Cross-hatched columns, free-living producers (F); solid columns, particle-associated producers (P).
α-Proteobacterial isolates were predominantly active against α-proteobacteria and members of the Bacteroidetes. The producers belonging to this group were predominately particle-associated bacteria (Fisher exact test, P = 0.04) which inhibited large numbers of the other isolates. When producers and nonproducers were placed in a phylogenetic tree, there was no clear clustering of producers and nonproducers (Fig. 1). In the case of individual interactions, attached α-proteobacteria were 13 times more likely to have antagonistic interactions than free-living α-proteobacteria were (Table 2).
The Bacteroidetes group had the lowest percentage of producers as determined by our assay system. The inhibitory activity was similar to that of the α-proteobacteria and was effective against α-proteobacteria, members of the Bacteroidetes, and two members of the Alteromonadales (Table 2). There was no discernible difference between the percentages of producers when attached and free-living members of the Bacteroidetes were compared (Table 2).
Sensitivity to inhibitory compounds.
Nearly 84% of all isolates were sensitive to one or more producers. There was no significant difference in the sensitivities of attached and free-living isolates in terms of the percentages of bacteria inhibited (79.5 and 88.1%, respectively) (Table 2) (Fisher exact test, P = 0.61). This general relationship was also true within each phylogenetic group.
Members of the Bacteroidetes were the most sensitive bacteria; they were inhibited by members of all four groups. On average, each Bacteroidetes isolate was sensitive to 8.2 producers. Free-living members of the Bacteroidetes were significantly more sensitive than attached members of the Bacteroidetes were; on average, members of these two groups were inhibited by 9.8 and 4.0 producers, respectively (two-sided t test with unequal variance, P = 0.045).
α-Proteobacteria were less sensitive than members of the Bacteroidetes. Similar percentages (90.1 and 85.7%) of free-living and attached α-proteobacteria were sensitive to one or more producers. However, attached α-proteobacteria were sensitive to fewer producers than free-living α-proteobacteria (2.0 and 6.4 producers, respectively; two-sided t test with unequal variance, P = 0.001).
The Alteromonadales and Vibrionales groups were the least sensitive groups; 73.7 and 75.0% of the isolates were sensitive. When the particle-attached members of these two groups were examined, 76.9 and 87.5% were sensitive to one or more isolates that produced an inhibitory compound. These values were not significantly different from the values for the free-living members, 66.7 and 58.3% of which were sensitive (Fisher exact test, P > 0.05). The levels of sensitivity as determined by the number of isolates that inhibited a target showed that attached members of the Alteromonadales were more sensitive than free-living isolates (2.5 versus 1.3 isolates; t test, P = 0.005). There was no significant difference within the Vibrionales group. It should also be noted that none of the Bacteroidetes or α-proteobacterial isolates affected any member of the Vibrionales group (Table 2).
DISCUSSION
Comparison with previous studies.
A remarkably large fraction (53.5%) of our marine bacterial isolates exhibited antagonistic properties against other pelagic bacteria. Thus, bacterium-bacterium antagonism is potentially very common in the pelagic ocean. In contrast to our study, previous studies found much lower percentages of bacterial antagonism (5 to 8%) (30, 36). This may be because we used a wide array of marine bacterial isolates as target organisms, while in previous studies the initial screening was often limited to few target species. Also, unlike previous studies, we examined the distribution of antibiotic production and the sensitivity of marine bacteria in the context of 16S rRNA phylogenetic relationships. We also note that it is not necessary for an inhibitor to be an antibiotic. For instance, some inhibitors may be involved in bacterium-bacterium communication (e.g., quorum sensing) (38) that regulates metabolism or physiology (53) and can alter the species composition of the bacterial community on particles. Quorum- sensing molecules are known to exert not only intraspecies control but also interspecies control on growth and the expression of specific phenotypes (7). Inhibition may also reflect changes in the availability of nutrients (e.g., iron through production of siderophores) or alteration of the pH of the environment.
Particle-attached versus free-living bacteria.
Our findings suggest that particle-attached bacteria are more likely to produce inhibitory compounds than their free-living counterparts. This agrees qualitatively with the results of Nair and Simidu (36), who found that a greater percentage of particle-derived bacteria than of free-living bacteria produced inhibitory compounds (6.9 to 12.9 and 3.2 to 5.6%, respectively). However, we found 5- to 10-fold-higher frequencies of inhibition than those found in the study of Nair and Simidu (36). The high frequency of producers among the particle-attached isolates suggests that bacterium-bacterium antagonism in the pelagic ocean may be more common on particles. This possibility has implications for strategies for searching for new antibiotics of marine bacterial origin.
In addition to being more likely to produce inhibitory molecules, particle-attached isolates either produced more inhibitory compounds than free-living bacteria or produced compounds that inhibited more species than the compounds produced by free-living bacteria. While we have not characterized the inhibitory molecules, it is not uncommon for a single species to produce multiple inhibitory compounds (54). Thus, our attached bacterial isolates produced all of the broad-range inhibitors, and the mean number of isolates inhibited by the attached bacteria was greater than the mean number of isolates inhibited by the free-living bacteria.
Whether particle-attached bacteria use inhibitory molecules to prevent other bacteria from colonizing particles is an important ecological and biogeochemical question. Our results show that the ability of particle-attached bacteria to chemically inhibit free-living bacteria is greater than the ability of free-living bacteria to chemically inhibit particle-attached bacteria. Antibiotic production may be a mechanism used by particle specialists to dominate the particle phase by deterring other potential colonizers. This is consistent with the conclusion, based on 16S rRNA sequence analyses, that marine snow displays bacterial species dominance distinct from that in the surrounding seawater (15). Variations in bacterial species dominance can strongly influence the nature and rate of degradation of sinking organic matter (e.g., through variations in species-specific ectohydrolase profiles) (33). Thus, bacterium-bacterium antagonisms on particles may be important variables in the ecology of marine bacteria, as well as in oceanic carbon cycling.
The specific particle source from which the bacteria were isolated also influenced the level of bacterium-bacterium antagonism. While the majority of isolates from seawater enrichment mesocosms produced inhibitors, it was the SWAT isolates, which were obtained from particles collected directly from the ocean, that appeared to have the most potent arsenal. All of these isolates were inhibitory, and they accounted for the majority of the broad-range inhibitors. While interesting, our data are too limited to make a generalization or to offer an explanation for the differences.
Phylogenetic relationship.
A novel aspect of this study was that we used 16S rRNA-based phylogeny, as well as a large number of marine target isolates, to examine whether members of certain phylogenetic groups were more likely to be antibiotic producers or particularly sensitive to inhibition. The γ-proteobacteria (the Alteromonadales and Vibrionales groups) in our study were found to be the dominant producers. This is consistent with most previous studies which showed that production of antibiotics by marine bacterial isolates from seawater is due to γ-proteobacteria (26). Members of the Bacteroidetes were the least prolific producers in our study. Most noticeably absent among the producers in our study were members of the Actinobacteridae. While these organisms are common antibiotic producers in soil and marine sediment (26), they are not commonly found in the water column (21).
Members of the γ-proteobacterial groups Vibrionales and Alteromonadales (which were the dominant producers) were the least sensitive to inhibition in our study. Thus, they may dominate the particle phase and biofilms in the ocean. This observation may also be relevant to the discussion of the culturability of marine bacteria. Giovannoni and Rappé (21) noted that γ-proteobacteria are the dominant cultivable marine bacteria. Their resilience to antibiotics and their ability to produce antibiotics may account for the bias towards these groups observed in marine isolate libraries. One might speculate that the production of antibiotics on isolation plates by γ-proteobacteria may, in part, limit the formation of colonies of other bacteria. A number of mechanisms of bacterial resistance to antibiotics have been documented, including hydrolytic inactivation of inhibitors (e.g., β-lactamases) (11), alteration of the target molecule (46), prevention of inhibitor influx into the cell, and the efflux of inhibitor from the cell. Multidrug transporters are pumps that remove a variety of toxic molecules (for a review, see reference 41). While these mechanisms have not been studied in the marine environment sufficiently to understand their distribution or occurrence, the last two have been reported for the marine bacteria Vibrio parahaemolyticus and Vibrio cholerae (14, 34). Whether marine γ-proteobacteria are better equipped with these or other antibiotic resistance mechanisms has not been examined.
We did not expect to find that members of the Bacteroidetes are so sensitive to inhibition by other marine bacteria. Members of this taxon are typically selected based on their resistance to kanamycin, and marine members are also kanamycin resistant. Members of the Bacteroidetes are perceived to be particle specialists (15), and a recent study showed that they were a significant component of both the particle-attached and free-living communities during a dinoflagellate bloom (18). Thus, it is interesting that members of the Bacteroidetes manage to become significant in organically enriched microenvironments despite the fact that they are very sensitive to inhibition by other pelagic bacteria. Factors other than bacterium-bacterium antagonism (e.g., heavy metal accumulation, pH, and oxygen concentration) may play a role in structuring communities on particles. Members of the Bacteroidetes were found to accumulate and grow in the presence of high concentrations of zinc and cadmium (17), and pigmented bacteria (members of the Bacteroidetes are typically pigmented) were found to be more tolerant to heavy metals (e.g., Zn, Cd, and Hg) (35) than nonpigmented bacteria.
The particle-derived α-proteobacteria strongly inhibited free-living α-proteobacteria and members of the Bacteroidetes, yet none of these organisms was effective against any of the other groups. The strong inhibitory activity of α-proteobacteria against members of the Bacteroidetes may in part explain the observations of Gonzalez et al. (23), who found that there was an inverse abundance relationship between the two groups with depth during an algal bloom. However, many other ecological factors that affect bacterial community structure (25, 39; Long and Azam, submitted) could also have caused the pattern observed by Gonzalez et al. (23) and should be considered when our hypothesis is tested.
Understanding antibiosis at the phylogenetic level may allow a more focused search for antibiotics that are active against a bacterial species or group. Such an understanding may also help workers devise strategies for pathogen control in aquatic environments. One example is in aquaculture, where members of the Bacteroidetes and Vibrio spp. are often associated with a number of fish and shellfish diseases (5, 12, 45, 50, 57). One method employed to protect aquaculture stocks involves the use of probiotics; a bacterial species is added to inhibit growth or colonization of the pathogenic species of concern. Our study suggests that α-proteobacteria derived from particles might be potential sources of probiotic species that are active against members of the Bacteroidetes. On the other hand, members of the Vibrionales and Alteromonadales may be good probiotics that are active against pathogenic Vibrio spp., and this has been recognized previously (49).
The approach used to search for antibiotics derived from bacteria in the ocean was an approach used in soil ecology. Emphasis was initially placed on members of the Actinobacteridae from marine sediments, and this was fruitful (27, 28). However, the approaches now used may have reached their apex in terms of the rate of discovery of novel molecules (19), and the emphasis has shifted to symbiotic bacteria associated with eukaryotes. However, it is probable that the full wealth of microbial diversity in the sea is yet to be revealed. A large fraction of marine bacteria have not been cultured yet, and novel cultivation methods need to be developed in order to culture them. We emphasize that not only should new cultivation techniques be developed, but the complex nature of the microenvironments that exist in the marine water column (6; Long and Azam, submitted) should also be taken into account. These microenvironments, including the highly diverse particles in the ocean, are a rich potential source of antibiotic-producing bacteria and include phylogenetic branches that have not been well explored.
TABLE 1.
Sources of bacterial isolatesa
| Designation | Source | Particle or free |
|---|---|---|
| BBAT | Enrichment | Attached to particle |
| BB2AT | Enrichment | Attached to particle |
| BBFL | Enrichment | Free-living |
| BB2FL | Enrichment | Free-living |
| LHAT | Enrichment | Attached to particle |
| LHFL | Enrichment | Free-living |
| SWAT | Water column | Attached to particle |
| SWFL | Water column | Free-living |
| SB | Enrichment | Attached to particle |
Bacterial isolates were collected directly from the water column or in F/2 enrichments to induce diatom blooms. Particles were individually picked, rinsed with 0.22-μm-pore-size filtered seawater, and plated on ZoBell 2216E plates to isolate attached bacteria. Free-living bacteria were isolated from seawater filtrate (pore size, 1.0 μm) on ZoBell 2216E plates. Organisms whose designations are not listed were isolated from seawater with no visible particles.
TABLE 2.
Bacterium-bacterium inhibitory interactions at the phylogenetic levela
Eighty-six marine bacterial isolates belonging to the α-proteobacterial (Alpha) (n = 19), Bacteroidetes (Bactero.) (n = 19), Alteromonadales (Altero.) (n = 28), and Vibrionales (Vibrio.) (n = 20) groups were examined for production and sensitivity to inhibitory molecules against one another. Isolates were operationally defined as either free-living (F) or particle-attached (P) based on the isolation procedure (see Materials and Methods).
ACKNOWLEDGMENTS
This work was supported by NSF grant OCE-9819603 and NIAID grant A146600 to F. Azam.
We thank M. Haygood and J. M. Gonzalez for donating several of the isolates. We thank two anonymous reviewers for their comments and insight.
REFERENCES
- 1.Alldredge A L, Jackson G A. Aggregation in marine systems. Deep-Sea Res Part II Top Studies Oceanogr. 1995;42:1–7. [Google Scholar]
- 2.Alldredge A L, Passow U, Logan B E. The abundance and significance of a class of large, transparent organic particles in the ocean. Deep-Sea Res Part I Oceanogr Res Papers. 1993;40:1131–1140. [Google Scholar]
- 3.Alldredge A L, Silver M. Characteristics, dynamics and significance of marine snow. Prog Oceanogr. 1988;20:41–82. [Google Scholar]
- 4.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Austin B, Day J G. Inhibition of prawn pathogenic Vibrio spp. by a commercial spray-dried preparation of Tetraselmis suecica. Aquaculture. 1990;90:389–392. [Google Scholar]
- 6.Azam F. Microbial control of oceanic carbon flux: the plot thickens. Science. 1998;280:694–696. [Google Scholar]
- 7.Bassler B L, Greenberg E P, Stevens A M. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bactereriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bidle K D, Fletcher M. Comparison of free-living and particle-associated bacterial communities in the Chesapeake Bay by stable low-molecular-weight RNA analysis. Appl Environ Microbiol. 1995;61:944–952. doi: 10.1128/aem.61.3.944-952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruehl G, Millar R, Cunfer B. Significance of antibiotic production by Cephalosporium gramineum to its saprophytic survival. Can J Plant Sci. 1969;49:235–246. [Google Scholar]
- 10.Burkholder P, Pfister R, Leitz F. Production of a pyrrole antibiotic by a marine bacterium. Appl Microbiol. 1966;14:649–653. doi: 10.1128/am.14.4.649-653.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carson J, Schmidtke L M, Munday B L. Cytophaga johnsonae—a putative skin pathogen of juvenile farmed barramundi, Lates calcarifer bloch. J Fish Dis. 1993;16:209–218. [Google Scholar]
- 13.Chin W C, Orellana M V, Verdugo P. Spontaneous assembly of marine dissolved organic matter into polymer gels. Nature. 1998;391:568–572. [Google Scholar]
- 14.Colmer J A, Fralick J A, Hamood A N. Isolation and characterization of a putative multidrug tesistance pump from Vibrio cholerae. Mol Microbiol. 1998;27:63–72. doi: 10.1046/j.1365-2958.1998.00657.x. [DOI] [PubMed] [Google Scholar]
- 15.DeLong E F, Franks D G, Alldredge A L. Phylogenetic diversity of aggregate-attached vs free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- 16.Don R H, Cox P T, Wainwwright B J, Baker K, Mattick J S. “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donocik A, Ledin M, Pedersen K, Allard B. Accumulation of zinc and cadmium by Cytophaga johnsonae. Biometals. 1996;9:169–175. [Google Scholar]
- 18.Fandino L B, Riemann L, Steward G F, Long R A, Azam F. Bacterial succession during a dinoflagellate bloom analyzed by DGGE and rDNA sequencing. Aquat. Microb. Ecol. . 2001;23:119–130. [Google Scholar]
- 19.Faulkner D J. Marine natural products. Nat Prod Rep. 2000;17:7–55. doi: 10.1039/a809395d. [DOI] [PubMed] [Google Scholar]
- 20.Fisher R A. Statistical methods for research workers. 13th ed. New York, N.Y: Hafner; 1958. [Google Scholar]
- 20a.Garrity G M, Winters M, Searles D B. Taxonomic outline of the procaryotic genera: Bergey's manual of systematic bacteriology. 2nd ed. New York, N.Y: Springer-Verlag; 2001. [Google Scholar]
- 21.Giovannoni S, Rappé M. Evolution, diversity, and molecular ecology of marine prokaryotes. In: Kirchman D L, editor; Kirchman D L, editor. Microbial ecology of the ocean. New York, N.Y: Wiley-Liss; 2000. pp. 47–84. [Google Scholar]
- 22.Gonzalez J M, Moran M A. Numerical dominance of a group of marine bacteria in the alpha subclass of the class Proteobacteria in coastal seawater. Appl Environ Microbiol. 1997;63:4237–4242. doi: 10.1128/aem.63.11.4237-4242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez J M, Simo R, Massana R, Covert J S, Casamayor E O, Pedros-Alio C, Moran M A. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 2000;66:4237–4246. doi: 10.1128/aem.66.10.4237-4246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guillard R R L, Ryther J H. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacea Cleve. Can J Microbiol. 1962;8:229. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- 25.Hennes K P, Suttle C A, Chan A M. Fluorescently labeled virus probes show that natural virus populations can control the structure of marine microbial communities. Appl Environ Microbiol. 1995;61:3623–3627. doi: 10.1128/aem.61.10.3623-3627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen P, Fenical W. Marine microorganisms and drug discovery: current status and future potential. In: Fusetani N, editor; Fusetani N, editor. Drugs from the sea. Basel, Switzerland: Karger; 2000. pp. 6–29. [Google Scholar]
- 27.Jensen P R, Fenical W. Marine bacterial diversity as a resource for novel microbial products. J Ind Microbiol Biotechnol. 1996;17:346–351. [Google Scholar]
- 28.Jensen P R, Fenical W. Strategies for the discovery of secondary metabolites from marine bacteria—ecological perspectives. Annu Rev Microbiol. 1994;48:559–584. doi: 10.1146/annurev.mi.48.100194.003015. [DOI] [PubMed] [Google Scholar]
- 29.Koike I, Shigemitsu H, Kazuki T, Kogure K. Role of sub-micrometre particles in the ocean. Nature. 1990;345:242–244. [Google Scholar]
- 30.Krasilnokova D N. On antibiotic properties of microorganisms isolated from various depths of the world ocean. Microbiology. 1961;30:545–550. [Google Scholar]
- 31.Li J, Yie J, Foo R W T, Ling J M L, Xu H S, Woo N Y S. Antibiotic resistance and plasmid profiles of Vibrio isolates from cultured silver sea bream, Sparus sarba. Mar Pollut Bull. 1999;39:245–249. [PubMed] [Google Scholar]
- 32.Long R A, Azam F. Abundant protein-containing particles in the sea. Aquat Microb Ecol. 1996;10:213–221. [Google Scholar]
- 33.Martinez J, Smith D C, Steward G F, Azam F. Variability in ectohydrolytic enzyme activities of pelagic marine bacteria and its significance for substrate processing in the sea. Aquat Microb Ecol. 1996;10:223–230. [Google Scholar]
- 34.Morita Y, Kodama K, Shiota S, Mine T, Kataoka A, Mizushima T, Tsuchiya T. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother. 1998;42:1778–1782. doi: 10.1128/aac.42.7.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nair S, Chandramohan D, Bharathi P A L. Differential sensitivity of pigmented and non-pigmented marine bacteria to metals and antibiotics. Water Res. 1992;26:431–434. [Google Scholar]
- 36.Nair S, Simidu U. Distribution and significance of heterotrophic marine bacteria with antibacterial activity. Appl Environ Microbiol. 1987;53:2957–2962. doi: 10.1128/aem.53.12.2957-2962.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okami Y. Marine microorganisms as a source of bioactive agents. Microb Ecol. 1986;12:65–78. doi: 10.1007/BF02153223. [DOI] [PubMed] [Google Scholar]
- 38.Pesci E C, Milbank J B J, Pearson J P, McKnight S, Kende A S, Greenberg E P, Iglewski B H. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinhassi J, Azam F, Hemphala J, Long R A, Martinez J, Zweifel U L, Hagstrom A. Coupling between bacterioplankton species composition, population dynamics, and organic matter degradation. Aquat Microb Ecol. 1999;17:13–26. [Google Scholar]
- 40.Ploug H, Grossart H P, Azam F, Jorgensen B B. Photosynthesis, respiration, and carbon turnover in sinking marine snow from surface waters of Southern California Bight: implications for the carbon cycle in the ocean. Mar Ecol Prog Ser. 1999;179:1–11. [Google Scholar]
- 41.Putman M, van Veen H W, Degener J E, Konings W N. Antibiotic resistance: era of the multidrug pump. Mol Microbiol. 2000;36:772–773. doi: 10.1046/j.1365-2958.2000.01871.x. [DOI] [PubMed] [Google Scholar]
- 42.Seymour J R, Mitchell J G, Pearson L, Waters R L. Heterogeneity in bacterioplankton abundance from 4.5 millimetre resolution sampling. Aquat Microb Ecol. 2000;22:143–153. [Google Scholar]
- 43.Smith D C, Simon M, Alldredge A L, Azam F. Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature. 1992;359:139–142. [Google Scholar]
- 44.Smith D C, Steward G F, Long R A, Azam F. Bacterial mediation of carbon fluxes during a diatom bloom in a mesocosm. Deep-Sea Res Part II Top Studies Oceanogr. 1995;42:75–97. [Google Scholar]
- 45.Speare D J, Markham R J F, Despres B, Whitman K, Macnair N. Examination of gills from salmonids with bacterial gill disease using monoclonal antibody probes for Flavobacterium branchiophilum and Cytophaga columnaris. J Vet Diagn Invest. 1995;7:500–505. doi: 10.1177/104063879500700413. [DOI] [PubMed] [Google Scholar]
- 46.Spratt B G. Resistance to antibiotics mediated by target alterations. Science. 1994;264:388–393. doi: 10.1126/science.8153626. [DOI] [PubMed] [Google Scholar]
- 47.Thomashow L S, Weller D M, Bonsall R F, Pierson L S. Production of the antibiotic phenazine-1-carboxylic acid by fluorescent Pseudomonas species in the rhizosphere of wheat. Appl Environ Microbiol. 1990;56:908–912. doi: 10.1128/aem.56.4.908-912.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson J D, Higgins D G, Gibson T G. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verschuere L, Heang H, Criel G, Sorgeloos P, Verstraete W. Selected bacterial strains protect Artemia spp. from the pathogenic effects of Vibrio proteolyticus CW8T2. Appl Environ Microbiol. 2000;66:1139–1146. doi: 10.1128/aem.66.3.1139-1146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wakabayashi H, Toyama T, Iida T. A study on serotyping of Cytophaga psychrophila isolated from fishes in Japan. Fish Pathol. 1994;29:101–104. [Google Scholar]
- 51.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wells M L, Goldberg E D. Occurrence of small colloids in sea water. Nature. 1991;353:342–344. [Google Scholar]
- 53.Wilcox M H, Finch R G, Smith D G E, Williams P, Denyer S P. Effects of carbon dioxide and sub-lethal levels of antibiotics on adherence of coagulase-negative staphylococci to polystyrene and silicone rubber. J Antimicrob Chemother. 1991;27:577–587. doi: 10.1093/jac/27.5.577. [DOI] [PubMed] [Google Scholar]
- 54.Wratten S J, Wolfe M S, Andersen R J, Faulkner D J. Antibiotic metabolites from a marine pseudomonad. Antimicrob Agents Chemother. 1977;11:411–414. doi: 10.1128/aac.11.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright J. The production of antibiotics in soil. IV. Production of antibiotics in coats of seeds sown in soil. Ann Appl Biol. 1956;44:561–566. [Google Scholar]
- 56.Wright J. The production of antibiotics in soil. III. Production of glixootin in wheatstraw buried in soil. Ann Appl Biol. 1956;44:461–466. [Google Scholar]
- 57.Zhao G J, Chung K T, Milow K, Wang W X, Stevens S E. Antibacterial properties of tannic acid and related compounds against the fish pathogen Cytophaga columnaris. J Aquat Anim Health. 1997;9:309–313. [Google Scholar]