Abstract
Salmonella enterica serovar Enteritidis, a major cause of food poisoning, can be transmitted to humans through intact chicken eggs when the contents have not been thoroughly cooked. Infection in chickens is asymptomatic; therefore, simple, sensitive, and specific detection methods are crucial for efforts to limit human exposure. Suppression subtractive hybridization was used to isolate DNA restriction fragments present in Salmonella serovar Enteritidis but absent in other bacteria found in poultry environments. Oligonucleotide primers to candidate regions were used in polymerase chain reactions to test 73 non-Enteritidis S. enterica isolates comprising 34 different serovars, including Dublin and Pullorum, two very close relatives of Enteritidis. A primer pair to one Salmonella difference fragment (termed Sdf I) clearly distinguished serovar Enteritidis from all other serovars tested, while two other primer pairs only identified a few non-Enteritidis strains. These primer pairs were also useful for the detection of a diverse collection of clinical and environmental Salmonella serovar Enteritidis isolates. In addition, five bacterial genera commonly found with Salmonella serovar Enteritidis were not detected. By treating total DNA with an exonuclease that degrades sheared chromosomal DNA but not intact circular plasmid DNA, it was shown that Sdf I is located on the chromosome. The Sdf I primers were used to screen a Salmonella serovar Enteritidis genomic library and a unique 4,060-bp region was defined. These results provide a basis for developing a rapid, sensitive, and highly specific detection system for Salmonella serovar Enteritidis and provide sequence information that may be relevant to the unique characteristics of this serovar.
In the last few decades, Salmonella enterica serovar Enteritidis has emerged as a major cause of food-borne illness worldwide. This pathogen is distinguished from its many close relatives also found in poultry environments by its ability to infect chicken ovaries before the eggshell is formed, allowing transmission through intact eggs. Once established in the human host from raw or undercooked eggs or egg products, this bacterium causes gastroenteritis similar to other S. enterica serovars. Infection in poultry flocks, which is asymptomatic, was first noticed in the late 1970s and in the 1980s spread rapidly throughout the United Kingdom, the United States, South America, and other areas. During this period, the proportion of salmonellosis cases attributed to Salmonella serovar Enteritidis increased substantially, showing a 275-fold increase in Argentina and becoming the predominant cause of this disease in the United States (10, 11, 14). Baumler et al. suggested that this rapid increase of Salmonella serovar Enteritidis may have been due to successful campaigns to eradicate the Salmonella serovars Pullorum and Gallinarum, the causative agents in chickens of bacillary white diarrhea and fowl typhoid, respectively (2). It is hypothesized that these avian-adapted salmonellae provided cross-immunity against Salmonella serovar Enteritidis because of important similarities in lipopolysaccharide structures. Therefore, these campaigns may have opened an ecological niche that has since been occupied by Salmonella serovar Enteritidis. This view remains controversial, however, since serovars Gallinarum and Pullorum remain prevalent in many developing countries where serovar Enteritidis has nevertheless increased dramatically, and turkey flocks in developed countries, now free of serovars Gallinarum and Pullorum, have not been colonized by serovar Enteritidis (12, 15). Unlike the avian-adapted salmonellae, rodents serve as an animal reservoir for Salmonella serovar Enteritidis, suggesting that culling would not be an effective method of control. It is possible that the use of Salmonella serovar Enteritidis as a rodenticide may have contributed to the current prevalence of this serovar, and it is also likely that infected rodents are currently a source of disease. In addition to the health risks, this pathogen has had a significant economic impact on the egg industry through decreased consumer confidence following well-publicized outbreaks.
Although Salmonella serovar Enteritidis is closely related to other pathogenic S. enterica serovars, several characteristics of this serovar appear to distinguish it from many others. For example, the fimbria Sef14 is found in a limited number of S. enterica serovars, including Enteritidis. This surface structure appears to be required for macrophage uptake and survival in intraperitoneal infections (6) in contrast to other Salmonella fimbriae that promote binding to host epithelial cells (5). There is also evidence that quorum sensing plays an important role in the life cycle of Salmonella serovar Enteritidis. Virulence correlates with a strain's ability to produce high-molecular-weight lipopolysaccharide and the ability of subpopulations to grow to high cell densities (8).
Because of the increased prevalence of Salmonella serovar Enteritidis and its complex life cycle, rapid and effective detection methods are important as a basis of control. Traditional culture methods require several days. More rapid methods have been developed but are often based on the sef operon, which is also found in other serogroup D salmonellae. One such test relies on recombinant Sef14 antigen to detect Salmonella serovar Enteritidis antibodies in chickens, but this test requires that serum samples be collected and also detects antibodies against S. enterica serovar Dublin (13). The gene encoding Sef14, sefA, is also found in the serovars Blegdam, Gallinarum, Pullorum, Rostock, Seremban, and Typhi (20). Several PCR-based assays have also been reported. One is based on sefA (18), and another is based on plasmid-borne sequences (17, 23). While the latter test appears quite specific for Salmonella serovar Enteritidis, the diversity of the isolates used for this study, normally based on phage typing, is unclear. Furthermore, since plasmids are often mobilizable and unstable, a plasmid-based test might not detect the occurrence of plasmid-less strains that could rapidly acquire virulence by plasmid transfer. Rapid plasmid transfer to plasmid-less strains is an important aspect of virulence in the plant pathogen Agrobacterium tumefaciens and may be important in other pathogens (7, 21, 24).
Here we describe the identification of a novel S. enterica serovar Enteritidis locus that serves as a marker for DNA-based identification of this bacterium. In contrast to other tests, this marker is not found in a wide range of closely related serovars, including Dublin and Pullorum, the two closest relatives of Enteritidis (19). Thus, this test allows highly specific detection of Salmonella serovar Enteritidis. Evidence is presented supporting a chromosomal location for the locus, thus circumventing the potential problems associated with plasmid-borne markers. An extensive array of Salmonella serovar Enteritidis phage types from around the world was tested by PCR for the presence of this DNA region, and all phage types associated with human infections were detected. An ∼7-kb region was isolated by PCR-based screening of a Salmonella serovar Enteritidis library and subsequently sequenced. The region of this clone that does not match the S. enterica serovar Typhi or Paratyphi complete genomes contains six short open reading frames (ORFs). The putative proteins show either weak or no similarity to database sequences. Two other primer pairs were developed that are also quite effective at detecting Salmonella serovar Enteritidis. The combined use of these primer pairs provides tools for developing rapid and specific detection methods for S. enterica serovar Enteritidis.
MATERIALS AND METHODS
Strains.
Strains used for these studies are listed in Tables 1, 2, and 3. Some strains, as indicated in the text, were obtained from the American Type Culture Collection (ATCC), Rockville, Md. Serotyping was verified or performed by the California Animal Health and Food Safety Laboratory (CAHFS) by using standard procedures. The National Veterinary Services Laboratory (NVSL), Ames, Iowa, performed phage typing by standard methods (9).
TABLE 1.
PCR results for selected S. enterica serovars
| Serovar (no. of strains) | Serogroup | Region evaluated (no. of strains positive)
|
||
|---|---|---|---|---|
| Sdf I | Sdf II | Sdf III | ||
| Agona (2) | B | 0 | 2 | 0 |
| Derby (2) | B | 0 | 0 | 0a |
| Heidelberg (3) | B | 0 | 0 | 0 |
| Reading (1) | B | 0 | 0 | 0a |
| Typhimurium (4) | B | 0 | 0 | 0 |
| Infantis (2) | C1 | 0 | 0 | 0 |
| Lille (1) | C1 | 0 | 0 | 0 |
| Livingstone (3) | C1 | 0 | 1 | 0 |
| Mbandaka (2) | C1 | 0 | 0 | 0a |
| Montevideo (3) | C1 | 0 | 0 | 0 |
| Ohio (2) | C1 | 0 | 0 | 0 |
| Oranienburg (3) | C1 | 0 | 0 | 0 |
| Tennessee (2) | C1 | 0 | 0 | 0 |
| Thompson (1) | C1 | 0 | 0 | 0 |
| Blockley (1) | C2 | 0 | 0 | 0a |
| Newport (2) | C2 | 0 | 0 | 0 |
| Corvallis (1) | C3 | 0 | 0 | 0 |
| Kentucky (4) | C3 | 0 | 0 | 0a |
| Berta (1) | D1 | 0 | 1 | 0a |
| Dublin (7) | D1 | 0 | 1 | 0 |
| Lomalinda (2) | D1 | 0 | 0 | 0a |
| Panama (1) | D1 | 0 | 0 | 0 |
| Pullorum (1) | D1 | 0 | 1 | 0 |
| Fresno (1) | D2 | 0 | 0 | 0 |
| Anatum (1) | E1 | 0 | 0 | 0 |
| Give (2) | E1 | 0 | 0 | 0 |
| Muenster (2) | E1 | 0 | 0 | 0 |
| Cambridge (1) | E2 | 0 | 0 | 0 |
| Newbrunswick (2) | E2 | 0 | 0 | 0 |
| Newington (2) | E2 | 0 | 0 | 0 |
| Menhaden (3) | E3 | 0 | 0 | 0 |
| Senftenberg (2) | E4 | 0 | 0 | 0 |
| Worthington (2) | G2 | 0 | 1 | 0 |
| Cerro (4) | K | 0 | 0 | 0 |
Products other than the expected size detected in one or more strains of this serovar.
TABLE 2.
PCR results for selected S. enterica serovar Enteritidis strains from various environmental sources
| Isolate no. | Source | State, district, or country of origin | Phage type | Region evaluateda
|
||
|---|---|---|---|---|---|---|
| Sdf I | Sdf II | Sdf III | ||||
| D1850 | Human, fecesb | Texas | 4 | Pos | Pos | Pos |
| D0918 | Human, fecesb | Mexico | 4 | Pos | Pos | Pos |
| D1832 | Human, fecesb | Italy | 7 | Pos | Pos | Pos |
| D0119 | Human, fecesb | Spain | 4 | Pos | Pos | Pos |
| 8-154 | Human, fecesc | California | 13 | Pos | Pos | Pos |
| D0927-CDC | Human, fecesd | Mexico | 4 | Pos | Pos | Pos |
| D1045-CDC | Human, fecesd | Texas | 4 | Pos | Pos | Pos |
| D3784-CDC | Human, fecesd | Hawaii | 4 | Pos | Pos | Pos |
| D4485-CDC | Human, fecesd | Minnesota | 4 | Pos | Pos | Pos |
| D4911-CDC | Human, fecesd | Connecticut | 4 | Pos | Pos | Pos |
| H4707 | Human, fecese | California | 6A | Pos | ND | ND |
| H5133 | Human, fecese | Oregon | 6A | Pos | ND | ND |
| H5134 | Human, fecese | Oregon | 6A | Pos | ND | ND |
| H6787 | Human, fecese | Arizona | 6A | Pos | ND | ND |
| D0144-CDC | Human, fecesb | Switzerland | 9A | Pos | ND | ND |
| D1760-CDC | Human, fecesd | Texas | 9A | Pos | ND | ND |
| CAHFS-525 | Chicken, environmentf | California | 13A | Pos | Pos | Pos |
| CAHFS-526 | Chicken, environmentf | California | 4B | Pos | Pos | Pos |
| 97-1866 | Porcine, tissueg | Indiana | 13A | Pos | Pos | Pos |
| 99-11315 | Chicken, egg beltg | Nebraska | 13A | Pos | Pos | Pos |
| 95-16526 | Chicken, egg poolg | Washington, D.C. | 14B | Pos | Pos | Pos |
| 95-13141 | Chicken, egg poolg | Maryland | 14B | Pos | Pos | Pos |
| 97-637 | Porcine, fecesg | Iowa | 8 | Pos | Pos | Pos |
| 97-604 | Turkey, cloacag | Ohio | 8 | Pos | Pos | Pos |
| CAHFS-5 | Environment, creekf | California | 8 | Pos | Pos | Pos |
| CAHFS-184 | Chicken, liverf | California | 4 | Pos | Pos | Pos |
| CAHFS-285 | Environment, creekf | California | 4 | Pos | Pos | Pos |
| CAHFS-320 | Chicken, environmentf | California | 4 | Pos | Pos | Pos |
| CAHFS-546 | Bovine, milk filterf | California | 8 | Pos | Pos | Pos |
| 426 93-0675 | Poultryb | France | 4 | Pos | Pos | Pos |
| CAHFS-435 | Chicken, environmentf | California | 6A | Pos | ND | ND |
| CAHFS-436 | Chicken, environmentf | California | 6A | Pos | ND | ND |
| CAHFS-739 | Chicken, environmentf | California | 6B | Pos | ND | ND |
Pos, positive; ND, not determined.
R. Gast, Southeast Poultry Laboratory, USDA, Agricultural Research Service, Athens, Ga.
S. Nunez, Tulare County Public Health, Tulare, California.
T. Barrett, Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, Ga.
B. Holland, Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, Ga.
H. Kinde, California Animal Health and Food Safety Laboratory, San Bernardino Laboratory, San Bernardino, Calif.
K. Ferris, NVSL, Ames, Iowa.
TABLE 3.
PCR results for S. enterica serovar Enteritidis phage type reference strains
| Phage typeb | Region evaluateda
|
||
|---|---|---|---|
| Sdf I | Sdf II | Sdf III | |
| 1 | Pos | Pos | Pos |
| 2 | Pos | Pos | Pos |
| 3 | Pos | Pos | Pos |
| 4 | Pos | Pos | Pos |
| 4A | Pos | Pos | Pos |
| 5 | Pos | Pos | Pos |
| 5A | Pos | Pos | Pos |
| 6 | Pos | Pos | Pos |
| 6A | Neg | Pos | Neg |
| 7 | Pos | Pos | Pos |
| 8 | Pos | Pos | Pos |
| 9 | Pos | Pos | Pos |
| 9A | Neg | Pos | Neg |
| 9B | Pos | Pos | Pos |
| 10 | Pos | Pos | Pos |
| 11 | Neg | Pos | Neg |
| 11A | Pos | Pos | Pos |
| 12 | Pos | Pos | Pos |
| 13 | Pos | Pos | Pos |
| 13A | Pos | Pos | Pos |
| 14 | Pos | Pos | Pos |
| 15 | Pos | Pos | Pos |
| 16 | Neg | Pos | Neg |
| 17 | Pos | Pos | Pos |
| 18 | Pos | Pos | Pos |
| 19 | Pos | Pos | Pos |
| 20 | Neg | Pos | Neg |
| 20A | Pos | Pos | Pos |
| 22 SC2 | Pos | Pos | Pos |
| 23 | Pos | Pos | Pos |
| 24 | Pos | Pos | Pos |
| 27 | Neg | Pos | Neg |
| 28 | Pos | Pos | Pos |
| 31 | Pos | Pos | Pos |
| 32 | Pos | Pos | Pos |
| 34 | Pos | Pos | Pos |
| 40 SC2 | Pos | Pos | Neg |
Pos, positive; Neg, negative.
NVSL, Ames, Iowa.
DNA preparation.
DNA was isolated from 3-ml cultures after overnight growth in Luria-Bertani medium (Sigma, St. Louis, Mo.). Either of two methods was used to purify total DNA; both methods yielded consistent results. DNA STAT-60 isolation reagent (Tel-Test, Friendswood, Tex.) was used according to the manufacturer's recommendations (1 ml per culture). Alternatively, cell pellets were resuspended in 200 μl of TE buffer (10 mM Tris HCl, 1 mM EDTA; pH 8.0) and treated with 2.5 μg of proteinase K/ml for 30 min at 37°C. Successive extractions were performed with saturated phenol, phenol-chloroform, (1:1, vol/vol) and chloroform-isoamyl alcohol (24:1, vol/vol). DNA was precipitated with 0.5 ml of cold 95% ethanol and 75 μl of 3 M sodium acetate (pH 5.2), dried under vacuum in a desiccator, and resuspended in water.
DNA amplification for strain testing.
Oligonucleotide primers (Sigma-Genosys, The Woodlands, Tex.) at a 400 nM final concentration were combined with 200 pg of genomic DNA template and amplified with Advantage 2 Polymerase (ClonTech, Palo Alto, Calif.). After an initial denaturation at 94°C for 1 min, the samples were subjected to 27 cycles of 94°C for 30s, 58°C for 30s, and 72°C for 1 min, followed by a final 7-min incubation at 72°C. Samples were fractionated by 2% agarose gel electrophoresis and visualized by ethidium bromide staining. A primer pair to either the 23S or 16S rRNA gene was used as a positive control for the amplification of each DNA sample, or a primer pair to the rplI gene (encoding the L9 ribosomal protein) was used as an internal control.
Suppression subtraction hybridization, DNA sequencing, and analysis.
Genome comparisons by suppression subtraction hybridization were performed essentially as described by Akopyants et al. (1) with the following exceptions. For subtractions with Sau3AI-digested DNA, adapter 1 was formed by annealing the adapter 1 long oligonucleotide with the oligonucleotide 5′ GATCACCTGCCCGG to form an adapter with appropriate cohesive ends. Similarly, adapter 2 was formed by annealing the adapter 2 long oligonucleotide with the oligonucleotide 5′-GATCCAATCGGCCG. Ligase (New England Biolabs, Beverly, Mass.) was inactivated by incubation at 72°C for 20 min. Unpurified PCR products were cloned using the pGEM-T Easy TA cloning kit (Promega, Madison, Wis.). Recombinant clones were picked by using a BioRobotics (Woburn, Mass.) BioPick automated colony picker, and plasmid templates were prepared by boiling lysis and magnetic bead capture with a high-throughput procedure (16). Sequencing of plasmid templates was performed by using the Applied Biosystems (Foster City, Calif.) BigDye Terminator system and either ABI 377 or 3700 automated sequencers. The sequencing primers used were 5′-TGTAAAACGACGGCCAGT (forward) and 5′-CAGGAAACAGCTATGACC (reverse). Sequences were assembled and oligonucleotide primers were designed by using the Consed software package (University of Washington, Seattle). Sequence comparisons with the GenBank databases were performed using the BLAST (basic local alignment search tool) server at the Baylor College of Medicine (Houston, Tex.) or the server at the National Center for Biotechnology Information (Bethesda, Md.). Both the nonredundant and the unfinished microbial databases were used for comparisons.
Oligonucleotide primers.
The sequences of the primer pairs used (Sigma-Genosys) for DNA amplification were as follows: spvC, 5′-CTCTGCATTTCACCACCATCACG and 5′ CTTGCACAACCAAATGCGGAAGAT; rplI, 5′ GGGTGATCAGGTTAACGTTAAAG and 5′CTTCGTGTTCGCCAGTGGTACGC; 23S, 5′-CTACCTTAGGACCGTTATAGTTAC and 5′-GAAGGAACTAGGCAAAATGGTGCC; 16S, 5′AGAGTTTGATCCTGGCTCAG and 5′-GGTTACCTTGTTACGACTT; Sdf I, 5′-TGTGTTTTATCTGATGCAAGAGG and 5′-CGTTCTTCTGGTACTTACGATGAC; Sdf II, 5′-GCGAATATCATTCAGGATAAC and 5′-GCATGTCATACCGTTGTGGA; and Sdf III, 5′-GCTGACTCACACAGGAAATCG and 5′-TCTGATAAGACTGGGTTTCACT.
DNase assays.
Plasmids were prepared from Salmonella serovar Enteritidis CAHFS-285, by a standard alkaline lysis method (4), except that proteins and cell debris were precipitated with 7.5 M ammonium acetate (1/2 volume) instead of sodium acetate. The DNA from a 10-ml culture was resuspended in 40 μl of TE, and 10 μl was digested with Plasmid-Safe DNase (Epicentre Technologies, Madison, Wis.) in a 250-μl reaction with 50 U of enzyme for 5 h according to the manufacturer's recommendations. Then, 5 μl of this reaction was used as a template in PCRs (30 cycles of 1 min of annealing at 65°C, 1 min of extension at 72°C, and 30 s of denaturation at 94°C).
Library construction and screening.
To construct a genomic library of Salmonella serovar Enteritidis strain CAHFS-285, 100 μg of total DNA was partially digested with 100 U Sau3AI (New England Biolabs) for 10 min. The DNA was fractionated by electrophoresis, and 4- to 6-kb fragments were excised and gel purified by electroelution. These fragments were ligated to pUC9 (22) that had been digested with BamHI (New England Biolabs), gel purified, and treated with shrimp alkaline phosphatase (U.S. Biochemicals, Cleveland, Ohio). Products were introduced into Escherichia coli DH10B cells (Gibco-BRL, Rockville, Md.) by electroporation (Gene-Pulser; Bio-Rad, Richmond, Calif.), and transformants were selected with 50 μg of ampicillin (Sigma, St. Louis, Mo.)/ml on agar plates with Luria-Bertani (LB) medium. By using a BioPick automated colony picker, white colonies (total of 6,528) were used to inoculate 384-well microtiter plates (Nalge Nunc, Rochester, N.Y.) containing LB medium with 7.5% (vol/vol) glycerol, followed by overnight incubation at 37°C. The library was replicated with a 384-pin tool and stored at −70°C. Screening was performed with the Sdf I primers by amplification of combined cultures, followed by amplification of single cultures. For each row, 5 μl of each culture was combined, and 1 μl of the mixture was PCR tested. For rows with a positive signal, the individual clones were then tested. One clone consistently yielded positive results in PCRs and was selected for sequencing.
DNA sequencing of the Sdf I region.
The library clone identified by PCR with the Sdf I primers was purified by alkaline lysis and anion-exchange chromatography with a Qiagen (Valencia, Calif.) Plasmid Preparation Kit. The plasmid DNA was digested with EcoRI and HindIII and separated by electrophoresis, and the two insert fragments were gel purified using a Qiaex II kit (Qiagen). The purified fragments were first treated with the Klenow fragment of DNA polymerase I (New England Biolabs) and deoxynucleoside triphosphates, followed by digestion with AluI, HaeIII, and RsaI in separate reactions. Then, the products from each of the three reactions were separately cloned into pPA9 that had been digested with EcoRV and treated with shrimp alkaline phosphatase. The plasmid pPA9 was constructed by annealing the oligonucleotides 5′ AGCTTGGAATTCGATATCAGGCCTCG and 5′ GATCCGAGGCCTGATATCGAATTCCA, which were then cloned between the HindIII and BamHI sites of pUC9 (22). We sequenced 32 clones from each enzyme sublibrary (96 total) as described above and assembled overlapping sequences with the Consed program to generate the complete sequence of the insert. The assembly was corroborated with restriction mapping based on the sequence.
Nucleotide sequence accession numbers.
The sequences for Salmonella difference fragments (Sdf) I to IX from S. enterica serovar Enteritidis CAHFS-5 have been submitted to GenBank with accession numbers AF370707 to AF370715, respectively. The sequence for Salmonella difference region I (Sdr I) from S. enterica serovar Enteritidis CAHFS-285 has been submitted to GenBank with accession number AF370716.
RESULTS
Isolation of DNA fragments unique to S. enterica serovar Enteritidis.
Suppression subtractive hybridization (SSH) was used to identify Salmonella serovar Enteritidis-specific sequences that could serve as diagnostic markers. SSH is a PCR-based technique that enriches for restriction fragments that are present in one strain, termed the tester, but absent in another, termed the driver. Salmonella serovar Enteritidis strain CAHFS-5 was used as the tester (phage type 8), and the closely related serovar Dublin (strain CAHFS-9008117D), also in serogroup D1, was used as the driver. This way, any true SSH products would be likely to distinguish serovar Enteritidis from serovar Dublin and its close relatives. Four restriction enzymes were used in separate SSH experiments: RsaI, AluI, Sau3AI, and HaeIII. We sequenced 48 clones from each subtraction (192 total) and synthesized PCR primers for 98 of the products. Ninety-four clones with high similarity to available non-Enteritidis database sequences were not studied further.
PCR amplifications were then performed by using the driver and tester DNAs as templates to identify true subtraction products. Nine primer pairs showed amplification with Salmonella serovar Entertitidis but not with serovar Dublin. These unique restriction fragments from which the primers were designed were designated Sdf I to Sdf IX (Salmonella difference fragment). One of the nine fragments was from an SSH experiment using Sau3AI (Sdf I), one was an AluI fragment, five were HaeIII fragments (including Sdf II and Sdf III), and two were RsaI fragments. The primer pairs (referred to as “Sdf I primer pair,” etc.) based on these nine sequences were selected for further analysis.
Characterization of DNA fragments unique to Salmonella serovar Enteritidis.
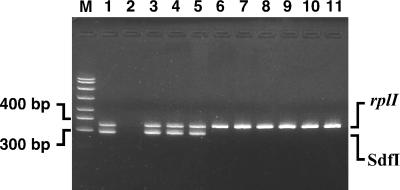
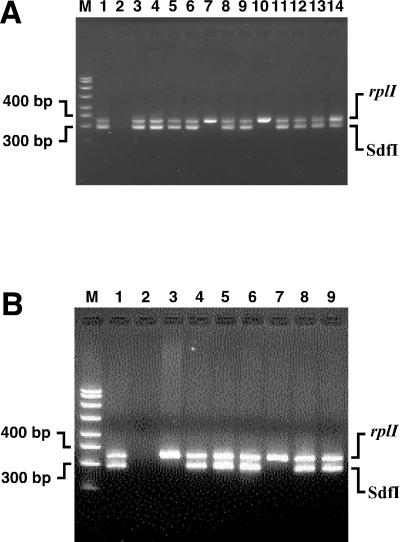
The nine primer pairs that amplified sequences from Salmonella serovar Enteritidis but not serovar Dublin were PCR tested with several other serovars commonly found in the poultry environment to eliminate those primer pairs that were not serovar Enteritidis specific. Amplification of sequences from one isolate each of Salmonella serovars Typhimurium, Heidelberg, Montevideo, and another isolate of Salmonella serovar Enteritidis (CAHFS-285, a phage type 4 strain) were used for this purpose. In addition, the two strains used in the subtraction (Salmonella serovar Dublin CAHFS-9008117D and Salmonella serovar Enteritidis CAHFS-5) were included as controls. Three of the nine primer pairs detected both strains of serovar Enteritidis but none of the other serovars. These three primer pairs were further evaluated using an extensive collection of S. enterica serovars available at the CAHFS. We also tested 81 additional S. enterica isolates, including 30 additional serovars (for a total of 34 non-Enteritidis serovars, including those described above [Table 1]) and 12 additional serovar Enteritidis environmental and poultry isolates (Table 2). Most of the 34 non-Enteritidis serovars are encountered at egg production facilities and therefore complicate diagnostic efforts to detect serovar Enteritidis. The Sdf II primer pair identified 7 of the 73 non-Enteritidis isolates, representing six non-Enteritidis serovars. Interestingly, one of the strains was an isolate of Salmonella serovar Dublin, even though this primer pair does not detect the strain of serovar Dublin used for the subtraction experiments. Also, this primer pair detected one isolate of Salmonella serovar Worthington, while another isolate of the same serovar was not detected. This indicates that there is some degree of diversity within serovars that can be detected by primers from SSH experiments. It is not known if these differences are due to nucleotide differences in the 3′ end of a primer-binding site or whether larger differences are responsible. Another primer pair, to Sdf III, amplified a specific product of the predicted size only with the serovar Enteritidis isolates but amplified other products in six non-Enteritidis isolates (serovars Lomalinda, Mbandaka, Blockley, Derby, Reading, and Kentucky) and produced a smear with one isolate (serovar Berta). Clear positive results were obtained with all 14 serovar Enteritidis environmental, poultry, and other animal isolates tested in this panel (Table 2). The third primer pair that was tested with this panel of strains was that of Sdf I, which yielded remarkably clear results. No products were amplified from the 73 non-Enteritidis isolates, but all 14 serovar Enteritidis isolates showed a clear band of the expected size. Figure 1 shows an Sdf I amplification product for three of the most common phage types of Salmonella serovar Enteritidis (lanes 3 to 5), while four other Salmonella serovars found in the poultry environment do not show this amplicon (lanes 6 to 9). In addition, two other enteric bacteria, E. coli ATCC 25922 and Citrobacter freundii ATCC 43864 (lanes 10 and 11) are not detected with this primer pair. The Sdf I primer pair was also tested with other bacteria common in poultry environments, namely, Proteus mirabilis ATCC 33946, Proteus vulgaris ATCC 13315, Enterobacter aerogenes ATCC 13048, Enterobacter cloacae ATCC 13047, and Providencia rettgeri ATCC 29944, and did not show any amplification. The Sdf I, Sdf II, and Sdf III primer pairs were then used to test 37 NVSL phage type reference strains of Salmonella serovar Enteritidis (Table 3). The Sdf I sequence was present in all but 6 of 37 phage types (phage types 6A, 9A, 11, 16, 20, and 27). No clinical isolates for phage types 11, 16, 20, and 27 are available from the Centers for Disease Control and Prevention (B. Holland, unpublished data), suggesting that infections from these phage types are exceedingly rare. A subset of these data is presented in Fig. 2A. Amplification of 12 different phage types is shown, with only phage type 6A (lane 7) and 9A (lane 10) showing negative results. Although these results suggest that the Sdf I primers cannot detect other isolates of phage type 6A or 9A, two clinical isolates of phage type 6A (lanes 4 and 5 of Fig. 2B) and phage type 9A (Fig. 2B, lanes 8 and 9) are readily detected with the Sdf I primer pair. Four additional isolates of phage type 6A were also tested and were detected with the Sdf I primers (Table 2). In addition, one isolate of phage type 6B was also detected (Fig. 2B, lane 6). These results suggest that strains that are clearly infectious are detected with the Sdf I primers. Interestingly, the Sdf I and Sdf III primers showed the same pattern when tested with the NVSL strains, except for the NVSL phage type 40 reference strain, raising the possibility that the Sdf I and Sdf III difference fragments may be linked in the Salmonella serovar Enteritidis genome. An important difference between the Sdf I and Sdf III primer pairs is that the Sdf III primers generate other products for several of the templates that are not the expected size. The Sdf II primer pair showed amplification with all 37 phage types.
FIG. 1.
Specificity of S. enterica serovar Enteritidis detection determined using the Sdf I primer pair in PCRs. Lane M, size standards; lane 1, Salmonella serovar Enteritidis CAHFS-546 (phage type 8); lane 2, no template; lane 3, Salmonella serovar Enteritidis CAHFS-184 (phage type 4); lane 4, Salmonella serovar Enteritidis 97-6371A (phage type 8); lane 5, Salmonella serovar Enteritidis 97-1866IN (phage type 13A); lane 6, Salmonella serovar Pullorum; lane 7, Salmonella serovar Typhimurium; lane 8, Salmonella serovar Heidelberg; lane 9, Salmonella serovar Montevideo; lane 10, E. coli; lane 11, C. freundii. Amplicons produced by the Sdf I primers (293 bp) and the rplI primers (343 bp) are indicated.
FIG. 2.
Specificity of detection of selected Salmonella serovar Enteritidis phage type reference strains determined using the Sdf I primer pair in PCRs. The strains used are from the NVSL unless indicated by a specific designation. (A) Detection of Sdf I in phage type reference strains. Lane M, size markers; lane 1, CAHFS-546 (positive control); lane 2, no template; lane 3, phage type 2; lane 4, phage type 3; lane 5, phage type 4; lane 6, phage type 6; lane 7, phage type 6A; lane 8, phage type 8; lane 9, phage type 9; lane 10, phage type 9A; lane 11, phage type 13A; lane 12, 95-13141 (phage type 14B); lane 13, phage type 24; lane 14, phage type 34. (B) Detection of Sdf I in phage type reference strains and clinical strains of phage types 6A, 6B, and 9A. Lane M, size markers; lane 1, CAHFS-546 (positive control); lane 2, no template; lane 3, NVSL 9 (phage type 6A); lane 4, CAHFS-435 (phage type 6A); lane 5, CAHFS-436 (phage type 6A); lane 6, CAHFS-739 (phage type 6B); lane 7, NVSL 13 (phage type 9A); lane 8, D0144-CDC (phage type 9A); lane 9, D01760-CDC (phage type 9A). Amplicons produced by the Sdf I primers (293 bp) and rplI primers (343 bp) are indicated.
The three primer pairs were also tested against 10 additional serovar Enteritidis clinical isolates taken from stool samples of afflicted humans (Table 2). Eight were phage type 4, one was phage type 7, and one was phage type 13. These 10 samples are geographically diverse, having been collected in Spain, Italy, Mexico, and across the United States from Connecticut to Hawaii. All three primer pairs detected the 10 strains.
Combined with the testing of the phage type 6A, 6B, and 9A strains discussed above, 16 clinical isolates were tested, and all were detected with the Sdf I primers. Thus, these data suggest one highly specific marker for Salmonella serovar Enteritidis (Sdf I) has been developed, as well as two other markers that are useful for narrowing S. enterica to just a few serovars.
Database searches with the sequences of Sdf I (333 bp), Sdf II (731 bp), and Sdf III (846 bp) showed that positions 5 to 274 of Sdf III, when translated, showed high similarity to the deduced amino acid sequence of a hypothetical protein of the putative cryptic phage CP-933R of E. coli O157:H7 strain EDL933 (expected probability of a fortuitous match [E] value 4 × 10−39). Sdf I and Sdf II showed no similarity to database sequences.
Chromosomal localization of the Sdf I locus.
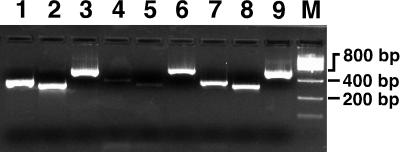
To determine whether the Sdf I marker is located on the chromosome or located on a circular plasmid, we developed the following novel assay (Fig. 3). Plasmid-Safe exodeoxyribonuclease from Epicentre Technologies was used to treat plasmid preparations of Salmonella serovar Enteritidis CAHFS-285, a phage type 4 isolate. The enzyme digests contaminating chromosomal DNA present in all plasmid preparations but does not affect covalently closed or nicked circular DNAs, i.e., circular plasmids. In addition to the Sdf I primer pair, a primer pair to a known chromosomal gene encoding the L9 ribosomal protein (rplI), and a primer pair to a known Salmonella plasmid-borne gene, spvC, were used as controls. Lanes 1 to 3 show that these primer pairs readily amplify products from total cellular DNA. As expected, all three amplicons were observed in the untreated plasmid preparations (lanes 7 to 9). In exonuclease-treated samples, however, the spvC product (lane 6) showed significant amplification, whereas the rplI (lane 4) and the Sdf I (lane 5) products were only faintly visible. Because the Sdf I signal was reduced similarly to a known chromosomal sequence, this suggests that Sdf I is located on the chromosome.
FIG. 3.
The Salmonella serovar Enteritidis Sdf I region is located on the chromosome. Lane 1, total DNA amplified with rplI primers (343-bp amplicon); lane 2, total DNA amplified with the Sdf I primers (293-bp amplicon); lane 3, total DNA amplified with the spvC primers (565-bp amplicon); lane 4, plasmid preparation treated with exo-DNase amplified with rplI primers; lane 5, plasmid preparation treated with exo-DNase and amplified with Sdf I primers; lane 6, plasmid preparation treated with exo-DNase and amplified with spvC primers. Lanes 7, 8, and 9 are the same as lanes 4, 5, and 6, respectively, but without exo-DNase treatment before amplification. Strain CAHFS-285 (phage type 4) was used for these experiments.
Cloning of the Sdf I locus.
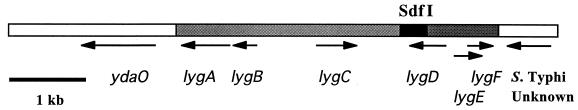
To define the region of the chromosome containing the 333-bp Sdf I SSH product (Salmonella difference region I [Sdr I]), a library was constructed using total DNA from the phage type 4 Salmonella serovar Enteritidis strain CAHFS-285. A total of 6,528 E. coli colonies containing plasmids with 4- to 6-kb inserts, representing >99% of the cellular DNA (assuming a genome size of 5 Mbp), were screened by PCR in pools by using the Sdf I primer pair. One clone was identified, and the complete sequence of its 6,907-bp insert was determined (Fig. 4). There was no similarity to the sequence of Sdf III, which was detected in a similar pattern to that of Sdf I. Sdf I, a Sau3AI fragment isolated by SSH, is found between positions 4928 and 5260 of the genomic clone (black region of Fig. 4). Nucleotide sequence comparisons with database sequences showed a near-perfect match at each end to the complete S. enterica serovar Typhi genome. On the left end as shown, the match extends from positions 1 to 2101, and on the right end it extends from positions 6160 to 6907. On the left end is a copy of a gene with near-perfect identity to E. coli ydaO. Surprisingly, the matches were to two widely separated regions of the serovar Typhi genome (1361375 to 1363475 on the left and 1920934 to 1920189 on the right), suggesting that this region is the site of a major rearrangement with respect to serovar Enteritidis. Overlapping PCR amplifications were used to confirm that the 6,907-bp region of the library clone is contiguous in Salmonella serovar Enteritidis and not the result of the ligation of two or more unrelated fragments (data not shown). There are six ORFs of >100 codons in the 4,060-bp novel region (gray and black bars in Fig. 4). We have designated these six ORFs lygA to lygF for “linked to the ydaO gene.” These six ORFs encode possible proteins of 207, 105, 173, 155, 119, and 110 amino acids for lygA to F, respectively. Using a protein BLAST search of the nonredundant database, LygA (positions 2161 to 2784) shows similarity to Exonuclease VIII of Salmonella serovar Typhimurium (E value 2 × 10−18). LygC (positions 3867 to 4388) exhibits weak similarity to phage superinfection exclusion protein B of E. coli (E value 6 × 10−5), while LygD (positions 5036 to 5503) shows even weaker similarity to phage λ repressor cI (E value 10−4). LygF shows some similarity to a hypothetical protein of prophage CP-933R of E. coli O157:H7, an enterohemorrhagic strain (E value 10−22). LygE and F overlap to a large extent, which may indicate that one, the other, or both are not genes. The deduced amino acid sequences of lygB and lygE do not show any similarity to database sequences with a protein BLAST search.
FIG. 4.
Chromosomal context of Sdf I. Schematic representation of the Sdf I region from Salmonella serovar Enteritidis CAHFS-285 (phage type 4). Open boxes indicate sequence with identity to S. enterica serovar Typhi. Gray and black boxes indicate novel sequences. Sdf I, bounded by Sau3AI sites, is shown in black. All ORFs of more than 100 codons are indicated with black arrows.
Amplification by PCR was used to examine other areas of the unique region defined by comparison to Salmonella serovar Typhi. Primer pairs to lygA, lygC, and lygD were used to amplify sequences from a Salmonella serovar Enteritidis phage type 8 strain (CAHFS-546), and a Salmonella serovar Dublin strain (CAHFS-9008117D), as well as the library strain, Salmonella serovar Enteritidis CAHFS-285 as a positive control. Products of the expected size were observed in the Enteritidis strains but not in the Dublin strain, a finding consistent with the view that the entire unique region is present in Enteritidis strains but absent in Dublin strains (data not shown). Interestingly, when primers to the nonunique flanking sequences, which would generate an ∼4.5-kb amplicon comprising the serovar Enteritidis unique region, were used with the serovar Dublin strain mentioned above, an ∼600-bp product was observed. This may indicate that all or most of the unique region is missing in serovar Dublin, and the locus is otherwise colinear. Sequencing the 600-bp amplicon will help to define the precise nature of the difference between the two serovars.
DISCUSSION
Using suppression subtractive hybridization, we found three loci that are restricted to S. enterica serovar Enteritidis or are found in a few close relatives. Remarkably, one region, Sdf I, appears to only be found in serovar Enteritidis strains, including a wide range of clinical and environmental samples, and has yielded clear results in laboratory testing. This makes this region an excellent candidate for the detection of serovar Enteritidis within complex samples. Given the wide range of other Salmonella serovars and other enteric bacteria found in poultry environments, it is desirable to have markers that will distinguish serovar Enteritidis strains from these bacteria. The Sdf I region appears to satisfy this criterion. It is important to note that in addition to making an excellent marker for nucleic acid detection, this region may also allow the development of an antibody-based test that relies on the detection of one or more putative protein products of the unique ORFs. The extent to which the cloned region varies within serovar Enteritidis strains will be an important question to answer in order to confirm that other areas of this region are useful for detection purposes. A phage type 8 strain was tested with three primer pairs spanning the unique region based on the nucleotide sequence from a phage type 4 strain. The expected products were observed, indicating that these regions were also present in this strain. An ongoing project at the University of Illinois to sequence the genome of the phage type 8 strain LK5 will allow a direct sequence comparison of the Sdf I region from two different strains.
Phage typing is currently the standard method for distinguishing subgroups of serovar Enteritidis (9). This technique has been exploited to ensure that a diverse collection of Enteritidis strains was tested with the diagnostic primer pairs in this study. Using the NVSL reference collection, all 37 phage types were detected with the Sdf I primer pair except phage types 6A, 9A, 11, 16, 20, and 27. Clinical samples for phage types 11, 16, 20, and 27 are not available, indicating that they are not a significant cause of human infections. Although the phage type 6A and 9A reference strains were not detected with the Sdf I primers, two clinical phage type 9A strains and four clinical phage type 6A strains were unambiguously identified by PCR with the Sdf I primer pair. In summary, the Sdf I primer pair clearly detects all strains of a diverse collection of clinical isolates, in addition to detecting all of the environmental isolates tested.
These results demonstrate the lack of a clear relationship between phage typing and the presence of Sdf I. It is possible that subtle differences such as point mutations in the primer binding sites could explain these results. PCR with a primer pair internal to Sdf I, however, showed the same results (data not shown), suggesting that this is not the case. It is also possible that in some reference strains the Sdf I region has been lost during laboratory passage but that selection maintains this region in the natural environment. Cloning and sequencing of the region corresponding to Sdf I from these aberrant strains could help to define the strain differences and perhaps provide insight into this question. Targeted deletion of the region defined by strain comparisons would allow otherwise isogenic strains to be tested to assign a functional role. It is possible that the Enteritidis-specific Sdf I region could be related to one or more of the unique properties of this serovar. Similarity to database sequences is not high enough to provide strong enough evidence to ascribe functions to the putative proteins encoded by this region. The similarity of the lygF deduced amino acid sequence to a hypothetical protein of an E. coli cryptic phage may suggest, however, that the sequences described here are those of phage remnants. Although Sdf III also showed some similarity to the same cryptic prophage, no Sdf III sequences are present in the 6,907-bp Sdr I in which Sdf I lay, and the degrees of similarity are quite different, so these data do not necessarily imply that these sequences are linked. Another interesting question is whether the phage type 6A and 9A NVSL reference strains, which are Sdf I negative, have the same functional properties, such as chicken colonization, egg infection, and virulence, compared to the phage type 6A and 9A clinical strains, which are Sdf I positive. Importantly, testing of the NVSL reference collection also showed that the most common phage types, i.e., phage types 4, 8, 13, and 13A, were all detected with the Sdf I primer pair. Taken together, the results presented here suggest that Sdf I is a robust marker for pathogenic Salmonella serovar Enteritidis strains.
A DNA-based test offers the potential for a significant improvement over current methods of S. enterica serovar Enteritidis detection. DNA detection offers the possibility of greater speed, sensitivity, and ease. An important extension of these studies will be their application to detection in samples taken directly from poultry environments and comparisons to current methods. Combined with improved technology (see, for example, reference 3), it may be possible to perform tests on-site, thus greatly facilitating detection and regular monitoring for serovar Enteritidis.
ACKNOWLEDGMENTS
This work was performed under the auspices of the U.S. Department of Energy by the University of California, Lawrence Livermore National Laboratory, under contract no. W-7405-Eng-48 and was funded by the U.S. Department of Energy, NN-20, Chemical and Biological Non-Proliferation Program. The California Egg Commission provided additional funds.
We thank Silvia Gamez-Chin, Anne Marie Erler, and Warren Regala for excellent technical assistance. We also thank the NVSL for providing the phage type reference collection.
REFERENCES
- 1.Akopyants N S, Fradkov A, Diatchenko L, Hill J E, Siebert P D, Lukyanov S A, Sverdlov E D, Berg D E. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:13108–13113. doi: 10.1073/pnas.95.22.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bäumler A J, Hargis B M, Tsolis R M. Tracing the origins of Salmonella outbreaks. Science. 2000;287:50–52. doi: 10.1126/science.287.5450.50. [DOI] [PubMed] [Google Scholar]
- 3.Belgrader P, Benett W, Hadley D, Richards J, Stratton P, Mariella R, Jr, Milanovich F. PCR detection of bacteria in seven minutes. Science. 1999;284:449–450. doi: 10.1126/science.284.5413.449. [DOI] [PubMed] [Google Scholar]
- 4.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dibb-Fuller M P, Allen-Vercoe E, Thorns C J, Woodward M J. Fimbriae- and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology. 1999;145:1023–1031. doi: 10.1099/13500872-145-5-1023. [DOI] [PubMed] [Google Scholar]
- 6.Edwards R A, Schifferli D M, Maloy S R. A role for Salmonella fimbriae in intraperitoneal infections. Proc Natl Acad Sci USA. 2000;97:1258–1262. doi: 10.1073/pnas.97.3.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuqua W C, Winans S C. A LuxR-LuxI type regulatory system activates Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guard-Petter J. Variants of smooth Salmonella enterica serovar Enteritidis that grow to higher cell density than the wild type are more virulent. Appl Environ Microbiol. 1998;64:2166–2172. doi: 10.1128/aem.64.6.2166-2172.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hickman-Brenner F W, Stubbs A D, Farmer J J., III Phage typing of Salmonella enteritidis in the United States. J Clin Microbiol. 1991;29:2817–2823. doi: 10.1128/jcm.29.12.2817-2823.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hogue A, White P, Guard-Petter J, Schlosser W, Gast R, Ebel E, Farrar J, Gomez T, Madden J, Madison M, McNamara A M, Morales R, Parham D, Sparling P, Sutherlin W, Swerdlow D. Epidemiology and control of Salmonella enteritidis in the United States of America. Rev Sci Tech. 1997;16:542–553. doi: 10.20506/rst.16.2.1045. [DOI] [PubMed] [Google Scholar]
- 11.Morales R A, McDowell R M. Economic consequences of Salmonella enterica serovar Enteritidis infection in humans and the U.S. egg industry. In: Saeed A M, Gast R K, Potter M E, Wall P G, editors. Salmonella enterica serovar Enteritidis in humans and animals. Ames: Iowa State University Press; 1999. pp. 271–290. [Google Scholar]
- 12.Pomeroy B S, Nagaraja K V. Fowl typhoid. In: Calnek B W, Barnes H J, Beard C W, Reid W M, Yoder H W, editors. Diseases of poultry. Ames: Iowa State University Press; 1991. pp. 87–99. [Google Scholar]
- 13.Rajashekara G, Wanduragala D, Halvorson D A, Nagaraja K V. A rapid strip immunoblot assay for the specific detection of Salmonella enteritidis infection in chickens. Int J Food Microbiol. 1999;53:53–60. doi: 10.1016/s0168-1605(99)00148-8. [DOI] [PubMed] [Google Scholar]
- 14.Rodrigue D C, Tauxe R V, Rowe B. International increase in Salmonella enteritidis: a new pandemic? Epidemiol Infect. 1990;105:21–27. doi: 10.1017/s0950268800047609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silva E N. Salmonella gallinarum problem in Central and South America. In: Snoyenbos G H, editor. Proceedings of International Symposium on Salmonella, New Orleans. Kennett Square, Pa: La. American Association of Avian Pathologists; 1985. pp. 150–156. [Google Scholar]
- 16.Skowronski E W, Armstrong N, Andersen G, Macht M, McCready P M. Magnetic, microplate-format plasmid isolation protocol for high-yield, sequencing-grade DNA. BioTechniques. 2000;29:786–792. doi: 10.2144/00294st05. [DOI] [PubMed] [Google Scholar]
- 17.Soumet C, Ermel G, Rose N, Rose V, Drouin P, Salvat G, Colin P. Evaluation of a multiplex PCR assay for simultaneous identification of Salmonella spp. Salmonella enteritidis and Salmonella typhimurium from environmental swabs of poultry houses. Lett Appl Microbiol. 1999;28:113–117. doi: 10.1046/j.1365-2672.1999.00488.x. [DOI] [PubMed] [Google Scholar]
- 18.Soumet C, Ermel G, Rose V, Rose N, Drouin P, Salvat G, Colin P. Identification by a multiplex PCR-based assay of Salmonella typhimurium and Salmonella enteritidis strains from environmental swabs of poultry houses. Lett Appl Microbiol. 1999;29:1–6. doi: 10.1046/j.1365-2672.1999.00559.x. [DOI] [PubMed] [Google Scholar]
- 19.Stanley J, Baquar N. Phylogenetics of Salmonella enteritidis. Int J Food Microbiol. 1994;21:79–87. doi: 10.1016/0168-1605(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 20.Turcotte C, Woodward M J. Cloning, DNA nucleotide sequence and distribution of the gene encoding the SEF14 fimbrial antigen of Salmonella enteritidis. J Gen Microbiol. 1993;139:1477–1485. doi: 10.1099/00221287-139-7-1477. [DOI] [PubMed] [Google Scholar]
- 21.van Larebeke N, Genetello C, Schell J, Schilperoort R A, Hermans A K, Hernalsteens J P, van Montagu M. Acquisition of tumor-inducing ability by non-oncogenic agrobacteria as a result of plasmid transfer. Nature. 1975;255:742–743. doi: 10.1038/255742a0. [DOI] [PubMed] [Google Scholar]
- 22.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- 23.Wood M W, Mahon J, Lax A J. Development of a probe and PCR primers specific to the virulence plasmid of Salmonella enteritidis. Mol Cell Probes. 1994;8:473–479. doi: 10.1006/mcpr.1994.1068. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Murphy P J, Kerr A, Tate M E. Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature. 1993;362:446–448. doi: 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]