Abstract
Organoids are complex multicellular three-dimensional (3D) in vitro models that are designed to allow accurate studies of the molecular processes and pathologies of human organs. Organoids can be derived from a variety of cell types, such as human primary progenitor cells, pluripotent stem cells, or tumor-derived cells and can be co-cultured with immune or microbial cells to further mimic the tissue niche. Here, we focus on the development of 3D lung organoids and their use as disease models and drug screening tools. We introduce the various experimental approaches used to model complex human diseases and analyze their advantages and disadvantages. We also discuss validation of the organoids and their physiological relevance to the study of lung diseases. Furthermore, we summarize the current use of lung organoids as models of host-pathogen interactions and human lung diseases such as cystic fibrosis, chronic obstructive pulmonary disease, or SARS-CoV-2 infection. Moreover, we discuss the use of lung organoids derived from tumor cells as lung cancer models and their application in personalized cancer medicine research. Finally, we outline the future of research in the field of human induced pluripotent stem cell-derived organoids.
Keywords: induced pluripotent stem cells, lung organoids, cystic fibrosis - CF, human disease modelling, 3D structure, chronic obstructive pulmonary disease, lung cancer
1 Introduction
Respiratory disorders such as asthma, chronic obstructive pulmonary disease (COPD), and idiopathic pulmonary fibrosis (IPF) affect millions of people worldwide, accounting for approximately 8% of global mortality (Dwyer-Lindgren et al., 2017). Lung tumors and airborne infections add to the burden of lung disease-associated morbidity and mortality, with the ongoing COVID-19 pandemic demonstrating the potential impact of viral lung disease on the worldwide population (Pollard et al., 2020; Kumar et al., 2021). Despite intensive research efforts spanning decades, our knowledge of lung biology and its interaction with the disease process remains incomplete. For example, key questions remain around the nature of lung stem cells, their roles in tissue regeneration and their therapeutic potential. A better understanding of the lung stem cell progenitor’s capacity to generate the various cell types of the adult lung and unraveling the pathways involved in their responses to injury could lead to the discovery of more efficient treatments for numerous pulmonary diseases. Thus, there is a persistent and urgent need to develop more relevant in vitro models of the human lung that authentically recapitulate the organ’s physiology and can be used to elucidate the mechanisms and biomarkers of pulmonary disease, as well as to test and guide the development of new treatments for conditions affecting the airways.
Studies of lung biology have, until recently, been limited by the unique structural and cellular complexity of the organ. The human lung comprises over 40 different types of cells (Franks et al., 2008; Cunniff et al., 2021; Varghese et al., 2022), arranged in a complex three-dimensional (3D) architecture that is designed to withstand the continuous dynamic mechanical stress associated with respiratory movements (Nossa et al., 2021) while existing and functioning at the air-tissue-interface, in the presence of a varied and interactive microbiota (Paolicelli et al., 2019; Barcik et al., 2020). Studies of primary tissue ex vivo cannot provide dynamic information, while the relative inaccessibility of the lung and its constant movement have precluded the widespread use of intra-vital imaging in pre-clinical models. Together, these challenges have driven researchers to develop and explore the use of lung organoids (LOs). These 3D in vitro models of the lung incorporate multiple cell types derived from induced pluripotent stem cells (iPSCs) (Clevers, 2016; Paolicelli et al., 2019; Jose et al., 2020; Wang et al., 2020a) and offer the potential to answer long-standing questions about lung biology in health and disease.
In this review we outline the unique features of the lung that make studying the organ and its pathologies such a challenge, then summarize the pre-organoid era leading to the development of these latest models. We then review the different approaches used to generate LOs, and report the latest findings made in the field, before finally looking at the direction of pulmonary research in the future.
2 The Pre-Organoid era: Early Attempts to Model the Human Lung
While all organs are, to some degree, complex, the human lung is remarkably so. While the stomach (Busslinger et al., 2021) and liver (Ramachandran et al., 2020), for example, are comprised of four or five main cell types, the lung contains more than 40 (Franks et al., 2008; Cunniff et al., 2021; Varghese et al., 2022), varying in proportion depending on the region of the lung and exhibiting clear patterns of polarization at the air-liquid interface (Castellani et al., 2018). Thus, the development of reliable in vitro lung models is a challenge on a grand scale (Castellani et al., 2018).
Accordingly, for many years, studies of lung tissue either employed a reductionist approach based on 2D culture of human cells, or relied on animal models, in which the 3D structure of the lung is intact. Together with increasing use of tools such as genetic engineering and inducible gene expression, these approaches have enabled us to better understand the molecular basis of some lung diseases (Guilbault et al., 2007; Williams and Roman, 2016; Lambrecht et al., 2019). However, 2D cultures of human cells have clear drawbacks for the study of an organ with a function that is tightly linked to its 3D organization and multiple studies have shown that the overall ability of rodent models in particular to accurately replicate human pulmonary diseases is limited (Shmidt and Nitkin, 2004; Matute-Bello et al., 2008; Wang et al., 2008). Thus, there is mounting evidence for the need to develop human cell-based systems to study pulmonary disease.
2.1 Basic 2D Cultures of Human Lung Cells
Prior to the advent of 3D tissue culture methods, many studies of the lung used human primary cells and/or cell lines derived from healthy pulmonary or tumor tissues, grown in monolayer cultures (Gottschling et al., 2012). The 2D models are ideal for high-throughput drug screening and their generation from cell lines with histological and genetic changes allows modeling of different clinical conditions and drug responses (Huang et al., 2021). Gazdar et al. (2010) reviewed the important discoveries made using 2D models such as, the spectrum of TP53 gene mutations in lung cancer (Takahashi et al., 1989), mechanisms of resistance in EGFR mutant cells (Kwak et al., 2005; Pao et al., 2005; Engelman et al., 2007) and the description of BRAF mutations in various tumors, including lung (Davies et al., 2002; Pratilas et al., 2008).
While these models have the advantage of human origin, they are constrained by the culture’s limited amount of extracellular matrix and the absence of different lung cell subtypes—including progenitors—and their inability to replicate the morphology or structural features of the in vivo lung architecture. The significance of these factors has become even more apparent in recent years: for example, we now know that lung-infiltrating immune cells are fundamental to the biology of several lung diseases, including COPD (Huang et al., 2019; David et al., 2021), IPF (Tanabe et al., 2020; van Geffen et al., 2021; Zhu et al., 2021), and non-small cell lung cancer (Tamminga et al., 2020). Furthermore, recent studies have illustrated the central role of tissue polarity in lung conditions, which can alter the functions of native cells in terms of spreading, migrating, and sensing soluble factors and other ligands (Duval et al., 2017). This implies limited relevance of findings from 2D cell cultures or isolated primary cells.
2.2 Rodent Models of Human Pulmonary Disorders
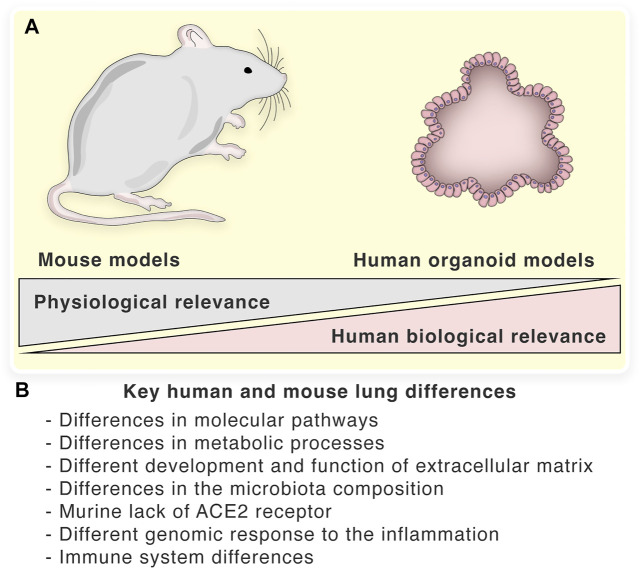
Although the etiology of human lung diseases is complicated and multi-factorial, there are three broad underlying forces: genetic mutation/variation/predisposition, environmental exposure/influences, and infection by a pathogen. Animal models have been used widely to investigate the mechanisms of each of these forces. While such studies have generated important insights into human lung disorders, there is a growing body of evidence that indicates their lack of ability to achieve progress in the field. Molecular characterization of human lungs using proteomic, transcriptomic, and in silico approaches, such as the Molecular Atlas of Lung Development Program (LungMAP), revealed major molecular differences between mouse and human lung epithelial tissues (Ardini-Poleske et al., 2017; Pan et al., 2019). Recent studies also showed that human lungs significantly differ from those of mouse models in several key metabolic processes as well as molecular pathways regulating the development and function of the lung extracellular matrix (Wang et al., 2015; Aichler et al., 2018). The main differences between human and mouse lungs are summarized in Figure 1. Below, we note key examples of the types of animal models commonly in use, acknowledging the advances and also noting their limitations that are driving the design of ever better LOs.
FIGURE 1.
Key human and mouse lung differences: (A)—Biological and physiological relevance of mouse and organoid models. (B)—The main differences between human and mouse lungs.
2.2.1 Models of Genetic Diseases Affecting the Lungs
CF is among the most common autosomal recessive diseases in humans and is caused by mutation of the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Genetically modified rodents bearing one or several of the more than 2,000 CFTR mutations that can result in CF in humans (De Boeck, 2020) have been used to model the condition, as reviewed by Semaniakou et al. (2018). However, although these models bear mutations known to be associated with the onset of CF in humans, the disease phenotypes in these mice vary in severity and pathology. These differences have been attributed to the rodent’s lack of specific cell types, receptors, and mediators involved in CF (Grubb and Boucher, 1999; Pan et al., 2019). Recent studies indicate that the differences in the microbial composition of the murine and human lung could also be a key factor (Semaniakou et al., 2018). Taken together, the distinct microbiota, alongside the molecular and physiological differences between mouse and human lungs, mean that murine models of CF are limited in their ability to advance our knowledge.
The same pattern emerges for the study of IPF—a chronic progressive disease characterized by scarring of the lung tissue—which has incompletely understood environmental and genetic components to its etiology (Chanda et al., 2019). The main mouse model of IPF involves the induction of pulmonary fibrosis by direct administration of bleomycin into the lungs of rodents (Walters and Kleeberger, 2008), with more recent developments including transgenic overexpression of pro-fibrotic cytokines such as TGF-β, TNF-α, IL-1β, and IL-13 in mice (Tashiro et al., 2017). However, despite showing fibrotic features, mouse models of IPF often display heterogeneous disease kinetics, especially in terms of the severity and extent of pulmonary lesions and levels of different cytokines (Moeller et al., 2008). Humanized mice have also been used as recipients of injected IPF patient lung biopsy material or tissue explants, which has allowed researchers to study the biology of the resulting lung remodeling and fibrotic lung phenotype (Habiel et al., 2018). As a result of murine studies, two IPF drugs have been developed, pirfenidone (King et al., 2014) and nintedanib (Richeldi et al., 2014), but neither are able to cure the disease and patient survival remains poor, with a median lifespan of 3.8 years post-diagnosis (Raghu et al., 2014).
2.2.2 Models of Pulmonary Infections
Another challenge is the development of relevant preclinical models of human pulmonary infections. Rodents and humans differ in their susceptibility to many pulmonary pathogens. This is particularly clear in mouse models of lung conditions that, in humans, develop through recurrent exacerbations of pulmonary infections including those by Staphylococcus aureus, Haemophilus influenzae, Pseudomonas aeruginosa, and Burkholderia cepacian (Semaniakou et al., 2018). Again, the difference in the microbes colonizing laboratory mice and humans is likely a major factor limiting the applicability of these models, and it is not yet clear whether this challenge can be overcome, although some attempts are being made [reviewed in Chang et al. (Chang and Sharma, 2020)]. Human viral lung infections are also difficult to study in murine models. For instance, investigations of human infection with the pathogenic avian influenza virus H5N1 in these models is impeded by the absence of the epithelial receptor for viral entry in mice (Ibricevic et al., 2006). Similarly, mice lack the ACE2 receptor required for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, initially dramatically limiting their applicability in this field. Although the development of a mouse model expressing the human ACE2 receptor was recently reported (Cleary et al., 2020), it is still unclear whether the response of mice to the virus mimics that of humans, and whether they express other as yet unidentified co-factors involved in infection/pathology.
Sepsis is a condition characterized by life-threatening organ dysfunction, and is caused by the host response to infection, most commonly by bacteria (Minasyan, 2019; Reyes et al., 2020) but also by some viruses (Singer et al., 2016; Hortova-Kohoutkova et al., 2020; Hortova-Kohoutkova et al., 2021). In the respiratory system, sepsis manifests clinically as acute lung injury (ALI) or, the more severe acute respiratory distress syndrome (ARDS) (Sevransky et al., 2009; Angus and van der Poll, 2013). Sepsis-induced ALI is marked by a “cytokine storm” that triggers inflammation and is followed by uncontrolled infiltration of pulmonary tissue by neutrophils (Aziz et al., 2013), while ARDS also involves pulmonary edema caused by alveolar injury that results in dangerously low oxygen levels in the blood (Sweeney and McAuley, 2016). Respiratory-infection-linked sepsis deaths totaled 1.8 million globally in 2017 (Rudd et al., 2020) driving urgent research into the condition. Sepsis has been experimentally induced in rodents by the administration of microbial components or living pathogens (e.g., Escherichia coli, or S. aureus) into the bloodstream, peritoneal cavity, or trachea (Karzai et al., 2003; Wang et al., 2004; Charavaryamath et al., 2006; Stahl et al., 2013; Kapicibasi et al., 2020). These models have shown that sepsis induced in mice by injection of peptidoglycan causes organ injury, organ, and systemic inflammation (Wang et al., 2004). Similarly, injection of a high dose of LPS and alphatoxin induces a systemic inflammatory response syndrome in rodents (Stahl et al., 2013). However, despite extensive animal studies, clinical translation has been largely unsuccessful. In this case, one of the reasons for the poor translation of the mouse data to humans is likely due to their markedly different genomic response to inflammation. A study comparing mouse and human endotoxemia showed no correlation between the top 10 most downregulated signaling pathways in the two species. In contrast to mouse models, the response to injury in humans was dominated by upregulation of genes related to innate immune response and downregulation of pathways related to adaptive immunity (Seok et al., 2013). Therefore, one must conclude that the balance of harm to the animal—in this case significant suffering is induced during sepsis modeling—and the low potential for valuable clinical insight, renders the use of such models highly undesirable for future research (Nandi et al., 2020).
2.2.3 Animal Models of Human Lung Cancer
Since the first animal model of leukemia was reported in 1950, many types of animal models of cancer have been developed to understand tumor biology and test drug efficacy. One of the simplest human tumor models is created by injecting tumor cell lines into immunodeficient mice. However, the pressure to develop more standardized model emerged due to unanticipated and unpredictable phenotype alterations compared to the original tumor. The patient-derived xenograft (PDX) model represents a reliable translation research tool that accurately mimics parental tumor tissue. It is generated by implanting a small tissue sample into highly immunodeficient mice. The great advantages of PDX models are in preserving the histologic features, genetic aberrations and gene expression profiles of the original tumor, as well as matching the characteristics of inflammation and the responses to chemotherapeutics and other anti-tumor drugs of the parental tissue. Therefore, PDX models are a valuable tool in tumor biology research, development of novel anti-cancer therapeutics, and pre-clinical drug screening (Lai et al., 2017; Bleijs et al., 2019; Yoshida, 2020). Although PDX models represent highly complex systems that accurately replicate tumor tissue in vivo, they also have several drawbacks, for example, the low success rate (30%–40%, lack of efficacy and the high cost of their establishment in terms of time and money, as well as their limited potential for application in high-throughput studies (Kim et al., 2019; Li et al., 2020). These features render PDX models unsuitable for use in the personalized treatment industry and other models suitable for high-throughput research are urgently required.
In lung cancer modeling, PDXs were reported in development of a novel anti-tumor therapeutic approaches, for example, in evaluation of microwave hyperthermia therapy (Motomura et al., 2010). Furthermore, PDX models of lung cancer have been used in pre-clinical drug testing (Hai et al., 2020; Li et al., 2020; Shi et al., 2020; Wang et al., 2020b), which is discussed in the following sections.
3 The Birth of Human Lung Organoids: Physiologically Relevant Models of Pulmonary Tissues
To overcome the limitations of classical 2D cultures and inter-species models, the use of lung tissue organoids has been pioneered in the field of lung research. Organoids are 3D “organ in a dish” models that aim to recreate key aspects of the in vivo structure of tissues using a mixture/variety of cell types from the species of interest to generate a relevant microenvironment in which cells within the organoid exhibit key aspects of the function of that organ (Lancaster and Huch, 2019).
Research into organoids was initially pioneered in the field of cancer studies, with cultured 3D structures based on simple air-medium interface system used to characterize the histological features of tumors (Kondo and Inoue, 2019). A significant advance in our ability to model non-malignant cells was the innovative demonstration by Sato et al. (2009) that individual Lgr5+ stem cells isolated from small intestinal crypts of adult mice could be grown and maintained in 3D cultures in which they regenerated the intestinal niche, comprising villus-like structures and fully differentiated epithelial cells. Since then, the use of tissue organoids in disease modeling research has expanded rapidly, with more than 3,000 scientific articles now published in this field alone (Lancaster and Huch, 2019).
It is now possible to generate LOs from cultures of pluripotent stem cells (PSCs) (including embryonic stem cells), induced PSCs (iPSCs), or adult stem cells (ASCs). Regardless of the source cell type, a key procedure common across all protocols for organoid derivation is the embedding of cells in extracellular matrix, which serves as the basal lamina for tissue culture and supports the development of the 3D architecture (Schutgens and Clevers, 2020). Fully differentiated organoids can then be passaged, further expanded, and used for basic tissue research (Tian et al., 2021), cell interaction studies (Nikolic and Rawlins, 2017), and cancer drug testing (Clevers, 2016; Drost and Clevers, 2018; Kim et al., 2019).
3.1 Different Methods for Generating Lung Organoids
As mentioned above, human LOs can now be derived through several routes and different biological materials, each with its own strengths and limitations. Selecting the most appropriate method for each application is of key importance, but with careful application of the methodologies mentioned below, valuable insights into lung biology and disease pathology can be made.
3.1.1 Generation of Primary Lung Progenitor Cell-Derived Organoids
Early protocols for LO differentiation were based on the use of primary lung progenitor cells (Leeman et al., 2019; Rabata et al., 2020; Schutgens and Clevers, 2020), or transformed lung cancer cells (Neal and Kuo, 2016; Sato et al., 2017). Several methods have been published describing the isolation of these cells from lung tissue that was first subjected to mechanical disruption followed by digestion using enzymes (Katsura et al., 2020; Salahudeen et al., 2020; Lamers et al., 2021; Tindle et al., 2021). Once a single-cell suspension has been generated, the cells are either directly seeded into Matrigel droplets for organoid culture or sorted based on the expression of cell-type-specific markers for organoid cultures that require a more homogeneous population of a particular cell type (Katsura et al., 2020).
Importantly, it is also possible to generate LOs from specific lung regions to model their physiology more accurately. For example, human lung basal cells can be isolated from either the trachea or upper bronchia, after which they are expanded ex vivo and stem cells are isolated for embedding into the matrix (Schutgens and Clevers, 2020). Similarly, alveolar-like organoids can be generated by sorting of type II alveolar cells identified as CD45−CD31−Epcam+HTII+ cells from dissociated lung tissue dissociation and mixing them with MRC5 fibroblasts before embedding in Matrigel (Barkauskas et al., 2013). A second method of alveolar organoid generation involves co-culture of epithelial progenitors with stromal fibroblasts (Tan et al., 2017), although this approach is less commonly used. More advanced LOs exhibiting a limited lung-like structure have been generated by mixing epithelial and fibroblastic cells using multi-layered microfluidic devices that enable the formation of differentiated LOs with morphological and secretory cellular phenotypes similar to those in human lungs (Hegab et al., 2015; Gkatzis et al., 2018). Similarly, organoids derived from alveolar or small airway epithelial cells have been co-cultured with mesenchymal stromal cells (MSCs) to promote alveolar organoid formation (Leeman et al., 2019). Leeman et al. (2019) also showed that ASC-derived LOs exhibit increased alveolar differentiation and decreased self-renewal when cultured with MSC-conditioned medium, thus demonstrating that MSC-secreted factors are important for the self-organization of lung epithelial organoids. Despite the achievements in the field of primary lung progenitor cell-derived organoid research, some limitations of these in vitro models remain. First, primary cell-derived LOs lack the mesenchymal cells that produce factors to support LO growth and differentiation (Leeman et al., 2019). Therefore, the successful generation of LOs requires either co-cultivation with mesenchymal cells or supplementation of the medium with these factors. Furthermore, cell-derived organoids are formed from only a limited number of cell types, which does not accurately mirror the complex structures of the human lung. Finally, primary cell-derived organoids require lung tissue biopsies, which provide only a limited amount of material for LO differentiation.
3.1.2 Generation of Induced Pluripotent Stem Cells-Derived Lung Organoids
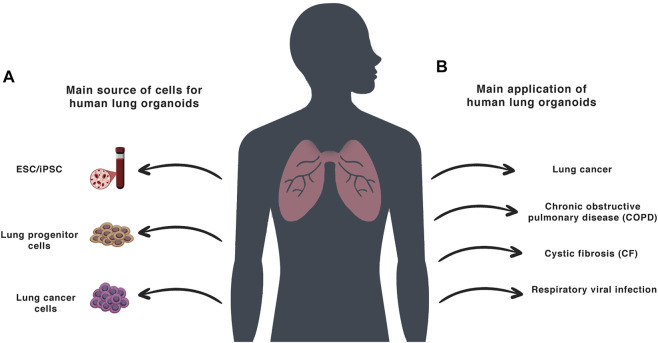
iPSC-based organoids can be generated by reprogramming somatic cells from patients (often easily accessible skin fibroblasts or peripheral blood mononuclear cells), making them a uniquely valuable and versatile tool for clinical research (Figure 2). This approach has revolutionized the study of genetic disorders in particular, superseding the previous ASC-based organoids, that were limited by their lack of MSCs and fibroblastic stromal cells, which are required for tissue-like structures and functions (Schutgens and Clevers, 2020). Thus, the use of PSCs to generate LOs with both epithelial and mesenchymal components has been a breakthrough in the field, and these organoids are now being utilized to study lung development and lung diseases across a broad range of fields (Choi et al., 2016).
FIGURE 2.
Lung organoids: (A)—The main sources for the creation of the human LOs. (B)—The main application of human LO models.
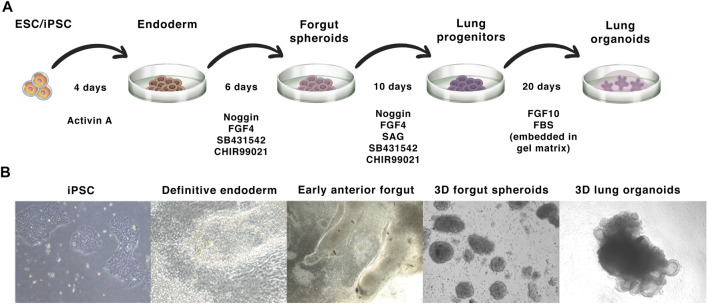
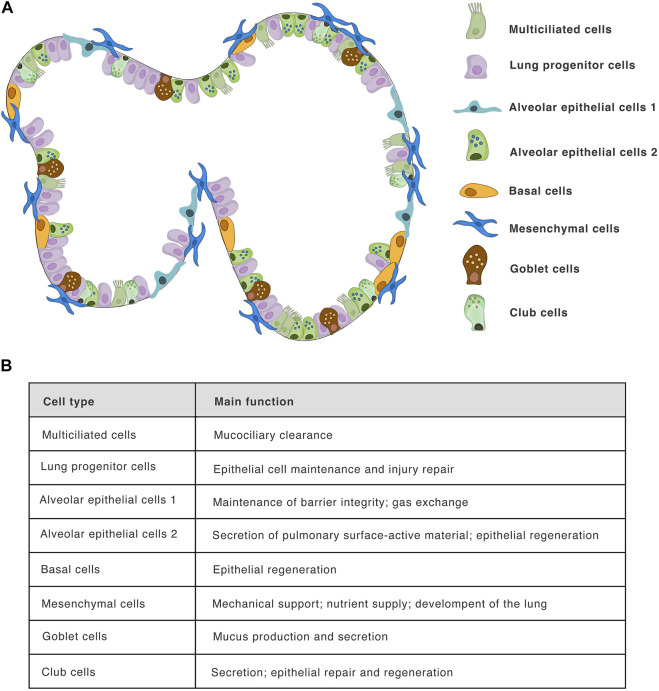
The process of LO differentiation follows that of normal embryogenesis, in which fetal lungs arise from the anterior foregut endoderm, forming the bronchial, and alveolar tissues (Schittny, 2017). In vitro, this process is driven/directed by sequential activation and inhibition of different signaling pathways using growth factors and small molecules. The iPSC-derived LO differentiation protocol was firstly described by Dye et al. (2015). This protocol involves the initial differentiation of either ESCs or iPSCs into definitive endoderm by treatment with activin A for a period of 4 days (Figure 3, days 1–4). This endodermal layer is then differentiated to form anterior foregut spheroids by using a combination of noggin, the TGFβ/activin/NODAL pathway inhibitor SB431542, FGF4, and the GSK3 inhibitor CHIR99021 (Figure 3, days 4–10). These signals are sufficient to induce both mesenchymal and epithelial cell populations within the anterior foregut spheroids. The activation of the WNT and FGF pathway and the simultaneous inhibition of the BMP/TGFβ signalling in PSCs was found to be optimal for the generation of lung tissue expressing NKX2.1+, a key transcription factor that regulates pulmonary development (Minoo et al., 1995; Choi et al., 2016). For the generation of NKX2.1+ E-cadherin+ foregut spheroids, the sonic hedgehog activator SAG is also added on days 4–10. Finally, NKX2.1+ SOX9+ spheroids are embedded into a gel matrix and expanded in an FGF10-containing medium, leading to the development of 3D LOs comprising both P63+ basal cells and FOXJ1+ ciliated epithelial cells (Dye et al., 2015; De Luca et al., 2017). Importantly, these organoids also exhibit tissue polarity, evidenced by the localization of acetyl-tubulin and E-cadherin markers (Jose et al., 2020). As previously mentioned, human lung tissue comprises more then 40 cell types (Franks et al., 2008; Cunniff et al., 2021; Varghese et al., 2022), including infiltrating immune cells. However, iPSC-derived organoids do not consist of any cells of hematopoietic origin, which can be an experimental advantage allowing controlled cocultures, such as Jose et al. (2020) showed for monocytes. Moreover, distinct protocols differ in the cell types composition within the organoids. The composition of several cell types forming iPSC-derived organoids is shown in Figure 4, the overall complexity of iPSCs model indeed does not reach completely the normal lungs.
FIGURE 3.
iPSC-derived lung organoid differentiation: (A)—The protocol starts with the induction of iPSC colonies to form the definitive endoderm, which is then differentiated into foregut spheroids. These spheroids are then embedded in a gel basal matrix and further differentiated into LOs. (B)—Bright-field images of the different stages of LO development from iPSCs.
FIGURE 4.
Lung iPSC-derived organoid composition: (A)—The structure of human LOs and their cellular composition. The scheme shows selected major cell types forming in published iPSC-derived LOs models, including club cells, goblet cells, basal cells, alveolar epithelial cells 1 and 2, multiciliated epithelial cells, lung progenitor cells, and mesenchymal cells. While human lung tissue comprises more than 40 different cell type, including infiltrating immune cells, the iPSC-derived organoids models provide unique opportunity to study the “tissue” without the infiltrating hematopoietic cells. Although, iPSC-derived organoids represent excellent in vitro model, they do not fully mimic the human lung tissue complexity. (B)—Main function of mentioned cell types in human the lung.
A recent advance in LO research is represented by reporter iPSCs, which allows the differentiation of different cell types to be tracked within the organoid. Pioneered by Sharma et al. (2018), this method involves fluorescent tagging of endogenous proteins in a stable human iPSC line using CRISPR/Cas9 genome editing technology (Sharma et al., 2018). Since almost any intracellular protein can be tagged, this system represents a powerful tool with the potential for application in many fields of research. Notably, this technique has recently been used in our own laboratory to study the activation of transcriptional factors such as NF-κB during LO differentiation and stimulation (Jose et al., 2020). Hawkins et al. (2021) published the protocol for directed differentiation of human iPSCs into airway basal cells allowing use of the iPSC NKX2-1GFP/TP63tdTomato dual reporter system. This protocol facilitates tracking of the differentiation state of the cells and sorting of the positive population to obtain pure culture of lung progenitor cells (Hawkins et al., 2021). In another study, the fluorescent reporter NKX2-1 and SFTPC iPSCs line were used to generate and purify alveolar epithelial type 2 cells (AEC2s) (Jacob et al., 2017). Importantly, this approach provides opportunities to study AEC dysfunction, which has been implicated in the pathogenesis of many lung disorders (Hawkins et al., 2021).
Advances in iPSC-derived LO generation have led to the generation of organoids with close transcriptional similarities to human fetal lung (Dye et al., 2016); however, their ability to recapitulate the characteristics of the adult tissue remains to be fully defined.
4 The Application of Lung Organoids
The special characteristics of organoids make them an excellent model for a wide range of basic and translational investigations, including drug testing, genetic screening, and disease modeling (Table 1). The major advantage of LO cultures is the interdependent existence of epithelial and mesenchymal cell populations, which—alongside epithelial polarization—is required for proper pulmonary function in vivo (Tan et al., 2017).
TABLE 1.
Lung organoids used for disease investigation: Disease studies where 3D LOs were used as experimental models.
| Disease modeled | Source cell type | References |
|---|---|---|
| Lung cancer | Human primary tumor tissue/cells | Jung et al. (2019); Kim et al. (2019) |
| Patient-derived xenograft models | Shi et al. (2020) | |
| Human lung cancer cell lines | Ramamoorthy et al. (2019) | |
| Human ESCs | Chen et al. (2017) | |
| COPD | Primary human lung epithelial cells | Ng-Blichfeldt et al. (2018) |
| Human lung epithelial cell lines | Tan et al. (2017) | |
| CF | Patient-derived iPSCs | Mou et al. (2012); Wong et al. (2012); Firth et al. (2015) |
| Lung cell pellet from the broncho‐alveolar lavage fluid of patients | Sachs et al. (2019) | |
| IPF | Patient-derived iPSCs | Wilkinson et al. (2017); Strikoudis et al. (2019) |
| Human ESCs | Chen et al. (2017) | |
| Neonatal respiratory distress syndrome | Patient-derived iPSCs | Jacob et al. (2017) |
| Interstitial lung disease | Patient-derived iPSCs | Leibel et al. (2019) |
| Bronchopulmonary dysplasia | Human fetal lung fibroblasts | Sucre et al. (2016) |
| Pulmonary metaplasia | Normal primary human epithelial cells | Danahay et al. (2015) |
| Pulmonary edema | Human pulmonary epithelial cells and microvascular endothelial cells used to form 3D-lung organoids on a chip | Huh et al. (2012) |
| Lung inflammation | Human 3D differentiated airway epithelium cultured on-chip (inflammation induced by IL-13) | Benam et al. (2016) |
| Mouse lung tissue (inflammation induced by bacterial flagellar hooks stimulation) | Shen et al. (2017) | |
| Mouse type 2 alveolar epithelial cells (inflammation induced by IL-1β and TNFα) | Katsura et al. (2019) | |
| Lung tissue injury and regeneration | Primary mouse lung epithelial cells, endothelial cells, and MSCs | Leeman et al. (2019); Riemondy et al. (2019) |
| Respiratory viral infection | IRF7 mutant patient-derived iPSCs (influenza virus infection) | Ciancanelli et al. (2015) |
| Mouse epithelial stem/progenitor cells (influenza virus infection) | Quantius et al. (2016) | |
| Human ESCs (respiratory syncytial virus infection) | Chen et al. (2017) | |
| Human ESCs (parainfluenza virus infection) | Porotto et al. (2019) | |
| Human airway epithelial cell cultures (parechovirus infection) | Karelehto et al. (2018) | |
| Human alveolar epithelial type II of KRT5+ basal cells (severe acute respiratory syndrome coronavirus 2) | Salahudeen et al. (2020) | |
| Human alveolar type 2 cells/pneumocytes (severe acute respiratory syndrome coronavirus 2) | Katsura et al. (2020) | |
| Human epithelial progenitor cells (severe acute respiratory syndrome coronavirus 2) | Xu et al. (2021) | |
| Human alveolar type 2 cells (severe acute respiratory syndrome coronavirus 2) | Ebisudani et al. (2021) | |
| Primary human lung tissue (enterovirus infection) | van der Sanden et al. (2018) |
Used abbreviations: COPD, chronic obstructive pulmonary disease; CF, cystic fibrosis; IPF, idiopathic pulmonary fibrosis; ESCs, embryonal stem cells; iPSCs, induced pluripotent stem cells; MSCs, mesenchymal stromal cells.
4.1 Lung Organoids as Models of Genetic Diseases Affecting the Lungs
LOs have been used successfully to investigate the pathogenesis of CF and protocols have been published for generating disease-specific lung progenitor cells from human CF patient-derived iPSCs (Mou et al., 2012; Wong et al., 2012). The value of this approach lies in the use of a human model, as the murine models may not be exact phenocopy of the human lung disease (Clarke et al., 1992; Snouwaert et al., 1992; Guilbault et al., 2007; Mou et al., 2012). Moreover, patient-derived iPSCs can be used to study the clinical variability of CF without the need for prior analysis of the genetic background of patients (Mou et al., 2012). Furthermore, Mou et al. (2012) used modified RNAs as a replacement for viral vectors to establish iPSC lines that are not genetically modified and therefore, provide an advantage for potential clinical use. Wong et al. (2012) validated the applicability of iPSC-derived epithelial cells as an in vitro model for the drug screening. In this study, CF patient-specific iPSC-derived lung epithelial cells were used to test a novel small molecule compound called a “corrector”, which is able to restore the trafficking of a mutant CFTR protein to the plasma membrane. “Corrector” treatment of cells with the F508del CF mutation resulted in the enhanced plasma membrane localization of the CFTR protein (Wong et al., 2012). Finally, this approach provides the ability to test drugs for personalized medicine and the possible future development of regenerative medicine for lung disorders. An additional advantage of iPSC-derived LOs in the study of lung fibrosis lies in their capacity for expansion of lung stem cell populations and the induction of differentiated cells from very limited amounts of starting material. In one study, Firth et al. (2015) took advantage of this feature to generate iPSC-derived LOs from CF patient fibroblasts. These organoids exhibited fibrotic characteristics in vitro, which were abrogated by CRISPR-mediated CFTR correction, ultimately giving rise to normal LOs (Firth et al., 2015). Alongside these disease-modelling successes, LOs have been instrumental in advances in CF drug testing (Kim et al., 2021). In one study, fibrotic response and collagen accumulation were ameliorated in a LO model of CF by treatment with NP-011, a novel potential anti-fibrotic drug (Kim et al., 2021). In the future, the use of patient-derived organoids may hold the potential for improved diagnostics (Dekkers et al., 2013), with the hope of achieving greater insight into disease processes, and perhaps even personalized therapies for CF (Berkers et al., 2019).
The limitations of mouse models of IPF also make LOs the natural choice for advancing knowledge in the field. This is especially so given the important role of altered extracellular matrix (ECM) composition in IPF pathology, which is readily studied in organoid models of the IPF lung (Kim et al., 2021; Suezawa et al., 2021). These models have yielded important insights, including the identification of new inhibitors of fibrinogenesis, such as NP-011, which was shown to ameliorate fibrosis induced by TGFβ (Kim et al., 2021). Moreover, similar results were obtained using in a mouse model of pulmonary fibrosis, suggesting the important potential of LOs for respiratory disease modeling and drug testing (Kim et al., 2021). Another condition extensively studied using LOs is fibrotic lung injury and the associated repair mechanisms. For instance, Chen et al. (2017) recapitulated fibrotic lung disease in vitro in normal iPSC line-derived LOs by the introduction of a mutation in the HPS1 gene. ECM and MSCs accumulated in affected LOs, in processes similar to those observed in HPS1 mutant-driven lung fibrosis in vivo (Chen et al., 2017).
Cytokine stimulation of organoids has been investigated in the field of pulmonary research. IL-1β is produced by lung interstitial macrophages in mouse lungs following bleomycin-induced injury (Choi et al., 2020). In the alveolar region, IL-1β induces differentiation of AT2 cells into damage-associated transition progenitors (DAPTs), which further differentiate into AT1 and AT2 cells to regenerate the alveolar compartment (Choi et al., 2020). This also occurs in IL-1β-stimulated mouse AT2 organoids (Choi et al., 2020), indicating the suitability of cytokine treatment of organoids for the study of inflammation and regeneration after injury. Choi et al. (2020) also demonstrated that chronic inflammation can be mimicked by IL-1β treatment of LOs, with sustained stimulation with IL-1β found to impair the terminal differentiation of AT1 cells and cause accumulation of DAPTs. Such an increase in the number of cells with expression profiles similar to DAPTs also occurs in the lung tissue of patients with IPF (Choi et al., 2020). Interestingly, withdrawing IL-1β after 14 days of LO stimulation resulted in increased terminal differentiation of AT1 cells (Choi et al., 2020), suggesting the potential benefits of therapies targeting IL-1β in the treatment of IPF. These examples of organoid models of immune responses in various tissues indicate a promising trajectory for basic and preclinical immunological research.
4.2 Lung Organoids to Model Lung Infections—The Model of SARS-CoV-2 Infection
The inflammatory response plays a crucial role in pathology as well as in the regeneration of lung tissue (Lechner et al., 2017). For example, cytokine storm and cytokine-release syndrome are life-threatening systemic inflammatory events involving elevated levels of circulating cytokines and immune cell hyperactivation (Fajgenbaum and June, 2020). In experimental settings, the induction of cytokine storm-like environments can be achieved by infection of lung tissue with a pathogenic agent or by direct stimulation with relevant cytokines. The cytokine approach has the advantage of mimicking the inflammatory niches in culture, even in relatively simplified in vitro tissue models, which lack the complete milieu of naturally occurring cell types.
Airway epithelial cells serve as a first line of defense against pathogen attack or inflammatory stimuli. Accordingly, LOs have been exploited to help understand how epithelial cell function/dysfunction contributes to the pathogenesis of various inflammatory lung diseases and infections (Gkatzis et al., 2018).
Several organoid-based in vitro models of lung infection have been established, which have provided valuable insights into the underlying host-pathogen interactions at the cellular and molecular levels. The recent SARS-CoV-2 pandemic has increased the demand for, and focus on, in vitro models of human lung tissue to facilitate disease pathology investigations and drug testing experiments. A seminal work in the field of LO infection models was published by Chen et al. (2017), who presented a protocol to generate PSC-derived lung bud organoids consisting of mesodermal (Vim+, CD90+) and pulmonary endodermal (FOXA2+, NKX2.1+, EPCAM+, SOX9+) cells. These organoids were shown to undergo branch morphogenesis when cultured in 3D Matrigel or transplanted into a mouse, rendering the model highly relevant as branching morphogenesis is a critical step in lung tissue development (Schittny, 2017). The researchers also infected the organoids with respiratory syncytial virus (RSV), which causes small airway obstruction and bronchiolitis in infants. They revealed a process of shedding of swollen, infected cells similar to that seen in infected human lungs (Chen et al., 2017). Therefore, RSV-infected LOs represent a useful model for RSV infection research, especially as most commonly used mouse models are limited by crucial differences between human and murine physiology, especially in metabolism, which may also be due to the fact that model organisms develop faster than humans (Kim et al., 2020).
A system for the culture of organoids derived from a single adult human alveolar epithelial type II cell (AT2) has also been developed. The resulting organoids successfully supported the differentiation of AT2 cells into AT1 cells (Salahudeen et al., 2020), which together form the human lung epithelium, in which AT2 cells produce pulmonary surfactant proteins, and AT1 cells cover most of the surface area of the alveoli and perform the function of gas exchange (Cunniff et al., 2021). These organoids have also been used to model human SARS-CoV-2 infection (Salahudeen et al., 2020). Around 10% of AT2-derived organoids (specifically the SFPTC+ cells) and 10% of basal cell-derived organoids (specifically SCGB1A1+ club cells) were infected with SARS-CoV-2. These results indicate that AT2 cells are directly infected by SARS-CoV-2 and that club cells are a distal lung target population (Salahudeen et al., 2020).
Xu et al. (2021) used LOs to examine the innate cellular immune response of lung tissue during SARS-CoV-2 infection. They found that expression of receptor interacting serine/threonine-protein kinase 1 (RIPK1), an important mediator of inflammation and cell death (Mifflin et al., 2020), is upregulated in patients with COVID-19 who experience a cytokine storm, and is also activated in SARS-CoV-2-infected primary LOs. Interestingly, treating infected LOs with the RIPK1-inhibitor Nec-1s reduced the transcription of proinflammatory cytokines, as well as ACE2 and the epidermal growth factor receptor (EGFR), which mediate viral entry (Xu et al., 2021).
Clinical trials of the immunomodulatory and clinical effects of another RIPK1 inhibitor, SAR443122, in patients with severe COVID-19 are currently in progress. Interestingly, these studies conducted in LOs, show the applicability of these organoids as suitable models to dissect the pathological mechanisms underlying infectious diseases. Moreover, LOs can be used for rapid screening of the infectivity of emerging human airway pathogens and to test disease-specific therapeutics. Using LO models to test the safety and efficacy of potential drugs instead of time-consuming and costly clinical trials can greatly contribute to faster and cheaper development of new drugs. Indeed, an LO model has been used to test drugs for SARS-CoV-2 infection (Ebisudani et al., 2021). In this model, infected LOs were treated with physiologically relevant concentrations of lopinavir, nelfinavir, and remdesivir (Ebisudani et al., 2021). Lopinavir and nelfinavir are protein inhibitors commonly used in combination with other drugs for the treatment of HIV-1 infection in adults, adolescents and children (Croxtall and Perry, 2010). Remdesivir is an adenosine nucleotide analogue with broad-spectrum activity against viruses from various families (Lamb, 2020). Although lopinavir and nelfinavir significantly decreased the viral titer, SARS-CoV-2 viral replication was not halted. Interestingly, remdesivir inhibited the replication of the virus in infected organoids (Ebisudani et al., 2021). These results correlate with clinical data of patients infected by SARS-CoV-2 (Beigel et al., 2020; Grein et al., 2020; Ebisudani et al., 2021) and thus, suggest that LOs represent appropriate models for the study of infectious lung diseases.
To conclude, organoids have been already used successfully as models of human disease caused by infections with viruses such SARS-CoV-2 and RSV. These models represent effective and relatively cheap tools for preclinical and clinical trials of potential treatment strategies. Furthermore, LO models of human lung tissue and infectious disease offer possibilities for comparison with, and validation of, serological data from the patients in the clinic.
4.3 Lung Organoids in Cancer Research
Compared to conventional 2D monolayers and suspension cell cultures, LOs used in cancer research provide a valuable opportunity to develop physiologically, genetically, and histologically relevant tumor models. LOs can mimic some of the high cellular organization, tumor microenvironment, and cell interactions of in vivo tumor tissue. As such, LOs have been used in various contexts from carcinogenesis, to personalized medicine and drug development (Gunti et al., 2021).
Lung cancer organoids (tumoroids) precisely reflect the histological features of primary lung tumors and maintain their genomic abnormalities during long-term expansion in vitro. Recognizing that these models represent a valuable resource, Kim et al. (2019) created a biobank of 80 lung tumoroids from the five most frequent subtypes of lung cancers, and five normal bronchial organoids as controls, which replicate the unique histological features of the primary tissues. Such biobanks serve as a beneficial platform for drug testing and pre-clinical research to advance personalized medicine approaches.
Similar to LOs derived from primary lung progenitors, organoids established from lung cancer cell lines or primary tumor cells contain only a few cell types and so fail to completely replicate the tumor in vitro (Fiorini et al., 2020). Nevertheless, these systems have been used successfully in drug sensitivity studies. In one such study, high-throughput screening of anticancer agents on liver organoids indicated that patient-derived organoids can mimic a patient’s response to a particular therapy. Thus, these types of organoids could potentially be used for the development of personalized medicine approaches or as part of clinical trials (Vlachogiannis et al., 2018). Indeed, others have shown that cancer-cell-derived LOs can serve as a tool for pre-treatment drug screening and personalized medicine. Hai et al. (2020) developed genetically modified mouse LOs to mimic human lung squamous cell carcinoma (LSCC) and used this model to demonstrate that treatment with the WEE1 inhibitor (AZD1775) and the adjuvant PD-1 inhibitor enhanced T-cell anti-tumour activity. This demonstrates the importance of drug efficacy screening for identifying more efficient therapeutic drug combinations for evaluation in future clinical trials (Hai et al., 2020). Shi et al. (2020) published a protocol for in vitro generation of non-small cell lung cancer (NSCLC) organoids that mimic the histological attributes of the original tumor tissue. Moreover, these NSCLC organoids retained the molecular profile of matching tumor tissue long-term, even after multiple passages (>3 months, >10 passages). Next, they and others showed that NSCLC organoids and PDX models from the same parental tissue exhibit similar drug responses and proposed that organoid cancer models are a valid pre-clinical model to explore novel therapeutic options for NSCLC disease (Hai et al., 2020; Li et al., 2020; Shi et al., 2020; Wang et al., 2020b). Li et al. (2020) established organoid models of lung adenocarcinoma (LADC), the most common subtype of NSCLC. They also showed that the in vitro generated organoids preserve the key tumor features and therefore, can be used as a valuable pre-clinical tool. Using LADC organoids derived from 12 cancer lines they performed high-throughput drug response screening and showed that the sensitivity to a particular drug was consistent throughout individual passages and also that the response to the drug varied among different organoid lines (Li et al., 2020). Moreover, they discovered unpredicted drug sensitivity regardless of genetic markers. For example, the ACI-3_O line and SOL-4_O line responded gefitinib despite the lack of EGFR mutations. Again, their conclusions highlight the benefits of drug response screening on organoids (Li et al., 2020). Kim et al. (2019) used LOs generated from patient-derived cells to dissect the genetic and phenotypic basis of heterogeneous responses to anticancer therapy. They demonstrated that variability in drug responses correlated with particular genomic mutations and persisted during multiple organoid passages. For example, organoids with a BRCA2 gene mutation showed increased sensitivity to the PARP inhibitor, Olaparib (Kim et al., 2019). Moreover, PDX derived from these organoids also responded to Olaparib treatment. On the other hand, although two organoids derived from different tissues expressed the same EGFR mutation, their responses to erlotinib and crizotinib differed due to secondary mutations. Overall, these studies highlight the critical importance of the LO biobank mentioned above for anti-tumor drug screening to predict individual patient drug responses (Kim et al., 2019).
The results mentioned above demonstrate that organoids are suitable for high-throughput screening while providing the advantages of 3D cancer models which, combined with clinical evaluation of specific genetic alterations, can serve as a tool for personalized therapy. Indeed, another recent study by Hu et al. (2021) proved that LOs successfully mimic original tumor tissue features and preserve them after in vitro cultivation. Moreover, they developed an integrated superhydrophobic microwell array chip (InSMAR-chip) suitable for high-throughput testing. With this set-up, they analyzed drugs administered during patient treatment and obtained results showing 100% accuracy and specificity in 10 of 21 samples. The remaining 11 on-chip samples could not be compared to relative patient responses due to differences in subsequent treatment. Nevertheless, this study showed a strong correlation of the drug response with genetic mutation and clinical outcomes and therefore, confirmed this as a promising approach for predicting the most effective drugs for individual patients (Hu et al., 2021).
As a result of such successes, LOs derived from patients with various lung pathologies have now entered clinical trials to establish their efficacy in predicting individual patients’ responses to a specific therapy (Table 2).
TABLE 2.
Clinical trials in lung organoids: List of the clinical trials using 3D LOs as in vitro models.
| Disease | Model | Source of the cells | Purpose of the study | ClinicalTrials.gov Identifier |
|---|---|---|---|---|
| Lung cancer | Spheroids | Lung tumor biopsies | Characterization of the consistency and accuracy of the organoids derived from patient lung biopsies to predict clinical response to the chemotherapy | NCT03979170 |
| Patient-derived LOs | ||||
| 3D model OncoCilAir™ (OncoTheis) | ||||
| Lung cancer | Patient-derived normal and cancer LOs | Biopsies from endobronchial tumors or lymph nodes | Biobanking of normal and primary lung cancer organoids. Analysis of microvesicles secreted by lung cancer cells in organoid-derived culture supernatants and patient blood samples. Comparison of the response to the drugs in normal and cancer LOs | NCT05092009 |
| Blood samples | ||||
| Lung cancer | Patient-derived organoids | Tumor tissue biopsies | Comparison of the xenografts with donor tissue. Testing novel anti-cancer treatment. Developing assays to predict tumor response to the drug | NCT04859166 |
| Xenografts | ||||
| CF | Organoids derived from the tissue of patients with the R334W-CFTR mutation | Rectal biopsies | Study of the response of organoids to CFTR modulators, which will be compared to the patients’ response to the same drug in the next study | NCT04254705 |
| Lung cancer | Patient-derived LOs | Lung tumor biopsies | Biobanking of organoids derived from stage I–IV lung cancer patients | NCT03655015 |
| Lung cancer | Patient-derived LOs | Non-small cell lung cancer patient biopsies | Use of organoids for the drug sensitivity testing and comparison with clinical treatment data | NCT03453307 |
| COPD and IPF | Patient-derived LOs | Lung tissue biopsies from patients with emphysema or pulmonary fibrosis | Characterization of the stem cell niche in different tissues (healthy, emphysematous and fibrotic pulmonary tissue). Further use of the organoids for drug screening and personalized medicine | NCT02705144 |
| Lung cancer | Patient-derived LOs | Non-small cell lung cancer patient biopsies | Testing of different drugs in vitro using organoids. Evaluation of the responders to Osimertinib and screening of alternative therapies for non-responders in vitro | NCT05136014 |
| Lung cancer | Patient-derived LOs | Tumor biopsies | Use of organoids and a microfluidic system as an innovative model of the tumor microenvironment and HUVECS or endothelial cells as a model of tumor vascularization, to create a tool for personalized medicine | NCT04826913 |
| Microfluidic system | Blood samples | |||
| Lung cancer | Patient-derived organoids from lung tumors or other solid tumors | Lung cancer tissue or solid tumor biopsies | Co-cultivation of organoids with lymphocytes to screen for tumor-responsive T-cells, which will be further expanded and used as immunotherapy for the patient | NCT03778814 |
| TILs or/and peripheral T-cells | ||||
| CF | Patient-derived organoids | Not specified | Use of an ex vivo organoid model to establish the correlation between the clinical response of CF patients to Vx-770 (Ivacaftor) | NCT03390985 |
Used abbreviations: COPD, chronic obstructive pulmonary disease; CF, cystic fibrosis; IPF, idiopathic pulmonary fibrosis; LOs, lung organoids; 3D, three-dimensional; CFTR, cystic fibrosis transmembrane conductance regulator.
These studies of high-throughput screening have highlighted the potential of cancer organoids as a promising tool for personalized treatment. Using such progressive methods, organoids can be generated for each individual patient and used to screen drug responses based on the initial clinical tests of specific genetic mutations and gene expression profiles. This approach holds great potential for achieving better clinical outcomes and prolonged survival of patients. High-throughput testing also offers a great advantage for screening samples currently available in biobanks. Furthermore, as more patient-derived samples are contributed, the collection of unique genetic alterations and their specific combinations will expand. Thus, we expect that this approach will provide a standardized tool for rapid development and testing of new anti-tumor treatments.
5 Lung Organoids: Future Prospects and Remaining Challenges
So far, we have discussed the great progress made using primary lung progenitor cell-derived and iPSC-derived LOs in modeling human diseases such as CF, IPF, lung cancer, and SARS-CoV-2 infection. However, certain challenges still need to be addressed to improve the relevance of these models to in vivo conditions. Several approaches have been developed to further enhance the physiological resemblance of in vitro 3D models to lung tissue in vivo. One is based on the cultivation of lung epithelial cells at an air-liquid interface, where the basolateral side of the epithelial cells is immersed in culture medium while the apical side is exposed to humidified air. This approach was used by Mas et al. (2016) to develop OncoCilAir™ tumor-stroma airway model by co-culture of lung adenocarcinoma cells, human primary bronchial cells, and lung fibroblasts. This model provides an alternative means of testing drug efficacy and toxicity that could replace animal models by overcoming the disadvantages of differences in rodent cancer physiology and genetic features. Specifically, the major drawback of animal lung models is the diversity in progression of lung disease and response to therapy (Mas et al., 2016).
The newly developed cultures that incorporate an extracellular scaffold represent a further advance that enables standardized long-term cultivation conditions and reproducibility (Zscheppang et al., 2018). Moreover, this approach facilitates exploration of the interaction between the ECM and lung cells in the context of aberrant repair or regeneration. The scaffolds are generated by decellularization of healthy human lung tissue or tissue derived from patients with conditions such as IPF. These models will contribute greatly to our understanding of the pathogenesis of lung disease and evaluation of the role of ECM in the onset and course of these pathologies (Zscheppang et al., 2018).
While 3D models have many advantages compared to conventional 2D models, many still fail to fully represent even a small fraction of the dynamic features of human lungs, such as the processes of nutrient and gas exchange, the mechanical forces created by breathing movements, and dynamic flow conditions. The tumor-on-a-chip model however, was designed to overcome these limitations (Barros et al., 2021). These microfluidic models of lung tumors are often derived from established lung cancer cell lines, which form a spheroid in the microchannel, adding a further 3D aspect to these studies (Mehta et al., 2022). Microfluidic devices have been used to study drug resistance, the efficiency of photodynamic therapy, the influence of mechanical forces in the lungs on tumor progression and drug resistance, and characterization of cell-cell communication in the tumor microenvironment (Del Piccolo et al., 2021). For example, Xu et al. (2013) developed a microfluidic device to study drug sensitivity of a co-cultured human non-small cell lung cancer cell line (SPCA-1) and stromal cells. They also established a culture of fresh lung cancer tissues from eight patients in their microchip device, in order to identify personalized medical treatments. This approach accomplished replication of the actual condition of the solid tumor in vivo. Microfluidic devices provide an easy way to administer single or combined anti-cancer treatments in high-throughput assays, which is beneficial for the development of personalized medicine. Further advantages are that these devices require only small amounts of samples and reagents, short assay time and high sensitivity. Microfluidic devices can also be used with various tissue set-ups, such as cell line monolayers or co-culture of cancer cell lines and stromal cells, to more accurately recapitulate tumor tissue in vivo, or even primary fresh tissue. This approach revealed significant differences in treatment sensitivity, with the poorest detected in fresh tissue (Xu et al., 2013). Thus, further studies are warranted to develop methods for cultivation of patient tumor tissue in vitro while closely maintaining the physiological features of the human body that can be used to exploit the potential of organoids in the development of personalized therapy.
Sepsis and septic shock greatly contribute to the mortality of human population (Rudd et al., 2020). Therefore, the development of an in vitro model of sepsis represents another prospect for the LO approach for defining the exact pathogenesis of the disease and identifying potential therapeutic strategies and new biomarkers. However, an in vitro model of sepsis-induced lung injury based on LO technology may require a complex set-up due to the number of different cell types and their specific effects in septic reactions. For instance, hypoxemia, which is an important factor in the pathophysiology of ALI/ARDS, is caused by neutrophil entrapment in the pulmonary microvasculature (Park et al., 2019). Therefore, to create a relevant in vitro model of sepsis might warrant culture under hypoxic conditions. Nevertheless, similar to the situation for infectious diseases, a great advantage of LOs as a model of human sepsis is in the possibility of comparison with, and validation of, serological data from patients in the clinic.
The use of iPSCs to derive LOs has rapidly advanced the field in recent years; however, these models are not yet perfect. LOs developed from iPSCs resemble fetal tissues rather than adult lungs and, therefore, transplantation into a living organism may be required to fully recapitulate the adult lung phenotype (Dye et al., 2015). This engraftment procedure has been successfully achieved using humanized/immunodeficient mouse recipients to develop mature intestinal (Cortez et al., 2018; Poling et al., 2018; Boyle et al., 2021), brain (Dong et al., 2021), and kidney (van den Berg et al., 2018) organoids, but it is not so commonly practiced with LOs. An early study by Dye et al. (2016) demonstrated that human ESC-derived LOs can be maintained for up to 15 weeks after engraftment into the epididymal fat pad of male NOD-scid IL2Rgnull mice. After the engraftment, LOs improved their cellular differentiation of secretory lineages and formed airway-like structures that were similar to adult human lungs, including vasculature and smooth muscles. However, a bioartificial microporous poly (lactide-co-glycolide) (PLG) scaffold niche was necessary for proper engraftment (Dye et al., 2016), which requires advanced techniques for its fabrication. Nevertheless, alternative approaches such as decellularized lung scaffolds are also available to provide relevant physical and environments for the generation of organized lung structures (Gilpin et al., 2014).
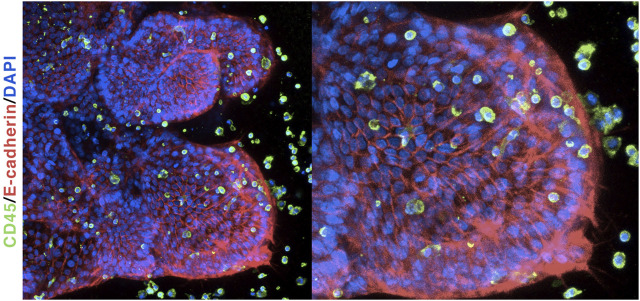
A notable drawback of iPSC-derived LOs is the lack of a pulmonary immune cell populations. The human lung comprises both tissue-resident immune cells (such as pulmonary macrophages) as well as infiltrating immune cells, such as neutrophils, monocytes, and T-cells, which are not found in LOs. This immune cell-deprived scenario renders such a model unsuitable for studying diseases in which immune cells have a major role—including microbial infections and inflammation. However, recent studies have demonstrated that immune cells can be co-cultured with organoids to generate a more physiologically relevant model. Our own findings show that iPSC-derived LOs, although deprived of immune cell populations, can respond to microbial ligands and are able to recruit human primary monocytes in vitro (Jose et al., 2020). Interestingly, co-cultured monocytes interacted closely with the organoid tissue and significantly changed their phenotype (Figure 5). Similarly, another study showed that RSV-infected airway organoids were able to recruit and interact with human neutrophils (Sachs et al., 2019). Taken together, these results suggest that co-culturing LOs with primary immune cells will allow a better dissection of the interactions of immune cells with human tissues and thus, contribute to a better understanding of complex pathologies.
FIGURE 5.
Lung organoids co-cultured with primary monocytes: Immunofluorescent labeling of human LOs shows tissue polarity and recruitment of human monocytes.
As already mentioned, the local microbiota plays a crucial role in the pathology and homeostasis of the lung tissue (Paolicelli et al., 2019; Barcik et al., 2020). Although many studies have focused on the role of the microbiota in the gastrointestinal tract using human intestinal organoids (Min et al., 2020), few studies using LOs have been reported. Therefore, future research should focus on dissection of the cell-cell interaction of the microbiota with lung tissue and the molecular mechanisms by which the microbiota influences the development, pathology, and homeostasis of human lung.
6 Discussion
Using 3D organoids as a tool to understand lung development and function holds great potential for generating important insights not only into genetic lung disorders, but also into diseases such as COPD, CF, and lung infections. The generation of LOs from human PSCs is crucial for overcoming the obstacles to in vitro studies of human cells, which for many years have relied on the scant availability of post-mortem tissues. LOs can bridge the knowledge gap between lung disease pathways identified in rodent models and therapeutic possibilities in their human counterparts. The end of our long-term dependence on rodent models for genetic manipulation studies may also be heralded by the emergence of novel genome editing techniques such as CRISPR, which could be used to generate genetic models of pulmonary disease using organoids. This approach would facilitate mechanistic studies in a physiological setting that closely resembles the human respiratory system without the need for presumably costly and time-consuming animal research. The recent advances in the use of iPSC-derived LO in clinical trials are also promising in terms of drug screening for personalized treatment or transplantation of healthy tissues, especially allogenic tissues, in patients, and hints at a future that includes “organoid-based treatment” of pulmonary diseases.
In summary, we believe that organoids represent the future of lung disease modeling as they allow long-term cultivation of human lung tissue, maintaining many of its phenotypical, and functional characteristics. LOs represent one of the best tools for translational research, due to the comparatively inexpensive and increasingly biologically-relevant methodology. The past 10 years have seen an increasing number of studies leveraging organoids to shed light on previously unexplained pathologies of diseases such as IPF (Chen et al., 2017; Kim et al., 2021; Suezawa et al., 2021), CF (Dekkers et al., 2013; Firth et al., 2015; Kim et al., 2021), and lung cancer (Hai et al., 2020; Li et al., 2020; Shi et al., 2020; Wang et al., 2020b; Gunti et al., 2021; Hu et al., 2021). Harnessing the potential that LOs have to offer will promote the design of new treatments and diagnostic methodologies and thus, finally improve the quality of life of people affected by various pulmonary diseases.
Acknowledgments
Authors would like to thank Lucy Robinson and Jessica Tamanini of Insight Editing London for critically reviewing the manuscript before submission.
Author Contributions
VB prepared the figures and table and wrote the manuscript. MZ wrote and critically reviewed the manuscript. LS and ZG wrote the manuscript. SJ prepared the table and wrote the manuscript. MH and TZ critically reviewed the manuscript. JF conceptualized, wrote, and critically reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The authors were supported by the European Social Fund and European Regional Development Fund—Project MAGNET (No. CZ.02.1.01/0.0/0.0/15_003/0000492) and ENOCH (CZ.02.1.01/0.0/0.0/16_019/0000868), and by the Ministry of Health of the Czech Republic—DRO (Institute of Hematology and Blood Transfusion—IHBT, 00023736) and grant nr. NU22-A-121. All rights reserved. MZ was supported by the European Regional Development Fund—Project Support of MSCA IF fellowships at FNUSA-ICRC (No. CZ.02.2.69/0.0/0.0/19_074/0016274).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Aichler M., Kunzke T., Buck A., Sun N., Ackermann M., Jonigk D., et al. (2018). Molecular Similarities and Differences from Human Pulmonary Fibrosis and Corresponding Mouse Model: MALDI Imaging Mass Spectrometry in Comparative Medicine. Lab. Invest. 98, 141–149. 10.1038/labinvest.2017.110 [DOI] [PubMed] [Google Scholar]
- Angus D. C., van der Poll T. (2013). Severe Sepsis and Septic Shock. N. Engl. J. Med. 369, 840–851. 10.1056/nejmra1208623 [DOI] [PubMed] [Google Scholar]
- Ardini-Poleske M. E., Clark R. F., Ansong C., Carson J. P., Corley R. A., Deutsch G. H., et al. (2017). LungMAP: The Molecular Atlas of Lung Development Program. Am. J. Physiology-Lung Cell. Mol. Physiology 313, L733–L740. 10.1152/ajplung.00139.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz M., Jacob A., Yang W. L., Matsuda A., Wang P. (2013). Current Trends in Inflammatory and Immunomodulatory Mediators in Sepsis. J. Leukoc. Biol. 93, 329–342. 10.1189/jlb.0912437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcik W., Boutin R. C. T., Sokolowska M., Finlay B. B. (2020). The Role of Lung and Gut Microbiota in the Pathology of Asthma. Immunity 52, 241–255. 10.1016/j.immuni.2020.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas C. E., Cronce M. J., Rackley C. R., Bowie E. J., Keene D. R., Stripp B. R., et al. (2013). Type 2 Alveolar Cells Are Stem Cells in Adult Lung. J. Clin. Invest. 123, 3025–3036. 10.1172/jci68782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros A. S., Costa A., Sarmento B. (2021). Building Three-Dimensional Lung Models for Studying Pharmacokinetics of Inhaled Drugs. Adv. drug Deliv. Rev. 170, 386–395. 10.1016/j.addr.2020.09.008 [DOI] [PubMed] [Google Scholar]
- Beigel J. H., Tomashek K. M., Dodd L. E., Mehta A. K., Zingman B. S., Kalil A. C., et al. (2020). Remdesivir for the Treatment of Covid-19 - Final Report. N. Engl. J. Med. 383, 1813–1826. 10.1056/nejmoa2007764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benam K. H., Villenave R., Lucchesi C., Varone A., Hubeau C., Lee H. H., et al. (2016). Small Airway-On-A-Chip Enables Analysis of Human Lung Inflammation and Drug Responses In Vitro . Nat. Methods 13, 151–157. 10.1038/nmeth.3697 [DOI] [PubMed] [Google Scholar]
- Berkers G., van Mourik P., Vonk A. M., Kruisselbrink E., Dekkers J. F., de Winter-de Groot K. M., et al. (2019). Rectal Organoids Enable Personalized Treatment of Cystic Fibrosis. Cell Rep. 26, 1701–1708. e3. 10.1016/j.celrep.2019.01.068 [DOI] [PubMed] [Google Scholar]
- Bleijs M., van de Wetering M., Clevers H., Drost J. (2019). Xenograft and Organoid Model Systems in Cancer Research. EMBO J. 38, e101654. 10.15252/embj.2019101654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle M. A., Sequeira D. J., McNeill E. P., Criss Z. K., 2nd, Shroyer N. F., Speer A. L. (2021). Vivo Transplantation of Human Intestinal Organoids Enhances Select Tight Junction Gene Expression. J. Surg. Res. 259, 500–508. 10.1016/j.jss.2020.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger G. A., Weusten B. L. A., Bogte A., Begthel H., Brosens L. A. A., Clevers H. (2021). Human Gastrointestinal Epithelia of the Esophagus, Stomach, and Duodenum Resolved at Single-Cell Resolution. Cell Rep. 34, 108819. 10.1016/j.celrep.2021.108819 [DOI] [PubMed] [Google Scholar]
- Castellani S., Di Gioia S., di Toma L., Conese M. (2018). Human Cellular Models for the Investigation of Lung Inflammation and Mucus Production in Cystic Fibrosis. Anal. Cell Pathol. (Amst) 2018, 3839803. 10.1155/2018/3839803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda D., Otoupalova E., Smith S. R., Volckaert T., De Langhe S. P., Thannickal V. J. (2019). Developmental Pathways in the Pathogenesis of Lung Fibrosis. Mol. Aspects Med. 65, 56–69. 10.1016/j.mam.2018.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D., Sharma L. (2020). Harnessing Murine Microbiome Models to Study Human Lung Microbiome. Chest 157, 776–778. 10.1016/j.chest.2019.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charavaryamath C., Janardhan K. S., Caldwell S., Singh B. (2006). Pulmonary Intravascular Monocytes/macrophages in a Rat Model of Sepsis. Anat. Rec. A Discov. Mol. Cell Evol. Biol. 288, 1259–1271. 10.1002/ar.a.20401 [DOI] [PubMed] [Google Scholar]
- Chen Y. W., Huang S. X., de Carvalho A., Ho S. H., Islam M. N., Volpi S., et al. (2017). A Three-Dimensional Model of Human Lung Development and Disease from Pluripotent Stem Cells. Nat. Cell Biol. 19, 542–549. 10.1038/ncb3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Iich E., Lee J. H. (2016). Organogenesis of Adult Lung in a Dish: Differentiation, Disease and Therapy. Dev. Biol. 420, 278–286. 10.1016/j.ydbio.2016.10.002 [DOI] [PubMed] [Google Scholar]
- Choi J., Park J. E., Tsagkogeorga G., Yanagita M., Koo B. K., Han N., et al. (2020). Inflammatory Signals Induce AT2 Cell-Derived Damage-Associated Transient Progenitors that Mediate Alveolar Regeneration. Cell Stem Cell 27, 366–382 e7. 10.1016/j.stem.2020.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciancanelli M. J., Huang S. X., Luthra P., Garner H., Itan Y., Volpi S., et al. (2015). Infectious Disease. Life-Threatening Influenza and Impaired Interferon Amplification in Human IRF7 Deficiency. Science 348, 448–453. 10.1126/science.aaa1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L. L., Grubb B. R., Gabriel S. E., Smithies O., Koller B. H., Boucher R. C. (1992). Defective Epithelial Chloride Transport in a Gene-Targeted Mouse Model of Cystic Fibrosis. Science 257, 1125–1128. 10.1126/science.257.5073.1125 [DOI] [PubMed] [Google Scholar]
- Cleary S. J., Pitchford S. C., Amison R. T., Carrington R., Robaina Cabrera C. L., Magnen M., et al. (2020). Animal Models of Mechanisms of SARS-CoV-2 Infection and COVID-19 Pathology. Br. J. Pharmacol. 177, 4851–4865. 10.1111/bph.15143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. (2016). Modeling Development and Disease with Organoids. Cell 165, 1586–1597. 10.1016/j.cell.2016.05.082 [DOI] [PubMed] [Google Scholar]
- Cortez A. R., Poling H. M., Brown N. E., Singh A., Mahe M. M., Helmrath M. A. (2018). Transplantation of Human Intestinal Organoids into the Mouse Mesentery: A More Physiologic and Anatomic Engraftment Site. Surgery 164, 643–650. 10.1016/j.surg.2018.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxtall J. D., Perry C. M. (2010). Lopinavir/Ritonavir: a Review of its Use in the Management of HIV-1 Infection. Drugs 70, 1885–1915. 10.2165/11204950-000000000-00000 [DOI] [PubMed] [Google Scholar]
- Cunniff B., Druso J. E., van der Velden J. L. (2021). Lung Organoids: Advances in Generation and 3D-Visualization. Histochem Cell Biol. 155, 301–308. 10.1007/s00418-020-01955-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danahay H., Pessotti A. D., Coote J., Montgomery B. E., Xia D., Wilson A., et al. (2015). Notch2 Is Required for Inflammatory Cytokine-Driven Goblet Cell Metaplasia in the Lung. Cell Rep. 10, 239–252. 10.1016/j.celrep.2014.12.017 [DOI] [PubMed] [Google Scholar]
- David B., Bafadhel M., Koenderman L., De Soyza A. (2021). Eosinophilic Inflammation in COPD: from an Inflammatory Marker to a Treatable Trait. Thorax 76, 188–195. 10.1136/thoraxjnl-2020-215167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H., Bignell G. R., Cox C., Stephens P., Edkins S., Clegg S., et al. (2002). Mutations of the BRAF Gene in Human Cancer. Nature 417, 949–954. 10.1038/nature00766 [DOI] [PubMed] [Google Scholar]
- De Boeck K. (2020). Cystic Fibrosis in the Year 2020: A Disease with a New Face. Acta Paediatr. 109, 893–899. 10.1111/apa.15155 [DOI] [PubMed] [Google Scholar]
- De Luca A., Pariano M., Cellini B., Costantini C., Villella V. R., Jose S. S., et al. (2017). The IL-17F/IL-17RC Axis Promotes Respiratory Allergy in the Proximal Airways. Cell Rep. 20, 1667–1680. 10.1016/j.celrep.2017.07.063 [DOI] [PubMed] [Google Scholar]
- Dekkers J. F., Wiegerinck C. L., de Jonge H. R., Bronsveld I., Janssens H. M., de Winter-de Groot K. M., et al. (2013). A Functional CFTR Assay Using Primary Cystic Fibrosis Intestinal Organoids. Nat. Med. 19, 939–945. 10.1038/nm.3201 [DOI] [PubMed] [Google Scholar]
- Del Piccolo N., Shirure V. S., Bi Y., Goedegebuure S. P., Gholami S., Hughes C. C. W., et al. (2021). Tumor-on-chip Modeling of Organ-specific Cancer and Metastasis. Adv. drug Deliv. Rev. 175, 113798. 10.1016/j.addr.2021.05.008 [DOI] [PubMed] [Google Scholar]
- Dong X., Xu S. B., Chen X., Tao M., Tang X. Y., Fang K. H., et al. (2021). Human Cerebral Organoids Establish Subcortical Projections in the Mouse Brain after Transplantation. Mol. psychiatry 26, 2964–2976. 10.1038/s41380-020-00910-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J., Clevers H. (2018). Organoids in Cancer Research. Nat. Rev. Cancer 18, 407–418. 10.1038/s41568-018-0007-6 [DOI] [PubMed] [Google Scholar]
- Duval K., Grover H., Han L.-H., Mou Y., Pegoraro A. F., Fredberg J., et al. (2017). Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology 32, 266–277. 10.1152/physiol.00036.2016 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Dwyer-Lindgren L., Bertozzi-Villa A., Stubbs R. W., Morozoff C., Shirude S., Naghavi M., et al. (2017). Trends and Patterns of Differences in Chronic Respiratory Disease Mortality Among US Counties, 1980-2014. Jama 318, 1136–1149. 10.1001/jama.2017.11747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye B. R., Dedhia P. H., Miller A. J., Nagy M. S., White E. S., Shea L. D., et al. (2016). A Bioengineered Niche Promotes In Vivo Engraftment and Maturation of Pluripotent Stem Cell Derived Human Lung Organoids. Elife 5. 10.7554/eLife.19732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye B. R., Hill D. R., Ferguson M. A., Tsai Y. H., Nagy M. S., Dyal R., et al. (2015). In Vitro generation of Human Pluripotent Stem Cell Derived Lung Organoids. Elife 4. 10.7554/eLife.05098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebisudani T., Sugimoto S., Haga K., Mitsuishi A., Takai-Todaka R., Fujii M., et al. (2021). Direct Derivation of Human Alveolospheres for SARS-CoV-2 Infection Modeling and Drug Screening. Cell Rep. 35, 109218. 10.1016/j.celrep.2021.109218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman J. A., Zejnullahu K., Mitsudomi T., Song Y., Hyland C., Park J. O., et al. (2007). MET Amplification Leads to Gefitinib Resistance in Lung Cancer by Activating ERBB3 Signaling. Science 316, 1039–1043. 10.1126/science.1141478 [DOI] [PubMed] [Google Scholar]
- Fajgenbaum D. C., June C. H. (2020). Cytokine Storm. N. Engl. J. Med. 383, 2255–2273. 10.1056/nejmra2026131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorini E., Veghini L., Corbo V. (2020). Modeling Cell Communication in Cancer with Organoids: Making the Complex Simple. Front. Cell Dev. Biol. 8, 166. 10.3389/fcell.2020.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth A. L., Menon T., Parker G. S., Qualls S. J., Lewis B. M., Ke E., et al. (2015). Functional Gene Correction for Cystic Fibrosis in Lung Epithelial Cells Generated from Patient iPSCs. Cell Rep. 12, 1385–1390. 10.1016/j.celrep.2015.07.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks T. J., Colby T. V., Travis W. D., Tuder R. M., Reynolds H. Y., Brody A. R., et al. (2008). Resident Cellular Components of the Human Lung: Current Knowledge and Goals for Research on Cell Phenotyping and Function. Proc. Am. Thorac. Soc. 5, 763–766. 10.1513/pats.200803-025hr [DOI] [PubMed] [Google Scholar]
- Gazdar A. F., Girard L., Lockwood W. W., Lam W. L., Minna J. D. (2010). Lung Cancer Cell Lines as Tools for Biomedical Discovery and Research. J. Natl. Cancer Inst. 102, 1310–1321. 10.1093/jnci/djq279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin S. E., Ren X., Okamoto T., Guyette J. P., Mou H., Rajagopal J., et al. (2014). Enhanced Lung Epithelial Specification of Human Induced Pluripotent Stem Cells on Decellularized Lung Matrix. Ann. Thorac. Surg. 98, 1721–1729. 10.1016/j.athoracsur.2014.05.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkatzis K., Taghizadeh S., Huh D., Stainier D. Y. R., Bellusci S. (2018). Use of Three-Dimensional Organoids and Lung-On-A-Chip Methods to Study Lung Development, Regeneration and Disease. Eur. Respir. J. 52. 10.1183/13993003.00876-2018 [DOI] [PubMed] [Google Scholar]
- Gottschling S., Jauch A., Kuner R., Herpel E., Mueller-Decker K., Schnabel P. A., et al. (2012). Establishment and Comparative Characterization of Novel Squamous Cell Non-small Cell Lung Cancer Cell Lines and Their Corresponding Tumor Tissue. Lung cancer 75, 45–57. 10.1016/j.lungcan.2011.05.020 [DOI] [PubMed] [Google Scholar]
- Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., et al. (2020). Compassionate Use of Remdesivir for Patients with Severe Covid-19. N. Engl. J. Med. 382, 2327–2336. 10.1056/nejmoa2007016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb B. R., Boucher R. C. (1999). Pathophysiology of Gene-Targeted Mouse Models for Cystic Fibrosis. Physiol. Rev. 79, S193–S214. 10.1152/physrev.1999.79.1.s193 [DOI] [PubMed] [Google Scholar]
- Guilbault C., Saeed Z., Downey G. P., Radzioch D. (2007). Cystic Fibrosis Mouse Models. Am. J. Respir. Cell Mol. Biol. 36, 1–7. 10.1165/rcmb.2006-0184tr [DOI] [PubMed] [Google Scholar]
- Gunti S., Hoke A. T. K., Vu K. P., London N. R., Jr. (2021). Organoid and Spheroid Tumor Models: Techniques and Applications. Cancers 13. 10.3390/cancers13040874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habiel D. M., Espindola M. S., Coelho A. L., Hogaboam C. M. (2018). Modeling Idiopathic Pulmonary Fibrosis in Humanized Severe Combined Immunodeficient Mice. Am. J. Pathology 188, 891–903. 10.1016/j.ajpath.2017.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai J., Zhang H., Zhou J., Wu Z., Chen T., Papadopoulos E., et al. (2020). Generation of Genetically Engineered Mouse Lung Organoid Models for Squamous Cell Lung Cancers Allows for the Study of Combinatorial Immunotherapy. Clin. Cancer Res. 26, 3431–3442. 10.1158/1078-0432.ccr-19-1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins F. J., Suzuki S., Beermann M. L., Barilla C., Wang R., Villacorta-Martin C., et al. (2021). Derivation of Airway Basal Stem Cells from Human Pluripotent Stem Cells. Cell stem Cell 28, 79–95 e8. 10.1016/j.stem.2020.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegab A. E., Arai D., Gao J., Kuroda A., Yasuda H., Ishii M., et al. (2015). Mimicking the Niche of Lung Epithelial Stem Cells and Characterization of Several Effectors of Their In Vitro Behavior. Stem Cell Res. 15, 109–121. 10.1016/j.scr.2015.05.005 [DOI] [PubMed] [Google Scholar]
- Hortova-Kohoutkova M., De Zuani M., Laznickova P., Bendickova K., Mrkva O., Andrejcinova I., et al. (2021). Polymorphonuclear Cells Show Features of Dysfunctional Activation during Fatal Sepsis. Front. Immunol. 12, 741484. 10.3389/fimmu.2021.741484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortova-Kohoutkova M., Tidu F., De Zuani M., Sramek V., Helan M., Fric J. (2020). Phagocytosis-Inflammation Crosstalk in Sepsis: New Avenues for Therapeutic Intervention. Shock 54, 606–614. 10.1097/shk.0000000000001541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Sui X., Song F., Li Y., Li K., Chen Z., et al. (2021). Lung Cancer Organoids Analyzed on Microwell Arrays Predict Drug Responses of Patients within a Week. Nat. Commun. 12, 2581. 10.1038/s41467-021-22676-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Feng H., Zhuge D. (2019). M1 Macrophage Activated by Notch Signal Pathway Contributed to Ventilator-Induced Lung Injury in Chronic Obstructive Pulmonary Disease Model. J. Surg. Res. 244, 358–367. 10.1016/j.jss.2019.06.060 [DOI] [PubMed] [Google Scholar]
- Huang Y., Huang Z., Tang Z., Chen Y., Huang M., Liu H., et al. (2021). Research Progress, Challenges, and Breakthroughs of Organoids as Disease Models. Front. Cell Dev. Biol. 9, 740574. 10.3389/fcell.2021.740574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D., Leslie D. C., Matthews B. D., Fraser J. P., Jurek S., Hamilton G. A., et al. (2012). A Human Disease Model of Drug Toxicity-Induced Pulmonary Edema in a Lung-On-A-Chip Microdevice. Sci. Transl. Med. 4, 159ra147. 10.1126/scitranslmed.3004249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibricevic A., Pekosz A., Walter M. J., Newby C., Battaile J. T., Brown E. G., et al. (2006). Influenza Virus Receptor Specificity and Cell Tropism in Mouse and Human Airway Epithelial Cells. J. virology 80, 7469–7480. 10.1128/jvi.02677-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob A., Morley M., Hawkins F., McCauley K. B., Jean J. C., Heins H., et al. (2017). Differentiation of Human Pluripotent Stem Cells into Functional Lung Alveolar Epithelial Cells. Cell stem Cell 21, 472–488 e10. 10.1016/j.stem.2017.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose S. S., De Zuani M., Tidu F., Hortová Kohoutková M., Pazzagli L., Forte G., et al. (2020). Comparison of Two Human Organoid Models of Lung and Intestinal Inflammation Reveals Toll-like Receptor Signalling Activation and Monocyte Recruitment. Clin. Transl. Immunol. 9, e1131. 10.1002/cti2.1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D. J., Shin T. H., Kim M., Sung C. O., Jang S. J., Jeong G. S. (2019). A One-Stop Microfluidic-Based Lung Cancer Organoid Culture Platform for Testing Drug Sensitivity. Lab a chip 19, 2854–2865. 10.1039/c9lc00496c [DOI] [PubMed] [Google Scholar]
- Kapicibasi H. O., Kiraz H. A., Demir E. T., Adali Y., Elmas S. (2020). Pulmonary Effects of Ozone Therapy at Different Doses Combined with Antibioticotherapy in Experimental Sepsis Model. Acta Cir. Bras. 35, e202000604. 10.1590/s0102-865020200060000004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karelehto E., Cristella C., Yu X., Sridhar A., Hulsdouw R., de Haan K., et al. (2018). Polarized Entry of Human Parechoviruses in the Airway Epithelium. Front. Cell. Infect. Microbiol. 8, 294. 10.3389/fcimb.2018.00294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai W., Cui X., Mehlhorn B., Straube E., Hartung T., Gerstenberger E., et al. (2003). Protection with Antibody to Tumor Necrosis Factor Differs with Similarly Lethal Escherichia coli versus Staphylococcus aureus Pneumonia in Rats. Anesthesiology 99, 81–89. 10.1097/00000542-200307000-00016 [DOI] [PubMed] [Google Scholar]
- Katsura H., Kobayashi Y., Tata P. R., Hogan B. L. M. (2019). IL-1 and TNFalpha Contribute to the Inflammatory Niche to Enhance Alveolar Regeneration. Stem Cell Rep. 12, 657–666. 10.1016/j.stemcr.2019.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura H., Sontake V., Tata A., Kobayashi Y., Edwards C. E., Heaton B. E., et al. (2020). Human Lung Stem Cell-Based Alveolospheres Provide Insights into SARS-CoV-2-Mediated Interferon Responses and Pneumocyte Dysfunction. Cell Stem Cell 27, 890–904. e8. 10.1016/j.stem.2020.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., An G. H., Kim J. Y., Rasaei R., Kim W. J., Jin X., et al. (2021). Human Pluripotent Stem-Cell-Derived Alveolar Organoids for Modeling Pulmonary Fibrosis and Drug Testing. Cell Death Discov. 7, 48. 10.1038/s41420-021-00439-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Koo B. K., Knoblich J. A. (2020). Human Organoids: Model Systems for Human Biology and Medicine. Nat. Rev. Mol. Cell Biol. 21, 571–584. 10.1038/s41580-020-0259-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Mun H., Sung C. O., Cho E. J., Jeon H. J., Chun S. M., et al. (2019). Patient-derived Lung Cancer Organoids as In Vitro Cancer Models for Therapeutic Screening. Nat. Commun. 10, 3991. 10.1038/s41467-019-11867-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King T. E., Jr., Bradford W. Z., Castro-Bernardini S., Fagan E. A., Glaspole I., Glassberg M. K., et al. (2014). A Phase 3 Trial of Pirfenidone in Patients with Idiopathic Pulmonary Fibrosis. N. Engl. J. Med. 370, 2083–2092. 10.1056/nejmoa1402582 [DOI] [PubMed] [Google Scholar]
- Kondo J., Inoue M. (2019). Application of Cancer Organoid Model for Drug Screening and Personalized Therapy. Cells 8. 10.3390/cells8050470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Singh R., Kaur J., Pandey S., Sharma V., Thakur L., et al. (2021). Wuhan to World: The COVID-19 Pandemic. Front. Cell. Infect. Microbiol. 11, 596201. 10.3389/fcimb.2021.596201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak E. L., Sordella R., Bell D. W., Godin-Heymann N., Okimoto R. A., Brannigan B. W., et al. (2005). Irreversible Inhibitors of the EGF Receptor May Circumvent Acquired Resistance to Gefitinib. Proc. Natl. Acad. Sci. U.S.A. 102, 7665–7670. 10.1073/pnas.0502860102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Wei X., Lin S., Qin L., Cheng L., Li P. (2017). Current Status and Perspectives of Patient-Derived Xenograft Models in Cancer Research. J. Hematol. Oncol. 10, 106. 10.1186/s13045-017-0470-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb Y. N. (2020). Remdesivir: First Approval. Drugs 80, 1355–1363. 10.1007/s40265-020-01378-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrecht B. N., Hammad H., Fahy J. V. (2019). The Cytokines of Asthma. Immunity 50, 975–991. 10.1016/j.immuni.2019.03.018 [DOI] [PubMed] [Google Scholar]
- Lamers M. M., van der Vaart J., Knoops K., Riesebosch S., Breugem T. I., Mykytyn A. Z., et al. (2021). An Organoid-Derived Bronchioalveolar Model for SARS-CoV-2 Infection of Human Alveolar Type II-like Cells. EMBO J. 40, e105912. 10.15252/embj.2020105912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M. A., Huch M. (2019). Disease Modelling in Human Organoids. Dis. Model Mech. 12. 10.1242/dmm.039347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner A. J., Driver I. H., Lee J., Conroy C. M., Nagle A., Locksley R. M., et al. (2017). Recruited Monocytes and Type 2 Immunity Promote Lung Regeneration Following Pneumonectomy. Cell stem Cell 21, 120–134 e7. 10.1016/j.stem.2017.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeman K. T., Pessina P., Lee J. H., Kim C. F. (2019). Mesenchymal Stem Cells Increase Alveolar Differentiation in Lung Progenitor Organoid Cultures. Sci. Rep. 9, 6479. 10.1038/s41598-019-42819-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibel S. L., Winquist A., Tseu I., Wang J., Luo D., Shojaie S., et al. (2019). Reversal of Surfactant Protein B Deficiency in Patient Specific Human Induced Pluripotent Stem Cell Derived Lung Organoids by Gene Therapy. Sci. Rep. 9, 13450. 10.1038/s41598-019-49696-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Qian Y., Li W., Liu L., Yu L., Liu X., et al. (2020). Human Lung Adenocarcinoma-Derived Organoid Models for Drug Screening. iScience 23, 101411. 10.1016/j.isci.2020.101411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas C., Boda B., Caul Futy M., Huang S., Wisniewski L., Constant S. (2016). Establishment of a Tumour-Stroma Airway Model (OncoCilAir) to Accelerate the Development of Human Therapies against Lung Cancer. Altern. Lab. Anim. 44, 479–485. 10.1177/026119291604400509 [DOI] [PubMed] [Google Scholar]
- Matute-Bello G., Frevert C. W., Martin T. R. (2008). Animal Models of Acute Lung Injury. Am. J. Physiology-Lung Cell. Mol. Physiology 295, L379–L399. 10.1152/ajplung.00010.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P., Rahman Z., Ten Dijke P., Boukany P. E. (2022). Microfluidics Meets 3D Cancer Cell Migration. Trends Cancer S2405-8033 (22), 00072–00073. 10.1016/j.trecan.2022.03.006 [DOI] [PubMed] [Google Scholar]
- Mifflin L., Ofengeim D., Yuan J. (2020). Receptor-interacting Protein Kinase 1 (RIPK1) as a Therapeutic Target. Nat. Rev. Drug Discov. 19, 553–571. 10.1038/s41573-020-0071-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S., Kim S., Cho S. W. (2020). Gastrointestinal Tract Modeling Using Organoids Engineered with Cellular and Microbiota Niches. Exp. Mol. Med. 52, 227–237. 10.1038/s12276-020-0386-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minasyan H. (2019). Sepsis: Mechanisms of Bacterial Injury to the Patient. Scand. J. Trauma Resusc. Emerg. Med. 27, 19. 10.1186/s13049-019-0596-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoo P., Hamdan H., Bu D., Warburton D., Stepanik P., deLemos R. (1995). TTF-1 Regulates Lung Epithelial Morphogenesis. Dev. Biol. 172, 694–698. 10.1006/dbio.1995.8080 [DOI] [PubMed] [Google Scholar]
- Moeller A., Ask K., Warburton D., Gauldie J., Kolb M. (2008). The Bleomycin Animal Model: a Useful Tool to Investigate Treatment Options for Idiopathic Pulmonary Fibrosis? Int. J. Biochem. Cell Biol. 40, 362–382. 10.1016/j.biocel.2007.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motomura T., Ueda K., Ohtani S., Hansen E., Ji L., Ito K., et al. (2010). Evaluation of Systemic External Microwave Hyperthermia for Treatment of Pleural Metastasis in Orthotopic Lung Cancer Model. Oncol. Rep. 24, 591–598. 10.3892/or_00000896 [DOI] [PubMed] [Google Scholar]
- Mou H., Zhao R., Sherwood R., Ahfeldt T., Lapey A., Wain J., et al. (2012). Generation of Multipotent Lung and Airway Progenitors from Mouse ESCs and Patient-specific Cystic Fibrosis iPSCs. Cell stem Cell 10, 385–397. 10.1016/j.stem.2012.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi M., Jackson S. K., Macrae D., Shankar-Hari M., Tremoleda J. L., Lilley E. (2020). Rethinking Animal Models of Sepsis - Working towards Improved Clinical Translation whilst Integrating the 3Rs. Clin. Sci. 134, 1715–1734. 10.1042/cs20200679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal J. T., Kuo C. J. (2016). Organoids as Models for Neoplastic Transformation. Annu. Rev. Pathol. 11, 199–220. 10.1146/annurev-pathol-012615-044249 [DOI] [PubMed] [Google Scholar]
- Ng-Blichfeldt J. P., Schrik A., Kortekaas R. K., Noordhoek J. A., Heijink I. H., Hiemstra P. S., et al. (2018). Retinoic Acid Signaling Balances Adult Distal Lung Epithelial Progenitor Cell Growth and Differentiation. EBioMedicine 36, 461–474. 10.1016/j.ebiom.2018.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic M. Z., Rawlins E. L. (2017). Lung Organoids and Their Use to Study Cell-Cell Interaction. Curr. Pathobiol. Rep. 5, 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nossa R., Costa J., Cacopardo L., Ahluwalia A. (2021). Breathing In Vitro: Designs and Applications of Engineered Lung Models. J. Tissue Eng. 12, 20417314211008696. 10.1177/20417314211008696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Deutsch G. H., Deutsch G. H., Wert S. E., Consortium N. M. A. o. L. D. P. (2019). Comprehensive Anatomic Ontologies for Lung Development: A Comparison of Alveolar Formation and Maturation within Mouse and Human Lung. J. Biomed. Semant. 10, 18. 10.1186/s13326-019-0209-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W., Miller V. A., Politi K. A., Riely G. J., Somwar R., Zakowski M. F., et al. (2005). Acquired Resistance of Lung Adenocarcinomas to Gefitinib or Erlotinib Is Associated with a Second Mutation in the EGFR Kinase Domain. PLoS Med. 2, e73. 10.1371/journal.pmed.0020073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli G., Luca A. D., Jose S. S., Antonini M., Teloni I., Fric J., et al. (2019). Using Lung Organoids to Investigate Epithelial Barrier Complexity and IL-17 Signaling during Respiratory Infection. Front. Immunol. 10, 323. 10.3389/fimmu.2019.00323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I., Kim M., Choe K., Song E., Seo H., Hwang Y., et al. (2019). Neutrophils Disturb Pulmonary Microcirculation in Sepsis-Induced Acute Lung Injury. Eur. Respir. J. 53. 10.1183/13993003.00786-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poling H. M., Wu D., Brown N., Baker M., Hausfeld T. A., Huynh N., et al. (2018). Mechanically Induced Development and Maturation of Human Intestinal Organoids In Vivo . Nat. Biomed. Eng. 2, 429–442. 10.1038/s41551-018-0243-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard C. A., Morran M. P., Nestor-Kalinoski A. L. (2020). The COVID-19 Pandemic: a Global Health Crisis. Physiol. genomics 52, 549–557. 10.1152/physiolgenomics.00089.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porotto M., Ferren M., Chen Y. W., Siu Y., Makhsous N., Rima B., et al. (2019). Authentic Modeling of Human Respiratory Virus Infection in Human Pluripotent Stem Cell-Derived Lung Organoids. mBio 10. 10.1128/mBio.00723-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratilas C. A., Hanrahan A. J., Halilovic E., Persaud Y., Soh J., Chitale D., et al. (2008). Genetic Predictors of MEK Dependence in Non-small Cell Lung Cancer. Cancer Res. 68, 9375–9383. 10.1158/0008-5472.can-08-2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quantius J., Schmoldt C., Vazquez-Armendariz A. I., Becker C., El Agha E., Wilhelm J., et al. (2016). Influenza Virus Infects Epithelial Stem/Progenitor Cells of the Distal Lung: Impact on Fgfr2b-Driven Epithelial Repair. PLoS Pathog. 12, e1005544. 10.1371/journal.ppat.1005544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabata A., Fedr R., Soucek K., Hampl A., Koledova Z. (2020). 3D Cell Culture Models Demonstrate a Role for FGF and WNT Signaling in Regulation of Lung Epithelial Cell Fate and Morphogenesis. Front. Cell Dev. Biol. 8, 574. 10.3389/fcell.2020.00574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghu G., Chen S.-Y., Yeh W.-S., Maroni B., Li Q., Lee Y.-C., et al. (2014). Idiopathic Pulmonary Fibrosis in US Medicare Beneficiaries Aged 65 Years and Older: Incidence, Prevalence, and Survival, 2001-11. Lancet Respir. Med. 2, 566–572. 10.1016/s2213-2600(14)70101-8 [DOI] [PubMed] [Google Scholar]
- Ramachandran P., Matchett K. P., Dobie R., Wilson-Kanamori J. R., Henderson N. C. (2020). Single-cell Technologies in Hepatology: New Insights into Liver Biology and Disease Pathogenesis. Nat. Rev. Gastroenterol. Hepatol. 17, 457–472. 10.1038/s41575-020-0304-x [DOI] [PubMed] [Google Scholar]
- Ramamoorthy P., Thomas S. M., Kaushik G., Subramaniam D., Chastain K. M., Dhar A., et al. (2019). Metastatic Tumor-In-A-Dish, a Novel Multicellular Organoid to Study Lung Colonization and Predict Therapeutic Response. Cancer Res. 79, 1681–1695. 10.1158/0008-5472.can-18-2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes M., Filbin M. R., Bhattacharyya R. P., Billman K., Eisenhaure T., Hung D. T., et al. (2020). An Immune-Cell Signature of Bacterial Sepsis. Nat. Med. 26, 333–340. 10.1038/s41591-020-0752-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richeldi L., du Bois R. M., Raghu G., Azuma A., Brown K. K., Costabel U., et al. (2014). Efficacy and Safety of Nintedanib in Idiopathic Pulmonary Fibrosis. N. Engl. J. Med. 370, 2071–2082. 10.1056/nejmoa1402584 [DOI] [PubMed] [Google Scholar]
- Riemondy K. A., Jansing N. L., Jiang P., Redente E. F., Gillen A. E., Fu R., et al. (2019). Single Cell RNA Sequencing Identifies TGFbeta as a Key Regenerative Cue Following LPS-Induced Lung Injury. JCI Insight 5. 10.1172/jci.insight.123637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd K. E., Johnson S. C., Agesa K. M., Shackelford K. A., Tsoi D., Kievlan D. R., et al. (2020). Global, Regional, and National Sepsis Incidence and Mortality, 1990-2017: Analysis for the Global Burden of Disease Study. Lancet 395, 200–211. 10.1016/s0140-6736(19)32989-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs N., Papaspyropoulos A., Zomer-van Ommen D. D., Heo I., Bottinger L., Klay D., et al. (2019). Long-term Expanding Human Airway Organoids for Disease Modeling. EMBO J. 38. 10.15252/embj.2018100300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salahudeen A. A., Choi S. S., Rustagi A., Zhu J., van Unen V., de la O. S., et al. (2020). Progenitor Identification and SARS-CoV-2 Infection in Human Distal Lung Organoids. Nature 588, 670–675. 10.1038/s41586-020-3014-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Morita M., Tanaka R., Inoue Y., Nomura M., Sakamoto Y., et al. (2017). Ex Vivo model of Non-small Cell Lung Cancer Using Mouse Lung Epithelial Cells. Oncol. Lett. 14, 6863–6868. 10.3892/ol.2017.7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Vries R. G., Snippert H. J., van de Wetering M., Barker N., Stange D. E., et al. (2009). Single Lgr5 Stem Cells Build Crypt-Villus Structures In Vitro without a Mesenchymal Niche. Nature 459, 262–265. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- Schittny J. C. (2017). Development of the Lung. Cell Tissue Res. 367, 427–444. 10.1007/s00441-016-2545-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutgens F., Clevers H. (2020). Human Organoids: Tools for Understanding Biology and Treating Diseases. Annu. Rev. Pathol. 15, 211–234. 10.1146/annurev-pathmechdis-012419-032611 [DOI] [PubMed] [Google Scholar]
- Semaniakou A., Croll R. P., Chappe V. (2018). Animal Models in the Pathophysiology of Cystic Fibrosis. Front. Pharmacol. 9, 1475. 10.3389/fphar.2018.01475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok J., Warren H. S., Cuenca A. G., Mindrinos M. N., Baker H. V., Xu W., et al. (2013). Host Response to Injury, Genomic Responses in Mouse Models Poorly Mimic Human Inflammatory Diseases. Proc. Natl. Acad. Sci. U. S. A. 110, 3507–3512. 10.1073/pnas.1222878110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevransky J. E., Martin G. S., Shanholtz C., Mendez-Tellez P. A., Pronovost P., Brower R., et al. (2009). Mortality in Sepsis versus Non-sepsis Induced Acute Lung Injury. Crit. care 13, R150. 10.1186/cc8048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Toepfer C. N., Ward T., Wasson L., Agarwal R., Conner D. A., et al. (2018). CRISPR/Cas9-Mediated Fluorescent Tagging of Endogenous Proteins in Human Pluripotent Stem Cells. Curr. Protoc. Hum. Genet. 96, 21 11 1–21 11 20. 10.1002/cphg.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Chen L., Wang M., Lin D., Liang Z., Song P., et al. (2017). Flagellar Hooks and Hook Protein FlgE Participate in Host Microbe Interactions at Immunological Level. Sci. Rep. 7, 1433. 10.1038/s41598-017-01619-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Radulovich N., Ng C., Liu N., Notsuda H., Cabanero M., et al. (2020). Organoid Cultures as Preclinical Models of Non-small Cell Lung Cancer. Clin. Cancer Res. 26, 1162–1174. 10.1158/1078-0432.ccr-19-1376 [DOI] [PubMed] [Google Scholar]
- Shmidt E. N., Nitkin A. Y. (2004). Pathology of Mouse Models of Human Lung Cancer. Comp. Med. 54, 23–26. [PubMed] [Google Scholar]
- Singer M., Deutschman C. S., Seymour C. W., Shankar-Hari M., Annane D., Bauer M., et al. (2016). The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 315, 801–810. 10.1001/jama.2016.0287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snouwaert J. N., Brigman K. K., Latour A. M., Malouf N. N., Boucher R. C., Smithies O., et al. (1992). An Animal Model for Cystic Fibrosis Made by Gene Targeting. Science 257, 1083–1088. 10.1126/science.257.5073.1083 [DOI] [PubMed] [Google Scholar]
- Stahl O., Loffler B., Haier J., Mardin W. A., Mees S. T. (2013). Mimicry of Human Sepsis in a Rat Model-Pprospects and Limitations. J. Surg. Res. 179, e167–75. 10.1016/j.jss.2012.01.042 [DOI] [PubMed] [Google Scholar]
- Strikoudis A., Cieslak A., Loffredo L., Chen Y. W., Patel N., Saqi A., et al. (2019). Modeling of Fibrotic Lung Disease Using 3D Organoids Derived from Human Pluripotent Stem Cells. Cell Rep. 27, 3709–3723. e5. 10.1016/j.celrep.2019.05.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucre J. M., Wilkinson D., Vijayaraj P., Paul M., Dunn B., Alva-Ornelas J. A., et al. (2016). A Three-Dimensional Human Model of the Fibroblast Activation that Accompanies Bronchopulmonary Dysplasia Identifies Notch-Mediated Pathophysiology. Am. J. physiology. Lung Cell. Mol. physiology 310, L889–L898. 10.1152/ajplung.00446.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suezawa T., Kanagaki S., Moriguchi K., Masui A., Nakao K., Toyomoto M., et al. (2021). Disease Modeling of Pulmonary Fibrosis Using Human Pluripotent Stem Cell-Derived Alveolar Organoids. Stem Cell Rep. 16, 2973–2987. 10.1016/j.stemcr.2021.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney R. M., McAuley D. F. (2016). Acute Respiratory Distress Syndrome. Lancet 388, 2416–2430. 10.1016/s0140-6736(16)00578-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Nau M. M., Chiba I., Birrer M. J., Rosenberg R. K., Vinocour M., et al. (1989). p53: a Frequent Target for Genetic Abnormalities in Lung Cancer. Science 246, 491–494. 10.1126/science.2554494 [DOI] [PubMed] [Google Scholar]
- Tamminga M., Hiltermann T. J. N., Schuuring E., Timens W., Fehrmann R. S., Groen H. J. (2020). Immune Microenvironment Composition in Non-small Cell Lung Cancer and its Association with Survival. Clin. Transl. Immunol. 9, e1142. 10.1002/cti2.1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q., Choi K. M., Sicard D., Tschumperlin D. J. (2017). Human Airway Organoid Engineering as a Step toward Lung Regeneration and Disease Modeling. Biomaterials 113, 118–132. 10.1016/j.biomaterials.2016.10.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe N., McDonough J. E., Vasilescu D. M., Ikezoe K., Verleden S. E., Xu F., et al. (2020). Pathology of Idiopathic Pulmonary Fibrosis Assessed by a Combination of Microcomputed Tomography, Histology, and Immunohistochemistry. Am. J. pathology 190, 2427–2435. 10.1016/j.ajpath.2020.09.001 [DOI] [PubMed] [Google Scholar]
- Tashiro J., Rubio G. A., Limper A. H., Williams K., Elliot S. J., Ninou I., et al. (2017). Exploring Animal Models that Resemble Idiopathic Pulmonary Fibrosis. Front. Med. 4, 118. 10.3389/fmed.2017.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L., Gao J., Garcia I. M., Chen H. J., Castaldi A., Chen Y. W. (2021). Human Pluripotent Stem Cell-Derived Lung Organoids: Potential Applications in Development and Disease Modeling. Wiley Interdiscip. Rev. Dev. Biol. 10, e399. 10.1002/wdev.399 [DOI] [PubMed] [Google Scholar]
- Tindle C., Fuller M., Fonseca A., Taheri S., Ibeawuchi S. R., Beutler N., et al. (2021). Adult Stem Cell-Derived Complete Lung Organoid Models Emulate Lung Disease in COVID-19. Elife 10. 10.7554/eLife.66417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C. W., Ritsma L., Avramut M. C., Wiersma L. E., van den Berg B. M., Leuning D. G., et al. (2018). Renal Subcapsular Transplantation of PSC-Derived Kidney Organoids Induces Neo-Vasculogenesis and Significant Glomerular and Tubular Maturation In Vivo . Stem Cell Rep. 10, 751–765. 10.1016/j.stemcr.2018.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sanden S. M. G., Sachs N., Koekkoek S. M., Koen G., Pajkrt D., Clevers H., et al. (2018). Enterovirus 71 Infection of Human Airway Organoids Reveals VP1-145 as a Viral Infectivity Determinant. Emerg. microbes Infect. 7, 84. 10.1038/s41426-018-0077-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Geffen C., Deissler A., Quante M., Renz H., Hartl D., Kolahian S. (2021). Regulatory Immune Cells in Idiopathic Pulmonary Fibrosis: Friends or Foes? Front. Immunol. 12, 663203. 10.3389/fimmu.2021.663203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese B., Ling Z., Ren X. (2022). Reconstructing the Pulmonary Niche with Stem Cells: a Lung Story. Stem Cell Res. Ther. 13, 161. 10.1186/s13287-022-02830-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachogiannis G., Hedayat S., Vatsiou A., Jamin Y., Fernandez-Mateos J., Khan K., et al. (2018). Patient-derived Organoids Model Treatment Response of Metastatic Gastrointestinal Cancers. Science 359, 920–926. 10.1126/science.aao2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters D. M., Kleeberger S. R. (2008). Mouse Models of Bleomycin-Induced Pulmonary Fibrosis. Curr. Protoc. Pharmacol. Chapter 5, Unit 5.46. 10.1002/0471141755.ph0546s40 [DOI] [PubMed] [Google Scholar]
- Wang H. M., Bodenstein M., Markstaller K. (2008). Overview of the Pathology of Three Widely Used Animal Models of Acute Lung Injury. Eur. Surg. Res. 40, 305–316. 10.1159/000121471 [DOI] [PubMed] [Google Scholar]
- Wang J. E., Dahle M. K., Yndestad A., Bauer I., McDonald M. C., Aukrust P., et al. (2004). Peptidoglycan of Staphylococcus aureus Causes Inflammation and Organ Injury in the Rat. Crit. Care Med. 32, 546–552. 10.1097/01.ccm.0000109775.22138.8f [DOI] [PubMed] [Google Scholar]
- Wang J., Li X., Chen H. (2020). Organoid Models in Lung Regeneration and Cancer. Cancer Lett. 475, 129–135. 10.1016/j.canlet.2020.01.030 [DOI] [PubMed] [Google Scholar]
- Wang L., Liu H., Jiao Y., Wang E., Clark S., Postlethwaite A., et al. (2015). Differences between Mice and Humans in Regulation and the Molecular Network of Collagen, Type III, Alpha-1 at the Gene Expression Level: Obstacles that Translational Research Must Overcome. Int. J. Mol. Sci. 16, 15031–15056. 10.3390/ijms160715031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., McCauley K. B., Kotton D. N., Hawkins F. (2020). Differentiation of Human Airway-Organoids from Induced Pluripotent Stem Cells (iPSCs). Methods Cell Biol. 159, 95–114. 10.1016/bs.mcb.2020.03.008 [DOI] [PubMed] [Google Scholar]
- Wilkinson D. C., Alva-Ornelas J. A., Sucre J. M., Vijayaraj P., Durra A., Richardson W., et al. (2017). Development of a Three-Dimensional Bioengineering Technology to Generate Lung Tissue for Personalized Disease Modeling. Stem cells Transl. Med. 6, 622–633. 10.5966/sctm.2016-0192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K., Roman J. (2016). Studying Human Respiratory Disease in Animals - Role of Induced and Naturally Occurring Models. J. Pathol. 238, 220–232. 10.1002/path.4658 [DOI] [PubMed] [Google Scholar]
- Wong A. P., Bear C. E., Chin S., Pasceri P., Thompson T. O., Huan L. J., et al. (2012). Directed Differentiation of Human Pluripotent Stem Cells into Mature Airway Epithelia Expressing Functional CFTR Protein. Nat. Biotechnol. 30, 876–882. 10.1038/nbt.2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Li Y., Zhang S., Peng H., Wang Y., Li D., et al. (2021). SARS-CoV-2 Promotes RIPK1 Activation to Facilitate Viral Propagation. Cell Res. 31, 1230–1243. 10.1038/s41422-021-00578-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Gao Y., Hao Y., Li E., Wang Y., Zhang J., et al. (2013). Application of a Microfluidic Chip-Based 3D Co-culture to Test Drug Sensitivity for Individualized Treatment of Lung Cancer. Biomaterials 34, 4109–4117. 10.1016/j.biomaterials.2013.02.045 [DOI] [PubMed] [Google Scholar]
- Yoshida G. J. (2020). Applications of Patient-Derived Tumor Xenograft Models and Tumor Organoids. J. Hematol. Oncol. 13, 4. 10.1186/s13045-019-0829-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F., Zuo L., Hu R., Wang J., Yang Z., Qi X., et al. (2021). Effect of Immune Cell Infiltration on Occurrence of Pulmonary Hypertension in Pulmonary Fibrosis Patients Based on Gene Expression Profiles. Front. Med. 8, 671617. 10.3389/fmed.2021.671617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zscheppang K., Berg J., Hedtrich S., Verheyen L., Wagner D. E., Suttorp N., et al. (2018). Human Pulmonary 3D Models for Translational Research. Biotechnol. J. 13. 10.1002/biot.201700341 [DOI] [PMC free article] [PubMed] [Google Scholar]