Abstract
Alkane monooxygenases in Nocardioides sp. strain CF8 were examined at the physiological and genetic levels. Strain CF8 can utilize alkanes ranging in chain length from C2 to C16. Butane degradation by butane-grown cells was strongly inhibited by allylthiourea, a copper-selective chelator, while hexane-, octane-, and decane-grown cells showed detectable butane degradation activity in the presence of allylthiourea. Growth on butane and hexane was strongly inhibited by 1-hexyne, while 1-hexyne did not affect growth on octane or decane. A specific 30-kDa acetylene-binding polypeptide was observed for butane-, hexane-, octane-, and decane-grown cells but was absent from cells grown with octane or decane in the presence of 1-hexyne. These results suggest the presence of two monooxygenases in strain CF8. Degenerate primers designed for PCR amplification of genes related to the binuclear-iron-containing alkane hydroxylase from Pseudomonas oleovorans were used to clone a related gene from strain CF8. Reverse transcription-PCR and Northern blot analysis showed that this gene encoding a binuclear-iron-containing alkane hydroxylase was expressed in cells grown on alkanes above C6. These results indicate the presence of two distinct monooxygenases for alkane oxidation in Nocardioides sp. strain CF8.
A number of bacteria have been isolated for their ability to utilize gaseous or liquid alkanes as growth substrates (5, 7, 44). It is convenient to recognize three classes of alkane-utilizing bacteria, namely, those that grow on methane, those that grow optimally on gaseous alkanes, and those that grow optimally on liquid alkanes. The methane utilizers (methanotrophs) have been well characterized. Methane metabolism is initiated by the oxidation of methane to methanol in a reaction catalyzed by methane monooxygenase (MMO). Two forms of MMO have been characterized, a particulate MMO (pMMO) that contains copper, and a soluble MMO (sMMO) that contains a binuclear iron cluster at the catalytic site (27, 50). Both pMMO and sMMO can catalyze the oxidation of several alkanes in addition to methane but cannot grow on any of these compounds (10, 11). Long-chain, liquid alkanes are utilized by a wide variety of bacterial species, including both gram-negative and gram-positive bacteria (8, 47). Among these bacteria, the alkane hydroxylase system in Pseudomonas oleovorans has been characterized most thoroughly. P. oleovorans can grow on n-alkanes ranging from C6 to C12 (5). It harbors a large plasmid (OCT plasmid) encoding the enzymes required to oxidize n-alkanes to the corresponding terminal acyl coenzyme A derivatives, which then enter the β-oxidation cycle (46). Alkane hydroxylase consists of three polypeptides: a hydroxylase (AlkB; 41 kDa), a rubredoxin (AlkG; 19 kDa), and a rubredoxin reductase (AlkT; 48 kDa) (13–15). The alkane hydroxylase component contains a binuclear iron cluster, which seems to be a common motif among bacteria that harvest alkanes, alkenes, and aromatic hydrocarbons as growth substrates (40). Several different long-chain alkane oxidation pathways have been described for strains of Acinetobacter spp.: alkane dioxygenase is involved in degradation of alkanes ranging from C13 to C44 in Acinetobacter sp. strain M-1 (28, 37), some strains utilize a P-450 monooxygenase system (3), and an alkane hydroxylase homologous to that of P. oleovorans has been found in Acinetobacter sp. strain ADP1 (34). Gaseous alkanes have not been shown to serve as growth substrates for these bacteria.
Although a wide variety of microorganisms can utilize long-chain, liquid alkanes, the ability to utilize short-chain, gaseous alkanes is mostly restricted to the Corynebacterium-Nocardia-Mycobacterium-Rhodococcus group of gram-positive bacteria (2, 29). In addition, some gram-negative Pseudomonas spp. were reported to grow on short-chain alkanes other than methane (25, 44). There have been neither descriptions of the purification to homogeneity of a monooxygenase from short-chain-alkane-utilizing bacteria nor any isolations of genes that code for this group of monooxygenases. We have recently isolated a butane-utilizing bacterium, Nocardioides sp. strain CF8, and characterized a butane monooxygenase in this organism at the physiological level (22, 24). Butane degradation by strain CF8 was strongly inhibited by allylthiourea (ATU) and inactivated by light and acetylene. Both ATU and light are known inhibitors and inactivators of copper-containing monooxygenases such as pMMO and ammonia monooxygenase (AMO) (6). Acetylene serves as an inactivator of butane monooxygenase in strain CF8, and incubation with [14C]acetylene results in the covalent binding of 14C label to a specific polypeptide with a molecular mass of 30 kDa (24). This result supports the similarity between butane monooxygenase of strain CF8 and the copper-containing monooxygenases, pMMO and AMO, which contain an acetylene-binding protein with a similar molecular mass (ca. 27 kDa) (26, 33). Therefore, it was suggested that butane monooxygenase in strain CF8 is a third example of the copper-containing monooxygenases, which include pMMO and AMO.
Bacteria that grow on gaseous alkanes often include some liquid alkanes in their range of growth substrates. This observation also extends to Nocardioides sp. strain CF8 (22). In this work, we extended the characterization of the alkane monooxygenase system in Nocardioides sp. strain CF8 to include longer-chain alkanes (C6 to C10). The possibility of two distinct monooxygenases for alkane degradation in Nocardioides sp. strain CF8 was examined.
MATERIALS AND METHODS
Growth conditions.
Cells of Nocardioides sp. strain CF8 were grown as previously described (23). Strain CF8 was grown in the Xanthobacter Py2 medium (48) except that NH4Cl replaced NaNO3, yeast extract was removed, and the pH was adjusted to 7.5. Cells were incubated in 150-ml sealed vials containing 50 ml of medium and alkanes as growth substrates. Gaseous alkanes (50 ml) were added as an overpressure to the gas phase. Liquid alkanes were added to a total amount of 500 μmol. All cultures were incubated at 30°C with constant shaking. Cell growth was monitored by removing a portion of the cultures (1 ml) and measuring the optical density at 600 nm (OD600). To examine the effect of ATU and 1-hexyne on growth, ATU (200 μM) and 1-hexyne (4.5 μmol) were added at the beginning of growth assays. The extent of growth of each culture was determined by measuring the OD600 at selected times (4 days for C4-, C6-, and C8-grown cells; 5 days for C10-grown cells). No significant increase in OD600 was observed in any of the experimental setups after 5 days. Experiments were repeated at least three times.
Butane degradation assay.
Cells were harvested from cultures by centrifugation (6,000 × g for 10 min), washed twice with the same phosphate buffer as the growth medium, and resuspended to a constant cell density (based on OD). Butane degradation assays were conducted as previously described (24). For ATU inhibition assays, butane degradation was monitored with 200 μM ATU in the reaction vials. Experiments were repeated at least three times.
[14C]acetylene labeling assays.
Cells of C4-, C6-, C8-, and C10-grown strain CF8 were radiolabeled with [14C]acetylene as described previously (24). C8- and C10-grown cells were also grown in the presence of 1-hexyne (4.5 μmol) and subjected to [14C]acetylene labeling. The labeled cells were disrupted with a Mini-beadbeater (Biospec Products, Bartlesville, Okla.). Protein samples (50 μg) were then separated on a sodium dodecyl sulfate–12% polyacrylamide gel at a constant current of 15 mA. The gel was stained with Coomassie blue, dried onto filter paper, and radioactive polypeptides were visualized by exposure to phosphor screens (Molecular Dynamics, Sunnyvale, Calif.) for 5 days.
Protein determinations.
Protein content was determined by using the biuret assay (19) after cells were solubilized in 3 N NaOH for 30 min at 65°C. Bovine serum albumin was used as the standard.
PCR, sequence analysis, and library screening to characterize alkB in strain CF8.
Chromosomal DNA was isolated from strain CF8 by the CTAB (hexadecyltrimethylammonium bromide) method described previously (4). PCRs were performed with degenerate oligonucleotide primers TS2S (5′-AAYAGAGCTC AYGARYTRGG TCAYAAG-3′) and deg1RE (5′-GTGGAATTCG CRTGRTGRTC IGARTG-3′) (42), and the program (4 min at 95°C; 25 cycles of 45 s at 95°C, 1 min at 40°C, and 1 min at 72°C; 5 min at 72°C; indefinitely at 4°C) described by Smits et al. (42) was used. The PCR products were cloned into the pGEM-T Easy vector (Promega, Madison, Wis.) following the directions of the manufacturer.
A genomic library was constructed in Lambda FIX II (Stratagene, La Jolla, Calif.) using Gigapack III XL-11 packaging extract (Stratagene) following the direction of the manufacturer. The genomic library was screened as described (38) using a DNA probe generated by PCR with TS2S and deg1RE primers. DNA probes were labeled by random priming using a Prime-a-gene kit (Promega) and [α-32P]dCTP (3,000 Ci/mmol; DuPont NEN products, Wilmington, Del.). Lambda phage DNA isolation, restriction digests, agarose gel electrophoresis, Southern hybridization, and cloning were performed as standard procedures (38). DNA fragments (0.6, 1, and 2.7 kb) of a XhoI digest were cloned into pBluescript II KS (Stratagene) and sequenced. A total of 3.2 kb, including the complete alkB, was sequenced in both directions at least three times by a combination of primer walking with custom oligonucleotides and primers complementary to cloning vectors.
DNA sequencing and oligonucleotide synthesis were performed at the Center for Gene Research and Biotechnology at Oregon State University (Corvallis, Oreg.). The nucleotide and the predicted amino acid sequences were compared with the EMBL, SWISSPROT, and GenBank databases using BLAST (1). DNA sequencing data was analyzed and assembled using the Genetics Computer Group (Madison, Wis.) software package (Wisconsin Package, version 10.0).
Reverse transcription (RT)-PCR and Northern blotting.
Cells of strain CF8 were grown on n-alkanes ranging from C4 to C8 in 50 ml of medium as described earlier. Cells were grown to late exponential phase (OD600 of 0.5 to 0.6), and 25 ml of each culture was harvested by centrifugation (6,000 × g for 10 min) in a rotor chilled to 4°C. Total cellular RNA was isolated as described by Brzostowicz et al. (9) with slight modifications. The pellets were resuspended in 0.7 ml of an ice-cold lysis solution (1% sodium dodecyl sulfate, 100 mM sodium acetate at pH 5) and transferred to a 2-ml conical screw-cap Microtube (Biospec Products) containing 0.7 ml of aqueous phenol (pH 5) and 0.3 ml of 0.5-mm-diameter zirconia beads (Biospec Products). The tubes were placed in a Mini-beadbeater (Biospec Products) and disrupted at 4,600 beats/min for 2 min. The liquid phase was transferred to microcentrifuge tubes and centrifuged for 3 min at 14,000 × g. The supernatant was extracted twice with phenol (pH 5) and twice with phenol-chloroform solution (pH 7.5). Nucleic acids were precipitated from the aqueous phase with ethanol, resuspended in 200 μl of diethyl pyrocarbonate-treated water, and treated with 5 U of RNase-free DNase (Promega Co.) at 37°C for 2 h. Subsequently, the solution was extracted twice with phenol-chloroform solution (pH 7.5). RNA was recovered by ethanol precipitation and resuspended in 200 μl of diethyl pyrocarbonate-treated water.
The extracted RNA was subjected to RT and subsequent PCR amplification. Primers, revalk (5′-AGTGTCGCTG CAGGTGGTA-3′) and cfalkF (5′-AGAAGGAGAG CCACGAACG-3′), were designed based on the nucleotide sequence of the PCR product amplified with TS2S and deg1RE primers. The RT reaction mixtures (20 μl) contained 200 U of Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega Co.), 1 mM each deoxynucleoside triphosphate, 1 U of RNase inhibitor (Promega Co.), 30 pmol of primer revalk, and 100 ng of extracted RNA in 1× M-MLV buffer provided by the manufacturer. Control reactions were performed without addition of M-MLV reverse transcriptase to verify the absence of DNA in the RNA preparations. RT was carried out at 42°C for 15 min, 95°C for 1 min, and 5°C for 5 min. Aliquots from each RT reaction (5 μl) were used as templates in subsequent PCR mixtures (25 μl) containing deoxynucleoside triphosphates (0.2 mM each), MgCl2 (2 mM), revalk and cfalkF primers (30 pmol each), and 2.5 U of Taq polymerase (Promega Co.). The following temperature profile was used: 4 min at 94°C and 35 cycles of 1 min at 94°C, 1 min at 62°C, and 1 min at 72°C. The PCR products (10 μl) were separated by electrophoresis on a 1.2% (wt/vol) agarose gel in Tris-borate-EDTA buffer and visualized with ethidium bromide staining.
Northern blot analysis was carried out as described (39). DNA probes specific for alkB and 16S rRNA were generated by PCR with revalk and cfalkF primers and a set of primers described by Giovannoni (18), respectively. The hybridization signals were visualized using a PhosphorImager (Molecular Dynamics).
Nucleotide sequence accession number.
The nucleotide sequence reported in this study has been deposited in the GenBank database under accession number AF350429.
RESULTS
Nocardioides sp. strain CF8 was grown on n-alkanes of various chain lengths and tested for butane degradation activity. Cells grown on butane, hexane, octane, and decane (C4, C6, C8, and C10 chain length, respectively) readily degraded butane at similar rates (Table 1). In our previous study, it was shown that butane degradation by butane-grown strain CF8 was strongly inhibited by ATU, a copper-selective chelator (24). Along with other lines of evidence, it was suggested that butane-grown strain CF8 contains a copper-containing butane monooxygenase that is responsible for butane oxidation in this organism (22). We have now examined the effect of ATU on butane degradation by cells grown on longer alkanes. As previously shown, 200 μM ATU inhibited butane degradation by C4-grown cells to below detectable levels (1.5 nmol min−1 mg of protein−1). In contrast, C6-, C8-, and C10-grown cells of strain CF8 showed detectable levels of butane degradation (10 to 30% relative to the absence of ATU) in the presence of the same concentration of ATU (Table 1). These low levels of activity could be due to an incomplete inhibition of the copper-containing butane monooxygenase by ATU, although this explanation would require different inhibitory effects of ATU on cells grown on various alkanes. Alternatively, it is possible that in addition to copper-containing butane monooxygenase, a second monooxygenase which degrades butane and is not inhibited by ATU is expressed when cells are grown on C6, C8, and C10 but not C4 alkanes.
TABLE 1.
Effect of allylthiourea on butane degradation by strain CF8 grown on various alkanes
| Growth substratea | Butane degradation rateb (nmol min−1 mg of protein−1)
|
|
|---|---|---|
| No additions | +ATU (200 μM) | |
| C4 | 30.8 ± 2.1 | <1.5 |
| C6 | 28.2 ± 5.1 | 3.0 ± 3.8 |
| C8 | 30.9 ± 4.9 | 6.4 ± 3.4 |
| C10 | 23.9 ± 1.9 | 7.5 ± 2.4 |
Butane was added as 50% (vol/vol [gas phase]) in the gas phase as an overpressure. Liquid alkanes were added to a total amount of 500 μmol.
Data are expressed as experimental means ± standard deviations.
To further investigate the latter possibility, we examined the inhibitory effects of ATU and 1-hexyne on the growth of strain CF8 with C4, C6, C8, and C10 alkanes as the growth substrate (data not shown). As expected, growth on C4 was greatly inhibited (∼80%) by 200 μM ATU. However, ATU (200 μM) did not inhibit growth on C6, C8, and C10 alkanes. In the presence of 1-hexyne (4.5 μmol), growth was not observed on C4 and C6 alkanes. Growth on C8 and C10 was unaffected by the presence of 1-hexyne (4.5 μmol) (data not shown). These results are consistent with the presence of a second monooxygenase that is not inhibited by ATU and is less sensitive to 1-hexyne inhibition (inactivation). The different response of C6-grown cells to 1-hexyne inhibition compared to those of C8- and C10-grown cells may reflect the higher affinities of the second monooxygenase toward longer-chain alkanes. Because ATU is a reversible inhibitor, butane-oxidizing activity of the copper-containing butane-monooxygenase can be recovered in cells grown in the presence of ATU. Indeed, cells grown on C6, C8, and C10 in the presence of ATU (200 μM) and subsequently washed free of ATU had similar butane degradation rates to those grown in the absence of ATU (data not shown). In contrast, the cells grown on C8 and C10 in the presence of 1-hexyne (4.5 μmol) and washed as above showed much lower butane degradation rates (5.2 ± 2.7 and 4.3 ± 3.1 nmol min−1 mg of protein−1, respectively [means ± standard deviations]) than those grown in the absence of 1-hexyne (Table 1). These results support the ideas that 1-hexyne irreversibly inactivates the copper-containing monooxygenase and that the residual butane degradation activity of the cells grown on C8 and C10 in the presence of 1-hexyne is due to the second monooxygenase.
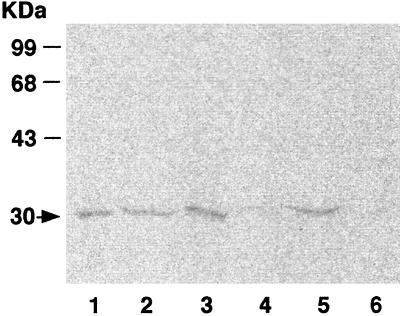
We have previously shown that the incubation of strain CF8 with [14C]acetylene results in the covalent binding of 14C label to a specific polypeptide with a molecular mass of 30 kDa and that the incorporation of 14C label correlates with butane monooxygenase activity (24). Thus, cells grown on various chain length alkanes either with or without 1-hexyne (4.5 μmol) were treated with [14C]acetylene (Fig 1). The 14C-labeled 30-kDa polypeptide was present in C4-, C6-, C8-, and C10-grown cells without 1-hexyne. In contrast, 14C label was not incorporated into cellular polypeptides of cells grown on C8 and C10 in the presence of 1-hexyne. Since labeling of the 30-kDa polypeptide with 14C from acetylene requires active butane monooxygenase, the 30-kDa polypeptide of the copper-containing butane monooxygenase is most likely being produced and then irreversibly inactivated by the 1-hexyne in the growth medium. These results further confirm that the cells grown in the presence of 1-hexyne do not possess an active copper-containing butane-monooxygenase. Again, production of a second monooxygenase would support growth under these conditions. Interestingly, there were no additional labeled polypeptides observed other than the 30-kDa polypeptide under any of the growth conditions tested. This result indicates that acetylene might act either as a conventional reversible inhibitor or as an unusually weak mechanism-based inactivator for the second monooxygenase.
FIG. 1.
Incorporation of 14C from [14C]acetylene into cellular proteins of strain CF8 grown on C4, C6, C8, and C10 alkanes (lanes 1, 2, 3, and 5, respectively) and cells grown with C8 or C10 in the presence of 1-hexyne (lanes 4 and 6, respectively). Incorporation of 14C into polypeptides was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized with a phosphorimager as described in Materials and Methods. The sizes of molecular mass markers and the apparent molecular mass of the labeled polypeptide (arrow) are shown on the left side of the gel. Each gel lane contains approximately 50 μg of cell extract protein.
Among the long-chain-alkane-oxidizing enzymes, alkane hydroxylase in P. oleovorans has been characterized most thoroughly (13–15). Recent study has shown that a large proportion of bacteria able to grow on long-chain alkanes possess genes related to the alkane hydroxylase gene in P. oleovorans (42). Based on biochemical analyses and sequence comparisons, alkane hydroxylase from P. oleovorans belongs to a family of integral-membrane, binuclear-iron hydrocarbon oxygenases including alkane hydroxylase from Acinetobacter sp. strain ADP1 and xylene monooxygenase from Pseudomonas putida (34, 40, 41). These enzymes all contain an eight-histidine motif as iron-binding ligand (41). This motif is also conserved among the soluble, binuclear-iron hydrocarbon oxygenases, such as sMMO and toluene 2-monooxygenase from Burkholderia cepacia G4 (17). It was shown that acetylene is a weak inactivator of toluene 2-monooxygenase, while longer-chain alkynes are more effective inactivators (49). These previous studies drew our attention to the binuclear-iron monooxygenases as potential candidates for the second monooxygenase in strain CF8.
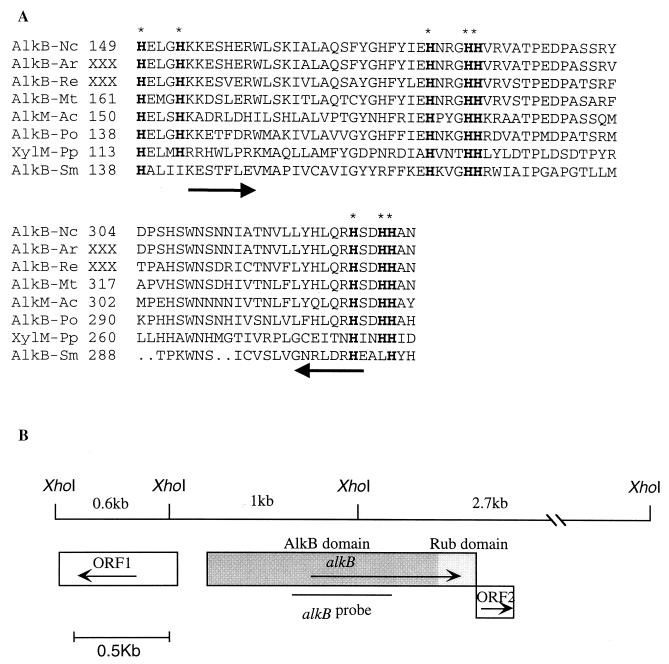
In order to examine the possible presence of a binuclear-iron monooxygenase gene in strain CF8, we employed the PCR method developed recently by Smits et al. (42) which uses degenerate primers based on the sequence alignment of the conserved histidine motif (42). By using this PCR method, a PCR product of the expected size (557 bp) was obtained from strain CF8. The fragment was cloned and sequenced. The peptide sequence encoded by the PCR product was compared with the known alkane hydroxylase sequences, including some of the PCR fragments obtained by Smits et al. (42), and the sequence of xylene monooxygenase (Fig. 2A). The sequence alignment showed that the regions around the eight-histidine motif were well conserved in the sequence from strain CF8. The deduced peptide fragment (186 amino acids) from strain CF8 showed high sequence identities to putative AlkB fragments from other gram-positive bacteria, including Amycolatopsis rugosa (86%) and Rhodococcus erythropolis (69%), and an alkane hydroxylase homologue present on the chromosome of Mycobacterium tuberculosis H37Rv (63%). Lower sequence identities were observed to the corresponding sequences from gram-negative bacteria, including AlkM from Acinetobacter sp. strain ADP1 (54%), AlkB from P. oleovorans (49%), and XylM from P. putida (27%).
FIG. 2.
(A) Part of a multiple sequence alignment of AlkB from strain CF8 with other known integral-membrane, binuclear-iron hydrocarbon oxygenases. Eight conserved His residues are indicated by letters in boldface type and marked by asterisks. The positions of the PCR primers used for RT-PCR are shown by arrows. Peptide numbering refers to the position in the full-length enzyme (XXX is shown for the sequences whose full length sequences are not available). Abbreviations: Nc, Nocardioides sp. strain CF8; Ar, A. rugosa; Re, R. erythropolis; Mt, M. tuberculosis; Ac, Acinetobacter sp. strain ADP1; Po, P. oleovorans; Pp, P. putida; Sm, Stenotrophomonas maltophilia. (B) Gene organization and restriction map of the cloned regions including complete AlkB gene. Location of the PCR fragment used as a probe for alkB is shown. Each box indicates an ORF, and the arrows indicate the orientation of each gene.
The complete AlkB gene (alkB) in strain CF8 was isolated by screening a genomic library with the cloned PCR fragment as a probe. The nucleotide sequences of three contiguous XhoI restriction fragments totaling 3.2 kb and including alkB were determined, and sequence analysis revealed three open reading frames (ORFs) (Fig. 2B). alkB was located in an ORF that continuously contains a domain similar to rubredoxin (deduced amino acid sequence showed 55% identity to rubredoxin 2 in P. oleovorans, 52% identity to rubredoxin in Clostridium butyricum, and 51% identity to rubredoxin 2 of P. putida). The entire ORF consists of 1,452 bp and potentially encodes a unique alkane hydroxylase fused to rubredoxin, although further biochemical characterization of this enzyme is required to confirm this idea. The stop codon of the alkB-rub ORF overlaps with the start codon of a second ORF (ORF2) which consists of 192 bp. The deduced amino acid sequence of ORF2 shows 30 to 40% identity to the ferredoxins of Rhodococcus fascians, Thermotoga maritima, and Archaeoglobus fulgidus. ORF1 is located 150 bp upstream of the alkB-rub ORF in the reverse orientation. The putative product of this gene, encoding 208 amino acid residues, shows ∼30% identity to the TetR-family of transcriptional regulators and contains the tetR DNA-binding helix-turn-helix motif.
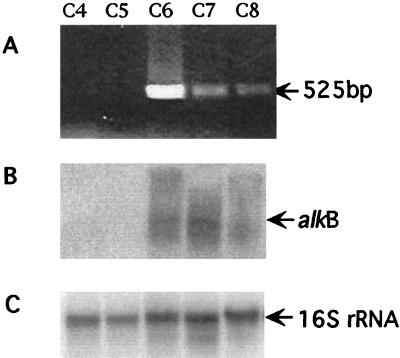
We investigated whether this alkB encodes the second monooxygenase that was proposed to be expressed when cells of strain CF8 were grown on alkanes above C6 in chain length. The expression of alkB in strain CF8 was examined by RT-PCR using primers designed from the cloned 550 bp PCR fragment, and by Northern hybridization using the same PCR fragment as a probe. Total RNA was prepared from cells grown on alkanes ranging from C4 to C8 as growth substrates. RT-PCR showed that a single product of the expected size (525 bp) was amplified from RNA prepared from cells grown on alkanes above C6 in chain length, while no products were detected from RNA prepared from C4 and C5-grown cells (Fig. 3A). The absence of alkB transcripts from C4 and C5-grown cells was also confirmed by the Northern blot (Fig. 3B). The probe hybridized to one major transcript of approximately 2 kb. These transcripts were absent from RNA from C4 and C5-grown cells. Hybridization with the 16S rRNA gene probe showed that all lanes contained similar amounts of RNA (Fig. 3C). These results indicate that alkB in strain CF8 is expressed when cells are growing on long-chain alkanes (C6) but not on the short-chain alkanes. Therefore, it seems likely that expression of alkB leads to production of the second monooxygenase in strain CF8.
FIG. 3.
RT-PCR (A) and Northern blot analysis (B and C) of alkB expression in strain CF8 grown on alkanes of various chain length. Total RNA was prepared from cells grown on C4 to C8 alkanes. RT-PCR was performed using primers shown in Fig. 2A, and the expected product size (525 bp) is indicated (A). Northern blot analysis was performed with the alkB probe shown in Fig. 2B, and the hybridizing band (about 2 kb) is indicated (B). The same blot was stripped and hybridized to the 16S rRNA probe (C).
DISCUSSION
Our results indicate that Nocardioides sp. strain CF8 can produce two distinct monooxygenases for oxidation of alkanes: a copper-containing monooxygenase and an integral-membrane, binuclear-iron monooxygenase. In this study, we took advantage of the sensitivity of the copper-containing monooxygenase to ATU and to alkynes in order to reveal the presence of the binuclear iron monooxygenase which is not inhibited by ATU or inactivated by alkynes. Expression of the copper-containing monooxygenase occurred on all the alkanes tested, while the binuclear iron monooxygenase was observed only in cells grown on alkanes C6 and above.
The ability of alkane-grown cells to express more than one monooxygenase is not without precedent. Several methanotrophs can produce two methane monooxygenases (pMMO and sMMO). The two situations are similar in that the copper-containing butane monooxygenase from strain CF8 is similar to pMMO. Like sMMO, the second alkane monooxygenase in strain CF8 apparently contains a binuclear-iron cluster. A distinct difference between this system and that of the methanotrophs is that in strain CF8, both monooxygenases can be expressed simultaneously, whereas, in methanotrophs, expression is limited to one monooxygenase at a time (32, 43). Another difference is in the apparent protein composition and cellular location of the binuclear-iron monooxygenases. While sMMO is a soluble enzyme and the hydroxylase component contains three polypeptides (16), the hydroxylase component of the binuclear-iron monooxygenase from strain CF8 is expected to be membrane-associated and to contain only a single polypeptide based on the similarity of the deduced amino acid sequence to that of the alkane hydroxylase (13, 46). Recently, the presence of two alkane hydroxylases in long-chain alkane-utilizing Acinetobacter sp. strain M-1 was reported (45). In this case, two alkane hydroxylases are both integral-membrane, binuclear-iron enzymes with high sequence similarity to the sequence of alkane hydroxylase (alkM) of Acinetobacter sp. strain ADP1. The expression of two alkane hydroxylase-encoding genes is regulated by the chain length of alkanes: alkMa expression is induced by solid alkanes (>C22), while alkMb expression is preferentially induced by liquid alkanes (C16 to C22) (45).
Sequence analysis of the complete alkB in strain CF8 revealed putative AlkB and rubredoxin domains in one continuous ORF that potentially encodes a fusion protein. The presence of a monooxygenase as a fused polypeptide is not without precedent. The cytochrome P-450 fatty acid monooxygenase from Bacillus megaterium was shown to consist of hydroxylase and reductase components on a single polypeptide encoded by a single continuous ORF (31, 36). This enzyme can be cleaved by trypsin into the respective domains (30). Further biochemical characterization of the alkane hydroxylase in strain CF8 is required to elucidate the function of the fused polypeptide and the possible involvement of the adjacent ferredoxin gene product.
The genetic organization of alkB in strain CF8 showed that genes encoding the enzymes in alkane metabolism are not clustered with alkB. This organization is unlike the alk genes in P. oleovorans, where the genes encoding alkane hydroxylase, two rubredoxins, an aldehyde dehydrogenase, an alcohol dehydrogenase, an acyl coenzyme A synthetase, and an outer membrane protein constitute a single operon (alkBFGHJKL) on the OCT plasmid (46). In the case of alk genes in Acinetobacter sp. strain ADP 1, the essential genes for alkane degradation are separately located on the chromosome where alkM and alkR are located about 369 kb from the rubA and rubB genes, encoding rubredoxin and rubredoxin reductase, respectively (20). The genetic organization of alkB in strain CF8 is rather similar to that of M. tuberculosis, in which the putative alkB is followed by the putative rubredoxin A and B, and no other genes encoding enzymes of alkane metabolism are nearby (12).
Our results suggest that the chain length of the alkane growth substrate plays a major role in the regulation of the expression of the binuclear iron monooxygenase in Nocardioides sp. strain CF8. In strain CF8, expression of the binuclear-iron monooxygenase was only observed in cells grown on alkanes C6 and above. In contrast, the substrate range of the enzyme appeared to extend to the gaseous alkane, butane. This response is reminiscent of alkB regulation in P. oleovorans, where the alkane specificity of the transcriptional regulator is more restrictive than the range of alkanes oxidized by the monooxygenase. In the case of AlkB in P. oleovorans, expression of the alkane hydroxylase operon (alkBFGHJKL) is regulated by a LuxR-UhpA-like transcriptional regulator, AlkS (15). AlkS induces the expression of the alkBFGHJKL operon in the presence of alkanes that are used as growth substrates (21). In contrast to AlkB, AlkM, the alkane hydroxylase from Acinetobacter sp. strain ADP1, can be induced by a variety of alkanes, including non-growth-supporting alkanes (35). AlkR, an AraC-XylS-like transcriptional regulator, induces the expression of alkM in the presence of alkanes ranging from C7 to C11, which do not support growth of ADP1, as well as alkanes ranging from C12 to C18, which are used as growth substrate (35). Approximately 150 bp upstream of the putative alkB sequence in strain CF8, there is an ORF that shows low similarity to TetR-like transcriptional regulators. This ORF may be a transcriptional regulator for alkB in strain CF8.
In conclusion, Nocardioides sp. strain CF8 possesses two distinct monooxygenases for alkane oxidation. To our knowledge, this is the first example of an alkane-utilizing bacterium that contains both copper- and binuclear-iron-containing monooxygenases specific for different chain length alkanes. The expression of the binuclear iron monooxygenase is influenced by the alkane chain length.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health grant GM56128 to D.J.A. and the Oregon Agricultural Experimental Station.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myersand E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ashraf W, Mihdhir A, Murrell J C. Bacterial oxidation of propane. FEMS Microbiol Lett. 1994;122:1–6. doi: 10.1111/j.1574-6968.1994.tb07134.x. [DOI] [PubMed] [Google Scholar]
- 3.Asperger O, Naumann A, Kleber H-P. Inducibility of cytochrome P-450 in Acinetobacter calcoaceticus by n-alkanes. Appl Microbiol Biotechnol. 1984;19:3948–4403. [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons; 1994. [Google Scholar]
- 5.Baptist J N, Gholson R K, Coon M J. Hydrocarbon oxidation by a bacterial enzyme system. I. Products of octane oxidation. Biochim Biophys Acta. 1963;69:40–47. doi: 10.1016/0006-3002(63)91223-x. [DOI] [PubMed] [Google Scholar]
- 6.Bédard C, Knowles R. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol Rev. 1989;53:68–84. doi: 10.1128/mr.53.1.68-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blevins W T, Perry J J. Metabolism of propane. n-propylamine, and propionate by hydrocarbon-utilizing bacteria. J Bacteriol. 1972;112:513–518. doi: 10.1128/jb.112.1.513-518.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britton L N. Microbial degradation of aliphatic hydrocarbons. In: Gibson D T, editor. Microbiology series. Microbial degradation of organic compounds. Vol. 13. New York, N.Y: Marcel Dekker; 1984. pp. 89–129. [Google Scholar]
- 9.Brzostowicz P C, Gibson K L, Thomas S M, Blasko M S, Rouvière P E. Simultaneous identification of two cyclohexanone oxidation genes from an environmental Brevibacterium isolate using mRNA differential display. J Bacteriol. 2000;182:4241–4248. doi: 10.1128/jb.182.15.4241-4248.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burrows K J, Cornish A, Scott D, Higgins I J. Substrate specificities of the soluble and particulate methane mono-oxygenases of Methylosinus trichosporium OB3b. J Gen Microbiol. 1984;130:3327–3333. [Google Scholar]
- 11.Colby J, Stirling D I, Dalton H. The soluble methane mono-oxygenase of Methylococcus capsulatus (Bath). Its ability to oxygenate n-alkanes. n-alkenes, ethers, and alicyclic, aromatic and heterocyclic compounds. Biochem J. 1977;165:395–402. doi: 10.1042/bj1650395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E I, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyed S, Hornsby T, Jagels K, Krrogh A, McLeah J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Soeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrett B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 13.Eggink G, van Lelyveld P H, Arnberg A, Arfman N, Witteveen C, Witholt B. Structure of the Pseudomonas putida alkBAC operon. Identification of transcription and translation products. J Biol Chem. 1987;262:6400–6406. [PubMed] [Google Scholar]
- 14.Eggink G, Engel H, Vriend G, Terpstra P, Witholt B. Rubredoxin reductase of Pseudomonas oleovorans. Structural relationship to other flavoprotein oxidoreductases based on one NAD and two FAD fingerprints. J Mol Biol. 1990;212:135–142. doi: 10.1016/0022-2836(90)90310-I. [DOI] [PubMed] [Google Scholar]
- 15.Eggink S, Engel H, Meijer W G, Otten J, Kingma J, Witholt B. Alkane utilization in Pseudomonas oleovorans. J Biol Chem. 1988;263:13400–13405. [PubMed] [Google Scholar]
- 16.Fox B G, Froland W A, Dege J E, Lipscomb J D. Methane monooxygenase from Methylosinus trichosporium OB3b. Purification and properties of a three-component system with high specific activity from a type II methanotroph. J Biol Chem. 1989;264:10023–10033. [PubMed] [Google Scholar]
- 17.Fox B G, Shanklin J, Ai J, Loehr T M, Sanders-Loehr J. Resonance raman evidence for an Fe-O-Fe center in stearoyl-ACP desaturase. Primary sequence identity with other diiron-oxo proteins. Biochemistry. 1994;33:12776–12786. doi: 10.1021/bi00209a008. [DOI] [PubMed] [Google Scholar]
- 18.Giovannoni S J. The polymerase chain reaction. In: Stackebrandt E, Goodfellow M, editors. Sequencing and hybridization techniques in bacterial systematics. New York, N.Y: John Wiley & Sons; 1991. pp. 177–201. [Google Scholar]
- 19.Gornall A G, Bardawill C J, David M M. Determination of serum proteins by means of the Biuret reaction. J Biol Chem. 1949;177:751–766. [PubMed] [Google Scholar]
- 20.Gralton E M, Campbell A L, Neidle E L. Directed introduction of DNA cleavage sites to produce a high-resolution genetic and physical map of the Acinetobacter sp. strain ADP1 (BD413UE) chromosome. Microbiology. 1997;143:1345–1357. doi: 10.1099/00221287-143-4-1345. [DOI] [PubMed] [Google Scholar]
- 21.Grund A, Shapiro J, Fennewald M, Bacha P, Leahy F, Markbreiter K, Nieder M, Toepfer M. Regulation of alkane oxidation in Pseudomonas putida. J Bacteriol. 1975;123:546–556. doi: 10.1128/jb.123.2.546-556.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamamura N, Arp D J. Isolation and characterization of alkane-utilizing Nocardioides sp. strain CF8. FEMS Microbiol Lett. 2000;186:21–26. doi: 10.1111/j.1574-6968.2000.tb09076.x. [DOI] [PubMed] [Google Scholar]
- 23.Hamamura N, Page C, Long T, Semprini L, Arp D J. Chloroform cometabolism by butane-grown CF8, Pseudomonas butanovora, and Mycobacterium vaccae JOB5 and methane-grown Methylosinus trichosprium OB3b. Appl Environ Microbiol. 1997;63:3607–3613. doi: 10.1128/aem.63.9.3607-3613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamamura N, Storfa R T, Semprini L, Arp D J. Diversity in butane monooxygenases among butane-grown bacteria. Appl Environ Microbiol. 1999;65:4586–4593. doi: 10.1128/aem.65.10.4586-4593.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou C T, Patel R, Laskin A I, Barnabe N, Barist I. Production of methyl ketones from secondary alcohols by cell suspensions of C2 to C4 n-alkane-grown bacteria. Appl Environ Microbiol. 1983;46:178–184. doi: 10.1128/aem.46.1.178-184.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyman M R, Wood P M. Suicidal inactivation and labeling of ammonia monooxygenase by acetylene. Biochem J. 1985;227:719–725. doi: 10.1042/bj2270719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lipscomb J D. Biochemistry of the soluble methane monooxygenase. Annu Rev Microbiol. 1994;48:371–399. doi: 10.1146/annurev.mi.48.100194.002103. [DOI] [PubMed] [Google Scholar]
- 28.Maeng J H, Sakai Y, Tani Y, Kato N. Isolation and characterization of a novel oxygenase that catalyzes the first step of n-alkane oxidation in Acinetobacter sp. strain M-1. J Bacteriol. 1996;178:3695–3700. doi: 10.1128/jb.178.13.3695-3700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLee A G, Kormendy A C, Wayman M. Isolation and characterization of n-butane-utilizing microorganisms. Can J Microbiol. 1972;18:1191–1195. doi: 10.1139/m72-186. [DOI] [PubMed] [Google Scholar]
- 30.Narhi L O, Fulco A J. Identification and characterization of two functional domains in cytochrome P-450BM-3, a catalytically self-sufficient monooxygenase induced by barbiturates in Bacillus megaterium. J Biol Chem. 1987;262:6683–6690. [PubMed] [Google Scholar]
- 31.Narhi L O, Fulco A J. Characterization of a catalytically self-sufficient 119,000-Dalton cytochrome P-450 monooxygenase induced by barbiturates in Bacillus megaterium. J Biol Chem. 1986;261:7160–7169. [PubMed] [Google Scholar]
- 32.Nielsen A K, Gerdes K, Murrell J C. Copper-dependent reciprocal transcriptional regulation of methane monooxygenase genes in Methylococcus capsulatus and Methylosinus trichosporium. Mol Microbiol. 1997;25:399–409. doi: 10.1046/j.1365-2958.1997.4801846.x. [DOI] [PubMed] [Google Scholar]
- 33.Prior S D, Dalton H. Acetylene as a suicide substrate and active site probe for methane monooxygenase from Methylococcus capsulatus (Bath) FEMS Microbiol Lett. 1985;29:105–109. [Google Scholar]
- 34.Ratajczak A, Geißdörfer W, Hillen W. Alkane hydroxylase from Acinetobacter sp. strain ADP1 is encoded by alkM and belongs to a new family of bacterial integral-membrane hydrocarbon hydroxylases. Appl Environ Microbiol. 1998;64:1175–1179. doi: 10.1128/aem.64.4.1175-1179.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ratajczak A, Geißdörfer W, Hillen W. Expression of alkane hydroxylase from Acinetobacter sp. strain ADP1 is induced by a broad range of n-alkanes and requires the transcriptional activator AlkR. J Bacteriol. 1998;180:5822–5827. doi: 10.1128/jb.180.22.5822-5827.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruettinger R T, Wen L-P, Fulco A J. Coding nucleotide, 5′ regulatory, and deduced amino acid sequences of P-450BM-3, a single peptide cytochrome P-450:NADPH-P-450 reductase from Bacillus megaterium. J Biol Chem. 1989;264:10987–10995. [PubMed] [Google Scholar]
- 37.Sakai Y, Maeng J H, Tani Y, Kato N. Use of long-chain n-alkanes (C13-C44) by an isolate, Acinetobacter sp. M-1. Biosci Biotechnol Biochem Biotechnol Biochem. 1994;58:2128–2130. [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Springs Harbor, N.Y: Cold Springs Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Sayavedra-Soto L A, Hommes N G, Alzerreca J J, Arp D J, Norton J M, Klotz M G. Transcription of the amoC, amoA, and amoB genes in Nitrosomonas europaea and Nitrosospira sp. NpAV FEMS Microbiol Lett. 1998;167:81–88. doi: 10.1111/j.1574-6968.1998.tb13211.x. [DOI] [PubMed] [Google Scholar]
- 40.Shanklin J, Achim C, Schmidt H, Fox B G, Münck E. Mössbauer studies of alkane ω-hydroxylase: evidence for a diiron cluster in an integral-membrane enzyme. Proc Natl Acad Sci USA. 1997;94:2981–2986. doi: 10.1073/pnas.94.7.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shanklin J, Whittle E, Fox B G. Eight histidine residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry. 1994;33:12787–12794. doi: 10.1021/bi00209a009. [DOI] [PubMed] [Google Scholar]
- 42.Smits T H M, Röthlisberger M, Witholt B, van Beilen J B. Molecular screening for alkane hydroxylase genes in gram-negative and gram-positive strains. Environ Microbiol. 1999;1:307–317. doi: 10.1046/j.1462-2920.1999.00037.x. [DOI] [PubMed] [Google Scholar]
- 43.Stanley S H, Prior D J, Leak D J, Dalton H. Copper stress underlines the fundamental change in intracellular location of methane mono-oxygenase in methane utilizing organisms: studies in batch and continuous cultures. Biotechnol Lett. 1983;5:487–492. [Google Scholar]
- 44.Takahashi J. Production of intracellular protein from n-butane by Pseudomonas butanovora sp. nov. Adv Appl Microbiol. 1980;26:117–127. [Google Scholar]
- 45.Tani A, Ishige T, Sakai Y, Kato N. Gene structures and regulation of the alkane hydroxylase complex in Acinetobacter sp. strain M-1. J Bacteriol. 2001;183:1819–1823. doi: 10.1128/JB.183.5.1819-1823.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Beilen J B, Wubbolts M G, Witholt B. Genetics of alkane oxidation by Pseudomonas oleovorans. Biodegradation. 1994;5:161–174. doi: 10.1007/BF00696457. [DOI] [PubMed] [Google Scholar]
- 47.Watkinson R J, Morgan P. Physiology of aliphatic hydrocarbon-degrading microorganisms. Biodegradation. 1990;1:79–92. doi: 10.1007/BF00058828. [DOI] [PubMed] [Google Scholar]
- 48.Wiegant W W, de Bont J A M. A new route for ethylene glycol metabolism in Mycobacterium E44. J Gen Microbiol. 1980;120:325–331. [Google Scholar]
- 49.Yeager C M, Bottomley P J, Arp D J, Hyman M R. Inactivation of toluene 2-monooxygenase in Burkholderia cepacia G4 by alkynes. Appl Environ Microbiol. 1999;65:632–639. doi: 10.1128/aem.65.2.632-639.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zahn J A, DiSpirito A. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath) J Bacteriol. 1996;178:1018–1029. doi: 10.1128/jb.178.4.1018-1029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]