Abstract
Neural activity is costly and requires continuous ATP from aerobic metabolism. Brainstem motor function of American bullfrogs normally collapses after minutes of ischaemia, but following hibernation, it becomes ischaemia-tolerant, generating output for up to 2 h without oxygen or glucose delivery. Transforming the brainstem to function during ischaemia involves a switch to anaerobic glycolysis and brain glycogen. We hypothesized that improving neural performance during ischaemia involves a transcriptional program for glycogen and glucose metabolism. Here we measured mRNA copy number of genes along the path from glycogen metabolism to lactate production using real-time quantitative PCR. The expression of individual genes did not reflect enhanced glucose metabolism. However, the number of co-expressed gene pairs increased early into hibernation, and by the end, most genes involved in glycogen metabolism, glucose transport and glycolysis exhibited striking linear co-expression. By contrast, co-expression of genes in the Krebs cycle and electron transport chain decreased throughout hibernation. Our results uncover reorganization of the metabolic transcriptional network associated with a shift to ischaemia tolerance in brain function. We conclude that modifying gene co-expression may be a critical step in synchronizing storage and use of glucose to achieve ischaemia tolerance in active neural circuits.
Keywords: metabolism, hypoxia tolerance, ischaemia tolerance, brain, gene co-expression
1. Introduction
Brain function is energetically expensive due to the high cost of ion regulation and synaptic transmission [1]. To provide ATP for these processes, glucose is oxidized through glycolysis and mitochondrial respiration. Compared to glycolysis alone, oxidative metabolism produces roughly 15-times more ATP per glucose molecule [2] and is, therefore, essential for providing consistent energy to the brain. Brief disruptions in the delivery of oxygen and glucose impair energy homeostasis, resulting in lethal outcomes commonly observed in stroke, as well as other energetic pathologies of the brain including drug overdose and starvation. Although the brain typically requires oxidative metabolism, some organisms have evolved the ability to survive in hypoxic environments without neural injury. Notable examples include turtles, goldfish, naked mole-rats and hooded seals, among others. Adaptations in these species can shed light on strategies for human neuroprotection [3,4].
Although the brain of frogs undergoes a slower death during hypoxia compared to mammals [5], most frogs are not generally counted among the champion hypoxia and ischaemia-tolerant vertebrates. Accordingly, motor circuit activity in the brainstem of frogs stops after a few minutes without oxygen, with tissue death following shortly after the removal of glucose [6,7]. In frogs that had recently emerged from hibernation, however, this same brainstem circuit not only survives ischaemic insults, but it transforms to produce coordinated motor activity for 25–30-fold longer (figure 1a) [6,7]. Mechanistically, this transformation involves a shift from dominant use of aerobic metabolism to anaerobic glycolysis fueled by local brain glycogen stores [6], which increases in frogs during winter [8]. Active circuits typically require oxidative phosphorylation to meet the demands of synaptic function and action potential firing, with little ability to run on anaerobic glycolysis and glycogen alone [9–11]. Thus, the frog brainstem can provide direct insight into endogenous mechanisms that remodel circuits of ischaemia-intolerant species into a state of ischaemia tolerance, with clear implications for humans who could benefit from modifying brain metabolism to function and survive during ischaemia.
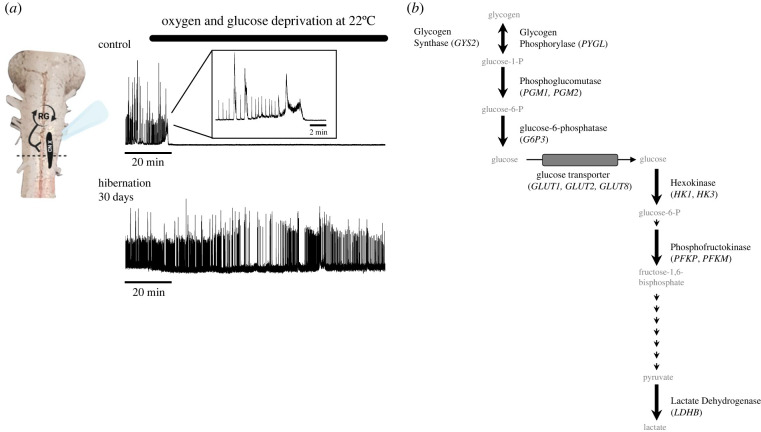
Figure 1.
Transformation from ischaemia intolerance to ischaemia tolerance in brainstem circuit function. (a) Example motor nerve recordings of output from the brainstem respiratory network before (top) and after (bottom) 30 days in a simulated overwintering environment (hibernation). The activity of this network is generated by a group of rhythm-generating neurons (RG) that activate motor neurons (black oval) to drive lung ventilation. After hibernation, motor circuit function gains the ability to operate for dramatically longer without oxygen and glucose delivery. (b) Metabolic enzymes assessed in this study are indicated in bold, with the gene name italicized. Small arrows indicate enzymatic steps not assessed in this study. (Online version in colour.)
Improving hypoxia and ischaemia tolerance in the bullfrog brainstem appears to represent a strategy to fuel high-cost circuit activity during emergence rather than a survival strategy during hibernation characteristic of other aquatic hibernators [12–14]. Frogs from northern latitudes overwinter in ice-covered ponds and rely solely on cutaneous gas exchange without lung ventilation. The arterial partial pressure of O2 falls as low as 1–3 mmHg, but the animal remains aerobic due to reduced metabolic rates in the cold [15]. However, as body temperature rises upon emergence, brainstem motor circuits must quickly restart to generate adaptive behaviors like breathing. This presents a problem because output from the respiratory network in frogs cannot occur with low O2 tensions associated with submergence [6,7]. Thus, if the respiratory motor network did not become capable of functioning during severe hypoxia at warming temperatures, lung ventilation could not restart, disrupting the recovery of whole-body O2 homeostasis in the spring.
Here we aim to gain insight into the regulation scheme through which glycogen metabolism and anaerobic glycolysis are enhanced to fuel circuit function following hibernation. As anaerobic glucose metabolism is a multi-step process, the regulation of rate-limiting enzymes at any step, or interactions among steps, have the potential to enhance glucose metabolism to support network activity [16–23]. Therefore, we hypothesized that glucose transport and metabolic processes along the path from glycogen metabolism to lactate production may either be upregulated or interact to permit anaerobic support of network function after winter. The metabolic phenotype of a variety of tissue and cell types is often reflected at the level of mRNA expression [24–27]; thus, we manually curated the bullfrog genome to identify genes involved in glycogen metabolism, glucose transport, glycolysis, as well as genes involved in metabolic regulation, such as AMPK and HIF-1α (AMP-activated protein kinase and hypoxia-inducible factor 1α). To compare with glycolysis and glycogen processing, we also assessed the expression of several genes involved in the Krebs cycle and electron transport chain. We then measured mRNA expression using absolute real-time quantitative PCR (RT-qPCR) in control, 2 and 30 day cold-acclimated brainstem samples. Finally, we compared mRNA abundances to assess up or downregulation of gene expression and changes in the co-expression of gene pairs to infer interactions across different aspects of metabolism.
2. Methods
(a) . Animals
Adult female American bullfrogs (Lithobates catesbeianus) were purchased from Rana Ranch (Twin Falls, ID, USA) and housed in plastic tanks with dechlorinated, aerated tap water in a light/dark cycle of 12/12 h. Control frogs (n = 8) were placed in tubs at 22°C with access to both wet and dry areas, and they were fed pellets provided by Rana Ranch once per week. Animals were acclimated to laboratory conditions for at least one week before experiments. To simulate overwintering conditions associated with hibernation, we placed other groups of animals in low-temperature incubators (Thermo Fisher Scientific, Waltham, MA, USA). One group of animals (n = 7) was placed in incubators that were gradually cooled from 20°C to 4°C over 7 days and then were kept at 4°C for 30 days before use (long-term cold acclimation). The other group was gradually cooled from 20°C to 4°C over 2 days and held at this temperature for 2–3 days (n = 7; short-term cold acclimation). Once the temperature reached 4°C, plastic screens were placed directly below water level to prevent frogs from accessing the surface. Water was bubbled with room aim in all treatments.
(b) . Brainstem dissection
The brainstem–spinal cord was dissected according to the established methods [7]. First, frogs were deeply anaesthetized with approximately 1 ml of isoflurane in a 1 l container until loss of the toe-pinch reflex, followed by rapid decapitation. The head was submerged in cold (2–4°C) artificial cerebrospinal fluid (aCSF) containing (in mM) 104 NaCl, 4 KCl, 1.4 MgCl2, 7.5 d-glucose, 40 NaHCO3, 2.5 CaCl2 and 1 NaH2PO4, all purchased from Fisher Scientific (Waltham, MA, USA). The aCSF was gassed with 1.5% CO2/98.5% O2 (pH = 7.85) during dissection. The skull was quickly removed. The animal was decerebrated, and then the brainstem–spinal cord was carefully removed. The dura covering the brain was removed and the brainstem was placed in TRIzol (Ambion by life technologies, CA, USA) and homogenized by strong vortexing.
(C) . RNA extraction and cDNA synthesis
Total RNA was isolated using a phenol–chloroform extraction according to the TRIzol protocol provided by the manufacturer (Invitrogen). The aqueous phase containing the RNA was then run through a column-based liquid clean-up protocol (Quick-RNA MicroPrep Kit, Zymo Research). Following RNA isolation, 1000 ng of total RNA underwent genomic DNA digestion using DNase 1 according to the manufacturer's instructions (DNase I, RNase-free, Thermo Scientific). cDNA was generated from 1000 ng total RNA following DNAse I digestion using qScript cDNA Supermix according to the protocol from the manufacturer (Quantabio, MA, USA). Samples were diluted fivefold in RNase-free molecular grade water (final volume 100 µl) and used as template in real-time quantitative PCR (RT-qPCR) analyses.
To identify candidate genes for RT-qPCR, we either used previously annotated coding sequences for Lithobates catesbeianus or identified unannotated genes via the homologous protein in other related amphibian species with well-annotated genomes (e.g. Nanorana parkeri or Rana temporaria). We then used NCBI BLAST to search the protein database for sequences for Lithobates catesbeianus that were similar to the queried target. As the Lithobates catesbeianus does not have a well-annotated genome, hits with the greatest homology were often unannotated ‘hypothetical proteins’. We confirmed the identity of the hypothetical protein by using this sequence from Lithobates catesbeianus as a query against the entire non-redundant protein database. All sequences used in this study shared close sequence identity with Rana temporaria and Nanorana parekeri. Accession numbers for each amino acid sequence were then used to identify the coding DNA sequence (CDS).
Primers for RT-qPCR were designed based on the CDS using Primer-BLAST [28]. Each set of primers were validated with fourfold serial dilutions of cDNA generated from frog brain, encompassing the range of cDNA inputs for this study. RT-qPCR reactions consisted of primer pairs at a final concentration of 2.5 µM, diluted cDNA template (5/32 of the total reaction volume), RNase-free molecular grade water and 2X SYBR master mix (Applied Biosystems by Thermo Fischer Scientific) and were run according to the manufacturer's instructions. Ten microlitre reactions for each target were run in triplicate on QuantStudio6 Real-Time PCR system (Applied Biosystems by Thermo Fischer Scientific) with a four-step cycle of 50°C for 2 minutes, 95°C for 10 minutes, 95°C for 15 seconds, 60°C for 1 minute. Following 40 cycles of PCR (95°C for 15 seconds, 60°C 1 minute), melt curves for all PCR products were acquired by increasing the temperature in increments of 0.3°C for 5 seconds from 60°C to 95°C. All melt curves had a single peak in every reaction well. All RT-qPCR reactions were run in triplicate, and the average Ct (cycle threshold) was determined for each sample. We then estimated absolute copy number per 400 ng total RNA by interpolating the Ct value into standard curves of known copy numbers, from 108–103 copies of DNA. We adjusted the estimate of absolute copy number by a normalization factor to account for the potential of sample-to-sample variation in efficiency of the cDNA synthesis reaction as has been done previously for absolute RT-qPCR in bulk tissue [29]. Briefly, the normalization factor was defined for each sample as 2−ΔCt18S, where ΔCt18S is the difference between the sample 18 S Ct value and population average, and 2 represents the doubling at each cycle of PCR assuming 100% efficiency, which is near the efficiency of your 18 S rRNA RT-qPCR assay (approx. 101%).
(d) . Electrophysiological recordings
To illustrate the transformation to ischaemia tolerance, sample electrophysiological recordings of respiratory motor activity of one brainstem preparation from a control frog and one from a frog after 30 days of cold submergence are shown in figure 1a. Recordings where were obtained exactly as described in [6].
(e) . Statistics
All datasets followed an approximately normal distribution based on the Shapiro–Wilk test. Analysis of group means was carried out using a one-way ANOVA test followed by Holm–Sidak's multiple comparison test. Pairwise co-expression relationships among genes were assessed using computing Pearson correlation coefficients (r) and p-values for the linear regression of each co-expression relationship. For correlation analyses, individual outliers can force artificial correlations. One sample in the 2 day cold group showed expression that was roughly double the mean for several mitochondrial genes, and therefore, produced correlations that were likely to be artificial. Thus, correlation analysis of the mitochondrial genes for 2 days of cold acclimation was run without this point for genes indicated on the available data spreadsheet. Group shifts in the distribution of Pearson correlation coefficients were assessed using Kolmogorov–Smirnov test. Analyses were ran using GraphPad Prism 9.0 (San Diego, CA, USA), and plots of multiple correlations were plotted using the ‘pairs’ function in R. Significance was accepted when p < 0.05. Data are presented as box plots, with dots representing data points for individual samples.
3. Results
Patterned activity of the brainstem respiratory network in the bullfrog can switch from low to high resistance to hypoxia and ischaemia after hibernation [6] (figure 1a). Therefore, we assessed gene expression profiles for all steps from glycogen synthesis to lactate production in control brainstem samples, as well as short-term (2 days) and longer-term (30 days) cold acclimation (figure 1b). To identify candidate genes for rate-limiting steps of glucose metabolism and glycogen processing, we manually curated the Lithobates catesbeianus genome [30]. We identified 13 metabolic enzymes and glucose transporters with brainstem expression that shared close amino acid sequence identity with two other frog species Nanorana parkeri and Rana temporaria (table 1).
Table 1.
qPCR primer sequences, efficiencies, NCBI accession numbers for bullfrog and homologous sequences.
| targets | forward primer sequence | reverse primer sequence | PCR efficiency (%) | NCBI accession number Lithobates catesbeianus | NCBI accession number Rana temporaria | NCBI accession number Nanorana parkeri |
|---|---|---|---|---|---|---|
| GLUT2 | GCAAGAAAACTGGGCCTGGTG | TGAAGACACTAAACCCCCAAGAGA | 97.808 | PIO25754.1 | XP_040205273.1 | XP_018409257.1 |
| GLUT3 | TGCCCTGCTGCCATTCTGTC | TCCGCTCCCCTTCCTCCTTC | 90.737 | PIO23325.1 | XP_040217157.1 | XP_018408713.1 |
| GLUT8 | GCTGGCTTGCTGTTCTCTGC | GGTCTACATCTGGCCCCCTC | 95.389 | PIO37827.1 | XP_040179949.1 | XP_018417566.1 |
| PFK-Muscle | GTCACATGCAGCAGGGAGGA | CCATAGCACGGCTTTTGCACC | 98.04 | PIO34539.1 | XP_040196734.1 | XP_018420044.1 |
| PFK-Platelet | GCTGTGCGCTCTGCAGTAAGA | GTCCAGCCACCAACATCTCCC | 86.601 | PIO12465.1 | XP_040208600.1 | XP_018425173.1 |
| LDH-B | GCTGCCATGGCTACGTTTTGG | GAAGGGCGACTCCAGCAACA | 92.093 | PIO23073.1 | XP_040207849.1 | XP_018413570.1 |
| GP | AGGCCTCTGGCACCGGTAAT | TCCGGCCTCTTCTGCCATCT | 102.41 | PIN97109.1 | XP_040206956.1 | XP_018428677.1 |
| G6P3 | CAGGTCCTGGCTGGCATTCT | GAGCCCCGAGCATCAGGAAC | 87.294 | PIN99572.1 | XP_040185933.1 | XP_018408213.1 |
| PGM1 | TTTTGGAGGGCACCACCCTG | ATCGAACGCTGCCCCAAAGT | 89.483 | PIO25062.1 | XP_040216711.1 | XP_018409357.1 |
| PGM2 | GGAGTGCGTGACCTAACCACC | TCATTGTGGCCACTCCTCCA | 85.876 | PIO31462.1 | XP_040191078.1 | XP_018422734.1 |
| GS2 | CGGCCGTCCAGTCCTCATTC | CCGGGCCCACACCAAATGAC | 108.561 | PIO24940.1 | XP_040201375.1 | XP_018425820.1 |
| Hexokinase1 | GCAAGGCACTGGAGAGGAGC | GCCGGGCTCCTTTAATCCCC | 95.033 | PIO33088.1 | XP_040218120.1 | XP_018422592.1 |
| Hexokinase3 | CGCGCTACGCCAGATGGAAC | GCTCTTCATCTGCACGCCAC | 81.684 | PIO02597.1 | XP_040200339.1 | XP_018407907.1 |
| HIF-1α | CTGCGAACCCATCCCTCACC | TCATGTCAAGGCTGTGGCGG | 108.699 | PIO38914.1 | XP_040188507.1 | XP_018418129.1 |
| AMPK (PRKAA1) | GGAGGTGCTCAGCTGCTTGT | AATGAATCGGGCGGGCTTGT | 94.923 | PIO34006.1 | XP_040194696.1 | XP_018416671.1 |
| 18S rRNA | CAGGCCGGTCGCCTGAATAC | GGCCCCAGTTCCGAAAACCA | 101.068 | KV934453.1 | XR_005744676.1 | ▪ |
| CS | GCCGTGCTCCGAAAGACAGA | GGGGTTCTTGGCTTTGCCCT | 82.139 | PIO22545.1 | XP_040197062 | XP_018429365.1 |
| ACO | AGCTGTCTGGCTGGTCGTCA | TGGCTCCTGTGCCACCTTTC | 92.896 | PIO31261.1 | XP_040215395.1 | XP_018423465.1 |
| IDH | TCTGGTGGCTTTGTGTGGGC | TGTTCCATGGGCTGCCTCAG | 83.307 | PIO36173.1 | XP_040198393.1 | XP_018427329.1 |
| SDHA | TGGGCCACAGGAGCTAAGGA | CAGGTCTGGCTGCCTGAGAC | 81.401 | PIO34735.1 | XP_040184340.1 | XP_018420689.1 |
| MDH | GCCAAGGAGGGAAGGGATGG | TTGCAGGATTGCCCACCACA | 84.083 | ACO51684.1 | XP_040207155.1 | XP_018411095.1 |
| NDUFA1 | TTGCTACCGCTTTCACCGCA | GGATGGAGCCTAGGGCGAGA | 82.133 | YP_008757908.1 | QLY89590.1 | YP_009130298.1 |
| CYCS | GGTGCCTGAGCCGGGATAGT | CCAAGGAGGGTTCCTGGCTG | 86.674 | YP_008757900.1 | QQM17989.1 | YP_009130290.1 |
| ATP6 | AGTTCGACTCACCGCCAACC | AGGGAGGCCGAGATTGATAAAGG | 82.62 | YP_008757903.1 | YP_009631379.1 | YP_009130293.1 |
| ATP5F1C | ATGTTGACGCTGGTTTCCCATT | TGGGCGGCTTTCTACCAACA | 83.89 | PIO33875.1 | XP_040199283.1 | XP_018421711.1 |
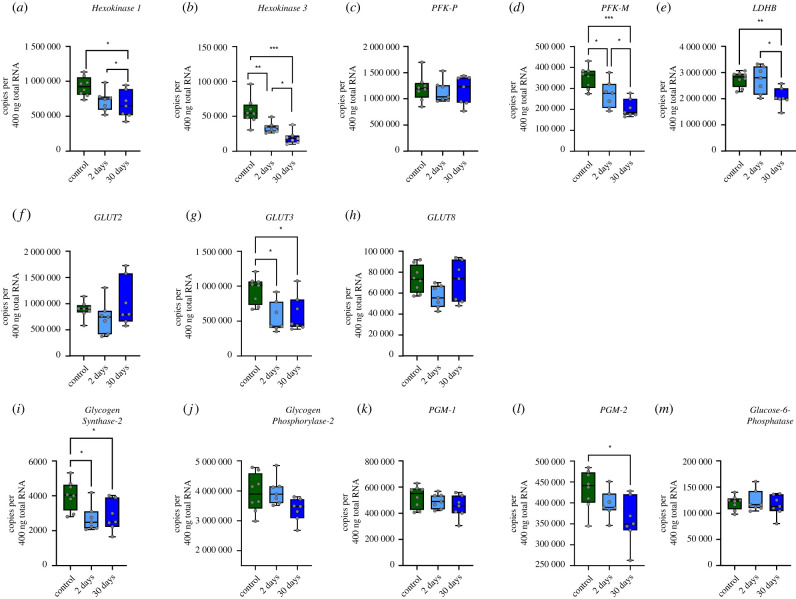
Given that a large upregulation of glycolytic metabolism contributes to the function of the brainstem during ischaemia [6], we first assessed changes in mRNA abundance for metabolic enzymes from glycogen processing to lactate production. Out of 13 genes, seven showed statistically significant changes in expression (figure 2). For glycolysis, Hexokinase I (figure 2a), Hexokinase 3 (figure 2b), Phosphofructokinase—Muscle (PFK-M, figure 2d), and Lactate Dehydrogenase—B (LDH-B, figure 2e) were reduced in both the 30-day and 2-day hibernation groups. One of the three glucose transporters, GLUT3 decreased in both the 2-day and 30-day hibernation groups (figure 2g). For glycogen metabolism, Glycogen Synthase—2 (figure 2i) and Phosphoglucomutase—2 (PGM—2, figure 2l) decreased by 30-days, with Glycogen Synthase—2 also showing a decrease in the 2-day hibernation group. Altogether, contrary to the expectation for a brainstem with activity that can be fueled solely by anaerobic glycolysis, the expression of all genes involved in glycolysis and glycogen processing showed decreases in expression or did not change.
Figure 2.
Mean expression of genes involved in glycolysis, glycogen metabolism, and glucose transport does not increase throughout hibernation. Boxplots for steady-state mRNA copy numbers for glucose metabolism genes. The control group is represented by green. The 2-day hibernation group is light blue, and the 30-day group are represented by darker blue. In each boxplot, the whiskers extend to the maximum and minimum values. (a–e) Genes for glycolysis, (f–h) genes for glucose transport, and (i–m) genes for glycogen synthesis and breakdown. The box spans from the 25th to 75th percentiles, and the median is presented as a horizontal line. Each individual data point is denoted by open circles. Significant differences between control and experimental groups are denoted with asterisks and the number of asterisk indicate the strength of the significance (one-way ANOVA test followed by Holm–Sidak's multiple comparisons test). *p < 0.05, **p <0.01,***p < 0.001. (Online version in colour.)
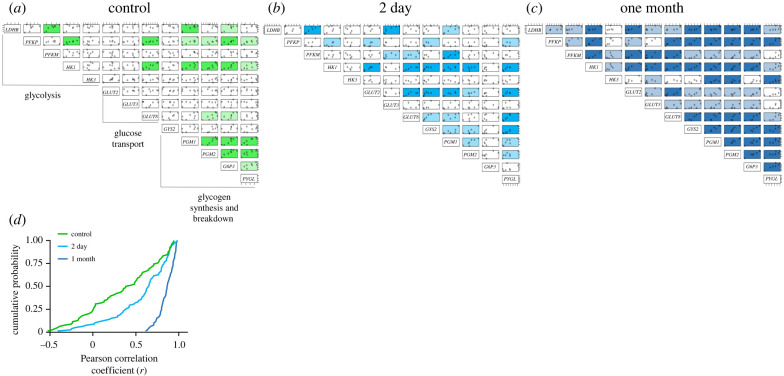
Given that anaerobic glycolysis supports network activity during oxygen and glucose deprivation after hibernation (figure 1, [6]), reduced or maintained glycolytic gene expression appears inconsistent with the large role of anaerobic glycolysis in fueling circuit function. Rather than upregulating rate-limiting enzymes, interactions across metabolic processes may contribute to the metabolic phenotype [31]. These interactions often exist as coupling of metabolite fluxes which are preserved as mRNA co-expression relationships for metabolic enzymes [32]. Therefore, we explored the dynamics of mRNA co-expression throughout cold acclimation. In the control group, only a few pairs of genes were significantly correlated, mostly concentrated within glycogen metabolism. Interestingly, we observed an increase in the number of gene pairs with significant correlation by 2 days of cold acclimation, particularly between glycolytic genes and glycogen metabolism, as well as glycolytic genes and glucose transporters (figure 3b). By the end of cold acclimation, most gene pairs became significantly correlated (figure 3c). When comparing the cumulative distribution of Pearson correlation coefficients for all pairwise correlations in control, 2-day, and 30-day groups, we observed a strong, progressive right-shift in correlation strength of population throughout hibernation. These results indicate that genes involved in glycolysis, glycogen metabolism and glucose transport become strongly co-expressed at fixed ratios over the course of 30-days of cold submergence (figure 3d).
Figure 3.
Hibernation progressively induces strong co-expression relationships across genes involved in glycolysis and glycogen processing. Correlation matrices showing co-expression for all pairs of metabolic genes in control (a), 2-day cold (b), and 30-day cold (c) groups. Each box shows a scatter plot of the pairwise expression relationships of two genes obtained for each sample. For example, the box on the top left of each matrix shows the mRNA copy number for LHDB (plotted on the y axis) and PFKP (plotted on the x axis) across the population of samples in the group. Pairwise correlations that reached statistical significance are shaded light when p < 0.05 and dark when p < 0.01. (d) The cumulative distribution of all Pearson r correlation coefficients obtained for all pairwise co-expression relationship in each group. There is a progressive increase in correlation coefficient at 2 days compared to control (p = 0.007; Kolmogorov–Smirnov test), with significantly greater amounts of co-expression at 30-days compared to control and 2-days (p < 0.0001 for both analyses; Kolmogorov–Smirnov test). (Online version in colour.)
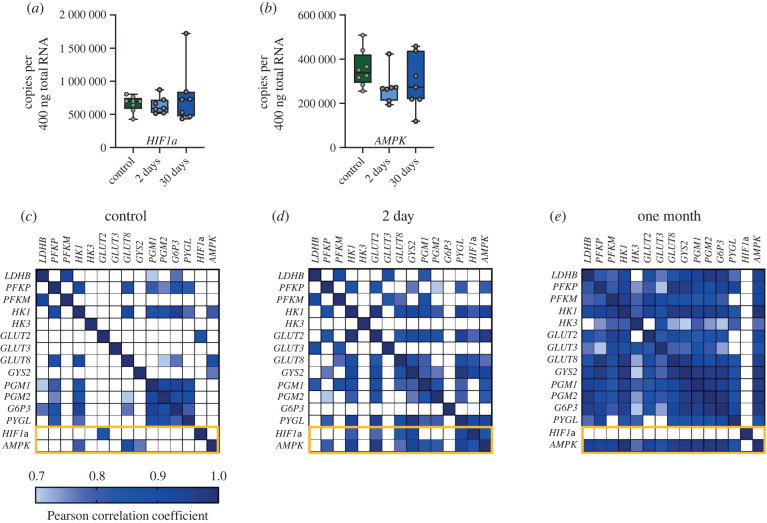
Metabolism is controlled by various transcriptional and signalling pathways. Hypoxia-inducible factor-1α (HIF-1α) is a transcription factor that can bind to the promoter/enhancer elements of genes for multiple glycolytic enzymes and glucose transporters [33]. In addition, AMPK is an energy sensor that can influence transcription to facilitate energy homeostasis [34,35]. Therefore, we measured the mRNA expression of AMPK and HIF-1α, as well as their co-expression with other metabolic genes. Neither AMPK nor HIF-1α showed differential expression across groups (figure 4a,b). However, both increased the number of co-expressed gene pairs early into the hibernation, outlined with an orange rectangle in the heat maps (figure 4c,d). Late into hibernation, AMPK became strongly co-expressed with most metabolic genes, whereas HIF-1α becomes decoupled from all other genes (figure 4e). In sum, these results point to widespread changes in mRNA co-expression for glucose metabolism throughout hibernation, with dynamic interactions with master regulators of metabolism, AMPK and HIF-1α.
Figure 4.
Metabolic regulators, AMPK and HIF-1α, change their association with the co-expression network across hibernation. (a,b) The mean mRNA expression for HIF-1α and AMPK. These genes did not show changes in mean abundance. (c–e) The same correlation matrices as in figure 3 drawn heat maps, highlighting co-expression relationships with HIF-1α and AMPK (orange rectangles). Each heat map pixel displays the Pearson correlation coefficient (r) for each pairwise comparison. Strong correlations are shown in dark blue (r of 1) and the light boxes represent the lowest Pearson r that showed statistical significance in the data set (0.7). White boxes have a Pearson r of less than 0.7 and were not statistically significant. The diagonal row represents the autocorrelation for each pair. Few correlations exist between metabolic regulators and all other metabolic genes in the control group. By 2 days, both AMPK and HIF-1α become correlated with metabolic enzymes and glucose transporters. At 30-days, AMPK becomes tightly coupled to all other genes except for HIF-1α, and HIF-1α becomes decoupled from every other gene. (Online version in colour.)
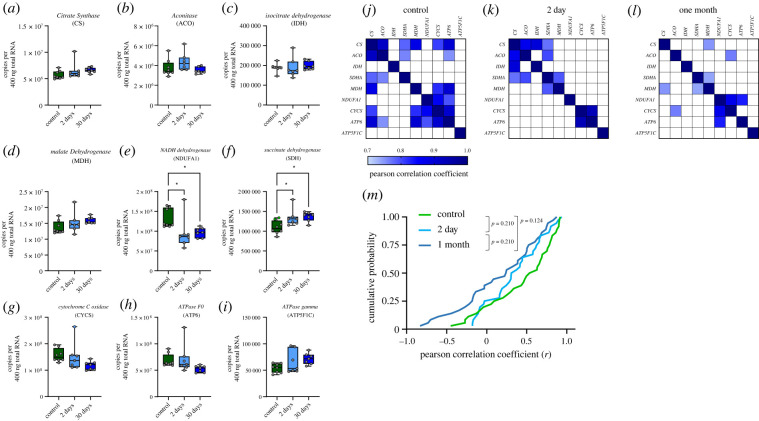
We last sought to assess mRNA co-expression for genes involved in aspects of aerobic metabolism, the Krebs cycle and electron transport chain (ETC; table 1). This would inform whether the strengthening of co-expression relationships is specific to anaerobic processes versus a general transcriptional strategy induced by hibernation. Of the nine Krebs cycle and ETC genes measured, NADH dehydrogenase (a subunit of complex I of the ETC) decreased expression at 2 and 30 days, while succinate dehydrogenase (part of the Krebs cycle and complex II of the ETC) increased expression at both time points (figure 5e,f). We observed several instances of co-expression across Krebs cycle and ETC genes in controls, with 12/36 strong correlations. (figure 5j). Strikingly—and in contrast to the trends observed for glycolysis/glycogen processing—co-expression relationships were reduced to 6/36 pairs at 2 days and 5/36 pairs at 30 days (figure 5k,l). Although the number of strong correlations decreased by greater than half at 30 days, there was no significant difference in the distribution of Pearson r values across groups (figure 5m). These results demonstrate a selective shift towards co-expression of genes for glycolysis and glycogen metabolism, and away from those involved in aerobic metabolism, in association with functional hypoxia and ischaemia tolerance [6].
Figure 5.
Genes involved in aerobic metabolism reduce co-expression during hibernation. (a–i) Mean copy number of genes involved in the Krebs cycle and electron transport chain (ETC). The box spans from the 25th to 75th percentiles, and the median is presented as a horizontal line. Each individual data point is denoted by open circles. Significant differences between control and experimental groups are denoted with asterisks (one-way ANOVA test followed by Holm–Sidak's multiple comparison test). (j–l) Changes in co-expression across Krebs cycle and ETC genes throughout hibernation, with (m) illustrating the cumulative distribution in Pearson correlation coefficients. *p < 0.05. (Online version in colour.)
4. Discussion
Function of the brainstem in adult American bullfrogs transitions to become remarkably ischaemia-tolerant following emergence from hibernation. In the present study, we investigated the mRNA expression profiles for a panel of genes that form the path from glycogen metabolism to lactate production, as well as the Krebs cycle and electron transport chain. Our results introduce the possibility that widespread alterations in gene co-expression across anaerobic and aerobic metabolism correspond to the transformation to ischaemia tolerance of brainstem function.
(a) . mRNA co-expression relationships for glucose metabolism in ischaemia tolerance
Considering the highly glycolytic and glycogenolytic phenotype of the brainstem following hibernation [6], we hypothesized that genes coding for rate-limiting enzymes may be upregulated to supply ATP needed for circuit function. By contrast, the expression of these genes decreased or did not change throughout simulated overwintering. These results suggest a more complicated relationship between metabolic gene expression and the highly glycolytic phenotype of the brainstem following hibernation. Although counterintuitive, this discrepancy between expected increases in gene expression and the hypoxia-tolerant phenotype is not uncommon. For example, the brain of hooded seals has far greater glycogen concentration and glycogenolytic capacity compared to mice, yet they have lower glycogen phosphorylase mRNA expression in the cerebellum and no significant difference in the neocortex [36]. For enzyme activity, turtle and rat brain has similar hexokinase (HK) and lactate dehydrogenase (LDH) activities despite vastly different capacities to survive on glycolysis alone [37]. In fish, expression of rate-limiting glycolytic enzymes in the brain does not correlate with hypoxia-tolerance across multiple species [38]. Although elevated mRNA expression and enzyme activity can correspond to enhanced glycolytic capacity in some cases [24,39], our results point to more complex control over brain glucose metabolism than simply upregulating rate-limiting steps.
Metabolic flux may be regulated by multiple enzymes rather than by any single rate-limiting enzyme [31]. The coupling of multiple metabolite fluxes are reflected in the co-expression of mRNA for various metabolic processes [32,40]. Therefore, understanding changes in mRNA co-expression within and across pathways may provide insight into critical metabolic interactions that contribute to ischaemia tolerance beyond that which can be easily observed through the mean expression of individual genes. Interestingly, we found that the number of co-expressed gene pairs increased after short-term cold submergence. Long-term cold acclimation elicited even a stronger effect, showing linear co-expression in most genes involved in glycogen metabolism, glucose transport and glycolysis (figure 3a–c). Thus, there appears to be regulation occurring that reorganizes the expression of genes at fixed ratios relative to one another even though the mean abundance decreases or remains the same. Some intriguing relationships with potential functional implications are the glucose transporters. In the control group, neither GLUT2 nor GLUT3 exhibited correlations with any other genes. However, both became more correlated with other enzymes in the 2-day hibernation group, and by one month, both GLUTs formed correlations with most metabolic enzymes. GLUT2 is often expressed in brainstem glial cells, and GLUT3 is expressed largely by neurons [41–45], suggesting coregulation of glucose transporters with other metabolic processes to facilitate glucose transport from glia to neurons. In addition, most genes involved in glycogen metabolism became correlated with those involved in glycolysis. These results are in direct contrast to the patterns observed for select genes from the Krebs cycle and electron transport chain, where we observed an elimination of over half of the co-expressed gene pairs by 30 days of hibernation (figure 5j–m). Together, these observations align with our functional data whereby glycogen can support neural activity during an ischaemic insult after hibernation, reducing its reliance on aerobic metabolism for circuit function [6].
These results lead us to suggest that the metabolic state of the brain can be read out through the co-expression of metabolic genes. Intriguingly, modification of mRNA co-expression plays a role in the metabolic phenotype of other highly glycolytic models, such as cancer cells. Indeed, some genes in cancer cells demonstrate a high degree of differential co-expression without differences in mean expression [46]. Additionally, disturbances in the co-expression of four genes involved in the electron transport chain occur in seven types of cancers [46]. Furthermore, multiple cancer cell types have disturbed gene co-expression networks for different pathways including folate, glutamine and glycine metabolism compared to normal tissue [47]. Similarly, mRNAs that encode proteins involved in oxidative phosphorylation (OXPHOS) also co-express in humans and mice [48–50], with subunits of each complex showing stronger ‘within complex’ than ‘across complex’ co-expression [51]. These results emphasize the potential for coregulation of gene expression to contribute to a tissue's metabolic phenotype, consistent with our results. Altogether, the data suggest that dynamics in mRNA co-expression across different aspects of anaerobic and aerobic metabolism may contribute to shifting the metabolic fuel source to support brainstem function during hypoxia and ischaemia.
(b) . Potential mechanisms that form mRNA correlations
The mechanisms underlying gene co-expression and their dynamic regulation remain elusive. One hypothesis for the control of gene co-expression involves a ‘master molecule’, such as transcription factors, that regulate the expression of multiple genes through shared enhancer elements and transcription factor binding sites [52]. For instance, Yin et al. [53] found that highly correlated gene pairs tend to share at least one common cis regulatory site. In Drosophila, genes that share transcription factor binding sites have a higher degree of co-expression than those that do not [54]. HIF-1α can bind to enhancer sites and has been characterized in amphibians [33,55,56], making it a potential regulator of mRNA co-expression in response to hibernation. Our data are consistent with this possibility because several metabolic genes became co-expressed with HIF-1α by 2 days of cold acclimation. However, these correlations vanish by 30 days, while the number of genes that co-express with AMPK increase throughout hibernation. Given that transcription factors controlling tissue-level processes, such as muscle growth, have been identified through mRNA co-expression without obvious changes in mean expression [57], our results suggest that HIF-1α and AMPK may contribute to reorganizing the metabolic network to favour glycolysis and glycogen breakdown. Since HIF-1α correlations emerge and disappear, we speculate that HIF-1α may respond to acute drops in tissue PO2, while AMPK contributes to the sustained reorganization of the glycolytic and glycogen metabolic network over longer timescales. These outcomes align with data showing that acute decreases in tissue PO2—consistent with the low arterial PO2 reported in cold-submerged frogs [58]—activates HIF-1α but is degraded under chronic low oxygen conditions [59,60]. Overall, we provide evidence for the rearrangement of the transcriptional network that regulates glucose metabolism and the possible involvement of AMPK and HIF-1α.
(c) . Limitations
Although there is a strong precedent for mRNA co-expression to, in part, explain metabolite fluxes [32,40], we must acknowledge a few limitations. First, enzyme concentration, post-translational modifications, allosteric modulation and protein–protein interactions can influence enzyme activity and metabolic fluxes. Therefore, dynamics in mRNA co-expression is only one level of regulation that has the potential to alter the brain metabolic phenotype. It will be important for future work to determine if mRNA correlations scale to enzyme activity, and if not, how the emergence of co-expression at the mRNA level influences the ability of brainstem to function without oxygen and glucose delivery. Second, by using whole brainstem samples, our results represent mRNA from a heterogeneous population of cells. Therefore, we cannot conclude which cell types are involved in the responses and how that relates to the emergence of co-expression relationships. That said, we expect most of the glycogen processing genes and GLUT2 may be expressed in glia, while glycolysis, ETC, and Krebs cycle genes are likely to be expressed globally. Finally, our original study assessed ischaemia tolerance of circuit function after 30-days of cold submergence [6]. Thus, we do not yet know whether the emergence of gene co-expression at 2 days are associated with functional ischaemia tolerance or if this represents an initial step in transforming the metabolic state of the brainstem. Further investigation using higher throughput methods, such as RNA-seq and metabolomics, are needed to determine how mRNA correlations relate to other biological scales and interactions between cell types.
5. Conclusion
In sum, our data indicate that transforming brain function to become ischaemia-tolerant involves dynamic changes in co-expression of metabolic genes for anaerobic (increases) and aerobic (decreases) metabolism. These results introduce the possibility that reorganization of gene co-expression networks may modify different aspects of metabolism to induce, or contribute to, hypoxia and ischaemia tolerance of brain function. Our data suggest that gene co-expression spanning multiple aspects of metabolism may contribute to hypoxia tolerance in other models and explain evolutionarytrends in hypoxia tolerance where rate-limiting steps for glucose metabolism do not obviously differ across species [36,38]. Much of the interest in natural models of hypoxia tolerance arises from their ability to avoid brain dysfunction and damage during hypoxia and ischaemia. Understanding the triggers and organizing mechanisms of gene co-expression may inform new design principles for inducible neuroprotection approaches, highlighting the value identifying multiple solutions for hypoxia tolerance across species.
Acknowledgements
We thank Nikolaus Bueschke, Lara do Amaral-Silva and Sarah Pellizzari for assistance with tissue sample collection.
Ethics
All experiments have been approved by the Institutional Animal Care and Use Committee at The University of North Carolina at Greensboro (protocol no. 19-006).
Data accessibility
Raw data generated for this study are available in the electronic supplemental material [61].
Authors' contributions
M.H.: data curation, formal analysis, investigation, writing—original draft, writing—review and editing; J.M.S.: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, supervision, visualization, writing—original draft, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
Funding was providing by the National Institutes of Health (R15NS112920), the US Department of Defense (W911NF2010275), and laboratory start-up funds from the University of North Carolina at Greensboro to J.M.S.
References
- 1.Harris JJ, Jolivet R, Attwell D. 2012. Synaptic energy use and supply. Neuron 75, 762-777. ( 10.1016/j.neuron.2012.08.019) [DOI] [PubMed] [Google Scholar]
- 2.Dienel GA. 2019. Brain glucose metabolism: integration of energetics with function. Physiol. Rev. 99, 949-1045. ( 10.1152/physrev.00062.2017) [DOI] [PubMed] [Google Scholar]
- 3.Dave KR, Christian SL, Perez-Pinzon MA, Drew KL. 2012. Neuroprotection: lessons from hibernators. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 162, 1-9. ( 10.1016/j.cbpb.2012.01.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larson J, Drew KL, Folkow LP, Milton SL, Park TJ. 2014. No oxygen? No problem! Intrinsic brain tolerance to hypoxia in vertebrates. J. Exp. Biol. 217, 1024-1039. ( 10.1242/jeb.085381) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milton SL, Manuel L, Lutz PL. 2003. Slow death in the leopard frog Rana pipiens: neurotransmitters and anoxia tolerance. J. Exp. Biol. 206, 4021-4028. ( 10.1242/jeb.00647) [DOI] [PubMed] [Google Scholar]
- 6.Bueschke N, Do Amaral-Silva L, Adams S, Santin JM. 2021. Transforming a neural circuit to function without oxygen and glucose delivery. Curr. Biol. 31, R1564-R1565. ( 10.1016/j.cub.2021.11.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams S, Zubov T, Bueschke N, Santin JM. 2021. Neuromodulation or energy failure? Metabolic limitations silence network output in the hypoxic amphibian brainstem. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 320, R105-R116. ( 10.1152/ajpregu.00209.2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mcdougal DB Jr, Holowach J, Howe MC, Jones EM, Thomas CA. 1968. The effects of anoxia upon energy sources and selected metabolic intermediates in the brains of fish, frog and turtle. J. Neurochem. 15, 577-588. ( 10.1111/j.1471-4159.1968.tb08956.x) [DOI] [PubMed] [Google Scholar]
- 9.Hall CN, Klein-Flügge MC, Howarth C, Attwell D. 2012. Oxidative phosphorylation, not glycolysis, powers presynaptic and postsynaptic mechanisms underlying brain information processing. J. Neurosci. 32, 8940-8951. ( 10.1523/JNEUROSCI.0026-12.2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rangaraju V, Calloway N, Ryan TA. 2014. Activity-driven local ATP synthesis Is required for synaptic function. Cell 156, 825-835. ( 10.1016/j.cell.2013.12.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trevisiol A, Saab AS, Winkler U, Marx G, Imamura H, Möbius W, Kusch K, Nave K-A, Hirrlinger J. 2017. Monitoring ATP dynamics in electrically active white matter tracts. Elife 6, e24241. ( 10.7554/eLife.24241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buck LT, Land SC, Hochachka PW. 1993. Anoxia-tolerant hepatocytes: model system for study of reversible metabolic suppression. Am. J. Physiol. 265, R49-R56. ( 10.1152/ajpregu.1993.265.1.R49) [DOI] [PubMed] [Google Scholar]
- 13.Herbert CV, Jackson DC. 1985. Temperature effects on the responses to prolonged submergence in the turtle Chrysemys picta bellii. II. Metabolic rate, blood acid–base and ionic changes, and cardiovascular function in aerated and anoxic water. Physiol. Zool. 58, 670-681. ( 10.1086/physzool.58.6.30156071) [DOI] [Google Scholar]
- 14.Jackson DC. 2002. Hibernating without oxygen: physiological adaptations of the painted turtle. J. Physiol. 543, 731-737. ( 10.1113/jphysiol.2002.024729) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ultsch GR, Reese SA, Stewart ER. 2004. Physiology of hibernation in Rana pipiens: metabolic rate, critical oxygen tension, and the effects of hypoxia on several plasma variables. J. Exp. Zool. A Comp. Exp. Biol. 301, 169-176. ( 10.1002/jez.a.20014) [DOI] [PubMed] [Google Scholar]
- 16.Tanner LB, Goglia AG, Wei MH, Sehgal T, Parsons LR, Park JO, White E, Toettcher JE, Rabinowitz JD. 2018. Four key steps control glycolytic flux in mammalian cells. Cell Syst. 7, 49-62.e8. ( 10.1016/j.cels.2018.06.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Birsoy K, et al. 2014. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature 508, 108-112. ( 10.1038/nature13110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ros S, Schulze A. 2013. Balancing glycolytic flux: the role of 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatases in cancer metabolism. Cancer Metab. 1, 8. ( 10.1186/2049-3002-1-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yi W, Clark PM, Mason DE, Keenan MC, Hill C, Goddard WA, Peters EC, Driggers EM, Hsieh-Wilson LC. 2012. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 337, 975-980. ( 10.1126/science.1222278) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu H, et al. 2016. Phosphoinositide 3-kinase regulates glycolysis through mobilization of aldolase from the actin cytoskeleton. Cell 164, 433-446. ( 10.1016/j.cell.2015.12.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shestov AA, et al. 2014. Quantitative determinants of aerobic glycolysis identify flux through the enzyme GAPDH as a limiting step. Elife 3, e03342. ( 10.7554/eLife.03342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dayton TL, Jacks T, Vander Heiden MG. 2016. PKM2, cancer metabolism, and the road ahead. EMBO Rep. 17, 1721-1730. ( 10.15252/embr.201643300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV. 1997. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc. Natl Acad. Sci. USA 94, 6658-6663. ( 10.1073/pnas.94.13.6658) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park TJ, et al. 2017. Fructose-driven glycolysis supports anoxia resistance in the naked mole-rat. Science 356, 307-311. ( 10.1126/science.aab3896) [DOI] [PubMed] [Google Scholar]
- 25.Chandrasekaran S, Price ND. 2010. Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 107, 17 845-17 850. ( 10.1073/pnas.1005139107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cakir T, Kirdar B, Ulgen KO. 2004. Metabolic pathway analysis of yeast strengthens the bridge between transcriptomics and metabolic networks. Biotechnol. Bioeng. 86, 251-260. ( 10.1002/bit.20020) [DOI] [PubMed] [Google Scholar]
- 27.Liao B-Y, Weng M-P. 2015. Unraveling the association between mRNA expressions and mutant phenotypes in a genome-wide assessment of mice. Proc. Natl Acad. Sci. USA 112, 4707-4712. ( 10.1073/pnas.1415046112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 13, 134. ( 10.1186/1471-2105-13-134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia VB, Garcia ML, Schulz DJ. 2014. Quantitative expression profiling in mouse spinal cord reveals changing relationships among channel and receptor mRNA levels across postnatal maturation. Neuroscience 277, 321-333. ( 10.1016/j.neuroscience.2014.07.012) [DOI] [PubMed] [Google Scholar]
- 30.Hammond SA, et al. 2017. The North American bullfrog draft genome provides insight into hormonal regulation of long noncoding RNA. Nat. Commun. 8, 1433. ( 10.1038/s41467-017-01316-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pierce VA, Crawford DL. 1997. Phylogenetic analysis of glycolytic enzyme expression. Science 276, 256-259. ( 10.1126/science.276.5310.256) [DOI] [PubMed] [Google Scholar]
- 32.Ihmels J, Levy R, Barkai N. 2004. Principles of transcriptional control in the metabolic network of Saccharomyces cerevisiae. Nat. Biotechnol. 22, 86-92. ( 10.1038/nbt918) [DOI] [PubMed] [Google Scholar]
- 33.Semenza GL, Roth PH, Fang HM, Wang GL. 1994. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J. Biol. Chem. 269, 23 757-23 763. ( 10.1016/S0021-9258(17)31580-6) [DOI] [PubMed] [Google Scholar]
- 34.Hardie DG, Ross FA, Hawley SA. 2012. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 13, 251-262. ( 10.1038/nrm3311) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bungard D, et al. 2010. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science 329, 1201-1205. ( 10.1126/science.1191241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Czech-Damal NU, Geiseler SJ, Hoff MLM, Schliep R, Ramirez J-M, Folkow LP, Burmester T. 2014. The role of glycogen, glucose and lactate in neuronal activity during hypoxia in the hooded seal (Cystophora cristata) brain. Neuroscience 275, 374-383. ( 10.1016/j.neuroscience.2014.06.024) [DOI] [PubMed] [Google Scholar]
- 37.Suarez RK, Doll CJ, Buie AE, West TG, Funk GD, Hochachka PW. 1989. Turtles and rats: a biochemical comparison of anoxia-tolerant and anoxia-sensitive brains. Am. J. Physiol. 257, R1083-R1088. ( 10.1152/ajpregu.1989.257.5.R1083) [DOI] [PubMed] [Google Scholar]
- 38.Nilsson GE, Ostlund-Nilsson S. 2008. Does size matter for hypoxia tolerance in fish? Biol. Rev. Camb. Philos. Soc. 83, 173-189. ( 10.1111/j.1469-185X.2008.00038.x) [DOI] [PubMed] [Google Scholar]
- 39.Murphy B, Zapol WM, Hochachka PW. 1980. Metabolic activities of heart, lung, and brain during diving and recovery in the Weddell seal. J. Appl. Physiol. Respir. Environ. Exerc. Physiol. 48, 596-605. ( 10.1152/jappl.1980.48.4.596) [DOI] [PubMed] [Google Scholar]
- 40.Notebaart RA, Teusink B, Siezen RJ, Papp B. 2008. Co-regulation of metabolic genes is better explained by flux coupling than by network distance. PLoS Comput. Biol. 4, e26. ( 10.1371/journal.pcbi.0040026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leloup C, Arluison M, Lepetit N, Cartier N, Marfaing-Jallat P, Ferré P, Pénicaud L. 1994. Glucose transporter 2 (GLUT 2): expression in specific brain nuclei. Brain Res. 638, 221-226. ( 10.1016/0006-8993(94)90653-x) [DOI] [PubMed] [Google Scholar]
- 42.Mcewen BS, Reagan LP. 2004. Glucose transporter expression in the central nervous system: relationship to synaptic function. Eur. J. Pharmacol. 490, 13-24. ( 10.1016/j.ejphar.2004.02.041) [DOI] [PubMed] [Google Scholar]
- 43.Gerhart DZ, Broderius MA, Borson ND, Drewes LR. 1992. Neurons and microvessels express the brain glucose transporter protein GLUT3. Proc. Natl Acad. Sci. USA 89, 733-737. ( 10.1073/pnas.89.2.733) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leino RL, Gerhart DZ, Van Bueren AM, Mccall AL, Drewes LR. 1997. Ultrastructural localization of GLUT 1 and GLUT 3 glucose transporters in rat brain. J. Neurosci. Res. 49, 617-626. () [DOI] [PubMed] [Google Scholar]
- 45.Mantych GJ, James DE, Chung HD, Devaskar SU. 1992. Cellular localization and characterization of Glut 3 glucose transporter isoform in human brain. Endocrinology 131, 1270-1278. ( 10.1210/endo.131.3.1505464) [DOI] [PubMed] [Google Scholar]
- 46.Reznik E, Sander C. 2015. Extensive decoupling of metabolic genes in cancer. PLoS Comput. Biol. 11, e1004176. ( 10.1371/journal.pcbi.1004176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang D, Guo Y, Xie N. 2020. Prognostic value and co-expression patterns of metabolic pathways in cancers. BMC Genom. 21, 860. ( 10.1186/s12864-020-07251-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P. 2004. Coexpression analysis of human genes across many microarray data sets. Genome Res. 14, 1085-1094. ( 10.1101/gr.1910904) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mootha VK, et al. 2003. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115, 629-640. ( 10.1016/s0092-8674(03)00926-7) [DOI] [PubMed] [Google Scholar]
- 50.Shyamsundar R, et al. 2005. A DNA microarray survey of gene expression in normal human tissues. Genome Biol. 6, R22. ( 10.1186/gb-2005-6-3-r22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van Waveren C, Moraes CT. 2008. Transcriptional co-expression and co-regulation of genes coding for components of the oxidative phosphorylation system. BMC Genom. 9, 18. ( 10.1186/1471-2164-9-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ribeiro DM, Rubinacci S, Ramisch A, Hofmeister RJ, Dermitzakis ET, Delaneau O. 2021. The molecular basis, genetic control and pleiotropic effects of local gene co-expression. Nat. Commun. 12, 4842. ( 10.1038/s41467-021-25129-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yin W, Mendoza L, Monzon-Sandoval J, Urrutia AO, Gutierrez H. 2021. Emergence of co-expression in gene regulatory networks. PLoS ONE 16, e0247671. ( 10.1371/journal.pone.0247671) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marco A, Konikoff C, Karr TL, Kumar S. 2009. Relationship between gene co-expression and sharing of transcription factor binding sites in Drosophila melanogaster. Bioinformatics 25, 2473-2477. ( 10.1093/bioinformatics/btp462) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Beaucourt A, Coumailleau P. 2007. Molecular cloning and characterization of the Xenopus hypoxia-inducible factor 1alpha (xHIF1alpha). J. Cell. Biochem. 102, 1542-1552. ( 10.1002/jcb.21376) [DOI] [PubMed] [Google Scholar]
- 56.Bergeron M, Yu AY, Solway KE, Semenza GL, Sharp FR. 1999. Induction of hypoxia-inducible factor-1 (HIF-1) and its target genes following focal ischaemia in rat brain. Eur. J. Neurosci. 11, 4159-4170. ( 10.1046/j.1460-9568.1999.00845.x) [DOI] [PubMed] [Google Scholar]
- 57.Hudson NJ, Reverter A, Dalrymple BP. 2009. A differential wiring analysis of expression data correctly identifies the gene containing the causal mutation. PLoS Comput. Biol. 5, e1000382. ( 10.1371/journal.pcbi.1000382) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tattersall GJ, Ultsch GR. 2008. Physiological ecology of aquatic overwintering in ranid frogs. Biol. Rev. 83, 119-140. ( 10.1111/j.1469-185X.2008.00035.x) [DOI] [PubMed] [Google Scholar]
- 59.Holmquist-Mengelbier L, et al. 2006. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell 10, 413-423. ( 10.1016/j.ccr.2006.08.026) [DOI] [PubMed] [Google Scholar]
- 60.Uchida T, Rossignol F, Matthay MA, Mounier R, Couette S, Clottes E, Clerici C. 2004. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression in lung epithelial cells: implication of natural antisense HIF-1alpha. J. Biol. Chem. 279, 14 871-14 878. ( 10.1074/jbc.M400461200) [DOI] [PubMed] [Google Scholar]
- 61.Hu M, Santin JM. 2022. Transformation to ischaemia tolerance of frog brain function corresponds to dynamic changes in mRNA co-expression across metabolic pathways. Figshare. ( 10.6084/m9.figshare.c.6080795) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Hu M, Santin JM. 2022. Transformation to ischaemia tolerance of frog brain function corresponds to dynamic changes in mRNA co-expression across metabolic pathways. Figshare. ( 10.6084/m9.figshare.c.6080795) [DOI] [PMC free article] [PubMed]
Data Availability Statement
Raw data generated for this study are available in the electronic supplemental material [61].