Abstract
Pandemics originating from non-human animals highlight the need to understand how natural hosts have evolved in response to emerging human pathogens and which groups may be susceptible to infection and/or potential reservoirs to mitigate public health and conservation concerns. Multiple zoonotic coronaviruses, such as severe acute respiratory syndrome-associated coronavirus (SARS-CoV), SARS-CoV-2 and Middle Eastern respiratory syndrome-associated coronavirus (MERS-CoV), are hypothesized to have evolved in bats. We investigate angiotensin-converting enzyme 2 (ACE2), the host protein bound by SARS-CoV and SARS-CoV-2, and dipeptidyl-peptidase 4 (DPP4 or CD26), the host protein bound by MERS-CoV, in the largest bat datasets to date. Both the ACE2 and DPP4 genes are under strong selection pressure in bats, more so than in other mammals, and in residues that contact viruses. Additionally, mammalian groups vary in their similarity to humans in residues that contact SARS-CoV, SARS-CoV-2 and MERS-CoV, and increased similarity to humans in binding residues is broadly predictive of susceptibility to SARS-CoV-2. This work augments our understanding of the relationship between coronaviruses and mammals, particularly bats, provides taxonomically diverse data for studies of how host proteins are bound by coronaviruses and can inform surveillance, conservation and public health efforts.
Keywords: SARS-CoV-2, COVID-19, ACE2, DPP4, molecular evolution, Chiroptera
1. Introduction
The COVID-19 pandemic has highlighted the disastrous impacts of zoonotic spillovers and underscores the need to understand how pathogens and hosts evolve in response to one another. Evolutionary analyses of host proteins targeted by infections reveal the pressures that hosts have faced from pathogens and how they have evolved to resist disease, informing predictions about spread of infections and how to counter them. Bats have been suggested to be ‘special’ reservoirs of emerging infectious viruses [1], particularly coronaviruses [2]. Three significant zoonotic coronaviruses, severe acute respiratory syndrome-associated coronavirus (SARS-CoV), SARS-CoV-2 and Middle Eastern respiratory syndrome-associated coronavirus (MERS-CoV), likely have their origins in bats [3–5]. However, often this diverse clade is treated as a homogeneous group, represented by few species, particularly when considering the interaction of SARS-CoV, SARS-CoV-2 and MERS-CoV with host proteins, though some studies consider multiple species [6–9]. The varied ecologies and evolutionary histories of bat species have likely driven differences in their infections and immunity [2,10,11]; it is therefore important to examine many species to determine which species are at greatest risk of transmitting infections to humans or vice versa and not to treat bats as a monolith.
Examination of host proteins bound by potential zoonoses can be used not only to infer past and current evolutionary pressure but to inform the likelihood of cross-species transmission. One major barrier to cross-species transmission is the ability of the virus, adapted to one host protein, to bind another species' protein [12,13]. Accordingly, there have been many studies attempting to understand how different viral strains bind different species’ angiotensin-converting enzyme 2 (ACE2) and dipeptidyl-peptidase 4 (DPP4) and where zoonotic spillover may have originated (e.g. [6,10,12,14]). The prevailing hypothesis, and one that we proposed early in the pandemic [15], is that increased similarity in the residues that contact viruses between people and other species will be correlated with increased susceptibility to viral binding and/or infection (e.g. [16]). The proliferation of research and the devastating scale of the pandemic allows us the ability to test whether, at broad scales, similarity in host receptors is indeed predictive of infection.
Here, we investigate how ACE2, the host protein bound by SARS-CoV and SARS-CoV-2 [5,17,18], and DPP4, the host protein bound by MERS-CoV [19,20], have evolved, using the largest bat genetic datasets to date and a large suite of other mammal species. Specifically, we ask (i) are bats, or some bats, special, compared to other mammals, in their evolutionary pressure to adapt to coronaviruses? (ii) how can these evolutionary and genomic insights inform predictions about disease transmission between humans and other animals, and risk factors and projections of future pandemics?
2. Results and discussion
To better understand the impact of coronaviruses on mammalian, particularly bat, evolution, we generated new ACE2 and DPP4 sequences from 55 bat species representing five families and 37 genera, more than doubling the taxonomic diversity of described bat ACE2 and DPP4 sequences.
(a) . Mammals (particularly bats) are diverse at ACE2 contact residues for SARS-CoV and SARS-CoV-2 and DPP4 contact residues for MERS-CoV
To understand the similarity between the residues that contact viruses in humans and other species, we analysed a total of 270 ACE2 sequences from 206 species (98 bat; 108 non-bat) and 248 DPP4 sequences from 235 species (92 bat; 143 non-bat), representing 18 and 21 mammalian orders, respectively (electronic supplementary material, tables S1 and S2). Twenty-four ACE2 amino acid sites stabilize the binding of ACE2 with the receptor-binding domain of SARS-CoV (22 sites; figure 1c; electronic supplementary material, table S3) and/or SARS-CoV-2 (21 sites; figure 1c; electronic supplementary material, table S3) [6,12,14,17–19,21]. Across these 24 sites, which we refer to by their position in the human ACE2, we found a minimum of 137 unique amino acid combinations, 78 in bats (n = 159) and 68 in non-bat mammals (n = 111); when considering one individual per species, we found 67 combinations each in bats and other mammals (electronic supplementary material, table S1). Across a subset of seven amino acids identified as virus-contacting residues or important for the maintenance of salt bridges by most studies [6,12,18,21], we found at least 91 unique amino acid sequences, 64 in bats and 38 in non-bat mammals; when considering only one individual per species, there were 55 unique combinations in bats and 38 in non-bat mammals despite the larger number of non-bat mammal species (electronic supplementary material, table S1). The examination of ACE2 within species revealed little intraspecific diversity in ACE2 in most species, though most species were represented by only two sequences; 12 species (10 bat) were identical across individuals and sites, while Canis lupus showed one amino acid difference across two individuals (electronic supplementary material, table S1). By contrast, rhinolophid bats showed dramatic differences in diversity; Rhinolophus ferrumequinum (n = 4) was identical within species; Rhinolophus pearsonii (n = 2) showed two amino acid differences; Rhinolophus affinis (n = 23) had only three haplotypes; by contrast Rhinolophus sinicus (n = 26) had at least eight haplotypes, consistent with the observation that R. sinicus is a particularly diverse host species in its ACE2 [22] (electronic supplementary material, table S1). In DPP4, 15 residues are important for binding [7,19]; electronic supplementary material, table S2). Across these 15 sites, we found at least 108 unique amino acid sequences, 43 in bats (n = 104) and 69 in non-bat mammals (n = 144). Nine species (eight bats) had identical residues across individuals and sites. Rhinolophus ferrumequinum, and Saccopteryx bilineata, both bats, showed one amino acid difference across two individuals each (electronic supplementary material, table S2).
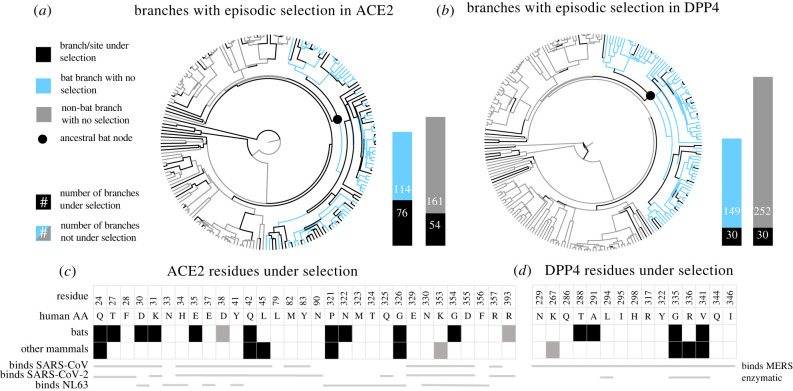
Figure 1.
ACE2 and DPP4 are under positive selection more frequently in bats compared to other mammals, and in residues that interact with viruses. (a,b) Phylogenetic hypotheses are coloured for (a) ACE2 and (b) DPP4 according to whether the branch was inferred to be under selection by aBSREL (p < 0.05 without correction). Black branches indicate detection of episodic positive selection; blue (lighter grey in print) and grey branches indicate bat and not-bat branches not under selection, respectively. Note that the basal branch of bats is under selection in both genes (black), denoted with a black circle. Bar charts next to the phylogenies show the number of branches in the bat clade (left) and other mammals (right) under selection (black) or not (blue or grey); length of bars is proportional to the number of branches in each category. (c,d) Charts depict the residues that viruses bind in (c) ACE2 and (d) DPP4. Numbers indicate the amino acid position in the human sequence and amino acids below depict the human sequence. Black (p < 0.05) and grey (p < 0.1) blocks indicate the residue was found to be under selection in bats (top row) or other mammals (bottom row). Below the ACE2 chart (c), residues that bind SARS-CoV (top), SARS-CoV-2 (middle) or NL63 (bottom) are indicated by the presence of a grey line. Below the DPP4 chart (d) residues that bind MERS (top) and/or are involved in the native enzymatic function of DPP4 (bottom) are noted by grey lines. Only the DPP4 residues that interact with MERS are depicted. (Online version in colour.)
Across all ACE2 sequences, the 24 amino acid sites varied from monomorphic across all examined sequences (e.g. Phe28 and Arg357) to having 10 or more possible amino acids. Of the 22 sites with more than one amino acid, bats were more diverse (Shannon's diversity index) than other mammals at 14 and were more even at 15 (electronic supplementary material, table S3). All residues in DPP4 were polymorphic across species, though two residues, 294 and 298, were monomorphic in bats, while 229 was monomorphic in non-bat mammals. In contrast with ACE2, bats were more diverse than other mammals at only one DPP4 site (229) and more even at only two (229 and 295).
Bats demonstrate a similar diversity in their ACE2 across the loci that contact SARS-CoV and SARS-CoV-2 and greater diversity in some sites than that observed across the rest of mammals suggesting they may be particularly diverse in their ACE2 [6], though some species, e.g. R. sinicus, are more diverse within species than others [22]. By contrast, bats do not seem to be particularly diverse in their DPP4 residues compared to other mammals—both bats and non-bat mammals have roughly half as many genotypes as sequences. Coronaviruses related to MERS-CoV have been found in multiple bat families across the globe, but not generally other mammals [2]. We might predict high diversity in the residues that contact MERS-CoV in bats but not other mammals; instead, we observe similar levels of diversity in both. Viral pressure on these residues may be balanced or eclipsed by other pressures on the protein (e.g. constraints on its function, see below). Alternatively, bats may resist merbecovirus infections through other mechanisms, such as modifications to host proteases. The host protease plays a major role in mediating MERS-CoV infection and the lack of an appropriate protease has been suggested as the explanation for the lack of MERS-CoV infection in sheep and cattle, even though the virus can bind their DPP4 [19].
(b) . Bats (but not other mammals) are under widespread selection due to SARS-CoV and SARS-CoV-2
We also conducted a series of selection analyses on a recent mammalian maximum clade credibility (MCC) tree [23]. In bats, a greater proportion of residues that contact SARS-CoV (p = 0.048; electronic supplementary material, table S4) and SARS-CoV-2 (p = 0.010; electronic supplementary material, table S4) were under selection than other residues in the gene, whereas residues that contact SARS-CoV (p = 0.48; electronic supplementary material, table S4) or SARS-CoV-2 (p = 0.46; electronic supplementary material, table S4) were not more likely to be under selection in non-bat mammals. Increased sampling can improve the ability of MEME to detect selection at individual sites [24]. Because our dataset of bat sequences is smaller than our mammalian dataset, it further strengthens our conclusion that bats are under positive selection in contact residues. Across all mammals, positions 24 and 42 were under selection (figure 1c; electronic supplementary material, table S3; MEME, p < 0.05), but in bats positions 27, 30, 31, 35 and 354 (figure 1c; electronic supplementary material, table S3, MEME, p < 0.05) and 38 and 393 (figure 1c; electronic supplementary material, table S3, MEME, p < 0.1) were additionally under positive selection, while positions 45 (figure 1c; electronic supplementary material, table S3, MEME, p < 0.05) and 353 (figure 1c; electronic supplementary material, table S3, MEME, p < 0.1) were under selection in non-bat mammals but not bats. These residues do not overlap with the active site residues of ACE2, Arg273, His345 and His505, suggesting selection is due to coronaviruses [25].
Using aBSREL, we tested two a priori hypotheses, (i) that bats are under positive selection in ACE2 and (ii) that the family Rhinolophidae, the bat family in which the progenitors of SARS-CoV and SARS-CoV-2 are hypothesized to have evolved [5,26], specifically, is under positive selection. Both bats (p = 0.0078) and Rhinolophidae (p = 0.0002) are under positive selection in ACE2 (electronic supplementary material, table S5). When we conducted an adaptive branch-site test of positive selection on all branches without specifying a foreground branch, branches in the bat clade were more likely to be selected than branches in other parts of the mammalian phylogeny (p = 0.0019; figure 1a; electronic supplementary material, table S5), and the branch at the base of Rhinolophidae was under positive selection (pHolm-Bonferroni correction = 0.03, electronic supplementary material, table S5). We found that bat branches are more likely to be under positive selection than other branches, even though these branches are a subset of the total phylogeny and branch-site tests of positive selection have reduced power to detect selection on shorter branches, making our test conservative [27,28].
Two bat families, Rhinolophidae and Hipposideridae, have been associated with SARS-related betacoronaviruses [2]. Interestingly, we found widespread selection across bats in ACE2 (figure 1a). Branches in the rhinolophid/ hipposiderid clade were not more likely to be under selection than other branches within bats (p = 0.81; electronic supplementary material, table S5) and bat lineages that live outside the predicted range of these viruses (e.g. in the Americas [2]) are also under positive selection. Therefore, there are still aspects of the bat-coronavirus relationship that we do not fully understand.
At least one other coronavirus uses ACE2 to gain entry into the host cell, HCoV-NL63, which may have its origin in bats [10]; we found evidence for increased selection in the residues that contact this virus in bats (p = 0.034; electronic supplementary material, table S4), but not in non-bat mammals (p = 0.66; electronic supplementary material, table S4). Many ACE2 residues that interact with HCoV-NL63 also interact with one or both of SARS-CoV and SARS-CoV-2 [29,30], which may be driving the evidence of selection in these residues. However, we did find evidence of selection in residues 321 and 326 in both bats and non-bat mammals (figure 1c; electronic supplementary material, table S3, MEME, p < 0.05), as well as selection in bats in residue 322 (figure 1c; electronic supplementary material, table S3, MEME, p < 0.05); these three residues contact HCoV-NL63 but not SARS-CoV or SARS-CoV-2. This finding contrasts with the findings of a smaller dataset of bats mostly from Europe, Asia and Africa which found no evidence of selection due to HCoV-NL63 [9] and may result from our greater power to detect signal or signal originating from bats in different regions than previously tested [31,32]. Recent evidence suggests that some MERS-CoV-related viruses use ACE2 as their host receptor [31], so this signal could be driven by coronaviruses from outside the SARS-related clade. Given that least three coronaviruses, distributed broadly across the viral family [2], bind ACE2 it is probable that there have been other CoVs driving positive selection across the evolutionary history of bats.
(c) . Bats are probably under selection due to MERS-CoV, but DPP4 is also under selection across mammals, probably for functional reasons
Conducting similar analyses to those conducted on ACE2 on DPP4 revealed comparable but less definite results. In bats, a greater proportion of residues that contact MERS-CoV (p = 0.012; figure 1d; electronic supplementary material, table S4) were under selection than other residues in the gene; non-bat mammals showed a similar but weaker trend (figure 1d; p = 0.096; electronic supplementary material, table S4). When running aBSREL with the basal bat branch as the foreground, we detected positive episodic selection (p = 0.0002), and a greater proportion of branches in the bat clade were under selection than in other mammals (p = 0.065; figure 1b; electronic supplementary material, table S5). As in ACE2, selection in DPP4 is widespread across the bat clade; widespread selection is consistent with evidence that MERS-related coronaviruses are found in multiple bat families found around the globe [2] and findings that MERS-CoV can infect many species [7].
Residues 335 and 341 are under selection across mammals, but bats were additionally under selection in 288 and 291 (MEME p < 0.05), while non-bat mammals were under selection at 336 (MEME p < 0.05) and 267 (MEME p < 0.1) (figure 1d). Residue 288 was also found to be under selection in a smaller study of bat DPP4 [32]. However, much of this signal may be due to selection on other functions of the protein. Residues 335–341 fall within the DPP4 adenosine deaminase-binding region [33], and residues that are important for peptidase function are more likely to be under selection than other residues in both bats (p = 0.0014; electronic supplementary material, table S4) and non-bat mammals (p = 0.016, electronic supplementary material, table S4). Selection on residues 288 and 291, which contact MERS but are not involved in the enzymatic function, suggests viruses may be driving selection in bats more than other mammals, but functional data on how exactly these residue changes affect binding would improve our understanding of the likely drivers of this selection.
Increased positive selection in bats in ACE2 and DPP4 compared with other mammals is consistent with their status as rich hosts of coronaviruses [2], and studies that have found selection in ACE2 and DPP4 in bats [9,16,32], and in aminopeptidase N in response to coronaviruses in mammals [27].
(d) . Similarity of host residues to humans is different from phylogenetic similarity between host orders and between genes
We quantified similarity between each species and humans in their ACE2 and DPP4 residues that bind CoVs (see Methods). All of the apes and most of the Old World monkeys we examined were identical to humans across all amino acid sites that contact coronavirus in ACE2; those that were not identical differed by only 1 or 2 amino acids (figure 2; electronic supplementary material, table S1). Similarly, all the primates had identical DPP4 sequences to humans except the gorilla, which differed by one amino acid. This consistent with predictions of significant capacity for viral exchange between humans and other primates (e.g. [32,34]).
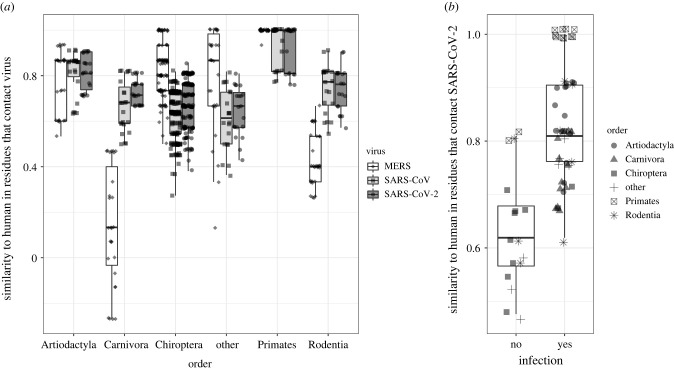
Figure 2.
Mammals vary in the similarity of their ACE2 or DPP4 residues contacting SARS-CoV, SARS-CoV-2 or MERS to humans, predicting SARS-CoV-2 infection. (a) Similarity of residues was calculated based on the number of residues that were identical or highly similar in binding properties to those found in human ACE2 or DPP4 with penalties for residues that would likely disrupt binding (see Methods). Scores of 1 indicate residues that contact the virus are identical (or highly similar) to humans. Boxes cover the interquartile range with a line indicating the median and whiskers extending to the largest value less than 1.5 times the interquartile range. Each point indicates a single sequence; only sequences with data for at least 22 of 24 ACE2 or 13 of 15 DPP4 residues are shown. Sequences are grouped by mammalian order; ‘other’ includes all orders with fewer than four sequences. Data points have been jittered for clarity. (b) Similarity of residues that bind SARS-CoV-2 to humans was compared between species that were infected with SARS-CoV-2 in vivo or in vitro or whose ACE2 proteins were shown to bind SARS-CoV-2 in vitro. On average susceptible species have more similar residues to humans. Symbols indicate the mammalian order of the species plotted; as in (a), orders with fewer than four representatives were grouped into ‘other’.
The similarity of ACE2 and DPP4 residues that contact coronaviruses to human residues varied both within and between the other mammalian orders for which we had many data points (Artiodactyla, Carnivora, Chiroptera and Rodentia). Artiodactyls were generally more similar to humans than other orders of mammals both in ACE2 and DPP4, though we observed greater spread within DPP4 than ACE2. Many ruminants (e.g. cows, deer, sheep and goats) and cetaceans were as or more similar to humans than New World monkeys in ACE2 residues that contact SARS-CoV and SARS-CoV-2. Camels, the intermediate hosts of MERS-CoV [19], were also highly similar to human DPP4 residues (electronic supplementary material, table S2). Rodents and carnivores were not particularly like humans in ACE2, with some exceptions (figure 2a). Two rodents (Mesocricetus auratus and Peromyscus leucopus) were like humans in all but two ACE2 sites for SARS-CoV and SARS-CoV-2. Domestic and big cats were among the species most like humans in ACE2 residues that contact SARS-CoV and SARS-CoV-2 and can shed both viruses [35,36]. However, carnivores and rodents are strikingly dissimilar to humans in DPP4 residues that contact MERS-CoV (figure 2a; electronic supplementary material, figure S1).
Bats were not very similar to humans in sites that bind SARS-CoV and SARS-CoV-2, some with as many as five changes that would likely reduce virus binding, the most observed across mammals. Additionally, most bat sequences (100 of 154) showed that at least one of the two salt bridges (Lys31-Glu35; Asp38-Lys353 in humans) within ACE2 would be disrupted (electronic supplementary material, table S1). In Rhinolophidae, 27 of the 58 sequences examined had a change in position 31 or 35 that would result in two clashing charged amino acids. This family was particularly diverse in its similarity to humans, especially R. sinicus (electronic supplementary material, figure S2), and numerous SARS-related CoVs that use ACE2 have been isolated from this group [22,37], suggesting further investigations into the implications within species ACE2 variation in this group may have important public health implications [22,38]. By contrast, bats were on average very similar to humans in their DPP4 residues that contact MERS-CoV (figure 2). No bat species demonstrated the dissimilar residues found in carnivores and rodents, and 23 bat species had identical DPP4-binding residues to humans (figure 2a; electronic supplementary material, table S2).
Because of the large overlap in residues that contact SARS-CoV and SARS-CoV-2 (19 residues), generally species were as similar to humans in residues that contact SARS-CoV as in residues that contact SARS-CoV-2 (figure 2a). However, bats (paired Wilcoxon signed-rank test, V = 8, p < 10−15) and carnivores (paired Wilcoxon signed-rank test, V = 21, p < 0.001), particularly mustelids including ferrets, were more similar to humans in residues that contact SARS-CoV-2 than residues that contact SARS-CoV (figure 2a). European mink, congenerics of ferrets, have been linked to significant zoonotic infections of SARS-CoV-2 [39]. Together these findings suggest susceptibility to one coronavirus infection does not necessarily inform predictions of susceptibility to another, and each pandemic threat must be treated individually.
(e) . Similarity of ACE2 yields predictions of susceptible hosts but cannot completely determine host range of SARS-CoV-2
Examination of ACE2 sequences across mammals and the similarity between distantly related groups at key residues for interaction with viruses may allow us to make predictions about potential spillover hosts or other susceptible species (e.g. [16,35]). We compared our human similarity scores in ACE2 residues that bind SARS-CoV-2 to experimental ACE2/ SARS-CoV-2 binding data and infection case reports. Overall, the species that are more like humans are more susceptible (Student's t-test, t = −6.24, d.f. = 26, p < 10−5; meanno infection = 0.64; meaninfection = 0.83; figure 2b), consistent with the predictions of a machine-learning based method of identifying spillover hosts [40]. Old World primates were identical to humans across all 24 residues and SARS-CoV replicates in multiple macaque species [41]. Pangolins were as similar in their ACE2 residues to humans as cats, lending support for the idea that a virus that can bind pangolin ACE2 might be able to transmit to humans. Accordingly, people should exercise precautions when interacting with species whose ACE2 is like to humans in the contact residues for CoVs, especially domestic animals such as cats. Care should also be taken with wild animals; for example, interactions between people with macaques or visitation of mountain gorillas by tourists could lead to cross-species transmission, endangering the health of humans and wildlife.
However, it can be hard to predict susceptibility to SARS-CoV or SARS-CoV-2 infection based on overall similarity of ACE2 residues, or even depending on the criteria considered. Pigs seem resistant to infection [36], but their ACE2 allows SARS-CoV-2 entry in vitro [42]. Squirrel ACE2 is bound by SARS-CoV-2 RBD at levels less than 40% of human ACE2 but the expression of squirrel ACE2 allows similar levels of SARS-CoV-2 pseudovirus transduction as human ACE2 expression [38]. Also, a single amino acid change can impact the binding of a virus to ACE2. In position 24, a diverse, even and selected site across all mammals that contacts both viruses, mutation from Gln (human) to Lys (18 bat species and Rattus norvegicus) reduced association between the SARS-CoV spike protein and ACE2 [34]. Position 27, a selected site in bats with many amino acid variants, is a Thr in humans; when mutated to a Lys (as in some bats), the interaction disfavoured SARS-CoV binding, while mutation to Ile, found in other bats, increased the ability of the virus to infect cells [6].
Some rodents, including Mesocricetus auratus and Peromyscus leucopus, which were among the most similar species to humans, have a glycosylated Asn in position 82 of ACE2 that disrupts the hydrophobic contact with Leu472 on SARS-CoV, reducing association and binding between the spike protein and ACE2 [18,19,34]. We predict this glycosylated Asn is in some rhinolophid bats (R. ferrumequinum and some R. sinicus), though not all (R. affinis, R. pearsoni, R. shameli, R. pusillus, R. macrotis and some R. sinicus) which may impact the potential of these species to act as reservoir or spillback hosts. Similarly, glycosylation of DPP4 on human residue 334 prevents MERS-CoV infection in mice [43]. We observed glycosylation in this position in many rodents and bats, and other glycosylated residues in a variety of species in residues 331 to 338 (positions 335, 336 and 341 contact MERS-CoV), making it probable that many species are less susceptible to infection than predicted from amino acid sequence level similarity alone (electronic supplementary material, table S2). Additionally, not all individuals in a species are equally susceptible to infection, complicating the identification of reservoirs, e.g. [22,38].
Spillover potential is not regulated solely by host receptor sequence. Compatibility of the host protease with the virus is important for determining host range for both SARS-CoV and MERS-CoV [13] and viral strains, including SARS-CoV-2 variants, vary in their binding properties to different species [18,44], and in their contacts with ACE2 residues [45]. MERS-CoV can rapidly evolve to exploit other DPP4 variants [7]. Viruses may also bind divergent receptors, e.g. a single SARS-like CoV that binds human, rhinolophid bat and civet ACE2 [26]. Both SARS-CoV and SARS-CoV-2 replicated well in ferrets, whose ACE2 ranked among the least similar to humans in their contact residues for SARS-CoV, though more similar for SARS-CoV-2 [35,36]. And species whose ACE2 sequence is not very similar to humans can be experimentally infected with SARS-CoV [19,46]. Natural variation in host receptors may create strong selective pressure on the viruses to bind a diverse array of host–receptor sequences that allow these viruses to infect spillover hosts such as humans. Indeed, Guo et al. [22] demonstrated variation in ACE2 within R. sinicus has probably driven the evolution of diverse SARS-related coronavirus strains, some of which infect cells displaying human ACE2 more effectively than those with R. sinicus ACE2.
(f) . What are the implications for this and future pandemics?
Approaches that incorporate the examination of evolutionary selection and similarity of host residues that contact viruses are useful for identifying broad groups of animals to target for viral surveillance, whether to identify probable sources of zoonotic infection (e.g. the hosts that viruses have evolved in or that infected humans), potential secondary reservoir species (e.g. domestic animals that can be infected by humans and reinfect humans) and/or species of conservation concern for which viral infection is a significant threat (e.g. [16,35]). The variation between species within an order, between orders and between residues interacting with different viruses highlights the lack of universal patterns in predicting the path of viral zoonoses, even within a single family of viruses coming from a single mammalian host order. For SARS-related coronaviruses, direct transmission from and to bats may be of less concern than for MERS-related coronaviruses, while carnivores may show the opposite patterns. Interestingly, we, like others (e.g. [35]), observe that many domestic animals have similar ACE2 to humans and are susceptible to SARS-CoV-2. Of 11 species of domestic hoof stock, carnivores and rabbits, plus Mus musculus and the Norway rat, 12 species are susceptible to infection. Domestic animals are one of the most important sources of zoonotic disease [47]. This reinforces the need to take precautions to not infect these animals for human and animal health.
While these analyses are useful on a broad scale, they have variable utility in predicting the reservoir potential of any individual host. The pangolin, an early suspect as the proximate spillover host of SARS-CoV-2 into humans [48], shows more similarity in its ACE2 residues to humans than most New World monkeys. Similarly, camels, the proximate host for MERS-CoV are very similar to humans, but the two bat species in which the closest related coronaviruses to MERS-CoV have been found, Tylonycteris pachypus and Pipistrellus abramus [49,50], are two of the bat species that are least like human in the DPP4 contact residues. And the civet, thought to be the intermediate host of SARS-CoV [51], fell in the middle of mammals in its similarity to humans in residues that contact either or both SARS-CoV and SARS-CoV-2.
3. Conclusion
Taken together, our data suggest that mammals, particularly bats, are evolving in response to coronaviruses with a diverse suite of ACE2, and to a lesser extent, DPP4, sequences that likely confer differing susceptibility to various coronavirus strains. This reinforces other findings of exceptional selection in bat lineages in ACE2 and DPP4 [9,16,32]. It seems that selection is widely distributed in the bat radiation and not restricted to a subset of ‘special’ species, suggesting that coronaviruses have been circulating in bats throughout much of their evolution.
Predicting which species will expose humans to infections, or suffer from infections transmitted by humans, is difficult. Genomic assessment of host receptors can make meaningful contributions to risk assessments, yielding deep evolutionary information about potential reservoir groups and complementing contemporary viral surveillance efforts. Such analyses also create predictions about likely susceptibility of different groups that can be further interrogated in functional [7,8] and in vivo [36,52] studies and used to train machine-learning analyses [40]. The idiosyncratic patterns in receptor similarity between humans and other mammals, and the differences in these patterns across host proteins, highlight the difficulty in applying data gleaned from one pandemic to another. However, with the increasing availability of genomic data and the tools to rapidly assess susceptibility and infection patterns, we are more empowered than ever before to protect human and animal health.
4. Methods
(a) . Alignment of mammalian ACE2 and DPP4 sequences
Sequences for ACE2 and DPP4 were obtained through GenBank or sequenced for this study. Mammalian ACE2 and DPP4 orthologues were downloaded from GenBank on 21 February 2020 and 17 December 2021, respectively [53]. We also sought out all bat sequences of ACE2 and DPP4, and the palm civet ACE2 sequence because of their putative role as SARS-CoV reservoirs. Because of the increased interest in ACE2 since 2020 and our focus on bats, we conducted an additional search on GenBank on 15 April 2022 for bat ACE2 sequences and included all new sequences in our analyses of ACE2. If the species for which we had data were not present in Upham et al. [23], we used the closest relative in the phylogeny (or a congeneric if no other species in the genus was in our dataset), retaining the same tree topology and distances. Artibeus [Dermanura] watsoni, Artibeus [Dermanura] phaeotis and Equus przewalskii were included in amino acid identity and diversity analyses but excluded from the molecular evolution analyses because they could not be placed accurately in the phylogeny (electronic supplementary material, tables S1 and S2). Only one sequence per species was used in molecular evolution analyses, noted in the electronic supplementary material, tables S1 and S2.
We augmented our data on the ACE2 and DPP4 diversity in bats using field-collected samples and tissues granted from museums (55 species; summarized in the electronic supplementary material, tables S1 and S2; see Ethics statement). Briefly, DNA was isolated from tissue using the Qiagen DNeasy Blood and Tissue kit (Valencia, CA, USA) and genomic libraries created with the NEBNext Ultra II kit (New England BioLabs, Ipswich, MA, USA), according to manufacturer's instructions. ACE2 and DPP4 were either captured as part of a targeted capture using genomic libraries and a custom target enrichment kit (Arbor Biosciences, Ann Arbor, MI, USA) according to the manufacturer's instructions with modifications [54], or isolated from total genomic sequence. Genomic reads were mapped to the nearest bat genome of Rousettus aegyptiacus, Pteropus alecto, Desmodus rotundus, Myotis lucifugus or Eptesicus fuscus using LAST [55] to generate a consensus sequence and the coding regions were extracted using a translated DNA search in BLAT [56] and the coding sequences from Myotis lucifugus [57]. Sequences are available from GenBank (MT333480–MT333534; ON714432–ON714486; electronic supplementary material, tables S1 and S2).
Sequences were aligned in Geneious [58] and corrected by hand to remove sequences outside the coding region and adjust gaps to be in frame using the human mRNA as a guide. Missing sequence, gaps and premature stop codons were converted to Ns for downstream analyses and comparison of residues with the human coding region.
(b) . Investigation of important residues for CoV binding
To understand how the residues important for coronavirus binding are conserved across mammals to determine the probable host range of MERS-CoV, SARS-CoV and SARS-CoV-2, we compared amino acid sequences across 24 ACE2 positions important for binding of SARS-CoV and/or SARS-CoV-2 [6,12,14,17–19] and 15 sites in DPP4 important for the binding of MERS-CoV [7,19]. To determine which amino acid positions were the most variable, we calculated Shannon's diversity index (which accounts for the number and evenness of amino acids), number of unique amino acids and evenness for each of the amino positions using the vegan [59] package (version 2.5-6) in R [60] (version 3.6.2). We also calculated how ‘human-like’ a species was in residues contacting each CoV by scoring each amino acid residue. Residues that were identical or equivalent to humans were given a score of 1; amino acids were deemed equivalent if they had similar properties and abilities to participate in hydrogen bonds, Van der Waals forces or salt bridges. Residues likely be worse at binding were given scores of −1; reduced binding was inferred when amino acid properties were dramatically altered from that of the human amino acid motif (e.g. replacement of a positively charged amino acid with a negatively charged amino acid in a salt bridge). In general, asparagine and glutamine were considered similar, as were amino acids with the same charge and amino acids with small hydrophobic side chains (valine, leucine, isoleucine and methionine). Other amino acids were given scores of zero; exact assignments are in the electronic supplementary material, table S3. The human-like score was calculated as a sum of each amino acid score divided by the total amino acids observed across all sites that contacted a given virus. When calculating differences between similarity in residues that bind SARS-CoV versus SARS-CoV-2 across mammalian orders, we only considered sequences for which we had data on at least 22 of the 24 ACE2 contact residues and for residues that bind MERS-CoV sequences for which we had data on at least 13 of 15 DPP4 contact residues. Wilcoxon signed-rank tests were performed on a single individual per species to avoid bias from well-sampled species. We predicted the N-linked glycosylation of Asn in the motif N-X-S/T where X is not a proline [61]. Glycosylation was not considered when calculating the human-like score.
(c) . Molecular evolution analyses
To determine whether coronaviruses are driving the evolution of ACE2 and DPP4, we used MEME [24] to infer the residues under selection across the mammal phylogeny, in bats and in non-bat mammals and used a Fisher's exact test to determine whether residues that interact with MERS, SARS-CoV, SARS-CoV-2 or HCoV-NL63 [29] were more likely to be under selection than other residues in DPP4 or ACE2. Only residues that showed variation (e.g. more than one amino acid across all species) and that were present in humans were considered in the Fisher's exact test. We report results using a p < 0.05 cutoff for inferring selection at each site via MEME but some results were sharper when using a p < 0.1 cutoff, probably due to the reduction in loss of statistical power (electronic supplementary material, table S4).
To determine whether bats, and specifically the family Rhinolophidae in the case of ACE2, are under strong selection to adapt to viruses, we used the adaptive branch-site random effects model test of positive selection, aBSREL [28], as implemented in HyPhy, version 2.5.8 [62], using the MCC phylogenetic hypothesis from a recent mammalian analysis [23]. A single sequence was used for each species (electronic supplementary material, tables S1 and S2). We tested three conditions: (i) in which the branch leading to Rhinolophidae was considered the foreground branch (ACE2 only); (ii) in which the branch leading to the common ancestor of all bats was considered the foreground branch; and (iii) in which all branches were tested without a priori specification of background and foreground branches. We used Fisher's exact tests to test whether an excess of branches in the bat lineage was under selection compared to the rest of the phylogeny. We used p < 0.05 as our cutoff for a branch being under selection without any HolmBonferroni correction because bat branches were unlikely to be more susceptible to false positives than any other branch, and all our comparisons were between branches within the same aBSREL analysis. When accepting significance at p < 0.05 with HolmBonferroni correction, a very stringent requirement, the general trends in ACE2 remain but some results lose statistical significance (electronic supplementary material, table S5).
It is possible that the sequences we generated through target capture and genomic sequence were less complete than the reference genomes. The number of ACE2 residues covered by our sequences and publicly available sequences was similar; mean ± s.e.this study = 770 ± 6.6 residues; mean ± s.e.publicly available = 784 ± 7.5 residues; DPP4 sequences were shorter; mean ± s.e.this study = 700 ± 5.5 residues; mean ± s.e.publicly available = 746 ± 4.1 residues. Although our sequences are distributed across the bat clade, to guard against bias, we removed the bat sequences we generated and examined the remaining terminal branches. A greater proportion of bat branches were under selection in ACE2 than non-bat branches (Fisher's exact test; p = 0.0075; electronic supplementary material, table S5). A similar but weaker pattern was observed for DPP4 (Fisher's exact test; p = 0.19; electronic supplementary material, table S5). Limiting our analyses to terminal branches and removing the sequences, we generated yields datasets less than 40% as large as the original datasets and therefore reduced power.
(d) . Determination of SARS-CoV-2 susceptibility
To test our predictions about susceptibility, we searched the literature for evidence of natural or in vitro infection of SARS-CoV-2 in the species in our study. We considered reports of natural or experimental in vivo infection or in vitro experiments on whether the SARS-CoV-2 bound host ACE2 to determine whether the host was susceptible to infection or not. In silico predictions were removed. In total, we were able to consider 65 species summarized in the electronic supplementary material, table S6. Correlation between how similar residues were to humans and infection susceptibility was tested with a Student's t-test.
Acknowledgements
We thank Julianna Gilson and Ellie Armstrong for research assistance and Sandra Nielsen and two anonymous reviewers for insightful comments. Thanks to CONAGEBIO and the Organization for Tropical Studies for assistance and access to Costa Rican genetic resources and the following museums for grants of tissue: Field Museum, Museum of Southwestern Biology, University of Alaska Museum, Museum of Vertebrate Zoology, University of Kansas Museum and Texas Tech Museum.
Ethics
For samples collected in the field, bats were captured in mist nets and a wing biopsy sample was collected. Bats were released immediately after sampling. Tissue samples were collected and analysed under the following Costa Rican permits: RT-019-2013-OT-CONAGEBIO, RT-044-2015-OTCONAGEBIO, RT-042-2015-OT-CONAGEBIO and R-054-2019-OT-CONAGEBIO. Research was approved by the Stanford Institutional Animal Care and Use Committee (protocols 26 920 and 29 978).
Data accessibility
All of the data (alignments, phylogenetic trees, metadata) and relevant R code required to reproduce the study are available as electronic supplementary material [63]. The DNA sequences generated in this study are available from GenBank with the primary accession codes MT333480-MT333534 (ACE2) and ON714432-ON714486 (DPP4).
Authors' contributions
H.K.F.: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, validation, visualization, writing—original draft and writing—review and editing; D.E.: conceptualization, formal analysis, methodology, resources and writing—review and editing; S.D.B.: conceptualization, funding acquisition, resources, supervision and writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
We declare we have no competing interests.
Funding
We are grateful to the following organizations for funding the work: National Science Foundation IOS (2032157; HKF), National Science Foundation Doctoral Dissertation Improvement Grant (1404521; HKF), Life Sciences Research Foundation Fellowship (HKF), Open Philanthropy Project, Stanford Woods Environmental Venture Program, Bing-Mooney Fellowship in Environmental Science, Stanford Center for Computational, Evolutionary and Human Genomics Postdoctoral Fellowship (HKF), National Institutes of Health grants 'Molecular and Cellular Immunobiology' (5 T32 AI07290), Stanford School of Medicine Dean’s Postdoctoral Fellowship (HKF), and endowment from the Crown Family foundation (SDB).
References
- 1.Brook CE, Dobson AP. 2015. Bats as ‘special’ reservoirs for emerging zoonotic pathogens. Trends Microbiol. 23, 172-180. ( 10.1016/j.tim.2014.12.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony SJ, et al. 2017. Global patterns in coronavirus diversity. Virus Evol. 3, vex012. ( 10.1093/ve/vex012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. 2020. The proximal origin of SARS-CoV-2. Nat. Med. 26, 450-452. ( 10.1038/s41591-020-0820-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu R, et al. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565-574. ( 10.1016/S0140-6736(20)30251-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P, et al. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270-273. ( 10.1038/s41586-020-2012-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou Y, Peng C, Yu M, Li Y, Han Z, Li F, Wang LF, Shi Z. 2010. Angiotensin-converting enzyme 2 (ACE2) proteins of different bat species confer variable susceptibility to SARS-CoV entry. Arch. Virol. 155, 1563-1569. ( 10.1007/s00705-010-0729-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Letko M, Miazgowicz K, McMinn R, Seifert SN, Sola I, Enjuanes L, Carmody A, Van Doremalen N, Munster V. 2018. Adaptive evolution of MERS-CoV to species variation in DPP4. Cell Rep. 24, 1730-1737. ( 10.1016/j.celrep.2018.07.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan H, et al. 2021. ACE2 receptor usage reveals variation in susceptibility to SARS-CoV and SARS-CoV-2 infection among bat species. Nat. Ecol. Evol. 5, 600-608. ( 10.1038/s41559-021-01407-1) [DOI] [PubMed] [Google Scholar]
- 9.Demogines A, Farzan M, Sawyer SL. 2012. Evidence for ACE2-utilizing coronaviruses (CoVs) related to severe acute respiratory syndrome CoV in bats. J. Virol. 86, 6350-6353. ( 10.1128/JVI.00311-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banerjee A, Kulcsar K, Misra V, Frieman M, Mossman K. 2019. Bats and coronaviruses. Viruses 11, 41. ( 10.3390/v11010041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang LF, Gamage AM, Chan WOY, Hiller M, Teeling EC. 2021. Decoding bat immunity: the need for a coordinated research approach. Nat. Rev. Immunol. 21, 269-271. ( 10.1038/s41577-021-00523-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan Y, Shang J, Graham R, Baric RS, Li F. 2020. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J. Virol. 94, e00127-20. ( 10.1128/JVI.00127-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Letko M, Marzi A, Munster V. 2020. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 5, 562-569. ( 10.1038/s41564-020-0688-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, et al. 2020. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 92, 595-601. ( 10.1002/jmv.25726) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frank HK, Enard D, Boyd SD. 2020. diversity and selection pressure on SARS-CoV and SARS-CoV-2 host receptor in bats compared to other mammals. bioRxiv. 2020.04.20.051656. ( 10.1101/2020.04.20.051656) [DOI]
- 16.Damas J, et al. 2020. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl Acad. Sci. USA 117, 22 311-22 322. ( 10.1073/pnas.2010146117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. 2020. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science 367, 1444-1448. ( 10.1126/science.abb2762) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li F, Li W, Farzan M, Harrison SC. 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309, 1864-1868. ( 10.1126/science.1116480) [DOI] [PubMed] [Google Scholar]
- 19.Lu G, Wang Q, Gao GF. 2015. Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 23, 468-478. ( 10.1016/j.tim.2015.06.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raj VS, et al. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495, 251-254. ( 10.1038/nature12005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lan J, et al. 2020. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, 215-220. ( 10.1038/s41586-020-2180-5) [DOI] [PubMed] [Google Scholar]
- 22.Guo H, Hu BJ, Yang XL, Zeng LP, Li B, Ouyang S, Shi ZL. 2020. Evolutionary arms race between virus and host drives genetic diversity in bat severe acute respiratory syndrome-related coronavirus spike genes. J. Virol. 94, e00902-20. ( 10.1128/jvi.00902-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Upham NS, Esselstyn JA, Jetz W. 2019. Inferring the mammal tree: species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biol. 17, e3000494. ( 10.1371/journal.pbio.3000494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8, e1002764. ( 10.1371/journal.pgen.1002764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guy JL, Jackson RM, Jensen HA, Hooper NM, Turner AJ. 2005. Identification of critical active-site residues in angiotensin-converting enzyme-2 (ACE2) by site-directed mutagenesis. FEBS J. 272, 3512-3520. ( 10.1111/j.1742-4658.2005.04756.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge XY, et al. 2013. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503, 535-538. ( 10.1038/nature12711) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enard D, Cai L, Gwennap C, Petrov DA. 2016. Viruses are a dominant driver of protein adaptation in mammals. Elife 5, e12469. ( 10.7554/eLife.12469) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Kosakovsky Pond SL. 2015. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol. Biol. Evol. 32, 1342-1353. ( 10.1093/molbev/msv022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu K, Li W, Peng G, Li F. 2009. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl Acad. Sci. USA 106, 19 970-19 974. ( 10.1073/pnas.0908837106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W, Sui J, Huang IC, Kuhn JH, Radoshitzky SR, Marasco WA, Choe H, Farzan M. 2007. The S proteins of human coronavirus NL63 and severe acute respiratory syndrome coronavirus bind overlapping regions of ACE2. Virology 367, 367-374. ( 10.1016/j.virol.2007.04.035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan H, et al. 2022. Close relatives of MERS-CoV in bats use ACE2 as their functional receptors. bioRxiv. See https://www.biorxiv.org/content/early/2022/01/25/2022.01.24.477490.
- 32.Cui J, Eden JS, Holmes EC, Wang LF. 2013. Adaptive evolution of bat dipeptidyl peptidase 4 (dpp4): implications for the origin and emergence of Middle East respiratory syndrome coronavirus. Virol. J. 10, 304. ( 10.1186/1743-422X-10-304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thoma R, Löffler B, Stihle M, Huber W, Ruf A, Hennig M. 2003. Structural basis of proline-specific exopeptidase activity as observed in human dipeptidyl peptidase-IV. Structure 11, 947-959. ( 10.1016/S0969-2126(03)00160-6) [DOI] [PubMed] [Google Scholar]
- 34.Li W, et al. 2005. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 24, 1634-1643. ( 10.1038/sj.emboj.7600640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martina BEE, Haagmans BL, Kuiken T, Fouchier RAM, Rimmelzwaan GF, Van Amerongen G, Peiris JSM, Lim W, Osterhaus ADME. 2003. SARS virus infection of cats and ferrets. Nature 425, 915. ( 10.1038/425915a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi J, et al. 2020. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science 368, 1016-1020. ( 10.1126/science.abb7015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo H, et al. 2021. Identification of a novel lineage bat SARS-related coronaviruses that use bat ACE2 receptor. Emerg. Microbes Infect. 10, 1507-1514. ( 10.1080/22221751.2021.1956373) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li P, et al. 2021. The Rhinolophus affinis bat ACE2 and multiple animal orthologs are functional receptors for bat coronavirus RaTG13 and SARS-CoV-2. Sci. Bull. (Beijing) 66, 1215-1227. ( 10.1016/j.scib.2021.01.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oreshkova N, et al. 2020. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance 25, 2001005. ( 10.2807/1560-7917.ES.2020.25.23.2001005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischhoff IR, Castellanos AA, Rodrigues JPGLM, Varsani A, Han BA. 2021. Predicting the zoonotic capacity of mammals to transmit SARS-CoV-2. Proc. R. Soc. B 288, 20211651. ( 10.1098/rspb.2021.1651) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McAuliffe J, et al. 2004. Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology 330, 8-15. ( 10.1016/j.virol.2004.09.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, et al. 2021. Functional and genetic analysis of viral receptor ACE2 orthologs reveals a broad potential host range of SARS-CoV-2. Proc. Natl Acad. Sci. USA 118, e2025373118. ( 10.1073/pnas.2025373118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peck KM, Cockrell AS, Yount BL, Scobey T, Baric RS, Heise MT. 2015. Glycosylation of mouse DPP4 Plays a role in inhibiting middle east respiratory syndrome coronavirus infection. J. Virol. 89, 4696-4699. ( 10.1128/JVI.03445-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peacock TP, et al. 2022. The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. bioRxiv. See https://www.biorxiv.org/content/early/2022/01/03/2021.12.31.474653.
- 45.Koehler M, Ray A, Moreira RA, Juniku B, Poma AB, Alsteens D. 2021. Molecular insights into receptor binding energetics and neutralization of SARS-CoV-2 variants. Nat. Commun. 12, 6977. ( 10.1038/s41467-021-27325-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi Z, Hu Z. 2008. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 133, 74-87. ( 10.1016/j.virusres.2007.03.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson CK, Hitchens PL, Pandit PS, Rushmore J, Evans TS, Young CCW, Doyle MM. 2020. Global shifts in mammalian population trends reveal key predictors of virus spillover risk. Proc. R. Soc. B 287, 20192736. ( 10.1098/rspb.2019.2736) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang T, Wu Q, Zhang Z. 2020. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 30, 1346-1351. ( 10.1016/j.cub.2020.03.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frutos R, Serra-Cobo J, Pinault L, Lopez Roig M, Devaux CA. 2021. Emergence of bat-related betacoronaviruses: hazard and risks. Front. Microbiol. 12, 591535. ( 10.3389/fmicb.2021.591535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woo PCY, et al. 2006. Molecular diversity of coronaviruses in bats. Virology 351, 180-187. ( 10.1016/j.virol.2006.02.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu D, et al. 2005. Civets are equally susceptible to experimental infection by two different severe acute respiratory syndrome coronavirus isolates. J. Virol. 79, 2620-2625. ( 10.1128/JVI.79.4.2620-2625.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlottau K, et al. 2020. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. Lancet Microbe 1, e218-25. ( 10.1016/S2666-5247(20)30089-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.National Library of Medicine (US), National Center for Biotechnology Information. 2004. ACE2 - angiotensin I converting enzyme 2. See https://www.ncbi.nlm.nih.gov/gene/59272/ortholog/?scope=7776.
- 54.Portik DM, Smith LL, Bi K. 2016. An evaluation of transcriptome-based exon capture for frog phylogenomics across multiple scales of divergence (Class: Amphibia, Order: Anura). Mol. Ecol. Res. 16, 1069-1083. ( 10.1111/1755-0998.12541) [DOI] [PubMed] [Google Scholar]
- 55.Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. 2011. Adaptive seeds tame genomic sequence comparison. Genome Res. 21, 487-493. ( 10.1101/gr.113985.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kent WJ. 2002. BLAT - The BLAST-like alignment tool. Genome Res. 12, 656-664. ( 10.1101/gr.229202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smedley D, et al. 2015. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 43(W1), W589-W598. ( 10.1093/nar/gkv350) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kearse M, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647-1649. ( 10.1093/bioinformatics/bts199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oksanen J, et al. 2019. vegan: commmunity ecology package. R package version 2.5-6. See https://CRAN.R-project.org/package=vegan.
- 60.R Core Team. 2019. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 61.UniProt Consortium. 2018. Glycosylation. See https://www.uniprot.org/help/carbohyd. [DOI] [PMC free article] [PubMed]
- 62.Kosakovsky Pond SL, et al. 2020. HyPhy 2.5 - A customizable platform for evolutionary hypothesis testing using phylogenies. Mol. Biol. Evol. 37, 295-299. ( 10.1093/molbev/msz197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frank HK, Enard D, Boyd SD. 2022. Exceptional diversity and selection pressure on coronavirus host receptors in bats compared to other mammals. FigShare. ( 10.6084/m9.figshare.c.6080908) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Frank HK, Enard D, Boyd SD. 2022. Exceptional diversity and selection pressure on coronavirus host receptors in bats compared to other mammals. FigShare. ( 10.6084/m9.figshare.c.6080908) [DOI] [PMC free article] [PubMed]
Data Availability Statement
All of the data (alignments, phylogenetic trees, metadata) and relevant R code required to reproduce the study are available as electronic supplementary material [63]. The DNA sequences generated in this study are available from GenBank with the primary accession codes MT333480-MT333534 (ACE2) and ON714432-ON714486 (DPP4).