Keywords: PARP inhibitors, cancer, synthetic lethality, biomarkers, drug resistance
Abstract
PARP inhibitors (PARPi) have been demonstrated to exhibit profound anti-tumour activity in individuals whose cancers have a defect in the homologous recombination DNA repair pathway. Here, we describe the current consensus as to how PARPi work and how drug resistance to these agents emerges. We discuss the need to refine the current repertoire of clinical-grade companion biomarkers to be used with PARPi, so that patient stratification can be improved, the early emergence of drug resistance can be detected and dose-limiting toxicity can be predicted. We also highlight current thoughts about how PARPi resistance might be treated.
1. What do we know?
1.1. PARP1 function
The target of clinical PARPi, PARP1 (poly[ADP-ribose] polymerase 1), is classically known for its role as a sensor of DNA damage and mediator of DNA repair. PARP1 mediates these effects via its ability to synthesise branched poly(ADP-ribose) (PAR) chains on substrate proteins (PARylation) and also itself (autoPARylation) [1,2]. For example, PARP1 plays a major role in promoting the repair of single-strand DNA breaks (SSB), including unligated Okazaki fragments that escape processing by the DNA repair enzymes FEN1 and LIG1 [3,4]. As part of its repair functions, PARP1 binds to damaged DNA, including SSBs, via N-terminal zinc-finger (ZnF) domains [5,6]. DNA binding invokes a conformational change in PARP1 that causes the release of an autoinhibitory interaction between the helical domain (HD) and the catalytic ADP-ribosyl transferase domain (ART); this, in turn, allows the PARP1 cofactor, NAD+ to access ART. The subsequent PARylation of substrate proteins involved in DNA repair enables their retention at the site of DNA damage, the relaxation of chromatin structure to increase access for DNA repair machinery and the repair of damaged DNA [7]. For example, PARylation leads to the recruitment of XRCC1, which in turn leads to the XRCC1-mediated recruitment of single-strand break repair (SSBR) proteins including DNA ligase 3 (LIG3) and DNA polymerase β (Polβ) [8,9]. PARP1 also autoPARylates, an event that drives its dissociation from DNA [1,2,10]. The PARylation status of PARP1 is also controlled by PAR glycohydrolase (PARG) and ARH3 whose activity enhances the retention of PARP1 on DNA [11,12].
PARP1 is also activated upon binding to double-strand DNA breaks (DSBs) and plays a role in the rapid recruitment and activation of the DNA-damage-sensing MRE11 and NBS1 components of the MRN (MRE11-RAD50-NBS1) complex to DSBs; the MRN complex generates 3′ single-stranded DNA (ssDNA) overhangs required for homologous recombination (HR) [13]. The activity of Ataxia telangiectasia mutated (ATM), a major activator of DSB repair pathways, is also controlled by an interaction with PAR chains [14,15]. In addition, the BRCT domain of BRCA1 recognises PAR chains and PARP1 activity plays an important role in the recruitment of BRCA1 to DSBs [16]. Finally, PARP1 has also been implicated in the suppression of DSB repair by non-homologous end joining (c-NHEJ); PARP1 PARylates the Ku70/80 NHEJ complex, decreasing its affinity for DNA [17]. Additionally, by competing with the Ku70/80 complex for access to the DNA ends, PARP1 may also act to suppress c-NHEJ and promote alternative NHEJ (alt-NHEJ, also known as microhomology end joining or theta-mediated end joining) [18].
1.2. PARP1 inhibitors and cancer synthetic lethality
Figure 1 illustrates the mechanisms of anti-tumour activity of PARPi. Existing clinical PARP1 inhibitors bind the catalytic domain of PARP1 and prevent PARylation by structurally mimicking nicotinamide, the by-product of the PARylation reaction [19–21]. In addition to preventing PARylation, clinically-approved PARPi (such as olaparib, niraparib, rucaparib and talazoparib) extend the retention of PARP1 at the site of DNA damage (PARP1 trapping), an effect probably caused by PARPi inducing a conformational change in the structure of PARP1 that increases the avidity of the ZnF for DNA [22–24]. Similar to other ‘trapped’ DNA-associated proteins, trapped PARP1, can be removed from DNA by the p97 segregase [25]. The importance of PARP1 trapping in PARPi-induced cytotoxicity is perhaps best illustrated by the observation that genetic deletion of PARP1 or mutation of PARP1 ZnF domains causes profound PARPi resistance [23,26,27].
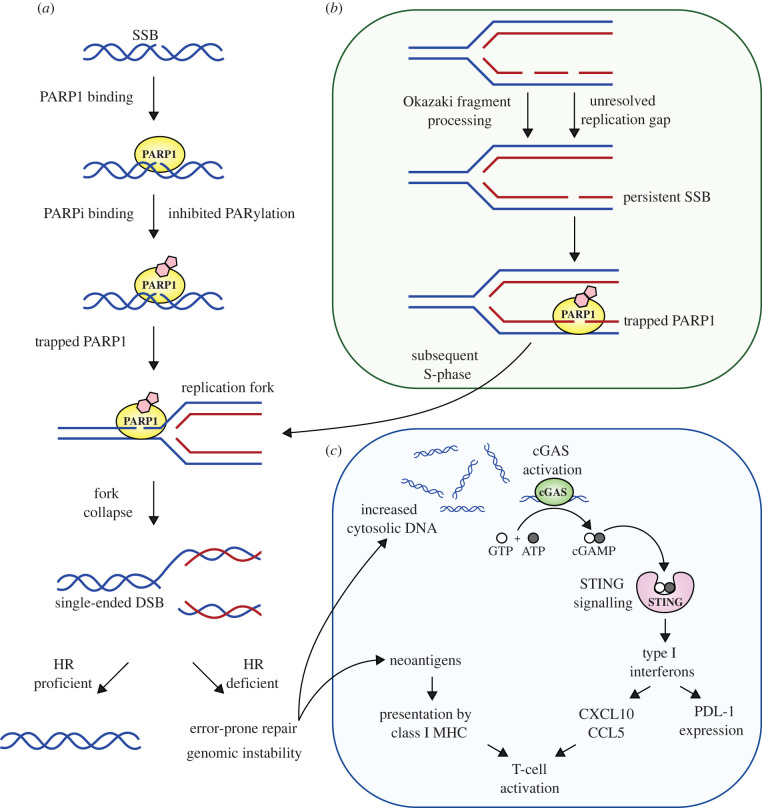
Figure 1.
Mechanisms of anti-tumour activity of PARPi. (a) PARP1 recognizes DNA lesions, such as single-stranded breaks (SSBs). DNA binding induces conformational changes in PARP1, including a change in an autoinhibitory interaction between the helical (HD) and catalytic (ART) domains; this in turn enables NAD+ to access the catalytic site where it initiates PARylation. PARPi bind the ART domain and inhibit catalytic activity but also alter the conformation of PARP1, trapping PARP1 at the site of DNA damage. Trapped PARP1 forms a replication barrier, leading to fork stalling and collapse. When there is a homologous recombination (HR) defect, error-prone DNA repair pathways are used to repair and restart the replication fork, events that can lead to increased genomic instability and loss of fitness. (b) Recent work suggests that unligated Okazaki fragments can form persistent SSBs that are bound by PARP1. In addition, defects in BRCA1/2 also lead to an increase in post-replicative SSBs. When PARP1 is trapped at post-replicative SSBs, this eventually poses an obstacle for the replication fork during the subsequent S phase. (c) The DNA damage induced by PARPi causes the generation of cytosolic DNA, which activates the cyclic GMP-AMP (cGAMP) synthetase (cGAS) DNA sensor. This in turn activates stimulator of interferon genes (STING) signalling and the production of type-I interferons and pro-inflammatory chemokines (e.g. CXCL10, CCL5) which, alongside the presentation of neoantigens created upon genomic instability, results in the activation of CD4+ and CD8+ T cells.
One model to explain the cytotoxicity caused by PARPi suggests that PARP1 becomes trapped by PARPi at unligated Okazaki fragments, with the result that cells undergo mitosis with persistent SSBs and trapped PARP1 [3]. In the subsequent S phase, trapped PARP1 forms a replication barrier, leading to fork stalling and collapse, an event which normally requires homologous recombination (HR) for successful repair [28]. HR is controlled by a series of tumour suppressor proteins including BRCA1, BRCA2, PALB2, RAD51C and RAD51D. A further iteration of this mechanistic model suggests that defects in BRCA1/2 themselves (and their associated DNA recombinase, RAD51) cause an accumulation of post-replicative ssDNA gaps; when combined with the increase in post-replicative ssDNA gaps caused by PARPi, the RPA exhaustion that ensues causes cell death [29,30]. In addition, other DNA lesions enhance PARPi sensitivity, such as those caused by the processing of genomic uracil [31] or DNA alkylation [32]. These DNA lesions are processed to form PARP1 binding sites, which in the presence of PARPi, cause increased PARP1 trapping and enhanced PARPi sensitivity [24].
Recent work has also highlighted a contribution of the immune system to the antitumour efficacy of PARPi. For example, the DNA damage that PARPi elicit has been shown to cause the generation of cytosolic DNA, which in turn is recognized by the cyclic GMP-AMP (cGAMP) synthetase (cGAS) DNA sensor; cGAS recognition of cytosolic DNA activates stimulator of interferon genes (STING) signalling, type-I interferon and pro-inflammatory chemokine production and CD4+ and CD8+ T cells [33–37]. Importantly, the anti-tumour activity of PARPi in tumour-bearing mice is impaired by CD8+ T-cell depletion, or by neutralization with an anti-CD8 antibody [34,35], suggesting that the adaptive immune system also plays a role in PARPi efficacy.
The synthetic lethality between BRCA1, BRCA2 and PARPi seen in pre-clinical models [38,39], also extends to clinical synthetic lethality [40], with PARPi now forming part of the standard-of-care approaches for the treatment of breast, ovarian, prostate or pancreatic cancers with defects in DNA repair by homologous recombination [41]. For example, in gynaecological cancers, the PARPi olaparib is approved for use according to four criteria: (i) as a maintenance treatment for patients with deleterious or suspected germline or somatic BRCA1/2-mutated advanced cancer patients who are in complete or partial response to first-line platinum-based chemotherapy (a clinical indication that HR is defective); (ii) as a combination maintenance treatment used in combination with the VEGF inhibitor bevacizumab, in those with HR defective cancer who are in complete or partial response to first-line platinum-based chemotherapy, where homologous recombination deficiency (HRD) is defined by either a deleterious or suspected deleterious BRCA1/2 mutation and/or an FDA-approved diagnostic that estimates the presence of cancer-associated genomic rearrangements normally associated with HRD; (iii) for the maintenance treatment of patients who are in complete or partial response to platinum-based chemotherapy and (iv) for the treatment of adult patients with deleterious or suspected deleterious germline BRCA1/2-mutated (gBRCAm) advanced ovarian cancer who have been treated with three or more prior lines of chemotherapy [42]. The use of olaparib is slightly distinct in breast, prostate and pancreatic cancers, but still focuses on patients who have HR defective cancers, defined either by the presence of deleterious BRCA1 or BRCA2 mutations or in the case of prostate cancers, by the presence of deleterious mutations in any one of a panel of genes that control HR [42]. For example, the recently reported OlympiA phase III trial demonstrated the utility of olaparib when used as an adjuvant treatment following standard-of-care chemotherapy in women with BRCA1 or BRCA2 mutant, HER2-negative, early breast cancer [43]. Additional PARPi (talazoparib (Pfizer), rucaparib (Clovis Oncology), niraparib (GlaxoSmithKline) and pamiparib (BeiGene) [44]) are also approved for the treatment of cancer, while others (e.g. AZD5305 (AstraZeneca) [45]) are still in clinical development. Future improvements to PARPi could include increased PARP1 specificity to circumvent off-target toxicity [45], decreased PgP efflux to decrease likelihood of this resistance mechanism [46] (see below) and modifications that increase tumour cell-selective PARP1 trapping [22].
1.3. PARP1 inhibitor resistance
Despite PARPi being able to elicit significant and sustained anti-tumour responses in some patients, PARPi resistance is a growing clinical problem, particularly in patients with advanced disease [41]. For example, in the Study 10 trial (NCT01482715 – Rucaparib in Patients With gBRCA Mutation Ovarian Cancer), 59.5% of germline BRCA1/2-mutant high grade ovarian cancer patients achieved an investigator-assessed confirmed RECIST response to rucaparib, while 40.5% of patients exhibited de novo resistance [47]. As well as de novo resistance (e.g. no detectable clinical response to PARPi in patients expected to respond due to the presence of a BRCA1/2 mutation), acquired PARPi resistance is also an issue. In some patients, the cause of PARPi resistance is reversion mutation in either BRCA1, BRCA2, RAD51C, RAD51D or PALB2 [48–54]. These reversion mutations, originally identified in patients with platinum-salt resistance [55,56], are secondary, additional, mutations (i.e. mutations other than the original pathogenic mutation in the gene) that restore the normal open reading frame of the tumour suppressor gene and encode somewhat functional proteins that are able to repair the DNA damage caused by PARPi [48]. Pre-clinical studies have also identified other candidate mechanisms of PARPi resistance. For example, in the absence of BRCA1, HR functionality can be restored by the further loss of DNA end resection inhibitors (e.g. 53BP1, REV7, Shieldin), enabling the resection of DNA ends necessary to initiate RAD51 recruitment [46,57,58]. BRCA1 and BRCA2 also play important roles in protecting stalled DNA replication forks and in the absence of their function, stalled forks are extensively degraded by nucleases such as MRE11 and MUS81, leading to fork collapse [59,60]. PARPi resistance can also be caused by inhibited recruitment of MRE11 and MUS81 [61,62], or increased fork stability via FANCD2 overexpression [63,64].
PARP inhibitor resistance can also occur via upregulation of ABC-family drug efflux pumps that reduce the cellular concentration of PARPi [65]. Although originally identified in a mouse model of BRCA1-mutant cancer [65], ABCB1 gene fusions that enhance activity have been observed in treatment-refractory breast and ovarian cancers, implying this could also be a cause of clinical PARPi resistance [66,67].
2. What don't we know?
Both pre-clinical and clinical investigation have taught us much about how PARPi work and where they might be best used. However, there are still a number of issues that if addressed, could further refine the clinical use of this class of drugs. Two of the key issues, which focus on biomarkers and how PARPi resistance might be targeted, are discussed below.
3. Which biomarkers are required to refine the best use of PARPi?
At present the biomarkers used to direct the use of PARPi focus on the presence of a deleterious BRCA1 or BRCA2 mutation, a deleterious mutation in other tumour suppressor genes that have been implicated in homologous recombination [68–70], prior platinum sensitivity or the presence in the tumour DNA of a genomic scar of HRD. While these biomarkers have utility, there is a clear need to refine the full package of biomarkers that are used to direct the use of this class of drugs (figure 2). We highlight five areas where additional biomarkers could be of use.
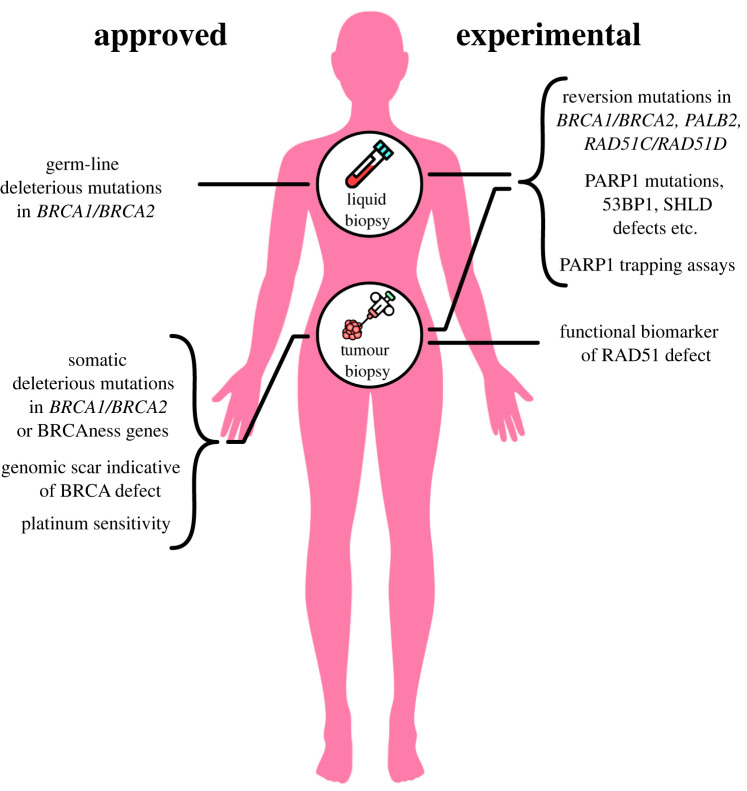
Figure 2.
Approved and experimental biomarkers for use with PARPi. Currently approved PARPi companion biomarkers primarily detect either (i) germ-line or somatic mutations in BRCA1/2 or other homologous recombination pathway ‘BRCAness’ genes, or (ii) the presence of HRD-associated genomic scars. Prior platinum salt sensitivity in gynaecological cancers is also used to select patients for subsequent PARPi treatment. Experimental biomarkers that could refine how PARPi are used clinically include the detection of PARPi resistance-causing reversion mutations in BRCA1, BRCA2, PALB2, RAD51C, RAD51D, mutations in PARP1 that impair PARP1 trapping and mutations or reduced expression in genes such as 53BP1, REV7, SHLD1/2/3 that restore HR in BRCA1 mutant tumour cells. These could be used alongside existing biomarkers and also a functional biomarker of the RAD51 defect that characterises a HR defect. There are currently no biomarkers used to predict dose-limiting toxicity and, given the association between PARP1 trapping and the haematological toxicities of PARPi, assays to measure the extent of PARP1 trapping may predict the magnitude of dose-limiting toxicity before it occurs. Figure modified from Lord & Ashworth [41].
3.1. Making the distinction between different BRCA1 and BRCA2 mutations and different HR-associated genes
At present, the presence of any deleterious mutation in either BRCA1 or BRCA2 is sufficient to select a patient for PARPi therapy. Already there is pre-clinical evidence to suggest some pathogenic BRCA1 mutations are hypomorphs and cause less cellular PARPi sensitivity than others [71–74], although it is not yet known whether the distinctive effect of different BRCA1 or BRCA2 mutations extends to distinct clinical responses. Furthermore, there is also the suggestion that some pathogenic BRCA1 or BRCA2 mutations are less likely to revert than others (particularly those in splice sites or those that are pathogenic missense mutations [26]). This implies that giving all deleterious BRCA1/2 mutations equal weight in terms of predicting response and resistance might be short-sighted. Further work (both clinical and pre-clinical) is clearly required to clarify the relative impact each has on PARPi sensitivity and the possibility of resistance. Likewise, although the concept of BRCAness (cancers that phenocopy cancers with BRCA1/2 mutations [68,75,76]) has proven useful in extending the use of PARPi beyond those individuals with BRCA1 or BRCA2 mutant cancers to those with a homologous recombination defect caused by some other means (e.g. mutation in PALB2, RAD51C, RAD51D), there is the implicit assumption that each of these other defects causes a similar extent of PARPi sensitivity as for a deleterious BRCA1 or BRCA2 mutation. Understanding whether this is the case or not might also allow a refinement of the effectiveness of biomarkers used to direct the use of PARPi.
3.2. Clinical-grade assays for identifying reversion mutations
There is already evidence from the study of gynaecological cancers that reversion mutations that are selected for by platinum treatment predict a poorer subsequent response to PARPi [50] and therefore understanding the presence of reversions prior to PARPi treatment is important. Reversions have been detected in circulating tumour DNA from individuals with clinical PARPi resistance [53,54,77,78]; if shown to be present prior to the emergence of clinical resistance (e.g. prior to the detection of a treatment-refractory lesion), detecting reversions in ctDNA could be used to adapt therapy so that resistant tumour cell clones could be targeted before they start to dominate the tumour cell population. We therefore see the development of clinical-grade tests that assay reversions as being critical. These could build on existing DNA sequencing-based assays already applied in the retrospective analysis of clinical trials involving platinum salts or PARPi [48–50,79].
3.3. Biomarkers that detect non-reversion mechanisms of PARPi resistance
Although tumour-associated reversions are associated with many cases of PARPi resistance, these are not detected in all cases. Other mechanisms of PARPi resistance have been identified from pre-clinical studies (as detailed above) but as yet, there is only anecdotal evidence for their existence in the clinical disease [26,80]. As such, robust, clinical grade biomarkers that allow these candidate resistance mechanisms to be detected are required. Ideally functional biomarkers (e.g. of 53BP1-Shieldin pathway function or PARP1 trapping) will also be available to interrogate non-mutational loss of these pathways.
3.4. Biomarkers of HR function
With the exception of prior platinum sensitivity, all of the existing clinically approved biomarkers used to stratify patients for PARPi treatment seek to identify that there has been an HR defect at some point in the history of cancer. What these biomarkers do not indicate is that the HR defect is present at the time of treatment. For example, HR defects arising via promoter methylation of HR genes (e.g. of BRCA1) may be reversed during the course of the disease. Such tumours retain the historical mutational signature of HRD, but have restored HR function and thus are not sensitive to PARP or platinum treatment [81–84]. Efforts are being made to convert a research-used assay of functional HRD, nuclear RAD51, into a clinical grade test which could estimate HR function [85,86]. It seems reasonable to think that such a biomarker might not be used in isolation but would be most powerful when used as part of an algorithm that also includes information from the other aforementioned biomarkers.
3.5. Biomarkers that predict dose-limiting toxicity
At present, biomarkers that predict dose-limiting toxicity do not exist. It seems possible that PARP1 trapping in myeloid cells explains (at least in part) the haematological toxicities seen with PARPi [45,87]. Given this, biomarkers that allow PARP1 trapping to be measured in patient samples might be of use and could predict those more likely to eventually show dose limiting toxicity before it occurs (e.g. those with elevated PARP1 trapping in lymphocytes).
4. How can we prevent or delay PARPi resistance?
At present, the treatment options for patients who develop PARP inhibitor resistance are limited and tend not to involve a targeted approach based upon the particular molecular make-up of resistant disease. Furthermore, approaches that are proven to delay the emergence of resistance do not exist. Here we highlight three areas of research that could inform how PARPi resistance is managed clinically.
4.1. Targeting PARPi resistance when caused by reversion
The study of reversions has suggested that some of the new DNA sequences formed by reversions could encode antigenic neopeptides [48]. This could suggest that therapeutic approaches that activate immune responses to these neopeptides could target reversion-mediated resistance. To this end, future work should establish whether neopeptide antigens are presented by class I MHC, and whether a robust and specific T-cell response can be induced against these neopeptides. If this proves to be the case, it may be possible to design vaccines based on candidate neopeptides predicted to occur upon reversion and to use these to prevent or delay BRCA1/2 revertant tumour outgrowth.
4.2. Targeting PARPi resistance when caused by non-reversion-based mechanisms
Pre-clinical studies have suggested that PARPi resistance in BRCA1 mutant cancers could emerge via loss of DNA end resection inhibitors (e.g. 53BP1, REV7, Shieldin) and the restoration of DNA resection (see earlier). Recent work suggests that while loss of these end resection inhibitors causes PARPi resistance, it also imparts upon tumour cells enhanced sensitivity to either ATR inhibitors [88], ionizing radiation [57], Polθ inhibitors [89,90] or LIG3 inhibition [30]. To fully realise the potential of using these approaches to either delay or target PARPi once it has occurred, biomarkers that identify these non-reversion-based mechanisms of resistance are required, so that patients can receive an appropriate treatment (see earlier).
4.3. Using drug combination approaches to target PARPi resistance
One approach to improving the overall efficacy of cancer treatments is to use these in combination with other treatments. Historically, PARPi have generally been combined with other drugs that target DDR defects, including DNA damaging chemotherapies [91–95]. However, the clinical experience with such combinations has not been wholly positive, with dose-limiting toxicity being an issue [96–100]. What has been more profitable has been to combine PARPi with drugs that have different mechanism of action. When the drugs involved in combinations have different mechanisms of action, the possibility of resistance-mechanisms emerging that cause resistance to both agents (cross resistance) is minimized and thus the overall efficacy of treatment could be improved [101]. It is possible that this is the case with the approved bevacizumab/olaparib combination used in ovarian cancers, although some have suggested that this combination works because VEGF inhibition causes a HR defect [102–104].
Whether the combination of PARPi with additional agents that target other, mechanistically independent, drivers in cancer turn out to be effective remains to be seen. To this end, a series of clinical trials are currently underway where PARPi are used in combination with agents that target PI3 K/AKT (NCT04729387, NCT02208375, NCT04586335, NCT03586661, NCT02338622, NCT01623349), MEK1/2 (NCT03162627) signalling, and other pathways. For example, in metastatic, castration-resistant, prostate cancer (mCRPC), where PARPi are already approved for use in those with tumoural mutations in a series of BRCAness genes [69,105], androgen signalling inhibitors such as abiraterone acetate (a CYP17 inhibitor) and enzalutamide (an androgen receptor antagonist) are also part of the standard-of-care [106]. A phase I trial has already established the safety and tolerable dose of the PARPi niraparib when used in combination with abiraterone acetate plus prednisone (AAP) [107]. This combination is now being investigated further in a larger randomized placebo-controlled, phase 3 study in patients with mCRPC (MAGNITUDE; NCT03748641). Similarly, recent data from the PROpel Phase III trial indicate that the combination of olaparib plus abiraterone acetate deliver an improvement in radiographic progression-free survival (rPFS) as a first line treatment for men mCRPC, when compared to standard-of-care abiraterone [108]. Whether the drugs in this combination act synergistically or independently on different vulnerabilities in the same cancer remains to be seen. HR defects are relatively common in mCRPC as is the addiction to androgen signalling, suggesting that if the two drug types act independently, clinical benefit could be achieved as tumour clones with resistance mechanisms to one agent (such as reversion mutation in a HR gene) might still exhibit sensitivity to the second agent (targeting the androgen signalling addiction). Alternatively, it is possible that some synergistic interaction between the two drug classes contributes to the therapeutic effect; some studies have shown that using AR signalling inhibitors cause reduced expression of HR-associated genes, including BRCA1, RAD54 L and RMI2 [109] and that loss of AR signalling reduces ATM signalling and MRE11 foci formation [110], effects that could be synthetic lethal with PARPi.
5. Concluding remarks
Over 50 years have passed since the first description of PARP1 function, 20 years since clinical trials using PARPi were initiated and close to eight years have passed since the first clinical approval of a PARPi [111–114]. Yet still, there is much to be discovered about PARPi that could refine how these drugs are used clinically. Some of these discoveries will no doubt come from pre-clinical work, but we also foresee a greater contribution to this field coming from ‘reverse translation’ where clinical observations made in people receiving PARPi as part of their cancer treatment tells us much about how these drugs work, and where we might better use these in the future. While we have summarized some of the key questions pertaining to refining the clinical use of PARPi, there are also other areas of research we have not covered. For example, how PARPi treatment influences the behaviour of patients' immune systems is very likely to have an impact on the overall clinical efficacy of these drugs [115,116] and no doubt further studies of those receiving PARPi will highlight how other bodily systems influence therapeutic responses.
Acknowledgements
The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care.
Contributor Information
Stephen J. Pettitt, Email: stephen.pettitt@icr.ac.uk.
Andrew N. J. Tutt, Email: andrew.tutt@icr.ac.uk.
Christopher J. Lord, Email: chris.lord@icr.ac.uk.
Data accessibility
This article has no additional data.
Authors' contributions
A.J.W.: conceptualization, data curation, visualization, writing—original draft; D.B.K.: conceptualization, supervision, writing—review and editing; S.J.P.: conceptualization, supervision, writing—review and editing; A.N.J.T.: conceptualization, funding acquisition, project administration, resources, supervision, writing—review and editing; C.J.L.: conceptualization, funding acquisition, project administration, resources, supervision, writing—review and editing.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Conflict of interest declaration
C.J.L. makes the following disclosures. Receives and/or has received research funding from: AstraZeneca, Merck KGaA, Artios. Received consultancy, SAB membership or honoraria payments from: Syncona, Sun Pharma, Gerson Lehrman Group, Merck KGaA, Vertex, AstraZeneca, Tango, 3rd Rock, Ono Pharma, Artios, Abingworth, Tesselate, Dark Blue Therapeutics. Has stock in: Tango, Ovibio, Enedra Tx., Hysplex, Tesselate. C.J.L. is also a named inventor on patents describing the use of DNA repair inhibitors and stands to gain from their development and use as part of the ICR ‘Rewards to Inventors' scheme. A.N.J.T. reports personal honoraria from Pfizer, Vertex, Prime Oncology, Artios, MD Anderson, Medscape Education, EM Partners, GBCC conference, Cancer Panel, Research to Practise, honoraria to either the Institute of Cancer Research or King's College research accounts from SABCS, VJ oncology, GE healthcare, Gilead, AZ ESMO symposium, IBCS conference, AstraZeneca Ad boards, honoraria and stock in InBioMotion, honoraria and financial support for research from AstraZeneca, Medivation, Myriad Genetics, Merck Serono. Travel expenses covered by AstraZeneca for any trial-related meetings or trial commitments abroad. A.N.J.T. reports benefits from ICR's Inventors Scheme associated with patents for PARP inhibitors in BRCA1/2 associated cancers, paid into research accounts at the Institute of Cancer Research and to A.N.J.T.'s personal account. The remaining authors declare no competing interests.
Funding
The work in our laboratories is funded by Programme Grants from Breast Cancer Now as part of Programme Funding to the Breast Cancer Now Toby Robins Research Centre, via Programme Grant funding from Cancer Research UK and via project grant funding from the Basser Foundation. This work represents independent research supported by the National Institute for Health Research (NIHR) Biomedical Research Centre at The Royal Marsden NHS Foundation Trust and the Institute of Cancer Research, London.
References
- 1.Hilz H, Stone P. 1976. Poly(ADP-ribose) and ADP-ribosylation of proteins. Rev. Physiol. Biochem. Pharmacol. 76, 1-58, 177. [DOI] [PubMed] [Google Scholar]
- 2.Purnell MR, Stone PR, Whish WJ. 1980. ADP-ribosylation of nuclear proteins. Biochem. Soc. Trans. 8, 215-227. ( 10.1042/bst0080215) [DOI] [PubMed] [Google Scholar]
- 3.Hanzlikova H, Kalasova I, Demin AA, Pennicott LE, Cihlarova Z, Caldecott KW. 2018. The importance of poly(ADP-ribose) polymerase as a sensor of unligated Okazaki fragments during DNA replication. Mol. Cell 71, 319-331. ( 10.1016/j.molcel.2018.06.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maya-Mendoza A, Moudry P, Merchut-Maya JM, Lee M, Strauss R, Bartek J. 2018. High speed of fork progression induces DNA replication stress and genomic instability. Nature 559, 279-284. ( 10.1038/s41586-018-0261-5) [DOI] [PubMed] [Google Scholar]
- 5.Langelier M-F, Planck JL, Roy S, Pascal JM. 2012. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science 336, 728-732. ( 10.1126/science.1216338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. 1999. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342, 249-268. ( 10.1042/bj3420249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ray Chaudhuri A, Nussenzweig A. 2017. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 18, 610-621. ( 10.1038/nrm.2017.53) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH. 1994. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell Biol. 14, 68-76. ( 10.1128/mcb.14.1.68-76.1994) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marintchev A, Robertson A, Dimitriadis EK, Prasad R, Wilson SH, Mullen GP. 2000. Domain specific interaction in the XRCC1-DNA polymerase beta complex. Nucleic Acids Res. 28, 2049-2059. ( 10.1093/nar/28.10.2049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zahradka P, Ebisuzaki K. 1982. A shuttle mechanism for DNA-protein interactions: the regulation of poly(ADP-ribose) polymerase. Eur. J. Biochem. 127, 579-585. ( 10.1111/j.1432-1033.1982.tb06912.x) [DOI] [PubMed] [Google Scholar]
- 11.Lautier D, Lagueux J, Thibodeau J, MéNard L, Poirier GG. 1993. Molecular and biochemical features of poly (ADP-ribose) metabolism. Mol. Cell. Biochem. 122, 171-193. ( 10.1007/BF01076101) [DOI] [PubMed] [Google Scholar]
- 12.Gogola E, et al. 2018. Selective loss of PARG restores PARylation and counteracts PARP inhibitor-mediated synthetic lethality. Cancer Cell 33, 1078-1093.e12. ( 10.1016/j.ccell.2018.05.008) [DOI] [PubMed] [Google Scholar]
- 13.Haince JF, McDonald D, Rodrigue A, Déry U, Masson JY, Hendzel MJ, Poirier GG. 2008. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 283, 1197-1208. ( 10.1074/jbc.M706734200) [DOI] [PubMed] [Google Scholar]
- 14.Haince JF, Kozlov S, Dawson VL, Dawson TM, Hendzel MJ, Lavin MF, Poirier GG. 2007. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J. Biol. Chem. 282, 16 441-16 453. ( 10.1074/jbc.M608406200) [DOI] [PubMed] [Google Scholar]
- 15.Aguilar-Quesada R, et al. 2007. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol. Biol. 8, 29. ( 10.1186/1471-2199-8-29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, Yu X. 2013. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell 23, 693-704. ( 10.1016/j.ccr.2013.03.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Navarro S, Kasahara N, Comai L. 2004. Identification and biochemical characterization of a Werner's syndrome protein complex with Ku70/80 and poly(ADP-ribose) polymerase-1. J. Biol. Chem. 279, 13 659-13 667. ( 10.1074/jbc.M311606200) [DOI] [PubMed] [Google Scholar]
- 18.Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. 2006. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 34, 6170-6182. ( 10.1093/nar/gkl840) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durkacz BW, Omidiji O, Gray DA, Shall S. 1980. (ADP-ribose)n participates in DNA excision repair. Nature 283, 593-596. ( 10.1038/283593a0) [DOI] [PubMed] [Google Scholar]
- 20.Bowman KJ, Newell DR, Calvert AH, Curtin NJ. 2001. Differential effects of the poly (ADP-ribose) polymerase (PARP) inhibitor NU1025 on topoisomerase I and II inhibitor cytotoxicity in L1210 cells in vitro. Br J Cancer 84, 106-112. ( 10.1054/bjoc.2000.1555) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curtin NJ, Szabo C. 2020. Poly(ADP-ribose) polymerase inhibition: past, present and future. Nat Rev Drug Discov 19, 711-736. ( 10.1038/s41573-020-0076-6) [DOI] [PubMed] [Google Scholar]
- 22.Zandarashvili L, et al. 2020. Structural basis for allosteric PARP-1 retention on DNA breaks. Science 368, eaax6367. ( 10.1126/science.aax6367) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murai J, Huang SYN, Das BB, Renaud A, Zhang YP, Doroshow JH, Ji JP, Takeda S, Pommier Y. 2012. Trapping of PARP1 and PARP2 by Clinical PARP inhibitors. Cancer Res. 72, 5588-5599. ( 10.1158/0008-5472.CAN-12-2753) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krastev DB, Wicks AJ, Lord CJ. 2021. PARP Inhibitors - Trapped in a Toxic Love Affair. Cancer Res. 81, 5605-5607. ( 10.1158/0008-5472.CAN-21-3201) [DOI] [PubMed] [Google Scholar]
- 25.Krastev DB, et al. 2022. The ubiquitin-dependent ATPase p97 removes cytotoxic trapped PARP1 from chromatin. Nat. Cell Biol. 24, 62-73. ( 10.1038/s41556-021-00807-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pettitt SJ, et al. 2018. Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat. Commun. 9, 1-4. ( 10.1038/s41467-018-03917-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettitt SJ, et al. 2013. A genetic screen using the PiggyBac transposon in haploid cells identifies Parp1 as a mediator of olaparib toxicity. PLoS ONE 8, e61520. ( 10.1371/journal.pone.0061520) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Wietmarschen N, Nussenzweig A. 2018. Mechanism for synthetic lethality in BRCA-deficient cancers: no longer lagging behind. Mol. Cell 71, 877-878. ( 10.1016/j.molcel.2018.08.045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cong K, et al. 2021. Replication gaps are a key determinant of PARP inhibitor synthetic lethality with BRCA deficiency. Mol. Cell 81, 3128-3144.e7. ( 10.1016/j.molcel.2021.06.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dias P, et al. 2021. Loss of nuclear DNA ligase III reverts PARP inhibitor resistance in BRCA1/53BP1 double-deficient cells by exposing ssDNA gaps. Mol. Cell 81, 4692-4708.e9. ( 10.1016/j.molcel.2021.09.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zimmermann M, et al. 2018. CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature 559, 285-289. ( 10.1038/s41586-018-0291-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boulton S, Pemberton L, Porteous J, Curtin N, Griffin R, Golding B, Durkacz B. 1995. Potentiation of temozolomide-induced cytotoxicity: a comparative study of the biological effects of poly(ADP-ribose) polymerase inhibitors. British J. Cancer 72, 849-856. ( 10.1038/bjc.1995.423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parkes EE, et al. 2017. Activation of STING-dependent innate immune signaling by S-phase-specific DNA damage in breast cancer. J. Natl Cancer Inst. 109, djw199. ( 10.1093/jnci/djw199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding L, et al. 2018. PARP Inhibition elicits STING-dependent antitumor immunity in Brca1-deficient ovarian cancer. Cell Rep. 25, 2972-2980.e5. ( 10.1016/j.celrep.2018.11.054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pantelidou C, et al. 2019. PARP inhibitor efficacy depends on CD8+ T-cell recruitment via intratumoral STING pathway activation in BRCA-deficient models of triple-negative breast cancer. Cancer Disc. 9, 722-737. ( 10.1158/2159-8290.CD-18-1218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chopra N, et al. 2020. Homologous recombination DNA repair deficiency and PARP inhibition activity in primary triple negative breast cancer. Nat. Commun. 11, 1-2. ( 10.1038/s41467-020-16142-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chabanon RM, et al. 2019. PARP inhibition enhances tumor cell-intrinsic immunity in ERCC1-deficient non-small cell lung cancer. J. Clin. Invest. 129, 1211-1228. ( 10.1172/JCI123319) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farmer H, et al. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917-921. ( 10.1038/nature03445) [DOI] [PubMed] [Google Scholar]
- 39.Bryant HE, et al. 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913-917. ( 10.1038/nature03443) [DOI] [PubMed] [Google Scholar]
- 40.Fong PC, et al. 2009. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl J. Med. 361, 123-134. ( 10.1056/NEJMoa0900212) [DOI] [PubMed] [Google Scholar]
- 41.Lord CJ, Ashworth A. 2017. PARP inhibitors: synthetic lethality in the clinic. Science 355, 1152-1158. ( 10.1126/science.aam7344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.AstraZeneca Pharmaceuticals LP 2021. AstraZeneca Pharmaceuticals LP. LYNPARZA (olaparib) [package insert]. See https://www.accessdata.fda.gov/drugsatfda_docs/label/2021/208558s019s020lbl.pdf. Revised March 2021 (accessed 11 February 2022).
- 43.Tutt ANJ, et al. 2021. Adjuvant Olaparib for patients with BRCA1- or BRCA2-mutated breast cancer. N. Engl. J. Med. 384, 2394-2405. ( 10.1056/NEJMoa2105215) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Markham A. 2021. Pamiparib: first approval. Drugs 81, 1343-1348. ( 10.1007/s40265-021-01552-8) [DOI] [PubMed] [Google Scholar]
- 45.Johannes JW, et al. 2021. Discovery of 5-{4-[(7-ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}-N-m ethylpyridine-2-carboxamide (AZD5305): a PARP1-DNA trapper with high selectivity for PARP1 over PARP2 and other PARPs. J. Med. Chem. 64, 14 498-14 512. ( 10.1021/acs.jmedchem.1c01012) [DOI] [PubMed] [Google Scholar]
- 46.Jaspers JE, et al. 2013. Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer Disc. 3, 68-81. ( 10.1158/2159-8290.CD-12-0049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kristeleit R, et al. 2017. A phase I–II study of the oral PARP inhibitor rucaparib in patients with germline BRCA1/2-mutated ovarian carcinoma or other solid tumors. Clin. Cancer Res. 23, 4095-4106. ( 10.1158/1078-0432.CCR-16-2796) [DOI] [PubMed] [Google Scholar]
- 48.Pettitt SJ, et al. 2020. Clinical BRCA1/2 reversion analysis identifies hotspot mutations and predicted neoantigens associated with therapy resistance. Cancer Disc. 10, 1475-1488. ( 10.1158/2159-8290.CD-19-1485) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swisher EM, et al. 2021. Molecular and clinical determinants of response and resistance to rucaparib for recurrent ovarian cancer treatment in ARIEL2 (Parts 1 and 2). Nat. Commun. 12, 1-3. ( 10.1038/s41467-021-22582-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin KK, et al. 2019. BRCA reversion mutations in circulating tumor DNA predict primary and acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Disc. 9, 210-219. ( 10.1158/2159-8290.CD-18-0715) [DOI] [PubMed] [Google Scholar]
- 51.Kondrashova O, et al. 2017. Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Disc. 7, 984-998. ( 10.1158/2159-8290.CD-17-0419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tobalina L, Armenia J, Irving E, O'Connor MJ, Forment JV. 2021. A meta-analysis of reversion mutations in BRCA genes identifies signatures of DNA end-joining repair mechanisms driving therapy resistance. Ann. Oncol. 32, 103-112. ( 10.1016/j.annonc.2020.10.470) [DOI] [PubMed] [Google Scholar]
- 53.Quigley D, et al. 2017. Analysis of circulating cell-free DNA identifies multiclonal heterogeneity of BRCA2 reversion mutations associated with resistance to PARP inhibitors. Cancer Disc. 7, 999-1005. ( 10.1158/2159-8290.CD-17-0146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goodall J, et al. investigators 2017. Circulating cell-free DNA to guide prostate cancer treatment with PARP inhibition. Cancer Discov. 7, 1006-1017. ( 10.1158/2159-8290.CD-17-0261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A. 2008. Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451, 1111-1115. ( 10.1038/nature06548) [DOI] [PubMed] [Google Scholar]
- 56.Sakai W, et al. 2008. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 451, 1116-1120. ( 10.1038/nature06633) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu G, et al. 2015. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 521, 541-544. ( 10.1038/nature14328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dev H, et al. 2018. Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nature Cell Biology 20, 954-965. ( 10.1038/s41556-018-0140-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529-542. ( 10.1016/j.cell.2011.03.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schlacher K, Wu H, Jasin M. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 22, 106-116. ( 10.1016/j.ccr.2012.05.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rondinelli B, et al. 2017. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nature Cell Biology 19, 1371-1378. ( 10.1038/ncb3626) [DOI] [PubMed] [Google Scholar]
- 62.Ray Chaudhuri R, et al. 2016. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535, 382-387. ( 10.1038/nature18325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kais Z, Rondinelli B, Holmes A, O'Leary C, Kozono D, D'Andrea AD, Ceccaldi R. 2016. FANCD2 maintains fork stability in BRCA1/2-deficient tumors and promotes alternative end-joining DNA repair. Cell Rep. 15, 2488-2499. ( 10.1016/j.celrep.2016.05.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Michl J, Zimmer J, Buffa FM, Mcdermott U, Tarsounas M. 2016. FANCD2 limits replication stress and genome instability in cells lacking BRCA2. Nat. Struct. Mol. Biol. 23, 755-757. ( 10.1038/nsmb.3252) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rottenberg S, et al. 2008. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc. Natl Acad. Sci. USA 105, 17 079-17 084. ( 10.1073/pnas.0806092105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patch A-M, et al. 2015. Whole–genome characterization of chemoresistant ovarian cancer. Nature 521, 489-494. ( 10.1038/nature14410) [DOI] [PubMed] [Google Scholar]
- 67.Christie EL, et al. 2019. Multiple ABCB1 transcriptional fusions in drug resistant high-grade serous ovarian and breast cancer. Nat. Commun. 10, 1-10. ( 10.1038/s41467-019-09312-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mccabe N, et al. 2006. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 66, 8109-8115. ( 10.1158/0008-5472.CAN-06-0140) [DOI] [PubMed] [Google Scholar]
- 69.De Bono J, et al. 2020. Olaparib for metastatic castration-resistant prostate cancer. N. Engl. J. Med. 382, 2091-2102. ( 10.1056/NEJMoa1911440) [DOI] [PubMed] [Google Scholar]
- 70.Bajrami I, et al. 2014. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res. 74, 287-297. ( 10.1158/0008-5472.CAN-13-2541) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y, et al. 2016. The BRCA1-Δ11q alternative splice isoform bypasses germline mutations and promotes therapeutic resistance to PARP inhibition and cisplatin. Cancer Res. 76, 2778-2790. ( 10.1158/0008-5472.CAN-16-0186) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y, et al. 2016. RING domain–deficient BRCA1 promotes PARP inhibitor and platinum resistance. J. Clin. Invest. 126, 3145-3157. ( 10.1172/JCI87033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Drost R, et al. 2016. BRCA1185delAG tumors may acquire therapy resistance through expression of RING-less BRCA1. J. Clin. Invest. 126, 2903-2918. ( 10.1172/JCI70196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Johnson N, et al. 2013. Stabilization of mutant BRCA1 protein confers PARP inhibitor and platinum resistance. Proc. Natl Acad. Sci. USA 110, 17 041-17 046. ( 10.1073/pnas.1305170110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turner N, Tutt A, Ashworth A. 2004. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat. Rev. Cancer 4, 814-819. ( 10.1038/nrc1457) [DOI] [PubMed] [Google Scholar]
- 76.Lord CJ, Ashworth A. 2016. BRCAness revisited. Nat. Rev. Cancer 16, 110-120. ( 10.1038/nrc.2015.21) [DOI] [PubMed] [Google Scholar]
- 77.Christie EL, Fereday S, Doig K, Pattnaik S, Dawson S-J, Bowtell DDL. 2017. Reversion of BRCA1/2 germline mutations detected in circulating tumor DNA from patients with high-grade serous ovarian cancer. J. Clin. Oncol. 35, 1274-1280. ( 10.1200/JCO.2016.70.4627) [DOI] [PubMed] [Google Scholar]
- 78.Weigelt B, et al. 2017. Diverse BRCA1 and BRCA2 reversion mutations in circulating cell-free DNA of therapy-resistant breast or ovarian cancer. Clin. Cancer Res. 23, 6708-6720. ( 10.1158/1078-0432.CCR-17-0544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lai Z, Brosnan M, Sokol ES, Xie M, Dry JR, Harrington EA, Carl Barrett J, Hodgson D. 2022. Landscape of homologous recombination deficiencies in solid tumours: analyses of two independent genomic datasets. BMC Cancer 22, 1-13. ( 10.1186/s12885-021-09033-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Waks AG, et al. 2020. Reversion and non-reversion mechanisms of resistance to PARP inhibitor or platinum chemotherapy in BRCA1/2-mutant metastatic breast cancer. Ann. Oncol. 31, 590-598. ( 10.1016/j.annonc.2020.02.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hurley RM, et al. 2021. Characterization of a RAD51C-silenced high-grade serous ovarian cancer model during development of PARP inhibitor resistance. NAR Cancer 3, zcab028. ( 10.1093/narcan/zcab028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nesic K, et al. 2021. Acquired RAD51C promoter methylation loss causes PARP inhibitor resistance in high-grade serous ovarian carcinoma. Cancer Res. 81, 4709-4722. ( 10.1158/0008-5472.CAN-21-0774) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kondrashova O, et al. 2018. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat. Commun. 9, 1-6. ( 10.1038/s41467-018-05564-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tutt A, et al. 2018. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat. Med. 24, 628-637. ( 10.1038/s41591-018-0009-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Castroviejo-Bermejo M, et al. 2018. A RAD51 assay feasible in routine tumor samples calls PARP inhibitor response beyond BRCA mutation. EMBO Mol. Med. 10, e9172. ( 10.15252/emmm.201809172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cruz C, et al. 2018. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann. Oncol. 29, 1203-1210. ( 10.1093/annonc/mdy099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hopkins TA, et al. 2019. PARP1 Trapping by PARP inhibitors drives cytotoxicity in both cancer cells and healthy bone marrow. Mol. Cancer Res. 17, 409-419. ( 10.1158/1541-7786.MCR-18-0138) [DOI] [PubMed] [Google Scholar]
- 88.Yazinski SA, et al. 2017. ATR inhibition disrupts rewired homologous recombination and fork protection pathways in PARP inhibitor-resistant BRCA-deficient cancer cells. Genes Dev. 31, 318-332. ( 10.1101/gad.290957.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zatreanu D, et al. 2021. Poltheta inhibitors elicit BRCA-gene synthetic lethality and target PARP inhibitor resistance. Nat. Commun. 12, 3636. ( 10.1038/s41467-021-23463-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou J, et al. 2021. A first-in-class polymerase theta inhibitor selectively targets homologous-recombination-deficient tumors. Nat. Cancer 2, 598-610. ( 10.1038/s43018-021-00203-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dent RA, et al. 2013. Phase I trial of the oral PARP inhibitor olaparib in combination with paclitaxel for first- or second-line treatment of patients with metastatic triple-negative breast cancer. Breast Cancer Res. 15, R88. ( 10.1186/bcr3484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Balmaña J, et al. 2014. Phase I trial of olaparib in combination with cisplatin for the treatment of patients with advanced breast, ovarian and other solid tumors. Ann. Oncol. 25, 1656-1663. ( 10.1093/annonc/mdu187) [DOI] [PubMed] [Google Scholar]
- 93.Oza AM, et al. 2015. Olaparib combined with chemotherapy for recurrent platinum-sensitive ovarian cancer: a randomised phase 2 trial. Lancet Oncol. 16, 87-97. ( 10.1016/S1470-2045(14)71135-0) [DOI] [PubMed] [Google Scholar]
- 94.Kunos C, et al. 2015. A phase I–II evaluation of Veliparib (NSC #737664), Topotecan, and Filgrastim or Pegfilgrastim in the treatment of persistent or recurrent carcinoma of the uterine cervix. Int. J. Gynecol. Cancer 25, 484-492. ( 10.1097/IGC.0000000000000380) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Han HS, et al. 2018. Veliparib with temozolomide or carboplatin/paclitaxel versus placebo with carboplatin/paclitaxel in patients with BRCA1/2 locally recurrent/metastatic breast cancer: randomized phase II study. Ann. Oncol. 29, 154-161. ( 10.1093/annonc/mdx505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bang Y-J, et al. 2017. Olaparib in combination with paclitaxel in patients with advanced gastric cancer who have progressed following first-line therapy (GOLD): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 18, 1637-1651. ( 10.1016/S1470-2045(17)30682-4) [DOI] [PubMed] [Google Scholar]
- 97.Somlo G, et al. 2017. Efficacy of the PARP inhibitor veliparib with carboplatin or as a single agent in patients with germline BRCA1- or BRCA2-associated metastatic breast cancer: California Cancer Consortium Trial NCT01149083. Clin. Cancer Res. 23, 4066-4076. ( 10.1158/1078-0432.CCR-16-2714) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wilson RH, et al. 2017. A phase I study of intravenous and oral rucaparib in combination with chemotherapy in patients with advanced solid tumours. British J. Cancer 116, 884-892. ( 10.1038/bjc.2017.36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pietanza MC, et al. 2018. Randomized, double-blind, phase II study of temozolomide in combination with either veliparib or placebo in patients with relapsed-sensitive or refractory small-cell lung cancer. J. Clin. Oncol. 36, 2386-2394. ( 10.1200/JCO.2018.77.7672) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moore KN, et al. 2020. A phase I study of intravenous or intraperitoneal platinum based chemotherapy in combination with veliparib and bevacizumab in newly diagnosed ovarian, primary peritoneal and fallopian tube cancer. Gynecol. Oncol. 156, 13-22. ( 10.1016/j.ygyno.2019.10.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dréan A, Lord CJ, Ashworth A. 2016. PARP inhibitor combination therapy. Critical Rev. Oncology/Hematol. 108, 73-85. ( 10.1016/j.critrevonc.2016.10.010) [DOI] [PubMed] [Google Scholar]
- 102.Ivy SP, Liu JF, Lee J-M, Matulonis UA, Kohn EC. 2016. Cediranib, a pan-VEGFR inhibitor, and olaparib, a PARP inhibitor, in combination therapy for high grade serous ovarian cancer. Expert Opin. Investig. Drugs 25, 597-611. ( 10.1517/13543784.2016.1156857) [DOI] [PubMed] [Google Scholar]
- 103.Bindra RS, Gibson SL, Meng A, Westermark U, Jasin M, Pierce AJ, Bristow RG, Classon MK, Glazer PM. 2005. Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res. 65, 11 597-11 604. ( 10.1158/0008-5472.CAN-05-2119) [DOI] [PubMed] [Google Scholar]
- 104.Bindra RS, et al. 2004. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol. Cell. Biol. 24, 8504-8518. ( 10.1128/MCB.24.19.8504-8518.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hussain M, et al. 2020. Survival with olaparib in metastatic castration-resistant prostate cancer. N. Engl. J. Med. 383, 2345-2357. ( 10.1056/NEJMoa2022485) [DOI] [PubMed] [Google Scholar]
- 106.Derleth CL, Yu EY. 2013. Targeted therapy in the treatment of castration-resistant prostate cancer. Oncology (Williston Park) 27, 620-628. [PubMed] [Google Scholar]
- 107.Saad F, et al. 2021. Niraparib with androgen receptor-axis-targeted therapy in patients with metastatic castration-resistant prostate cancer: safety and pharmacokinetic results from a phase 1b study (BEDIVERE). Cancer Chemother. Pharmacol. 88, 25-37. ( 10.1007/s00280-021-04249-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.AstraZeneca PLC 2021. Lynparza in combination with abiraterone significantly delayed disease progression in all-comers in PROpel Phase III trial in 1st-line metastatic castration-resistant prostate cancer [Press release]. See https://www.astrazeneca.com/media-centre/press-releases/2021/lynparza-propel-trial-meets-primary-endpoint.html. Published 24 September 2021 (accessed 24 February 2022).
- 109.Asim M, et al. 2017. Synthetic lethality between androgen receptor signalling and the PARP pathway in prostate cancer. Nat. Commun. 8, 374. ( 10.1038/s41467-017-00393-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li L, et al. 2017. Androgen receptor inhibitor-induced ‘BRCAness’ and PARP inhibition are synthetically lethal for castration-resistant prostate cancer. Sci. Signal. 10, eaam7479. ( 10.1126/scisignal.aam7479) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Virag L. 2013. 50Years of poly(ADP-ribosyl)ation. Mol. Aspects Med. 34, 1043-1045. ( 10.1016/j.mam.2013.05.002) [DOI] [PubMed] [Google Scholar]
- 112.Chambon P, Weill JD, Mandel P. 1963. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem. Biophys. Res. Commun. 11, 39-43. ( 10.1016/0006-291X(63)90024-X) [DOI] [PubMed] [Google Scholar]
- 113.Plummer ER, et al. 2005. Temozolomide pharmacodynamics in patients with metastatic melanoma: dna damage and activity of repair enzymes O6-alkylguanine alkyltransferase and poly(ADP-ribose) polymerase-1. Clin. Cancer Res. 11, 3402-3409. ( 10.1158/1078-0432.CCR-04-2353) [DOI] [PubMed] [Google Scholar]
- 114.Deeks ED. 2015. Olaparib: first global approval. Drugs 75, 231-240. ( 10.1007/s40265-015-0345-6) [DOI] [PubMed] [Google Scholar]
- 115.Pilger D, Seymour LW, Jackson SP. 2021. Interfaces between cellular responses to DNA damage and cancer immunotherapy. Genes Dev. 35, 602-618. ( 10.1101/gad.348314.121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chabanon RM, Rouanne M, Lord CJ, Soria JC, Pasero P, Postel-Vinay S. 2021. Targeting the DNA damage response in immuno-oncology: developments and opportunities. Nat. Rev. Cancer 21, 701-717. ( 10.1038/s41568-021-00386-6) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.