Abstract
We cloned and sequenced a cluster of genes involved in the biosynthesis of rhizobitoxine, a nodulation enhancer produced by Bradyrhizobium elkanii. The nucleotide sequence of the cloned 28.4-kb DNA region encompassing rtxA showed that several open reading frames (ORFs) were located downstream of rtxA. A large-deletion mutant of B. elkanii, USDA94Δrtx::Ω1, which lacks rtxA, ORF1 (rtxC), ORF2, and ORF3, did not produce rhizobitoxine, dihydrorhizobitoxine, or serinol. The broad-host-range cosmid pLAFR1, which contains rtxA and these ORFs, complemented rhizobitoxine production in USDA94Δrtx::Ω1. Further complementation experiments involving cosmid derivatives obtained by random mutagenesis with a kanamycin cassette revealed that at least rtxA and rtxC are necessary for rhizobitoxine production. Insertional mutagenesis of the N-terminal and C-terminal regions of rtxA indicated that rtxA is responsible for two crucial steps, serinol formation and dihydrorhizobitoxine biosynthesis. An insertional mutant of rtxC produced serinol and dihydrorhizobitoxine but no rhizobitoxine. Moreover, the rtxC product was highly homologous to the fatty acid desaturase of Pseudomonas syringae and included the copper-binding signature and eight histidine residues conserved in membrane-bound desaturase. This result suggested that rtxC encodes dihydrorhizobitoxine desaturase for the final step of rhizobitoxine production. In light of results from DNA sequence comparison, gene disruption experiments, and dihydrorhizobitoxine production from various substrates, we discuss the biosynthetic pathway of rhizobitoxine and its evolutionary significance in bradyrhizobia.
Rhizobitoxine [2-amino-4-(2-amino-3-hydropropoxy)-trans-but-3-enoic acid] is synthesized by the legume symbiont Bradyrhizobium elkanii (37) and the plant pathogen Burkholderia andropogonis (29). Because it induces foliar chlorosis of soybeans, rhizobitoxine has been regarded as a plant toxin (18, 36, 57). In terms of biochemical functions, rhizobitoxine inhibits β-cystathionase in the methionine biosynthesis pathway (39, 57) and 1-aminocyclopropane-1-carboxylate (ACC) synthase in the ethylene biosynthesis pathway (59).
Recently, a beneficial role for rhizobitoxine in Rhizobium-legume symbiosis has been revealed. Using a rhizobitoxine mutant, Yuhashi et al. (60) found that rhizobitoxine production by B. elkanii enhances nodulation and competitiveness in the legume Macroptilium atropurpureum (siratro), probably via the inhibition of endogenous ethylene production in the host plant. Duodu et al. (7) reported that rhizobitoxine mutants formed fewer mature nodules on Vigna radiata (mung bean) than the wild-type strain. In addition, application of ethylene inhibitors to the rhizobitoxine mutants partly restored the nodulation phenotype. Therefore, rhizobitoxine is a nodulation enhancer rather than a phytotoxin for siratro and mung bean, although it is unlikely that rhizobitoxine exerts this positive effect in nodulation of soybean cultivars (28, 43, 60).
The biosynthetic pathway for rhizobitoxine has not been elucidated fully. Ruan et al. (43–45) obtained two Tn5-induced rhizobitoxine null mutants of B. elkanii USDA61 and isolated the rtxA gene, which is responsible for rhizobitoxine biosynthesis in culture and in planta. The N-terminal region of the amino acid sequence of rtxA has a motif that is homologous to an aminotransferase, whereas one similar to O-acetylhomoserine sulfhydrolase is found in the C-terminal portion (43, 44); however, there is some confusion about frameshift in rtxA genes. Proposed as a precursor of rhizobitoxine, serinol is abundant in soybean nodules formed by B. elkanii (23, 26, 30). A mutant with a Tn5 insertion in the portion of the rtxA gene corresponding to the N-terminal part of the protein was defective in serinol accumulation in soybean nodules, suggesting that the N-terminal part functions as an aminotransferase in serinol production, but this function has not yet been verified in pure culture.
Dihydrorhizobitoxine [O-(2-amino-3-hydroxypropyl) homoserine] has been found in cultures and nodules of B. elkanii (38), and it is less potent than rhizobitoxine as an inhibitor of ACC synthase and β-cystathionase (59). Mitchell and Coddington (30) suggested that dihydrorhizobitoxine is an end product that lacks biological activity. However, in light of the homology with sulfhydrylase (44, 45), the C-terminal portion of the rtxA product may be involved in dihydrorhizobitoxine formation as an intermediate in rhizobitoxine biosynthesis. The absence of unequivocal, systematic determination of dihydrorhizobitoxine and serinol has complicated efforts to study the rhizobitoxine biosynthetic pathway, although two bioassay systems have been developed for rhizobitoxine (46, 59).
The aim of the present work was to investigate the rhizobitoxine biosynthetic pathway of B. elkanii in culture by an approach that combines mutagenesis of the rtxA gene and its flanking regions with unequivocal determination of the rhizobitoxine intermediates in culture by using liquid chromatography and mass spectroscopy. To this end, we chose the B. elkanii strain USDA94, which produces high concentrations of rhizobitoxine in culture; however, this strain is rather difficult to manipulate genetically because of increased resistance to antibiotics, particularly tetracycline.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. B. elkanii cultures were grown aerobically at 30°C in HM salt medium (5) supplemented with 0.1% arabinose and 0.025% yeast extract (Difco, Detroit, Mich.) or in Tris-YMRT medium (25). Escherichia coli cells were grown at 30°C in Luria-Bertani medium (47). Antibiotics were added to media at the following concentrations: spectinomycin and streptomycin at 250 μg/ml and kanamycin at 150 μg/ml for B. elkanii, and tetracycline at 12.5 μg/ml, ampicillin at 100 μg/ml, and kanamycin at 100 μg/ml for E. coli.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| B. elkanii | ||
| USDA94 | Wild-type strain producing high concn of rhizobitoxine in culture; Tcr Kms | H. H. Keyserb |
| USDA94Δrtx::Ω1 | USDA94, nodPQ rtxA rtxC (ORF1), ORF2, and ORF3::del/ins Ω cassette; Smr, Spr | This study |
| E. coli | ||
| JM109 | recA; cloning strain | Toyobo Inc. |
| DH5α | recA; cloning strain | Toyobo Inc. |
| HB101 | recA hsdR hsdM pro Smr | 3 |
| Plasmids | ||
| pRK2013 | ColE1 replicon carrying RK2 transfer genes; Kmr | 9 |
| pUC118 | Cloning vector for shotgun sequence; Apr | Takara Shuzo Co. |
| pBluescript II SK(+) | Cloning vector; Apr | Takara Shuzo Co. |
| pBS IIΔSacI | pBluescript II SK(+) lacking SacI site; Apr | This study |
| pHP45Ω | Plasmid carrying 2.1-kb Ω cassette; Smr Spr Apr | 8 |
| pSUP202 | pBR325 carrying oriT from RP4; Apr Cmr Tcr | 51 |
| pBS13.4 | pBSII ΔSacI carrying a 13.4-kb ApaI-NotI fragment from pRTF1 | This study |
| pBS3.6::Ω | pBS13.4 derivatives containing 9.8-kb SacI fragment deletion and Ω cassette insertion | This study |
| pSUP3.6::Ω | pSUP202 carrying a 5.7-kb ApaI-NotI fragment from pBS3.6::Ω | This study |
| pLAFR1 | Broad-host-range cosmid; IncP Tcr | 12 |
| pRTB7, pRTE1, pRTF1, pRTF3, pRTL1, pRTN2, pRTS1 | pLAFR1 containing in rtxA and flanking regions from B. elkanii USDA94 (Fig. 1) | This study |
| pGPS1.1 | Transprimer donor plasmid; Kmr Tcr | New England BioLabs |
| pRTF1-C1 | pRTF1 containing KM cassette insertion at 631-bp upstream region of rtxA | This study |
| pRTF1-C3 | pRTF1 containing KM cassette insertion at 1.3-kb upstream region of rtxA | This study |
| pRTF1-D2 | pRTF1 containing KM cassette insertion at N-terminal domain of rtxA | This study |
| pRTF1-D3 | pRTF1 containing KM cassette insertion at ORF1 (rtxC) | This study |
| pRTF1-D5 | pRTF1 containing KM cassette insertion at C-terminal domain of rtxA | This study |
| pRTF1-D8 | pRTF1 containing KM cassette insertion at 224-bp upstream region of rtxA | This study |
| pRTF1-E2 | pRTF1 containing KM cassette insertion at ORF3 | This study |
| pRTF1-E8 | pRTF1 containing KM cassette insertion at ORF3 | This study |
| pRTF1-E9 | pRTF1 containing KM cassette insertion at 183-bp downstream region of ORF2 | This study |
| pRTF1-E10 | pRTF1 containing KM cassette insertion at 337-bp downstream region of ORF2 | This study |
| pRTF1-E26 | pRTF1 containing KM cassette insertion at ORF2 | This study |
| pRTF1-F1 | pRTF1 containing KM cassette insertion at 5.7-kb downstream region of ORF4 | This study |
| pRTF1-F6 | pRTF1 containing KM cassette insertion at ORF4 | This study |
Apr, ampicillin resistant; Tcr, tetracycline resistant; Kmr, kanamycin resistant; Kms, kanamycin sensitive; Smr, streptomycin resistant; Spr, spectinomycin resistant; Cmr, chloramphenicol resistant; KM, kanamycin.
U.S. Department of Agriculture, Beltsville, Md.
DNA isolation and manipulations.
Isolation of plasmid DNA, restriction enzyme digestion, DNA ligation, bacterial transformation of E. coli, and Southern hybridization were performed as described by Sambrook et al. (47). Total DNA of B. elkanii was prepared as described previously (21). To isolate the DNA regions flanking rtxA, we used a cosmid library of B. elkanii USDA94 in the pLAFR1 vector (12) that had been constructed previously (61). E. coli HB101 cells containing the library were plated on Luria-Bertani agar (1.5%) with tetracycline. The B. elkanii USDA94 rtxA gene was PCR amplified as described previously (60) and was used as a colony hybridization probe. The probe was labeled by using digoxigenin-dUTP random priming (Boehringer Mannheim, Mannheim, Germany), and hybridization signals were detected by using the digoxigenin nucleic acid detection kit (Boehringer Mannheim). Cosmid clones from colonies showing a positive hybridization signal were isolated, digested with EcoRI, and subjected to electrophoresis in 0.8% agarose–TAE (47) to make a restriction map of the regions flanking rtxA of B. elkanii USDA94.
DNA sequencing.
The B. elkanii USDA94 genome cosmid clones pRTN2, pRTF1, and pRTS1, which cover a 40- to 45-kb DNA region containing rtxA, were digested into 4- to 8-kb fragments by using several restriction enzymes. These DNA fragments were ligated into pBluescript SK(+) (Takara Shuzo Co., Ltd., Kusatsu, Japan) and transformed into E. coli JM109. The cloned plasmid DNAs were isolated and digested into 0.5- to 4-kb fragments by sonication. The 1- to 2-kb DNA fragments were purified by 1% agarose gel electrophoresis, blunt ended by using KOD polymerase (Toyobo Inc., Tokyo, Japan), ligated into HincII-digested pUC118 (Takara Shuzo Co.), and transformed into E. coli DH5α. Consequently, more than 300 plasmids containing portions of rtxA and its flanking regions were isolated.
Sequencing was performed by using dye primer technology and a model 373A sequencer (Perkin-Elmer Applied Biosystems, Warrington, United Kingdom). Gaps in the DNA sequence were closed by sequencing PCR-generated fragments with primers based on the sequence flanking the gaps. Sequence data were assembled and analyzed with Malign and Mac Vector software (Oxford Molecular Ltd., Oxford, United Kingdom). BLAST similarity searches were done by using the National Center for Biotechnology Information database. The secondary structure analysis for hydropathy index (56) was carried out by Genetyx-Mac software (Software Development Co. Ltd., Tokyo, Japan).
Construction of a B. elkanii mutant with a large deletion of rtxA and its flanking region.
For the construction of a large deletion mutant for the putative rhizobitoxine gene, the 13.4-kb ApaI-NotI fragment from pRTF1 was cloned into pBSIIΔSacI. The resulting plasmid (pBS13.4) was digested by SacI, and a 6.4-kb fragment was isolated. This 6.4-kb fragment was blunt ended and then ligated with the 2.1-kb Ω cassette from pHP45Ω (42). The resulting plasmid (pBS3.6::Ω) was double digested with ApaI and NotI, and a 5.7-kb ApaI-NotI fragment containing noeE, the Ω cassette, a partial open reading frame (ORF3), and ORF4 was cloned into pSUP202 (51), yielding pSUP3.6::Ω. pSUP3.6::Ω was introduced into B. elkanii USDA94 by triparental mating using pRK2013 as a helper plasmid (9). Crossover mutants were selected by screening for resistance to streptomycin and spectinomycin, and a double-crossover mutant was identified by Southern hybridization with the 5.7-kb ApaI-NotI fragment from pSUP3.6::Ω as a probe. The relevant characteristics of the double-crossover mutant USDA94Δrtx::Ω 1 and plasmids used for the construction are listed in Table 1.
Construction and complementation of pRTF1 derivatives with kanamycin cassette insertion.
To examine the function of rtxA and its flanking region in rhizobitoxine biosynthesis, we used the genome priming system kit (GPS-1; New England BioLabs, Inc., Beverly, Mass.) according to the manufacturer's instructions to randomly insert a kanamycin cassette into these sequences. After mutation of 0.08 μg of pRTF1 and 0.02 μg of pGPS1.1 and transformation, kanamycin-resistant colonies were isolated, and plasmids were purified, digested with EcoRI, and electrophoresed to broadly specify the sites of insertion and eliminate mutants with multiple cassettes. The exact insertion points were determined by DNA sequencing using outward primers of the kanamycin cassette, as described in the manufacturer's instructions. These pRTF1 insertion mutant derivatives were introduced into B. elkanii USDA94Δrtx::Ω1 by triparental mating with pRK2013 (9). Transconjugants were selected by screening for kanamycin resistance and were assayed for serinol, dihydrorhizobitoxine, and rhizobitoxine production.
LC/MS analysis of serinol, dihydrorhizobitoxine, and rhizobitoxine.
We simultaneously determined the serinol, dihydrorhizobitoxine, and rhizobitoxine concentrations in cultures of B. elkanii by using liquid chromatography and mass spectrometry (LC/MS) to quantitate their phenylthiocarbamyl derivatives. A 15-ml aliquot of a stationary-phase culture of B. elkanii grown in Tris-YMRT medium was centrifuged at 10,000 × g for 10 min. The resulting supernatant was loaded on a Dowex 50 column (H+ type; resin size, 50 to 100 mesh; column volume, 5 ml; Muromachi Chemicals, Tokyo, Japan). The column was washed with 10 column volumes of deionized water. Serinol, dihydrorhizobitoxine, and rhizobitoxine were eluted with 3 column volumes of 2 M NH4OH, and evaporated in vacuo. Pellets were dissolved in 500 μl of deionized water, and 10 nmol of aminoethoxyvinylglycine (a structural analogue of rhizobitoxine) was added as an internal standard before phenylthiocarbamyl derivatization.
Phenylthiocarbamyl derivatization was carried out according to the method of Yamaya and Matsumoto (58). A 50-μl aliquot of the sample solution was evaporated in vacuo in a 1.5-ml tube, and the pellet was dissolved in 20 μl of ethanol-triethylamine-water (2:1:2). After evaporation, the pellet was dissolved in 10 μl of ethanol-triethylamine-water-phenylisothiocyanate (PITC) (7:1:1:1), incubated for 20 min at room temperature, and then evaporated to dryness. Each pellet of PITC derivative was dissolved in 100 μl of deionized water and passed through a 0.2-μm cellulose nitrate filter prior to LC/MS analysis.
A JMS-LCmate (JEOL, Tokyo, Japan) equipped with an electrospray ionization system and high-performance liquid chromatograph (HP-1100, Hewlett Packard, Waldbronn, Germany) was used for analysis of PITC-serinol, -dihydrorhizobitoxine, and -rhizobitoxine under the following conditions: column, Inertsil ODS-2 (1.5 by 150 mm; GL Sciences Inc., Tokyo, Japan); column temperature, 40°C; flow rate, 0.1 ml/min; mobile phase, a linear gradient from 30% solvent B (100% MeCN) in solvent A (0.1% HCOOH) to 100% solvent B for 15 min. Under these conditions the retention times of PITC-serinol, -dihydrorhizobitoxine, -rhizobitoxine, and -aminoethoxyvinylglycine (internal standard) were 3.8, 10.4, 10.4, and 12.4 min, respectively. The concentrations of serinol, dihydrorhizobitoxine, and rhizobitoxine in the cultures and buffers were calculated according to the ratio between the peak area of the PITC derivative of each compound (m/z = 227, 463, and 461, respectively) and the peak area of PITC-aminoethoxyvinylglycine (m/z = 431). Authentic dihydrorhizobitoxine and rhizobitoxine were isolated and purified from cultures of B. elkanii USDA94 as described previously (25); serinol and aminoethoxyvinylglycine were purchased from Sigma Chemical Co. (St. Louis, Mo.).
Conversion of serinol and various compounds to dihydrorhizobitoxine.
A 500-ml aliquot of a stationary-phase culture of B. elkanii USDA94 in HM medium was centrifuged at 10,000 × g for 10 min at 20°C. The cells were washed twice with 300 ml of 50 mM potassium phosphate buffer (KP; pH 6.8), resuspended in 10 ml of 50 mM KP (pH 6.8), and aliquoted into Eppendorf tubes (1.5 ml). Then the amount of cells was adjusted to 60 mg (wet weight)/tube. After centrifugation, the cells were resuspended in 1 ml of 20 mM KP (pH 6.8) containing 1 mM homoserine, O-acetylhomoserine, cysteine, cystathionine, homocysteine, or methionine. O-Acetylhomoserine was synthesized from l-homoserine and acetic anhydride according to the method of Nagai and Flavin (33); the other compounds were purchased from Wako Pure Chemical Industries Ltd. (Osaka, Japan), Nacalai Tesque Inc. (Kyoto, Japan), and Sigma Chemical Co. The cell suspensions were incubated with shaking at 30°C for 1 h in the dark. After centrifugation, the supernatants were analyzed by LC/MS as described above.
Nucleotide sequence accession number. The nucleotide sequence determined in this study appears in the DDBJ/GenBank/EMBL databases under accession number AB062279.
RESULTS
Selection of pLAFR1 cosmids containing rtxA from B. elkanii USDA94 library.
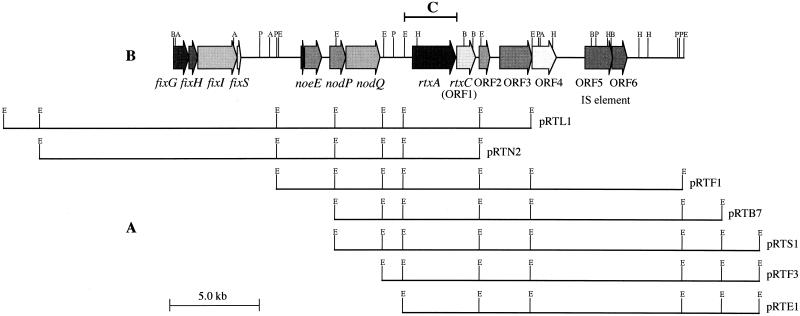
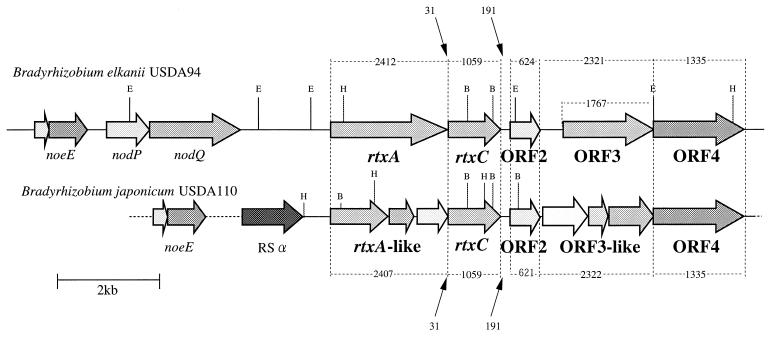
In previous work, the rtxA gene of B. elkanii USDA94, a high rhizobitoxine producer, was PCR amplified by using two primers designed from published rtxA sequence of B. elkanii USDA61, a low rhizobitoxine producer in culture (60). Using a PCR-derived fragment of rtxA in B. elkanii USDA94 as a probe (Fig. 1), we carried out colony hybridization against the pLAFR1 cosmid library of B. elkanii USDA94 (61). Consequently, we were able to align seven independent cosmids containing rtxA in light of their EcoRI restriction sites (Fig. 1). The identity of the 4.3-kb EcoRI fragment containing rtxA in the seven cosmids was verified by Southern hybridization with the PCR-derived fragment of rtxA from B. elkanii USDA94 as a probe (Fig. 1).
FIG. 1.
Physical map of the flanking regions of rtxA. (A) Seven independent overlapping pLAFR1 cosmids containing rtxA from a genomic library of B. elkanii USDA94. (B) In the 28,401 bp of sequence obtained, an fixGHIS cluster, noeE, nodPQ, rtxA, and six ORFs were found. Restriction sites: A, ApaI; B, BamHI; E, EcoRI; H, HindIII; P, PstI. C, probe used for colony and Southern hybridization.
Nucleotide sequence around the rtxA gene.
The DNA encompassing the rtxA gene of B. elkanii USDA94 was sequenced, and we identified 14 ORFs, all of which were in the same orientation. Of these, seven ORFs upstream of rtxA are homologous to fixGHIS, noeE, and nodPQ and appear to be involved in symbiotic functions. Another four ORFs are downstream of rtxA, suggesting that they are involved in rhizobitoxine biosynthesis. A potential promoter for sigma 70 was found 0.5 kb upstream of rtxA, although the other potential promoters recognized by NodD, FixK, and sigma 54 were not found in the DNA regions by their consensus sequences (15). Detailed descriptions of each gene and ORF follow.
The deduced amino acid sequence of rtxA in B. elkanii USDA94.
The deduced amino acid sequence of rtxA (803 amino acid residues) in B. elkanii USDA94 was 95% similar to that of B. elkanii USDA61, a low rhizobitoxine producer (44, 45). The 346 N-terminal residues had 24% identity and 40% similarity to the aminotransferase of Methanobacterium thermoautotrophicum (52). The 443 C-terminal residues had 41% identity and 56% similarity to the O-acetylhomoserine sulfhydrylase of Leptospira meyer (2). O-Acetylhomoserine sulfhydrylase synthesizes sulfur-containing amino acids from O-acetylhomoserine and sulfide. Generally, the enzyme shows O-alkylhomoserine synthase activity from O-acetylhomoserine and alcohol as well (31, 32), whose reaction mode resembles that of dihydrorhizobitoxine synthesis from O-acetylhomoserine and serinol. These amino acid homologies of rtxA product in B. elkanii USDA94 are similar to those of B. elkanii USDA61, although the rtxA gene was formally separated into rtxA and rtxB genes in B. elkanii USDA61 because of a sequencing error (44, 45). The predicted amino acid sequences for rtxA suggested that their possible enzymatic functions might be involved in serinol formation (1) and dihydrorhizobitoxine synthesis.
Deduced amino acid sequence of ORF1 (rtxC).
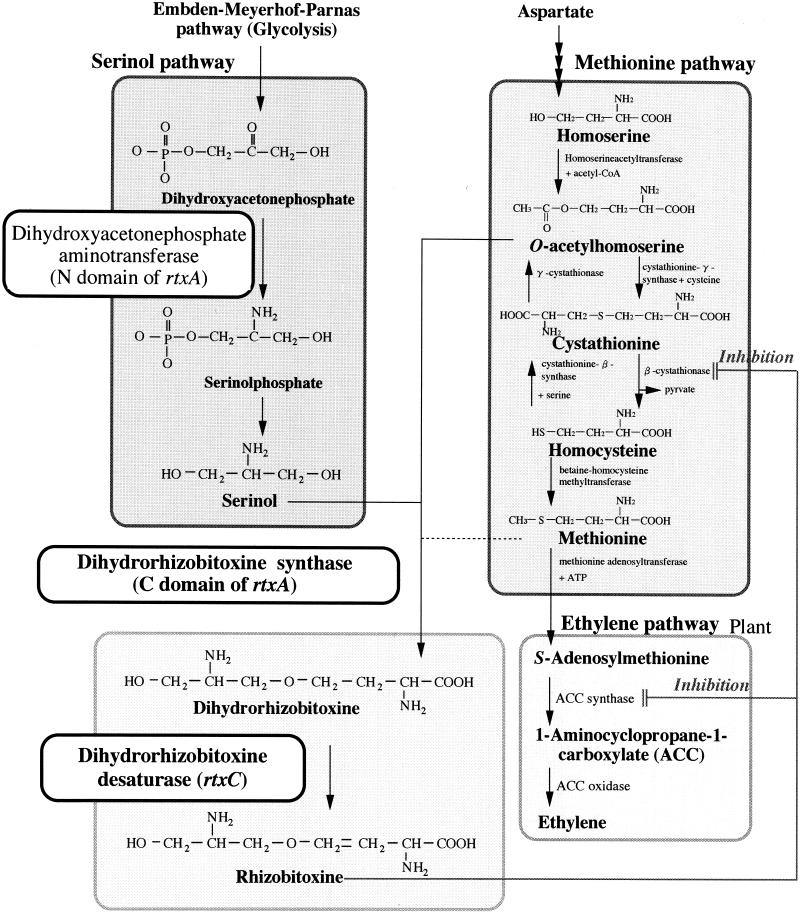
The deduced amino acid sequence of ORF1 (352 amino acid residues) had 19% identity and 31% similarity to the fatty acid desaturase of Pseudomonas syringae (62) (Fig. 2). Although the predicted amino acid sequence of ORF1 indicated low similarity to other desaturases, alignment of these sequences revealed the presence of two regions conserved among membrane-bound desaturases (49, 62): a copper-binding signature and eight histidine residues. Analysis of the deduced secondary structure showed two potential transmembrane regions in the ORF1 product (56) (Fig. 2). Rhizobitoxine possesses a double bond between C-3 and C-4 (37). Therefore, we hypothesized that the ORF1 product catalyzes the introduction of the carbon double bond into dihydrorhizobitoxine. We consequently designated ORF1 as rtxC, which begins 32 bp downstream of rtxA and lacks an upstream promoter-like sequence. Therefore, rtxA and rtxC (at least) probably form an operon.
FIG. 2.
Alignment of the amino acid sequence of rtxC product with those of various desaturases. PsDES, P. syringae desaturase (U27310); AtDES, Arabidopsis thaliana desaturase (Al022198); EgDES, Euglena gracilis Δ-8 fatty acid desaturase (AF139720); CrDES, Chlamydomonas reinhardtii desaturase (AB007640). Conserved among membrane-bound desaturases, the copper-binding signature (●) and eight histidine residues (★) of rtxC product were highly homologous to those of fatty acid desaturases from several sources. Putative membrane-spanning regions were deduced by hydropathy analysis and are indicated by asterisks.
Deduced amino acid sequences of ORF2, ORF3, and ORF4.
ORF2 began 192 bp downstream of rtxC, and its deduced amino acid sequence (207 amino acid residues) had 45% identity and 61% similarity to the amidotransferase subunit of Pseudomonas aeruginosa PAO1 (53). ORF3 began 555 bp downstream of ORF2, and the deduced amino acid sequence of ORF3 (588 amino acid residues) lacked homology to any known protein. ORF4 began in the termination codon of ORF3 (TGA-TG). The deduced amino acid sequence of ORF4 (444 amino acid residues) had 34% identity and 53% similarity to the glutamine synthetase of Mycobacterium tuberculosis (6).
Deduced amino acid sequences of the remaining ORFs.
The deduced amino acid sequence of ORF5 had 84% similarity to the transposase of Rhizobium sp. strain NGR234 (11), and that of ORF6 had 79% similarity to the ATP-binding helper protein of Rhizobium sp. strain NGR234. The insertion sequence (IS) element NGR IS5 belongs to the IS21/IS1162 family. ORF5 and ORF6 probably form an IS element belonging to the IS21/IS1162 family, which has been identified as IS1631 in Bradyrhizobium japonicum (17).
The deduced amino acid sequences of four ORFs located 14 kb upstream of rtxA showed 81, 70, 82, and 62% similarity to the sequences of the 3′ end of fixG and to the complete sequences of fixH, fixI, and fixS of B. japonicum USDA110, respectively (34). In B. japonicum USDA110, the FixGHIS complex is necessary for symbiotic nitrogen fixation and might play a role in the uptake and metabolism of copper required for cbb3-type heme-copper oxidase (41).
The deduced amino acid sequences of the remaining three ORFs upstream of rtxA showed 83, 90, and 82% similarity to the sequences of the interrupted noeE of Rhizobium sp. strain NGR234, the complete nodP of Azospirillum brasilense, and nodQ of Rhizobium sp. strain N33 (11, 55). Numerous reports have shown that the nod, nol, and noe gene products are required for the synthesis of variant Nod factors. In Rhizobium sp. strain NGR234, NoeE transferred sulfate from 3′-phosphoadenosine 5′-phosphosulfate to fucosylated lipochitin oligosaccharides (16). In Sinorhizobium meliloti and Rhizobium tropici, the nodPQ genes were required for sulfation of Nod factor (10, 50). However, the Nod factor of B. elkanii was not modified by those sulfational adjunctions (4, 48). A consensus nod box sequence (15) was not found in this region as well. These genes probably do not function in Nod factor synthesis.
Establishment of a complementation system for rhizobitoxine biosynthesis.
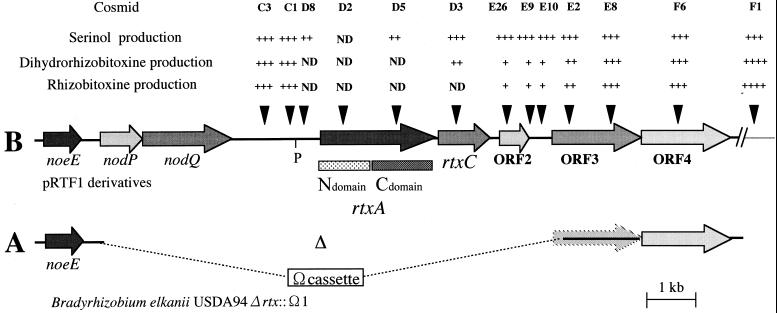
The efficiency of homologous recombination in B. elkanii is lower than that in B. japonicum (22). Therefore, to evaluate their functions in rhizobitoxine biosynthesis, we adopted a shortcut strategy in which cosmids mutagenized by insertion of a kanamycin cassette complement a B. elkanii USDA94 mutant lacking a putative DNA region for rhizobitoxine biosynthesis. We first constructed the large-deletion mutant USDA94Δrtx::Ω1, which lacks a 9.8-kb region (nodQP, rtxA, rtxC, ORF2, and truncated ORF3) of the B. elkanii USDA94 chromosome (Fig. 3A). We could not delete the entire ORF3 sequence or ORF4 because of the absence of appropriate restriction sites.
FIG. 3.
Serinol, dihydrorhizobitoxine, and rhizobitoxine production by B. elkanii USDA94Δrtx::Ω1 complemented with pRTF1 cosmid derivatives created by insertion of a kanamycin cassette. (A) Large-deletion mutant USDA94Δrtx::Ω1, which lacks a 9.8-kb region (nodQP, rtxA, rtxC, ORF2, and truncated ORF3) of B. elkanii USDA94. Δ, 9.8-kb deleted SacII fragment. (B) The insertion point of each kanamycin cassette is indicated by the arrowheads. The N and C domains of rtxA show the regions where their deduced amino acid sequences are homologous to aminotransferase and O-acetylhomoserine sulfhydrylase, respectively (see the text). P, putative promoter (according to the sequence). For serinol production, +, ++, and +++ indicate 0 to 50, 50 to 100, and >100 μM, respectively; for dihydrorhizobitoxine production, +, ++, +++, and ++++ indicate 0 to 2, 2 to 5, 5 to 10, and >10 μM, respectively; for rhizobitoxine production, +, ++, +++, and ++++ indicate 0 to 0.5, 0.5 to 1.0, 1.0 to 10, and >10 μM, respectively. ND, not detected.
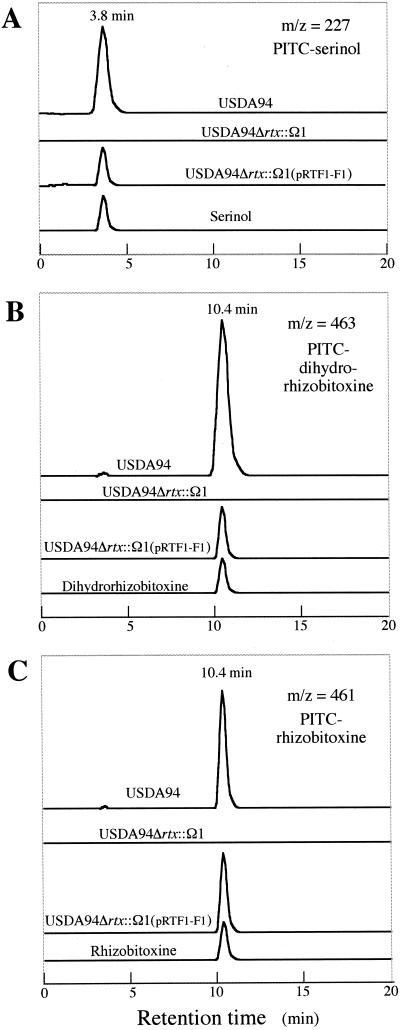
The serinol, dihydrorhizobitoxine, and rhizobitoxine concentrations in culture supernatants were analyzed as phenylthiocarbamyl derivatives by LC/MS (Fig. 4). High concentrations of serinol, dihydrorhizobitoxine, and rhizobitoxine (280, 120 and 15 μM, respectively) were detected in cultures of wild-type USDA94. However, as expected, USDA94Δrtx::Ω1 did not produce rhizobitoxine, dihydrorhizobitoxine, or serinol.
FIG. 4.
LC/MS chromatograms of cultures of B. elkanii USDA94 (wild-type), USDA94Δrtx::Ω1, and USDA94Δrtx::Ω1(pRTF1-F1). (A) PITC derivatives of serinol were detected at m/z 227 and eluted at a retention time of 3.8 min. (B) PITC-dihydrorhizobitoxine was detected at m/z 463 and eluted at a retention time of 10.4 min. (C) PITC-rhizobitoxine was detected at m/z 461 and eluted at a retention time of 10.4 min. The large-deletion mutant USDA94Δrtx::Ω1 ceased to produce serinol, dihydrorhizobitoxine, and rhizobitoxine. However, these compounds were again produced after introduction of pRTF1-F1 into the mutant.
Because B. elkanii USDA94 possesses high resistance to tetracycline (22), a pLAFR1 cosmid with a tetracycline selection marker could not be used for transconjugation. Therefore, we conferred kanamycin resistance on pRTF1. The resulting cosmid, pRTF1-F1, contains the kanamycin cassette on the 3′ side of the cloned region (1.7 kb downstream from the end of ORF6) (Fig. 1). Serinol, dihydrorhizobitoxine, and rhizobitoxine production was recovered when pRTF1-F1 was introduced into USDA94Δrtx::Ω1 (Fig. 4). These results indicate that the complementation system using pRTF1 and USDA94Δrtx::Ω1 was established successfully, thereby enabling us to examine the functions of various genes and ORFs in rhizobitoxine biosynthesis.
Production of rhizobitoxine, dihydrorhizobitoxine, and serinol in USDA94Δrtx::Ω1 complemented by various pRTF1 derivatives.
To examine the functions of rtxA and its associated ORFs in rhizobitoxine biosynthesis, we constructed 12 independent pRTF1 derivatives in which the kanamycin cassette was inserted into this DNA region and then analyzed the serinol, dihydrorhizobitoxine, and rhizobitoxine in the culture supernatants of the pRTF1 derivatives (Fig. 3B). The derivatives were named by adding the abbreviations C3, C1, D8, D2, D5, D3, E26, E9, E10, E2, E8, and F6 to the designation pRTF1 (Table 1), and the corresponding positions of kanamycin cassette insertions are shown in Fig. 3. Mutants with insertions of the cassette at positions C3 and C1, which are located 1.3 and 0.6 kb upstream of rtxA, continued to produce serinol (C3, 142 μM; C1, 116 μM), dihydrorhizobitoxine (C3, 7 μM; C1, 6 μM), and rhizobitoxine (C3, 4 μM; C1, 5 μM) in culture. However, the D8 insertion (located 0.2 kb upstream of rtxA) stopped dihydrorhizobitoxine and rhizobitoxine production and reduced serinol production (24 μM). Because we found a putative promoter sequence (5′-TTGAAA-cgcacctaacgtcaagttg-TACGAT-3′) 0.5 kb upstream of rtxA, the loss of or decrease in the ability to produce these compounds probably is due to a polar effect of the D8 insertion downstream of the rtxA promoter.
The derivative with the D2 insertion (located in the N-terminal region of the rtxA amino acid sequence) completely turned off the production of serinol, dihydrorhizobitoxine, and rhizobitoxine in culture. The D5 insertion (in the C-terminal region) also eliminated the ability to produce dihydrorhizobitoxine and rhizobitoxine but resulted in partial recovery of serinol production (40 μM). These results indicate that the N-terminal region of the rtxA product is responsible for serinol biosynthesis in culture, which is supported by the fact that the DNA sequence of rtxA corresponding to this region is homologous to that for aminotransferase. This function is in accordance with the lack of serinol production in soybean nodules inoculated with a Tn5 rtxA mutant of B. elkanii USDA61 (43).
The D3 insertion (in rtxC) construct provided recovery of dihydrorhizobitoxine (2 μM) as well as serinol (202 μM) production but not rhizobitoxine production. In light of the polar effect of the insertion, the C-terminal region of the rtxA product is probably involved in dihydrorhizobitoxine biosynthesis. This idea is supported by the DNA homology of the rtxA sequence corresponding to the C-terminal domain to that for O-acetylhomoserine sulfhydrylase, which catalyzes the combination of O-acetylhomoserine with alcohol (31, 32). Therefore, this finding suggests that the rtxA gene product is responsible for two crucial steps in rhizobitoxine biosynthesis.
Recovery of rhizobitoxine production after complementation with the E26 insertion (in ORF2)-containing derivative compared with the D3 insertion suggests that rtxC is involved in an enzymatic conversion from dihydrorhizobitoxine to rhizobitoxine. Moreover, the nucleotide sequence of rtxC has similarity to that of fatty acid desaturase, which introduces a carbon double bond in molecules. Therefore, it seems reasonable to postulate that the rtxC product catalyzes the introduction of a carbon double bond into the C3 position of dihydrorhizobitoxine to produce rhizobitoxine.
The derivatives with the E26, E9, and E10 insertions, which are located within ORF2 and between ORF2 and ORF3, led to low concentrations of dihydrorhizobitoxine and rhizobitoxine (dihydrorhizobitoxine, 0.8 to 1.1 μM; rhizobitoxine, 0.4 to 0.5 μM), although serinol production (serinol, 200 to 222 μM) had reached the original level after complementation with the derivatives containing the C3 and F1 insertions. The E2 insertion in ORF3 retained a low level of dihydrorhizobitoxine and rhizobitoxine production (serinol, 240 μM; dihydrorhizobitoxine, 3.9 μM; rhizobitoxine, 0.9 μM). Therefore, the ORF2 and ORF3 products might be involved in the synthesis of other intermediates or in the secretion or regulation of rhizobitoxine biosynthesis. Their functions cannot be inferred just from their homology and the previous literature.
Effect of homoserine-like compounds on dihydrorhizobitoxine production in B. elkanii USDA94.
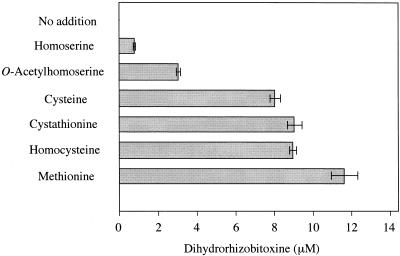
The C-terminal portion of the rtxA product is homologous to O-acetylhomoserine sulfhydrylase, and the results of our disruption experiments suggested that its enzymatic function is involved in dihydrorhizobitoxine synthesis. However, O-acetylhomoserine is located within the methionine biosynthetic pathway (31, 32). To identify possible substrates for this putative dihydrorhizobitoxine synthase, we investigated the dihydrorhizobitoxine production by B. elkanii USDA94 cell suspensions in the presence of various compounds in the methionine biosynthetic pathway (Fig. 5). Dihydrorhizobitoxine production was increased dramatically by the addition of these compounds, in particular sulfur-containing compounds such as methionine.
FIG. 5.
Effect of homoserine, O-acetylhomoserine, and sulfur-containing amino acids on dihydrorhizobitoxine production in B. elkanii USDA94. B. elkanii cells were incubated in 20 mM potassium phosphate buffer (pH 6.8) containing homoserine, O-acetylhomoserine, or sulfur-containing amino acids at 30°C for 1 h in the dark. Values are means and standard errors from two separate experiments.
Comparison of the rtx regions of B. elkanii and B. japonicum.
The 410-kb DNA region related to symbiosis in B. japonicum USDA110 was sequenced and analyzed by Göttfert et al. (15). Interestingly, they found rtxA-like genes in this region, even though B. japonicum could not produce rhizobitoxine (13, 27). Therefore, we compared the sequence of rtxA and its flanking regions of B. elkanii USDA94 with those of B. japonicum USDA110 (Fig. 6). The comparison of the 8,641-bp DNA sequence from rtxA to ORF4 shows that this region of the B. japonicum USAD110 gene is 79% homologous to that of the B. elkanii USDA94 gene. In contrast, the comparison of DNA upstream of rtxA and downstream of ORF4 in these two species of Bradyrhizobium revealed no noteworthy similarities. Furthermore, the ORFs corresponding to rtxA, rtxC, ORF2, ORF3, and ORF4 were also found in B. japonicum, and their order was well conserved between the two species. The DNA sequences of rtxC, ORF2, and ORF4 of B. japonicum showed 93, 87, and 89% similarity to those of B. elkanii, respectively. However, the ORFs corresponding to rtxA and ORF3 were fragmented into three ORFs in B. japonicum. In particular, the fragmentation of the C domain of the rtxA product appeared to abolish the ability to synthesize dihydrorhizobitoxine.
FIG. 6.
Comparison of the rtx gene regions of B. elkanii USDA94 and B. japonicum USDA110. The nucleotide sequence of the rtx gene region of B. japonicum USAD110 (8,641 bp) is 79% homologous to that of B. elkanii USDA94. The amino acid sequences of rtxC, ORF2, and ORF4 of B. japonicum USAD110 show 96, 87, and 89% identity to those of B. elkanii USDA94, respectively. The rtxA and ORF3 regions of B. japonicum USAD110 are interrupted and fragmented compared with those of B. elkanii USDA94. Numbers indicate lengths (in base pairs) of genes and intergenic regions.
DISCUSSION
Here we demonstrated that at least the rtxA and rtxC genes are responsible for rhizobitoxine biosynthesis in free-living B. elkanii, in light of results from mutagenesis experiments and the determination of concentrations of rhizobitoxine intermediates in culture by using LC/MS. The biosynthetic route to rhizobitoxine and the biological activities of the various genes are summarized in Fig. 7.
FIG. 7.
Proposed biosynthetic pathway of rhizobitoxine and relevant metabolism.
Whether dihydrorhizobitoxine is an end product (30) or intermediate (44) of rhizobitoxine biosynthesis has been a source of discussion. Our work indicates that the rtxC gene is responsible for dihydrorhizobitoxine desaturation at the final step of rhizobitoxine biosynthesis, and this gene product catalyzes the conversion of dihydrorhizobitoxine to rhizobitoxine by creating a double bond between C-3 and C-4. Therefore, we confirmed that dihydrorhizobitoxine is a key intermediate in rhizobitoxine biosynthesis (Fig. 7).
Ruan et al. (44, 45) isolated the rtxA (formally rtxA and rtxB) gene from USDA61 of B. elkanii and observed that rtxA mutants do not accumulate serinol in nodules and do not produce rhizobitoxine in culture or nodules. Those authors speculated that rtxA is involved in serinol formation and dihydrorhizobitoxine synthesis in light of DNA homology and results from other studies (44, 45). In our work, determination of serinol and dihydrorhizobitoxine concentrations in culture revealed that the N-terminal region of rtxA product is responsible for serinol formation and that the C-terminal portion is involved in dihydrorhizobitoxine biosynthesis—two crucial steps in rhizobitoxine biosynthesis. However, it is still unclear whether the predicted protein of 90 kDa mediated both activities simultaneously.
Serinol probably is a precursor of dihydrorhizobitoxine, because the one rtxA gene has two functions: serinol formation and dihydrorhizobitoxine biosynthesis. However, the substrates of the homoserine moiety in dihydrorhizobitoxine biosynthesis remain ambiguous. The addition of methionine and its intermediates (including O-acetylhomoserine, cysteine, cystathionine, and homocysteine) dramatically increased dihydrorhizobitoxine production in culture (Fig. 5). This suggests that the sulfur-containing intermediates are candidate precursors for dihydrorhizobitoxine as well as O-acetylhomoserine, although substrate specificity ultimately should be determined by using the purified enzyme derived from the rtxA gene. If so, it would be interesting if β-cystathionase is subject to feedback inhibition by rhizobitoxine in B. elkanii (Fig. 7).
In B. japonicum, most genes concerned with nodulation and symbiotic nitrogen fixation are clustered within an approximately 410-kb region on the 8.7-Mb chromosome (15, 20). In Mesorhizobium loti strain ICMP3153, a symbiotic cluster, termed the symbiosis island, can be transferred to other strains when it is integrated into a phenylalanine-specific tRNA gene (54). Further, this island structure is well conserved on a similar chromosome of another M. loti strain, MAFF303099 (19). Therefore, it is generally accepted that symbiosis genes in rhizobia have evolved by horizontal gene transfer and genomic rearrangements thereafter.
The rtxA and rtxC genes of B. elkanii were located in a DNA region that includes nodulation and symbiotic nitrogen fixation genes (Fig. 1). Interestingly, the rtx cluster and noeE gene upstream of the cluster were almost completely conserved in B. japonicum USDA110 (Fig. 6) (15), which does not synthesize rhizobitoxine (24, 25, 27). A possible explanation is that the loss of the ability to synthesize rhizobitoxine is due to the fragmentation in B. japonicum USDA110 of the C-terminal region of the rtxA gene (Fig. 6). The transfer of cosmids containing B. elkanii rtxA and rtxC genes to B. japonicum would help answer the question of whether the loss of production of rhizobitoxine in B. japonicum was brought about only by the fragmentation of rtxA gene or by other mechanisms as well.
Experiments using hypernodulation legume mutants (40) and ethylene inhibitor applications (35) indicate that detection of ethylene by host legumes is involved in the control of nodulation. Ethylene has been reported to reduce nodulation of several legumes, with the exception of soybeans (Glycine max) (35). B. japonicum preferentially nodulates soybean cultivars in a multistrain environment (28). Therefore, perhaps after an ancestor of bradyrhizobia acquired rhizobitoxine biosynthetic genes as well as various symbiotic genes, B. japonicum lost the ability to synthesize rhizobitoxine in the absence of selection pressure because of the ethylene insensitivity of soybean nodulation. The partial collapse of the rtx region in B. japonicum USDA110 supports this idea (Fig. 6).
So far, the ability to synthesize rhizobitoxine is confined to the slow-growing B. elkanii (25, 27, 37) and Burkholderia andropogonis (29). The question arises whether fast-growing rhizobia other than Bradyrhizobium spp. produce another inhibitor for ethylene biosynthesis of host plants, because it could enhance nodulation. To test this possibility, we sought potential enzymes and compounds for reducing ethylene biosynthesis from the entire genome of the fast-growing M. loti represented in a database (http://www.kazusa.or.jp/en/) and identified the ACC deaminase gene as a candidate. The ACC deaminase gene is located within a 611-kb symbiosis island (downstream of the nifDK genes) on the 7.0-Mb chromosome (19). The plant growth-promoting Pseudomonas spp. possess ACC deaminase and reduce the amount of plant ethylene by degrading ACC into α-ketobutylate and ammonia (14). It is, therefore, a fascinating hypothesis that rhizobia have two strategies for fulfilling nodulation enhancement by ethylene inhibition in host plants: rhizobitoxine biosynthesis in the slow-growing bradyrhizobia and ACC deaminase in fast-growing rhizobia.
ACKNOWLEDGMENTS
We thank M. Göttfert (Dresden University) for kindly providing DNA sequences before their publication and for valuable advice regarding genetic strategy, Y. Kiyota (Tohoku University) for synthesizing O-acetylhomoserine, Y. Murooka (Osaka University) for valuable discussions on O-acetylhomoserine metabolism, and M. Sugawara (Tohoku University) for alignment of rtx genes between B. elkanii USDA94 and B. japonicum USDA110.
We thank PROBRAIN (Japan) for supporting the research of K. Yuhashi and K. Minamisawa. This work was supported in part by grants from the Ministry of Education, Science, Sports and Culture of Japan (no. 11556012) and the Joint Research Program of the Institute of Genetic Ecology, Tohoku University (no. 981002).
REFERENCES
- 1.Babczinski P, Matern U, Strobel G A. Serinol phosphate as an intermediate in serinol formation in sugarcane. Plant Physiol. 1978;61:46–49. doi: 10.1104/pp.61.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourhy P, Martel A, Margarita D, Saint G I, Belfaiza J. Homoserine O-acetyltransferase, involved in the Leptospira meyeri methionine biosynthetic pathway, is not retroinhibited. J Bacteriol. 1997;179:4396–4398. doi: 10.1128/jb.179.13.4396-4398.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 4.Carlson R W, Sanjuan J, Bhat U R, Glushka J, Spaink H P, Wijfjes A H M, van Brussel A A N, Stokkermans T J W, Peters N K, Stacey G. The structures and biological activities of the lipo-oligosaccharide nodulation signals produced by type I and II strains of Bradyrhizobium japonicum. J Biol Chem. 1993;268:18372–18381. [PubMed] [Google Scholar]
- 5.Cole M A, Elkan G H. Transmissible resistance to penicillin G, neomycin, and chloramphenicol in Rhizobium japonicum. Antimicrob Agents Chemother. 1973;4:248–253. doi: 10.1128/aac.4.3.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas III S, Barry C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver S, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seeger K, Skelton S, Squares S, Suares R, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 7.Duodu S, Bhuvaneswari T V, Stokkermans T J W, Peters N K. A positive role for rhizobitoxine in Rhizobium-legume symbiosis. Mol Plant-Microbe Interact. 1999;12:1082–1089. [Google Scholar]
- 8.Fallay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertion mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 9.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans . Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Folch-Mallol J L, Marroqui S, Sousa C, Manyani H, Lopez-Lara I M, van der Drift K M G M, Haverkamp J, Quinto C, Gil-Serrano A, Thomas-Oates J, Spaink H P, Megias M. Characterization of Rhizobium tropici CIAT899 nodulation factors: the role of nodH and nodPQ genes in their sulfation. Mol Plant-Microbe Interact. 1996;9:151–163. doi: 10.1094/mpmi-9-0151. [DOI] [PubMed] [Google Scholar]
- 11.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 12.Friedman A M, Long S R, Brown S R, Buikema W J, Ausubel F M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 13.Fuhrmann J. Symbiotic effectiveness of indigenous soybean bradyrhizobia as related to serological, morphological, rhizobitoxine, and hydrogenase phenotypes. Appl Environ Microbiol. 1990;56:224–229. doi: 10.1128/aem.56.1.224-229.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glick B R, Patten C L, Holguin G, Penrose D M. Biochemical and genetic mechanisms used by plant growth promoting bacteria. London, United Kingdom: Imperial College Press; 1999. [Google Scholar]
- 15.Göttfert M, Röthlisberger S, Kündig C, Beck C, Marty R, Hennecke H. Potential symbiosis-specific genes uncovered by sequencing a 410-kilobase DNA region of the Bradyrhizobium japonicum chromosome. J Bacteriol. 2001;183:1405–1412. doi: 10.1128/JB.183.4.1405-1412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanin M, Jabbouri S, Quesada-Vincens D, Freiberg C, Perret X, Prome J-C, Broughton W J, Fellay R. Sulphation of Rhizobium sp. NGR234 nod factors is dependent on noeE, a new host-specificity gene. Mol Microbiol. 1997;24:1119–1129. doi: 10.1046/j.1365-2958.1997.3981777.x. [DOI] [PubMed] [Google Scholar]
- 17.Isawa T, Sameshima R, Mitsui H, Minamisawa K. IS1631 occurrence in Bradyrhizobium japonicum highly reiterated sequence-possessing strains with high copy numbers of repeated sequences RSa and RSb. Appl Environ Microbiol. 1999;65:3493–3501. doi: 10.1128/aem.65.8.3493-3501.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson H W, Means U M, Clark F E. Responses of seedlings to extracts of soybean nodules bearing selected strains of Rhizobium japonicum. Nature. 1959;183:308–309. [Google Scholar]
- 19.Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S, Watanabe A, Idesawa K, Ishikawa A, Kawashima K, Kimura T, Kishida Y, Kiyokawa C, Kohara M, Matsumoto M, Matsuno A, Mochizuki Y, Nakayama S, Shimpo S, Sugimoto M, Takeuchi C, Yamada M, Tabata S. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 2000;7:331–338. doi: 10.1093/dnares/7.6.331. [DOI] [PubMed] [Google Scholar]
- 20.Kündig C, Hennecke H, Göttfert M. Correlated physical and genetic map of Bradyrhizobium japonicum 110 genome. J Bacteriol. 1993;175:613–622. doi: 10.1128/jb.175.3.613-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minamisawa K. Division of rhizobitoxine-producing and hydrogen-uptake positive strains of Bradyrhizobium japonicum by nifDKE sequence divergence. Plant Cell Physiol. 1990;31:81–89. [Google Scholar]
- 22.Minamisawa K, Mitsui H. Genetic ecology of soybean bradyrhizobia. Soil Biochem. 2000;10:349–377. [Google Scholar]
- 23.Minamisawa K, Watanabe H. Serinol (2-amino-1,3-propanediol) and 3-amino-1,2-propanediol in soybean nodules. Plant Cell Physiol. 1986;27:1109–1116. [Google Scholar]
- 24.Minamisawa K, Fukai K. Production of indole-3-acetic acid by Bradyrhizobium japonicum: a correlation with genotype grouping and rhizobitoxine production. Plant Cell Physiol. 1991;32:1–9. [Google Scholar]
- 25.Minamisawa K, Fukai K, Asami T. Rhizobitoxine inhibition of hydrogenase synthesis in free-living Bradyrhizobium japonicum. J Bacteriol. 1990;172:4505–4509. doi: 10.1128/jb.172.8.4505-4509.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minamisawa K, Kume N. Determination of rhizobitoxine and dihydrorhizobitoxine in soybean plants by amino acid analyzer. Soil Sci Plant Nutr. 1987;33:645–649. [Google Scholar]
- 27.Minamisawa K, Seki T, Onodera S, Kubota M, Asami T. Genetic relatedness of Bradyrhizobium japonicum field isolates as revealed by repeated sequences and various other characteristics. Appl Environ Microbiol. 1992;58:2832–2839. doi: 10.1128/aem.58.9.2832-2839.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minamisawa K, Onodera S, Tanimura Y, Kobayashi N, Yuhashi K, Kubota M. Preferential nodulation of Glycine max, Glycine soja, Macroptilium atropurpureum by two Bradyrhizobium species japonicum and elkanii. FEMS Microbiol Ecol. 1997;24:49–56. [Google Scholar]
- 29.Mitchell R E, Frey E J, Benn M K. Rhizobitoxine and 1-threo-hydroxythreonine production by the plant pathogen Pseudomonas andropogonis. Phytochemistry. 1986;25:2711–2715. [Google Scholar]
- 30.Mitchell R E, Coddington J M. Biosynthetic pathway to rhizobitoxine in Pseudomonas andropogonis. Phytochemistry. 1991;30:1809–1814. [Google Scholar]
- 31.Murooka Y, Kakihara K, Miwa T, Koji K, Harada T. O-Alkylhomoserine synthesis catalyzed by O-acetylhomoserine sulfhydrylase in microorganisms. J Bacteriol. 1977;130:62–73. doi: 10.1128/jb.130.1.62-73.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murooka Y, Seto K, Harada T. O-Alkylhomoserine synthesis from O-acetylhomoserine and alcohol. Biochem Biophys Res Commun. 1970;41:407–414. doi: 10.1016/0006-291x(70)90519-x. [DOI] [PubMed] [Google Scholar]
- 33.Nagai S, Flavin M. Synthesis of O-acetylhomoserine. Methods Enzymol. 1971;17:423–424. [Google Scholar]
- 34.Nellen-Anthamatten D, Rossi P, Presig O, Kullik I, Babst M, Fisher H M, Hennecke H. Bradyrhizobium japonicum FixK2, a crucial distributor in the FixLJ-dependent regulatory cascade for the control of genes inducible by low oxygen level. J Bacteriol. 1998;180:5251–5255. doi: 10.1128/jb.180.19.5251-5255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nukui N, Ezura H, Yuashi K-I, Yasuta T, Minamisawa K. Effects of ethylene precursor and inhibitors for ethylene biosynthesis and perception on nodulation in Lotus japonicus and Macroptilium atropurpureum. Plant Cell Physiol. 2000;41:893–897. doi: 10.1093/pcp/pcd011. [DOI] [PubMed] [Google Scholar]
- 36.Owen L D, Wright D A. Rhizobial-induced chlorosis in soybean: isolation, production in nodules, and varietal specificity of the toxin. Plant Physiol. 1965;40:927–930. doi: 10.1104/pp.40.5.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owen L D, Thompson J F, Pitcher R G, Williams T. J. Chem. Sci. Chem. Commun. 1972:714. 1972. Structure of rhizobitoxine, an antimetabolic enol-ether amino-acid from Rhizobium japonicum. [Google Scholar]
- 38.Owen, L. D., J. F. Thompson, and V. F. Paul. 1972. Dihydrorhizobitoxine, a new ether amino-acid from Rhizobium japonicum. Chem. Sci. Chem. Commun. 1972:715.
- 39.Owen L D, Guggenheim S, Hilton J L. An inhibitor of β-cystathionase in Salmonella typhimurium. Biochim Biophys Acta. 1968;158:219–225. doi: 10.1016/0304-4165(68)90134-7. [DOI] [PubMed] [Google Scholar]
- 40.Penmetsa R V, Cook D R. A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science. 1997;275:527–530. doi: 10.1126/science.275.5299.527. [DOI] [PubMed] [Google Scholar]
- 41.Preisig O, Anthamatten D, Hennecke H. The Bradyrhizobium japonicum fixGHIS genes are required for the formation of the high-affinity cbb3-type cytochrome oxidase. Arch Microbiol. 1996;165:297–305. doi: 10.1007/s002030050330. [DOI] [PubMed] [Google Scholar]
- 42.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 43.Ruan X, Peters N K. Isolation and characterization of rhizobitoxine mutants of Bradyrhizobium japonicum. J Bacteriol. 1992;174:3467–3473. doi: 10.1128/jb.174.11.3467-3473.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruan X, Zhang C, Peters N K. Bradyrhizobium japonicum rhizobitoxine genes and putative enzyme functions: expression requires a translational frameshift. Proc Natl Acad Sci USA. 1993;90:2641–2645. doi: 10.1073/pnas.90.7.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruan X, Zhang C, Peters N K. Authors collection. Proc Natl Acad Sci USA. 1993;90:12055. [Google Scholar]
- 46.Ruan X, Peters N K. Rapid and sensitive assay for the phytotoxin rhizobitoxine. Appl Environ Microbiol. 1991;57:2097–2101. doi: 10.1128/aem.57.7.2097-2100.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Sanjuan J, Carlson R W, Spaink H P, Bhat U R, Mark Barborr W, Glushka J, Stacey G. A 2-O-methylfucose moiety is present in the lipo-oligosaccharide nodulation signal of Bradyrhizobium japonicum. Proc Natl Acad Sci USA. 1992;89:8789–8793. doi: 10.1073/pnas.89.18.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sato N, Fujiwara S, Kawaguchi A, Tsuzuki M. Cloning of gene chloroplast w6 desaturase of green alga, Chlamydomonas reinhardtii. J Biochem. 1997;122:1224–1232. doi: 10.1093/oxfordjournals.jbchem.a021885. [DOI] [PubMed] [Google Scholar]
- 50.Schwedock J S, Liu C X, Leyh T S, Long S R. Rhizobium meliloti NodP and NodQ form a multifunctional sulfate-activating complex requiring GTP for activity. J Bacteriol. 1994;176:7055–7064. doi: 10.1128/jb.176.22.7055-7064.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:789–791. [Google Scholar]
- 52.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H-M, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicare R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nolling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stover T C K, Pham X-Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S L, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrook-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbig K, Lim R M, Smith K A, Spencer D H, Wong G K-S, Wu Z, Paulsen I T, Reizer J, Saier M H, Hancock R E W, Lory S, Olson M V. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 54.Sullivan J T, Ronson C W. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci USA. 1998;95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vieille C, Elmerich C. Characterization of two Azospirillum brasilense Sp7 plasmid genes homologous to Rhizobium meliloti nodPQ. Mol Plant-Microb Interact. 1990;3:389–400. doi: 10.1094/mpmi-3-389. [DOI] [PubMed] [Google Scholar]
- 56.Wada H, Gombos Z, Murata N. Enhancement of chilling tolerance of a cyanobacterium by genetic manipulation of fatty acid desaturation. Nature. 1990;347:200–203. doi: 10.1038/347200a0. [DOI] [PubMed] [Google Scholar]
- 57.Xiong K, Fuhrmann J J. Comparison of rhizobitoxine-induced inhibition of β-cystathionase from different bradyrhizobia and soybean genotypes. Plant Soil. 1996;186:53–61. [Google Scholar]
- 58.Yamaya T, Matsumoto H. Analysis of phenylthiocarbamyl-amino acids at pico-mole level by high performance liquid chromatography and application to plant materials. Soil Sci Plant Nutr. 1988;34:297–302. [Google Scholar]
- 59.Yasuta T, Satoh S, Minamisawa K. New assay for rhizobitoxine based on inhibition of 1-aminocyclopropane-1-carboxylate synthase. Appl Environ Microbiol. 1999;65:849–852. doi: 10.1128/aem.65.2.849-852.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuhashi K I, Ichikawa N, Ezura H, Akao S, Minakawa Y, Nukui N, Yasuta T, Minamisawa K. Rhizobitoxine production by Bradyrhizobium elkanii enhances nodulation and competitiveness on Macroptilium atropurpureum. Appl Environ Microbiol. 2000;66:2658–2663. doi: 10.1128/aem.66.6.2658-2663.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuhashi K I, Akao S, Fukuhara H, Tateno E, Chun J Y, Stacy G, Hara H, Kubota M, Asami T, Minamisawa K. Bradyrhizobium elkanii induces outer root swelling in soybean. Plant Cell Physiol. 1995;36:1571–1577. [Google Scholar]
- 62.Zhang Y X, Patil S S. The phtE locus in the phaseolotoxin gene cluster has ORFs with homologies to genes encoding amino acid transferases, the AraC family of transcriptional factors, and fatty acid desaturases. Mol Plant-Microbe Interact. 1997;10:947–960. doi: 10.1094/MPMI.1997.10.8.947. [DOI] [PubMed] [Google Scholar]