Summary
Background
Ebola virus (EBOV) vaccines containing glycoprotein (GP) provide protection against severe Ebola virus disease (EVD). EBO vaccinations elicit antibodies that are detectable in Ebola serodiagnostic tests, as EBOV GP is a major target antigen. This vaccine-induced seropositivity presents issues with early detection of natural EBOV infections, following vaccination and during surveillance, leading to ‘uninfected’ vaccine trial participants being falsely diagnosed as ‘EBOV infected’ potentially resulting in long-term social and economic distress. Since mass vaccinations are being employed to curtail the recurrent EBOV epidemics in multiple African countries, it is, therefore, essential to differentiate vaccine-induced from natural infection–induced antibodies by a differential serodiagnosis assay for accurate detection of Ebola virus infections.
Methods
To develop a serodiagnostic test that can differentiate between individuals with EBOV infection-induced antibodies and individuals with EBOV vaccine-induced antibodies, we analysed peptides of EBOV viral protein 40 (VP40), viral protein 35 (VP35) and nucleocapsid protein (NP) using an ELISA with a panel of 181 human sera collected from healthy controls, EBO vaccinees, and EBOV-infected survivors. Receiver Operating Characteristic (ROC) curve analysis was used to calculate sensitivity and specificity of the assay. A simple peptide-based serodiagnostic assay was used to evaluate detection of breakthrough EBOV infections in vaccinated non-human primates (NHP) in EBOV challenge studies.
Findings
We identified conserved peptide sequences in EBOV VP40, VP35 and NP, produced soon after EBOV infection that are not part of the current EBO vaccine target antigens. The new ELISA-based differential serodetection assay termed ‘EBOV-Detect’ demonstrated >94% specificity and 96% sensitivity for diagnosis of EBOV infection. Importantly, the uninfected vaccine-trial participants scored negative in ‘EBOV-Detect’ assay. The results from the NHPs EBOV challenge study established that post-EBO vaccination serum scored negative in ‘EBOV-Detect’ and all NHPs with Ebola breakthrough infections, following EBOV challenge, were serodiagnosed positively with EBOV-Detect.
Interpretation
The new ‘EBOV-Detect’ is a simple and sensitive serodiagnostic assay that can specifically differentiate between natural Ebola virus infected and those with vaccine-induced immunity. This could potentially be implemented as a robust diagnostic tool for epidemiology and surveillance of EBOV infections during and after outbreaks, especially in countries with mass Ebola vaccinations.
Funding
The antibody characterization work described in this manuscript was supported by FDA Office of Counterterrorism and Emerging Threats (OCET) - Medical Countermeasures initiative (MCMi) grant- OCET 2019-1018 and Defense Threat Reduction Agency (HDTRA1930447) funds to S.K.
Keywords: Ebola, Diagnosis, Vaccine, Glycoprotein, VP40, Virus
Research in context.
Evidence before this study
EBO vaccines have been deployed at a large-scale in some African countries to contain the spread of EBOV outbreaks. However, the extent of breakthrough EBOV infections following vaccination is unknown due to the lack to availability of serological diagnostic tests that can differentiate between EBOV infection induced antibodies from vaccination-induced antibodies. Since EBO vaccines induce antibodies that target GP, vaccinated but uninfected individuals will be seropositive in a GP-based EBOV serodiagnostic test, resulting in vaccine-induced seropositivity (VISP), wherein, uninfected vaccine trial participants can be falsely diagnosed as ‘EBOV infected’.
Added value of this study
We developed a peptide based differential serodiagnostic ELISA assay termed ‘EBOV-Detect’ that demonstrates >95% specificity and sensitivity for detection of EBOV infection induced-antibodies. Importantly, uninfected vaccine-trial participants scored negative using ‘EBOV-Detect’. Similarly, serum from vaccinated non-human primate tested negative in EBOV-Detect, whereas serum from only EBOV infected animals tested positive. EBOV-Detect can be used as an effective serodiagnostic assay to differentiate host immune response following natural EBOV infection vs. ERVEBO or ChAd/MVA-EBOV vaccine-induced seropositivity in GP-based assays, thereby reducing false positives and enabling diagnosis of true breakthrough EBOV infections in resource-limited settings.
Implications of all the available evidence
‘EBOV-Detect’ could be implemented as a robust diagnostic tool for epidemiology and surveillance of EBOV infections, especially in countries with mass Ebola vaccination campaigns.
Alt-text: Unlabelled box
Introduction
The Ebola virus disease (EVD) outbreak in West Africa (2014-15) and more recent outbreaks in the Democratic Republic of Congo and Côte d'Ivoire have highlighted the critical need for a rapid serodiagnostic assay for detection of EBOV infection for accurate surveillance. The development of simple and rapid diagnostic assays that can be conducted with minimal personnel training, in resource-limited settings, is crucial to overcome challenges with existing assays such as Polymerase Chain Reaction (PCR)-based diagnosis.1,2 The PCR-based diagnostic tests are only sensitive towards detection of virus genomic material during active virus replication and suffer from several bottlenecks including limit of detection and lower sensitivity for long-term EBOV persistence in immune-privileged sites (e.g., semen, eyes, etc). Serological tests such as the enzyme-linked immunosorbent assay (ELISA) are used for rapid detection of antibody responses associated with filovirus infection.1,3 Rapid point-of-care serodiagnosis assays, including antigen detection tests have been developed and are being used in the field at various national reference laboratories with high specificity of >95%. However, their ability to detect EBOV infection has been limited with sensitivity ranging between 38 to 87% that does not achieve the desired sensitivity and specificity per the World Health Organization (WHO) target product profile.4 While PCR-based detection tests have routinely been used during the EBOV epidemics,5 however, they require significant infrastructure and with operational and technical expertise, thus complicating deployment in resource-limited settings.3,6,7 A simple ELISA or another serodiagnostic assays for the detection of EBOV-infection induced antibodies offers many advantages, as they are rapid and can be performed with limited resources. In addition, such assays can be made compatible with inactivation methods like gamma-irradiation, detergent-treatment or solvent-extraction that are used to inactivate infectious components in samples prior to handling and thus minimizing impact on measurable levels of anti-EBOV antibodies.1,3,8 Moreover, post-EBOV infection antibodies targeting structural proteins, apart from GP, can be detected in EVD survivors even years after their infection, including in 1995 Kikwit outbreak or 2014 Makona outbreak.8,9 This unique quality of antibodies makes their detection useful for epidemiological and surveillance studies.
With the licensure of two EBO vaccines, one in the US (ERVEBO, rVSV-ZEBOV) and another in Europe (Ad26.ZEBOV/MVA-BN-Filo), there have been efforts for large-scale vaccination campaigns in African countries to curtail the spread of EBOV outbreaks. Even though the vaccines have shown good effectiveness in clinical trials, there have been observations of breakthrough EBOV infections following vaccination in western Africa.10, 11, 12, 13 However, the scale of these breakthrough infections during vaccination campaigns is unknown due to the lack of availability of serological diagnostic tests that can differentiate EBOV infection from vaccinated individuals. Current EBOV serodiagnostic assays uses either the recombinant EBOV GP or GP-containing viral lysates as the detecting antigen. However, these assays are prone to false-positive results, increased cross-reactivity and hence decreased specificity.1,14 Since most EBO vaccines currently licensed or under development induce antibodies that target GP, and therefore serum/plasma from vaccinated but uninfected individuals will be seropositive in a GP-based EBOV serodiagnostic test, resulting in false-positive results and confounding the interpretation of EBOV GP-based or the whole virus based serodiagnostic assays. This, in turn, will lead to phenomenon called vaccine-induced seropositivity (VISP), wherein, uninfected vaccine trial participants could be falsely diagnosed as ‘EBOV infected’, similar to that observed for the HIV vaccine trial participants in earlier clinical studies.15, 16, 17, 18 As a result, this can lead to social stigma, and uninfected, seropositive vaccinees may encounter long-term social and economic harm. With EBO vaccines being used for large-scale vaccinations in Africa, it is critical to develop simple point-of-care serodiagnostic assays that can be designed for the differential detection of true EBOV infections or breakthrough EBOV infections in the face of vaccine-induced antibodies.
Therefore, in this study, synthetic peptides representing non-GP antigenic sites identified in EVD survivors8 (and unpublished studies) using whole EBOV genome fragment phage display library (GFPDL), were used as antigens for development of a simple ELISA-based serological test to differentiate EBO vaccine-induced antibodies from antibodies generated following EBOV-infection. Human serum samples from rVSV-EBOV or ChAd3/MVA-EBOV vaccinated, or EBOV-infected, or control sera from unexposed adults were analyzed for presence of IgG antibodies against EBOV peptides spanning NP, VP35, VP30, VP40 and VP24, for development of ELISA-based differential serodiagnosis of EBOV breakthrough infection in the face of vaccine-induced antibodies.
Methods
Ethics
The study at CBER, FDA was conducted with de-identified samples (ClinicalTrials.gov Identifier: NCT02363322; NIH IRB protocol #15-I-0083) under Research Involving Human Subjects (RIHSC) exemption #15-0B; and all assays performed fell within the permissible usages in the original consent.
Human samples
A panel of 181 human serum/plasma samples obtained from 33 EBOV-infected, 33 ERVEBO-vaccinated but uninfected, 2 ChAd3/MVA vaccinated-uninfected, and 44 H1N1pdm09 influenza infected, and 69 healthy control adults were tested in ELISA. The clinical samples from EBOV survivors or ChAd3/MVA prime-boost vaccinated adults were obtained from National Institute for Biological Standards and Control (NIBSC) as part of a “WHO collaborative study to assess the suitability of an interim standard for antibodies to Ebola virus” 19,20 for determining the reactivity of the EBOV-infection induced antibodies in ELISA. Briefly, plasma was obtained from convalescent patients between time-period of 2 months to 2 years post-recovery from Ebola virus disease when they were negative for Ebola virus RNA and other blood viral markers. The PCR-negative plasma samples were solvent-detergent-extracted using a method validated at NIBSC. Serum samples from additional EBOV survivors were obtained from Dr. Miles Carroll (PHE UK). For testing of EBO-vaccination induced antibodies, serum samples from adults vaccinated with rVSV-ZEBOV (ERVEBO; 20 million pfu vaccine dose) at 1-month post-vaccination were obtained from Drs. Richard Davey and John Beigel at National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) from a clinical study performed in the US.19,20,21 Serum from 69 healthy US adults (18-45 years) and 44 influenza H1N1pdm09-infected adults (18-45 years) were used as controls.22 All samples were aliquoted and kept frozen at -80°C until usage to ensure avoidance of freeze-thaw cycles. Serum or plasma were used based on the sample availability.
Vaccinated non-human primate EBOV challenge study samples
Longitudinal serum samples of 11 cynomolgus macaque's either prior to vaccination (Pre-Vac), or following vaccination with rVSV expressing the EBOV-Makona GP (Post-Vac), or after EBOV challenge (Post-EBOV) were obtained from Drs. Andrea Marzi and Heinz Feldmann at NIAID, NIH.23 The rVSV-GP vaccine dose ranged from 101 to 106 plaque-forming units. The NHP samples were used to determine the capacity of the assay to diagnose breakthrough EBOV infection in presence of vaccine-induced antibodies.
Enzyme-linked immunosorbent assay (ELISA)
Nine biotinylated peptides selected from whole-genome GFPDL analysis of post-EBOV infected human serum/plasma were chemically synthesized. The 9 biotinylated individual peptides were evaluated in an ELISA. The chosen peptides are as follows: NP 1-41 (1); NP 453-514 (2); VP35 742-815 (3); VP35- 895-934 (4); VP35 930-965 (5); VP40 1084-1133 (6); VP40 1313-1387 (7); VP30 2088-2114 (8); VP24 2560-2612 (9). Consequently, ELISA was performed using different combinations of peptides namely, EBOV-Detect-1 (VP35 930-965 + VP40 1313-1387) and EBOV-Detect-2 (VP35 930-965+VP40 1084-1133+ VP40 1313-1387+ VP24 2560-2612). 96-well ELISA plates (Immulon 2HB, Fisher) were coated overnight at 4℃ with 200 ng Streptavidin (NEB) in 100 µL PBS in each well. After washing the plates three times with PBST (PBS with 0.05% Tween-20), each of the biotinylated peptides (50 ng each) or peptide mixtures (equal amount of each peptide for total content of 50 ng) were added to each well and incubated for 1 hour. Recombinant EBOV-GP (without the transmembrane domain obtained from Sino Biologicals, catalog number 40442-V08H1; 50 ng/well) was directly coated as a control. Plates were then washed three times with PBST and blocked for 2 hours at RT with 5% BSA-PBST. The serum/plasma samples were diluted 100-fold in 2% BSA-PBST, added to the plates, and incubated for 1 hour. After washing the plates with PBST, 5000-fold dilution of HRP-conjugated anti-human IgG-Fc antibodies (Jackson Research, Cat #709-035-098; for human samples) or HRP-conjugated anti-monkey IgG for NHP sera (Brookwood, Cat #SAB1303) were added and incubated for 1 hour. After washing with PBST, the bound antibodies were developed with O-phenylenediamine substrate solution (Thermofisher). The reaction was stopped with 3.3 M H2SO4, and plates were read at 492 nm. Cut-off value for each antigen was determined by Receiver Operating Characteristic (ROC) curve analysis24 in Graphpad Prism. Specimens with an Absorbance/Cut-off ratios of ≥ 1 are considered EBOV seropositive and those with ratios < 1 are considered EBOV seronegative. Area under the ROC curve was used to calculate sensitivity (%) and specificity (%) values with 95% CI (confidence interval) by comparing ELISA reactivity for post-EBOV infection samples vs. un-infected samples (from healthy controls or post-vaccinations) by GraphPad Prism (v. 9.0). No significant differences were observed in reactivity of using serum vs plasma with no impact of detergent treatment or gamma irradiation of samples in the peptide ELISA.
Statistics
All statistical analyses were performed in GraphPad Prism (v. 9.0) using one-way ANOVA with a Tukey post-hoc analysis for multiple comparisons.
Experiments were performed based on sample availability during the study and hence sample size calculations were not done a priori. Samples were allocated randomly to each test group and tested in blinded fashion (researcher was blinded to sample identity) to minimize selection bias or detection bias. There were no exclusion criteria. All samples and data were used for analysis and are presented in the study.
Role of funders
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Results
Vaccine-induced seropositivity and identification of EBOV non-GP serodiagnostic targets
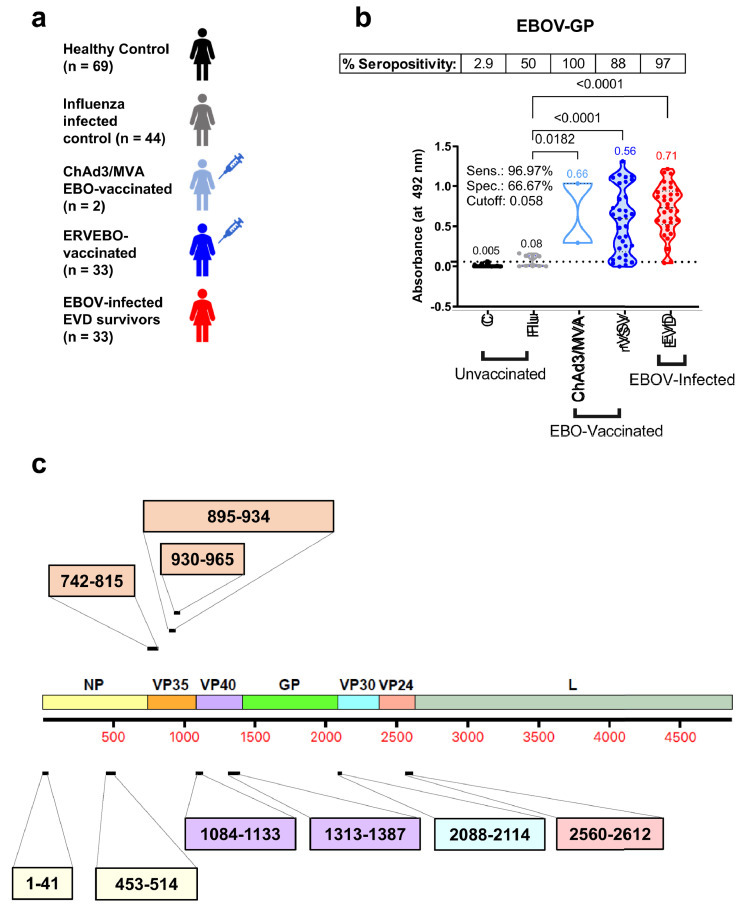
Human serum/plasma samples collected from 69 healthy control adults, 44 H1N1pdm09 influenza-infected adults, 2 ChAd3/MVA-vaccinated and uninfected vaccinees, 33 ERVEBO-vaccinated but uninfected adults, and 33 EBOV-infected EVD survivors, were evaluated in this study by IgG-ELISA (Figure 1a).
Figure 1.
Demographics, GP reactivity and selected peptides used for development of Ebola-Detect. (A) Overview of 181 serum/plasma samples used in the study including unvaccinated [healthy controls (C, n = 69) and H1N1-influenza infected (Flu, n = 44)], EBO-vaccinated [ChAd3/MVA-vaccinated but uninfected (ChAd3/MVA, n = 2) and rVSV-ZEBOV (ERVEBO) vaccinated (rVSV, n = 33)], and EBOV-infected convalescent sera from EVD survivors (EVD, n = 33). (B) Serum IgG absorbance values at a 100-fold serum dilution for each sample group to EBOV Makona GP. Mean absorbance values for each group are color-coded and indicated above each group. Cut-off value for GP in ELISA was determined by ROC curve analysis and is represented as the dotted line on Y-axis. Specimens with an absorbance greater than Cut-off value are considered EBOV seropositive and those with absorbance less than Cut-off value are considered EBOV seronegative. Percent seropositive are shown for each sample cohort above the panel. Area under the ROC curve was performed to calculate sensitivity (%) and specificity (%) values with 95% CI (confidence interval) by comparing ELISA reactivity for 33 EVD survivors vs. 148 uninfected samples. Statistical differences among groups were analyzed by one-way ANOVA and Tukey-adjusted p values using a pairwise multiple comparison. Statistically significant differences between cohorts are shown. (C) A whole proteome map of EBOV showing different encoded proteins: NP, VP35, VP40, GP, VP30, VP24 and L. Peptides representing immunodominant epitopes from different proteins identified using EBOV-GFPDL approach on post-EBOV infected sera are indicated with their amino acid positions. The amino acid labels of the peptides are color-coded corresponding to the color of the EBOV protein on the proteome map. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
EBO vaccines primarily induce antibodies that target GP, and serum from ChAd3/MVA vaccinated and 30 of the 33 ERVEBO-vaccinated but uninfected individuals reacted with EBOV GP in IgG-ELISA (Figure 1b). Also, four of the 44 H1N1-infected samples showed a low non-specific reactivity with the EBOV-GP. As expected, serum/plasma from all 33 EVD survivors showed antibody binding against EBOV-GP. The GP-reactivity of most uninfected EBO vaccinees confirm the phenomenon of VISP in current GP-based serodiagnostic tests. We, therefore, focused on identifying differential antigenic sites in EBOV proteins other than GP that were recognized in EBOV-infected patients8 (and unpublished studies) but not in healthy controls using EBOV GFPDL analysis. Nine EBOV peptides representing immunodominant antigenic sites up to 74 amino acid residues derived from EBOV NP, VP35, VP40, VP30 and VP24 proteins were chemically synthesized (Figure 1c) to develop a serodiagnostic ELISA.
Specificity and sensitivity of EBOV GFPDL-identified non-GP antigenic site peptides
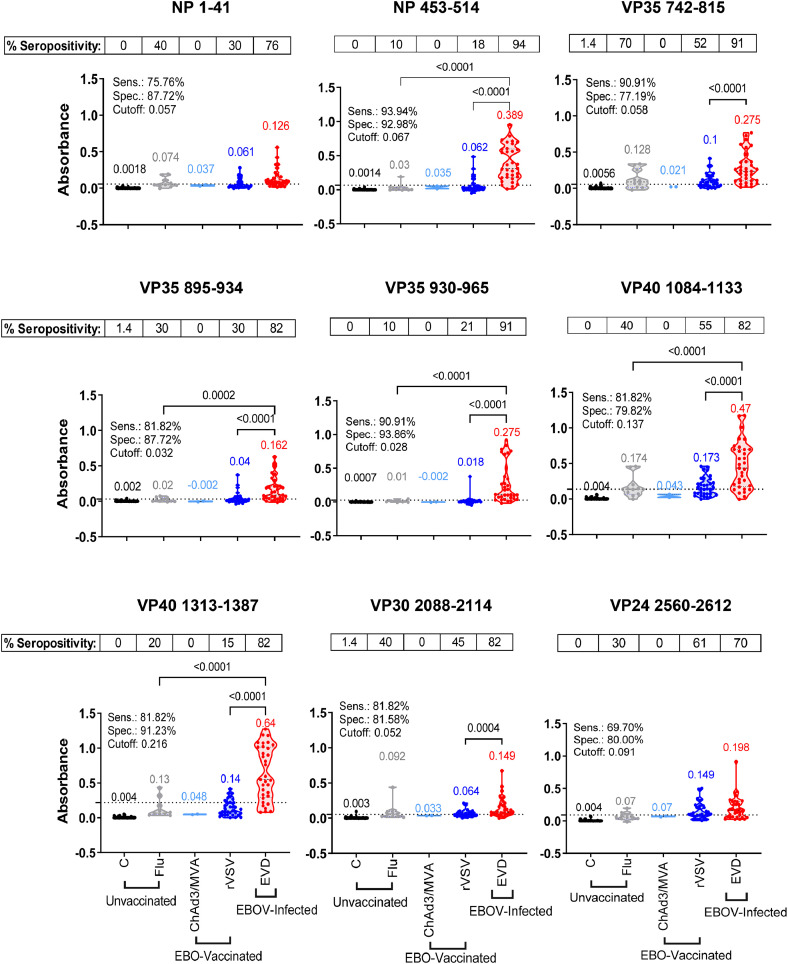
First, the 9 peptides were evaluated individually with serum/plasma from 69 healthy adults to establish cut-off values and determine specificity of ELISA (Figure 2). Each of these 9 peptides displayed very low seroreactivity with 69 healthy control samples. Then, antibody binding of these individual peptides with serum samples from 33 post-EBOV-infected EVD survivors were compared with 44 unvaccinated H1N1-influenza infected adults (ages 18-45 years, collected in 2009), 33 ERVEBO and 2 ChAd3-MVA vaccinated serum samples (Figure 2). Percent sensitivity and specificity (true seropositive and true seronegative, respectively) values were calculated for each peptide using the cut-off value as determined using Receiver Operating Characteristic (ROC) curve analysis.24 The individual peptides demonstrated sensitivity and specificity values above 77% in ELISA. Except for the low non-specific reactivity of some ERVEBO-vaccinated samples to a few peptides, minimal IgG binding was observed with ChAd3/MVA vaccinated samples or H1N1-infected samples. For individual peptides, highest specificity of 91% was observed for the VP35 930-965 peptide.
Figure 2.
Reactivity of human samples to individual EBOV peptides in ELISA. IgG absorbance values at a 1:100 serum/plasma dilution for each sample group namely, unvaccinated [healthy controls (C, black, n = 69) and H1N1-infected (Flu, grey, n = 44)], EBO-vaccinated [ChAd3/MVA-vaccinated (ChAd3/MVA, light blue, n = 2) and rVSV-ZEBOV (ERVEBO) vaccinated (rVSV, blue, n = 33)], and EBOV-infected convalescent sera from EVD survivors (EVD, red, n = 33) were plotted based on the reactivity to each 9 individual peptides. Mean absorbance values for each group are color-coded and indicated above each group. Cut-off value for each individual peptide was determined separately by ROC curve analysis and represented as a dotted line on the Y-axis. Specimens with an absorbance greater than Cut-off value are considered EBOV seropositive and those with absorbance less than Cut-off value are considered EBOV seronegative. Percent seropositive are shown for each sample cohort above the panel. Area under the ROC curve was performed to calculate sensitivity (%) and specificity (%) values with 95% CI (confidence interval) by comparing ELISA reactivity for 33 EVD survivors vs. 148 uninfected samples. Statistical differences among groups were analyzed by one-way ANOVA and Tukey-adjusted p values using a pairwise multiple comparison. Statistically significant differences between cohorts are shown. All ELISA experiments were performed twice and the researcher performing the assay was blinded to sample identity. The variation for each sample in duplicate ELISA runs was <7%. The data shown are the average value of two experimental runs. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
EVD survivors demonstrated a broad range of antibody reactivity to the nine individual peptides (Figure 2). Convalescent serum samples from EVD survivors showed a statistically significant higher reactivity to NP 453-514 (p<0.0001), three VP35 peptides (p<0.01), two VP40 peptides (p<0.0001) and VP30 (p<0.05) peptides compared with ERVEBO-vaccinated samples (Figure 2). The highest antibody reactivity for post-EBOV-infection samples was observed against the peptides derived from VP40 and the NP 453-514, with lowest IgG binding against the NP 1-41 peptide. Highest sensitivity of 94% was observed for NP 453-514 for these EVD survivors in ELISA.
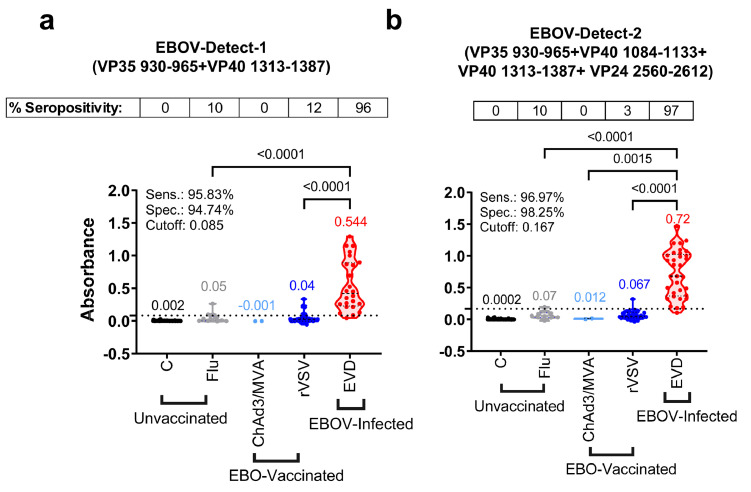
Establishment of differential serodiagnostic test termed EBOV-Detect
Based on the observed specificity and sensitivity of these 9 individual peptides, we evaluated different combinations of high-reactivity peptides to develop a differential serodiagnostic assay, termed EBOV-detect. The reactivity pattern of different peptide mixtures and assay optimization led to the down-selection of two different mixtures of fewer peptides. These two peptide combinations were evaluated further with the complete panel of 181 human samples in IgG-ELISA. EBOV-Detect-1 consists of an equal mix of only 2 peptides (VP35 930-965 and VP40 1313-1387), and EBOV-Detect-2 consists of an equivalent mix of 4 peptides (VP35 930-965 + VP40 1084-1133 + VP40 1313-1387 + VP24 2560-2612). The sensitivity of two peptide combinations was >95% for the detection of 33 post-EBOV-infected samples. A significantly (p<0.0001) high reactivity was observed for EVD survivors when compared with ERVEBO-vaccinated or control H1N1 influenza infected samples (p<0.0001) and achieved >94% specificity (Figure 3). EBOV-Detect-1 and EBOV-Detect-2 (Figure 3) demonstrated higher sensitivity compared with those of individual peptides (Figure 2) possibly due to the additive reactivity of EBOV infection-specific IgG-antibodies against multiple epitopes contained in the mixture of peptides rather than the individual peptides, resulting in the greater sensitivity for detection of IgG antibodies elicited by EBOV infection in EBOV-Detect.
Figure 3.
Reactivity of human serum samples in EBOV-Detect assay. Combination of the EBOV peptides were used in ELISA. EBOV-Detect-1 consists of equal mix of 2 peptides (VP35 930-965 and VP40 1313-1387) and EBOV-Detect-2 consist of equivalent mix of 4 peptides (VP35 930-965 + VP40 1084-1133 + VP40 1313-1387 + VP24 2560-2612). Serum IgG absorbance values at a 1:100 serum dilution for each sample group namely unvaccinated; healthy controls (C, black, n = 69) and H1N1-infected (Flu, grey, n = 44), EBO-vaccinated; ChAd3/MVA-vaccinated (ChAd3/MVA, light blue, n = 2) and rVSV-ZEBOV (ERVEBO) vaccinated (rVSV, blue, n = 33), and EBOV-infected convalescent sera from EVD survivors (EVD, red, n = 33) were plotted for reactivity to EBOV-Detect-1 and EBOV-Detect-2. Mean values for each group are color-coded and indicated above each group. Cut-off value for EBOV-Detect-1 and EBOV-Detect-2 was determined separately by ROC curve analysis and represented as a dotted line on the Y-axis. Specimens with an absorbance greater than Cut-off value are considered EBOV seropositive and those with absorbance less than Cut-off value are considered EBOV seronegative. Percent seropositive are shown for each sample cohort above the panel. Area under the ROC curve was performed to calculate sensitivity (%) and specificity (%) values with 95% CI (confidence interval) by comparing ELISA reactivity for 33 EVD survivors vs. 148 uninfected samples. Statistical differences among groups were analyzed by one-way ANOVA and Tukey-adjusted p values using a pairwise multiple comparison. Statistically significant differences between cohorts are shown. All ELISA experiments were performed twice and the researcher performing the assay was blinded to sample identity. The variation for each sample in duplicate ELISA runs was <6%. The data shown are the average value of two experimental runs. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Serodiagnosis of breakthrough EBOV-infection in the presence of vaccine-induced antibodies by EBOV-Detect
To further evaluate the potential of EBOV-Detect assay to detect breakthrough EBOV infection following EBO vaccination, we evaluated serum reactivity of 11 NHPs either prior to vaccination (Pre-Vac), 1-month after two vaccine doses of rVSV expressing the EBOV-Makona GP (Post-Vacc) and following a month after EBOV-Makona challenge (Post-EBOV)23 against EBOV-GP and EBOV-Detect (Figure 4). Pre-vaccination NHP samples did not react either with EBOV GP or EBOV-Detect. All post-vaccination NHP serum showed moderate to strong reactivity (100% seropositivity) to EBOV-GP, confirming the phenomenon of VISP (Figure 4a). Importantly, negligible reactivity (0% seropositivity) was observed with post-rVSV-Makona-GP vaccinated (Post-Vacc) NHP samples in either EBOV-Detect-1 or EBOV-Detect-2 serodiagnostic assay (Figure 4b-c). In contrast, all the post-EBOV infection samples following EBOV viral challenge (Post-EBOV) reacted positively (100% seropositive) in the EBOV-Detect-1 and EBOV-Detect-2 (Figure 4b-c). The level of antibody binding among the infected animals in EBOV-Detect assay correlated with outcome in this NHP challenge study.23 The strong IgG reactivity of the post-challenge EBOV-infected NHP serum samples but not post-vaccination samples resulting in 100% sensitivity and 100% specificity demonstrated the potential of EBOV-Detect for differential serodiagnosis of true EBOV infection in presence of vaccine-induced antibody response and surveillance during or after outbreaks.
Figure 4.
Immunoreactivity of vaccinated and EBOV-infected NHP serum samples in EBOV-Detect. Serum IgG absorbance values at a 1:100 serum dilution for samples collected prior to vaccination (pre-vac), one month after two doses of rVSV-Makona GP vaccination (post-vacc) or following EBOV challenge (Post-EBOV) from 11 cynomolgus macaque's against EBOV-GP (A) or in EBOV-Detect-1 (B) and EBOV-Detect-2 (C). Mean values for each group are indicated and color coded. Cut-off value for EBOV-Detect-1 and EBOV-Detect-2 was determined separately by ROC curve analysis and represented as a dotted line on the Y-axis. Specimens with an absorbance greater than Cut-off value are considered EBOV seropositive and those with absorbance less than Cut-off value are considered EBOV seronegative. Percent seropositive are shown for each sample cohort above the panel. Area under the ROC curve was performed to calculate sensitivity (%) and specificity (%) values with 95% CI (confidence interval) by comparing ELISA reactivity for post-EBOV infection vs. un-infected (pre-vacc and post-vacc) samples. Statistical differences among groups were analyzed by one-way ANOVA and Tukey-adjusted p values using a pairwise multiple comparison. Statistically significant differences between cohorts are shown. All ELISA experiments were performed twice and the researcher performing the assay was blinded to sample identity. The variation for each sample in duplicate ELISA runs was <7%. The data shown are the average value of two experimental runs. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Discussion
Taken holistically, our data demonstrates that an EBOV infection mounts an IgG response, which recognizes diverse epitopes in multiple EBOV proteins, in addition to EBOV-GP. These immunodominant epitopes can serve as differential serodiagnostic targets for detection of EBOV infection in the presence of vaccine-induced antibodies. Previous studies have used recombinant NP and VP35 antigens in ELISAs, as these proteins were thought to be major determinants of immune responses following infection in humans and primates.25, 26, 27, 28 We have, however, observed high antibody titers against VP35, VP40, VP30 and VP24 up to 2 years post-EBOV exposure8 (and unpublished studies). The ChAd3/MVA and ERVEBO-vaccinated serum samples showed minimal reactivity to individual peptides within these EBOV proteins. While the samples from EBOV survivors showed a statistically significant high IgG reactivity to various individual peptides derived from EBOV proteins excluding EBOV-GP. The use of entire EBOV protein in place of peptides can potentially result in higher background reactivity with serum/plasma from healthy controls as well as post-vaccination individuals leading to more non-specific reactivity. The complete protein may also occlude an immunodominant epitope within the corresponding protein. Conclusively, carefully selected immunodominant peptides provide an advantage over using the entire EBOV protein for serodiagnostic assay.
The sensitivity and specificity of individual peptide ELISA was further improved by testing various combination of VP35 and VP40 derived peptides as mixture. The EBOV-infected samples showed a statistically significant higher response compared with all controls and ChAd3/MVA- or ERVEBO-vaccinated samples in this differential serodiagnosis assay termed EBOV-Detect-1. Both the two peptide-mix (EBOV-Detect-1) and four peptide-mix (EBOV-Detect-2) assays show >95% sensitivity and >94% specificity, similar or superior to other published EBOV serodiagnostic assays.1,4,19,29 The high specificity of EBOV-Detect-1 and EBOV-Detect-2 for vaccinated individuals compared with current GP-based serodiagnostic assays would ensure that very few EBO-vaccinated individuals will be diagnosed false positives. The false-seropositive diagnosis of EBOV vaccinated but uninfected individuals can be further mitigated by testing in a secondary confirmatory assay like a western blot assay, similar to the two-pronged testing approach for the participants in HIV vaccine clinical trials.15, 16, 17, 18 The 100% sensitivity and 100% specificity of the EBOV-Detect-1 (two peptide-mix assay) to diagnose breakthrough EBOV infections in the presence of vaccine-induced antibodies (Figure 4) can be simple to implement in resource-limited settings as a stand-alone assay, rather than the four peptide-mix assay (EBOV-Detect-2). All advanced/licensed vaccines contain GP as the primary antigen, we, therefore, focused on non-GP peptides for development of EBOV-Detect assays. Other vaccine approaches are being evaluated; and in future, if EBO vaccines containing non-GP antigens are used for mass vaccination programs, the EBOV-Detect can be easily modified to replace peptides in either EBOV-Detect-1 or EBOV-Detect-2 while continuing to retain sensitivity to diagnose breakthrough Ebola virus infections. A potential caveat in this study is the low number of the ChAd3/MVA samples due to limited availability of these samples.
To summarize, EBOV-Detect can be used as an effective serodiagnostic assay to differentiate host immune response following natural EBOV infection vs. vaccine-induced seropositivity in GP-based assays induced following ERVEBO or ChAd/MVA-EBOV vaccination, thereby minimizing false positives and diagnosing true breakthrough EBOV infections in the face of mass vaccination campaigns. Hence, this allows for the potential use of EBOV-Detect as a differential assay for detection of true EBOV-infection in a population of vaccinated and unvaccinated individuals using a simple ELISA-based approach that can be performed in resource-limited settings during or after the EBOV outbreak. These simple peptide assays can be further developed into a rapid point-of-care differential serodiagnostic assay like lateral flow, biosensor or lab-on-a-chip assays for detection of EBOV infection.
Contributors
Conceptualisation, funding acquisition and supervision: S.K. Performed assays: S.R. and S.K. verified the underlying data. Contributed to Writing: S.R. and S.K. verified the underlying data. All authors read and approved the final version of the manuscript.
Data sharing statement
All data are present in the manuscript. The materials generated during the current study are available from the corresponding author under a material transfer agreement on reasonable request.
Declaration of interests
The authors have nothing to declare.
Acknowledgements
We thank Hana Golding and Keith Peden at FDA for review of the manuscript. We thank NIBSC and Dr. Miles Caroll (PHE UK) for providing clinical samples from EBOV survivors or ChAd3/MVA prime-boost vaccinated group or healthy controls. We thank Drs. Richard Davey and John Beigel at NIAID, NIH for providing rVSV-ZEBOV (ERVEBO) vaccinated human serum samples and Drs. Andrea Marzi and Heinz Feldmann at NIAID, NIH for sharing samples from the NHP EBOV challenge study. The antibody characterization work described in this manuscript was supported by FDA Office of Counterterrorism and Emerging Threats (OCET) - Medical Countermeasures initiative (MCMi) grant- OCET 2019-1018 and Defense Threat Reduction Agency (HDTRA1930447) funds to S. K. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
References
- 1.Paweska JT, Moolla N, Storm N, et al. Evaluation of diagnostic performance of three indirect enzyme-linked immunosorbent assays for the detection of IgG antibodies to Ebola virus in human sera. Viruses. 2019;11(8):678. doi: 10.3390/v11080678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayouba A, Toure A, Butel C, et al. Development of a sensitive and specific serological assay based on luminex technology for detection of antibodies to Zaire Ebola virus. J Clin Microbiol. 2017;55(1):165–176. doi: 10.1128/JCM.01979-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broadhurst MJ, Brooks TJ, Pollock NR. Diagnosis of Ebola virus disease: past, present, and future. Clin Microbiol Rev. 2016;29(4):773–793. doi: 10.1128/CMR.00003-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukadi-Bamuleka D, Bulabula-Penge J, De Weggheleire A, et al. Field performance of three Ebola rapid diagnostic tests used during the 2018-20 outbreak in the eastern Democratic Republic of the Congo: a retrospective, multicentre observational study. Lancet Infect Dis. 2022 doi: 10.1016/S1473-3099(21)00675-7. [DOI] [PubMed] [Google Scholar]
- 5.Semper AE, Broadhurst MJ, Richards J, et al. Performance of the GeneXpert Ebola assay for diagnosis of Ebola virus disease in Sierra Leone: a field evaluation study. PLoS Med. 2016;13(3) doi: 10.1371/journal.pmed.1001980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Presser LD, Coffin J, Koivogui L, et al. The deployment of mobile diagnostic laboratories for Ebola virus disease diagnostics in Sierra Leone and Guinea. Afr J Lab Med. 2021;10(1):1414. doi: 10.4102/ajlm.v10i1.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erickson BR, Sealy TK, Flietstra T, et al. Ebola virus disease diagnostics, Sierra Leone: analysis of real-time reverse transcription-polymerase chain reaction values for clinical blood and oral swab specimens. J Infect Dis. 2016;214(suppl 3):S258–S262. doi: 10.1093/infdis/jiw296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khurana S, Ravichandran S, Hahn M, et al. Longitudinal human antibody repertoire against complete viral proteome from Ebola virus survivor reveals protective sites for vaccine design. Cell Host Microbe. 2020;27(2):262–276.e4. doi: 10.1016/j.chom.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowe AK, Bertolli J, Khan AS, et al. Clinical, virologic, and immunologic follow-up of convalescent Ebola hemorrhagic fever patients and their household contacts, Kikwit, Democratic Republic of the Congo. Commission de Lutte contre les Epidemies a Kikwit. J Infect Dis. 1999;179(suppl 1):S28–S35. doi: 10.1086/514318. [DOI] [PubMed] [Google Scholar]
- 10.Kasereka MC, Ericson AD, Conroy AL, Tumba L, Mwesha OD, Hawkes MT. Prior vaccination with recombinant Vesicular Stomatitis Virus - Zaire Ebolavirus vaccine is associated with improved survival among patients with Ebolavirus infection. Vaccine. 2020;38(14):3003–3007. doi: 10.1016/j.vaccine.2020.02.044. [DOI] [PubMed] [Google Scholar]
- 11.Mulangu S, Mbala-Kingebeni P, Mbaya OT. Antibody use during an outbreak of Ebola virus disease in the Democratic Republic of Congo, 2020. N Engl J Med. 2022;386(12):1188–1191. doi: 10.1056/NEJMc2113505. [DOI] [PubMed] [Google Scholar]
- 12.Mbala-Kingebeni P, Pratt C, Mutafali-Ruffin M, et al. Ebola virus transmission initiated by relapse of systemic Ebola virus disease. N Engl J Med. 2021;384(13):1240–1247. doi: 10.1056/NEJMoa2024670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulangu S, Dodd LE, Davey RT, Jr., et al. A randomized, controlled trial of Ebola virus disease therapeutics. N Engl J Med. 2019;381(24):2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groen J, van den Hoogen BG, Burghoorn-Maas CP, et al. Serological reactivity of baculovirus-expressed Ebola virus VP35 and nucleoproteins. Microbes Infect. 2003;5(5):379–385. doi: 10.1016/s1286-4579(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 15.Enterprise VWGoGHV. Voronin Y, Zinszner H, et al. HIV vaccine-induced sero-reactivity: a challenge for trial participants, researchers, and physicians. Vaccine. 2015;33(10):1243–1249. doi: 10.1016/j.vaccine.2014.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khurana S, Norris PJ, Busch MP, et al. HIV-Selectest enzyme immunoassay and rapid test: ability to detect seroconversion following HIV-1 infection. J Clin Microbiol. 2010;48(1):281–285. doi: 10.1128/JCM.01573-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khurana S, Needham J, Mathieson B, et al. Human immunodeficiency virus (HIV) vaccine trials: a novel assay for differential diagnosis of HIV infections in the face of vaccine-generated antibodies. J Virol. 2006;80(5):2092–2099. doi: 10.1128/JVI.80.5.2092-2099.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooper CJ, Metch B, Dragavon J, Coombs RW, Baden LR, Force NHVTNV-IST Vaccine-induced HIV seropositivity/reactivity in noninfected HIV vaccine recipients. JAMA. 2010;304(3):275–283. doi: 10.1001/jama.2010.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkinson DE, Page M, Mattiuzzo G, et al. Comparison of platform technologies for assaying antibody to Ebola virus. Vaccine. 2017;35(9):1347–1352. doi: 10.1016/j.vaccine.2016.11.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuentes S, Ravichandran S, Coyle EM, Klenow L, Khurana S. Human antibody repertoire following Ebola virus infection and vaccination. iScience. 2020;23(3) doi: 10.1016/j.isci.2020.100920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khurana S, Fuentes S, Coyle EM, Ravichandran S, Davey RT, Jr., Beigel JH. Human antibody repertoire after VSV-Ebola vaccination identifies novel targets and virus-neutralizing IgM antibodies. Nat Med. 2016;22(12):1439–1447. doi: 10.1038/nm.4201. [DOI] [PubMed] [Google Scholar]
- 22.Verma N, Dimitrova M, Carter DM, et al. Influenza virus H1N1pdm09 infections in the young and old: evidence of greater antibody diversity and affinity for the hemagglutinin globular head domain (HA1 Domain) in the elderly than in young adults and children. J Virol. 2012;86(10):5515–5522. doi: 10.1128/JVI.07085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marzi A, Reynolds P, Mercado-Hernandez R, et al. Single low-dose VSV-EBOV vaccination protects cynomolgus macaques from lethal Ebola challenge. EBioMedicine. 2019;49:223–231. doi: 10.1016/j.ebiom.2019.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem. 1993;39(4):561–577. [PubMed] [Google Scholar]
- 25.Becker S, Feldmann H, Will C, Slenczka W. Evidence for occurrence of filovirus antibodies in humans and imported monkeys: do subclinical filovirus infections occur worldwide? Med Microbiol Immunol. 1992;181(1):43–55. doi: 10.1007/BF00193395. [DOI] [PubMed] [Google Scholar]
- 26.Elliott LH, Sanchez A, Holloway BP, Kiley MP, McCormick JB. Ebola protein analyses for the determination of genetic organization. Arch Virol. 1993;133(3-4):423–436. doi: 10.1007/BF01313780. [DOI] [PubMed] [Google Scholar]
- 27.Lucht A, Grunow R, Moller P, Feldmann H, Becker S. Development, characterization and use of monoclonal VP40-antibodies for the detection of Ebola virus. J Virol Methods. 2003;111(1):21–28. doi: 10.1016/s0166-0934(03)00131-9. [DOI] [PubMed] [Google Scholar]
- 28.Niikura M, Ikegami T, Saijo M, Kurane I, Miranda ME, Morikawa S. Detection of Ebola viral antigen by enzyme-linked immunosorbent assay using a novel monoclonal antibody to nucleoprotein. J Clin Microbiol. 2001;39(9):3267–3271. doi: 10.1128/JCM.39.9.3267-3271.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brangel P, Sobarzo A, Parolo C, et al. A serological point-of-care test for the detection of IgG antibodies against Ebola virus in human survivors. ACS Nano. 2018;12(1):63–73. doi: 10.1021/acsnano.7b07021. [DOI] [PubMed] [Google Scholar]