Abstract
Previous attempts to express the toxin complex genes of Photorhabdus luminescens W14 in Escherichia coli have failed to reconstitute their oral toxicity to the model insect Manduca sexta. Here we show that the combination of three genes, tcdA, tcdB, and tccC, is essential for oral toxicity to M. sexta when expression in E. coli is used. Further, when transcription from native toxin complex gene promoters is used, maximal toxicity in E. coli cultures is associated with the addition of mitomycin C to the growth medium. In contrast, the expression of tcdAB (or the homologous tcaABC operon) with no recombinant tccC homolog in a different P. luminescens strain, K122, is sufficient to confer oral toxicity on this strain, which is otherwise not orally toxic. We therefore infer that P. luminescens K122 carries a functional tccC-like homolog within its own genome, a hypothesis supported by Southern analysis. Recombinant toxins from both P. luminescens K122 and E. coli were purified as high-molecular-weight particulate preparations. Transmission electron micrograph (TEM) images of these particulate preparations showed that the expression of tcdAB (either with or without tccC) in E. coli produces visible ∼25-nm-long complexes with a head and tail-like substructure. These data are consistent with a model whereby TcdAB constitutes the majority of the complex visible under TEM and TccC either is a toxin itself or is an activator of the complex. The implications for the potential mode of action of the toxin complex genes are discussed.
The toxin complex (tc) genes of Photorhabdus luminescens belong to a family of genes found across different members of the Enterobacteriaceae (10). Photorhabdus spp. are bacteria symbiotic with entomopathogenic nematodes of the family Heterorhabditidae, and homologs of the toxin complex genes are also found in a second genus of bacteria, Xenorhabdus, that are symbiotic with nematodes of a different family, Steinernematidae (4, 8). As well as being found in nematode-symbiotic bacteria, toxin complex gene homologs have recently been reported for Serratia entomophila (6). In this species, they are present on a 115-kb plasmid associated with amber disease in the grass grub Costelytra zealandica, where they cause cessation of feeding, clearance of the gut, amber coloration, and eventual insect death (6). We have also located a tca-like operon in the unfinished genome sequence of the human pathogen Yersinia pestis (10), which is the causative agent of bubonic plague (9). However, the role of this operon in either the insect vector (the flea) or the mammalian host remains unknown (10).
P. luminescens strain W14 produces several high-molecular-weight toxin complexes with oral toxicity to insects (2). Two of these, Tca and Tcd, have previously been shown to be responsible for the majority of oral toxicity to the lepidopteran insect Manduca sexta via gene knockout studies (2). Tca also shows characteristic, midgut-specific histopathology in M. sexta (1). Previous attempts to express recombinant toxins by the introduction of tca and tcd plasmids into Escherichia coli have failed (2). In these experiments, although the full-length toxin complex polypeptides were produced, they were not exported from the bacterial cell. In order to define the essential subset of W14 genes required for oral toxicity and to verify conditions enhancing the release of active toxin, we expressed tca, tcd, and tcc gene components in E. coli and in a second strain of P. luminescens, K122, that shows no oral toxicity to M. sexta.
Here we report that the coexpression of tccC with the tcdAB operon is sufficient for oral toxicity in E. coli XL1Blue. In contrast, the expression of either tcdAB or the homologous tcaABC operon alone is sufficient to confer oral toxicity on P. luminescens K122 in the absence of W14-derived tccC expression. Maximal oral toxicity in E. coli also requires the addition of mitomycin C to the culture medium. Particulates of high molecular weight purified from either recombinant bacterial species were toxic when fed to larvae of M. sexta. Examination of these toxin preparations by transmission electron microscopy (TEM) showed that tcdAB encodes a visually distinct complex with a head and tail-like substructure. These data are discussed in relation to a model whereby tcdAB encodes the visually distinct complex, whose synthesis and/or release is stimulated by mitomycin C, and tccC encodes either an actual toxin or an activator of the complex.
MATERIALS AND METHODS
Bacterial strains and plasmids.
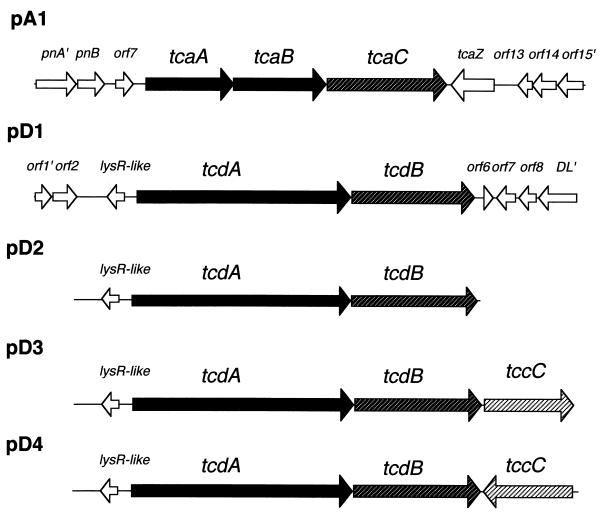
P. luminescens strains W14 (3) and K122 (isolated by D. Clarke from a nematode collected in the Republic of Ireland) were cultured at 30°C with aeration in 2% Proteose Peptone 3 (PP3) and Luria-Bertani (LB) broth, respectively. When necessary, agar powder was added to 1.5% (wt/vol). The five plasmids used in this study are diagrammed in Fig. 1. Plasmid pA1 represents a portion of the tca locus containing the tcaA, tcaB, and tcaC genes. Plasmid pD1 contains the equivalent genes, tcdA and tcdB, from the tcd locus. The three remaining plasmids represent variants of the tcd locus either with the lysR-like regulator and promoter region alone upstream of tcdA (pD2) or with different orientations of tccC downstream (pD3 and pD4). Note that, with the exception of the tccC gene in pD4, which is transcribed by the pBR322 tet gene promoter, transcription in these plasmids relies upon native Photorhabdus promoters.
FIG. 1.
Diagram of toxin complex genes present in the recombinant plasmids used in this study. These are derived from either the tca (pA) or the tcd (pD) locus of P. luminescens W14. Plasmids pA1 and pD1 are Sau3A restriction fragments cloned into the BamHI site of Bluescript pBCKS(+). Plasmid pD2 is an SphI-BamHI fragment of pD1 cloned into pBR322. Plasmids pD3 and pD4 represent the pD2 backbone with a PCR-generated copy of tccC cloned into the BamHI site. In pD4, tccC is transcribed by the tetracycline resistance gene promoter on pBR322.
Plasmids were transformed into P. luminescens K122 using electroporation. For antibiotic selection, 15 μg of chloramphenicol/ml was added to the growth medium to select for plasmids pA1 and pD1, while 100 μg of ampicillin/ml was used for pD2, pD3, and pD4. Briefly, a fresh culture of P. luminescens K122 was grown overnight. Cells were harvested by centrifugation and washed in an equivalent volume of ice-cold sterile water. Cells were washed twice again in equivalent volumes of ice-cold 10% glycerol before final resuspension in 1/200 the original volume of 10% glycerol. Concentrated plasmid DNA prepared from E. coli by the Qiagen miniprep kit protocol was added to 50 μl of cells in a 0.2-cm electroporation cuvette. After a 2.1-kV pulse (100 Ω of resistance), the cells were resuspended in ice-cold LB medium, incubated at 30°C for 1 h, and then plated on LB agar supplemented with the relevant antibiotic. Plates were incubated for 2 days at 30°C.
Oral bioassays.
For bioassays, 100 μl of either bacterial culture supernatants or toxin preparations was applied to 1-cm3 disks of artificial wheat germ diet. The treatment was allowed to soak into the food block and dry for approximately 20 min under laminar flow. Three first-instar M. sexta neonate larvae were then placed on each food block before incubation at 25°C for 7 days. After this time, the percent mortality of larvae and the weight of surviving larvae were recorded. Oral toxicity was assessed as the relative weight gain of animals on the treated diet in comparison to control animals fed nonrecombinant K122 or E. coli supernatants. Due to difficulties in assessing and comparing the levels of protein expression of the different toxin complex components in the various strains, supernatants were compared using only the relative toxicity of standardized dilutions. The effective supernatant concentration which reduced relative weight gain by 50% (EC50) was calculated using nonlinear regression analysis, and t tests of EC50s were performed to test for significant differences.
Particulate toxin preparations.
Prior to either DEAE-Sepharose or CsCl gradient purification of recombinant toxins, crude preparations of particulate material were made. Briefly, 300 ml of a 3-day-old stationary-phase Photorhabdus culture (grown at 30°C) in either LB medium or 2% PP3 medium was centrifuged at 8,000 rpm in a GS3 rotor for 30 min at 4°C. The supernatant was decanted to remove the cell pellet, and the centrifugation was repeated to remove any remaining cells. The cell-free supernatant was centrifuged at 150,000 × g for 90 min at 4°C in a Sorvall ultracentrifuge to harvest particulate material. The supernatant was discarded, and the pellet was washed by gentle resuspension in 50 ml of 50 mM HEPES buffer (pH 6.8) before a second centrifugation at 150,000 × g for 90 min at 4°C to pellet particulate material. The pellet was finally resuspended in 600 μl of ice-cold phosphate-buffered saline (PBS) and stored at 4°C.
DEAE-Sepharose purification.
As a second step in recombinant toxin purification, the particulate preparations were further separated by DEAE-Sepharose chromatography. For DEAE-Sepharose chromatography, the particulate material recovered after the first high-speed centrifugation described above was resuspended in 10 ml of ice-cold PBS, and an equivalent volume of DEAE–Sepharose CL-6B anion exchanger (in PBS) was added. This mixture was incubated at room temperature for 15 min. The Sepharose resin was harvested by low-speed centrifugation (3,000 × g), and the supernatant was discarded. The resin was resuspended in 40 ml of ice-cold PBS and again harvested by centrifugation. This wash was repeated three more times, and the resin finally was resuspended in 10 ml of elution buffer (0.5 M NaCl, 50 mM phosphate buffer [pH 7.4]). The resin was removed by centrifugation, and the supernatant containing toxins was centrifuged at 150,000 × g for 90 min at 4°C to pellet particulate material. Particulate material was finally resuspended in 600 μl of ice-cold PBS and stored at 4°C.
CsCl density gradient column purification.
Particulate material containing the toxins prepared using the DEAE-Sepharose method was further fractionated on a CsCl density gradient. A 300-μl portion of the sample was brought to a volume of 5.4 ml at a density of 1 g of CsCl/ml and centrifuged for 24 h at 14°C in a Beckman VTi65 rotor. Fractions of approximately 200 μl each were sequentially drawn off the bottom of the column and examined by sodium dodecyl sulfate (SDS)–10% polyacrylamide gel electrophoresis (PAGE) for protein content. Fractions containing the same protein profiles were pooled and dialyzed against 5 liters of PBS at 4°C.
Native gel electrophoresis.
Samples from particulate preparations were brought to 30 μl with 50 mM Tris-HCl buffer (pH 7). Native gel electrophoresis was performed as described elsewhere (3) with modifications. Briefly, samples were separated by electrophoresis in an agarose gel consisting of a 5-cm-long 1.5% (wt/vol) agarose “stacking” region in 100 mM Tris-HCl (pH 7) and a 15-cm-long 1.9% agarose “resolving” region in 200 mM Tris-borate (pH 8.3). The anode and cathode buffers consisted of 1 M Tris-HCl (pH 8.3) and 0.025 M Tris–0.192 M glycine (pH 8.3), respectively. The gel was run for 4 to 5 h at 120 mA on ice. Duplicate lanes were run on either side of the gel. Half was then cut off and stained with Coomassie brilliant blue to visualize the positions of the proteins. The unstained half of the gel was then laid on top of this portion in order to localize and excise the unstained native protein complexes from the gel. Protein was electroeluted from these excised agar blocks by electrophoresis inside dialysis membranes suspended in Tris-borate-EDTA running buffer (on ice). Electroeluted protein was concentrated by phenol precipitation for visualization by SDS–10% PAGE and Coomassie brilliant blue staining.
Electron microscopy.
For TEM, pioloform-covered 300-mesh copper grids, coated with a fine layer of carbon, were used as substrates for the protein fractions in the microscope. Four different aqueous negative stains were tested with the protein samples: 1% uranyl acetate, 3% ammonium molybdate, 3% methylamine tungstate, and 2% sodium silicotungstate. The preferred stain, 3% methylamine tungstate, produced acceptable contrast and minimum artifacts and was subsequently used for all samples viewed in the microscope.
The coated grids were exposed to UV light for 16 h immediately prior to use to ensure adequate wetting of the substrate. A 10-μl drop of sample was applied to the TEM grid, and the protein was allowed to settle for 5 min. The liquid was absorbed with filter paper from the edge of the grid and substituted immediately with 10 μl of filtered negative stain solution. This drop was partially removed with filter paper, and the grid was allowed to air dry thoroughly before viewingwas done with a Jeol (Tokyo, Japan) 1200EX transmission electron microscope operating at 80 kV.
RESULTS
Expression of toxin genes in E. coli.
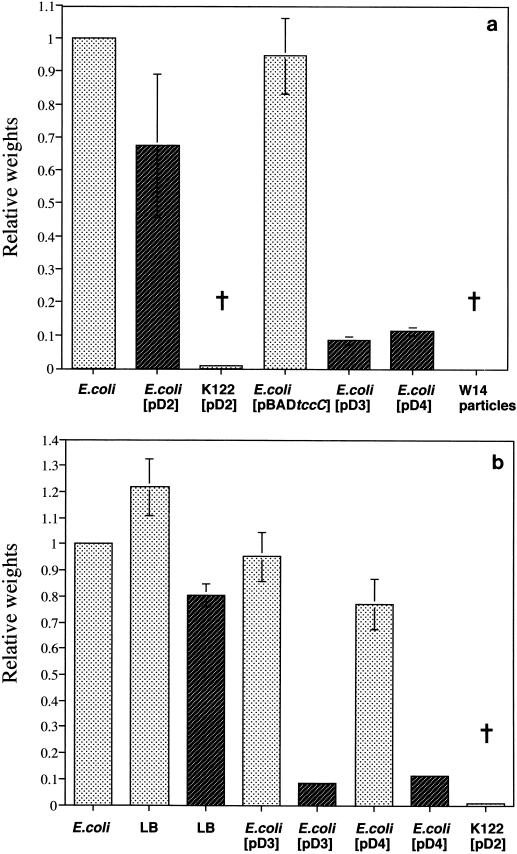
The expression of different combinations of genes from the P. luminescens W14 tcd and tcc loci shows that two factors are necessary and sufficient for high levels of oral toxicity to M. sexta. First, coexpression of tccC with tcdAB (plasmid pD3 or pD4) in E. coli is necessary to confer oral toxicity, while the expression of tcdAB alone (plasmid pD2) is not sufficient (Fig. 2a). Second, in addition to the coexpression of all three genes, to achieve full toxicity, mitomycin C must be added to the growth medium (Fig. 2b); alternatively, in its absence, cultures must be allowed to stand for more than 48 h for toxicity to become apparent. The expression of tccC alone in the arabinose-inducible expression plasmid pBAD30 (5) was insufficient to confer oral toxicity.
FIG. 2.
Effect of the presence of TccC and mitomycin C induction on the oral toxicity of recombinant P. luminescens K122 and E. coli. (a) Histogram showing relative weight gain (mean and standard error for six larvae per treatment) of M. sexta fed on a diet treated with whole cultures of strains containing plasmids with or without tccC. Note that plasmids containing tccC alone [E. coli(pBADtccC)] do not inhibit weight gain, whereas plasmids carrying tcdAB in P. luminescens K122 [K122(pD2)] or tcdAB and tccC in E. coli [E. coli(pD3) and E. coli(pD4)] inhibit growth to 10% or less of the control weight. (b) Relative weight gain of larvae fed on supernatants grown in the presence (dark shading) or absence (light shading) of mitomycin C. Note that although some toxicity can be observed without mitomycin C [e.g., the 20% reduction seen with E. coli(pD4)], toxicity is substantially increased (∼90% weight reduction) by the addition of mitomycin C to cultures containing plasmids coexpressing both tcdAB and tccC [E. coli(pD3) and E. coli(pD4)]. The dagger represents a sample in which the mortality of the insects was 100%.
Interestingly, low levels of oral toxicity to M. sexta are seen in exponentially growing cultures but are putatively cell associated (i.e., absent from the growth medium). Further, oral toxicity is not increased via physical lysis (sonication) of cells from these cultures. The fact that purified Tcd particles are still orally toxic after sonication (data not shown) suggests that release from the cytoplasm is not limiting toxicity in these cultures. One possible explanation for these observations is that the toxin complexes are surface associated and that either aging of the culture or the addition of mitomycin C promotes their detachment (either directly or via disruption of the cell membranes).
Expression of toxin genes in P. luminescens K122.
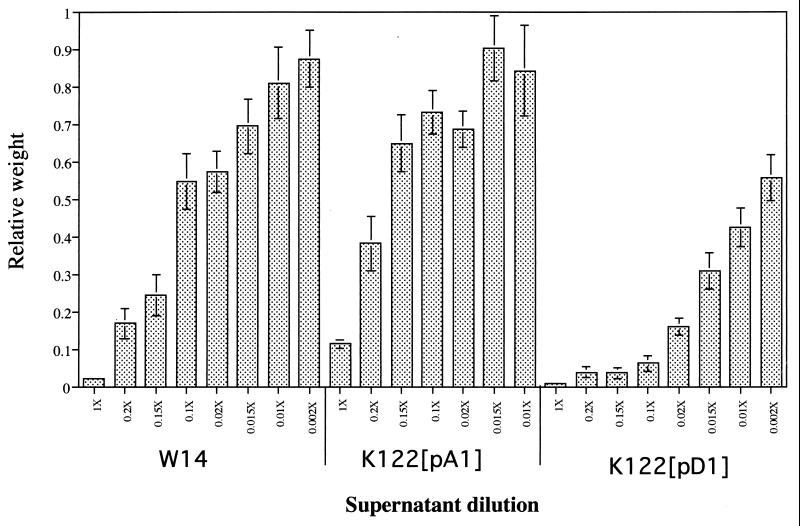
Both the tcaABC and the tcdAB expression plasmids (pA1 and pD1, respectively) conferred the oral toxicity of the supernatants on recombinant K122, with relative weight gain EC50s of 0.23- and 0.003-fold, respectively (Fig. 3). Notably, the EC50 for K122 expressing tcdAB (pD1) was significantly lower (P > 0.001) than that for the original strain, W14, itself (0.003- versus 0.05-fold). Further, supernatants from K122 expressing tcdAB (pD1) were 38.6 times more potent than the equivalent supernatants from K122 expressing tcaABC (pA1). These comparisons do not take into account the specific amount of toxin complex protein produced by each recombinant strain; however, they do illustrate that it is possible to make recombinant P. luminescens strains which are more orally toxic than the original P. luminescens strain, W14, itself.
FIG. 3.
Relative weight gain of M. sexta larvae fed on different dilutions (1× to 0.002×) of P. luminescens supernatants. Strains used were orally toxic P. luminescens W14 and P. luminescens K122 (lacking oral toxicity) expressing recombinant Tca or Tcd from the W14-derived recombinant plasmid pA1 or pD1 (Fig. 1), respectively. Note that the expression of Tcd alone in strain K122 produces a more orally toxic supernatant than does the expression of strain W14 (which contains a mixture of Tca and Tcd). Weights (mean and standard error for nine larvae) are relative to the mean weight for larvae fed on a supernatant from nonrecombinant K122 (which is set at 1.0). The means and standard errors for three independent experiments are shown. The estimated EC50s are as follows: W14, 0.05×; Tca, 0.23×; and Tcd, 0.003× (see Discussion).
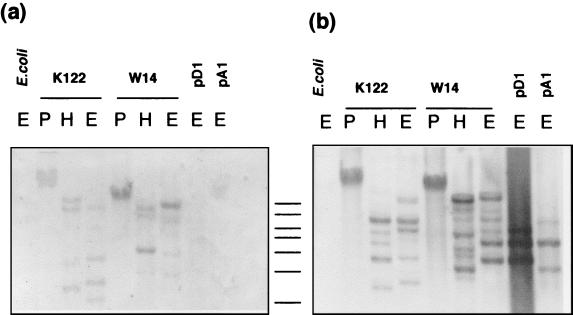
In contrast to the results obtained in the experiments with E. coli, both pA1 and pD1 lack a copy of tccC but can still confer oral toxicity on P. luminescens strain K122. This result suggests that a functional tccC homolog may be present already in P. luminescens strain K122. Support for this hypothesis was obtained by Southern analysis of K122 DNA with a tccC probe derived from W14 by PCR. This blot reveals the presence of several tccC-like sequences (Fig. 4a), supporting the hypothesis that P. luminescens K122 indeed carries tccC-like homologs. A similar Southern analysis also provides evidence for P. luminescens K122 carrying tcdA- and/or tcdB-like genes as well (Fig. 4b). These results infer that, in these experiments, oral toxicity is conferred on P. luminescens K122 specifically by the tcdA and tcdB genes from strain W14 but that the tccC-like gene function is provided by the genetic background of K122 itself.
FIG. 4.
Southern analysis supports the hypothesis that P. luminescens K122 carries homologs of W14 tccC-like and tcdA-like genes. The blots were probed with the highly conserved core region of the tccC gene (10) (a) and the P. luminescens W14 tcdAB operon (b). Probes were derived by PCR from the W14 genome. The DNA was digested and electrophoresed on 0.7% agarose gels and blotted using standard techniques. Probe hybridization was visualized using the Boehringer digoxigenin system. The locations of 1-kb marker DNA fragments are shown between the two gels (from top to bottom, 10, 8, 6, 5, 4, 3, and 2 kb). Digests were as follows: E, EcoRI; P, PstI; and H, HindIII.
Purification of recombinant toxins.
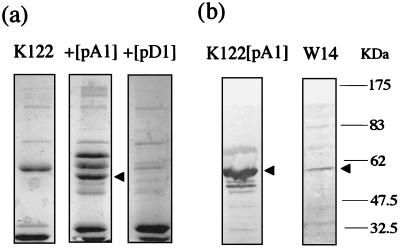
In order to determine the particulate nature of the recombinant toxin complexes and as a prelude to electron microscopy, we enriched the toxin complexes via DEAE column chromatography. SDS-PAGE analysis of these particulate preparations was then used to confirm the presence of novel polypeptides associated with the recombinant toxin complexes. Particulate preparations of host strain P. luminescens K122 show two predominant species, of ∼30 and 60 kDa. Expression of either the tcaABC or the tcdAB operon results in the appearance of additional protein bands putatively corresponding to cleaved toxin complex fragments (Fig. 5a). The identities of these recombinant fragments were confirmed by Western blotting with anti-TcdA and anti-Tca antibodies (Fig. 5b). Note that the cleaved Tca fragment highlighted by the anti-Tca antibody is the same as that identified in the particulate preparation from P. luminescens W14.
FIG. 5.
(a) Denaturing gel electrophoresis (SDS-PAGE) of particulate preparations from P. luminescens wild-type K122 and K122 expressing recombinant Tca (pA1) and Tcd (pD1). Note that expression from the pA1 and pD1 plasmids leads to the production of additional protein species, presumably toxin components. (b) Western analysis of P. luminescens K122 expressing recombinant Tca with an anti-Tca antibody. Note the detection of an ∼60-kDa cross-reacting species (arrowhead), putatively TcaBii. Note also that the antibody recognizes the same species in a preparation from P. luminescens W14.
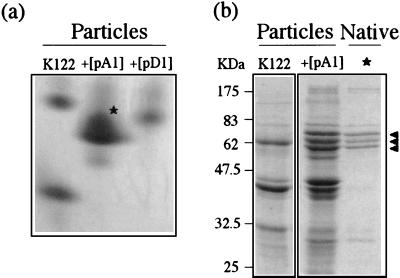
To further examine the compositions of the recombinant toxin complexes, we used native nondenaturing gel electrophoresis. Native gel analysis of particulate preparations from host strain K122 resolves two complexes. However, recombinant expression of either tcaABC or tcdAB causes the resulting particulate preparation to migrate as a single native complex with a mobility different from that seen in the K122 parent strain (Fig. 6a). This result suggests that heterologous expression of tca and tcd genes from strain W14 produces gene products that coassemble with complexes from host strain K122 or that alter the synthesis of certain native K122 polypeptides. Subsequent analysis of these complexes by denaturing SDS-PAGE confirmed the composition of each native complex. Excision of the predominant native band associated with recombinant tca expression (Fig. 6a), electroelution, and reanalysis by SDS-PAGE (Fig. 6b) confirmed that it consists of a specific subset of Tca-derived polypeptides clustered at ∼60 kDa (compare Fig. 5 with Fig. 6). It was not possible to determine exactly which toxin complex polypeptide subfragments are represented without N-terminal protein sequencing of each fragment.
FIG. 6.
(a) Native gel of particulate preparations from P. luminescens K122 and K122 expressing recombinant Tca (pA1) and Tcd (pD1). Note that preparations from the acceptor strain P. luminescens K122 migrate as two clearly separable complexes, whereas coexpression of W14 Tca or Tcd results in a single native complex being produced by recombinant P. luminescens K122. The asterisk represents the recombinant Tca complex excised from the gel and reanalyzed by SDS-PAGE. (b) Denaturing gel electrophoresis (SDS-PAGE) of the same K122 and K122-Tca preparations as those analyzed in panel a. Note that the native Tca complex excised from the gel in panel a contains three predominant polypeptides (arrowheads). Note also that this native complex forms visible particles under TEM examination (see Fig. 8d).
SDS-PAGE analysis of recombinant Tcd particulate preparations from E. coli shows that mature Tcd components are more abundant in recombinant E. coli than in P. luminescens K122. Thus, large polypeptides, presumably corresponding to unprocessed TcdA and TcdB, can clearly be seen in recombinant E. coli preparations (Fig. 7) but are not visible in similar preparations from host strain P. luminescens K122 (Fig. 5a). Importantly, TcdA and TcdB appear to be produced at similar levels either in the presence (pD4) or in the absence (pD2) of TccC. However, as noted above (Fig. 2b), the tcdA and tcdB gene products become orally toxic only after coexpression with tccC. These findings confirm that TcdA and TcdB are necessary but not sufficient for toxicity and that TccC is required to confer full oral toxicity to M. sexta.
FIG. 7.
Denaturing gel electrophoresis (SDS-PAGE) of particulate preparations from E. coli expressing recombinant TcdAB from three plasmids, pD2, pD3, and pD4. Species corresponding to TcdA and TcdB are indicated by arrowheads.
Electron microscopy.
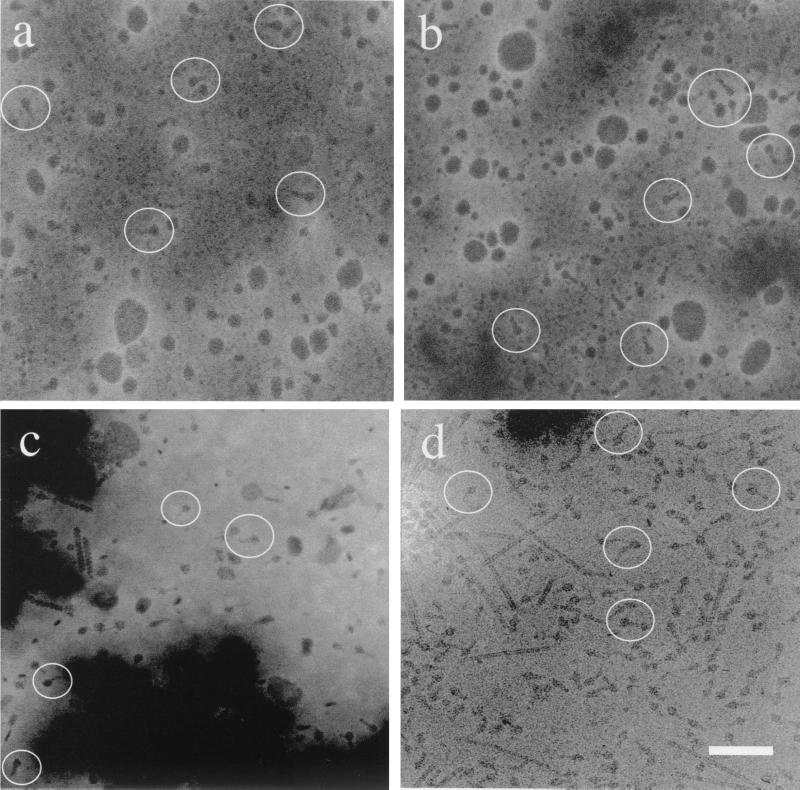
To identify the supramolecular structures associated with native and recombinant toxin complexes, we performed TEM on particulate preparations from both recombinant E. coli and P. luminescens K122 containing toxin complex gene-carrying plasmids and also from the original P. luminescens strain, W14. Particulate preparations from E. coli carrying tcd-containing plasmids show the presence of distinct supramolecular complexes (Fig. 8a and b), which are absent from control preparations of E. coli lacking the recombinant plasmids. These complexes are ∼25 nm long and have a distinct head and protruding tail-like substructure. Similar structures are also present in particulate preparations from P. luminescens strain W14 (Fig. 8c) and in preparations from recombinant P. luminescens strain K122 (Fig. 8d). These complexes are visible both in strains expressing tcdAB (Fig. 8a) and in strains coexpressing tcdAB together with tccC (Fig. 8b). These results suggest that only the tcdA and tcdB gene products are necessary to form the visibly distinct complexes. Further, coexpression of tccC does not change the visible structure of the complexes (at this resolution) but does confer on recombinant bacterial strains expressing all three genes oral toxicity to M. sexta. These data suggest the alternative hypotheses that either TccC activates the TcdA-TcdB complex or TccC is itself a toxic component required for high levels of oral toxicity to M. sexta.
FIG. 8.
Transmission electron micrographs of particulate preparations from wild-type and recombinant strains of E. coli and P. luminescens showing the supramolecular structure of the toxin complexes. (a) E. coli expressing TcdAB (pD2). Note the presence of visible complexes (circled); however, these preparations are not orally toxic (Fig. 2a). (b) E. coli coexpressing TcdAB and TccC. Note the presence of the same visible particles, which are now orally toxic (Fig. 2a). (c) Preparation from wild-type P. luminescens W14 showing visible toxin complexes. (d) Native gel preparation (asterisk in Fig. 6a) from K122 expressing TcaABC, again showing the same particles. Bar, 50 nm. These data show that TcdA and TcdB together are sufficient to produce visible particles but that coexpression of TccC is essential for oral toxicity to M. sexta (see Discussion).
DISCUSSION
Three genes are required for oral toxicity.
High levels of recombinant Tcd-based oral toxicity to M. sexta require the presence of three distinct genetic elements, i.e., TcdA, TcdB, and TccC. Further, the data presented here suggest that tccC itself is an essential part of this high-level oral toxicity. We note that in the plasmids used here, the copy of tccC was derived from the original (first-described) tcc locus. However, it is important to realize that our recent extended sequencing of the original four toxin complex loci shows that further tccC-like sequences are also present both upstream and downstream of the original tcd locus (10). This result suggests that physical linkage in the genome between tcdAB genes and tccC-like elements is not necessary in order for them to confer oral toxicity when cloned together on the same recombinant plasmid. This hypothesis is supported by our recent isolation of a P. luminescens W14 cosmid that encompasses tcdA, tcdB, and one of the neighboring tccC-like elements and that confers oral toxicity on the E. coli strain that carries it (unpublished results).
The suggestion that three genetic elements are necessary for high levels of oral toxicity to insects is consistent with the work of others on toxin complex gene homologs in the free-living bacterium S. entomophila. In this species, all three of the equivalent genes, sepA, sepB, and sepC, are necessary and sufficient to confer the amber disease phenotype (which includes attack of the insect gut) in the New Zealand grass grub on recombinant E. coli (6). Further, Morgan and others, working with further toxin complex gene homologs from the nematode symbiont Xenorhabdus nematophilus, recently indicated that high-level oral toxicity of recombinant E. coli also requires tcdA, tcdB, and tccC homologs (xptA1, xptB1, and xptC1) (8). Interestingly, however, these workers also reported that high-level expression of the tcdA homolog alone can confer lower levels of oral toxicity on E. coli. In our experiments, however, plasmids encoding P. luminescens W14 tcdA but lacking tcdB could not be recovered due to consistent rearrangements in the resulting plasmid constructs. To date, we have therefore been unable to test the level of oral toxicity to M. sexta associated with recombinant expression of tcdA alone.
When either the tcdAB or the tcaABC operon was expressed in a different P. luminescens host, strain K122, the presence of W14 tccC was not required for oral toxicity. This result suggests that a tccC-like homolog already present in P. luminescens K122 is able to confer oral toxicity on recombinant W14 TcdAB. This hypothesis is supported by Southern analysis, which shows that K122 carries tcdA- and/or tcdB-like sequences (Fig. 4b). If, as proposed, P. luminescens K122 therefore carries both tcd- and tccC-like gene homologs, either they are not expressed in this strain under the conditions tested or they do not confer oral toxicity to M. sexta. As oral toxicity can be conferred on P. luminescens K122 simply by the addition of either the tcdAB or the tcaABC operon from strain W14, these genes may themselves encode the factors essential for lepidopteran gut toxicity. Further, these data also raise the formal possibility that toxin complex homologs in other P. luminescens strains that (like strain K122) show no observable oral toxicity to M. sexta may not be effective on the lepidopteran gut.
Finally, the observations presented here also have potential implications for the putative mechanism(s) of secretion (and/or display) of the toxin complex proteins from bacteria. First, oral toxicity of the P. luminescens W14 supernatant is seen only in stationary phase when the bacteria are cultured in vitro (3). Second, and similarly, the oral toxicity of Tcd-producing E. coli supernatants increases either with the age of the bacterial culture or with the addition of mitomycin C. One potential explanation for these observations is that recombinant toxin complexes are cell associated and are released into the culture supernatant only when mitomycin C is added (presumably resulting in increased cell lysis and associated membrane disruption). As the total level of toxicity (combined cell- and supernatant-associated oral toxicity) also increases with the addition of mitomycin C, this compound also may stimulate increased Tcd production via stress-induced activation of the tcd promoter itself. At this stage, it is still not clear how such large toxin complexes are exported from the bacterial cell without any obvious secretion device. However, the hypotheses presented above can be tested directly and should allow researchers to gain an understanding of the relative contributions of bacterial cell lysis and culture stress in the production and release of toxin complex proteins.
In summary, here we have shown via TEM analysis that both recombinant TcdAB complexes from E. coli and native complexes (of unknown composition) from P. luminescens W14 are ∼25 nm long and have a distinct head and tail-like substructure. Coexpression of TccC with TcdAB does not change the visual nature of the complex in TEM analysis but is required for full levels of oral toxicity to our model insect, M. sexta. At present, we are uncertain about the significance of either the structure of the toxin complexes or the molecular role of the genetic component tccC. However, we note two salient features that may form the basis for further investigation. First, the observation of phage-related holin-lysin genes within some toxin complex loci (10) raises the possibility that they are released from the bacteria via cell lysis. Second, although the mode of action of the toxin complex proteins remains obscure, we note that the two TccC homologs presently identified for Y. pestis both contain putative active sites for protein tyrosine phosphatases (10), enzymes that are absent from bacteria and that are typically associated with bacterial effectors, such as SptP (7).
ACKNOWLEDGMENTS
This work was supported by a grant to R.F.-C. from the BBSRC. Nucleotide sequencing was performed by P. Wilkinson with an ABI3700 sequencer supported by a Wellcome/JIF grant to R.F.-C.
We thank D. Bowen and T. Rocheleau, formerly in the laboratory of R.F.-C. at the University of Wisconsin—Madison, for providing plasmids.
REFERENCES
- 1.Blackburn M, Golubeva E, Bowen D, ffrench-Constant R H. A novel insecticidal toxin from Photorhabdus luminescens: histopathological effects of toxin complex A (Tca) on the midgut of Manduca sexta. Appl Environ Microbiol. 1998;64:3036–3041. doi: 10.1128/aem.64.8.3036-3041.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowen D, Rocheleau T A, Blackburn M, Andreev O, Golubeva E, Bhartia R, ffrench-Constant R H. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science. 1998;280:2129–2132. doi: 10.1126/science.280.5372.2129. [DOI] [PubMed] [Google Scholar]
- 3.Bowen D J, Ensign J C. Purification and characterization of a high-molecular-weight insecticidal protein complex produced by the entomopathogenic bacterium Photorhabdus luminescens. Appl Environ Microbiol. 1998;64:3029–3035. doi: 10.1128/aem.64.8.3029-3035.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.ffrench-Constant R, Bowen D. Photorhabdus toxins: novel biological insecticides. Curr Opin Microbiol. 1999;2:284–288. doi: 10.1016/s1369-5274(99)80049-6. [DOI] [PubMed] [Google Scholar]
- 5.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurst M R, Glare T R, Jackson T A, Ronson C W. Plasmid-located pathogenicity determinants of Serratia entomophila, the causal agent of amber disease of grass grub, show similarity to the insecticidal toxins of Photorhabdus luminescens. J Bacteriol. 2000;182:5127–5138. doi: 10.1128/jb.182.18.5127-5138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaniga K, Uralil J, Bliska J B, Galan J E. A secreted protein tyrosine phosphatase with modular effector domains in the bacterial pathogen Salmonella typhimurium. Mol Microbiol. 1996;21:633–641. doi: 10.1111/j.1365-2958.1996.tb02571.x. [DOI] [PubMed] [Google Scholar]
- 8.Morgan J A, Sergeant M, Ellis D, Ousley M, Jarrett P. Sequence analysis of insecticidal genes from Xenorhabdus nematophilus PMFI296. Appl Environ Microbiol. 2001;67:2062–2069. doi: 10.1128/AEM.67.5.2062-2069.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perry R D, Fetherston J D. Yersinia pestis—etiologic agent of plague. Clin Microbiol Rev. 1997;10:35–66. doi: 10.1128/cmr.10.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waterfield N R, Bowen D J, Fetherston J D, Perry R D, ffrench-Constant R H. The toxin complex genes of Photorhabdus: a growing gene family. Trends Microbiol. 2001;9:185–191. doi: 10.1016/s0966-842x(01)01978-3. [DOI] [PubMed] [Google Scholar]