Abstract
During sporulation, many Bacillus thuringiensis subspecies synthesize several related δ-endotoxins which are packaged into bipyramidal intracellular inclusions. These inclusions are solubilized in the alkaline, reducing conditions of the midguts of susceptible insect larvae and are converted by proteolysis to active toxins. The toxins insert into the membranes of cells lining the midgut and form cation-selective channels, which results in lethality. There are three δ-endotoxins, Cry1Ab3, Cry1Ca1, and Cry1Da1, present in the inclusions produced by a B. thuringiensis subsp. aizawai cell. While the ratio of the steady-state mRNAs for these three protoxins has been shown to differ (cry1Ab3/cry1Ca1/cry1Da1 mRNA ratio, 4:2:1), the half-lives of the cry1Da1 and cry1Ab3 mRNAs were found to be similar, indicating that there were differences in the transcription rates. The relative contents of these δ-endotoxins in purified inclusions from B. thuringiensis subsp. aizawai have been measured previously, and an even greater relative deficiency of the Cry1Da1 protoxin (ratio, 20:12:1) was found. In order to account for this deficiency, other steps which could be involved in inclusion formation, such as translation and packaging, were examined. The three cry genes have the same dual overlapping promoters, but the ribosome binding sequence for the cry1Da1 gene was not the consensus sequence. Translation was enhanced about fourfold by changing to the consensus sequence. In addition, the relative amount of Cry1Da1 protoxin in inclusions was twofold lower when cells were sporulated in Luria-Bertani (LB) medium than when cells were sporulated in a glucose-yeast extract medium. This difference was attributable to packaging since the relative amounts of Cry1Da1 antigen in cells sporulating in the two media were the same. Some factor(s) required for packaging of the Cry1Da1 protoxin in inclusions is apparently limiting in LB medium. Differences in the initial transcription rates, translation efficiencies, and packaging all contribute to the δ-endotoxin composition of an inclusion.
A large number of Bacillus thuringiensis subspecies contain several plasmid-encoded δ-endotoxin genes (cry genes) that lead to production of intracellular inclusions, each of which is comprised of related δ-endotoxins (2, 24). These proteins are synthesized as protoxins, and the members of a major class, designated Cry1 (molecular mass, about 130 kDa), contain 15 to 17 cysteine residues in their carboxyl halves (18). Apparently, all of these cysteines form intermolecular disulfide bonds when they are packaged in inclusions (11). For example, B. thuringiensis subsp. aizawai HD133 contains the cry1Ca1 and cry1Da1 genes in close proximity on a 120-MDa plasmid plus the cry1Ab3 gene on a 45-MDa plasmid (6). The protoxins encoded by these three cry genes have very similar sequences in their carboxyl halves, the portions of the protoxins involved in cross-linking. They differ from each other in specific regions in their amino halves and thus have unique but overlapping specificities for target insects (18).
When an inclusion is ingested by a susceptible insect larva (certain Lepidoptera in this case), it is solubilized in the alkaline, reducing conditions of the larval midgut. Trypsin-like enzymes remove the carboxyl halves of the protoxins, releasing ca. 60-kDa toxins which have a well-conserved three-domain structure (24). The toxins bind reversibly but with high affinity to receptors on cells lining the larval midgut. This binding is rather specific and is attributable to certain toxin-specific sequences in loops connecting the β sheets in domains II and III (24). Subsequently, there is an irreversible binding step involving a close association of most of the toxin with the membrane and insertion of certain amphipathic α-helices in domain I (7). The toxin molecules aggregate (either before or after insertion) and form cation-selective channels that result in osmotic lysis of the cells and larval death (20).
It has been known for some time that both the overall toxicity and the specificity profile of a particular B. thuringiensis subspecies for target insects vary substantially depending on the medium used for growth and sporulation (14). The medium-dependent differences, especially the specificity profile differences, imply that there is regulation of the amount of each of the protoxins produced. In support of this possibility, differences in the steady-state levels of mRNAs for the cry1Ab3, cry1Ca1, and cry1Da1 genes in B. thuringiensis subsp. aizawai HD133 were determined by measuring each level relative to the gene content (4). While the gene content did differ, probably due to differences in plasmid copy number, the steady-state mRNA levels relative to the gene content were about 1.00:0.50:0.25.
In a subsequent study, the δ-endotoxin contents of purified inclusions from this organism were compared by quantifying unique peptides produced by trypsin digestion of the proteins solubilized from purified inclusions (22). A ratio of 1.00:0.60:0.05 was obtained, which was substantially different from the steady-state mRNA ratio, especially in terms of the relatively low recovery of the Cry1Da1 protoxin. One difference in these experiments, aside from the fact that δ-endotoxin-specific peptides were measured instead of steady-state mRNAs, was the media used for growth and sporulation.
In order to resolve the apparent discrepancy, the relative contributions of transcription, translation, and packaging to the δ-endotoxin content of an inclusion were determined. Differences in the initial transcription rates of the genes were established by measuring mRNA half-lives. A lower rate of translation of the cry1Da1 mRNA due to a suboptimal ribosome binding sequence was also found. In addition, a medium-dependent difference in packaging of the Cry1Da1 δ-endotoxin in inclusions contributed to the relatively low level of this protoxin. The latter finding was unexpected and indicates that a cellular component(s) involved in packaging can be limiting, which influences the protoxin content of an inclusion and thus the toxicity profile of a B. thuringiensis subspecies.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. thuringiensis subsp. aizawai HD133 and HD68 were obtained from the H. Dulmage culture collection maintained by L. Nakamura at the USDA Northern Regional Research Center, Peoria, Ill. Strain #5 is a derivative of HD133 spontaneously cured of the 45-MDa plasmid containing the cry1Ab3 gene (6). Strain 80-21 is a derivative of B. thuringiensis subsp. kurstaki HD1 spontaneously cured of a 44-MDa plasmid containing the cry1Ab3 gene (3). These strains were transformed by electroporation with various lacZ fusion plasmids as described below. Cells were grown and sporulated either in Luria-Bertani (LB) medium or in a glucose-yeast extract medium (G-tris medium) (5). Liquid cultures comprising 10 to 15% of the flask volume were grown at 30°C in a New Brunswick shaker at 250 rpm.
Escherichia coli DH5α, which was used for plasmid construction, was grown at 37°C in LB medium containing ampicillin (50 μg/ml) when necessary.
Plasmid construction.
A 940-bp oligonucleotide from the upstream region of the cry1Da1 gene extending through the first five codons was synthesized by PCR by using a plasmid containing a 1.6-kb fragment of the cry1Da1 gene as the template (21, 23). The primers were 5′-GCTATGATCTAGATTACGAATTCGAGCTCG-3′ from the T7 primer region and the multiple cloning site of pUC18 contained in vector pHT3101 (21) and 5′-CATTGGTCTAGATTATTTATTTCCATAAACTATCCCCTA-3′ starting in the cry1Da1 coding region and extending through the ribosome binding site (boldface type indicates XbaI sites). The PCR conditions used have been described previously (25). Both primers contained XbaI sites, so the PCR product was digested with XbaI and ligated into pUC18. The orientation was established by exploiting a BglII site in the cloned fragment. The cry1Da1 fragment was then excised with HindIII and BamHI for ligation into a double-digested B. thuringiensis-E. coli shuttle vector, pHT304-18Z (1), to form a transcriptional fusion with lacZ (15). The resulting 10.7-kb plasmid contained both the cry1Da1 and Bacillus subtilis spoVG ribosome binding sites separated by 31 bp. The latter site was not optimal for translation in B. subtilis (9). In addition, changing the cry1Da1 ribosome binding sequence from AAGGGGAT (Fig. 1) to an optimal B. subtilis sequence, AAGGAGGT (9), by PCR performed with the second oligonucleotide described above but with the opitmal ribosome binding sequence resulted in enhanced expression of β-galactosidase (see Results). It is likely, therefore, that this ribosome binding site rather than the spoVG site was used for translation in the lacZ fusions.
FIG. 1.
Sequence of the cry1Da1 upstream region, including the overlapping promoters and the initiation of translation. The upstream oligonucleotide used to amplify this region was complementary to the T7 primer region and extended into the multiple cloning site of pUC18 which was present in the cloned cry1Da1 fragment used for PCR (see Materials and Methods). The downstream oligonucleotide extended from within the cry1Da1 coding region through the ribosome binding site. The ribosome binding sequence is overlined and was changed to AAGGAGGT by modifying the oligonucleotide primer as described in Materials and Methods. The positions of the overlapping promoter regions (−10 and −35) for the ςE and ςK forms of RNA polymerase are indicated. The GenBank accession number of the sequence is AF337948.
The plasmids were isolated from transformed E. coli DH5α, and constructs were confirmed by restriction enzyme digestion profiles and by sequencing. Plasmids were introduced into various B. thuringiensis strains by electroporation, with plating on G-tris medium containing erythromycin (25 μg/ml) for the pHT304-18Z constructs. Each resistant colony was then streaked onto the same medium onto which 0.1 ml of a 10% (wt/vol) solution of methylumbelliferyl-β-d-galactoside (Sigma) in 50% dimethyl formamide had been spread to confirm expression of the lacZ gene (12). The transformed cells were grown in G-tris medium containing the appropriate antibiotic at 30°C. Duplicate samples were removed at hourly intervals starting at the end of growth and were frozen at −70°C. Assays for β-galactosidase activity were performed as previously described (12), and the results are reported below in Miller units per unit of optical density at 600 nm (OD600).
Immunoblotting.
Twenty milliliters of cells was harvested at two stages of sporulation, when about 30% of the cells contained phase-dull endospores and when about 70 to 80% of the cells contained phase-white to bright endospores. The values were somewhat lower for LB medium, in which the sporulation efficiency was only 60 to 70%, compared to >95% for G-tris medium. The cells were harvested by centrifugation at 10,000 × g in a Sorvall SS4 rotor, washed, and lysed as previously described (12). The amount of cell extract protein loaded onto sodium dodecyl sulfate-polyacrylamide gels was adjusted to compensate for the difference in sporulation frequency.
The remainder of each culture was incubated for a total of 40 h until primarily free spores and inclusions were present. These spores and inclusions were harvested and washed, and the inclusions were purified with Renografin-76 (66% diatrizoate meglumine–10% diatrizoate sodium) (Solvay) gradients as previously described (6). Inclusion proteins were solubilized by incubating suspensions in 30 to 50 μl of 0.03 M Na2CO3–1% β-mercaptoethanol (pH 9.8) for 20 min at 37°C. Following centrifugation for 5 min in an Eppendorf microcentrifuge, the pellets were reextracted with 0.5 volume of the same buffer, and the supernatants were pooled.
The inclusion and cell extracts were dialyzed for 20 h at 4°C versus 2,000 volumes of 0.03 M NaHCO3. The dialyzed preparations were then treated with tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin (Sigma) at a ratio of 1:50 for 90 min at 37°C; then the same amount of trypsin was added and the preparations were incubated for an additional 90 min. The specificity of each of the Cry antibodies was for the toxin rather than for the protoxin, and thus the trypsin treatment prior to electrophoresis was necessary. The preparations were then dialyzed as described above by using 50-cut dialysis tubing (Spectrum). Aliquots were removed for protein determinations (BCA reagent; Pierce Chemical Co.), and the remaining portions of the preparations were frozen at −70°C.
Portions of each preparation (25 to 50 μg of extract protein and 5 to 10 μg of inclusion protein) were fractionated on sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis gels and transferred to polyvinylidene difluoride membranes (Immobilon P; Millipore) in a Hoffer semidry apparatus. The additional treatments used have been described previously (12). The developed blots were scanned, and major bands were quantified with NIH Image software. Experiments were repeated with the same extracts (sometimes the protein concentrations were varied) at least two additional times with different cell and inclusion preparations. The average densities of defined stained areas from two experiments were determined (the standard deviations were ±10%), and the relative values for the three toxin antigens were determined.
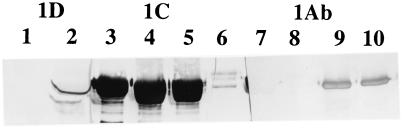
A monoclonal antibody specific for the Cry1Ab3 toxin (23) and a polyclonal rabbit antibody specific for the Cry1Ca1 toxin have been described previously (4). An antibody reacting with both the Cry1Ca1 and Cry1Da1 δ-endotoxins was kindly provided by D. Dean. An affinity column was prepared by coupling purified Cry1Ca1 toxin (12) to React-Gel (1,1′carbonyldiimidazole cross-linked to 6% agarose; Pierce) as described previously (17). The effluent from the mixed antibody was collected and passed through a second affinity column. As discussed below, this second effluent did not react with the Cry1Ca1 toxin but did react with a mixture of Cry1Ca1 and Cry1Da1 toxins (see Fig. 3).
FIG. 3.
Immunoblots obtained with 200 ng of purified Cry1Ab3 toxin (lanes 6, 9, and 10), 500 ng of purified Cry1Ca1 toxin (lanes 1, 4, 5, and 8), and a mixture of the Cry1Ca1 and Cry1Da1 toxins (500 ng) from purified inclusions from strain #5 (lanes 2, 3, and 7). Lanes 1 and 2 were treated with Cry1Da antibody, lanes 3 to 6 were treated with Cry1Ca1 antibody, and lanes 7 to 10 were treated with Cry1Ab3 antibody.
Determination of mRNA stability.
Cells (300 ml) were grown at 30°C in G-tris medium in 2-liter flasks. At the two stages of sporulation described above, 30-ml portions of cells were transferred to tubes containing 20 ml of frozen, crushed G-tris medium. The cells were harvested immediately by centrifugation in a Sorvall SS34 rotor at 12,000 × g for 10 min. The pellets were washed once with 15 ml of 0.05 M sodium acetate–0.1 M NaCl–0.001 M EDTA (pH 5.5). The cells were suspended in 1 ml of this buffer plus 0.2 ml of a 30% bentonite suspension in deionized water and lysed by passage through a French press at 9,000 lb/in2. The lysed cells were collected directly in an equal volume of phenol-chloroform (24:1). RNA was purified as described previously (8). A solution of rifampin (10 mg per ml in 50% ethanol) was added to the remaining culture to obtain a final concentration of 100 μg/ml. Thirty-milliliter samples were removed after 10, 20, and 30 min and used for RNA extraction as described above. The cry1Ab3 and cry1Da1 mRNAs in each sample were quantified following electrophoresis, transfer, and hybridization with gene-specific 32P-labeled oligonucleotides (4). The hybridizing bands on X-ray film were quantified with a phosphorimager, and decay curves were plotted with Kaleidagraph.
RESULTS
mRNA stability.
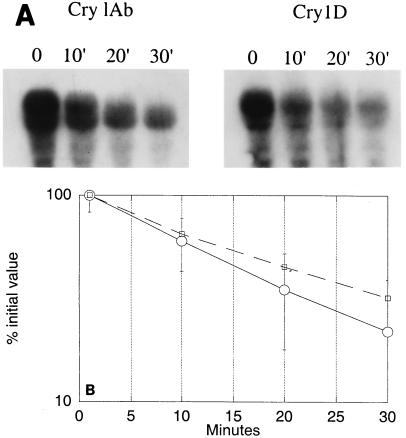
Differences in the steady-state amounts of the three cry mRNAs reported previously (4) could have been due to differences in rates of transcription and/or turnover. The stabilities of the cry1Ab3 and cry1Da1 mRNAs in B. thuringiensis subsp. aizawai HD133 were measured (Fig. 2). The half-lives were 14 to 18 min, and there were no significant differences. This range is comparable to that reported for cry1Aa mRNA (28). Therefore, the smaller amount of steady-state Cry1Da1 mRNA cannot be attributed to a less stable mRNA and must be due to differences in the initial transcription rates.
FIG. 2.
(A) Autoradiograms of Northern gels of RNAs from sporulating cells sampled after the addition of rifampin at the times indicated (see Materials and Methods). Hybridization was to a 32P-labeled cry1Da1 (Cry1D) or 32P-labeled cry1Ab3 (Cry1Ab) probe. (B) X-ray film was quantified with a phosphorimager, and the percentages of the initial values were plotted on a log scale versus time in order to calculate mRNA half-lives of cry1Ab3 mRNA (○) and cry1Da1 mRNA (□). Standard errors are indicated by error bars.
Alteration of the ribosome binding sequence improved expression.
The two- or fourfold differences in transcription were not as great as the differences in the relative amounts of the two protoxins in inclusions produced in LB medium (ratio, 1:20) (22). Control at the level of translation was a possibility since the ribosome binding sequence for the cry1Da1 gene is AAGGGGAT (Fig. 1) rather than the optimal B. subtilis consensus sequence, AAGGAGGT (9), which is present in the cry1Ab3 and cry1Ac1 genes. When the cry1Da1 sequence was changed to the latter sequence in a cry1Da1-lacZ fusion plasmid, the β-galactosidase specific activity increased from 300 to 1,400 Miller units per OD600 unit in strain 80-21 and from 450 to 1,800 Miller units per OD600 unit in strain #5.
Relative accumulation of the protoxins in sporulating cells and inclusions.
Antibodies specific to each of the three toxins were prepared either in rabbits (Cry1Ca1 and Cry1Da1) or as a monoclonal antibody (Cry1Ab3). The specificity of each antibody is shown in Fig. 3. Since purified Cry1Da1 toxin was not available, the specificity of the absorbed Cry1Da1 antibody (see Materials and Methods) was determined by its reaction with toxins from strain #5 inclusions. This strain contains only the cry1Ca1 and cry1Da1 genes and thus reacts with the Cry1Ca1 and Cry1Da1 antibodies but not with the Cry1Ab3 antibody. The Cry1Da1 antibody did not react with purified Cry1Ca1 toxin (Fig. 3) or with the Cry1Ab3 toxin (data not shown). In a given immunoblot, the reactions with all three antibodies increased as the amounts of the toxins increased.
The relative amounts of the toxins in extracts of sporulating B. thuringiensis subsp. aizawai HD133 and in purified inclusions produced in LB or G-tris medium were determined (Fig. 4 and 5). The ratio of the three antigens in extracts of sporulating cells in both LB medium and G-tris medium (Fig. 4) was 1.0:2.1:0.8. These values are averages based on two separate experiments, and the standard deviations were ±10%. Samples were also taken when more than 70% of the cells contained phase-white endospores, and similar results were obtained. There did not appear to be medium-dependent translational impairment of the cry1Da1 gene, nor is it likely that the Cry1Da1 protoxin was less stable in cells sporulating in LB medium.
FIG. 4.
Immunoblots of trypsin-treated extracts from B. thuringiensis subsp. aizawai HD133 grown in G-tris medium (G) (25 μg of protein) or LB medium (LB) (30 μg of protein) until 30% of the cells in G-tris medium or ca. 25% of the cells in LB medium contained phase-gray endospores. Samples were prepared as described in Materials and Methods. Lanes 1 and 2 were treated with the Cry1D antibody, lanes 3 and 4 were treated with the Cry1Ca1 antibody, and lanes 5 and 6 were treated with the Cry1Ab3 antibody. The blots were scanned and quantitated as described in Materials and Methods.
FIG. 5.
Immunoblots of trypsin-treated extracts from purified inclusions prepared from B. thuringiensis subsp. aizawai HD133 grown and sporulated in G-tris medium (G) or LB medium (LB). Preparations were treated with the Cry1Ab3, Cry1Ca1, and Cry1Da1 antibodies as described in Materials and Methods. Lanes 1 to 4 were treated with Cry1Da1 antibody, lanes 5 to 8 were treated with Cry1Ca1 antibody, and lanes 9 and 10 were treated with Cry1Ab3 antibody. Lanes 1, 2, 5, 6, and 9 contained inclusions from G-tris medium; lanes 1 and 2 were duplicates, as were lanes 5 and 6. Lanes 3, 4, 7, 8, and 10 contained inclusions from LB medium; lanes 3 and 4 were duplicates, as were lanes 7 and 8.
Inclusions from the two media, however, did differ in terms of the relative content of the Cry1Da1 toxin (Fig. 5). The Cry1Da1/Cry1Ca1/Cry1Ab3 ratios for G-tris medium inclusions and LB medium inclusions were 2.0:3.3:1.4 and 1.0:3.3:1.4, respectively. These values are averages based on two separate experiments, and the standard deviations were ±10%. There was twofold less Cry1Da1 toxin in inclusions from LB medium than in inclusions from G-tris medium.
B. thuringiensis subsp. aizawai HD68 is the original source of the cloned cry1Da1 gene (19). The relative content of this toxin in inclusions formed by this subspecies in LB medium was also twofold less than the relative content in inclusions formed in G-tris medium (R. Grant, unpublished data).
DISCUSSION
The results presented here account for the relatively low content of Cry1Da1 δ-endotoxin in inclusions. In a previous study (4), the steady-state ratio of the cry1D, cry1C, and cry1Ab mRNAs was reported to be 1:2:4. The ratios were about the same for cells that sporulated in G-tris medium and cells that sporulated in LB medium. These steady-state values are attributable to differences in the rates of transcription since the half-lives of the cry1Ab3 and cry1Da1 mRNAs were the same (14 to 18 min) (Fig. 2). The half-life of cry1Aa mRNA was reported to be about 12 min (28), which is much greater than the half-lives of most mRNAs in vegetative cells (2 to 3 min). The stability of these cry mRNAs is in part due to a large stem-loop structure at the 3′ end (28).
The cry1D, cry1C, and cry1Ab δ-endotoxin genes in B. thuringiensis subsp. aizawai HD133 have very similar dual overlapping promoters (10), and yet the steady-state amounts of the three mRNAs differ. The differences may be due to sequences upstream of the promoters which differ for ca. 1 kb in the three genes. There is some evidence that this region is involved in transcriptional regulation (27).
A second factor contributing to the relatively small amount of the Cry1Da1 protoxin is the fact that the ribosome binding sequence differs from the B. subtilis consensus sequence (9), which results in fourfold-lower translation of a cry1Da1-lacZ fusion. Transcription and translation thus account for ca. 10- to 15-fold less Cry1Da1 than Cry1Ab3 toxin. A third factor which augments this difference in inclusions is a medium-dependent limitation in packaging. Overall, a 20- to 30-fold difference in the amounts of these two protoxins in inclusions formed in LB medium would be expected, which is very similar to the ratio of these protoxins in purified inclusions (22).
Many B. thuringiensis subspecies produce a mixture of related Cry1 protoxins which are packaged together in a crystalline inclusion. The sequences of the carboxyl halves of these protoxins are well conserved and include 15 to 19 cysteines. Apparently, all of these cysteines participate in intermolecular disulfide bond formation (11). Such interactions among δ-endotoxins to form a regular crystalline array probably require other factors. For example, there is a stabilizing protein, P20, which enhances accumulation of a cytolytic toxin produced by B. thuringiensis subsp. israelensis (26, 29). There may be a functionally similar protein encoded by an open reading frame in the operon which also contains a cry2 δ-endotoxin gene (13). While these examples suggest some specificity for δ-endotoxins, packaging or stabilizing factors which are probably not specific for the δ-endotoxins may also have functions. Crystalline inclusions are produced by Bacillus cereus transformants containing clones of only a cry gene (8), implying that plasmid-encoded packaging functions are not essential.
Other cellular components involved in such packaging are not known but could include one or more enzymes involved in disulfide interchange reactions, such as protein disulfide isomerase (16) and perhaps some of the chaperone(s). The smaller amount of the Cry1Da1 δ-endotoxin in inclusions formed in LB medium than in inclusions formed in G-tris medium indicates that there is a limitation in one or more of these packaging factors in LB medium. There also appeared to be a limitation when the Cry1Ac1 δ-endotoxin was synthesized at a very high and apparently excessive rate due to promoter-up mutations (25). The excess protoxin was rapidly turned over, which resulted in smaller inclusions. There appears to be a close coupling between δ-endotoxin synthesis and packaging in inclusions. The Cry1Da1 δ-endotoxin which is produced in relatively small amounts may not bind efficiently to one of the limiting factors; i.e., there may be some specificity of the packaging factors for the various δ-endotoxins.
However, it is puzzling that the Cry1Da1 δ-endotoxin is produced at all given the limited capacity of sporulating cells to synthesize δ-endotoxins (3). In addition to its relatively small quantity, it is not very active against the array of insects that have been tested, nor is its specificity profile unique (18). It could function synergistically with other toxins and thus may be required only in small amounts. This would allow the presence in inclusions of larger amounts of the more active Cry1Ab3 and Cry1Ca1 protoxins.
ACKNOWLEDGMENTS
Lan Wu assisted in some of the experiments. We thank D. Dean, Ohio State University, for providing the Cry1Ca1/Cry1Da1 antibody.
Lily Chang was a visiting scientist supported by the Ministry of Agriculture, People's Republic of China.
REFERENCES
- 1.Agaisse H, Lereclus D. Structural and functional analysis of the promoter region involved in full expression of the cryIIIA toxin gene of Bacillus thuringiensis. Mol Microbiol. 1994;13:97–107. doi: 10.1111/j.1365-2958.1994.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 2.Aronson A I. The two faces of Bacillus thuringiensis: insecticidal proteins and post-exponential survival. Mol Microbiol. 1993;7:489–496. doi: 10.1111/j.1365-2958.1993.tb01139.x. [DOI] [PubMed] [Google Scholar]
- 3.Aronson A I. Flexibility in the protoxin composition of Bacillus thuringiensis. FEMS Microbiol Lett. 1994;117:21–28. doi: 10.1111/j.1574-6968.1994.tb06737.x. [DOI] [PubMed] [Google Scholar]
- 4.Aronson A I. The protoxin composition of Bacillus thuringiensis insecticidal inclusions affects solubility and toxicity. Appl Environ Microbiol. 1995;61:4057–4060. doi: 10.1128/aem.61.11.4057-4060.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aronson A I, Angelo N, Holt S C. Regulation of extra-cellular protease production in B. cereus T: characterization of mutants producing altered amounts of protease. J Bacteriol. 1971;106:1016–1025. doi: 10.1128/jb.106.3.1016-1025.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aronson A I, Han E-S, McGaughey W, Johnson D. The solubility of inclusion proteins from Bacillus thuringiensis is dependent upon protoxin composition and is a factor in toxicity to insects. Appl Environ Microbiol. 1991;57:981–986. doi: 10.1128/aem.57.4.981-986.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aronson A I, Shai Y. Why Bacillus thuringiensis insecticidal toxins are so effective: unique features of their mode of action. FEMS Microbiol Lett. 2001;195:1–8. doi: 10.1111/j.1574-6968.2001.tb10489.x. [DOI] [PubMed] [Google Scholar]
- 8.Arvidson H, Dunn P E, Strnad S, Aronson A I. Specificity of Bacillus thuringiensis for lepidopteran larvae: factors involved in vivo and in the structure of a purified protoxin. Mol Microbiol. 1989;3:1533–1543. doi: 10.1111/j.1365-2958.1989.tb00139.x. [DOI] [PubMed] [Google Scholar]
- 9.Band L, Henner D J. Bacillus subtilis requires a stringent Shine-Dalgarno sequence for gene expression. DNA. 1984;3:17–21. doi: 10.1089/dna.1.1984.3.17. [DOI] [PubMed] [Google Scholar]
- 10.Baum J A, Malvar T. Regulation of insecticidal crystal protein production in Bacillus thuringiensis. Mol Microbiol. 1995;18:1–12. doi: 10.1111/j.1365-2958.1995.mmi_18010001.x. [DOI] [PubMed] [Google Scholar]
- 11.Bietlot H P L, Vishmulhatla J, Carey P R, Pozsgay M, Kaplan H. Characterization of the cysteine residues and disulphide linkages in the protein crystal of Bacillus thuringiensis. Biochem J. 1990;267:309–315. doi: 10.1042/bj2670309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng P, Wu L, Aronson A. Subspecies-dependent regulation of Bacillus thuringiensis protoxin genes. Appl Environ Microbiol. 1999;65:1849–1853. doi: 10.1128/aem.65.5.1849-1853.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crickmore N, Ellar D J. Involvement of a possible chaperonin in the efficient expression of a cloned Cry IIA δ-endotoxin gene in Bacillus thuringiensis. Mol Microbiol. 1992;6:1533–1537. doi: 10.1111/j.1365-2958.1992.tb00874.x. [DOI] [PubMed] [Google Scholar]
- 14.Dulmage H T. Production of the spore–δ-endotoxin complex by variants of Bacillus thuringiensis in two fermentation media. J Invert Pathol. 1970;16:385–389. doi: 10.1016/0022-2011(70)90157-6. [DOI] [PubMed] [Google Scholar]
- 15.Errington J. A general method for fusion of the Escherichia coli lacZ gene to chromosomal genes in Bacillus subtilis. J Gen Microbiol. 1986;132:2953–2966. doi: 10.1099/00221287-132-11-2953. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert H F. Protein disulfide isomerase and assisted protein folding. J Biol Chem. 1997;272:29399–29402. doi: 10.1074/jbc.272.47.29399. [DOI] [PubMed] [Google Scholar]
- 17.Harlow E, David L. Antibodies—a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 18.Hofte H, Whiteley H R. Insecticidal crystal proteins of Bacillus thuringiensis. Microbiol Rev. 1989;53:242–255. doi: 10.1128/mr.53.2.242-255.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofte H, Soetaert P, Jansens S, Peferoen M. Nucleotide sequence and deduced amino acid sequence of a new lepidopteran-specific crystal protein gene from Bacillus thuringiensis. Nucleic Acids Res. 1990;18:5545. doi: 10.1093/nar/18.18.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knowles B H, Ellar D J. Colloid osmotic lysis is a general feature of the mechanism of action of Bacillus thuringiensis delta-endotoxins with different insect specificities. Biochim Biophys Acta. 1987;924:509–518. [Google Scholar]
- 21.Lereclus D, Arantes O, Chaufaux J, Lecadet M-M. Transformation and expression of a cloned δ-endotoxin gene in Bacillus thuringiensis. FEMS Microbiol Lett. 1989;60:211–218. doi: 10.1016/0378-1097(89)90511-9. [DOI] [PubMed] [Google Scholar]
- 22.Masson L, Erlandson M, Puzstai-Carey M, Brousseau R, Juarez-Perez V, Frutos R. A holistic approach for determining the entomopathogenic potential of Bacillus thuringiensis strains. Appl Environ Microbiol. 1998;64:4782–4788. doi: 10.1128/aem.64.12.4782-4788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minnich S A, Aronson A I. Regulation of protoxin synthesis in Bacillus thuringiensis. J Bacteriol. 1984;158:447–454. doi: 10.1128/jb.158.2.447-454.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schnepf E, Crickmore N, VanRie J, Lereclus D, Baum J, Feitelson J, Zeigler R, Dean D H. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev. 1998;62:775–806. doi: 10.1128/mmbr.62.3.775-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sedlak M, Walter T, Aronson A. The function of overlapping promoters in the regulation of Bacillus thuringiensis protoxin genes. J Bacteriol. 1998;182:734–741. doi: 10.1128/jb.182.3.734-741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Visick J E, Whiteley H R. Effect of a 20-kilodalton protein from Bacillus thuringiensis subsp. israelensis on production of the CytA protein by Escherichia coli. J Bacteriol. 1991;173:1748–1756. doi: 10.1128/jb.173.5.1748-1756.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walter T, Aronson A. Specific binding of the E2 subunit of pyruvate dehydrogenase to the upstream regions of Bacillus thuringiensis protoxin genes. J Biol Chem. 1998;274:7901–7906. doi: 10.1074/jbc.274.12.7901. [DOI] [PubMed] [Google Scholar]
- 28.Wong H C, Schnepf H E, Whiteley H R. Transcriptional and translational start sites for the Bacillus thuringiensis crystal protein gene. J Biol Chem. 1983;258:1960–1967. [PubMed] [Google Scholar]
- 29.Wu D, Federici B A. A 20-kilodalton protein preserves cell viability and promotes CytA crystal formation during sporulation in Bacillus thuringiensis. J Bacteriol. 1993;175:5276–5280. doi: 10.1128/jb.175.16.5276-5280.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]