Summary
The four dengue virus serotypes (DENV1-4) are mosquito-borne flaviviruses of humans. Several live attenuated tetravalent DENV vaccines are at different stages of clinical development and approval. In children with no baseline immunity to DENVs, a leading vaccine (Dengvaxia™) was efficacious against vaccine-matched DENV4 genotype II (GII) strains but not vaccine-mismatched DENV4 genotype I (GI) viruses. We used a panel of recombinant DENV4 viruses displaying GI or GII envelope (E) proteins to map Dengvaxia™ induced neutralizing antibodies (NAb) linked to protection. The vaccine stimulated antibodies that neutralized the DENV4 GII virus better than the GI virus. The neutralization difference mapped to 5 variable amino acids on E protein located within a region targeted by DENV4 NAbs, supporting a mechanistic role for these epitope-specific NAbs in protection. In children with no baseline immunity to DENVs, levels of DENV4 serotype- and genotype-specific NAbs induced by vaccination are predictive of vaccine efficacy.
Introduction
Dengue virus (DENV) is a single-stranded positive sense RNA virus, transmitted by Aedes mosquitoes. Over a third of the world is at risk for DENV infection, with an estimated 390 million infections annually (Bhatt et al., 2013). There are four distinct DENV serotypes (DENV1-4). Natural infection with one serotype results in durable serotype-specific (TS) protective immunity but limited cross protective immunity to new serotypes. Within each DENV serotype, there are multiple distinct genotypes (Holmes and Twiddy, 2003, Weaver and Vasilakis, 2009, Gallichotte et al., 2018b). Historically, genetic and antigenic differences between DENVs belonging to the same serotype have not been considered significant enough to impact protective immunity and leading DENV vaccines have been formulated under the assumption that the envelope (E) protein from a single strain will stimulate broadly protective antibody (Ab) responses to all genotypes within a serotype. There is growing evidence in the literature to challenge this assumption. Several studies have established that natural antigenic variation in the E protein of DENV strains within a serotype can have a large impact on the efficiency of neutralization by monoclonal Abs and immune sera from people exposed to DENV infections or vaccines (Gallichotte et al., 2018b, Wahala et al., 2010, Katzelnick et al., 2015, Dowd et al., 2015). Additionally, reinfection with homologous serotypes is rare, but does occasionally occur, especially after clade replacements altering the circulating genotype (Forshey et al., 2016, Waggoner et al., 2016). These observations challenge established dogma about human immunity to DENV serotypes and highlight the need to study how E protein variation impacts Ab neutralization and protective immunity. DENV4 contains five genotypes, with genotypes I and II as the dominant strains currently circulating in human populations (Gallichotte et al., 2018b). In a recent DENV vaccine clinical trial, baseline seronegative children were reliably protected from DENV4 genotype II (GII) but not genotype I (GI) strains (Juraska et al., 2018, Rabaa et al., 2017). Here we test if amino acid differences at critical sites in the envelope protein of DENV4 GI and II strains lead to differences in neutralization by vaccine immune sera and vaccine efficacy.
Sanofi Pasteur’s live-attenuated, chimeric yellow fever-dengue, tetravalent DENV vaccine (Dengvaxia) contains equal amounts of each DENV serotype component, and is given in three doses, six months apart (Capeding et al., 2014, Hadinegoro et al., 2015). Dengvaxia recently completed phase III clinical trials in Southeast Asia (CYD14), and Latin America (CYD15) among children who were DENV seronegative or seropositive at baseline (Poo et al., 2011, Morrison et al., 2010, Leo et al., 2012, Lanata et al., 2012, Guy et al., 2015). In seropositive children, the vaccine stimulated pre-existing DENV-specific memory B and T cells, which led to secondary immune responses and significant clinical efficacy (10). In seronegative children, overall efficacy was low or non-existent (Dayan et al., 2020a, Dayan et al., 2020b, Sridhar et al., 2018). Poor efficacy in seronegative children was unexpected because most children developed neutralizing antibodies (NAbs), widely considered to be a correlate of protection for flaviviruses.
The poor efficacy of Dengvaxia in baseline seronegative children is, most likely, due to the unbalanced replication of the 4 attenuated viruses in the vaccine, specifically the DENV4 vaccine component outcompeting and replicating to higher levels than the other three components (Barban et al., 2012, Guirakhoo et al., 2004, Thomas and Yoon, 2019). We recently demonstrated that, in seronegative individuals, the vaccine stimulated higher levels of type-specific neutralizing antibodies to DENV4 compared to the other three serotypes, which is consistent with the DENV4 component replicating better than the other components (Henein et al., 2017). While significant overall vaccine efficacy (combined across the 4 serotypes) was not observed in seronegative children who were vaccinated, the vaccine did provide significant protection against DENV4 but not the other three serotypes in that population, further supporting the replication and immunodominance of the DENV4 vaccine component (Sridhar et al., 2018).
Another level of complexity in DENV vaccine development was revealed by investigators who analyzed sequences of DENVs infecting children enrolled in Dengvaxia clinical trials (Rabaa et al., 2017, Juraska et al., 2018). When a genetic sieve analysis was performed to compare viral sequences from vaccine and placebo arms, investigators observed high vaccine efficacy (VE) against vaccine matched DENV4 GII viruses (VE 76%) and no efficacy against vaccine mismatched DENV4 GI viruses (VE = 23.9%) among children aged 2-8 years (Rabaa et al., 2017, Juraska et al., 2018). Importantly, this phenomenon was not observed in older children (≥9 years) (GII VE = 89.8%; GI VE = 85.5%), who are mainly DENV pre-immune at baseline, suggesting a difference in genotypic breadth and protection by vaccine-elicited Abs in seronegative versus seropositive populations.
Here we performed experiments to identify the mechanism by which minor differences in the E proteins of DENV4 GI and GII viruses drives large differences in vaccine efficacy. We observed that in baseline seronegative children, the vaccine elicited DENV4 TS Abs that neutralized the vaccine-matched GII viruses better than vaccine-mismatched GI viruses. The difference in neutralization mapped to a few variable E protein residues located close to sites targeted by DENV4 neutralizing MAbs. We propose that in seronegative individuals, the dominant DENV4 vaccine component stimulates an effective but highly focused NAb response to the vaccine matched genotype, increasing the probability for breakthrough infections with a vaccine mismatched genotype.
Results
Contemporary DENV4 genotypes in circulation during Dengvaxia clinical trials
DENV4 is sub-divided into five genotypes (Holmes and Twiddy, 2003). Genotypes III and V are former human epidemic strains that have not been detected in humans recently. Genotype IV is restricted to a sylvatic cycle in non-human primates. DENV4 genotypes I and II are contemporary epidemic strains responsible for cases during CYD14 and 15 Dengvaxia clinical trials (Juraska et al., 2018, Rabaa et al., 2017) (Fig. 1A). The DENV4 E protein in Dengvaxia is derived from a historic GII isolate (Strain 1228, Indonesia 1978) (Fig. 1A). In younger children (ages 2-8), which included many individuals that were seronegative at baseline, Dengvaxia was efficacious against contemporary DENV4 GII but not GI strains (Juraska et al., 2018, Rabaa et al., 2017). To determine if vaccine induced antibodies were sensitive to naturally occurring genetic variation in the E protein of DENV4 GI and II viruses, we generated 2 recombinant viruses containing the envelope (E) protein sequence of each genotype (I, II), on a DENV4 GII infectious clone (infectious clone – Sri Lanka 1992, GI - Cambodia 2010, and GII - French Polynesia 2009) (Fig. 1B, Table S1) (Gallichotte et al., 2018b). The viruses are genetically identical in all but the E sequences, allowing one to determine phenotypes driven entirely by E protein variation.
Figure 1. DENV4 GI and II phylogeny and virus design.
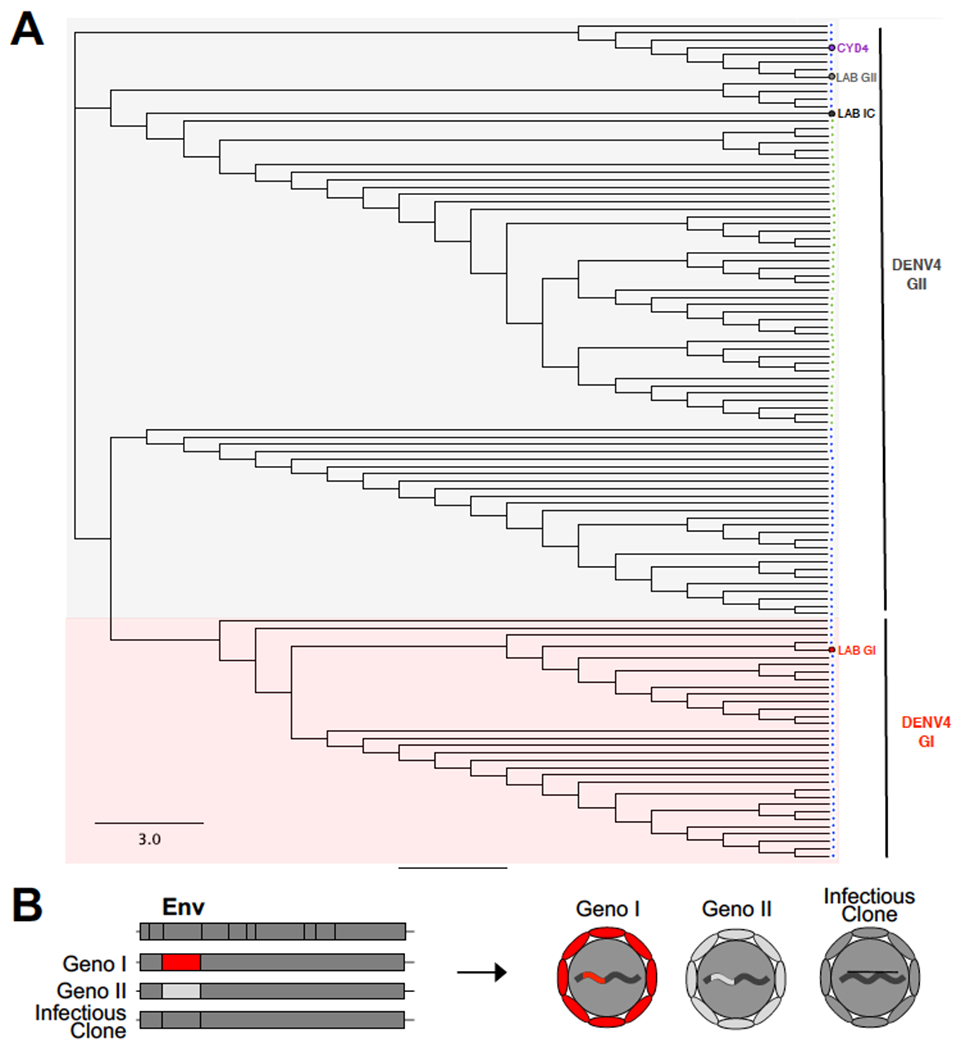
A) Maximum likelihood phylogeny of envelope protein sequences of DENV4 viruses detected during CYD14 (blue dots) and CYD15 trial (green dots). The tree also depicts the positions of the DENV4 vaccine virus envelope (CYD4, purple), and the DENV4 infectious clone (Lab IC), DENV4 GI (Lab GI) and DENV4 GII (Lab GII) envelopes on recombinant viruses used in the current study. The tree was created using Geneious version 2019.0 B) Design of recombinant GI and II viruses.
The impact of baseline DENV serostatus on the properties of vaccine induced DENV4 NAbs
We compared the ability of immune sera from individuals who had received 3 doses of the vaccine to neutralize the recombinant DENV4 GI and II viruses. Vaccine immune sera from individuals who were seronegative at baseline neutralized the vaccine-matched GII better than the vaccine-mismatched GI viruses (p<0.001) (Fig. 2A). Immune sera from individuals who were seropositive at baseline equally cross-neutralized both genotypes, demonstrating broader neutralization breadth in this population (Fig. 2B). DENV4 GII is widely distributed and consists of 2 clades designated GIIa and IIb. Both clades were similarly neutralized by baseline seronegative and seropositive subjects demonstrating the vaccine was response was not influenced by variation between DENV4 GII clades (SFig. 1).
Figure 2. Baseline serostatus impacts genotype cross-neutralization.
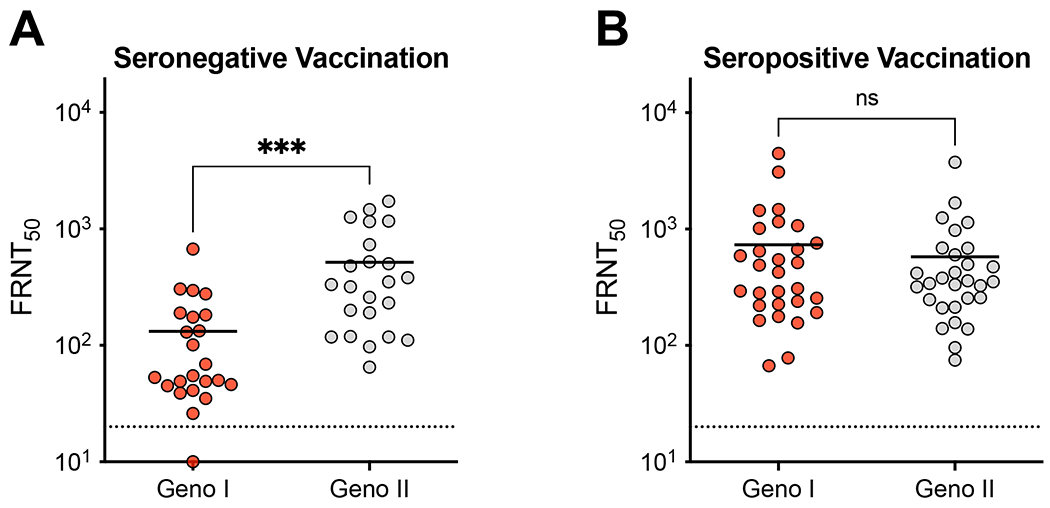
DENV4 GI and II neutralization titers by A) seronegative and B) seropositive post-vaccination immune sera. FRNT50 represents sera dilution factor required to neutralize 50% of virus. Dotted line represents limit of detection (20). Samples not neutralized are graphed at ½ the limit of detection (10). Neutralization titers against GI and GII viruses were compared by unpaired t-test (***p<0.001).
Individuals exposed to natural DENV infections or vaccines develop NAbs that bind to TS (unique to serotype) and CR (conserved between serotypes) epitopes on the viral E protein (Gallichotte et al., 2018a). In general, primary DENV infections stimulate neutralizing and protective Abs directed against TS epitopes, whereas secondary infections activate pre-existing DENV-specific memory, which leads to high levels of neutralizing and protective Abs targeting epitopes that are conserved between serotypes (Dejnirattisai et al., 2015b, Gallichotte et al., 2018a, Patel et al., 2017, Tsai et al., 2013). Using a standard approach (Gallichotte et al., 2018b), we depleted all DENV1, 2 and 3 Abs (DENV1+2+3 depletion) in each vaccine serum sample to measure the contribution of DENV4-TS Abs to neutralization of DENV4 GI and GII strains (Fig. 3A). As controls, each sample was processed without removing any DENV-specific Ab (control depletion) and by depleting all DENV binding Abs (DENV1+2+3+4 depletion). Depletion of relevant Ab populations was confirmed by ELISA before performing neutralization assays with GI and II viruses (Fig. 3A). We found that neutralization of DENV4 by immune sera from individuals who were naturally infected with DENV4 is driven primarily (~95%) by DENV4-TS Abs that were equally effective against DENV4 GI and II strains. (Fig. 3B). In baseline seronegative subjects, the vaccine induced DENV4-TS response was narrow and significantly more effective against vaccine matched DENV4 GII strain compared to GI strain (p<0.005) (Fig. 3B and C). Additionally, the low level of DENV4 GI neutralization seen in baseline seronegative vaccinees is mainly driven by serotype CR Abs, because most individuals no longer neutralized the GI strain after removal of CR Abs (Fig. 2A vs. Fig. 3C). DENV4 neutralization following vaccination of baseline seropositive individuals was mainly driven by CR Abs that were equally effective against GI and II strains (Fig. 3B and C).
Figure 3. Quality of Ab response is impacted by pre-vaccination serostatus.
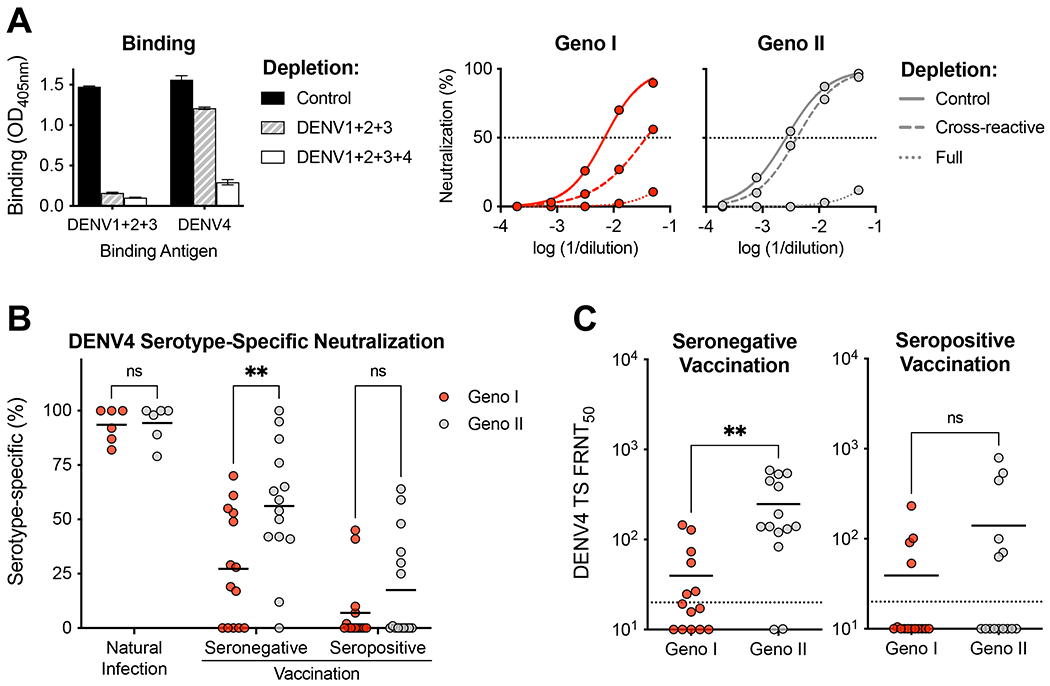
A) Representative example of the impact of depleting cross-reactive (DENV1, 2 and 3 binding) or total (DENV1, 2, 3, 4 binding) DENV binding antibodies from a baseline seronegative subject who was vaccinated. Control or specific antibody depleted samples were tested for binding to a mix DENV1+2+3 antigens to confirm depletion of antibodies binding to epitopes conserved (cross-reactive) between serotypes. The samples were tested for binding to DENV4 to detect any DENV4 type-specific antibodies. The difference in binding between sera depleted with a mix of DENV1,2 and 3 antigens and a mix of DENV1,2,3 and 4 antigens is a measure of DENV4 type-specific binding antibodies. The antibody depleted samples were also used to perform neutralization assays with DENV4 GI and II strains to measure levels of DENV4 type-specific neutralizing antibodies. The difference in neutralizing antibody levels after removing cross-reactive (DENV1,2,3 depletion) and total (DENV1,2,3,4) DENV-specific antibodies is a measure of DENV4 type-specific neutralizing antibodies. B) Percentage of DENV4 GI and II neutralization by TS specific Abs, calculated as = (CR depletion FRNT50 – full depletion FRNT50)/(control depletion FRNT50 – full depletion FRNT50) x 100. The proportions of DENV4 TS NAbs against GI and GII strains were compared using a 2-way ANOVA with Šidák’s multiple comparison test (**p<0.005). C) GI and II DENV4-specific neutralization titers, FRNT50 represents sera dilution factor required to neutralize 50% of virus. Dotted line represents limit of detection (20). Samples not neutralized are graphed at ½ the limit of detection (10). The levels of DENV4 TS NAbs against GI and II strains were compared by unpaired T-test.
Differential genotypic neutralization is driven by signature amino acids
To map the E protein amino acids responsible for DENV4 GI and GII neutralization differences, we used an infectious clone of DENV4 to create chimeras exchanging sequences between GI and GII strains (Fig. 4A, Table S2). The DENV4 GI E segment contains 15 amino acid differences relative to the GII E segment, distributed evenly throughout all three domains (Fig. 4A Table S2). In a previous sieve analysis of the sequences of DENV4 viruses infecting placebo and Degvaxia recipients, five amino acids (blue asterisks) that differ between DENV4 GI and GII were linked to vaccine efficacy (Juraska et al., 2018). Importantly, these amino acids map to epitopes of DENV4 TS strongly neutralizing MAbs (Fig. 4B) (Nivarthi et al., 2017). To directly test if these 5 variant amino acids were responsible for the ability of Dengvaxia immune sera to neutralize the vaccine matched GII genotype better than the GI genotype, we generated chimeric viruses that contained the GI residues on a GII envelope, and vice versa (Fig. 4A & C, Table S2).
Figure 4. Design of GI and II chimeric viruses for mapping key residues.
A) Schematic of E protein amino acid differences between DENV4 GI virus relative to GII virus. The figure depicts signature amino acids (blue asterisks), and design of chimeric viruses, aligned with envelope protein domains (red = EDI, yellow = EDII, blue = EDIII). B) Signature amino acids and DENV4 MAb epitopes shown on one monomer within the E protein dimer. C) Amino acid differences of GI (red) relative to DEVN4 GII, and signature amino acids transplanted in chimeric viruses (black and red) (PDB = 1OAN).
Before using the DENV4 WT GI, GII and chimeric envelope G/I/II and GII/I viruses to map vaccine responses, we compared the sensitivity of these viruses to a panel of DENV4-specific neutralizing MAbs (5H2, D4-126, D4-131) isolated from humans and non-human primates. Each MAb neutralized the GII viruses better than GI (SFig. 2A). The GI/II virus containing the signature amino acids from GII gained sensitivity to neutralization by TS MAbs 5H2 and D4-131 (SFig. 2A & B). Conversely, the GII/I viruses required higher concentrations of MAbs 5H2 and D4-126 to neutralize when the signature amino acids were replaced with those from GI (SFig. 2A & B). These results indicate that signature amino acids defined by the sieve analysis are located within epitopes targeted by DENV4-TS NAbs and impact the neutralization potency of DENV4 TS MAbs. Importantly, DENV CR MAbs (C10, B7, 1M7) similarly neutralize all wildtype and chimeric viruses (SFig. 2C & D), suggesting the genotype differential neutralization and importance of signature amino acids is specific to DENV4-TS Abs.
Next, we tested if differences in the ability of Dengvaxia immune sera to neutralize DENV4 GI and II viruses mapped to the 5 amino acids linked to DENV4 vaccine efficacy. As demonstrated in Figure 2, vaccinated individuals who were seronegative at baseline developed higher levels of NAbs to vaccine matched DENV4 GII viruses compared to the vaccine mismatched GI virus. Vaccine immune sera from baseline seronegative individuals poorly neutralized the DENV4 GII/I chimera compared to the parental GII virus (Fig. 5A & B). Sera from these subjects neutralized the GI/II chimeric virus better than the parental GI virus (Fig. 5B). These results demonstrate that the ability of Dengvaxia induced antibodies to neutralize DENV4 GII better than GI map to the same signature amino acids linked to DENV4 genotype-specific vaccine efficacy (Fig. 5A & 5B). Individuals who were seropositive at baseline mainly developed DENV serotype CR NAbs that were equally effective against both DENV4 genotypes and the chimeric viruses (Fig. 5A & B), confirming that the signature amino acids define epitopes that are the target of DENV4 TS but not CR NAbs.
Figure 5. Pre-vaccination serostatus alters tracking with signature amino acids.
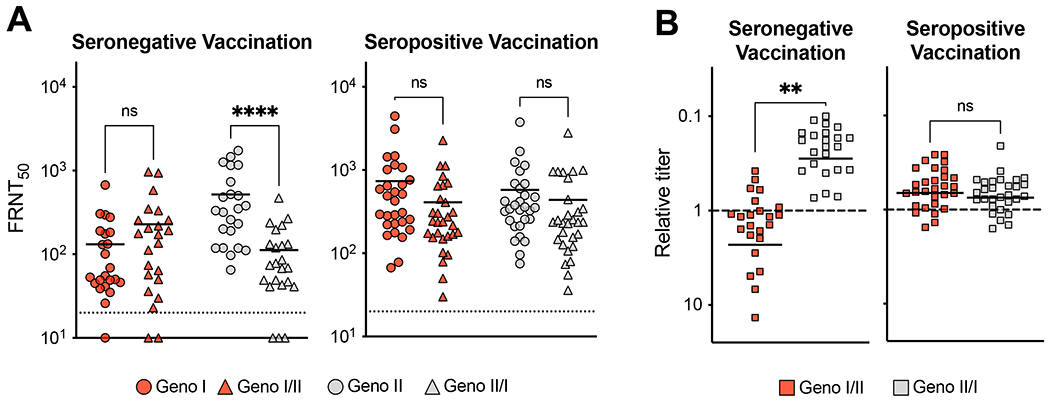
A) Seronegative and seropositive vaccination sera neutralization of wild-type and chimeric GI and II viruses. FRNT50 represents sera dilution factor required to neutralize 50% of virus. Dotted line represents limit of detection (20). Samples not neutralized are graphed at ½ the limit of detection (10). One-way ANOVA with Tukey’s multiple comparison test (****p<0.0001). B) Seronegative and seropositive vaccination sera chimeric virus relative titer = (chimeric FRNT50 / parental wild-type FRNT50). Relative titer < 1 = parental wild-type virus better neutralized, relative titer > 1 = chimeric better neutralized. Relative titers were compared with an unpaired t-test (**p<0.005).
Discussion
Dengvaxia was predicted to be effective against the four DENV serotypes because in most people, including children with no immunity to DENVs, the vaccine elicited Abs that neutralized all 4 serotypes (Guy et al., 2015). As our understanding of the adaptive immune response to DENV infection evolves, the metrics used to evaluate vaccine responses has evolved as well. While CYD-TDV neutralization titers have been correlated with vaccine efficacy, these studies are based on a pooled analysis of baseline seropositive and seronegative children that mainly consisted of seropositive children (Moodie et al., 2017). When the analysis was stratified by baseline serostatus, total NAb levels were an imprecise and weak correlate, at best, in baseline seronegative (Carpp et al., 2020, Moodie et al., 2017). Given fundamental differences in how adaptive immunity is activated in individuals who are DENV seronegative and seropositive at the time of vaccination, these two populations must be studied separately when evaluating immune correlates, vaccine efficacy and safety (Gilbert et al., 2019, Huang et al., 2020, Lam et al., 2019).
Future development efforts would ideally focus on a vaccine that can be administered to young children (who are primarily DENV seronegative) living in regions with active transmission. Our in-depth examination of the humoral immune responses in vaccinated individuals strongly support the premise that neutralizing Abs directed to unique (serotype-specific) epitopes are a mechanistic correlate of DENV4 protection in this population. Studies in animal models and humans have established that the DENV4 component in Dengvaxia replicates to higher levels than the other three components (Barban et al., 2018, Thomas and Yoon, 2019). In baseline seronegative individuals, the vaccine readily stimulated DENV4 TS NAbs, whereas the other three serotypes were mainly neutralized by CR Abs, most likely derived from the replication dominant DENV4 vaccine component (Henein et al., 2017, Henein et al., 2021). CR NAbs derived from the replication of one DENV serotype are known to be transient and not correlated with long-term protection after natural infection or vaccination (Dayan et al., 2020b, de Silva and Harris, 2018). Among children who were seronegative at baseline, significant efficacy of Dengvaxia against serotypes 1, 2 and 3 was not observed and there was no evidence for genotype-specific sieve effects (Juraska et al., 2018, Rabaa et al., 2017). In contrast, a sieve effect of high efficacy against vaccine matched DENV4 GII viruses and minimal efficacy against vaccine mismatched DENV4 GI viruses were observed in this population (Juraska et al., 2018, Rabaa et al., 2017). Genotype-specific vaccine efficacy is only possible if the protective Ab response is directed to TS epitopes that vary between genotypes. Indeed, our results establish that just 5 amino acids located at known DENV4 TS NAb epitopes have a strong impact on the TS NAb titer. We propose that DENV4 genotype specific NAbs are the major mechanism of protective immunity in baseline seronegative individuals in addition to being a strong correlate of vaccine efficacy. Additionally, this reveals the need for more detailed studies into the role of genotype-specific immunity in regions experiencing genotype replacements within a single serotype as a potential mechanisms of serotype persistence in DENV endemic regions.
Our results highlight the fundamental differences in correlates and mechanisms of protection in baseline seropositive and seronegative individuals who receive tetravalent DENV vaccines (de Silva and Harris, 2018). Protective immunity in seronegative individuals will require the robust and balanced replication of each vaccine component to induce independent, TS neutralizing and protective Abs to each serotype (de Silva and Harris, 2018). In baseline DENV seropositive individuals, even unbalanced replication dominated by one or two vaccine components will activate immune memory and induce serotype cross protective B and T cell responses that are analogous to cross-protective immune responses observed in people who have recovered from a second DENV infection with a new serotype (Dejnirattisai et al., 2015a, Patel et al., 2017). In fact, human MAbs that bind to conserved epitopes that strongly neutralize the 4 serotypes in cell culture and cross protect in animal models have been isolated from people exposed to secondary DENV infections (Tsai et al., 2013). We observed that baseline seropositive vaccinees, who were protected from both DENV4 GI and GII strains, had high levels of serotype CR NAbs that were effective against both DENV4 GI and GII viruses.
Our results also highlight the importance of mapping the targets of neutralizing and protective antibodies and considering natural E protein variation among contemporary DENV serotypes and genotypes when developing and evaluating DENV vaccines (Katzelnick et al., 2017). As previously reported, wild type DENV4 infections elicit DENV4 type-specific antibodies that effectively neutralize multiple current, past, and sylvatic genotype (Gallichotte et al., 2018b). Our results indicate that live attenuated vaccines induce NAbs that are more narrowly focused to the vaccine matched genotype, at least, in the case of DENV4. As new DENV vaccines are developed, we need to identify major epitopes targeted by vaccine induced protective antibodies, determine the variability of these epitopes across genotypes, and select vaccine E glycoproteins that match the major antigenic sites on circulating serotypes and genotypes.
Limitations of Study
This study has mapped the differences in Dengvaxia induced antibody neutralization of DENV4 GI and II viruses to a subset of 5 amino acids that differ between these genotypes. Four of the residues (46, 120, 160, 203) cluster on either side of the hinge between E protein domains I and II and the 5th residue (329) is located at a greater distance on domain III. Given the limited quantities of vaccine sera available for the study, we were unable to create and test additional recombinant viruses to determine contribution of each residue to the neutralization phenotype.
STAR METHODS
Resource Availability
Lead Contact:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Aravinda Desilva (desilva@med.unc.edu)
Materials Availability:
The recombinant DENV4 viruses created for this study are available to other investigators and institutions authorized for handling DENVs. The transfer of the material will require a standard academic materials transfer agreement between UNC and the requesting institution.
Data and Code Availability:
This paper does not report original code. All data generated for the project, including vaccine antibody titers, are archived in the Desilva laboratory. This data is available for review or reanalysis upon request from the Lead Author.
Study approvals and Immune sera.
DENV4 natural infection human immune sera were obtained from a previously described and approved (protocol 08-0895) traveler study at the University of North Carolina (Gallichotte et al., 2018b). Dengvaxia clinical trial protocols for CYD14 (ClinicalTrials.gov ID NCT01373281) and CYD15 (ClinicalTrials.gov ID NCT01374516) have been approved by all relevant ethics review boards (Capeding et al., 2014, Villar et al., 2015). The Institutional Review Board of the University of North Carolina at Chapel Hill reviewed and approved the receipt and analysis of anonymized CYD14 and CYD15 clinical specimens at the University of North Carolina at Chapel Hill for the current study (protocol-08-0895). All vaccine sera used in the current study were collected one month following the third dose of vaccine.
Cells.
Vero cells were maintained in DMEM with 5% fetal bovine serum (FBS) at 37°C. C6/36 cells were maintained in MEM with 10% FBS and non-essential amino acids at 32°C. All media were supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL amphotericin B and 5% CO2.
Viruses.
Recombinant DENV4 viruses were constructed using a DENV4 quadripartite infectious clone as described previously (Gallichotte et al., 2018b). Our DENV4 infectious clone (Sri Lanka, 1992 isolate) belongs to genotype II. We used this infectious clone to produce new recombinant viruses that were genetically identical with the exception of the envelope gene, which was derived from genotype I, IIA, IIB and I/II envelope chimeras using standard methods. In brief, plasmid DNA encoding viral genomes were digested, ligated together, and in vitro transcribed with T7 polymerase. Infectious, genome-length viral mRNA transcripts were electroporated into C6/36, supernatant was passaged onto C6/36 cells once, harvested and stored at −80°C. The recombinant viruses used for the current study, Genbank accession numbers and E protein amino acid differences between viruses are listed in supplementary material (Table S1 and S2).
Depletion Assays.
Immune sera were depleted of Abs populations as previously described (Gallichotte et al., 2018b, Henein et al., 2017). Briefly, polystyrene beads were coated with BSA, purified DENV1+2+3 or purified DENV1+2+3+4 antigen. Immune sera were incubated with bead:antigen complexes at 37°C for 45 minutes, and repeated two additional times with new bead:antigen mix. Confirmation of depletion of Abs was performed using an ELISA as previously described (Gallichotte et al., 2018b, Henein et al., 2017). Briefly, 96-well plates were coated with anti-DENV antibodies (4G2 and 2H2), blocked in normal goal serum, then purified DENV antigen was captured for 1 hour at 37°C. Depleted human sera added and incubated for an additional hour at 37°C, then secondary anti-human-alkaline phosphatase antibody incubated for an additional hour. P-nitrophenyl phosphate substrate was added, and absorbance was measured at OD 405nm.
Focus reduction neutralization assay (FRNT).
Neutralization assays were performed as previously described (Gallichotte et al., 2018b). Briefly, Vero cells were plated one day prior to infection. MAbs or immune sera were diluted and mixed with virus, Ab:virus complex was incubated at 37°C for one hour, then added to confluent cells. After one hour, 1% carboxymethycellulose overlay was added and cells were incubated for 4-5 days. Cells were fixed in 20% methanol, blocked in non-fat dried milk, and immunostained using mouse 4G2 and 2H2 antibodies, secondary anti-mouse-horseradish peroxidase conjugated antibody, and developed using HRP substrate. Foci were counted manually.
Supplementary Material
Supplementary Figure 1. Baseline serostatus impacts DENV4 genotype I and II cross-neutralization. DENV4 GI, GIIa and GIIb neutralization titers by A) seronegative and B) seropositive post-vaccination immune sera. FRNT50 represents sera dilution factor required to neutralize 50% of virus. Dotted line represents limit of detection (20). Samples not neutralized are graphed at ½ the limit of detection (10). Neutralization titers against the different viruses were compared by one way ANOVA (*p<0.05, **p<0.005).
Supplementary Figure 2. MAb neutralization of DENV4 genotype I, II and chimeric envelope viruses. Wild-type (DENV GI, GII) and chimeric (GI/II, GII/I) virus neutralization by A) DENV4-specific and C) DENV CR MAbs. FRNT50 represents MAb concentration [ng/μl] required to neutralize 50% of virus. B) DENV4-specific and D) DENV CR MAb chimeric virus relative titer = (chimeric FRNT50 / parental wild-type FRNT50). Relative titer > 1 = parental wild-type virus better neutralized, relative titer < 1 = chimeric better neutralized.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse 4G2 | Novus Biologicals | NBP2-52709 |
| Mouse 2H2 | Sigma Aldrich | MAB8705 |
| Secondary anti-mouse-horseradish peroxidase conjugated antibody | ThermoFisher | 62-6520 |
| Secondary anti-human-alkaline phosphatase conjugated antibody | Sigma Aldrich | A9544 |
| Bacterial and virus strains | ||
| DENV4 infectious clone | Gallichotte et. al. 2018 | N/A |
| Recombinant DENV4 viruses | This paper | N/A |
| Biological samples | ||
| Human immune sera | Gallichotte et al. 2018 | N/A |
| Human vaccine sera | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Purified DENV1, DENV2, DENV3, DENV4 | This paper | N/A |
| Experimental models: Cell lines | ||
| Vero cell | ATCC | CCL-81 |
| C6/36 | ATCC | CCL-1660 |
| Recombinant DNA | ||
| DENV4 infectious clone | Gallichotte et al. 2018 | N/A |
| Software and algorithms | ||
| MacPyMOL | https://pymol.org/2/ | N/A |
| GraphPad Prism Version 9.2.1 | https://www.graphpad.com/ | N/A |
| Other | ||
| T7 RNA polymerase | New England Biolabs | M0251 |
| Polystyrene microspheres | Polysciences | 17135 |
| Bovine Serum Albumin | Sigma Aldrich | A9418 |
| P-nitrophenyl phosphate substrate | ThermoFisher | 34047 |
| Normal Goat Serum | abcam | ab7481 |
| TrueBlue HRP substrate | Seracare | 5510-0030 |
Acknowledgments
This research was supported by U.S. National Institute of Allergy and Infectious Diseases (NIAID) grants R01s AI107731 and AI125198 (PI: de Silva).
Footnotes
Declaration of Interests
Matthew Bonaparte, Janice Moser, and Alina Munteanu are employees of the company (Sanofi Pasteur) that developed Dengvaxia. The other authors have declared that no conflict of interest exists.
References
- Barban V, Mantel N, De Montfort A, Pagnon A, Pradezynski F, Lang J & Boudet F 2018. Improvement of the Dengue Virus (DENV) Nonhuman Primate Model via a Reverse Translational Approach Based on Dengue Vaccine Clinical Efficacy Data against DENV-2 and −4. J Virol, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barban V, Munoz-Jordan JL, Santiago GA, Mantel N, Girerd Y, Gulia S, Claude JB & Lang J 2012. Broad neutralization of wild-type dengue virus isolates following immunization in monkeys with a tetravalent dengue vaccine based on chimeric yellow fever 17D/dengue viruses. Virology, 429, 91–8. [DOI] [PubMed] [Google Scholar]
- Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ & Hay SI 2013. The global distribution and burden of dengue. Nature, 496, 504–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capeding MR, Tran NH, Hadinegoro SR, Ismail HI, Chotpitayasunondh T, Chua MN, Luong CQ, Rusmil K, Wirawan DN, Nallusamy R, Pitisuttithum P, Thisyakorn U, Yoon IK, van der Vliet D, Langevin E, Laot T, Hutagalung Y, Frago C, Boaz M, Wartel TA, Tornieporth NG, Saville M, Bouckenooghe A & Group CYDS 2014. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. Lancet, 384, 1358–65. [DOI] [PubMed] [Google Scholar]
- Carpp LN, Fong Y, Bonaparte M, Moodie Z, Juraska M, Huang Y, Price B, Zhuang Y, Shao J, Zheng L, Chambonneau L, Small R, Sridhar S, DiazGranados CA & Gilbert PB 2020. Microneutralization assay titer correlates analysis in two phase 3 trials of the CYD-TDV tetravalent dengue vaccine in Asia and Latin America. PLoS One, 15, e0234236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayan GH, Langevin E, Forrat R, Zambrano B, Noriega F, Frago C, Bouckenooghe A, Machabert T, Savarino S & DiazGranados CA 2020a. Efficacy after 1 and 2 doses of CYD-TDV in dengue endemic areas by dengue serostatus. Vaccine, 38, 6472–6477. [DOI] [PubMed] [Google Scholar]
- Dayan GH, Langevin E, Gilbert PB, Wu Y, Moodie Z, Forrat R, Price B, Frago C, Bouckenooghe A, Cortes M, Noriega F & DiazGranados CA 2020b. Assessment of the long-term efficacy of a dengue vaccine against symptomatic, virologically-confirmed dengue disease by baseline dengue serostatus. Vaccine, 38, 3531–3536. [DOI] [PubMed] [Google Scholar]
- de Silva AM & Harris E 2018. Which Dengue Vaccine Approach Is the Most Promising, and Should We Be Concerned about Enhanced Disease after Vaccination? The Path to a Dengue Vaccine: Learning from Human Natural Dengue Infection Studies and Vaccine Trials. Cold Spring Harb Perspect Biol, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinski A, Jumnainsong A, Edwards C, Quyen NT, Duangchinda T, Grimes JM, Tsai WY, Lai CY, Wang WK, Malasit P, Farrar J, Simmons CP, Zhou ZH, Rey FA, Mongkolsapaya J & Screaton GR 2015a. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol, 16, 170–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinski A, Jumnainsong A, Edwards C, Quyen NTH, Duangchinda T, Grimes JM, Tsai WY, Lai CY, Wang WK, Malasit P, Farrar J, Simmons CP, Zhou ZH, Rey FA, Mongkolsapaya J & Screaton GR 2015b. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol, 16, 170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd KA, DeMaso CR & Pierson TC 2015. Genotypic Differences in Dengue Virus Neutralization Are Explained by a Single Amino Acid Mutation That Modulates Virus Breathing. MBio, 6, e01559–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forshey BM, Reiner RC, Olkowski S, Morrison AC, Espinoza A, Long KC, Vilcarromero S, Casanova W, Wearing HJ, Halsey ES, Kochel TJ, Scott TW & Stoddard ST 2016. Incomplete Protection against Dengue Virus Type 2 Re-infection in Peru. PLoS Negl Trop Dis, 10, e0004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallichotte EN, Baric RS & de Silva AM 2018a. The Molecular Specificity of the Human Antibody Response to Dengue Virus Infections. Adv Exp Med Biol, 1062, 63–76. [DOI] [PubMed] [Google Scholar]
- Gallichotte EN, Baric TJ, Nivarthi U, Delacruz MJ, Graham R, Widman DG, Yount BL, Durbin AP, Whitehead SS, de Silva AM & Baric RS 2018b. Genetic Variation between Dengue Virus Type 4 Strains Impacts Human Antibody Binding and Neutralization. Cell Rep, 25, 1214–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert PB, Huang Y, Juraska M, Moodie Z, Fong Y, Luedtke A, Zhuang Y, Shao J, Carpp LN, Jackson N, Chambonneau L, Bouckenooghe A, Zambrano B, Frago C, Pallardy S & Noriega F 2019. Bridging Efficacy of a Tetravalent Dengue Vaccine from Children/Adolescents to Adults in Highly Endemic Countries Based on Neutralizing Antibody Response. Am J Trop Med Hyg, 101, 164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirakhoo F, Pugachev K, Zhang Z, Myers G, Levenbook I, Draper K, Lang J, Ocran S, Mitchell F, Parsons M, Brown N, Brandler S, Fournier C, Barrere B, Rizvi F, Travassos A, Nichols R, Trent D & Monath T 2004. Safety and efficacy of chimeric yellow Fever-dengue virus tetravalent vaccine formulations in nonhuman primates. J Virol, 78, 4761–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy B, Briand O, Lang J, Saville M & Jackson N 2015. Development of the Sanofi Pasteur tetravalent dengue vaccine: One more step forward. Vaccine, 33, 7100–11. [DOI] [PubMed] [Google Scholar]
- Hadinegoro SR, Arredondo-Garcia JL, Capeding MR, Deseda C, Chotpitayasunondh T, Dietze R, Ismail HI, Reynales H, Limkittikul K, Rivera-Medina DM, Tran HN, Bouckenooghe A, Chansinghakul D, Cortes M, Fanouillere K, Forrat R, Frago C, Gailhardou S, Jackson N, Noriega F, Plennevaux E, Wartel TA, Zambrano B, Saville M & Group, C.-T. D. V. W. 2015. Efficacy and Long-Term Safety of a Dengue Vaccine in Regions of Endemic Disease. N Engl J Med. [DOI] [PubMed] [Google Scholar]
- Henein S, Adams C, Bonaparte M, Moser JM, Munteanu A, Baric R & Desilva AM 2021. Dengue vaccine breakthrough infections reveal properties of neutralizing antibodies linked to protection. J Clin Invest. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henein S, Swanstrom J, Byers AM, Moser JM, Shaik SF, Bonaparte M, Jackson N, Guy B, Baric R & de Silva AM 2017. Dissecting Antibodies Induced by a Chimeric Yellow Fever-Dengue, Live-Attenuated, Tetravalent Dengue Vaccine (CYD-TDV) in Naive and Dengue-Exposed Individuals. J Infect Dis, 215, 351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC & Twiddy SS 2003. The origin, emergence and evolutionary genetics of dengue virus. Infect Genet Evol, 3, 19–28. [DOI] [PubMed] [Google Scholar]
- Huang Y, Moodie Z, Juraska M, Fong Y, Carpp LN, Chambonneau L, Coronel DL, Dayan GH, DiazGranados CA & Gilbert PB 2020. Immunobridging efficacy of a tetravalent dengue vaccine against dengue and against hospitalized dengue from children/adolescents to adults in highly endemic countries. Trans R Soc Trop Med Hyg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juraska M, Magaret CA, Shao J, Carpp LN, Fiore-Gartland AJ, Benkeser D, Girerd-Chambaz Y, Langevin E, Frago C, Guy B, Jackson N, Duong Thi Hue K, Simmons CP, Edlefsen PT & Gilbert PB 2018. Viral genetic diversity and protective efficacy of a tetravalent dengue vaccine in two phase 3 trials. Proc Natl Acad Sci U S A, 115, E8378–E8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzelnick LC, Fonville JM, Gromowski GD, Bustos Arriaga J, Green A, James SL, Lau L, Montoya M, Wang C, VanBlargan LA, Russell CA, Thu HM, Pierson TC, Buchy P, Aaskov JG, Munoz-Jordan JL, Vasilakis N, Gibbons RV, Tesh RB, Osterhaus AD, Fouchier RA, Durbin A, Simmons CP, Holmes EC, Harris E, Whitehead SS & Smith DJ 2015. Dengue viruses cluster antigenically but not as discrete serotypes. Science, 349, 1338–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzelnick LC, Harris E & Participants in the Summit on Dengue Immune Correlates of, P. 2017. Immune correlates of protection for dengue: State of the art and research agenda. Vaccine, 35, 4659–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam HM, Phuong HT, Thao Vy NH, Le Thanh NT, Dung PN, Ngoc Muon TT, Van Vinh Chau N, Rodriguez-Barraquer I, Cummings DAT, Wills BA, Boni MF, Rabaa MA & Clapham HE 2019. Serological inference of past primary and secondary dengue infection: implications for vaccination. J R Soc Interface, 16, 20190207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanata CF, Andrade T, Gil AI, Terrones C, Valladolid O, Zambrano B, Saville M & Crevat D 2012. Immunogenicity and safety of tetravalent dengue vaccine in 2-11 year-olds previously vaccinated against yellow fever: randomized, controlled, phase II study in Piura, Peru. Vaccine, 30, 5935–41. [DOI] [PubMed] [Google Scholar]
- Leo YS, Wilder-Smith A, Archuleta S, Shek LP, Chong CY, Leong HN, Low CY, Oh ML, Bouckenooghe A, Wartel TA & Crevat D 2012. Immunogenicity and safety of recombinant tetravalent dengue vaccine (CYD-TDV) in individuals aged 2-45 y: Phase II randomized controlled trial in Singapore. Hum Vaccin Immunother, 8, 1259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodie Z, Juraska M, Huang Y, Zhuang Y, Fong Y, Carpp LN, Self SG, Chambonneau L, Small R, Jackson N, Noriega F & Gilbert PB 2017. Neutralizing Antibody Correlates Analysis of Tetravalent Dengue Vaccine Efficacy Trials in Asia and Latin America. J Infect Dis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D, Legg TJ, Billings CW, Forrat R, Yoksan S & Lang J 2010. A novel tetravalent dengue vaccine is well tolerated and immunogenic against all 4 serotypes in flavivirus-naive adults. J Infect Dis, 201, 370–7. [DOI] [PubMed] [Google Scholar]
- Nivarthi UK, Kose N, Sapparapu G, Widman D, Gallichotte E, Pfaff JM, Doranz BJ, Weiskopf D, Sette A, Durbin AP, Whitehead SS, Baric R, Crowe JE Jr. & de Silva AM 2017. Mapping the Human Memory B Cell and Serum Neutralizing Antibody Responses to Dengue Virus Serotype 4 Infection and Vaccination. J Virol, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel B, Longo P, Miley MJ, Montoya M, Harris E & de Silva AM 2017. Dissecting the human serum antibody response to secondary dengue virus infections. PLoS Negl Trop Dis, 11, e0005554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poo J, Galan F, Forrat R, Zambrano B, Lang J & Dayan G 2011. Live-attenuated Tetravalent Dengue Vaccine in Dengue-naive Children, Adolescents, and Adults in Mexico City: Randomized Controlled Phase 1 Trial of Safety and Immunogenicity. Pediatr Infect Dis J, 30, e9–17. [DOI] [PubMed] [Google Scholar]
- Rabaa MA, Girerd-Chambaz Y, Duong Thi Hue K, Vu Tuan T, Wills B, Bonaparte M, van der Vliet D, Langevin E, Cortes M, Zambrano B, Dunod C, Wartel-Tram A, Jackson N & Simmons CP 2017. Genetic epidemiology of dengue viruses in phase III trials of the CYD tetravalent dengue vaccine and implications for efficacy. Elife, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar S, Luedtke A, Langevin E, Zhu M, Bonaparte M, Machabert T, Savarino S, Zambrano B, Moureau A, Khromava A, Moodie Z, Westling T, Mascarenas C, Frago C, Cortes M, Chansinghakul D, Noriega F, Bouckenooghe A, Chen J, Ng SP, Gilbert PB, Gurunathan S & DiazGranados CA 2018. Effect of Dengue Serostatus on Dengue Vaccine Safety and Efficacy. N Engl J Med, 379, 327–340. [DOI] [PubMed] [Google Scholar]
- Thomas SJ & Yoon IK 2019. A review of Dengvaxia(R): development to deployment. Hum Vaccin Immunother, 15, 2295–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WY, Lai CY, Wu YC, Lin HE, Edwards C, Jumnainsong A, Kliks S, Halstead S, Mongkolsapaya J, Screaton GR & Wang WK 2013. High-avidity and potently neutralizing cross-reactive human monoclonal antibodies derived from secondary dengue virus infection. J Virol, 87, 12562–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar L, Dayan GH, Arredondo-Garcia JL, Rivera DM, Cunha R, Deseda C, Reynales H, Costa MS, Morales-Ramirez JO, Carrasquilla G, Rey LC, Dietze R, Luz K, Rivas E, Miranda Montoya MC, Cortes Supelano M, Zambrano B, Langevin E, Boaz M, Tornieporth N, Saville M, Noriega F & Group, C. Y. D. S. 2015. Efficacy of a tetravalent dengue vaccine in children in Latin America. N Engl J Med, 372, 113–23. [DOI] [PubMed] [Google Scholar]
- Waggoner JJ, Balmaseda A, Gresh L, Sahoo MK, Montoya M, Wang C, Abeynayake J, Kuan G, Pinsky BA & Harris E 2016. Homotypic Dengue Virus Reinfections in Nicaraguan Children. J Infect Dis, 214, 986–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahala WM, Donaldson EF, de Alwis R, Accavitti-Loper MA, Baric RS & de Silva AM 2010. Natural strain variation and antibody neutralization of dengue serotype 3 viruses. PLoS Pathog, 6, e1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC & Vasilakis N 2009. Molecular evolution of dengue viruses: contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect Genet Evol, 9, 523–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Baseline serostatus impacts DENV4 genotype I and II cross-neutralization. DENV4 GI, GIIa and GIIb neutralization titers by A) seronegative and B) seropositive post-vaccination immune sera. FRNT50 represents sera dilution factor required to neutralize 50% of virus. Dotted line represents limit of detection (20). Samples not neutralized are graphed at ½ the limit of detection (10). Neutralization titers against the different viruses were compared by one way ANOVA (*p<0.05, **p<0.005).
Supplementary Figure 2. MAb neutralization of DENV4 genotype I, II and chimeric envelope viruses. Wild-type (DENV GI, GII) and chimeric (GI/II, GII/I) virus neutralization by A) DENV4-specific and C) DENV CR MAbs. FRNT50 represents MAb concentration [ng/μl] required to neutralize 50% of virus. B) DENV4-specific and D) DENV CR MAb chimeric virus relative titer = (chimeric FRNT50 / parental wild-type FRNT50). Relative titer > 1 = parental wild-type virus better neutralized, relative titer < 1 = chimeric better neutralized.
Data Availability Statement
This paper does not report original code. All data generated for the project, including vaccine antibody titers, are archived in the Desilva laboratory. This data is available for review or reanalysis upon request from the Lead Author.


