Abstract
The long-term benefit of insecticidal products based on Cry toxins, either in sprays or as transgenic crops, is threatened by the development of resistance by target pests. The models used to predict evolution of resistance to Cry toxins most often are monogenic models in which two alleles are used. Moreover, the high-dose/refuge strategy recommended for implementation with transgenic crops relies on the assumption that the resistance allele is recessive. Using selection experiments, we demonstrated the occurrence in a laboratory colony of diamondback moth of two different genes (either allelic or nonallelic) that confer resistance to Cry1Ab. At the concentration tested, resistance was dominant in one selection line and partially recessive in the other. Resistant insects from the two selection lines also differed in their cross-resistance patterns. The diamondback moth colony was derived from a field population from the Philippines, which originally showed a different resistance phenotype. This is the first time that an insect population has been directly shown to carry more than one gene conferring resistance to the same Cry toxin.
Resistance to insecticides is a key issue in agriculture and in public health (with respect to control of insect-transmitted diseases) because of the capacity of insects to develop resistance to any pesticide to which they are exposed. More than 500 species of insects and mites have been reported to have developed resistance to one or more pesticides (12), and cases of resistance to pesticides, including biological insecticides such as those based on the bacterium Bacillus thuringiensis, continue to appear (11, 29).
At the onset of the sporulation phase, B. thuringiensis produces proteinaceous crystalline parasporal bodies (1). Some proteins in the crystals are active against insects, and for this reason they are generically called insecticidal crystal proteins, δ-endotoxins, Cry proteins, or Cry toxins. There are many formulations based on a mixture of spores and crystals from different B. thuringiensis strains. These formulations have been used for many years as alternatives to chemical insecticides when resistance to other insecticides is severe, when natural enemies need to be preserved, when application just before harvest is necessary, or when organic farming methods are used. Since 1987 some B. thuringiensis genes coding for Cry proteins have been transferred to the genomes of plants, which have become resistant to insects (for reviews see references 16 and 19). Despite the high number of plant species transformed to date with B. thuringiensis genes, only two transformed crops (corn and cotton) are planted widely in the United States and, on a smaller scale, in other parts of the world. In 2000, a total of 11.5 million hectares was dedicated to these crops (including plants with both B. thuringiensis and herbicide tolerance); this represents 26% of total transgenic area (15).
Sooner or later, extensive use of B. thuringiensis-based insecticide sprays and particularly the high selection pressure exerted by B. thuringiensis cultivars will lead to insect populations that develop resistance to Cry toxins. In fact, there are a number of insect species that have already developed resistance to single Cry toxins or mixtures of toxins in laboratory selection experiments (11, 29). So far, the diamondback moth, Plutella xylostella, is the only pest that has developed resistance to B. thuringiensis in the field. Genetic and biochemical studies with resistant insects belonging to different species have allowed workers to draw the following general conclusions concerning B. thuringiensis resistance: (i) in all cases this resistance is autosomally inherited; (ii) in most cases resistance is due to a recessive allele; and (iii) high levels of resistance and cross-resistance are generally related to a lack of toxin binding to midgut receptors. Resistance due to dominant (14, 22, 28) alleles and high levels of resistance not explained by receptor binding alteration (31) have been reported in a few cases.
A strategy that has been widely recommended to delay resistance to B. thuringiensis in insect populations in the field is to combine the high-dose strategy (expression by plants of a level of toxin sufficient to kill all heterozygous insects) with the use of refuges (plots containing non-B. thuringiensis-treated plants) (21; http://www.epa.gov/pesticides/biopesticides). However, for this strategy to be effective, resistance has to be recessive, random mating must occur between susceptible and resistant individuals, and the frequency of resistance alleles must be low (23). Models used to predict evolution of resistance to a given Cry toxin most often assume that resistance is due to one gene with two alleles, one susceptible allele and one resistant allele. A few studies have suggested that more than one gene conferring resistance to a given Cry toxin is present in the same population (13, 24, 27, 28), but direct evidence of this is not yet available.
In this study we used the PHI colony, which was derived from insects that were collected in the Philippines from B. thuringiensis-treated fields and originally showed high levels of resistance to Cry1Ab (3, 4). After selection in the laboratory, this colony was shown to have independent genetic control of Cry1Aa and Cry1Ab resistance (28). By selecting two sample lines of the PHI colony with different selective agents and with different larval instars, we obtained evidence that there are two different genes (either allelic or nonallelic) that confer resistance to Cry1Ab (we use the term gene in this paper to indicate genetic variants, regardless of whether they occur at the same locus or at different loci). Resistant insects obtained from the two selection lines differed in their patterns of cross-resistance and in their inheritance of resistance. Along with the findings of previous studies showing that the PHI colony contains other genes for resistance to Cry1A toxins, our findings indicate the high degree of variability in B. thuringiensis resistance genes that can be present in field populations.
MATERIALS AND METHODS
Insects.
The PHI colony was derived from 130 pupae collected in the Philippines in 1993 (4). During the 7 years that this colony has been maintained in the laboratory, insects were subjected to selection with Cry1Ab and then with MYX 03604, a product containing chimeric Cry1Ab-Cry1Ac protoxin (domain I and almost all domain II from Cry1Ac plus a small part of domain II, domain III, and the C-terminal half of the protein from Cry1Ab) expressed in recombinant Pseudomonas fluorescens (Mycogen Corporation, San Diego, Calif.) (3). After selection was discontinued, the resistance values reverted to values close to those of the control strain. The LAB-V strain, which originated from The Netherlands, was used as the susceptible control and had never been exposed to B. thuringiensis (10). All insects were reared on fresh cabbage leaves at 25°C with 60% relative humidity and a photoperiod consisting of 16 h of light and 8 h of darkness.
Cry toxins.
Cry1Aa, Cry1Ab, Cry1Ac, Cry1F, and Cry1J were obtained from recombinant B. thuringiensis strains EG1273, EG7077, EG11070, EG11069, and EG7279, respectively (Ecogen Inc.). Bacteria were grown for 48 h in CCY medium (25) supplemented with the appropriate antibiotic. Spores and crystals were collected by centrifugation at 9,700 × g for 10 min at 4°C. Each pellet was washed four times with a 1 M NaCl–10 mM EDTA solution, and then it was thoroughly suspended in 10 mM KCl. Purification and activation of toxins were carried out by alkaline solubilization and trypsin activation as previously described (24).
Toxins used for labeling and binding experiments were chromatographically purified by using a MonoQ HR 5/5 anion-exchange column (fast protein liquid chromatography system; Pharmacia, Uppsala, Sweden) (24).
Protein concentrations in solutions of activated toxins were determined by the method of Bradford (8). The concentrations of spore-crystal suspensions were expressed in units of optical density at 600 nm (OD600 units).
Bioassays.
Mortality was scored after 48 h. Groups of 10 third-instar larvae were placed on cabbage leaf discs that previously had been dipped in a test solution containing the surfactant 0.2% Triton AG-98. Dilutions of toxins were prepared with 50 mM carbonate buffer (pH 10.5). Dilutions of spore-crystal mixtures were prepared with distilled water. Control leaves were dipped in distilled water containing 0.2% Triton AG-98. Five dilutions of toxins were used to estimate the concentrations that killed 50% of the larvae tested (LC50) with the Polo-PC program (17). The LC50s reported below are means based on two independent experiments. Single-point mortality tests with Cry1F and Cry1J were performed with 50 larvae for each concentration of toxin. These tests were performed twice.
Selection.
Selection experiments were performed with two samples of the PHI colony. For the first sample, approximately 1,000 eggs were transferred to cabbage leaves that previously had been dipped in a mixture of spores and crystals of Cry1Aa (0.074 OD600 unit). Two days after hatching, additional treated leaves were added, and larvae were allowed to feed for two more days. Then, fresh untreated leaves were added until pupation. The emerged adults were pooled to produce progeny for the next generation. The selection process was continued until generation 13, although selective pressure was applied only in 10 generations (selective pressure was not applied in generations 8, 9, and 12). The resulting selection line was called Sel-A.
The other sample was selected with activated Cry1Ab. Close to 300 third-instar larvae were placed on leaf discs that previously had been dipped in a solution containing 50 mg of Cry1Ab per liter. After 2 days, the survivors were transferred to fresh untreated leaves until pupation. The emerged adults were pooled to produce progeny for the next generation. Selection was applied for three generations, and the resulting selection line was called Sel-B.
Evaluation of dominance.
Bioassays to determine the type of inheritance were carried out with a solution containing 50 mg of Cry1Ab per liter by crossing resistant individuals (Sel-A or Sel-B) with susceptible LAB-V individuals. For single-pair crosses, one virgin male was caged together with one virgin female for mating and egg production. The sexes of the parents were selected randomly. Only single pairs that produced enough progeny were used in Cry1Ab bioassays. Before genetic analysis, Sel-A individuals went through one generation without selection, and then they were treated with a solution containing 50 mg of Cry1Ab per liter to eliminate individuals susceptible to this concentration of toxin.
Effective dominance (DML) was calculated from mortality values at a single concentration (6), as follows: DML = (MLRS − MLSS)/(MLRR − MLSS), where MLRR, MLRS, and MLSS are the mortality values at a particular toxin concentration for the resistant line, the F1 progeny, and the susceptible strain, respectively. The DML values range from 0 (completely recessive resistance) to 1 (completely dominant resistance).
Binding assays.
Binding assays were performed with brush border membrane vesicles (BBMV) prepared from whole fourth-instar Sel-A, Sel-B, and LAB-V larvae (9). Total protein concentrations of BBMV preparations were determined by the method of Bradford (8). Activated Cry1Ab and Cry1Ac toxins were labeled with 125I by the chloramine-T method (30). Binding experiments were conducted as described previously (31), except that BBMV were incubated with labeled toxin for 30 min. At the highest concentration of BBMV used, the levels of total binding of 125I-labeled Cry1Ab were 9.5% for LAB-V, 1.9% for Sel-A, and 1.7% for Sel-B, and the levels of total binding of 125I-labeled Cry1Ac were 35% for LAB-V and 2.4% for Sel-B.
RESULTS
Response to selection and cross-resistance.
Two samples of the PHI colony were selected with different B. thuringiensis products. Selection line Sel-A was derived from a PHI sample selected with a mixture of spores and crystals containing only Cry1Aa protoxin. Before selection, the LC50 for the spore-crystal mixture was 0.074 OD600 unit (95% fiducial limits [FL95], 0.006 to 0.176 OD600 unit) (Table 1). After 10 generations of selection, Sel-A did not show any significant response to the selective agent (LC50, 0.23 OD600 unit; FL95, 0.08 to 0.50 OD600 unit). The other selection line, Sel-B, was derived from a second PHI sample and was selected with solubilized and trypsin-activated Cry1Ab toxin. In this case, we observed a strong response to the selective agent. The LC50 of Cry1Ab changed from 2.08 mg/liter (FL95, 1.44 to 3.20 mg/liter) to 143 mg/liter (FL95, 119 to 186 mg/liter) in just three generations of selection (Table 1).
TABLE 1.
Toxicities of activated Cry1A toxins and spores and crystals of Cry1Aa
| Insects | Toxin | LC50 (FL95)a | Resistance ratiob |
|---|---|---|---|
| Sel-A | Cry1Aa (S+C) | 0.23 (0.08–0.50) | 6.4 |
| Cry1Aa | 4.47 (2.66–6.18) | 1.3 | |
| Cry1Ab | 36.2 (23.14–55.17) | 60.3 | |
| Cry1Ac | 26.1 (14.7–43.45) | 118.6 | |
| Sel-B | Cry1Aa (S+C) | 0.15 (0.06–0.38) | 4.2 |
| Cry1Aa | 2.70 (1.16–4.53) | 0.8 | |
| Cry1Ab | 143 (119–186) | 238 | |
| Cry1Ac | 22.7 (12.0–62.1) | 103.2 | |
| PHI | Cry1Aa (S+C) | 0.074 (0.006–0.176) | 2.1 |
| Cry1Aa | 11.73 (5.54–24.69) | 3.5 | |
| Cry1Ab | 2.08 (1.44–3.20) | 3.5 | |
| Cry1Ac | 0.93 (0.56–1.40) | 4.2 | |
| LAB-V | Cry1Aa (S+C) | 0.036 (0.019–0.056) | |
| Cry1Aa | 3.32 (1.53–6.98) | ||
| Cry1Ab | 0.60 (0.40–0.86) | ||
| Cry1Ac | 0.22 (0.14–0.31) |
Values for spores and crystals (S+C) of Cry1Aa are expressed in OD600 units. Other values are expressed in milligrams per liter.
Resistance ratio = LC50 of resistant strain/LC50 of LAB-V.
Despite the differences in the responses to the selective agents described above, both selection lines showed very similar patterns of cross-resistance to the Cry1A toxins (Table 1). In both cases there was a significant increase in resistance to Cry1Ab and Cry1Ac and no increase in resistance to Cry1Aa in either form (solubilized activated toxin or crystallized protoxin with spores). However, it is important to note that the responses to Cry1Ab were significantly different in the two selection lines.
Bioassays performed with Cry1F and Cry1J also revealed differences between the two selection lines (Table 2). Sel-B developed cross-resistance to Cry1F, whereas Sel-A was even more susceptible than the control LAB-V strain. For Cry1J, the susceptibilities of LAB-V and Sel-B were essentially the same, but Sel-A had significantly higher mortality at the two concentrations tested.
TABLE 2.
Toxicities of activated Cry1F and Cry1J toxins
| Insects | Toxin | Concn (mg/liter) | % Mortality (FL95)a |
|---|---|---|---|
| Sel-A | Cry1F | 1 | 46 (33–60) |
| 10 | 88 (76–95) | ||
| Cry1J | 1 | 73 (59–83) | |
| 10 | 98 (88–100) | ||
| Sel-B | Cry1F | 1 | 0 (0–9) |
| 10 | 0 (0–9) | ||
| Cry1J | 1 | 2 (0–12) | |
| 10 | 38 (26–52) | ||
| LAB-V | Cry1F | 1 | 8 (3–20) |
| 10 | 47 (34–61) | ||
| Cry1J | 1 | 8 (3–20) | |
| 10 | 44 (31–58) |
FL95 were estimated by the modified exact Wald method (2).
Inheritance of resistance to Cry1Ab.
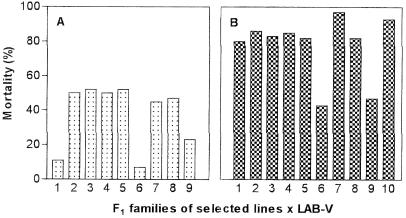
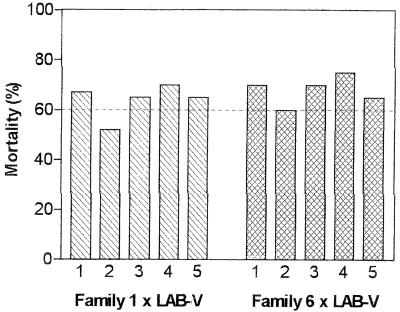
Analysis of the F1 progeny from single-pair crosses between resistant insects and susceptible strain LAB-V insects clearly showed that there were differences in the mode of inheritance of Cry1Ab resistance in the two selection lines at the test concentration used (50 mg/liter). Bioassays of the progeny from the cross between Sel-A and LAB-V suggested that resistance to Cry1Ab was due to an autosomal dominant gene (A) at a single locus (Fig. 1A). Cry1Ab produced around 50% mortality in six of nine F1 families (families 2, 3, 4, 5, 7, and 8), around 10% mortality in two families (families 1 and 6), and 23% mortality in one family (family 9). The sex of the parental insects did not have any effect on the results. The results obtained for the six families with mortality values around 50% corresponded to the segregation expected if the Sel-A parents were heterozygous (AS). The results obtained for the two families with mortality values around 10% are consistent with crosses with homozygous Sel-A parents (AA). If this occurred, the F1 progeny from these two families should have been heterozygous (AS) and, when crossed with LAB-V insects (SS), should produce 50% resistant offspring and 50% susceptible offspring. To test this hypothesis, we crossed the family 1 and 6 survivors with LAB-V insects in single-pair mating experiments. Five crosses for each family produced enough offspring, and in all cases mortality was close to 60% (the expected level of mortality, since the resistant parents had 10% mortality at the test concentration) when the insects were exposed to Cry1Ab (Fig. 2). Because mortality in the presence of Cry1Ab was not determined with the Sel-A parents used in the initial crosses, we could not tell whether this was a case of partial dominance or a case of complete dominance. Finally, the 23% mortality for family 9 seemed to have been strongly affected by environmental conditions, and therefore this result is not informative.
FIG. 1.
Mortalities of F1 progeny from single-pair crosses between insects from selected lines and LAB-V insects in the presence of 50 mg of Cry1Ab per liter. (A) Sel-A × LAB-V; (B) Sel-B × LAB-V. Assays were performed with an average of 38 larvae.
FIG. 2.
Mortalities of the offspring from two test crosses of F1 insects (families 1 and 6 in Fig. 1A) with LAB-V insects in the presence of 50 mg of Cry1Ab per liter.
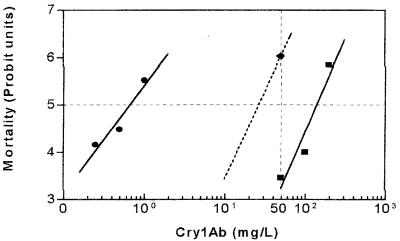
The pattern of resistance to Cry1Ab in the Sel-B line was different. Analysis of the F1 progeny from the cross between Sel-B and LAB-V insects suggested that resistance was due to an autosomal partially recessive gene (b) at a single locus (Fig. 1B). Cry1Ab produced mortalities ranging from 80 to 97% (mean, 86%) in 8 of the 10 families tested and of around 45% (43 and 47%) in the other two families. No effect of the sex of the parents on the progeny was detected. The results obtained for the eight families with a mean mortality value of 86% corresponded to results expected for progeny of homozygous Sel-B parents (bb) crossed with homozygous susceptible LAB-V insects (ss). Since at the test concentration the resistant parents had a mortality of 4% and the susceptible parents had a mortality of 100% (Fig. 3), we calculated that the effective dominance of resistance (DML) was 0.15, which corresponded to partially recessive inheritance. The simplest explanation for the two families with mortalities around 45% is that the LAB-V insects used in these crosses were heterozygous (bs) for the resistance gene. The progeny of a homozygous Sel-B insect with a heterozygous LAB-V insect would be 50% bb and 50% bs, which would give a global mortality of [(0.5 × 0.04) + (0.5 × 0.86)] × 100% or 45%. To test this hypothesis, survivors in these two F1 families were allowed to mate among themselves (within each family), and the F2 progeny were exposed to Cry1Ab and Cry1F. If our hypothesis was correct, the contributions to the F2 offspring of the different parental genotypes would be f(bb) = (0.5 × 0.96/0.55) = 0.873 and f(bs) = (0.5 × 0.14/0.55) = 0.127. The allele frequencies in the parental F1 generation would then be f(b) = 0.873 + (0.5 × 0.127) = 0.9365 and f(s) = 0.5 × 0.127 = 0.0635. Assuming that random mating occurred, Hardy-Weinberg equilibrium can be applied, and the expected frequencies of genotypes in the F2 generation would be f(bb) = 0.877, f(bs) = 0.119, and f(ss) = 0.004. If the empirical mortality data for these three genotypes at the Cry1Ab concentration tested (4% for bb, 86% for bs, and 100% for ss) were used, the overall expected mortality with this toxin would be 14%. The actual mortality observed in the F2 generation was 20% (n = 41 or 49, depending on the family), which is not significantly different from the calculated value (P < 0.05). In the case of the Cry1F toxin, the mortalities for the bb and ss genotypes were 0 and 47%, respectively (Table 2). If it was assumed that the mortality for the bs genotype at the concentration tested (10 mg/liter) was 47%, the overall expected mortality would be 5.8%. Our actual results with Cry1F gave a mortality for the F2 generation of 10% (n = 50 for each family), which again was close to the calculated value and consistent with the hypothesis that two individuals of the LAB-V strain were heterozygous for the resistant gene.
FIG. 3.
Concentration-mortality responses of LAB-V (●) and Sel-B (▪) insects when they were tested with activated Cry1Ab. The dashed line indicates the expected response of the F1 generation (Sel-B × LAB-V) based on the mortality observed in the presence of 50 mg/liter (⧫), assuming that the slope was the same as that of the regression line for Sel-B.
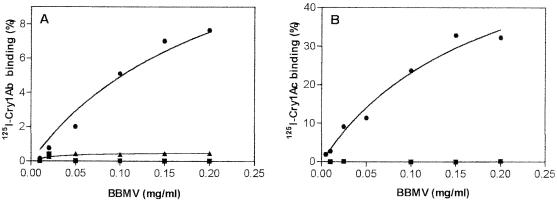
Binding assays.
BBMV from LAB-V, used as a control susceptible strain, exhibited specific binding of 125I-labeled Cry1Ab and 125I-labeled Cry1Ac (Fig. 4). In contrast, BBMV from Sel-B exhibited no specific binding at all with either labeled toxin. BBMV from Sel-A were tested only with 125I-labeled Cry1Ab and exhibited highly reduced binding.
FIG. 4.
Specific binding of 125I-labeled Cry1Ab (A) and 125I-labeled Cry1Ac (B) as a function of BBMV protein concentration in the resistant lines and the susceptible control strain. Symbols: ▴, Sel-A; ▪, Sel-B; ●, LAB-V.
DISCUSSION
The analysis of susceptibility to Cry1Ab, Cry1F, and Cry1J and the genetic analysis showed that the types of resistance obtained in the two selection experiments were substantially different. Since at the concentration of Cry1Ab used (50 mg/liter), resistance in the Sel-A line is dominant, we could not perform complementation tests to determine whether the mutations that confer resistance are alleles of the same locus. At this concentration the two selection lines clearly differed in terms of the type of inheritance of Cry1Ab resistance, which is dominant in Sel-A and partially recessive in Sel-B.
In the selection experiments described here, similar patterns of resistance to Cry1A toxins were obtained when Cry1Aa protoxin plus spores and activated Cry1Ab were used as selective agents. It seems that the initial frequency of genes conferring resistance to Cry1Ab and Cry1Ac in the PHI colony must have been much higher than the initial frequency of genes conferring resistance to Cry1Aa, and thus the former were selected even when Cry1Aa was used as the selective agent. Previously, Tabashnik et al. (28) showed that in the PHI colony, there is evidence of independent genetic control of resistance to Cry1Aa and Cry1Ab. The fact that Cry1Ab selected for Cry1Ac resistance indicates that there are mechanisms of resistance which are shared by these toxins and can be explained by alteration of a common binding site in the midgut receptor, as shown by the binding analysis. Why selection with Cry1Aa selected for Cry1Ab and Cry1Ac resistance but not for Cry1Aa resistance is more difficult to explain. Any mechanism related to crystal solubilization or protoxin processing would be irrelevant when the insects are tested with activated Cry1A toxins. Moreover, any mechanism related to the activated toxin should have selected for Cry1Aa as well. It is possible that because the selection procedure was performed with neonate larvae and the bioassays were performed with third-instar larvae, a Cry1Aa-resistant phenotype in neonates was missed. In addition, because Sel-A went through important bottlenecks during the selection process (in generation 11 the number of parents was limited to 10 individuals), genetic drift might have influenced the final resistance phenotype of Sel-A.
The PHI colony was derived from a field population from the Philippines. The first time that bioassays were carried out, insects showed high resistance to Cry1Ab but not to Cry1Aa or Cry1Ac (4), indicating that there was a gene that conferred resistance to Cry1Ab but did not provide protection against the other Cry1A toxins. This was in agreement with the finding that BBMV from the resistant insects did not bind Cry1Ab, whereas they bound Cry1Aa and Cry1Ac (5, 28). To maintain or even increase the level of resistance, this colony was subjected to selection with pure Cry1Ab for several generations, and the resistance pattern did not change. However, resistance to Cry1Aa and Cry1Ac started to build up when the colony was exposed to MYX 03604, a product containing a chimeric Cry1Ac-Cry1Ab protoxin (3). After selection with this product, the PHI colony became resistant to Cry1Aa, Cry1Ab, and Cry1Ac but not to Cry1F (28). In the present study we obtained a new phenotype (Sel-B) with resistance to Cry1Ab, Cry1Ac, and Cry1F, most likely caused by alteration of the common Cry1Ab/Cry1Ac/Cry1F receptor (5). Therefore, the PHI population has been shown to carry genes conferring resistance to (i) just Cry1Ab (4); (ii) Cry1Aa (28); (iii) Cry1Ab and Cry1Ac but not Cry1F, with a loss of Cry1Ab binding but not of Cry1Ac binding (28); and (iv) Cry1Ab, Cry1Ac, and Cry1F, with a loss of Cry1Ab and Cry1Ac binding (Sel-B in this study). We cannot completely eliminate the possibility that the resistance gene in the Sel-A line is the same gene that was previously reported for the PHI colony (28). In both cases the insects were resistant to Cry1Ab and Cry1Ac but not to Cry1F. Although PHI insects were found to be resistant to Cry1Aa as well, resistance to this toxin was shown to be independent of resistance to Cry1Ab and Cry1Ac. Finally, the finding that resistance to Cry1Ab in Sel-A is dominant whereas it was previously found to be recessive in the PHI colony might have been due to differences in the concentration or the toxin form used (7). In summary, the PHI population must have carried at least one gene for resistance to Cry1Aa, one gene for resistance to Cry1Ab, one gene for resistance to Cry1Ac (not affecting receptor binding), and one gene conferring resistance to Cry1Ab, Cry1Ac, and Cry1F that altered binding. In the latter case, if binding of Cry1F was not affected, at least one additional mutation would be required for resistance to Cry1F. The PHI population is not unique, because other populations have been shown to exhibit substantial genetic variation in resistance to Cry1A toxins (20, 26, 27).
The presence of heterozygous individuals in the LAB-V strain is not completely surprising, since the presence of resistance genes has been reported in other susceptible control strains (18, 27). Furthermore, we cannot eliminate the possibility that there was contamination of the susceptible colony by resistant PHI insects. An alternative explanation for the two families with 45% mortality in the F1 generation resulting from the cross between Sel-B and LAB-V is that the selection that led to the Sel-B line also selected for dominant A genes. These two families would be the result of the cross of two AS insects from the Sel-B line with susceptible homozygotes from the LAB-V line. However, in this case we would expect most members of the F2 generation to be susceptible to Cry1F, which does not agree with the actual results.
Binding of Cry1Ab was strongly reduced in the two selection lines. Since the Sel-A line is not homozygous for the resistance gene, it is expected to produce susceptible homozygotes once selection is discontinued. Because this was the case when the binding analysis was performed, the residual binding observed with BBMV from Sel-A insects must have been due to the contribution of susceptible individuals. Therefore, absence of Cry1Ab binding seems to have been the cause of resistance to this toxin in both selection lines. In addition, resistance to Cry1Ac, at least in Sel-B insects, seemed to be also due to an absence of binding. Since Sel-B insects are also resistant to Cry1F, it is likely that the resistance mechanism is an alteration in the common receptor affecting binding of Cry1Ab, Cry1Ac, and Cry1F (5).
The two selection procedures applied to insects from the PHI colony in the present study had important differences. With regard to the selective agents used, besides the presence of spores, the differences were restricted not only to the primary structure of the toxin but also to the level of processing (protoxin versus activated toxin) and its physical state (crystal versus solubilized). Moreover, in Sel-A the selective pressure was exerted on neonate larvae, whereas in Sel-B it was exerted on third-instar larvae. It is worth bearing in mind that under field conditions insects can encounter either soluble Cry toxins in transgenic plants or B. thuringiensis spores and crystalline inclusions in sprayed plants or both and that different instars may be exposed to the selective agent. All these variables have an influence on the final resistance outcome.
Another result of our work is that, based on different cross-resistance patterns and types of inheritance, we found evidence of at least two distinct genes conferring resistance to the same Cry toxin (Cry1Ab) in the same insect population. Although the presence of more than one gene conferring resistance to the same Cry toxin has been suggested in other studies, the evidence reported here is more direct than the evidence provided previously (13, 24, 27, 28). Our results indicate that insect populations may carry, more frequently than has been assumed, more than one gene involved in resistance to a given Cry toxin or even to a set of toxins if cross-resistance appears. Our results have important implications for resistance management strategies, since they show the high variability of B. thuringiensis resistance genes present in field populations and since they stress the effect of the selective agent and/or larval instar on the final resistance outcome. Models to predict the evolution of resistance in hypothetical scenarios and especially when management strategies are designed should consider high genetic variability to be not a rare phenomenon but a very common phenomenon in field populations.
ACKNOWLEDGMENTS
We thank Bruce E. Tabashnik for his thoughtful comments on the manuscript. We also thank Luis Calzada for his technical assistance and Ecogen for providing B. thuringiensis strains.
This work was supported by Agencia Española de Cooperación Internacional (AECI) grant AECI99-02-1°. J.G.C. was funded by an AECI fellowship.
REFERENCES
- 1.Agaisse H, Lereclus D. How does Bacillus thuringiensis produce so much insecticidal crystal protein? J Bacteriol. 1995;177:6027–6032. doi: 10.1128/jb.177.21.6027-6032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agresti A, Coull B A. Approximate is better than “exact” for interval estimation of binomial proportion. Am Stat. 1998;52:119–126. [Google Scholar]
- 3.Ballester V. Resistencia a δ-endotoxinas de Bacillus thuringiensis en poblaciones naturales de Plutella xylostella. Ph.D. thesis. Valencia, Spain: University of Valencia; 1997. [Google Scholar]
- 4.Ballester V, Escriche B, Ménsua J L, Riethmacher G W, Ferré J. Lack of cross-resistance to other Bacillus thuringiensis crystal proteins in a population of Plutella xylostella highly resistant to CryIA(b) Biocontrol Sci Technol. 1994;4:437–443. [Google Scholar]
- 5.Ballester V, Granero F, Tabashnik B E, Malvar T, Ferré J. Integrative model for binding of Bacillus thuringiensis toxins in susceptible and resistant larvae of the diamondback moth (Plutella xylostella) Appl Environ Microbiol. 1999;65:1413–1419. doi: 10.1128/aem.65.4.1413-1419.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourguet D, Genissel A, Raymond M. Insecticide resistance and dominance levels. J Econ Entomol. 2000;93:1588–1595. doi: 10.1603/0022-0493-93.6.1588. [DOI] [PubMed] [Google Scholar]
- 7.Bourguet D, Prout M, Raymond M. Dominance of insecticide resistance presents a plastic response. Genetics. 1996;143:407–416. doi: 10.1093/genetics/143.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Escriche B, Silva F J, Ferré J. Testing suitability of brush border membrane vesicles prepared from whole larvae from small insects for binding studies with Bacillus thuringiensis CryIA(b) crystal protein. J Invertebr Pathol. 1995;65:318–320. [Google Scholar]
- 10.Ferré J, Real M D, Van Rie J, Jansens S, Peferoen M. Resistance to the Bacillus thuringiensis bioinsecticide in a field population of Plutella xylostella is due to a change in a midgut membrane receptor. Proc Natl Acad Sci USA. 1991;88:5119–5123. doi: 10.1073/pnas.88.12.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frutos R, Rang C, Royer M. Managing insect resistance to plants producing Bacillus thuringiensis toxins. Crit Rev Biotechnol. 1999;19:227–276. [Google Scholar]
- 12.Georghiou G P, Lagunes-Tejeda A. The occurrence of resistance to pesticides in arthropods. Rome, Italy: Food and Agriculture Organization of the United Nations; 1991. [Google Scholar]
- 13.Herrero S, Oppert B, Ferré J. Different mechanisms of resistance to Bacillus thuringiensis toxins in the Indianmeal moth. Appl Environ Microbiol. 2001;67:1085–1089. doi: 10.1128/AEM.67.3.1085-1089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang F, Buschman L L, Higgins R A, McGaughey W H. Inheritance of resistance to Bacillus thuringiensis toxin (Dipel ES) in the European corn borer. Science. 1999;284:965–967. doi: 10.1126/science.284.5416.965. [DOI] [PubMed] [Google Scholar]
- 15.James C. Global status of commercialized transgenic crops: 2000. Preview. Ithaca, N.Y: International Service for the Acquisition of Agri-Biotech Applications; 2000. [Google Scholar]
- 16.Jenkins J N. Transgenic plants expressing toxins from Bacillus thuringiensis. Biopesticides. 1999;5:211–232. [Google Scholar]
- 17.LeOra Software. POLO-PC: a user's guide to probit or logit analysis. Berkeley, Calif: LeOra Software; 1987. [Google Scholar]
- 18.Liu Y-B, Tabashnik B E. Elimination of a recessive allele conferring resistance to Bacillus thuringiensis from a heterogeneous strain of diamondback moth (Lepidoptera: Plutellidae) J Econ Entomol. 1998;91:1032–1037. [Google Scholar]
- 19.Mazier M, Pannetier C, Tourneur J, Jouanin L, Giband M. The expression of Bacillus thuringiensis genes in plant cells. Biotechnol Annu Rev. 1997;3:313–347. [Google Scholar]
- 20.McGaughey W H, Johnson D E. Influence of crystal protein composition of Bacillus thuringiensis strains on cross-resistance in Indian- meal moth (Lepidoptera: Pyralidae) J Econ Entomol. 1994;87:535–540. [Google Scholar]
- 21.Mellon M, Rissler J, editors. Now or never: serious new plans to save a natural pest control. Cambridge, Mass: Union of Concerned Scientists; 1998. [Google Scholar]
- 22.Rahardja U, Whalon M E. Inheritance of resistance to Bacillus thuringiensis subsp. tenebrionis CryIIIA delta-endotoxin in Colorado potato beetle (Coleoptera: Chrysomelidae) J Econ Entomol. 1995;88:21–26. doi: 10.1093/jee/88.1.21. [DOI] [PubMed] [Google Scholar]
- 23.Roush R T. Managing pests and their resistance to Bacillus thuringiensis: can transgenic plants be better than sprays? Biocontrol Sci Technol. 1994;4:501–516. [Google Scholar]
- 24.Sayyed A H, Haward R, Herrero S, Ferré J, Wright D J. Genetic and biochemical approach for characterization of resistance to Bacillus thuringiensis toxin Cry1Ac in a field population of the diamondback moth, Plutella xylostella. Appl Environ Microbiol. 2000;66:1509–1516. doi: 10.1128/aem.66.4.1509-1516.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart G S A B, Johnstone K, Hagelberg E, Ellar D J. Commitment of bacterial spores to germinate. Biochem J. 1981;198:101–106. doi: 10.1042/bj1980101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tabashnik B E, Finson N, Marshall W J, Heckel D G. Prolonged selection affects stability of resistance to Bacillus thuringiensis in diamondback moth (Lepidoptera: Plutellidae) J Econ Entomol. 1995;88:219–224. [Google Scholar]
- 27.Tabashnik B E, Liu Y-B, Finson N, Masson L, Heckel D G. One gene in diamondback moth confers resistance to four Bacillus thuringiensis toxins. Proc Natl Acad Sci USA. 1997;94:1640–1644. doi: 10.1073/pnas.94.5.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabashnik B E, Liu Y-B, Malvar T, Heckel D G, Masson L, Ballester V, Granero F, Ménsua J L, Ferré J. Global variation in the genetic and biochemical basis of diamondback moth resistance to Bacillus thuringiensis. Proc Natl Acad Sci USA. 1997;94:12780–12785. doi: 10.1073/pnas.94.24.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Rie J, Ferré J. Insect resistance to Bacillus thuringiensis crystal proteins. In: Charles J F, Delecluse A, Nielsen-LeRoux C, editors. Entomopathogenic bacteria: from laboratory to field applications. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2000. pp. 219–237. [Google Scholar]
- 30.Van Rie J, Jansens S, Höfte H, Degheele D, Van Mellaert H. Receptors on the brush border membrane of the insect midgut as determinants of the specificity of Bacillus thuringiensis delta-endotoxins. Appl Environ Microbiol. 1990;56:1378–1385. doi: 10.1128/aem.56.5.1378-1385.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao J-Z, Collins H L, Tang J D, Cao J, Earle E D, Roush R T, Herrero S, Escriche B, Ferré J, Shelton A M. Development and characterization of diamondback moth resistance to transgenic broccoli expressing high levels of Cry1C. Appl Environ Microbiol. 2000;66:3784–3789. doi: 10.1128/aem.66.9.3784-3789.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]