Abstract
Trichoderma virens (synonym, Gliocladium virens), a deuteromycete fungus, suppresses soilborne plant diseases caused by a number of fungi and is used as a biocontrol agent. Several traits that may contribute to the antagonistic interactions of T. virens with disease-causing fungi involve the production of peptide metabolites (e.g., the antibiotic gliotoxin and siderophores used for iron acquisition). We cloned a 5,056-bp partial cDNA encoding a putative peptide synthetase (Psy1) from T. virens using conserved motifs found within the adenylate domain of peptide synthetases. Sequence similarities with conserved motifs of the adenylation domain, acyl transfer, and two condensation domains support identification of the Psy1 gene as a gene that encodes a peptide synthetase. Disruption of the native Psy1 gene through gene replacement was used to identify the function of this gene. Psy1 disruptants produced normal amounts of gliotoxin but grew poorly under low-iron conditions, suggesting that Psy1 plays a role in siderophore production. Psy1 disruptants cannot produce the major T. virens siderophore dimerum acid, a dipetide of acylated Nδ-hydroxyornithine. Biocontrol activity against damping-off diseases caused by Pythium ultimum and Rhizoctonia solani was not reduced by the Psy1 disruption, suggesting that iron competition through dimerum acid production does not contribute significantly to disease suppression activity under the conditions used.
Trichoderma virens (synonym, Gliocladium virens [30]) is an effective biological control agent for plant diseases caused by soilborne fungi (26). The disease-suppressive ability of T. virens is presumably related to its antagonistic behavior towards other fungi. Multiple traits may contribute to this antagonistic activity, particularly since the modes of interaction appear to differ for different target fungi (17). Production of gliotoxin, an epidithiodiketopiperazine antibiotic, is associated with T. virens suppression of damping-off caused by Pythium ultimum (24, 25, 37). Production of additional antibiotics, competition for resources in the soil or rhizosphere, and hyperparasitism of target fungi are traits that may account for other antagonistic interactions (7). Cloning of genes associated with antagonistic traits could permit manipulation of the genes in T. virens and could increase the biocontrol activity of this organism.
Small peptides are associated with a number of the traits that might contribute to T. virens biocontrol activity. Gliotoxin is a modified cyclic phenylalanine-serine dipeptide (19). Fungal siderophores involved in iron uptake, a potential means of competition in the soil, are commonly short peptides containing nonprotein amino acids (22). T. virens produces three types of hydroxamate siderophores: a monohydroxamate (cis- and trans-fusarinines), a dipeptide of trans-fusarinine (dimerum acid), and a trimer disdepsipeptide (copragen) (18). Peptide toxins and siderophores in bacteria and fungi often are produced nonribosomally by large multifunctional peptide synthetases. The enzymes are organized into repetitive synthase units, each of which has the functions required to complete a different single amino acid elongation step in the synthesis of the peptide product (20). The number and linear order of the repetitive synthase units correspond to the amino acid sequence of the peptide product. The functions of each synthase unit include ATP-dependent activation of the amino acid, transfer of the acyl adenylates to specific thiols located on enzyme-bound cofactors (4′-phosphopantetheine), and condensation to form a peptide bond; separate domains within the synthase unit perform these functions (the acylation domain, the acyl carrier domain, and the condensation domain, respectively) (20).
The conservation of consensus signature sequence motifs, along with their spatial separation (20, 32, 34), provides a tool for cloning peptide synthetases in other organisms. In this study our objectives were to clone a peptide synthetase from T. virens and to use specific gene replacement to identify the function of the cloned gene and its role, if any, in biocontrol activity.
MATERIALS AND METHODS
Fungal isolates and inoculum.
T. virens G20-4VIB is a single-spore isolate of strain G20 (= GL21) (24). Non-gliotoxin-producing UV-induced mutants were produced in a previous study (37). T. virens strains, P. ultimum PuZS1, and Rhizoctonia solani Rs-23A (24) were maintained on V8 juice agar. (200 ml of V8 juice per liter, 3 g of CaCO3 per liter, 20 g of agar per liter).
Isolation of a peptide synthase gene fragment by PCR.
A PCR was performed with a T. virens G20-4VIB genomic DNA template primed with two degenerate oligonucleotides corresponding to conserved amino acid residues of peptide synthetases (34). Primer A was based on the GKPKG sequence in core C (5′-GGNAAPCCNAAPGG, where N is A, G, C, or T, P is A or G, and Y is C or T). Primer B was based on the YKTGD sequence in core F (5′-PTCNCCNGTYTTPTA). The PCR was carried out in a 50-μl (total volume) mixture containing 100 ng of genomic DNA, 200 pmol of each primer, 1.5 mM MgCl2, and 2.5 U Taq of DNA polymerase. The thermal program was 40 cycles of 1 min at 94°C, 1.5 min at 37°C, and 2 min at 72°C. The final cycle was followed by 7 min of incubation at 72°C. An approximately 700-bp product was cloned into the pCRII vector (Invitrogen, Carlsbad, Calif.) by using T/A overhang ligation according to the manufacturer's directions, resulting in pPCR675.
cDNA library construction and screening.
Total RNA was isolated from individual 100-ml cultures of G20-4VIB grown in malt extract medium (1 × 105 conidia/ml of inoculum) at 160 rpm and 24°C in the dark and harvested 24, 28, 33, 39, and 49 h after inoculation. RNA isolation was carried out essentially as described by Chomczynski and Sacchi (6). Polyadenylated mRNA was isolated with oligo(dT)-cellulose spin columns (mRNA separator kit; Clontech, Palo Alto, Calif.) by following the manufacturer's instructions. A custom directional, deoxyribosylthymine-primed cDNA library was constructed by Clontech in the λZAPII cloning vector. The custom library consisted of 2 × 106 independent clones with an insert size range of 0.6 to 3.5 kb (average insert size, ∼1.5 kb).
A cDNA clone was isolated by hybridization of the 675-bp amplified fragment from pCR675 to 75,000 PFU of the unamplified cDNA library in Escherichia coli XL1-Blue MRF′. The 675-bp clone was used to generate a high-specific-activity riboprobe (∼1.6 × 109 cpm/μg). Positive plaques in replicate filters were subjected to secondary screening to isolate a single hybridizing plaque. In vivo excision using ExAssist/SOLR (Stratagene, La Jolla, Calif.) produced the cDNA insert in a pBluescript vector, pPsy1.
Southern blotting.
Fungal DNA was isolated (35) for Southern blot analysis (29) by capillary transfer to MagnaGraph nylon membranes (MSI, Westboro, Mass.) and UV cross-linking in a Strata-linker (Stratagene). Hybridizations were performed in a solution containing 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 2× Denhardt's reagent, 0.5% sodium dodecyl sulfate (SDS), 100 μg of single-stranded DNA per ml, and 50% formamide at 42°C. Membranes were prehybridized for 3 h prior to addition of fresh hybridization solution containing (per milliliter) 1 × 106 to 5 × 106 cpm of radioactively labeled DNA probe prepared by using a random oligonucleotide labeling kit (Pharmacia, Piscataway, N.J.) as recommended in the manufacturer's protocol. Following overnight hybridization at 42°C, the membranes were washed at 65°C in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 30 min, in 2× SSC–1% SDS for 30 min, and in 0.1× SSC–0.1% SDS for 10 to 30 min.
T. virens transformation.
Protoplasts were prepared as described by Thomas and Kenerley (33), except that conidia (1 × 106 conidia/ml) were germinated in a malt extract broth for 12 h at 28°C. This procedure generally yielded 1 × 108 to 2 × 108 protoplasts, which was adequate for two to four individual transformations at a level of 5 × 107 protoplasts per transformation.
The transforming construct for gene disruptions was prepared by inserting a selectable marker into pPsy1. A 3.3-kb NarI fragment of pCPGHY1 (5), containing the E. coli hygromycin B phosphotransferase gene (hygB) flanked by the Cryphonectria parasitica glyceraldehyde-3-phosphate dehydrogenase gene (Gpd1) promoter and terminator, was inserted into a ClaI site of Psy1 (between the C and D motifs in the adenylate domain). The resulting construct was confirmed by restriction analysis of plasmid DNA and was designated pPS-HYG1. The DNA used for transformation was a linear fragment produced by PCR amplification from pPS-HYG1 with primers located near the termini of the cloned Psy1 cDNA, which eliminated the bacterial vector sequences.
Transformation of protoplasts was performed by using a modification of the method of Thomas and Kenerley (33), as follows. A total of 1 × 107 to 5 × 107 protoplasts in an MES [2-(N-morpholino)ethanesulfonic acid]-buffered osmoticum (50 mM MES, 50 mM CaCl2, 0.5 M mannitol) were mixed with 1 μg of the linear transforming construct in a 0.6-ml preparation and incubated for 20 min on ice and then for 20 min at room temperature. An equal volume of 40% polyethylene glycol 8000 in MES osmoticum was added, and aliquots of the suspension were transferred to 20 petri plates, each containing 25 ml of molten regeneration medium (1 g of yeast extract per liter, 1 g of casein hydrolysate per liter, 342 g of sucrose per liter, 16 g of agar per liter). After incubation at 28°C for 6 to 12 h, each of the plates was overlaid with 25 ml of molten water agar containing 600 μg (215 U) of hygromycin B (Calbiochem, La Jolla, Calif.) per ml. Hygromycin B-resistant colonies were recovered after 2 to 3 days of incubation at 28°C. Single-spore isolates were recovered from hygromycin B-resistant colonies. Transformant stability was determined by four successive passages of conidia on nonselective potato dextrose agar, followed by plating of 100 conidia from each isolate onto potato dextrose agar plates with and without 500 μg of hygromycin B per ml.
Siderophore production and determination.
Spores were inoculated into the following low-iron medium (2), which is similar to the Grimm-Allen medium used previously for induction of siderophore production in T. virens (18). Autoclaved base medium (1 g of K2SO4 per liter, 1 g of K2HPO4 per liter, 3 g of ammonium acetate per liter, 1 g of citric acid per liter, 2 mg of ZnSO4 · 7H2O per liter, 80 mg of MgSO4 · 7H2O per liter) was supplemented with (per liter) 20 g of sucrose, 1 ml of a trace element solution (carbon-free trace elements [2] minus iron), and 1 ml of a stock vitamin solution (0.1 mg of biotin per ml, 2 mg of myo-inositol per ml, 2 mg of thiamine HCl per ml, 1 mg of p-aminobenzoic acid per ml, 1 mg of pantothenic acid per ml, 1 mg of nicotinamide per ml, 1 mg of pyridoxine HCl per ml), and then the pH was adjusted to 6.8 with NH4OH. High-iron medium was prepared by supplementing this base medium with FeSO4 · 7H2O (55.6 mg/liter).
Siderophore contents in culture filtrates were measured by measuring total hydroxamate contents (2). In this assay, 0.2 ml of a culture filtrate was mixed with 1 ml of 5 mM Fe(ClO4)3, and the optical density at 500 nm was determined by using a blank prepared from sterile medium. Values are expressed below in millimolar equivalents of ethylenediamine-di(o-hydroxyphenylacetic acid) (Sigma Chemical Co., St. Louis, Mo.), a synthetic hydroxamate standard, as measured with a standard curve. For partial purification and thin-layer chromatography (TLC) analysis of dimerum acid (18) we utilized 18 ml of culture filtrate evaporated in vacuo. The remaining gum was extracted in 6 ml of methanol overnight at room temperature and filtered through miracloth (Calbiochem) to remove undissolved material. Six hundred milligrams of XAD-2 resin (Supelco, Bellefonte, Pa.) was added to the sample, which shaken in an orbital shaker for 24 h at room temperature (25 to 28°C). The solvent was collected by filtration through miracloth, and more XAD-2 resin (600 mg) was added to the solvent. After a second filtration, the XAD-2 resin from the two extractions was combined in a 3-ml column, washed with 12 ml of distilled water, and eluted with 12 ml of methanol. Dimerum acid was released in the methanol extraction, while trans- and cis-fusarines were partially lost in the water washes (18). The methanol extract was dried, resuspended in 20 ml of methanol, and subjected to TLC on silica plates (60A; Whatman) with a chloroform-methanol-water (35:12:2) solvent. Hydroxamates were visualized by spraying the plates with 5 mM Fe(ClO4)3.
Biological control assays.
The biological control assays used were assays that were modified from the assays described by Lumsden and Locke (24). The antagonist inoculum used for biological control assays was prepared by transferring the conidia from one V8 juice agar slant (18-mm test tube) into 60 g of autoclaved bran (ratio of bran to H2O, 1:1 [wt/vol]) and incubating the preparation in the light at room temperature for 3 days. An R. solani inoculum was prepared by mixing autoclaved RE/CM (20 g of RediEarth [W.R. Grace & Co, Columbia, Md.], 0.4 g of 80-mesh cornmeal, 60 ml of water) with a 3-day-old agar plate culture and incubating the mixture at room temperature for 10 days. A P. ultimum inoculum was prepared by mixing autoclaved SCM (200 g of coarse sand, 6 g of coarse-ground cornmeal, 40 ml of water) with an agar plate culture and incubating the mixture at room temperature for 2 weeks. For each treatment, soilless mix (360 g of RediEarth, 720 ml of water containing 1.5 g of 20-10-20 N-P-K fertilizer) was amended with antagonist and pathogenic isolates (10 g of T. virens-colonized wheat bran and 1.8 g of R. solani RediEarth-cornmeal inoculum or 8.0 g of P. ultimum sand-cornmeal inoculum). All ingredients were mixed in large plastic bags and incubated for 10 to 13 days at 30°C. The bags were shaken at the end of the incubation period, and then the mixture in each bag was subdivided and placed into three planting flats (12 by 16 cm; 300 g each) to obtain a total of three replicates per treatment. Three rows (10 seeds/row) of eggplant seeds were planted in each flat, and the seedlings were examined after 2 weeks of incubation at 26 to 30°C (R. solani) or after 3 weeks of incubation at 24°C (P. ultimum) for disease incidence. The healthy control contained no fungal inoculum, whereas the disease control was treated with the pathogen and sterile bran. Disease incidence was scored by determining the percentage of seedlings standing compared to the number of seeds planted (percentage of plant stand). A statistical analysis was carried out as described by Lumsden and Locke (24).
The pH of RediEarth was modified to decrease iron availability by adding Ca(OH)2 dispersed in water at the rates indicated in Table 1 4 days before fungal isolates were added. No fertilizer was added to the RediEarth to avoid introducing contaminating iron present in the fertilizer.
TABLE 1.
Relative biocontrol activities of T. virens strains carrying the Psy1 disruption compared to the activity of parental isolate G20-M37
| Straina | Relative biocontrol activityb
|
||||
|---|---|---|---|---|---|
|
R. solani
|
P. ultimum
|
||||
| Expt 1 | Expt 2 | Expt 3 | Expt 1 | Expt 2 | |
| G20-M37 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| M37-T14 | 0.98 | 0.97 | 0.97 | ||
| M37-T18 | 0.83 | 1.3 | 1.1 | 1.0 | 1.1 |
| M37-T24 | 1.2 | 0.91 | 1.1 | ||
| M37-T42 | 0.17∗ | 1.1 | 1.0 | 1.1 | 0.94 |
| M37-T53 | 0.82 | 1.2 | 0.94 | ||
| M37-T62 | 0∗ | 1.1 | 1.0 | 1.1 | 0.90 |
| M37-T71 | 0∗ | 0.74 | 1.0 | 1.2 | 0.94 |
R. solani or P. ultimum was incubated with a T. virens strain for 10 to 13 days before eggplant seeds were planted. The T. virens strains used were recipient strain G20-M37 (glx mutant) and Psy1 disruptants (T strains).
Relative increases in the plant stand values compared to the disease control values. All values are relative to the data obtained for G20-M37, the parental isolate, which was assigned a relative biocontrol activity of 1.00 for each experiment. Relative biocontrol activity was calculated as follows: (percent plant stand after treatment − percent plant stand for R. solani-inoculated control)/(percent plant stand with G20-M37 treatment − percent plant stand for R. solani-inoculated control). Values that are significantly different (P = 0.05) from the G20-M37 value as determined by Duncan's multiple-range test are indicated by an asterisk. The plant stand values for control treatments in which neither pathogen nor T. virens was used were 87 to 90%. The plant stand values for pathogen and uninfested bran treatments were 0% for R. solani in all three experiments and 30 and 0% for P. ultimum in experiments 1 and 2, respectively. The plant stand values for the G20-M37 treatment were 33 and 60% for R. solani in experiments 1 and 2, respectively, and 77% for P. ultimum in both experiments.
Nucleotide sequence accession number.
The nucleotide sequence of Psy1 has been deposited in the GenBank database under accession number AF351825.
RESULTS
Cloning and sequence analysis of Psy1.
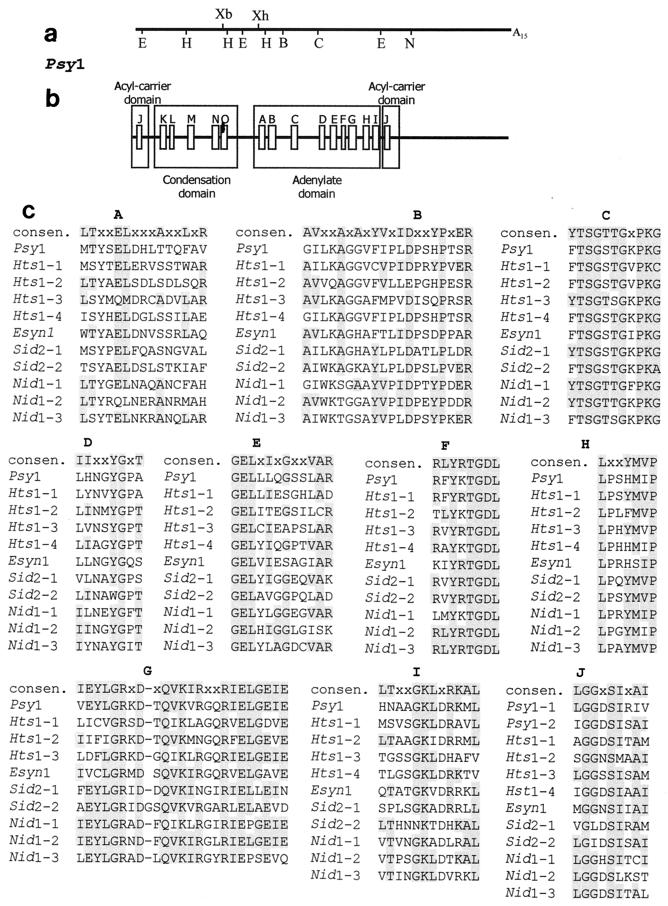
We used degenerate oligonucleotides corresponding to portions of conserved cores C and F of peptide synthetases to prime a PCR performed with T. virens genomic DNA. A fragment that was 675 bp long, which is consistent with the expected size, was cloned and sequenced. The predicted amino acid sequence encoded by this fragment exhibited extended sequence similarity with the sequence of the HC peptide synthetase from Cochliobolus carbonum (31). The 675-bp fragment was used to screen 75,000 independent clones of a polydeoxythymidylic acid-primed cDNA library prepared from T. virens poly(A)+ RNA. This screening yielded one cDNA clone (pPsy1) with a 5,056-bp insert. The cDNA insert possessed an open reading frame from the 5′ terminus corresponding to a 1,609-amino-acid protein fragment with a molecular mass of 178 kDa. Sequence similarity was observed in core consensus regions (Fig. 1) found in the adenylate domains of bacterial peptide synthetases, including the C motif believed to be characteristic of peptide synthetase adenylate domains (36). In addition, domains for cofactor binding (acyl carrier domain) and condensation which are unique to peptide synthetases (20) were identified (Fig. 1). The organization of the domains comprising the synthetase unit (condensation-adenylation-cofactor binding) is similar to the organization of gramicidin S synthetase and tyrocidine synthetase 2 from Bacillus brevis and surfactin synthetases 1, 2, and 3 from Bacillus subtilis (20). A second region with sequence similarity to the acyl carrier domain was located towards the N terminus of the Psy1-encoded fragment, which suggests that there is a second (and incomplete) synthase unit encoded upstream of the cloned gene fragment. Optimized protein alignments of the Psy1-encoded fragment with known peptide synthetase domains of fungal origin, obtained by using CLUSTAL W (European Bioinformatics Institute [http://www2.ebi.ac.uk/clustalw]), revealed similarity to the consensus core sequences defining these domains (Fig. 1). The deviations from the consensus core sequences derived from bacterial genes were similar to those observed for the sequences derived from other fungal genes and usually involved hydrophobic amino acid substitutions. The level of amino acid sequence identity between the Psy1-encoded fragment and the consensus sequence derived from bacterial genes was 62% for the adenylate domain, compared to 63 to 88% for Nid1 and Hts1; the level of amino acid sequence identity was 71% for the acyl carrier domain, compared to 57 to 86% for Nid1 and Hts1 units; and for the condensation domain the level of identity was 31% and the level of similarity was 60%, compared to 21 to 43% identity and 45 to 55% similarity among Nid1 and Hts1 units.
FIG. 1.
Restriction map and modular organization of Psy1 with regions of high sequence similarity to conserved adenylate domain motifs. (a) Restriction map of the 5,056-bp cloned cDNA of Psy1. Abbreviations: B, BamHI; C, ClaI; E, EcoRI; H, HindIII; S, SalI; N, NarI; Xb, XbaI; Xh, XhoI. (b) Regions of similarity to consensus sequence motifs of bacterial peptide synthetases (20). (c) Sequences in the motifs compared for the adenylate domains of Psy1 and other fungal peptide synthetases. Shading indicates sequence identity with the bacterial consensus sequence (consen.). Sequence comparisons were carried out with CLUSTALW (European Bioinformatics Institute [http://www2.ebi.ac.uk/clustalw]), and the entire adenylate domains and acyl carrier domains were used to preserve relative spacing between blocks. The genes used were the genes for ferrichrome siderophore synthetase (Sid2) from U. maydis (GenBank accession no. U62738), ACV synthetase (Nid1) from Aspergillus nidulans (GenBank accession no. X54853), HC toxin synthetase (Hts1) from C. carbonum (GenBank accession no. M98024), and enniatin synthetase (Esyn1) from Fusarium avenaceum (GenBank accession no. Z18755). Each adenylate domain/acyl carrier domain in a gene is numbered in the order of its appearance in the gene. However, Psy1 acyl carrier domain 2 (Psy1-2) is the acyl carrier domain located at the 5′ end of the cDNA and potentially is part of another incomplete synthetase unit. The relative organization of motifs is conserved in the adenylate domain; however, the organization of the condensation, adenylate, and acyl carrier domains is not the same for all genes.
Disruption of the Psy1 gene.
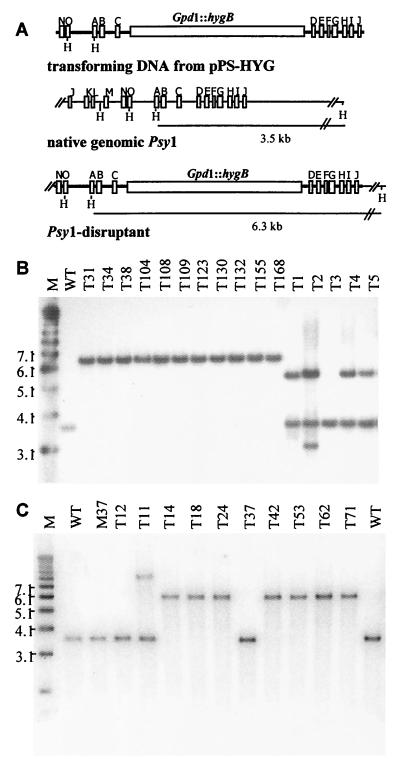
Wild-type biocontrol strain G20-4VIB was transformed with gene disruption construct pPS-HYG1, in which the selectable marker (hygB) was inserted into the middle of the cloned Psy1 gene (Fig. 2). Following production of single-spored hygromycin B-resistant isolates, we noticed that 11 of 168 isolates recovered germinated better (80 to 100% germination) on the hygromycin B-containing medium than the majority of the transformants germinated (<0.1% germination). These 11 isolates, six additional transformants, and G20-4VIB (untransformed) were analyzed by Southern blotting (Fig. 2B) for the expected integration. The untransformed G20-4VIB strain had only one copy of Psy1 and produced a single HindIII fragment (3.5 kb) when it was hybridized to the probe located at the 3′ end of the gene. The G20-4VIB transformants that were unstable in the presence of hygromycin B, T1 to T5 and T9, produced both the native 3.5-kb fragment from the native gene and one or more bands representing ectopic integrations of the transforming construct. All 11 isolates that were stable in the presence of hygromycin B, T31, T34, T38, T104, T108, T109, T123, T130, T132, T155, and T168, lacked the 3.5-kb fragment associated with the native gene and instead contained a single 6.3-kb fragment that was consistent with the expected gene replacement event resulting in disruption of the Psy1 gene (Fig 2B).
FIG. 2.
Disruption of the Psy1 gene in G20-4VIB (wild type) and non-gliotoxin-producing mutant G20-M37. (A) Gene disruption construct (pPS-HYGI) obtained by insertion of HygB (hygromycin phosphotransferase) expressed from the C. parasitica Gpd1 promoter (Gpd1::hygB) into the ClaI restriction site of the Psy1 cDNA insert in pPsy1. The positions of nucleotides corresponding to adenylate domain motifs A to O are indicated. PCR amplification with primers located at the termini of the Psy1 insert was used to produce a linear DNA fragment used for transformation to eliminate vector sequences. Insertion of the hygB cassette into native Psy1 resulted in a 2.8-kb increase in the size of the HindIII restriction fragment. (B) Southern blot analysis of HindIII-digested chromosomal DNA of T. virens wild-type strain G20-4VIB (lane WT) and transformants for detection of gene disruption events. Lane M contained molecular weight markers. (C) Southern blot analysis of HindIII-digested chromosomal DNA of T. virens wild-type strain G20-4VIB (lanes WT), gliotoxin mutant G20-M37 (lane M37), and G20-M37 transformants for detection of gene disruption events. Blots were probed with a 32P-labeled PCR fragment corresponding to the 3′ end of Psy1. Lane M contained molecular weight markers.
Effect of the Psy1 disruption on gliotoxin production.
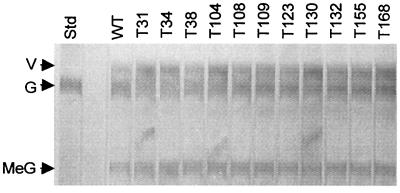
The abilities of the Psy1-disrupted isolates to produce gliotoxin were determined by TLC analysis of filtrates from 2-day-old malt extract broth (Difco, Detroit, Mich.) cultures. The Psy1 disruptants displayed no decrease in gliotoxin production compared to the wild-type strain, as determined by visual comparison of the bands on the plates by UV shadowing (Fig. 3) or after staining for gliotoxin with silver nitrate (37) (data not shown). The lack of a change in gliotoxin levels was confirmed by gas chromatography-mass spectrometry analysis of culture filtrates for gliotoxin. The heights and elution times of the gliotoxin peak were similar in the Psy1 disruptant and wild-type strains (data not shown).
FIG. 3.
Gliotoxin production in Psy1 disruptants of G20-4VIB. Filtrates from malt broth cultures grown for 2 days were extracted with chloroform and subjected to TLC on a plate with a fluorescent background (37). Dark bands indicate UV-absorbing compounds. Lane Std contained a gliotoxin standard. Culture filtrates of wild-type strain G20-4VIB (lane WT) and Psy1disruptants (T strains) derived from this strain were also included. The arrows indicate the positions of gliotoxin (G) and viridin (V). Viridin is a sterol metabolite that is not related to gliotoxin. Band MeG is dimethyl gliotoxin, an inactivated derivative of gliotoxin formed in culture.
Effect of Psy1 disruption on iron-dependent growth.
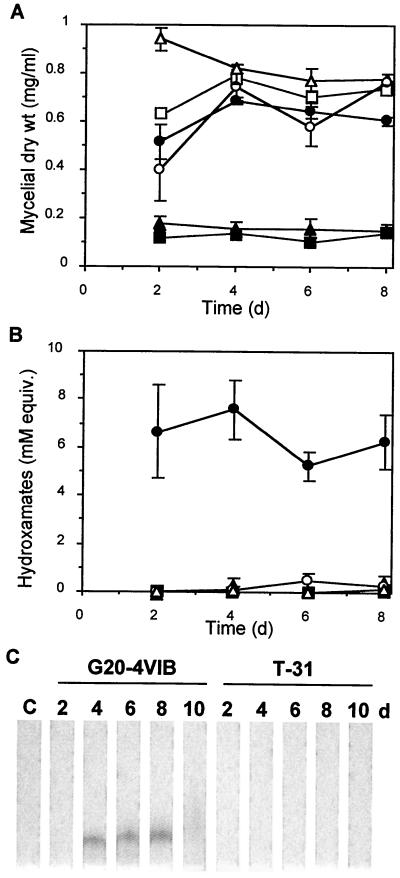
When we cultured the transformants on Weindling's modified medium (a defined T. virens culture medium [37]) lacking the usual 3 μM Fe-EDTA supplement, we observed that the colonies of all 11 Psy1 disruptants were smaller than the colonies of wild-type T. virens or five ectopic transformants (T1 to T5). In contrast, the colony sizes were similar to the colony sizes for these other strains when the organisms were grown on Weindling's modified medium containing the iron supplement. High iron levels (e.g., micromolar) in the growth medium normally repress siderophore production due to sufficient nonspecific iron transport, while low iron concentrations normally induce siderophore production (22). We more fully assessed the effect of Psy1 disruption on siderophore production by examining two representative disruptants, T31 and T155. Siderophore production was measured by determining (i) the ability to grow (as measured by mycelial dry weight) in a liquid medium that minimizes levels of trace iron compared to the ability to grow in the same medium supplemented with iron and (ii) the amount of hydroxamate siderophore produced in the low-iron medium. Both growth and hydroxamate levels were monitored for 8 days in culture, after the rapid growth phase.
Compared to the wild type, both Psy1 disruptants grew poorly under low-iron conditions (Fig. 4A) but normally under high-iron conditions. This iron-dependent growth defect was not observed in six T. virens transformants that carried only an ectopic copy of the disruption construct and contained an intact Psy1 gene (data not shown). A general hydroxamate siderophore assay was performed with partially purified culture medium collected during growth. In low-iron medium, in which siderophore production is normally induced, hydroxamate siderophore was detected in Psy1 disruptant T31, but the levels were 30-fold lower than the wild-type levels (Fig. 4B). Although part of the loss of siderophore activity could be attributed to the decreased growth in the low-iron medium, the amount of siderophore was at least eightfold lower in the disruptants than in the G20-4VIB recipient strain when the values were normalized to the dry weights of mycelia recovered from the same cultures. The other Psy1 disruptant, T155, displayed no detectable siderophore production (Fig. 4B). TLC after XAD-2 resin purification was used to detect the hydroxamate dimerum acid in culture filtrates. Although dimerum acid was clearly detected in wild-type cultures and was present on days 4 to 8, it was not found in the Psy1 disruptant cultures (Fig. 4C and data not shown) in any of the samples collected through day 10. Copragen was not detected in this assay, as expected from the relatively low levels previously reported (18). Fusarinines were not detected, as expected from their tendency to wash from the XAD-2 resin used in the purification process (18). TLC analysis of the methanol extracts without XAD-2 resin purification (data not shown) indicated that a ninhydrin-stained compound remained at the origin, which was consistent with the presence of trans- or cis-fusarinine. Such staining was observed with G20-4VIB, T31, and T155.
FIG. 4.
Mycelial growth, siderophore production, and dimerum acid levels in G20-4VIB and Psy1 disruptants. Spores of G20-4VIB (wild type) (circles) or Psy1 disruptants T31 (triangles) and T155 (squares) were inoculated into a base medium used to limit trace iron (<0.1 μM Fe), and the resulting cultures were divided into equivalent 10-ml cultures. Supplemental iron was added to one-half of the flasks to create high-iron conditions (100 μM Fe) (open symbols), while the other cultures were not supplemented in order to create low-iron conditions (solid symbols). Triplicate cultures were harvested by filtration at different times after inoculation for the wild-type and T31 strains. Duplicate cultures were harvested for strain T155. The dry weight of the mycelium in each 10-ml culture (A) and the total hydroxamate siderophore yield for each culture filtrate (B) are shown. To determine the total amount of hydroxamate utilized, we performed an Fe(ClO4)3 assay with samples of culture filtrate that were not purified. The error bars indicate standard errors of the mean. (C) Dimerum acid levels in the culture filtrates analyzed by methanol extraction, purification on XAD-2 resin, and TLC. The only band was the Fe(ClO4)3-stained band on TLC chromatograms, and this band comigrated with a dimerum acid standard. Lane C contained a control extraction of uninoculated culture medium.
Effect of the Psy1 disruption on biocontrol activity.
We also generated a set of Psy1 disruptants in a gliotoxin mutant (glx) background G20-M37, a UV-induced glx mutant of G20-4VIB (37), in order to focus the biocontrol assays on differences in siderophore production. Eight of the 80 single-spore transformants possessed the stable hygromycin B-resistant phenotype, and seven of these had the Psy1 disruption (Fig. 2C).
Biocontrol activities of the disruptants were tested by performing standard assays for suppression of pythium and rhizoctonia damping-off (Table 1). One experiment showed that two Psy1 disruptants reduced R. solani damping-off to some extent, but for replicates of R. solani or P. ultimum there was no consistent reduction in biocontrol activity compared to controls. We also tested biocontrol activity under conditions that decreased the availability of iron by increasing the pH of the soilless mix (Table 2). For one Psy1 disruptant of G20-4VIB, T155, there was a small but significant reduction at the highest pH, but there was not a consistent trend toward less biocontrol activity in all replicates.
TABLE 2.
Relative biocontrol activities of G20-4VIB and T. virens Psy1 disruptants against pythium and rhizoctonia damping-off with different pH values
| Straina | Relative biocontrol activityb
|
||
|---|---|---|---|
| pH 5.0 | pH 5.6 | pH 6.6 | |
| G20-4VIB | 1.00 | 1.00 | 1.00 |
| T31 | 0.97 | 1.14 | 0.97 |
| T155 | 1.04 | 0.92 | 0.74∗ |
R. solani was incubated with T. virens for 10 days before eggplant seeds were planted. RediEarth (pH 5.0) was amended 4 days prior to use with Ca(OH)2 at concentrations of 0.25 and 0.7 g/liter to obtain pHs 5.6 and 6.6, respectively.
Relative increases in the plant stand values compared to the disease control values. All values are relative to the data obtained for G20-4VIB, the recipient isolate, which was assigned a relative biocontrol activity of 1.00 for each experiment. Relative biocontrol activity was calculated as follows: (percent plant stand after treatment − percent plant stand for R. solani-inoculated control)/(percent plant stand after G20-4VIB treatment − percent plant stand for R. solani-inoculated control). A value that is significantly different (P = 0.05) from the G20-4VIB value as determined by Duncan's multiple-range test is indicated by an asterisk.
DISCUSSION
The T. virens Psy1 gene that we cloned appears to encode a segment of a peptide synthetase. This gene is not involved in biosynthesis of the dipeptide antibiotic gliotoxin but instead appears to be associated with production of a major siderophore, dimerum acid. We used Psy1 disruptants to test the role of siderophores in fungal biocontrol activity. The retention of full biocontrol activity by the disruptants indicates that dimerum acid siderophore production and perhaps iron competition are not limiting factors in the suppression of rhizoctonia and pythium damping-off by T. virens under the conditions tested.
The Psy1 gene probably encodes a peptide synthetase. There is extended sequence similarity to the conserved motif of the adenylate domain of peptide synthetases, particularly peptide synthetases from other filamentous fungi, which are different from related adenylate-forming enzymes (34). The Psy1-encoded sequence also contains regions similar to two acyl carrier domains and a condensation domain flanking the adenylate domain. The positions of these domains are conserved in peptide synthetases (20), and this suggests that one complete synthase unit and part of a second synthase unit are located near the N terminus of the translated peptide. The Psy1-encoded sequence also contains a region that matches the thioesterase/transferase consensus region (GXSXG) often found near the C termini of other peptide synthetases, including the peptide synthetases for ACV, gramicidin S, and surfactin (20). Although the open reading frame fragment encoding 1,609 amino acids is only a partial coding region, as expected from the larger sizes of other peptide synthetases, the cloned fragment is large enough to permit manipulation of the gene through targeted gene disruption (15).
Assuming that there are no redundant genes or partial activity of the disrupted gene, the continued production of gliotoxin by the Psy1 disruptants indicated that PSY1 is not involved in biosynthesis of gliotoxin. Peptide synthetases are also involved in synthesis of siderophores for iron uptake. T. virens produces siderophores that are monomers and multimers of the monohydroxamates cis-fusarinine and trans-fusarinine (18), which are Nδ-acylated derivatives of Nδ-hydroxylated ornithine, a nonproteinaceous amino acid. The fusarinines and the dihydroxamate (dimerum acid) constitute the majority of siderophores in T. virens, and each accounts for 45% of the total siderophore yield (18). The trihydroxamates copragen and copragen B account for about 6% of the total siderophores.
The Psy1 disruptants display several traits that are consistent with a large reduction in siderophore production. First, they grow poorly in low-iron medium, and the defect appears to be a defect in iron acquisition. Second, the amount of total hydroxamate siderophores is markedly decreased in the Psy1 disruptants, and dimerum acid is not present in culture filtrates. We infer from these findings that PSY1 is involved specifically in dimerum acid synthesis. Previous studies with mycelial extracts of Fusarium cubensis, another producer of dimerum acid, detected ATP-pyrophosphate exchange in the presence of trans-fusarinine. These results suggest that a peptide synthetase utilizes trans-fusarinine as a substrate (1). The loss of dimerum acid and the apparent presence of the monohydroxamates in the Psy1 disruptants are consistent with PSY1 being a peptide synthetase, presumably forming the cyclic peptide bond from two trans-fusarinine molecules. The role of PSY1 in synthesis of copragen or copragen B, which adds a third trans-fusarinine in an ester linkage to dimerum acid, remains to be determined. Such linkages can be produced by peptide synthetases (e.g., the depsipeptide enniatin [12]), but a peptide synthetase other than that encoded by Psy1 may be responsible for copragen synthesis.
If the role of PSY1 is to produce a fungal hydroxamate siderophore, then it is surprising that the sequence of the PSY1 adenylate domain is more similar to the sequences of the adenylate domains of the C. carbonum HC toxin synthetase (47 to 54% similarity) than to the sequences of the adenylate domains of the Ustilago maydis ferrichrome synthetase (42 to 47% similarity), the only other cloned fungal peptide synthetase that produces a siderophore. The differences may be attributable to the large differences in the structures of these siderophores. Ferrichromes have other acyl modifications of ornithine and additional neutral amino acids in head-to-tail linkages as hexapeptides (22).
Competition for nutrients is thought to be one mechanism for antagonistic interactions between biocontrol agents and plant pathogens, and it has been proposed that siderophore production is an important mode of competition for iron in the soil (23). Studies of bacterial biocontrol agents in which non-siderophore-producing mutants have been used have provided conflicting results in tests of this hypothesis. Pseudomonas aeruginosa mutants that lack siderophores have reduced biocontrol activity against Fusarium oxysporum (11), while similar Pseudomonas fluorescens and Pseudomonas putida mutants have full biocontrol activity against Pythium aphanadermatum, P. ultimum, and Gaeumannomyces graminis var. tritici (21, 27, 28). Manipulation of multiple siderophores in P. aeruginosa (4) and Enterobacter cloacae (8) has provided similarly disparate results, indicating that the roles of siderophores may depend on the pathogen-bacterial biocontrol agent pair. In our test of a fungal biocontrol agent, T. virens, we did not observe any consistent loss of biocontrol activity against either P. ultimum or R. solani, even when the soil pH was increased to reduce iron availability. The biocontrol assay used, which involved preincubating T. virens with the pathogen for 10 days prior to introduction of the plant, should have increased competition for iron in the soilless mix. Production of dimerum acid does not appear to be a significant factor in the ability of T. virens to suppress pythium or rhizoctonia damping-off. We cannot, however, rule out the possibility that conditions in the biocontrol assay enhanced production of the monohydroxamate siderophores in the Psy1 disruptants, compensating for the loss of dimerum acid. Cloning and disruption of a gene or genes encoding ornithine hydroxylation and acylation enzymes in T. virens are needed to test the role of fusarinine in biocontrol activity. The clustering of ornithine-N5-oxygenase activity and the peptide synthetase for ferrichrome synthesis in U. maydis (22) suggests that a similar gene may be located near Psy1 in T. virens.
Psy1 should be a useful tool for molecular definition of the diverse siderophores produced by fungi, particularly the ascomycetes, in which no other genes have been cloned (22). A range of fungi produce copragen and dimerum acid; these fungi include the human pathogen Histoplasma capsulatum (3, 16) and the plant pathogens Verticillium dahliae (13) and G. graminis var. tritici (10). Homologs of Psy1 may be present in these fungi and have a role in animal or human pathogenicity, while the same siderophores produced by Penicillium chrysogenum increase iron utilization by plants (14) and may have a role in plant growth promotion by this strain of T. virens (9). An intact Psy1 clone may be useful in engineering hydroxamate siderophore production in plants or in providing plant growth enhancement by T. virens beyond suppression of disease-causing fungi.
ACKNOWLEDGMENTS
We thank Richard van der Helm for providing a dimerum acid standard and useful discussions and Donald Nuss for providing plasmid pCPGHY1.
We thank the Maryland Agriculture Experiment Station for partial support.
REFERENCES
- 1.Anke T, Diekmann H. Incorporation of delta-N-hydroxy-l-ornithine and delta-N-acyl-delta-N-hydroxy-l-ornithine into sideramines of fungi. Arch Microbiol. 1974;95:227–236. [Google Scholar]
- 2.Atkin C L, Neilands J B, Phaff H J. Rhodotorulic acid from species of Leucosporidium, Rhodosporidium, Rhodotorula, Sporidiobolus, and Sporobolomyces, and a new alanine-containing ferrichrome from Cryptococcus melibiosum. J Bacteriol. 1970;103:722–733. doi: 10.1128/jb.103.3.722-733.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burt W R. Identification of coprogen B and its breakdown products from Histoplasma capsulatum. Infect Immun. 1982;35:990–996. doi: 10.1128/iai.35.3.990-996.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buysens S, Heungens K, Poppe J, Hofte M. Involvement of pyochelin and pyoverdin in suppression of Pythium-induced damping-off of tomato by Pseudomonas aeruginosa NSK2. Appl Environ Microbiol. 1996;62:865–871. doi: 10.1128/aem.62.3.865-871.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi G, Nuss D. Nucleotide sequence of the glyceraldehyde-3-phosphate dehydrogenase gene from Cryphonectria parasitica. Nucleic Acids Res. 1990;18:5566. doi: 10.1093/nar/18.18.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Biochemistry. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 7.Cook R J, Baker K F. The nature and practice of biological control of plant pathogens. St. Paul, Minn: American Phytopathological Society; 1983. [Google Scholar]
- 8.Costa J, Loper J. Characterization of siderophore production by the biological control agent Enterobacter cloacae. Mol Plant-Microbe Interact. 1994;7:440–448. [Google Scholar]
- 9.De Silva A, Patterson K, Rothrock C, Moore J. Growth promotion of highbush blueberry by fungal and bacterial inoculants. HortScience. 2000;35:1228–1230. [Google Scholar]
- 10.Dori S, Solel Z, Kashman Y, Barash I. Characterization of hydroxamate siderophores and siderophore-mediated iron uptake in Gaeumannomyces graminis var. tritici. Physiol Mol Plant Pathol. 1990;37:95–106. [Google Scholar]
- 11.Duijff B J, Bakker A H M, Schippers B. Suppression of fusarium wilt of carnation by Pseudomonas putida WCS358 at different levels of disease incidence and iron availability. Biocontrol Sci Technol. 1994;4:279–288. [Google Scholar]
- 12.Haese A, Schubert M, Herrmann M, Zocher R. Molecular characterization of the enniatin synthetase gene encoding a multifunctional enzyme catalysing N-methyldepsipeptide formation in Fusarium scirpi. Mol Microbiol. 1993;7:905–914. doi: 10.1111/j.1365-2958.1993.tb01181.x. [DOI] [PubMed] [Google Scholar]
- 13.Harrington G J, Neilands J B. Isolation and characterization of dimerum acid from Verticillium dahliae, a fungal phytopathogen. In: Nelson S D, editor. Iron nutrition and interactions in plants. New York, N.Y: Marcel Dekker; 1982. pp. 675–682. [Google Scholar]
- 14.Hördt W, Römheld V, Winkelmann G. Fusarinines and dimerum acid, mono- and dihydroxamate siderophores from Penicillium chrysogenum, improve iron utilization by strategy I and strategy II plants. Biometals. 2000;13:37–46. doi: 10.1023/a:1009234612486. [DOI] [PubMed] [Google Scholar]
- 15.Hoskins J, O'Callaghan N, Queener S, Cantwell C, Wood J, Chen V, Skatrud P. Gene disruption of the pcbAB gene encoding ACV synthetase in Cephalosporium acremonium. Curr Genet. 1990;18:523–530. doi: 10.1007/BF00327023. [DOI] [PubMed] [Google Scholar]
- 16.Howard D, Rafie R, Tiwari A, Faull K. Hydroxamate siderophores of Histoplasma capsulatum. Infect Immun. 2000;68:2338–2343. doi: 10.1128/iai.68.4.2338-2343.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howell C R. Relevance of mycoparasitism in the biological control of Rhizoctonia solani by Gliocladium virens. Phytopathology. 1987;77:992–994. [Google Scholar]
- 18.Jalal M A, Love S K, van der Helm D. Siderophore mediated iron(III) uptake in Gliocladium virens. 1. Properties of cis-fusarinine, trans-fusarinine, dimerum acid, and their ferric complexes. J Inorg Biochem. 1986;28:417–430. doi: 10.1016/0162-0134(86)80027-7. [DOI] [PubMed] [Google Scholar]
- 19.Kirby G W, Patrick G L, Robins D J. Cyclo-(l-phenyl-l-seryl) as an intermediate in the biosynthesis of gliotoxin. J Chem Soc Trans I. 1978;11:1336–1338. [Google Scholar]
- 20.Kleinkauf H, Von Dohren H. A nonribosomal system of peptide biosynthesis. Eur J Biochem. 1996;236:335–351. doi: 10.1111/j.1432-1033.1996.00335.x. [DOI] [PubMed] [Google Scholar]
- 21.Kraus J, Loper J E. Lack of evidence for a role of antifungal metabolite production by Pseudomonas fluorescens Pf-5 in biological control of pythium damping-off of cucumber. Phytopathology. 1992;82:264–271. [Google Scholar]
- 22.Leong S A, Winkelmann G. Molecular biology of iron transport in fungi. Met Ions Biol Syst. 1998;35:147–186. [PubMed] [Google Scholar]
- 23.Loper J E, Buyer J S. Siderophores in microbial interactions on plant surfaces. Mol Plant-Microbe Interact. 1991;4:5–13. [Google Scholar]
- 24.Lumsden R D, Locke J C. Biological control of damping-off caused by Pythium ultimum and Rhizoctonia solani with Gliocladium virens in soilless mix. Phytopathology. 1989;79:361–366. [Google Scholar]
- 25.Lumsden R D, Locke J C, Adkins S T, Walter J F, Ridout C J. Isolation and localization of the antibiotic gliotoxin produced by Gliocladium virens from alginate prill in soil and soilless media. Phytopathology. 1992;82:230–235. [Google Scholar]
- 26.Lumsden R D, Walter J F, Baker C P. Development of Gliocladium virens for damping-off disease control. Can J Plant Pathol. 1996;18:463–468. [Google Scholar]
- 27.Ongena M, Daayf F, Jacques P, Thonart P, Thonart P, Benhamou N, Paulitz T C, Cornelis P, Koedam N, Belanger R R. Protection of cucumber against Pythium root rot by fluorescent pseudomonads: predominant role of induced resistance over siderophores antibiosis. Plant Pathol. 1998;48:66–76. [Google Scholar]
- 28.Ownley B, Weller D, Thomashow L. Influence of in situ and in vitro pH on suppression of Gaeumannomyces graminis var. tritici by Pseudomonas fluorescens 2-79. Phytopathology. 1992;82:178–184. [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Samuels G J, Rehner S A. Toward a concept of genus and species in Trichoderma. In: Lumsden R D, Vaughn J L, editors. Pest management: biologically based technologies. Washington, D.C.: American Chemical Society; 1993. pp. 186–188. [Google Scholar]
- 31.Scott-Craig J S, Panaccione D G, Pocard J A, Walton J D. The cyclic peptide synthetase catalyzing HC-toxin production in the filamentous fungus Cochliobolus carbonum is encoded by a 15.7-kilobase open reading frame. J Biol Chem. 1992;267:26044–26049. [PubMed] [Google Scholar]
- 32.Stachelhaus T, Marahiel M. Modular structure of genes encoding multifunctional peptide synthetases required for non-ribosomal peptide synthesis. FEMS Microbiol Lett. 1995;125:3–14. doi: 10.1111/j.1574-6968.1995.tb07328.x. [DOI] [PubMed] [Google Scholar]
- 33.Thomas M D, Kenerley C M. Transformation of the mycoparasite Gliocladium. Curr Genet. 1989;15:415–420. [Google Scholar]
- 34.Turgay K, Krause M, Marahiel M. Four homologous domains in the primary structure of GrsB are related to domains in a superfamily of adenylate-forming enzymes. Mol Microbiol. 1992;6:529–546. doi: 10.1111/j.1365-2958.1992.tb01498.x. [DOI] [PubMed] [Google Scholar]
- 35.Turgeon G B, Garber R C, Yoder O C. Transformation of the fungal maize pathogen Cocliobolus heterostrophus using the Aspergillus nidulans amdS gene. Mol Gen Genet. 1985;201:450–453. [Google Scholar]
- 36.von Döhren H, Keller U, Vater J, Zocher R. Multifunctional peptide synthetases. Chem Rev. 1997;97:2675–2705. doi: 10.1021/cr9600262. [DOI] [PubMed] [Google Scholar]
- 37.Wilhite S E, Lumsden R D, Straney D C. Mutational analysis of gliotoxin production by the biocontrol fungus Gliocladium virens in relation to suppression of pythium damping-off. Phytopathology. 1994;84:816–821. [Google Scholar]