Abstract
Background
Transcription factors GATAs were a member of zinc finger protein, which could bind DNA regulatory regions to control expression of target genes, thus influencing plant growth and development either in normal condition or environmental stresses. Recently, GATA genes have been found and functionally characterized in a number of plant species. However, little information of GATA genes were annotated in wheat.
Results
In the current study, 79 GATA genes were identified in wheat, which were unevenly located on 21 chromosomes. According to the analysis of phylogenetic tree and functional domain structures, TaGATAs were classified into four subfamilies (I, II, III, and IV), consist of 35, 21, 12, and 11 genes, respectively. Meanwhile, the amino acids of 79 TaGATAs exhibited apparent difference in four subfamilies according to GATA domains comparison, gene structures and conserved motif analysis. We then analyze the gene duplication and synteny between the genomes of wheat and Arabidopsis, rice and barley, which provided insights into evolutionary characteristics. In addition, expression patterns of TaGATAs were analyzed, and they showed obvious difference in diverse tissues and abiotic stresses.
Conclusion
In general, these results provide useful information for future TaGATA gene function analysis, and it helps to better understand molecular breeding and stress response in wheat.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-022-03733-3.
Keywords: Wheat (Triticum aestivum L.), GATA,Genome-wide, Abiotic stress, Expression patterns
Introduction
Plants face many environmental challenges during development, and they must optimize growth to adapt to all kinds of environmental condition including abiotic and biotic stress. During the long-term evolution, many kinds of plants have evolved a range of protective mechanisms in response to various environmental stress, among which transcriptional regulations play a dominant role [1]. Transcription factors (TFs) are vital modulators to control gene expression level via specifically binding to promoter region of the downstream gene, thus influencing or regulating a lot of important biological processes, including cellular morphogenesis, signaling transduction, and environmental stress responses [2, 3]. In plants, many well-known transcription factor families have been found, such as GATA (GATA-binding factor) [4], WRKY [5], MYB [6], DREB (Dehydration-responsive element-binding protein) [7], bZIP (Basic region-leucine zipper) [8], and MADS-box [9].
GATAs are a class of DNA binding proteins widely existed in fungi, animals and plants, which belongs to a member of type IV zinc finger, and the DNA binding domain of which is consist of a basic region with C-X2-C-X17–20-C-X2−C form [4]. The GATA proteins could modulate the transcription level of their target genes by recognizing and binding to the (T/A)GATA(A/G) sequences of genes promoter. The first GATA factor NTL1 was found in tobacco (Nicotiana tabacum), which was a homolog of NIT2 form Neurospora crassa [10]. The GATA family gene was successively identified in a number of plants, such as Arabidopsis thaliana [4], Oryza sativa [11, 12], Glycine max [13, 14], Brachypodium distachyon [15, 16], Capsicum annuum [17], Cucumis sativus [18], Gossypium genus [19] and so on. In plants, all GATA factors are featured with one single zinc finger domain with 18 or 20 residues. In general, the GATA family were classified into four subfamilies in Arabidopsis thaliana, as subfamily I, II, III and IV, in terms of phylogenetic analysis, DNA binding domains and gene structures [4].
With the rapid development of Next Generation Sequencing (NGS), GATAs family have been found both in monocots and dicots. There were 30 GATA genes in Arabidopsis, 28 in rice, 35 in apple, and 64 in soybean according to Genome-wide analyses [4, 13, 20]. Plant GATA TFs have various roles like the chloroplast development [21], photosynthesis and growth [22, 23], seed dormancy [24], host immune response [25], Grain shape [12], and abiotic stress [18, 26, 27]. In Arabidopsis, GATA12 are involved in primary seed dormancy [24]. SWI2/SNF2 ATPase BRM could associate with GNC (GATA, NITRATE-INDUCIBLE, Carbon metabolism Involved) to regulate SOC1 (Suppressor of Overexpression of Constans 1) expression and control bloom time in Arabidopsis [28]. In rice, Over-expressed OsGATA12 lead to reduction of leaf and tiller numbers, thus affecting yield-related characters [23]. OsGATA7 regulated architecture and grain shape by mediating brassinosteroids content [12]. In poplar, PdGATA19 was responsible for photosynthesis and growth [22]. In soybean, low nitrogen treatment led to the obvious repression of GATA44 and GATA58 in seedlings [13]. Given the importance of GATA in plants, the above reports manifest that GATA TFs are needed to conduct a comprehensive assessment in development, growth and stress response.
Wheat is the second major cereal crop in the world. Hence, it is important to conduct genetic and physiological research. Many wheat GATA genes have been found and functionally characterized. For example, over-expressing TaZIM-A1 postponed flower time and led to the reduction of thousand seed weight [29]. Liu et al. [25] reported that plants of over-expressed TaGATA1 showed high resistance to Rhizoctonia cerealis in wheat. In spite of this, the function of GATA factors defined remains very little in wheat. In the current study, 79 candidate TaGATA genes were identified based on the bioinformatic analysis of wheat genome. Generally, we performed a genome-wide analysis in wheat GATA genes, such as phylogeny, conserved motifs, gene structures, chromosomal distribution, and expression profiles of GATA genes in different tissues and diverse abiotic stresses.
Materials and methods
Identification of TaGATA genes in wheat
Gene and protein sequences were obtained from the Ensemble Plants database (http://plants.ensembl.org) [30]. To identify the candidate TaGATA genes, we used a Hidden Markov Model (HMM) to search the protein database in wheat genome by HMMER3.0 [31], in which the profiles of the GATA protein domain, PF00320, were used as queries with default parameters. Within the same gene ID, we left the longest transcript sequence, and incomplete sequence without start or termination codon were discarded. Then, we used Pfam tool with e-value <e− 20 [32] and Conserved Domain Database (CDD) to analyzed the left sequence. Ultimately, 79 TaGATA genes were identified. Furthermore, ExPASy tool (http://www.expasy.ch/tools/pi_tool.html) was used to calculate amino acids number, molecular weights (MW) and isoelectric point (pI) [33].
Phylogenetic analysis of TaGATAs
Sequence alignment of 79 TaGATA protein was conducted using ClustalW [34]. We used MEGA 7.0 to construct Evolutionary tree by the Neighbor-Joining (NJ) method [35], with the following parameters: poisson model, pairwise deletion and 1000 bootstrap replications. The phylogenetic tree was further beautified using the iTOL (https://itol.embl.de/) [36].
Chromosomal location and gene duplication
TaGATA genes localization on chromosome was visualized by MapChart tools (v2.3.2) [37]. Syntenic relationship of the orthologous GATA genes between Triticum aestivum and other species, including Arabidopsis thaliana, Oryza sativa and Hordeum vulgare, were analyzed by the MCScanX software [38]. We then used KaKs_Calculator 2.0 to calculate non-synonymous (Ka) and synonymous (Ks) substitution of each duplicated TaGATA gene [39]. Formula T = Ks/2R was used to assess Divergence time, where R is 1.5 × 10–8 synonymous substitutions per site per year [39].
Gene structures and protein motifs analysis
The exon/intron organization of TaGATA genes was identified using the Gene Structure Display Server (GSDS) tool (http://gsds.cbi.pku.edu.cn/) [40]. The Multiple Expectation Maximization for Motif Elicitation (MEME) online program (http://meme.sdsc.edu/ meme/itro.html) was performed to identify conserved motifs of TaGATA proteins [41]. The exon-intron structure and conserved motif of TaGATA was examined by TBtools [42] and GFF3 database obtained from Ensemble Plants.
Cis-elements in the promoter of TaGATA genes
Promoter sequences (− 1500 bp) of TaGATA gene was obtained from the wheat genome sequence, and cis-element of promoter region was analyzed using PlantCARE software (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [43]. The full graphics of Cis-elements were annotated by TBtools [42].
Gene expression analysis
The specific expression patterns of GATA gene from various tissues in the wheat Chinese spring were obtained from Wheatomics (http://202.194.139.32/) [44]. The gene expression values are represented by transcript per Kilobase of exon per million reads mapped (TPM). The average expression level of three biological replicates was calculated and used to show their expression patterns in each tissue. The data were normalized to expression level in roots. Furthermore, transcriptome data under abiotic stress were also obtained from Wheatomics. The genes with log2 ratio ≥ 0.5 and log2 ratio ≤ − 0.5 were regarded as differentially expressed genes (DEGs). A heatmap of expression pattern profile on log2 (TPM + 1) and log2 fold change scale was conducted by TBtools [42, 45].
Plant growth and treatment
The hydroponic experiment was conducted in a greenhouse at Qingdao Agricultural University, Qingdao, China. Wheat cultivar “Chinese Spring” were used in this study. Chinese Spring is a well-known wheat variety, which has been widely used in wheat genetics research [44]. All the testing wheat seeds were harvested in the summer of 2020 at the Jiaozhou Experimental Station, Qingdao Agricultural University, Jiaozhou, Shandong, China. Seeds of wheat were treated with 3% H2O2 for 10 min, rinsed seven times with distilled water. The seeds were sown in a controlled environment with a day-night temperature of 22 ± 3 °C on moist filter papers. After germination, uniform seedlings were transferred to 2 L pots containing 1.5 L basic nutrient solution (BNS). On the seventh day after transplanting, PEG and NaCl were added to the containers to form three treatments: BNS (control), BNS plus 10% PEG and BNS plus 100 mM NaCl. The composition of BNS was (mg L− 1): (NH4)2SO4, 48.2; MgSO4, 65.9; K2SO4, 15.9; KNO3, 18.5; Ca(NO3)2, 59.9; KH2PO4, 24.8; Fe-citrate, 5; MnCl2 4H2O, 0.9; ZnSO4 7H2O, 0.11; CuSO4·5H2O, 0.04; HBO3, 2.9; H2MoO4, 0.01. The solution pH was adjusted to 5.8 ± 0.1 with NaOH or HCl, as required. Plant leaves and stems were sampled on the seventh day after transplanting. Plant roots samples for RNA isolation were collected 6 h after PEG and NaCl treatment. All samples were stored at − 80 °C for downstream analysis.
Quantitative RT-PCR validation
Quantitative real-time PCR (qRT-PCR) was performed by a QuantStudio3 PCR system (Thermo, USA) [46]. First-strand cDNA synthesis was performed using the PrimeSciptTM RT reagent Kit (Takara, Japan), followed by qRT-PCR using a SYBR Green Supermix (Takara, Japan) with TaActin as a reference. The total PCR volume was 10 μl. The reaction process was 94 °C denaturation for 30 s, followed by 40–45 cycles of 94 °C for 5 s, 58 °C for 15 s, and 72 °C for 10 s. Experiments were replicated three times with 2-ΔΔCq relative quantification method. Primers were listed in Table S13.
Results
Identification of TaGATAs in wheat
In total, 79 GATA family members were identified in wheat. The detailed information of genes and proteins were listed in Tables S1. For example, the amino acid length of 79 TaGATA proteins ranges from 146 to 499. Meanwhile, the molecular weight is ranged from 16.1 to 54.1 kDa. The GATA domain sequences were listed in Table S2.
Phylogenetic analysis of TaGATA proteins
To figure out the phylogenetic relationship of the GATA proteins, we constructed a evolutionary tree in terms of the alignment of 79 wheat TaGATAs and 30 Arabidopsis AtGATAs (Fig. 1). The AtGATAs protein sequence were listed in Table S3. It was reported that 30 AtGATA proteins could be categorized into four clusters [4]. On the basis of classification standard used for Arabidopsis, the wheat GATA proteins were classified into four group. Group I, II, III, and IV consist of 35, 21, 12, and 11 TaGATA proteins, respectively (Figs. 1 and 2A).
Fig. 1.
Phylogenetic tree of full-length TaGATA and AtGATA proteins. The different-colored arcs indicate subfamilies of the GATA proteins. The tree was constructed using identified 79 TaGATAs (asterisks) in wheat, 30 AtGATAs (triangle) from Arabidopsis. The unrooted Neighbour-Joining phylogenetic tree was constructed using MEGA7 with full-length amino acid sequences and the bootstrap test replicate was set as 1000 times
Fig. 2.
Phylogenetic relationships, architecture of conserved protein motifs and gene structure in GATA genes from wheat. A The phylogenetic tree was constructed based on the full-length sequences of wheat GATA proteins using MEGA 7 software. B Exon-intron structure of wheat GATA genes. Blue boxes indicate untranslated 5′- and 3′- regions; yellow boxes indicate exons; black lines indicate introns. C The motif composition of wheat GATA proteins. The motifs, numbers 1–10, are displayed in different colored boxes. The sequence information for each motif is provided in Supplementary Files. The length of protein can be estimated using the scale at the bottom
Gene structure and protein motif analysis of TaGATA
We used the web server GSDS to analyze TaGATA genes structures. The results showed that TaGATA genes contained one to eight exons unevenly (Fig. 2B). Protein motifs were determined by MEME. In general, 10 conserved motifs were found in TaGATA proteins and considered motifs 1–10 (Figs. 2C). The detailed information of conserved motif were listed in Table S4. In total, 19 of 79 TaGATAs only contain motif 1. Thirty five of 79 TaGATAs contain motif 1 and 2. The motif 1 were primarily presented in subfamily I and II, and the motif 3–10 were detected in the members of group II and IV. In a word, similar gene structures and conserved motifs in the same subfamily forcefully back up phylogenetic analysis for subfamily classifications.
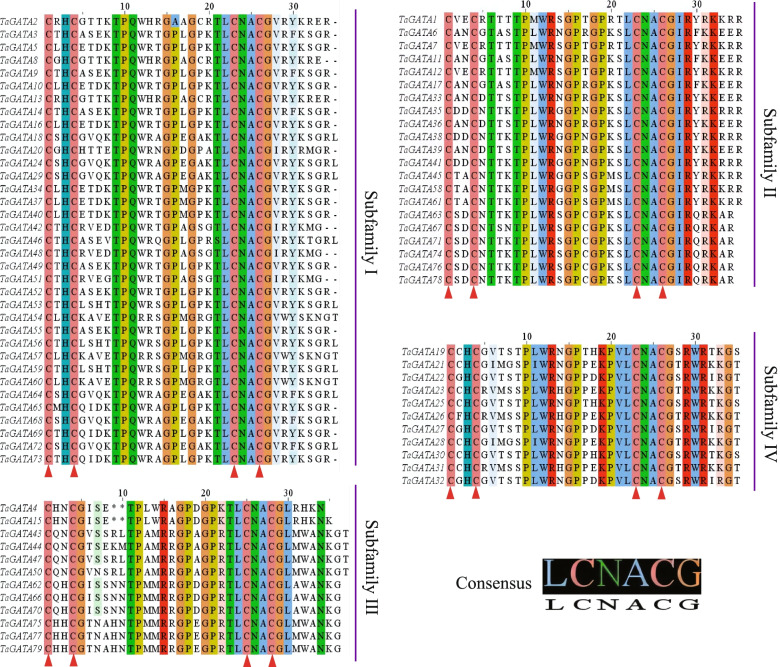
In addition, GATA domain analysis showed that TaGATAs in the subfamilies I, II and IV comprised 18 residues in the zinc finger loop between the second and the third Cys residues, while TaGATAs in the subfamily III comprised 20 residues, with the exception of TaGATA4 and TaGATA15 comprised 18 residues. In the GATA domains, many amino acid sites exhibited high conservation, such as LCNACG residues (Fig. 3).
Fig. 3.
Alignments of GATA domain sequences of the GATA family members in wheat. Highly conserved amino acid positions are marked with letters and triangles at the bottom
Chromosomal location and genome Synteny of TaGATA genes
The chromosomal distribution of TaGATA gene were analyzed. In total, 79 TaGATAs were mapped to the wheat genome (Fig. 4). The TaGATA genes were evenly located among A (29), B (25), D (25) subgenomes. This was consistent with the finding that a large proportion of TaGATAs have three homoeologous sequences distributed on three subgenomes. There were three TaGATA genes located on chromosome 3, 5. Six TaGATAs could be found on each of chromosomes 1 and 2. Four TaGATA genes were located on chromosome 6. Five TaGATA genes were distributed on chromosome 4A and three TaGATA genes were located on chromosome 4B and 4D. Chromosome 7 carried 2 TaGATAs which was the minimum number. With approach of BLAST and MCScanX, we detected 96 segmental duplication events in TaGATAs (Fig. 6; Table S5). All events were almost happened between the different chromosomes. Furthermore, 4 duplication events happened on the AA subgenome, 3 events on the BB subgenome, 4 events on the DD subgenome, and 85 events across AA/BB/DD subgenomes. The above results demonstrate that a number of TaGATA genes are likely to appear in the course of gene duplication, and the segmental duplication events could be of great importance in the expansion of TaGATA genes in wheat.
Fig. 4.
Distribution of TaGATA genes in wheat chromosomes. The chromosome numbers are indicated at the top of each chromosome image
Fig. 6.
the synteny analysis of TaGATA family in wheat. Gray lines indicate all synteny blocks in the wheat genome, and the red lines indicate duplicated TaGATA gene pairs. The chromosome number is indicated at the bottom of each chromosome
The colinearity of TaGATA gene pairs between Hordeum vulgare genome, Arabidopsis thaliana genome and Oryza sativa genome was compared. The result exhibited that three and ten TaGATA genes exhibited syntenic relationship with AtGATA and OsGATA genes, respectively (Fig. 7; Table S6 and S7). For example, AT2G45050 showed syntenic relationship with TaGATA3, TaGATA9 and TaGATA14 (Table S6). However, 54 TaGATAs showed syntenic relationship with GATAs in barley (Table S8), implying that these genes may be responsible for the evolution of TaGATAs family.
Fig. 7.
Synteny analysis of GATA genes between wheat, Arabidopsis, rice and barley. Gray lines in the background indicate the collinear blocks within wheat and other plant genomes, while the red lines highlight the syntenic gata gene pairs. The specie names with the prefixes, Hv, Ta, At and Os indicate barley, wheat, Arabidopsis and rice, respectively
To assess the evolutionary constraints acting, we calculated Ks values, Ka values, Ka/Ks ratios and divergence time of paralogous and orthologous on GATA family genes (Tables S9). Ka/Ks ratios were less than 1 in several segmental duplicated TaGATA gene pairs, while TaGATA26/TaGATA31 were more than 1. The results demonstrated that TaGATAs family probably have suffered strong purifying selective stress in the course of evolution.
Cis-elements analysis in TaGATAs promoters
To explore the underlying function of TaGATA genes, we used Plant-CARE to detect the cis-elements in these genes promoter. 79 TaGATAs were estimated with cis-elements, such as ABRE, circadian, G-box, LTR, MSA, P-box, TCA, TGA TGACC-motif and MBSI involving in ABA responses, circadian control, light response, low-temperature response, cell cycle regulation, gibberellin response, salicylic acid response, auxin response, MeJA response, drought-inducibility and flavonoid biosynthetic genes regulation (Fig. 5, Table S10). In general, 69 TaGATA genes (87.3%) carried ABRE cis-elements, 75 TaGATA genes (94.9%) had G-box cis-elements, and 63 TaGATA genes (79.7%) carried TGACC cis-elements. In a word, the cis-elements analysis implied that a large portion of TaGATA genes are likely to be responded to various environmental stresses.
Fig. 5.
Predicted cis-elements in TaGATA promoters. Promoter sequences (− 1500 bp) of 79 TaGATA genes were analyzed by PlantCARE. The upstream length to the translation starting site can be inferred according to the scale at the bottom
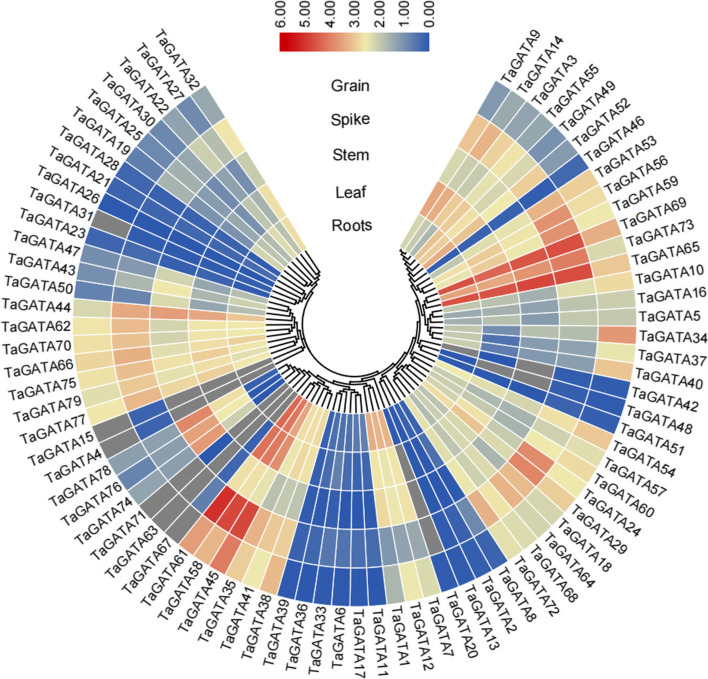
Expression analysis of TaGATAs in wheat tissues
The expression patterns of 79 TaGATAs in 5 tissues of Chinese spring, including roots, leaves, stems, spikes, and grains, were compared (Fig. 8; Table S11). On the basis of different expression pattern of these genes, they could be classified into two groups. Group 1 include 9 genes, and they were only expressed in some tissues. For example, TaGATA4 were only expressed in spike, and no expressed in other tissues. Group 2 includes 70 genes, which displayed expression in all tissues analyzed in the current study. Group 2 can be divided into two subgroup. Twelve TaGATAs were assigned to the subgroup 1 with high expression levels (log2TPM + 1 > 2) in all tissues. 10 TaGATAs were assigned to the subgroup 2 with low expression levels (log2TPM + 1 < 0.5) in all tissues. The rest of 48 genes of 70 genes were belong to the subgroup 3. These results implied that TaGATAs showed different expression level and genes in the same subfamily also displayed different expression profile.
Fig. 8.
Expression profiles of the TaGATA genes in different tissues. Expression data were processed with log2 normalization. The color scale represents relative expression levels from high (red) to low (blue)
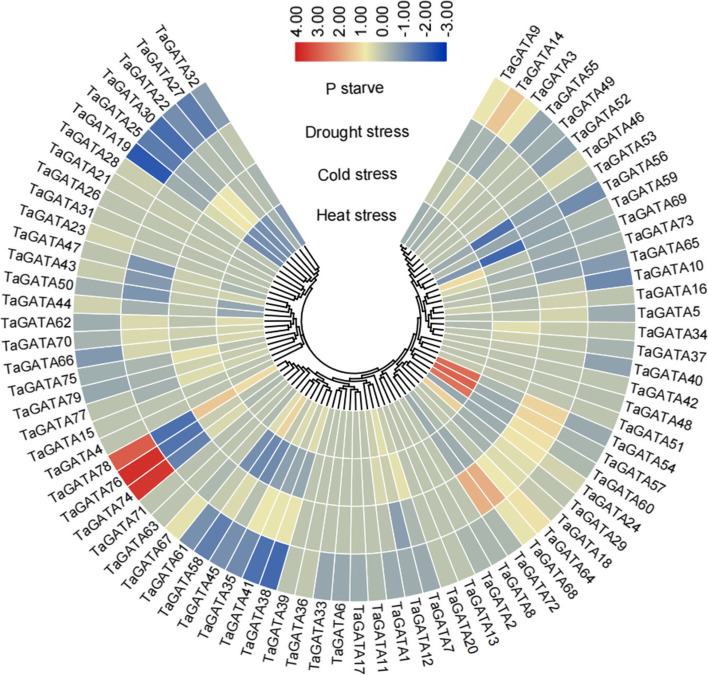
Expression patterns of TaGATAs under abiotic stress
We analyzed the expression level of TaGATA genes under different abiotic stress using the wheat transcriptome data recently published, such as drought, heat, cold stresses and P starvation. Overall, the expression level of TaGATA genes significantly changed under diverse abiotic stresses (Fig. 9; Table S12). Several TaGATA genes were in response to heat stress or P starvation. For example, the expression level of TaGATA74, TaGATA76 and TaGATA78 were extremely increased by P starvation. TaGATA54, TaGATA57 and TaGATA60 showed high expression level responding to heat stress. Meanwhile, some TaGATA genes were repressed by cold stress, such as TaGATA53 and TaGATA59, or by P starvation, such as TaGATA19. In contrast, several TaGATAs were not induced by any abiotic stresses. For example, TaGATA4 and TaGATA20 displayed almost no expression alteration in response to all analyzed treatments. Instead, several genes displayed opposite expression patterns under different abiotic stress. For instance, TaGATA78 was extremely induced by all treatments, which showed down-regulation in drought stress, but up-regulation in other treatments. In addition, several TaGATA genes were chosen for qRT-PCR to verify the reliability of transcriptome data, and the results were uniform to the sequencing data (Fig. S1, S2).
Fig. 9.
Expression profiles of the TaGATA genes under different abiotic stresses. Expression data were the ratio to control values. The color scale represents expression levels from upregulation (red) to downregulation (blue)
Discussion
Transcription factors take a vital regulatory role in plant growth and development. They are the key links in modulating many kinds of physiological activities. Thus far, the GATA family has been reported in a number of plant species, such as Arabidopsis, rice [4], maize [47], apple [20], and Brassica napus [48]. The gene structures, expression profile, characteristic features and functions have already been reported in some GATA genes. Nevertheless, a genome-wide analysis of the GATA family genes have not yet been reported in wheat (Triticum aestivum L.). In the present study, 79 members of TaGATA genes were found in the Triticum aestivum genome, which were identified as TaGATA1 to TaGATA79 based on their chromosome location (Fig. 1; Table S1). TaGATAs classified into four subfamilies showed obvious difference in genetic structures and expression patterns (Fig. 1 and Fig. 2; Table S11 and Table S12). The current study gives a valuable information for future functional characterization of GATA genes and it contributes to increase adaptive capacity when plants subjected to abiotic stress.
In plants, GATA genes showed low conservation in their exon/introns structures. In wheat, exons number in TaGATA genes ranges from 1 to 8 (Fig. 2), which is very similar to that of Brassica napus (1 to 9) [48] and Arabidopsis (2 to 8), and rice (2 to 9) [4]. Except for the zinc finger, the low level of similarity in flanking sequences suggested that the different subfamilies have appeared by modular evolution through shuffling of exons encoding the zinc finger domains [4]. Large divergences in TaGATA gene and protein structures could cause functional differences. For instance, the GATA domain featured with 20 residues in the zinc finger in subfamily III, while other three subfamilies showed 18 residues (Fig. 3; Table S4). The CCT and TIFY domains were particularly existed in the subfamily III, which were found to be responsible for flowering, hypocotyl and root development in Arabidopsis thaliana [49]. For instance, AtGATA23 modulates the auxin response factors ARF7 and ARF19 and influences the lateral root initiation cell differentiation and root branching pattern [50]. The first GATA factor is identified on the basis of the light and circadian clock-related cis-elements in its promoters [51]. Moreover, the Arabidopsis GATA factors AtGATA1, AtGATA2, and AtGATA4 have been reported to be associated with light regulation of gene expression and photomorphogenesis [52, 53]. Thus, the function of the GATA factors can be predicted according to the identification of cis-elements from their promoter. In this study, TaGATA75 and TaGATA77 showed high expression level in most tissues of wheat (Fig. 8, Table S11). Meanwhile, the promoter of these genes had TGA-element involved in auxin-responsive. It suggested the importance of these genes in root development. The subfamily I genes were reported to be associated with plant growth and in response to environmental stresses. In Arabidopsis, BME3 (ortholog of TaGATA24) could enhance seed germination capacity [54]. In comparison with wild-type plants, seeds in knockout of BME3 plants were more prone to dormancy and more vulnerable to cold stress. In this study, TaGATA24 were highly expressed in all tissues, and the promoter of TaGATA24 had ABRE-element and G-box involved in abscisic acid responsiveness and light responsiveness (Fig. 5; Table S10), which are consistent with our results concerning expression pattern under heat and drought stresses (Fig. 8 and Fig. 9; Table S11 and Table S12). Meanwhile, Ravindran et al. [24] found that RGL2-DOF6 complex modulates GATA12 (GATA subfamily I) to promote seed dormancy in Arabidopsis. GATA genes in subfamily II may be associated with flowering and also in response to abiotic stresses. Expression pattern analysis exhibit that GATA genes respond to diverse abiotic stresses, such as high temperature, salinity, cold, and drought treatments in rice, Brassica juncea, Brassica napus, Cucumis sativus, and pepper [17, 18, 48, 55, 56]. In Arabidopsis, GNC and GNL (ortholog of TaGATA38) were associated with germination, bloom and cold stress [49]. In Brassica napus, the expression level of BnGATA2.5 (ortholog of TaGATA38) was highly depressed under ABA, drought and cold stresses [48]. In this study, TaGATA38 was expressed across many tissues, and was down-regulated in cold stress and P starve, and up-regulated in heat and drought stress (Fig. 8 and Fig. 9; Table S11 and Table S12). Meanwhile, the promoter of TaGATA38 had ABRE-element, G-box, MBS involved in abscisic acid responsiveness, light responsiveness and drought-inducibility (Fig. 5; Table S10), thus showing its strong response to environmental stresses. Moreover, over-expressing BdGATA13 in Arabidopsis led to darker green leaves, more delayed flowering, and more increased drought tolerance [16]. In rice, over-expressing OsGATA16 and OsGATA8 enhances cold and drought tolerance, respectively [27, 57]. Over-expression of SlGATA17 increases drought tolerance in tomato [26]. In this study, TaGATA54, TaGATA57 and TaGATA60 were increased in heat stress, but decreased in cold stress and P starve. Our qRT-PCR analysis showed that the expression of TaGATA60 were up-regulated under salt stress, but no response under drought stress (Fig. S1), suggesting that TaGATA60 could be a functional gene in response to salt stress. However, GATA subfamily IV have known very little so far. Here, Expression pattern analysis showed that TaGATA19, TaGATA22 and TaGATA25 were down-regulated in response to heat stress and P starve (Fig. 9; Table S12). Salicylic acid and jasmonic acid have been reported to play important roles in plants responding to abiotic stress [58, 59]. In this study, the promoter of TaGATA19, TaGATA22 and TaGATA25 had TGACG-motif and TCA-element involved in MeJA-responsiveness and salicylic acid responsiveness (Fig. 5; Table S10), suggesting subfamily IV of TaGATA may be also associated with abiotic stress.
In general, we conducted a comprehensive characterization of GATA family genes in wheat. All these results provide a basis information for manipulating GATA genes and facilitate marker-assisted breeding in wheat. Nevertheless, functional identification is necessary for further study to uncover the exact functional characteristic of TaGATA genes.
Supplementary Information
Acknowledgements
We thank Dr. Lei Ge for his insightful advising and contribution in manuscript revision.
Authors’ contributions
XF and QY conceived and designed the research. XF, QY, XH and JZ performed the experiments and data analyses. WL and XF wrote the article. All authors read and approved the final article.
Funding
This work was supported by the National Natural Science Foundation of China [grant No. 32101660; 32001449], and the Natural Science Foundation of Shandong Province [grant No. ZR2021QC052].
Availability of data and materials
All data analyzed during this study are included in this article and its Additional files.
Declarations
Ethics approval and consent to participate
All experimental research and field studies on plants in our study complies with Chinese institutional, national, and international guidelines and legislation. The planting and management of experimental materials are permitted by the Experimental Station of Qingdao Agricultural University (Qingdao,Shandong Province, China).
Consent for publication
Not applicable.
Competing interests
Authors have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Xu L, Yang L, Huang H. Transcriptional, post-transcriptional and post-translational regulations of gene expression during leaf polarity formation. Cell Res. 2007;17:512–519. doi: 10.1038/cr.2007.45. [DOI] [PubMed] [Google Scholar]
- 2.Jin J, He K, Tang X, Li Z, Lv L. An Arabidopsis transcriptional regulatory map reveals distinct functional and evolutionary features of novel transcription factors. Molecular Biology Evolution. 2015;32:1767–1773. doi: 10.1093/molbev/msv058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci U S A. 2014;111:2367–2372. doi: 10.1073/pnas.1316278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reyes JC, Muro-Pastor MI, Florencio FJ. The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 2004;134:1718–1732. doi: 10.1104/pp.103.037788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H, Zou S, Li Y, et al. An ankyrin-repeat and WRKY-domain-containing immune receptor confers stripe rust resistance in wheat. Nat Commun. 2020;11:1353. doi: 10.1038/s41467-020-15139-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hao L, Shi S, Guo H, Zhang J, Li P, Feng Y. Transcriptome analysis reveals differentially expressed MYB transcription factors associated with silicon response in wheat. Sci Rep. 2021;11:4320. doi: 10.1038/s41598-021-83761-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niu X, Luo T, Zhao H, Su Y, Li H. Identification of wheat dreb genes and functional characterization of tadreb3 in response to abiotic stresses. Gene. 2020;740:144514. doi: 10.1016/j.gene.2020.144514. [DOI] [PubMed] [Google Scholar]
- 8.Li X, Gao S, Tang Y, Li L, Zhang F, Feng B, Fang Z, Ma L, Zhao C. Genome-wide identification and evolutionary analyses of bZIP transcription factors in wheat and its relatives and expression profiles of anther development related TabZIP genes. BMC Genomics. 2015;16:976. doi: 10.1186/s12864-015-2196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li K, Debernardi JM, Li C, Huiqiong L, Chaozhong Z, Judy J, von Korff M, Jinshun Z, Jorge D. Interactions between SQUAMOSA and SHORT VEGETATIVE PHASE MADS-box proteins regulate meristem transitions during wheat spike development. Plant Cell. 2021;33:3621–3644. doi: 10.1093/plcell/koab243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel-Vedele F, Caboche M. A tobacco cDNA clone encoding a GATA-1 zinc fifinger protein homologous to regulators of nitrogen metabolism in fungi. Mol Gen Genet. 1993;240:365–373. doi: 10.1007/BF00280388. [DOI] [PubMed] [Google Scholar]
- 11.He P, Wang X, Zhang X, Jiang Y, Tian W, Zhang X, Li Y, Sun Y, Xie J, Ni J, He G, Sang X. Short and narrow flag leaf1, a GATA zinc finger domain-containing protein, regulates flag leaf size in rice (Oryza sativa) BMC Plant Biol. 2018;18:273. doi: 10.1186/s12870-018-1452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Zhang Y, Zhang L, Huang H, Yang B, Luan S, Xue H, lin W. OsGATA7 modulates brassinosteroids-mediated growth regulation and influences architecture and grain shape. Plant Biotechnol J. 2018;16:1261–1264. doi: 10.1111/pbi.12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C, Hou Y, Hao Q, Chen H, Chen L, Yuan S, Shan Z, Zhang X, Yang Z, Qiu D, Zhou X, Huang W. Genome-wide survey of the soybean GATA transcription factor gene family and expression analysis under low nitrogen stress. PLoS One. 2015;10:e0125174. doi: 10.1371/journal.pone.0125174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C, Huang Y, Xiao Z, Yang H. A GATA transcription factor from soybean (Glycine max) regulates chlorophyll biosynthesis and suppresses growth in the transgenic Arabidopsis thaliana. Plants-Basel. 2020;9:1036. doi: 10.3390/plants9081036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng W, Li W, Song N, Tang Z, Liu J, Wang Y, Pan S, Dai L, Wang B. Genome-wide characterization, evolution, and expression profile analysis of GATA transcription factors in Brachypodium distachyon. International Journal of Molecular Science. 2021;22:2026. doi: 10.3390/ijms22042026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji G, Bai X, Dai K, Yuan X, Guo P, Zhou M, Shi W, Hao C. Identification of GATA transcription factors in Brachypodium distachyon and functional characterization of BdGATA13 in drought tolerance and response to gibberellins. Front Plant Sci. 2021;12:2386. doi: 10.3389/fpls.2021.763665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu C, Li N, Yin Y, Wang F, Gao S, Jiao C, Yao M. Genome-wide identification and function characterization of GATA transcription factors during development and in response to abiotic stresses and hormone treatments in pepper. J Appl Genet. 2021;62:265–280. doi: 10.1007/s13353-021-00618-3. [DOI] [PubMed] [Google Scholar]
- 18.Zhang K, Jia L, Yang D, Hu Y, Njogu MK, Wang P, Lu X, Yan C. Genome wide identifification, phylogenetic and expression pattern analysis of GATA family genes in cucumber (Cucumis sativus L.) Plants-Basel. 2021;10:1626. doi: 10.3390/plants10081626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, Zou X, Huang Z, Fan S, Qun G, Liu A, Gong J, Li J, Gong W, Shi Y, et al. Genome-wide identifification and analysis of the evolution and expression patterns of the GATA transcription factors in three species of Gossypium genus. Gene. 2019;680:72–83. doi: 10.1016/j.gene.2018.09.039. [DOI] [PubMed] [Google Scholar]
- 20.Chen H, Shao H, Li K, Zhang D, Fan S, Li Y, Han M. Genome wide identification, evolution, and expression analysis of GATA transcription factors in apple (Malus× domestica Borkh.) Gene. 2017;627:460–472. doi: 10.1016/j.gene.2017.06.049. [DOI] [PubMed] [Google Scholar]
- 21.Hudson D, Guevara DR, Hand AJ, Xu ZH, Hao LX, Chen X, Zhu T, Bi YM, Rothstein SJ. Rice cytokinin GATA transcription Factor1 regulates chloroplast development and plant architecture. Plant Physiol. 2013;162:132–144. doi: 10.1104/pp.113.217265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.An Y, Zhou Y, Han X, Shen C, Wang S, Liu C, Yin W, Xia X. The GATA transcription factor GNC plays an important role in photosynthesis and growth in poplar. J Exp Bot. 2020;71:1969–1984. doi: 10.1093/jxb/erz564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu G, Casaretto JA, Ying S, Mahmood K, Liu F, Yong B, Rothstein SJ. Overexpression of OsGATA12 regulates chlorophyll content, delays plant senescence and improves rice yield under high density planting. Plant Mol Biol. 2017;94:215–227. doi: 10.1007/s11103-017-0604-x. [DOI] [PubMed] [Google Scholar]
- 24.Ravindran P, Verma V, Stamm P, Kumar PP. 2017. A novel RGL2-DOF6 complex contributes to primary seed dormancy in Arabidopsis thaliana by regulating a GATA transcription factor. Mol Plant 10, 1307–1320. [DOI] [PubMed]
- 25.Liu X, Zhu X, Wei X, Lu C, Shen F, Zhang X, Zhang Z. The wheat LLM-domain-containing transcription factor TaGATA1 positively modulates host immune response to Rhizoctonia cerealis. J Exp Bot. 2020;71:344–355. doi: 10.1093/jxb/erz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao T, Wu T, Pei T, Wang Z, Yang H, Jiang J, Zhang H, Chen X, Li J, Xu X. Over-expression of SlGATA17 promotes drought tolerance in transgenic tomato plants by enhancing activation of the phenylpropanoid biosynthetic pathway. Front Plant Sci. 2021;12:634888. doi: 10.3389/fpls.2021.634888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nutan KK, Singla-Pareek SL, Pareek A. The Saltol QTL-localized transcription factor OsGATA8 plays an important role in stress tolerance and seed development in Arabidopsis and rice. J Exp Bot. 2020;71:684–698. doi: 10.1093/jxb/erz364. [DOI] [PubMed] [Google Scholar]
- 28.Yang J, Xu YC, Jianhao W, Sujuan G, Yisui H, Hung F-Y, Tao L, Qing L, Yue Lin W, Keqiang YS. The chromatin remodelling ATPase BRAHMA interacts with GATA-family transcription factor GNC to regulate flowering time in Arabidopsis. J Exp Bot. 2021;73:835–847. doi: 10.1093/jxb/erab430. [DOI] [PubMed] [Google Scholar]
- 29.Liu H, Li T, Wang YM, Zheng J, Li HF, Hao CY, Zhang XY. TaZIM-A1 negatively regulates flowering time in common wheat (Triticum aestivum L.) Journal of Integrated Plant Biology. 2019;61:359–376. doi: 10.1111/jipb.12720. [DOI] [PubMed] [Google Scholar]
- 30.Kersey PJ, Allen JE, Allot A, Barba M, Boddu S, Bolt BJ, Carvalho-Silva D, Christensen M, Davis P, Grabmueller C, Kumar N, Liu Z, Maurel T, Moore B, McDowall MD, Maheswari U, Naamati G, Newman V, Ong CK, Paulini M, Pedro H, Perry E, Russell M, Sparrow H, Tapanari E, Taylor K, Vullo A, Williams G, Zadissia A, Olson A, Stein J, Wei S, Tello-Ruiz M, Ware D, Luciani A, Potter S, Finn RD, Urban M, Hammond-Kosack KE, Bolser DM, De Silva N, Howe KL, Langridge N, Maslen G, Staines DM, Yates A. Ensemble genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res. 2018;46:802–808. doi: 10.1093/nar/gkx1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:279–285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov D, Liechti R, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:597–603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 35.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Letunic I, Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:256–259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Tang H, DeBarry JD, Tan X, Li J, Wang X, Lee T, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D, Zhang Z, Zhang Z, Zhu J, Yu J. KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinformatics. 2010;8:77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu B, Jin J, Guo A, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 43.Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma S, Wang M, Wu J, Guo W, Chen Y, Li G, Wang Y, Shi W, Xia G, Gu D, Kang Z, Ni F. WheatOmics: a platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol Plant. 2021;14:1965–1968. doi: 10.1016/j.molp.2021.10.006. [DOI] [PubMed] [Google Scholar]
- 45.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview version 2 multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feng X, Liu W, Dai H, Qiu Y, Zhang G, Chen Z, Wu F. HvHOX9, a novel homeobox leucine zipper transcription factor revealed by root miRNA and RNA sequencing in Tibetan wild barley, positively regulates Al tolerance. J Exp Bot. 2020;19:6057–6073. doi: 10.1093/jxb/eraa290. [DOI] [PubMed] [Google Scholar]
- 47.Long J, Yu X, Chen D, Hu F, Li J. Identification, phylogenetic evolution and expression analysis of GATA transcription factor family in maize (Zea mays) Int J Agric Biol. 2020;23:637–643. [Google Scholar]
- 48.Zhu W, Guo Y, Chen Y, Wu D, Jiang L. Genome-wide identification, phylogenetic and expression pattern analysis of GATA family genes in Brassica napus. BMC Plant Biol. 2020;20:543. doi: 10.1186/s12870-020-02752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richter R, Bastakis E, Schwechheimer C. Cross-repressive interactions between SOC1 and the GATAs GNC and GNL/CGA1 in the control of greening, cold tolerance, and flowering time in Arabidopsis. Plant Physiol. 2013;162:1992–2004. doi: 10.1104/pp.113.219238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rybel BD, Vassileva V, Parizot B, et al. A novel aux/IAA28 signaling Cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr Biol. 2010;20:1697–1706. doi: 10.1016/j.cub.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 51.Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. doi: 10.1146/annurev.pp.46.060195.002305. [DOI] [Google Scholar]
- 52.Luo X, Lin W, Zhu S, Zhu J, Sun Y, Fan X, Cheng M, Hao Y, Oh E, Tian M, Liu L, Zhang M, Xie Q, Chong K, Wang Z. Integration of light-and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Development Cell. 2010;19:872–883. doi: 10.1016/j.devcel.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H, Wang H, Zhu Q, Gao Y, Wang H, Zhao L, Wang Y, Xi F, Wang W, Yang Y, Lin C, Gu L. Transcriptome characterization of moso bamboo (Phyllostachys edulis) seedlings in response to exogenous gibberellin applications. BMC Plant Biol. 2018;18:125. doi: 10.1186/s12870-018-1336-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu P, Koizuka N, Martin RC, Nonogaki H. Te BME3 (blue Micropylar end 3) GATA zinc fnger transcription factor is a positive regulator of Arabidopsis seed germination. Plant J. 2005;44:960–971. doi: 10.1111/j.1365-313X.2005.02588.x. [DOI] [PubMed] [Google Scholar]
- 55.Bhardwaj AR, Joshi G, Kukreja B, Malik V, Arora P, Pandey R, et al. Global insights into high temperature and drought stress regulated genes by RNA-Seq in economically important oilseed crop Brassica juncea. BMC Plant Biol. 2015;15:9. doi: 10.1186/s12870-014-0405-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gupta P, Nutan KK, Singla-Pareek S, Pareek A. Abiotic stresses cause differential regulation of alternative splice forms of GATA transcription factor in rice. Front Plant Sci. 2017;8:1944. doi: 10.3389/fpls.2017.01944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang H, Wu T, Li Z, Huang K, Kim NE, Ma Z, Kwon SW, Jiang W, Du X. OsGATA16, a GATA transcription factor, confers cold tolerance by repressing OsWRKY45-1 at the seedling stage in rice. Rice. 2021;14:42. doi: 10.1186/s12284-021-00485-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Janda T, Szalai G, Pál M. Salicylic acid signalling in plants. Int J Mol Sci. 2020;21:2655. doi: 10.3390/ijms21072655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jang G, Yoon Y, Choi YD. Crosstalk with jasmonic acid integrates multiple responses in plant development. Int J Mol Sci. 2020;21:305. doi: 10.3390/ijms21010305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data analyzed during this study are included in this article and its Additional files.