ABSTRACT
Influenza virus infections are thought to be initiated in a small number of cells; however, the heterogeneity across the cellular responses of the epithelial cells during establishment of disease is incompletely understood. Here, we used an H1N1 influenza virus encoding a fluorescent reporter gene, a cell lineage-labeling transgenic mouse line, and single-cell RNA sequencing to explore the range of responses in a susceptible epithelial cell population during an acute influenza A virus (IAV) infection. Focusing on multiciliated cells, we identified a subpopulation that basally expresses interferon-stimulated genes (ISGs), which we hypothesize may be important for the early response to infection. We subsequently found that a population of infected ciliated cells produce most of the ciliated cell-derived inflammatory cytokines, and nearly all bystander ciliated cells induce a broadly antiviral state. From these data together, we propose that variable preexisting gene expression patterns in the initial cells targeted by the virus may ultimately affect the establishment of viral disease.
IMPORTANCE Influenza A virus poses a significant threat to public health, and each year, millions of people in the United States alone are exposed to the virus. We do not currently, however, fully understand why some individuals clear the infection asymptomatically and others become severely ill. Understanding how these divergent phenotypes arise could eventually be leveraged to design therapeutics that prevent severe disease. As a first step toward understanding these different infection states, we used a technology that allowed us to determine how thousands of individual murine lung epithelial cells behaved before and during IAV infection. We found that small subsets of epithelial cells exhibited an antiviral state prior to infection, and similarly, some cells made high levels of inflammatory cytokines during infection. We propose that different ratios of these individual cellular responses may contribute to the broader antiviral state of the lung and may ultimately affect disease severity.
KEYWORDS: single cell, interferon-stimulated genes, gene expression, RNA sequencing, transgenic mouse, reporter virus
INTRODUCTION
Influenza viruses cause significant morbidity and mortality each year, with an estimated 290,000 to 650,000 deaths globally (1). With relatively limited available therapeutics, significant effort has been focused on defining the mechanisms of viral pathogenesis to explore alternate avenues of intervention. One area of particular interest is in defining why seemingly similar individuals exhibit different courses of disease. While viral and host genetics certainly play a role, some models suggest that stochastic variability within early innate responses may also be a contributing factor (2). A limitation to testing this hypothesis is that the field currently has an incomplete understanding of the heterogeneity in cellular responses after the initiation of viral infection.
Since its introduction, single-cell RNA sequencing (scRNA-seq) has been used to resolve tissue heterogeneity in a variety of environments (3–5). It has also been used to study host-influenza interactions both in vitro and in vivo (2, 6–17). Previous in vitro studies in immortalized cell lines have primarily focused on understanding how coinfection, genetic variability in viral populations, and infection with different strains of influenza virus influence the cellular innate immune response (2, 6, 7, 10, 15). In vivo studies profiling the entire lung have identified changes in inflammatory responses throughout infection, described broad viral tropism, and found interferon-dependent and independent gene modules (8, 9). Few, mostly in vitro studies, have focused on the responses of epithelial cells, the main targets of IAV infection (12, 13, 16). Even less understood is the heterogeneity across responses by genetically defined subsets of epithelial cells in vivo.
A complicating factor in defining the responses of epithelial cell subsets using scRNA-seq is that their transcriptional profiles are dramatically altered by infection, often due to high viral burden and degradation of host mRNAs (9, 18, 19). Additionally, damage to the lung results in the differentiation of progenitor populations or dedifferentiation of cells to repair and regenerate the respiratory epithelium, often beginning within the first several days of injury (20–23). Consequently, it can be difficult to assign transcriptional profiles to specific cell populations with certainty by detecting cell type-specific marker gene transcripts alone (24).
As a first step in understanding how differences in cellular responses may ultimately influence infection outcomes, we explored how a single, highly susceptible epithelial cell population responded to IAV infection. We generated a fluorescent reporter virus and identified ciliated cells as a major target of the pdmH1N1 A/California/04/2009 (Cal09) influenza strain in vivo. We then used a transgenic mouse model to genetically label ciliated cells prior to infection and track them throughout the establishment of viral disease. Finally, we used scRNA-seq to interrogate gene expression in mock-infected, bystander, and influenza virus-infected ciliated cells from matched infected animals. Interestingly, we found expression of type I and type III interferons in a subset of ciliated cells, which we hypothesize may be linked to the basal expression of antiviral genes prior to infection. Additionally, there was notable expression of strong interferon-stimulated gene (ISG) transcriptional profiles in almost all bystander cells within 48 h of infection, indicating the sensitivity of these cells to inflammatory cytokines. These data support the growing literature showing variability in the response to infection, and we propose that divergent gene expression before exposure to influenza virus could potentially contribute to disease pathogenesis.
RESULTS
Epithelial ciliated cells are a main target of a reporter H1N1 virus in the upper respiratory tract.
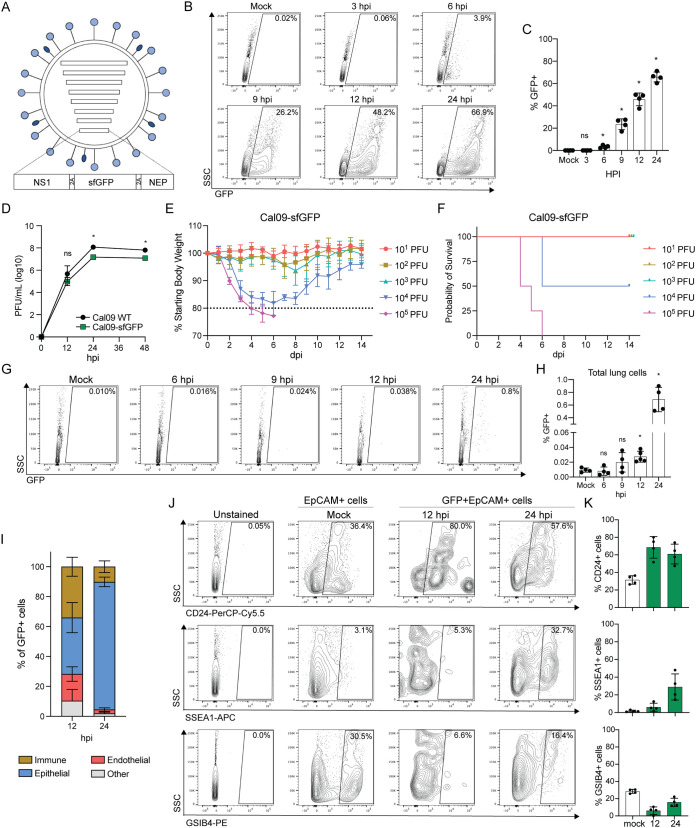
Our initial aim was to understand the potential heterogeneity of responses by a single population of epithelial cells to a nonlaboratory-adapted influenza virus. We therefore generated a fluorescent reporter virus in the prototype pdmH1N1 A/California/04/2009 genetic background (Cal09-superfolder green fluorescent protein [sfGFP]) to allow for the identification of infected cells based on fluorescence. We modified the eighth viral segment to eliminate splicing and encode NS1, sfGFP, and NEP as a single open reading frame (ORF), with 2A autoproteolytic cleavage sites separating the proteins cotranslationally (Fig. 1A).
FIG 1.
Ciliated cells are a target of a Cal09 reporter virus in the murine upper respiratory tract. (A) Schematic of A/California/04/2009 reporter virus expressing sfGFP from segment 8, which also encodes nonstructural proteins NS1 and NEP. 2A, porcine teschovirus 2A sequence. (B) Representative flow cytometry of GFP+ cells collected at the indicated time points following infection with Cal09-sfGFP (MDCK cells, MOI = 0.5, single-cycle infection). hpi, hours postinfection. (C) Quantification of flow cytometry in panel B. Means with SDs are shown; n = 4 independent experiments; Mann-Whitney U test. (D) Viral titer after replication in embryonated chicken eggs for the time indicated for wild-type Cal09 or Cal09-sfGFP (1,000 PFU), measured using plaque assays. PFU, plaque-forming units. Means with SDs are shown; n = 4 eggs per sample; Mann-Whitney U test. (E) Body weights of mice infected with the indicated doses of Cal09-sfGFP. Means with SDs are shown; n = 4 to 6 mice per dose. Dashed line represents humane endpoint. dpi, days postinfection. (F) Kaplan-Meier survival curve of mice infected with the indicated doses of Cal09-sfGFP. n = 4 to 6 mice per dose. (G) Representative flow cytometry of GFP+ cells from the lungs of mice infected with Cal09-sfGFP at the indicated times postinfection. (H) Quantification of flow cytometry in panel G. Means with SDs are shown; n = 4 to 5 mice per time point; Mann-Whitney U test. (I) Quantification of GFP+ cell surface expression of CD45, EpCAM, or CD31. Immune cells, CD45+; epithelial cells, CD45-EpCAM+; endothelial, CD45-CD31+. Cells without expression of either three were categorized as “other.” Cells were isolated from tracheas of Cal09-sfGFP-infected mice at the indicated times postinfection. Means with SDs are shown; n = 4 mice per time point. (J) Representative flow cytometry of unstained, EpCAM+ (Mock) or GFP+ EpCAM+ (12 hpi, 24 hpi) cells isolated from the tracheas of mice infected with Cal09-sfGFP at the indicated times postinfection stained for surface expression of CD24 (ciliated cells), SSEA1 (secretory cells), and GSIB4 (basal cells). (K) Quantification of flow cytometry in panel J (EpCAM+ cells, white bars; GFP+ EpCAM+ cells, green bars). Means with SDs are shown; n = 4 mice per time point. For all panels, ns, not significant, and *, P ≤ 0.05.
Although we and others have previously used this genetic strategy to generate experimentally useful reporter viruses (25, 26), genetically modified viruses can fail to express the reporter protein or be significantly attenuated (27, 28). To determine reporter expression and the degree of attenuation for our reporter virus, we performed single-cycle infections and multi-cycle growth curves of Cal09-sfGFP and wild-type Cal09. We could detect a statistically significant increase in GFP expression in infected MDCK cells relative to background by 6 h postinfection (hpi), indicating this virus functions as a sensitive reporter of viral infection (Fig. 1B and C). We also found our reporter virus grew to similar titers and with similar kinetics as wild-type Cal09 in embryonated chicken eggs, suggesting only slight attenuation (Fig. 1D). As we ultimately wanted to investigate IAV infection in a mouse model, we next asked whether Cal09-sfGFP could establish disease in vivo. We infected mice and found a dose-dependent decrease in body weight and survival following infection (Fig. 1E and F). Finally, we wanted to determine the earliest time points at which we could reliably identify GFP-positive (GFP+) cells in vivo. To do so, we infected mice and then collected lung and tracheal cells at several time points following infection for flow cytometry analysis. We could detect a significant increase in GFP signal above the background beginning 12 hpi, with an additional 25-fold increase in fluorescent cells by 24 hpi (Fig. 1G and H).
With our reporter virus functioning appropriately, we next turned our attention to identifying the types of cells targeted by the virus during the establishment of infection. Although the tropism of pdmH1N1 viruses has been described previously, there are conflicting reports depending on the method used for assessment (29–31). We therefore collected cells from the upper respiratory tract of our Cal09-sfGFP-infected animals and stained them for markers of immune (CD45+), epithelial CD45− EpCAM+), and endothelial (CD45− CD31+) cells (32–34). As expected, epithelial cells represent most of the infected cells at both 12 (average, 37%) and 24 hpi (average, 85%) (Fig. 1I). We detected a sizeable population of apparently infected immune cells at 12 hpi; however, because our assay cannot distinguish between infected immune cells and those that have phagocytosed GFP, we focused on the epithelial compartment. To determine the types of epithelial cells infected, we repeated this experiment but stained cells for markers of the major respiratory epithelial cell types, ciliated (EpCAM+ CD24+), secretory (EpCAM+ SSEA1+), and basal (EpCAM+ GSIB4+) (35–38). At both 12 and 24 hpi, we found that ciliated cells accounted for more than 50% of infected epithelial cell population (Fig. 1J and K). We therefore concluded that ciliated cells were a relevant in vivo target during murine infection with Cal09-sfGFP and thus decided to focus additional study on this population of the epithelium during influenza virus infection.
A transgenic model to genetically label epithelial ciliated cells for scRNA-seq.
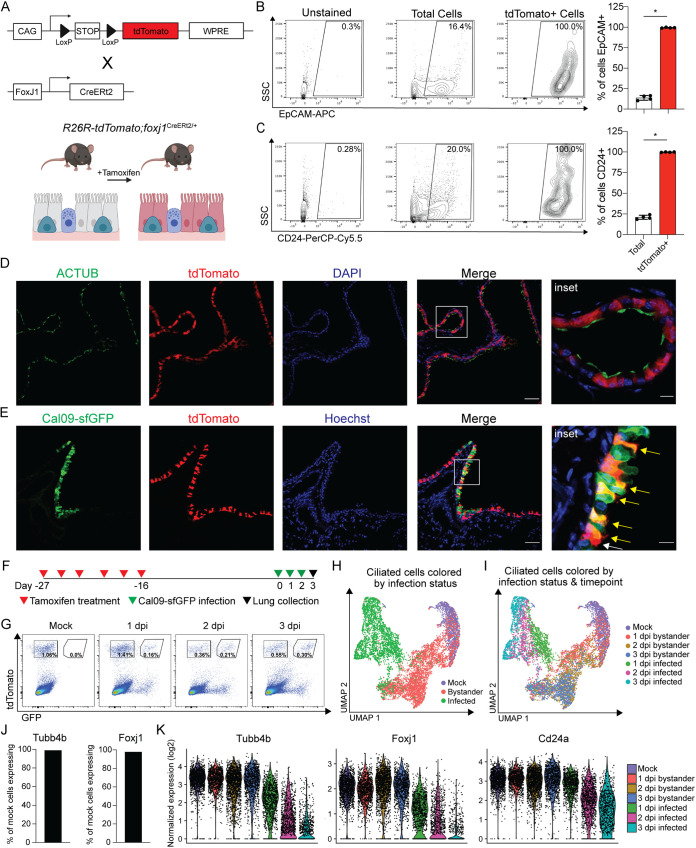
To understand heterogeneity in the ciliated cell response to IAV infection, we wanted to perform scRNA-seq on mock-infected, influenza virus-infected, and bystander ciliated cells. However, we and others have documented that ciliated cells lose expression of canonical markers during acute viral infection (35, 39–41), which could affect our ability to collect or analyze them during infection. Furthermore, progenitor cells could begin to differentiate into ciliated cells during infection experiments, and the intermediate gene expression profile could confound our capacity to define virally induced gene signatures. To circumvent these challenges, we employed an inducible lineage tracing strategy to fluorescently label ciliated cells prior to infection. We crossed mice that contained a Cre-responsive reporter cassette (tdTomato mice) to mice that expressed tamoxifen-inducible Cre recombinase under the promoter of the ciliated cell-specific transcription factor Foxj1 (foxj1CreERt2/+ mice) (42). When tdTomato;foxj1CreERt2/+ mice (further referred to as tdT;foxj1 mice) are treated with tamoxifen, Cre recombinase expressed in ciliated cells enters the nucleus and induces recombination, allowing for constitutive tdTomato expression (Fig. 2A). Importantly, in this system, cells are only labeled during the tamoxifen treatment window, and expression of tdTomato in labeled cells is permanent.
FIG 2.
A transgenic mouse model genetically labels ciliated cells for identification during infection. (A) Schematic of transgenic mouse model. Mice containing a Cre-responsive reporter cassette were crossed to mice expressing Cre recombinase under the promoter for the ciliated cell-specific transcription factor Foxj1. When tdT;foxj1 mice are treated with tamoxifen, ciliated cells permanently express tdTomato. WPRE, woodchuck hepatitis virus posttranscriptional regulatory element. (B) Representative flow cytometry and quantification of total and tdTomato+ cell surface EpCAM expression. Cells isolated from the lungs of tamoxifen-treated tdT;foxj1 mice. Means with SDs are shown. n = 4; Mann-Whitney U test. (C) Representative flow cytometry and quantification of total and tdTomato+ cell surface CD24 expression. Cells isolated from the lungs of tamoxifen treated tdT;foxj1 mice. Means with SDs are shown. n = 4; Mann-Whitney U test. (D) Cross-sectioned microscopy of lung epithelial cells from tamoxifen treated tdT;foxj1 mice. ACTUB (green), cilia; tdTomato (red), Foxj1 activity; DAPI (blue), DNA. Scale bar, 50 μm, 10-μm inset. Image representative of two independent experiments with at least 2 mice each. (E) Cross-sectioned microscopy of lung epithelial cells from tamoxifen-treated tdT;foxj1 mice 24 hpi with Cal09-sfGFP. Cal09-sfGFP (green), GFP; tdTomato (red), Foxj1 activity; Hoechst (blue), DNA. Yellow arrows, GFP+ ciliated cells. White arrows, GFP- ciliated cells. Scale bar, 50 μm; 10-μm inset. Image representative of two independent experiments with at least 2 mice each. (F) Schematic of scRNA-seq experiment design. TdT;foxj1 mice were treated with tamoxifen six times over 2 weeks followed by a 2-week recovery period before infection with Cal09-sfGFP. Total lung cells were isolated at the indicated times postinfection for FACS. (G) Flow cytometry of FACS-sorted tdTomato+ ciliated cells isolated from tamoxifen-treated tdT;foxj1 mice at the indicated times postinfection with Cal09-sfGFP or mock infected with PBS. (H) UMAP dimensionality reduction plot showing ciliated cells clustered based on their transcriptomes and relation to one another. Ciliated cells are colored based on their infection status. (I) UMAP dimensionality reduction plot showing ciliated cells clustered based on their transcriptomes and relation to one another. Ciliated cells are colored based on their infection status and time point. (J) The percentage of mock tdTomato+ cells with at least one transcript of ciliated cell markers Tubb4b and Foxj1. (K) Violin plot of normalized expression of ciliated cell markers Tubb4b, Foxj1, and CD24a by sample. The width of each violin represents the frequency of that expression level. For all panels, ns, not significant, and *, P ≤ 0.05.
To validate that this model specifically labeled ciliated cells, we stained lung cells from tamoxifen-treated tdT;foxj1 mice for the ciliated cell markers EpCAM and CD24. We found nearly 100% of tdTomato+ cells were positive for EpCAM or CD24 compared to less than 25% of total lung cells (Fig. 2B and C). We then cross-sectioned lungs from tdT;foxj1 mice and found tdTomato+ cells to be positive for expression of acetylated tubulin, a component of cilia (Fig. 2D). Based on these data and the timing of tamoxifen treatment, we considered any cell with tdTomato expression to be a bona fide ciliated cell prior to infection. To verify our previous results of viral infection of ciliated cells in this model, we collected the lungs from tdT;foxj1 mice 1 day postinfection (dpi) with Cal09-sfGFP. Using confocal microscopy, we identified distinct populations of tdTomato+ GFP+ cells in infected mice (Fig. 2E).
To perform single-cell RNA sequencing of ciliated cells, we infected tamoxifen-treated tdT;foxj1 mice with Cal09-sfGFP or phosphate-buffered saline (PBS) as a mock control and then collected the lungs 1, 2, and 3 dpi for ciliated cell isolation using fluorescence-activated cell sorting (FACS) (Fig. 2F and G). We prepared the sorted cells for scRNA-seq using the 10× Genomics platform and then sequenced the resulting cDNA libraries. As a first test of the quality of our data, we explored the dimensionality of our data set with Uniform Manifold Approximation and Projection (UMAP) dimension reduction; as expected, cells grouped predominantly by their infection status (Fig. 2H). We also found that within bystander or infected cell groupings, cells from the same time point tended to group with each other (Fig. 2I). We next quantified expression of ciliated cell markers to confirm that we had collected ciliated cells and found that 98.98% and 97.88% of tdTomato+-sorted cells from uninfected animals had at least one transcript detected of Tubb4b and Foxj1, respectively (Fig. 2J). Consistent with previous work showing ciliated cells downregulate genes related to ciliogenesis or ciliated cell morphology during infection, expression of Tubb4b and Foxj1 decreased over time in infected cells but was stable in bystander cells (Fig. 2K) (35, 39–41). Although expression of Cd24a decreased, it remained detectable in infected cells, which suggested CD24 can be used as a marker for ciliated cells throughout infection (Fig. 2K).
Identification of a subpopulation of ciliated cells with basal ISG expression.
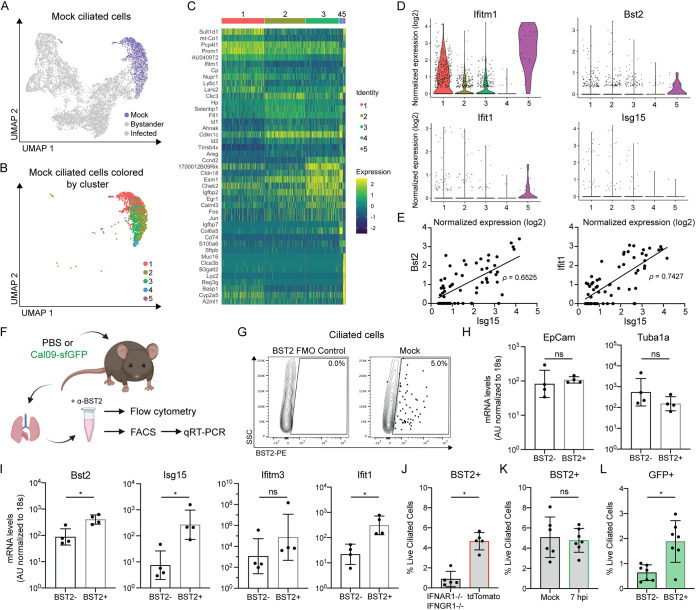
Next, we performed unbiased clustering on ciliated cells from uninfected animals (Fig. 3A) but restricted our analyses to mock cells containing at least one transcript of Foxj1 to exclude any potentially missorted cells (Fig. 3B). These cells clustered into five discrete groups, although most uninfected ciliated cells displayed similar gene expression profiles with varied expression of genes related to negative regulation of the cell cycle and cytoskeletal and tight junction maintenance (e.g., Ccnd2, Cdkn1c, Tmsb4x, and Cldn18) (Fig. 3B and C). However, we observed a subpopulation of ciliated cells (cluster 5) which displayed high levels of the antiviral interferon-stimulated gene (ISG) Ifitm1 (Fig. 3C and D). ISGs are a set of genes expressed by cells in response to interferon signaling and induce an antiviral state within the cell (43–45). More in-depth analysis revealed that additional ISGs were disproportionately expressed by this cluster of ciliated cells (e.g., Bst2 and Ifit1), but in all ciliated cell clusters, we could identify cells with basal ISG expression (Fig. 3D). Thus, basal ISG expression can be detected in some uninfected ciliated cells; however, the magnitude of expression of these genes is insufficient to drive clustering, at least under our analysis parameters.
FIG 3.
Identification of a subpopulation of ciliated cells with basal ISG expression prior to infection. (A) UMAP dimensionality reduction plot showing ciliated cells clustered by their transcriptome similarity. Mock ciliated cells are colored purple. (B) UMAP dimensionality reduction plot showing clusters of mock ciliated cells containing at least one transcript of Foxj1. (C) Heat map of the top 10 most variably expressed genes by each mock ciliated cell cluster. Genes are ordered based on P value. The scale of the heat map shows the expression of a gene by each cell relative to the mean expression by all cells in the sample. (D) Violin plots showing the normalized expression of each ISG by the cells in each cluster. The width of each violin represents the frequency of that expression level. (E) Normalized expression of the ISGs Bst2 and Ifit1 plotted against normalized expression of Isg15 in mock cells containing at least one transcript of Isg15. Pearson’s correlation coefficient, ρ, was used to measure the linear correlation of expression. (F) Schematic of experiments used to evaluate basal ISG expression in ciliated cells in panels G to L. (G) Representative flow cytometry of ciliated cells (CD45− CD31− CD24+) isolated from mock-infected mice stained for BST2/tetherin. The BST2 gate was set using fluorescence minus one (FMO) control as shown on the left side of the panel. (H and I) qRT-PCR of mRNA isolated from FACS-sorted BST2+ or BST2− ciliated cells (CD45− CD31− CD24+) was used to quantify transcripts for ciliated cell marker genes (EpCam, Tuba1a) (H) and ISGs (Bst2, Isg15, Ifitm3, and Ifit1) (I). Gene expression was quantified based on a standard curve and normalized to endogenous 18s expression. Means with SDs are shown. n = four independent mice; Mann-Whitney U test. AU, arbitrary units. (J) Quantification of flow cytometry. Ciliated cells (CD45− CD31− CD24+) were isolated from untreated IFNAR1−/− IFNGR1−/− or tdTomato mice and stained for BST2. Means with SDs are shown; n = 5 mice per group; Mann-Whitney U rest. (K) Quantification of flow cytometry. Ciliated cells (CD45− CD31− CD24+) were isolated from mock-infected or Cal09-sfGFP-infected (1.4e5 PFU) mice 7 hpi and stained for BST2. Means with SDs are shown; n = at least 6 mice per group from two independent experiments; Mann-Whitney U test. (L) Quantification of flow cytometry. GFP expression by BST2+ or BST2− ciliated cells (CD45− CD31− CD24+) isolated from mice 7 hpi with Cal09-sfGFP (1.4e5 PFU) was measured. Means with SDs are shown. n = 7 mice from two independent experiments; Mann-Whitney U test. For all panels, ns, not significant, and *, P ≤ 0.05.
In order to understand if the ISG expression in these cells was stochastic or if they have a more broadly active antiviral state, we performed a correlation analysis of cells expressing either Bst2 or Ifit1 and another ISG, Isg15. A strong correlation between the ISG transcripts (Fig. 3E) is consistent with the idea that these ciliated cells are likely to have upregulated a canonical antiviral response. To experimentally validate the existence of this subpopulation of cells, we collected ciliated cells from mock-infected mice to analyze ISG expression (Fig. 3F). By performing flow cytometry for the surface-expressed ISG BST2-tetherin, we identified a small population (~3 to 6%) of ciliated cells with BST2 expression (Fig. 3G). This proportion was consistent with the percentage of mock ciliated cells with >1 transcript for Bst2 (6.5%) in our single-cell data set. To verify that this preinfection BST2+ subpopulation also expressed the other ISGs, we isolated BST2+ and BST2− ciliated cells from mock-infected mice for reverse transcription-quantitative PCR (qRT-PCR) analysis of key transcripts. While ciliated cell markers were unchanged between the two populations (Fig. 3H), the levels of the other ISGs tested were higher in BST2+ cells (Fig. 3I).
This population of ISG-expressing cells could be explained by a low level of authentic IFN signaling in the epithelium, likely produced by lung-resident immune cells (46). To test this hypothesis, we collected epithelial cells from untreated animals deficient in the type I and type II interferon receptors, the two classes of interferon predominantly produced by immune cells (47–53). We observed a significant decrease in basal ciliated cell BST2 expression in INFAR1−/− IFNGR1−/− mice relative to tdTomato mice, suggesting that these interferons drive most of this phenotype (Fig. 3J). Finally, we wanted to understand how these basally innate immune-activated cells could affect viral infection. We hypothesized that they could either be resistant to infection (and contribute to “bystander” cell phenotypes) or they could be infected by IAV and potentially contribute to heterogeneity in the infected cell responses. We therefore infected animals with Cal09-sfGFP, and at a time point before BST2 expression was significantly induced (Fig. 3K), we looked for GFP signal in the BST2+ cells. Relative to the BST2− ciliated cells, we were unable to observe a decrease in infection (Fig. 3L). Because we cannot formally rule out that some of the detected GFP+ BST2+ cells may have resulted from rapid antiviral protein expression after infection, we can only interpret these data to indicate that basal ISG expression in ciliated cells is likely insufficient to make these cells completely refractory to infection. Further, any gene expression changes resulting from the basal transcriptional profiles of these cells likely manifest phenotypically in the infected cell population.
Bystander ciliated cells robustly, and mostly uniformly, respond to interferon signaling.
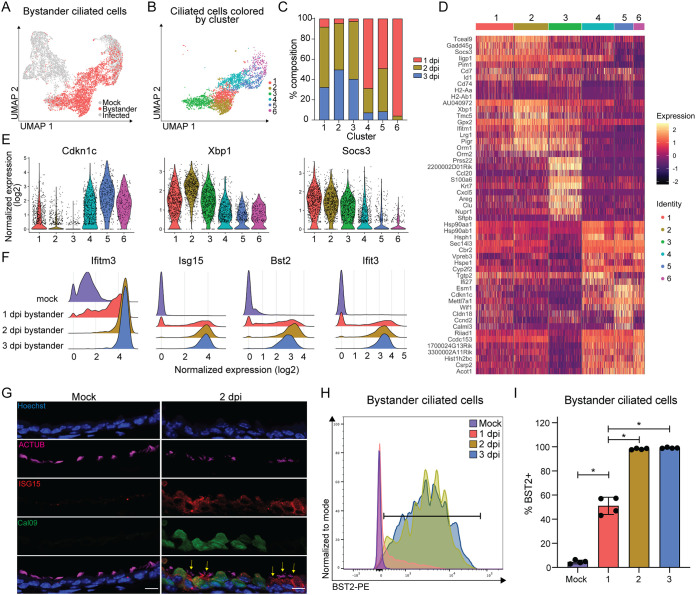
Next, we wanted to understand the potential spread of gene expression in the uninfected bystander ciliated cell population from our broader experiment (Fig. 4A). We performed unbiased clustering of GFP− cells harboring less than 10 viral transcripts at all three infection time points to focus on truly uninfected cells. This strategy yielded 6 individual clusters (Fig. 4B), and further analysis revealed that clusters 1 to 3 were largely composed of ciliated cells from 2 and 3 dpi, while clusters 4 to 6 were mostly made of cells from 1 dpi (Fig. 4C). This suggested that gene expression differences across bystander cells were likely primarily driven by the inflammatory state of the lung rather than basal heterogeneity in the ciliated cell population. Consistent with this idea, a heat map of the top 10 most variably expressed genes in each cluster showed that clusters 4 to 6 were dominated by expression of genes found in mock cells (e.g., Cdkn1c, Cldn18, Calml3, and Esm1), while clusters 1 to 3 are marked by expression of genes related to stress response (Xbp1, Nupr1) and interferon signaling (Socs3, Ifitm1, and Ccl20) (Fig. 4D and E).
FIG 4.
Uninfected bystander ciliated cells respond homogenously to pulmonary inflammation during IAV infection. (A) UMAP dimensionality reduction plot showing bystander cells. (B) UMAP dimensionality reduction plot showing unbiased clustering of bystander GFP− ciliated cells. GFP− ciliated cells were considered bystander cells if they had less than 10 viral transcripts. (C) Stacked bar graph plotting the percentage of each cluster that belongs to each time point. (D) Heat map of the top 10 most variably expressed genes by each cluster of bystander GFP− ciliated cells. Genes are ordered based on P value. The scale of the heat map shows the expression of a gene by each cell relative to the mean expression by all cells in the sample. (E) Violin plot showing the normalized expression of Cdkn1c (negative regulation of cell cycle), Xbp1 (stress response), and Socs3 (interferon signaling). The width of each violin represents the frequency of that expression level. (F) Ridge plots depicting normalized expression of ISGs, grouped by sample. Ridge height indicates the frequency of expression level. (G) Microscopy of cross-sectioned lung epithelial cells from mock- or Cal09-sfGFP-infected mice (2 dpi) stained for ACTUB and ISG15. Scale bar, 10 μm. Yellow arrows indicate ISG15+ bystander ciliated cells. Image representative of sections from three mice from two independent experiments. (H) Flow cytometry of bystander ciliated cells (CD45− CD31− CD24+ GFP−) stained for BST2. Ciliated cells were isolated from the lungs of mock-infected or Cal09-sfGFP-infected mice at the indicated times postinfection. Gate for BST2 was set using FMO control. (I) Quantification of flow cytometry in panel H. Means with SDs are shown; n = 4 mice per group; Mann-Whitney U test. For all panels, ns, not significant, and *, P ≤ 0.05.
We were surprised to see, however, how few of the “classic” ISGs appeared in the heat map, as these cells were presumably in a highly inflammatory environment. Specific interrogation of ISGs revealed widespread expression, as nearly all bystander cells at 2 and 3 dpi expressed ISGs, including Ifitm3, Isg15, Bst2, and Ifit3 (Fig. 4F). This broad expression pattern explained why they did not appear in the list of differentially expressed genes (Fig. 4D). To experimentally validate what our analysis predicted would essentially be the ubiquity of ISG expression in bystander cells, we stained lung sections from mock-infected and Cal09sfGFP-infected mice for ISG15 and found robust expression in bystander (GFP−) ciliated cells, even more so than in infected (GFP+) cells (Fig. 4G). For a more quantitative approach, we used flow cytometry to measure BST2 expression in bystander (GFP−) ciliated cells and found over 97% were BST2+ at both 2 and 3 dpi (Fig. 4H and I). Thus, despite some minor differences in the response profiles of bystander ciliated cells during infection, the innate antiviral response is mostly homogenous and consistent with the idea that basal gene expression differences in ciliated cells had little to no effect on their responses.
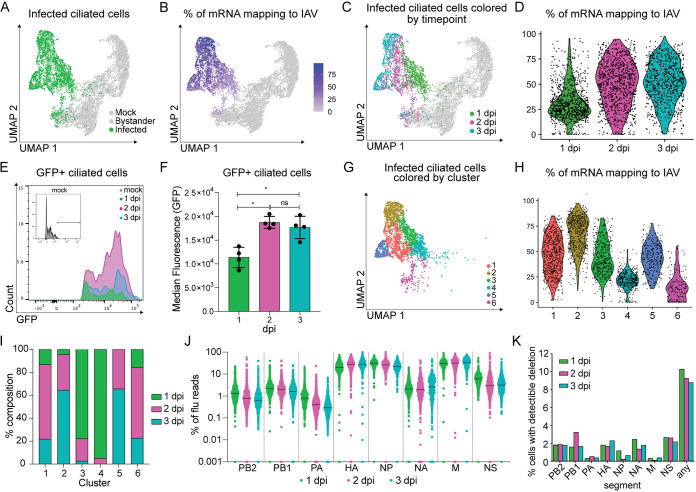
Viral burden in infected ciliated cells increases throughout infection.
We next turned our attention to the influenza virus-infected ciliated cells collected during our time course experiment (Fig. 5A). It has been reported that lung epithelial cells, both in vitro and in vivo, can have a high viral burden during IAV infection (6, 9, 15). In agreement with previous studies, we found viral mRNAs comprised from less than 1 to over 90% of the total cell transcriptome (Fig. 5B). Interestingly, we could observe a “gradient” of viral gene expression across our population of infected cells (Fig. 5B); labeling infected cells by time point revealed that viral RNA levels were generally associated with the infection time point (Fig. 5C and D). We experimentally validated this phenomenon by infecting mice with Cal09-sfGFP and measuring fluorescence of infected cells at different time points. Consistent with the scRNA-seq, we saw a larger amount of virally expressed reporter protein per cell at days 2 to 3 compared to day 1 (Fig. 5E and F).
FIG 5.
Viral replication characteristics across ciliated cells during the first 3 days of infection. (A) UMAP dimensionality reduction plot depicting infected ciliated cells. (B) UMAP dimensionality reduction plot of total ciliated cells shaded based on the fraction of total mRNA transcripts that are viral. (C) UMAP dimensionality reduction plot depicting GFP+ ciliated cells 1, 2, or 3 days post-Cal09-sfGFP infection. (D) Violin plot showing the fraction of total mRNA transcripts that are viral for each GFP+ cell grouped by sample. Violin width indicates the frequency of the percentage level. (E) Representative flow cytometry histogram of GFP+ ciliated cells (CD45− CD31− CD24+). Ciliated cells were isolated from mock-infected or Cal09-sfGFP-infected mice at the indicated times postinfection. (F) Quantification of the median fluorescence (GFP) of GFP+ ciliated cells in panel E. Means with SDs are shown; n = 4 mice per time point; Mann-Whitney U test. (G) UMAP dimensionality reduction plot depicting clusters of infected cells (GFP+, ≥10 viral transcripts), which are colored by cluster identity. (H) Violin plot showing the fraction of total mRNA transcripts that are viral for each GFP+ cell grouped by cluster identity. Violin width indicates the frequency of the percentage level. (I) Quantification of the fraction of cells from each time point that make up each cluster. Cells were clustered based on host and viral gene expression. (J) Plot showing the fraction of total viral transcripts belonging to each influenza segment in infected GFP+ cells, grouped by time point. GFP+ cells were classified as infected based on the presence of 10 or more viral transcripts. If a fraction for a gene was 0, it was set to 0.001 for inclusion in the plot. (K) Bar graph depicting the percentage of infected cells (GFP+, ≥10 viral transcripts) with detectable deletions (>15 reads, >75 nt deletion, final product > 150 nt) in each flu segment or any segment.
We next performed unbiased clustering on the infected cells (Fig. 5G). This analysis revealed that, in contrast to the bystander cells, clusters did not appear to predominately group by infection time points. We found viral burden between clusters of infected cells varied and that clusters spanned multiple time points (Fig. 5H and I). Additionally, the relative expression of viral genes was similar to those seen in influenza virus-infected A549 cells (M > NP ~ HA > NS > NA > PB1 ~ PB2 ~ PA), apart from NS, which may be due to our genetic manipulations in this segment (Fig. 5J) (6, 7).
Next, we performed an analysis for deletions within the IAV genomic segments. While the scRNA-seq methodology we employed sequenced the 3′ end of viral transcripts and thus poorly sampled total viral genetic diversity, some variants were nevertheless captured. One form of variation that can still be partially assessed in our analysis is the presence of large internal deletions in any given segment, a hallmark of defective influenza A virus populations and whose presence can be associated with altered infection outcome (54, 55). Setting an empirical threshold of deletion abundance and size based on rarefaction curves, we found deletions throughout all eight viral segments (Fig. 5K; see Table S1 in the supplemental material), although the majority of infections did not meet our thresholds for positive identification of large deletions.
Rare production of interferons by infected ciliated cells at later time points.
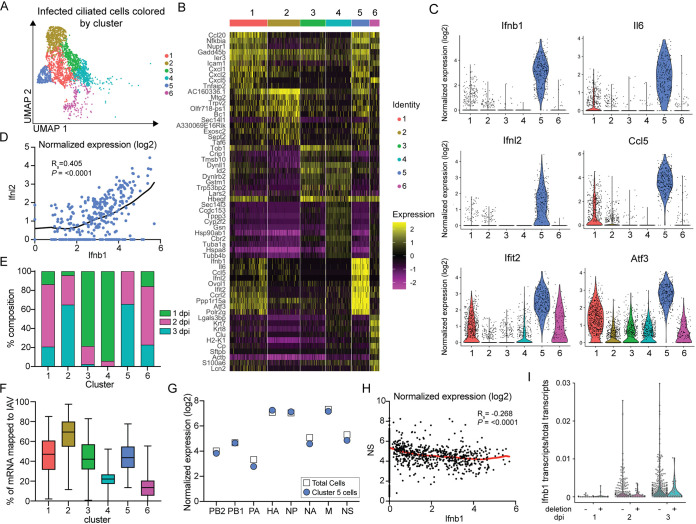
Finally, we turned our attention to understanding the variability in the cell response to IAV infection. We performed unbiased clustering on the GFP+ populations of cells with 10 or more viral transcripts across all three time points. For this analysis, we generated a new gene expression matrix in which viral genes were removed to ensure viral RNA levels would not confound our analysis of the host response. However, this strategy revealed clusters (Fig. 6A) that were highly similar to those generated by the matrix, including both host and viral genes (Fig. 5G).
FIG 6.
Inflammatory antiviral responses are restricted to a subpopulation of infected ciliated cells during infection. (A) UMAP dimensionality reduction plot showing unbiased clustering of infected GFP+ cells. GFP+ cells were classified as infected based on the presence of 10 or more viral transcripts. Clustering was based on the expression of host genes only. (B) Heat map of the top 10 most variably expressed genes by each cluster of infected GFP+ ciliated cells. Cells are clustered based on host gene expression only. Genes are ordered based on P value. The scale of the heat map shows the expression of a gene by each cell relative to the mean expression by all cells in the sample. (C) Violin plots of normalized expression for 6 of the top 10 most variably expressed genes in cluster 5. Violin width indicates the frequency of the expression level. (D) Plot of the normalized expression (log2) of Ifnb1 versus Ifnl2 by cells in cluster 5. A locally weighted scatterplot smoothing (LOWESS) curve was fitted to visualize the relationship of expression. Spearman’s rank order correlation, P < 0.0001; we reject the null hypothesis that there is no correlation between Ifnb1 and Ifnl2 expression. (E) Stacked bar graph showing the percentage of each cluster in panel A that belongs to each of the three GFP+ time points. (F) Box-and-whisker plot showing the fraction of total mRNA transcripts that are virally grouped by cluster. Clustering was based on the expression of host genes only. Box extends from 25th to 75th percentiles with a line representing the median value for each cluster. Whiskers extend to the minimum and maximum value. (G) Plot of average normalized expression of each viral gene for total infected GFP+ cells and cluster 5 cells. (H) Plot of the normalized expression of Ifnb1 versus NS for total infected GFP+ cells. A LOWESS curve was fitted to visualize the relationship of expression. Spearman’s rank order correlation, P < 0.0001; we reject the null hypothesis that there is no correlation between Ifnb1 and NS expression. (I) Distribution of Ifnb1 transcripts normalized to total cellular transcripts in GFP+ infections in which deletions were (+) and were not (−) detected according to our thresholds. For all panels: ns, not significant, and *, P ≤ 0.05.
Analysis of the top 10 most variably expressed genes by cluster revealed a number of cellular responses after viral infection, including one subpopulation (cluster 1) which expressed multiple chemokines (Cxcl1, Cxcl2, and Cxcl5) and another (cluster 6) which expressed genes involved in epithelial cell integrity (Lgals3bp, Krt7, and Krt8) (Fig. 6B). However, one subpopulation of ciliated cells (cluster 5) displayed high expression of both type I and type III IFN in addition to other inflammatory cytokines such as Il6 and Ccl5 (Fig. 6B and C). Further analysis of gene expression in this cluster revealed a correlation between type I and type III IFN expression (Spearman’s correlation coefficient (rs) = 0.405, P < 0.0001), which had been previously reported during IAV infection in vitro but not in vivo (Fig. 6D) (2). Interestingly, cluster 5 was comprised of cells from 2 and 3 dpi, but not 1 dpi (Fig. 6E). Thus, a time-dependent subpopulation of infected ciliated cells has the potential to not only respond to, but also contribute to, the inflammatory cytokines in the infected lung.
We next asked what could be driving the expression of cytokines in this IFN+ subpopulation. We found average expression levels of viral genes in the IFN+ cluster, which suggested IFN expression was not due to high or low viral burden (Fig. 6F). We therefore compared the average expression of each segment in this cluster to total infected cells, as the IAV protein NS1 is a known interferon antagonist, and the absence of NS in single cells has been shown to be associated with interferon expression (2, 7). We found a slight decrease in NS expression along with PA and NA (Fig. 6G) and measured a statistically significant negative correlation between NS and Ifnb1 expression (rs = −0.268, P < 0.0001) (Fig. 6H). However, the IFN+ cells were not predominantly derived from NS-negative viral particles, as we did detect NS expression in nearly every Ifnb1+ cell. This population of NS-negative cells may, in fact, exist but would be missed in our experimental system, as our fluorescent infection reporter was encoded in this segment. Finally, we explored whether Ifnb1+ cells contained viral genes with internal deletions, as infection with defective viruses can induce IFN (15, 56–59). While we cannot definitively claim any given infection did not contain a segment with an internal deletion, in those infections in which we identified such variants, we did not see any enrichment for Ifnb1 expression (Fig. 6I). Taken together, these results suggest that interferon expression by infected ciliated cells cannot be entirely explained by differences in viral gene expression and is likely dictated at least in part by the specific transcriptional state of the infected cell.
DISCUSSION
In this study, we sought to understand variability in the host response to IAV infection. To do so, we used a fluorescent H1N1 virus, a transgenic mouse model, and scRNA-seq to explore the heterogeneity of gene expression in response to influenza virus infection in a single, highly susceptible epithelial cell population. Our analyses support several findings from prior single-cell studies focused on IAV infection but provide further insights into the dynamic interplay between virus and host in vivo. Our data set revealed that a small population of ciliated cells expressed ISGs before infection, which we were able to validate using qRT-PCR and flow cytometry. During acute infection, we found a small population of ciliated cells not only produce the majority of cytokines but also multiple cytokines at once. Finally, we found that ciliated cells are highly responsive to interferon signaling, as nearly all bystander ciliated cells had induced expression of ISGs by 2 dpi.
Despite our in-depth analysis of the ciliated cell-specific response to IAV infection, there remain several outstanding questions. For example, in the infected (GFP+) ciliated cell samples, we identified a subpopulation of ciliated cells with high expression of inflammatory cytokines, including type I and III IFN, Il6, and Ccl5 (Fig. 6B and C). These data support the concept that epithelial cells in vivo may be relevant producers of cytokines during IAV infection (60–63). We do not know, however, whether cytokine expression was a stable characteristic of this population of cells or if at any given time, infected cells have the potential to transiently upregulate these genes. Additionally, the mechanism of the observed increased viral “burden” in infected cells over time remains unclear (Fig. 5D to F). We favor a model in which the initiation of lytic cell death is delayed in vivo relative to cell culture after infection, allowing for more complex patterns of viral protein accumulation. Future studies will be required to determine if the “rates” of virally induced cell death are different across different experimental systems, however, or if a completely different mechanism may explain this observation.
We also identified a subpopulation of ciliated cells with basal ISG expression prior to infection (Fig. 3G to I), but the biological significance of this population currently remains unknown. Despite the upregulation of Ifitm3 and Ifitm1, genes encoding proteins known to prevent viral entry (64–66), we were unable to detect a block in the infection of these cells (Fig. 3L). We therefore hypothesize that this subpopulation of cells is primed to become the cells that produce interferon and other inflammatory mediators after infection. Along these lines, an independent study recently showed that epithelial cells in the upper respiratory tract of uninfected children expressed higher basal levels of viral-sensing and antiviral genes than adults (67). The authors hypothesized this may be linked to basal expression of type II IFN in the airways and could explain why children have reduced replication and increased clearance of SARS-CoV-2 relative to adults. Further investigation will be required, however, to understand if there is truly any functional link between basal gene expression signatures in epithelial cells and phenotypically differential responses to infection.
We also made some experimental analysis choices that may be important for interpretation of our data. First, we captured contaminating lymphocytes and cells which concurrently expressed both ciliated cells and other epithelial cell markers in our single-cell samples. This is not uncommon, as cells with expression of markers for multiple epithelial cell types have been described in single-cell data sets (12, 24, 68). We excluded cells with high expression of secretory cell marker Scgba3a and alveolar type I/II cell markers Pdpn and Ager, which could have led us to exclude rare, but real, populations of ciliated cells. Along similar lines, in our infected cell analysis, we defined infected cells as those that were GFP+ and contained 10 or more transcripts of viral genes. This likely resulted in some truly infected cells being thrown out, possibly due to errors in sorting or failure to express GFP.
Additionally, we did not perform full virus genotyping in our infected cells. Although we identified Ifnb1+ cells that lacked observable viral gene deletions (Fig. 6I), it is possible there were deletions or mutations in NS in cells with high Ifnb1 expression that we were unable to detect. Further, the distribution of deletions across viral segments did not match their anticipated abundance from prior studies, predominantly in the three polymerase segments. However, we rarely observed the same deletion between time points, suggesting these may be generated de novo over the course of these infections, absent, or at least rare in our initial inoculum. Recent work suggests de novo deletion frequencies may be more evenly distributed across the eight segments (69), whereupon selection thereafter leads to their observed distribution (59).
In conclusion, our study provides an in-depth characterization of the ciliated cell response to IAV infection and highlights the power of scRNA-seq to identify small but discrete subpopulations of cells. We also propose that our data are consistent with the idea that the cell state prior to infection may influence how a cell ultimately responds to infection. Further, we demonstrate the range of gene expression changes that can occur in “bystander” cell populations. These data represent a step toward unraveling the complexities during the establishment of viral replication beginning in a small number of cells after a transmission event. Future studies have the potential to link these types of changes to outcomes of infection and may ultimately allow for a better understanding or prediction of individuals who will have mild or severe influenza disease.
MATERIALS AND METHODS
Cell culture.
MDCK cells (ATCC) were grown at 37°C and in 5% CO2 and maintained in minimal essential medium (MEM) supplemented with 5% fetal bovine serum (FBS), GlutaMAX, HEPES, sodium bicarbonate, and penicillin-streptomycin.
Viruses.
Cal09-sfGFP (70) was generated by inserting the gene that encodes sfGFP into segment 8 using the same strategy previously used to encode Cre recombinase in A/Puerto Rico/8/1934 (26). The sequence for each segment matches A/California/04/2009 GenBank accession numbers FJ966079 to FJ966086 with the following nucleotide substitutions noted: PB2, C1170T; PA, G1953A; HA, G6A, G368T, A640G, A716G; NP, T9C, C865T.
Growth curve.
Wild-type Cal09 or Cal09-sfGFP virus (1,000 PFU) was injected into 10-day-old embryonated chicken eggs and then incubated until the indicated time points at 37°C to allow for viral replication. Allantoic fluid was collected, serially diluted, and then plaqued on MDCK cells. Plaques were stained using sera from Cal09-infected mice and sheep anti-mouse IgG peroxidase-linked whole antibody (Cytiva; catalog no. NXA931V) and then visualized with KPL TrueBlue peroxidase substrate (SeraCare; catalog no. 5510-0030).
Time course of single-cycle infection in MDCK cells.
Confluent MDCK cells were mock infected or infected with Cal09-sfGFP with a multiplicity of infection (MOI) of 0.5 for 1 h at 37°C. Virus was removed, and maintenance medium was added to cells. Cells were collected at the indicated time points and fixed with 2% paraformaldehyde (PFA) before being analyzed.
Mouse lines and tamoxifen treatment.
Wild-type C57BL/6, B6.Cg-Gt(ROSA)26Sortm14(CAGtdTomato)Hze/J (tdTomato), and B6.Cg-Ifngr1tm1Agt Ifnar1tm1.2Ees/J (IFNAR1−/− IFNGR1−/−) mice were purchased from The Jackson Laboratory. Foxj1CreERt2/+ mice were developed by Chay T. Kuo (42). tdT;foxj1CreERt2/+ litters were genotyped using the following primers: F, TGGCTTGCAGGTACAGGAGG, and R, CCTCATTGTGGCCAACAAACCC. To induce reporter expression, tdT;foxj1 mice were treated with 2 mg of tamoxifen (Sigma-Aldrich; catalog no. T5648-1G) resuspended in corn oil via oral gavage 5 to 6 times over a 2-week period.
Mouse infections.
For all infections, mice were anesthetized with 100 μL of ketamine-xylazine and infected intranasally with 40 μL of virus (2.8e4 PFU Cal09-sfGFP for all experiments except Fig. 3K and L (1.4e5 PFU)) diluted in pharmaceutical-grade PBS. The humane endpoint was reaching 80% of starting body weight. Body weight was monitored once daily for 14 dpi or until collection time point. All experiments used a minimum of 3 mice per condition. Male and female mice were used and randomly assigned to experimental and control groups. All experiments were conducted in accordance with Duke IACUC.
Flow cytometry.
To isolate murine lung cells for flow cytometry analysis, lungs were perfused with 7 mL PBS, inflated with 0.75 mL Dispase (Corning, catalog no. 354235, or Sigma, catalog no. 4942078001) and 0.75 mL 1% low-melt NuSieve GTG agarose (Lonza; catalog no. 50081) dissolved in water, and covered with an ice pack for 30 s. The lungs were removed and then incubated in 1 mL Dispase for at least 45 min at room temperature before being minced by razor blade in 7 mL Dulbecco’s modified Eagle medium (DMEM) with DNase I (Sigma-Aldrich; catalog no. D4527-200KU). For tracheal cell isolation only (Fig. 1I to K), the trachea was removed and incubated in 400 μL of Dispase for at least 45 min before continuing the same protocol. The cells were further homogenized by vigorous pipetting and filtered using a 70-μm cell strainer (Olympus; catalog no. 25-376). Red blood cells were lysed by incubating cells in 1× Pharm Lyse buffer (BD Biosciences; catalog no. 555899) for 10 min at room temperature. The remaining cells were neutralized in PBS with 2% bovine serum albumin (BSA) (Lampire; catalog no. 7500855) before antibody staining. The following primary antibodies were used: CD45 (BioLegend; clone 30-F11; 1:200), CD31 (BioLegend; clone 390; 1:200), CD326/EpCAM (BD Biosciences; clone G8.8; 1:100), CD24 (BD Biosciences; clone M1/69; 1:200), CD317/BST2 (BioLegend; clone 129C1; 1:100), CD15/SSEA1 (BioLegend; clone MC-480; 1:50), and isolectin B4 (Enzo; catalog no. ALX-650-001B-MC05; 1:1,000). Cells were incubated with primary antibodies for 1 h at 4°C in the dark. For collection of live BST2− or BST2+ cells, cells were incubated with Live/Dead stain (Thermo Fisher Scientific; catalog no. L34964) prior to primary antibody staining. Cells were analyzed using the FACSCanto II or LSRFortessa X-20 using standard laser and filter combinations. Gates were set using fluorescence minus one (FMO) or unstained controls. Data were analyzed using FlowJo software. If cells were not run the same day as isolation, cells were fixed with 2% PFA after antibody staining and kept at 4°C in the dark for up to 48 h.
Microscopy.
Lungs were inflated with 1.5 mL of 1:1 Tissue-Tek OCT (VWR; catalog no. 25608-930) and 8% PFA before the trachea was tied off with suture string. The lungs were fixed overnight at 4°C in 4% PFA before incubation in 30% sucrose in PBS at 4°C for at least 24 h. The lungs were then embedded in OCT and cryosectioned (8-μm sections). Sections were thawed and allowed to dry for 2 h at room temperature before blocking with 3% BSA in PBST (PBS with 0.1% Triton X-100) and 5% normal goat serum (Thermo Fisher Scientific; catalog no. 31872) for 1 h at room temperature. The sections were then stained with primary antibodies overnight at 4°C and secondary antibodies at room temperature for 2 h (1:1,000). Primary antibodies used include ACTUB (Abcam, clone EPR16772, catalog no. ab179484, 1:1,000, or Sigma-Aldrich, clone 6-11B-1, catalog no. T7451, 1:1,000) and ISG15 (Invitrogen; clone 1H9L21; catalog no. 703132; 1:100). Following incubation with Hoechst 33342 (1:10,000) for 5 min at room temperature and 2 PBST washes, the sections were mounted in Prolong Diamond antifade mountant (Invitrogen; catalog no. P36965). If Hoechst 33342 was not used (Fig. 2D), nuclei were stained using Prolong Diamond antifade mountant with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen; catalog no. P36962). Imaging was performed using a Zeiss 780 upright confocal microscope with consistent settings, and raw images were processed using Fiji software (NIH).
Isolation of ciliated cells for scRNA-seq.
Tamoxifen-treated tdT;foxj1 mice were infected with Cal09-sfGFP or mock infected with PBS, and the lungs were collected 1, 2, and 3 dpi (2 mice per condition). Single-cell suspensions were generated as described above, and cells from mice of the same condition were pooled. Single tdTomato+, tdTomato+ GFP−, and tdTomato+ GFP+ cells were isolated using a BD FACSDiVa. The cell suspensions were then counted using a Nexcelom Cellometer K2. The cells were diluted to capture 3,000 cells per sample and loaded into a 10× Genomics Chromium Next GEM chip. cDNA libraries were generated from the GEMs according to the manufacturer’s instructions (Chromium Next GEM single-cell 3′ library kit v3.1) and run on an Agilent 4200 tape station for quality control. Libraries were quantified using Qubit high-sensitivity DNA assay (Thermo Fisher; catalog no. Q32854) and then sequenced on an Illumina NovaSeq. The Illumina base call files (BCLs) were demultiplexed into FASTQs, which were then aligned to a custom-concatenated mm10 and Cal09 genome reference file and counted using 10× Genomics CellRanger pipeline (version 3.1.0).
Single-cell data analysis.
(i) Data QC.
The raw and filtered output files from the CellRanger pipeline were read into SoupX (71) (v1.5.2) to filter reads from lysed cells using the command Load10x. Contaminating reads from lysed cells were estimated using the command autoEstCounts, and the adjusted count table (whole integers) was generated using adjustCounts. The output was then used to make a Seurat (v4.0.4) object using CreateSeuratObject. The count matrices for the 7 samples were merged, and initial QC was performed. Cells that contained fewer than 500 or more than 6,000 genes were excluded in addition to cells whose mitochondrial genes made up more than 20% of the total transcriptome. Cells with fewer than 1,500 or more than 80,000 unique molecular identifiers (UMIs) were also removed.
Following an initial round of unbiased clustering using the standard Seurat workflow (72), clusters with high expression of lymphocyte markers (Cd52, Ctla2a, Cd8a, Lyz2, and Ptprc/CD45) were removed, along with cells with high expression of secretory cell marker Scgb3a2 (>5 UMIs) and alveolar type I/II markers Ager (>5 UMIs) and Pdpn (>5 UMIs). We did not positively select cells based on expression of ciliated cell markers for bystander and infected cell analysis, as expression of these genes decreases dramatically during infection (35, 39). However, the remaining cells were considered ciliated based on >97.88% expression of ciliated cell markers Foxj1 and Tubb4b in the mock population.
(ii) Mock sample analysis.
The mock sample was subsetted to make a new Seurat object. The percentage of cells expressing Tubb4b and Foxj1 was calculated using the function PrctCellExpringGene (Ryan-Zhu, Github). For clustering, we restricted our analyses to cells with at least 1 transcript of Foxj1.
(iii) Infected sample analysis.
The three GFP-positive samples (1, 2, and 3 dpi) were subsetted to make a new Seurat object. To remove GFP+ cells that were considered “uninfected,” the raw UMI counts of each flu gene per cell were exported to an Excel sheet, and viral counts were summed. Cells with fewer than 10 total viral transcripts were considered uninfected and removed from the object. To cluster infected GFP+ cells based on host gene expression, a new count matrix with viral genes removed was created. The GFP+ object was then subjected to the standard Seurat workflow for unbiased clustering. The cluster ID for each cell was added to the meta data of the original object containing viral genes to explore the relationship between host and viral gene expression.
(iv) Bystander sample analysis.
The three GFP− samples (1, 2, and 3 dpi) were subsetted to make a new Seurat object. To remove GFP− cells that were considered “infected,” the raw UMI counts of each flu gene per cell were exported to an Excel sheet, and viral counts were summed. Cells with 10 or more total viral transcripts were considered infected and removed from the object. The GFP− object was then subjected to the standard Seurat workflow for unbiased clustering.
Deletion analysis.
All gapped reads mapping to influenza A virus from cellranger-generated BAM files were extracted based on their CIGAR string and converted to FASTA. Critically, these gapped reads use splice scoring and are inappropriate for calling bona fide deletion junctions. Therefore, similar to previous work (59), we remapped these reads using BLASTn against an empirically verified Cal09-sfGFP genome using the following parameters: a percent identity of 90, a word size of 10, a gap open penalty of 5, extend penalty of 2, and an E value cutoff of 0.000001 (73). Files were then parsed to identify potential deletion junctions with the following rules: such reads must map completely with no “missing” bases and must map with the same polarity with no inversion, and repetitive elements, such as 2 nucleotides (nt) repeated at the 5′ and 3′ ends of the junction, which could, in theory, be assigned to either, were assigned to the 5′ end arbitrarily. Each junction-spanning read was compared to the original BAM file, and identical UMIs were collapsed to a single transcript supporting the presence of a junction and thereafter enumerated on a per-cell barcode basis. To set a threshold read support for deletion calling, a rarefaction analysis was performed looking at the fraction of deletions called that were lost as we increased the read depth required for support; a depth of 15 was chosen as the point at which the curve flattened. This presumably will help to reduce template-switching artifacts or lysis-derived deletions, although it will likely come at the cost of missing low-depth deletions and bias our data set toward deletion calling in high-burden infections. A similar rarefaction curve on deletion size revealed a large set of small deletions <75 nt, which comprised 20% of deletions present in our data set. We excluded these deletions as potentially exhibiting divergent behavior from large internal deletions. Lastly, a small, <150-nt species was observed in the M segment, which may have been either a novel splice product or a template-switching artifact, which was excluded from further analysis, as its relative abundance across many barcodes and incredibly small size made its identity as a large internal deletion unlikely.
qRT-PCR.
Live BST2+ and BST2− ciliated cells (CD45− CD31− CD24+ or EpCAM+ CD24+) from tdTomato mice were sorted using the Sony SH800S cell sorter. At least 180 cells were collected for each sample per mouse. cDNA libraries were prepared for each sample using SMART-Seq v4 Ultra Low Input RNA kit for sequencing (TaKaRa Bio; catalog no. 634888). qPCR was performed using TaqMan Universal PCR master mix (Applied Biosystems; catalog no. 43-044-37) and the following TaqMan assays: EpCam (Mm00493214_m1), Tuba1a (Mm01300524_g1), Bst2 (Mm01340357_g1), Isg15 (Mm01705338_s1), Ifitm3 (Mm00847057_s1), and Ifit1 (Mm00515153_m1).
Data analysis.
For all experiments, the statistical tests used are noted in the figure legends and were performed in GraphPad Prism (version 9.0.1). In vitro experiments were conducted 4 times with 3 technical replicates each. Egg or animal studies used a minimum of n = 3 for all groups and were conducted at least twice except for the scRNA-seq experiment (n = 2, conducted once).
Data availability.
Raw and processed sequencing data are available to download from NCBI GEO via accession number GSE203018.
ACKNOWLEDGMENTS
We thank Mike Cook and Bin Li from the Duke Cancer Institute Flow Cytometry Core for their assistance and support. Confocal microscopy was performed at the Duke Light Microscopy Core Facility with guidance and training from Yasheng Gao. The 10× Genomics single-cell libraries were prepared by Stephanie Arvai, Emily Hocke, and Karen Abramson at the Duke University Molecular Genomics Core. Next-generation sequencing was performed by the Duke Center for Genomic & Computational Biology.
We thank Alfred Harding for rescuing Cal09-sfGFP and Heather Froggatt for helpful discussion.
A.B.R. is partially supported by a grant from NIAID: AI141678. N.S.H. is partially supported by grants from NIAID and NHLBI, grant numbers R01-HL142985 and R01-AI137031.
Conceptualization, C.E.H and N.S.H.; methodology, C.E.H., A.B.R., and N.S.H.; software, A.B.R.; validation, C.E.H.; formal analysis, C.E.H., A.B.R., and N.S.H.; investigation, C.E.H.; resources, N.S.H.; data curation, C.E.H. and A.B.R., writing – original draft, C.E.H. and N.S.H.; writing – review & editing, C.E.H., A.B.R., N.S.H.; visualization, C.E.H. and A.B.R.; supervision, N.S.H.; funding acquisition, N.S.H.
We declare no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Nicholas S. Heaton, Email: nicholas.heaton@duke.edu.
Stacey Schultz-Cherry, St. Jude Children's Research Hospital.
REFERENCES
- 1.Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, Cohen C, Gran JM, Schanzer D, Cowling BJ, Wu P, Kyncl J, Ang LW, Park M, Redlberger-Fritz M, Yu H, Espenhain L, Krishnan A, Emukule G, van Asten L, Pereira da Silva S, Aungkulanon S, Buchholz U, Widdowson M-A, Bresee JS, Azziz-Baumgartner E, Cheng P-Y, Dawood F, Foppa I, Olsen S, Haber M, Jeffers C, MacIntyre CR, Newall AT, Wood JG, Kundi M, Popow-Kraupp T, Ahmed M, Rahman M, Marinho F, Sotomayor Proschle CV, Vergara Mallegas N, Luzhao F, Sa L, Barbosa-Ramírez J, Sanchez DM, Gomez LA, Vargas XB, Acosta Herrera A, Llanés MJ, et al. 2018. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391:1285–1300. 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russell AB, Elshina E, Kowalsky JR, Te Velthuis AJW, Bloom JD. 2019. Single-cell virus sequencing of influenza infections that trigger innate immunity. J Virol 93:e00500-19. 10.1128/JVI.00500-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papalexi E, Satija R. 2018. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol 18:35–45. 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- 4.Paik DT, Cho S, Tian L, Chang HY, Wu JC. 2020. Single-cell RNA sequencing in cardiovascular development, disease and medicine. Nat Rev Cardiol 17:457–473. 10.1038/s41569-020-0359-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carraro G, Langerman J, Sabri S, Lorenzana Z, Purkayastha A, Zhang G, Konda B, Aros CJ, Calvert BA, Szymaniak A, Wilson E, Mulligan M, Bhatt P, Lu J, Vijayaraj P, Yao C, Shia DW, Lund AJ, Israely E, Rickabaugh TM, Ernst J, Mense M, Randell SH, Vladar EK, Ryan AL, Plath K, Mahoney JE, Stripp BR, Gomperts BN. 2021. Transcriptional analysis of cystic fibrosis airways at single-cell resolution reveals altered epithelial cell states and composition. Nat Med 27:806–814. 10.1038/s41591-021-01332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell AB, Trapnell C, Bloom JD. 2018. Extreme heterogeneity of influenza virus infection in single cells. Elife 7:e32303. 10.7554/eLife.32303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun J, Vera JC, Drnevich J, Lin YT, Ke R, Brooke CB. 2020. Single cell heterogeneity in influenza A virus gene expression shapes the innate antiviral response to infection. PLoS Pathog 16:e1008671. 10.1371/journal.ppat.1008671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Liu J, Yuan Y, Huang F, Ma R, Luo B, Xi Z, Pan T, Liu B, Zhang Y, Zhang X, Luo Y, Wang J, Zhao M, Lu G, Deng K, Zhang H. 2020. Two waves of pro-inflammatory factors are released during the influenza A virus (IAV)-driven pulmonary immunopathogenesis. PLoS Pathog 16:e1008334. 10.1371/journal.ppat.1008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steuerman Y, Cohen M, Peshes-Yaloz N, Valadarsky L, Cohn O, David E, Frishberg A, Mayo L, Bacharach E, Amit I, Gat-Viks I. 2018. Dissection of influenza infection in vivo by single-cell RNA sequencing. Cell Syst 6:679–691.e4. 10.1016/j.cels.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramos I, Smith G, Ruf-Zamojski F, Martínez-Romero C, Fribourg M, Carbajal EA, Hartmann BM, Nair VD, Marjanovic N, Monteagudo PL, DeJesus VA, Mutetwa T, Zamojski M, Tan GS, Jayaprakash C, Zaslavsky E, Albrecht RA, Sealfon SC, García-Sastre A, Fernandez-Sesma A. 2019. Innate immune response to influenza virus at single-cell resolution in human epithelial cells revealed paracrine induction of interferon lambda 1. J Virol 93:e00559-19. 10.1128/JVI.00559-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kupke SY, Ly LH, Börno ST, Ruff A, Timmermann B, Vingron M, Haas S, Reichl U. 2020. Single-cell analysis uncovers a vast diversity in intracellular viral defective interfering RNA content affecting the large cell-to-cell heterogeneity in influenza A virus replication. Viruses 12:71. 10.3390/v12010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao Y, Guo Z, Vangala P, Donnard E, Liu P, McDonel P, Ordovas-Montanes J, Shalek AK, Finberg RW, Wang JP, Garber M. 2020. Single-cell analysis of upper airway cells reveals host-viral dynamics in influenza infected adults. bioRxiv. 10.1101/2020.04.15.042978:2020.04.15.042978. [DOI]
- 13.Kelly JN, Laloli L, V’kovski P, Holwerda M, Portmann J, Thiel V, Dijkman R. 2020. Comprehensive single cell analysis of pandemic influenza A virus infection in the human airways uncovers cell-type specific host transcriptional signatures relevant for disease progression and pathogenesis. bioRxiv. 10.1101/2020.04.03.014282:2020.04.03.014282. [DOI] [PMC free article] [PubMed]
- 14.Kudo E, Song E, Yockey LJ, Rakib T, Wong PW, Homer RJ, Iwasaki A. 2019. Low ambient humidity impairs barrier function and innate resistance against influenza infection. Proc Natl Acad Sci USA 116:10905–10910. 10.1073/pnas.1902840116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang C, Forst CV, Chou TW, Geber A, Wang M, Hamou W, Smith M, Sebra R, Zhang B, Zhou B, Ghedin E. 2020. Cell-to-cell variation in defective virus expression and effects on host responses during influenza virus infection. mBio 11:e02880-19. 10.1128/mBio.02880-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medaglia C, Kolpakov I, Zhu Y, Constant S, Huang S, Zwygart AC-A, Cagno V, Dermitzakis ET, Stellacci F, Xenarios I, Tapparel C. 2021. A novel anti-influenza combined therapy assessed by single cell RNA-sequencing. bioRxiv 10.1101/2021.07.27.453967. [DOI] [PMC free article] [PubMed]
- 17.O'Neill MB, Quach H, Pothlichet J, Aquino Y, Bisiaux A, Zidane N, Deschamps M, Libri V, Hasan M, Zhang SY, Zhang Q, Matuozzo D, Cobat A, Abel L, Casanova JL, Naffakh N, Rotival M, Quintana-Murci L. 2021. Single-cell and bulk RNA-sequencing reveal differences in monocyte susceptibility to influenza A virus infection between Africans and Europeans. Front Immunol 12:768189. 10.3389/fimmu.2021.768189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bercovich-Kinori A, Tai J, Gelbart IA, Shitrit A, Ben-Moshe S, Drori Y, Itzkovitz S, Mandelboim M, Stern-Ginossar N. 2016. A systematic view on influenza induced host shutoff. Elife 5:e18311. 10.7554/eLife.18311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beloso A, Martinez C, Valcarcel J, Santaren JF, Ortin J. 1992. Degradation of cellular mRNA during influenza virus infection: its possible role in protein synthesis shutoff. J Gen Virol 73:575–581. 10.1099/0022-1317-73-3-575. [DOI] [PubMed] [Google Scholar]
- 20.Kumar PA, Hu Y, Yamamoto Y, Hoe NB, Wei TS, Mu D, Sun Y, Joo LS, Dagher R, Zielonka EM, Wang de Y, Lim B, Chow VT, Crum CP, Xian W, McKeon F. 2011. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell 147:525–538. 10.1016/j.cell.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tata PR, Mou H, Pardo-Saganta A, Zhao R, Prabhu M, Law BM, Vinarsky V, Cho JL, Breton S, Sahay A, Medoff BD, Rajagopal J. 2013. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature 503:218–223. 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong KU, Reynolds SD, Watkins S, Fuchs E, Stripp BR. 2004. In vivo differentiation potential of tracheal basal cells: evidence for multipotent and unipotent subpopulations. Am J Physiol Lung Cell Mol Physiol 286:L643-9. 10.1152/ajplung.00155.2003. [DOI] [PubMed] [Google Scholar]
- 23.Pardo-Saganta A, Law BM, Tata PR, Villoria J, Saez B, Mou H, Zhao R, Rajagopal J. 2015. Injury induces direct lineage segregation of functionally distinct airway basal stem/progenitor cell subpopulations. Cell Stem Cell 16:184–197. 10.1016/j.stem.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiege JK, Thiede JM, Nanda HA, Matchett WE, Moore PJ, Montanari NR, Thielen BK, Daniel J, Stanley E, Hunter RC, Menachery VD, Shen SS, Bold TD, Langlois RA. 2021. Single cell resolution of SARS-CoV-2 tropism, antiviral responses, and susceptibility to therapies in primary human airway epithelium. PLoS Pathog 17:e1009292. 10.1371/journal.ppat.1009292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reuther P, Gopfert K, Dudek AH, Heiner M, Herold S, Schwemmle M. 2015. Generation of a variety of stable influenza A reporter viruses by genetic engineering of the NS gene segment. Sci Rep 5:11346. 10.1038/srep11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chambers BS, Heaton BE, Rausch K, Dumm RE, Hamilton JR, Cherry S, Heaton NS. 2019. DNA mismatch repair is required for the host innate response and controls cellular fate after influenza virus infection. Nat Microbiol 4:1964–1977. 10.1038/s41564-019-0509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiege JK, Langlois RA. 2015. Investigating influenza A virus infection: tools to track infection and limit tropism. J Virol 89:6167–6170. 10.1128/JVI.00462-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breen M, Nogales A, Baker SF, Martinez-Sobrido L. 2016. Replication-competent influenza A viruses expressing reporter genes. Viruses 8:179. 10.3390/v8070179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Childs RA, Palma AS, Wharton S, Matrosovich T, Liu Y, Chai W, Campanero-Rhodes MA, Zhang Y, Eickmann M, Kiso M, Hay A, Matrosovich M, Feizi T. 2009. Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray. Nat Biotechnol 27:797–799. 10.1038/nbt0909-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soundararajan V, Tharakaraman K, Raman R, Raguram S, Shriver Z, Sasisekharan V, Sasisekharan R. 2009. Extrapolating from sequence–the 2009 H1N1 'swine' influenza virus. Nat Biotechnol 27:510–513. 10.1038/nbt0609-510. [DOI] [PubMed] [Google Scholar]
- 31.Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, Gustin KM, Pearce MB, Viswanathan K, Shriver ZH, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. 2009. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 325:484–487. 10.1126/science.1177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, García-Sastre A. 2010. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc Natl Acad Sci USA 107:11531–11536. 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pusztaszeri MP, Seelentag W, Bosman FT. 2006. Immunohistochemical expression of endothelial markers CD31, CD34, von Willebrand factor, and Fli-1 in normal human tissues. J Histochem Cytochem 54:385–395. 10.1369/jhc.4A6514.2005. [DOI] [PubMed] [Google Scholar]
- 34.Teijaro JR, Walsh KB, Cahalan S, Fremgen DM, Roberts E, Scott F, Martinborough E, Peach R, Oldstone MB, Rosen H. 2011. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell 146:980–991. 10.1016/j.cell.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dumm RE, Fiege JK, Waring BM, Kuo CT, Langlois RA, Heaton NS. 2019. Non-lytic clearance of influenza B virus from infected cells preserves epithelial barrier function. Nat Commun 10:779. 10.1038/s41467-019-08617-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balasooriya GI, Johnson JA, Basson MA, Rawlins EL. 2016. An FGFR1-SPRY2 signaling axis limits basal cell proliferation in the steady-state airway epithelium. Dev Cell 37:85–97. 10.1016/j.devcel.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao R, Fallon TR, Saladi SV, Pardo-Saganta A, Villoria J, Mou H, Vinarsky V, Gonzalez-Celeiro M, Nunna N, Hariri LP, Camargo F, Ellisen LW, Rajagopal J. 2014. Yap tunes airway epithelial size and architecture by regulating the identity, maintenance, and self-renewal of stem cells. Dev Cell 30:151–165. 10.1016/j.devcel.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pardo-Saganta A, Tata PR, Law BM, Saez B, Chow RD, Prabhu M, Gridley T, Rajagopal J. 2015. Parent stem cells can serve as niches for their daughter cells. Nature 523:597–601. 10.1038/nature14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinot R, Hubert M, de Melo GD, Lazarini F, Bruel T, Smith N, Levallois S, Larrous F, Fernandes J, Gellenoncourt S, Rigaud S, Gorgette O, Thouvenot C, Trebeau C, Mallet A, Dumenil G, Gobaa S, Etournay R, Lledo PM, Lecuit M, Bourhy H, Duffy D, Michel V, Schwartz O, Chakrabarti LA. 2021. SARS-CoV-2 infection induces the dedifferentiation of multiciliated cells and impairs mucociliary clearance. Nat Commun 12:4354. 10.1038/s41467-021-24521-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Look DC, Walter MJ, Williamson MR, Pang L, You Y, Sreshta JN, Johnson JE, Zander DS, Brody SL. 2001. Effects of paramyxoviral infection on airway epithelial cell Foxj1 expression, ciliogenesis, and mucociliary function. Am J Pathol 159:2055–2069. 10.1016/S0002-9440(10)63057-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravindra NG, Alfajaro MM, Gasque V, Huston NC, Wan H, Szigeti-Buck K, Yasumoto Y, Greaney AM, Habet V, Chow RD, Chen JS, Wei J, Filler RB, Wang B, Wang G, Niklason LE, Montgomery RR, Eisenbarth SC, Chen S, Williams A, Iwasaki A, Horvath TL, Foxman EF, Pierce RW, Pyle AM, van Dijk D, Wilen CB. 2021. Single-cell longitudinal analysis of SARS-CoV-2 infection in human airway epithelium identifies target cells, alterations in gene expression, and cell state changes. PLoS Biol 19:e3001143. 10.1371/journal.pbio.3001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abdi K, Lai CH, Paez-Gonzalez P, Lay M, Pyun J, Kuo CT. 2018. Uncovering inherent cellular plasticity of multiciliated ependyma leading to ventricular wall transformation and hydrocephalus. Nat Commun 9:1655. 10.1038/s41467-018-03812-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoggins JW. 2019. Interferon-stimulated genes: what do they all do? Annu Rev Virol 6:567–584. 10.1146/annurev-virology-092818-015756. [DOI] [PubMed] [Google Scholar]
- 45.Iwasaki A, Pillai PS. 2014. Innate immunity to influenza virus infection. Nat Rev Immunol 14:315–328. 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bradley KC, Finsterbusch K, Schnepf D, Crotta S, Llorian M, Davidson S, Fuchs SY, Staeheli P, Wack A. 2019. Microbiota-driven tonic interferon signals in lung stromal cells protect from influenza virus infection. Cell Rep 28:245–256.e4. 10.1016/j.celrep.2019.05.105. [DOI] [PubMed] [Google Scholar]
- 47.Kallfass C, Lienenklaus S, Weiss S, Staeheli P. 2013. Visualizing the beta interferon response in mice during infection with influenza A viruses expressing or lacking nonstructural protein 1. J Virol 87:6925–6930. 10.1128/JVI.00283-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshimoto T, Okamura H, Tagawa YI, Iwakura Y, Nakanishi K. 1997. Interleukin 18 together with interleukin 12 inhibits IgE production by induction of interferon-gamma production from activated B cells. Proc Natl Acad Sci USA 94:3948–3953. 10.1073/pnas.94.8.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puddu P, Fantuzzi L, Borghi P, Varano B, Rainaldi G, Guillemard E, Malorni W, Nicaise P, Wolf SF, Belardelli F, Gessani S. 1997. IL-12 induces IFN-gamma expression and secretion in mouse peritoneal macrophages. J Immunol 159:3490–3497. [PubMed] [Google Scholar]
- 50.Ohteki T, Fukao T, Suzue K, Maki C, Ito M, Nakamura M, Koyasu S. 1999. Interleukin 12-dependent interferon gamma production by CD8alpha+ lymphoid dendritic cells. J Exp Med 189:1981–1986. 10.1084/jem.189.12.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cella M, Jarrossay D, Facchetti F, Alebardi O, Nakajima H, Lanzavecchia A, Colonna M. 1999. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat Med 5:919–923. 10.1038/11360. [DOI] [PubMed] [Google Scholar]
- 52.Bach EA, Aguet M, Schreiber RD. 1997. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol 15:563–591. 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- 53.Hogner K, Wolff T, Pleschka S, Plog S, Gruber AD, Kalinke U, Walmrath HD, Bodner J, Gattenlohner S, Lewe-Schlosser P, Matrosovich M, Seeger W, Lohmeyer J, Herold S. 2013. Macrophage-expressed IFN-beta contributes to apoptotic alveolar epithelial cell injury in severe influenza virus pneumonia. PLoS Pathog 9:e1003188. 10.1371/journal.ppat.1003188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dimmock NJ, Dove BK, Scott PD, Meng B, Taylor I, Cheung L, Hallis B, Marriott AC, Carroll MW, Easton AJ. 2012. Cloned defective interfering influenza virus protects ferrets from pandemic 2009 influenza A virus and allows protective immunity to be established. PLoS One 7:e49394. 10.1371/journal.pone.0049394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vignuzzi M, Lopez CB. 2019. Defective viral genomes are key drivers of the virus-host interaction. Nat Microbiol 4:1075–1087. 10.1038/s41564-019-0465-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tapia K, Kim WK, Sun Y, Mercado-Lopez X, Dunay E, Wise M, Adu M, Lopez CB. 2013. Defective viral genomes arising in vivo provide critical danger signals for the triggering of lung antiviral immunity. PLoS Pathog 9:e1003703. 10.1371/journal.ppat.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boergeling Y, Rozhdestvensky TS, Schmolke M, Resa-Infante P, Robeck T, Randau G, Wolff T, Gabriel G, Brosius J, Ludwig S. 2015. Evidence for a novel mechanism of influenza virus-induced type I interferon expression by a defective RNA-encoded protein. PLoS Pathog 11:e1004924. 10.1371/journal.ppat.1004924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Te Velthuis AJW, Long JC, Bauer DLV, Fan RLY, Yen HL, Sharps J, Siegers JY, Killip MJ, French H, Oliva-Martin MJ, Randall RE, de Wit E, van Riel D, Poon LLM, Fodor E. 2018. Mini viral RNAs act as innate immune agonists during influenza virus infection. Nat Microbiol 3:1234–1242. 10.1038/s41564-018-0240-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mendes M, Russell AB. 2021. Library-based analysis reveals segment and length dependent characteristics of defective influenza genomes. PLoS Pathog 17:e1010125. 10.1371/journal.ppat.1010125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galani IE, Triantafyllia V, Eleminiadou EE, Koltsida O, Stavropoulos A, Manioudaki M, Thanos D, Doyle SE, Kotenko SV, Thanopoulou K, Andreakos E. 2017. Interferon-lambda mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity 46:875–890.e6. 10.1016/j.immuni.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 61.Crotta S, Davidson S, Mahlakoiv T, Desmet CJ, Buckwalter MR, Albert ML, Staeheli P, Wack A. 2013. Type I and type III interferons drive redundant amplification loops to induce a transcriptional signature in influenza-infected airway epithelia. PLoS Pathog 9:e1003773. 10.1371/journal.ppat.1003773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hsu AC, Parsons K, Barr I, Lowther S, Middleton D, Hansbro PM, Wark PA. 2012. Critical role of constitutive type I interferon response in bronchial epithelial cell to influenza infection. PLoS One 7:e32947. 10.1371/journal.pone.0032947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jewell NA, Vaghefi N, Mertz SE, Akter P, Peebles RS, Jr., Bakaletz LO, Durbin RK, Flano E, Durbin JE. 2007. Differential type I interferon induction by respiratory syncytial virus and influenza A virus in vivo. J Virol 81:9790–9800. 10.1128/JVI.00530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feeley EM, Sims JS, John SP, Chin CR, Pertel T, Chen LM, Gaiha GD, Ryan BJ, Donis RO, Elledge SJ, Brass AL. 2011. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog 7:e1002337. 10.1371/journal.ppat.1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254. 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Desai TM, Marin M, Chin CR, Savidis G, Brass AL, Melikyan GB. 2014. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog 10:e1004048. 10.1371/journal.ppat.1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loske J, Rohmel J, Lukassen S, Stricker S, Magalhaes VG, Liebig J, Chua RL, Thurmann L, Messingschlager M, Seegebarth A, Timmermann B, Klages S, Ralser M, Sawitzki B, Sander LE, Corman VM, Conrad C, Laudi S, Binder M, Trump S, Eils R, Mall MA, Lehmann I. 2022. Pre-activated antiviral innate immunity in the upper airways controls early SARS-CoV-2 infection in children. Nat Biotechnol 40:319–324. 10.1038/s41587-021-01037-9. [DOI] [PubMed] [Google Scholar]
- 68.Ruiz García S, Deprez M, Lebrigand K, Cavard A, Paquet A, Arguel MJ, Magnone V, Truchi M, Caballero I, Leroy S, Marquette CH, Marcet B, Barbry P, Zaragosi LE. 2019. Novel dynamics of human mucociliary differentiation revealed by single-cell RNA sequencing of nasal epithelial cultures. Development 146:dev177428. 10.1242/dev.177428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alnaji FG, Reiser WK, Rivera-Cardona J, Te Velthuis AJW, Brooke CB. 2021. Influenza A virus defective viral genomes are inefficiently packaged into virions relative to wild-type genomic RNAs. mBio 12:e02959-21. 10.1128/mBio.02959-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Froggatt HM, Harding AT, Chaparian RR, Heaton NS. 2021. ETV7 limits antiviral gene expression and control of influenza viruses. Sci Signal 14:eabe1194. 10.1126/scisignal.abe1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Young MD, Behjati S. 2020. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. Gigascience 9:giaa151. 10.1093/gigascience/giaa151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, III, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R. 2021. Integrated analysis of multimodal single-cell data. Cell 184:3573–3587.e29. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download jvi.00505-22-s0001.pdf, PDF file, 0.3 MB (269.8KB, pdf)
Data Availability Statement
Raw and processed sequencing data are available to download from NCBI GEO via accession number GSE203018.