ABSTRACT
Respiratory coronaviruses cause serious health threats to humans and animals. Porcine respiratory coronavirus (PRCoV), a natural transmissible gastroenteritis virus (TGEV) mutant with partial spike deletion, causes mild respiratory disease and is an interesting animal respiratory coronavirus model for human respiratory coronaviruses. However, the absence of robust ex vivo models of porcine airway epithelium hinders an understanding of the pathogenesis of PRCoV infection. Here, we generated long-term porcine airway organoids (AOs) derived from basal epithelial cells, which recapitulate the in vivo airway complicated epithelial cellularity. Both 3D and 2D AOs are permissive for PRCoV infection. Unlike TGEV, which established successful infection in both AOs and intestinal organoids, PRCoV was strongly amplified only in AOs, not intestinal organoids. Furthermore, PRCoV infection in AOs mounted vigorous early type I and III interferon (IFN) responses and upregulated the expression of overzealous inflammatory genes, including pattern recognition receptors (PRRs) and proinflammatory cytokines. Collectively, these data demonstrate that stem-derived porcine AOs can serve as a promising disease model for PRCoV infection and provide a valuable tool to study porcine respiratory infection.
IMPORTANCE Porcine respiratory CoV (PRCoV), a natural mutant of TGEV, shows striking pathogenetic similarities to human respiratory CoV infection and provides an interesting animal model for human respiratory CoVs, including SARS-CoV-2. The lack of an in vitro model recapitulating the complicated cellularity and structure of the porcine respiratory tract is a major roadblock for the study of PRCoV infection. Here, we developed long-term 3D airway organoids (AOs) and further established 2D AO monolayer cultures. The resultant 3D and 2D AOs are permissive for PRCoV infection. Notably, PRCoV mediated pronounced IFN and inflammatory responses in AOs, which recapitulated the inflammatory responses associated with PRCoV in vivo infection. Therefore, porcine AOs can be utilized to characterize the pathogenesis of PRCoV and, more broadly, can serve as a universal platform for porcine respiratory infection.
KEYWORDS: coronavirus, PRCoV, TGEV, airway organoids, PRRs, interferons
INTRODUCTION
Human coronaviruses (HCoVs), including three highly pathogenic CoVs and four low-pathogenic CoVs, have a significant impact on health worldwide and cause respiratory diseases ranging from the common cold to serious respiratory diseases, including severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) (1–3). Porcine respiratory coronavirus (PRCoV), a naturally occurring spike deletion mutant of highly enteropathogenic transmissible gastroenteritis virus (TGEV), is a surrogate to study the pathogenesis of human respiratory coronaviruses, including SARS-CoV and SARS-CoV-2, in humans (4–6). PRCoV contains a large deletion (200 to 230 amino acids [aa]) near the N terminus of the spike protein and alters tissue tropism from the intestinal to the respiratory tract (6, 7). PRCoV largely replicates in the epithelial cells of the nasal mucosa, trachea, bronchi, and bronchioles (8). PRCoV spreads mainly by aerogenic routes and causes mild respiratory disease, such as coughing, but no watery diarrhea, unlike the parental TGEV (7). PRCoV is commonly found in pigs and shows many of the same features as seen with SARS-CoV-2 infection, including a similar tissue tropism and overlapping, although milder, clinical signs and lesions (6, 8, 9). Similar to SARS-CoV-2, most PRCoV infections are clinically mild or asymptomatic, and PRCoV primarily causes extensive lung lesions with atypical interstitial pneumonia (6, 9). PRCoV provides a promising animal model for the study of human coronavirus respiratory disease given that the anatomy, physiology, and immune system of the porcine respiratory tract closely resemble those of humans. An understanding of PRCoV infections in the natural host is critical to provide insights into SARS-CoV-2 pathogenesis and the potential viral or host factors that contribute to the severity of COVID-19. In addition, it is well recognized that the emergence of PRCoV reduces the spread of TGEV in pork herds due to cross-protective immunity to TGEV (7, 10). Therefore, PRCoV is also a promising model to investigate the contributions of respiratory versus gastrointestinal mucosal immunity to local protection.
The respiratory tract and gastrointestinal tract are the main target tissues for most coronaviruses and are the first line of defense against the infection of these viruses. The respiratory airway is covered with a continuous epithelial layer mainly consisting of four major types of cells in humans (ciliated, basal, secretory goblet, and columnar club cells) (11, 12). The airway epithelia are dynamic tissue and undergo continuous and slow turnover (12). The ciliated, goblet, and club cells constitute the primary host defense barrier. A better understanding of the cellular intrinsic responses of airway epithelia to respiratory coronavirus infection could contribute to the clarification of coronavirus pathogenesis and the development of novel therapies for respiratory coronavirus diseases. Most studies of porcine respiratory infection have been performed using nonphysiological cell lines or short-term cultures of porcine lung explants and primary airway epithelial cells (13, 14). None of the approaches allow continued, long-term expansion of porcine airway epithelia in vitro, thus limiting their practical use.
3D tissue organoids can mimic key structural and functional properties of organs and have been widely applied to model in vivo organ development and various infectious or noninfectious diseases (11, 15, 16). Airway basal cells account for approximately 30% of the pseudostratified mucociliary epithelium of the respiratory tracts and function as epithelial stem cells, which play essential roles in organ homeostasis and repair after injury (12, 15). Respiratory airway basal cells can generate airway or lung organoids in vitro (15). Since murine bronchiolar lung organoid culture derived from adult basal cells was first reported in 2010 (17), airway or lung organoids of multiple species, including humans and mice, have been successfully established and used to clarify viral infections, such as influenza virus and SARS-CoV-2 (18, 19). Airway or lung organoids provide valuable models of respiratory disease and facilitate the therapeutic development for respiratory disorders. However, porcine airway organoids have not yet been reported.
In this study, we developed long-term and passible porcine 3D airway organoids derived from airway basal stem/progenitor cells and further established a 2D monolayer culture of airway organoids, which recapitulated the airway epithelial surface in vivo. We demonstrate the susceptibility of airway organoids to PRCoV and TGEV. Furthermore, substantial interferon (IFN)/IFN-stimulated gene (ISG) induction in airway organoids derived from neonatal piglets profoundly impacts the susceptibility of airway organoids to PRCoV, which provides novel insights into PRCoV pathogenesis.
RESULTS
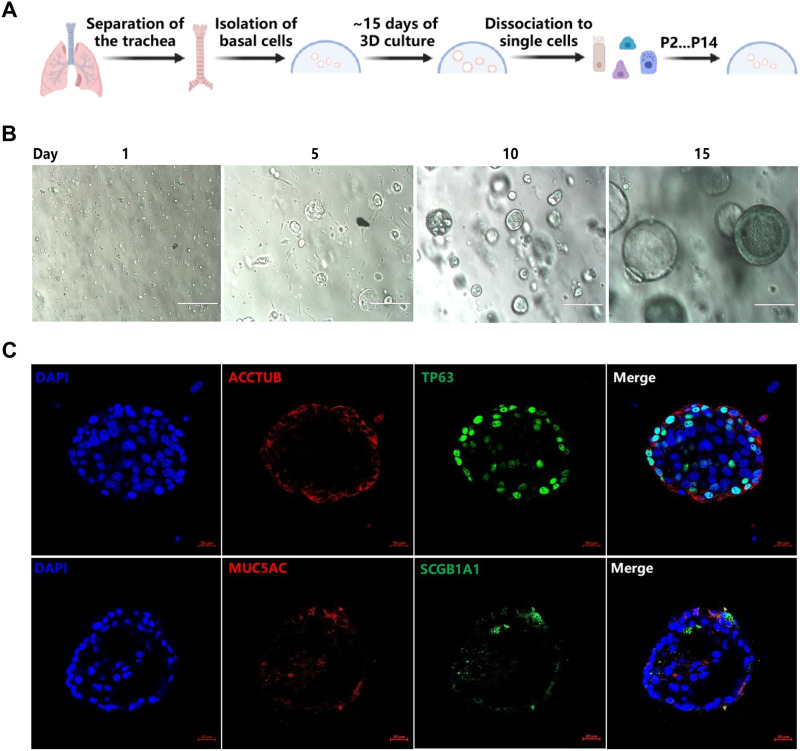
Establishing 3D cultures of porcine 3D airway organoids.
We collected small pieces of tracheal tissues from 2-day-old neonatal specific-pathogen-free (SPF) piglets and isolated epithelial cells through mechanical and enzymatic tissue disruption as described previously (20). Modified from the recently reported protocol, tracheal epithelial cells were cultured in semisolid Matrigel matrix and differentiated for 15 to 20 days as described in detail in Materials and Methods (Fig. 1A). Individual tracheal epithelial cells gave rise to “spheres” by day 10 and grew into clonal spheres with visible lumen called 3D airway organoids (AOs) after approximately 3 weeks of differentiation (Fig. 1B). The immature 3D AOs could be passaged by mechanical disruption, frozen-thawed, and maintained for at least 7 months. Immunostaining shows that 3D AOs consist of the four major types of airway epithelial cells, including ciliated cells (ACCTUB+), goblet cells (MUC5AC+), basal cells (TP63+), and club cells (SCGB1A1+) (Fig. 1C). ACCTUB-labeled ciliated cells have two orientations: toward or reverse of the lumen, as previously reported by Zhou et al. (21). Consistent with 3D human airway organoids (21), the ciliated cells, goblet cells, and club cells observed were also mainly distributed on the surface of “tracheaespheres” (Fig. 1C). Collectively, these results demonstrated that the porcine AOs we generated can simulate the pseudostratified airway epithelium.
FIG 1.
3D cultures of porcine airway organoids (AOs) were established. (A) Diagram of 3D porcine airway organoid isolation and culture. (B) Representative images of AOs differentiation derived from the trachea of 2-day-old piglets. The AOs were cultured in Matrigel matrix with organoid growth medium and monitored daily under a microscope to verify the formation diameter of the organoids. The developed organoids were subjected to freeze-thaw cycling, and their daily growth was observed under a microscope. Images were obtained with an Evos XL microscope. Scale bar = 200 μm. (C) Identification of distinct cell lineages in 3D airway organoids. 3D airway organoids were fixed and labeled with DAPI (nuclear staining, blue) and cell marker antibodies: ACCTUB for ciliated cells (red), MUC5AC for goblet cells (red), SCGB1A1 for club cells (green), and TP63 for basal cells (green). Images were obtained with an LSM980-ZEISS confocal laser scanning microscope. Scale bar = 20 μm.
Porcine 3D airway organoids support PRCoV replication.
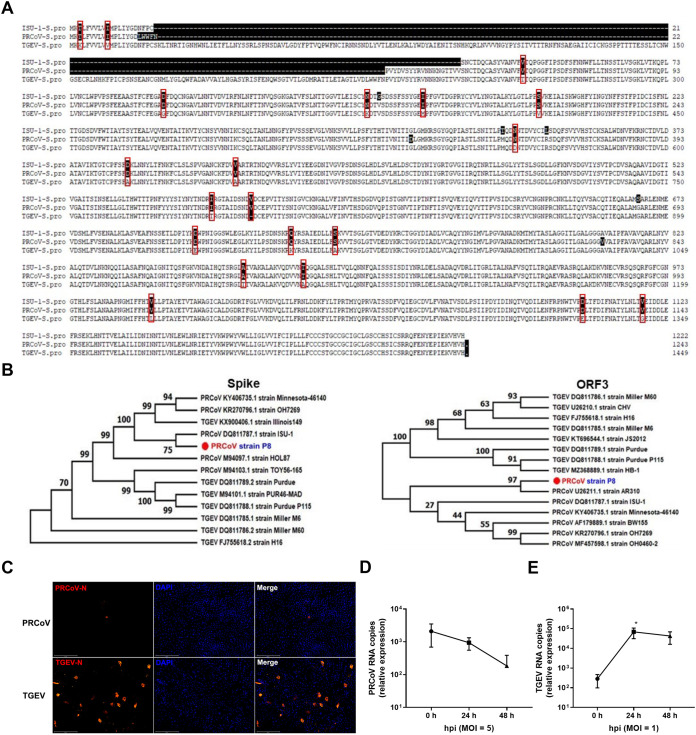
To assess whether porcine AOs can be infected by PRCoV, we initially confirmed that the PRCoV P8 strain stocked by our group is a mutant of TGEV with a 16- to 225-aa partial deletion at the N-terminal spike protein by spike gene sequencing (Fig. 2A). Phylogenetic trees of the complete S gene and ORF3 gene further revealed that PRCoV P8 is most closely related to the classical American PRCoV ISU-1 strain for the spike gene and AR310 strain, respectively (Fig. 2B). These results demonstrate that PRCoV P8 is a PRCoV strain derived from classical U.S. PRCoV strains. Recent studies have suggested that the N-terminal partial deletion of the TGEV spike protein is necessary but not sufficient for the tissue tropism switch of TGEV from intestinal to respiratory tropism (6, 22, 23). Next, we explored the tissue tropism alteration of the PRCoV P8 strain (PRCoV). As expected, TGEV robustly replicated in porcine intestinal organoid monolayers, a good ex vivo model for enteric coronavirus infection (16) (Fig. 2C and E). Unlike TGEV, PRCoV did not successfully establish a productive infection in porcine intestinal organoids measured by nucleocapsid protein (NP) immunofluorescence assay (IFA) (Fig. 2C) and viral genomes by quantitative PCR (qPCR) (Fig. 2D and E), even though a higher multiplicity of infection (MOI) was applied for PRCoV than with TGEV (Fig. 2D and E), indicating that the PRCoV P8 strain does lose intestinal tropism.
FIG 2.
PRCoV loses the tropism of porcine intestinal epithelia. (A) Sequence alignment of the S protein of the PRCoV P8 strain, PRCoV ISU-1 strain (GenBank no. DQ811787.1), and TGEV H88 strain. (B) Phylogenetic tree of the spike and ORF3 genes of PRCoV P8 and other major groups of PRCoV and TGEV. (C) Detection of PRCoV or TGEV infection in porcine 2D intestinal enteroids by IFA. Porcine 2D intestinal enteroids were infected with PRCoV at an MOI of 5 and TGEV at an MOI of 1, and the levels of viral infection at 24 hpi were detected by staining with anti-N monoclonal antibody (MAb) (scale bar = 200 μm). (D and E) The kinetic curve of PRCoV or TGEV replication in porcine 2D intestinal enteroids. Porcine 2D intestinal enteroids derived from 2-day-old piglets were infected with PRCoV at an MOI of 5 and TGEV at an MOI of 1. The kinetics of PRCoV (D) and TGEV (E) amplification were determined by RT-qPCR. The relative fold change of PRCoV and TGEV RNA was calculated by normalizing the mock control of 2D intestinal enteroids. Error bars denote the deviations indicated by *: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
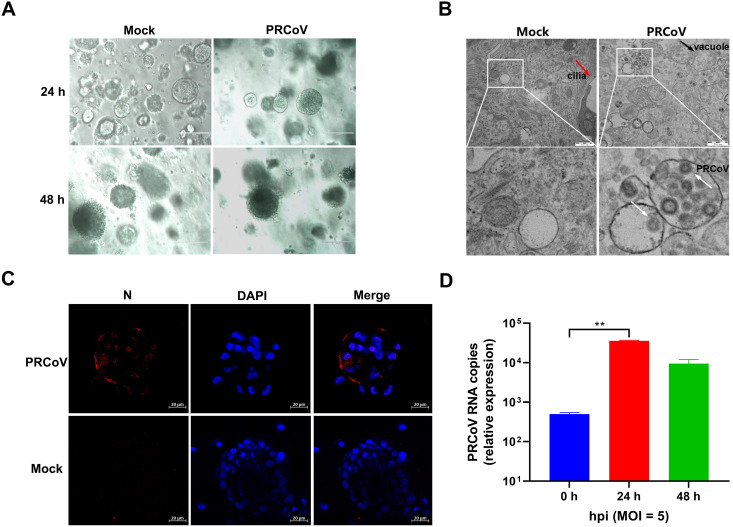
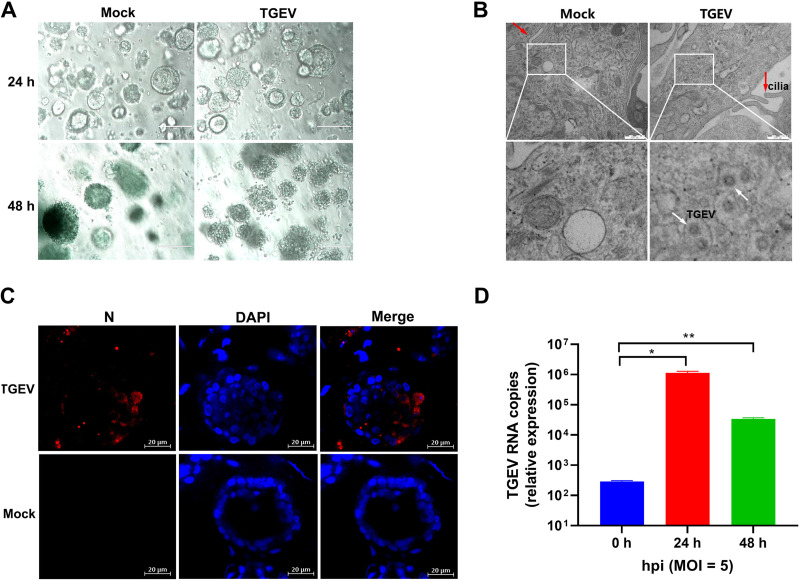
To assess the permissiveness of 3D AOs for PRCoV infection, we removed Matrigel matrix from AO culture and inoculated 3D AOs differentiated from the tracheal epithelia of day 2 newborn piglets (AOs) with PRCoV. PRCoV infection caused the morphological changes of AOs and disrupted AOs at 24 h postinfection (hpi) (Fig. 3A). To further determine the successful infection in porcine AOs, we evaluated PRCoV infection at 24 hpi by transmission electron microscopy (TEM) analysis at higher resolution. Cilia made up of microtubules coated by the plasma membrane were observed in the epithelia of AOs (Fig. 3B). Compared with uninfected AOs, infected porcine AOs had more massive vacuoles (Fig. 3B). The viral particles approximately 100 nm in diameter were either dispersed or encapsulated in cytoplasmic vacuoles in PRCoV-infected 3D AOs instead of the mock-control AOs (Fig. 3B), suggesting the establishment of productive PRCoV infection in 3D AOs. The successful infection of PRCoV in porcine AOs was further confirmed by NP IFA. PRCoV NP was easily detected in PRCoV-infected AOs at 24 hpi (Fig. 3C). These results show that porcine 3D AOs are permissive to PRCoV infection. To further confirm the susceptibility of AOs to PRCoV, we monitored the viral replication kinetics of PRCoV in AOs (Fig. 3D). After 3 h of incubation and washing to remove nonspecifically bound residual viral particles with phosphate-buffered saline (PBS), PRCoV particles associated with AOs were marked as the input 0 hpi. PRCoV viral genomes in AOs increased 71- and 19-fold at 24 hpi and 48 hpi, respectively, after normalizing the viral input at 0 hpi (Fig. 3D). Altogether, these results demonstrate that porcine 3D AOs are permissible for PRCoV infection.
FIG 3.
The permissiveness of porcine 3D airway organoids to PRCoV infection. (A) The morphological changes of PRCoV-infected or mock-infected 3D AOs derived from 2-day-old piglets were observed under a bright-field microscope at 24 hpi and 48 hpi. Scale bars = 200 μm. (B) Detection of PRCoV infection in porcine 3D airway organoids by transmission electron microscopy (TEM). Porcine 3D AOs mock-inoculated or inoculated with PRCoV were harvested at 24 hpi, and then the 3D AOs were fixed in extracellular matrix (ECM) with 2.5% glutaraldehyde and analyzed by H-7650 electron microscopy (Hitachi, Japan). The red arrow points to cilia, the black arrow points to the vacuole, and the white arrows point to PRCoV particles. Scale bars = 500 nm. (C) Monitoring PRCoV infection in 3D AOs derived from a 2-day-old piglet (D2) by IFA. 3D AOs were infected with PRCoV at an MOI of 5, and the levels of infection at 24 hpi were determined by anti-NP IFA staining (scale bar = 20 μm). (D) The kinetics of PRCoV replication in 3D AOs. 3D AOs were infected with PRCoV at an MOI of 5, and samples were collected at the indicated time points for PRCoV viral load detection by RT-qPCR compared with mock AOs. Error bars denote the deviations indicated by *: *, P < 0.05; **, P < 0.01; ***; P < 0.001.
Transcriptome changes at single-cell resolution in 3D airway organoids after PRCoV infection.
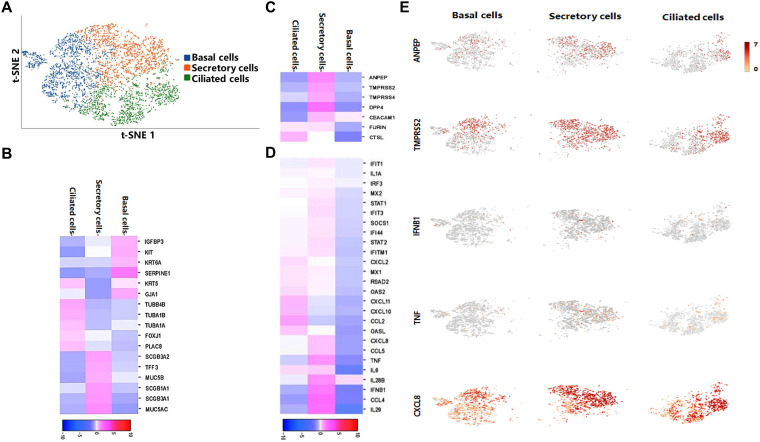
To further explore the cellular complexity of porcine 3D AOs and characterize the transcriptional profiles in AOs following PRCoV infection, we performed single-cell transcriptome analysis on PRCoV-infected 3D AOs at 24 hpi by a 10× Genomics single-cell RNA sequencing (RNA-seq) platform. We captured a total of 6,010 single cells, with 4,145 detectable genes per cell by 21,853 median unique molecular identifiers (UMIs). We conducted integration (count normalization, feature selection, dimension reduction, and visualization) through t-distributed stochastic neighbor embedding (t-SNE) (Fig. 4A) through Seurat and SC3 packages. According to Loupe Browser software analysis, the Seurat clusters consisted of 3 clusters: cluster 1 for basal epithelial cells, cluster 2 for secretory cells, including goblet and club cells, as reported by Carraro et al. (24), and cluster 3 for ciliated epithelial cells (Fig. 4A). Among them, basal cells accounted for 35.66%, secretory cells accounted for 34.51%, and ciliated cells accounted for 29.83%. Based on previously reported cell markers (20, 24) and our data, we evaluated specific gene expression in these cell types: basal cells (GJA1, IGFBP3/6, SERPINE1, KIT, and KRT6A), goblet cells (SCGB3A1/2, TFF3, MUC5B, and MUC5AC), club cells (SCGB1A1), and ciliated cells (TUBB, TUBA1A/B, FOXJ1, TUBB4B, and PLAC8). As shown in Fig. 4B, and 4D AOs included basal cells, secretory cells (goblet cells and club cells), and ciliated cells. The single-cell RNA sequencing (scRNA-seq) results combined with IFA results (Fig. 1) demonstrate that porcine 3D AOs contain the major subpopulations of airway epithelial cells.
FIG 4.
Single-cell sequencing of PRCoV-infected 3D AOs. (A) The t-SNE plot of individual cells demonstrating basal cell, secretory cell, and ciliated cell populations in PRCoV-infected 3D AOs at 24 hpi. (B to D) The heatmap shows highly enriched different gene sets in each cluster in basal cells, secretory cells, and ciliated cells: the enriched expression of cell lineage marker genes (B), coronavirus entry factor genes (C), and type I and III IFN-related genes (D). (E) The tSNE plots of normalized expression of ANPEP (APN), TMPRSS2, IFNB1, TNF, and CXCL8 based on similarity of host transcriptional patterns.
To elucidate the potential specific cellular targets of PRCoV in AOs, we also evaluated the expression of known coronavirus entry factors in these three subpopulations by clustering analysis of the data. Notably, APN (ANPEP), transmembrane serine protease 2 and 4 (TMPRSS2 and TMPRSS4), and DPP4 are mainly expressed in secretory cells; FURIN and cathepsin L (CTSL) are expressed in ciliated cells, and these coronavirus entry receptors or factors are less expressed in basal cells (Fig. 4C and E), indicating that PRCoV potentially primarily infects secretory cells and ciliated cells in porcine 3D AOs given that APN is the viral receptor of TGEV and PRCoV (13, 25). Consistently, the virus-mediated induction of type I and type III IFNs and IFN-stimulated genes (ISGs) and inflammatory cytokines (tumor necrosis factor [TNF] and interleukin-6 [IL-6]) were mainly enriched in secretory cells, followed by ciliated cells and basal cells (Fig. 4D and E). Altogether, porcine AOs recapitulate respiratory airway epithelia and are permissible for PRCoV infection.
Porcine 3D airway organoids are susceptible to TGEV infection.
TGEV possesses both enteric and respiratory tropisms and can replicate in the trachea, although the enteric tract tropism of TGEV directly correlates with TGEV virulence (23, 26). To evaluate the susceptibility of porcine 3D airway organoids to TGEV infection, we infected AOs with TGEV at an MOI of 5, as we did for PRCoV infection above. TGEV infection significantly disrupted the structure of AOs at 24 hpi (Fig. 5A). TGEV viral particles were widely present in cytosolic vacuoles at 24 hpi by TEM analysis, suggesting a productive infection of TGEV in 3D AOs (Fig. 5B). TGEV infection was further confirmed by NP IFA staining and displayed a substantial positive level of TGEV NP staining (Fig. 5C). Next, we performed a time course analysis of TGEV infection in 3D AOs. Time-course analysis of TGEV growth kinetics showed that TGEV amplified robustly and peaked at 24 hpi, followed by a sharp decline at 48 hpi in AOs (Fig. 5D), which is in contrast with the plateau kinetics of TGEV from 24 to 48 hpi in intestinal organoids (Fig. 2D). Collectively, these results demonstrate that TGEV can also infect porcine 3D AOs, similar to PRCoV.
FIG 5.
Porcine 3D airway organoids are susceptible to TGEV infection. (A) The morphological changes of TGEV-infected or mock-infected AOs were monitored under a bright-field microscope at 24 hpi or 48 hpi. Scale bars = 200 μm. (B) Determination of TGEV infection in porcine 3D AOs by TEM. Porcine 3D AOs were mock-inoculated or inoculated with TGEV and harvested at 24 hpi, and then the 3D AOs were fixed in ECM with 2.5% glutaraldehyde and evaluated by H-7650 electron microscopy (Hitachi, Japan). The red arrows indicate cilia, and the white arrows indicate TGEV particles. Scale bars = 500 nm. (C) Detection of TGEV infection in 3D AOs by IFA. 3D AOs were infected with TGEV at an MOI of 5, and TGEV infection at 24 hpi was determined by IFA (scale bar = 20 μm). (D) The kinetic curve of TGEV replication in 3D AOs. The samples of 3D AOs were collected at the indicated time points for TGEV viral load detection by RT-qPCR compared with mock AOs. Error bars denote the deviations indicated by *: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
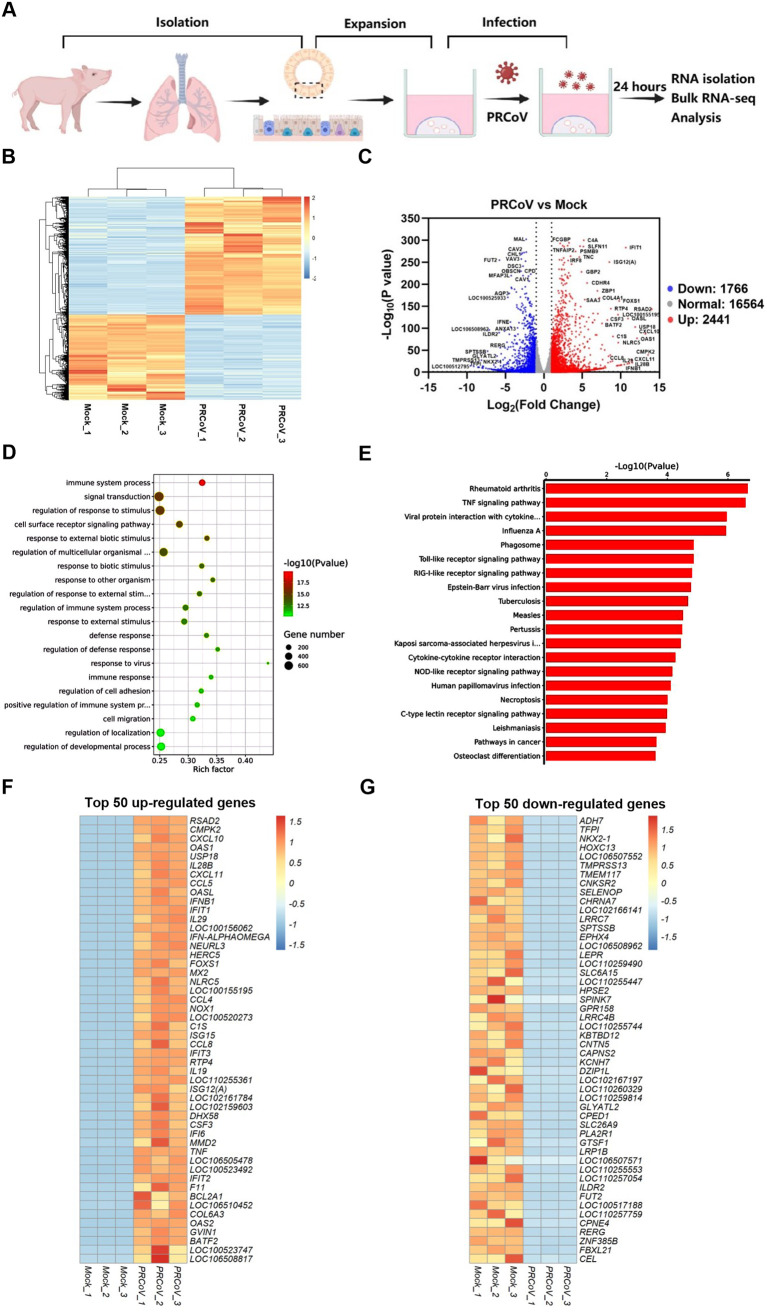
The gene transcriptional profiles of airway organoids following PRCoV infection.
The cellular intrinsic responses of airway epithelia to virus infection could contribute to the clarification of coronavirus pathogenesis and play critical roles in modifying the airway mucosal infection of respiratory coronaviruses. We next explored the cell-intrinsic responses in AOs after PRCoV infection. To this end, strand-specific Illumina mRNA libraries were constructed from AOs with or without PRCoV infection at 24 hpi, and bulk RNA-seq was performed (Fig. 6A). Bulk RNA-seq results showed that PRCoV infection elicited a unique gene expression profile in AOs (Fig. 6B). PRCoV infection upregulated the expression of 2,441 genes and downregulated expression of 1,766 genes in AOs relative to the mock control (Fig. 6C). To further clarify the functional consequences of the differentially expressed genes in AOs elicited by PRCoV infection, we performed gene ontology (GO) and KEGG pathway enrichment analyses. The dominant functions of the differentially expressed genes (DEGs) were the immune system process and response to viruses (Fig. 6D), which mainly concentrated on TNF, viral protein interaction with cytokines, and the pattern recognition receptor (PRR) signaling pathway (Fig. 6E). Heatmaps of the top 50 genes among the 2,411 upregulated genes and 1,766 downregulated genes in PRCoV-infected AOs are shown in Fig. 6F and G, respectively. Interestingly, most of the top 50 upregulated genes were IFN response-associated genes (such as IFN-β, IFN-λ, and RSAD2) in PRCoV-infected AOs, suggesting that PRCoV infection elicits a pronounced IFN response in AOs (Fig. 6F). Most of the 50 downregulated genes were associated with intracellular material transport and metabolism (such as ADH7, TMEM117, and SELENOP) in PRCoV-infected AOs (Fig. 6G). These results indicate that PRCoV infection induces vigorous cellular innate immunity in AOs.
FIG 6.
Transcriptional profiles in AOs following PRCoV infection. (A) Schematic for bulk RNA-seq of PRCoV-infected AO samples. (B) Heatmap of common differentially expressed genes in 3D AOs after PRCoV infection. (C) Volcano plot showing differentially expressed genes in PRCoV-infected AOs. (D and E) GO (D) and KEGG pathway (E) enrichment analyses of differentially expressed genes in PRCoV-infected AOs. (F and G) Heatmap showing the top 50 up- (F) or downregulated (G) genes modified by PRCoV in AOs. 3D AOs derived from the trachea of 2-day-old piglets were infected with or without PRCoV for 24 hpi. Total RNA was isolated from AOs, and the changes in gene expression were determined by bulk RNA-seq. Data are based on log(RPKM) values from three independent biological preparations.
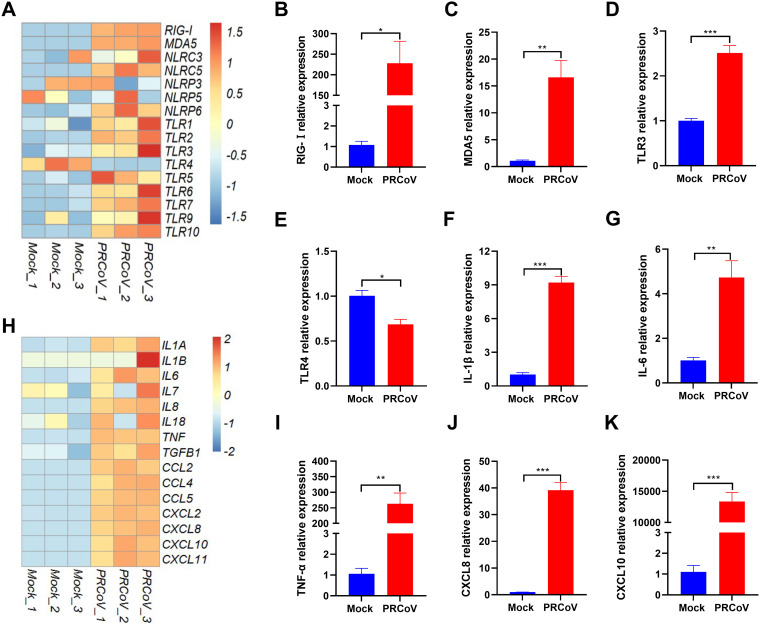
Significantly upregulated expression of PRRs and proinflammatory cytokines in PRCoV-infected AOs.
PRCoV infection can induce inflammatory responses in the respiratory tract and results in mild bronchointerstitial pneumonia in vivo (26, 27). Host pattern recognition receptors (PRRs) initiate the cellular response to viral infection by recognizing viral genomes and proteins, leading to the production of multiple inflammatory responses (28, 29). To explore whether PRCoV can induce inflammatory responses in AOs, we analyzed the transcriptional expression of PRRs in AOs following PRCoV infection, including RIG-I-like receptors (RLRs), toll-like receptors (TLRs), and nucleotide-binding oligomerization domain-like receptors (NLRs). The heatmap results indicated that the expression of RLRs (RIG-I and MDA5) and TLRs (TLR-3, -7, and -9) mainly responsible for sensing RNA virus infection in AOs was induced by PRCoV (Fig. 7A). This finding is consistent with the enrichment of upregulated IFN-related genes in PRCoV-infected AOs (Fig. 6F). The increased transcriptional levels of several typical PRR genes by PRCoV were further confirmed by reverse transcriptase quantitative PCR (RT-qPCR), which showed a similar trend as that observed in RNA-seq (Fig. 7A to 7E). In line with the in vivo observations, we observed that the transcriptional levels of proinflammatory cytokines and recruiting chemokines were largely upregulated in PRCoV-infected AOs compared with the mock control (Fig. 7H). Moreover, we randomly selected several typical proinflammatory cytokine representatives, including interleukin-1β (IL-1β), IL-6, tumor necrosis factor-α (TNF-α), CXCL8, and CXCL10, to be verified by RT–qPCR (Fig. 7F, G, and I to K). In agreement with the RNA-seq results, the increased mRNA levels of these proinflammatory factors in AOs after PRCoV infection were confirmed by RT-qPCR (Fig. 7F, G, and I to K). Collectively, PRCoV infection upregulated the expression of PRRs and induced substantial proinflammatory cytokines, leading to inflammatory responses.
FIG 7.
Enriched analysis of pattern recognition receptors (PRRs) and proinflammatory cytokines in AOs after PRCoV infection. (A and H). Heatmaps showing the expression of PRRs (A) and proinflammatory cytokines (H) in AOs following PRCoV infection. (B to E) RT-qPCR verification of the expression of PRRs (RIG-I, MDA5, TLR3, and TLR4) in AOs. (F, G, I to K). Verification of the expression of proinflammatory cytokines (IL-1β, IL-6, TNF-α, CXCL8, and CXCL10) in AOs by RT-qPCR. Data represent the means ± SEMs of results from three wells and three independent organoid preparations. Error bars denote the deviations indicated by *: *, P < 0.05; **, P < 0.01; ***, P < 0.001. The results are presented as the means ± SEMs (n = 3).
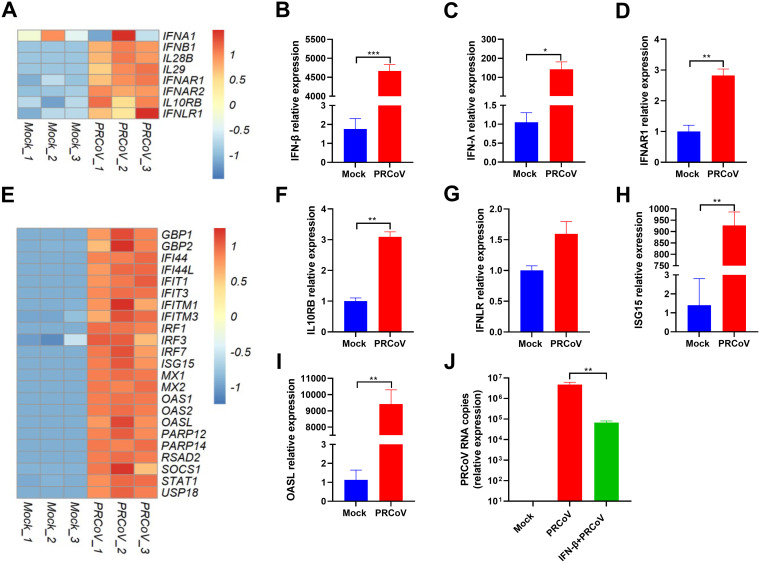
Robust induction of IFN and ISG responses in AOs following PRCoV infection.
Type I and III IFNs are the initial antiviral cytokines in response to virus infection in mucosal epithelia and play crucial roles against respiratory virus infection. Next, we evaluated IFN production and ISG induction following PRCoV infection of airway epithelial organoids. In agreement with previous in vivo studies (9, 27), PRCoV infection induced type I IFNs (IFN-α, -β) and type III IFNs (IL-29, IL-28) (Fig. 8A). The substantial induction of type I and type III IFNs by PRCoV infection in AOs was confirmed by in vitro RT-qPCR (Fig. 8B and C). The amplitude and kinetics of the response of IFN-elicited ISG responses can be imprinted by the expression levels of IFN receptors on the cellular surface except for the amount of IFNs. We next explored the receptor expression of type I and III IFNs in AOs. Notably, PRCoV infection substantially increased the expression of both types of IFN receptors, including type I IFN receptors (IFNAR1 and IFNAR2) (Fig. 8D) and type III receptors (IFNLR1 and IL-10RB) in AOs after PRCoV infection (Fig. 8F and G).
FIG 8.
The divergent responses of IFN and ISGs induced by PRCoV infection in 3D AOs. (A) Heatmap showing the expression of IFN and IFN receptors in 3D AOs infected with or without PRCoV. (B to D, F to I). Verification of the expression of the listed differentially expressed genes between PRCoV-infected AOs by RT-qPCR: IFN-β (B), IFN-λ (C), IFNAR1 (D), IL10RB (F), IFNLR (G), ISG15 (H), and OASL (I). (E) Heatmap showing the expression of randomly selected upregulated ISGs in PRCoV-infected 3D AOs. (J) PRCoV is sensitive to exogenous IFN-β. All the data were calculated using the 2-ΔΔCT method, normalized to GAPDH expression, and compared with mock AOs. Error bars denote the deviations indicated by *: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Consistent with the substantial production of IFNs, PRCoV-infected AOs showed substantial induction of ISG expression in comparison with the mock control (Fig. 8D). The pronounced ISG expression induced by PRCoV infection was verified by in vitro RT-qPCR of randomly selected ISG15 and OASL (Fig. 8H and I). Additionally, porcine AOs respond to exogenous IFN-β priming, which significantly inhibited PRCoV infection in AOs (Fig. 8J), indicating that PRCoV is sensitive to the antiviral activity of IFNs. Together, these data suggest that PRCoV infection elicits pronounced IFN production and ISG responses in AOs.
Establishing porcine 2D airway organoid monolayers from 3D AOs to assess the infectivity and innate immunity of PRCoV.
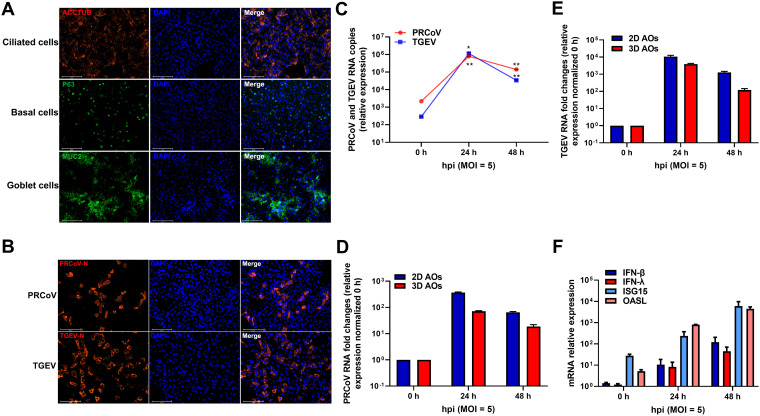
One of the limitations of 3D AOs for studying respiratory virus infections is to access the apical surface for pathogens since most airway organoids are a spheroid structure inside the lumen. To model natural respiratory virus infections through the apical surface, we established 2D airway organoid monolayers from 3D AOs as we described previously for monolayer enteroids (16, 30). To this end, 3D AOs were enzymatically dissociated into a single-cell suspension, seeded in Matrigel-coated plates, and then submerged in growth medium for another 3 days to grow into 2D AO monolayers (2D AOs). Differentiated 2D AOs also included ciliated cells (ACCTUB+), goblet cells (MUC2+), and basal cells (TP63+) (Fig. 9A). The substantial signal of ACCTUB and lower signal of TP63 indicated that the 2D monolayers might represent more differentiated appreciable AOs compared with 3D AOs.
FIG 9.
Porcine 2D AO monolayers were established from 3D AOs to assess the infectivity and innate immunity of PRCoV. (A) Identification of different cell lineages in 2D AO monolayers. 2D airway organoid monolayers were fixed and labeled with DAPI (nuclear staining, blue) and cell marker antibodies: ACCTUB for ciliated cells (red), MUC2 for goblet cells (green), and TP63 for basal cells (green) (scale bar = 125 μm). (B) Detection of 2D AO monolayers infected with PRCoV or TGEV by anti-NP IFA. 2D AO monolayers were infected with PRCoV or TGEV (MOI = 5) and fixed at 24 hpi for staining with anti-NP antibody (red). DAPI was used for nuclear staining. Images were obtained with an Evos FL Auto2 fluorescence microscope. Scale bars = 125 μm. (C) The kinetic curve of PRCoV or TGEV replication in 2D airway monolayers. 2D AO monolayers were infected with PRCoV or TGEV (MOI = 5), and the cultures were collected at the indicated time points for PRCoV or TGEV quantification by RT-qPCR compared with mock AO monolayers. (D and E) Infection comparison of PRCoV (D) and TGEV (E) in 2D AOs and 3D AOs. (F) Induction of ISG responses by PRCoV infection quantified by RT-qPCR. All the data were calculated using the 2-ΔΔCT method, normalized to GAPDH expression, and compared with mock AOs. Error bars denote the deviations indicated by *: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To assess the ability of 2D AOs to support the infection of porcine respiratory coronaviruses, we monitored PRCoV and TGEV replication kinetics in 2D AOs after challenge with the virus at an MOI of 5. PRCoV and TGEV peaked at 24 hpi and increased approximately 368- and 3,924-fold, respectively, relative to that of input viruses, indicating that 2D AOs are permissible to both TGEV and PRCoV infection (Fig. 9C to E). The successful infection of both PRCoV and TGEV in 2D AOs was confirmed by NP IFA (Fig. 9B). More PRCoV infection was observed in 2D AOs than in 3D AOs, which might be due to the more accessible apical respiratory epithelia formed in the 2D monolayers compared with 3D AOs (Fig. 9D). As expected, PRCoV infection elicited substantial production of type I and type III IFNs and the initiation of ISG responses (Fig. 9F). Altogether, these findings show that 2D AOs support TGEV and PRCoV infection and are a suitable ex vivo model for porcine respiratory infection.
DISCUSSION
Porcine respiratory coronavirus (PRCoV) is a counterpart in pigs to human respiratory CoV infections and a valuable model for human respiratory CoV infections, including SARS-CoV-2 (6, 8). In this study, we established long-term cultures of porcine airway organoids derived from basal epithelial cells, which recapitulate the in vivo airway complicated epithelial cellularity. We demonstrated that 3D AOs and 2D AOs are susceptible to both PRCoV and TGEV despite the variable extent of permissiveness. We show that PRCoV infection induced vigorous IFN and inflammatory responses in AOs, mimicking the events associated with PRCoV infection in vivo (9, 26, 27).
The tissue distribution of coronavirus receptors is one of the determinants in the pathogenesis of coronavirus infections. PRCoV, a natural mutant of TGEV, has a large deletion of the S gene and minor deletions in genes 3/3a and 3-1/3b, which results in the tissue tropism switch from enteric to respiratory tropism (7). Consistent with previous work, our results show that PRCoV did not establish a productive infection in intestinal organoids even though the MOI of PRCoV is 5-fold that of TGEV (Fig. 2). The S1 region of coronavirus spike mediates attachment of CoV particles to cell surface molecules and is the major determinant of coronavirus tropism (31). With the large deletion of the PRCoV N-terminal spike, both PRCoV and TGEV use APN as the major CoV cell entry receptor (13). The results of AO single-cell RNA-seq demonstrated that porcine APN was largely expressed on nonciliated secretory cells in AOs (Fig. 4C). This is consistent with the virus-mediated expression profiles of IFN-β and inflammatory cytokines among the three clustered major cell types (Fig. 4D and E). These results indicate that PRCoV primarily infects secretory cells, which is consistent with a recent elegant study showing that PRCoV cellular tropism in respiratory epithelia is strongly determined by the abundance of APN on the cell surface (13).
Mounting a potent early antiviral IFN response following infection is critical to limiting virus infection and disease severity (32, 33). Notably, we demonstrated that the AOs derived from newborn piglets exhibited pronounced IFN production (types I and III) and ISG induction in response to PRCoV infection (Fig. 4 and 8). In agreement with the substantial induction of IFN responses, PRCoV infection strongly increased the expression of most pattern recognition receptors (PRRs) sensing coronavirus infection, including RIG-1, MDA5, TLR3, TLR7, and TLR9 (Fig. 7). The substantial induction of IFN responses by PRCoV infection in AOs is in line with previous in vivo studies of PRCoV infection (9, 27, 34). Consequently, the pronounced IFN responses together with the high IFN sensitivity of PRCoV may result in mild clinical severity after PRCoV infection in vivo. Notably, PRCoV infection in AOs elicited significant expression of proinflammatory cytokines (IL-1β, IL-6, and TNF) and chemokines (CXCL8, CXCL10, and CXCL11) (Fig. 7), which is consistent with mild pneumonia characterized by bronchiolar exudation and neutrophil and macrophage infiltration within the alveoli after PRCoV infection (9, 26). The specific protective or pathological roles of PRCoV-elicited innate responses need further investigation.
There are several limitations to this study. One of the limitations of this study is that the airway organoid model contains only epithelial cells. The interactions between airway epithelial cells and immune cells could not be explored in this study due to the lack of immune cells in our system. Introducing immune cells into the culture system by coculturing airway organoids with immune cells obtained from the same donor will likely advance our understanding of the pathogenesis of respiratory coronavirus infection. In addition, we cannot exclude the possibility that our findings only reflect varieties in cellular composition or differentiation of AOs.
In conclusion, we established porcine 3D and 2D airway organoids derived from airway epithelial basal cells, which simulate the complicated cellularity and structure of airway epithelia in vivo. The 3D and 2D AOs are susceptible to both PRCoV and TGEV and can produce a pronounced IFN and inflammatory response to virus infection, which well recapitulates the events associated with PRCoV infection in vivo. Taken together, long-term porcine airway organoids provide an interesting model for respiratory coronavirus infection and other porcine respiratory infections.
MATERIALS AND METHODS
Viruses.
The TGEV strain H88 derived from the virulent strain H16 (GenBank accession no. FJ755618) was passaged and stocked by our laboratory. PRCoV strain P8 was isolated, passaged, and stocked by our laboratory.
Porcine intestinal enteroid cultures.
3D porcine intestinal enteroids were previously isolated from specific-pathogen-free (SPF) 2-day-old piglets, stored by our laboratory (30), cultured in IntestiCult organoid growth medium (Stemcell, Canada), and mixed with Matrigel (BD Biosciences, USA) in an incubator with 5% CO2 at 37°C. 2D porcine intestinal enteroid monolayers were prepared from differentiated 3D enteroids as previously described (16). Briefly, 3D porcine intestinal enteroids were separated from the Matrigel after 5 to 7 days of culture, processed into single cells, and seeded in Matrigel-pretreated cell culture plates. After further differentiation for 3 days, the 2D enteroid monolayers were ready for the next experiments. The Institutional Animal Care and Use Committee of the Harbin Veterinary Research Institute approved all the protocols used for animal experiments in this study.
3D airway organoid isolation and culture.
Porcine airway organoids were prepared from specific-pathogen-free (SPF) 2-day-old landrace swine using previously described protocols (20, 21) with minor modifications. Briefly, the complete trachea was isolated from the piglet by dissection and then cut longitudinally. The epithelium of the inner wall of the trachea was carefully peeled off and cut into 2-mm segments. The segments were washed with 15 to 20 mL prechilled PBS several times in a 50-mL centrifuge tube until the supernatant was clear. The pieces were incubated in collagenase IV (1 to 2 mg/mL) at 37°C for 1 h. Then, collagenase IV was removed from the pieces. The airway debris was dissociated with 0.25% EDTA-Trypsin at 37°C for 0.5 h. The debris was harvested by centrifugation at 400 × g at 4°C for 5 min, suspended in ice-cold PBS with 0.1% bovine serum albumin (BSA), and then passed through a 70-μm filter to obtain the airway pellets. The airway pellets were resuspended with PneumaCult-Ex Plus medium (Stemcell, Canada), mixed with Matrigel (BD Biosciences, USA), and seeded into a 48-well plate in an incubator at 37°C for 10 min. After solidification, Matrigel droplets were cultured with PneumaCult-Ex Plus medium at 37°C in an incubator with 5% CO2 at 37°C. The culture medium was exchanged every 4 days. The airway organoids were passaged every 15 to 20 days according to a cell density of 1:5 to 1:10. The Institutional Animal Care and Use Committee of the Harbin Veterinary Research Institute approved all the protocols used for animal experiments in this study.
2D airway organoid monolayer cultures.
2D airway organoid monolayer cultures were prepared from differentiated 3D airway organoids as previously described (30). First, after culturing for 15 to 20 days, 3D airway organoids were washed by using ice-cold Dulbecco’s modified Eagle’s medium (DMEM)-F12 and dissociated with 0.25% trypsin-EDTA (Gibco, USA). Trypsin was then inactivated by adding DMEM-F12 with 10% (vol/vol) fetal bovine serum (FBS). Single-cell suspensions were prepared for dissociation by repeated pipetting. Single-cell suspensions were centrifuged and cultured in PneumaCult-Ex Plus medium and then seeded at 100 airway organoid cells in each well of a 96-well plate precoated with Matrigel. After culturing for 3 days, the 2D airway organoid monolayers were ready for the experiments.
Virus infection in 3D AOs.
After culturing for 15 to 20 days, 3D airway organoids were washed gently with ice-cold 0.5% EDTA-PBS and harvested in a 15-mL centrifuge tube at 400 × g at 4°C for 5 min. The 3D AOs were then washed with ice-cold DMEM-F12 to ensure that the Matrigel was completely removed by 400 × g at 4°C for 5 min. After gently mixing the above-described cell precipitate with the viral dilution, the mixture was added to a 24-well plate and incubated for 3 h at 37°C. 3D AOs were washed three times with DMEM-F12 and recultured under the above-described 3D AO culture conditions, and samples were collected at various time points for subsequent experiments.
Virus infection in 2D AOs.
2D AO monolayers were mixed with virus dilution and incubated at 37°C for 2 h. Unbound virus was washed away with DMEM-F12, and then PneumaCult-Ex Plus medium was added to each well. Samples were collected at the indicated times to quantify viral infection.
Immunofluorescence assay.
AOs were infected with the virus at an MOI of 5 for 24 h. The levels of virus infection and virus-targeted specific cell types were analyzed by IFA as described previously (16, 35). AOs were washed with DMEM-F12 and fixed with 4% paraformaldehyde (PFA) at room temperature, followed by 0.2% Triton X-100 to permeabilize cell membranes, blocked for 2 h at 37°C in PBS containing 5% FBS and 5% skim milk (Sigma-Aldrich, USA), incubated with the primary antibodies for 2 h at 37°C, and then incubated with the secondary antibodies for 1 h at 37°C. Expression of the PRCoV or TGEV nucleocapsid protein (NP) was detected using a mouse anti-N monoclonal antibody stocked in our laboratory. AOs were then labeled with secondary antibody conjugated to Alexa Fluor 488 donkey polyclonal antibody against rabbit IgG (Thermo Fisher Scientific, USA) or Alexa Fluor 546 goat anti-mouse IgG antibody (Thermo Fisher Scientific, USA). DAPI (4′,6-diamidino-2-phenylindole) (Sigma, USA) was used to stain cellular nuclei. The stained cells were visualized using an Evos FL Auto2 fluorescence microscope and confocal laser scanning microscopy with fast Airyscan LSM980-ZEISS.
The expression of differentiation markers of airway epithelia was detected using primary anti-SCGB1A1 (Abcam, USA) for club cells, anti-α-tubulin (ACCTUB) (Novus Biologicals, USA) for ciliated cells, anti-mucin 2 (Abcam, USA) or anti-mucin 5AC (Santa Cruz, USA) for goblet cells, and anti-TP63 (Abcam, USA) for basal cells.
RNA isolation and reverse transcriptase quantitative PCR.
Total RNA was extracted using the RNAiso plus reagent (TaKaRa, Japan) following the manufacturer’s instructions. Total RNA (1 μg) was reverse-transcribed with the PrimeScript II first-strand cDNA synthesis kit (TaKaRa, Japan) following the manufacturer’s instructions. Three repetitions of qPCR were performed on the LightCycler 480 II system (Roche, Switzerland) using SYBR green PCR mix (Life Science, USA). The relative gene expression levels were determined based on the cycle threshold (ΔΔCT) method, normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression, and compared with mock airway organoids or intestinal enteroids. The qPCR primers for TGEV or PRCoV targeted conserved regions of the spike gene (3,422 to 3,630 nucleotides [nt]). All qPCR primer sequences used in the study are listed in Table 1.
TABLE 1.
Primers for qPCR
| Gene name | Primer direction | Primer sequences (5′–3′) |
|---|---|---|
| ISG15 | Forward | AGCATGGTCCTGTTGATGGTG |
| Reverse | CAGAAATGGTCAGCTTGCACG | |
| OASL | Forward | TCCCTGGGAAGAATGTGCAG |
| Reverse | CCCTGGCAAGAGCATAGTGT | |
| IFN-β | Forward | CTGCTGCCTGGAATGAGAGCC |
| Reverse | TGACACAGGCTTCCAGGTCCC | |
| IFNAR1 | Forward | ACATCACCTGCCTTCACCAG |
| Reverse | CATGGAGCCACTGAGCTTGA | |
| IFNLR1 | Forward | GGGGCTTCTCATCACCAC |
| Reverse | ATTCGCTCCCACCCTCTT | |
| APN | Forward | TGTTTGACCCACAGTCCT |
| Reverse | TCCACATAGGAGGCAAAG | |
| Furin | Forward | GGTGGCCCTTCCCCGTCCCTCTAA |
| Reverse | CCTGCCTGGCCTTGGTCCCTTCCT | |
| TMPRSS2 | Forward | GGCCGGCGTGAGCTGCTTTTACAA |
| Reverse | RCCCCGAGGCCCCCATCTTCT | |
| IL10RB | Forward | CATGGGCTTATCATGTGC |
| Reverse | GCTCGGACTTGAACACAG | |
| IFN-λ1 | Forward | CCACGTCGAACTTCAGGCTT |
| Reverse | ATGTGCAAGTCTCCACTGGT | |
| PRCoV | Forward | GCTTGATGAATTGAGTGCTGATG |
| Reverse | CCTAACCTCGGCTTGTCTGG | |
| TGEV | Forward | GCTTGATGAATTGAGTGCTGATG |
| Reverse | CCTAACCTCGGCTTGTCTGG | |
| GAPDH | Forward | CCTTCCGTGTCCCTACTGCCAAC |
| Reverse | GACGCCTGCTTCACCACCTTCT | |
| RIG-I | Forward | GGCTGAAGCCACAGAATA |
| Reverse | TCAGTGGTCCGTAATTCC | |
| MDA5 | Forward | TCTCCGGGAAACAGGCAAC |
| Reverse | CAAAGGATGGAGAGGGCAAGT | |
| TLR3 | Forward | AGTAAATGAATCACCCTGCCTAGCA |
| Reverse | GCCGTTGACAAAACACATAAGGAC | |
| TLR4 | Forward | GCCATCGCTGCTAACATCATC |
| Reverse | CTCATACTCAAAGATACACCATCGG | |
| TLR10 | Forward | CCTGTCCAACTGCCTCATTTG |
| Reverse | CTAAGTGTTCTAAGGATGTGTTTCT | |
| IL-1β | Forward | TGTGCAAGGAGATGACAGTG |
| Reverse | CATCACACAAGACAGGTACAG | |
| IL-6 | Forward | TACATCCTCGGCAAAATC |
| Reverse | TCTCATCAAGCAGGTCTCC | |
| TNF-α | Forward | GACAGCTCCAATGGCAGAGTG |
| Reverse | GCTGATGGTGTGAGTGAGGAA | |
| CXCL8 | Forward | GCAGTTCTGGCAAGAGTAAGT |
| Reverse | CAGGCAGACCTCTTTTCCATT | |
| CXCL10 | Forward | GCCGTCATTTTCTGCCTCATC |
| Reverse | TAGGCTCGCAGGGATGATTTC |
Single-cell RNA sequencing.
SPF 2-day-old 3D airway organoids were infected with or without PRCoV (MOI = 5) at 37°C for 24 h and then prepared for single-cell suspensions to be handed over to Shanghai Jiayin Co., Ltd., for single-cell RNA sequencing. Single cells were captured in 10× Genomics chromium single cell 3′ solution, and RNA-seq libraries were prepared following the manufacturer’s protocol (10× Genomics). The libraries were subjected to high-throughput sequencing on an Illumina HiSeq X Ten PE150 platform, and 150-bp paired-end reads were generated. The integration (count normalization, feature selection, dimension reduction, and visualization through t-distributed stochastic neighbor embedding [t-SNE]) and unsupervised clustering of single cells were performed using the Seurat and SC3 packages. Single-cell RNA sequencing data were analyzed with cell subsets using Loupe Browser software.
RNA-seq analysis.
SPF 2-day-old 3D airway organoids were infected with or without PRCoV (MOI = 5) at 37°C for 24 h with three biological replicates and then were prepared for RNA sequencing. Total RNA was extracted using the RNAiso Plus reagent (TaKaRa, Japan) following the manufacturer’s instructions. The samples were handed over to Shanghai Jiayin Co., Ltd., for RNA-seq experiments and postdata analysis based on sequencing data.
RNA sequencing data were processed and mapped to the Sus scrofa reference genome and PRCoV genome. Differential expression was analyzed using the DESeq2 R package at a significance cutoff of P < 0.05. Differentially expressed genes were identified by the screening criteria based on |log2(fold change)| >1 and a false-discovery rate (FDR) of <0.05.
Statistical analysis.
All statistical analyses were performed using Prism (GraphPad Software, Inc.). The experiment was performed three times independently, as shown in all the legend. The scale data of viral genomes and titers were taken as log (fold change) values and expressed as a power of 10. Data are presented as the means ± the standard errors of the mean (SEMs). Differences were considered significant when the P value was <0.05. The P values are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Data availability.
All data are in the manuscript and/or supporting information files, except for raw RNA-seq data, which can be available upon request.
ACKNOWLEDGMENTS
We are grateful to Zhaohui Qian (Chinese Academy of Medical Sciences and Peking Union Medical College) for discussion about the manuscript.
Conceptualization: Pinghuang Liu. Funding acquisition: Pinghuang Liu, Li Feng. Investigation: Chengfan Jiang, Liang Li, Liyuan Zhao, Mei Xue, Xiang Liu, Wenzhe Wang, Li Feng, Pinghuang Liu. Methodology: Chengfan Jiang, Liang Li, Liyuan Zhao, Mei Xue, Pinghuang Liu. Resources: Pinghuang Liu, Li Feng, Mei Xue. Supervision: Pinghuang Liu, Li Feng, Mei Xue. Writing: Pinghuang Liu, Chengfan Jiang, Liang Li.
This work was supported by the National Key Research and Development Program of China (2021YFD1801104 and 2021YFD1801105), the National Natural Science Fund (32192453 and 32172865), and China Postdoctoral Science Foundation (2020M670556). The funders had no role in the study design, data collection, analysis, decision to publish, or preparation of the manuscript.
We declare that no competing interests exist.
Contributor Information
Li Feng, Email: fengli@caas.cn.
Pinghuang Liu, Email: liupinghuang@cau.edu.cn.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Hu B, Guo H, Zhou P, Shi ZL. 2021. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol 19:141–154. 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.V’Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. 2021. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol 19:155–170. 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong LR, Perlman S. 2022. Immune dysregulation and immunopathology induced by SARS-CoV-2 and related coronaviruses: are we our own worst enemy? Nat Rev Immunol 22:47–56. 10.1038/s41577-021-00656-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pensaert M, Callebaut P, Vergote J. 1986. Isolation of a porcine respiratory, non-enteric coronavirus related to transmissible gastroenteritis. Vet Q 8:257–261. 10.1080/01652176.1986.9694050. [DOI] [PubMed] [Google Scholar]
- 5.Heegaard PMH, Sturek M, Alloosh M, Belsham GJ. 2020. Animal models for COVID-19: more to the picture than ACE2, rodents, ferrets, and non-human primates. A case for porcine respiratory coronavirus and the obese Ossabaw pig. Front Microbiol 11:573756. 10.3389/fmicb.2020.573756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saif LJ, Jung K. 2020. Comparative pathogenesis of bovine and porcine respiratory coronaviruses in the animal host species and SARS-CoV-2 in humans. J Clin Microbiol 58:e01355-20. 10.1128/JCM.01355-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim L, Hayes J, Lewis P, Parwani AV, Chang KO, Saif LJ. 2000. Molecular characterization and pathogenesis of transmissible gastroenteritis coronavirus (TGEV) and porcine respiratory coronavirus (PRCV) field isolates co-circulating in a swine herd. Arch Virol 145:1133–1147. 10.1007/s007050070114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox E, Hooyberghs J, Pensaert MB. 1990. Sites of replication of a porcine respiratory coronavirus related to transmissible gastroenteritis virus. Res Vet Sci 48:165–169. 10.1016/S0034-5288(18)30984-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Alekseev K, Jung K, Vlasova A, Hadya N, Saif LJ. 2008. Cytokine responses in porcine respiratory coronavirus-infected pigs treated with corticosteroids as a model for severe acute respiratory syndrome. J Virol 82:4420–4428. 10.1128/JVI.02190-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyazaki A, Fukuda M, Kuga K, Takagi M, Tsunemitsu H. 2010. Prevalence of antibodies against transmissible gastroenteritis virus and porcine respiratory coronavirus among pigs in six regions in Japan. J Vet Med Sci 72:943–946. 10.1292/jvms.09-0377. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Wu Q, Sun X, Shen J, Chen H. 2020. Organoids as a powerful model for respiratory diseases. Stem Cells Int 2020:5847876. 10.1155/2020/5847876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller AJ, Yu Q, Czerwinski M, Tsai YH, Conway RF, Wu A, Holloway EM, Walker T, Glass IA, Treutlein B, Camp JG, Spence JR. 2020. In vitro and in vivo development of the human airway at single-cell resolution. Dev Cell 53:117–128.e116. 10.1016/j.devcel.2020.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng JY, Punyadarsaniya D, Shin DL, Pavasutthipaisit S, Beineke A, Li G, Wu NH, Herrler G. 2020. The cell tropism of porcine respiratory coronavirus for airway epithelial cells is determined by the expression of porcine aminopeptidase N. Viruses 12:1211. 10.3390/v12111211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mora-Diaz JC, Pineyro PE, Rauh R, Nelson W, Sankoh Z, Gregg E, Carrillo-Avila JA, Shen H, Nelli RK, Zimmerman JJ, Gimenez-Lirola LG. 2021. Porcine hemagglutinating encephalomyelitis virus infection in vivo and ex vivo. J Virol 95:e02335-20. 10.1128/JVI.02335-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hild M, Jaffe AB. 2016. Production of 3-D airway organoids from primary human airway basal cells and their use in high-throughput screening. Curr Protoc Stem Cell Biol 37:IE.9.1–IE.9.15. 10.1002/cpsc.1. [DOI] [PubMed] [Google Scholar]
- 16.Li L, Fu F, Guo S, Wang H, He X, Xue M, Yin L, Feng L, Liu P. 2019. Porcine intestinal enteroids: a new model for studying enteric coronavirus porcine epidemic diarrhea virus infection and the host innate response. J Virol 93:e01682-18. 10.1128/JVI.01682-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McQualter JL, Yuen K, Williams B, Bertoncello I. 2010. Evidence of an epithelial stem/progenitor cell hierarchy in the adult mouse lung. Proc Natl Acad Sci USA 107:1414–1419. 10.1073/pnas.0909207107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tindle C, Fuller M, Fonseca A, Taheri S, Ibeawuchi SR, Beutler N, Katkar G, Claire A, Castillo V, Hernandez M, Russo H, Duran J, Crotty Alexander LE, Tipps A, Lin G, Thistlethwaite PA, Chattopadhyay R, Rogers TF, Sahoo D, Ghosh P, Das S. 2021. Adult stem cell-derived complete lung organoid models emulate lung disease in COVID-19. bioRxiv. 10.1101/2020.10.17.344002. [DOI] [PMC free article] [PubMed]
- 19.Lamers MM, van der Vaart J, Knoops K, Riesebosch S, Breugem TI, Mykytyn AZ, Beumer J, Schipper D, Bezstarosti K, Koopman CD, Groen N, Ravelli RBG, Duimel HQ, Demmers JAA, Verjans G, Koopmans MPG, Muraro MJ, Peters PJ, Clevers H, Haagmans BL. 2021. An organoid-derived bronchioalveolar model for SARS-CoV-2 infection of human alveolar type II-like cells. EMBO J 40:e105912. 10.15252/embj.2020105912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sachs N, Papaspyropoulos A, Zomer-van Ommen DD, Heo I, Bottinger L, Klay D, Weeber F, Huelsz-Prince G, Iakobachvili N, Amatngalim GD, de Ligt J, van Hoeck A, Proost N, Viveen MC, Lyubimova A, Teeven L, Derakhshan S, Korving J, Begthel H, Dekkers JF, Kumawat K, Ramos E, van Oosterhout MF, Offerhaus GJ, Wiener DJ, Olimpio EP, Dijkstra KK, Smit EF, van der Linden M, Jaksani S, van de Ven M, Jonkers J, Rios AC, Voest EE, van Moorsel CH, van der Ent CK, Cuppen E, van Oudenaarden A, Coenjaerts FE, Meyaard L, Bont LJ, Peters PJ, Tans SJ, van Zon JS, Boj SF, Vries RG, Beekman JM, Clevers H. 2019. Long-term expanding human airway organoids for disease modeling. EMBO J 38:e100300. 10.15252/embj.2018100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou J, Li C, Sachs N, Chiu MC, Wong BH, Chu H, Poon VK, Wang D, Zhao X, Wen L, Song W, Yuan S, Wong KK, Chan JF, To KK, Chen H, Clevers H, Yuen KY. 2018. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc Natl Acad Sci USA 115:6822–6827. 10.1073/pnas.1806308115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang G, Liang R, Liu Z, Shen Z, Shi J, Shi Y, Deng F, Xiao S, Fu ZF, Peng G. 2019. The N-terminal domain of spike protein is not the enteric tropism determinant for transmissible gastroenteritis virus in piglets. Viruses 11:313. 10.3390/v11040313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez CM, Pascual-Iglesias A, Sola I, Zuniga S, Enjuanes L. 2019. Minimum determinants of transmissible gastroenteritis virus enteric tropism are located in the N-terminus of spike protein. Pathogens 9:2. 10.3390/pathogens9010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carraro G, Langerman J, Sabri S, Lorenzana Z, Purkayastha A, Zhang G, Konda B, Aros CJ, Calvert BA, Szymaniak A, Wilson E, Mulligan M, Bhatt P, Lu J, Vijayaraj P, Yao C, Shia DW, Lund AJ, Israely E, Rickabaugh TM, Ernst J, Mense M, Randell SH, Vladar EK, Ryan AL, Plath K, Mahoney JE, Stripp BR, Gomperts BN. 2021. Transcriptional analysis of cystic fibrosis airways at single-cell resolution reveals altered epithelial cell states and composition. Nat Med 27:806–814. 10.1038/s41591-021-01332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delmas B, Gelfi J, Sjostrom H, Noren O, Laude H. 1993. Further characterization of aminopeptidase-N as a receptor for coronaviruses. Adv Exp Med Biol 342:293–298. 10.1007/978-1-4615-2996-5_45. [DOI] [PubMed] [Google Scholar]
- 26.Keep S, Carr BV, Lean FZX, Fones A, Newman J, Dowgier G, Freimanis G, Vatzia E, Polo N, Everest H, Webb I, McNee A, Paudyal B, Thakur N, Nunez A, MacLoughlin R, Maier H, Hammond J, Bailey D, Waters R, Charleston B, Tuthill T, Britton P, Bickerton E, Tchilian E. 2022. Porcine respiratory coronavirus as a model for acute respiratory coronavirus disease. Front Immunol 13:867707. 10.3389/fimmu.2022.867707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Reeth K, Labarque G, Nauwynck H, Pensaert M. 1999. Differential production of proinflammatory cytokines in the pig lung during different respiratory virus infections: correlations with pathogenicity. Res Vet Sci 67:47–52. 10.1053/rvsc.1998.0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin L, Liu X, Hu D, Luo Y, Zhang G, Liu P. 2021. Swine enteric coronaviruses (PEDV, TGEV, and PDCoV) induce divergent interferon-stimulated gene responses and antigen presentation in porcine intestinal enteroids. Front Immunol 12:826882. 10.3389/fimmu.2021.826882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKernan DP. 2020. Pattern recognition receptors as potential drug targets in inflammatory disorders. Adv Protein Chem Struct Biol 119:65–109. 10.1016/bs.apcsb.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 30.Yin L, Chen J, Li L, Guo S, Xue M, Zhang J, Liu X, Feng L, Liu P. 2020. Aminopeptidase N expression, not interferon responses, determines the intestinal segmental tropism of porcine deltacoronavirus. J Virol 94:e00480-20. 10.1128/JVI.00480-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reguera J, Santiago C, Mudgal G, Ordono D, Enjuanes L, Casasnovas JM. 2012. Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies. PLoS Pathog 8:e1002859. 10.1371/journal.ppat.1002859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Channappanavar R, Fehr AR, Zheng J, Wohlford-Lenane C, Abrahante JE, Mack M, Sompallae R, McCray PB, Jr, Meyerholz DK, Perlman S. 2019. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J Clin Invest 129:3625–3639. 10.1172/JCI126363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loske J, Rohmel J, Lukassen S, Stricker S, Magalhaes VG, Liebig J, Chua RL, Thurmann L, Messingschlager M, Seegebarth A, Timmermann B, Klages S, Ralser M, Sawitzki B, Sander LE, Corman VM, Conrad C, Laudi S, Binder M, Trump S, Eils R, Mall MA, Lehmann I. 2022. Pre-activated antiviral innate immunity in the upper airways controls early SARS-CoV-2 infection in children. Nat Biotechnol 40:319–324. 10.1038/s41587-021-01037-9. [DOI] [PubMed] [Google Scholar]
- 34.Charley B, Riffault S, Van Reeth K. 2006. Porcine innate and adaptative immune responses to influenza and coronavirus infections. Ann N Y Acad Sci 1081:130–136. 10.1196/annals.1373.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Chen J, Liu Y, Da S, Shi H, Zhang X, Liu J, Cao L, Zhu X, Wang X, Ji Z, Feng L. 2020. Pathogenicity of porcine deltacoronavirus (PDCoV) strain NH and immunization of pregnant sows with an inactivated PDCoV vaccine protects 5-day-old neonatal piglets from virulent challenge. Transbound Emerg Dis 67:572–583. 10.1111/tbed.13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are in the manuscript and/or supporting information files, except for raw RNA-seq data, which can be available upon request.