ABSTRACT
Integration of the reverse-transcribed genome is a critical step of the retroviral life cycle. Strand-transfer inhibitors (INSTIs) used for antiretroviral therapy inhibit integration but can lead to resistance mutations in the integrase gene, the enzyme involved in this reaction. A significant proportion of INSTI treatment failures, particularly those with second-generation INSTIs, show no mutation in the integrase gene. Here, we show that replication of a selected dolutegravir-resistant virus with mutations in the 3′-PPT (polypurine tract) was effective, although no integrated viral DNA was detected, due to the accumulation of unintegrated viral DNA present as 1-LTR circles. Our results show that mutation in the 3′-PPT leads to 1-LTR circles and not linear DNA as classically reported. In conclusion, our data provide a molecular basis to explain a new mechanism of resistance to INSTIs, without mutation of the integrase gene and highlights the importance of unintegrated viral DNA in HIV-1 replication.
IMPORTANCE Our work highlights the role of HIV-1 unintegrated viral DNA in viral replication. A virus, resistant to strand-transfer inhibitors, has been selected in vitro. This virus highlights a mutation in the 3'PPT region and not in the integrase gene. This mutation modifies the reverse transcription step leading to the accumulation of 1-LTR circles and not the linear DNA. This accumulation of 1-LTR circles leads to viral replication without integration of the viral genome.
KEYWORDS: HIV-1, integrase, integration, resistance, unintegrated viral DNA
INTRODUCTION
HIV-1 generally selects mutations in the integrase (IN) gene to counteract IN inhibitors (raltegravir, RAL; elvitegravir, EVG; and dolutegravir, DTG) (1, 2). Interestingly, it has been reported that mutations in another part of the genome, more particularly in the env gene, can confer resistance to ISTI (3). Recently, we reported the in vitro selection of a virus highly resistant to these INSTIs, with mutations in the 3′-PPT (polypurine tract) and not the target integrase gene (4). The PPT is highly conserved in most retroviruses and used as the site of plus-strand initiation involved in viral reverse transcription (5). Mutations in the 3′-PPT have been confirmed in vivo in a patient failing to respond to DTG without mutations in the integrase gene (6). Studies about the impact of the mutations in the 3′-PPT on INSTI resistance have highlighted conflicted results. These apparent discrepancies could be explained by the different settings of the experiments involved (7–9). These observations of mutations outside the integrase gene raise the mechanism leading to the viral replication, if any, and resistance to integrase inhibitors. We therefore explored the impact of the mutations we have previously selected in the 3′-PPT named (5′-GCAGT) (4). These mutations consist in a replacement of the cytidine by a thymine in position 9053, change of the GGGGGG sequence by the GCAGT sequence from position 9069 to 9073 including a deletion of a guanine in position 9073 (4).
We found that these mutations lead to the replication of the virus, even at a less extent compared with the WT. Importantly, we demonstrate that replication is concomitant to 1-LTR circles accumulation and the absence of integrated viral DNA. We highlight, using biochemical experiments that mutation in the 3′-PPT leads to the modification of the reverse transcription step that generates a 1-LTR circle and not linear viral DNA. Taken together, our data demonstrate the replication of the virus encompassing the mutation in the 3′-PPT region involving unintegrated viral DNA (1-LTR circles), explaining the resistance to INSTI.
RESULTS AND DISCUSSION
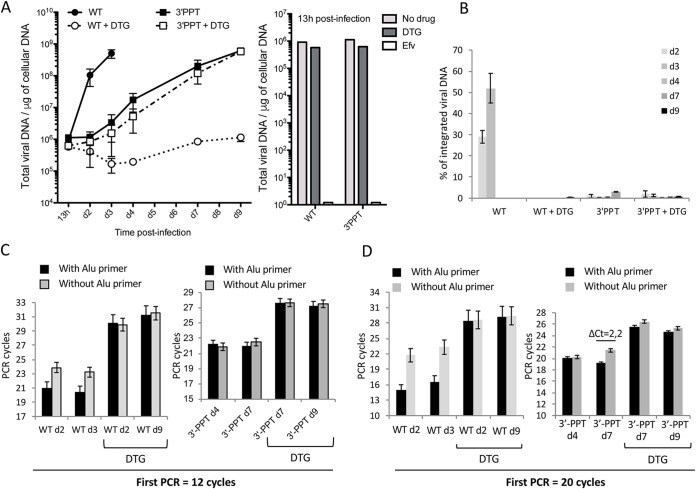
MT4 cells were infected, with or without DTG, and total viral DNA quantified by quantitative PCR. DTG was maintained throughout the experiment in the medium to ensure that unintegrated viral DNA was not integrated into the cellular genome, such reversibility having been previously described for RAL (10). The WT virus was sensitive to DTG, as shown by reduced DNA synthesis, up to the end of the experiment (d9 postinfection), compared to the condition without DTG. (Fig. 1A, left panel). These data indicate that DTG is efficient to mostly inhibit WT replication even if a faint DNA synthesis can be observed late after infection (from 7 days postinfection). This observation is similar to the weak replication of a catalytic mutant of integrase reported in some studies (11, 12). In contrast, the 3′-PPT mutant virus showed an increase in total viral DNA, with or without DTG, confirming its resistance to DTG. Moreover, the reverse transcription step was not affected since the amount of viral DNA at 13 h postinfection was similar for both viruses and not affected by DTG (Fig. 1A, right panel). This observation clearly shows that DTG has no influence on viral DNA synthesis either in the case of the 3′-PPT mutant or the WT. Quantification of integrated HIV-1 DNA was calculated by subtracting the signal obtained with Alu primers from that obtained without (13). We confirmed the presence of integrated DNA only for the WT virus, representing 28% and 52% of total viral DNA at d2 and d3, respectively (Fig. 1B). The presence of integrated viral DNA led to a significant difference between the two signals (with or without the Alu primers), 2.9 cycles on average (ΔCt = 2.9), and was observed only for the WT infection and not the other conditions (WT+DTG and 3′-PPT+/−DTG) (Fig. 1C, left panel). Indeed, for the 3′-PPT mutant, there was no difference between the two signals (with or without the Alu primers), with or without DTG, (Fig. 1C, right panel), demonstrating that no viral DNA integration occurred, as for the WT virus with DTG. We increased the number of cycles of the first integrated PCR from 12 to 20 to increase the sensitivity of integrated viral DNA detection. In this context, we were able to detect the integrated viral genome, but its quantification was not reliable due to the loss of linearity of the experiment. For the WT virus, these new amplification conditions made it possible to observe a large increase between the two signals from 2.9 to 6.7 (Fig. 1D, left panel), validating the possibility to increase the detection of integrated viral DNA. The difference between signals did not increase significantly, with or without DTG, in most 3′-PPT mutant samples. Only one sample without DTG (Fig. 1D, right panel) showed a significant ΔCt (ΔCt = 2.2) at d7 postinfection, making it impossible to exclude the presence of a very low level of integrated viral DNA under this specific condition.
FIG 1.
Replication of the WT and 3′-PPT mutant viruses in MT4 cells. MT4 T-cells were infected with WT or 3′-PPT mutant (100 ng of p24 per one million cells) in the absence or presence of 1 μM DTG. Quantitative-PCR (qPCR) was performed to quantify total viral DNA (A) and integrated viral DNA (B–D) during the course of the experiment. (A) Left panel: total viral DNA is expressed in copies per μg of DNA during the course of infection. The graph shows one representative experiment or three independent experiments. Right panel: total viral DNA at 13h postinfection. (B) Percentage of integrated viral DNA over total viral DNA. Results are shown for three independent experiments (mean ± standard deviation (SD)). (C) The ΔCt obtained for each condition of amplification with 12 cycles (with the Alu primer (black column) and without the Alu primer (gray column)) is shown for the WT and 3′-PPT mutant samples, with or without DTG. (D) The number of PCR cycles for the first ALU-LTR reaction was increased (20 cycles instead of 12 (B)). The ΔCt obtained for each condition of amplification of the same samples described in (B) (with Alu primer (black column) and without Alu primer (gray column)) is shown. Results are shown for three independent experiments (mean ± SD). Statistical differences were calculated using Student's t test (*, P < 0.05; **, P < 0.005; ***, P < 0.001; ns P > 0.05).
We evaluated the sensitivity of the detection of integrated viral DNA among nonintegrated viral DNA by diluting a known amount of integrated viral DNA (4 × 107 copies) in a constant amount of DNA from cells infected with the 3′-PPT mutant in the presence of DTG (4 × 107 copies) (Fig. S1A in the supplemental material). The amount of cellular DNA was kept constant. Integrated viral DNA PCR was performed using 20 cycles for the first PCR and the ΔCt calculated. The ΔCt decreased as the proportion of integrated viral DNA decreased in the mixture but was still significant after dilution of the initial integrated DNA by 1,000-fold. This data show that if integrated DNA is present in the case of the 3′-PPT for which we did not see significant ΔCt, its proportion over total viral DNA is less than 0,1%. These data were supported by the nondetection of viral insertion in the host cell genome by the sequencing of integration sites, excluding a bias of the quantification by PCR (Fig. S1B). Insertions were clearly detectable for the WT virus, whereas the signal was at the level of the background noise for the 3′-PPT mutant, with or without DTG. Taken together, we demonstrate a defect in the integration process of the 3′-PPT mutant.
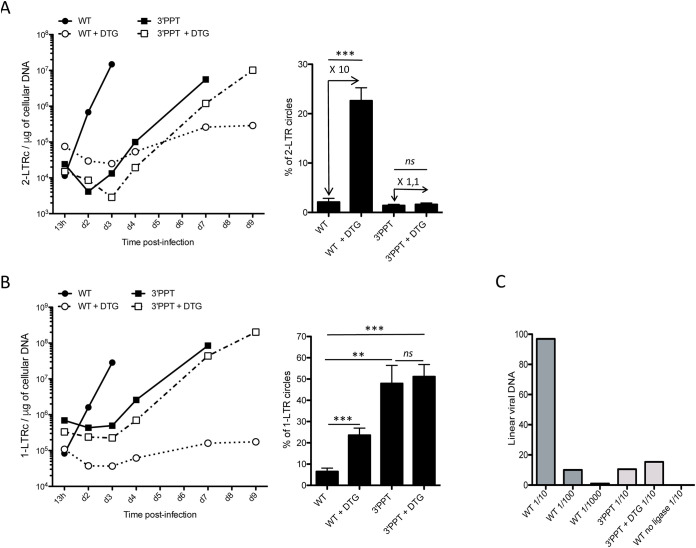
Use of the 3′-PPT mutant virus resulted in an increase in the absolute number of 2-LTR circles during the course of the experiment, with or without DTG, as shown for the WT virus without DTG (Fig. 2A) concomitant with the increase of total viral DNA (Fig. 1A) . Infection of the cells with the WT virus in the presence of DTG led to strong accumulation of 2-LTR circles, concomitant with the inhibition of integration, as already reported (14). Such accumulation of 2-LTR circles results from the ligation of the two LTRs of the linear viral DNA (15). 2-LTR circles represented 1% of the total viral DNA when the cells were infected with the 3′-PPTmutant, with or without DTG, which was equivalent to the level found for WT infection (without inhibitor) (Fig. 2A, right panel). In a first approach, the nonaccumulation of 2-LTR circles as well as the absence of integrated viral DNA for the 3′-PPT mutant seems controversial. This apparent discrepancy can be ruled out if the substrate involved in 2-LTR formation, i.e., linear viral DNA, is not available suggesting a modification of the reverse transcription mechanism. Another form of unintegrated viral DNA, called a 1-LTR circle, is formed mainly in the nucleus by homologous recombination or during reverse transcription (16, 17). This episomal form increased during the course of infection, except for the WT+DTG condition (Fig. 2B). 1-LTR circles strongly accumulated using the 3′-PPT mutant virus and represented approximately 50% of the total viral DNA, regardless of whether or not DTG was present, unlike the WT virus, for which the proportion of DNA in the 1-LTR form represented only 6% of the total viral DNA in the absence of DTG and 21% in its presence (Fig. 2B, right panel). Such accumulation, greater by a factor of 10, occurred during replication, even though the amount of total viral DNA was similar for the mutant and WT viruses. Quantification of linear viral DNA in cells infected with the 3′-PPT mutant virus showed only a small amount of linear viral DNA, with or without DTG, showing that the 1-LTR circles were formed directly during reverse transcription instead of the linear viral DNA, as already shown for PPT mutants in the retroviral vector context (Fig. 2C) (17).
FIG 2.
Characterization of unintegrated viral DNA (linear and 2-LTR and 1-LTR circles) genomes following infection with the WT or 3′-PPT mutant virus. Samples from the previously depicted experiment (Fig. 1) using MT4 T-cells infected with the WT or 3′-PPT mutant virus, in the presence or absence of 1 μM DTG, were analyzed by qPCR to quantify circular genomes: (A) Left panel: time course of 2-LTR circle formation. Right panel: average percentage of 2-LTR circles for all time points analyzed. The fold of 2-LTR circle accumulation due to DTG treatment is indicated in the Figure. (B) Left panel: time course of 1-LTR circle formation. Right panel: average percentage of 1-LTR circles for all time points analyzed. (C) Quantification of linear viral DNA at 13h postinfection by ligation-mediated PCR. Several dilutions have been tested for each sample (1/10, 1/100, 1/1000 for the WT and 1/10 for the 3′-PPT and 3′-PPT + DTG). In addition, as a negative control, a sample of WT was processed without the ligase. All results were obtained from three independent experiments (mean ± SEM). Statistical differences were calculated using Student’s test (*, P < 0.05; **, P < 0.005; ***, P < 0.001; ns P > 0.05).
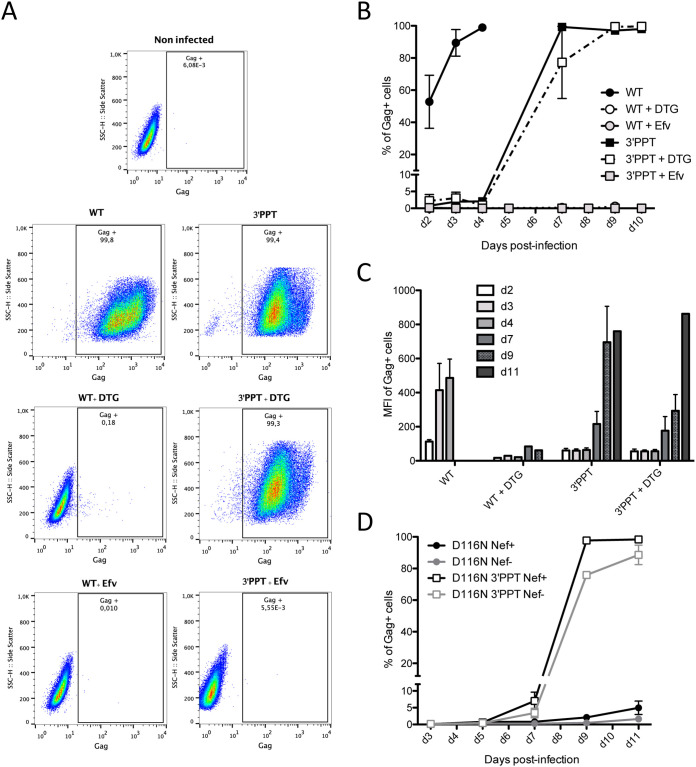
We next studied expression of the viral DNA genomes from WT and 3′-PPT mutant infections by quantifying the production of a late structural protein, the Gag protein (Fig. 3). MT4 cells were infected with the WT and 3′-PPT mutant viruses, in the presence or absence of DTG. To follow the percentage of cells able to express viral proteins, a Gag intracellular immunostaining was performed at different times postinfection (Fig. 3). No Gag-positive cells were detected until d10 when the cells were infected with the WT virus in the presence of DTG. At d10, only 0.8% of the cells were positive (Fig. 3A and B). In this setting, the DNA was in the nonintegrated form, as already described. These data show that nonintegrated DNA of the WT virus was not expressed and therefore not involved in Gag synthesis. However, when the WT DNA was integrated in the genome, 53% of the cells were positive for Gag at d2, showing that integrated viral DNA sustained a high level of viral protein expression as soon as 2 days postinfection. Infection of the cells with the 3′-PPT mutant virus resulted in a continuous increase in the percentage of Gag+ cells from d2 (1% Gag+ cells) to d10 (100% Gag+ cells), in the presence or absence of DTG. This result confirmed that the nonintegrated viral DNA from the 3′-PPT mutant virus was competent for Gag synthesis. We wanted to ensure that the quantified Gag proteins resulted from de novo synthesized proteins and not the remaining viral input used for infection. Thus, we performed the same experiment in the presence of efavirenz, an inhibitor of reverse transcriptase, which inhibits early replication of the virus. Indeed, no Gag antigen was detected when efavirenz was used, showing that the detection of Gag was due to novel synthesis of the protein (Fig. 3A and B). The mean of fluorescence intensity (MFI) of the Gag+ cells was high for the WT infection, as soon as d3 postinfection, revealing strong Gag synthesis, i.e., from integrated viral DNA (Fig. 3C). In the case of the 3′-PPT mutant, the synthesis of gag protein is delayed, and the mean of fluorescence intensity is weaker (at least until d7) compared with the mean of gag positive cells from the WT suggesting that the synthesis of viral protein is weaker from nonintegrated DNA compared to integrated viral DNA (Fig. 3C). Importantly, supernatants from WT or 3′-PPT infected cells were collected and the quantity of p24 antigen was quantified by ELISA (Fig. S2B in the supplemental material). Then, these supernatants were used for de novo infection of fresh MT4-cells in presence or absence of DTG (Fig. S2). Infection monitoring by intracellular staining of gag (Fig. S2c) or by quantification of total viral DNA (Fig. S2D) shows that MT4-cells are productively infected by the 3′-PPT supernatant, demonstrating that infection of the 3′-PPT virus leads to production of infectious viral particles. Sequences on newly infected cells as well as viral RNA from infectious viral particles highlights the presence of the 3′-PPT mutation showing that no reversion has occurred after infection of MT-4 cells with 3′-PPT virus.
FIG 3.
Expression from viral DNA of the 3′-PPT mutant virus is higher than that from nonintegrated viral DNA of the WT virus but lower than that of integrated viral DNA. MT4 T cells were infected with the WT and 3′-PPT mutant virus, with or without 1 μM DTG. (A) Gating strategy for flow cytometry analysis of intracellular gag synthesis after infection with the WT (left panel) or 3′-PPT mutant virus (right panel), with or without 1 μM DTG. Efavirenz (Efv) was used as a control to ensure that gag detection was not due to the initial infection. Data are representative of three independent experiments. Immunofluorescent staining was performed using an anti-Gag antibody (recognizing p55, p39, p33 and p24 proteins) conjugated to phycoerythrin (PE). Analysis was performed at d7 postinfection except for the WT virus (d2 postinfection). (B) The amount of intracellular gag antigen was quantified at various time points by immunofluorescent staining using an anti-Gag antibody conjugated to phycoerythrin (PE). Results are expressed as the percentage of Gag positive cells during the course of infection. Efavirenz (Efv) was used as a control to ensure that gag detection was due to de novo synthesis and not the persistence of gag antigen from the initial infection. Graph shows the mean ± (SEM) of 3 independent experiments. (C) The mean fluorescence intensity of phycoerythrin (PE) in Gag-PE+ cells during the course of infection is shown. Graph shows the mean ± (SEM) of 3 independent experiments. (D) MT4 T cells were infected with an integrase mutant virus (D116N), with an intact 3′-PPT region or with the mutated 3′-PPT (3′-PPT). Moreover, since mutations of the 3′-PPT destroyed the Nef gene, the Nef gene was rescued (Nef+) or not (Nef-). At different times postinfection, the percentage of cells expressing the Gag antigen was analyzed by immunofluorescent staining using an anti-Gag antibody conjugated to phycoerythrin (PE). Graph shows the mean ± (SEM) of 3 independent experiments.
We studied catalytic mutant of integrase, another way to prevent integration. We performed experiment on MT4-cells using viruses inactivated in the integrase gene (D116N mutation). The 3′-PPT mutations (5′-GCAGT) lead to the disruption of the Nef open reading frame. To prevent a potential effect of Nef in combination with the 3′-PPT mutation, we used different viral constructions with or without a functional Nef open reading frame (Nef + and Nef −, respectively) in the context of D116N. Our data clearly show that replication of the D116N virus is only efficient in combination with the 3′-PPT mutation (Fig. 3D), whatever the Nef context. Taken together, using DTG and catalytic mutant, our data demonstrate that replication of the 3′-PPT virus does not require integration activity.
We performed the same experiments using CEMss cells, in which HTLV-1 was absent, to ensure that the effect of the 3′-PPT mutation on HIV-1 replication was not influenced by HTLV-1 virus. Indeed, several studies report a role of Tax in the replication of the virus without integration using mutants in the integrase gene (11, 12). The results obtained with the CEMss cells were identical to those obtained with the MT-4 cells (Fig. S3 in the supplemental material). More particularly, we observed the same viral DNA synthesis (with and without DTG) (Fig. S3B) in which the proportion of 1-LTR circles is identical to the one found in MT4 cells. These data clearly show that replication of the 3′-PPT mutant in MT4 cells is not due to Tax.
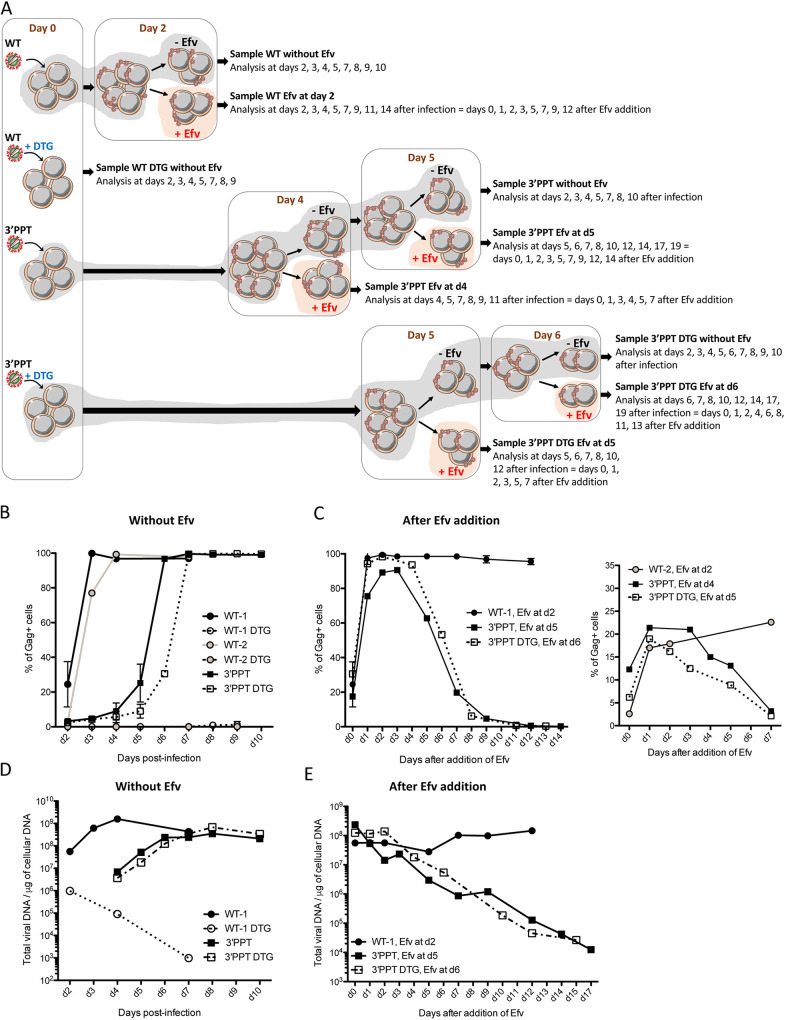
To ensure that the source of viral proteins produced during the infection by the 3′-PPT virus was indeed nonintegrated DNA, we next infected cells with the WT (with two amounts of viral particles) or 3′-PPT viruses, with or without DTG, and added efavirenz to the medium at various times postinfection to block the reverse-transcription step of newly synthesized viral particles (Fig. 4A). We assume that the blocking of virus propagation with efavirenz would lead progressively to the loss of nonintegrated viral genome while stably integrated viral DNA would be maintained. Thus, if the expression of viral proteins observed during the 3′-PPT infection is due to nonintegrated DNA, the loss of this nonintegrated genome following Efavirenz addition would lead to the loss of protein expression. Without efavirenz, the percentage of infected cells expressing the gag antigen (Fig. 4B), as well as the quantity of total viral DNA (Fig. 4D), increased during WT, 3′-PPT, and 3′-PPT+DTG infections confirming that the 3′-PPT mutant virus is highly resistant to DTG. As mentioned previously, supernatants from 3′-PPT infected cells contain infectious viral particles able to lead to de novo infection of fresh MT4-cells in the presence or absence of DTG (Fig. S2). Importantly, no gag antigen was detected following infection with the WT virus in the presence of DTG, showing that unintegrated viral DNA from the WT virus (WT+DTG infection) does not lead to viral production (Fig. 4B). As we assumed, the addition of efavirenz (at different times postinfection), led to different consequences depending on the virus used for infection (Fig. 4C and 4e). The percentage of gag positive cells remained constant after the addition of efavirenz when cells are infected with the WT virus independently of the initial quantity of virus used for the infection (Fig. 4C, left panel: WT-1, infection with 50 ng of p24 per million of cells, or right panel: WT-2, infection with 10 ng of p24 per million cells) showing that the stable signal for gag positive cells in the WT condition was not due to a high multiplicity of infection (several proviruses per cell) but rather to the nature of the viral genome (integrated vs nonintegrated). In this way, the amount of total viral DNA also remains constant (Fig. 4e, WT-1). Taken together, these results highlight the stability of the genome (integrated viral DNA) responsible of the viral protein expression in the WT condition. Alternatively, after the addition of efavirenz at different times postinfection (see figure legend), the percentage of gag positive cells, as well as the quantity of total viral DNA, decreased markedly for cells infected with the 3′-PPT mutant, with or without DTG (Fig. 4C and 4e). Conversely to the WT condition, the decrease of the percentage of Gag positive cells, concomitant to the decrease of total viral DNA, observed for the 3′-PPT mutant (+/− DTG) shows the instability of the viral genome, which is progressively lost after the blocking of the reverse-transcription of newly synthesized virus by Efavirenz. Therefore, these data confirm that nonintegrated viral DNA is responsible of the expression of viral proteins in the case of the 3′-PPT infection in both the presence and absence of DTG. Overall, our data highlight that the main genome involved in WT replication is integrated viral DNA and nonintegrated viral DNA in the case of the 3′-PPT mutant.
FIG 4.
Instability of the 3′-PPT mutant genome relative to that of the WT virus. (A) Experiment workflow. MT4 T cells were infected with the WT or 3′-PPT mutant virus, with or without 200 nM DTG. Two different quantities of virus were used for the WT virus (50 ng per million cells and 10 ng per million cells, WT-1 and WT-2, respectively). 100 ng of p24 per million cells were used for the 3′-PPT mutant infection. At 2 days postinfection, cells infected with WT-1 or WT-2 were split in two conditions: 100 nM Efavirenz (Efv) was added on part of cells in order to prevent further rounds of replication (samples WT-1, Efv at d2 and WT-2, Efv at d2) while the other part of cells remained without Efv (samples WT-1 and WT-2). For cells infected with the 3′-PPT virus, cells were split and 100 nM Efv was added at day 4 (sample 3′-PPT, Efv at d4). Cells without Efv have been again split at d5 for Efv addition (sample 3′-PPT, Efv at d5). For cells infected with the 3′-PPT virus + DTG, cells were split and 100 nM Efv was added at day 5 (sample 3′-PPT DTG, Efv at d5). Cells without Efv have been again split at d6 for Efv addition (sample 3′-PPT DTG, Efv at d6). Efv was added at different times postinfection, depending on the condition of infection, to add Efv when the percentage of gag+ cells was similar for all conditions. All samples were analyzed at different times points postinfection by intracellular gag antigen staining in order to measure the percentage of gag+ cells (B–C) or by qPCR analysis in order to quantify the total viral DNA (D–E). (B) Immunofluorescent staining was performed using an anti-Gag antibody and the percentage of gag+ cells is shown for the samples remained without Efv. The graph shows the mean ± SD of two independents experiments. (C) Left panel: Immunofluorescent staining was performed using an anti-Gag antibody and the percentage of gag+ cells is shown for the samples WT-1,Efv at d2, 3′-PPT, Efv at d5 and 3′-PPT DTG, Efv at d6. For these 3 samples, the percentage of Gag+ was between 20 and 30% when Efv was added. Right panel: Immunofluorescent staining was performed using an anti-Gag antibody and the percentage of gag+ cells is shown for the samples WT-2,Efv at d2, 3′-PPT, Efv at d4 and 3′-PPT DTG, Efv at d5. For these 3 samples, the percentage of Gag+ was between 3 and 12% when Efv was added. (D–E) Total viral DNA was quantified during the course of the experiment for samples remained without Efv (D) or for those in which Efv was added (E).
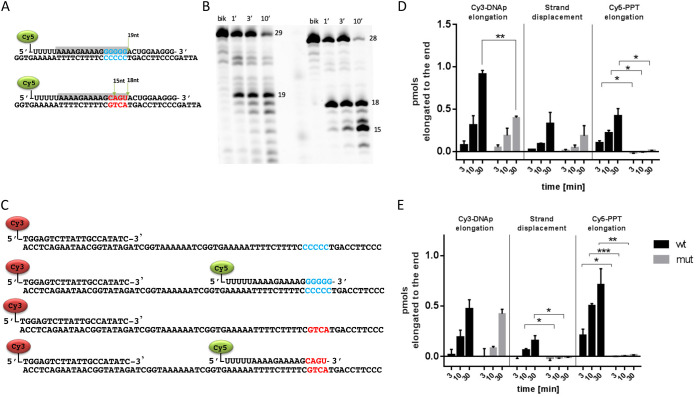
The behavior of the 3′-PPT mutant virus could be explained by hypothesizing that the mutated 3′-PPT sequence is not functional as a primer to initiate the plus strand cDNA synthesis. Since this priming event is critical for the second strand transfer reaction, its abrogation may cause the formation and accumulation of 1-LTRc. To verify such hypothesis we designed a set of experiments to dissect the impact of 3′-PPT mutations on the reverse transcription process. Firstly, we asked whether the mutant 3′-PPT was recognized and cleaved by RT. Results showed that the mutated sequence is recognized and cleaved at -18 and further cleaved at -15 nt (Fig. 5A and 5B), while the WT 3′-PPT is mainly cut only at -19 nt. Hence, one main cleavage takes place at -10 nt for both WT and mutated 3′-PPT, while the further −3 nt cleavage takes mainly place in the mutated 3′-PPT. In any case, the cleavage generates a fragment that could potentially serve as primer, that, to be functional, needs to be firstly elongated and then removed by strand displacement synthesis. Secondly, to verify whether these two events take place simultaneously, we designed an experiment using three oligonucleotides: an oligonucleotide DNA template annealed either with a Cy3-labeled DNA primer alone (to verify its elongation) or with both Cy3- labeled DNA primer and a Cy5-labeled RNA PPT, WT, or mutated (to verify both the strand displacement and PPT elongation events) (Fig. 5C). The substrate was incubated with wt RT enzyme, in presence and in the absence of dNTP mix, then the reaction product was detected with a dual channel imager (Fig. S4 in the supplemental material). Results showed that the Cy3- labeled DNA primer is fully elongated with both WT and mutated PPT, although the latter has a significant decrease in efficiency (Fig. 5D). Importantly, the Cy5-labeled RNA mutated PPT was not elongated by RT, demonstrating that it cannot serve as primer for plus strand cDNA synthesis (Fig. 5D). Differently the strand displacement synthesis occurred in the presence of both WT and mutated PPT, with no significant variations among the two (Fig. 5D). Noteworthy, the mutated PPT is further cleaved in a shorter fragment at -3nt.
FIG 5.
Selection and elongation of WT and mutant 3′-PPT. Substrates used for PPT cleavage assay (A). Reaction products resolved by high voltage denaturing 15% polyacrylamide gel electrophoresis (Acrylamide/bisacrylamide ratio 19:1, 7M Urea in Tris Borate EDTA buffer 1X) (B). Substrates used for PPT elongation and strand-displacement synthesis (C). PPT elongation and displacement by WT HIV-1 RT. Quantitative analysis of reaction products. (D) PPT elongation and displacement by RNase H defective E478Q HIV-1 RT. Quantitative analysis of reaction products (E). Bands were analyzed and quantified by ImageLab version 6.0.1. Mean ± standard deviation of two independent experiments; P values were calculated by paired, two-tailed t tests using GraphPad Prism 6.01 software (GraphPad Software, Inc.; San Diego, CA, USA). Figures were drawn with GraphPad Prism 6 version 6.01.
To ascertain the role of RT-associated RNase H activity in this process, the same experiment was performed with an RNase H deficient RT enzyme, carrying the E478Q substitution (Fig. S5 in the supplemental material). Results showed that the strand displacement DNA synthesis is completely abrogated for the substrate containing the mutated PPT, while still present and efficient for the WT PPT. This suggests that the RNase H cleavage of mutated PPT is required for its efficient displacement during the reverse transcription. Finally, we analyzed the independent effect of the C to U substitution at -6 form the 3′-PPT motif in a one-nucleotide longer Cy5-labeled oligo RNA (Fig. S6). Results showed that a longer Cy5-labeled PPT was internally cleaved at -7 with either C or U, in both the WT (Fig. S6a) or the mutated 3′-PPT (Fig. S6b). Quantitative analysis showed that the substitution did not affect the strand-displacement synthesis rate using the mutated 3′-PPT but increased significantly the strand-displacement synthesis for the WT PPT (Fig. S6c). The elongation of the longer mutated 3′-PPT was possible, even if it was significantly less efficient than the longer WT PPT, with or without the C to U substitution (Fig. S6d). Hence, the C to U substitution per se did not affect the elongation of mutated 3′-PPT.
This result suggests that the altered sequence and hence geometry of the mutated 3′-PPT compromises its efficient extension preventing the start of (+) strand synthesis from this primer and the correct accomplishment of canonical reverse transcription. Most probably, the (+) strand synthesis starts form that cPPT and the length of the so formed DNA fragment impedes the occurrence of the (+) strand transfer needed for the formation of the complete LTRs at both ends, required for an integration-competent cDNA.
Overall, we clearly show that replication of the 3′-PPT mutant is efficient and not impaired by DTG. Importantly, the replication of this mutant does not require integration but rather involves the 1-LTR circles that have been accumulated. Our results concerning 1-LTR circle accumulation are similar to those of Kantor et al., who reported increased 1-LTR circle formation of 3′-PPT mutants in the context of viral vectors (17). The authors reported that the reverse transcription process of certain 3′-PPT mutants directly led to an increase in the formation of 1-LTR circles, due to the fact that no strand displacement occurred due to disruption of the 3′-PPT.
Unintegrated viral DNA has been clearly shown to be involved in the expression of viral proteins (18–22). Its role in the synthesis of infectious viral particles and thus viral spread is a subject of debate. Indeed, it has been shown that, under specific conditions, nonintegrated genomes can lead to low-level viral replication (11, 22, 23).
Even though DTG is a highly potent integrase inhibitor, because of its stability on the integrase/DNA complex and its high genetic barrier, our data could explain why mutations in the integrase gene could not be detected in patients failing DTG-based treatment (24). Mutations in the 3′-PPT region led to a modification in the reverse transcription mechanism Thus, 1-LTR circles are not formed by recombination in the nucleus from linear viral DNA as in WT but directly from viral RNA. This synthesis leads to a stronger accumulation of 1-LTR circles in the case of the 3′-PPT mutant compared to the WT. This explains that the replication of the 3′-PPT mutant is more important compared to the WT with DTG and that DTG has no influence on the replication of the 3′-PPT virus. It is likely that resistance acquired against strand-transfer inhibitors can be due to mutations in the integrase gene, as well as other regions of the viral genome as previously suggested (25, 26), suggesting an important role of nonintegrated 1-LTR circles in the overall replication of the virus in patients treated with DTG-based regimens.
MATERIALS AND METHODS
Cells and viruses.
MT4 (from NIH AIDS Reference and Reagent Program) and CEMss cells were cultured in RPMI 1640 medium purchased from Gibco (Life Technologies Co.) supplemented with 10% fetal bovine serum (PAA Laboratories GmbH, Pasching, Austria) and 1% penicillin/streptomycin (100 units/mL). Cells were incubated at 37°C in a 5% C02 atmosphere. HIV-1 stocks WT (Laï strain) and 3′-PPT mutant, previously described (4), were prepared by amplification in MT4 cells.
Molecular clones of HIV.
MT-4 cells were infected by the WT (Laï) and the 3′-PPT viruses. DNA was extracted and the different part of the viral genomes were amplified by PCR using the Q5 Hot Start High-Fidelity Master Mix (New England Biolabs). The NEBuilder HiFi DNA Assembly was used in order to obtain the two viral constructions: the WT (Laï) and 3′-PPT viruses according to the manufacturer’s instructions (New England Biolabs). The D116N mutation was then introduced into the WT and 3′-PPT viruses using the QuikChange II XL site-directed mutagenesis kit (Agilent) leading to the D116N Nef+ and D116N 3′-PPT Nef- viruses, respectively. Since the 3′-PPT mutation leads to the disruption of the Nef Open reading frame, we restore the Nef open in the D116N construction leading to the D116N 3′-PPT Nef+ virus (5′-GCAGTT). Conversely, we disrupted the Nef open reading frame by removing one base (cytosine in position 11) leading to the D116N Nef- virus (5′-GGGGGGACTGGAAGGG-T). All constructions were fully sequenced.
The HIV-1 p24gag antigen content in viral inocula was determined by enzyme-linked immunosorbent assay (Perkin-Elmer Life Sciences).
Quantification of HIV-1 DNA and viral RNA extraction.
In total, 2-5 × 106 cells were collected during the course of infection. The cell pellet was washed in PBS. DNA and viral RNA were purified with the QIAamp DNA blood minikit (Qiagen) and RNeasy minikit (Qiagen), respectively, according to the manufacturer’s instructions. All DNA quantifications were performed by real-time PCR on the same apparatus. They were performed on nucleic-acid amounts equivalent to ≈200,000 cells. The sequences of the primers and probes used for qPCR for total HIV-1, 2-LTRc, 1-LTRc, linear viral DNA, and integrated viral DNA absolute quantification have been described previously (16). The copy number of the various nucleic-acid forms was determined from calibration curves obtained by amplification of pre-determined amounts (from 10 to 105 copies) of a plasmid carrying the matching sequences. The copy number of integrated viral DNA (iDNA) was first determined from a calibration curve obtained by concomitant two-stage PCR amplification of serial dilutions of iDNA contained in HeLa R7 Neo cells (16). The number of cell equivalents in each amplification was deduced by amplifying the β-globin gene. Integrated viral DNA quantification was improved by increasing the number of PCR cycles during the first Alu-LTR PCR (20 cycles instead of 12). In this setting, absolute quantification was not possible. This setting only allowed the detection of integrated viral DNA. Linear viral DNA was quantified using ligation-mediated PCR, as previously reported (16). Briefly, a linker compatible with the 3′-processed end of the linear DNA was used. After ligation, the DNA was purified and quantified using quantitative PCR with primers hybridizing in the linker and LTR.
Intracellular Gag staining for flow cytometry analysis.
WT and 3′-PPT replications were followed by gag intracellular immunostaining: after different times postinfection, infected cells were collected and were fixed with 4% paraformaldehyde 10 min at room temperature, washed with PBS, permeabilized with PBS-0.5% BSA-0.05% Saponin for 5 min at room temperature and stained with the anti-gag antibody KC57-RD1 (phycoerythrin (PE)) (Beckman Coulter 6604667) 1 h at 4°C. Fluorescence of PE positive cells was acquired using the flow cytometer FACSCalibur (BD Biosciences) and further analyzed using FlowJo software.
Integration site analysis.
To remove any nonintegrated viral DNA (one-, or two-LTR circles), 5 μg of genomic DNA (gDNA) samples isolated from infected MT4 cells were subjected to 0.6% agarose gel electrophoresis and high-molecular gDNA was isolated from the gel using the Zymoclean Large Fragment DNA Recovery Kit (Zymo Research). The eluents were sonicated to an average fragment length of 600 bp in screw-cap cuvettes with a Covaris M220 ultrasonicator with the following settings: peak power: 50.0, duty factor: 20, cycle/burst: 200, duration: 28 s. After bead purification, the DNA was end-repaired and 5′-phosphorylated with the NEBNext End Repair Module (New England Biolabs, (NEB)). The DNA was prepared for ligation with the NEBNextdA-Tailing Module (NEB) and eluted after bead purification in 10 μL water. Ligation with double-stranded linkers was performed in 15 μL for 15 min at room temperature using the Blunt/TA Ligase Master Mix (NEB). After purification with 0.8 volumes of AMPure XP beads (Beckman Coulter), the ligated DNA was eluted in 20 μL 10 mM Tris/HCl, pH 8.0 and the entire DNA solution used for multiple PCRs to amplify the virus-gDNA junctions with HIV1 primers and a linker primer using NEBNext High-Fidelity 2x PCR Master Mix (NEB) with the following cycling conditions: 98°C for 30 s; 20 cycles of 98°C for 10 s, 68°C for 30 s, and 72°C for 30 s; 10 cycles of 98°C for 10 s; ramp to 63°C at 1°C/s for 30 s; 72°C for 30 s; 72°C for 3 min. The PCR products were isolated using one volume of AMPure XP beads (Beckman Coulter), eluted in 20 μL 10 mM Tris/HCl, pH 8.0 and 2 μL of the eluents were used as template for five parallel PCRs with the primers: SIN-HIV-BC-N-Ill and PE-nest ind-N (where N stands for the sequences of Illumina TrueSeq indexes or their corresponding reverse-complement sequences) using the following cycling conditions: 98°C for 30 s; 20 cycles of 98°C for 10 s, 67°C for 30 s, and 72°C for 30 s; 72°C for 3 min. The 200–500 bp size range of the indexed libraries were agarose gel-isolated and mixed equimolarly for 100-base, single-end Illumina sequencing on a HiSeq 2000 instrument using a 40% PhiX DNA spike-in at Genewiz (USA).
Enzymes.
HIV-1 RT group M subtype B. Heterodimeric RT was expressed and purified essentially as described (27). Briefly, E. coli strain M15 containing the p6HRT-prot vector was grown to an optical density at 600 nm of 0.7 and induced with 1.7 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 4 h. Protein purification was carried out with a BioLogic LP system (Bio-Rad), using a combination of immobilized metal affinity and ion-exchange chromatography. Cell pellets were resuspended in lysis buffer (50 mM sodium phosphate buffer pH 7.8, containing 0.5 mg/mL lysozyme), incubated on ice for 20 min, and, after adding NaCl to a final concentration of 0.3 M, were sonicated and centrifuged at 30,000 × g for 1 h. The supernatant was loaded onto a Ni2+−NTA-Sepharose. RT was eluted with an imidazole gradient in wash buffer (0 – 0.5 M). Enzyme-containing fractions were pooled and diluted 1:1 with 50 mM sodium phosphate buffer pH 7.0, containing 10% glycerol; and then loaded into a Hi-trap heparin HP GE (Healthcare Lifescience) RT was eluted with Elute Buffer 2 (50 mM Sodium Phosphate pH 7.0, 10% glycerol, 1 M NaCl), protein was dialyzed and stored in buffer containing 50 mM Tris HCl pH 7.0, 25 mM NaCl, 1 mM EDTA, and 50% glycerol. Catalytic activities and protein concentrations were determined. Enzyme-containing fractions were pooled, and aliquots were stored at −80°C. Amino acidic substitution E478Q was introduced in the p6HRT-prot vector QuikChange mutagenesis kit by following the manufacturer’s instructions (Agilent Technologies Inc., Santa Clara, CA).
Assays.
RNA and DNA oligos were purchased form METABION, HPLC purified (oligo1 5′-Cy5-uuu uaa aag aaa agg ggg gac ugg aag gg-3′, oligo2 5′-ATT AGC CCT TCC AGT CCC CCC TTT TCT TTT AAA AAG TGG C-3′) (oligo 3 5′-Cy5-uuu uaa aag aaa agcagu ac ugg aag gg-3′, oligo 4 5′-ATT AGC CCT TCC AGT ACTGC TTT TCT TTT AAA AAG TGG C-3′; oligo 5 5′-Cy5-uuu uaa aag aaa agg ggg g-3′; oligo 6 5′-Cy5-u uuu uaa aag aaa agc agu-3′; oligo 7-TGGAGTCTTATTGCCATATC-3′; oligo 8 5′-CCC TCC AGT ACT 5′-Cy3GCT TTT CTT TTA AAA AGT GGC TAA AAA ATG GCT AGA TAT GGC AAT AAG ACT CCA-3′; oligo 9 5′-CCC TCC AGT CCC CCC TTT TCT T TT AAA AAG TGG CTA AAAA AGT GGC TAG ATA TGG CAA TAA GAC TCC A-3′; oligo 10 5′-Cy5 CUU UUU AAA AGA AAA GGG GGG-3′; oligo 11 5′-Cy5 UUU UUU AAA AGA AAA GGG GGG-3′; oligo 12 5′-Cy5- CUU UUU AAA AGA AAA GCA GU-3′; 5′-Cy5-UUU UUU AAA AGA AAA GCA GU-3′). The annealing was performed by warming at 95°C for 2 min and cool to storage temperature of 4°C. The reaction was initiated by adding 1 μL of 60 mM MgCl2 to 9 μL of mixture containing 100 ng of enzyme, 200 nM substrate, 50 mM Tris, pH 7.8, 80 mM KCl, and 2 mM DTT, with or without 0.2 mM dNTP mix, at 37°C. Reaction was quenched at selected time point with an equal volume of 8 M urea in Tris borate-EDTA buffer. Reaction products were resolved by high voltage denaturing 15% polyacrylamide gel electrophoresis (Acrylamide/bisacrylamide ratio 19:1, 7M Urea in Tris Borate EDTA buffer 1X) and visualized by fluorescent dual channel imaging (ChemiDoc Bio-Rad). Bands were analyzed and quantified by ImageLab version 6.0.1.
Statistics.
Data, error bars (standard deviation, standard error of the mean), and statistical tests were analyzed and calculated using Microsoft Excel (Microsoft, Redmond, WA) and Prism (GraphPad Software; La Jolla, CA).
Footnotes
Supplemental material is available online only.
Contributor Information
Olivier Delelis, Email: delelis@lbpa.ens-cachan.fr.
Guido Silvestri, Emory University.
REFERENCES
- 1.Quashie PK, Mesplède T, Wainberg MA. 2013. Evolution of HIV integrase resistance mutations. Curr Opin Infect Dis 26:43–49. 10.1097/QCO.0b013e32835ba81c. [DOI] [PubMed] [Google Scholar]
- 2.Quashie PK, Mesplède T, Wainberg MA. 2013. HIV drug resistance and the advent of integrase inhibitors. Curr Infect Dis Rep 15:85–100. 10.1007/s11908-012-0305-1. [DOI] [PubMed] [Google Scholar]
- 3.Hikichi Y, Van Duyne R, Pham P, Groebner JL, Wiegand A, Mellors JW, Kearney MF, Freed EO. 2021. Mechanistic analysis of the broad antiretroviral resistance conferred by HIV-1 envelope glycoprotein mutations. mBio 12:e03134-20. 10.1128/mBio.03134-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malet I, Subra F, Charpentier C, Collin G, Descamps D, Calvez V, Marcelin A-G, Delelis O. 2017. Mutations located outside the integrase gene can confer resistance to HIV-1 integrase strand transfer inhibitors. mBio 8:e00922-17. 10.1128/mBio.00922-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rausch JW, Le Grice SFJ. 2004. ‘Binding, bending and bonding’: polypurine tract-primed initiation of plus-strand DNA synthesis in human immunodeficiency virus. Int J Biochem Cell Biol 36:1752–1766. 10.1016/j.biocel.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 6.Wijting I, Rokx C, Boucher C, van Kampen J, Pas S, de Vries-Sluijs T, Schurink C, Bax H, Derksen M, Andrinopoulou E-R, van der Ende M, van Gorp E, Nouwen J, Verbon A, Bierman W, Rijnders B. 2017. Dolutegravir as maintenance monotherapy for HIV (DOMONO): a phase 2, randomised non-inferiority trial. Lancet HIV 4:e547–e554. 10.1016/S2352-3018(17)30152-2. [DOI] [PubMed] [Google Scholar]
- 7.Hachiya A, Kubota M, Shigemi U, Ode H, Yokomaku Y, Kirby KA, Sarafianos SG, Iwatani Y. 2022. Specific mutations in the HIV-1 G-tract of the 3’-polypurine tract cause resistance to integrase strand transfer inhibitors. J Antimicrob Chemother 77:574–577. 10.1093/jac/dkab448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith SJ, Ferris A, Zhao X, Pauly G, Schneider JP, Burke TR, Hughes SH. 2021. INSTIs and NNRTIs potently inhibit HIV-1 polypurine tract mutants in a single round infection assay. Viruses 13:2501. 10.3390/v13122501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei Y, Sluis-Cremer N. 2021. Mutations in the HIV-1 3’-polypurine tract and integrase strand transfer inhibitor resistance. Antimicrob Agents Chemother 65:e02432-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thierry S, Munir S, Thierry E, Subra F, Leh H, Zamborlini A, Saenz D, Levy DN, Lesbats P, Saïb A, Parissi V, Poeschla E, Deprez E, Delelis O. 2015. Integrase inhibitor reversal dynamics indicate unintegrated HIV-1 dna initiate de novo integration. Retrovirology 12:24. 10.1186/s12977-015-0153-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irwan ID, Karnowski HL, Bogerd HP, Tsai K, Cullen BR. 2020. Reversal of epigenetic silencing allows robust HIV-1 replication in the absence of integrase function. mBio 11:e01038-20. 10.1128/mBio.01038-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakajima N, Lu R, Engelman A. 2001. Human immunodeficiency virus type 1 replication in the absence of integrase-mediated DNA recombination: definition of permissive and nonpermissive T-cell lines. J Virol 75:7944–7955. 10.1128/JVI.75.17.7944-7955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brussel A, Delelis O, Sonigo P. 2005. Alu-LTR real-time nested PCR assay for quantifying integrated HIV-1 DNA. Methods Mol Biol Clifton NJ 304:139–154. [DOI] [PubMed] [Google Scholar]
- 14.Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA, Espeseth A, Gabryelski L, Schleif W, Blau C, Miller MD. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287:646–650. 10.1126/science.287.5453.646. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Olvera JM, Yoder KE, Mitchell RS, Butler SL, Lieber M, Martin SL, Bushman FD. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J 20:3272–3281. 10.1093/emboj/20.12.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munir S, Thierry S, Subra F, Deprez E, Delelis O. 2013. Quantitative analysis of the time-course of viral DNA forms during the HIV-1 life cycle. Retrovirology 10:87. 10.1186/1742-4690-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kantor B, Bayer M, Ma H, Samulski J, Li C, McCown T, Kafri T. 2011. Notable reduction in illegitimate integration mediated by a PPT-deleted, nonintegrating lentiviral vector. Mol Ther 19:547–556. 10.1038/mt.2010.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Y, Marsh JW. 2001. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science 293:1503–1506. 10.1126/science.1061548. [DOI] [PubMed] [Google Scholar]
- 19.Cara A, Cereseto A, Lori F, Reitz MS. 1996. HIV-1 protein expression from synthetic circles of DNA mimicking the extrachromosomal forms of viral DNA. J Biol Chem 271:5393–5397. 10.1074/jbc.271.10.5393. [DOI] [PubMed] [Google Scholar]
- 20.Sloan RD, Wainberg MA. 2011. The role of unintegrated DNA in HIV infection. Retrovirology 8:52. 10.1186/1742-4690-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trinité B, Ohlson EC, Voznesensky I, Rana SP, Chan CN, Mahajan S, Alster J, Burke SA, Wodarz D, Levy DN. 2013. An HIV-1 replication pathway utilizing reverse transcription products that fail to integrate. J Virol 87:12701–12720. 10.1128/JVI.01939-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan CN, Trinité B, Lee CS, Mahajan S, Anand A, Wodarz D, Sabbaj S, Bansal A, Goepfert PA, Levy DN. 2016. HIV-1 latency and virus production from unintegrated genomes following direct infection of resting CD4 T cells. Retrovirology 13:1. 10.1186/s12977-015-0234-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irwan ID, Cullen BR. 2021. Tax induces the recruitment of NF-κB to unintegrated HIV-1 DNA to rescue viral gene expression and replication. J Virol 95:e0028521. 10.1128/JVI.00285-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molina J-M, Clotet B, van Lunzen J, Lazzarin A, Cavassini M, Henry K, Kulagin V, Givens N, de Oliveira CF, Brennan C, FLAMINGO study team . 2015. Once-daily dolutegravir versus darunavir plus ritonavir for treatment-naive adults with HIV-1 infection (FLAMINGO): 96 week results from a randomised, open-label, phase 3b study. Lancet HIV 2:e127–e136. 10.1016/S2352-3018(15)00027-2. [DOI] [PubMed] [Google Scholar]
- 25.Darcis G, Berkhout B. 2018. Human immunodeficiency virus resistance to dolutegravir: are we looking in the wrong place? J Infect Dis 218:2020. 10.1093/infdis/jiy474. [DOI] [PubMed] [Google Scholar]
- 26.Thierry E, Deprez E, Delelis O. 2016. Different pathways leading to integrase inhibitors resistance. Front Microbiol 7:2165. 10.3389/fmicb.2016.02165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corona A, Di Leva FS, Thierry S, Pescatori L, Cuzzucoli Crucitti G, Subra F, Delelis O, Esposito F, Rigogliuso G, Costi R, Cosconati S, Novellino E, Di Santo R, Tramontano E. 2014. Identification of highly conserved residues involved in inhibition of HIV-1 RNase H function by Diketo acid derivatives. Antimicrob Agents Chemother 58:6101–6110. 10.1128/AAC.03605-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S6. Download jvi.00676-22-s0001.pdf, PDF file, 4.4 MB (4.4MB, pdf)