ABSTRACT
Human mannose receptor 1 (MRC1) is a cell surface receptor expressed in macrophages and other myeloid cells that inhibits human immunodeficiency virus type 1 (HIV-1) particle release by tethering virions to producer cell membranes. HIV-1 counteracts MRC1 expression by inhibiting mrc1 transcription. Here, we investigated the mechanism of MRC1 downregulation in HIV-1-infected macrophages. We identified the myeloid cell-specific transcription factor PU.1 as critical for regulating MRC1 expression. In the course of our study, we recognized a complex interplay between HIV-1 Tat and PU.1 transcription factors: Tat upregulated HIV-1 gene expression but inhibited mrc1 transcription, whereas PU.1 inhibited HIV-1 transcription but activated MRC1 expression. Disturbing this equilibrium by silencing PU.1 resulted in increased HIV-1 gene expression and reduced MRC1 promoter activity. Our study identified PU.1 as a central player in transcriptional control, regulating a complex interplay between viral and host gene expression in HIV-infected macrophages.
IMPORTANCE HIV-1 replication in primary human cells depends on the activity of virus-encoded proteins but also involves cellular factors that can either promote (viral dependency factors) or inhibit (host restriction factors) virus replication. In previous work, we identified human MRC1 as a macrophage-specific host restriction factor that inhibits the detachment of viral particles from infected cells. Here, we report that HIV-1 counteracts this effect of MRC1 by imposing a transcriptional block on cellular MRC1 gene expression. The transcriptional inhibition of the MRC1 gene is accomplished by Tat, an HIV-1 factor whose best-described function actually is the enhancement of HIV-1 gene expression. Thus, HIV-1 has evolved to use the same protein for (i) activation of its own gene expression while (ii) inhibiting expression of MRC1 and other host factors.
KEYWORDS: HIV-1, PU.1 transcription factor, Tat, transcriptional repression, mannose receptor
INTRODUCTION
Secretion of virus particles from human immunodeficiency virus type 1 (HIV-1)-infected monocyte-derived macrophages (MDMs) is inhibited by two mechanisms, (i) tethering of virus particles to the cell surface by BST-2 (1, 2), and (ii) tethering of progeny virions to the cell surface through an interaction with human mannose receptor 1 (MRC1) (3). While retention of mature and fully infectious virions at the surface of infected cells is likely to promote cell-to-cell transmission of HIV-1 and thus may be advantageous for localized spread within a tissue or organ, the virus makes a serious effort to counteract the effects of BST-2 and MRC1, presumably to allow efficient spread into the peripheral organs. Indeed, different lentiviruses employ different strategies to accomplish that goal. HIV-1, for instance, employs Vpu to counteract the antiviral activity of BST-2. Vpu downregulates BST-2 from the cell surface by interfering with the cycling of BST-2 from internal membranes to the plasma membrane (reviewed in references 4 and 5). On the other hand, HIV-2 and some simian immunodeficiency virus (SIV) strains lack Vpu. Instead, they encode Env glycoproteins with the ability to antagonize BST-2 (6–10). Finally, some SIV strains employ Nef to accomplish this goal (8, 11, 12). Regarding MRC1-based antiviral activity, we previously reported that HIV-1 counteracts the inhibitory effect of MRC1 by reducing its intracellular steady-state expression. Indeed, we found that reduction of MRC1 protein expression in HIV-infected MDMs was paralleled by a reduction of MRC1 mRNA levels; however, the underlying mechanism remained unclear (3, 13–15).
The regulation of the mannose receptor 1 promoter (p-mrc1) in uninfected MDMs is poorly understood even though the mrc1 gene was among a group of more than 100 cellular genes found to be regulated by the myeloid-specific transcription factor PU.1 (16, 17). PU.1 is a member of the Ets family of transcription factors and is expressed exclusively in cells of the hematopoietic lineage (18). It is a critical regulator of hematopoiesis, and mice homozygous for mutation of the PU.1 DNA binding domain die within a few days after birth due to septicemia (16–19). PU.1 was discovered in 1988 as a putative oncogene encoded by the Spi-1 locus, which was associated with virally induced murine erythroleukemia (20). During hematopoiesis, PU.1 is required for the development of both lymphoid and myeloid lineages (reviewed in references 21 and 22). The 272-residue PU.1 protein is made up of a 118-residue transcriptional activation domain at its N terminus, a 42-residue PEST motif in the center, and a 110-residue DNA-binding domain (ETS domain) at its C terminus (23). Phosphorylation at a conserved serine residue (S148 in the PEST domain) was reported to be important for protein-protein interactions and transcriptional activity of PU.1 (24). Recently, PU.1 was identified as a target for caspase-3 that can cleave the protein at two sites, thereby separating its DNA binding domain from the transactivating and PEST domains (25). PU.1 interacts with DNA as a monomer and recognizes DNA sequences containing the “PU-box,” a GGAA or AGAA nucleotide motif (19, 26, 27). Indeed, the sequence of the human MRC1 promoter (28) predicts two potential PU.1 binding sites; however, transcriptional regulation of the MRC1 promoter by PU.1 has not been experimentally tested to date.
Previous studies have implicated the viral Tat, Nef, and Vpr proteins in MRC1 downmodulation, although the relative contribution of each of these factors remains under debate (13, 14). In particular, Nef does not affect cellular steady-state levels of MRC1, and its effect is limited to the internalization of cell surface MRC1 (14). On the other hand, HIV-1 Tat was reported to act at the transcriptional level by inhibiting the basal activity of the rat mannose receptor promoter, which is about 60% identical to the human MRC1 promoter (13). However, the impact of Tat on the regulation of human mannose receptor was found to be insignificant by others, even though they did observe a transcriptional component in inhibition of MRC1 expression (15). That same study implicated Nef and Vpr in the inhibition of MRC1 expression via a concerted action that involved an interaction of Vpr with DCAF1, which was indicative of proteasome-mediated MRC1 degradation (15).
The goal of the current study was to further investigate the mechanism of MRC1 downregulation in HIV-1-infected macrophages. We identified a complex interplay between HIV-1 Tat and the myeloid-specific transcription factor PU.1, which upregulates the MRC1 promoter. Interestingly, while Tat activates HIV-1 gene expression through a positive feedback loop, PU.1 is involved in a negative feedback loop that acts on the HIV-1 LTR promoter and inhibits viral gene expression, including Tat. Tat, on the other hand, inhibits the activity of PU.1, thereby antagonizing the inhibitory effect of PU.1 on long terminal repeat (LTR)-based transcription. Thus, we found that there is a complex equilibrium between viral and host gene expression in HIV-infected macrophages. Indeed, disturbing this equilibrium, for instance, by silencing PU.1, had opposing effects and resulted in increased HIV-1 gene expression while reducing MRC1 promoter activity. Our work has important implications for understanding how HIV-1 gene expression is regulated in HIV-infected macrophages.
RESULTS
HIV-1 infection of MDMs reduces expression of MRC1 and PU.1.
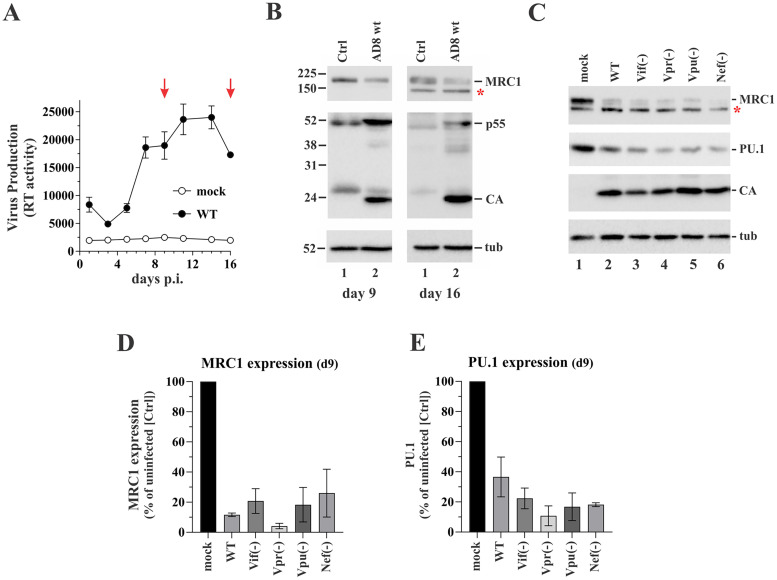
We initially analyzed the impact of HIV-1 infection on the expression of MRC1 in the course of a spreading infection of primary human MDMs (Fig. 1A). Protein expression was assessed by immunoblotting on days 9 and 16 postinfection (Fig. 1B). Consistent with our previous report (3), we found that MRC1 expression was downregulated relative to uninfected cells at both time points. To evaluate which HIV-1 gene products are involved in MRC1 downmodulation, we infected MDMs with recombinant vesicular stomatitis virus G protein (VSVg)-pseudotyped AD8 variants carrying knockout mutations in individual accessory genes (Fig. 1C). The use of VSVg-pseudotyped viruses allowed us to achieve comparable efficiency of infection by all viral mutants, irrespective of their ability to initiate a spreading infection in MDMs. As expected, probing viral Gag expression on day 9 postinfection revealed comparable amounts of cell-associated Gag protein (Fig. 1C, CA). Surprisingly, inactivation of individual accessory genes, including vpr and nef, previously reported to be involved in inhibition of MRC1 expression (15) did not affect the ability of HIV-1 to downmodulate MRC1 expression in two independent experiments (Fig. 1D). Consistent with the possible role of PU.1 in regulating MRC1 expression, downmodulation of endogenous PU.1 was evident in all infected samples as well (Fig. 1E). Thus, our results indicate that HIV-1 infection of MDMs leads to the silencing of both MRC1 and PU.1 through a mechanism that does not depend on expression of individual viral accessory proteins.
FIG 1.
HIV-1 replication in MDM induces the downmodulation of mannose receptor 1 (MRC1) and PU.1. (A) Terminally differentiated human monocyte-derived macrophages (MDMs) were infected in triplicate with the R5-tropic HIV-1 isolate AD8 (WT). Virus replication was monitored for 16 days by measuring the virus-associated reverse transcriptase (RT) assay. Uninfected MDMs were cultured in parallel (mock). On days 9 and 16 (marked by red arrows), parts of the cells were removed and processed for immunoblotting. Mean and error bars representing standard error of the mean (SEM) calculated from triplicate infections are shown. (B) Immunoblotting was performed on days 9 and 16 on whole-cell extracts using antibodies to MRC1, HIV-1 (CA, p55), and tubulin (tub). Molecular mass markers are on the left. Proteins are identified on the right. (C) MDMs were infected with concentrated, VSVg-pseudotyped stocks of AD8 WT or AD8 variants lacking expression of individual viral proteins as indicated at the top. Cells were harvested 9 days postinfection and processed for immunoblot analysis using antibodies to MRC1, PU.1, HIV (CA), and tubulin (tub). A representative blot of two independent experiments is shown. A nonspecific band below the MRC1 band (marked by a star in panels B and C) is seen in some experiments. Its intensity varies in a donor-dependent manner. (D and E) MRC- and PU.1-specific bands from panel C were quantified by image analysis and expressed relative to signals observed in the uninfected (mock) cells. Mean and error bars representing SEM calculated from two independent infections are shown.
Viruses expressing Tat86 or Tat101 do not differ in their ability to reduce MRC1 expression.
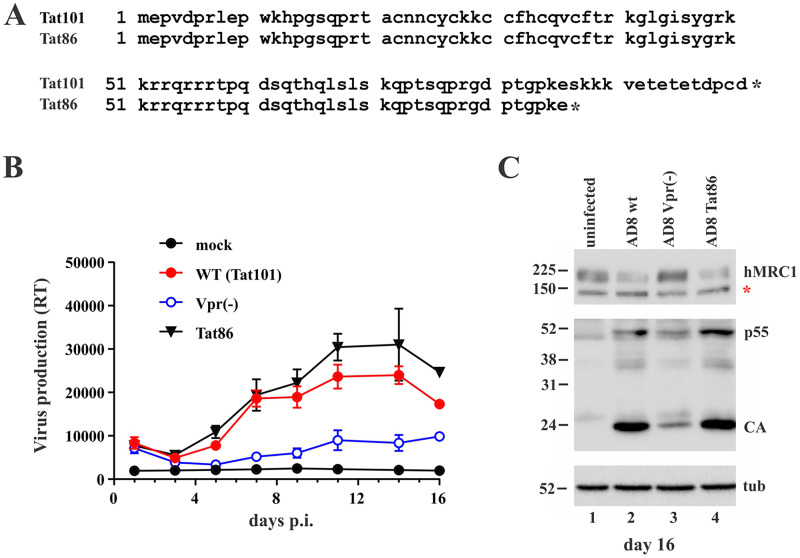
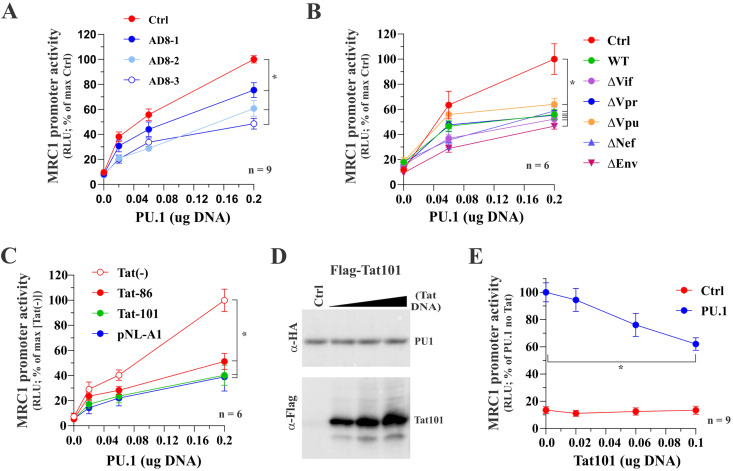
The experiment shown in Fig. 1 did not address the possible impact of HIV-1 Tat on MRC1 downmodulation. Indeed, Tat was previously implicated in the silencing of the rat MRC1 promoter (13), although others were unable to verify an independent function of Tat (15). HIV-1 Tat exists in two isoforms: the R5-tropic isolate AD8, for instance, expresses a 101-residue protein (Tat101), whereas the X4-tropic NL43 isolate encodes a shorter, 86-amino-acid form (Tat86) due to the presence of a stop codon in the second coding exon of Tat (Fig. 2A). It is conceivable that the highly charged C-terminal 15 residues in Tat101 are important for the replication of the R5-tropic AD8 in MDMs and/or the ability to downmodulate MRC1. To test that possibility, we inserted a stop codon in the Tat ORF of pAD8, resulting in an AD8 variant expressing the shorter Tat86. Importantly, changing Ser87 (TCG) to TAG (stop) in the Tat ORF did not affect the amino acid sequence of the overlapping Env and Rev ORFs. A Vpr-defective variant of AD8 was included as a reference for attenuated virus replication. In this experiment, infection of MDMs was initiated with nonpseudotyped virus stocks to allow for spreading of infection in the culture. We found that eliminating the highly charged C-terminal domain of Tat101 did not impair the replication fitness of the AD8 Tat86 variant in MDMs. In fact, the AD8 Tat86 virus replicated slightly better than the parental virus (Fig. 2B). Of note, immunoblot analysis of cell extracts on day 16 postinfection showed comparable levels of viral Gag proteins for both Tat variants and revealed comparable downmodulation of MRC1 (Fig. 2C, lanes 2 and 4). As expected, Vpr-defective AD8 virus replicated poorly in MDMs, as evident by the low replication profile (Fig. 2B, open blue circles) and the low Gag signal (Fig. 2C, lane 3) and, consequently, did not cause downmodulation of MRC1 (Fig. 2C, lane 3). Thus, our data confirm that productive HIV-1 infection of MDMs leads to the downmodulation of MRC1. This effect is, however, independent of the respective Tat isoform.
FIG 2.
Replication of HIV-1 AD8 in MDMs is not impacted by different Tat isoforms. (A) AD8 encodes a 101-residue Tat protein (Tat101), while HIV-1 NL-43 encodes a shorter, 86-residue protein due to a premature stop codon in the 2nd coding exon (Tat86). (B) MDMs were infected with wild-type AD8 (Tat101, red circles) or AD8 Tat86 (black triangles). A Vpr-defective AD8 variant (encoding Tat101) known to exhibit an attenuated replication phenotype in MDMs was included as a reference (blue open circles). Mock-infected cells (black circles) were cultured in parallel. Virus replication was monitored for 16 days by RT assay. Mean and error bars representing SEM from triplicate infections are shown. (C) Cells from panel B were collected on day 16 and processed for immunoblotting using antibodies to human MRC1 (hMRC1), HIV-1 (p55, CA), and tubulin (tub). The red star in the hMRC1 blot indicates a nonspecific background band as noted in Fig. 1B and C.
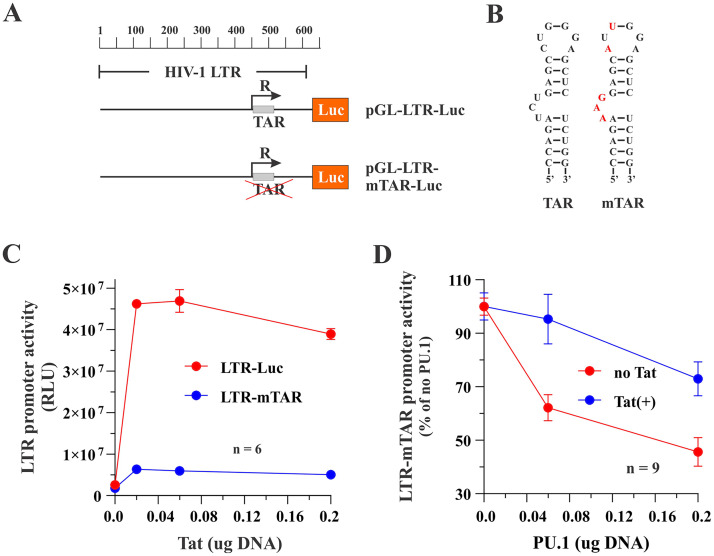
Tat does not affect the basal activity of the human MRC1 promoter.
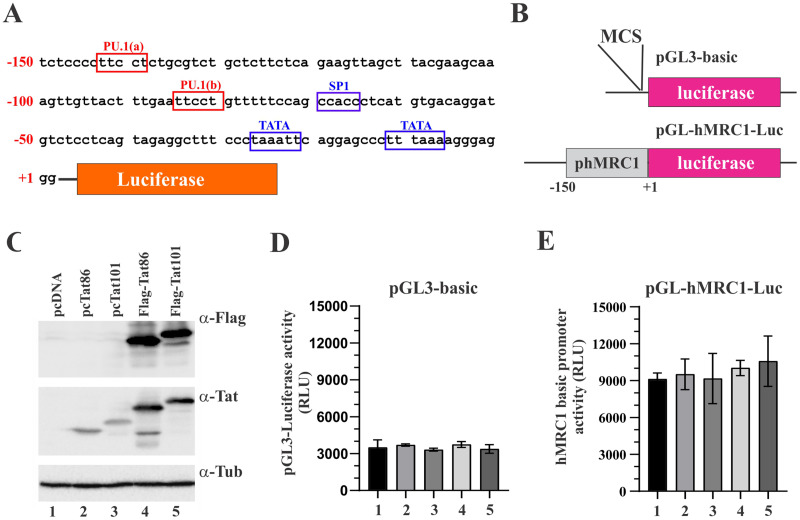
We previously reported that the reduction of MRC1 protein levels in infected MDMs is at least in part due to transcriptional silencing of the MRC1 promoter (3). Since none of the HIV-1 accessory proteins assayed in Fig. 1C showed an impact on the virus-induced silencing of MRC1 expression in infected MDMs, we wanted to assess the possible impact of the HIV-1 Tat protein. HIV-1 Tat is a well-studied transcriptional activator, but it has also been implicated in additional functions (reviewed in references 29 to 32). To assess a possible impact of HIV-1 Tat on the transcriptional activity of the MRC1 promoter, we cloned a 150-bp synthetic DNA fragment (IDT; gBlock) (Fig. 3A) containing the human mannose receptor promoter (28) into the promoter-trap vector pGL3 (Promega) carrying a luciferase indicator gene downstream of a multicloning site (MCS) (Fig. 3B). The MRC1 promoter sequence encoded in the resulting pGL-hMRC1-Luc vector contains several known transcriptional regulatory elements, including two TATA boxes and one SP1 site, as well as two potential PU.1 binding sites (TTCCT). When tested in transfected HEK293T cells, the presence of the MRC1 promoter sequence only very modestly (about 3-fold) upregulated luciferase activity compared to the empty pGL3-basic vector, indicating that, in 293T cells, the hMRC1 sequence has very poor promoter activity (Fig. 3D and E, column 1). Coexpression of Tat86 or Tat101, either in untagged or Flag-tagged form (Fig. 3C), did not appear to have an impact on hMRC1 promoter activity (Fig. 3D and E, compare columns 2 to 5 to column 1; for statistics, see Fig. S1). We conclude that HIV-1 Tat neither activated nor inhibited the basal activity of the MRC1 promoter in HEK293T cells.
FIG 3.
Human mannose receptor promoter has low basal activity in HEK293T cells. (A) A 150-bp fragment of the hMRC1 promoter (positions −1 to −150) was cloned into the promoter trap vector pGL3-basic (Promega). Known (blue) or predicted (red) transcriptional elements are boxed. (B) The structures of pGL3-basic and pGL-hMRC1-Luc are shown schematically. (C) Tat expression vectors used in this study. Tat86 and Tat101 were cloned into pcDNA3.1 either in untagged form (lanes 2 and 3) or carrying an N-terminal triple-Flag epitope (lanes 4 and 5). Expression of Tat was assessed by immunoblotting using antibodies to the Flag epitope (α-Flag) or Tat (α-Tat). Tubulin expression was monitored as a loading control. (D and E) Expression of luciferase from pGL3-basic (D) and pGL-hMRC1-Luc (E) in the presence or absence of Tat. HEK293T cells were transfected in 24-well plates with pGL3-basic (0.25 μg) or pGL-hMRC1-Luc (0.25 μg) together with empty vector (pUC19; 0.25 μg) or vectors encoding Tat variants (0.25 μg each) as indicated in panel C. Luciferase production was measured 48 h later. Mean and error bars reflecting SEM calculated from three independent transfections are shown. Differences between samples in columns 2 to 5 were not statistically significant from those in column 1 in both panels D and E (see Fig. S1 in the supplemental material).
The myeloid-specific transcription factor PU.1 activates the MRC1 promoter.
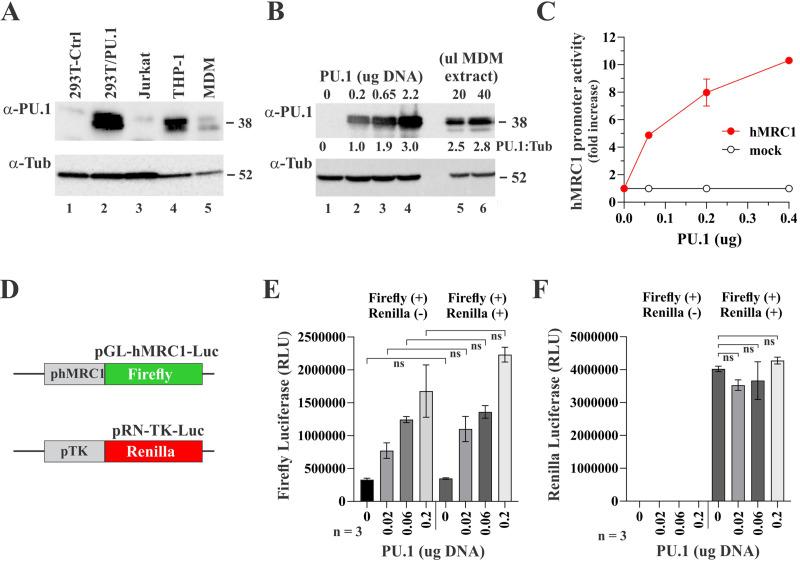
As noted above, the sequence for the MRC1 promoter reveals the presence of two potential PU.1 binding sites [identified as PU.1(a) and PU.1(b) in Fig. 3A]. PU.1 is a myeloid-specific transcription factor that is involved in macrophage proliferation (33) and also directs the tissue-specific expression of the macrophage colony-stimulating factor (MCSF) receptor (34). Furthermore, PU.1 has the ability to either repress or activate gene expression during T cell development by redirecting partner transcription factor binding (35). PU.1 expression was confirmed in MDMs as well as the monocytic cell line THP-1 (Fig. 4A, lanes 4 and 5). Most other cell types tested, including HEK293T and Jurkat cells, did not express significant levels of PU.1 (Fig. 4A, lanes 1 and 3). We cloned human PU.1 into pcDNA3.1. Expression of PU.1 was confirmed by immunoblotting (Fig. 4A, lane 2). To compare the levels of PU.1 in transiently transfected 293T cells to levels of endogenous PU.1 in uninfected human MDMs, 293T cells were transfected with increasing amounts of PU.1 vector and analyzed by immunoblotting together with cell extract of uninfected MDMs by using tubulin as internal reference (Fig. 4B). Band intensities were used to calculate PU.1/tubulin ratios (Fig. 4B, shown below the PU.1 blot). We found that levels of exogenously expressed PU.1 were in the general range of the endogenous PU.1 in MDMs. We tested the same PU.1 vector concentrations shown in lanes 1 to 4 of Fig. 4B in our luciferase induction assay. Note that amounts of transfected DNA had to be lowered to account for the smaller cell number in 24-well plates relative to 25-cm2 flasks used for the immunoblot analysis in Fig. 4B. We observed strong dose-dependent activation of the MRC1 promoter with increasing amounts of PU.1 (Fig. 4C). To ascertain that the effect of PU.1 on the MRC1 promoter was specific, we performed a dual-reporter assay where firefly luciferase was controlled by the MRC1 promoter and Renilla luciferase was under the control of the thymidine kinase (TK) promoter (Fig. 4D). Thus, constant amounts of firefly and Renilla luciferase indicator vectors were cotransfected with increasing amounts of PU.1 vector. As predicted, firefly luciferase was activated at increasing amounts of PU.1 irrespective of the presence or absence of the Renilla luciferase vector (Fig. 4E; for statistics, see Fig. S1). On the other hand, the PU.1 amount had no effect on the expression of the TK promoter-driven Renilla luciferase (Fig. 4F), attesting to the specificity of the PU.1-mediated activation of the MRC1 promoter. Taken together, these results indicate that the low basal activity of the MRC1 promoter in Fig. 3E is likely due to the absence of PU.1 in HEK293T cells. Our results also raise the possibility that MRC1 expression in macrophages is under the control of the myeloid-specific PU.1 transcription factor and that the HIV-1 induced reduction of MRC1 expression directly or indirectly involves PU.1.
FIG 4.
PU.1 is expressed in myeloid cells and activates the MRC1 promoter. (A) Immunoblot analysis of various cell types for expression of PU.1. A sample of HEK293T cells transfected with pcDNA-PU.1 (lane 2) was included as control. Tubulin expression was used as internal reference. (B) Comparison of exogenous and endogenous PU.1 expression. HEK293T cells (3 × 106) were transfected with increasing amounts (0, 0.2, 0.65, or 2.2 μg) of a vector encoding untagged PU.1 (lanes 1 to 4). DNA concentrations were chosen to match the DNA concentrations (i.e., μg DNA per 106 cells) of panel C. All samples were adjusted to 5 μg total transfected DNA using empty vector DNA (pUC19). Lane 1 is a mock-transfected control. Cell extract (20 and 40 μL) from uninfected MDMs was included as reference. Whole-cell extracts were prepared 24 h posttransfection and processed for immunoblotting using antibodies to PU.1 (α-PU.1) or tubulin (α-Tub). Relative expression of PU.1 was calculated by dividing the signal obtained for PU.1 by the signal for tubulin in each lane. Results are indicated below the PU.1 blot. (C) HEK293T cells were plated in 24-well plates (1 × 105 cells/well) and transfected in triplicate with 0.1 μg each of pGL-hMRC1-Luc (hMRC1; red line) or promoterless pGL3-basic (mock; black line) together with increasing amounts (0.06 μg, 0.2 μg, and 0.4 μg) of pcDNA-PU.1 DNA. Total amounts of transfected DNA were adjusted to 0.5 μg in each sample using empty vector DNA as needed. Production of luciferase was measured 24 h later. Means and error bars representing SEM calculated from triplicate transfections are shown. (D) Cartoon of constructs employed in a dual-reporter assay. Firefly luciferase is under the control of the PU.1 responsive hMRC1 promoter; Renilla luciferase is under the control of the HSV-thymidine kinase (TK) promoter. (E and F) Dual-reporter assay to assess the specificity of PU.1 activation of the MRC1 promoter. HEK293T cells were plated in 24-well plates (1 × 105 cells/well) and transfected in triplicate with 0.1 μg of pGL-hMRC1-Luc with or without 0.1 μg of pRN-TK-Luc, together with increasing amounts (0, 0.02, 0.06, and 0.2 μg) of pcDNA-PU.1. After 24 h, samples were analyzed for firefly and Renilla expression using the Promega dual-luciferase reporter assay system (Promega; catalog no. E1910). (E) Results of firefly luciferase activity. There were no statistically significant differences in firefly luciferase activities between samples with and without transfection of the Renilla luciferase regardless of PU.1 amounts (all comparisons were nonsignificant [ns]). (F) Results of Renilla luciferase activity. Renilla luciferase activities at increased PU.1 amounts were not statistically significantly different from no PU.1. Mean and error bars reflecting the SEM calculated from three analyses are shown. For details of the statistical analyses, see Fig. S1 in the supplemental material.
HIV-1 Tat inhibits activation of the MRC1 promoter by PU.1.
Our observation in Fig. 4 that PU.1 upregulates the MRC1 promoter activity in a dose-dependent manner raises the possibility that the inhibition of MRC1 expression in HIV-1-infected MDMs is due to interference by HIV-1 Tat with the function of PU.1. To test this hypothesis, we first assessed the ability of the full-length AD8 molecular clone to affect PU.1 function. For this purpose, constant amounts of pGL-hMRC1-Luc vector were cotransfected with increasing amounts of PU.1 vector in the presence of increasing amounts of pAD8. As expected, increasing the amounts of PU.1 in the absence of AD8 resulted in increased activation of the MRC1 promoter (Fig. 5A, red line); however, MRC1 promoter activity at the largest amount of PU.1 was reduced compared to control in all three samples with increasing amounts of cotransfected pAD8 DNA (Fig. 5A, blue lines; for statistics, see Fig. S1).
FIG 5.
Tat inhibits the PU.1-dependent activation of the hMRC1 promoter. HEK293T cells were plated into 24-well plates (5 × 104 cells/well) and transfected in triplicate with a total of 0.5 μg DNA mixture each. Luciferase production was determined 2 days posttransfection as described in Materials and Methods. (A) Full-length AD8 inhibits PU.1-mediated activation of the MRC1 promoter. Cells were transfected with pGL-hMRC1-Luc (0.1 μg) and increasing amounts of PU.1 (0.02 μg, 0.06 μg, and 0.2 μg) as well as pAD8 WT (AD8-1, 0.02 μg; AD8-2, 0.06 μg; AD8-3, 0.2 μg). The effect of PU.1 in the absence of AD8 was assessed in parallel (red circles). Mean and error bars representing SEM from nine independent transfections (n = 9) are shown. MRC1 promoter activity was statistically significantly different between the control sample and all three AD8-transfected samples at 0.2 μg PU.1 (indicated by an asterisk). (B) Inactivation of individual HIV-1 genes has no effect on the ability of AD8 to interfere with the function of PU.1. Cells were transfected with pGL-hMRC1-Luc (0.1 μg) and increasing amounts of PU.1 (0.06 μg, 0.2 μg) as well as 0.2 μg each of pAD8 WT or pAD8 mutants as indicated. A control lacking AD8 proviral DNA was included as reference (red circles). Mean and error bars representing SEM from six independent transfections (n = 6) are shown. The MRC1 promoter activity of all virus-transfected samples was statistically significantly different from the control sample at 0.2 μg PU.1 (indicated by an asterisk). (C) Tat expression is sufficient to inhibit PU.1 activity. Cells were transfected with pGL-hMRC1-Luc (0.1 μg) and increasing amounts of PU.1 (0.02 μg, 0.06 μg, and 0.2 μg) either in the absence of Tat [Tat(−)] or the presence of 0.1 μg of pTat86 or pTat101 or 0.2 μg of pNL-A1 DNA. Mean and error bars representing the SEM from six independent transfections (n = 6) are shown. MRC1 promoter activity of all three transfected samples was statistically significantly different from that of the [Tat(−)] at 0.2 μg PU.1 (indicated by an asterisk). (D) Tat does not affect the stability of PU.1 protein. HEK293T cells were transfected in 25-cm2 flasks with 2 μg of pcDNA-PU.1-HA and increasing amounts (1 μg, 2 μg, and 3 μg) of p3xFlag-Tat101. The total amount of transfected DNA was adjusted in all samples to 5 μg each using empty vector DNA as appropriate. Cells were collected 24 h later, and whole-cell extracts were subjected to immunoblot analysis using antibodies to HA (PU.1) and Flag (Tat101) epitope tags. Expression of PU.1 in the absence of Tat served as reference (Ctrl). (E) Inhibition of the hMRC1 promoter by Tat. Cells were transfected with pGL-hMRC1-Luc (0.1 μg) and increasing amounts of pTat101 (0, 0.02, 0.06, and 0.1 μg) either in the absence of PU.1 (Ctrl) or in the presence of 0.1 μg of PU.1 DNA (PU.1). Means and error bars representing SEM from nine independent transfections (n = 9) are shown. In the presence of PU.1, MRC1 promoter activity was statistically significantly different between 0 μg and 0.2 μg Tat (P < 0.001); this was not the case in the absence of PU.1 (P = 0.985). The slope of Tat on MRC1 promoter activity was significant only in the presence of PU.1, and the slope of the curve in each group was statistically significantly different from each other. For details of the statistical analyses, see Fig. S1 in the supplemental material.
Next, we compared the accessory protein mutants of AD8 employed in Fig. 1C for their effect on MRC1 promoter activity. In this experiment, only one concentration of each proviral vector, corresponding to the largest amount of pAD8 used in panel A (see Fig. 5A, open blue circles), was tested against increasing amounts of PU.1. Consistent with the results from infected macrophages (Fig. 1C), we found that inactivation of Vif, Vpr, Vpu, and Nef did not have an impact on the HIV-1-mediated inhibition of PU.1 activity in our in vitro assay (Fig. 5B). Additionally, we tested an Env-deficient variant of pAD8 and found that it was capable of interfering with the function of PU.1 as well (Fig. 5B, ΔEnv). At the largest amount of PU.1, all variants had significantly reduced MRC1 promoter activity compared to control (for statistics, see Fig. S1).
To further narrow down the region in HIV-1 required for the inhibition of PU.1, we tested the effect of the HIV-1 NL43-based vector pNL-A1. This vector is derived from a Vif-cDNA clone and encodes and expresses all HIV-1 proteins except for Gag and Pol (36). The results indicate that Gag and Pol are dispensable for the HIV-induced inhibition of PU.1 (Fig. 5C). Additionally, we directly tested the ability of HIV-1 Tat to inhibit the activity of PU.1. Here, we tested the full-length AD8 Tat (Tat101), as well as the early termination product (Tat86). Interestingly, both Tat101 and Tat86 efficiently inhibited the PU.1-mediated activation of the MRC1 promoter (Fig. 5C); at the largest amount of PU.1, MRC1 promoter activity was significantly reduced in these samples relative to control (for statistics, see Fig. S1). Of note, Tat protein expression had no impact on PU.1 protein stability (Fig. 5D), indicating that Tat targets PU.1 function via a degradation-independent mechanism. However, attempts to demonstrate a physical interaction of Tat and PU.1 failed (data not shown). Finally, doing the reverse experiment by testing the effects of increasing amounts of Tat101 in the absence or presence of PU.1 revealed that Tat inhibited the PU.1-activated MRC1 promoter activity in a dose-dependent manner (Fig. 5E). Consistent with the results from Fig. 3D and E, the basal activity of the MRC1 promoter in the absence of PU.1 was too low to reliably measure any effects of Tat. Taken together, we conclude that HIV-1 Tat101 inhibited the PU.1-dependent activation of the MRC1 promoter. Thus, our data suggest that the reduction of MRC1 expression observed in HIV-infected MDMs (Fig. 1 and 2) is at least in part due to interference by HIV-1 Tat with the PU.1-dependent regulation of the MRC1 promoter activity.
Tat transactivator function is not required for inhibition of PU.1 activity.
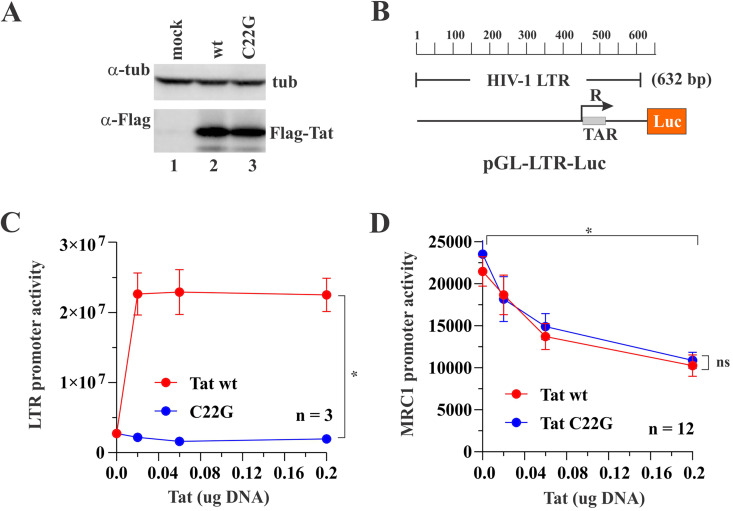
It was previously reported that Tat transactivator function is dependent on conserved cysteine residues present in the first exon of Tat. Indeed, a Tat variant carrying a C22G mutation was found to have lost more than 90% of its transactivator activity (37). To test whether Tat transactivator function was required for interference with PU.1 activity, we created a catalytically inactive variant (C22G) in the context of AD8 Tat101. We confirmed that Tat wild type (WT) and Tat C22G were expressed equally well (Fig. 6A). To verify loss of transactivator function by Tat C22G, we made use of an indicator vector, pGL-LTR-Luc, containing the 603 bp HIV-1 LTR in the backbone of pGL3-Luc (Fig. 6B). Loss of transactivator function of Tat C22G was confirmed by transfecting pGL-LTR-Luc together with increasing amounts of Tat WT or Tat C22G (C22G). As expected, at the largest amount of Tat, Tat WT strongly activated the HIV-1 LTR promoter (Fig. 6C, red line), whereas Tat C22G had lost its ability to activate the HIV-1 LTR (Fig. 6C, blue line). Interestingly, both Tat WT and Tat C22C retained the ability to inhibit the PU.1-induced MRC1 promoter activity (Fig. 6D, compare red and blue lines; for statistics, see Fig. S1). This indicates that the inhibitory effect of Tat on the MRC1 promoter activity is independent of its transactivator function.
FIG 6.
Inhibition of PU.1-activated MRC1 promoter activity does not require Tat transactivator function. (A) HEK293T cells were transfected with 1 μg each of N-terminally Flag-tagged Tat WT (lane 2) or Tat C22G (lane 3) or were mock transfected (lane 1). After 24 h, whole-cell extracts were prepared, and samples were processed for immunoblotting using a Flag epitope-specific antibody (α-Flag). Tubulin levels were determined by probing with an antibody to alpha-tubulin (α-tub). (B) The 632-bp 5′ HIV-1 LTR fragment from pNL4-3 was cloned into the backbone of pGL3-basic, resulting in pGL-LTR-Luc. The construct is shown schematically. (C) To measure the transactivator function of Tat C22G, HEK293T cells were transfected in triplicate in a 24-well plate with 0.1 μg of pGL-LTR-Luc together with increasing amounts (0.02, 0.06, and 0.2 μg) of Flag-tagged Tat WT (red line) or Tat C22G (blue line). Total amounts of transfected DNA were adjusted to 0.5 μg for each sample using empty vector (pUC19) as needed. LTR-driven luciferase activity was determined 24 h later. Mean and error bars representing SEM from three transfections are shown. LTR activity at 0.2 μg Tat induced by Tat WT or Tat C22G is statistically significantly different (P < 0.001). (D) Inhibition of the PU.1-stimulated MRC1 promoter activity by Tat C22G was measured by transfecting HEK293T cells in triplicate in a 24-well plate with 0.1 μg of pGL-hMRC1-Luc and 0.1 μg of pcDNA-PU.1, together with increasing amounts (0.02, 0.06, and 0.2 μg) of Flag-tagged Tat WT (red line) or Tat C22G (blue line). Total amounts of transfected DNA were adjusted to 0.5 μg for each sample using empty vector (pUC19) as needed. LTR-driven luciferase activity was determined 24 h later. Mean and error bars reflecting SEM from 12 transfections are shown. MRC1 promoter activity by both Tat WT and Tat C22G was statistically significantly reduced between 0 μg Tat and 0.2 μg Tat (P < 0.001 for both). There is no significant difference between Tat WT and Tat C22G (P = 0.562) at 0.2 μg Tat. The effect of Tat is significant regardless of group, and the slopes of the line in each group are not statistically significantly different from each other. For details on the statistical analyses, see Fig. S1 in the supplemental material.
Silencing of PU.1 enhances HIV-1 gene expression in infected macrophages.
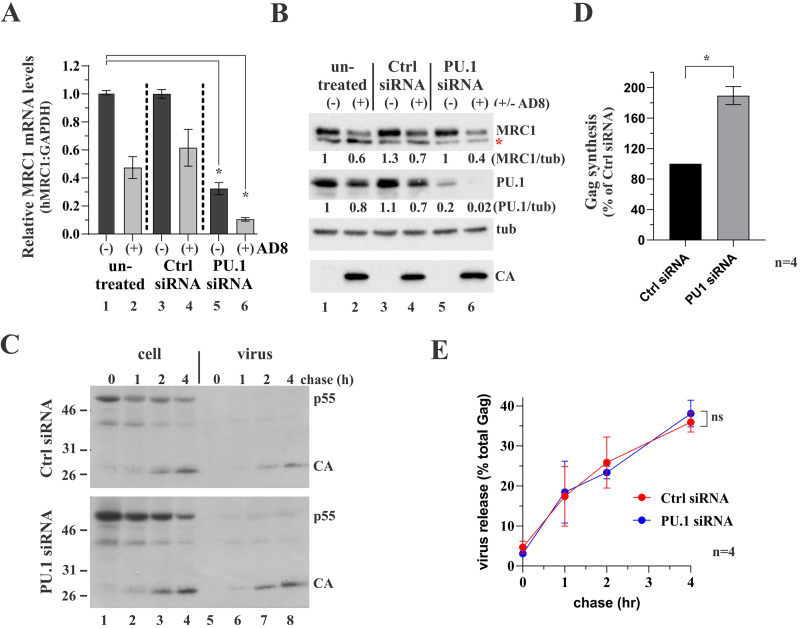
If reduction of MRC1 expression by HIV-1 Tat in HIV-infected cells is linked to interference with PU.1 activity, then artificially reducing endogenous PU.1 levels through small interfering RNA (siRNA)-mediated silencing should have the same effect. To test this hypothesis, we determined the effect of siRNA silencing of PU.1 on MRC1 expression in uninfected or HIV-1 AD8-infected MDMs (Fig. 7). MDMs were infected with VSVg-pseudotyped AD8 virus stock (AD8+) or left uninfected (AD8−). Beginning on day 9 postinfection, one-third of each of the uninfected and infected cultures was either left untreated (untreated) or were treated with control siRNA (Ctrl siRNA) or PU.1-specific siRNA (PU.1 siRNA). The siRNA treatment was repeated 2 days later (day 11). On day 12, cells were harvested, and two-thirds of each sample were processed for reverse transcriptase quantitative PCR (RT-qPCR) to determine MRC1 mRNA levels (Fig. 7A), while the remaining samples were processed for immunoblotting to assess the effect of PU.1 siRNA on protein expression (Fig. 7B). We found that MRC1 mRNA levels were reduced by about 50% following infection by AD8. This effect was amplified in cells treated with PU.1 siRNA where MRC1 mRNA levels were reduced to about 10% (Fig. 7A, compare column 1 to column 6), supporting our above-stated hypothesis of the involvement of PU.1 in the regulation of MRC1 mRNA expression.
FIG 7.
Silencing of PU.1 upregulates gene expression in HIV-1-infected MDMs. (A and B) Monocytes from a healthy donor were plated in 6-well plates (4 × 106/well; 18 wells total) and allowed to differentiate into MDMs for 6 days. Nine wells were then infected with VSVg-pseudotyped HIV-1 AD8 WT virus stock; the other nine wells remained uninfected. On days 9 and 11, cells were treated as follows: 3 wells each of infected and uninfected cells were left untreated (lanes 1 and 2) or were treated with nontargeting siRNA (lanes 3 and 4) or PU.1 siRNA (lanes 5 and 6). (A) One day later, cells from two of the three wells were processed for RT-qPCR as detailed in Materials and Methods. RT-qPCR was done twice in triplicate sets. MRC1 mRNA levels were plotted relative to GAPDH mRNA levels. Mean and error bars representing SEM from replicate data sets are shown. Differences in relative MRC1 mRNA levels between column 1 and columns 2, 3, and 4 were not statistically significant. Differences in relative MRC1 mRNA levels between column 1 and columns 5 and 6 are statistically significant (P = 0.022 and P = 0.001, respectively). For details on the statistical analyses, see Fig. S1 in the supplemental material. (B) Cells from the third well of each set were processed for immunoblotting and probed for expression of MRC1, PU.1, tubulin (tub), and HIV-1 Gag (CA). The red star in the MRC1 blot indicates a nonspecific background band as noted in Fig. 1. (C) MDMs (4 wells of a 6-well plate) were infected with VSVg-pseudotyped HIV-1 AD8 WT virus stock as in panels A and B. On days 9 and 11 postinfection, cultures were treated with nontargeting control siRNA (Ctrl siRNA) or with PU.1-specific siRNA (PU.1 siRNA). Cells were collected on day 12 and labeled for 25 min with [35S]-Expre35S35S-label and chased for up to 4 h as indicated on the top. Detergent extracts were immunoprecipitated with HIV-1 IgG, separated by SDS-PAGE, and visualized by fluorography. Proteins are identified on the right. Representative gels from one of four independent pulse-chases are shown. (D) Signals for p55gag at the end of the pulse (lane 1 in panel C) were quantified by phosphoimage analysis. The signal obtained with the control samples (nontargeting siRNA treated) was defined as 100% in each of the four replicate experiments. The signal obtained with the PU.1 siRNA-treated samples was calculated relative to the control. Mean and error bars representing SEM from four independent experiments are shown. Differences are statistically significant (P = 0.005). For details on the statistical analyses, see Fig. S1. (E) Silencing of PU.1 does not affect virus assembly and release. Gag-specific bands from panel C were quantified by phosphoimage analysis, and the fraction of extracellular Gag present at each time point was calculated as percentage of the total intra- and extracellular Gag. Means and error bars representing SEM calculated from four independent experiments are shown. Differences between the two samples at 4 h are not statistically significant (P = 0.623). For details on the statistical analyses, see Fig. S1.
Consistent with the results from Fig. 1C, HIV infection of MDMs resulted in the partial reduction of both MCR1 and PU.1 at the protein level in untreated and Ctrl-siRNA treated samples (Fig. 7B, compare lane 1 to lanes 2 and 4). Surprisingly, despite the approximate 70% silencing of MRC1 mRNA in uninfected MDMs following PU.1 siRNA treatment (Fig. 7A, column 5), MRC1 protein levels remained relatively stable (Fig. 7B, lane 5). The reason for this is unclear; however, it is possible that due to the long half-life of MRC, which was reported to be 32 h (38), changes at the mRNA level were slow to translate into changes at the protein level. At any rate, treatment of HIV-1-infected MDMs with PU.1 siRNA resulted in additional downregulation of MRC1 protein levels (Fig. 7B, compare lanes 2 and 4 to lane 6). Of note, metabolic labeling revealed that the kinetics of viral particle assembly and release were not affected by PU.1 siRNA treatment and that, after 4 h, virus release was not significantly different between groups (Fig. 7C and E; for statistics, see Fig. S1). Interestingly, however, metabolic labeling of infected MDMs revealed a noticeable increase in de novo Gag protein synthesis in PU.1 siRNA-treated samples compared to the Ctrl siRNA-treated sample (Fig. 7D; for statistics, see Fig. S1). Thus, silencing of PU.1 increases HIV-1 LTR-driven de novo viral protein synthesis in infected MDMs.
PU.1 inhibits HIV-1 LTR promoter activity.
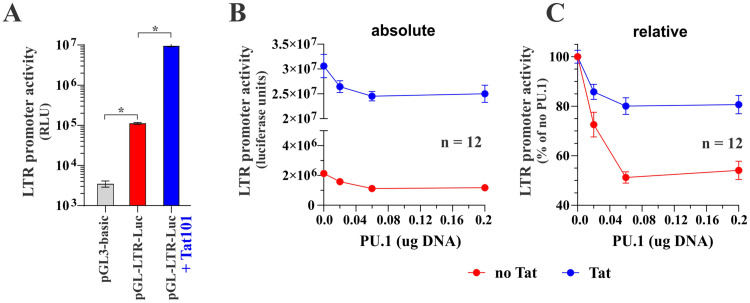
Since PU.1 is a transcriptional regulator, the observation that PU.1 silencing in infected MDMs increases HIV-1 gene expression points toward a PU.1-dependent inhibition of the HIV-1 LTR promoter activity. To test this hypothesis, we assessed the impact of PU.1 on Tat-dependent and Tat-independent HIV-1 LTR promoter activity in an in vitro assay using the pGL-LTR-Luc indicator vector (see Fig. 6B). The basal HIV-1 LTR promoter activity in HEK293T cells in the absence of Tat was significantly above the background (Fig. 8A, compare gray and red columns; for statistics, see Fig. S1). As expected, Tat expression caused an additional 100-fold increase in LTR promoter activity on top of its basal activity (Fig. 8A, compare red and blue columns; for statistics, see Fig. S1). Interestingly, coexpression of PU.1 caused a dose-dependent reduction in LTR promoter activity both in the presence and absence of Tat (Fig. 8B and C; for statistics, see Fig. S1). In fact, the effect of PU.1 on LTR promoter activity was significant only in the absence of Tat (see relative effect in Fig. 8C), although this may be caused by the overall higher absolute luciferase levels in the absence of PU.1, which are harder to neutralize (Fig. 8B). At any rate, we conclude that the inhibition of LTR promoter activity by PU.1 is largely, if not entirely, Tat independent.
FIG 8.
PU.1 inhibits the basal activity of the HIV-1 LTR in a Tat-independent manner. (A) HEK293T cells were seeded in 24-well plates and transfected with pGL3-basic (0.1 μg), pGL-LTR-Luc (0.1 μg), or pGL-LTR-Luc (0.1 μg) plus pTat101 (0.1 μg). Total amounts of transfected DNA were adjusted to 0.5 μg using empty vector DNA (pUC19) as appropriate. Luciferase production was measured 2 days posttransfection. Means and error bars representing SEM from triplicate transfections are shown. The P value for the test comparing promoter activity between pGL3-basic and pGL-LTR-Luc is 0.001. The P value for the test comparing log10 LTR promoter activity between pGL-LTR-Luc and pGL-LTR-Luc plus Tat101 is <0.001. (B and C) HEK293T cells were transfected in 24-well plates with pGL-LTR-Luc (0.1 μg) in the absence or presence of Tat (0.1 μg) as well as increasing amounts of pcDNA-PU.1 (0 μg, 0.02 μg, 0.06 μg, and 0.2 μg). Luciferase production was measured 24 h later. Results are shown as absolute luciferase units (B) or as relative units (C) with the signal obtained in the absence of PU.1 being defined as 100%. Means and error bars representing SEM from 12 transfections are shown. Statistical analyses demonstrate that the effect of PU.1 on LTR promoter activity is significant in the control group (no Tat) only. For details on the statistical analyses, see Fig. S1 in the supplemental material.
To further support this conclusion, we repeated the experiment shown in Fig. 8 using an LTR-Luc construct carrying mutations in the TAR stem-loop structure (Fig. 9A and B) that dramatically reduced, but did not completely abolish, the ability of Tat to activate the HIV-1 LTR promoter (Fig. 9C, blue line). Consistent with the results from Fig. 8, we found that PU.1 inhibited the LTR-mTAR basal activity in a dose-dependent manner (Fig. 9D, red line; for statistics, see Fig. S1). Coexpression of Tat partially inhibited the effect of PU.1 on LTR promoter activity (Fig. 9D; compare blue and red lines). This is likely due to residual promoter activity of Tat (Fig. 9C, blue line) rather than direct interference of Tat with PU.1 activity.
FIG 9.
Tat binding to the TAR stem-loop structure of the HIV-1 LTR is not required for PU.1-mediated inhibition of the HIV-1 LTR promoter activity. (A) Schematic representation of the constructs used in this experiment. (B) To minimize binding of Tat to the LTR, the TAR stem-loop structure in pGL-LTR-mTAR-Luc was mutated in the bulge-and-loop region, as previously reported, (52) using site-directed mutagenesis. Mutated residues are shown in red. (C) HEK293T cells were transfected in 24-well plates with pGL-LTR-Luc (0.1 μg) or pGL-LTR-mTAR-Luc (0.1 μg) together with increasing amounts of pTat101 (0 μg, 0.02 μg, 0.06 μg, and 0.2 μg). Production of luciferase was assessed 2 days later. Means and error bars reflecting SEM calculated from six independent transfections are shown. LTR promoter activity was statistically significantly different between 0 μg and 0.2 μg Tat for both LTR-Luc (P = 0.001) and LTR-mTAR (P = 0.009). (D) HEK293T cells were transfected in 24-well plates with pGL-LTR-mTAR-Luc (0.1 μg) in the absence (red circles) or presence (blue circles) of 0.1 μg pTat101 and increasing amounts of PU.1 DNA (0 μg, 0.06 μg, and 0.2 μg). Production of luciferase was assessed 2 days later. Means and error bars reflecting SEM calculated from nine independent transfections are shown. LTR-mTAR-driven luciferase activity by PU.1 was statistically significantly different between 0 μg and 0.2 μg PU.1 both in the presence (P = 0.006) and absence (P < 0.001) of Tat. The slope of the inhibition curves was significant in the presence or absence of Tat, although the curves were not statistically significantly different from each other. For details on the statistical analyses, see Fig. S1 in the supplemental material.
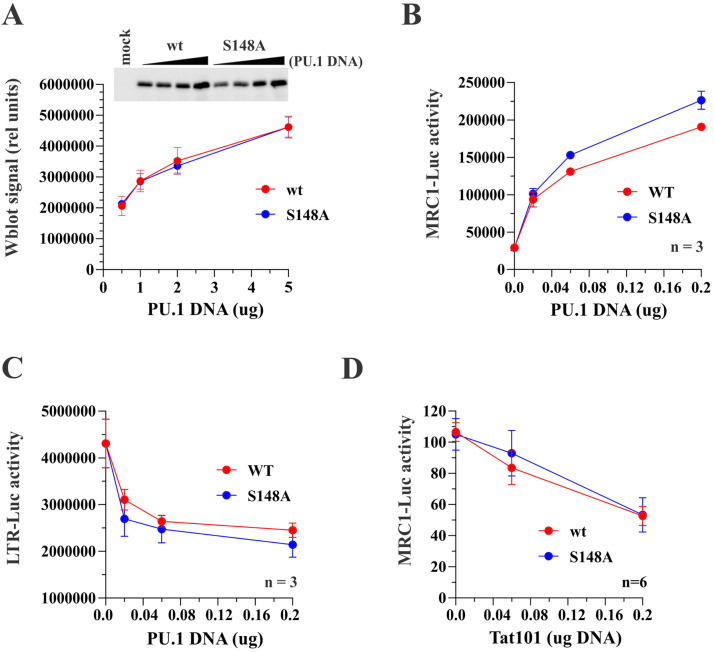
Inhibition of phosphorylation at S148 does not affect the functional properties of PU.1.
PU.1 is subject to posttranslational phosphorylation by multiple kinases that is required for the regulation of a variety of promoters by PU.1 (39). In particular, the interaction of PU.1 with another B cell-restricted nuclear factor, EM5, was shown to be dependent on PU.1 phosphorylation at Ser148 (24). Unphosphorylated PU.1 was able to bind DNA but unable to interact with EM5 (24). To test the possible impact of PU.1 phosphorylation at S148 in our assays, we constructed an S148A mutant of PU.1 in the context of C-terminally hemagglutinin (HA)-tagged PU.1 (Fig. 10). Transfection of increasing amounts of PU.1 WT and PU.1 S148A vector into HEK293T cells verified that mutation of S148A had no impact on PU.1 expression in HEK293T cells (Fig. 10A). Analysis of the ability of PU.1 S148A to activate the MRC1 promoter did not reveal a requirement of S148 phosphorylation for this function of PU.1 (Fig. 10B). Similarly, the ability of the PU.1 S148 mutant to inhibit the activity of the HIV-1 LTR was not affected (Fig. 10C). Importantly, HIV-1 Tat was able to inhibit the activation of the MRC1 promoter by both PU.1 WT and PU.1 S148A (Fig. 10D; for statistics, see Fig. S1). These results suggest that lack of PU.1 phosphorylation at S148 does not abolish its sensitivity to HIV-1 Tat.
FIG 10.
Phosphorylation at S148 is not critical for PU.1 function or sensitivity to Tat. (A) S148 in PU.1 was mutated to alanine in the backbone of pcDNA-PU.1-HA. Expression of the resulting mutant relative to the wild-type protein was analyzed by immunoblotting. For that, HEK293T cells were transfected with increasing amounts (0.5, 1.0, 2.0, and 5.0 μg) of pcDNA-PU.1-HA WT or pcDNA-PU.1-HA S148A. Cells were harvested 24 h later and processed for immunoblotting using HA tag-specific antibodies. Band intensities were quantified using Bio-Rad Image Lab software and blotted as a function of transfected PU.1 DNA. Quantitation of PU.1 WT is shown in red; quantitation of the S148A mutant is shown in blue. Means and error bars representing SEM calculated from three independent experiments are shown. (B and C) The ability of PU.1 S148A to activate the MRC1 promoter (B) and to inhibit the HIV-1 LTR promoter (C) was tested in HEK293T cells. Cells were transfected with pGL-HMRC1-Luc (0.1 μg) or pGL-LTR-Luc (0.1 μg) together with increasing amounts (0, 0.02, 0.06, and 0.2 μg) of pcDNA-PU.1-HA (red lines) or pcDNA-PU.1-HA S148A (blue lines). (D) The ability of Tat to interfere with the inhibition of the MRC1 promoter by PU.1 S148A was tested by transfecting HEK293T cells with pGL-MRC1-Luc (0.1 μg) and either pcDNA-PU.1-HA WT (0.1 μg; red line) or pcDNA-PU.1-HA S148A (0.1 μg; blue line), along with increasing amounts (0, 0.02, 0.06, and 0.2 μg) of pTat101. (B to D) Luciferase activity was measured 24 h later. Means and error bars representing the SEM calculated from 3 to 6 experiments as indicated in the figure panels are shown. Statistical analyses demonstrate that while the slope (i.e., the change in outcome between the lowest and largest amounts of PU.1 [panels B and C] or Tat DNA [panel D]) for each group is significant, there are no significant differences between the curves for PU.1 wild type and PU.1 S148A in all three assays. Thus, at 0.2 μg, there are no statistically significant differences between PU.1 wild type and PU.1 S148A in all three assays. For details on the statistical analyses, see Fig. S1 in the supplemental material.
DISCUSSION
Macrophages are long-lived cells that are susceptible to productive infection in vivo by R5-tropic or dual-tropic strains of HIV-1. Indeed, HIV-1 species circulating in a patient typically become more macrophage tropic during the course of infection (40–42). Thus, the ability to infect macrophages appears to be of crucial importance to HIV-1. Infected macrophages are relatively resistant to HIV-induced cytopathicity and therefore have the potential to act as a long-lasting reservoir. Compared to productive infection of CD4-positive (CD4+) T lymphocytes, virus output from infected macrophages is comparatively low (43). It is therefore conceivable that viral gene expression in differentiated macrophages is moderated by cell type-specific transcription factors. As such, our observation that the myeloid cell-specific transcription factor PU.1 acts as an inhibitor of HIV-1 LTR-driven gene expression could support such a hypothesis. Given the inhibitory effect of PU.1 on HIV-1 gene expression combined with the regulatory feedback loops identified in our study, it is even conceivable that PU.1 is involved in regulating HIV-1 latency. This will be the subject of a future study.
The mechanistic details of how PU.1 activates the MRC1 promoter while inhibiting the HIV-1 LTR promoter have yet to be elucidated. However, preliminary results from electrophoretic mobility shift assays (EMSAs) indicate that PU.1 binds directly to the PU.1 elements identified in the MRC1 promoter (unpublished data). The finding that PU.1 has opposing effects with regard to the regulation of the HIV-1 LTR and the MRC1 promoter in itself is not surprising since such opposing functions had already been reported for other PU.1-regulated promoter elements (35). In those situations, it was proposed that the inhibitory effect of PU.1 is caused by its recruitment of other transcription factors to PU.1 binding sites on promoters that are activated by PU.1 (e.g., the MRC1 promoter).
Interestingly, phosphorylation of PU.1 at Ser148, which was previously reported to affect its interaction with other cellular factors (24), did not seem to affect the ability of HIV-1 Tat to inhibit PU.1 function. Thus, while it is currently unclear exactly how PU.1 inhibits HIV-1 LTR-driven gene expression, the fact that the HIV-1 LTR lacks a canonical TTCCT PU.1 binding site could be consistent with such a competitive binding model. On the other hand, the presence of two potential PU.1 binding sites in the human MRC1 promoter (Fig. 3A) may explain the activating effect of PU.1 on the MRC1 promoter. It is interesting to note that an earlier study reported a modest activating effect of PU.1 on the HIV-1 LTR as opposed to the inhibitory effect reported here (44). However, that study focused on the effects of lipopolysaccharide (LPS) stimulation on HIV-1 promoter activity, and the study neither tested the effect of Tat on PU.1 function nor did it offer other mechanistic insights into the PU.1 activation of the HIV-1 LTR.
One of the key findings of our study is that HIV-1 Tat inhibits the transcriptional activity of the MRC1 promoter (Fig. 5). Our experiments were facilitated by the fact that HEK293T cells do not express PU.1, resulting in very low background activity of the MRC1 promoter indicator vector. Thus, the results from our experiments involving full-length infectious AD8 (Fig. 5A) or a variety of individual accessory gene mutants (Fig. 5B) demonstrate the inhibitory effect of HIV-1 on the MRC1 promoter and suggest that none of the viral accessory proteins on their own, including Vpr, are responsible for the inhibition of the MRC1 promoter. Indeed, expression of Tat alone was sufficient to inhibit the PU.1-activated MRC1 promoter (Fig. 5C and E). Somewhat surprisingly, the function of Tat was not affected by the presence or absence of a 15-residue highly charged C-terminal domain that is present in the 101-residue Tat variant encoded by the R5-tropic AD8 isolate but is absent in the 86-residue variant encoded by the X4-tropic NL4-3 isolate. Also, interestingly, Tat transactivator activity was not required for the interference with PU.1 function. Taken together, our results demonstrate that PU.1 has a strong activating effect on the MRC1 promoter, which is counteracted efficiently by the HIV-1 Tat protein.
Our finding of Tat-dependent transcriptional repression of the MRC1 promoter is not unique. Indeed, an earlier comprehensive chromatin immunoprecipitation sequencing (ChIP-seq) analysis of the effects of Tat on the epigenetic landscape of the host cell showed that Tat both upregulates and downregulates cellular genes (45). For instance, Tat was found to act as a transcriptional inhibitor of MHC class I genes (46). Interestingly, the inhibition of MHC class I genes was dependent on the second exon of Tat since the one-exon form of Tat (Tat72) failed to inhibit transcription of MHC class I genes (47). The observation that HIV-1 Tat repressed the human MRC1 promoter is in line with the previous report on Tat’s inhibitory effect on the rat promoter (13). However, our data clearly indicate that the effect of Tat is not mediated by a direct effect on the MRC1 promoter but, instead, is exerted indirectly by inhibiting the activity of PU.1 (Fig. 5). Indeed, the MRC1 promoter has very low basal activity in PU.1-deficient HEK293T cells, and providing Tat in trans neither increased nor reduced this basal activity (Fig. 3E). Moreover, we failed to detect a direct physical interaction between Tat and PU.1 (data not shown). Thus, exactly how Tat inhibits PU.1 activity remains to be investigated. One possible mechanism could involve competitive binding to shared cellular cofactors. Finally, while HIV infection of MDMs resulted in the inhibition of PU.1 expression (Fig. 1C and E), we can rule out Tat-induced proteolytic degradation of PU.1 (Fig. 5D). Preliminary data also argue against a role of Vpr in the inhibition of PU.1 expression since WT Vpr or its DCAF1 binding mutant have similar effects on transiently expressed PU.1 (data not shown), and deletion of Vpr did not reverse the ability of HIV-1 AD8 to downmodulate PU.1 steady-state levels (Fig. 1C and E). It is therefore likely that the reduction of PU.1 expression in HIV-1-infected macrophages is mediated by a degradation-independent mechanism. We are in the process of cloning the PU.1 promoter in order to study factors affecting PU.1 promoter activity.
Our data do not formally rule out a contribution of Vpr to the reduction of MRC1 expression in infected MDMs as reported by others (15) since our work focused primarily on the role of Tat. It is indeed possible that the discrepant results regarding the contribution of Vpr come from the use of different HIV-1 strains since we used the R5-tropic AD8 isolate, while Lubow et al. employed the dual-tropic 89.6 isolate. The Vpr proteins of these isolates are only 88% identical, and it will be interesting to perform a side-by-side comparison of their biological activities in the future. However, at least under the experimental conditions used in our study, the absence of Vpr did not adversely affect the ability of the AD8 virus to reduce MRC1 protein expression (Fig. 1C and D). Furthermore, our in vitro assessment of the MRC1 promoter did not provide any evidence for a contribution of Vpr (Fig. 5B). Finally, the inability of Vpr-defective AD8 to repress MRC1 expression under the experimental conditions used in Fig. 2C is probably not caused by the absence of Vpr but is more likely the result of inefficient replication of AD8 that presumably left most of the cells in the culture uninfected.
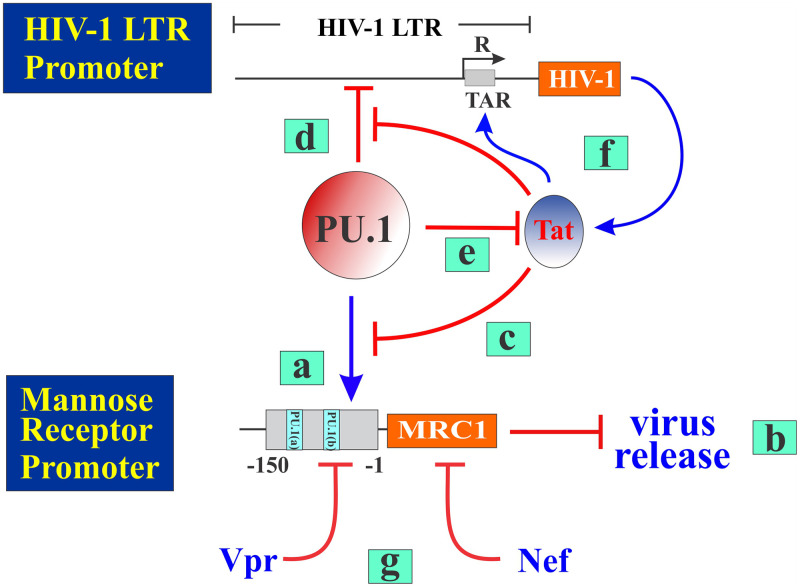
In summary, our data reveal a complex interplay between Tat and PU.1 proteins, as well as the HIV-1 LTR and MRC1 promoters, as summarized in Fig. 11. Individual steps of the model are explained in the legend to the figure. Our current work established that PU.1 activates the MRC1 promoter but inhibits the HIV-1 LTR promoter. Inhibition of the LTR promoter limits expression of HIV-1-encoded proteins, including Tat. However, HIV-1 Tat ensures continued expression of virus-encoded proteins (including Tat) via a positive feedback loop involving its interaction with the TAR stem-loop structure on the HIV-1 RNA. Continued production of Tat therefore increasingly inhibits PU.1 activity, which results in reduced expression of MRC1 and thus in increased virus release. Aside from Tat, Nef and Vpr have been implicated in regulating MRC1 expression in macrophages. However, Nef does not significantly affect steady-state expression of MRC1 but, instead, triggers cell surface downmodulation of MRC1 in HIV-infected cells (14, 15). In contrast, Vpr was reported to reduce expression of MRC1 through an incompletely defined transcriptional mechanism (15). Thus, HIV-1 appears to employ multiple pathways to control the expression of MRC1 in infected macrophages.
FIG 11.
PU.1 inhibits transcription from the HIV-1 LTR but activates the MRC1 promoter. PU.1 is a myeloid-specific transcription factor that regulates expression of multiple genes, including mannose receptor (MRC1). PU.1-mediated upregulation (blue arrow) of MRC1 (a) (see Fig. 4) accounts for the previously observed inhibition of HIV-1 virus release (b) (3). We now report that this effect is counteracted by the HIV-encoded Tat protein that inhibits PU.1 function (c) (see Fig. 5). Inhibition of PU.1 function by Tat reduces MRC1 expression, which contributes to enhanced virus release. While PU.1 upregulates MRC1 expression, it has an inhibitory effect on HIV-1 LTR promoter activity (d) (see Fig. 8 and 9). This leads to reduced Tat protein synthesis (e), which, in turn, increases the inhibition of HIV-1 LTR transcription by PU.1 (d). However, a positive feedback loop ensures continued Tat synthesis via TAR-dependent activation of HIV-1 transcription (f) and thus control of PU.1 expression (see Fig. 1C and E and Fig. 8B). Aside from Tat, HIV-1 Vpr (15) and Nef (14, 15) have been implicated in the regulation of MRC1 (h).
MATERIALS AND METHODS
Cells.
HEK293T cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 4.5 g/L glucose (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 mg/mL streptomycin in a 37°C and 5.0% CO2 environment. Elutriated human monocytes from multiple anonymous healthy donors were obtained from the NIH Blood Bank. For some experiments, elutriated monocytes from healthy donors were obtained from Chunling Gao (FDA, White Oak, MD). Monocytes were precultured as adherent cell monolayers by a modification of the procedure of Lazdins et al. (48). Monocytes (4 × 106 cells/well in a 6-well plate) were first cultured in 2 mL of serum-free DMEM for 90 min. The medium was then changed to macrophage medium (complete DMEM supplemented with 1 mM sodium pyruvate and 10% pooled human serum [Gemini Bio-Products, West Sacramento, CA]). Cells were cultured for 5 to 7 days to allow differentiation into monocyte-derived macrophages (MDMs). Replication studies were performed in 24-well plates containing 0.75 × 106 cells/well in 1 mL of macrophage medium. For protein analyses (immunoblotting and pulse-chase), cells were cultured in 6-well plates containing 4 × 106 cells per well in 2 mL of macrophage medium.
Plasmids and viral vectors.
An hMRC1 cDNA clone (GenBank accession no. NM_002438) in the backbone of pCMV6-Entry was purchased from OriGene (OriGene Technologies, Inc., Rockville. MD; catalog no. SC303200). A PU.1 (SPI1) cDNA (GenBank accession no. BC111379) clone was obtained from Dharmacon (catalog no. OHS5898-202627596) and was subcloned into vectors for expression of untagged or C-terminally HA-tagged proteins. A phosphorylation mutant (S148A) was created by PCR-based mutagenesis. Infectious molecular clones pNL4-3 (49) and pAD8 (50) have been reported previously. An Env mutant of pAD8 was created by QuikChange (Stratagene, La Jolla, CA) mutagenesis (3). The Vif open reading frame (ORF) was interrupted by introducing a stop codon after residue 23 using QuikChange mutagenesis. The Vpr ORF was interrupted by filling in a unique EcoRI site, resulting in a translational frameshift after residue 62. A Vpu-defective variant, pAD8-Udel2, carrying an out-of-frame deletion, was reported previously (51). Nef-deficient AD8 was created by introducing a stop codon after residue 6 by using QuikChange mutagenesis. To create pAD8-Tat86, Ser86 (TCG) was converted to a premature stop codon (TAG). For expression of Tat in the absence of other viral proteins, the two-exon Tat101 gene was synthesized (IDT; gBlock) and subcloned into pcDNA3.1 with or without an N-terminal Flag tag. Tat86 was created by inserting a stop codon in the pcDNA-Tat101 construct using site-directed mutagenesis. The mannose receptor promoter vector pGL-hMRC1-Luc was constructed using a synthetic DNA fragment (IDT; gBlock) corresponding to positions −1 to −150 in the human mannose receptor promoter (sequence based on reference 28) and subcloning it into the promoter trap vector pGL3-Basic (Promega; GenBank accession no. U47295). The HIV-1 LTR vector pGL-LTR-Luc was created by subcloning a 632-bp 5′-LTR fragment from pNL4-3. The Tat binding mutant pGL-LTR-mTAR-Luc was constructed by introducing five point mutations in the bulge and loop of the TAR stem-loop structure (52) by using site-directed mutagenesis. The Renilla luciferase control vector, pRL-TK, encoding Renilla luciferase under the control of the herpes simplex virus (HSV)-thymidine kinase promoter was purchased from Promega (GenBank accession no. AF025846) and used in dual-reporter assays.
Transient transfection of HEK293T cells for protein analyses.
For transient transfection of HEK293T cells, 3 × 106 cells were plated in a 25-cm2 flask and grown overnight. The following day, cells were transfected using Lipofectamine Plus (Invitrogen Corp., Carlsbad, CA) according to the manufacturer’s instructions. Total amounts of plasmid DNA in all samples were adjusted to 5 μg with empty vector DNA as appropriate. After 24 h, whole-cell extracts were produced as follows: cells were scraped, washed with phosphate-buffered saline (PBS), suspended in PBS (100 μL/106 cells), and mixed with an equal volume of 2× sample buffer. Samples were then heated for 10 to 15 min at 95°C with occasional vortexing to shear cellular DNA. Samples were then processed for immunoblot analysis.
Transient transfection of HEK293T cells for luciferase assays.
For transient transfection of HEK293T cells, cells were plated into 24-well plates (1 × 105 cells per well in 1 mL). The following day, cells were transfected using Lipofectamine Plus as follows. The medium was removed from 24-well plates and replaced by 0.5 mL of serum-free DMEM. DNA samples (2.5 μg total DNA) were mixed with 150 μL of DMEM containing 6 μL of Plus reagent. After 15 min, 150 μL of DMEM containing 6.5 μL of Lipofectamine reagent was added to the DNA-containing samples for a total volume of 312.5 μL. After 15 min incubation at room temperature, 50 μL of transfection mix was added to each of three wells. The remaining transfection mix was discarded. After 24 h, medium was aspirated from plates, 250 μL of 1× Promega lysis buffer (Promega; catalog no. E397A) was added, and plates were transferred to a −80°C freezer for at least 30 min. Then, plates were warmed to 37°C for 30 min, and 15 μL of lysate from each well was mixed with 50 μL of Steady-Glo substrate (Promega; catalog no. E2510). After 5 min incubation at room temperature, light emission was measured in a Promega GloMax Explorer.
Virus replication in MDMs.
Virus stocks for the infection of MDMs were prepared as follows. For low-titer infections, HEK293T cells (in 75-cm2 flasks) were transfected with 15 μg of pAD8 DNA using Lipofectamine Plus according to the manufacturer’s instructions. To produce high-titer VSVg-pseudotyped virus stocks, 1.5 μg of pCMV-VSVg vector DNA was cotransfected with 13.5 μg of pAD8 DNA. Virus-containing supernatants were harvested 2 days later, and cellular debris was removed by centrifugation (5 min, 1,500 rpm), followed by filtration (0.45 μm). Virus was then concentrated about 10-fold by ultracentrifugation.
For infection of MDMs, 70% of the culture medium was removed from the cells, and virus-containing supernatants or concentrated virus stocks were added. Virus was allowed to adsorb for 4 h at 37°C before the medium was replaced by macrophage medium. For determination of replication kinetics, half of the culture medium was replaced by fresh medium every 3 days. Virus production was monitored by determining the reverse transcriptase (RT) activity in the culture supernatants (53). For immunoblot analysis, MDMs were detached using a cell scraper, washed once with PBS, suspended in PBS (200 μL per well of a 6-well plate), mixed with an equal volume of 2× sample buffer (4% sodium dodecyl sulfate, 125 mM Tris-HCl [pH 6.8], 10% 2-mercaptoethanol, 10% glycerol, 0.002% bromophenol blue), and heated at 95°C for 5 to 10 min with occasional vortexing.
Metabolic labeling and pulse-chase analysis.
For pulse-chase analysis, infected siRNA-treated MDMs from 2 wells each of a 6-well plate were harvested by scraping, pooled, washed with PBS, and suspended in 5 mL of labeling medium (methionine- and cysteine-free RPMI [MP Biomedical, Solon, OH] containing 5% fetal calf serum [FCS]). Samples were incubated for 20 min at 37°C in labeling medium to deplete the intracellular methionine-cysteine pool. Cells were then labeled for 25 min at 37°C in 200 μL of labeling medium, supplemented with 30 μL (300 μCi) of [35S]-Expre35S35S-label (product no. NEG072; PerkinElmer, Waltham, MA). After the labeling period, cells were pelleted, and supernatant containing unincorporated isotope was removed. The cell pellet was then suspended in 850 μL of complete DMEM, and aliquots of 200 μL each were distributed into 4 tubes containing 1 mL of prewarmed complete DMEM (except for the tube designated time zero, which was kept on ice) and chased for the times indicated in the text. At each time point, cells were pelleted, and virus-containing supernatants were stored separately on dry ice until all samples had been collected. For immunoprecipitation of intracellular and virus-associated Gag proteins, cells and virus-containing supernatants were lysed in 1 mL of Triton X-100-based lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, and 10% glycerol) and incubated on ice for 20 min. After lysis, insoluble material from the cellular fractions was pelleted at 13,000 rpm for 10 min, and the clarified supernatants were added to protein A Sepharose beads (Sigma-Aldrich, Inc.; catalog no. P-3391) that had been preadsorbed with HIV-1 Ig. Beads were incubated for 1 h at 4°C on a rotator to keep the Sepharose beads in suspension. Beads were then extensively washed with lysis buffer. Precipitated proteins were eluted by boiling in sample buffer and separated by SDS-PAGE. Gels were soaked in 1 M sodium salicylic acid and dried. Finally, gels were exposed to Kodak XMR film, and proteins were visualized by fluorography. For protein quantitation, gels were exposed to imaging plates, and analysis of the relevant bands was performed using a Typhoon FLA 9500 phosphoimager (GE Healthcare).
siRNA knockdown of PU.1 in MDMs.
Elutriated human monocytes were plated in 6-well plates (4 × 106 cells per well in 2 mL medium) and cultured for 5 to 7 days to allow differentiation into MDMs. Then, MDMs were infected as described in the text. On day 9 and day 11 postinfection, MDMs were transfected either with 100 nM ON-TARGETplus nontargeting control siRNAs (catalog no. D-001810-01; Horizon Discovery, Cambridge, UK) or 100 nM ON-TARGETplus Human SPI1 (PU.1) siRNA SMARTpool (catalog no. L-010537-00; Horizon Discovery, Cambridge, UK) using TransIT-TKO transfection reagent (Mirus Bio, Madison, WI) following the manufacturer’s recommendations. One day after the second siRNA treatment, cells were harvested by scraping and processed for immunoblot analysis or metabolic labeling (pulse-chase analysis).
Immunoblot analysis.
Cells were washed once with PBS, suspended in PBS (100 μL/106 cells), and mixed with an equal volume of 2× sample buffer. Samples were heated at 95°C with occasional vortexing until samples were completely dissolved. Samples were subjected to SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes, and reacted with primary antibodies as described in the text. To enhance sensitivity, SignalBoost immunoreaction enhancer kit (CalBiochem; catalog no. 407207-1KIT) was employed in some of the experiments. Human MRC1 was identified using a rabbit monoclonal antibody to hMRC1 (Abcam, Inc., Cambridge MA; catalog no. ab125028). HIV-1 Gag was identified using pooled HIV Ig (NIH Research and Reference Reagent Program; catalog no. 3957), and tubulin was identified using a mouse monoclonal antibody to alpha-tubulin (catalog no. T9026; Sigma-Aldrich Inc.). A rabbit monoclonal antibody to PU.1 was from Abcam (catalog no. ab76543), and a rabbit polyclonal antibody to Tat was from Bryan Cullen through the NIH Research and Reference Reagent Program (catalog no. 705). Mouse anti-HA antibodies were obtained from Sigma (catalog no. H3663; Sigma-Aldrich, Inc.). Membranes were then incubated with horseradish peroxidase-conjugated secondary antibodies (GE Healthcare, Piscataway, NJ), and proteins were visualized by enhanced chemiluminescence (Clarity Western ECL substrate; catalog no. 170-5061; Bio-Rad Laboratories, Hercules, CA).
RNA isolation and RT-qPCR.
Total RNA was extracted from MDM using Direct-zol RNA miniprep kit (Zymo Research, Irvine, CA) according to the manufacturer’s instructions, and the RNA concentration was quantified by NanoDrop (Thermo Fisher Scientific, Inc., Waltham, MA). RT-qPCR was performed with 20 ng of total RNA and specific primer sets for hMRC1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using iTaq Universal SYBR Green one-step kit (Bio-Rad). Primers for RT-qPCR were as follows: hMRC1 sense, 5′-AAAGCTGCCA ACAACAGAAC GCTGAG-3′; hMRC1 antisense, 5′-ATATAGCCCA GTTTCTGAAC ACATTCC-3′; PU.1 sense, 5′-AGCTCAGATG AGGAGGAGGG-3′; PU.1 antisense, 5′-AACAGGAACT GGTACAGGCG-3′; GAPDH sense, 5′-AAGGTCGGAG TCAACGGATT-3′, and GAPDH antisense, 5′-CTCCTGGAAG ATGGTGATGG-3′. RT-qPCR was carried out using the CFX96 Touch real-time PCR detection system (Bio-Rad). The housekeeping gene GAPDH was used as an internal control, and relative mRNA levels were determined using the threshold cycle (ΔΔCT) quantification method.
Quantitation of extracellular virus by RT assay.
Extracellular virus was quantified by measuring the amounts of virus-associated reverse transcriptase using a 32P-based assay essentially as described (53). We mixed 10 μL of virus-containing culture supernatant with 50 μL of an RT reaction cocktail, which contained, as the template, poly(A) (5 μg/mL) and, as primer, oligo(dT) [oligo(dT)12-18; 0.16 μg/mL] in 50 mM Tris, pH 7.8, 7.5 mM KCl, 2 mM dithiothreitol, 5 mM MgCl2, 0.05% Nonidet P-40, and 1 μCi/mL cocktail of [32P]dTTP (800 Ci/mmol). Following a 90-min incubation at 37°C, 10 μL of the reaction mixture was spotted onto DEAE ion exchange paper (Whatman) and washed three times in 2× SSC (l× SSC is 0.15 M NaCl plus 0.015 M sodium chloride) to remove unincorporated [32P]dTTP. Spots were counted in a scintillation counter.
Statistical analysis.
Comparisons of continuous variables between two groups were done using an unpaired two-tailed t test with the assumption of unequal variances between groups. If outliers were present (Fig. 8C), a Wilcoxon test was used to compare outcomes between groups. To evaluate dose response, linear regression was performed to predict the outcome from microgram DNA, group, and an interaction between DNA and group. When absolute values were used as the outcome, values were log10 transformed. Analyses were done in R version 3.6.3, and P values of <0.05 were considered significant (indicated by an asterisk). P values were not adjusted for multiple comparisons and should be interpreted cautiously. Details of the statistical analyses are in Fig. S1 in the supplemental material.
ACKNOWLEDGMENTS
We thank Chunling Gao for help with elutriation of monocytes and Kathryn Brittain for helpful discussions.
The following reagents were obtained through the NIH HIV Reagent Program, Division of AIDS, NIAID, NIH: (i) polyclonal anti-HIV-1 Tat antibody (catalog no. 705), contributed by Bryan Cullen, and (ii) HIV-1 immunoglobulin (catalog no. 3957), contributed by NABI and the National Heart Lung and Blood Institute (Luiz Barbosa).
This work was supported by the Intramural Research Program of the NIH, NIAID (1 Z01 AI000669; K.S.). This project has been funded in whole or in part with federal funds from the National Institute of Allergy and Infectious Diseases and the National Cancer Institute, National Institutes of Health, under contract no. 75N91019D00024. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products, or organizations imply endorsement by the United States Government.
We all declare no conflict of interest.
E.M. and K.S. designed the experiments. S.K., E.M., R.M., H.S., D.F., H.F., and K.S. performed the experiments. A. Mukherji, R.M., K.C., and S.S. provided critical reagents. A. Mateja. performed the statistical analysis. K.S. wrote the manuscript. All authors discussed results and commented on the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Klaus Strebel, Email: kstrebel@nih.gov.
Frank Kirchhoff, Ulm University Medical Center.
REFERENCES
- 1.Neil SJ, Sandrin V, Sundquist WI, Bieniasz PD. 2007. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe 2:193–203. 10.1016/j.chom.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252. 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sukegawa S, Miyagi E, Bouamr F, Farkasova H, Strebel K. 2018. Mannose receptor 1 restricts HIV particle release from infected macrophages. Cell Rep 22:786–795. 10.1016/j.celrep.2017.12.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strebel K, Luban J, Jeang KT. 2009. Human cellular restriction factors that target HIV-1 replication. BMC Med 7:48. 10.1186/1741-7015-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arias JF, Iwabu Y, Tokunaga K. 2012. Sites of action of HIV-1 Vpu in BST-2/tetherin downregulation. Curr HIV Res 10:283–291. 10.2174/157016212800792423. [DOI] [PubMed] [Google Scholar]
- 6.Le Tortorec A, Neil SJ. 2009. Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J Virol 83:11966–11978. 10.1128/JVI.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen CY, Shingai M, Welbourn S, Martin MA, Borrego P, Taveira N, Strebel K. 2016. Antagonism of BST-2/tetherin is a conserved function of the Env glycoprotein of primary HIV-2 isolates. J Virol 90:11062–11074. 10.1128/JVI.01451-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang SJ, Lopez LA, Hauser H, Exline CM, Haworth KG, Cannon PM. 2010. Anti-tetherin activities in Vpu-expressing primate lentiviruses. Retrovirology 7:13. 10.1186/1742-4690-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hauser H, Lopez LA, Yang SJ, Oldenburg JE, Exline CM, Guatelli JC, Cannon PM. 2010. HIV-1 Vpu and HIV-2 Env counteract BST-2/tetherin by sequestration in a perinuclear compartment. Retrovirology 7:51. 10.1186/1742-4690-7-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta RK, Mlcochova P, Pelchen-Matthews A, Petit SJ, Mattiuzzo G, Pillay D, Takeuchi Y, Marsh M, Towers GJ. 2009. Simian immunodeficiency virus envelope glycoprotein counteracts tetherin/BST-2/CD317 by intracellular sequestration. Proc Natl Acad Sci USA 106:20889–20894. 10.1073/pnas.0907075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia B, Serra-Moreno R, Neidermyer W, Rahmberg A, Mackey J, Fofana IB, Johnson WE, Westmoreland S, Evans DT. 2009. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog 5:e1000429. 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang F, Wilson SJ, Landford WC, Virgen B, Gregory D, Johnson MC, Munch J, Kirchhoff F, Bieniasz PD, Hatziioannou T. 2009. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6:54–67. 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caldwell RL, Egan BS, Shepherd VL. 2000. HIV-1 Tat represses transcription from the mannose receptor promoter. J Immunol 165:7035–7041. 10.4049/jimmunol.165.12.7035. [DOI] [PubMed] [Google Scholar]
- 14.Vigerust DJ, Egan BS, Shepherd VL. 2005. HIV-1 Nef mediates post-translational down-regulation and redistribution of the mannose receptor. J Leukoc Biol 77:522–534. 10.1189/jlb.0804454. [DOI] [PubMed] [Google Scholar]
- 15.Lubow J, Virgilio MC, Merlino M, Collins DR, Mashiba M, Peterson BG, Lukic Z, Painter MM, Gomez-Rivera F, Terry V, Zimmerman G, Collins KL. 2020. Mannose receptor is an HIV restriction factor counteracted by Vpr in macrophages. Elife 9. 10.7554/eLife.51035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turkistany SA, DeKoter RP. 2011. The transcription factor PU.1 is a critical regulator of cellular communication in the immune system. Arch Immunol Ther Exp (Warsz) 59:431–440. 10.1007/s00005-011-0147-9. [DOI] [PubMed] [Google Scholar]
- 17.De Smedt J, van Os EA, Talon I, Ghosh S, Toprakhisar B, Furtado Madeiro Da Costa R, Zaunz S, Vazquez MA, Boon R, Baatsen P, Smout A, Verhulst S, van Grunsven LA, Verfaillie CM. 2021. PU.1 drives specification of pluripotent stem cell-derived endothelial cells to LSEC-like cells. Cell Death Dis 12:84. 10.1038/s41419-020-03356-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, Maki RA. 1996. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J 15:5647–5658. 10.1002/j.1460-2075.1996.tb00949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klemsz MJ, McKercher SR, Celada A, Van Beveren C, Maki RA. 1990. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell 61:113–124. 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 20.Moreau-Gachelin F, Tavitian A, Tambourin P. 1988. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature 331:277–280. 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- 21.Fisher RC, Scott EW. 1998. Role of PU.1 in hematopoiesis. Stem Cells 16:25–37. 10.1002/stem.160025. [DOI] [PubMed] [Google Scholar]
- 22.Imperato MR, Cauchy P, Obier N, Bonifer C. 2015. The RUNX1-PU.1 axis in the control of hematopoiesis. Int J Hematol 101:319–329. 10.1007/s12185-015-1762-8. [DOI] [PubMed] [Google Scholar]
- 23.Klemsz MJ, Maki RA. 1996. Activation of transcription by PU.1 requires both acidic and glutamine domains. Mol Cell Biol 16:390–397. 10.1128/MCB.16.1.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pongubala JM, Van Beveren C, Nagulapalli S, Klemsz MJ, McKercher SR, Maki RA, Atchison ML. 1993. Effect of PU.1 phosphorylation on interaction with NF-EM5 and transcriptional activation. Science 259:1622–1625. 10.1126/science.8456286. [DOI] [PubMed] [Google Scholar]
- 25.Zhao M, Duan XF, Wen DH, Chen GQ. 2009. PU.1, a novel caspase-3 substrate, partially contributes to chemotherapeutic agents-induced apoptosis in leukemic cells. Biochem Biophys Res Commun 382:508–513. 10.1016/j.bbrc.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 26.Kodandapani R, Pio F, Ni CZ, Piccialli G, Klemsz M, McKercher S, Maki RA, Ely KR. 1996. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature 380:456–460. 10.1038/380456a0. [DOI] [PubMed] [Google Scholar]
- 27.Ray-Gallet D, Mao C, Tavitian A, Moreau-Gachelin F. 1995. DNA binding specificities of Spi-1/PU.1 and Spi-B transcription factors and identification of a Spi-1/Spi-B binding site in the c-fes/c-fps promoter. Oncogene 11:303–313. [PubMed] [Google Scholar]
- 28.Rouleux F, Monsigny M, Legrand A. 1994. A negative regulatory element of the macrophage-specific human mannose receptor gene represses its expression in nonmyeloid cells. Exp Cell Res 214:113–119. 10.1006/excr.1994.1239. [DOI] [PubMed] [Google Scholar]
- 29.Jeang KT. 1998. Tat, Tat-associated kinase, and transcription. J Biomed Sci 5:24–27. 10.1007/BF02253352. [DOI] [PubMed] [Google Scholar]
- 30.Jones KA. 1997. Taking a new TAK on Tat transactivation. Genes Dev 11:2593–2599. 10.1101/gad.11.20.2593. [DOI] [PubMed] [Google Scholar]
- 31.Clark E, Nava B, Caputi M. 2017. Tat is a multifunctional viral protein that modulates cellular gene expression and functions. Oncotarget 8:27569–27581. 10.18632/oncotarget.15174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rice AP. 2017. The HIV-1 Tat protein: mechanism of action and target for HIV-1 cure strategies. Curr Pharm Des 23:4098–4102. 10.2174/1381612823666170704130635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Celada A, Borras FE, Soler C, Lloberas J, Klemsz M, van Beveren C, McKercher S, Maki RA. 1996. The transcription factor PU.1 is involved in macrophage proliferation. J Exp Med 184:61–69. 10.1084/jem.184.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang DE, Hetherington CJ, Chen HM, Tenen DG. 1994. The macrophage transcription factor PU.1 directs tissue-specific expression of the macrophage colony-stimulating factor receptor. Mol Cell Biol 14:373–381. 10.1128/mcb.14.1.373-381.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hosokawa H, Ungerback J, Wang X, Matsumoto M, Nakayama KI, Cohen SM, Tanaka T, Rothenberg EV. 2018. Transcription factor PU.1 represses and activates gene expression in early T cells by redirecting partner transcription factor binding. Immunity 48:1119–1134.e7. 10.1016/j.immuni.2018.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strebel K, Daugherty D, Clouse K, Cohen D, Folks T, Martin MA. 1987. The HIV 'A' (sor) gene product is essential for virus infectivity. Nature 328:728–730. 10.1038/328728a0. [DOI] [PubMed] [Google Scholar]
- 37.Rice AP, Carlotti F. 1990. Mutational analysis of the conserved cysteine-rich region of the human immunodeficiency virus type 1 Tat protein. J Virol 64:1864–1868. 10.1128/JVI.64.4.1864-1868.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vigerust DJ, Vick S, Shepherd VL. 2012. Characterization of functional mannose receptor in a continuous hybridoma cell line. BMC Immunol 13:51. 10.1186/1471-2172-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gross SA, Zheng JH, Le AT, Kerzic PJ, Irons RD. 2006. PU.1 phosphorylation correlates with hydroquinone-induced alterations in myeloid differentiation and cytokine-dependent clonogenic response in human CD34(+) hematopoietic progenitor cells. Cell Biol Toxicol 22:229–241. 10.1007/s10565-006-0128-7. [DOI] [PubMed] [Google Scholar]
- 40.Ellery PJ, Tippett E, Chiu YL, Paukovics G, Cameron PU, Solomon A, Lewin SR, Gorry PR, Jaworowski A, Greene WC, Sonza S, Crowe SM. 2007. The CD16+ monocyte subset is more permissive to infection and preferentially harbors HIV-1 in vivo. J Immunol 178:6581–6589. 10.4049/jimmunol.178.10.6581. [DOI] [PubMed] [Google Scholar]
- 41.Gorry PR, Churchill M, Crowe SM, Cunningham AL, Gabuzda D. 2005. Pathogenesis of macrophage tropic HIV-1. Curr HIV Res 3:53–60. 10.2174/1570162052772951. [DOI] [PubMed] [Google Scholar]
- 42.Igarashi T, Brown CR, Endo Y, Buckler-White A, Plishka R, Bischofberger N, Hirsch V, Martin MA. 2001. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): implications for HIV-1 infections of humans. Proc Natl Acad Sci USA 98:658–663. 10.1073/pnas.98.2.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koppensteiner H, Brack-Werner R, Schindler M. 2012. Macrophages and their relevance in human immunodeficiency virus type I infection. Retrovirology 9:82–82. 10.1186/1742-4690-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lodie TA, Reiner M, Coniglio S, Viglianti G, Fenton MJ. 1998. Both PU.1 and nuclear factor-kappa B mediate lipopolysaccharide-induced HIV-1 long terminal repeat transcription in macrophages. J Immunol 161:268–276. [PubMed] [Google Scholar]
- 45.Reeder JE, Kwak YT, McNamara RP, Forst CV, D'Orso I. 2015. HIV Tat controls RNA Polymerase II and the epigenetic landscape to transcriptionally reprogram target immune cells. Elife 4:e08955. 10.7554/eLife.08955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weissman JD, Brown JA, Howcroft TK, Hwang J, Chawla A, Roche PA, Schiltz L, Nakatani Y, Singer DS. 1998. HIV-1 Tat binds TAFII250 and represses TAFII250-dependent transcription of major histocompatibility class I genes. Proc Natl Acad Sci USA 95:11601–11606. 10.1073/pnas.95.20.11601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howcroft TK, Strebel K, Martin MA, Singer DS. 1993. Repression of MHC class I gene promoter activity by two-exon Tat of HIV. Science 260:1320–1322. 10.1126/science.8493575. [DOI] [PubMed] [Google Scholar]
- 48.Lazdins JK, Woods-Cook K, Walker M, Alteri E. 1990. The lipophilic muramyl peptide MTP-PE is a potent inhibitor of HIV replication in macrophages. AIDS Res Hum Retroviruses 6:1157–1161. 10.1089/aid.1990.6.1157. [DOI] [PubMed] [Google Scholar]
- 49.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol 59:284–291. 10.1128/JVI.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Theodore TS, Englund G, Buckler-White A, Buckler CE, Martin MA, Peden KW. 1996. Short communication: construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res Hum Retroviruses 12:191–194. 10.1089/aid.1996.12.191. [DOI] [PubMed] [Google Scholar]
- 51.Schubert U, Clouse KA, Strebel K. 1995. Augmentation of virus secretion by the human immunodeficiency virus type 1 Vpu protein is cell type independent and occurs in cultured human primary macrophages and lymphocytes. J Virol 69:7699–7711. 10.1128/JVI.69.12.7699-7711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mueller N, Pasternak AO, Klaver B, Cornelissen M, Berkhout B, Das AT. 2018. The HIV-1 Tat protein enhances splicing at the major splice donor site. J Virol 92:e01855-17. 10.1128/JVI.01855-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willey RL, Smith DH, Lasky LA, Theodore TS, Earl PL, Moss B, Capon DJ, Martin MA. 1988. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol 62:139–147. 10.1128/JVI.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Download jvi.00652-22-s0001.pdf, PDF file, 0.2 MB (250.5KB, pdf)