ABSTRACT
Bats are natural reservoirs of numerous coronaviruses, including the potential ancestor of SARS-CoV-2. Knowledge concerning the interaction between coronaviruses and bat cells is sparse. We investigated the ability of primary cells from Rhinolophus and Myotis species, as well as of established and novel cell lines from Myotis myotis, Eptesicus serotinus, Tadarida brasiliensis, and Nyctalus noctula, to support SARS-CoV-2 replication. None of these cells were permissive to infection, not even the ones expressing detectable levels of angiotensin-converting enzyme 2 (ACE2), which serves as the viral receptor in many mammalian species. The resistance to infection was overcome by expression of human ACE2 (hACE2) in three cell lines, suggesting that the restriction to viral replication was due to a low expression of bat ACE2 (bACE2) or the absence of bACE2 binding in these cells. Infectious virions were produced but not released from hACE2-transduced M. myotis brain cells. E. serotinus brain cells and M. myotis nasal epithelial cells expressing hACE2 efficiently controlled viral replication, which correlated with a potent interferon response. Our data highlight the existence of species-specific and cell-specific molecular barriers to viral replication in bat cells. These novel chiropteran cellular models are valuable tools to investigate the evolutionary relationships between bats and coronaviruses.
IMPORTANCE Bats are host ancestors of several viruses that cause serious disease in humans, as illustrated by the ongoing SARS-CoV-2 pandemic. Progress in investigating bat-virus interactions has been hampered by a limited number of available bat cellular models. We have generated primary cells and cell lines from several bat species that are relevant for coronavirus research. The various permissivities of the cells to SARS-CoV-2 infection offered the opportunity to uncover some species-specific molecular restrictions to viral replication. All bat cells exhibited a potent entry-dependent restriction. Once this block was overcome by overexpression of human ACE2, which serves at the viral receptor, two bat cell lines controlled well viral replication, which correlated with the inability of the virus to counteract antiviral responses. Other cells potently inhibited viral release. Our novel bat cellular models contribute to a better understanding of the molecular interplays between bat cells and viruses.
KEYWORDS: SARS-CoV-2, bat cells, coronavirus, innate immunity
INTRODUCTION
Bats are natural hosts of numerous coronaviruses, including members of the Betacoronavirus genus, which comprises viruses belonging to the severe acute respiratory syndrome coronavirus (SARS-CoV) 1 and 2 lineages (1). RNAs from SARS-CoV-2 related coronaviruses (SC2r-CoVs) have been recently detected in different species of Rhinolophus living in Northern Laos. One of these viruses, named BANAL-52, was found in R. malayanus and is, to date, the closest relative to SARS-CoV-2, with an overall sequence identity of 96.8% (2). Other viruses belonging to this lineage have been identified in Rhinolophus bats sampled in China (3, 4) Thailand (5), Cambodia (6), and Japan (7). SC2r-CoVs are thus probably widely distributed in South-East Asia. In addition, numerous other bat species worldwide are infected with betacoronaviruses, including species of the Myotis, Nyctalus, Tadarida, and Eptesicus genera (8–14).
The risk of spillback transmission of SARS-CoV-2 from humans to domestic animals or wildlife remains a major concern, since this reverse zoonotic transmission has been already documented in pet animals and farmed minks, as well as in tigers and gorillas in zoos (15, 16). Infection and transmission of SARS-CoV-2 has also been reported in white-tailed deer (17). Given the likely bat origin of SARS-CoV-2, bats could be putatively at risk of spillback transmission (18). The establishment of novel bat reservoirs would have a severe impact on wildlife conservation and public health measures.
SARS-CoV, SARS-CoV-2, and one of the BANAL viruses use the surface receptor angiotensin-converting enzyme 2 (ACE2) to enter human cells (2, 4, 19–21). Viral binding to ACE2 is followed by the proteolytic cleavage of the viral spike (S) proteins by either the plasma-membrane resident transmembrane protease serine 2 (TMPRSS2) or the endosomal cathepsin L (CTSL) (22). This cleavage is mandatory for the fusion between the viral and cellular membranes. Thus, localization and expression of TMPRSS2 and CTSL dictate whether the virus enters cells by fusing at the cell surface or in endosomes (22, 23).
Several approaches have been used to predict the ability of ACE2 from phylogenetically diverse bat species to promote SARS-CoV-2 entry. First, comparison of ACE2 protein sequences from 37 bat species, including species of the genus Rhinolophus, predicted a low or very low ability to interact with viral S proteins (24). Second, ectopic expression of ACE2 from several bat species in nonpermissive mammalian cells, followed by infection with genuine viruses or pseudoviruses carrying SARS-CoV-2 S proteins, revealed that ACE2 from Rhinolophus, Myotis, and Eptesicus species allowed viral entry, albeit often less efficiently than human ACE2. However, these approaches using in silico analysis or ectopic expression of bat ACE2 in human or hamster cells do not allow to draw conclusions as to which bat species might actually support SARS-CoV-2 replication. Other factors unique to bat cells may potentially modulate viral entry and replication. Indeed, experiments performed with cells derived from lung tissue of Rhinolophus alcyone and Myotis daubentonii showed that they were not susceptible to infection with vesicular stomatitis viruses (VSVs) bearing SARS-CoV-2 S proteins (20). Cells originating from lung and kidney tissue of Rhinolophus sinicus and Eptesicus fuscus were not permissive to SARS-CoV-2 either (25, 26). These studies underline the limitation of predicting the ability of S proteins to interact with ACE2 orthologs based on computational models or ectopic expression. These findings also highlight the limitation of predicting viral host tropism based on cellular studies.
Only a few models are available to study the replication of betacoronaviruses in bat cells. Viral replication was detected in Rhinolophus sinicus lung and brain cells, as well as in Pipistrellus abramus kidney cells (27), but viral titers were very low. In contrast, SARS-CoV-2 replicated efficiently in R. sinicus intestinal organoids (28), confirming further the ability of Rhinolophus cells to support viral replication. Intranasal inoculation of SARS-CoV-2 in Rousettus aegypticus resulted in transient infection of their respiratory tract and oral shedding of the virus (29), indicating that bats unrelated to the Rhinolophus genus can be productively infected with the virus. Since the manipulation of bat organoid and animal models remains challenging, there is a need to develop cell lines from various organs and species to gain deeper insights into bat-virus coevolution (30). Here, we developed novel cellular models derived from understudied bat species widely circulating in Europe and Asia. The various susceptibilities and permissivities of the cells to SARS-CoV-2 infection offered the opportunity to uncover species-specific molecular restrictions to viral replication.
RESULTS
Resistance to SARS-CoV-2 infection in selected bat cell lines.
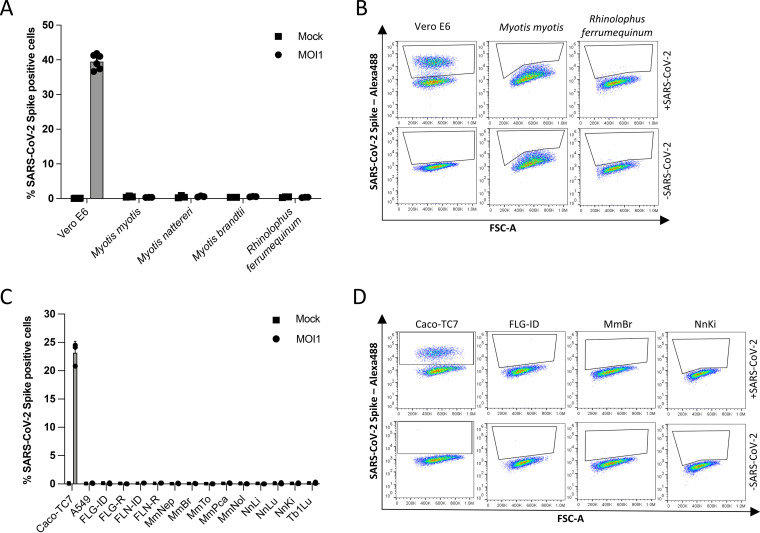
Species belonging to the Rhinolophus genus are known natural hosts for numerous SARS-CoV-related betacoronaviruses (1). Alphacoronaviruses (10, 31, 32), and possibly betacoronaviruses (1, 9), circulate in species belonging to the Myotis genera. Primary cells generated from wing biopsy specimens of R. ferrumequinum, M. myotis, M. nattereri, and M. brandtii (Table 1) were subjected to infection by SARS-CoV-2 at a multiplicity of infection (MOI) of 1. Flow cytometry analyses were performed using anti-S antibodies at 24 h postinfection (hpi) to assess viral antigen expression. Vero E6 cells, which are African green monkey kidney cells known to be permissive to SARS-CoV-2 (33), were used as positive controls. Around 40% of Vero E6 cells were positive for the viral S protein (Fig. 1A and B). Neither R. ferrumequinum nor Myotis spp. primary cells were permissive for productive infection (Fig. 1A and B).
TABLE 1.
Overview of the primary and immortalized bat cells used in the study
| Cell line | Bat species | Common name | Family | Organ | Transformation method | Source or reference |
|---|---|---|---|---|---|---|
| 16104 | Rhinolophus ferrumequinum | Greater Horseshoe Bat | Rhinolophidae | Skin (patagium) | None (primary cell) | This study |
| 29B | Myotis myotis | Common serotine bat | Vespertilionidae | Skin (patagium) | None (primary cells) | This study |
| 19PL50 | Myotis nattereri | Natterer’s bat | Vespertilionidae | Skin (patagium) | None (primary cells) | This study |
| MBra10 | Myotis brandtii | Brandt’s bat | Vespertilionidae | Skin (patagium) | None (primary cells) | This study |
| FLG-ID | Eptesicus serotinus | Common serotine bat | Vespertilionidae | Brain | Immortalized FLG-R cells with SV40 large T antigen | CCLV-RIE 1152 |
| FLG-R | Eptesicus serotinus | Common serotine bat | Vespertilionidae | Brain | Natural | CCLV-RIE 1093 |
| FLN-ID | Eptesicus serotinus | Common serotine bat | Vespertilionidae | Kidney | Immortalized FLN-R cells with SV40 large T antigen | CCLV-RIE 1134 |
| FLN-R | Eptesicus serotinus | Common serotine bat | Vespertilionidae | Kidney | Natural | CCLV-RIE 1091 |
| MmBr | Myotis myotis | Greater mouse-eared bat | Vespertilionidae | Brain | SV40 large T antigen | 35 |
| MmNep | Myotis myotis | Greater mouse-eared bat | Vespertilionidae | Nasal epithelium | SV40 large T antigen | 35 |
| MmNol | Myotis myotis | Greater mouse-eared bat | Vespertilionidae | Nerve | SV40 large T antigen | 35 |
| MmPca | Myotis myotis | Greater mouse-eared bat | Vespertilionidae | Macrophage | SV40 large T antigen | 35 |
| MmTo | Myotis myotis | Greater mouse-eared bat | Vespertilionidae | Tonsil | SV40 large T antigen | 35 |
| NnKi | Nyctalus noctula | Common noctule | Vespertilionidae | Kidney | SV40 large T antigen | This study |
| NnLi | Nyctalus noctula | Common noctule | Vespertilionidae | Liver | SV40 large T antigen | This study |
| NnLu | Nyctalus noctula | Common noctule | Vespertilionidae | Lung | SV40 large T antigen | This study |
| Tb1Lu | Tadarida brasiliensis | Mexican/Brazilian free-tailed bat | Molossidae | Lung | Natural? | CCLV-RIE 0072 |
FIG 1.
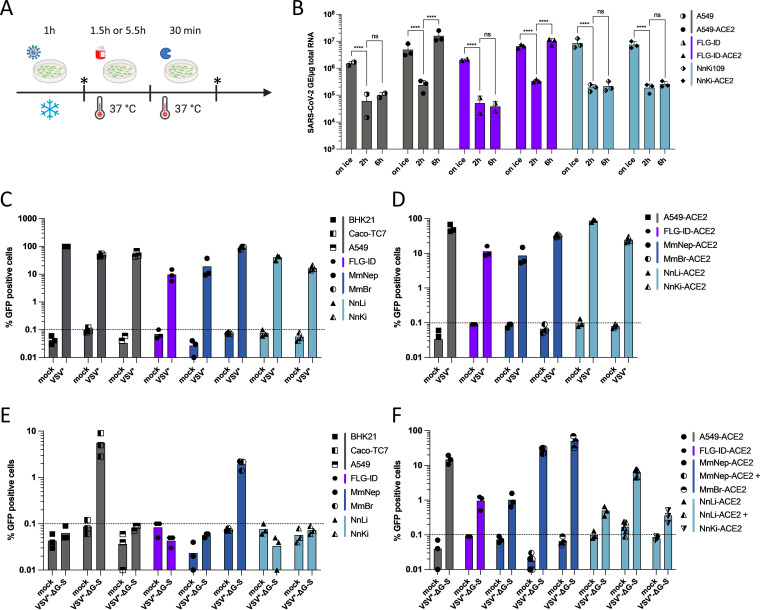
Resistance to SARS-CoV-2 infection in selected bat cell lines. (A) Primary bat cells derived from wing tissues from four different species, as well as Vero E6 cells, were left uninfected (Mock) or were infected with SARS-CoV-2 at an MOI of 1 for 24 h and then analyzed via flow cytometry for viral spike (S) protein expression. (B) Representative dot plots of selected primary bat cells and Vero E6 cells. Data points represent three technical replicates. (C) Immortalized bat cell lines from four different species, as well as Caco-TC7 human intestine and A549 human lung epithelial cells, were left uninfected (Mock) or were infected with SARS-CoV-2 at an MOI of 1 for 24 h and then analyzed via flow cytometry for S expression. (D) Representative dot plots of selected immortalized cells. Data points represent three independent experiments with the exception of A549, FLN-ID, and FLN-R cells, where data points represent three technical replicates.
We then tested the permissivity of previously described, immortalized cell lines generated from Eptesicus serotinus (34), Myotis myotis (35), and Tadarida brasiliensis (Table 1) to SARS-CoV-2. E. serotinus cells were isolated from brain (FLG) and kidney (FLN) (34) (Table 1). M. myotis cells were established from brain (MmBr), tonsil (MmTo), peritoneal cavity (MmPca), nasal epithelium (MmNep), and nervus olfactorius (MmNol) (35) (Table 1). Tb1Lu cells are T. brasiliensis lung cells. We also generated immortalized Nyctalus noctula cell lines from lung (NnLu), liver (NnLi), and kidney (NnKi) (Table 1). Betacoronaviruses have been sampled in species belonging to all 4 bat genera (9–14). Human intestinal Caco-TC7 cells and human lung A549 cells, which are both representative of tissues targeted by the virus in infected patients (36), were used as controls. All cells were infected with SARS-CoV-2 at an MOI of 1. Around 23% of Caco-TC7 cells were positive for the viral S protein at 24 hpi (Fig. 1C and D). None of the other selected cells were permissive for productive infection (Fig. 1C and D).
The lack of production of viral protein in the primary and immortalized bat cells, as well as in A549 cells, might be explained by the absence of one or several key proviral factor(s) and/or the presence of potent antiviral factor(s).
Expression of endogenous ACE2 and ectopically expressed hACE2 in bat cell lines.
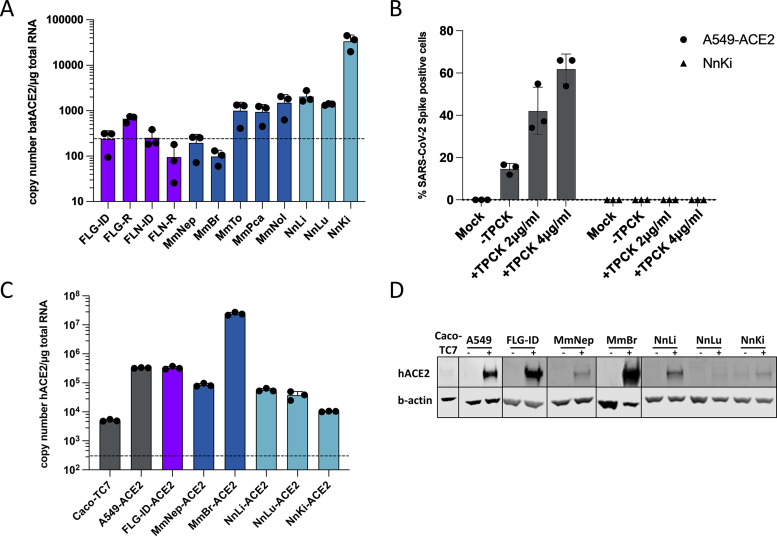
To determine whether the absence or low expression of ACE2 was the main limiting factor for SARS-CoV-2 replication in the selected bat cell lines, we evaluated the level of ACE2 expression by RT-qPCR analysis. The levels of ACE2 were above the detection limit in FLG-R, MmTo, MmPca, MmNol cells and in the three Nn cells (Fig. 2A). These cells did not, however, support viral replication (Fig. 1C and D). Thus, the SARS-CoV-2 S protein may have a low affinity for ACE2 expressed in these cells. An alternative explanation might be that the cells analyzed were deficient in expression of both TMPRSS2 and CTSL. To test this hypothesis, viral input was treated with the serine-protease trypsin to activate the S protein and allow viral fusion in a TMPRSS2- and CTSL-independent manner (22). The assay was performed with NnKi cells since they express the highest level of ACE2 of all bat cells (Fig. 2A). A549-ACE2 cells, which stably overexpressed ACE2, served as positive-control cells as they express low levels of TMPRSS2 and moderate levels of CTSL (22). Preactivation of viruses with trypsin increased the percentage of A549-ACE2 cells that were expressing the viral protein S, in a concentration-dependent manner (Fig. 2B). NnKi cells were resistant to productive infection with virions treated or not with trypsin (Fig. 2B), suggesting that cleavage of the S protein is not the factor limiting viral infection in NnKi cells.
FIG 2.
Expression of endogenous ACE2 or ectopically expressed hACE2 in bat cell lines. (A) Quantification of copy numbers per μg of total cellular RNA of endogenously expressed ACE2 in indicated bat cell lines via qPCR analysis. (B) A549-ACE2 and NnKi cells were left uninfected (Mock) or were infected with SARS-CoV-2 at an MOI of 1 in the absence of FBS and TPCK (–TPCK) or in the absence of FBS and the presence of trypsin TPCK at 2 or 4 μg/mL. The percentages of S-positive cells were determined by flow cytometric analysis. Data points represent three biological replicates. (C and D) Indicated bat and human cell lines were stably transduced with a lentivirus vector expressing the hACE2 gene and selected with blasticidin treatment. Human Caco-TC7 intestine cell line served as nontransduced control. (B) The amount of ectopically expressed hACE2 gene in each cell line was measured by qPCR analysis and is indicated as the gene copy number per μg of total cellular RNA. (C) Whole-cell lysates were analyzed by Western blotting with antibodies against the indicated proteins. Western blots are representative of two independent experiments. In panels A and C, a dashed line indicates the limit of detection in the qPCR assays.
To bypass entry-mediated restriction(s) and subsequently investigate the ability of SARS-CoV-2 to replicate in bat cells, we used lentiviral transduction to stably express hACE2 in bat cells. A549-ACE2 served as positive-control cells. Six of the thirteen bat cell lines, representing three species (Myotis myotis, Nyctalus noctula, and Eptesicus serotinus) tolerated the lentiviral transduction and antibiotic selection. We used RT-qPCR and Western blot analyses to evaluate hACE2 expression in these cell lines (Fig. 2C and D). RT-qPCR analysis revealed that hACE2 mRNA abundances in all transduced cells were higher than in Caco-TC7 cells (Fig. 2C), which support SARS-CoV-2 replication (Fig. 1C and D). This suggests that the transduced cells expressed hACE2 at levels that should permit viral entry. In line with the RT-qPCR analysis (Fig. 2C), Western blot analysis showed that MmBr-ACE2 cells expressed the highest level of hACE2 among all transduced cell lines (Fig. 2D). ACE2 was barely detectable in Caco-TC7 cells (Fig. 2D). A faint band was also detected in nontransduced NnKi cells, likely representing endogenous bACE2. This suggests that N. noctula ACE2 is recognized by the antibody raised against hACE2 in this assay and that NnKi cells expressed higher levels of ACE2 than lung and liver cells from the same bat. These data are in line with the RT-qPCR analysis of endogenous ACE2 expression (Fig. 2A). These assays revealed that the level of hACE2 expression varied considerably among transduced bat cell lines. Expression of hACE2 and antibiotic resistance are under the control of two different promoters in the bicistronic lentiviral vector used. Variable strength of the two promoters in the different cell lines could generate cells that survived the antibiotic treatment but did not express hACE2 (or very little of it). Nevertheless, despite expressing differential levels of hACE2, the transduced cells offered the opportunity to investigate the interaction between viruses belonging to the SARS-CoV-2 lineage and bat cells.
Expression of hACE2 allows efficient replication of SARS-CoV-2 in Eptesicus serotinus and Myotis myotis brain cells.
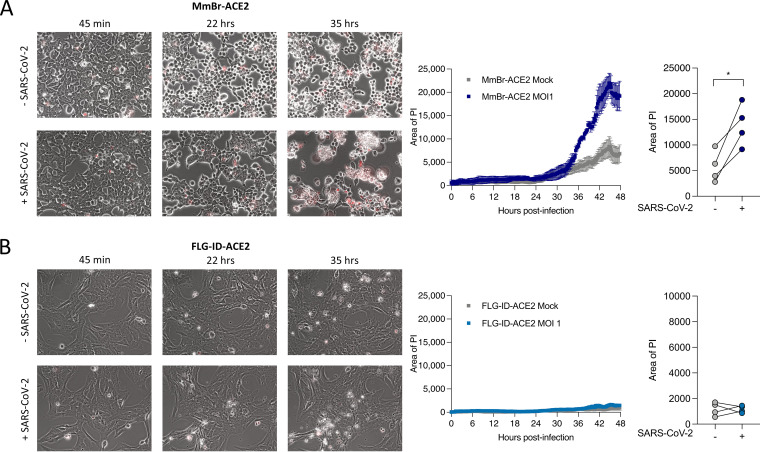
The six transduced bat cell lines and A549-ACE2 cells were infected with SARS-CoV-2 for 24 h at an MOI of 1. Cytopathic effects (CPEs) were observed in MmBr-ACE2 cells. To illustrate this, we performed time-lapse microscopy of MmBr-ACE2 cells, infected or not, in the presence of propidium iodide (PI) for 48 h. Cells were rapidly forming syncytia (around 12 h). Cell death was observed as early as 34 h, as assessed by the PI uptake due to the loss of membrane integrity (Fig. 3A and Movies S1 and S2 in the supplemental material). Syncytia represent cell-to-cell fusing events mediated by the interaction between cell-surface expressed S protein and ACE2 (37). Neither CPE nor syncytium formation were observed in the other cells, as illustrated by the video of infected FLG-ID-ACE2 cells (Fig. 3B and movies 3 and 4).
FIG 3.
Time-lapse microscopy of Myotis myotis and Eptesicus serotinus brain cells during SARS-CoV-2 infection. MmBr-ACE2 (A) and FLG-ID-ACE2 (B) cells were left uninfected (Mock) or infected with SARS-CoV-2 at an MOI of 1 in medium containing propidium iodide (PI) as a cell death marker. Images were taken every 10 min. Quantification of cell death (area of PI) displayed on the right of corresponding video cutouts. The results indicate means ± the SD from three fields per condition.
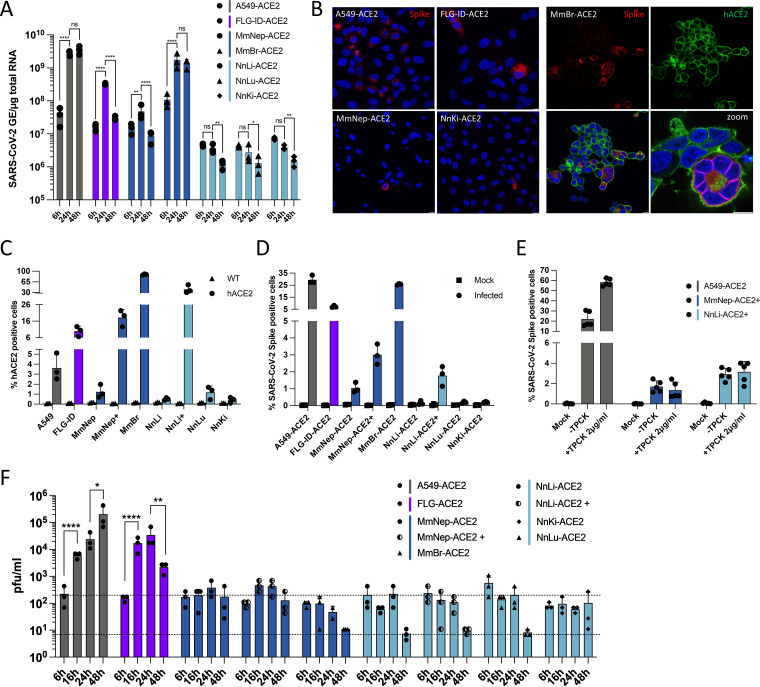
To avoid cell death, MmBr-ACE2 cells were infected with 25 times lower virus doses (MOI of 0.04) than the other cells (Fig. 4). Assessment of viral replication by RT-qPCR revealed that viral RNA yields increased between 6 and 24 hpi in A549-ACE2 cells, and subsequently reached a plateau (Fig. 4A). Viral RNA yields also increased between 6 and 24 hpi in FLG-ID-ACE2 cells but then dropped back to their 6-h levels at 48 hpi (Fig. 4A), suggesting that these cells efficiently controlled viral replication. Viral RNA abundance slightly increased between 6 and 24 hpi in MmNep-ACE2 cells (Fig. 4A), suggesting a low level of viral RNA production, before decreasing at 48 hpi. The profile of viral RNA yield was similar in MmBr-ACE2 cells than in A549-ACE2 cells (Fig. 4A), indicating a robust viral replication, especially when considering that the cells were infected with 25 times less viruses than the others. No increase in viral RNA yield was observed in the three Nn cell lines between 6 hpi and later time points (Fig. 4A), suggesting an absence of viral replication.
FIG 4.
Expression of hACE2 allows efficient replication of SARS-CoV-2 in Myotis myotis and Eptesicus serotinus cells. Transduced bat cell lines were left uninfected (Mock) or were infected with SARS-CoV-2 at an MOI of 1, with the exception of MmBr cells that were infected at an MOI of 0.04 (A, B, and E). (A) The relative amounts of cell-associated viral RNA were determined by qPCR analysis and are expressed as genome equivalents (GE) per μg of total cellular RNA at different times postinfection. All results are expressed as fold increases relative to uninfected cells. (B) Infected cells were stained at 24 hpi with anti-SARS-CoV-2 S protein (red) and/or anti-hACE2 antibodies (green). Nuclei were stained with Nucblue (blue). Scale bar, 10 μm. (C and D) The percentages of the indicated cells that expressed hACE2 (C) or SARS-CoV-2 S proteins (D) were determined by flow cytometric analysis at 24 hpi. (E) A549-ACE2, MmNep-ACE2+, and NnLi-ACE2+ cells were left uninfected (Mock) or were infected with SARS-CoV-2 at an MOI of 1 in the absence of FBS and TPCK (–TPCK) or in the absence of FBS and presence of trypsin TPCK at 2 μg/mL. The percentages of S-positive cells were determined by flow cytometric analysis. Data points represent two independent experiments (containing two or three technical replicates). (F) The presence of extracellular infectious viruses in the culture medium of the indicated cells was determined by TCID50 assays with Vero E6 cells at 6, 16, 24, and 48 hpi. The lower dashed line indicates the limit of detection, and the upper dashed line indicates the viral input (based on the average titration values at 6 h). (A, D, E, and F) Data points represent three independent experiments. Statistical tests: (a) Dunnett’s multiple-comparison test on a two-way ANOVA (n.s: not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001); (e and f) Tukey’s multiple-comparison test on a two-way ANOVA (ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
A549-ACE2, FLG-ID-ACE2, MmNep-ACE2 and MmBr-ACE2 cells, which are competent for viral RNA production (Fig. 4A), were analyzed for the expression of viral proteins through immunofluorescence imaging using antibodies specific for S and hACE2 at 24 hpi. NnKi-ACE2 cells were included in the analysis for comparison. Cells positive for S protein were observed in all cell lines (Fig. 4B). However, the proportion of positive cells seemed to vary considerably among cell lines (Fig. 4B), which agrees with the disparities in the ability of the cells to sustain a productive infection (Fig. 4A). As expected (Fig. 4A), almost none of the NnKi-ACE2 cells were expressing the S protein (Fig. 4B). An hACE2 signal was only detected in MmBr-ACE2 cells (Fig. 4B). These cells expressed the highest levels of hACE2 among the transduced cell lines (Fig. 2C and D). Thus, the selected anti-hACE2 antibodies appeared to allow detection by immunofluorescence analysis only when the protein was expressed at high levels. The confocal images also confirmed the presence of syncytia in MmBr-ACE2 infected cells (Fig. 4B).
Flow cytometry analyses were used to further evaluate hACE2 expression in transduced bat cells. They revealed that around 80% of MmBr-ACE2 cells were positive for hACE2 (Fig. 4C), which is in line with the RT-qPCR and Western blot and analyses (Fig. 2C and D). About 10% of FLG-ID-ACE2 brain cells were positive for hACE2 (Fig. 4C). On average, 1 to 4% of A549-ACE2 and MmNep-ACE2 cells were positive for hACE2 and even lower percentages of Nn cells were expressing hACE2 (Fig. 4C). These low percentages were surprising in light of the RT-qPCR and Western blot data (Fig. 2C and D). Cells counted as negative for hACE2 signal may, however, express levels that are under the detection limit of the assay. Alternatively, anti-ACE2 antibodies may recognize only a subpopulation of the protein by cytometry, such as, for instance, glycosylated and/or truncated forms (38, 39). To reduce the disparities between cell lines and to obtain cell populations that were enriched for hACE2 expression (ACE2+), live MmNep-ACE2 and Nn-ACE2 cells were sorted based on hACE2 cell surface expression. Our attempts to produce NnKi-ACE2+ and NnLu-ACE2+ cells remained unsuccessful. In contrast, flow cytometric analysis revealed a significant enrichment of cells expressing hACE2 in MmNep-ACE2+ and NnLi cells-ACE2+ cells (Fig. 4C). On average, 20% of MmNep-ACE2+ cells and 40% of NnLi-ACE2+ cells were positive for hACE2 (Fig. 4C).
Flow cytometry analyses were performed to estimate the number of transduced cells positive for S. On average 25% of A549-ACE2 cells were positive for S proteins when infected for 24 h at an MOI of 1 (Fig. 4D). This was surprising since only 3% of them appeared hACE2-positive (Fig. 4C). Knowing that these cells are not permissive to viral replication in the absence of hACE2 overexpression (Fig. 1C), these results further suggest that the anti-hACE2 antibodies recognized only a subpopulation of hACE2. Around 25% of MmBr-ACE2 cells and 5% of FLG-ID-ACE2 were positive for S protein when infected for 24 h at an MOI of 0.04 or 1, respectively (Fig. 4D). Very little MmNep-ACE2 cells (around 1%) were expressing the S protein upon infection. Despite being significantly more enriched in ACE2-positive cells (Fig. 4C), only 3% of MmNep-ACE2+ cells were infected (Fig. 4D). Similarly, NnLi-ACE2+ cells, which expressed significantly more ACE2 than the initial NnLi-ACE2 population (Fig. 4C), were poorly infected (Fig. 4D). This suggests that expression of hACE2 was not sufficient to allow robust viral replication in NnLi-ACE2 and MmNep-ACE2 cells. Less than 0.2% of the NnLu-ACE2 and NnKi-ACE2 cells were positive for the S protein (Fig. 4D). These flow cytometry data were consistent with viral RNA yield (Fig. 4A). Trypsin activation did not enhanced infection of MmNep-ACE2+ or NnLi-ACE2+ cells (Fig. 4E), suggesting that cleavage of the S protein was not the factor limiting viral infection in these cells.
To assess whether infectious virions were released from transduced bat cells, all cells were all infected at an MOI of 1, and supernatants were collected at 6, 16, 24, and 48 hpi. Viral titers were evaluated on Vero E6 cells (Fig. 4F). Approximately 200 PFU/mL were collected from the supernatant of all cell lines at 6 hpi (Fig. 4F). These infectious viruses represent input viruses that were carried over from the inoculum. A549-ACE2 cells released around 8 × 103 and 2 × 104 PFU/mL at 16 and 24 hpi, respectively (Fig. 4F). Production of infectious viruses reached 2 × 105 PFU/mL at 48 hpi in these cells (Fig. 4F). Despite producing around 1 log less viral RNA than A549-ACE2 cells at 24 hpi (Fig. 4A), FLG-ID-ACE2 cells released similar amounts of infectious particles at 24 hpi (Fig. 4F). This suggests that A549-ACE2 cells produce more noninfectious defective genomes than FLG-ID-ACE2 cells. Significantly less infectious particles were produced by FLG-ID-ACE2 cells at 48 hpi than at 24 hpi (Fig. 4F), which is in accordance with a decrease of viral RNA production between 24 and 48 hpi (Fig. 4A) and further suggests a control of viral infection. Approximately 200 PFU/mL were collected from the supernatant of Mm and Nn transduced cell lines at all time point, including the two cell lines enriched in ACE2, which suggests that none of these cells release infectious particles. Based on viral RNA and viral protein quantification (Fig. 4A and D), transduced Nn cells were not expected to produced infectious particles. In contrast, it was surprising that MmBr-ACE2 cells did not release infectious virions since they produced viral RNA and proteins (Fig. 4A and D).
Together, these data revealed that expression of hACE2 allowed the virus to complete its replication cycle in E. serotinus FLG-ID brain cells, suggesting that once the ACE2-mediated refractory state to SARS-CoV replication is overcome, these cells are permissive for productive infection. Overcoming the ACE2-mediated restriction in M. myotis brain cells stably expressing hACE2 (MmBr-ACE2) renders the cells competent for production of viral RNA and proteins. Infectious particles, however, were not released from these cells, suggesting the existence of another cellular restriction at a late stage of the viral replication cycle. In MmNep-ACE2+ and NnLi-ACE2+ cells, high expression of hACE2 was not sufficient to allow robust viral replication, suggesting a deficiency in key proviral factor(s) and/or expression of potent antiviral factor(s) that act at an early stage or viral replication.
An abortive entry route exists in bat cells.
To investigate further ACE2-mediated restriction, we performed viral binding and entry assays on a selection of bat cells, transduced or not with hACE2 (Fig. 5A). Infected cells were kept on ice for 1 h, washed three times and then either lysed (“on ice”) or incubated at 37°C for 2 or 6 h. To remove potential residual bound particles, the warmed cells were treated with trypsin for 30 min prior to lysis (Fig. 5A). We performed the assays with A549, FLG-ID, and NnKi cells since they tolerated the three washes on ice without detaching from the plates. For each cell line, we compared viral RNA abundance in wild-type versus hACE2-expressing cells. Viral RNA detected in cells that were kept on ice represent input viruses bound to cellular membranes. In all six cell lines, we indeed detected viral RNA bound to cell membranes (Fig. 5B), suggesting that hACE2 expression is not required for viral attachment to the cell surface. Such ACE2-independent binding of the S protein could be mediated by heparan sulfate, as described in several human cell lines (40, 41), or by endogenous bat ACE2 when it is expressed at detectable levels (Fig. 2A). hACE2 expression may, however, enhance viral binding to A549 and FLG cell membranes since around 500 more viral genome copies per μg of total RNA were detected in cold transduced cells than in unmodified ones (Fig. 5B). The abundance of viral RNAs increased between 2 and 6 h both in A549-ACE2 and FLG-ACE2 cells but not in wild-type cells (Fig. 5B). These results confirm that viral replication occurred in hACE-2-expressing A549 and FLG cells (Fig. 4). No increase in viral RNA yield was observed between 2 and 6 h in NnKi-ACE2 cells (Fig. 5B), confirming the inability of the virus to replicate in these cells (Fig. 4). Viral RNA detected at 2 or 6 hpi in nontransduced cells (Fig. 5B) may represent viruses that remained attached to the cell surface despite the trypsin treatment or viruses that penetrated the cells via an hACE2-independent route.
FIG 5.
An abortive entry route exists in bat and human cells. (A) Scheme summarizing the experimental workflow. Cells were incubated with SARS-CoV-2 at an MOI of 1 for 1 h on ice to allow viral attachment. After extensive washing, a portion of the cells was lysed (“on ice”), and the remaining cells were incubated for 1.5 or 5.5 h at 37°C to permit viral internalization. After the incubation period, these cells were lysed after 30 min of trypsinization to remove bound viruses from the cell surface (“2 h” and “6 h”). (B) The relative amounts of cell-associated viral RNA were determined by qPCR analysis and are expressed as genome equivalents (GE) per μg of total cellular RNA as depicted in panel A. All results are expressed as fold increases relative to uninfected cells. Data points represent three independent experiments. Statistical test: Dunnett’s multiple-comparison test on a two-way ANOVA (ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (C to F) The indicated cell lines were infected with VSV* (at MOIs ranging from 0.001 to 5) or with VSV*ΔG-S at an MOI of 7. The percentages of infected cells were determined at around 16 to 18 hpi based on GFP expression and flow cytometry analysis. Data points represent independent experiments. Dotted lines represent the limit of sensibility of the assay. (C) BHK21, Caco-TC7, A549, and MmNep cells were infected at an MOI of 0.001, MmBr cells at an MOI of 0.05, FLG-ID cells at an MOI of 5, NnLi cells at an MOI of 0.5, and NnKi cells at an MOI of 0.1. (D) A549-ACE2 and MmBr-ACE2 cells were infected at an MOI of 0.001, MmNep-ACE2 at an MOI of 0.005, FLG-ID-ACE2 cells at an MOI of 5, NnLi cells at an MOI of 0.1, and NnKi cells at an MOI of 0.05.
To address whether SARS-CoV-2 enters and fuses with endosomal membranes of bat cells, we took advantage of a chimeric VSV*ΔG-SARS-CoV-2-SΔ21 (VSV*ΔG-S), which lacks its homotypic glycoprotein G but encodes for the spike protein of SARS-CoV-2 (Wuhan-Hu-1 strain), along with green fluorescent protein (GFP) instead (42). Human A549 and hamster BHK-21 cells, which are not susceptible to SARS-CoV-2 (20, 43), were used as negative controls, while human A549-ACE2 and Caco-TC7 cells served as positive controls. We first tested the ability of a GFP-recombinant VSV harboring its glycoprotein G (VSV*) to replicate in FLG-ID, MmNep, MmBr, NnLi, and NnKi cells. VSV* replicated in all transduced and nontransduced bat and human cell lines, as well as in BHK-21 cells (Fig. 5C and D), albeit at different levels. For instance, around 90% of MmBr cells were expressing GFP when infected with an MOI of 0.001 while only 10% of FLG cells were GFP positive when infected with an MOI of 5 (Fig. 5D).
As expected, Caco-TC7 cells were susceptible to VSV*ΔG-S, whereas BKH-21 cells were not (Fig. 5E). No GFP-positive A549 cells were identified following VSV*ΔG-S infection (Fig. 5E). However, detection of viral RNA in A549 cells at early times postinfection (Fig. 5B) and at 24 hpi (44) suggests that the virus can be internalized by these cells. The fusion step may thus be the step limiting viral replication in A549 cells. Nontransduced FLG-ID, MmNep, NnLi, and NnKi cells were not susceptible to S-mediated entry of the chimeric virus (Fig. 5E). In contrast, around 2% of nontransduced MmBr cells were GFP positive (Fig. 5E), indicating that at least a fraction of MmBr cells are competent for viral entry and fusion.
Around 15% of A549-ACE2 cells and 1% of FLG-ID-ACE2 cells were expressing GFP upon VSV*ΔG-S infection (Fig. 5F). This was surprising since both cell lines express similar level of hACE2 (Fig. 2C). This difference could be due to a poor replication of VSV in FLG-ID cells (Fig. 5C). The percentages of MmNep-ACE2, MmNep-ACE2+, MmBR-ACE2, NnLi-ACE2, NnLi-ACE2+, and NnKi-ACE2 cells that were expressing GFP varied considerably from one cell line to another (Fig. 5F). These percentages correlated with the level of hACE2 expression (Fig. 4C). For instance, around 30% of MmNep-ACE2+ cells and 10% of NnLi-ACE2+ cells were expressing GFP upon VSV*ΔG-S infection, while only around 1% of MmNep-ACE2 cells and NnLi-ACE2 were GFP positive (Fig. 5E). Thus, the level of hACE2 expression in transduced bat cells largely contribute to their susceptibility.
Infectious particles are produced by MmBr-ACE2 cells but are not released.
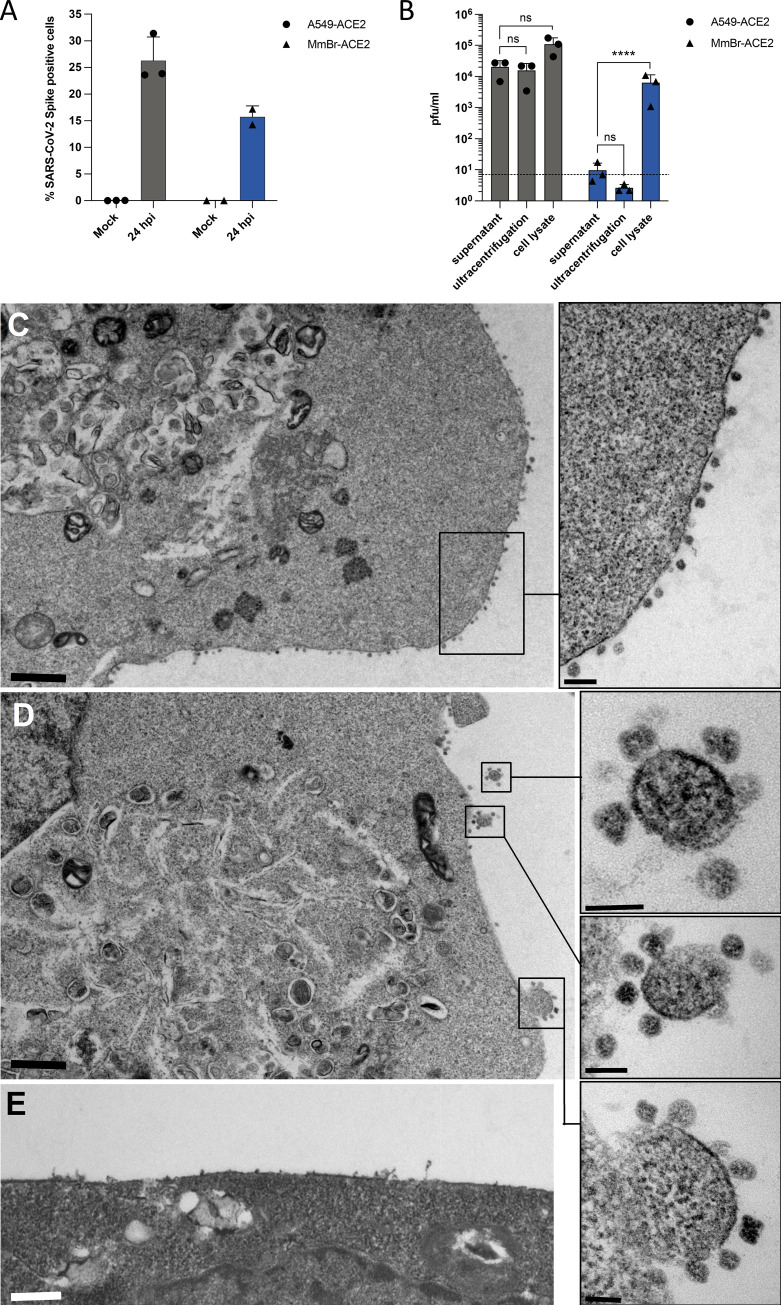
Since MmBr-ACE2 cells sustained the production of viral RNA and proteins (Fig. 4A and D), we were intrigued by the absence of infectious particle release (Fig. 4F). Despite infecting these cells with an MOI of 0.04 (Fig. 4A, B, and D) to reduce the CPEs observed at an MOI of 1 (Fig. 3), we wondered whether cytokines released by infected cells and/or dying cells may stimulate damage-associated molecular patterns (DAMPs) and thus trigger an antiviral response inhibiting viral replication in Vero E6 cells. Other possibilities include a defect in viral assembly and/or in viral transport through the secretory pathway in MmBr-ACE2 cells. Alternatively, these cells may only produce immature noninfectious viral particles. To investigate these hypotheses, supernatants collected from MmBr-ACE2 cells, and as controls, from A549-ACE2 cells, were titrated on Vero E6 cells. Prior to titration, supernatants were first clarified to remove cell debris and then subjected to ultracentrifugation to pellet the virus and remove potential cytokines. Flow cytometry analyses using anti-S antibodies were done on the same samples to verify that the cells were infected (Fig. 6A). Supernatants from A549-ACE2 cells contained around 104 PFU/mL infectious particles (Fig. 6B). In line with previous analysis (Fig. 4F), only infectious particles representing input viruses were recovered in supernatants, ultracentrifuged or not, of infected MmBr-ACE2 cells (Fig. 6B). These data suggest that immunostimulatory components, such as dying cells or cytokines, that may be present in infected MmBr-ACE2 cell culture supernatants did not affect the results of the titration assays. To assess the presence of intracellular infectious particles in MmBr-ACE2 cells, the titration assays were performed on crude cell lysates collected at 24 hpi. Around 1 log more infectious particles was retrieved from lysed A549-ACE2 cells than from their supernatants (Fig. 6B). About 104 PFU/mL were retrieved from lysed MmBr-ACE2 cells, which is around 3 logs more than in the culture supernatant (Fig. 6B), suggesting that viral assembly and maturation takes place in these cells. The absence of viral release may thus be due to a defect in viral transport through the secretory pathway.
FIG 6.
Infectious particles are retained at the surface of MmBr-ACE2 cells. A549-ACE2 and MmBr-ACE2 cells were left uninfected (Mock) or were infected for 24 h at an MOI of 1 or 0.04, respectively. (A) The percentages of cells that contained SARS-CoV-2 S proteins were determined by flow cytometric analysis. (B) The presence of extracellular infectious viruses in the culture medium of the cells was determined by TCID50 assays performed on Vero E6 cells. Supernatants were either clarified or clarified and purified by ultracentrifugation. Alternatively, cell-associated infectious virions were titrated on Vero E6 cells from whole-cell lysates. Data points represent three independent experiments. Statistical test: Dunnett’s multiple-comparison test on a two-way ANOVA (ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). (C to E) At 16 h postinfection, numerous viral particles were observed on the cell surfaces of MmBr-ACE2 cells, attached to the plasma membrane (C and D). Boxed areas in the low-magnification photographs in panels C and D are shown at higher magnification in the right panels. (E) Uninfected cells showed no viral particles on their surfaces. The scale bars are 1 mm in panels C and D, 0.5 mm in panel E, and 200 and 100 nm in the high-magnification panels extracted from images C and D, respectively.
To investigate the fate of infectious virions in MmBr-ACE2 cells, we performed a transmission electron microscopy analysis of cells infected at an MOI of 0,04 for 24 h (Fig. 6C and D). Noninfected cells served as negative-control cells (Fig. 6E). As previously described in SARS-CoV-2 infected cells (45, 46), viral replication factories, which consisted of double-membrane vesicles, were observed in infected MmBr-ACE2 cells (Fig. 6C and D). The virions were around 80 to 100 nm in diameter, which is the expected size for SARS-CoV-2 particles (46). The images showed that virions remained bound to the surface of infected MmBr-ACE2 cells (Fig. 6C and D). Such retention could be due to an endogenous expression of tetherin, an interferon (IFN)-induced gene known to retain SARS-CoV-2 at the cell surface of human cells (47) or to a high expression of hACE2.
Viral IFN counteraction mechanisms are species specific.
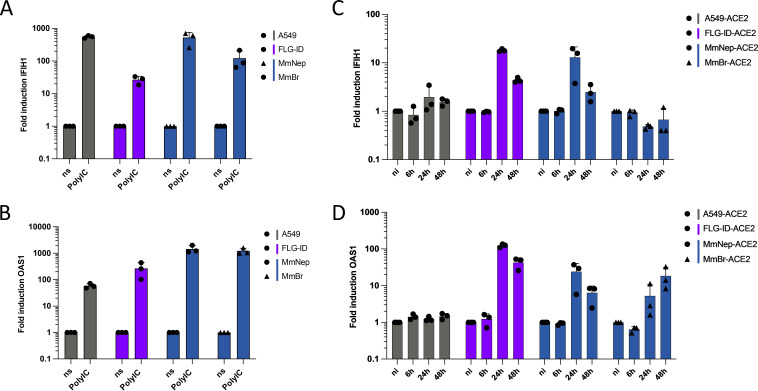
Quantification of intracellular viral RNA and titration assays revealed that FLG-ACE2 cells, to a lesser extent, MmNep-ACE2 cells, controlled viral replication over time (Fig. 4A). In contrast, viral RNA yield remained high between 24 and 48 hpi in A549-ACE2 and MmBr-ACE2 cells (Fig. 4A). To assess whether the IFN response could contribute to viral containment in FLG-ACE2 and MmNep-ACE2 cells, we compared mRNA abundance of two IFN-stimulated genes (ISGs) upon stimulation or infection in these four cell lines. We selected OAS1 and IFIH1, two ISGs that are conserved across vertebrate species (48). Moreover, OAS1 expression is associated with reduced COVID-19 death (49) and IFIH1 codes for Mda5, the protein responsible for sensing SARS-CoV-2 replication intermediates and thus initiating the IFN response, in human cells (50, 51). We first evaluated the expression of the two selected ISGs upon transfection with poly(I·C), a synthetic dsRNA analog. All four cell lines contained transcripts for these two ISGs and responded well to the stimulation (Fig. 7A and B), demonstrating that they possess intact IFN induction and signaling pathways.
FIG 7.
Viral IFN counteraction mechanisms are species-specific. (A and B) Nontransduced cell lines were transfected with 250 ng of low-molecular-weight poly(I·C) or treated with PBS for 16 h. The relative amounts of IFIH1 mRNA (a) and OAS1 mRNA (b) were determined by qPCR analysis. The results are expressed as fold increases relative to unstimulated PBS-treated cells. (C and D) Whole-cell lysates of infected cells (the same lysates used for the viral quantification in Fig. 4A) were analyzed via RT-qPCR assays for the relative amounts of IFIH1 mRNA (c) and OAS1 mRNA (d). The results are expressed as fold increases relative to uninfected cells. (A to D) Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) of corresponding species was used as a housekeeping gene. Data points represent three independent experiments.
We then evaluated the mRNA abundance of these two ISGs in cells infected for 6, 24, and 48 h (Fig. 7C and D). No increase of OAS1 and IFIH1 expression was observed in A549-ACE2 cells (Fig. 7C and D). This agrees with a previous report showing that infection of A549-ACE2 by SARS-CoV-2 is characterized by an absence of IFN response (52). In contrast, the abundance of OAS1 and IFIH1 transcripts increased between 6 and 24 hpi in FLG-ACE2 and MmNep-ACE2 cells (Fig. 7C and D) and remained elevated at 48 hpi in both cell lines (Fig. 7C and D). In MmBr-ACE2 cells, the infection triggered the induction of OAS1 expression but not of IFIH1 (Fig. 7C and D), suggesting that the virus is able to dampen the expression of at least one ISG in this cell type.
Together, our data confirmed that SARS-CoV-2 efficiently counteracts ISG induction in A549-ACE2 cells (52) and revealed that it is not the case in FLG-ACE2 and MmNep-ACE2 cells. The control of viral replication observed in these two cell lines (Fig. 5) could thus be due to the expression of a set of ISGs with potent antiviral functions. Interestingly, IFN-mediated barriers could also differ in bat cells derived from different organs since the virus seems to dampen IFIH1 expression in MmBr-ACE2 cells but not in MmNep-ACE2 cells.
DISCUSSION
The development of novel bat cellular models is essential to understand the molecular mechanisms underlying the ability of bats to serve as reservoirs for numerous viruses, including alpha- and betacoronaviruses. We first produced R. ferrumequinum, M. myotis, M. nattereri, and M. brandtii primary cells to evaluate their permissivity to infection with SARS-CoV-2. None of them supported viral replication, not even R. ferrumequinum cells, which have been isolated from bats belonging to the same genus as the bat host of BANAL-52, a potential ancestor of SARS-CoV-2 (2). These primary cells, which were generated from patagium biopsy specimens of living bats, exhibited a dermal-fibroblast phenotype. A single-cell transcriptomic analysis showed that R. sinicus skin cells express moderate levels of ACE2 and very little TMPRSS2 (53). The virus may thus be able to enter the skin primary cells, but the fusion between viral and membrane may not take place as the S protein is not cleaved. Further experiments will be required to identify at which step of the replication cycle the virus is stopped in these primary cells.
We established the first three immortalized Nyctalus noctula cell lines using liver, kidney, and lung tissues from a single bat. These organs may be physiologically relevant for bat infection since bat coronaviruses, such as MERS-CoV, infect the lungs and livers of fruit bats (1). In addition to coronaviruses, N. noctula carry other viruses with zoonotic potential such as paramyxoviruses and hantaviruses (54, 55). The Nn cells that we have developed represent thus novel opportunities to study bat-borne viruses. We found that Nn kidney cells expressed higher levels of ACE2 than Nn cells derived from the lung or liver. Likewise, ACE2 is expressed at high levels in R. sinicus kidney, as revealed by comparative single-cell transcriptomic (53) and in silico analysis of ACE2 expression pattern in various tissues. ACE2 is also highly expressed in human kidney (56). Thus, kidney cells appear relevant to study betacoronavirus replication.
Myotis myotis, Tadarida brasiliensis, Eptesicus serotinus, and Nyctalus noctula cells were resistant to infection. ACE2 from M. myotis and T. brasiliensis, as well as from a species of the Eptesicus genus, permitted S-mediated entry of pseudotyped VSV when ectopically expressed in human cells refractory to SARS-CoV-2 infection (57). This means that when expressed at high levels, ACE2 from these three species interacts with the viral S protein. As in human A549 cells, ACE2 may be expressed at a level which is too low to allow viral entry in our bat cell lines. The potential ability of N. noctula ACE2 to bind S protein has not been reported and the genome of this bat genus is yet to be sequenced. Hence, low affinity between S protein and ACE2 and/or low level of ACE2 expression may hamper viral replication in these cells. Our results highlight the importance of performing experiments in the context of genuine infection of bat cells to predict their ability to support viral replication.
SARS-CoV-2 RNA was detected in nontransduced A549, FLG-ID, and NnKi cells at early times postinfection. Since the virus could not be removed by trypsin treatment of the cells, it likely represents input virus that entered the cells via an ACE2-independent manner, as described previously for Vero E6 cells (22) and human H522 lung adenocarcinoma cells (58). The VSV-based entry assay clearly showed that A549, FLG-ID, and NnKi cells are not susceptible to S-mediated infection. On the contrary, the same assay demonstrated that a subset of MmBr cells is susceptible to S-mediated infection, suggesting that these cells express all the factors necessary for binding, entry, and viral fusion. However, since they were not permissive to SARS-CoV-2, they probably lack factors that are essential for viral replication. Alternatively, they may express potent viral restriction factors that remain to be identified.
To bypass entry-mediated restriction(s), we generated eight chimeric bat cell lines stably expressing hACE2. Four cell lines (MmNep-ACE2, NnLi-ACE2, NnLu-ACE2, and NnKi-ACE2) expressed too little hACE2 to draw conclusion about a potential rescue of infection. Four cell lines (FLG-ID-ACE2, MmBr-ACE2, MmNep-ACE2+, and NnLi-ACE2+) expressed more hACE2 than A549-ACE2, which were competent for viral replication. The virus completed its replication cycle in FLG-ID-ACE2 cells. MmBr-ACE2 cells were competent for production of viral RNA and proteins but not for infectious particles release. The virus replicated poorly in MmNep-ACE2+ and NnLi-ACE2+ cells, even when pretreated with trypsin. A protease-independent restriction may thus exist in these cells. The various permissivities of these four cell lines to SARS-CoV-2 infection offer opportunities to decipher species-specific and tissue-specific antiviral mechanisms that have evolved in bats. Efficient viral RNA and protein production in A549-ACE2, FLG-ID-ACE2, and MmBr-ACE2 cells suggest that they express proteases that cleave S proteins. It also shows that ACE2 alone was responsible for the lack of viral replication in nontransduced A549, FLG-ID and MmBr cells. This ACE2-mediated entry block might be rather due to a low or absent ACE2 expression than to an incompatibility between ACE2 and S protein since ectopic expression of Myotis sp. and Eptesicus sp. ACE2 facilitated S-mediated entry of pseudoviruses (43, 57). Infectious particles were produced in MmBr cells but were retained at the cell surface. This block may be mediated by the restriction factor tetherin, which retains numerous enveloped viruses, including SARS-CoV-2, at the surface of human cells (47). An analysis of the tetherin gene of 27 species of bats indicates that bats have undergone tetherin gene expansion and diversification relative to other mammals (59). Bats belonging to the genus Myotis possess five unique tetherin variants that may potently restrict SARS-CoV-2 release (59). Alternatively, infectious virion retention in MmBr cells could be due to an overexpression of hACE2 and may thus not be bat specific.
Since infection induced S-mediated syncytium formation in MmBr cells, viruses might spread from cell-to-cell via syncytia, as do other syncytium-forming viruses such as respiratory syncytial virus, parainfluenza viruses and measles virus (60). Syncytium-mediated intercellular spreading allows viruses to escape virus-neutralizing antibodies. Such mode of transport has been previously proposed in human cells infected with the Middle East respiratory syndrome coronavirus (MERS-CoV) (61), another betacoronavirus. Analysis of postmortem samples of patients that succumb of COVID-19 revealed the presence of syncytial pneumocytes positives for viral RNAs (62). However, the significance of syncytium formation for virus pathogenicity remains to be investigated.
SARS-CoV-2 has evolved numerous synergetic mechanisms to evade the IFN response in human cells (63), resulting in an absence of IFN and ISG expression in some cells, including A549 cells (52, 64). The virus was unable to counteract OAS1 and IFIH1 induction in Eptesicus serotinus kidney cells and in Myotis myotis nasal epithelial cells. This is especially intriguing in E. serotinus cells since the virus replicates to high levels in these cells and thus produce viral proteins with described IFN antagonist activities. Similarly, MERS-CoV suppresses the antiviral IFN response in human cells but not in E. fuscus cells (65). One can envisage that escape of IFN-mediated restriction by betacoronaviruses is species specific. For instance, SARS-CoV-2 Nsp14 targets human IFNAR1 for lysosomal degradation (63) but may be unable to degrade bat IFNAR1. This inability to evade the expression of two ISGs in E. serotinus kidney cells and in Myotis myotis nasal epithelial cells may contribute to the cellular control of infection in these cells, as in experimentally infected E. fuscus (66). Alternatively, the basal level of IFN may be high in these two cell lines, as reported in several other bat species (67–69). Expression of a mutated form of IRF3, which is a key transcription factor involved in the induction of the IFN signaling cascade, contributes to enhanced IFN responses in bat species, including E. fuscus, compared to humans (70). Investigation of IRF7, another transcription factor that mediates IFN expression, in Pteropus alecto cells revealed a more widespread tissue distribution in bats than in humans (71, 72). Bats may thus launch IFN-dependent measures against viruses in a faster and broader manner than humans (73). Expression of atypical ISGs has been reported for different bat species, including RNA-degrading RNase L (RNaseL) in P. alecto cells and RNA-binding Microrchidia 3 (MORC3) in Pteropus vampyrus and Eidolon helvum cells (74, 75). Pursuing the characterization of bat innate immunity in relevant in vitro models is essential to understand the mechanisms by which they control the replication of numerous unrelated viruses.
An obvious need to develop additional bat cell lines still remains (30). Particularly valuable cells would be cells derived from bat intestine, a tissue that expresses high level of proteins known to mediate or facilitate cellular entry of bat-borne betacoronaviruses, such as ACE2 and TMPRSS2, at least in R. sinicus (53). This tissue is relevant for coronavirus infection, as demonstrated by the detection of viral genomes in duodenum tissue of Rousettus aegypticus experimentally infected with SARS-CoV-2 (29) and in anal swabs of Rhinolophus bats infected with SC2r-CoVs (76).
MATERIALS AND METHODS
Bat primary cells.
M. myotis samples were collected in July 2020 from two bat colonies in Inca and Llucmajor on Mallorca (Balearic Islands, Spain) (agreement CEP 31/2020 delivered by the Ministry of the Environment and Territory, government of the Balearic Islands). R. ferrumequinum biopsy specimens were collected in France in 2020. Authorization for bat capture was delivered by the French Ministry of Ecology, Environment, and Sustainable development (approval C692660703 from the Departmental Direction of Population Protection, Rhone, France). All methods were approved by the Société Française pour l’Étude et la Protection des Mammifères. Patagium biopsy specimens were shipped in freezing medium Cryo-SFM (PromoCell), on dry ice or at 4°C with ice packs. Primary cells were obtained as previously described (77, 78). Briefly, skin biopsy specimens were washed twice with sterile phosphate-buffered saline (PBS), excised in small pieces and enzymatically digested, either with 500 μL of collagenase D (1 mg/mL) (Roche) and overnight incubation at 37°C without agitation, or with 100 to 200 μL of TrypLE Express Enzyme (Gibco) and incubation 10 min at 37°C under gentle agitation. Dissociated cells and remaining pieces of tissue were placed in a single well of a 6-well plate containing 2 mL of Dulbecco modified Eagle medium (DMEM; Gibco) containing 20% heat-inactivated fetal bovine serum (FBS; Eurobio), 1% penicillin/streptomycin (P/S; Gibco), and 50 μg/mL gentamicin (Gibco) and then incubated at 37°C under 5% CO2. Cell cultures were regularly checked to determine the need for medium refreshment or splitting. After 5 to 10 passages, cells were grown in DMEM supplemented with 10% FBS.
Cell lines.
FLG-ID, FLG-R, FLN-ID, FLN-R, and Tb1Lu cell lines (Table 1) were maintained in equal volumes of Ham F-12 and Iscove modified Dulbecco medium (Gibco), supplemented with 10% FBS and 1% P/S (Gibco) in nonvented flasks. Mm cells, which were obtained from a single common serotine bat (Eptesicus serotinus), were previously described (35). Nn kidney-, liver-, and lung-derived cell cultures were obtained from a common noctule bat (Nyctalus noctula) euthanized because of poor chance of survival associated with traumatic injuries sustained while a dead tree sheltering bat hibernaculum was cut. The decision to euthanize the specimen was made by a veterinarian following inspection of a group of noctule bats presented for examination and therapy in the rescue center at the University of Veterinary and Pharmaceutical Sciences Brno, Czech Republic, in November 2015 (79). The bat was anesthetized with isofluranum (Piramal Enterprises, Ltd.) and euthanized by quick decapitation. The cadaver was immersed into 96% ethanol for a few seconds and then subjected to necropsy under aseptic conditions to collect organs, which were loosened mechanically with scalpel blades, minced into small pieces, suspended in DMEM (Biosera) containing 1 mg/mL collagenase (Thermo Fisher Scientific) and 1 mg/mL trypsin (Sigma-Aldrich), and incubated at 37°C on a shaking Thermoblock for 45 min. The cells were then separated through a 100 μm nylon filter and washed twice in a medium supplemented with 10% FBS to stop enzymatic digestion. The cells yielded in this way were cultured in DMEM supplemented with 10% FBS and 1% P/S (Sigma). Primary cells were immortalized by transfection of pRSVAg1 plasmid expressing simian vacuolating virus 40 large T antigen (SV40T) with lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol, expanded and cryopreserved. Mm and Nn cell lines (Table 1), as well as African green monkey Vero E6 cells (ATCC CRL-1586), human lung epithelial A549 cells (a gift from Frédéric Tangy, Institut Pasteur, Paris, France) and human colorectal adenocarcinoma Caco-TC7 cells (ATCC HTB-37), were maintained in DMEM (Gibco), supplemented with 10% FBS and 1% P/S in vented flasks. All cells were maintained at 37°C in a humidified atmosphere with 5% CO2. Bat and A549 cells were modified to stably express hACE2 using the pLenti6-hACE2 lentiviral transduction as described previously (37). Briefly, 2 × 105 cells were resuspended in 150 μL of culture medium containing 15 μL of ultracentrifuged lentiviral vectors supplemented with 2 mM HEPES (Gibco) and 4 μg/mL Polybrene (Sigma). Cells were agitated for 30 s every 5 min for 2.5 h at 37°C in a Thermomixer and then plated. At 48 h after transduction, blasticidin (concentrations ranging from 7 to 15 μg/mL depending on cell lines) was added in the culture media.
Viruses and infections.
The SARS-CoV-2 strain BetaCoV/France/IDF0372/2020 (historical) and hCoV-19/France/PDL-IPP01065/2021 (20H/501Y.V2 or SA) were supplied by the French National Reference Centre for Respiratory Viruses hosted by Institut Pasteur (Paris, France) and headed by S. van der Werf. The human samples from which the historical and South African strains were isolated were provided by X. Lescure and Y. Yazdanpanah from the Bichat Hospital, Paris, France, and Vincent Foissaud, HIA Percy, Clamart, France, respectively. These strains were supplied through the European Virus Archive goes Global (EVAG) platform, a project that has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement 653316. The hCoV-19/Japan/TY7-501/2021 strain (20J/501Y.V3 or Brazil) was kindly provided by Jessica Vanhomwegen (Environment and Infectious Risks Research and Expertise Unit, Institut Pasteur). Viral stocks were produced by amplification on Vero E6 cells, for 72 h in DMEM supplemented with 2% FBS and 1% P/S. The cleared supernatant was stored at −80°C and titrated on Vero E6 cells by using standard plaque assays to measure PFU/mL. Cells were infected at the indicated MOIs in DMEM without FBS. Virus inoculum was either removed after 6 h and replaced or topped up with FBS containing culture medium to a final concentration of 2% FBS and 1% P/S. For infections with proteolytically activated SARS-CoV-2, cell monolayers were washed twice with PBS before adding virus inoculum in DMEM supplemented with 2 or 4 μg/mL of trypsin TPCK (Sigma) and no FBS. After 4h, DMEM containing FBS was added to a final concentration of 2%.
TCID50 assays.
Supernatants of infected cells were first cleared of cell debris by centrifugation at 3,500 rpm for 10 min at 4°C. They were 10-fold serially diluted in DMEM supplemented with 2% FBS and 1% P/S. For the “ultracentrifugation” condition in MmBr-ACE2 50% tissue culture infective dose (TCID50) assays, cleared supernatants were ultracentrifuged for 1 h at 45,000 rpm at 4°C. to remove cytokines and other proteins. Virus-containing pellets were resuspended in DMEM with 2% FBS and 1% P/S after 4 h of incubation at 4°C. For the “lysate” condition, infected MmBr-ACE2 or A459-ACE2 cells were lysed and scraped in ddH2O. After one freeze-thaw cycle, whole-cell lysates were cleared by centrifugation, supplemented with 10× PBS to a physiological condition and used for serial dilutions. Around 9 × 103 Vero E6 cells and 50-μL potions of serially diluted virus suspensions were deposited in 96-well plate in quintuplicate wells. Cells were fixed with 4% paraformaldehyde (PFA) for 30 min at room temperature and revealed with crystal violet 5 days later. CPEs were assessed by calculating the TCID50 values using the Spearman-Karber method (80).
Flow cytometry.
Cells were detached with trypsin or versene for hACE2 staining. Cells were then fixed in 4% PFA for 30 min at 4°C and staining was performed in PBS, 2% bovine serum albumin (BSA), 2 mM EDTA, and 0.1% saponin (FACS buffer). Cells were incubated with goat pAB anti-hACE2-647 (1:100, FAB933R; R&D Systems) and/or with antibodies recognizing the spike protein of SARS-CoV (anti-S, 1:1,000, GTX632604; Genetex) or anti-S mAb10 (1 μg/mL; kindly provided by Hugo Mouquet, Institut Pasteur, Paris, France) and subsequently with secondary antibodies anti-human Alexa Fluor-647 (1:1,000, A21455; Thermo), anti-mouse Alexa Fluor-488 (1:1000, A28175; Thermo) or Dylight488 (1:100, SA5-10166; Thermo) for 30 min at 4°C. Cells were acquired on an Attune NxT flow cytometer (Thermo Fisher), and data were analyzed using FlowJo software v10 (TriStar).
RNA extraction and RT-qPCR assays.
Total RNA was extracted from cells with the NucleoSpin RNA II kit (Macherey-Nagel) according to the manufacturer’s instructions. First-strand complementary DNA (cDNA) synthesis was performed with the RevertAid H Minus M-MuLV reverse transcriptase (Thermo Fisher Scientific) using random primers. For batACE2 determination, total RNA was treated with DNase I (DNase-free kit; Thermo Fisher Scientific) for 30 min at 37°C before cDNA synthesis with SuperScript IV reverse transcriptase. Quantitative real-time PCR was performed on a real-time PCR system (QuantStudio 6 Flex; Applied Biosystems) with Power SYBR green RNA-to-CT 1-Step kit (Thermo Fisher Scientific). Data were analyzed using the 2–ΔΔCT method, with all samples normalized to GAPDH. Genome equivalent concentrations were determined by extrapolation from a standard curve generated from serial dilutions of the pcDNA3.1-hACE2 plasmid (Addgene, 145033) or plasmids encoding a fragment of the RNA-dependent RNA polymerase/IP4 of SARS-CoV-2 or a fragment of the ACE2 genome of each bat species. Primers used for RT-qPCR analysis are listed in Table 2.
TABLE 2.
Primers used for RT-qPCR analysis
| Target gene | Primer sequence (5′–3′) |
|
|---|---|---|
| Forward | Reverse | |
| ACE2 human | GGACCCAGGAAATGTTCAGA | GGCTGCAGAAAGTGACATGA |
| ACE2 FLG | TGGGACTCTACCGTTCACTTA | GCTTCATCTCCCACCACTTT |
| ACE2 Mm | TGCTTATGTCAGGGCAAAGT | CCCACATATCACCAAGCAAATG |
| ACE2 Nn | CAGTCCTGGGATGCAGATAAG | TGGCTCAGTTAGCATGGATTTA |
| GAPDH human | GGTCGGAGTCAACGGATTTG | ACTCCACGACGTACTCAGCG |
| GAPDH FLG | TCATCAACGGAAAGTCCATCTC | ACATACTCAGCACCAGCATC |
| GAPDH Mm | GTAGTGAAGCAGGCATCAGAG | GGAGTGGGTGTCACTGTTAAA |
| GAPDH Nn | CCTGTTCGTCAGACAGCCTT | TTGATGGCGACAACTTGCAC |
| IFIH1 human | ACA CGT TCT TTG CGA TTT CC | ACC AAA TAC AGG AGC CAT GC |
| IFIH1 Mm/FLG | GGAGTCAAAGCCCACCATCT | TCCAGACCTTCTTCTGCCAC |
| IFIH1 Nn | TTTGCCAAGTGAGCCCAATG | AAGCGGTCTTTGCGATTTCC |
| OAS human | GAGCTCCTGACGGTCTATGC | TTCGTGAGCTGCCTTCTCAG |
| OAS pan-bat | GGAAGGAGGGCGAGTTCTC | GGTACCAGTGCTTGACCAGG |
| SARS-CoV-2 nsp12 polymerase | GGTAACTGGTATGATTTC | CTGGTCAAGGTTAATATAGG |
Cloning of qPCR amplicon.
To quantify the amounts of bat ACE2 in each cell line, plasmids containing the qPCR amplicon obtained with the primers described in Table 2 were generated via TOPO cloning. Briefly, total RNA was extracted from a cadaver of Myotis myotis stored at the University of Veterinary and Pharmaceutical Sciences in Brno. For the remaining two bat species, total RNA extracted from NnKi and FLG-R cells were used. RNA was treated for 30 min at 37°C with DNase I and cDNA synthesized with SuperScript IV reverse transcriptase. These cDNAs were then used as the template for PCR amplification of the qPCR bACE2 amplicon using the primers in Table 2 and Phusion high-fidelity DNA polymerase (Thermo). PCR products were gel-purified (NucleoSpin gel and PCR cleanup kit; Macherey-Nagel) and cloned into pCR-Blunt II-TOPO vectors using a Zero Blunt TOPO PCR cloning kit (Thermo). Inserts were verified via Sanger sequencing. Plasmids were then used as quantitative qPCR standards.
Western blot analysis.
Proteins extracted from cell lysates were resolved by SDS-PAGE on 4 to 12% NuPAGE Bis-Tris gel (Life Technologies) with MOPS running buffer and semidry transferred to a nitrocellulose membrane with Trans-Blot Turbo system (Bio-Rad). After blocking with 0.05% Tween 20 in PBS containing 5% dry milk powder for 1 h at room temperature, the membrane was incubated with goat pAB anti-hACE2-700 (1:200, FAB933N R&D Systems) and mouse monoclonal antibody anti-β-actin (1:5,000, A5316; Sigma) diluted in blocking buffer overnight at 4°C. The membranes were then incubated with DyLight800 secondary antibody (1:5,000, 46421; Thermo) diluted in blocking buffer for 1 h. Finally, the membranes were revealed using an Odyssey CLx infrared imaging system (LI-COR Bioscience).
Immunofluorescence microscopy and live cell imaging.
Cells grown on glass coverslips were fixed in 4% PFA for 30 min at room temperature and permeabilized with 0.2% Triton X-100 (Sigma/Merck) in PBS for 10 min at room temperature. After blocking with 3% BSA (Sigma) in PBS for 1 h at room temperature, cells were incubated with goat pAB anti-hACE2 (1:50, AF933; R&D Systems) and monoclonal antibody anti-SARS-CoV-2-spike (1:1,000, GTX632604; Genetex) in 1% BSA in PBS (AB buffer) for 1 h at room temperature or overnight at 4°C. Subsequently, cells were incubated with anti-goat Alexa 488 (A-11055; Thermo Fisher Scientific) and anti-mouse Alexa 555 (A21427; Thermo Fisher Scientific) secondary antibodies diluted 1:500 in antibody buffer for 30 min at room temperature. Finally, cells were stained with NucBlue Fixed Cell ReadyProbes reagent (Thermo) in PBS for 5 min at room temperature. Coverslips were washed with ultrapure water (Gibco) and mounted in ProLong Gold antifade (Life Technologies). Sample were visualized with a Leica TCS SP8 confocal microscope (Leica Microsystems), and a white light excitation laser and a 405-nm diode laser were used for excitation. Confocal images were taken with an automatically optimized pixel format, a 4× frame averaging, and a scan speed of 400 Hz through an HC PL APO CS2 63× NA 1.4 oil immersion objective. Overlay pictures of single channel images were digitally processed in Leica LAS X lite software. For live imaging, 5.4 × 104 to 105 cells were plated per quadrant in a μ‐Dish 35 mm Quad dish (80416; Ibidi). Cells were infected the next day with SARS‐CoV‐2 at an MOI of 1 in culture media supplemented with 2.5% FBS and 1% P/S containing propidium iodide. Transmission and fluorescence images were taken at 37°C every 15 min, for up to 48 h, using a Nikon BioStation IMQ, with three fields for each condition.
Attachment and entry assays.
Cells plated in monolayers were prechilled on ice for 10 min and washed once with cold PBS. Cells were then incubated with SARS-CoV-2 at an MOI of 1 for 1 h on ice. After three washes with cold PBS, half of the cells was lysed in RA1 lysis buffer (Macherey-Nagel) (“on ice”). The second half of the cells was trypsinized for 15 min on ice and 15 min at 37°C after washing the virus inoculum and then washed with PBS and lysed (“on ice + trypsin”). The remaining cells were directly transferred to 37°C after washing of the virus inoculum and incubated for 2 or 6 h in warm culture media supplemented with 2% FBS and 1% P/S. After this incubation period, these cells were trypsinized for 30 min at 37°C, washed with PBS, and lysed in RA1 buffer (“2 h” and “6 h”). Finally, the total RNA was extracted from all cell lysates using a NucleoSpin RNA II kit (Macherey-Nagel).
VSV-based entry assays.
VSV encoding GFP has been previously described as VSV* (81). The chimeric virus VSV*ΔG-SARS-CoV-2-SΔ21 (VSV*ΔG-S), which lacks the homotypic glycoprotein G but rather encodes the spike protein of SARS-CoV-2 (Wuhan-Hu-1 strain), along with GFP, has recently been described (42). Cells were seeded at 1 × 105 cells per well in 24-well plates in DMEM with 1% FBS. The next day, cells were infected with VSV* (at the indicated MOI) or VSV*ΔG-S (MOI of 7) in DMEM without FBS. The virus suspension was removed after 2 h and replaced with DMEM with 1% FBS. At 16 hpi, the cells were washed once with PBS, trypsinized, and subsequently fixed in 4% PFA. Fixed cells were washed once with PBS and analyzed by flow cytometry. The percentage of infected cells was identified based on GFP expression.
Transmission electron microscopy.
Infected and mock-infected MmBr-ACE2 cells were fixed at 16 hpi by incubation for 24 h in 1% glutaraldehyde/4% paraformaldehyde (Sigma, St. Louis, MO) in 0.1 M phosphate buffer (pH 7.2). Samples were then washed in PBS and postfixed by incubation for 1 h with 2% osmium tetroxide (Agar Scientific, Stansted, UK). The cells were then fully dehydrated in a graded series of ethanol solutions and propylene oxide. They were impregnated with a 1:1 mixture of propylene oxide/Epon resin (Sigma) and left overnight in pure resin. Samples were then embedded in Epon resin (Sigma), which was allowed to polymerize for 48 h at 60°C. Ultrathin sections (90 nm) of these blocks were obtained with a Leica EM UC7 ultramicrotome (Wetzlar, Germany). Sections were stained with 2% uranyl acetate (Agar Scientific) and 5% lead citrate (Sigma), and observations were made with a transmission electron microscope (JEM-1011; JEOL, Tokyo, Japan).
Poly(I·C) stimulation.
Cells were plated in monolayers in 24-well culture plates. The next day, they were transfected with 250 ng low-molecular-weight poly(I·C) (InvivoGen) or PBS, respectively, using INTERFERin (Polyplus Transfection) transfection reagent. Cells were lysed 16 h after transfection, and the total RNA was extracted using a NucleoSpin RNA II kit (Macherey-Nagel).
Statistical analysis.
Graphical representation and statistical analyses were performed using Prism version 9.0.2 software (GraphPad). Unless otherwise stated, results are shown as means ± the standard deviations (SD) from three independent experiments. Significance was calculated using either Dunnett’s multiple-comparison test with two-way analysis of variance (ANOVA) or Tukey’s multiple-comparison test with two-way ANOVA, as indicated. Statistically significant differences are indicated by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant).
ACKNOWLEDGMENTS
We thank Noémie Aurine (SBRI, U1208 INSERM, France) for help designing RT-qPCR primers for bat samples; Ondine Filippi-Codaccioni and Marc López-Roig (Université Lyon 1, France) for help in bat sampling; the French National Reference Centre for Respiratory Viruses hosted by Institut Pasteur (France) and headed by S. van der Werf for providing the historical viral strains; Hugo Mouquet and Cyril Planchais (Institut Pasteur) for providing anti-S antibodies; Françoise Porrot (Institut Pasteur) for lentiviral production; Florence Guivel-Benhassine (Institut Pasteur) for help in titration assays; and Matthias Lenk (Friedrich-Loeffler-Institut, Germany) for providing the E. serotinus and Tadarida brasiliensis cell lines. We are grateful to the members of our laboratories for helpful discussions. We acknowledge the UTechS Photonic BioImaging (Imagopole), C2RT, Institut Pasteur, supported by the French National Research Agency (France BioImaging; ANR-10-INBS-04; Investments for the Future) for the use of the confocal microscope.
This study was funded by the CNRS (N.J. and O.S.), Institut Pasteur (N.J., L.D., and O.S.), the “Urgence COVID-19” fundraising campaign of Institut Pasteur (N.J., L.D., and O.S.), the University of Veterinary Sciences Brno (FVHE/Pikula/ITA2021) (J.P.), the FINOVI Fondation (AO13) and Covid IDEX Université Lyon 1 (B.P.), LabEx Ecofect (ANR-11-LABX-0048) (Do.P.), Labex IBEID (ANR-10-LABX-62-IBEID) (O.S.), ANR/FRM Flash Covid PROTEO-SARS-CoV-2 (O.S.), and IDISCOVR (O.S.). S.-M.A. and De.P. are supported by the Pasteur-Paris University International Ph.D. Program and the Vaccine Research Institute (ANR-10-LABX-77), respectively. D.L. was funded by the Chinese Scholarship Council and Institut Pasteur. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We declare that no competing interests exist.
Footnotes
Supplemental material is available online only.
Contributor Information
Laurent Dacheux, Email: laurent.dacheux@pasteur.fr.
Nolwenn Jouvenet, Email: nolwenn.jouvenet@pasteur.fr.
Mark T. Heise, University of North Carolina at Chapel Hill
REFERENCES
- 1.Ruiz-Aravena M, McKee C, Gamble A, Lunn T, Morris A, Snedden CE, Yinda CK, Port JR, Buchholz DW, Yeo YY, Faust C, Jax E, Dee L, Jones DN, Kessler MK, Falvo C, Crowley D, Bharti N, Brook CE, Aguilar HC, Peel AJ, Restif O, Schountz T, Parrish CR, Gurley ES, Lloyd-Smith JO, Hudson PJ, Munster VJ, Plowright RK. 2022. Ecology, evolution and spillover of coronaviruses from bats. Nat Rev Microbiol 20:299–314. 10.1038/s41579-021-00652-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Temmam S, Vongphayloth K, Baquero E, Munier S, Bonomi M, Regnault B, Douangboubpha B, Karami Y, Chrétien D, Sanamxay D, Xayaphet V, Paphaphanh P, Lacoste V, Somlor S, Lakeomany K, Phommavanh N, Pérot P, Dehan O, Amara F, Donati F, Bigot T, Nilges M, Rey FA, van der Werf S, Brey PT, Eloit M. 2022. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature 604:330–336. 10.1038/s41586-022-04532-4. [DOI] [PubMed] [Google Scholar]
- 3.Zhou H, Chen X, Hu T, Li J, Song H, Liu Y, Wang P, Liu D, Yang J, Holmes EC, Hughes AC, Bi Y, Shi W. 2020. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol 30:2196–2203. 10.1016/j.cub.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wacharapluesadee S, Tan CW, Maneeorn P, Duengkae P, Zhu F, Joyjinda Y, Kaewpom T, Chia WN, Ampoot W, Lim BL, Worachotsueptrakun K, Chen VC-W, Sirichan N, Ruchisrisarod C, Rodpan A, Noradechanon K, Phaichana T, Jantarat N, Thongnumchaima B, Tu C, Crameri G, Stokes MM, Hemachudha T, Wang L-F. 2021. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat Commun 12:972. 10.1038/s41467-021-21768-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delaune D, Hul V, Karlsson EA, Hassanin A, Ou TP, Baidaliuk A, Gámbaro F, Prot M, Tu VT, Chea S, Keatts L, Mazet J, Johnson CK, Buchy P, Dussart P, Goldstein T, Simon-Lorière E, Duong V. 2021. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. Nat Commun 12:6563. 10.1038/s41467-021-26809-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murakami S, Kitamura T, Suzuki J, Sato R, Aoi T, Fujii M, Matsugo H, Kamiki H, Ishida H, Takenaka-Uema A, Shimojima M, Horimoto T. 2020. Detection and characterization of bat sarbecovirus phylogenetically related to SARS-CoV-2, Japan. Emerg Infect Dis 26:3025–3029. 10.3201/eid2612.203386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjee A, Kulcsar K, Misra V, Frieman M, Mossman K. 2019. Bats and coronaviruses. Viruses 11:41. 10.3390/v11010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frutos R, Serra-Cobo J, Pinault L, Lopez Roig M, Devaux CA. 2021. Emergence of bat-related betacoronaviruses: hazard and risks. Front Microbiol 12:591535. 10.3389/fmicb.2021.591535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Benedictis P, Marciano S, Scaravelli D, Priori P, Zecchin B, Capua I, Monne I, Cattoli G. 2014. Alpha and lineage C βCoV infections in Italian bats. Virus Genes 48:366–371. 10.1007/s11262-013-1008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee S, Jo S-D, Son K, An I, Jeong J, Wang S-J, Kim Y, Jheong W, Oem J-K. 2018. Genetic characteristics of coronaviruses from Korean bats in 2016. Microb Ecol 75:174–182. 10.1007/s00248-017-1033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falcón A, Vázquez-Morón S, Casas I, Aznar C, Ruiz G, Pozo F, Perez-Breña P, Juste J, Ibáñez C, Garin I, Aihartza J, Echevarría JE. 2011. Detection of alpha and betacoronaviruses in multiple Iberian bat species. Arch Virol 156:1883–1890. 10.1007/s00705-011-1057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lecis R, Mucedda M, Pidinchedda E, Pittau M, Alberti A. 2019. Molecular identification of Betacoronavirus in bats from Sardinia (Italy): first detection and phylogeny. Virus Genes 55:60–67. 10.1007/s11262-018-1614-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lelli D, Papetti A, Sabelli C, Rosti E, Moreno A, Boniotti MB. 2013. Detection of coronaviruses in bats of various species in Italy. Viruses 5:2679–2689. 10.3390/v5112679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patterson EI, Elia G, Grassi A, Giordano A, Desario C, Medardo M, Smith SL, Anderson ER, Prince T, Patterson GT, Lorusso E, Lucente MS, Lanave G, Lauzi S, Bonfanti U, Stranieri A, Martella V, Solari Basano F, Barrs VR, Radford AD, Agrimi U, Hughes GL, Paltrinieri S, Decaro N. 2020. Evidence of exposure to SARS-CoV-2 in cats and dogs from households in Italy. Nat Commun 11:6231. 10.1038/s41467-020-20097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koopmans M. 2021. SARS-CoV-2 and the human-animal interface: outbreaks on mink farms. Lancet Infect Dis 21:18–19. 10.1016/S1473-3099(20)30912-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cool K, Gaudreault NN, Morozov I, Trujillo JD, Meekins DA, McDowell C, Carossino M, Bold D, Mitzel D, Kwon T, Balaraman V, Madden DW, Artiaga BL, Pogranichniy RM, Sosa GR, Henningson J, Wilson WC, Balasuriya UBR, García-Sastre A, Richt JA. 2021. Infection and transmission of ancestral SARS-CoV-2 and its alpha variant in pregnant white-tailed deer. Emerg Microbes Infect:1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olival KJ, Cryan PM, Amman BR, Baric RS, Blehert DS, Brook CE, Calisher CH, Castle KT, Coleman JTH, Daszak P, Epstein JH, Field H, Frick WF, Gilbert AT, Hayman DTS, Ip HS, Karesh WB, Johnson CK, Kading RC, Kingston T, Lorch JM, Mendenhall IH, Peel AJ, Phelps KL, Plowright RK, Reeder DM, Reichard JD, Sleeman JM, Streicker DG, Towner JS, Wang L-F. 2020. Possibility for reverse zoonotic transmission of SARS-CoV-2 to free-ranging wildlife: a case study of bats. PLoS Pathog 16:e1008758. 10.1371/journal.ppat.1008758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. 2020. Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280.e8. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ge X-Y, Li J-L, Yang X-L, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, Zhang Y-J, Luo C-M, Tan B, Wang N, Zhu Y, Crameri G, Zhang S-Y, Wang L-F, Daszak P, Shi Z-L. 2013. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503:535–538. 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murgolo N, Therien AG, Howell B, Klein D, Koeplinger K, Lieberman LA, Adam GC, Flynn J, McKenna P, Swaminathan G, Hazuda DJ, Olsen DB. 2021. SARS-CoV-2 tropism, entry, replication, and propagation: considerations for drug discovery and development. PLoS Pathog 17:e1009225. 10.1371/journal.ppat.1009225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koch J, Uckeley ZM, Doldan P, Stanifer M, Boulant S, Lozach P-Y. 2021. TMPRSS2 expression dictates the entry route used by SARS-CoV-2 to infect host cells. EMBO J 40:e107821. 10.15252/embj.2021107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Damas J, Hughes GM, Keough KC, Painter CA, Persky NS, Corbo M, Hiller M, Koepfli K-P, Pfenning AR, Zhao H, Genereux DP, Swofford R, Pollard KS, Ryder OA, Nweeia MT, Lindblad-Toh K, Teeling EC, Karlsson EK, Lewin HA. 2020. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci USA 117:22311–22322. 10.1073/pnas.2010146117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu H, Chan JF-W, Yuen TT-T, Shuai H, Yuan S, Wang Y, Hu B, Yip CC-Y, Tsang JO-L, Huang X, Chai Y, Yang D, Hou Y, Chik KK-H, Zhang X, Fung AY-F, Tsoi H-W, Cai J-P, Chan W-M, Ip JD, Chu AW-H, Zhou J, Lung DC, Kok K-H, To KK-W, Tsang OT-Y, Chan K-H, Yuen K-Y. 2020. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe 1:e14–e23. 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J, Queen K, Tao Y, Paden CR, Zhang J, Li Y, Uehara A, Wang H, Goldsmith C, Bullock HA, Wang L, Whitaker B, Lynch B, Gautam R, Schindewolf C, Lokugamage KG, Scharton D, Plante JA, Mirchandani D, Widen SG, Narayanan K, Makino S, Ksiazek TG, Plante KS, Weaver SC, Lindstrom S, Tong S, Menachery VD, Thornburg NJ. 2020. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg Infect Dis 26:1266–1273. 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lau SKP, Wong ACP, Luk HKH, Li KSM, Fung J, He Z, Cheng FKK, Chan TTY, Chu S, Aw-Yong KL, Lau TCK, Fung KSC, Woo PCY. 2020. Differential tropism of SARS-CoV and SARS-CoV-2 in bat cells. Emerg Infect Dis 26:2961–2965. 10.3201/eid2612.202308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou J, Li C, Liu X, Chiu MC, Zhao X, Wang D, Wei Y, Lee A, Zhang AJ, Chu H, Cai J-P, Yip CC-Y, Chan IH-Y, Wong KK-Y, Tsang OT-Y, Chan K-H, Chan JF-W, To KK-W, Chen H, Yuen KY. 2020. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med 26:1077–1083. 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 29.Schlottau K, Rissmann M, Graaf A, Schön J, Sehl J, Wylezich C, Höper D, Mettenleiter TC, Balkema-Buschmann A, Harder T, Grund C, Hoffmann D, Breithaupt A, Beer M. 2020. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. Lancet Microbe 1:e218–e225. 10.1016/S2666-5247(20)30089-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banerjee A, Misra V, Schountz T, Baker ML. 2018. Tools to study pathogen-host interactions in bats. Virus Res 248:5–12. 10.1016/j.virusres.2018.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dominguez SR, O’Shea TJ, Oko LM, Holmes KV. 2007. Detection of group 1 coronaviruses in bats in North America. Emerg Infect Dis 13:1295–1300. 10.3201/eid1309.070491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendenhall IH, Kerimbayev AA, Strochkov VM, Sultankulova KT, Kopeyev SK, Su YCF, Smith GJD, Orynbayev MB. 2019. Discovery and characterization of novel bat coronavirus lineages from Kazakhstan. Viruses 11:356. 10.3390/v11040356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogando NS, Dalebout TJ, Zevenhoven-Dobbe JC, Limpens RWAL, van der Meer Y, Caly L, Druce J, de Vries JJC, Kikkert M, Bárcena M, Sidorov I, Snijder EJ. 2020. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J Gen Virol 101:925–940. 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richter M, Reimann I, Schirrmeier H, Kirkland PD, Beer M. 2014. The viral envelope is not sufficient to transfer the unique broad cell tropism of Bungowannah virus to a related pestivirus. J Gen Virol 95:2216–2222. 10.1099/vir.0.065995-0. [DOI] [PubMed] [Google Scholar]
- 35.He X, Korytář T, Zhu Y, Pikula J, Bandouchova H, Zukal J, Köllner B. 2014. Establishment of Myotis myotis cell lines: model for investigation of host-pathogen interaction in a natural host for emerging viruses. PLoS One 9:e109795. 10.1371/journal.pone.0109795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puelles VG, Lütgehetmann M, Lindenmeyer MT, Sperhake JP, Wong MN, Allweiss L, Chilla S, Heinemann A, Wanner N, Liu S, Braun F, Lu S, Pfefferle S, Schröder AS, Edler C, Gross O, Glatzel M, Wichmann D, Wiech T, Kluge S, Pueschel K, Aepfelbacher M, Huber TB. 2020. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med 383:590–592. 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buchrieser J, Dufloo J, Hubert M, Monel B, Planas D, Rajah MM, Planchais C, Porrot F, Guivel-Benhassine F, Van der Werf S, Casartelli N, Mouquet H, Bruel T, Schwartz O. 2021. Syncytium formation by SARS-CoV-2-infected cells. EMBO J 40:e107405. 10.15252/embj.2020107405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Onabajo OO, Banday AR, Stanifer ML, Yan W, Obajemu A, Santer DM, Florez-Vargas O, Piontkivska H, Vargas JM, Ring TJ, Kee C, Doldan P, Tyrrell DL, Mendoza JL, Boulant S, Prokunina-Olsson L. 2020. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. 12. Nat Genet 52:1283–1293. 10.1038/s41588-020-00731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allen JD, Watanabe Y, Chawla H, Newby ML, Crispin M. 2021. Subtle influence of ACE2 glycan processing on SARS-CoV-2 recognition. J Mol Biol 433:166762. 10.1016/j.jmb.2020.166762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clausen TM, Sandoval DR, Spliid CB, Pihl J, Perrett HR, Painter CD, Narayanan A, Majowicz SA, Kwong EM, McVicar RN, Thacker BE, Glass CA, Yang Z, Torres JL, Golden GJ, Bartels PL, Porell RN, Garretson AF, Laubach L, Feldman J, Yin X, Pu Y, Hauser BM, Caradonna TM, Kellman BP, Martino C, Gordts PLSM, Chanda SK, Schmidt AG, Godula K, Leibel SL, Jose J, Corbett KD, Ward AB, Carlin AF, Esko JD. 2020. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell 183:1043–1057.e15. 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Q, Chen CZ, Swaroop M, Xu M, Wang L, Lee J, Wang AQ, Pradhan M, Hagen N, Chen L, Shen M, Luo Z, Xu X, Xu Y, Huang W, Zheng W, Ye Y. 2020. Heparan sulfate assists SARS-CoV-2 in cell entry and can be targeted by approved drugs in vitro. Cell Discov 6:1–14. 1. 10.1038/s41421-020-00222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cramer J, Lakkaichi A, Aliu B, Jakob RP, Klein S, Cattaneo I, Jiang X, Rabbani S, Schwardt O, Zimmer G, Ciancaglini M, Abreu Mota T, Maier T, Ernst B. 2021. Sweet drugs for bad bugs: a glycomimetic strategy against the DC-SIGN-mediated dissemination of SARS-CoV-2. J Am Chem Soc 143:17465–17478. 10.1021/jacs.1c06778. [DOI] [PubMed] [Google Scholar]
- 43.Conceicao C, Thakur N, Human S, Kelly JT, Logan L, Bialy D, Bhat S, Stevenson-Leggett P, Zagrajek AK, Hollinghurst P, Varga M, Tsirigoti C, Tully M, Chiu C, Moffat K, Silesian AP, Hammond JA, Maier HJ, Bickerton E, Shelton H, Dietrich I, Graham SC, Bailey D. 2020. The SARS-CoV-2 Spike protein has a broad tropism for mammalian ACE2 proteins. PLoS Biol 18:e3001016. 10.1371/journal.pbio.3001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao Y, Xu X, Kitanovski S, Song L, Wang J, Hao P, Hoffmann D. 2021. Comprehensive comparison of RNA-Seq data of SARS-CoV-2, SARS-CoV and MERS-CoV infections: alternative entry routes and innate immune responses. Front Immunol 12:656433. 10.3389/fimmu.2021.656433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eymieux S, Rouillé Y, Terrier O, Seron K, Blanchard E, Rosa-Calatrava M, Dubuisson J, Belouzard S, Roingeard P. 2021. Ultrastructural modifications induced by SARS-CoV-2 in Vero cells: a kinetic analysis of viral factory formation, viral particle morphogenesis and virion release. Cell Mol Life Sci 78:3565–3576. 10.1007/s00018-020-03745-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cortese M, Lee J-Y, Cerikan B, Neufeldt CJ, Oorschot VMJ, Köhrer S, Hennies J, Schieber NL, Ronchi P, Mizzon G, Romero-Brey I, Santarella-Mellwig R, Schorb M, Boermel M, Mocaer K, Beckwith MS, Templin RM, Gross V, Pape C, Tischer C, Frankish J, Horvat NK, Laketa V, Stanifer M, Boulant S, Ruggieri A, Chatel-Chaix L, Schwab Y, Bartenschlager R. 2020. Integrative Imaging Reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe 28:853–866.e5. 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin-Sancho L, Lewinski MK, Pache L, Stoneham CA, Yin X, Becker ME, Pratt D, Churas C, Rosenthal SB, Liu S, Weston S, De Jesus PD, O’Neill AM, Gounder AP, Nguyen C, Pu Y, Curry HM, Oom AL, Miorin L, Rodriguez-Frandsen A, Zheng F, Wu C, Xiong Y, Urbanowski M, Shaw ML, Chang MW, Benner C, Hope TJ, Frieman MB, García-Sastre A, Ideker T, Hultquist JF, Guatelli J, Chanda SK. 2021. Functional landscape of SARS-CoV-2 cellular restriction. Mol Cell 81:2656–2668.e8. 10.1016/j.molcel.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaw AE, Hughes J, Gu Q, Behdenna A, Singer JB, Dennis T, Orton RJ, Varela M, Gifford RJ, Wilson SJ, Palmarini M. 2017. Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biol 15:e2004086. 10.1371/journal.pbio.2004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou S, Butler-Laporte G, Nakanishi T, Morrison DR, Afilalo J, Afilalo M, Laurent L, Pietzner M, Kerrison N, Zhao K, Brunet-Ratnasingham E, Henry D, Kimchi N, Afrasiabi Z, Rezk N, Bouab M, Petitjean L, Guzman C, Xue X, Tselios C, Vulesevic B, Adeleye O, Abdullah T, Almamlouk N, Chen Y, Chassé M, Durand M, Paterson C, Normark J, Frithiof R, Lipcsey M, Hultström M, Greenwood CMT, Zeberg H, Langenberg C, Thysell E, Pollak M, Mooser V, Forgetta V, Kaufmann DE, Richards JB. 2021. A Neanderthal OAS1 isoform protects individuals of European ancestry against COVID-19 susceptibility and severity. Nat Med 27:659–667. 10.1038/s41591-021-01281-1. [DOI] [PubMed] [Google Scholar]
- 50.Yin X, Riva L, Pu Y, Martin-Sancho L, Kanamune J, Yamamoto Y, Sakai K, Gotoh S, Miorin L, De Jesus PD, Yang C-C, Herbert KM, Yoh S, Hultquist JF, García-Sastre A, Chanda SK. 2021. MDA5 governs the innate immune response to SARS-CoV-2 in lung epithelial cells. Cell Rep 34:108628. 10.1016/j.celrep.2020.108628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rebendenne A, Valadão ALC, Tauziet M, Maarifi G, Bonaventure B, McKellar J, Planès R, Nisole S, Arnaud-Arnould M, Moncorgé O, Goujon C. 2021. SARS-CoV-2 triggers an MDA-5-dependent interferon response which is unable to control replication in lung epithelial cells. J Virol 95:e02415-20. 10.1128/JVI.02415-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blanco-Melo D, Nilsson-Payant BE, Liu W-C, Uhl S, Hoagland D, Møller R, Jordan TX, Oishi K, Panis M, Sachs D, Wang TT, Schwartz RE, Lim JK, Albrecht RA, tenOever BR. 2020. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181:1036–1045. 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ren L, Wu C, Guo L, Yao J, Wang C, Xiao Y, Pisco AO, Wu Z, Lei X, Liu Y, Shi L, Han L, Zhang H, Xiao X, Zhong J, Wu H, Li M, Quake SR, Huang Y, Wang J, Wang J. 2020. Single-cell transcriptional atlas of the Chinese horseshoe bat (Rhinolophus sinicus) provides insight into the cellular mechanisms which enable bats to be viral reservoirs. Cell Biol. https://www.biorxiv.org/content/10.1101/2020.06.30.175778v1. [Google Scholar]
- 54.Straková P, Dufkova L, Širmarová J, Salát J, Bartonička T, Klempa B, Pfaff F, Höper D, Hoffmann B, Ulrich RG, Růžek D. 2017. Novel hantavirus identified in European bat species Nyctalus noctula. Infect Genet Evol 48:127–130. 10.1016/j.meegid.2016.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurth A, Kohl C, Brinkmann A, Ebinger A, Harper JA, Wang L-F, Mühldorfer K, Wibbelt G. 2012. Novel paramyxoviruses in free-ranging European bats. PLoS One 7:e38688. 10.1371/journal.pone.0038688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hikmet F, Méar L, Edvinsson Å, Micke P, Uhlén M, Lindskog C. 2020. The protein expression profile of ACE2 in human tissues. Mol Syst Biol 16:e9610. 10.15252/msb.20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yan H, Jiao H, Liu Q, Zhang Z, Xiong Q, Wang B-J, Wang X, Guo M, Wang L-F, Lan K, Chen Y, Zhao H. 2021. ACE2 receptor usage reveals variation in susceptibility to SARS-CoV and SARS-CoV-2 infection among bat species. Nat Ecol Evol 5:600–609. 10.1038/s41559-021-01407-1. [DOI] [PubMed] [Google Scholar]
- 58.Puray-Chavez M, LaPak KM, Schrank TP, Elliott JL, Bhatt DP, Agajanian MJ, Jasuja R, Lawson DQ, Davis K, Rothlauf PW, Liu Z, Jo H, Lee N, Tenneti K, Eschbach JE, Shema Mugisha C, Cousins EM, Cloer EW, Vuong HR, VanBlargan LA, Bailey AL, Gilchuk P, Crowe JE, Diamond MS, Hayes DN, Whelan SPJ, Horani A, Brody SL, Goldfarb D, Major MB, Kutluay SB. 2021. Systematic analysis of SARS-CoV-2 infection of an ACE2-negative human airway cell. Cell Rep 36:109364. 10.1016/j.celrep.2021.109364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hayward JA, Tachedjian M, Johnson A, Irving AT, Gordon TB, Cui J, Nicolas A, Smith I, Boyd V, Marsh GA, Baker ML, Wang L-F, Tachedjian G. 2020. Unique evolution of antiviral tetherin in bats. bioRxiv. https://www.biorxiv.org/content/10.1101/2020.04.08.031203v3. [DOI] [PMC free article] [PubMed]
- 60.Cifuentes-Muñoz N, Dutch RE, Cattaneo R. 2018. Direct cell-to-cell transmission of respiratory viruses: the fast lanes. PLoS Pathog 14:e1007015. 10.1371/journal.ppat.1007015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qian Z, Dominguez SR, Holmes KV. 2013. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytium formation. PLoS One 8:e76469. 10.1371/journal.pone.0076469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bussani R, Schneider E, Zentilin L, Collesi C, Ali H, Braga L, Volpe MC, Colliva A, Zanconati F, Berlot G, Silvestri F, Zacchigna S, Giacca M. 2020. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EBioMedicine 61:103104. 10.1016/j.ebiom.2020.103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayn M, Hirschenberger M, Koepke L, Nchioua R, Straub JH, Klute S, Hunszinger V, Zech F, Bozzo CP, Aftab W, Christensen MH, Conzelmann C, Müller JA, Badarinarayan SS, Stürzel CM, Forne I, Stenger S, Conzelmann K-K, Münch J, Schmidt FI, Sauter D, Imhof A, Kirchhoff F, Johannes Sparrer KM. 2021. Systematic functional analysis of SARS-CoV-2 proteins uncovers viral innate immune antagonists and remaining vulnerabilities. Cell Rep 35:109126. 10.1016/j.celrep.2021.109126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stukalov A, Girault V, Grass V, Karayel O, Bergant V, Urban C, Haas DA, Huang Y, Oubraham L, Wang A, Hamad MS, Piras A, Hansen FM, Tanzer MC, Paron I, Zinzula L, Engleitner T, Reinecke M, Lavacca TM, Ehmann R, Wölfel R, Jores J, Kuster B, Protzer U, Rad R, Ziebuhr J, Thiel V, Scaturro P, Mann M, Pichlmair A. 2021. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature 594:246–252. 10.1038/s41586-021-03493-4. [DOI] [PubMed] [Google Scholar]
- 65.Banerjee A, Falzarano D, Rapin N, Lew J, Misra V. 2019. Interferon regulatory factor 3-mediated signaling limits Middle-East respiratory syndrome (MERS) coronavirus propagation in cells from an insectivorous bat. Viruses 11:152. 10.3390/v11020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hall JS, Knowles S, Nashold SW, Ip HS, Leon AE, Rocke T, Keller S, Carossino M, Balasuriya U, Hofmeister E. 2021. Experimental challenge of a North American bat species, big brown bat (Eptesicus fuscus), with SARS-CoV-2. Transbound Emerg Dis 68:3443–3452. 10.1111/tbed.13949. [DOI] [PubMed] [Google Scholar]
- 67.Zhou P, Tachedjian M, Wynne JW, Boyd V, Cui J, Smith I, Cowled C, Ng JHJ, Mok L, Michalski WP, Mendenhall IH, Tachedjian G, Wang L-F, Baker ML. 2016. Contraction of the type I IFN locus and unusual constitutive expression of IFN-α in bats. Proc Natl Acad Sci USA 113:2696–2701. 10.1073/pnas.1518240113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Banerjee A, Baker ML, Kulcsar K, Misra V, Plowright R, Mossman K. 2020. Novel insights into immune systems of bats. Front Immunol 11:26. 10.3389/fimmu.2020.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bondet V, Le Baut M, Le Poder S, Lécu A, Petit T, Wedlarski R, Duffy D, Le Roux D. 2021. Constitutive IFNα protein production in bats. Front Immunol 12:735866. 10.3389/fimmu.2021.735866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Banerjee A, Zhang X, Yip A, Schulz KS, Irving AT, Bowdish D, Golding B, Wang L-F, Mossman K. 2020. Positive selection of a serine residue in bat IRF3 confers enhanced antiviral protection. iScience 23:100958. 10.1016/j.isci.2020.100958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou P, Cowled C, Mansell A, Monaghan P, Green D, Wu L, Shi Z, Wang L-F, Baker ML. 2014. IRF7 in the Australian black flying fox, Pteropus alecto: evidence for a unique expression pattern and functional conservation. PLoS One 9:e103875. 10.1371/journal.pone.0103875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brook CE, Boots M, Chandran K, Dobson AP, Drosten C, Graham AL, Grenfell BT, Müller MA, Ng M, Wang L-F, van Leeuwen A. 2020. Accelerated viral dynamics in bat cell lines, with implications for zoonotic emergence. Elife 9:e48401. 10.7554/eLife.48401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Irving AT, Ahn M, Goh G, Anderson DE, Wang L-F. 2021. Lessons from the host defences of bats, a unique viral reservoir. Nature 589:363–370. 10.1038/s41586-020-03128-0. [DOI] [PubMed] [Google Scholar]
- 74.De La Cruz-Rivera PC, Kanchwala M, Liang H, Kumar A, Wang L-F, Xing C, Schoggins JW. 2018. The IFN response in bats displays distinctive IFN-stimulated gene expression kinetics with atypical RNASEL induction. J Immunol 200:209–217. 10.4049/jimmunol.1701214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Glennon NB, Jabado O, Lo MK, Shaw ML. 2015. Transcriptome profiling of the virus-induced innate immune response in Pteropus vampyrus and its attenuation by Nipah virus interferon antagonist functions. J Virol 89:7550–7566. 10.1128/JVI.00302-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Temmam S, Vongphayloth K, Salazar EB, Munier S, Bonomi M, Régnault B, Douangboubpha B, Karami Y, Chretien D, Sanamxay D, Xayaphet V, Paphaphanh P, Lacoste V, Somlor S, Lakeomany K, Phommavanh N, Pérot P, Donati F, Bigot T, Nilges M, Rey F, van der Werf S, Brey P, Eloit M. 2021. Coronaviruses with a SARS-CoV-2-like receptor-binding domain allowing ACE2-mediated entry into human cells isolated from bats of Indochinese peninsula. Biol Sci. 10.21203/rs.3.rs-871965/v1. [DOI]
- 77.Yohe LR, Devanna P, Davies KTJ, Potter JHT, Rossiter SJ, Teeling EC, Vernes SC, Dávalos LM. 2019. Tissue collection of bats for -omics analyses and primary cell culture. J Vis Exp 10.3791/59505. [DOI] [PubMed] [Google Scholar]
- 78.Aurine N, Baquerre C, Gaudino M, Jean C, Dumont C, Rival-Gervier S, Kress C, Horvat B, Pain B. 2019. Reprogrammed pteropus bat stem cells present distinct immune signature and are highly permissive for henipaviruses. bioRxiv. https://www.biorxiv.org/content/10.1101/846410v1.
- 79.Pikula J, Bandouchova H, Kovacova V, Linhart P, Piacek V, Zukal J. 2017. Reproduction of rescued vespertilionid bats (Nyctalus noctula) in captivity: veterinary and physiologic aspects. Vet Clin North Am Exot Anim Pract 20:665–677. 10.1016/j.cvex.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 80.Ramakrishnan MA. 2016. Determination of 50% endpoint titer using a simple formula. World J Virol 5:85–86. 10.5501/wjv.v5.i2.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hoffmann M, Wu Y-J, Gerber M, Berger-Rentsch M, Heimrich B, Schwemmle M, Zimmer G. 2010. Fusion-active glycoprotein G mediates the cytotoxicity of vesicular stomatitis virus M mutants lacking host shut-off activity. J Gen Virol 91:2782–2793. 10.1099/vir.0.023978-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Download jvi.00608-22-s0002.avi, AVI file, 15.5 MB (15.5MB, avi)
Movie S2. Download jvi.00608-22-s0003.avi, AVI file, 12.4 MB (12.4MB, avi)
Movie S3. Download jvi.00608-22-s0004.avi, AVI file, 10.6 MB (10.6MB, avi)
Movie S4. Download jvi.00608-22-s0005.avi, AVI file, 11.9 MB (11.9MB, avi)
Movie legends. Download jvi.00608-22-s0001.pdf, PDF file, 0.07 MB (68.4KB, pdf)