ABSTRACT
HIV-1 persistence in different cell types presents the main obstacle to an HIV-1 cure. We have previously shown that the renal epithelium is a site of HIV-1 infection and that the kidney represents a separate viral compartment from blood. Whether renal cells can harbor latent virus that can be reactivated upon treatment with latency reversing agents (LRAs) is unknown. To address this question, we developed an in vitro HIV-1 latency model in renal tubule epithelial (RTE) cells using a dual color HIV-1 reporter virus, R7/E-/GFP/EF1a-mCherry (R7GEmC), and evaluated the effect of LRAs, both as single agents and in combination, on viral reactivation. Our data show that HIV-1 can establish latency in RTE cells early postinfection. While the pool of latently infected cells expanded overtime, the percentage of productively infected cells declined. Following LRA treatment only a small fraction of latently infected cells, both T cells and RTE cells, could be reactivated, and the drug combinations more effective in reactivating HIV transcription in RTE cells differed from those more active in T cells. Our study demonstrates that HIV can establish latency in RTE cells and that current LRAs are only marginally effective in inducing HIV-1 reactivation. This suggests that further study of LRA dynamics in non-T cells may be warranted to assess the suitability of LRAs as a sterilizing cure strategy.
IMPORTANCE Anti-retroviral therapy (ART) has dramatically reduced HIV-related morbidity and mortality. Despite this success, a number of challenges remain, including the long-term persistence of multiple, clinically latent viral reservoirs capable of reactivation in the absence of ART. As efforts proceed toward HIV eradication or functional cure, further understanding of the dynamics of HIV-1 replication, establishment of latency and mechanisms of reactivation in reservoirs harboring the virus throughout the body is necessary. HIV-1 can infect renal epithelial cells and the expression of viral genes in those cells contributes to the development of HIV associated nephropathy (HIVAN) in untreated individuals. The significance of our work is in developing the first model of HIV-1 latency in renal epithelial cells. This model enhances our understanding of HIV-1 latency and persistence in the kidney and can be used to screen candidate latency reversing agents.
KEYWORDS: HIV-1, latency, renal epithelial cells, LRAs, reservoir
INTRODUCTION
Despite the profound successes of antiretroviral therapy (ART), HIV-1 remains both a significant health risk to infected individuals and a global disease burden. The use of antiretroviral drugs as the cornerstone of HIV-1 treatment presents clinical and public health challenges, including drug intolerance, development of resistance, high cost, and lack of access to health care and medication (1). In consideration of these issues, there remains a pressing need for the development of an effective HIV-1 cure. Though current drug therapies are capable of reducing viral load to undetectable levels, they fail to clear latently infected cells (2). The establishment of a latent infection allows lifelong persistence of HIV-1 in infected individuals that enables viral load to rebound upon cessation of therapy (3). A sterilizing cure would require the elimination of every latently infected cell from the body, a challenging proposition compounded by the fact that no high-specificity method exists to identify or target latently infected cells in vivo (4).
Prominent among potential HIV-1 cure strategies is the “shock-and-kill” approach. This method relies upon transcription-activating drugs to stimulate latently infected cells to emerge from quiescence and initiate HIV-1 gene expression, which renders them susceptible to the host immune response and vulnerable to the direct cytotoxic effects of HIV-1 transcription (5). Though significant progress is being made in the optimization of latency reversing agents (LRAs), the majority of research efforts investigate reactivation only in the peripheral CD4+ T-cell population. While CD4+ T cells represent the main viral reservoir, HIV can infect several tissues, including the gut-associated lymphoid tissues, genital tract, lymph nodes, central nervous system, spleen, liver, lungs, and kidney (3, 6). In fact, the source of reactivating virus is often not found within the circulating CD4+ pool (7–9). A sterilizing cure will require that these secondary HIV-1 compartments also be purged of latently infected cells. Given the functional and anatomic diversity among these compartments and their local T cell subsets, it is likely that LRAs will exhibit similarly diverse pharmacodynamic and kinetic profiles across compartments (6).
We have previously shown that the renal epithelium is a site of HIV-1 infection (10, 11) and that the kidney represents a separate viral compartment from blood (12). HIV-1 gains access to renal tubule epithelial (RTE) cells through cell-to-cell contact with infected T cells (13) or macrophages (14), and infected RTE cells support the full HIV-1 life cycle (15). Furthermore, HIV-1 can be found in the urine of infected individuals (16) and as previously observed through the phylogenetic analysis of viral quasispecies in renal biopsy specimens (12), the urine derived viruses cluster separately from blood (16) but are closely related to viral sequences from urine-derived renal epithelial cells (17), suggesting autonomous replication in the kidney of HIV-1 positive subjects.
Whether RTE cells can harbor latent virus that can be reactivated upon LRAs treatment is not known. To assess whether HIV-1 can establish latency in RTE cells we used a previously described dual color HIV-1 reporter virus, R7/E-/GFP/EF1a-mCherry (R7GEmC) (18). The R7GEmC virus allows the separation of latently infected cells (those expressing only mCherry) from productively infected cells (those expressing GFP) (18). We found that HIV-1 can establish latency in RTE cells early postinfection and that following LRA treatment only a small fraction of latently infected cells can be reactivated. Furthermore, we observed that while the percentage of productively infected RTE cells declined over time, the pool of latently infected cells continued to expand in culture and persisted for up to 12 weeks. Our model provides additional understanding of the dynamics of renal cell infection by HIV-1 and can be used to further advance the study of HIV-1 latency and reactivation in this unique cell type.
RESULTS
HIV-1 establishes latency in renal tubule epithelial cells.
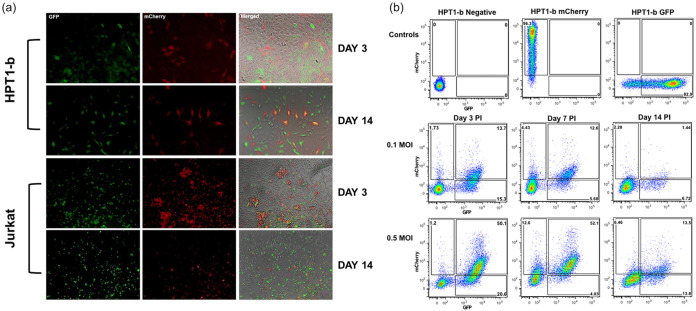
To determine whether HIV-1 can establish latency in RTE cells we infected the Hpt1-b RTE cell line with the VSV-G pseudotyped R7GEmCh virus at MOI 1. In parallel we also infected Jurkat T cells as a positive control. Productive infection (GFP+ and mCherry+/GFP+ cells) and latent infection (GFP−/mCherry+ cells) were monitored by fluorescence microscopy and flow cytometry for 14 days following virus inoculation. As shown in Fig. 1a, both productive and latent infections were detected early in RTE cells and persisted over time. While a small enrichment of the latent population (GFP−/mCherry+ cells) was observed between day 3 and 7 postinfection, the percentage of productively infected cells declined overtime (up to day 14 p.i.), likely due to the cytotoxic effects of HIV transcription (Fig. 1b). We also observed a decline in the percentage of latently infected cells (GFP−/mCherry+ cells) between day 7 and 14 p.i., suggesting silencing of the internal promoter.
FIG 1.
HIV-1 Latency is established early in renal cells. (a) The renal tubule epithelial Hpt1-b and the Jurkat T cell lines were infected with 1 multiplicity of infection (MOI) of the VSV-G pseudotyped R7GEmC construct and followed over time by fluorescence microscopy. (b) Time course FACS analysis of Hpt1-b cells infected with 0.1 or 0.5 MOI of R7GEmC. Expression of GFP and mCherry was analyzed at day 3, 7 and 14 postinfection. Representative of 3 independent experiments.
Latently infected renal tubule epithelial cells expand in culture and persist for a prolonged period of time.
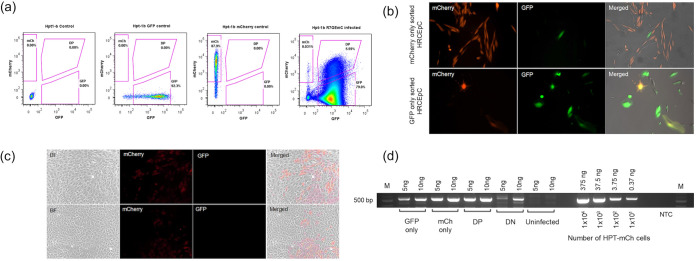
We next used the same virus to infect primary RTE cells (HRCEp cells) and performed flow cytometry sort (Fig. 2a) at 3 weeks postinfection to separate latently infected RTE cells (GFP−/mCherry+ cells) from productively infected RTE cells (GFP+ and mCherry+/GFP+ cells). The two different cell populations were then replated in separate wells and monitored overtime. In contrast to the productively infected HRCEp cells, we observed efficient expansion of the latent cell pool at 2 weeks post-sort (Fig. 2). Interestingly, spontaneous HIV-1 reactivation was also observed in some of the latent cells (Fig. 2b).
FIG 2.
Long term persistence of HIV-1 latency in renal epithelial cells. Gating strategy for flow cytometry sort of latently infected and actively infected Hpt1-b renal epithelial cells (day 5 postinfection). Uninfected cells were used as negative control. Positive controls include Hpt1-b cells stably transduced with a lentiviral vector expressing either GFP or mCherry. (b) Flow-sorted primary renal cells (HRCEp cells) were reseeded and the expression of mCherry and GFP was analyzed at 2 weeks post-sort in both the latent (top panel) and active cells (bottom panel). (c) Flow-sorted mCherry only (latent) Hpt1-b cells were cultured for 12 weeks postinfection. mCherry expression could be detected in about half of the cells, while the other half did not express any fluorescent marker. No GFP expression (active infection) was detected at this late time point. Each panel represents a separate experiment. (d) The presence of the HIV-R7GEmC construct DNA was assessed in the four populations of flow-sorted Hpt1-b cells: GFP only, GFP/mCherry double positive (DP), latently infected (mCherry only) and double negative (DN). Serial dilutions of genomic DNA extracted from Hpt-1b cells stably transduced with a lentiviral vector expressing mCherry were used to generate a standard curve. The amount of DNA and the corresponding number of cells used to generate the standard curve are indicated. Uninfected Hpt1-b cells were included as negative control. NTC = no template control.
To determine whether latent cells can persist in culture for a prolonged period of time we repeated the infection and flow cytometry sort on R7GEmC infected Hpt1-b cells and followed the latent cell population for a total of 12 weeks. As shown in Fig. 2c, the latent cell pool continued to expand in culture. We did not observe spontaneous reactivation of HIV-1 after such a prolonged time in culture, suggesting that the latent state of HIV-1 in these cells is stable. We observed two distinct phenotypes among the latently infected cell population, RTE cells GFP−/mCherry+ and cells that lost the expression of mCherry in addition to not expressing GFP (GFP−/mCherry−). To determine whether this GFP/mCherry double negative cell population comprised latent cells in which the R7GEmCh construct was completely silenced, as previously shown for T cells (19), we performed a semiquantitative PCR using a primer set specific for the mCherry gene. As shown in Fig. 2d, the DN population comprised both uninfected and R7GEmCh infected cells (~1/3 of the entire population based on similar intensity of the 10 ng band in the DN population and the 3.75 ng band in the standard curve), suggesting that cells in which both the LTR (driving GFP expression) and the internal promoter (driving mCherry expression) were transcriptionally silent were flow-sorted together with uninfected cells.
Moderate HIV-1 reactivation in latently infected renal cells following LRA treatment.
One approach currently under clinical evaluation for eradicating HIV-1 includes the use of pharmacologic agents known as latency-reversing agents (LRA) to reactivate viral gene expression and induce the elimination of these cells either through virus-induced apoptosis or by immune-mediated cytotoxic killing (20–22). The ability of several LRAs to reactivate HIV has been tested primarily on infected CD4+ T cells, therefore the effects of these compounds on viral gene expression reactivation in other cell types harboring the virus is not known.
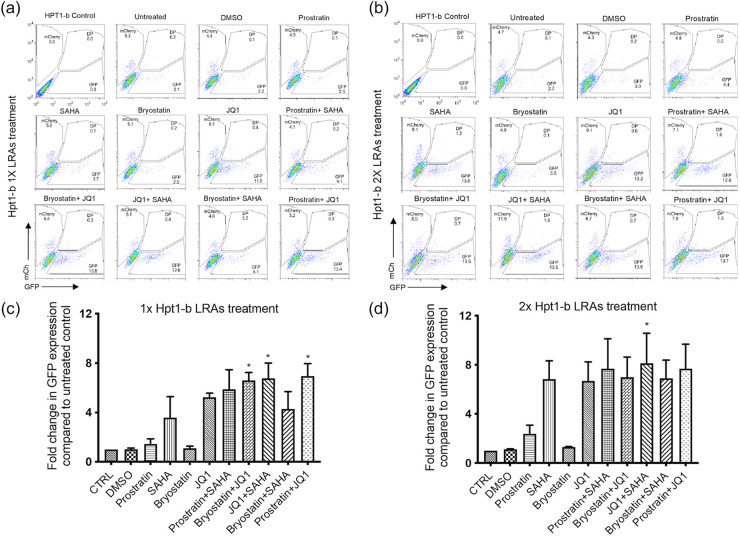
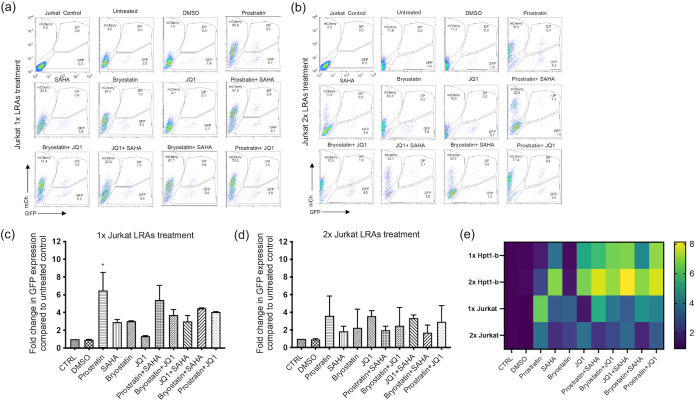
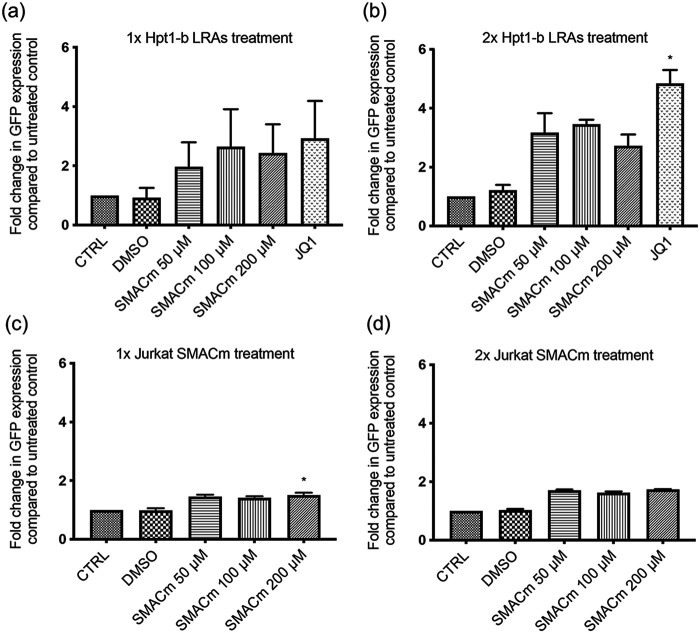
We therefore tested a panel of LRAs reported to reactivate latent HIV-1 both as single agents or in combination in our RTE cell latency model. Flow-sorted GFP−/mCherry+ Hpt1-b cells (Fig. 2a) were kept in culture for 1 week to expand the pool of latently infected cells and then treated either once or twice with the LRAs indicated in Fig. 3 (a to d) for 24 h. We performed the same experiment on latently infected Jurkat T cells for comparison (Fig. 4a to d). At the time of LRAs administration, approximately 5% of the mCherry+/GFP− sorted Hpt1-b cells expressed only the mCherry marker, while 2% of these cells, likely spontaneously reactivated cells, expressed only GFP (Fig. 3a). In the mCherry+/GFP− sorted Jurkat T cells, approximately 8% continued to express only the mCherry marker and 2% expressed only GFP (Fig. 4a). In RTE cells, a five to 7-fold increase in the number of GFP-expressing cells compared to the DMSO treated control was observed after one LRA treatment with JQ1 alone (5.5-fold – P value >0.05, nonparametric Kruskall-Wallis test), JQ1 in combination with either Bryostatin (6.5-fold), SAHA (~7-fold) or Prostratin (~7-fold) (Fig. 3c). A single treatment with the HDACi SAHA induced a 3.5-fold increase in GFP+ cells, while the combination of Prostratin and SAHA and Bryostatin and SAHA induced a 5.9 and a 4-fold increase in GFP+ cells, respectively (Fig. 3c). No reactivation was observed following a single treatment with either Prostratin or Bryostatin alone (Fig. 3a, c). A second LRA administration induced a ~7-fold reactivation in both the SAHA and SAHA/Bryostatin treated cells, suggesting that multiple treatments with SAHA can increase HIV-1 reactivation in RTE cells (Fig. 3b, d). Following a second treatment with either Prostratin or Bryostatin a ≤ 2-fold increase in GFP+ cells was observed (Fig. 3b, d), suggesting that PKC agonists are only minimally effective at inducing HIV-1 reactivation in RTE cells. On the other hand, when these two drugs were administered to latent Jurkat T cells, they induced a 3 to 6-fold increase in GFP+ cells (Fig. 4c–d). In contrast to RTE cells, a single treatment with the BET-inhibitor JQ1, did not induce measurable HIV-1 reactivation in Jurkat T cells (Fig. 4a, c), suggesting that different mechanisms might be involved in HIV-1 latency persistence in renal cells versus T cells. For both cell types, although significantly more pronounced in T cells, we observed a shift from double negative to mCherry only expressing cells after the second LRA treatment, indicating that a good proportion of the cells, originally sorted based on the expression of the latent marker mCherry, had silencing of both the HIV and EF1a promoters, probably due to the integration site of the provirus and/or the cell state. These results are in line with the microscopy data in Fig. 2c and the PCR data shown in Fig. 2d. While the second LRAs treatment induced a shift in the mCherry+/GFP− cell population, no further increase in the percentage of GFP+ and GFP+/mCherry+ cells was observed (Fig. 4b).
FIG 3.
Moderate HIV-1 reactivation following LRAs treatment of latently infected Hpt1-b renal cells. Latently infected renal cells were flow sorted 2 weeks postinfection and cultured for an additional week before LRAs treatment. Cells were then treated for 24 h with the indicated LRAs or DMSO either once (a) or twice (b). FACS analyses of representative single and double treatments are shown. Histograms represent the fold change in GFP expression compared to untreated controls for the single (c) or double treatments (d). Bars represent mean+SD of the percentage of GFP+ population in three independent experiments. Asterisks indicate a significant difference (P value <0.05; nonparametric Kruskall-Wallis test) between LRA treated cells and DMSO.
FIG 4.
Moderate HIV-1 reactivation following LRAs treatment of latently infected Jurkat T cells. Latently infected Jurkat T cells were flow sorted 2 weeks postinfection and cultured for an additional week before LRAs treatment. Cells were then treated for 24 h with the indicated LRAs or DMSO either once (a) or twice (b). FACS analyses of representative single and double treatments are shown. Histograms represent the fold change in GFP expression compared to untreated controls for the single (c) or double treatments (d). (e) Heatmap comparing fold change in GFP expression for both Hpt1-b and Jurkat cells. Bars represent mean+SD of the percentage of GFP+ population in three independent experiments. Asterisks indicate a significant difference (P value <0.05; nonparametric Kruskall-Wallis test) between LRA treated cells and DMSO.
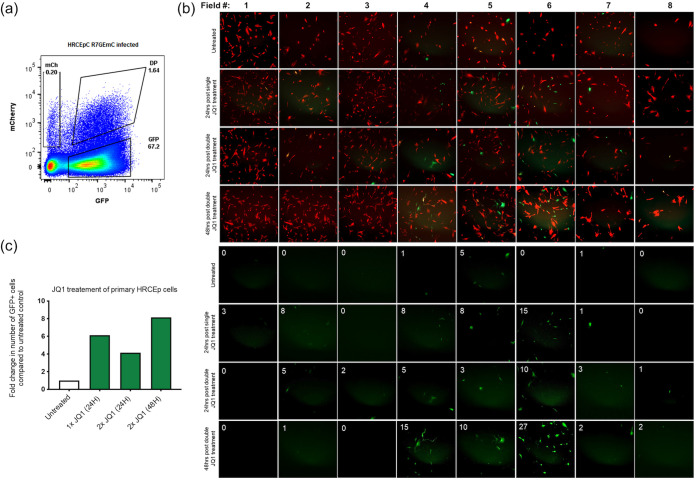
We next tested the ability of JQ1 to reactivate latent HIV-1 in primary HRCEp cells. Flow-sorted GFP−/mCherry+ HRCEp cells (Fig. 5a) were kept in culture for 1 week to expand the pool of latently infected cells and then treated either once or twice with JQ1 for 24 h. Treated cells were then imaged at 24 h after the first treatment and 24 and 48 h after the second treatment. As shown in Figure 5b, both the first and second treatment with JQ1 induced HIV reactivation in these cells, and the number of GFP positive cells was higher at 48 h after the second JQ1 treatment.
FIG 5.
HIV-1 reactivation following JQ1 treatment of latently infected primary HRCEp cells. Latently infected primary HRCEp cells were flow sorted 11 days postinfection and cultured for an additional 8 days before treatment with JQ1. (a) Gating strategy for flow cytometry sort of latently infected HRCEp cells (day 11 postinfection). (b) Latently infected (mCherry only) flow-sorted HRCEp cells were reseeded and the expression of mCherry and GFP was analyzed at 8 days post-sort. Cells were then treated twice with JQ1 for 24 h and imaged 24 h after the first treatment, and 24 and 48 h after the second treatment. (b) Shown are images of mCherry and GFP positive HRCEp cells from eight separate fields taken at the indicated time points from untreated and JQ1 treated cells. Number of GFP+ cells present in each field are indicated in the top left corner of each image. (c) Fold change in number of GFP+ cells in untreated and JQ1 treated cells at the indicated time points.
Recently, mimetics of the second mitochondrial-derived activator of caspases (SMAC) have gained attention for their potential to act as LRAs (23) with a lower chance of off-target effects and toxicity, relative to traditional LRAs (24, 25). AZD5582, is one such SMAC mimetic and has been reported to successfully reverse HIV latency in vitro as well as in vivo in humanized mice and rhesus macaques (25). As with most standard LRAs, these newer SMAC mimetic compounds have not been evaluated for their reactivation effects in renal cells. To investigate this new class of molecule we tested AZD5582 in our RTE latency model. Flow-sorted GFP−/mCherry+ Hpt1-b cells and Jurkat T cells were treated either once or twice with escalating doses of AZD5582, ranging from 50 to 200 nM, for 24 h. In line with previous reports in T cells (25), treatment with AZD5582 resulted in increased GFP expression in RTE cells (Fig. 6). We found that the 100 nM dose was slightly more effective at reversing latency in RTE cells (Fig. 6a) while the 200 nM dose was more effective in Jurkat T cells (Fig. 6c). Similar to what we observed with other LRAs, a second treatment with AZD5582 resulted in higher levels of reactivation compared to a single treatment (Fig. 6b, d). We compared the latency reversal activity of AZD5582 to that of the BET-inhibitor JQ1, which as shown in Fig. 3c was the most effective agent at reversing latency in RTE when administered alone. As shown in Fig. 6a, b, JQ1 was more efficient at reversing latency in RTE cells compared to AZD5582.
FIG 6.
Moderate HIV-1 reactivation following SMACm treatment of latently infected Hpt1-b renal cells and Jurkat T cells. Latently infected Hpt-1b renal cells or Jurkat T cells were flow sorted 2 weeks postinfection and cultured for an additional week before SMACm treatment. Cells were then treated for 24 h with SMACm, JQ1 or DMSO either once (a–c) or twice (b–d). Bars represent mean+SD of the percentage of GFP+ population in three independent experiments. Asterisks indicate a significant difference (P value <0.05; nonparametric Kruskall-Wallis test) between LRA treated cells and DMSO.
DISCUSSION
Reducing the persistent HIV reservoir remains an important goal for the development of a functional cure for HIV-1. This goal has been hindered by the challenge of identifying cells that harbor persistent replication-competent virus, as well as their anatomic location. Several cell populations have been proposed as HIV-1 reservoirs, including renal epithelial cells; however, the only population yet convincingly shown to be a long-term reservoir for HIV-1 are resting memory CD4+ T cells harboring latent but replication-competent HIV-1 (26). Due to the scarcity of HIV-1 latently infected cells in patients, in vitro latency models represent a good tool to study the HIV-1 reservoir in different cell types.
Here, we developed an HIV-1 latency model in renal tubule epithelial (RTE) cells and evaluated the dynamics of viral infection, latency, and reactivation in this unique cell type. As previously reported for T cells (19), we observed the establishment of latent infection as early as 3 days postinfection. The percentage of latently infected RTE cells was significantly lower than those harboring active virus. Actively infected RTE cells showed a hypertrophic phenotype, a cellular abnormality also observed in HIVAN biopsy specimens (27) as a result of viral gene expression, and did not efficiently expand in culture, while the latent cell pool demonstrated a phenotype more similar to uninfected cells and appeared to retain their proliferation capacity. These observations are in agreement with recent studies highlighting the possibility that proliferation of latently infected cells may be a major contributor in determining the composition and stability of the latent reservoir (28). Notably, we recently demonstrated that RTE cells that acquired HIV following cell-to-cell contact with infected macrophages can undergo multiple rounds of proliferation, with or without transcriptional silencing, providing a mechanism for HIV-1 persistence in the kidney (14). Furthermore, the proliferation of infected RTE cells might explain the pattern of renal tubule infection observed in HIVAN biopsy specimens, where all cells of a single tubule appear to be infected (10, 12, 29).
Spontaneous HIV-1 reactivation was also observed in some of the latent cells. This phenomenon has also been previously observed in T cell models of latency by a number of groups (30–32). Clonal populations of infected cells are not static and demonstrate spontaneous reactivation and transcriptional shutdown. Fluctuations in HIV-1 Tat expression, due to proviral integration into genomic positions that support a low basal expression rate, have been associated with spontaneous HIV reactivation (31, 33).
The possibility that part of the latent reservoir arises from the proliferation of a smaller number of infected cells renders the development of an HIV-1 cure even more challenging. A potential strategy to cure HIV-1-infection is the use of latency reversing agents (LRAs) to reactivate and eliminate infected cells. Here, we tested the ability of several LRAs, both as single agent or in combination to reactivate latent HIV-1 in both immortalized and primary RTE cells. In line with data from several T cell models of latency, the tested drugs were only marginally effective at viral reactivation in renal cells. Several possibilities have been proposed to explain the lack of viral reactivation by LRAs, including provirus integration into regions non permissive for viral gene expression (34, 35), transcriptional interference (36–38), or epigenetic modifications (39). Finally, stochastic features of the Tat transactivation mechanism may prevent induction of all intact proviruses (33). Identification of successful latency-reversing strategies requires a deeper understanding of these issues.
In summary, our in vitro model demonstrates that HIV-1 can establish latency in renal tubule epithelial cells and that latently infected RTE cells are stable over time. Because kidney samples are not readily available from HIV-1 individuals, unless there is a clinical indication for the biopsy specimen, it has been difficult to definitely conclude that RTE cells are a true reservoir for HIV-1 according to the reservoir definition of cells that harbor persistent, replication-competent HIV-1 (26) that can seed reactivation. Despite the inherent limitations of in vitro models relaying on cell lines and reporter viruses, the study presented here complements previous in vitro and in vivo studies from our group and others (13–17, 40), and provides additional evidence that the kidney might serve as a reservoir for HIV-1. Future studies will be focused on determining the reactivation potential of HIV-1 infected renal epithelial cells isolated from urine samples or kidney biopsy specimens of people with HIV-1, as well as mechanistic studies of HIV-latency in RTE cells. As novel cure strategies are developed, it will be important to understand their impact in both lymphoid and nonlymphoid reservoirs.
MATERIALS AND METHODS
Primary cells and cell lines.
The previously described human proximal tubular epithelial cell line Hpt1-b (13, 41) and the primary human renal cortical epithelial cells (HRCEp cells) were cultured in Lonza Renal Cell Growth Medium supplemented with Lonza SingleQuot Supplement and Growth Factors (catalog number CC-3190). Primary HRCEp cells were obtained from Promocell (catalog number C-12660). Jurkat, Clone E6-1 cells (ATCC TIB-152) were cultured in RPMI 1640 supplemented with 10% FBS, 100 mg/mL penicillin/streptomycin. To generate the Hpt-1b-mCherry and Hpt-1b-GFP cell line, constitutively expressing the mCherry or the GFP genes, Hpt-1b cells were transduced with 1 multiplicity of infection (MOI) of the SIV-based lentiviral vector (42) expressing both the mCherry and neomycin resistance gene (GAE-CMV-mCh-IRES-Neo) or the GFP and neomycin resistance gene under the CMV promoter (GAE-CMV-GFP-IRES-Neo). Stably transduced cells were selected by treatment with 800 μg/mL of G418 sulfate (Corning) for 2 weeks.
Virus preparation and cells infection.
The reporter HIV-1 construct R7/E-/GFP/EF1a-mCherry (pR7GEmC) was kindly provided by Eric Verdin (University of San Francisco, CA, USA). To generate viral particles pseudotyped with the vesicular stomatitis virus glycoprotein (VSV-G), 293T Lenti-X cells (Clontech) were transiently transfected with 10 μg of R7GEmC, and 2 μg pHCMV-VSV-G as previously described (42, 43) using the JetPRIME transfection kit from PolyPlus (cat number 114-75), according to the manufacturer’s instruction. Transduction efficiency for each virus stock was assessed on infected Hpt1-b and Jurkat T cells by flow cytometry as previously described (42, 43). RTE cells and Jurkat T cells were inoculated overnight with 1 multiplicity of infection (MOI) of VSV-G pseudotyped R7GEmCh.
Flow cytometry and cell sorting.
At 5, 9, 11, or 14 days postinfection, infected cells were analyzed by flow cytometry (Fluorescence-activated cell sorting [FACS] Calibur, BD Biosciences, Franklin Lakes, NJ) to determine the percentage of latent (mCherry only) versus active (GFP positive) cells. RTE cells were flow sorted into 4 populations based on mCherry and GFP expression (double Negative, mCherry-only, GFP-only, and double positive) and returned to culture for further analysis. Sorts were performed using a BD FACSAriaII (BD Biosciences, Franklin Lakes, NJ). All data were analyzed using FlowJo, LLC (Treestar, Ashland, OR).
PCR detection of HIV-1 construct in flow-sorted renal epithelial cell populations.
Genomic DNA was extracted from flow sorted populations of R7GEmC-infected Hpt1-b cells using QiAmp DNA micro-kit (Qiagen Cat number: 56304). The following primers, mCh_For: 5′-GAGGAGGATAACATGGCCAT-3′ and mCh_Rev: 5′-GGTGGTCTTGACCTCAGCGT-3′, were used to detect the presence of viral DNA using the following cycling conditions: 94°C for 3 min, 40 cycles of 94°C for 20 sec, 61°C for 30 sec, 72°C for 45 sec; followed by 72°C for 3 min. Amplicons of 540 bp were visualized on 2% agarose gel stained with SYBR safe. Serial dilutions of genomic DNA extracted from Hpt-1b cells stably transduced with a lentiviral vector expressing mCherry were used to generate the standard curve.
Latency-reversing agent assays.
Flow sorted R7GEmC-infected cells were expanded in culture for up to 21 days until a sufficient number of cells for each of the 4 populations was obtained. Double negative, mCherry-only, GFP-only, and double positive cells were seeded on 12-well plates at a density of 5 × 104 cells/well. All latency-reversing agents (LRAs) tested were obtained from Sigma-Aldrich (MilliporeSigma, St. Louis, MO, USA) and dissolved in dimethyl sulfoxide (DMSO). LRAs were diluted in culture media and applied to the cells at 24 and 48 h after plating, at the following concentrations: 5 μM Prostratin; 2.5 μM SAHA; 100 nM Bryostatin; 1 μM JQ1; 50 nM, 100 nM and 200 nM AZD5582. 24 h after final LRA treatment, cells were imaged via fluorescence microscopy and then collected and fixed in 1% paraformaldehyde for FACS analysis. Latently infected primary HRCEp cells were treated only with 1 μM JQ1 due to a low number of cells recovered after flow-cytometry sorting and were imaged via fluorescence microscopy at 24 and 48 h after final LRA treatment.
Fluorescence microscopy.
Fluorescence microscopy images were collected on a Nikon TE2000-E microscope and a Zeiss Axiovert A1 microscope.
Statistics.
Statistical analyses were performed using GraphPad Prism version 7.0 (GraphPad, La Jolla, CA).
ACKNOWLEDGMENTS
E.J.B. performed the majority of the experiments, analyzed the data and wrote the first draft of the manuscript. K.H. performed PCR and SMAC mimetic experiments. T.T. performed infection, flow cytometry sorting, imaging, and LRA treatment of primary renal epithelial cells. M.E.K. contributed to study design and interpretation of the data. M.B. contributed to study design, oversaw the planning and direction of the project, including analysis and interpretation of the data and editing of the manuscript. All authors read, revised, and approved the final manuscript.
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases (R01DK108367 and P01DK056492).
We have no conflict of interest to declare.
Contributor Information
Mary E. Klotman, Email: mary.klotman@duke.edu.
Maria Blasi, Email: maria.blasi@duke.edu.
Frank Kirchhoff, Ulm University Medical Center.
REFERENCES
- 1.Deeks SG, Lewin SR, Ross AL, Ananworanich J, Benkirane M, Cannon P, Chomont N, Douek D, Lifson JD, Lo Y-R, Kuritzkes D, Margolis D, Mellors J, Persaud D, Tucker JD, Barre-Sinoussi F, Alter G, Auerbach J, Autran B, Barouch DH, Behrens G, Cavazzana M, Chen Z, Cohen ÉA, Corbelli GM, Eholié S, Eyal N, Fidler S, Garcia L, Grossman C, Henderson G, Henrich TJ, Jefferys R, Kiem H-P, McCune J, Moodley K, Newman PA, Nijhuis M, Nsubuga MS, Ott M, Palmer S, Richman D, Saez-Cirion A, Sharp M, Siliciano J, Silvestri G, Singh J, Spire B, Taylor J, Tolstrup M, International AIDS Society Towards a Cure Working Group, et al. 2016. International AIDS Society global scientific strategy: towards an HIV cure 2016. Nat Med 22:839–850. 10.1038/nm.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shan L, Siliciano RF. 2013. From reactivation of latent HIV-1 to elimination of the latent reservoir: the presence of multiple barriers to viral eradication. Bioessays 35:544–552. 10.1002/bies.201200170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dahabieh MS, Battivelli E, Verdin E. 2015. Understanding HIV latency: the road to an HIV cure. Annu Rev Med 66:407–421. 10.1146/annurev-med-092112-152941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siliciano JD, Siliciano RF. 2014. Recent developments in the search for a cure for HIV-1 infection: targeting the latent reservoir for HIV-1. J Allergy Clin Immunol 134:12–19. 10.1016/j.jaci.2014.05.026. [DOI] [PubMed] [Google Scholar]
- 5.Xing S, Siliciano RF. 2013. Targeting HIV latency: pharmacologic strategies toward eradication. Drug Discov Today 18:541–551. 10.1016/j.drudis.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rasmussen TA, Tolstrup M, Sogaard OS. 2016. Reversal of latency as part of a cure for HIV-1. Trends Microbiol 24:90–97. 10.1016/j.tim.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 7.De Scheerder MA, Vrancken B, Dellicour S, Schlub T, Lee E, Shao W, Rutsaert S, Verhofstede C, Kerre T, Malfait T, Hemelsoet D, Coppens M, Dhondt A, De Looze D, Vermassen F, Lemey P, Palmer S, Vandekerckhove L. 2019. HIV rebound is predominantly fueled by genetically identical viral expansions from diverse reservoirs. Cell Host Microbe 26:347–358.e7. 10.1016/j.chom.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaillon A, Gianella S, Dellicour S, Rawlings SA, Schlub TE, De Oliveira MF, Ignacio C, Porrachia M, Vrancken B, Smith DM. 2020. HIV persists throughout deep tissues with repopulation from multiple anatomical sources. J Clin Invest 130:1699–1712. 10.1172/JCI134815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, Chung C, Hu BS, He T, Guo Y, Kim AJ, Skulsky E, Jin X, Hurley A, Ramratnam B, Markowitz M, Ho DD. 2000. Genetic characterization of rebounding HIV-1 after cessation of highly active antiretroviral therapy. J Clin Invest 106:839–845. 10.1172/JCI10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruggeman LA, Ross MD, Tanji N, Cara A, Dikman S, Gordon RE, Burns GC, D'Agati VD, Winston JA, Klotman ME, Klotman PE. 2000. Renal epithelium is a previously unrecognized site of HIV-1 infection. J Am Soc Nephrol 11:2079–2087. 10.1681/ASN.V11112079. [DOI] [PubMed] [Google Scholar]
- 11.Hughes K, Chang J, Stadtler H, Wyatt C, Klotman M, Blasi M. 2021. HIV-1 infection of the kidney: mechanisms and implications. AIDS 35:359–367. 10.1097/QAD.0000000000002753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marras D, Bruggeman LA, Gao F, Tanji N, Mansukhani MM, Cara A, Ross MD, Gusella GL, Benson G, D'Agati VD, Hahn BH, Klotman ME, Klotman PE. 2002. Replication and compartmentalization of HIV-1 in kidney epithelium of patients with HIV-associated nephropathy. Nat Med 8:522–526. 10.1038/nm0502-522. [DOI] [PubMed] [Google Scholar]
- 13.Chen P, Chen BK, Mosoian A, Hays T, Ross MJ, Klotman PE, Klotman ME. 2011. Virological synapses allow HIV-1 uptake and gene expression in renal tubular epithelial cells. J Am Soc Nephrol 22:496–507. 10.1681/ASN.2010040379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes K, Akturk G, Gnjatic S, Chen B, Klotman M, Blasi M. 2020. Proliferation of HIV-infected renal epithelial cells following virus acquisition from infected macrophages. AIDS 34:1581–1591. 10.1097/QAD.0000000000002589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blasi M, Balakumaran B, Chen P, Negri DR, Cara A, Chen BK, Klotman ME. 2014. Renal epithelial cells produce and spread HIV-1 via T-cell contact. AIDS 28:2345–2353. 10.1097/QAD.0000000000000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blasi M, Carpenter JH, Balakumaran B, Cara A, Gao F, Klotman ME. 2015. Identification of HIV-1 genitourinary tract compartmentalization by analyzing the env gene sequences in urine. AIDS 29:1651–1657. 10.1097/QAD.0000000000000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blasi M, Stadtler H, Chang J, Hemmersbach-Miller M, Wyatt C, Klotman P, Gao F, Wolfe C, Klotman M. 2020. Detection of donor's HIV strain in HIV-positive kidney-transplant recipient. N Engl J Med 382:195–197. 10.1056/NEJMc1910189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calvanese V, Chavez L, Laurent T, Ding S, Verdin E. 2013. Dual-color HIV reporters trace a population of latently infected cells and enable their purification. Virology 446:283–292. 10.1016/j.virol.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chavez L, Calvanese V, Verdin E. 2015. HIV latency is established directly and early in both resting and activated primary CD4 T cells. PLoS Pathog 11:e1004955. 10.1371/journal.ppat.1004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. 2012. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487:482–485. 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasmussen TA, Tolstrup M, Brinkmann CR, Olesen R, Erikstrup C, Solomon A, Winckelmann A, Palmer S, Dinarello C, Buzon M, Lichterfeld M, Lewin SR, Ostergaard L, Sogaard OS. 2014. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV 1:e13-21–e21. 10.1016/S2352-3018(14)70014-1. [DOI] [PubMed] [Google Scholar]
- 22.Spivak AM, Andrade A, Eisele E, Hoh R, Bacchetti P, Bumpus NN, Emad F, Buckheit R, 3rd, McCance-Katz EF, Lai J, Kennedy M, Chander G, Siliciano RF, Siliciano JD, Deeks SG. 2014. A pilot study assessing the safety and latency-reversing activity of disulfiram in HIV-1-infected adults on antiretroviral therapy. Clin Infect Dis 58:883–890. 10.1093/cid/cit813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pache L, Dutra MS, Spivak AM, Marlett JM, Murry JP, Hwang Y, Maestre AM, Manganaro L, Vamos M, Teriete P, Martins LJ, Konig R, Simon V, Bosque A, Fernandez-Sesma A, Cosford ND, Bushman FD, Young JA, Planelles V, Chanda SK. 2015. BIRC2/cIAP1 is a negative regulator of HIV-1 transcription and can be targeted by Smac mimetics to promote reversal of viral latency. Cell Host Microbe 18:345–353. 10.1016/j.chom.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fulda S. 2014. Molecular pathways: targeting death receptors and smac mimetics. Clin Cancer Res 20:3915–3920. 10.1158/1078-0432.CCR-13-2376. [DOI] [PubMed] [Google Scholar]
- 25.Nixon CC, Mavigner M, Sampey GC, Brooks AD, Spagnuolo RA, Irlbeck DM, Mattingly C, Ho PT, Schoof N, Cammon CG, Tharp GK, Kanke M, Wang Z, Cleary RA, Upadhyay AA, De C, Wills SR, Falcinelli SD, Galardi C, Walum H, Schramm NJ, Deutsch J, Lifson JD, Fennessey CM, Keele BF, Jean S, Maguire S, Liao B, Browne EP, Ferris RG, Brehm JH, Favre D, Vanderford TH, Bosinger SE, Jones CD, Routy JP, Archin NM, Margolis DM, Wahl A, Dunham RM, Silvestri G, Chahroudi A, Garcia JV. 2020. Systemic HIV and SIV latency reversal via non-canonical NF-kappaB signalling in vivo. Nature 578:160–165. 10.1038/s41586-020-1951-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Simonetti FR, Siliciano RF, Laird GM. 2018. Measuring replication competent HIV-1: advances and challenges in defining the latent reservoir. Retrovirology 15:21. 10.1186/s12977-018-0404-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wyatt CM, Rosenstiel PE, Klotman PE. 2008. HIV-associated nephropathy. Contrib Nephrol 159:151–161. 10.1159/000125831. [DOI] [PubMed] [Google Scholar]
- 28.Hosmane NN, Kwon KJ, Bruner KM, Capoferri AA, Beg S, Rosenbloom DI, Keele BF, Ho YC, Siliciano JD, Siliciano RF. 2017. Proliferation of latently infected CD4(+) T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. J Exp Med 214:959–972. 10.1084/jem.20170193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ross MJ, Bruggeman LA, Wilson PD, Klotman PE. 2001. Microcyst formation and HIV-1 gene expression occur in multiple nephron segments in HIV-associated nephropathy. J Am Soc Nephrol 12:2645–2651. 10.1681/ASN.V12122645. [DOI] [PubMed] [Google Scholar]
- 30.Pearson R, Kim YK, Hokello J, Lassen K, Friedman J, Tyagi M, Karn J. 2008. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J Virol 82:12291–12303. 10.1128/JVI.01383-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinberger LS, Burnett JC, Toettcher JE, Arkin AP, Schaffer DV. 2005. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 122:169–182. 10.1016/j.cell.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Singh A, Weinberger LS. 2009. Stochastic gene expression as a molecular switch for viral latency. Curr Opin Microbiol 12:460–466. 10.1016/j.mib.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Razooky BS, Pai A, Aull K, Rouzine IM, Weinberger LS. 2015. A hardwired HIV latency program. Cell 160:990–1001. 10.1016/j.cell.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jordan A, Bisgrove D, Verdin E. 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J 22:1868–1877. 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bisgrove D, Lewinski M, Bushman F, Verdin E. 2005. Molecular mechanisms of HIV-1 proviral latency. Expert Rev Anti Infect Ther 3:805–814. 10.1586/14787210.3.5.805. [DOI] [PubMed] [Google Scholar]
- 36.Han Y, Lin YB, An W, Xu J, Yang HC, O'Connell K, Dordai D, Boeke JD, Siliciano JD, Siliciano RF. 2008. Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell Host Microbe 4:134–146. 10.1016/j.chom.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lenasi T, Contreras X, Peterlin BM. 2008. Transcriptional interference antagonizes proviral gene expression to promote HIV latency. Cell Host Microbe 4:123–133. 10.1016/j.chom.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shan L, Yang HC, Rabi SA, Bravo HC, Shroff NS, Irizarry RA, Zhang H, Margolick JB, Siliciano JD, Siliciano RF. 2011. Influence of host gene transcription level and orientation on HIV-1 latency in a primary-cell model. J Virol 85:5384–5393. 10.1128/JVI.02536-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tyagi M, Pearson RJ, Karn J. 2010. Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and P-TEFb restriction. J Virol 84:6425–6437. 10.1128/JVI.01519-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Canaud G, Dejucq-Rainsford N, Avettand-Fenoel V, Viard JP, Anglicheau D, Bienaime F, Muorah M, Galmiche L, Gribouval O, Noel LH, Satie AP, Martinez F, Sberro-Soussan R, Scemla A, Gubler MC, Friedlander G, Antignac C, Timsit MO, Onetti Muda A, Terzi F, Rouzioux C, Legendre C. 2014. The kidney as a reservoir for HIV-1 after renal transplantation. J Am Soc Nephrol 25:407–419. 10.1681/ASN.2013050564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ross MJ, Wosnitzer MS, Ross MD, Granelli B, Gusella GL, Husain M, Kaufman L, Vasievich M, D'Agati VD, Wilson PD, Klotman ME, Klotman PE. 2006. Role of ubiquitin-like protein FAT10 in epithelial apoptosis in renal disease. J Am Soc Nephrol 17:996–1004. 10.1681/ASN.2005070692. [DOI] [PubMed] [Google Scholar]
- 42.Blasi M, Negri D, LaBranche C, Alam SM, Baker EJ, Brunner EC, Gladden MA, Michelini Z, Vandergrift NA, Wiehe KJ, Parks R, Shen X, Bonsignori M, Tomaras GD, Ferrari G, Montefiori DC, Santra S, Haynes BF, Moody MA, Cara A, Klotman ME. 2018. IDLV-HIV-1 Env vaccination in non-human primates induces affinity maturation of antigen-specific memory B cells. Commun Biol 1:134. 10.1038/s42003-018-0131-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Negri D, Blasi M, LaBranche C, Parks R, Balachandran H, Lifton M, Shen X, Denny T, Ferrari G, Vescio MF, Andersen H, Montefiori DC, Tomaras GD, Liao HX, Santra S, Haynes BF, Klotman ME, Cara A. 2016. Immunization with an SIV-based IDLV expressing HIV-1 Env 1086 clade C elicits durable humoral and cellular responses in rhesus macaques. Mol Ther. 10.1038/mt.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]