ABSTRACT
Human cytomegalovirus (HCMV) exhibits a complex host-pathogen interaction with peripheral blood monocytes. We have identified a unique, cell-type specific retrograde-like intracellular trafficking pattern that HCMV utilizes to gain access to the monocyte nucleus and for productive infection. We show that infection of primary human monocytes, epithelial cells, and fibroblasts leads to an increase in the amount of the trafficking protein Syntaxin 6 (Stx6). However, only knockdown (KD) of Stx6 in monocytes inhibited viral trafficking to the trans-Golgi network (TGN), a requisite step for nuclear translocation in monocytes. Conversely, KD of Stx6 in epithelial cells and fibroblasts did not change the kinetics of nuclear translocation and productive infection. Stx6 predominantly functions at the level of the TGN where it facilitates retrograde transport, a trafficking pathway used by only a few cellular proteins and seldom by pathogens. We also newly identify that in monocytes, Stx6 exhibits an irregular vesicular localization rather than being concentrated at the TGN as seen in other cell-types. Lastly, we implicate that viral particles that associate with both Stx6 and EEA1 early in infection are the viral population that successfully traffics to the TGN at later time points and undergo nuclear translocation. Additionally, we show for the first time that HCMV enters the TGN, and that lack of Stx6 prevents viral trafficking to this organelle. We argue that we have identified an essential cell-type specific regulator that controls early steps in efficient productive infection of a cell-type required for viral persistence and disease.
IMPORTANCE Human cytomegalovirus (HCMV) infection causes severe and often fatal disease in the immunocompromised. It is one of the leading infectious causes of birth defects and causes severe complications in transplant recipients. By uncovering the unique pathways used by the virus to infect key cells, such as monocytes, responsible for dissemination and persistence, we provide new potential targets for therapeutic intervention.
KEYWORDS: early endosomes, HCMV, monocytes, Syntaxin 6, trans-Golgi network, signal transduction, vesicular trafficking, viral trafficking
INTRODUCTION
Human cytomegalovirus (HCMV) is a betaherpesvirus that infects 50–90% of the global population and is a significant cause of morbidity and mortality in immunocompromised populations including AIDS patients, cancer patients, and transplant recipients (1–5). HCMV is also a leading cause of congenital infections, which results in varying degrees of neurological damage in developing fetuses and often leads to fetal demise (1–5). HCMV pathogenesis is directly linked to the widespread dissemination of the virus throughout the body. Dissemination to the various organ systems is hematogenous and facilitated by infection of peripheral blood monocytes (4–7). These circulating infected monocytes can freely access all organ systems, cross the blood brain barrier, and enter the bone marrow, which serves as the reservoir for lifelong viral persistence (1, 4, 8). In this manner, monocytes serve as the essential vehicle for widespread dissemination of the virus and for the establishment of latency following primary infection.
To successfully infect monocytes, HCMV must gain access to the monocyte nucleus. HCMV is a DNA virus and as such requires access to the host cell nucleus to begin viral gene expression and genomic replication (1). Thus, nuclear translocation is a critical facet of the viral life cycle of HCMV and of all herpesviruses. We have previously identified that HCMV nuclear translocation in monocytes is both temporally and physically unique relative to that seen in other infected cell-types (9–11). In fibroblasts and epithelial cells, HCMV can be detected in the nucleus beginning around 30 min postinfection (mpi) (9, 10), while in CD34+ hematopoietic progenitor cells, this process takes approximately 1–2 h (9). However, in monocytes, nuclear translocation begins around 3 days postinfection (dpi) (10, 11). We have also previously noted that intracellular trafficking of the virus is distinct in monocytes and is navigated in the absence of released tegument proteins or de novo viral gene expression. First, HCMV attaches to and activates the cellular surface receptors epidermal growth factor receptor (EGFR) and β1/β3 integrins via viral glycoproteins gB and the pentamer complex gH/gL/UL128-131, respectively. Following the engagement and activation of monocyte EGFR and β1/β3 integrins, HCMV is taken into the cell where it is rapidly found in EEA1+ vesicles (early endosomes or macropinosomes) (10–14). This is contrary to viral entry in fibroblasts and epithelial cells where viral envelope fusion and de-envelopment typically occur at the plasma membrane or early endosome. In monocytes, the virus then transitions from the early endosome to the trans-Golgi network (TGN) and then subsequently to Rab11+ recycling endosomes where viral de-envelopment occurs. De-envelopment is followed by nuclear translocation and delayed viral genome replication, ultimately leading to productive infection (1, 9–11). Additionally, we have previously published that viral passage through the TGN in monocytes is required for nuclear translocation, as disruption of the TGN significantly inhibits the ability of the virus to reach the nucleus (10). These necessary trafficking events are directed by the viral glycoprotein-induced activation of integrin-Src signaling and the chronic activation of the EGFR-Akt signaling axis (10, 11). Unlike that seen in other cell-types, gB/EGFR and pentamer/integrin engagement and activation is required for attachment and viral entry into monocytes, as well as successful nuclear translocation of the virus. In fact, absence of HCMV-induced EGFR or c-Src signaling pathways in monocytes post-entry inhibits the progression of the virus to the nucleus and can result in viral degradation (10, 11).
This distinct route of HCMV nuclear translocation in monocytes that we have uncovered bears the hallmarks of retrograde transport; in that the virus travels from the cell surface to early endosomes, and then to the TGN (15). In nonneuronal cells, retrograde transport is defined by the recycling of endocytosed lipids, transmembrane receptors, and extracellular proteins from the cell surface to the endosomal network, and subsequently to the TGN and/or endoplasmic reticulum (16–18). Retrograde transport is an essential pathway to maintain and control various critical cellular processes such as intracellular signaling, lipid and cellular homeostasis, and organelle identity (17–20). This is a highly selective and limited trafficking pathway restricted to only a few classes of cellular proteins including acid hydrolase receptors, cation independent/dependent mannose-6-phosphate receptors, soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs), and a select few transmembrane and soluble enzymes (i.e., EGFR and furin) (15, 17–19, 21). Because retrograde trafficking exhibits complex regulation and limited cargo capacity, few pathogens utilize this pathway for entry into host cells. Most notably, bacterial and plant toxins utilize the retrograde transport pathway to enter and toxify cells. For example, Shiga toxin, cholera toxin, and ricin bind to glycoproteins or glycosphingolipids on the cell surface, which allow the correct sorting of these toxins along the retrograde transport route to the Golgi and subsequently the ER from which they then promote cytotoxicity (15–17, 19, 21–23). Only three viruses (HPV, SV40, and AAV) have been documented to utilize the retrograde transport pathway for entry and infection, all of which are nonenveloped viruses (15, 16, 24). While HCMV, along with other enveloped viruses such as HIV and vaccinia virus, are known to exploit retrograde transport machinery to facilitate virus assembly and egress, they have not been reported to exploit retrograde transport for viral entry (15, 16, 24–27). To our knowledge, our new findings implicate HCMV as the initial example of the use of retrograde transport for an enveloped virus entry process, which likely makes this process of herpesvirus infection an extremely rare event, especially for an enveloped virus.
Retrograde trafficking is predominantly facilitated by specific combinations of lipids, small GTPases, tethering molecules, and SNAREs (15, 16, 18, 19, 21, 23, 28). SNAREs function to facilitate membrane fusion between transport vesicles and target cellular compartments by complexing with cognate receptors to create membrane fusion complexes for transport of cargo throughout the cell (21, 29, 30). A key SNARE in retrograde transport is syntaxin 6 (Stx6). Stx6 predominantly functions at the endosomal/TGN interface where it mediates the fusion of endosomes and the TGN (31–35). Although Stx6 is predominantly concentrated at the TGN in most cell-types, in can also be present on early endosomes, recycling endosomes, and the plasma membrane. In cells of the myeloid lineage, which typically exhibit more dynamic trafficking needs than other cell-types, Stx6 localization and function changes with the needs of the cell (29, 32, 35–41). For example, Stx6 is concentrated at the TGN in macrophages, but is predominantly found on EEA1+ vesicles in dendritic cells (32, 35–38). It has been documented that perturbations of Stx6 have a profound impact on retrograde transport. Decreases in the amount or inhibition of Stx6 function causes a concomitant decrease in retrograde trafficking from endosomes to the TGN, while overexpression of Stx6 has been shown to increase retrograde trafficking (35, 36, 39, 42–44).
As previously mentioned, we have observed that in monocytes, HCMV undergoes a retrograde-like trafficking route for entry (10, 11) that we argue is abnormal for viruses and especially for enveloped viruses. Because this transport route, particularly the transition of HCMV from early endosomes to the TGN, is required for nuclear translocation and productive infection in monocytes, we believe the identification of this process represents a new and important pathway required for HCMV infection in a clinically relevant cell-types. This retrograde-like trafficking event is thereby critical for the viral life cycle within monocytes and warrants further study. Because we have demonstrated that HCMV undergoes requisite trafficking from early endosomes/macropinosomes (EEA1+ vesicles) to the TGN prior to nuclear translocation in monocytes, we hypothesized that HCMV manipulated Stx6 to successfully traffic the viral particle to the TGN. In this study we show that HCMV infection significantly and rapidly affects both Stx6 transcript and protein levels. While we observed these changes in protein levels in HCMV-infected fibroblasts and epithelial cells, it is likely due to a common, non-cell-type-specific, element of the viral binding events. However, we excitedly report that only in monocytes is Stx6 functionally important for HCMV nuclear translocation. Further, we demonstrate that this phenotype is specific to Stx6 and that in the absence of Stx6, HCMV is retained in EEA1+ vesicles rather than moving forward along the viral nuclear translocation route to the TGN. We also show Stx6 is unexpectedly concentrated on EEA1+ vesicles in primary monocytes, rather than at the TGN. Here we report that in contrast to what is typically seen in other cells, Stx6 seems to function on the early endosome side of the endosome-to-TGN transition to facilitate retrograde trafficking and fusion of the viral particle into the TGN. Finally, we observed that the viral particles that associate with both the early endosomes and Stx6 early in infection are the likely population of viral particles that successfully traffic to the TGN, and subsequently to the nucleus. Together, our data documents what we argue is the initial description of an enveloped virus exploiting retrograde trafficking machinery for entry and through our mechanistic analysis highlights the distinct trafficking mechanics underlying HCMV nuclear translocation in monocytes.
RESULTS
HCMV infection alters Stx6 transcript and protein levels in infected cells.
As mentioned previously, retrograde transport is a selective and limited intracellular trafficking route generally classified as cargo transport from the endosomal network to the TGN and/or ER. While some bacterial and plant toxins, along with a small number of nonenveloped viruses, have been documented to commandeer the retrograde trafficking pathway for entry, this seems to be a rare event for enveloped viruses. However, we have documented that after being endocytosed by monocytes, HCMV first traffics to EEA1+ vesicles (early endosomes or macropinosomes), then to the TGN, and subsequently to Rab11+ recycling endosomes prior to de-envelopment and nuclear translocation (10, 11). Because this nuclear translocation route is reminiscent of the uncommon retrograde transport pathway, we hypothesized that HCMV was utilizing retrograde transport machinery to facilitate these specific intracellular trafficking events, specifically the transition of HCMV from early endosomes to the TGN. Thus, we wanted to better understand this possible new trafficking mechanism in monocytes.
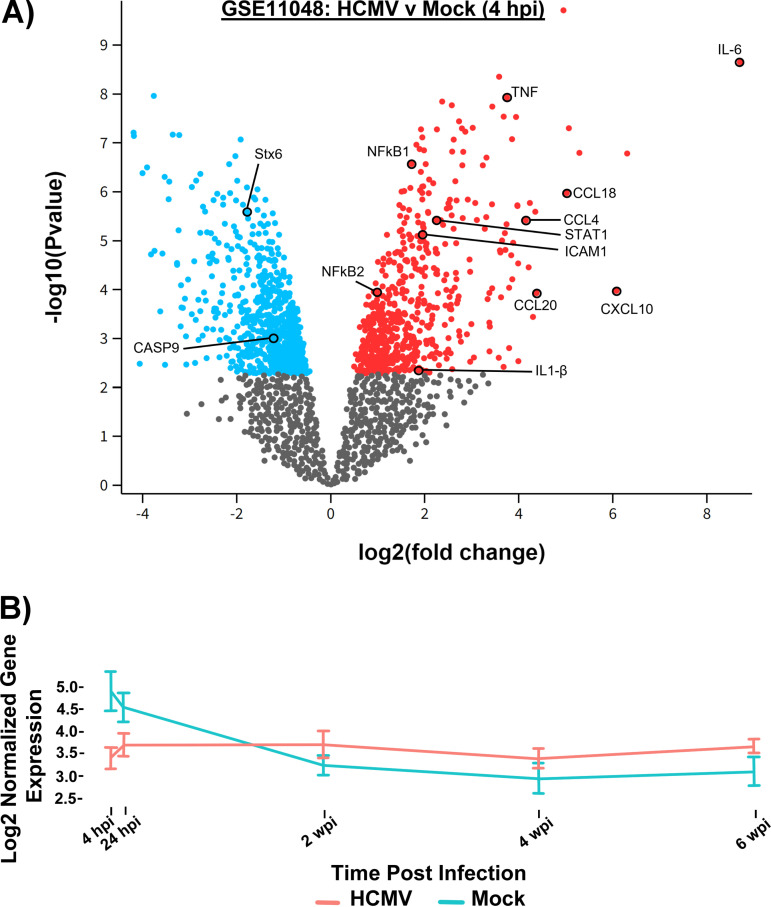
Our previous studies led to construction of a microarray database that compares the transcriptomes of mock and HCMV-infected primary human monocytes at 4 h postinfection (hpi) (45, 46). For the purposes of this study, we reexamined our established microarray transcriptome data to investigate transcriptional changes during early infection (4 hpi) (GEO accession number GSE11408) (Fig. 1A, Table 1). Utilizing this transcriptomic approach, we screened for trafficking protein transcripts significantly affected by viral infection. When analyzing the top 250 most significantly altered transcripts (Fig. 1A), we identified several transcripts of trafficking proteins that were increased or decreased relative to mock. The transcript of the major retrograde transport protein Stx6 was the most significantly dysregulated (P value: 2.62e−06) out of the transcripts examined (Table 1). Therefore, while all the proteins of the transcripts identified in our transcriptome analysis, such as Syntaxin 11 and VAMP5, are involved in vesicular transport in some capacity and will be investigated in the future, we focused on Stx6 due to its role in early endosome-to-TGN retrograde transport (19, 21, 28, 33–35, 37, 41, 42, 44). Surprisingly, as highlighted in the insert in Fig. 1A, transcripts of Stx6 were decreased relative to mock (-1.778 fold change). However, Stx6 transcripts are translationally repressed in P-bodies within the cytosol in order to be rapidly translated in response to the immediate needs of the cell (47–50). Furthermore, translation is coupled with RNA stability (51–53). That is, it has been reported that more frequently translated transcripts are associated with increased degradation (51–53). Therefore, a rapid significant drop of Stx6 transcripts in response to a stimulus, such as viral infection or cellular differentiation, we hypothesized would correlate to a rise in Stx6 protein levels. We confirmed the decrease in Stx6 transcripts during early infection via RNAseq analysis (4 hpi – 6 wpi [GSE206198]) (Fig. 1B). In this RNAseq analysis, we found that Stx6 transcripts increased over time and at approximately 1 week postinfection (wpi) overtook mock-infected cell Stx6 transcript levels, with a continued upward trend out to at least 6 wpi. This likely represents the virus-driven monocyte-to-macrophage differentiation (approximately 1–2 wpi [4, 54]), a process that is associated with a significant increase in trafficking proteins to accommodate the increased trafficking demands of the differentiated macrophage (4, 40).
FIG 1.
HCMV infection causes transcriptomic changes to trafficking proteins. (A) RNA was isolated from HCMV-infected primary human monocytes at 4 hpi from 6 donors and converted to cDNA (GEO accession number GSE11408). A microarray was then performed on whole cell transcripts, which were analyzed by GEO2R software and compared to mock infected cells. A volcano plot was constructed of transcripts upregulated (red) and downregulated (blue) relative to mock. P value versus fold change relative to mock is plotted. (B) RNA was isolated from mock and HCMV-infected primary human monocytes (3 donors) at 4 hpi, 24 hpi, 2, 4, and 6 wpi and prepared for RNAseq (GSE206198). Paired end sequencing was performed (>25 × 106 reads) and aligned to HCMV Lisa and human genome to determine differential expression (FDR=0.05). Transcripts per million (TPM) were log 2 normalized for gene level analysis. Stx6 gene expression in mock (blue) and HCMV-infected monocytes (red) are shown.
TABLE 1.
List of trafficking protein transcripts significantly altered by HCMV infectiona
| Gene title | Gene symbol | Function | P value |
|---|---|---|---|
| Kinesin family member 5B | KIF5B | Distribution of mitochondria and lysosomes | 3.52e-01 |
| Kinesin family member 5C | KIF5C | Endosomal recycling, compartment/slow Recycling | 1.65e-01 |
| Rab27A | RAB27A | Late endosome positioning, maturation, and secretion | 1.46e-01 |
| Synaptosome-associated protein 23 | SNAP23 | Endocytic recycling and phagosome maturation | 6.31e-03 |
| Syntaxin 6 | STX6 | Endosome-TGN retrograde transport | 2.62e-06 |
| Syntaxin 11 | STX11 | Late endosome-TGN protein transport | 1.10e-05 |
| Vesicle-associated membrane protein 5 | VAMP5 | Plasma membrane to early endosome vesicular transport | 3.07e-01 |
Transcript data from GSE11408 (4 hpi).
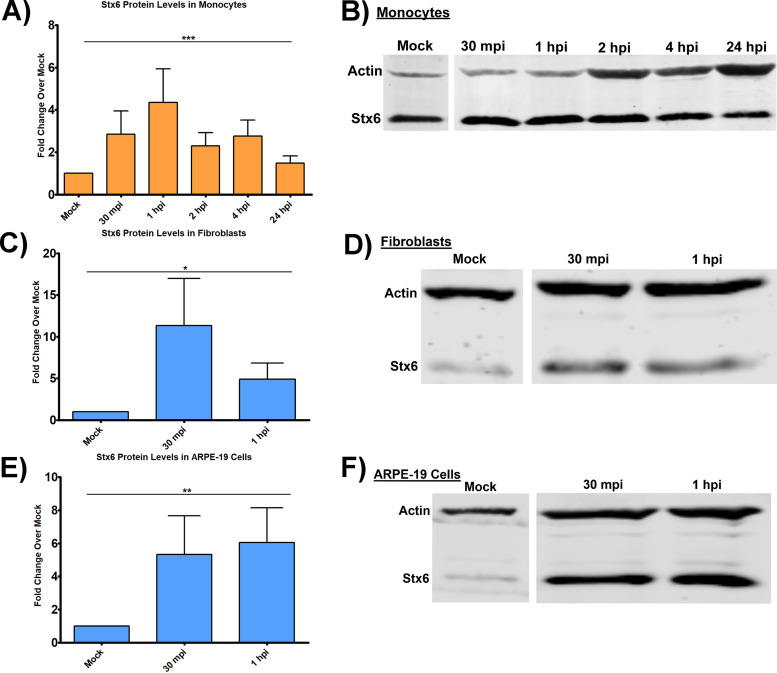
Because HCMV trafficking from EEA1+ endosomes to the TGN is a requisite step for nuclear translocation and is characteristic of retrograde transport, and because Stx6 is a central component of retrograde trafficking from early endosomes to the TGN, we next wanted to determine if HCMV infection impacted Stx6 protein levels. We infected primary human monocytes with HCMV TB40/E (multiplicity of infection [MOI] = 5) and immunoblotted for Stx6 at time points leading up to and including the time frame in which the virus traffics through the TGN (30 mpi – 24 hpi) (Fig. 2A and B). We also performed the same experiment in human embryonic lung (HEL) fibroblasts (Fig. 2C and D) and ARPE-19 epithelial cells (Fig. 2E and F). Because we and others have documented nuclear translocation to occur within 30 mpi in these cells (10, 55, 56), the infection time course was abridged to reflect a time point appropriate for nuclear translocation (1 hpi) (10). On average, HCMV infection led to a 5–15-fold increase in Stx6 protein levels in infected fibroblasts over mock-infected fibroblasts, a 5–8-fold increase in Stx6 levels in infected vs mock-infected epithelial cells, and a 2–6-fold increase in Stx6 levels in infected vs mock-infected monocytes at all time points examined. As de-envelopment of HCMV does not occur until approximately 1–3 dpi in monocytes (10, 11), these data indicate that upregulation of Stx6 protein is likely due to a commonality in receptor-ligand-mediated signaling events that follows viral binding. That is, EGFR and integrins are on all cell-types and once triggered, produce both common and cell-type-specific signaling. We have previously shown that EGFR and β1/β3 are the specific cellular receptors that mediate HCMV attachment and entry into monocytes (10, 13, 14), and that the signalosomes downstream of these receptors are broad and overlap with that of other known HCMV receptors on other cell-types such as PDGFR-α (1, 57, 58). Our data would argue that, in general, viral binding has some common signaling outcomes and this might be why Stx6 is upregulated in multiple cell-types.
FIG 2.
HCMV infection increases Stx6 protein levels. Primary human monocytes (A and B), HEL fibroblasts (C and D), and ARPE-19 epithelial cells (E and F) were synchronously infected with HCMV TB40/E UL32-GFP (MOI = 5). Protein was harvested 30 mpi, 1 hpi, and/or 2 hpi, 4 hpi, 24 hpi depending on the experiment. Protein lysates were immunoblotted for Stx6 and actin. Protein band densities were quantified by LI-COR Image Studio software, normalized to mock, and expressed as fold change over mock. Single factor ANOVA was performed for statistical significance. *, P < 0.05; **, P < 0.01; ***, P < 0.001. N = 3 per cell-type.
Knockdown of Stx6 disrupts HCMV trafficking to the TGN in monocytes.
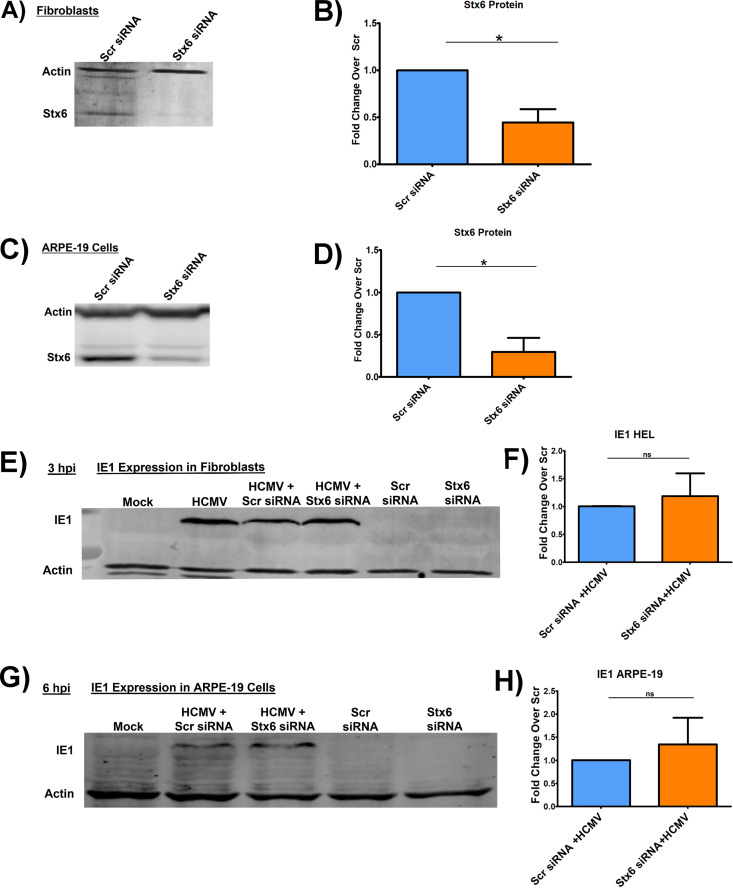
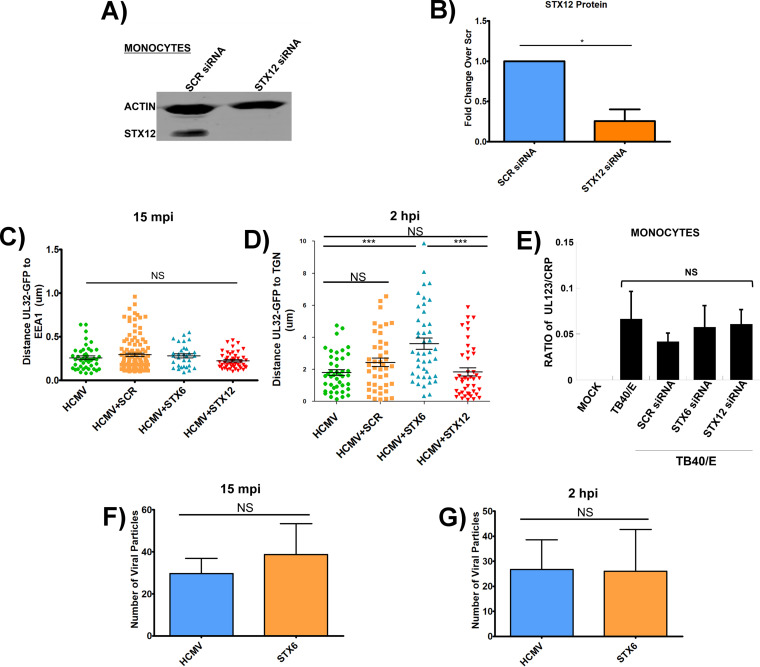
Because the HCMV-induced increase in the protein level of Stx6 was statistically significant in the cell-types tested, we next wanted to determine if the upregulation of Stx6 was functionally important for viral trafficking and/or nuclear translocation. Next, we treated primary human monocytes, ARPE-19 epithelial cells and fibroblasts with siRNA targeting Stx6 to knock down (KD) Stx6 gene expression, or a scrambled siRNA as a control. KD efficiency of Stx6 was measured by Western blot and quantified by densitometry (Fig. 3A–D and Fig. 4A–B). Knockdown efficiency of 60% or higher was consistently achieved in all cell-types. After a 24-h (ARPE-19 cells/fibroblasts) or 48-h (monocyte) siRNA incubation, the cells were then synchronously infected with HCMV TB40/E (MOI = 5 for immunoblots; MOI = 10 for immunofluorescence). Fibroblasts and epithelial cells are rapidly permissive for HCMV infection with viral DNA being found within the nucleus beginning approximately 30 mpi (10, 55, 56). Therefore, we immunoblotted for HCMV immediate early protein 1 (IE1), the first de novo expressed viral gene product at 3 hpi in fibroblasts (Fig. 3E and F) and 6 hpi in ARPE-19 cells (Fig. 3G and H), as a readout for successful nuclear translocation. In both fibroblasts and epithelial cells (Fig. 3), there was no significant difference in IE1 gene expression between cells treated with the siRNA targeting Stx6 or the scrambled control. Additionally, as HCMV exhibits similar viral entry into ARPE-19 cells relative to monocytes, we performed RT-qPCR to measure IE1 transcript levels at 1 hpi in HCMV-infected, Stx6 siRNA-treated ARPE-19 cells. We found no significant difference between IE1 gene expression in control HCMV-infected cells and infected cells treated with Stx6 siRNA (data not shown). This data indicated that although HCMV infection causes an increase in Stx6 protein in both fibroblasts and epithelial cells, Stx6 is not functionally important for HCMV nuclear translocation in fibroblasts and epithelial cells.
FIG 3.
Knockdown of Stx6 does not affect HCMV nuclear translocation in fibroblasts or epithelial cells. HEL fibroblasts (A and B) and ARPE-19 epithelial cells (B and D) were treated with siRNA targeting Stx6 or a scrambled (Scr) siRNA control. Treated cells were incubated for 24 h to ensure maximal knockdown. Immunoblots were performed on whole cell lysate and Stx6 band density was measured and normalized to quantify knockdown efficiency (B and D). The treated cells were then synchronously infected with HCMV TB40/E UL32-GFP (MOI = 5). Protein was harvested from infected and mock-infected fibroblasts (E and F) and ARPE-19 cells (G and H) at 3 hpi and 6 hpi, respectively. Lysates were immunoblotted for HCMV IE-1 and actin (E–H). Protein amount was quantified using LI-COR Image Studio software. Student’s t-tests were used to assess statistical significance. N = 3 per cell-type. (C) *, P < 0.05; **, P < 0.01; ***, P < 0.001.
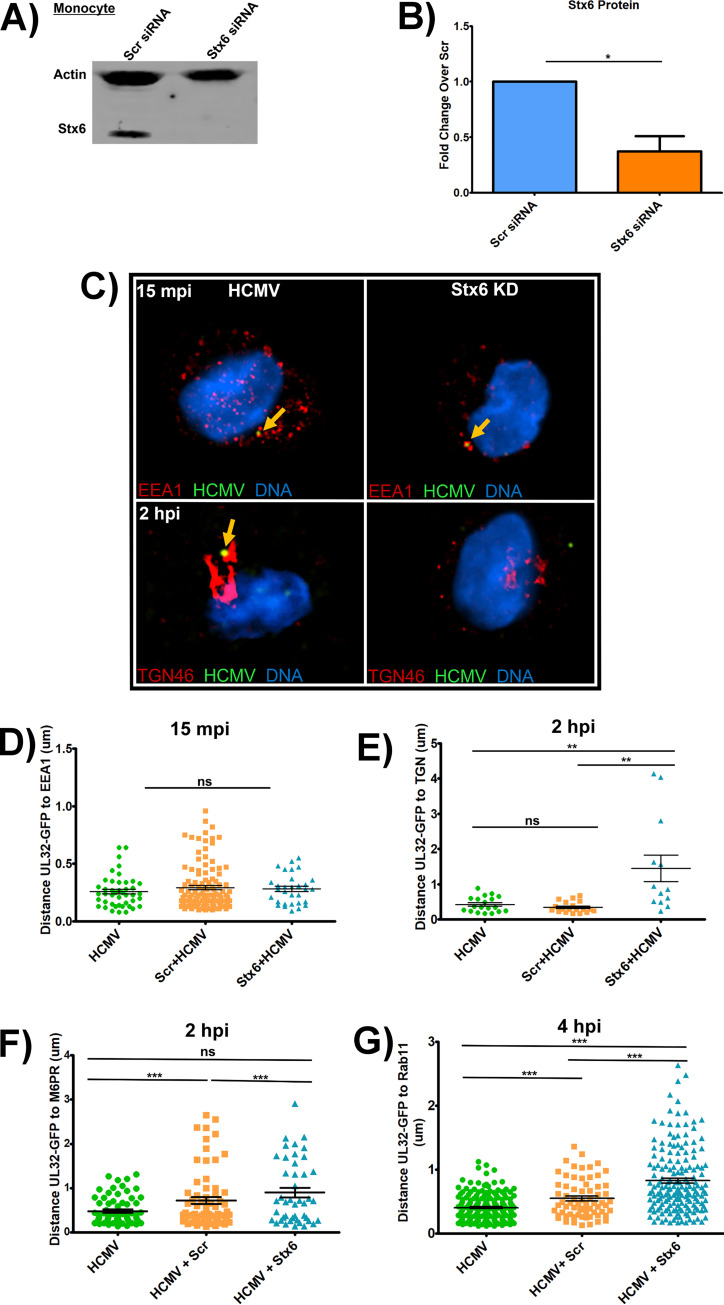
FIG 4.
Knockdown of Stx6 disrupts HCMV trafficking to the TGN in monocytes. Primary human monocytes were treated with siRNA targeting Stx6 or scrambled (Scr) siRNA control and incubated for 48 h for maximal knockdown. (A) An immunoblot was performed on whole cell lysate (representative blot shown). Stx6 band density was measured using LI-COR Image Studio software. Band density was normalized to Scr control to quantify knockdown efficiency (B). The cells were then synchronously infected with HCMV TB40/E UL32-GFP (MOI = 10), then fixed at 15 mpi, 2 hpi, and 4 hpi. Cells were then permeabilized and stained for markers of early endosome (EEA1), the TGN (TGN46), late endosome (M6PR), recycling endosome (Rab11), and HCMV (anti-GFP). Images were acquired on a Nikon N-SIM E Super Resolution microscope system (100x objective) in single slices (z stacks) using the same laser settings. Representative images are shown (C). Yellow arrows indicate co-localization. Viral particle distance to the nearest edge of the target organelle was quantified from at least 10 cells (D–G) using the microscope companion analysis software. Student’s t-tests were used to assess statistical significance. **, P < 0.01; ***, P < 0.001. A–E: N = 3; F–G: N = 2.
Monocytes, however, are not initially permissive for HCMV replication (10, 11, 55, 56). Therefore, to assess the impact of Stx6 on viral nuclear translocation, we used the Nikon N-SIM E Super Resolution microscope system (~100 nm limit of resolution) on Stx6 siRNA-treated primary human monocytes along with scrambled controls to track the progress of the viral particle to the nucleus by measuring viral particle colocalization with the specific cellular compartments we previously documented to be key for viral nuclear translocation in these cells (10, 11). Because Stx6 primarily functions in retrograde trafficking events from early endosomes to the TGN, we first focused on these compartments. We synchronously infected the siRNA-treated monocytes with HCMV TB40/E (MOI = 10). After incubation with the virus, the monocytes were then fixed at 15 mpi, when we expected the virus to be within early endosomes, and at 2 hpi, when we expected the virus to be associated with the TGN, as previously documented (10, 11). The fixed cells were then stained with antibodies to the viral particle and various cellular compartments. Specifically, a TB40/E UL32-GFP virus (9–11) was employed and an anti-GFP antibody was used to enhance the signal to the viral particle. The cellular compartments examined were early endosomes/macropinosomes (using an antibody to EEA1) and the TGN (using an antibody to TGN46). Distance between the center of the viral particle (the UL32-GFP signal) and nearest edge of target organelle (early endosome or TGN) was manually quantified by Nikon 3D reconstruction and line profiling, a technology we established previously (11). Our new data showed that HCMV trafficking to the early endosome at 15 mpi was unaffected by Stx6 KD (Fig. 4C, top left and right panels) with no significant difference in colocalization of viral particles with early endosomes between Stx6 KD cells, HCMV only, and scrambled siRNA controls (see quantified data in Fig. 4D). However, viral colocalization between HCMV and the TGN at 2 hpi was greatly affected by Stx6 KD, which resulted in a loss of colocalization (Fig. 4C, bottom left and right panels). On average, we observed a significant ~350% increase in the distance of the viral particles away from the TGN in Stx6 KD cells relative to HCMV only and scrambled siRNA controls (quantified in Fig. 4E). Because trafficking to the TGN is requisite for HCMV nuclear translocation in monocytes, this data suggested that Stx6 was functionally important for trafficking of HCMV to the TGN in monocytes and thus is important for nuclear translocation. We then wanted to determine the impact of Stx6 KD on viral degradation as well as subsequent trafficking from the TGN to recycling endosomes. To assess the impact of Stx6 KD on viral degradation, we performed Super Resolution microscopy to examine HCMV colocalization with late endosomes at 2 hpi, where Stx6 siRNA-treated, HCMV-infected monocytes were stained with antibodies targeting HCMV UL32-GFP (anti-GFP) and the late endosome (anti-M6PR) at 2 hpi (Fig. 4F). We observed no significant difference in virus colocalization with late endosomes between cells treated with Stx6 siRNA, HCMV only or scrambled siRNA controls, indicating that Stx6 KD does not influence viral degradation in monocytes. We previously showed that trafficking of the virus to the recycling endosomes is required for nuclear translocation and thus serves as a surrogate event for successful HCMV nuclear translocation (10, 11). To assess the impact of Stx6 KD on the late stage of the HCMV nuclear translocation pathway, and thereby successful nuclear translocation, we performed Super Resolution microscopy where Stx6 siRNA-treated, HCMV-infected cells were stained for HCMV UL32-GFP (anti-GFP) and recycling endosomes (Rab11) (Fig. 4G). We have previously published that HCMV is colocalized with recycling endosomes in monocytes beginning at approximately 2 hpi (10, 11). We chose to assess viral colocalization with recycling endosomes at 4 hpi rather than later time points in the viral life cycle for a few reasons: 1) 4 hpi is beyond the point of initial HCMV/Rab11+ vesicular co-localization; (2 hpi); and 3) we wanted to minimize the effect of other intracellular trafficking machinery compensating for Stx6 function, thereby confounding observable Stx6 defect-associated trafficking phenotypes. For example, Stx5 has been documented to compensate for Stx6 function during endosome-TGN retrograde trafficking (19–21). At 4 hpi, we observed that, similar to the viral trafficking phenotype observed at the TGN at 2 hpi, Stx6 KD significantly inhibited HCMV trafficking to the recycling endosome at 4 hpi relative to HCMV only and scrambled siRNA controls. This data suggests that Stx6 is an important regulator of not only HCMV early endosome-TGN transport, but also successful nuclear translocation of the virus.
Only Stx6 is associated with HCMV trafficking in monocytes.
Stx6 is known to be involved in the recycling of membranes and membrane receptors. Additionally, it has been documented that disruption of one syntaxin can disrupt entire portions of the syntaxin trafficking network (34, 35, 38, 42, 43, 59–61). Because these scenarios may have an impact on viral trafficking, we wanted to ensure that the viral trafficking defect observed in Stx6 KD cells was specific to Stx6. Syntaxin 12 (Stx12) is involved in trafficking events that functionally overlap with Stx6 (i.e., recycling of membranes from the TGN to the plasma membrane) and has previously been used as a control for understanding Stx6’s role in cells (38, 39, 60, 62). Therefore, we next treated primary human monocytes with siRNA targeting Stx12 (Fig. 5A and B). An average KD efficiency of approximately 75% was consistently achieved (Fig. 5B). After a 48-h incubation, the monocytes were synchronously infected with HCMV TB40/E UL32-GFP (MOI = 10) and subsequently fixed and stained for HCMV (anti-GFP) and organelles (EEA1 for early endosomes; TGN46 for TGN). Super resolution microscopy was then performed to measure the viral particles colocalization with the target cellular compartment (early endosome or TGN) as previously described (Fig. 5C and D). We observed no significant difference in the colocalization of the vial particle with early endosomes in Stx12 KD cells versus HCMV only, Stx6 or scrambled siRNA controls (Fig. 5C). Additionally, we observed no significant difference in colocalization of the viral particle with the TGN relative to HCMV only controls. We also observed a significant ~230% increase in distance of viral particles away from the TGN in Stx6 KD cells relative to Stx12 KD cells (Fig. 5D). Overall, HCMV trafficking in Stx12 siRNA-treated monocytes behaves similarly to that seen in untreated controls, suggesting that the viral trafficking defect observed in Stx6 KD cells is not due to a defect in syntaxin-driven lipid/membrane turnover or a general syntaxin network malfunction.
FIG 5.
Stx-associated trafficking disruption is specific to Stx6. Primary human monocytes were treated with siRNA targeting Stx12 or a scrambled control (Scr) (A). An immunoblot was performed on whole cell lysate and Stx12 band density was measured using LI-COR Image Studio software. Band density was normalized to Scr control to quantify knockdown efficiency. A representative blot is shown (A and B). (C and D) Primary human monocytes were treated with siRNA targeting Stx6, Stx12, or a Scr control. The cells were then synchronously infected with HCMV TB40/E UL32-GFP (MOI = 10). The cells were then fixed at 15 mpi and 2 hpi. The fixed cells were then permeabilized and stained for TGN (TGN46) and HCMV (measured via UL32-GFP). High resolution images were acquired on a Nikon N-SIM E Super Resolution microscope system (100x objective) in single slices (z stacks) using the same laser settings. Viral particle (measured via UL32-GFP) distance to the nearest edge of the TGN was quantified from at least 10 cells and 20 viral particles using the companion analysis software of the microscope. Quantification of images is shown. Student’s t-tests were used to assess statistical significance. *, P < 0.05; **, P < 0.01. (E) A viral entry assay (HCMV TB40/E UL32-GFP; MOI = 1) was performed on Stx6, Stx12, or Scr siRNA-treated primary human monocytes. DNA from these cells was then isolated and qPCR was performed with primers for HCMV UL123 and cellular CRP. (F–G) Total viral particles (measured via UL32-GFP) within infected and Stx6 siRNA-treated monocytes were counted at 15 mpi and 2 hpi using the NIKON N-SIM E Super Resolution microscope system (100× objective) in single slices (z stacks) using the same laser settings. Student’s t-tests were used to assess statistical significance. N = 3.
Furthermore, to determine if the observed viral trafficking defect could be due to impaired viral entry into the cell as a byproduct of the Stx6 or Stx12 KDs, we also performed a viral entry assay and qPCR to compare the amount of viral DNA detected within Stx6 and Stx12 siRNA-treated monocytes within 1 hpi (Fig. 5E). When calculating the ratio of viral genomes to the cellular housekeeping gene for C-reactive protein (CRP), we observed no significant difference in the amount of HCMV genome present within either Stx6 or Stx12 KD cells at 1 hpi relative to untreated and scrambled siRNA-treated control cells. In support of this data, we also used immunofluorescence microscopy to quantify the number of viral particles within Stx6 KD cells at 15 mpi and 2 hpi (Fig. 5F and G). We found that there was no significant difference between the total number of viral particles inside of untreated vs Stx6 siRNA-treated monocytes at either time point. Together, these data suggest that Stx6 KD does not affect in viral entry, but that it does play a significant role in the successful trafficking of the virus in monocytes. Furthermore, the data provides evidence that Stx6 is at least one of the key cell-types specific regulatory molecules controlling the retrograde transport of the virion in monocytes.
Stx6 exhibits vesicular localization in monocytes.
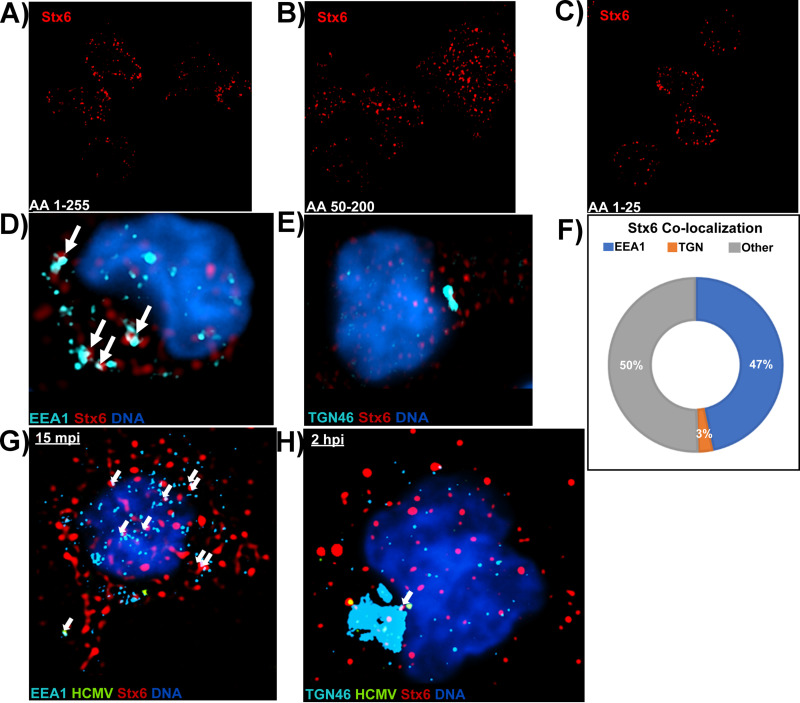
Next, we wanted to determine how Stx6 might be functioning in monocytes, and thereby how the virus may be utilizing Stx6 to facilitate retrograde trafficking from early endosomes to the TGN. As discussed previously, retrograde transport can be defined as the trafficking of cargo, lipids, and membrane-bound receptors from the endosomal network to the TGN and/or the ER; a process facilitated by retrograde trafficking proteins such as Stx6 (16–18). While predominantly distributed at the TGN in most cell-types, Stx6 is known to be differentially distributed in cells of the myeloid lineage, likely due to the more dynamic trafficking needs of these cells. For example, Stx6 is typically concentrated at the TGN in macrophages, while predominantly distributed on early endosomes in dendritic cells (32, 35–38). Because monocytes are also of the myeloid lineage and the location of Stx6 in monocytes would provide insight into where and how it might function in the HCMV-containing EEA1+ vesicle-to-TGN transition, we wanted to determine Stx6 localization in primary human monocytes. To address Stx6 localization, we first fixed uninfected primary human monocytes and subsequently stained the cells with antibodies against Stx6 (Fig. 6A–C). We utilized 3 different antibodies targeting 3 distinct C- and N-terminal residue sites of the Stx6 protein to ensure that observed staining was not due to potential antibody bias. Using super resolution immunofluorescence microscopy, we observed that all 3 antibodies used displayed similar punctate staining throughout the cells. Because Stx6 could potentially localize to either early endosomes or the TGN in monocytes, we also fixed and stained uninfected monocytes with antibodies against both Stx6 and the early endosome (EEA1) or TGN (TGN46) to determine Stx6 organelle localization (Fig. 6D and E). We observed that in uninfected monocytes, Stx6 exhibits punctate, vesicular staining that primarily overlaps with EEA1 rather than the TGN marker. Additionally, we manually quantified the colocalization of Stx6 with target organelles in these cells using the accompaniment software to the Nikon N-SIM E Super Resolution microscope system and found that approximately half (~47%) of Stx6 is colocalized with EEA1, while approximately 3% of Stx6 is colocalized with TGN46 (Fig. 6C). The remaining Stx6 is likely distributed between recycling endosomes and the plasma membrane. Therefore, the Stx6 distribution in primary human monocytes seems similar to that observed in dendritic cells, where the majority of Stx6 is located in the endosomal compartment with little Stx6 concentrated in the TGN as reported for macrophages and other cell-types (29, 32, 35–38, 40, 59). We also wanted to determine if HCMV infection altered the distribution of Stx6. Super resolution microscopy was performed next on triple-stained HCMV-infected monocytes at 15 mpi (Fig. 6G) and 2 hpi (Fig. 6H). While infection led to an observable increase in Stx6 at both time points relative to uninfected controls, supporting our previous immunoblot data (Fig. 2A), Stx6 staining remained punctate and predominantly associated with EEA1 throughout infection. These data suggest that HCMV infection does not alter Stx6 distribution during infection of monocytes. Based on the location we observe within the cell, it is likely that Stx6 functions on the endosomal side of the early endosome-to-TGN transition rather than at the TGN as reported in other cell-types (29, 35, 38, 59).
FIG 6.
Stx6 is localized on EEA1+ vesicles rather than the TGN in primary monocytes. Three distinct primary antibodies targeting Stx6 were used to assess the localization of Stx6 in uninfected primary human monocytes. (A) Anti-Stx6 targeting amino acids 1 to 255. (B) Anti-Stx6 targeting amino acids 50 to 200. (C) Anti-Stx6 targeting amino acids 1 to 25. Super resolution microscopy was performed as described in Fig. 4 (D–F). Uninfected primary human monocytes were fixed and stained with antibodies to EEA1 (A) or TGN46 (B) and Stx6. High resolution images were acquired on a Nikon N-SIM E Super Resolution microscope system (100× objective) in single slices (z stacks) using the same laser and lookup table (LUT) settings. Representative images are shown. Stx6 colocalization (~0.1 μm distance) with target organelle was manually quantified using the companion analysis software of the microscope (F). HCMV TB40/E (MOI = 10) -infected primary human monocytes were fixed at 15 mpi (G) and 2 hpi (H) and stained with antibodies against EEA1 or TGN46, HCMV UL32-GFP, and Stx6. High resolution images were acquired on a Nikon N-SIM E Super Resolution microscope system (100× objective) in single slices (z stacks) using the same laser and LUT settings. Representative images are shown. White arrows indicate co-localization between organelle and Stx6. N = 3 for all experiments.
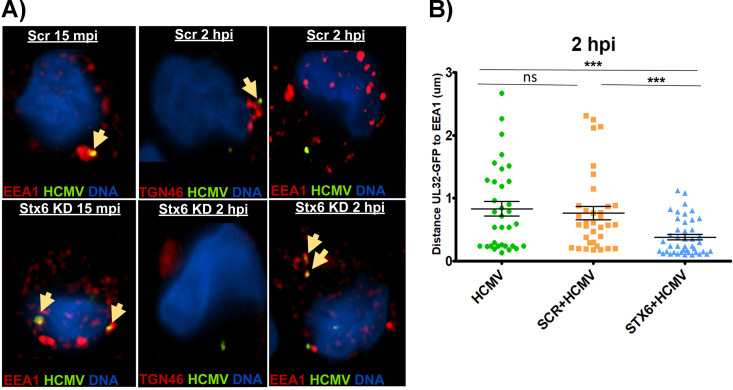
Lack of Stx6 leads to HCMV retention in EEA1+ endosomes.
Because the viral particle does not progress to the TGN during the expected time frame (45 mpi – 2 hpi) in the absence of Stx6, we next wanted to determine where the viral particle is trafficked when retrograde transport is disrupted via Stx6 KD. As our data indicates that Stx6 functions from the endosomal compartment, we hypothesized that in the absence of Stx6, HCMV cannot progress to the TGN and is retained in EEA1+ endosomes. To test this hypothesis, we treated primary human monocytes with siRNA targeting Stx6 or a scrambled siRNA as a control. The cells were then synchronously infected with HCMV TB40/E UL32-GFP (MOI = 10) following a 48-h siRNA incubation and then fixed at 15 mpi and 2 hpi, during which time the virus transitions from early endosomes to the TGN. Subsequently, the monocytes were stained with anti-GFP (mature viral particle), and anti-TGN46 (TGN) or anti-EEA1 (early endosome) (Fig. 7A). Nikon 3D reconstruction and line profiling was used to measure the distance between the viral particle and nearest target organelle as previously described (Fig. 7B). As expected, the viral particle successfully progressed to the early endosome around 15 mpi in both Stx6 siRNA and scrambled siRNA-treated cells. We also observed the expected failure of the viral particle to progress to the TGN at 2 hpi in Stx6 KD cells. However, when the infected, Stx6 siRNA-treated monocytes were stained for early endosomes rather than the TGN at 2 hpi, we found that there was significant viral colocalization with EEA1+ endosomes relative to that seen in untreated and scrambled siRNA control treated cells. This data is also supported by our previously published finding that indicates HCMV remains in EEA1+ vesicles when the TGN is chemically destroyed prior to infection (10), as many interacting partners for Stx6 are present on the TGN. Absence of the TGN would thereby restrict Stx6 function and lead to a similar outcome as a Stx6 KD. Thus, these data support that the absence of Stx6 inhibits the necessary movement of HCMV to the TGN by enforcing retention of mature viral particles within EEA1+ vesicles.
FIG 7.
HCMV is retained in EEA1+ vesicles in the absence of Stx6. Primary human monocytes were treated with siRNA targeting Stx6 or a scrambled (Scr) control and incubated for 48 h to ensure maximal knockdown. The cells were then synchronously infected with HCMV TB40/E (MOI = 10). Control and infected monocytes were then fixed at 2 hpi. The cells were then permeabilized and stained with antibodies against GFP (to stain the virus TB40/E UL32-GFP), early endosomes (EEA1) or the TGN (TGN46). High resolution images were acquired on a NIKON N-SIM E Super Resolution microscope system (100× objective) in single slices (z stacks) using the same laser settings. Representative images are shown. Yellow arrows indicate colocalization (A). Viral particle (measured by UL32-GFP) distance to the nearest edge of the target organelle was quantified from at least 10 cells and 20 viral particles using the companion analysis software of the microscope (B). Student’s t-tests were used to assess statistical significance. ***, P < 0.001. N = 3.
Stx6 facilitates fusion of HCMV-containing EEA1+ endosomes with the TGN.
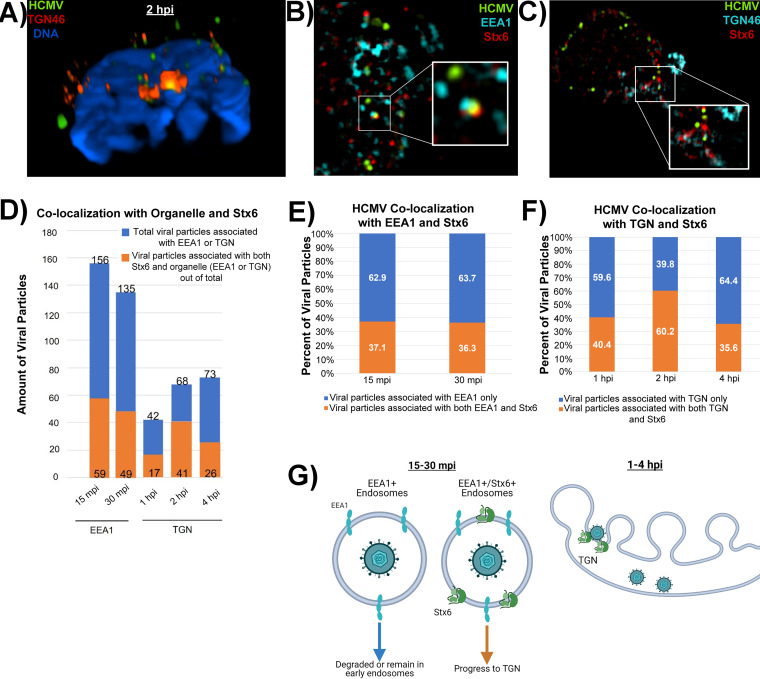
Stx6 functions as a fusion protein by nature, typically facilitating the fusion of early endosomes with the TGN. Because our new data indicates that Stx6 is located primarily within the endocytic compartment in monocytes, we wanted to determine if HCMV utilizes Stx6 on early endosomes as fusion machinery to enter the TGN in these leukocytes. To gain insight into the mechanical process that occurs during the transition of the virus from the early endosome to the TGN, we next wanted to determine if there is indeed a fusion event that occurs between HCMV-carrying EEA1+ vesicles and the TGN. To accomplish this goal, we synchronously infected primary human monocytes with HCMV TB40/E UL32-GFP (MOI = 10). The cells were then fixed at 2 hpi and stained with antibodies against HCMV (anti-GFP) and TGN (TGN46). We utilized the Nikon N-SIM E Super Resolution microscope system to determine if HCMV could be found within the TGN of infected monocytes. Specifically, we completed 3D volumetric reconstructions on infected cells and analyzed cross sections of the TGN to identify any viral particle signal surrounded by TGN signal on all sides, which we concluded to be present within the TGN (Fig. 8A). Indeed, we make the initial observation that HCMV particles can be consistently found within the TGN of monocytes at 2 hpi. While the nonenveloped viruses SV40, AAV, and HPV utilize retrograde transport for entry, only HPV is found in a Golgi-like structure (15, 16, 24). Thus, HCMV residing within the TGN appears to be a unique finding among viruses that utilize retrograde transport for entry. Because the Stx6 SNARE complex (Stx6-Stx16-Vti1a-VAMP4) is the major mediator of retrograde transport from early endosomes to the TGN as well as the fusion process between these compartments (19), these data indicate that the pool of Stx6 located predominantly on EEA1+ endosomes in monocytes functions in a retrograde manner to facilitate fusion of the virus-containing early endosome with the TGN.
FIG 8.
HCMV particles that associate with both Stx6 and EEA1 early in infection fuse with the TGN. Primary human monocytes were synchronously infected with HCMV TB40/E (MOI = 10). The cells were fixed at 15 mpi, 30 mpi, 1 hpi, 2 hpi, and 4 hpi. The fixed cells were then stained with antibodies against HCMV (UL32-GFP), TGN (TGN46) or the early endosome (EEA1) (A), and Stx6 (B–E). High resolution images were acquired on a Nikon N-SIM E Super Resolution microscope system (100× objective) in single slices (z stacks) using the same laser settings. Representative images are shown. (A) Representative image of an infected monocyte at 2 hpi. Volumetric 3D reconstruction was performed on the image. The cross-section is shown. Yellow staining indicates colocalization (~0.1 μm distance). (B and C) Representative images of triple-stained infected monocytes. White boxes indicate areas within the image where triple colocalization occurs. (D) Total viral particles associated with the target organelle (~0.1 μm distance; blue bars) were manually quantified using the companion software to the microscope. Out of that population, the number of viral particles colocalized with both target organelle and Stx6 were quantified (orange bars). (E) The number of viral particles associated with organelle only or both organelle and Stx6 was calculated as percentages. (F) Model of likely biological events happening between virus-containing early endosomes and the TGN at the observed time points. N = 3.
To gain more in-depth information as to how the biological processes involving Stx6 facilitate early endosome-to-TGN transition of HCMV, we synchronously infected primary human monocytes with HCMV TB40/E UL32-GFP (MOI = 10) over a time course reflecting this transitional trafficking period (15 mpi – 4 hpi). We subsequently fixed and triple-stained the cells with antibodies against HCMV (anti-GFP), Stx6, and EEA1 or TGN46 (Fig. 8B and C). Super resolution immunofluorescence microscopy was used to acquire 3D reconstructions of the stained cells. We then manually counted mature viral particles (via UL32-GFP staining) associated with both Stx6 and an organelle marker (EEA1 or TGN) (Fig. 8B–E). We expressed this data as both the number of viral particles (Fig. 8D) and the percentage of viral particles (Fig. 8E) associated with the target organelle. Out of the viral particles associated with the early endosome or TGN, we then quantified how many of those particles were also associated with Stx6. Initially, we observed that the total amount of viral particles within monocytes decreases throughout infection with approximately half of viral particles associated with EEA1 at 15 mpi being lost by 4 hpi. This observation further supports the “bottleneck” phenomenon surrounding viral trafficking through the TGN reported in our prior publications where few viral particles traffic through the TGN at a given time, causing a sharp decrease in the amount of virus found within the cells at later time points (10, 11). It is important to note that we have previously demonstrated some viral particles within late endosomes early in infection, which contributes to particle loss over time (10), but we now have mechanistic insights as to why and how that loss of viral particles over time might occur. Interestingly, the percentage of viral particles associated with both the target organelle and Stx6 remains constant throughout infection (approximately 36–40%), except for 2 hpi, where association of viral particles with both the TGN and Stx6 peaks at 60% before returning to 36% by 4 hpi. Because Stx6 predominantly functions on the outside of the TGN rather than inside, it is likely that from 1–4 hpi, the viral particles associated with both the TGN and Stx6 are those fixed mid-fusion event while the viral particles associated with the TGN only at these time points have likely already dissociated from Stx6 and lie within the TGN to be further transported to the recycling endosome. Because we have previously documented 2 hpi as the peak time point for viral trafficking to the TGN, we expected that this time point would be associated with the highest percentage of viral particles associated with both the TGN and Stx6.
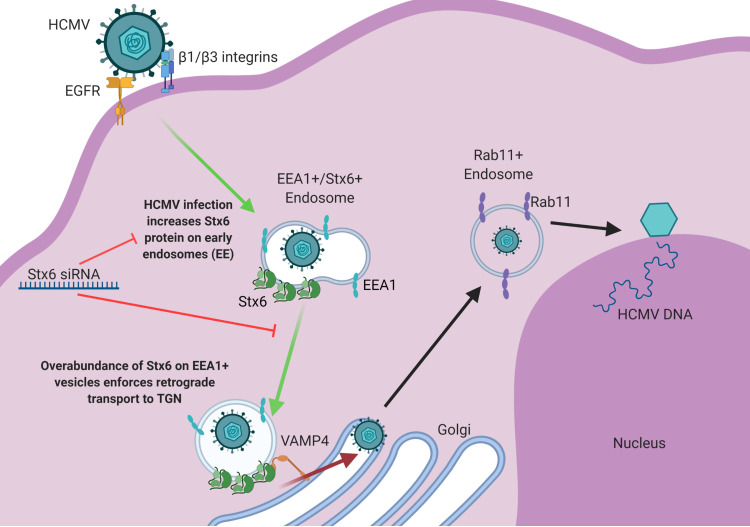
Surprisingly, similar amounts of viral particles associated with both EEA1 and Stx6 early in infection (15–30 mpi) comprise the approximate total amount of viral particles located at the TGN at later time points (1–4 hpi). Because of this trend, Stx6 localization to the endosomal compartment and its function on the exterior of the TGN, as well as the pattern of viral particle colocalization discussed above, we make the following proposal that early in infection (15–30 mpi), HCMV is taken into EEA1+ endosomes or EEA1+/Stx6+ endosomes (Fig. 8F). It is likely that viral particles in endosomes lacking Stx6 are either degraded or remain in early endosomes until Stx6 recruitment to that endosome occurs. Viral particles in EEA1+/Stx6+ endosomes can then successfully progress in a retrograde manner to the TGN. Once docked at the TGN (1–4 hpi), the HCMV-containing EEA1+/Stx6+ endosome can then fuse with the TGN, allowing the viral particle entrance to the lumen of the TGN.
While analysis is limited by the constraints of primary human monocytes, we propose that HCMV uses retrograde trafficking machinery to gain access to the TGN and that the population of viral particles in EEA1+/Stx6+ endosomes comprise the population of viral particles that successfully enter the TGN lumen via a Stx6-driven membrane fusion event. Further, we suggest that the EEA1+/Stx6+ viral particles comprise the population of HCMV that can subsequently traffic to Rab11+ recycling endosomes from the TGN and undergo successful nuclear translocation, thereby identifying Stx6 as a limiting factor for HCMV nuclear translocation in monocytes.
DISCUSSION
Monocytes are a critical cell-types for the infection cycle of HCMV within the host as they serve as essential vehicles of hematogenous dissemination throughout the body to all organ systems, as well as the bone marrow, which serves as the viral latency compartment (1, 5, 8). HCMV pathogenesis is, thus, directly associated with the ability of the virus to successfully infect monocytes. We have previously established that HCMV nuclear translocation in these cells, an essential part of the viral intracellular life cycle, follows a unique physical and temporal pathway to the nucleus relative to that seen in other cell-types and is primarily driven by virus-induced EGFR and integrin signaling events following viral binding (10, 11, 13, 14). However, the trafficking mechanics that physically underlie this pathway remain to be resolved. Because the nuclear translocation of HCMV is requisite for viral genome replication and productive viral infection, we began to investigate the specific trafficking mechanics of this pathway in monocytes.
HCMV utilizes a distinct nuclear translocation pathway in monocytes that bears the hallmarks of retrograde transport as it traffics from early endosomes to the TGN (10, 11). However, the retrograde transport pathway is restricted and highly selective even among cellular proteins. Because of this stringent regulation, it is uncommon for pathogens to utilize this pathway for entry (15–19, 21–23). However, in this study, we show that HCMV infection dysregulates Stx6, a SNARE protein that primarily functions in retrograde transport by facilitating the fusion of early endosomes with the TGN (31–35). We show that transcripts for Stx6 are significantly decreased during early infection while Stx6 protein is significantly increased during early infection. While this event seems counterintuitive, Stx6 transcripts are known to be stalled and held within cells for immediate translation in response to cellular trafficking demand (47–50). It is important to note that mRNA translation and mRNA stability are coupled, further supporting the observed transcript and protein kinetics of Stx6 (51–53). Because we see a significant decrease in Stx6 transcripts accompanied by a significant increase in Stx6 protein, and given the dynamic nature of monocytes, it is likely that HCMV infection causes the rapid translation of stalled Stx6 transcripts to accommodate the trafficking needs of the cell that change with viral infection.
While Stx6 transcript and protein was dysregulated by HCMV infection in monocytes, we also observed that Stx6 protein was significantly upregulated in all cell-types tested (monocytes, fibroblasts, and epithelial cells). However, this is most likely due to some common features of viral binding and ensuing signaling events that are not cell-types-specific. As previously mentioned, HCMV glycoproteins gB and pentamer complex bind to EGFR and β1/β3, respectively, on the monocyte cell surface and activate the subsequent PI3K/Akt and c-Src signaling cascades (10, 11). Even though there are unique aspects of binding of HCMV to EGFR and integrins in monocytes, there are also features that are common across various cell-types. That is, HCMV binding of these and other receptors on fibroblasts and epithelial/endothelial cells also initiate overlapping early downstream signaling events to promote cell-types-specific viral entry (63, 64). Therefore, we argue that the common overlapping viral binding and signaling events can result in some similar physiological outcomes across cell-types.
HCMV infection caused a significant increase of Stx6 protein expression throughout infection of all cell-types examined. While we observed an increase of Stx6 in fibroblasts and epithelial cells, loss of Stx6 did not impact the kinetics of the infection process in these cells, indicating a lack of functional importance during the early stage of infection. We also showed, importantly, that Stx6 is functionally required for nuclear translocation of HCMV in monocytes, demonstrating a cell-types-specific function. KD of Stx6 inhibited the ability of the virus to reach the TGN, which is a requisite trafficking event for nuclear translocation. Additionally, KD of Stx6 significantly inhibited progress of HCMV to recycling endosomes, a surrogate for successful nuclear translocation (10, 11), and did not significantly impact viral degradation. This biological effect seems to be specific to Stx6 rather than a defect in viral entry, lipid/membrane turnover, or defective syntaxin network due to off-target effects of the KD. This finding suggests that HCMV utilizes retrograde transport machinery for monocyte entry, which is uncommon for viruses and rare for enveloped viruses (15, 16, 24–27).
Additionally, we identified that Stx6 exhibits an unexpected localization in monocytes. In most cell-types, including macrophages, Stx6 is concentrated at the TGN where it functions primarily to facilitate endosome-TGN fusion events. However, in primary human monocytes, we found that Stx6 is instead vesicular and exhibits significant colocalization with EEA1+ early endosomes. While this distribution is uncharacteristic of macrophages, Stx6 is often associated with EEA1+ early endosomes in dendritic cells. Thus, Stx6 distribution in myeloid cells appears to change with the needs of the cell (29, 31, 32, 35–41). Further, while HCMV infection increases Stx6 protein, Stx6 localization within the cell appears to be unaffected and remains punctate and vesicle-associated throughout infection. This indicates to us that Stx6 functions on HCMV-containing early endosomes and facilitates the forward movement of the viral particle to the TGN. In fact, KD of Stx6 inhibits the progression of the viral particle to the TGN and simultaneously causes retention of viral particles within the endosomal compartment.
We have also shown here for the first time that HCMV enters the TGN in primary human monocytes. While we have previously published HCMV association with the TGN along the nuclear translocation route, we have now utilized super resolution microscopy and volumetric 3D reconstruction to determine that HCMV is definitively present within the TGN prior to nuclear translocation. This ability of the viral particle to enter the TGN further supports the distinct aspects of HCMV trafficking in monocytes and the distinct molecular mechanisms in this process. Because the mature viral particle is contained in EEA1+ endosomes prior to entering the TGN, we conclude that a fusion event occurs between the virus-containing early endosome and the membrane of the TGN, allowing the virus to cross into the lumen of the TGN. In the instance of retrograde-targeted cellular cargo, Stx6 predominantly performs this function (19, 29, 33, 34). There are no specific inhibitors of Stx6 function. However, this data, combined with the previously discussed data of Stx6 localization, as well as the loss of viral particle localization with the TGN concurrent with the loss of Stx6 lead us to conclude that Stx6 is responsible for the fusion event between the HCMV-containing endosome and the TGN. Furthermore, it has been demonstrated that overexpression of Stx6 protein alone is sufficient to drive dominant retrograde transport (35, 36, 39, 42–44). Although technologically limited within our primary cell system, we also have data indicating that the population of viral particles associated with both Stx6 and EEA1 early in infection is the population of viral particles that successfully navigate to the TGN.
With our combined data from this study, we propose a model in which HCMV infection causes an increase of Stx6 protein in primary human monocytes (Fig. 9). The excess Stx6 protein accumulates on EEA1+ endosomes that contain or will contain viral particles. This overabundance of Stx6 on EEA1+ endosomes enforces retrograde transport, allowing the virus-containing endosome to bypass the limiting retrograde trafficking proteins at the endosome-TGN interface and fuse directly with the TGN. This fusion event then gives the virus access to the TGN lumen, from which it can then progress to the recycling endosome and finally to the nucleus. Because the transition of the virus from the endosome to the TGN is a requisite trafficking event to achieve nuclear translocation, we argue that in turn, Stx6 is an important factor for HCMV nuclear translocation in monocytes. In this study we have shown for the first time that HCMV dysregulates Stx6 in primary human monocytes and is functionally important for viral entry events. We also show for the first time that Stx6 is primarily associated with endosomes in monocytes, a departure from canonical Stx6 localization at the TGN, and that loss of Stx6 prevents forward progress of the viral particle along the nuclear translocation pathway. Additionally, we have discovered that HCMV localizes within the TGN in primary human monocytes. Thus, this study significantly contributes to our understanding of how HCMV productively infects monocytes.
FIG 9.
Model: HCMV infection upregulates Stx6, which facilitates viral retrograde transport to the TGN, and thereby nuclear translocation. HCMV infection of monocytes triggers a significant increase in Stx6 protein, which accumulates on early endosomes. At approximately 15–45 mpi, HCMV is taken into early endosomes. A portion of viral particles are taken into vesicles containing both EEA1 and Stx6. Overabundance of Stx6 enforces or biases retrograde transport of these virus-containing vesicles to the TGN. At the TGN, the virus-containing endosome, laden with Stx6, can bypass retrograde transport-limiting complexes and bind directly to cognate receptors at the TGN surface such as VAMP4. This Stx6-dependent interaction mediates membrane fusion between the virus-carrying vesicle and the TGN. Once in the lumen of the TGN, the viral particles then subsequently travel on to recycling endosomes and then to the nucleus where viral genome replication and subsequent productive infection can occur. Stx6 siRNA inhibits the HCMV-driven upregulation of Stx6, which consequently inhibits viral trafficking to the TGN – a necessary trafficking event prior to nuclear translocation in monocytes. Image created with BioRender.com.
As previously mentioned, we have demonstrated that HCMV nuclear translocation within monocytes is directed by the binding of the viral glycoproteins gB and pentamer complex to cell surface EGFR and β1/β3 integrins (10, 11, 13, 14). Specifically, the engagement of these cellular receptors by HCMV, as well as the virus-induced activation of the respective PI3K/Akt and c-Src signaling cascades, drives successful intracellular trafficking of the virus (10, 11). Pentamer/integrin engagement and activation are critical for early trafficking events where virus-induced c-Src signaling prevents viral particle sorting to late endosomes and promotes progression of HCMV from early endosomes to the TGN, a required trafficking event to achieve nuclear translocation (10). Additionally, gB/EGFR engagement and chronic activation of the PI3K/Akt signaling pathway is important for progression through all organelles associated with the HCMV nuclear translocation pathway in monocytes (11). These post-entry trafficking events occur in the absence of de-enveloped viral effector proteins (~2 dpi) or de novo viral gene expression (~ 2 wpi) and are thus predominantly driven by virus-induced signaling (4, 7, 10, 11). We have shown here that Stx6 is an important factor within the nuclear translocation pathway of HCMV in monocytes. Therefore, it is likely that these virus-induced signaling events also manipulate Stx6 to facilitate trafficking of HCMV from early endosomes to the TGN. Thus, the role of virus-induced signaling on Stx6 in HCMV-infected monocytes will be assessed in future studies.
It is rare for viruses to utilize retrograde transport for entry and even rarer for enveloped viruses. We argue that we have at least made the initial discovery of this process during herpesvirus infections. That is, our current findings argue that HCMV utilizes retrograde trafficking machinery for post entry events prior to nuclear translocation in primary human monocytes. To our knowledge, this study may implicate HCMV as the first enveloped virus to utilize retrograde trafficking machinery for entry.
MATERIALS AND METHODS
Human primary monocyte isolation.
Human peripheral blood was collected via venipuncture and dual-density gradient centrifugation as previously described and in accordance with Institutional Review Board (H99-064) approval (65). Approval was granted by the Louisiana State University Health Shreveport Institutional Review Board (approval no. H99-064). All Health Insurance Portability Accountability Act guidelines were followed. Human subjects were informed of the current study as well as the potential risks involved within the venipuncture procedure. The subjects were also informed that their informed consent (gained prior to each blood donation) could be revoked at any time during their study participation.
Virus preparation.
The HCMV strain TB40/E containing a GFP fusion to UL32 (TB40/E UL32-GFP) was cultured using human embryonic lung (HEL) fibroblasts. The virus was harvested via ultracentrifugation through a sorbitol cushion (20% [wt/vol]) (6, 66, 67). The virus was resuspended in Gibco RPMI 1640 and subsequently used to infect monocytes at MOI = 5 unless stated otherwise.
Affymetrix gene array and analysis.
Whole transcriptome data of HCMV-infected primary human monocytes was generated and analyzed as previously described (45). The GEO accession information for these data is GSE11408.
RNASeq.
Primary human monocytes were cultured in 10% RPMI/human serum on tissue culture plates coated with fibronectin. Fresh media was added every other day to prolong cell viability. Total RNA was harvested from HCMV (TB40/E)-infected or mock-infected monocytes over the indicated time course. RNA quality was assessed, and cytoplasmic rRNA was depleted using the Illumina HMR Ribo-Zero rRNA removal kit. An mRNA sequencing library was prepared with the NEBNextUltra directional library kit and the TruSeq stranded mRNA kit (Illumina). Paired end sequencing (2 × 75 cycles) was performed on an Illumina NextSeq 500 obtaining over 25 × 106 reads per sample. Reads were aligned to the annotated HCMV Lisa reference strain (GenBank accession number KF297339.1) and human genome using STAR_2.4.2a and counted using RSEM 1.2.31. EBseq 1.2.0 was used to determine differential expression using an false-discovery rate (FDR) of 0.05. Transcripts per million (TPM) were log 2 normalized for gene level analysis.
Western blot analysis.
Fibroblasts (HEL), epithelial cells (ARPE-19), and primary human monocytes were left untreated or treated with siRNA targeting Stx6, Stx12, or a scrambled control as indicated in the text. The cells were then subsequently infected with HCMV TB40/E UL32-GFP. Protein was harvested over the designated time courses. Whole cell lysate was then resolved via SDS/PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were then blocked and incubated with primary antibodies (Stx6 [Cell Signaling], actin [Santa Cruz], or HCMV IE1 as previously described [68]). The immunoblots were subsequently incubated with IRDye 680RD and IRDye 800CW conjugated secondary antibodies (LI-COR). Bands were detected using a LI-COR fluorescent imaging system and accompanying Image Studio software.
siRNA transfection.
Primary human monocytes (3 × 106 per sample) were transfected with siRNA as previously described and incubated at 37°C for 24–48 h prior to HCMV infection (14, 54, 69, 70). Monocytes were treated with 100 nM Stx6, Stx12, and scrambled control siRNA (Sigma-Aldrich). Transfection was performed using an Amaxa Nucleofector (Lonza).
Immunofluorescence microscopy.
Primary human monocytes were plated onto coverslips coated with fibronectin and incubated overnight (10, 11). Cells were then infected with TB40/E UL32-GFP (MOI = 10) the following day. At each designated time point, the monocytes were fixed and permeabilized. Subsequently, the fixed cells were stained with primary antibodies to detect Stx6 (Abcam), EEA1 (Cell Signaling), TGN46 (Thermo Fisher Scientific), and GFP-488 (Life Technologies). A Nikon N-SIM E Super Resolution microscope system (100× objective) was used to capture high-resolution images. Images were acquired in single slices (z-stacks). Each image presented in the text is a representative single slice from a z-stack. Images were acquired using the same laser power settings. 488, 594, and 647 lasers of the super resolution system were used to visualize structures and proteins within the green, red, and far-red channels, respectively. Nuclei were stained with Hoescht and nuclei images were captured using the widefield function of the Nikon as the super resolution component of the microscope did not possess a laser for the blue channel. Widefield nuclear images were merged into the corresponding super resolution images using the companion analysis software of the microscope. Due to pandemic-related supply chain issues for the SIM component of the microscope, confocal microscopy on the same microscope system was used to capture images for Fig. 4F and G using settings similar to those discussed above.
Entry assays.
Entry assays for HCMV were performed as previously described (13, 14, 70). Primary human monocytes were treated with Stx6, Stx12, or scrambled control siRNA for 48 h prior to infection. Briefly, the treated cells were then cooled to 4°C for 1 h and subsequently infected on ice with HCMV TB40/E UL32-GFP (MOI = 1) to allow viral binding, but not entry. The cells were then shifted to 37°C (except 4°C binding controls) for 1 h to allow viral entry. Unbound virus was then washed with away with phosphate-buffered saline (PBS). HCMV-infected monocytes were then treated with 10 mg/mL proteinase K (Roche) in PBS-100 mM EDTA for 45 m on ice. DNA was subsequently isolated from the cells using the EZNA Tissue DNA Kit (Omega Bio-Tek) and prepared for qPCR.
qPCR.
Quantitative PCR was performed as previously described (10, 11, 13, 14) on isolated total cellular DNA. Primers to detect cellular CRP and HCMV UL123 were used in qPCR.
Data availability.
All data presented in this study in which large data sets were analyzed or reanalyzed are available in their entirety through submission of the data to the GEO repository (via NCBI). RNAseq data were submitted to GEO under accession number GSE206198. Accession numbers of previously existing data sets are individually discussed in the Materials and Methods section and/or are referenced for the methods employed in the generation of the data sets.
ACKNOWLEDGMENTS
We thank the LSUHS core microscopy facility for their help with our studies. We thank the CoBRE Center for Applied Immunology and Pathological Processes for their core support of the microscopy facilities and for their help with our omics analyses. We also thank M.S., R.S., J.W., M.B.-H., G.R.B. and V.Z. for helpful discussions and inspiration during the undertaking of our project.
This study was funded by National Institutes of Health Grants AI056077, AI127335, P20GM134974, P20GM121307, P20GM121288, and P30GM110703.
Contributor Information
Andrew D. Yurochko, Email: andrew.yurochko@lsuhs.edu.
Felicia Goodrum, University of Arizona.
REFERENCES
- 1.Mocarski ES, Jr, Shenk T, Griffiths PD, Pass RF. 2013. Cytomegaloviruses, p 1961–2010. In David M, Knipe PMH (ed), Fields Virology, 6th ed, vol II. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Azevedo LS, Pierrotti LC, Abdala E, Costa SF, Strabelli TMV, Campos SV, Ramos JF, Latif AZA, Litvinov N, Maluf NZ, Caiaffa Filho HH, Pannuti CS, Lopes MH, Santos VAd, Linardi CdCG, Yasuda MAS, Marques HHdS. 2015. Cytomegalovirus infection in transplant recipients. Clinics (Sao Paulo) 70:515–523. 10.6061/clinics/2015(07)09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jean Beltran PM, Cristea IM. 2014. The life cycle and pathogenesis of human cytomegalovirus infection: lessons from proteomics. Expert Rev Proteomics 11:697–711. 10.1586/14789450.2014.971116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith MS, Bentz GL, Alexander JS, Yurochko AD. 2004. Human cytomegalovirus induces monocyte differentiation and migration as a strategy for dissemination and persistence. J Virol 78:4444–4453. 10.1128/jvi.78.9.4444-4453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fulkerson HL, Nogalski MT, Collins-McMillen D, Yurochko AD. 2021. Overview of human cytomegalovirus pathogenesis. Methods Mol Biol 2244:1–18. 10.1007/978-1-0716-1111-1_1. [DOI] [PubMed] [Google Scholar]
- 6.Bentz GL, Jarquin-Pardo M, Chan G, Smith MS, Sinzger C, Yurochko AD. 2006. Human cytomegalovirus (HCMV) infection of endothelial cells promotes naive monocyte extravasation and transfer of productive virus to enhance hematogenous dissemination of HCMV. J Virol 80:11539–11555. 10.1128/JVI.01016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith MS, Bentz GL, Smith PM, Bivins ER, Yurochko AD. 2004. HCMV activates PI(3)K in monocytes and promotes monocyte motility and transendothelial migration in a PI(3)K-dependent manner. J Leukoc Biol 76:65–76. 10.1189/jlb.1203621. [DOI] [PubMed] [Google Scholar]
- 8.Goodrum F. 2016. Human cytomegalovirus latency: approaching the Gordian Knot. Annu Rev Virol 3:333–357. 10.1146/annurev-virology-110615-042422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JH, Collins-McMillen D, Buehler JC, Goodrum FD, Yurochko AD. 2017. Human cytomegalovirus requires epidermal growth factor receptor signaling to enter and initiate the early steps in the establishment of latency in CD34(+) human progenitor cells. J Virol 91. 10.1128/JVI.01206-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JH, Collins-McMillen D, Caposio P, Yurochko AD. 2016. Viral binding-induced signaling drives a unique and extended intracellular trafficking pattern during infection of primary monocytes. Proc Natl Acad Sci USA 113:8819–8824. 10.1073/pnas.1604317113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulkerson HL, Chesnokova LS, Kim JH, Mahmud J, Frazier LE, Chan GC, Yurochko AD. 2020. HCMV-induced signaling through gB-EGFR engagement is required for viral trafficking and nuclear translocation in primary human monocytes. Proc Natl Acad Sci USA 117:19507–19516. 10.1073/pnas.2003549117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins-McMillen D, Stevenson EV, Kim JH, Lee BJ, Cieply SJ, Nogalski MT, Chan GC, Frost RW, 3rd, Spohn CR, Yurochko AD. 2017. Human cytomegalovirus utilizes a nontraditional signal transducer and activator of transcription 1 activation cascade via signaling through epidermal growth factor receptor and integrins to efficiently promote the motility, differentiation, and polarization of infected monocytes. J Virol 91. 10.1128/JVI.00622-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nogalski MT, Chan GC, Stevenson EV, Collins-McMillen DK, Yurochko AD. 2013. The HCMV gH/gL/UL128-131 complex triggers the specific cellular activation required for efficient viral internalization into target monocytes. PLoS Pathog 9:e1003463. 10.1371/journal.ppat.1003463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan G, Nogalski MT, Yurochko AD. 2009. Activation of EGFR on monocytes is required for human cytomegalovirus entry and mediates cellular motility. Proc Natl Acad Sci USA 106:22369–22374. 10.1073/pnas.0908787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johannes L, Popoff V. 2008. Tracing the retrograde route in protein trafficking. Cell 135:1175–1187. 10.1016/j.cell.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Sivan G, Weisberg AS, Americo JL, Moss B. 2016. Retrograde transport from early endosomes to the trans-Golgi network enables membrane wrapping and egress of vaccinia virus virions. J Virol 90:8891–8905. 10.1128/JVI.01114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu Y, Zhao L, Billadeau DD, Jia D. 2020. Endosome-to-TGN trafficking: organelle-vesicle and organelle-organelle interactions. Front Cell Dev Biol 8:163. 10.3389/fcell.2020.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barlocher K, Welin A, Hilbi H. 2017. Formation of the legionella replicative compartment at the crossroads of retrograde trafficking. Front Cell Infect Microbiol 7:482. 10.3389/fcimb.2017.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laufman O, Hong W, Lev S. 2011. The COG complex interacts directly with Syntaxin 6 and positively regulates endosome-to-TGN retrograde transport. J Cell Biol 194:459–472. 10.1083/jcb.201102045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Personnic N, Barlocher K, Finsel I, Hilbi H. 2016. Subversion of retrograde trafficking by translocated pathogen effectors. Trends Microbiol 24:450–462. 10.1016/j.tim.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Bonifacino JS, Rojas R. 2006. Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol 7:568–579. 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- 22.Sandvig K, Skotland T, van Deurs B, Klokk TI. 2013. Retrograde transport of protein toxins through the Golgi apparatus. Histochem Cell Biol 140:317–326. 10.1007/s00418-013-1111-z. [DOI] [PubMed] [Google Scholar]
- 23.Chia PZ, Gleeson PA. 2011. The regulation of endosome-to-Golgi retrograde transport by tethers and scaffolds. Traffic 12:939–947. 10.1111/j.1600-0854.2011.01185.x. [DOI] [PubMed] [Google Scholar]
- 24.Spooner RA, Smith DC, Easton AJ, Roberts LM, Lord JM. 2006. Retrograde transport pathways utilised by viruses and protein toxins. Virol J 3:26. 10.1186/1743-422X-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cruz L, Buchkovich NJ. 2017. Rerouting the traffic from a virus perspective. Front Biosci (Landmark Ed) 22:1845–1866. 10.2741/4575. [DOI] [PubMed] [Google Scholar]
- 26.Close WL, Glassbrook JE, Gurczynski SJ, Pellett PE. 2018. Infection-induced changes within the endocytic recycling compartment suggest a roadmap of human cytomegalovirus egress. Front Microbiol 9:1888. 10.3389/fmicb.2018.01888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hook LM, Grey F, Grabski R, Tirabassi R, Doyle T, Hancock M, Landais I, Jeng S, McWeeney S, Britt W, Nelson JA. 2014. Cytomegalovirus miRNAs target secretory pathway genes to facilitate formation of the virion assembly compartment and reduce cytokine secretion. Cell Host Microbe 15:363–373. 10.1016/j.chom.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schindler C, Chen Y, Pu J, Guo X, Bonifacino JS. 2015. EARP is a multisubunit tethering complex involved in endocytic recycling. Nat Cell Biol 17:639–650. 10.1038/ncb3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wendler F, Tooze S. 2001. Syntaxin 6: the promiscuous behaviour of a SNARE protein. Traffic 2:606–611. 10.1034/j.1600-0854.2001.20903.x. [DOI] [PubMed] [Google Scholar]
- 30.Inoue H, Tani K, Tagaya M. 2016. SNARE-associated proteins and receptor trafficking. Receptors & Clinical Invest 3. [Google Scholar]
- 31.Aerbajinai W, Liu L, Zhu J, Kumkhaek C, Chin K, Rodgers GP. 2016. Glia maturation factor-gamma regulates monocyte migration through modulation of beta1-integrin. J Biol Chem 291:8549–8564. 10.1074/jbc.M115.674200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Angelis Rigotti F, De Gassart A, Pforr C, Cano F, N'Guessan P, Combes A, Camossetto V, Lehner PJ, Pierre P, Gatti E. 2017. MARCH9-mediated ubiquitination regulates MHC I export from the TGN. Immunol Cell Biol 95:753–764. 10.1038/icb.2017.44. [DOI] [PubMed] [Google Scholar]
- 33.Pfeffer SR. 2011. Entry at the trans-face of the Golgi. Cold Spring Harb Perspect Biol 3. 10.1101/cshperspect.a005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grimm C, Chen CC, Wahl-Schott C, Biel M. 2017. Two-pore channels: catalyzers of endolysosomal transport and function. Front Pharmacol 8:45. 10.3389/fphar.2017.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manickam V, Tiwari A, Jung JJ, Bhattacharya R, Goel A, Mukhopadhyay D, Choudhury A. 2011. Regulation of vascular endothelial growth factor receptor 2 trafficking and angiogenesis by Golgi localized t-SNARE syntaxin 6. Blood 117:1425–1435. 10.1182/blood-2010-06-291690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray RZ, Wylie FG, Khromykh T, Hume DA, Stow JL. 2005. Syntaxin 6 and Vti1b form a novel SNARE complex, which is up-regulated in activated macrophages to facilitate exocytosis of tumor necrosis Factor-alpha. J Biol Chem 280:10478–10483. 10.1074/jbc.M414420200. [DOI] [PubMed] [Google Scholar]
- 37.Weimershaus M, Mauvais FX, Saveanu L, Adiko C, Babdor J, Abramova A, Montealegre S, Lawand M, Evnouchidou I, Huber KJ, Chadt A, Zwick M, Vargas P, Dussiot M, Lennon-Dumenil AM, Brocker T, Al-Hasani H, van Endert P. 2018. Innate immune signals induce anterograde endosome transport promoting MHC class I cross-presentation. Cell Rep 24:3568–3581. 10.1016/j.celrep.2018.08.041. [DOI] [PubMed] [Google Scholar]
- 38.Enrich C, Rentero C, Hierro A, Grewal T. 2015. Role of cholesterol in SNARE-mediated trafficking on intracellular membranes. J Cell Sci 128:1071–1081. 10.1242/jcs.164459. [DOI] [PubMed] [Google Scholar]
- 39.Perera HK, Clarke M, Morris NJ, Hong W, Chamberlain LH, Gould GW. 2003. Syntaxin 6 regulates Glut4 trafficking in 3T3-L1 adipocytes. Mol Biol Cell 14:2946–2958. 10.1091/mbc.e02-11-0722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arango Duque G, Descoteaux A. 2014. Macrophage cytokines: involvement in immunity and infectious diseases. Front Immunol 5:491. 10.3389/fimmu.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bock JB, Klumperman J, Davanger S, Scheller RH. 1997. Syntaxin 6 functions in trans-Golgi network vesicle trafficking. Mol Biol Cell 8:1261–1271. 10.1091/mbc.8.7.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riggs KA, Hasan N, Humphrey D, Raleigh C, Nevitt C, Corbin D, Hu C. 2012. Regulation of integrin endocytic recycling and chemotactic cell migration by syntaxin 6 and VAMP3 interaction. J Cell Sci 125:3827–3839. 10.1242/jcs.102566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Du Y, Shen J, Hsu JL, Han Z, Hsu MC, Yang CC, Kuo HP, Wang YN, Yamaguchi H, Miller SA, Hung MC. 2014. Syntaxin 6-mediated Golgi translocation plays an important role in nuclear functions of EGFR through microtubule-dependent trafficking. Oncogene 33:756–770. 10.1038/onc.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lieu ZZ, Derby MC, Teasdale RD, Hart C, Gunn P, Gleeson PA. 2007. The golgin GCC88 is required for efficient retrograde transport of cargo from the early endosomes to the trans-Golgi network. Mol Biol Cell 18:4979–4991. 10.1091/mbc.e07-06-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan G, Bivins-Smith ER, Smith MS, Smith PM, Yurochko AD. 2008. Transcriptome analysis reveals human cytomegalovirus reprograms monocyte differentiation toward an M1 macrophage. J Immunol 181:698–711. 10.4049/jimmunol.181.1.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan G, Bivins-Smith ER, Smith MS, Yurochko AD. 2008. Transcriptome analysis of NF-kappaB- and phosphatidylinositol 3-kinase-regulated genes in human cytomegalovirus-infected monocytes. J Virol 82:1040–1046. 10.1128/JVI.00864-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hubstenberger A, Courel M, Benard M, Souquere S, Ernoult-Lange M, Chouaib R, Yi Z, Morlot JB, Munier A, Fradet M, Daunesse M, Bertrand E, Pierron G, Mozziconacci J, Kress M, Weil D. 2017. P-body purification reveals the condensation of repressed mRNA regulons. Mol Cell 68:144–157.e5. 10.1016/j.molcel.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 48.Eulalio A, Behm-Ansmant I, Izaurralde E. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8:9–22. 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 49.Shyu AB, Wilkinson MF, van Hoof A. 2008. Messenger RNA regulation: to translate or to degrade. EMBO J 27:471–481. 10.1038/sj.emboj.7601977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang C, Schmich F, Srivatsa S, Weidner J, Beerenwinkel N, Spang A. 2018. Context-dependent deposition and regulation of mRNAs in P-bodies. Elife 7. 10.7554/eLife.29815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Q, Medina SG, Kushawah G, DeVore ML, Castellano LA, Hand JM, Wright M, Bazzini AA. 2019. Translation affects mRNA stability in a codon-dependent manner in human cells. Elife 8. 10.7554/eLife.45396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan LY, Mugler CF, Heinrich S, Vallotton P, Weis K. 2018. Non-invasive measurement of mRNA decay reveals translation initiation as the major determinant of mRNA stability. Elife 7. 10.7554/eLife.32536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hanson G, Alhusaini N, Morris N, Sweet T, Coller J. 2018. Translation elongation and mRNA stability are coupled through the ribosomal A-site. RNA 24:1377–1389. 10.1261/rna.066787.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan G, Nogalski MT, Yurochko AD. 2012. Human cytomegalovirus stimulates monocyte-to-macrophage differentiation via the temporal regulation of caspase 3. J Virol 86:10714–10723. 10.1128/JVI.07129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collins-McMillen D, Kim JH, Nogalski MT, Stevenson EV, Chan GC, Caskey JR, Cieply SJ, Yurochko AD. 2015. Human cytomegalovirus promotes survival of infected monocytes via a distinct temporal regulation of cellular Bcl-2 family proteins. J Virol 90:2356–2371. 10.1128/JVI.01994-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stevenson EV, Collins-McMillen D, Kim JH, Cieply SJ, Bentz GL, Yurochko AD. 2014. HCMV reprogramming of infected monocyte survival and differentiation: a Goldilocks phenomenon. Viruses 6:782–807. 10.3390/v6020782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Regad T. 2015. Targeting RTK Signaling Pathways in Cancer. Cancers (Basel) 7:1758–1784. 10.3390/cancers7030860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Larsen M, Artym VV, Green JA, Yamada KM. 2006. The matrix reorganized: extracellular matrix remodeling and integrin signaling. Curr Opin Cell Biol 18:463–471. 10.1016/j.ceb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 59.Reverter M, Rentero C, Garcia-Melero A, Hoque M, Vila de Muga S, Alvarez-Guaita A, Conway JR, Wood P, Cairns R, Lykopoulou L, Grinberg D, Vilageliu L, Bosch M, Heeren J, Blasi J, Timpson P, Pol A, Tebar F, Murray RZ, Grewal T, Enrich C. 2014. Cholesterol regulates Syntaxin 6 trafficking at trans-Golgi network endosomal boundaries. Cell Rep 7:883–897. 10.1016/j.celrep.2014.03.043. [DOI] [PubMed] [Google Scholar]
- 60.Choudhury A, Marks DL, Proctor KM, Gould GW, Pagano RE. 2006. Regulation of caveolar endocytosis by syntaxin 6-dependent delivery of membrane components to the cell surface. Nat Cell Biol 8:317–328. 10.1038/ncb1380. [DOI] [PubMed] [Google Scholar]
- 61.Murray RZ, Stow JL. 2014. Cytokine secretion in macrophages: SNAREs, Rabs, and membrane trafficking. Front Immunol 5:538. 10.3389/fimmu.2014.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nozawa T, Minowa-Nozawa A, Aikawa C, Nakagawa I. 2017. The STX6-VTI1B-VAMP3 complex facilitates xenophagy by regulating the fusion between recycling endosomes and autophagosomes. Autophagy 13:57–69. 10.1080/15548627.2016.1241924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bentz GL, Yurochko AD. 2008. Human CMV infection of endothelial cells induces an angiogenic response through viral binding to EGF receptor and beta1 and beta3 integrins. Proc Natl Acad Sci USA 105:5531–5536. 10.1073/pnas.0800037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vanarsdall AL, Johnson DC. 2012. Human cytomegalovirus entry into cells. Curr Opin Virol 2:37–42. 10.1016/j.coviro.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mosher BS, Fulkerson HL, Yurochko AD. 2021. Collection and isolation of CD14(+) primary human monocytes via dual density gradient centrifugation as a model system to study human cytomegalovirus infection and pathogenesis. Methods Mol Biol 2244:103–113. 10.1007/978-1-0716-1111-1_6. [DOI] [PubMed] [Google Scholar]
- 66.Yurochko AD, Huang ES. 1999. Human cytomegalovirus binding to human monocytes induces immunoregulatory gene expression. J Immunol 162:4806–4816. [PubMed] [Google Scholar]
- 67.Yurochko AD, Hwang ES, Rasmussen L, Keay S, Pereira L, Huang ES. 1997. The human cytomegalovirus UL55 (gB) and UL75 (gH) glycoprotein ligands initiate the rapid activation of Sp1 and NF-kappaB during infection. J Virol 71:5051–5059. 10.1128/JVI.71.7.5051-5059.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Siddiquey MNA, Zhang H, Nguyen CC, Domma AJ, Kamil JP. 2018. The human cytomegalovirus endoplasmic reticulum-resident glycoprotein UL148 activates the unfolded protein response. J Virol 92. 10.1128/JVI.00896-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chan G, Nogalski MT, Bentz GL, Smith MS, Parmater A, Yurochko AD. 2010. PI3K-dependent upregulation of Mcl-1 by human cytomegalovirus is mediated by epidermal growth factor receptor and inhibits apoptosis in short-lived monocytes. J Immunol 184:3213–3222. 10.4049/jimmunol.0903025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nogalski MT, Chan G, Stevenson EV, Gray S, Yurochko AD. 2011. Human cytomegalovirus-regulated paxillin in monocytes links cellular pathogenic motility to the process of viral entry. J Virol 85:1360–1369. 10.1128/JVI.02090-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data presented in this study in which large data sets were analyzed or reanalyzed are available in their entirety through submission of the data to the GEO repository (via NCBI). RNAseq data were submitted to GEO under accession number GSE206198. Accession numbers of previously existing data sets are individually discussed in the Materials and Methods section and/or are referenced for the methods employed in the generation of the data sets.