Summary
The formation of nitrogen‐fixing nodules on legume hosts is a finely tuned process involving many components of both symbiotic partners. Production of the exopolysaccharide succinoglycan by the nitrogen‐fixing bacterium Sinorhizobium meliloti 1021 is needed for an effective symbiosis with Medicago spp., and the succinyl modification to this polysaccharide is critical. However, it is not known when succinoglycan intervenes in the symbiotic process, and it is not known whether the plant lysin‐motif receptor‐like kinase MtLYK10 intervenes in recognition of succinoglycan, as might be inferred from work on the Lotus japonicus MtLYK10 ortholog, LjEPR3. We studied the symbiotic infection phenotypes of S. meliloti mutants deficient in succinoglycan production or producing modified succinoglycan, in wild‐type Medicago truncatula plants and in Mtlyk10 mutant plants. On wild‐type plants, S. meliloti strains producing no succinoglycan or only unsuccinylated succinoglycan still induced nodule primordia and epidermal infections, but further progression of the symbiotic process was blocked. These S. meliloti mutants induced a more severe infection phenotype on Mtlyk10 mutant plants. Nodulation by succinoglycan‐defective strains was achieved by in trans rescue with a Nod factor‐deficient S. meliloti mutant. While the Nod factor‐deficient strain was always more abundant inside nodules, the succinoglycan‐deficient strain was more efficient than the strain producing only unsuccinylated succinoglycan. Together, these data show that succinylated succinoglycan is essential for infection thread formation in M. truncatula, and that MtLYK10 plays an important, but different role in this symbiotic process. These data also suggest that succinoglycan is more important than Nod factors for bacterial survival inside nodules.
Keywords: beneficial microbes, microbiome, exopolysaccharide, infection thread, LysM‐receptor‐like kinase, Medicago truncatula, root hair, Sinorhizobium meliloti, succinoglycan, symbiotic nitrogen fixation
Significance Statement
This work should be of interest across the field of plant–microbe endosymbioses by providing insights into key determinants of both bacteria and plants for successful microbial host invasion. It is timely with much current interest in symbiotic roles of plant LysM‐RLK proteins and the evolutionary origins of nitrogen‐fixing endosymbiosis. MtLYK10 is a Medicago truncatula component that specifically intervenes in rhizobial infection, independently of succinoglycan recognition.
Introduction
Sinorhizobium meliloti 1021 is a nitrogen‐fixing symbiont of the plants Medicago truncatula cv. Jemalong, M. truncatula ssp. tricycla R108, and Medicago sativa (alfalfa) (Hoffmann et al., 1997; Oldroyd et al., 2011). On roots of compatible hosts, S. meliloti induces nodule organogenesis, and invades and colonizes the developing nodule primordia, ultimately being endocytosed by cells of the nodule cortex (Oldroyd et al., 2011). Once internalized within so‐called ‘symbiosomes’, the bacteria differentiate into the nitrogen‐fixing ‘bacteroid’ form (Vasse et al., 1990), and fix dinitrogen gas, converting it to ammonia and providing it to the host (Hellriegel and Wilfarth, 1888; Burris, 1974). Invasion of host roots by rhizobia requires that compatible symbiont/host pairs exchange multiple signals that promote bacterial entry. S. meliloti produces a lipo‐chitooligosaccharide signal called Nod factor (NF) (Peters et al., 1986; Lerouge et al., 1990) responsible for inducing root hair curling around attached rhizobia. This traps S. meliloti microcolonies within an infection chamber (Fournier et al., 2015). NFs also induce cell division in the root cortex leading to formation of nodule primordia (Timmers et al., 1999; Xiao et al., 2014). Rhizobia access root cortical cell layers through structures called infection threads that initiate from the colonized curled root hairs (CCRHs). An infection thread is a progressive ingrowth of root hair cell membrane that forms a tube populated with S. meliloti and filled with a matrix of bacterial exopolysaccharide (EPS), secreted bacterial proteins, and plant cell wall material (Brewin, 2004; Gage, 2004). S. meliloti propagate in the infection thread during its extension, a process that requires bacterial production of both NFs and the EPS succinoglycan (Jones et al., 2007). Infection threads then extend through each successive cell layer until they reach the inner cortex, where bacteria are endocytosed by proliferating cells of the nodule primordium (Timmers et al., 1999; Xiao et al., 2014).
On the host M. truncatula, S. meliloti succinoglycan is required for symbiosis and cannot be substituted by other polysaccharides produced by this bacterium (Glazebrook and Walker, 1989). It is needed for infection thread progression on M. truncatula A17 (Jones et al., 2008) and on alfalfa (Cheng and Walker, 1998). Both M. truncatula A17 (Jones et al., 2008) and alfalfa (Niehaus et al., 1993) show signs of plant defense when inoculated with the succinoglycan‐deficient exoY mutant. Succinoglycan is a polymer of an octasaccharide composed of seven glucose and one galactose sugar that S. meliloti produces in both high molecular weight (HMW) and low‐molecular‐weight (LMW) forms (Reinhold et al., 1994; González et al., 1998). Production of the LMW form is dependent on cleavage by the glycanases encoded by the exoK and exsH genes (York and Walker, 1997; Mendis et al., 2016). Succinyl, pyruvyl, and acetyl groups are added to S. meliloti succinoglycan by, respectively, the products of the exoH, exoV, and exoZ genes (Reuber and Walker, 1993).
Not only is there a requirement for succinoglycan for host plant invasion, but the material that is produced must be properly modified by succinylation. If succinoglycan is not succinylated by ExoH, no functional nodules are formed by S. meliloti 1021 on M. truncatula A17 (Mendis et al., 2016) or on alfalfa (Leigh et al., 1987). Unsuccinylated succinoglycan produced by exoH mutants cannot be cleaved by exoK‐ or exsH‐encoded glycanases, leaving all the unsuccinylated polymer produced by exoH mutants in the HMW form (York and Walker, 1998). We determined that the strict requirement for the succinylated form of succinoglycan for functional nodule formation on M. truncatula A17 is independent of the effect that succinylation has on production of the LMW form by the ExoK and ExsH glycanases (Mendis et al., 2016). S. meliloti strains lacking both glycanases can form a successful symbiosis on M. truncatula A17 despite producing only HMW succinoglycan. However, these ‘double‐glycanase mutants’ have reduced symbiotic efficiency relative to control strains (c. 70% shoot fresh weight of wild‐type (WT)‐inoculated plants) (Mendis et al., 2016). Strikingly, strains that lack the exoH‐encoded succinyltransferase but are otherwise isogenic to the exoKdel/exsH double‐glycanase strains, have the same plant growth phenotype as non‐inoculated plants or plants inoculated with the succinoglycan‐deficient exoY mutant (c. 18% of shoot fresh weight of S. meliloti WT‐inoculated plants (Mendis et al., 2016)). Thus, loss of the LMW form of succinoglycan results in reduction of symbiotic efficiency, but the HMW form plays an even more critical role in the symbiosis as long as it is succinylated.
We set out to further characterize the symbiotic phenotypes of exoY succinoglycan‐deficient and exoH succinyltransferase‐deficient S. meliloti mutants. Specifically, to determine if these strains are equally deficient in invasion, and whether the defect occurs at the same stage in infection. We also aimed to determine how these rhizobial mutants and the LMW succinoglycan‐deficient strains interact with M. truncatula plants carrying a mutation in a LysM‐receptor‐like kinase (LysM‐RLK), MtLYK10 (Medtr5g033490). This plant protein is the ortholog of the exopolysaccharide receptor 3 (EPR3) protein from the determinate‐nodule‐forming legume Lotus japonicus (Buendia et al., 2018). In the symbiotic interaction between the L. japonicus ecotype 'Gifu' and its symbiont Mesorhizobium loti R7A, M. loti EPS facilitates more efficient infection thread formation and nodulation, but EPS is not strictly required for symbiosis (Kelly et al., 2013). Ljepr3 null mutants permit invasion of M. loti exoU mutants that produce a truncated, pentasaccharide form of EPS that blocks M. loti invasion on WT plants (Kawaharada et al., 2015). This suggests that LjEPR3 plays a role in EPS recognition and restricts infection of bacteria that produce inappropriate EPS (Kawaharada et al., 2015).
We identified a M. truncatula mutant in MtLYK10, and determined how this line interacts with WT S. meliloti and S. meliloti mutants that are completely succinoglycan deficient, those that do not produce LMW succinoglycan and those that produce only unsuccinylated HMW succinoglycan. Our results suggest that S. meliloti succinoglycan has a function in infection of M. truncatula that is independent of LYK10, but is entirely dependent on succinylation of the polysaccharide.
Results
Sinorhizobium meliloti succinoglycan‐deficient and succinoglycan‐succinyltransferase‐deficient mutants form arrested infection threads in both M. truncatula cv. Jemalong and M. truncatula ssp. tricycla R108 ecotypes
Analyses of infection threads formed by S. meliloti exo mutants have previously been performed in alfalfa cv. Iroquois (Leigh et al., 1987; Yang et al., 1992; Cheng and Walker, 1998), but the symbiosis between S. meliloti 1021 and M. truncatula has important differences from the symbiosis with alfalfa. Increased production of succinoglycan by S. meliloti 1021 enhances infection on M. truncatula Jemalong, but not on alfalfa (Jones, 2012). A second EPS (called EPSII or galactoglucan) produced by some strains of S. meliloti (but not S. meliloti 1021) can function in place of succinoglycan on alfalfa cv. Iroquois (González et al., 1996; Pellock et al., 2000), but not on M. truncatula (Glazebrook and Walker, 1989). Indeed in a similar comparison performed in our growth conditions, we confirmed that EPSII cannot substitute for succinoglycan on M. truncatula cv. Jemalong A17 (Figure S1). Therefore, we first examined infection threads formed on M. truncatula by S. meliloti exo mutants to determine how the different types of exo mutants are impaired in invasion.
In M. truncatula A17, plants inoculated with an exoY mutant or with S. meliloti 1021 WT look similar at 3 days post‐inoculation (dpi), with exoY‐inoculated plants not yet having produced the large numbers of failed infection threads that will accumulate over time (Jones et al., 2008). (See Table 1 and Figure S2 for strain descriptions.) However, using cerulean Cyan Fluorescent Protein (cCFP)‐expressing bacteria at the very low culture density of OD600 = 0.001, the near‐absence of background fluorescence makes it possible to see subtle differences in infection threads between exoY‐inoculated and WT‐inoculated roots. On M. truncatula Jemalong super numeric nodules‐2 mutant (sunn‐2) inoculated with S. meliloti 1021 WT expressing cCFP, infection threads can be observed at 3–5 dpi extended all the way to the base of a root hair cell (Figure 1a). (The M. truncatula sunn‐2 mutant is routinely used for infection thread analysis because it produces more infection events without any qualitative differences compared with M. truncatula WT plants (Schnabel et al., 2005, Fournier et al., 2015)). In contrast with WT S. meliloti 1021, the S. meliloti exoY mutant at the same time point has formed only short, probably aborted infection threads (Figure 1b–d single arrowheads). The microcolony‐containing infection chambers formed by exoY within the CCRHs are much larger than those usually formed by WT bacteria (Figure 1b–d, asterisks).
Table 1.
Sinorhizobium meliloti strains and plasmids*
| Strain name | Promoter controlling exoLAMON operon | Production of succinylated succinoglycan | Production of LMW succinoglycan | Successful symbiosis with M. truncatula | Reference |
|---|---|---|---|---|---|
| S. meliloti 1021 | native | + | + | + | Meade et al. (1982) |
| exoY | native | N/A | no succinoglycan | − | Cheng and Walker (1998) |
| modified wild‐type 961 | trp | + | + | + | Mendis et al. (2013) |
| exsH 1317 modified wild‐type with mutation in exsH glycanase | trp | + | + | + | Mendis et al. (2016) |
| exoKdel/exsH 1325 | trp | + | − | +, c. 70% shoot fresh weight of wild‐type | Mendis et al. (2016) |
| exoHKdel/exsH 1345 | trp | − | − | − | Mendis et al. (2016) |
| nodC (Rm5613) | native | + | + | − | Jacobs et al. (1985) |
| Plasmid | Description | Reference |
|---|---|---|
| pXLGD4 | hemA‐lacZ reporter plasmid, carrying tetracycline resistance (TcR) | Leong et al. (1985) |
| pHC60 | pHC41 constitutively expressing green fluorescent protein(GFP) with the S65T mutation, TcR | Cheng and Walker (1998) |
| pcCFP | pHC60 derivative constitutively expressing Cerulean cyan fluorescent protein (CFP), TcR | Fournier et al. (2015)) |
Figure 1.
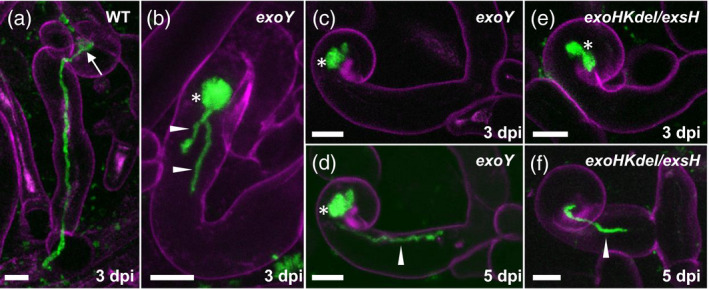
Infections formed by the succinoglycan‐deficient exoY mutant and the exoHKdel/exsH‐1345 mutant on Medicago truncatula Jemalong sunn‐2.
(a) WT Sinorhizobium meliloti 1021 expressing cCFP in a fully extended infection thread formed at 3 dpi on M. truncatula Jemalong sunn‐2. (b–d) CCRHs and short epidermal infection threads formed by the S. meliloti exoY mutant also expressing cCFP at 3 dpi (b, c) or 5 dpi (d). Note that in (c) and (d) the same site was imaged at two time‐points. (e, f) Infections formed by the exoHKdel/exsH‐1345 mutant at 3 dpi (e) or 5 dpi (f). Short, aborted infection threads (b, d, f) are labeled with arrowheads. The microcolony (infection chamber) is indicated in (a) (arrow). Excessively large root hair microcolonies (b–e) are labeled with asterisks. Images in (a–f) are z‐projections of confocal image stacks, combining the cCFP fluorescence of bacteria (green) and root hair cell wall autofluorescence (magenta). Results presented are representative of seven infection sites for S. meliloti WT 1021 (a), 24 sites for S. meliloti exoY (b–d) and 31 sites for S. meliloti exoHKdel/exsH‐1345 (e, f) recorded on three plants (six roots), five plants (11 roots) and seven plants (11 roots) respectively. Scale bars: 10 μm.
We next investigated the infection thread phenotypes of an S. meliloti strain that forms only HMW unsuccinylated succinoglycan. To this aim, we used the non‐polar exoHKdel/exsH‐1345 triple mutant whose symbiotic phenotype mainly results from the lack of the ExoH succinyltransferase (see next paragraph). In M. truncatula sunn‐2 at 3 dpi, the exoHKdel/exsH‐1345 mutant has formed only excessively large CCRHs (Figure 1e) or aborted infection threads (Figure 1f). Thus, the infection threads formed by exoY and exoHKdel/exsH‐1345 on the M. truncatula sunn‐2 line are quite similar in that they initiate some tubular growth but then elongate slowly and struggle to progress. No differences were observed in the infection thread phenotypes of these exo mutants on M. truncatula sunn‐2 versus M. truncatula Jemalong A17, and infections initiated by exoY or exoHKdel/exsH‐1345 are never observed penetrating a nodule primordium in M. truncatula Jemalong A17 (Figure S4). Note that the isogenic exoHKdel/exsH mutants 1345, 1343, and 1349 are lacking function of not only the exoH succinyltransferase, but also of the glycanases encoded by exoK and exsH (Figure S2). Complementation with a plasmid carrying exoH returns these strains to the level of symbiotic function of a non‐polar exoK mutant (70% shoot fresh weight of WT) (Figure S5).
exo mutant infection thread development and nodule formation were also analyzed on the host M. truncatula ssp. tricycla R108. This is the genotype for most Tnt1 retrotransposon‐induced plant mutants available for M. truncatula (d'Erfurth et al., 2003; Tadege et al., 2008). Two S. meliloti strains, 961 and exsH‐1317, which have the exoLAMON operon transcribed from a heterologous promoter and retain the WT copy of the exoH and exoK genes (Figures S2 and S3) were first tested. The exoLAMON operon has been placed under heterologous regulation in these strains to isolate it from polar effects from mutations in the upstream exoH and exoK (Mendis et al., 2013; Mendis et al., 2016). The control strains have the same altered genetic regulation as the exoK and exoHK deletion strains (Figures S2 and S3). Both strains form fully invaded nodules by 17 dpi on M. truncatula R108, similarly to WT S. meliloti 1021 (Figure 2). In addition, the double‐glycanase exoKdel/exsH‐1325 mutant, which only makes HMW succinoglycan (Mendis et al., 2016) also forms invaded nodules on M. truncatula R108, although significantly fewer nodules form compared with inoculation with the two WT 1021, 961 or single mutant exsH‐1317 strains (Figure 2). This is consistent with the fact that inoculation with the double‐glycanase exoKdel/exsH‐1325 mutant ultimately produces plants with c. 70% the shoot fresh weight of those inoculated with control strains (Mendis et al., 2016). In contrast with these strains, neither the S. meliloti exoHKdel/exsH‐1345 mutant nor the S. meliloti exoY mutant forms any invaded primordia or nodules on M. truncatula R108 (Figure 2). Instead, both strains have an arrested infection thread phenotype on M. truncatula R108 that closely resembles the phenotype on M. truncatula Jemalong. Thus, at 17 dpi, both exoHKdel/exsH‐1345 and exoY form excessively large microcolonies in CCRHs, and only very short infection threads that are blocked within root hairs and/or in the outer cortical cell layer above uninvaded nodule primordia (Figure 2). These infection blocks presumably result in the arrest of nodule development, such that only nodule primordia are formed. The complete lack of successful infection of nodule primordia explains the absence of shoot fresh weight gain by either strain (Mendis et al., 2016).
Figure 2.
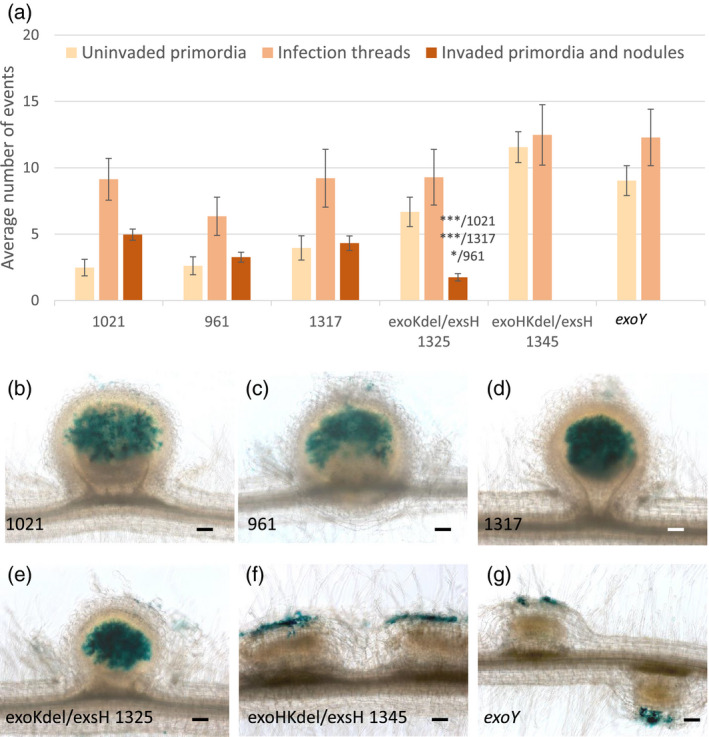
Infection and nodulation invasion phenotypes of the succinoglycan‐deficient exoY mutant and the exoHKdel/exsH‐1345 mutant on Medicago truncatula ssp. tricycla R108. Infection and nodule invasion phenotypes of M. truncatula WT R108 plants were scored 17 dpi following inoculation with six different Sinorhizobium meliloti strains; 1021, 961, exsH‐1317, exoKdel/exsH‐1325, exoHKdel/exsH‐1345 and exoY. The first three of these strains correspond to the unmodified WT strain (1021), a modified WT strain (961), which serves as a control for the expression of exoLAMON genes from a heterologous promoter, and exsH‐1317, which is identical to 961 except it lacks the exsH‐encoded glycanase. All three strains display an identical symbiotic phenotype. The double‐glycanase exoKdel/exsH‐1325 mutant (deficient for both the exoK and exsH glycanases) makes only HMW succinoglycan. The exoHKdel/exsH‐1345 mutant is isogenic to exoKdel/exsH‐1325, except that 1345 is missing the exoH succinyltransferase, and thus makes non‐succinylated HMW succinoglycan. exoY is completely deficient for production of succinoglycan.
(a) Quantification of uninvaded nodule primordia, infection threads, and invaded nodule primordia and nodules. Data are the averages of two independent experiments, n = 28 for each strain. Error bars = SEM. Statistically significant differences in invaded primordia and nodules between exoKdel/exsH‐1325 and other strains are indicated. *P < 0.05; ***P < 0.001.
(b–g) Photographs illustrating these infection phenotypes, all inoculated strains carrying a constitutive hemA‐lacZ reporter gene fusion (pXLGD4) for visualization of bacteria in blue. Fully invaded nodules were seen with 1021, 961, exsH‐1317 and exoKdelexsH‐1325 (b–e, respectively), while only uninvaded nodule primordia were observed with exoHKdel/exsH‐1345 and exoY (f and g, respectively). In these latter two cases, rhizobia appear restricted to the epidermis, but apparently continue to multiply in these unsuccessful infection compartments, resulting in excessively large root hair microcolonies. Scale bars: 100 µm.
M. truncatula lyk10 mutant plants show an early infection phenotype with WT S. meliloti
Having established these phenotypes on M. truncatula R108 plants, we studied an R108 line with a Tnt1 insertion in the LjEPR3 ortholog MtLYK10 (Medtr5g033490) (Cheng et al., 2014). Phylogenetic and synteny analysis confirmed the orthology between MtLYK10 and LjEPR3, and revealed that the genomes of some plants, including some Fabaceae species, encode two MtLYK10 homologs, but not M. truncatula (Figure S6). qRT‐PCR analysis confirmed that the Tnt1 insertion results in a null mutation in MtLYK10 (Figure S7).
WT S. meliloti 1021 was able to form infection threads with a normal appearance on Mtlyk10 mutant plants. However, compared with WT M. truncatula R108 plants, most infection threads remained in epidermal cells on Mtlyk10 mutant roots and significantly fewer infection threads progressed into the cortex at 11 dpi with WT S. meliloti 1021. Consequently, significantly fewer successfully invaded nodule primordia formed on mutant roots (Figure 3a–g). Numbers of uninvaded nodule primordia (Figure 3a) were not significantly different between genotypes (P = 0.0962), and more were seen on R108 compared with previous work on A17. However, a higher proportion of all nodules/primordia were uninvaded for Mtlyk10 (66.09%) compared with R108 (28.13%) (Figure 3a). Young nodules of Mtlyk10 plants appeared normal and well‐invaded (Figure 3d,g). We also found that nodulation was delayed on Mtlyk10, and that more developed nodules are also equivalently well‐invaded in mutant and WT plants (Figure 4).
Figure 3.
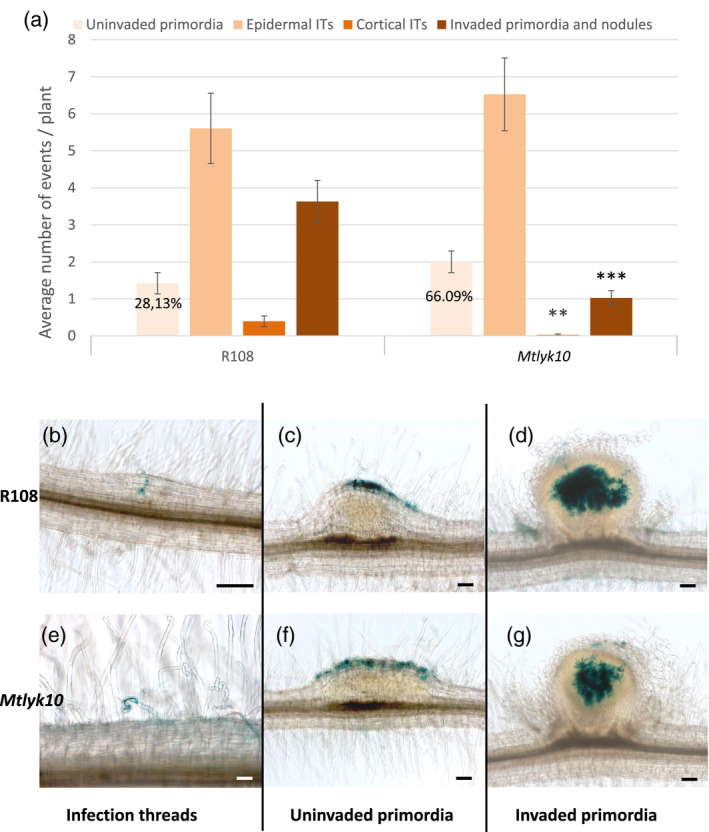
Early rhizobial infection and nodulation invasion phenotypes of Medicago truncatula lyk10 mutant plants with Sinorhizobium meliloti WT. Infection and nodulation invasion phenotypes of M. truncatula Mtlyk10 mutant and R108 WT plants were scored 11 dpi following inoculation with the S. meliloti WT strain 1021.
(a) Quantification of uninvaded nodule primordia, epidermal (restricted to the epidermis) and cortical (which have reached the cortex) infection threads (ITs) and invaded nodule primordia and nodules. Data are the averages of two independent experiments, n = 38 for each genotype. Error bars = SEM. On Mtlyk10 plants compared with R108, significantly fewer cortical infection threads, and significantly fewer invaded primordia and nodules formed. **P < 0.01; ***P < 0.001. The number of uninvaded primordia formed was not significantly different (P = 0.0962) between the two genotypes, but 28.13% (for R108) and 66.09% (for Mtlyk10) of all nodules and primordia were uninfected, as indicated.
(b–g) Photographs illustrating these infection phenotypes, the inoculated strain carrying a constitutive hemA‐lacZ reporter gene fusion (pXLGD4) for visualization of bacteria in blue. (b) a cortical infection thread in R108; (c) an uninvaded nodule primordium in R108; (d) an infected nodule in R108; (e) an epidermal infection thread in Mtlyk10; (f) an uninvaded nodule primordium in Mtlyk10; (g) an infected nodule in Mtlyk10. Scale bars: 100 µm.
Figure 4.
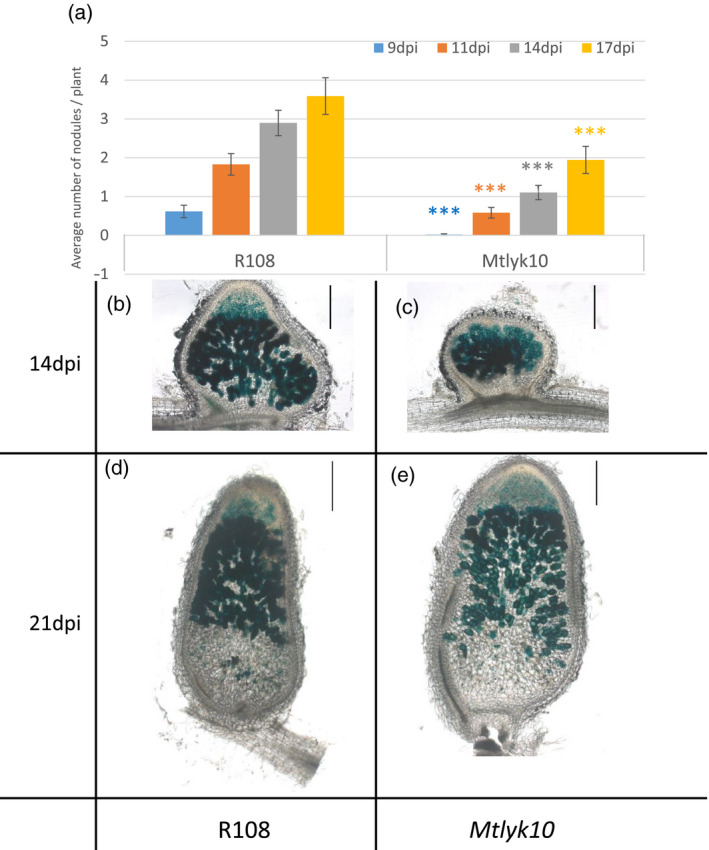
Kinetics of nodule appearance and invasion of well developed nodules on Medicago truncatula lyk10 mutant plants following inoculation with Sinorhizobium meliloti WT.
(a) The appearance of nodules was scored at 9, 11, 14 and 17 dpi following inoculation of M. truncatula Mtlyk10 mutant and R108 WT plants with the S. meliloti WT strain 1021. n = 35 for each genotype. ***P < 0.001. Error bars = SEM.
(b–e) Photographs of 70 μm nodule sections, colored to visualize bacteria in blue (the inoculated strain carries a constitutive hemA‐lacZ reporter gene fusion (pXLGD4)) after sectioning. (b) R108 at 14 dpi; (c) Mtlyk10 at 14 dpi; (d) R108 at 21 dpi; (e) Mtlyk10 at 21 dpi. Scale bars: 100 μm.
We also compared the nodulation ability of two independent lines carrying the Mtlyk10 mutation, selected from the backcross to WT R108 plants. Both Mtlyk10 lines similarly formed fewer invaded primordia and nodules, and fewer cortical infection threads than WT plants (Figure S8). We also showed that the infection and nodulation phenotype of a homozygous WT line selected from the F2 population of the backcross between the Mtlyk10 mutant and R108 was comparable to the R108 WT line (Figure S8). Taken together, these data show that MtLYK10 plays a role in the symbiotic process of rhizobial infection thread growth, and by consequence the rate of successful nodulation in M. truncatula.
M. truncatula lyk10 mutant plants show more severe infection phenotypes with S. meliloti succinoglycan mutants
To address the possible implication of MtLYK10 in EPS recognition, we next characterized the symbiotic phenotypes of Mtlyk10 mutant plants with S. meliloti exo mutants. First, we compared the double‐glycanase exoKdel/exsH‐1325 mutant (only makes HMW succinoglycan) with the appropriate modified WT S. meliloti 961 strain (makes both HMW and LMW succinoglycan), on Mtlyk10 and R108 plants at 14 dpi. As we had observed with the unmodified 1021 WT S. meliloti (Figure 3), the 961 WT S. meliloti strain induced fewer cortical infection threads and fewer successful infections on Mtlyk10 mutant plants compared with R108 plants (Figure 5). Also, no significant quantitative differences were noted in uninvaded nodule primordia (P = 0.06) induced by the 961 strain on Mtlyk10 mutant and WT plants (Figure 5), as previously observed with S. meliloti 1021.
Figure 5.
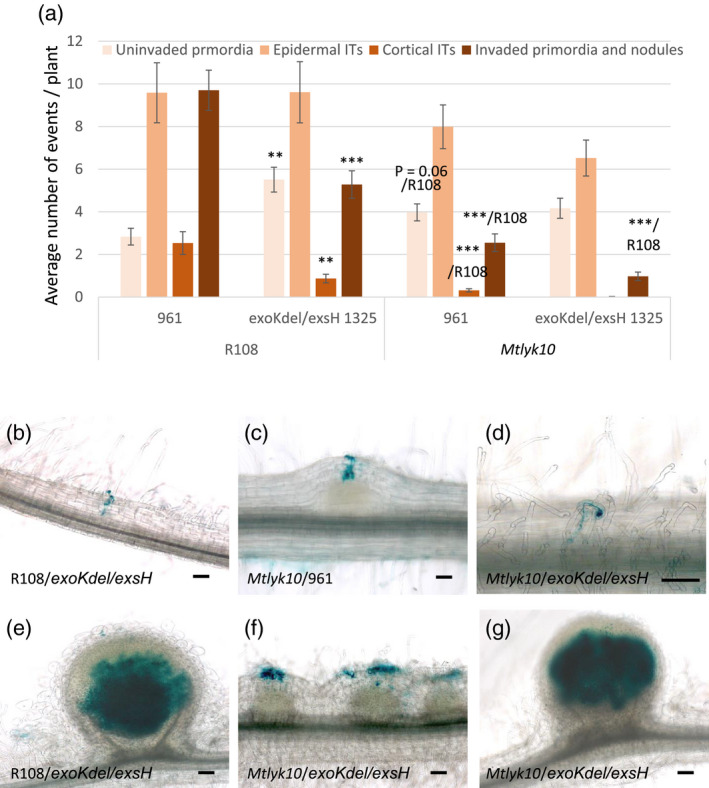
Infection and nodulation invasion phenotypes of the exoKexsH 1325 double‐glycanase Sinorhizobium meliloti mutant on M. truncatula lyk10 mutant plants. Infection and nodulation invasion phenotypes of M. truncatula Mtlyk10 mutant and R108 WT plants were scored 14 dpi following inoculation with the exoKdel/exsH‐1325 mutant that makes only HMW succinoglycan, or the S. meliloti WT strain 961.
(a) Quantification of uninvaded nodule primordia, epidermal (restricted to the epidermis) and cortical (which have reached the cortex) infection threads (ITs), and invaded nodule primordia and nodules. Data are the averages of four independent experiments, n = 60 for each genotype/strain combination. Error bars = SEM. Statistically significant differences are indicated as ***/R108 for comparisons for the same rhizobial strain (either 961 or exoKdelexsH‐1325) between plant genotypes (R108 versus Mtlyk10); and as ** or *** for comparisons between the two rhizobial strains (961 versus exoKdelexsH‐1325) for the same plant genotype (R108). **P < 0.01; ***P < 0.001. No long infection threads were observed with exoKdel/exsH‐1325 on Mtlyk10 plants. For the comparison of R108 and Mtlyk10 plants inoculated by exoKdel/exsH‐1325, no significant difference was observed (P = 0.06) for numbers of uninvaded nodule primordia (as indicated).
(b–g) Photographs illustrating these infection phenotypes, both inoculated strains carrying a constitutive hemA‐lacZ reporter gene fusion (pXLGD4) for visualization of bacteria in blue.
(b) A cortical infection thread on R108 inoculated by exoKdel/exsH‐1325. (c) A cortical infection thread on Mtlyk10 inoculated by 961. (d) An epidermal (restricted to the root hair) infection thread on Mtlyk10 inoculated by exoKdel/exsH‐1325. (e) A nodule on R108 inoculated by exoKdel/exsH‐1325. (f) Uninvaded primordia on Mtlyk10 inoculated by exoKdel/exsH‐1325. (g) A nodule on Mtlyk10 inoculated by exoKdel/exsH‐1325. Scale bars = 100 µm except for (d) = 20 µm.
Comparing the infection phenotype of the exoKdel/exsH‐1325 mutant on Mtlyk10 and WT roots, we observed no cortical infection threads on Mtlyk10 roots and significantly fewer invaded nodule primordia and nodules on Mtlyk10 roots compared with R108 (Figure 5). The different infection phenotypes are apparent in Figure 5(b) that shows an infection thread in the cortex with exoKdel/exsH‐1325/R108, Figure 5(c) that shows cortical infection with 961/Mtlyk10, and Figure 5(d) that shows an infection thread restricted to a root hair with exoKdel/exsH‐1325/Mtlyk10. As S. meliloti exoKdel/exsH‐1325 only makes HMW succinoglycan, this suggests that while LMW succinoglycan is not essential for infection, it enhances infection success in both R108 WT and Mtlyk10 mutant plants. Furthermore, the absence of LMW succinoglycan together with an MtLYK10 mutation creates a stronger infection defect than the absence of LMW succinoglycan alone.
WT R108 and Mtlyk10 mutant plants were next inoculated with succinoglycan‐deficient S. meliloti exoY, and the S. meliloti exoHKdel/exsH‐1345 mutant that only produces non‐succinylated HMW succinoglycan. In order to assess both infection and nodulation, we observed plants at 21 dpi. As previously shown (Figure 2) neither strain successfully infects R108, and in addition, we observed no successful infections with either strain on Mtlyk10 plants (Figure 6a–i). Indeed, on Mtlyk10 mutant plants both strains induced mainly uninfected bumps corresponding to uninvaded nodule primordia with large, abnormal colonies of rhizobia that remain superficial to these structures (Figure 6g,i). These infection events were scored as short epidermal infection threads (Figure 6a), illustrated in Figure 6(f) (exoY/Mtlyk10) and Figure 6(h) (exoHKdel/exsH‐1345/Mtlyk10). Some more developed, but nonetheless also aborted, infection threads (‘long epidermal infection threads’) were observed with both strains on WT R108 plants (for example, Figure 6d shows a long epidermal infection thread for exoHKdel/exsH‐1345/R108), but significantly fewer of such long, aborted infection threads were observed on Mtlyk10 mutant roots compared with WT R108, with both exo mutants (Figure 6a). Thus, the severe infection phenotype of exoY and exoHKdel/exsH‐1345 on WT plants was even stronger on Mtlyk10 mutant plants. Taken together, none of the rhizobial mutant phenotypes were suppressed by the mutation in MtLYK10, and in fact, the phenotypes were worse with each of the three S. meliloti exo mutants (exoKdel/exsH 1325, exoY and exoHKdel/exsH‐1345).
Figure 6.
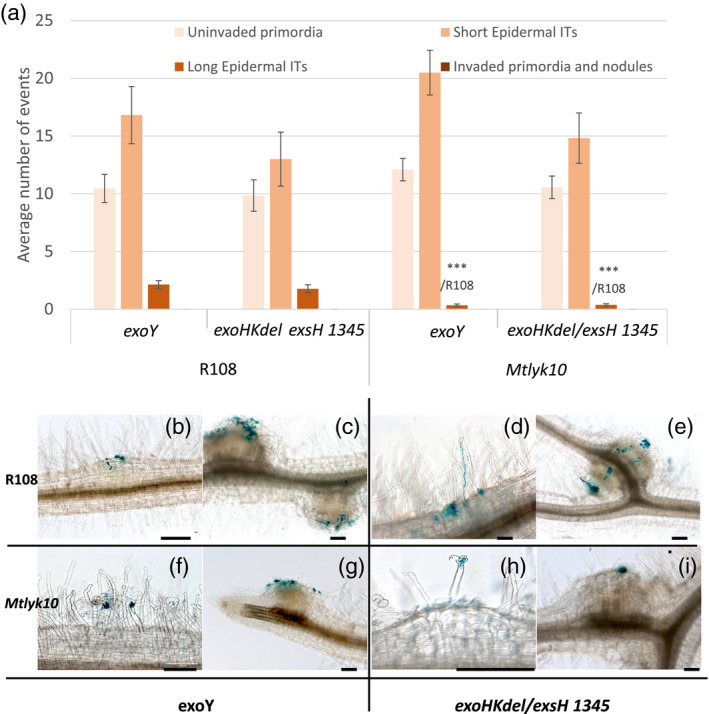
Infection and nodulation invasion phenotypes of the Sinorhizobium meliloti succinoglycan‐deficient exoY mutant and the exoHKdel/exsH‐1345 succinyltransferase mutant on M. truncatula lyk10 mutant plants. Infection and nodulation invasion phenotypes of M. truncatula Mtlyk10 mutant and R108 WT plants were scored 21 dpi following inoculation with the S. meliloti succinoglycan‐deficient exoY mutant or the S. meliloti exoHKdel/exsH‐1345 mutant, which makes non‐succinylated HMW succinoglycan.
(a) Quantification of uninvaded nodule primordia, and short and long epidermal infection threads. No invaded nodule primordia or invaded nodules were seen. Data are the averages of two independent experiments, n = 40 for each genotype/strain combination. Error bars = SEM. Statistically significant differences are indicated as ***/R108 to indicate that the comparison is to the R108 genotype inoculated with the same rhizobial strain. ***P < 0.001.
(b–g) Photographs illustrating these infection phenotypes, both inoculated strains carrying a constitutive hemA‐lacZ reporter gene fusion (pXLGD4) for visualization of bacteria in blue. (b, c) R108/exoY; (d, e) R108/exoHKdel/exsH‐1345; (f, g) Mtlyk10/exoY; (h, i) Mtlyk10/exoHKdel/exsH‐1345. Scale bars = 100 µm.
Rescue in trans by a strain producing normal succinoglycan indicates that succinoglycan‐defective mutants are at a disadvantage in mixed inoculation nodules, with non‐succinylated succinoglycan being even more of a disadvantage than the absence of succinoglycan
Rescue of failed infection by succinoglycan‐deficient exo mutants has previously been accomplished by providing WT succinoglycan by co‐inoculation with NF‐deficient mutants (Klein et al., 1988). Each strain individually is impaired in invasion, but when NF and succinoglycan are provided in trans within an infection thread, both strains succeed in invading. We used this in trans rescue method to investigate whether the non‐succinylated succinoglycan produced by exoHKdel/exsH‐1345 has a restrictive effect on the invasion of a strain that produces normal succinoglycan. To determine if a NF‐deficient S. meliloti mutant rescues the exoHKdel/exsH‐1345 mutant with the same efficiency with which it rescues the succinoglycan‐deficient exoY mutant, we performed co‐inoculations with a mutant in the NF N‐acetylglucosaminyltransferase gene, nodC (Jacobs et al., 1985; Atkinson and Long, 1992). If the nodC mutant rescues exoHKdel/exsH‐1345 less efficiently than it rescues exoY, it suggests that any restrictive interaction between non‐succinylated succinoglycan and the plant is dominant over the reaction with WT succinoglycan. If nodC rescues exoHKdel/exsH‐1345 equal to or more efficiently than it rescues exoY, it suggests that any restrictive interaction is not dominant.
M. truncatula A17 WT plants were inoculated with 1:1 mixtures of either nodC + exoY or nodC + exoHKdel/exsH‐1345, and nodule formation tracked over 28 days. As shown in Figure 7(a) both combinations induce nodule formation, in contrast with the negative controls of individual strain inoculations. Compared with nodC + exoY, the nodC + exoHKdel/exsH‐1345 mix was less efficient, with fewer plants forming nodules. In both cases relatively few nodules were observed (<1 nodule/plant) (Figures 7 and S9). To determine strain occupancy, mature nodules were surface sterilized, then crushed and bacterial growth was quantified on antibiotic selection plates (for nodC) or on Calcofluor (for exo strains). This showed that both types of nodules contained a clear majority of nodC bacteria (90–95%), and that exoY was significantly more present in mixed inoculation nodules compared with exoHKdel/exsH‐1345 (Figure 7b).
Figure 7.
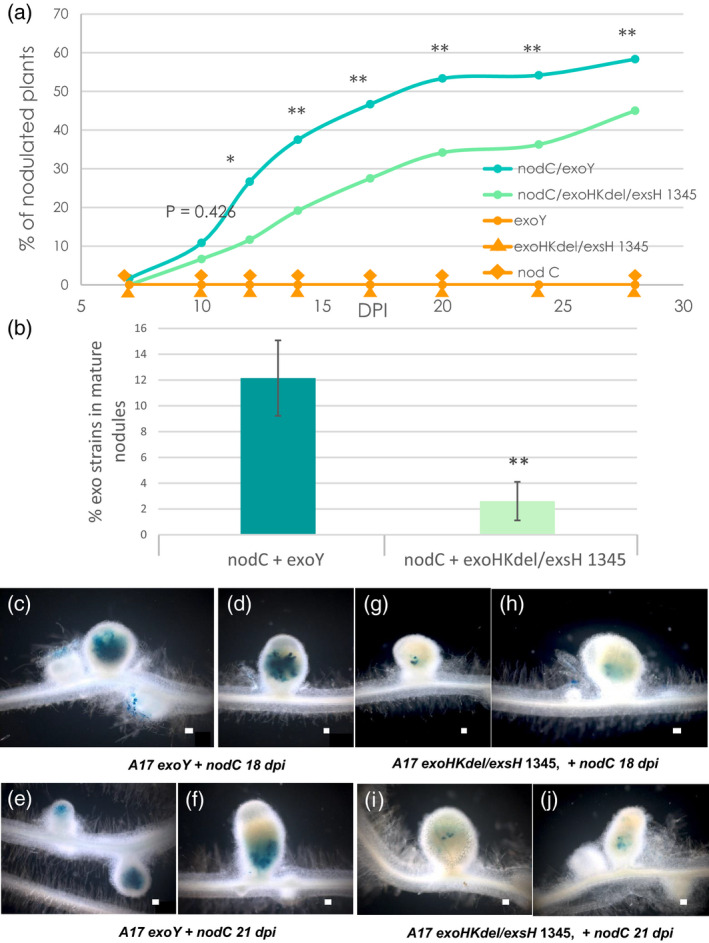
Nodulation defects of exo mutants can be rescued in trans by co‐inoculation with a nodC mutant. M. truncatula WT A17 plants were inoculated with 1:1 mixes of Sinorhizobium meliloti nodC + exoY or S. meliloti nodC + exoHKdel/exsH‐1345, or with individual control strains; exoY, exoHKdel/exsH‐1345 or S. meliloti nodC.
(a) Kinetics of nodule formation at 7, 10, 12, 14, 17, 20, 24, and 28 dpi expressed as percentages of nodulated plants at each time point. Data are the averages of two independent experiments n = 120 for each mixed inoculum. Statistical comparisons were performed between the two types of mixed inoculation on the average number of nodules per plant at each time point. *P < 0.05; **P < 0.01.
(b) Percentages of the exoY or exoHKdel/exsH‐1345 mutants recovered from mature nodules compared with total rhizobial counts recovered. Error bars = SEM. **P < 0.01.
(c–j) Photographs of mature nodules formed in the same experimental set‐up using the exoY and exoHKdel/exsH‐1345 mutants carrying a constitutive hemA‐lacZ reporter gene fusion (pXLGD4) for visualization of bacteria in blue. (c–f) nodules formed following co‐inoculation with S. meliloti 1021 nodC + exoY at 18 dpi (c, d) and 21 dpi (e, f). (g–j) nodules formed following co‐inoculation with S. meliloti 1021 nodC + exoHKdel/exsH‐1345 at 18 dpi (g, h) and 21 dpi (i, j). Scale bars = 100 µm.
To visualize nodule occupancy by the exo mutant strains, plants were co‐inoculated with the same mixes of S. meliloti strains, but using exo strain versions expressing beta‐galactosidase. Unlike for recoverable rhizobia, the in situ staining via lacZ of rhizobia inside nodules also enables visualization of terminally differentiated bacteria that are internalized within host cells. Coloration of well developed nodules of both types confirmed the presence of the exo strains in nodules, and again indicated that exoHKdel/exsH‐1345 is less present compared with exoY (Figure 7c–j). Thus, nodC + exoY nodules clearly showed the presence of exoY, usually throughout inner nodule tissue (Figure 7c–f). The dense staining of the exoY rhizobia within nodules suggests that they are internalized into cortical cells. In contrast, nodC + exoHKdel/exsH‐1345 nodules showed only quite limited, patchy areas of the exoHKdel/exsH‐1345 mutant within inner nodule tissue (Figure 7g–j). This suggests that the exoHKdel/exsH‐1345 mutant has a very limited, if any, ability to become terminally differentiated into bacteroids. In both cases, smaller nodule‐like structures that were visualized along with the mature nodules at 18–21 dpi, were not invaded by the exo strains. Co‐inoculation studies using R108 were only feasible in the presence of the ethylene biosynthesis inhibitor aminoethoxyvinylglycine (AVG) (extremely few nodules formed without AVG). Ethylene is a well known negative regulator of rhizobial infection, and symbiotic responses and nodulation are often better when AVG is present (Suzaki et al., 2015). In these conditions, co‐inoculated R108 plants did not show clear differences in nodulation ability by the two mixes of rhizobial strains. Nonetheless, coloration of nodules clearly showed exoHKdel/exsH‐1345 was less persistent in mixed inoculation nodules compared with exoY (Figure S10), as observed on M. truncatula A17. Taken together, these data indicate that early events of infection by trans‐complementation of NF‐deficient and succinoglycan‐deficient/aberrant rhizobial mutants lead to fully formed nodules in which the main occupant is the strain with normal succinoglycan (nodC). Furthermore, the exoHKdel/exsH‐1345 mutant producing non‐succinylated succinoglycan appears at even more of a disadvantage than the succinoglycan‐deficient exoY strain in mixed inoculation nodules.
Discussion
Our results show that the succinoglycan‐deficient exoY mutant of S. meliloti is completely blocked in invasion of M. truncatula, and only forms very short infection threads that do not elongate and do not reach beyond the epidermis. The exoHKdel/exsH‐1345 mutant that produces succinoglycan missing the succinyl group is as impaired in invasion as the succinoglycan‐deficient exoY mutant, with infections being similarly restricted to the epidermis. These infection phenotypes explain the previous finding that inoculation of M. truncatula A17 with either mutant results in the same plant growth phenotype as non‐inoculated plants (Jones et al., 2008; Mendis et al., 2016). Thus, no successful infections are possible without succinylated succinoglycan. Unlike these two non‐invading mutant strains, we had previously observed that the S. meliloti exoKdel/exsH‐1325 mutant, which only produces succinylated HMW succinoglycan, invades, but has a reduced symbiotic efficiency (Mendis et al., 2016). In this study, we now show that the S. meliloti exoKdel/exsH‐1325 mutant forms fewer invaded nodules than WT strains, so the enhancement that production of the LMW form of succinoglycan provides to the symbiotic productivity of S. meliloti on the host M. truncatula is due to increased infection thread progression, which results in more nitrogen‐fixing nodules. Thus, succinylated succinoglycan plays a critical and early role in infection thread progression in M. truncatula root hairs.
Rhizobial NFs are also essential for the infection process in M. truncatula, as in the majority of rhizobial host plants (Dénarié et al., 1996). Mutant M. truncatula plants expressing reduced levels of the M. truncatula NF receptor proteins MtNFP or MtLYK3 exhibit arrested infections with unusually large microcolonies of rhizobia and sac‐like infection events (Arrighi et al., 2006; Smit et al., 2007), which resemble to some extent, those we observed with S. meliloti exoY or exoHKdel/exsH‐1345. Thus, NF perception and the presence of succinylated succinoglycan are both needed for the correct initiation, maintenance, and direction of polar growth of infection threads, essential processes for successful infection of M. truncatula. In contrast with NFs, succinoglycan is not essential for root hair curling and rhizobial entrapment in root hair curls, neither for infection thread initiation nor the induction of nodule primordia formation, such that S. meliloti exoY or exoHKdel/exsH‐1345 mutants induce uninvaded nodule‐like structures, unlike NF‐deficient S. meliloti mutants (Dénarié et al., 1996). This suggests that an early symbiotic role of succinoglycan is probably restricted to a local function for infection thread elongation, unlike the pleiotropic roles of NFs.
The rescue of both S. meliloti exoY and exoHKdel/exsH‐1345 by the NF‐deficient nodC mutant shows that the prevention of epidermal penetration by both the succinoglycan‐deficient exoY mutant and the exoHKdel/exsH‐1345 mutant is relieved when normal succinoglycan is supplied. However, complementation efficiency was relatively low, and low percentages of exo rhizobia were recovered from within mature nodules. This suggests that these mutants were both at a disadvantage compared with the nodC mutant in nodules, and that once nodules are formed, the rhizobial production of NFs is less important than the extracellular rhizobial succinoglycan. Alternatively, differences in the quantities of NFs and succinoglycan could be required during nodulation. Laser‐capture microdissection RNA‐Seq experiments show that exo and nod genes are expressed in apices and nitrogen‐fixing zones of M. truncatula nodules (Roux et al., 2014). A disadvantage for internalization into and/or survival within symbiosomes relative to the nodC mutant might be due to greater sensitivity on the part of exo mutants to conditions within the symbiosome. As the dominance of nodC in exoHKdel/exsH‐1345 + nodC nodules (approximately 98%) was even more marked compared with exoY + nodC nodules (approximately 88%), we can hypothesize that the production of only non‐succinylated succinoglycan renders rhizobia even more sensitive than the complete absence of succinoglycan production. However, if the plant does actively resist invasion by strains producing structurally aberrant succinoglycan, this is only partially effective and the complete absence of succinoglycan is also resisted.
We have also shown that M. truncatula plants carrying a Tnt1 insertion in MtLYK10 have clear, early infection phenotypes, the severity of which depends on the S. meliloti strain tested. In response to WT rhizobia, Mtlyk10 plants form fewer cortical, successful infection threads and consequently fewer invaded nodules, compared with WT plants. Although Mtlyk10 mutant plants were clearly defective in infection, nodule primordia and some invaded nodules can be formed, indicating that the NF‐activated process of nodule organogenesis is independent of MtLYK10. Mtlyk10 mutants are thus symbiotic M. truncatula mutants that can uncouple infection and nodule organogenesis (Murray, 2011), revealing another plant component that is specifically involved in the infection process in M. truncatula. The expression profile of MtLYK10 is compatible with its role in infection, with strong induction in root hairs and nodules, and in response to either NFs or rhizobial inoculation, like many infection‐related genes (Breakspear et al., 2014; Roux et al., 2014; Larrainzar et al., 2015; van Zeijl et al., 2015; Damiani et al., 2016; Jardinaud et al., 2016; Liu et al., 2019). Within M. truncatula nodules, MtLYK10 is one of the most highly expressed LysM receptor genes, with strong expression in the distal infection zones of nodules, similarly to MtNFP and MtLYK3 (Roux et al., 2014; Bono et al., 2018).
In response to the S. meliloti exo mutants studied here, the infection phenotype of Mtlyk10 mutant plants was consistently more severe compared with S. meliloti WT strains on these plants. The S. meliloti exoKdel/exsH‐1325 mutant forms very few invaded nodules on Mtlyk10 mutant plants, while no invaded nodules and very few extended infection threads were observed with either the exoY or the exoHKdel/exsH‐1345 strains. Thus, the infection deficiencies conferred by defects in succinoglycan or mutation in MtLYK10 were apparently additive, indicating different mechanisms by which succinoglycan and MtLYK10 play a role in the infection process. Furthermore, if the infection deficiency of the S. meliloti exoY mutant is due to perception of bacterial molecular patterns exposed by the absence of succinoglycan, the restriction is not accomplished through MtLYK10.
In L. japonicus, the octasaccharide EPS produced by M. loti is also implicated in symbiosis. However, there are important differences between the two model nodulation systems in the role of EPS in infection thread formation. Notably, a M. loti R7A exoB mutant that does not make EPS can invade L. japonicus, albeit with reduced efficiency (Kelly et al., 2013; Kawaharada et al., 2015). Remarkably, in M. loti, structurally aberrant polysaccharide produced by the exoU mutant creates a much greater L. japonicus invasion defect than the complete absence of the EPS in an exoB mutant (Kelly et al., 2013; Kawaharada et al., 2015). It is not possible to perform a completely identical comparison between the two symbiont/host systems. There is no known mutant of S. meliloti that is comparable with the exoU mutant of M. loti in forming a truncated pentasaccharide EPS monomer. A glycosyltransferase that has been designated exoU in S. meliloti is known to function in the succinoglycan biosynthesis pathway, but this mutant does not secrete any succinoglycan (Reuber and Walker, 1993; Becker et al., 1993c). We have confirmed that this mutant has a succinoglycan‐deficient phenotype similar to an exoY mutant, in terms of succinoglycan production (Figure S11). Nonetheless, we can conclude that the interaction between the exoHKdel/exsH‐1345 mutant and M. truncatula is different from the interaction between the exoU mutant of M. loti that produces truncated pentasaccharide EPS and L. japonicus.
Another major difference between the two systems is that L. japonicus mutants in the MtLYK10 ortholog, LjEPR3, partially suppress the phenotype of a M. loti exoU mutant (Kawaharada et al., 2015), while none of the S. meliloti exo phenotypes we have characterized here are suppressed by the Mtlyk10 mutation. In fact, infection deficiencies are accumulated by the combination of plant (Mtlyk10) and rhizobial (exoKdel/exsH‐1325, exoHKdel/exsH‐1345 and exoY) mutations, indicating that each succinoglycan‐related infection phenotype is independent of MtLYK10. In other words, unlike the proposed role for LjEPR3, our data suggest that S. meliloti succinoglycan and MtLYK10 perform important, but different, roles in infection thread formation in M. truncatula. Thus, although we cannot exclude a role for MtLYK10 in recognizing a form of EPS not tested here, we favor the hypothesis that MtLYK10 does not intervene in EPS recognition. Despite the apparent difference between plant species for EPS recognition, Ljepr3 and Mtlyk10 mutants have similar phenotypes with WT rhizobia, both forming fewer invaded nodules than their corresponding WT plants, within 14 dpi. Also, these Ljepr3 and Mtlyk10 phenotypes are both weaker than those of the appropriate EPS‐minus rhizobial strains, M. loti exoB and S. meliloti exoY, respectively, on their WT host plants. This leaves open the possibility that an unknown EPS perception component exists in L. japonicus. So far, the few LysM‐RLKs that have been characterized in different plant species have mostly been linked to roles in recognizing chitin oligomers or lipo‐chitooligosaccharides, but as pointed out by Kawaharada et al. (2015), such proteins have LysM domains that are quite divergent from those of MtLYK10/LjEPR3 homologs. With the notable exception of Arabidopsis thaliana (Buendia et al., 2018), these MtLYK10/LjEPR3 homologs are widespread in Angiosperms. Interestingly, some dicot species have two copies of this gene as the result of ancient duplications (Figure S6), followed potentially by neofunctionalization or subfunctionalization. This could point toward different functions of MtLYK10‐type genes in different plant species and this gene is not always involved in nodulation, as shown recently in Parasponia andersonii (Gough et al., 2018; van Velzen et al., 2018). This may be linked to differences in rhizobial infection mechanisms among plant species.
It is not yet clear what role(s) succinylated succinoglycan plays in infection thread development. exoY, exoH, and exoK transposon‐insertion mutants are sensitive to acidic pH (Hawkins et al., 2017). However, acid sensitivity could not be the only cause of the symbiotic defect in these strains. exoK mutants are just as sensitive as exoH mutants to acid, yet the symbiotic defect of exoK mutants is much less severe than that of exoH mutants. There is some evidence for succinoglycan interaction with reactive oxygen species (ROS) produced in the infection thread. One possible role is protection of the bacteria from ROS. When tested in the free‐living state, succinoglycan‐overproducing strains are c. 1.5–2‐fold more resistant than WT S. meliloti to H2O2, whereas the exoY mutant has c. 0.4–0.5 the resistance to H2O2 of the WT (Lehman and Long, 2013). But the symbiotic phenotypes of S. meliloti mutants in genes involved in coping with reactive oxygen species do not closely resemble those of exo mutants. The extremely H2O2‐sensitive S. meliloti oxyR mutant has no symbiotic defect (Jamet et al., 2005). Also, an S. meliloti double‐mutant in two catalase‐encoding genes katB and katC has a symbiotic defect, but in contrast with exo mutants, it forms numerous infection threads that do not appear to fail until the time of S. meliloti release into symbiosomes (Jamet et al., 2003). Thus, it seems unlikely that sensitivity to ROS could be the only cause of the symbiotic defect of exo mutants.
Another recent article has shown that strains that produce an excess of succinoglycan are less sensitive to the NCR247 cationic peptide than is S. meliloti WT (Arnold et al., 2018). However, the exoY mutant was found to not be more sensitive than the WT to NCR247, at least in the free‐living state, and addition of non‐succinylated succinoglycan was as successful as wild‐type succinoglycan in enhancing survival of free‐living S. meliloti treated with NCR247 (Arnold et al., 2018). NCR peptides are secreted into the symbiosome by most legumes that form indeterminate nodules, including M. truncatula and alfalfa (Alunni and Gourion, 2016). These peptides play multiple roles in the process of bacteroid differentiation and in cell fate (Alunni and Gourion, 2016). A role has not yet been demonstrated for NCR peptides in root hairs and infection threads, and bacteria within M. truncatula infection threads do not show signs of bacteroid differentiation, but expression of NCR247 is strongly increased in root hairs by 3 dpi with S. meliloti (Breakspear et al., 2014). A difference in sensitivity to NCR peptides might explain the disadvantage in nodule occupancy that the exoY and exoHKdel/exsH‐1345 mutants appear to have relative to the nodC mutant in our co‐inoculation experiments. More than 600 NCR genes are induced in M. truncatula by S. meliloti, and in vitro tests with NCR peptides show increased sensitivity and different responses of a S. meliloti succinoglycan‐deficient mutant compared with WT bacteria (Montiel et al., 2017). Also, EPS‐minus mutants of Sinorhizobium fredii HH103 can form functional nodules on soybean and pigeonpea (Parada et al., 2006), species which lack NCR genes (Montiel et al., 2016), and also on Glycyrrhiza uralensis (Margaret‐Oliver et al., 2012), in which the expression of only a few NCR genes is detected (Montiel et al., 2017). Taken together, these data suggest a link between rhizobial EPS and the mediation of NCR peptide effects on differentiation and survival of endophytic rhizobia at different stages of nodulation.
Thus, the essential function of succinylated succinoglycan in Medicago infection might involve interaction with an as‐yet‐unidentified plant receptor, or with another component of the infection thread matrix. Alternatively, succinylated succinoglycan could have a role in protecting the bacteria from NCR peptides or an as‐yet‐unidentified stress within the infection thread.
Experimental Procedures
Bacterial strains, growth conditions, and plasmids
S. meliloti 1021 strains (Meade et al., 1982), were grown at 28–30°C with shaking in either Luria–Bertani Miller medium with 2.5 mm MgSO4 and 2.5 mm CaCl2 with streptomycin 200 or 500 µg/ml or glutamate–mannitol–salts (GMS) medium (York and Walker, 1997). S. meliloti mutants were grown in appropriate antibiotics at concentrations: neomycin 100 or 200 µg/ml; spectinomycin 50 µg/ml; gentamicin 25 µg/ml. For detail on strains, see Figure S2. Calcofluor polysaccharide indicator plates contained 0.02% Calcofluor white M2R (fluorescent brightener 28, Sigma, St. Louis, MO, USA) (Leigh et al., 1985). Strains carrying plasmids: pRF771 (Wells and Long, 2002); pExoH (this study, see Figure S2); pXLGD4 (hemA::lacZ reporter (Leong et al., 1985)); pHC60 (Cheng and Walker, 1998); or pcCFP (Fournier et al., 2015) were selected with 10 µg/ml tetracycline.
Plant genotypes
Two WT and two mutant genotypes of Medicago truncatula were used. Medicago truncatula cv. Jemalong A17 and M. truncatula ssp. tricycla R108 are WT lines. The M. truncatula Jemalong super numeric nodules‐2 mutant (sunn‐2) carries a mutation in the MtSUNN gene, leading to loss of the autoregulation of nodulation and consequently to increased infection events (Schnabel et al., 2005; Fournier et al., 2008). The M. truncatula ssp. tricycla Mtlyk10 (NF4929) mutant carries a Tnt1 retrotransposon in LysM1 of the MtLYK10 gene (Medtr5g033490). Two independent backcrossed lines of this mutant were used, most of the results presented were obtained using the Mtlyk10#46 line (see Figure S8). A line that is homozygous for the WT MtLYK10 gene was also selected after backcross and checked for its symbiotic phenotype (see Figure S8).
Real time quantitative RT PCR analysis on M. truncatula plants
qRT‐PCR expression analysis of MtLYK10 was performed on M. truncatula WT (R108) and the Mtlyk10 mutant. Expression was analyzed in the non‐inoculated and nodulated root segments for each genotype, using material pooled from 20 plants for each genotype inoculated or not with S. meliloti 1021. Nodulated material was collected from 14 to 25 dpi. Relative expression was calculated using two housekeeping genes, Medtr3g062450 and Medtr3g065110, and then fold changes were calculated for inoculated compared with non‐inoculated as the 2−ΔCT values. The primers for MtLYK10 were: F: GACCCAGTAGCTGCTTTGGAA; and R: TGACACTGCCACAACGATCTC. The RNA extraction protocol and qRT‐PCR conditions and primers for the reference genes have been previously described (Gibelin‐Viala et al., 2019).
Whole‐plant assay of nodulation and symbiotic efficiency
Medicago truncatula cv. Jemalong A17 was prepared for inoculation with S. meliloti as previously described (Jones et al., 2013). Seedlings were moved to individual Jensen’s medium microcosms and inoculated with S. meliloti of the appropriate strain. Plants were grown in a Percival AR‐36L incubator (Perry, IA) at 21°C, with 60–70% relative humidity and 100–175 µmol m−2 s−1 light. Shoot fresh weight was measured after 7 weeks.
Infection thread assays of cCFP‐expressing S. meliloti strains on M. truncatula Jemalong
Rhizobial infection sites were analyzed in M. truncatula Jemalong sunn‐2 or A17 root hairs by in vivo microscopy as previously described (Fournier et al., 2008; Fournier et al., 2015). In brief, plants were placed in 12 cm x 12 cm plates containing modified Fåhraeus medium (0.5% (w/v) Phytagel; Sigma‐Aldrich) supplemented with 50 nm aminoethoxyvinylglycine (AVG). Roots were covered with gas‐permeable plastic film (Lumox Film, Sarstedt, France), and plants were grown with the dishes slightly tilted to encourage the growth of the roots along the film. An aqueous suspension (OD600 = 0.001) of exponentially growing bacteria constitutively expressing cCFP was used for inoculation of nitrogen‐starved plants. Selected infection sites were imaged with a Leica TCS SP2 AOBS confocal laser scanning microscope (Figure 1) or a Zeiss epifluorescence microscope (Figure S4) equipped with long‐distance 40x HCX Apo L (numerical aperture, 0.80) water immersion objectives. For confocal images, the argon laser band of 458 nm was used to excite cCFP and a 561‐nm diode to observe cell wall autofluorescence, respectively. Specific emission windows used for cCFP and autofluorescence signals were 465–485 nm and 620–720 nm, respectively, and emitted fluorescence was false colored in green (cCFP) and magenta (wall autofluorescence). The composite images shown are maximal projections of selected planes of a z‐stack. For epifluorescence images, cCFP and cell wall autofluorescence were recorded using combinations of BP436/10 and BP450‐480 (for cCFP) or BP573/20 and BP625/30 (for autofluorescence) excitation and emission filters. Images were acquired and projected using Leica confocal software or MetaVue (Molecular Devices) and processed using Leica confocal software, metavue and fiji (http://imagej.nih.gov/ij/; Schindelin et al., 2012; Schindelin et al., 2015). Data are from two experiments in sunn‐2 and 2 experiments in A17.
Infection thread assays of β‐galactosidase‐expressing S. meliloti strains on M. truncatula tricycla R108
Seeds were surface sterilized, germinated, and grown in 12 cm x 12 cm plates containing slanting 0.2 mm NH4NO3‐Fåhraeus agar medium covered by growth paper for 6 days at 22°C with day/night cycles of 16/8 h. Each plant was inoculated with 2 × 103 bacteria.
Entire roots were collected 11, 14, 17 or 21 dpi, fixed with 2% (v/v) glutaraldehyde for 2 h under vacuum, rinsed 3× in Z′ buffer [0.1 m potassium phosphate buffer (pH 7.4), 1 mm MgSO4, and 10 mm KCl], and stained 1 h under vacuum then overnight at 28°C in Z′ buffer containing 0.08% 5‐bromo‐4‐chloro‐3‐indolyl‐β‐d‐galactoside (X‐gal), 5 mm K3Fe(CN)6, and 5 mm K4Fe(CN)6. Entire roots were observed under a light microscope after 3 min bleach clearing (3.2% active chloride).
Co‐inoculation assays
Plants grown as described above were inoculated with 100 µl per plant of a 1:1 mix of nodC + exo mutant at an OD600 = 0.5. Mature nodules were surface sterilized 5 min in bleach 2.4% active chloride, rinsed 5× in sterile water, crushed with a mortar in a 1.5 ml microtube, and spread on plates containing streptomycin and neomycin. Ten to 12 nodules of each combination were crushed and 52 randomly picked colonies/ nodule were characterized. M. truncatula A17 plants were grown without AVG, R108 plants were grown in the presence of 0.5 µm AVG.
Statistical analysis
Data are presented as means ± SEM. Data analyses were performed with r software (v.3.4.1). Statistical comparisons of phenotypic data were performed using a Wilcoxon test or a Student’s t‐test (when the normality assumption was followed).
Phylogeny analysis
MtLYK10 homologs were found from Buendia et al. (2018) and from NCBI blasts. Synteny was verified using phytozome (https://phytozome.jgi.doe.gov), soybase (https://www.soybase.org) and the Legume Information System, lis (http://legumeinfo.org). Protein or gene names are shown in the tree, together with the plant species name. The tree was generated using http://www.phylogeny.fr using the gblocks program to eliminate poorly aligned positions and divergent regions.
Author Contributions
FM, JF, CG, KMJ designed research and wrote the manuscript. FM, JF, HCM, KMJ performed experiments. FM, JF, HCM, CG, KMJ analyzed data. MT, JW, PR, KM provided resources.
Conflicts of interest
The authors have no conflicts of interest to declare.
Supporting information
Figure S1. Plant performance and nodulation phenotypes on alfalfa cv. Iroquois and M. truncatula A17 of S. meliloti strains that make EPSII (galactoglucan).
Figure S2. Information for all S. meliloti strains and plasmids, both those are discussed in the main text and in Supporting Information.
Figure S3. Information on the genetic modifications in strains 961, 1317, exoKdel/exsH‐1325 and exoHKdel/exsH‐1345 with the exoLAMON operon under the control of a heterologous promoter.
Figure S4. Infections formed by the exoY and exoHKdel/exsH‐1345 mutants on Medicago truncatula Jemalong A17.
Figure S5. Complementation of strains with an exoHK deletion by a plasmid carrying exoH.
Figure S6. Phylogenetic tree of MtLYK10 homologs from a variety of plant species, mostly Rosid dicots, but also two monocots, Oryza sativa and Brachypodium distachyon, and with MtLYK11 as an outgroup.
Figure S7. qRT‐PCR expression analysis of MtLYK10 in Medicago truncatula WT (R108) and the Mtlyk10 mutant.
Figure S8. Comparison of the infection and nodulation phenotype of two independent Mtlyk10 mutant lines (Mtlyk10#2 and Mtlyk10#46) and a homozygous MtLYK10‐WT line (MtLYK10‐WT), all selected from the backcross between the Mtlyk10 mutant and R108.
Figure S9. Kinetics of nodule formation when the nodulation defects of exo mutants are rescued in trans by co‐inoculation with a nodC mutant.
Figure S10. Nodules formed after mixed inoculation on Medicago truncatula WT R108 plants.
Figure S11. The exoU mutant of S. meliloti does not produce detectable succinoglycan.
Acknowledgements
The Mtlyk10 mutant, which is jointly owned by the Centre National de la Recherche Scientifique, was obtained from The Noble Research Institute and was created through research funding, in part, by grants from the US National Science Foundation, DBI‐0703285 and IOS‐1127155. This work was funded by INRA, the CNRS, the French Agence Nationale de la Recherche ‘AOI’ project (ANR‐15‐CE20‐0004‐01), the Agence Nationale de la Recherche ‘Laboratoire d’Excellence’ (LABEX) entitled TULIP (ANR‐10‐LABX‐41), and the TULIP Visiting Scientist Award and USDA, National Institute of Food and Agriculture award 2014‐67013‐21579. Thanks to TRI‐FR 3450 Imaging Platform Facilities, CNRS, France.
Contributor Information
Clare Gough, Email: clare.gough@inra.fr.
Kathryn M. Jones, Email: kmjones@bio.fsu.edu.
Data Availability Statement
Supporting Information is listed below and these figures are provided for this article. Image acquisition and modification information has been described in Experimental Procedures or in Figure legends.
References
- Alunni, B. and Gourion, B. (2016) Terminal bacteroid differentiation in the legume‐rhizobium symbiosis: nodule‐specific cysteine‐rich peptides and beyond. New Phytol. 211, 411–417. [DOI] [PubMed] [Google Scholar]
- Arnold, M.F.F. , Penterman, J. , Shabab, M. , Chen, E.J. and Walker, G.C. (2018) Important late‐stage symbiotic role of the Sinorhizobium meliloti exopolysaccharide succinoglycan. J. Bacteriol. 200, e00665–00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi, J.F. , Barre, A. , Ben Amor, B. et al . (2006) The Medicago truncatula lysin motif‐receptor‐like kinase gene family includes NFP and new nodule‐expressed genes. Plant Physiol. 142, 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson, E.M. and Long, S.R. (1992) Homology of Rhizobium meliloti NodC to polysaccharide polymerizing enzymes. Mol. Plant Microbe Interact. 5, 439–442. [DOI] [PubMed] [Google Scholar]
- Becker, A. , Kleickmann, A. , Kuster, H. , Keller, M. , Arnold, W. and Puhler, A. (1993c) Analysis of the Rhizobium meliloti genes exoU, exoV, exoW, exoT, and exoI involved in exopolysaccharide biosynthesis and nodule invasion: exoU and exoW probably encode glucosyltransferases. Mol. Plant Microbe Interact. 6, 735–744. [DOI] [PubMed] [Google Scholar]
- Bono, J.J. , Fliegmann, J. , Gough, C. and Cullimore, J. (2018) Expression and function of the Medicago truncatula lysin motif receptor‐like kinase (LysM‐RLK) gene family in the legume‐rhizobia symbiosis. In The Model Legume Medicago truncatula . ( de Bruijn, F. ed.). Hoboken, NJ: John Wiley and Sons. [Google Scholar]
- Breakspear, A. , Liu, C.W. , Roy, S. et al . (2014) The root hair “infectome" of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell, 26, 4680–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewin, N.J. (2004) Plant cell wall remodelling in the Rhizobium‐Legume symbiosis. Crit. Rev. Plant Sci. 23, 293–316. [Google Scholar]
- Buendia, L. , Girardin, A. , Wang, T.M. , Cottret, L. and Lefebvre, B. (2018) LysM Receptor‐like Kinase and LysM Receptor‐like protein families: an update on phylogeny and functional characterization. Front. Plant Sci. 9, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burris, R.H. (1974) Biological nitrogen fixation, 1924–1974. Plant Physiol, 54, 443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H.P. and Walker, G.C. (1998) Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti . J. Bacteriol. 180, 5183–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X.F. , Wang, M.Y. , Lee, H.K. , Tadege, M. , Ratet, P. , Udvardi, M. , Mysore, K.S. and Wen, J.Q. (2014) An efficient reverse genetics platform in the model legume Medicago truncatula . New Phytol. 201, 1065–1076. [DOI] [PubMed] [Google Scholar]
- Damiani, I. , Drain, A. , Guichard, M. et al . (2016) Nod Factor effects on root hair‐specific transcriptome of Medicago truncatula: Focus on plasma membrane transport systems and reactive oxygen species networks. Front Plant Sci. 7, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dénarié, J. , Debellé, F. and Promé, J.‐C. (1996) Rhizobium lipo‐chitooligosaccharide nodulation factors: signalling molecules mediating recognition and morphogenesis. Annu. Rev. Biochem. 65, 503–535. [DOI] [PubMed] [Google Scholar]
- d'Erfurth, I. , Cosson, V. , Eschstruth, A. , Lucas, H. , Kondorosi, A. and Ratet, P. (2003) Efficient transposition of the Tnt1 tobacco retrotransposon in the model legume Medicago truncatula . Plant J. 34, 95–106. [DOI] [PubMed] [Google Scholar]
- Fournier, J. , Timmers, A.C.J. , Sieberer, B.J. , Jauneau, A. , Chabaud, M. and Barker, D.G. (2008) Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol. 148, 1985–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier, J. , Teillet, A. , Chabaud, M. , Ivanov, S. , Genre, A. , Limpens, E. , de Carvalho‐Niebel, F. and Barker, D.G. (2015) Remodeling of the infection chamber before infection thread formation reveals a two‐step mechanism for rhizobial entry into the host legume root hair. Plant Physiol. 167, 1233–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage, D.J. (2004) Infection and invasion of roots by symbiotic, nitrogen‐fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68, 280–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibelin‐Viala, C. , Amblard, E. , Puech‐Pages, V. et al . (2019) The Medicago truncatula LysM receptor‐like kinase LYK9 plays a dual role in immunity and the arbuscular mycorrhizal symbiosis. New Phytol. 223, 1516–1529. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. and Walker, G.C. (1989) A novel exopolysaccharide can function in place of the calcofluor‐binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti . Cell, 56, 661–672. [DOI] [PubMed] [Google Scholar]
- Gonzalez, J.E. , Reuhs, B.L. and Walker, G.C. (1996) Low molecular weight EPS II of Rhizobium meliloti allows nodule invasion in Medicago sativa . Proc. Natl Acad. Sci. USA, 93, 8636–8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González, J.E. , Semino, C.E. , Wang, L.X. , Castellano‐Torres, L.E. and Walker, G.C. (1998) Biosynthetic control of molecular weight in the polymerization of the octasaccharide subunits of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti . Proc. Natl Acad. Sci. USA, 95, 13477–13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough, C. , Cottret, L. , Lefebvre, B. and Bono, J.J. (2018) Evolutionary history of plant LysM receptor proteins related to root endosymbiosis. Front. Plant Sci., 9, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins, J.P. , Geddes, B.A. and Oresnik, I.J. (2017) Succinoglycan production contributes to acidic pH tolerance in Sinorhizobium meliloti Rm1021. Mol. Plant Microbe Interact. 30, 1009–1019. [DOI] [PubMed] [Google Scholar]
- Hellriegel, H. and Wilfarth, H. (1888) Untersuchungen uber die Stickstoff‐nahrung der Gramineen und Leguminosen. D. R.: Beilageheft zu der Zeitschrift des Vereins f. d. Rübenzuckerindustrie d. [Google Scholar]
- Hoffmann, B. , Trinh, T.H. , Leung, J. , Kondorosi, A. and Kondorosi, E. (1997) A new Medicago truncatula line with superior in vitro regeneration, transformation, and symbiotic properties isolated through cell culture selection. Mol. Plant Microbe Interact. 10, 307–315. [Google Scholar]
- Jacobs, T.W. , Egelhoff, T.T. and Long, S.R. (1985) Physical and genetic map of a Rhizobium meliloti nodulation gene region and nucleotide sequence of nodC . J. Bacteriol. 162, 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet, A. , Sigaud, S. , Van de Sype, G. , Puppo, A. and Herouart, D. (2003) Expression of the bacterial catalase genes during Sinorhizobium meliloti‐Medicago sativa symbiosis and their crucial role during the infection process. Mol. Plant Microbe Interact. 16, 217–225. [DOI] [PubMed] [Google Scholar]
- Jamet, A. , Kiss, E. , Batut, J. , Puppo, A. and Herouart, D. (2005) The katA catalase gene is regulated by OxyR in both free‐living and symbiotic Sinorhizobium meliloti . J. Bacteriol. 187, 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardinaud, M.F. , Boivin, S. , Rodde, N. et al . (2016) A laser dissection‐RNAseq analysis highlights the activation of cytokinin pathways by Nod Factors in the Medicago truncatula root epidermis. Plant Physiol. 171, 2256–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, K.M. (2012) Increased production of the exopolysaccharide succinoglycan enhances Sinorhizobium meliloti 1021 symbiosis with the host plant Medicago truncatula . J. Bacteriol. 194, 4322–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, K.M. , Kobayashi, H. , Davies, B.W. , Taga, M.E. and Walker, G.C. (2007) How rhizobial symbionts invade plants: the Sinorhizobium‐Medicago model. Nat. Rev. Microbiol. 5, 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, K.M. , Sharopova, N. , Lohar, D.P. , Zhang, J.Q. , VandenBosch, K.A. and Walker, G.C. (2008) Differential response of the plant Medicago truncatula to its symbiont Sinorhizobium meliloti or an exopolysaccharide‐deficient mutant. Proc. Natl Acad. Sci. USA, 105, 704–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, K.M. , Mendis, H.C. and Queiroux, C. (2013) Single‐plant, sterile microcosms for nodulation and growth of the legume plant Medicago truncatula with the rhizobial symbiont Sinorhizobium meliloti . J. Vis. Exp. 80, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaharada, Y. , Kelly, S. , Nielsen, M.W. et al . (2015) Receptor‐mediated exopolysaccharide perception controls bacterial infection. Nature, 523, 308–312. [DOI] [PubMed] [Google Scholar]
- Kelly, S.J. , Muszynski, A. , Kawaharada, Y. , Hubber, A.M. , Sullivan, J.T. , Sandal, N. , Carlson, R.W. , Stougaard, J. and Ronson, C.W. (2013) Conditional requirement for exopolysaccharide in the Mesorhizobium‐Lotus symbiosis. Mol. Plant Microbe Interact. 26, 319–329. [DOI] [PubMed] [Google Scholar]
- Klein, S. , Hirsch, A.M. , Smith, C.A. and Signer, E.R. (1988) Interaction of nod and exo Rhizobium meliloti in alfalfa nodulation. Mol. Plant Microbe Interact. 1, 94–100. [DOI] [PubMed] [Google Scholar]
- Larrainzar, E. , Riely, B.K. , Kim, S.C. et al . (2015) Deep sequencing of the Medicago truncatula root transcriptome reveals a massive and early interaction between Nodulation factor and ethylene signals. Plant Physiol. 169, 233–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman, A.P. and Long, S.R. (2013) Exopolysaccharides from Sinorhizobium meliloti can protect against H2O2‐dependent damage. J. Bacteriol. 195, 5362–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh, J.A. , Signer, E.R. and Walker, G.C. (1985) Exopolysaccharide‐deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl Acad. Sci. USA, 82, 6231–6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh, J.A. , Reed, J.W. , Hanks, J.F. , Hirsch, A.M. and Walker, G.C. (1987) Rhizobium meliloti mutants that fail to succinylate their Calcofluor‐binding exopolysaccharide are defective in nodule invasion. Cell, 51, 579–587. [DOI] [PubMed] [Google Scholar]
- Leong, S.A. , Williams, P.H. and Ditta, G.S. (1985) Analysis of the 5' regulatory region of the gene for d‐aminolevulinic acid synthetase of Rhizobium meliloti . Nucleic Acids Res. 13, 5965–5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge, P. , Roche, P. , Faucher, C. , Maillet, F. , Truchet, G. , Prome, J.C. and Dénarié, J. (1990) Symbiotic host‐specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature, 344, 781–784. [DOI] [PubMed] [Google Scholar]
- Liu, C.W. , Breakspear, A. , Guan, D. et al . (2019) NIN acts as a network hub controlling a growth module required for rhizobial infection. Plant Physiol. 179, 1704–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaret‐Oliver, I. , Lei, W. , Parada, M. et al . (2012) Sinorhizobium fredii HH103 does not strictly require KPS and/or EPS to nodulate Glycyrrhiza uralensis, an indeterminate nodule‐forming legume. Arch. Microbiol. 194, 87–102. [DOI] [PubMed] [Google Scholar]
- Meade, H.M. , Long, S.R. , Ruvkun, G.B. , Brown, S.E. and Ausubel, F.M. (1982) Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149, 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendis, H.C. , Queiroux, C. , Brewer, T.E. , Davis, O.M. , Washburn, B.K. and Jones, K.M. (2013) The succinoglycan endoglycanase encoded by exoK is required for efficient symbiosis of Sinorhizobium meliloti 1021 with the host plants Medicago truncatula and Medicago sativa (Alfalfa). Mol. Plant Microbe Interact. 26, 1089–1105. [DOI] [PubMed] [Google Scholar]
- Mendis, H.C. , Madzima, T.F. , Queiroux, C. and Jones, K.M. (2016) Function of succinoglycan polysaccharide in Sinorhizobium meliloti host plant invasion depends on succinylation, not molecular weight. MBio, 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montiel, J. , Szucs, A. , Boboescu, I.Z. , Gherman, V.D. , Kondorosi, E. and Kereszt, A. (2016) Terminal bacteroid differentiation is associated with variable morphological changes in legume species belonging to the inverted repeat‐lacking clade. Mol. Plant Microbe Interact. 29, 210–219. [DOI] [PubMed] [Google Scholar]
- Montiel, J. , Downie, J.A. , Farkas, A. , Bihari, P. , Herczeg, R. , Balint, B. , Mergaert, P. , Kereszt, A. and Kondorosi, E. (2017) Morphotype of bacteroids in different legumes correlates with the number and type of symbiotic NCR peptides. Proc. Natl Acad. Sci. USA, 114, 5041–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, J.D. (2011) Invasion by invitation: rhizobial infection in legumes. Mol. Plant Microbe Interact. 24, 631–639. [DOI] [PubMed] [Google Scholar]
- Niehaus, K. , Kapp, D. and Puhler, A. (1993) Plant defence and delayed infection of alfalfa pseudonodules induced by an exopolysaccharide (EPS I)‐deficient Rhizobium meliloti mutant. Planta, 190, 415–425. [Google Scholar]
- Oldroyd, G.E. , Murray, J.D. , Poole, P.S. and Downie, J.A. (2011) The rules of engagement in the legume‐rhizobial symbiosis. Annu. Rev. Genet. 45, 119–144. [DOI] [PubMed] [Google Scholar]
- Parada, M. , Vinardell, J.M. , Ollero, F.J. et al . (2006) Sinorhizobium fredii HH103 mutants affected in capsular polysaccharide (KPS) are impaired for nodulation with soybean and Cajanus cajan. Mol. Plant Microbe Interact. 19, 43–52. [DOI] [PubMed] [Google Scholar]
- Pellock, B.J. , Cheng, H.P. and Walker, G.C. (2000) Alfalfa root nodule invasion efficiency is dependent on Sinorhizobium meliloti polysaccharides. J. Bacteriol. 182, 4310–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, N.K. , Frost, J.W. and Long, S.R. (1986) A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science, 233, 977–980. [DOI] [PubMed] [Google Scholar]
- Reinhold, B.B. , Chan, S.Y. , Reuber, T.L. , Marra, A. , Walker, G.C. and Reinhold, V.N. (1994) Detailed structural characterization of succinoglycan, the major exopolysaccharide of Rhizobium meliloti Rm1021. J. Bacteriol. 176, 1997–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber, T.L. and Walker, G.C. (1993) Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti . Cell, 74, 269–280. [DOI] [PubMed] [Google Scholar]
- Roux, B. , Rodde, N. , Jardinaud, M.F. et al . (2014) An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser‐capture microdissection coupled to RNA sequencing. Plant J. 77, 817–837. [DOI] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. et al . (2012) Fiji: an open‐source platform for biological‐image analysis. Nat. Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J. , Rueden, C.T. , Hiner, M.C. and Eliceiri, K.W. (2015) The ImageJ ecosystem: an open platform for biomedical image analysis. Mol. Reprod. Dev. 82, 518–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel, E. , Journet, E.P. , de Carvalho‐Niebel, F. , Duc, G. and Frugoli, J. (2005) The Medicago truncatula SUNN gene encodes a CLV1‐like leucine‐rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 58, 809–822. [DOI] [PubMed] [Google Scholar]
- Smit, P. , Limpens, E. , Geurts, R. , Fedorova, E. , Dolgikh, E. , Gough, C. and Bisseling, T. (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 145, 183–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki, T. , Yoro, E. and Kawaguchi, M. (2015) Leguminous plants: Inventors of root nodules to accommodate symbiotic bacteria. Int. Rev. Cell Mol. Biol. 316, 111–158. [DOI] [PubMed] [Google Scholar]
- Tadege, M. , Wen, J. , He, J. et al . (2008) Large‐scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula . Plant J. 54, 335–347. [DOI] [PubMed] [Google Scholar]
- Timmers, A.C. , Auriac, M.C. and Truchet, G. (1999) Refined analysis of early symbiotic steps of the Rhizobium‐Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development, 126, 3617–3628. [DOI] [PubMed] [Google Scholar]
- Vasse, J. , de Billy, F. , Camut, S. and Truchet, G. (1990) Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J. Bacteriol. 172, 4295–4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Velzen, R. , Holmer, R. , Bu, F.J. et al . (2018) Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen‐fixing rhizobium symbioses. Proc. Natl Acad. Sci. USA, 115, E4700–E4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zeijl, A. , den Camp, R.H.M.O. , Deinum, E.E. , Charnikhova, T. , Franssen, H. , den Camp, H.J.M.O. , Bouwmeester, H. , Kohlen, W. , Bisseling, T. and Geurts, R. (2015) Rhizobium lipo‐chitooligosaccharide signaling triggers accumulation of cytokinins in Medicago truncatula roots. Mol. Plant, 8, 1213–1226. [DOI] [PubMed] [Google Scholar]
- Wells, D.H. and Long, S.R. (2002) The Sinorhizobium meliloti stringent response affects multiple aspects of symbiosis. Mol. Microbiol. 43, 1115–1127. [DOI] [PubMed] [Google Scholar]
- Xiao, T.T. , Schilderink, S. , Moling, S. , Deinum, E.E. , Kondorosi, E. , Franssen, H. , Kulikova, O. , Niebel, A. and Bisseling, T. (2014) Fate map of Medicago truncatula root nodules. Development, 141, 3517–3528. [DOI] [PubMed] [Google Scholar]
- Yang, C. , Signer, E.R. and Hirsch, A.M. (1992) Nodules initiated by Rhizobium meliloti exopolysaccharide mutants lack a discrete, persistent nodule meristem. Plant Physiol. 98, 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York, G.M. and Walker, G.C. (1997) The Rhizobium meliloti exoK gene and prsD/prsE/exsH genes are components of independent degradative pathways which contribute to production of low‐molecular‐weight succinoglycan. Mol. Microbiol. 25, 117–134. [DOI] [PubMed] [Google Scholar]
- York, G.M. and Walker, G.C. (1998) The succinyl and acetyl modifications of succinoglycan influence susceptibility of succinoglycan to cleavage by the Rhizobium meliloti glycanases ExoK and ExsH. J. Bacteriol. 180, 4184–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Plant performance and nodulation phenotypes on alfalfa cv. Iroquois and M. truncatula A17 of S. meliloti strains that make EPSII (galactoglucan).
Figure S2. Information for all S. meliloti strains and plasmids, both those are discussed in the main text and in Supporting Information.
Figure S3. Information on the genetic modifications in strains 961, 1317, exoKdel/exsH‐1325 and exoHKdel/exsH‐1345 with the exoLAMON operon under the control of a heterologous promoter.
Figure S4. Infections formed by the exoY and exoHKdel/exsH‐1345 mutants on Medicago truncatula Jemalong A17.
Figure S5. Complementation of strains with an exoHK deletion by a plasmid carrying exoH.
Figure S6. Phylogenetic tree of MtLYK10 homologs from a variety of plant species, mostly Rosid dicots, but also two monocots, Oryza sativa and Brachypodium distachyon, and with MtLYK11 as an outgroup.
Figure S7. qRT‐PCR expression analysis of MtLYK10 in Medicago truncatula WT (R108) and the Mtlyk10 mutant.
Figure S8. Comparison of the infection and nodulation phenotype of two independent Mtlyk10 mutant lines (Mtlyk10#2 and Mtlyk10#46) and a homozygous MtLYK10‐WT line (MtLYK10‐WT), all selected from the backcross between the Mtlyk10 mutant and R108.
Figure S9. Kinetics of nodule formation when the nodulation defects of exo mutants are rescued in trans by co‐inoculation with a nodC mutant.
Figure S10. Nodules formed after mixed inoculation on Medicago truncatula WT R108 plants.
Figure S11. The exoU mutant of S. meliloti does not produce detectable succinoglycan.
Data Availability Statement
Supporting Information is listed below and these figures are provided for this article. Image acquisition and modification information has been described in Experimental Procedures or in Figure legends.


