Abstract
Radioproteomics is the integration of proteomics, the systematic study of the protein expression of an organism, with radiomics, the extraction and analysis of large numbers of quantitative features from medical images. This article examines this developing field, and it’s application in high grade serous ovarian carcinoma. Seminal proteomic studies in the area of ovarian cancer, such as the PROVAR and CPTA studies are discussed, along side recent research, such as that highlighting the central role of methyltransferase nicotinamide N-methyltransferase as the metabolic regulation of cancer progression in the tumour stroma. Finally, this article considers a novel, hypothesis generating approach to integrate CT-based qualitative and radiomic features with proteomic analysis, and the future direction of the field. Combined advances in radiomic, proteomic and genomic analysis has the potential to signal the age of true precision medicine, where treatment is centered specifically on the molecular profile of the tumour, rather than based on empirical knowledge, thus altering the course of a disease that has the highest mortality of all cancers of the female reproductive system.
Proteomics
Proteomics is the systematic study of the complete complement of proteins (proteome) of organisms. Research primarily focused on the discovery and validation of protein markers can be used for diagnostic and prognostic purposes in various diseases, including patients with high-grade serous ovarian cancer (HGSOC).1,2
Using mass-spectrometry (MS) or antibody-based assays of a tissue’s proteome, it is possible to identify, measure and validate the presence of therapeutically traceable pathways, which may not be represented or interpretable through genomic analysis alone, and this has been seen in combined proteomic and genomic analysis of the same tumour.3,4 Proteomics supplement genomic analysis by revealing the pathways and processes of ovarian cancer that correspond with clinical phenotypes.5 HGSOC, relative to other cancers, has a low mutation rate, which is associated with a high degree of copy number variations (CNV) and remarkable genomic disarray.6 Gonçalves et al7 analyzed the impact of CNVs on the proteome of tumour samples from patients with breast, ovarian, and colorectal cancers. They showed that disagreement existed between the CNV at a genomic level and protein abundance in the tumours, with 23–33% of proteins undergoing post-translational regulation to attenuate the impact of CNVs in changes that would not be visible through genomic analysis alone.7
Proteomic features of ovarian cancer
The PROVAR (PRotein-driven index of OVARian cancer) study used proteomic markers to predict recurrence and survival in 412 ovarian cancer cases from The Cancer Genome Atlas (TCGA).1 Five proteins (phosphorylated epidermal growth factor receptor (pEGFR); phosphorylated tafazzin (pTAZ); BH3-interacting domain death agonist (BID); 70 kilodalton heat shock protein (HSP70) and androgen receptor (AR)) were associated with longer progression free survival (PFS) and overall survival (OS).1 In contrast, four proteins (eukaryotic translation elongation factor 2 (EEF2), signal transducer and activator of transcription 5 α (STAT5 α); phosphorylated protein kinase C- α (pPKCα); and phosphorylated dual-specificity mitogen-activated protein kinase 1 (pMEK1)) were associated with shorter PFS and OS.1
TCGA is a invaluble resource in the study of ovarian cancer, and it’s potential for proteomic research was once again evident in work by the clinical proteomic tumour analysis consortium (CPTAC).5 Using mass spectrometry, they performed a comprehensive proteomic characterisation of 174 ovarian tumours, 169 of which were HGSOC. The proteomic data were integrated with the previously acquired genomic data to assess the effect of different copy number alterations (CNA) on the proteome, and those proteins associated with chromosomal instability and short overall survival. They observed that proteins associated with cell invasion and migration, and immune function, were enriched in association with multiple CNAs.5 Protein abundance in tumours of short survivors (<3 years) were compared with those of long survivors (>5 years). In short survivors, serum response factor (SRF), a proliferation-associated transcription factor and the proteins it regulates were found to be more abundant.5 Integrated proteomic, phosphoproteomic and transcriptomic analysis identified an association between pathways related to increased invasiveness and motility, and short survival; these pathways included PDGFR-beta signalling pathway (associated with angiogenesis), and rhoA-regulatory and integrin-like kinase pathways (both associated with cell mobility and invasion).5
Other proteins also recognized in CPTAC’s analysis of ovarian tumour samples have been previously shown to have an oncogenic association in other cancers. These include: catenin B2, a tumour suppressor whose loss is associated with increased migration and invasion properties in laryngeal cancer8; rho GDP dissociation inhibitor 2 (RhoGDI2), a protein involved in the regulation of cell migration and cell cycle progression, whose overexpression correlates with a poor prognosis in pancreatic carcinoma9; protein kinase C and casein kinase substrate in neurons (PACSIN2), a tumour suppressor involved in repressing cell migration associated with the oncogenic effects of cyclin D110; and RAN, a member of the rasGTPase super-family of proteins, highly expressed in high-grade and stage serous ovarian carcinoma, and associated with poor prognosis.11 The abundance of these cell migration and invasion factors corroborates the theory of haematogenous spread in the metastasis of ovarian cancer.12
The CPTAC’s database has proven to be a rich resource that research groups can access. One such team, Yu et al13, investigated the proteomic signatures of 130 patients from the CPTAC database using supervised machine-learning techniques. Their work showed that the expression of several proteins was highly associated with platinum resistance.13 Upregulation of KRT19 (a member of the type-1 cytokeratins) involved in tumour invasion and metastasis, which is present in many cancers, was also associated with early disease recurrence.13
Eckert et al14 analysed laser microdissected cancer cells, allowing compartment-resolved proteomic analysis of both tumour and stromal compartments from the whole spectrum of HGSOC, from serous tubal carcinoma in situ (STIC) to metastatic tumours. Their work showed that, between patients, there was a significant variation in the protein signatures of tumour compartments, emphasizing the molecular heterogeneity of HGSOC.14 Analysis also revealed that, although the tumour proteome remained remarkably unchanged as it progressed from STIC to metastatic disease, the metastasis-related stromal proteome had a distinct, highly conserved protein signature containing the methyltransferase nicotinamide N-methyltransferase (NNMT).14 The expression of NNMT was a crucial metabolic regulator of the cancer-associated fibroblast (CAF) and its associated cytokines.14 The presence of NNMT supported the migration of cancer cells, as well as proliferation and metastasis.14 Through the action of NNMT in CAFs, the level of S-adenosyl homocysteine (SAM—the universal methyl donor for histones, non-histone proteins, DNA, RNA, and others) was reduced.14 This resulted in a reduction in histone methylation and widespread gene expression changes in the tumour stroma.14
Although predominantly found in metastatic sites, a subset of the primary tumour sites did contain a high stromal expression of NNMT.14 Using the tissue microarrays employed to validate the presence of NNMT, an analysis of the potential of NNMTs to serve as a prognosticator in chemotherapy-naive HGSOC was conducted. It showed that high levels of NNMT in the stroma of the primary tumour correlated with platinum resistance and a significantly worse recurrence-free survival and OS.14 There were no proteomic distinctions between the protein environment of STIC or metastatic tumours, suggesting that STIC already possesses the machinery and capability to become metastatic.14
Integrating proteomic analysis with radiomic features
A recent study integrated CT-based qualitative and radiomic features with proteomic analysis.15 Tissue samples and images from 20 patients with HGSOC awaiting primary cytoreductive surgery were analyzed, with associations found between the presence of certain proteins and CT-based qualitative traits, specifically: Melanoma Antigen Gene A4 (MAGEA4a) which was positively correlated with supradiaphragmatic lymphadenopathy, and cysteine rich protein 2 (CRIP2), which was negatively correlated with mesenteric disease.15 In addition, proteomic data were integrated with radiomic features that capture tumour intra- and intersite heterogeneity (cluster-site entropy, cluster standard deviation, and cluster dissimilarity), and showed that low levels of argininosuccinate synthase 1 (ASS1) were associated with more heterogenous tumours.15 This is consistent with previous studies of ASS1 that show low levels of the protein correlate with more heterogenous tumours, platinum therapy resistance and ultimately a poor prognosis.16
Lu and colleagues developed a quantitative radiomics prognostic vector that predicts survival in patients with epithelial ovarian cancer and is able to identify patients with a poor overall survival of less than 2 years.17 The authors discovered a significant correlation between the radiomics prognostic vector and the tumour tissue fibronectin abundance,17 a protein known to promote tumour migration and invasion.18
Future directions
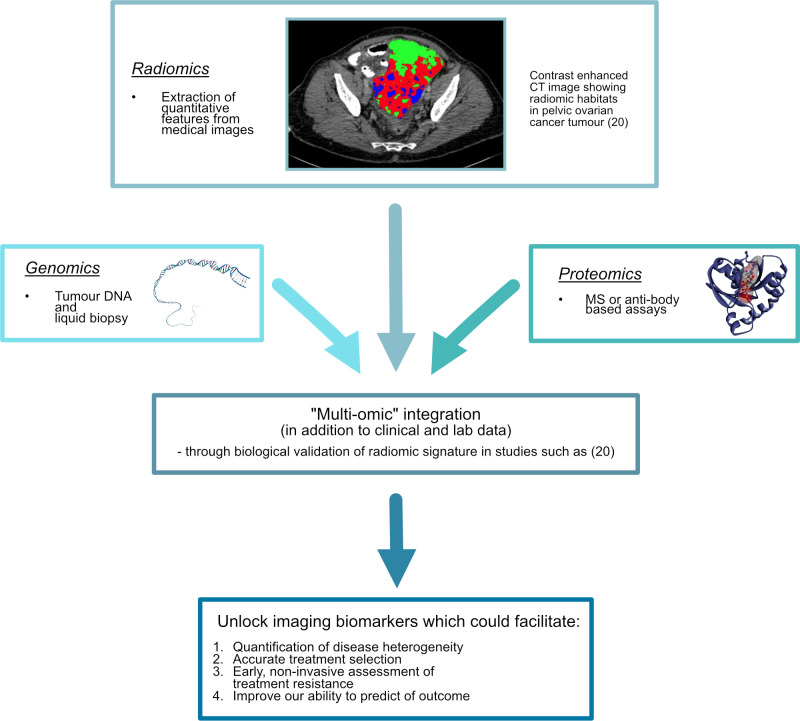
In the future, it is hoped that advances in the areas of proteomics, radiomics, and genomics will herald an age of precision medicine, where treatment is centered specifically on the molecular profile of the tumour, rather than being based on empirical knowledge.2,19 Precision tissue sampling techniques that use CT, MRI or PET-based radiomic tumour habitats for ultrasound-guided targeted biopsies will enable the prospective analysis of relevant tumour regions.20 Large-scale prospective trials like that of the APOLLO group (Applied Proteogenomics OrganizationaL Learning and Outcomes), which integrate these three disciplines (Figure 1), will give clearer insight into the pathways of ovarian cancer and unearth new therapeutic possibilities.19 Such studies could also allow the discovery and full validation of indicators of disease heterogeneity, treatment resistance, and predictors of outcome, thus altering the course of a disease that remains the most common cause of death from gynaecological cancer worldwide.19,21
Figure 1.
Schematic summary integrating proteomics, genomics and radiomics.
Footnotes
Acknowledgements: This work was partially supported by The Mark Foundation for Cancer Research and Cancer Research UK Cambridge Centre [C9685/A25177], the Wellcome Trust Innovator Award [RG98755] and the CRUK National Cancer Imaging Translational Accelerator (NCITA) [C42780/A27066].
Additional support was also provided by the National Institute of Health Research (NIHR) Cambridge Biomedical Research Centre (BRC-1215-20014).
Contributor Information
Cathal McCague, Email: cm2074@medschl.cam.ac.uk.
Lucian Beer, Email: lucian.beer@meduniwien.ac.at.
REFERENCES
- 1.Yang J-Y, Yoshihara K, Tanaka K, Hatae M, Masuzaki H, Itamochi H, et al. Predicting time to ovarian carcinoma recurrence using protein markers. J Clin Invest 2013; 123: 5410–50. doi: 10.1172/JCI74035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinker K, Chin J, Melsaether AN, Morris EA, Moy L. Precision medicine and Radiogenomics in breast cancer: new approaches toward diagnosis and treatment. Radiology 2018; 287: 732–47. doi: 10.1148/radiol.2018172171 [DOI] [PubMed] [Google Scholar]
- 3.Zhang B, Whiteaker JR, Hoofnagle AN, Baird GS, Rodland KD, Paulovich AG. Clinical potential of mass spectrometry-based proteogenomics. Nat Rev Clin Oncol 2019; 16: 256–68. doi: 10.1038/s41571-018-0135-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nougaret S, McCague C, Tibermacine H, Vargas HA, Rizzo S, Sala E. Radiomics and radiogenomics in ovarian cancer: a literature review. Abdom Radiol 2020;11 Nov 2020. doi: 10.1007/s00261-020-02820-z [DOI] [PubMed] [Google Scholar]
- 5.Zhang H, Liu T, Zhang Z, Payne SH, Zhang B, McDermott JE, et al. Integrated Proteogenomic characterization of human high-grade serous ovarian cancer. Cell 2016; 166: 755–65. doi: 10.1016/j.cell.2016.05.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network . Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474: 609–15. doi: 10.1038/nature10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonçalves E, Fragoulis A, Garcia-Alonso L, Cramer T, Saez-Rodriguez J, Beltrao P. Widespread post-transcriptional attenuation of genomic copy-number variation in cancer. Cell Syst 2017; 5: 386–98. doi: 10.1016/j.cels.2017.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fanjul-Fernández M, Quesada V, Cabanillas R, Cadiñanos J, Fontanil T, Obaya A, et al. Cell-Cell adhesion genes CTNNA2 and CTNNA3 are tumour suppressors frequently mutated in laryngeal carcinomas. Nat Commun 2013; 4: 2531. doi: 10.1038/ncomms3531 [DOI] [PubMed] [Google Scholar]
- 9.Yi B, Zhang Y, Zhu D, Zhang L, Song S, He S, et al. Overexpression of RhoGDI2 correlates with the progression and prognosis of pancreatic carcinoma. Oncol Rep 2015; 33: 1201–6. doi: 10.3892/or.2015.3707 [DOI] [PubMed] [Google Scholar]
- 10.Meng H, Tian L, Zhou J, Li Z, Jiao X, Li WW, et al. Pacsin 2 represses cellular migration through direct association with cyclin D1 but not its alternate splice form cyclin D1b. Cell Cycle 2011; 10: 73–81. doi: 10.4161/cc.10.1.14243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrès V, Ouellet V, Lafontaine J, Tonin PN, Provencher DM, Mes-Masson A-M. An essential role for Ran GTPase in epithelial ovarian cancer cell survival. Mol Cancer 2010; 9: 272. doi: 10.1186/1476-4598-9-272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pradeep S, Kim SW, Wu SY, Nishimura M, Chaluvally-Raghavan P, Miyake T, et al. Hematogenous metastasis of ovarian cancer: rethinking mode of spread. Cancer Cell 2014; 26: 77–91. doi: 10.1016/j.ccr.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu K-H, Levine DA, Zhang H, Chan DW, Zhang Z, Snyder M. Predicting ovarian cancer patients' clinical response to platinum-based chemotherapy by their tumor proteomic signatures. J Proteome Res 2016; 15: 2455–65. doi: 10.1021/acs.jproteome.5b01129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckert MA, Coscia F, Chryplewicz A, Chang JW, Hernandez KM, Pan S, et al. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature 2019; 569: 723–8. doi: 10.1038/s41586-019-1173-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beer L, Sahin H, Bateman NW, Blazic I, Vargas HA, Veeraraghavan H, et al. Integration of proteomics with CT-based qualitative and radiomic features in high-grade serous ovarian cancer patients: an exploratory analysis. Eur Radiol 2020; 30: 4306–16. doi: 10.1007/s00330-020-06755-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholson LJ, Smith PR, Hiller L, Szlosarek PW, Kimberley C, Sehouli J, et al. Epigenetic silencing of argininosuccinate synthetase confers resistance to platinum-induced cell death but collateral sensitivity to arginine auxotrophy in ovarian cancer. Int J Cancer 2009; 125: 1454–63. doi: 10.1002/ijc.24546 [DOI] [PubMed] [Google Scholar]
- 17.Lu H, Arshad M, Thornton A, Avesani G, Cunnea P, Curry E, et al. A mathematical-descriptor of tumor-mesoscopic-structure from computed-tomography images annotates prognostic- and molecular-phenotypes of epithelial ovarian cancer. Nat Commun 2019; 10: 764. doi: 10.1038/s41467-019-08718-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yousif NG. Fibronectin promotes migration and invasion of ovarian cancer cells through up-regulation of FAK-PI3K/Akt pathway. Cell Biol Int 2014; 38: 85–91. doi: 10.1002/cbin.10184 [DOI] [PubMed] [Google Scholar]
- 19.Fiore LD, Rodriguez H, Shriver CD. Collaboration to accelerate Proteogenomics cancer care: the Department of Veterans Affairs, department of defense, and the National cancer Institute's applied Proteogenomics organizational learning and outcomes (Apollo) network. Clin Pharmacol Ther 2017; 101: 619–21. doi: 10.1002/cpt.658 [DOI] [PubMed] [Google Scholar]
- 20.Beer L, Martin-Gonzalez P, Delgado-Ortet M, Reinius M, Rundo L, Woitek R, et al. Ultrasound-Guided targeted biopsies of CT-based radiomic tumour habitats: technical development and initial experience in metastatic ovarian cancer. Eur Radiol 2021; 31: 3765–72. doi: 10.1007/s00330-020-07560-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lheureux S, Gourley C, Vergote I, Oza AM. Epithelial ovarian cancer. Lancet 2019; 393: 1240–53. doi: 10.1016/S0140-6736(18)32552-2 [DOI] [PubMed] [Google Scholar]