Abstract
Binding interactions of the spike proteins of the severe acute respiratory syndrome corona virus 2 (SARS-CoV-2) to a peptide fragment derived from the human angiotensin converting enzyme 2 (hACE2) receptor are investigated. The peptide is employed as capture moiety in enzyme linked immunosorbent assays (ELISA) and quantitative binding interaction measurements that are based on fluorescence proximity sensing (switchSENSE). In both techniques, the peptide is presented on an oligovalent DNA nanostructure, in order to assess the impact of mono- versus trivalent binding modes. As the analyte, the spike protein and several of its subunits are tested as well as inactivated SARS-CoV-2 and pseudo viruses. While binding of the peptide to the full-length spike protein can be observed, the subunits RBD and S1 do not exhibit binding in the employed concentrations. Variations of the amino acid sequence of the recombinant full-length spike proteins furthermore influence binding behavior. The peptide was coupled to DNA nanostructures that form a geometric complement to the trimeric structure of the spike protein binding sites. An increase in binding strength for trimeric peptide presentation compared to single peptide presentation could be generally observed in ELISA and was quantified in switchSENSE measurements. Binding to inactivated wild type viruses could be shown as well as qualitatively different binding behavior of the Alpha and Beta variants compared to the wild type virus strain in pseudo virus models.
Subject terms: Nanostructures, Nanoscale biophysics, DNA nanostructures, Viral proteins, Biological physics
Introduction
The pandemic of the severe acute respiratory syndrome corona virus 2 (SARS-CoV-2) has hit the world severely, causing over 6 million deaths until May 20221. While the response was originally focused on prevention by individual precautions (e.g.2,3), the development of effective vaccines was a major step towards overcoming the coronavirus disease 19 (COVID-19)4. Still, there is evidence that the virus will become endemic5, therefore, any deeper understanding of the fundamental behavior of the virus is essential and SARS-CoV-2 will remain a relevant topic for science. While vaccination prevents severe cases of COVID-196–8, the search for effective treatment and therapeutics continues and remains challenging (e.g9). The understanding of binding interactions of the spike protein on the surface of the virions to the receptor on the surface cells in the respiratory tract is of essential importance. Here we characterize a peptide mimicking the targeted receptor and its binding to the spike protein under varying circumstances. Different subfragments of the spike protein as well as full-length proteins with and without amino acid modifications are tested for their binding behavior towards peptide-DNA nanostructures, which are used as structural scaffolds for investigating the impact of oligovalent binding modes. Furthermore, we investigate binding of inactivated wild type SARS-CoV-2 and pseudo viruses expressing the wild type and two mutations of the spike protein.
Peptide-based structures that bind virus surface proteins are valuable tools for investigating binding interactions and can potentially be used in therapeutics and diagnostics for viral infections such as COVID-19 as well10,11. In particular, peptide fragments that mimic the binding site of the viral protein on the targeted cellular structure can be used as “decoy” moieties to inhibit the attachment and fusion of virus particles with host cells12–16, or potentially serve as tools in analytical investigations of specific virus-host interactions17. In the simplest form, these comprise the amino acid sequence in the region of the targeted protein, which is directly involved in the binding interaction with the viral protein. For SARS-CoV, such peptides have been derived from the sequence of the human angiotensin converting enzyme 2 (hACE2) receptor. Several hACE2-derived sequences between 23 and 35 amino acids in length, derived by alanine scan mutagenesis analysis, were reported by Han et al.18 in 2006, which showed inhibitory activity against SARS-CoV, responsible for the 2002–2003 outbreak, with IC50 values ranging from 0.1 to 50 µM. For the currently relevant SARS-CoV-2, in-silico methods have been used to assist in the development of peptides19–22. The peptide SBP1 was developed by Zhang et al.21 from the alpha1 helix sequence of the N-terminal region of hACE2 and is made up of 23 amino acids. The hACE2 constitutes a known receptor for the spike protein of the new SARS-CoV-223,24. According to Zhang et al., SBP1 showed µM binding affinity to the receptor-binding domain (RBD) of SARS-CoV-2 from one supplier21. We characterize the peptide further and compare its binding to different subunits of the spike protein, as well as the full-length protein with and without modifications to the amino acid sequence.
Here, we utilize DNA nanostructures as structural scaffolds for enabling the presentation of multiple SBP1 peptides, arranged in an approximate geometric complement to the targeted binding sites on the spike protein. This strategy takes advantage of cooperative, oligovalent binding, which is a fundamental principle in both natural and engineered biological systems to enhance binding affinity between ligands and multimeric receptors25. Especially in the case of ligands that bind to virus surface proteins, small DNA-based nanostructures have been used to arrange carbohydrate26–29 and peptide14,15 moieties, enhancing the overall binding interaction beyond that provided by typically weak monovalent interactions. While in this work, the DNA structure is used as an analytical device to quantify oligovalent binding interactions, a hypothetic therapeutic application of oligovalent DNA nanostructures would require stabilization in order to avoid enzymatic digestion (e.g. Phosphorothioate-Oligonucleotides30).
Here a four-arm DNA nanostructure, formed by the hybridization of four, partially complementary DNA oligonucleotides, was employed as a structural scaffold. The DNA nanostructures are a modification of the ones presented by Moeser et al.31. In that case, a trimeric DNA structure was used to analyze oligovalent binding of a peptide to EphrinA2 receptors. The authors characterized the DNA structures showing assembly and stability, as well as atomic force microscopy images. Similar characteristics are expected for the adapted structure carrying the SBP1 peptide. The four-arm DNA structure has been previously employed by Kruse et al.17 in the case of an influenza A virus-binding peptide. The melting temperature of the fully double stranded structure was calculated to lie at ~ 84 °C.
In our binding assays, the DNA-peptide nanostructures are a substitute for the natural target, here the hACE2 receptor. One arm enables immobilization of the entire construct to a substrate (e.g. gold electrode or surface of an ELISA plate), while the three remaining arms carry the SBP1 peptide. This trimeric peptide presentation is meant to form an approximate geometric complement to the homo trimeric structure of the spike protein of SARS-CoV-2. The binding behavior of the trimer was determined and compared to the binding of single SBP1 peptide to the spike protein.
For the binding measurements we applied two measurement techniques, namely enzyme-linked immunosorbent assays (ELISA) and switchSENSE technology. By employing these two assays, we demonstrate how the concept of DNA-templated oligovalent interactions with virus proteins can be effectively integrated into both, a universally common measurement method available to nearly any biological facility (ELISA) as well as a more specialized, quantitative approach (switchSENSE). Generally, the methods here can be transferred to any analytical method that involves detecting a target analyte in solution using a surface-bound capture moiety, such as surface plasmon resonance (SPR) or even lateral flow assays.
In ELISA, the receptor, in this case the DNA-peptide nanostructure, is immobilized on a surface to act as capture molecule for binding the antigen, which is free in solution. An antibody is subsequently added that also binds the antigen at a different epitope. Then typically a secondary antibody that carries an enzyme is added and binds to the first antibody, if it is available. An enzymatic substance is then added to induce a visible color change. A microplate reader then measures the absorption. The results are interpreted semi-quantitatively. The absorbance signal should increase with increasing concentrations of the added analyte, in the case of a binding event. A higher absorbance signal is interpreted as an enhanced binding. ELISAs can in principle be used to determine quantitative values, but this requires a calibration curve32. This was not done here. ELISAs provide reliable, reproducible, economical and controllable results of binding, and are an ubiquitously employed analytical method, here primarily used for a semi-quantitative assessment of binding33.
switchSENSE technology on the other hand is a more quantitative technique, that measures association and dissociation rate constants34,35. Here the receptor or capture molecule is coupled to a single stranded DNA strand and then immobilized on gold electrodes by conjugate hybridization to an anchored complementary DNA strand. The anchor strand carries a fluorescent molecule at the distal end. A binding event is then detected in real time by a change in the fluorescence intensity upon binding36.
Results and discussion
Binding of DNA nanostructures to fragments of the SARS-CoV-2 spike protein and to full-length spike proteins with and without amino acid sequence variations
In order to functionalize the DNA strands with SBP1, copper-free click chemistry was utilized. It has proven to provide reliable receptor-mimicking constructs for peptide-virus interactions in previous studies17 and is in general favorable for the synthesis of oligovalent DNA-peptide nanostructures31,37. The DNA-SBP1 strands were assembled to DNA-SBP1 nanostructures by hybridization of four partially complementary DNA strands using a temperature ramp. Coupling of SBP1 to the DNA strands as well as correct folding of the structures was analyzed by native PAGE (see Supporting Information SI3 and SI4). The DNA nanostructure was optimized for the application in switchSENSE technology as it is made up of three peptide-coupled DNA strands and a fourth strand, that contains the complementary strand to the 48 base long anchor (NL-B48) on the gold electrodes of the measurement chip. The construct can therefore be immobilized by conjugate hybridization on the chip surface. After the hybridization, the protein was injected into the flow channel and binding was detected in the instrument’s static mode (fluorescence proximity sensing). In this measurement mode, the DNA strand is kept in an upright position by an applied negative direct voltage (U = − 0.1 V). The binding interaction is then observed by a change in the fluorescence intensity that is caused by the close chemical proximity of the binding partner36. In this case, the binding resulted in a strong decrease of the fluorescence intensity.
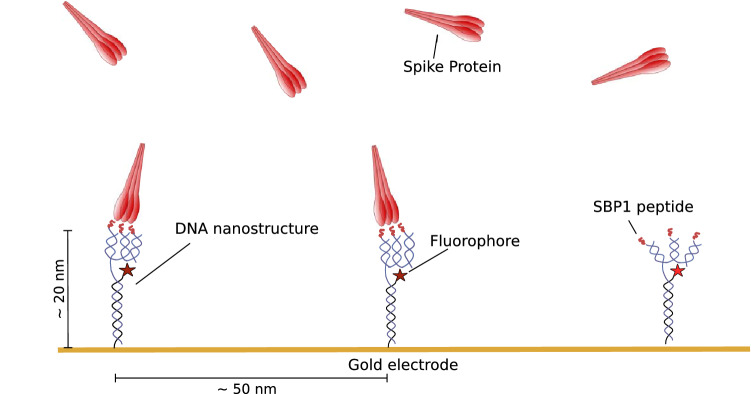
In a first step, we immobilized the four-arm DNA nanostructure that presents three SBP1 peptides and then observed the binding to the SARS-CoV-2 spike protein. The setup is presented schematically in Fig. 1.
Figure 1.
Schematic overview of the experimental setup. Oligovalent DNA structures are immobilized on a gold sensor surface and the binding of spike protein to the oligovalently presented SBP1 peptide is observed. A binding event leads to a reduction of detected fluorescent light intensity. The picture is not drawn to scale.
Two different subunits of the SARS-CoV-2 spike protein as well as the full-length protein itself were investigated. The protein has been studied intensively38 and relevant regions for binding have been determined. We focused our work primarily on the receptor-binding domain (RBD), subunit 1 (S1) and the full-length protein. Full-length protein in this case refers to the ectodomain (ECD). The spike glycoprotein is divided into the subunit 1 (S1) and the subunit 2 (S2). S1 contains the RBD that has been reported to bind to the hACE2-receptor39.
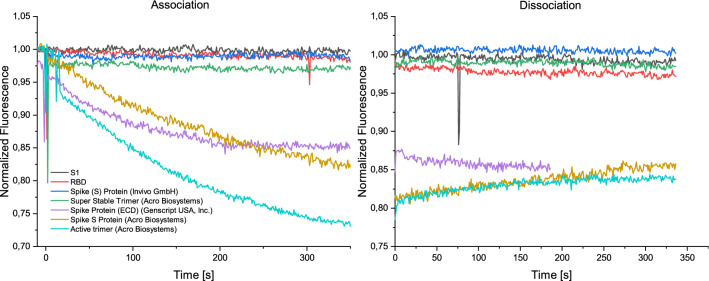
According to Zhang et al., the binding of the SBP1 peptide to the receptor binding domain of the spike protein is weak and for some samples nonexistent. Our experiments confirmed this observation. The peptide presented on the trimeric DNA nanostructure showed no binding to RBD from two different sources up to high concentrations, neither in switchSENSE, nor in ELISA. A similar observation was made for the S1 domain of the spike protein. However, the peptide did bind some of the full-length spike proteins as can be seen in Fig. 2.
Figure 2.
Binding measurements of spike protein and different subunits to SBP1 peptide. Association and dissociation of different subunits of the spike protein, as well as of full-length spike proteins with varying amino acid sequences were analyzed. All proteins were injected at a concentration of 100 nM. The SBP1-peptide-DNA-nanostructure bound the full-length spike proteins from Genscript USA, Inc. and the proteins “SARS-CoV-2 S protein” and “SARS-CoV-2 S protein, active trimer” by Acro Biosystems. The “Super Stable Trimer” by Acro Biosystms and the “Spike (S) Protein” by Invivo BioTech Services GmbH did not show any binding to the peptide. The two proteins have variations in their amino-acid sequences that seem to disturb the binding.
Only the full-length protein “SARS-CoV-2 Spike protein (ECD)” of Genscript USA, Inc. and the proteins “SARS-CoV-2 S protein” and “SARS-CoV-2 S protein, active trimer” by Acro Biosystems lead to a strong fluorescence intensity change upon injection.
According to the manufacturers, the proteins were produced in varying expression systems that do not correlate with binding or non-binding behavior of the particular fragments (see “Materials and methods” section, Table 1). The expression system therefore is most likely not responsible for whether a protein binds to the receptor.
Table 1.
Spike protein fragments and full-length spike proteins and their specifics.
| Name | AAS/MW | Modifications | Expression system | Producer |
|---|---|---|---|---|
| RBD |
AA 319–519, ~ 30 kDa |
His Tag, Avi Tag | Human | Genscript USA, Inc |
| RBD |
AA 329–538, ~ 30 kDa |
His Tag | Drosophila S2 | Fraunhofer IZI, Leipzig, Germany |
| S1 |
AA 16–685, ~ 79 kDa |
His Tag | Human | Genscript USA, Inc |
| S1 |
AA 15–682, ~ 100 kDa |
His Tag | Drosophila S2 | Fraunhofer IZI, Leipzig, Germany |
| SARS-CoV-2 spike protein (ECD) |
AA 16–1213, ~ 135 kDa |
His Tag, Flag Tag | Sf9 insect | Genscript USA, Inc |
| SARS-CoV-2 S protein |
AA 16 – 1213, ~ 135 kDa |
His Tag | HEK293 | Acro Biosystems |
| SARS-CoV-2 spike (S) protein |
AA 14–1213, ~ 137 kDa |
His Tag, Mutated polybasic/furin cleavage site to alanine & K986P/V987P | HEK293 | Invivo BioTech Services GmbH |
| Active trimer |
AA 16–1213, ~ 138 kDa |
His Tag, Trimer motif (26 AA linker) at C-terminus | HEK293 | Acro Biosystems |
| Super stable trimer |
AA 16–1213, ~ 138 kDa |
F817P, A892P, A899P, A942P, K986P, V987P & alanine substitutions R683A/R685A (furin cleavage site) | HEK293 | Acro Biosystems |
Curiously, two variations of full-length proteins did not bind to the SBP1-DNA nanostructure: the “SARS-CoV-2 S protein, Super Stable Trimer” by Acro Biosystems and the “Spike (S) protein” from Invivo BioTech Services GmbH. These two proteins have several variations in their amino acid sequences, with both containing identical alanine substitutions at the furin cleavage site (R683A and R685A) and two substitutions to proline at K986P and V987P.
The furin cleavage site is known to influence binding behavior40,41. Peacock et al. showed that the polybasic cleavage site is advantageous for human transmission. The variations done on the cleavage site in the two proteins might therefore be a contributing factor for the loss of binding to the hACE2-based peptide.
The amino acids 986 and 987 are located in the region of the Heptad-Repeat Domain 1 (HR-1)42. A substitution of these two consecutive amino acids to two prolines hinders a transformation from the prefusion state to the postfusion state42. The protein that carries these replacements is named S-2P43. The substitution has a stabilizing effect as it conserves the prefusion state of the protein. The stabilization can even be improved as has been done for the “Super Stable Trimer” by Acro Biosystems. Here, four additional amino acids are replaced by proline, namely F817P, A892P, A899P and A942P. This corresponds to the “HexaPro” version of the spike protein described by Hsieh et al.43. The reduced conformational flexibility of the stabilized spike protein might negatively influence the binding behavior.
The measurement results show that the variations at the furin cleavage site and the proline substitutions for stabilization of the prefusion state influence the binding of the proteins to the hACE2-derived SBP1-peptide. Since the proteins are modified in both of these aspects, it is not possible to determine, which of these variations causes the ceasing of the binding or if the combination of both variations is responsible. It would be interesting to investigate a protein that carries only one of the variations, either furin cleavage site variation or stabilization of the prefusion state.
The peptide SBP1 mimics the receptor and even though its alpha-helical conformation is supposed to be very similar to that of the original hACE2 binding domain, there can be small conformational differences influencing the binding behavior of the stabilized versions of the spike protein. A further hypothesis would be that the region of the hACE2 receptor, from which the SBP1 peptide was deduced, might play a role in the fusion process.
The results show that relatively small variations of the protein amino acid sequence can drastically affect binding behavior up to the point where no binding occurs.
DNA nanostructure carrying one or three peptides in comparison
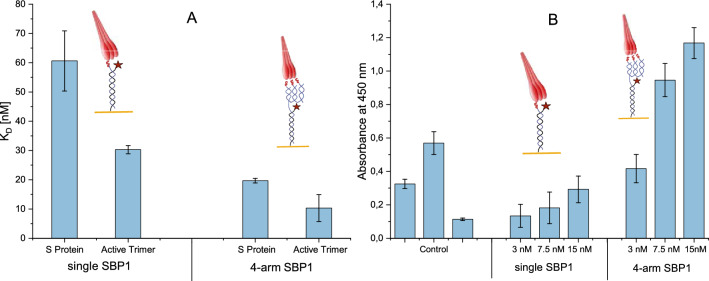
To determine the influence of the trimeric structure of the DNA-peptide-nanostructure, we compared the binding of the full-length spike proteins “SARS-CoV-2 S protein” and “SARS-CoV-2 S protein, active trimer” both by Acro Biosystems to the DNA nanostructure carrying three peptides or just a single peptide using the switchSENSE technology and ELISA. For the switchSENSE experiments, similar experimental parameters were applied to the interaction measurement between a mono- and trivalent SBP1 presentation on the DNA nanostructure as were chosen in “Binding of DNA nanostructures to fragments of the SARS-CoV-2 spike protein and to full-length spike proteins with and without amino acid sequence variations” section. Dissociation times were increased to t = 1200 s and the concentration of spike proteins was varied between 25 and 200 nM. After each binding experiment, the surface was regenerated with fresh DNA-peptide nanostructures since complete dissociation was not reached. As negative control, only the DNA nanostructure without any peptide conjugated was immobilized. The measurement parameters were identical for all measurements. The resulting graphs (see Supporting Information SI1) were then fit with a global mono-exponential function according to standard binding affinity theory44 in order to determine kinetic rates and dissociation constant KD as can be found in Fig. 3A.
Figure 3.
Spike protein binding to three or one peptide per DNA nanostructure. (A) Results from the switchSENSE measurements of the dissociation constants KD for the binding interaction of “SARS-CoV-2 S protein” and “SARS-CoV-2 S protein, active trimer” by Acro Biosystems to the monovalent peptide presenting DNA (single SBP1) and the trimeric peptide presenting DNA structure (4-arm SBP1). Trimeric peptide presentation reduces the dissociation constant by a third compared to the monovalent peptide presentation. The “SARS-CoV-2 S protein, active trimer” binds more strongly than the “SARS-CoV-2 S protein”, presumably due to variations in glycosylation and amino acid sequence variations. (B) Results from ELISA for a control, immobilized DNA with one peptide or three peptides and subsequent binding analysis of the “SARS-CoV-2 S protein, active trimer”. Here as well the trimeric presentation results in a significant increase of the signal compared to the monovalent peptide presentation. Although the experiment was repeated multiple times, the results shown here come from one well plate to allow for comparability.
The experiments were confirmed by ELISA for DNA nanostructures holding three, one and zero peptides, and varying concentrations of “SARS-CoV-2 S protein, active trimer” from Acro Biosystems (see Fig. 3B).
Figure 3A shows the results of the determination of dissociation constants KD for the binding of the “SARS-CoV-2 S protein” and “SARS-CoV-2 S protein, active trimer” by Acro Biosystems to the monovalent peptide presentation (single SBP1) and the trivalent peptide presentation (4-arm SBP1) measured in switchSENSE and ELISA.
As can be seen in Table 2, the dissociation constant of one SBP1-peptide per DNA nanostructure is around three times higher than that of the trivalent peptide-DNA nanostructure for both proteins. It was expected that the oligovalent peptide presentation on the four arm nanostructure would increase affinity (e.g.31,45).
Table 2.
Dissociation constants for monovalent and trivalent peptide presenting DNA nanostructures to two spike proteins.
| KD (nM) monovalent peptide presentation (single SBP1) | KD (nM) trivalent peptide presentation (4-arm SBP1) | |
|---|---|---|
| SARS-CoV-2 S protein | 60.1 ± 10.2 | 19.7 ± 0.8 |
| SARS-CoV-2 S protein, active trimer | 30.3 ± 1.4 | 10.3 ± 4.6 |
In the ELISA, the binding of “SARS-CoV-2 S protein, active trimer” by Acro Biosystems to the one and three peptide-carrying DNA was analyzed. For constructs carrying three peptides, the absorbance showed almost three times the signal strength as compared to the constructs holding one peptide. Nanostructures carrying zero peptides were used as control to confirm specific binding to the hACE2 derived peptide, and exclude unspecific binding to the DNA nanostructures.
The experiments were repeated several times. The spike proteins “SARS-CoV-2 S protein” and “SARS CoV-2 S protein, Active Trimer” were handled according to the manufacturer’s instructions. The lyophilized protein was first resuspended in ddH2O, then aliquoted and finally stored at − 80 °C until use. Nevertheless, it was observed that over time, the activity of the proteins seemed to degrade, which has also been reported in literature43. This degradation resulted in variations of the measured binding affinity and therefore effected the error. Still, we were able to differentiate between the one and three peptides per nanostructure. The trimeric presentation of the peptide always resulted in lower dissociation constants than the monomeric presentation.
DNA-SBP1 nanostructures binding to inactivated viruses
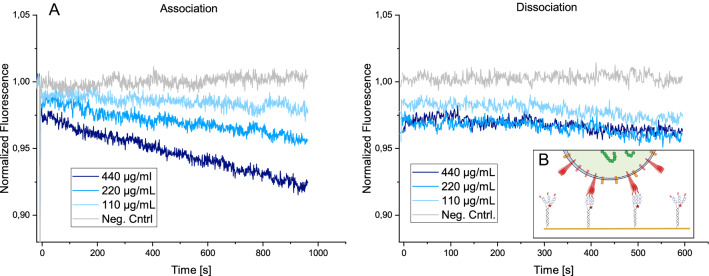
In order to prove that the DNA-peptide-nanostructures bind not only to the individual recombinant spike protein floating freely in solution, but also to the protein as it is found on the surface of the virus, the binding to whole SARS-CoV-2 particles was measured. Viruses were chemically inactivated by ß-propiolactone in order to adhere to biosafety requirements. The DNA construct that displayed three peptides was employed as it had shown to be the strongest binder. The DNA-SBP1 nanostructure was again immobilized by complementary hybridization. Subsequently inactivated SARS-CoV-2 sample was injected at varying concentrations. The virus particles have an average diameter of around 90 nm46, as was also verified by dynamic light scattering (see Supporting Information SI2). The average distance between the DNA nanostructures on the surface of the gold electrode is 50 nm36, therefore, one virus particle can bind to more than one DNA-peptide nanostructure (see Fig. 4B). The result of the binding experiment can be seen in Fig. 4A.
Figure 4.
SARS-CoV-2 virus material binding to SBP1-DNA nanostructure. (A) Association and dissociation of inactivated SARS-CoV-2 to the DNA nanostructure carrying three SBP1 peptides are presented. The association shows a concentration dependent decrease of the fluorescence intensity signal, indicating a correct binding interaction. The dissociation shows almost no signal change presumably due to multivalent binding interactions. (B) Schematic overview of binding process, not drawn to scale.
Figure 4A shows the association and dissociation of the inactivated virus from the DNA-peptide nanostructures. During association, a concentration dependent decrease of fluorescence intensity was observed. The signal change is more linear than exponential. This has been described before for multivalent virus-receptor interactions47. The dissociation shows almost no signal change over the measurement time of around 600 s. This indicates a strong binding interaction, which can be attributed to the high affinity of the individual DNA-peptide nanostructures to the spike protein as well as multivalent binding effects to multiple DNA-peptide nanostructures on the surface, which has been reported for previous virus-receptor interaction studies17. As a negative control, the ssDNA (cNLB48) that is complementary to the anchor strand (NL-B48) and that carries no peptide was hybridized onto the sensor surface and virus solution at a concentration of 220 µg/mL was injected. No signal change was observable upon injection.
DNA-SBP1 nanostructures binding to pseudo viruses
Further experiments were performed on pseudo virus models for SARS-CoV-2. These use Lenti-viruses as a basis, which are genetically modified to express the spike protein on their viral membrane. The pseudo viruses were also ß-propiolactone inactivated and purified prior to the measurement. Then the three SBP1-peptides carrying DNA structures were once again immobilized on the gold electrodes and the pseudo virus solution was injected. The results of the measurements can be found in Fig. 5.
Figure 5.
Pseudo virus—peptide interaction. Lenti-virus based pseudo viruses that carry the SARS-CoV-2 spike protein on their surface were injected and their binding to the DNA nanostructures with three SBP1 peptides was observed. Here as well, concentration dependent linear signal changes in the fluorescence intensity upon injection can be observed in the course of the association.
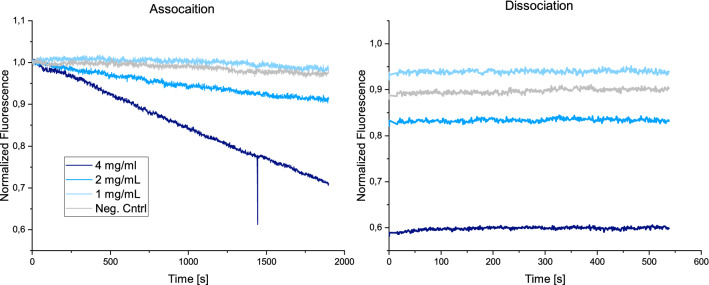
Figure 5 shows the association and dissociation of the pseudo viruses to and from the three-armed DNA-SBP1 nanostructure. Here as well, a strong linear fluorescence intensity change upon injection of the pseudo virus samples could be seen. The concentrations utilized here were higher than in the case of the inactivated original viruses in “DNA-SBP1 nanostructures binding to inactivated viruses” section. This might be due to differences in sample preparation and varying characteristics of the samples as for example the density of spike proteins on the viral surface or varying surface proteins for pseudo viruses and original viruses. After their production, the pseudo viruses were purified, but it is expected that proteins and other residues from the cell medium remained in the sample solution. For the original viruses, which were purified by ultracentrifugation and were delivered in buffer, the purity of the sample was expected to be higher.
The dissociation showed almost no signal change for any of the injected samples. This indicates again a strong binding over the measurement time. Similar to the original viruses, multivalent binding to multiple DNA nanostructures on the surface of the electrode was expected. As a negative control, DNA without peptide (cNLB48) was immobilized and 2 mg mL−1 pseudo virus solution was injected into the channel. There was almost no signal change for the association or the dissociation indicating that the pseudo viruses specifically bound the peptides on the nanostructure. Concentrations below 1 mg mL−1 virus solution resulted in signal changes that were no longer distinguishable from the control.
DNA-SBP1 nanostructure binding to pseudo virus variants
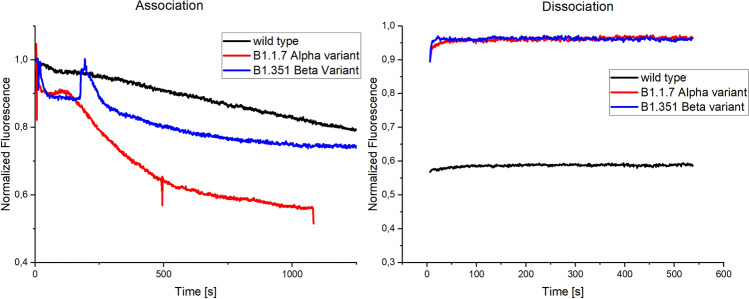
The spike protein on the pseudo viruses can be modified to existing variants of the SARS-CoV-2. The different binding behaviors of the various strains can therefore be analyzed. Here we tested three different spike protein variants namely the wild type (first detected in Wuhan, China), the B1.1.7 or Alpha variant (first detected in the United Kingdom) and the B1.351 or Beta variant (first detected in South Africa)48. The results of the binding interaction measurements indicate a substantially different binding behavior of the variants from the wild type. Although the measurements were not quantitative, a qualitative comparison was possible. The results can be seen in Fig. 6.
Figure 6.
SARS-CoV-2 pseudo virus variants binding to SBP1-peptide. The binding of pseudo viruses that carry spike proteins of three different variants of the SARS-CoV-2 virus to the DNA-peptide nanostructure was determined. Association behavior differed for all three variants. While the wild type showed the typical linear association behavior, Alpha and Beta presented an exponential binding behavior. Spikes in the signal can be attributed to air in the channel system. The Alpha variant induced the strongest signal change over the measurement time. The dissociation displayed almost no signal change for all three samples.
In Fig. 6, the association and the dissociation of the three pseudo virus samples and their binding to the DNA-SBP1 nanostructures is presented. The association showed strong signal changes for all three samples. The injection of the wild type leads to the typical linear binding behavior, while the injection of the Alpha and Beta variant resulted in exponential binding signals. The Alpha variant showed the fastest and strongest association of the three samples although the protein concentration (c) was the lowest (cwild ~ 4 mg mL−1, cAlpha ~ 3 mg mL−1, cBeta ~ 4 mg mL−1). The spikes in the graph can be attributed to air in the tubing. For the dissociation, almost no signal change was visible for all three samples. This again indicates a strong binding behavior.
While the measurements were repeated and the findings could be reproduced, the influence of production and sample differences has to be considered. The different binding behavior of the variants could be an indication for advantageous mutations for the SARS-CoV-2 variants. The findings need further investigations and should be expanded to other variants.
Conclusion
The measurements show that the SBP1 peptide specifically binds distinct full-length spike proteins of SARS-CoV-2. While subunits like the receptor binding domain (RBD) and S1 do not show any binding to the peptide, the unmodified full-length spike proteins bind strongly. A variation of the amino acid sequence at the C-Terminal as was done for one spike protein even increases binding. Substitutions to alanine at the furin cleavage site and variations for stabilization of the prefusion state of the protein on the other hand lead to reduced binding. The measurements suggest that even small variations of the amino acid sequence of the spike protein strongly influence the binding process to the hACE2-derived peptide.
The peptide was coupled to DNA nanostructures that mirror the homo trimeric structure of the spike protein. DNA nanostructures carrying three peptides showed the strongest binding affinity towards the full-length spike proteins “SARS-CoV-2 S protein” and “SARS-CoV-2 S protein, active trimer” by Acro Biosystems. Measurements with one peptide per construct resulted in binding of the protein, but, as expected, the affinity was lower. Influences of the protein degradation have to be considered and lead to measurement errors in the quantitative analysis.
The DNA nanostructure carrying three peptides was furthermore utilized as a ligand for measuring interactions with original SARS-CoV-2 and pseudo viruses. Strong concentration dependent binding interaction signals were visible for the associations. No dissociations were measurable presumably due to multivalent binding and rebinding effects of virus particles to the DNA-peptide nanostructures. Variations of the spike protein on the pseudo virus surface lead to a change in binding behavior. While the wild-type spike protein on Lentivirus based pseudo viruses lead to a linear signal change, spike proteins from the Alpha and Beta variants showed a more exponential behavior. While the findings are purely qualitative, they might point towards a change in binding behavior with possible consequences for the transmissibility and virulence of the variants.
Materials and methods
SBP1-peptide
The peptide SBP1 was developed by Zhang et al.21,22 from molecular dynamic simulations and cryo electron microscopy of the α1 helix sequence of the human angiotensin-converting enzyme 2 (hACE2). The peptide is 23 amino acids long (IEEQAKTFLDKFNHEAEDLFYQS). The peptide was adjusted to carry an azidobutyric acid and 16 PEG-linkers at the N-terminal and it was purchased from Peptide Speciality Laboratories GmbH (Heidelberg, Germany). SBP1 has a molecular mass of 3202 Da.
DNA-peptide nanostructure
DNA-peptide conjugate preparation is based on previously described protocols17,31,37. All DNA strands as presented in Table 3 were purchased from Biomers.net GmbH. Some strands were purchased with an amino group on the 5′ end. These DNA strands were then modified with a dibenzocyclooctyne group (DBCO)-NHS ester (Jena Bioscience GmbH, Jena, Germany) through NHS ester reaction. This allows for the azide-modified hACE2-derived peptide to be conjugated to the DBCO moiety on the oligonucleotide strands, through strain promoted azide-alkyne cycloaddition (SPAAC).
Table 3.
Oligonucleotide sequences to form DNA nanostructures.
| Name | Sequence 5′ → 3′ | Modification |
|---|---|---|
| p-cNL-B48 | acacacacacaactaatcagcgttcgatgcttccgactaatcagccatatcagcttacgacta | 5′ NH2 |
| n | atttagtttctatca | none |
| n*o | tgatagaaactaaatataatatgcgagcca | 5′ NH2 |
| o*p* | tggctcgcatattattagttgtgtgtgtgt | 5′ NH2 |
| NL-B48 | tagtcgtaagctgatatggctgattagtcggaagcatcgaacgctgat | 5′ Biotin |
Part n is complementary to part n*; part o to part o *; p to p* and cNL-B48 is complementary to NL-B48, which is already immobilized on the gold electrode of the switchSENSE device (without Biotin). For ELISAs, NL-B48 was ordered with 5´ Biotin to attach it to NeutrAvidin on the ELISA plate.
Concentration and purity of unmodified and modified ssDNA and DNA structures were determined using a NanoDrop UV/Vis spectrophotometer at 260 nm.
NHS ester reaction was carried out to attach a functional DBCO group to the DNA for later addition of the peptide through copper-free click chemistry reaction. DBCO-NHS ester was dissolved in DMSO to a concentration of 10 mM. 10% of total volume of 10 × PBS was added to amino-coupled oligonucleotides to a final concentration of 1 × PBS, followed by the addition of DBCO-NHS ester in 100 × molar excess compared to the DNA. NHS ester-activated compounds react with amino groups to yield stable amide bonds and the reaction releases N-hydroxysuccinimide (NHS). The solution was incubated overnight at room temperature and purified the next day through ethanol precipitation to remove excess DBCO-NHS ester.
For the conjugation of the DBCO modified oligonucleotides with the hACE2 derived peptide, SPAAC was performed. DBCO is a cyclooctyne derivative, which reacts with azide-coupled peptides to form a stable triazole. The peptide was dissolved in MilliQ water to a concentration of 2 mM. Afterwards, the peptide was added to DBCO-modified DNA in 1 × PBS at an excess between 15x and 30x. The conjugate solution was left to incubate overnight at room temperature and purified the next day through ethanol precipitation to remove excess peptide.
The DNA nanostructures were formed using partially complementary single stranded DNA, and folded in a thermocycler in 1 × PBS. First, the samples were incubated at 54 °C for 15 min, then incubated at 30 °C for 5 min, and then cooled to 4 °C.
Spike proteins
The spike proteins were all used according to the manufacturer’s instructions without further purification.
The molecular weights (MW) of the monovalent proteins as given in Table 1 were used to calculate protein concentrations for binding affinity measurements in “Binding of DNA nanostructures to fragments of the SARS-CoV-2 spike protein and to full-length spike proteins with and without amino acid sequence variations” and “DNA nanostructure carrying one or three peptides in comparison” sections.
Pseudo viruses
Pseudo viruses from preclinics GmbH (Potsdam, Germany) were used. An overview of the production process is given.
Cell culture
293T (ATCC, USA) and 293T-hACE2 (Integral Molecular Inc., Philadelphia, PA, USA) were used for pseudo virus creation and testing. Both strains were grown in DMEM (Corning, USA) supplemented with heat inactivated 10% FCS (Sigma-Aldrich, Darmstadt, Germany) and 1% penicillin/streptomycin (Corning). 293 T-hACE2 cells were additionally supplemented with 10 mM HEPES (Corning) and 0,5% puromycin (LEXSY). Cells were grown at 37 °C in a humidified incubator (Sanyo) with 5% CO2. They were removed and split with accutase (Corning) followed by centrifugation at 4000 rpm for 5 min with a Megafuge 1.0R (Heraeus, Hanau, Germany). Cells were counted with an automatic cell counter (EVE, NanoEnTek) and plated out according to the test conditions.
Generation of different pseudo-viruses
Various pseudo viruses were developed. Each of the pseudo viruses contains a GFP or firefly luciferase (fluc) reporter gene, a lentivirus backbone, and Sars-CoV-2 spike proteins. We followed the ViraSafeTm Lentiviral Packing System protocol from Biocat GmbH (Heidelberg, Germany) for pseudo virus generation.
The lentivirus backbone is from the third generation and can be processed under S2 conditions. Vectorbuilder GmbH (Neu-Isenburg, Germany) and SinoBiological Europe GmbH (Eschborn, Germany) delivered various plasmids containing the spike protein information. After transfection into 70–80% confluent 293 T cells, pseudo viruses were released into the supernatant and were harvested 36–48 h later and stored at 4 °C.
Pseudo virus concentration
All pseudo virus supernatant from the same spike plasmid were pooled and collected. All supernatants were centrifuged at 4000 rpm for 5 min to remove any remaining cell fragments, followed by sterile filtration with a 1.2 µm and a 0.2 µm filter (both filters: Sartorius GmbH, Göttingen, Germany). The sterile fraction was collected and concentrated with a 100 MWCO Spin-X UF centrifugal concentrators (Corning, USA) at 4000 rpm for 30 min.
Virus testing and titer determination
The virus titer was determined as relative activity via firefly luminescence measurement. 293 T-hsACE2 cells were seeded into a 96 well plate (Corning) at a density of 1*105 cells per well. After 5 h, all cells were adherent and were inoculated with the pseudo virus solution. Various dilutions from 1 to 10−9 were used to determine the virus concentration. 48 h post infection, the supernatants were removed and the cells were lysed inside the well with 25 µl of cell lysis buffer (PJK Biotech GmbH, Kleinbittersdorf, Germany) and incubated for 1 h at room temperature. All subsequent processing of samples was done using the beetle-juice luciferase assay firefly (PJK Biotech GmbH). All samples were analysed in a Mithras LB940 microplate reader (Berthold Technologies GmbH & Co.KG) at 560 nm.
Pseudo virus inactivation
The pseudo viruses were treated with ß-propiolactone (Serva Electrophoresis GmbH, Heidelberg, Germany) for inactivation. To validate the efficiency, a stock solution was divided into an untreated fraction (control) and a fraction, which was treated with 0.1% ß-propiolactone. After 16 h of incubation at 4 °C, samples were transferred to 37 °C for 2 h to hydrolyze all residual ß-propiolactone to prevent cytotoxicity to mammalian cells49. After hydrolysis, samples were inoculated onto 293T-hsACE2 cells, and incubated at 37 °C with 5% CO2 in a humidified incubator. 48 h after infection, the cells were observed via luminescence detection in order to determine the infection rate.
Inactivated SARS-CoV-2
Inactivated SARS CoV-2 were provided by Fraunhofer Institute for Cell Therapy and Immunology, Leipzig, Germany. They were ß-propiolactone inactivated and suspended in MES buffer. The concentration prior to inactivation was ~ 1.5 × 106 PFU/mL. The sample was kept at − 80 °C until further use.
SwitchSENSE assays
For binding affinity measurements, a DRX2 instrument (Dynamic Biosensors GmbH, Martinsried, Germany) was employed. The method was used according to standard operating procedure as described by Müller-Landau et al.36. The DNA-peptide nanostructures were immobilized via conjugate hybridization at a concentration of 500 nM over a period of 360 s. All binding affinity measurements were conducted in static mode. As running buffer, PE140 (10 mM Na2HPO4/NaH2PO4, 140 mM NaCl, 0.05% Tween20, 50 µM EDTA, 50 µM EGTA) was utilized and PE40 (10 mM Na2HPO4/NaH2PO4, 40 mM NaCl, 0.05% Tween20, 50 µM EDTA, 50 µM EGTA) was utilized as auxiliary buffer. All analytes were diluted in PE140 for measurements.
All measurements were carried out on standard multipurpose biochips MPC-48–1-R1-S (Dynamic Biosensors GmbH) at 25 °C system temperature. Prior to each interaction measurement, a passivation of the channel by injection of thiol-containing passivation solution (Dynamic Biosensors GmbH) was performed to avoid non-specific binding. Only electrodes that showed a fluorescence amplitude of > 50 kcps in the subsequently performed status tests were used for measurements.
For measurements with spike protein or fragments of the spike protein, an analyte concentration between 25 and 500 nM was chosen. After each interaction measurement the surface was regenerated. The association volume of the protein was chosen at 300 µL and the flow rate was set at 50 µL min−1 for 6 min. The dissociation volume was chosen at 20 mL and the flow rate at 1 mL min−1 for 20 min using the peristaltic pump.
For the measurements with inactivated virus material or pseudo viruses the parameters for the interaction were chosen differently. The association volume was 100 µL and was pumped at a flow rate of 5 µL min−1 for 20 min. The dissociation volume contained 10 mL and had a flow rate of 1 mL min−1 for a time of 10 min using the peristaltic pump. Here as well, the surface was regenerated after each interaction measurement.
ELISA assays
A functional ELISA was performed to analyze the binding parameters (KD value, Bmax value, linear dynamic range). First, a 96-well microtiter plate (Nunc maxisorp, transparent; Thermo Fisher Scientific, USA) was coated with 0.5 µg/well NeutrAvidin (Thermo Fisher Scientific, USA) in 0.1 M NaHCO3 buffer (pH 8.0) and incubated at 4 °C overnight. The plate was washed three times with wash buffer (1 × PBS, 1 mM MgCl2, 0.1% Tween 20, pH 7.4) and 1 µg/well (100 µL/well) of DNA nanostructures was added and incubated for 60 min at room temperature with gentle shaking. The supernatant was then discarded, and the plate was washed three times with 200 µL of blocking buffer (1 × PBS, 1 mM MgCl2, 0.1% Tween 20, 1% BSA, pH 7.4). A 1:2 dilution series of SARS-CoV 2 protein in blocking buffer, between 100 and 0.05 nM was then added and incubated for an additional 60 min at room temperature.
The supernatant was again discarded and the plate was washed three times with blocking buffer. Subsequently, a 6 × anti-His-Tag antibody (Thermo Fisher Scientific, USA) was diluted 1:500 in blocking buffer and 100 µL/well was added. The plate was incubated for 1 h at room temperature in the dark. It was finally washed three times with blocking buffer and 100 µL/well of TMB substrate solution (3,3',5,5'-Tetramethylbenzidine, Pierce™ TMB Substrate Kit—Thermo Fisher Scientific, USA) was added and incubated for 20 min at room temperature in the dark, during which the color of the solution changed from colorless to blue. The reaction was stopped by adding 50 µL/well of 2 NH2SO4, while the color changed from blue to yellow and the absorbance was measured in a microplate reader (Tecan, Germany) at 450 nm and at 520 nm; the latter value was subtracted as a reference.
Supplementary Information
Acknowledgements
This work was supported by the Fraunhofer Internal Programs under Grant No. Anti-Corona 840245. We thank Dr. Alexandra Rockstroh from Fraunhofer IZI (Leipzig, Germany) for providing spike protein and inactivated SARS-CoV-2 samples. We also thank Prof. Frank F. Bier and Dynamic Biosensors GmbH for their support and fruitful discussions.
Author contributions
M.K. wrote the manuscript text and prepared all figures with contributions from B.A., N.G., L.H., C.M. and D.M.S. All switchSENSE experiments and evaluations were performed by M.K. ELISA measurements and DNA-peptide nanostructure preparation were done by B.A., N.G. and C.M. L.H. prepared pseudo virus samples and advised with handling. E.M.L. analyzed and provided theoretical background research. Idea, project conceptualization, Grant acquisition and supervision by E.M.L., R.H. and D.M.S. All authors reviewed and edited the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The datasets generated during and analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
DMS and CM are co-inventors on patent applications WO2018215660A1 and WO2020212576A1 currently pending, which cover the multivalent presentation of virus-binding peptides on DNA scaffolds. All other authors declare no potential conflict of interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-16914-9.
References
- 1.World Health Organisation. Coronavirus (COVID-19) Dashboard. Available at https://covid19.who.int/ (2022).
- 2.Babino A, Magnasco MO. Masks and distancing during COVID-19: A causal framework for imputing value to public-health interventions. Sci. Rep. 2021;11:5183. doi: 10.1038/s41598-021-84679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galeazzi A, et al. Human mobility in response to COVID-19 in France, Italy and UK. Sci. Rep. 2021;11:13141. doi: 10.1038/s41598-021-92399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dagan N, et al. BNT162b2 mRNA Covid-19 vaccine in a nationwide mass vaccination setting. N. Engl. J. Med. 2021;384:1412–1423. doi: 10.1056/NEJMoa2101765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veldhoen M, Simas JP. Endemic SARS-CoV-2 will maintain post-pandemic immunity. Nat. Rev. Immunol. 2021;21:131–132. doi: 10.1038/s41577-020-00493-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polack FP, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voysey M, et al. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: A pooled analysis of four randomised trials. Lancet. 2021;397:881–891. doi: 10.1016/S0140-6736(21)00432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Sahly HM, et al. Efficacy of the mRNA-1273 SARS-CoV-2 vaccine at completion of blinded phase. N. Engl. J. Med. 2021;385:1774–1785. doi: 10.1056/NEJMoa2113017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le BL, et al. Transcriptomics-based drug repositioning pipeline identifies therapeutic candidates for COVID-19. Sci. Rep. 2021;11:12310. doi: 10.1038/s41598-021-91625-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pomplun S, et al. De novo discovery of high-affinity peptide binders for the SARS-CoV-2 spike protein. ACS Central Sci. 2021;7:156–163. doi: 10.1021/acscentsci.0c01309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panda SK, Sen Gupta PS, Biswal S, Ray AK, Rana MK. ACE-2-derived biomimetic peptides for the inhibition of spike protein of SARS-CoV-2. J. Proteome Res. 2021;20:1296–1303. doi: 10.1021/acs.jproteome.0c00686. [DOI] [PubMed] [Google Scholar]
- 12.Alhoot MA, Rathinam AK, Wang SM, Manikam R, Sekaran SD. Inhibition of dengue virus entry into target cells using synthetic antiviral peptides. Int. J. Med. Sci. 2013;10:719–729. doi: 10.7150/ijms.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Memczak H, et al. Anti-hemagglutinin antibody derived lead peptides for inhibitors of influenza virus binding. PLoS ONE. 2016;11:e0159074. doi: 10.1371/journal.pone.0159074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith, D. M. et al.Nanostructure with a Nucleic Acid Scaffold and Virus Binding Peptide Moieties. Patent, WO2018215660A1, 2018. Available at https://patentimages.storage.googleapis.com/49/75/27/5efe02b1905260/WO2018215660A1.pdf (2018).
- 15.Smith, D. M. et al.Biological and Synthetic Molecules Inhibiting Respiratory Syncytial Virus Infection. Patent, WO2020212576A1, 2020. Available at https://patentimages.storage.googleapis.com/38/57/28/c69c84866f848f/WO2020212576A1.pdf (2020).
- 16.Lauster D, et al. Multivalent peptide-nanoparticle conjugates for influenza-virus inhibition. Angewandte Chemie (Int. Ed. Engl.) 2017;56:5931–5936. doi: 10.1002/anie.201702005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kruse M, et al. Measuring influenza A virus and peptide interaction using electrically controllable DNA nanolevers. Adv. Mater. Technol. 2021 doi: 10.1002/admt.202101141. [DOI] [Google Scholar]
- 18.Han DP, Penn-Nicholson A, Cho MW. Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology. 2006;350:15–25. doi: 10.1016/j.virol.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freitas FC, Ferreira PHB, Favaro DC, de Oliveira RJ. Shedding light on the inhibitory mechanisms of SARS-CoV-1/CoV-2 spike proteins by ACE2-designed peptides. J. Chem. Inf. Model. 2021;61:1226–1243. doi: 10.1021/acs.jcim.0c01320. [DOI] [PubMed] [Google Scholar]
- 20.Baig MS, Alagumuthu M, Rajpoot S, Saqib U. Identification of a potential peptide inhibitor of SARS-CoV-2 targeting its entry into the host cells. Drugs R&D. 2020;20:161–169. doi: 10.1007/s40268-020-00312-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang, G. et al.Investigation of ACE2 N-Terminal Fragments Binding to SARS-CoV-2 Spike RBD (2020).
- 22.Pentelute, B. L., Zhang, G., Pomplun, S., Loftis, A. R. & Loas, A. SARS-CoV-2 Spike Protein Binding Peptides. Patent WO2021188966A1, 2021. Available at https://patentimages.storage.googleapis.com/04/d9/b0/46c6a576c7069e/WO2021188966A1.pdf
- 23.Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walls AC, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fasting C, et al. Multivalency as a chemical organization and action principle. Angewandte Chemie (Int. Ed. Engl.) 2012;51:10472–10498. doi: 10.1002/anie.201201114. [DOI] [PubMed] [Google Scholar]
- 26.Yamabe M, Fujita A, Kaihatsu K, Ebara Y. Synthesis of neuraminidase-resistant sialoside-modified three-way junction DNA and its binding ability to various influenza viruses. Carbohydr. Res. 2019;474:43–50. doi: 10.1016/j.carres.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Yamabe M, Kaihatsu K, Ebara Y. Sialyllactose-modified three-way junction DNA as binding inhibitor of influenza virus hemagglutinin. Bioconjug. Chem. 2018;29:1490–1494. doi: 10.1021/acs.bioconjchem.8b00045. [DOI] [PubMed] [Google Scholar]
- 28.Yamabe M, Kaihatsu K, Ebara Y. Binding inhibition of various influenza viruses by sialyllactose-modified trimer DNAs. Bioorg. Med. Chem. Lett. 2019;29:744–748. doi: 10.1016/j.bmcl.2018.12.064. [DOI] [PubMed] [Google Scholar]
- 29.Bandlow V, et al. Spatial screening of hemagglutinin on influenza A virus particles: Sialyl-LacNAc displays on DNA and PEG scaffolds reveal the requirements for bivalency enhanced interactions with weak monovalent binders. J. Am. Chem. Soc. 2017;139:16389–16397. doi: 10.1021/jacs.7b09967. [DOI] [PubMed] [Google Scholar]
- 30.Kilanowska A, Studzińska S. In vivo and in vitro studies of antisense oligonucleotides—A review. RSC Adv. 2020;10:34501–34516. doi: 10.1039/d0ra04978f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Möser C, Lorenz JS, Sajfutdinow M, Smith DM. Pinpointed stimulation of EphA2 receptors via DNA-templated oligovalence. Int. J. Mol. Sci. 2018 doi: 10.3390/ijms19113482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armston A. The Immunoassay Handbook. Elsevier; 2013. [Google Scholar]
- 33.Tighe PJ, Ryder RR, Todd I, Fairclough LC. ELISA in the multiplex era: Potentials and pitfalls. Proteom. Clin. Appl. 2015;9:406–422. doi: 10.1002/prca.201400130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rant U, et al. Switchable DNA interfaces for the highly sensitive detection of label-free DNA targets. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17364–17369. doi: 10.1073/pnas.0703974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langer A, et al. Protein analysis by time-resolved measurements with an electro-switchable DNA chip. Nat. Commun. 2013;4:2099. doi: 10.1038/ncomms3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller-Landau H, Varela PF. Standard operation procedure for switchSENSE DRX systems. Eur. Biophys. J. EBJ. 2021;50:389–400. doi: 10.1007/s00249-021-01519-3. [DOI] [PubMed] [Google Scholar]
- 37.Lorenz JS, et al. Synthetic transient crosslinks program the mechanics of soft, biopolymer-based materials. Adv. Mater. 2018;30:e1706092. doi: 10.1002/adma.201706092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harvey WT, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lan J, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 40.Peacock TP, et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021;6:899–909. doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]
- 41.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia X. Domains and functions of spike protein in Sars-Cov-2 in the context of vaccine design. Viruses. 2021 doi: 10.3390/v13010109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsieh C-L, et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodrich JA, Kugel JF. Binding and Kinetics for Molecular Biologists. CSH Press; 2007. [Google Scholar]
- 45.Bandlow V, et al. Sialyl-LacNAc-PNA⋅DNA concatamers by rolling-circle amplification as multivalent inhibitors of influenza A virus particles. Chembiochem Eur. J. Chem. Biol. 2019;20:159–165. doi: 10.1002/cbic.201800643. [DOI] [PubMed] [Google Scholar]
- 46.Klein S, et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Müller M, Lauster D, Wildenauer HHK, Herrmann A, Block S. Mobility-based quantification of multivalent virus-receptor interactions: New insights into influenza A virus binding mode. Nano Lett. 2019;19:1875–1882. doi: 10.1021/acs.nanolett.8b04969. [DOI] [PubMed] [Google Scholar]
- 48.Tao K, et al. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021;22:757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lei S, Gao X, Sun Y, Yu X, Zhao L. Gas chromatography-mass spectrometry method for determination of β-propiolactone in human inactivated rabies vaccine and its hydrolysis analysis. J. Pharm. Anal. 2018;8:373–377. doi: 10.1016/j.jpha.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and analyzed during the current study are available from the corresponding author on reasonable request.