Abstract
Growth and the production of acetone, butanol, and ethanol by Clostridium beijerinckii NCIMB 8052 on several polysaccharides and sugars were analyzed. On crystalline cellulose, growth and solvent production were observed only when a mixture of fungal cellulases was added to the medium. On lichenan growth and solvent production occurred, but this polymer was only partially utilized. To increase utilization of these polymers and subsequent solvent production, the genes for two new glycoside hydrolases, celA and celD from the fungus Neocallimastix patriciarum, were cloned separately into C. beijerinckii. To do this, a secretion vector based on the pMTL500E shuttle vector and containing the promoter and signal sequence coding region of the Clostridium saccharobutylicum NCP262 eglA gene was constructed and fused either to the celA gene or the celD gene. Stable C. beijerinckii transformants were obtained with the resulting plasmids, pWUR3 (celA) and pWUR4 (celD). The recombinant strains showed clear halos on agar plates containing carboxymethyl cellulose upon staining with Congo red. In addition, their culture supernatants had significant endoglucanase activities (123 U/mg of protein for transformants harboring celA and 78 U/mg of protein for transformants harboring celD). Although C. beijerinckii harboring either celA or celD was not able to grow, separately or in mixed culture, on carboxymethyl cellulose or microcrystalline cellulose, both transformants showed a significant increase in solvent production during growth on lichenan and more extensive degradation of this polymer than that exhibited by the wild-type strain.
The anaerobic conversion of carbohydrates into acetone, butanol, and ethanol, which is known as ABE fermentation, by several strains of Clostridium spp. was first described more than half a century ago. Interest in industrial applications of this process was lost following the development of the petrochemical industry, and currently ABE fermentation plants are operated only in the People's Republic of China (7). Recently, however, there has been increased interest in the production of chemicals and energy by using renewable resources as starting materials. Biomass is a widely available substrate, and utilization of biomass for production of energy carriers is considered an environmentally friendly process (3). For ABE fermentation the most interesting substrates appear to be agricultural or domestic organic wastes due to their low price, wide availability, and appropriate sugar compositions (3, 14). These materials contain mainly two major sugar polymers: cellulose, an insoluble, linear, unbranched homopolysaccharide consisting of glucose units linked by β-1,4 glycosidic bonds; and hemicellulose, which consists of noncellulosic polysaccharides, including xylans, mannans, and glucans. The cellulose fibrils are partially arranged in a crystalline structure, integrated with hemicellulose, and embedded in lignin (a complex polyphenolic compound). Therefore, in contrast to other substrates used in the past, such as starch or molasses, lignocellulosic substrates are very recalcitrant to degradation, and an expensive hydrolysis step is often necessary prior to fermentation.
Many solventogenic clostridia can utilize a wide range of carbohydrates, including mono- and disaccharides (such as glucose, cellobiose, fructose, maltose, or xylose), which are generated during degradation of plant polysaccharides. However, none of the known solventogenic species is able to utilize cellulose, although some of these species do show some cellulase activity (13). With respect to other polysaccharides, most of the solvent-producing clostridia can efficiently ferment starch, and Clostridium acetobutylicum ATCC 824 utilizes xylan, although not very efficiently (17). During the last decade, important advances in our knowledge concerning the physiology and genetics of solventogenic clostridia have been made. Genetic tools for transformation have been developed for a number of strains, including Clostridium beijerinckii NCIMB 8052 (23). Transformation of this strain with suitable genes coding for active extracellular hydrolytic enzymes would increase its substrate utilization range and, eventually, enable it to degrade cellulose and hemicellulose more efficiently. Cloning and expression of the Clostridium cellulovorans engB gene in C. acetobutylicum have been described. However, the recombinant strain exhibited increased extracellular endoglucanase activity but was not able to grow on cellulose as a sole carbon source (12). The engB gene from C. cellulovorans codes for a glycoside hydrolase that belongs to family 5 of glycoside hydrolases and exhibits endoglucanase and xylanase activities but not hydrolytic activity on crystalline cellulose (8).
To provide an alternative approach, we directed our attention to cloning glycoside hydrolase genes from eukaryotic microorganisms and focused on the genes from the anaerobic rumen fungus Neocallimastix patriciarum, which grows on crystalline cellulose as a sole carbon source and exhibits high cellulolytic and hemicellulolytic activities. Another reason why we chose this fungus was its low DNA G+C content and its codon usage, which is compatible with that of C. beijerinckii (30). Several cDNAs coding for cellulases from this fungus have been cloned and functionally expressed in Escherichia coli (6, 29, 30). These cDNAs include the celA gene coding for cellobiohydrolase (EC 3.2.1.91), an exoenzyme that releases cellobiose as a main product from crystalline cellulose (6). N. patriciarum CelA belongs to family 6 of the glycoside hydrolases (5; http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html), and its production is induced during fungal growth in the presence of cellulose. Another well-characterized gene is the N. patriciarum celD gene, which encodes an endoglucanase (EC 3.2.1.4) belonging to family 5 of the glycoside hydrolases (5, 6). CelD is constitutively produced by the fungus and exhibits activity towards carboxymethyl cellulose (CMC) and, to a lesser extent, towards Avicel (microcrystalline cellulose) and amorphous cellulose (29). It is generally assumed that enzymes with complementary activities, such as exoglucanases (cellobiohydrolases) and endoglucanases, act synergistically during degradation of crystalline cellulose.
In this study we describe the growth of and solvent production by C. beijerinckii NCIMB 8052 on different glucose polymers, including lichenan, CMC, and crystalline cellulose. In media with crystalline cellulose, growth and solvent production were observed only when a mixture of fungal cellulases was added to the medium. Subsequently, we cloned the N. patriciarum celA and celD genes individually into C. beijerinckii NCIMB 8052 using a secretion vector. Extracellular production of the fungal enzymes by C. beijerinckii transformants was observed, and substrate utilization by transformants was also studied in different combinations.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
C. beijerinckii NCIMB 8052 was kindly supplied by M. Young (University of Wales, Aberystwyth, United Kingdom). Clostridium saccharobutylicum NCP262 was kindly provided by S. R. Reid (University of Capetown, Capetown, South Africa). Stock cultures were maintained as spore suspensions in sterile 10% (vol/vol) glycerol at −20°C. Spore suspensions were heat shocked for 1 min at 100°C (for strain NCIMB 8052) or for 3 min at 75°C (for strain NCP262) in a water bath prior to inoculation. For production of precultures, vegetative cells were grown in a semisynthetic medium described previously (19) or in clostridial basal medium (CBM) (20) overnight at 37°C. For growth experiments the same media were supplemented with 1.5% (wt/vol) glucose (Merck, Darmstadt, Germany), 3% (wt/vol) cellobiose (Sigma), 6% (wt/vol) Avicel (Merck), 6% (wt/vol) Sigmacell (type 50; Sigma), 1 or 2% (wt/vol) CMC (low or high viscosity; Sigma), or 2% (wt/vol) lichenan (Sigma) as the carbon source, as indicated below. For simultaneous saccharification and fermentation experiments, media were supplemented with Cellulast 1.5L (Novozymes, Bagsvaerd, Denmark) at a concentration of 2% (weight of Celluclast 1.5L/weight of substrate), unless indicated otherwise, at the beginning of the fermentation (zero time). All experiments were performed anaerobically in a Coy anaerobic chamber (Coy Laboratory Products, Grass Lake, Mich.) under a 20% CO2–4% H2–76% N2 atmosphere unless indicated otherwise.
For vector construction E. coli XL1 blue (Stratagene) was used. This strain was usually grown in Luria-Bertani broth as described previously (25). When necessary, media were supplemented with ampicillin (50 μg/ml), isopropyl-β-thiogalactopyranoside (IPTG) (50 μg/ml), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (40 μg/ml), or erythromycin (10 μg/ml).
Plasmids pNPCA (6) and pBSFD (29) containing the cDNA sequences of the celA and celD genes, respectively, were kindly supplied by G.-P. Xue (CSIRO Tropical Agriculture, St. Lucia, Australia). Plasmid pMTL500E (21), used as cloning vector for C. beijerinckii, was kindly supplied by M. Young.
Transformation procedures, DNA manipulation, and PCR.
All general DNA manipulations in E. coli were carried out essentially as described previously (24). Transformation of C. beijerinckii NCIMB 8052 was performed by electroporation as described previously (21).
Restriction endonucleases and modification enzymes were purchased from Roche Diagnostics, Eurogentec, or Qiagen.
DNA isolation from E. coli was performed with a Wizard Plus SV miniprep kit (Promega Inc.). Plasmid DNA was isolated from C. beijerinckii by the modified alkaline lysis method described previously (21). Genomic DNA from C. saccharobutylicum NCP262 was isolated by the method of Pospiech and Neumann (22).
The oligonucleotides used for mutagenesis or PCR were purchased from Eurogentec. The DNA fragments containing the desired mutations were cloned into pGEMT-Easy (Promega Inc.). The mutations were verified by sequencing with an automated laser fluorescent ALF DNA sequencer (Amersham Pharmacia Biotech), using fluorescently labeled M13 universal and reverse primers. The promoter plus signal peptide fragment from the eglA gene of C. saccharobutylicum NCP262 was obtained by PCR as a 0.37-kb XhoI/NcoI fragment. The primers used were based on the sequence of the eglA gene (31) and included forward primer 5′GCCTCGAGCAAACTGCTTCCCCTAATTCCC3′ and reverse primer 5′GCCATGGTTGCAGCTTCAGCTTTATAA3′; XhoI and NcoI sites (underlined) were added at the 5′ and 3′ ends of the fragment, respectively. The NcoI site was added in such a way that it allowed the in-frame fusion of the genes to be cloned downstream. The celA gene was amplified from plasmid pNPCA by PCR performed with forward primer 5′GCACGCGTGTGGTGGTGCCTGGGCTCAATG3′ and reverse primer 5′GCTCTAGATTAAAATGATGGTCTAGC3′; this resulted in introduction of MluI and XbaI sites (underlined) at the 5′ and 3′ ends of the gene, respectively. The celA fragment was cloned after the promoter-signal sequence fragment in pMTL500E as an MluI/XbaI fragment, resulting in pWUR3 (Fig. 1). The celD gene was amplified from plasmid pBFSD by PCR performed with forward primer 5′GCCATGGAAGCTATACGATTTCGAACC3′ and reverse primer 5′GCTCTAGATTAGTTGGTTCTTCTGG3′; this resulted in introduction of NcoI and XbaI sites (underlined) at the 5′ and 3′ ends of the gene, respectively. The 1.2-kb celD fragment obtained was cloned as a NcoI/XbaI fragment after the promoter-signal sequence in pMTL500E, resulting in pWUR4.
FIG. 1.
Schematic representation of the constructions in pMTL500E that generated pWUR3 (A) and pWUR4 (B). Amino acids in boldface type are amino acids of the CelA or CelD protein. The position of the C. saccharobutylicum eglA promoter (P) is indicated, as is the position of the signal sequence (SP). The fungal cDNA is indicated by the striped arrow. The restriction sites in the fusions are underlined.
Analytical methods.
The concentrations of solvents (acetone, butanol, and ethanol) and acids (acetic and butyric acids) were determined in clear supernatants of centrifuged culture samples by high-performance liquid chromatography as described previously by using propionic acid as an internal standard (10). Separation was carried out by using a Shodex KC-311 (Shodex, Tokyo, Japan) column at 80°C and 3 mM H2SO4 as the eluent at a flow rate of 1 ml/min. A refractive index detector (Waters 410; Millipore) and a UV absorbance detector (model VWM2141; Pharmacia) were used in series. The concentrations of most of the metabolites were determined from the refractive index chromatograms; the only exception was the concentration of butyric acid, which was determined from UV chromatograms at 210 nm.
Preparation of culture samples for enzyme assays.
Cells were sedimented by centrifugation at 10,000 × g at 4°C for 15 min. The culture supernatant was collected and concentrated (approximately 30-fold) by ultrafiltration through a Millipore PM10 membrane at 4°C. Low-molecular-weight compounds and medium components were removed from the concentrated material by dilution with 4 volumes of ice-cold 20 mM sodium phosphate buffer (pH 6.0); this was followed by reconcentration by ultrafiltration. This process was repeated three times, and the resulting fractions were used for enzymatic activity determinations. C. beijerinckii cell extracts were prepared as follows. One milliliter of culture cells was harvested by centrifugation, resuspended in 0.1 ml of 50 mM Tris-HCl (pH 8.0) buffer, and sonicated with a Branson Sonifier. Cell debris was removed by centrifugation (10,000 × g for 10 min), and the resulting supernatant was used for enzymatic activity determinations.
Cellulase activity assays.
E. coli and C. beijerinckii transformants grown on agar plates were screened for endoglucanase activity by the Congo red staining method of Teather and Wood (26). Some modifications were made, as follows: CMC was added to the medium at a concentration of 0.1% (wt/vol), and carboxymethyl cellulase (CMCase) activity was detected after 2 to 4 days of incubation at 37°C by washing the cells out of the plates with sterile demineralized water and staining the CMC with Congo red.
Endoglucanase and exoglucanase activities were determined as described previously (14). The following substrates were used (final concentrations): 0.5% (wt/vol) CMC, 0.2% (wt/vol) lichenan (Sigma), 0.5% (wt/vol) laminarin (Sigma), and 0.5% (wt/vol) Avicel in 50 mM citrate buffer (pH 5.7). The substrates were incubated with the enzyme samples for 60 min at 39°C in a water bath. The reducing sugars formed were measured by the 3,5-dinitrosalicylic acid method (9). One unit of activity corresponded to the formation of 1 nmol of reducing sugar (d-glucose) per min. Protein concentrations in the samples were determined by the Bradford assay (Bio-Rad).
RESULTS
Growth of C. beijerinckii NCIMB 8052 on (hemi)cellulosic substrates.
To analyze the potential for acetone, butanol, and ethanol production on substrates other than the well-studied compounds starch and mono- and disaccharides (16), C. beijerinckii NCIMB 8052 was grown on various forms of cellulose and lichenan (Table 1). Because microcrystalline cellulose and lichenan were insoluble, growth on these substrates could not be quantified accurately, and growth was indicated as positive or negative. In media with microcrystalline cellulose (Avicel and Sigmacell) or soluble cellulose (CMC) there was virtually no growth. Very small amounts of solvents were produced, which resulted from utilization of residual glucose in the medium (concentration, less than 4 g/liter), which probably originated from impurities in the substrates added and from the inoculum (10% [vol/vol] from an overnight culture grown in the presence of 6% [wt/vol] glucose). In medium containing cellobiose as a carbon source, the growth and solvent production levels were similar to those in the same media containing glucose (Table 1). Growth and solvent production were also observed in medium containing lichenan, but a residue was left after fermentation, indicating that utilization of this polymer was incomplete.
TABLE 1.
Growth of and production of solvents (acetone and butanol) by C. beijerinckii on semisynthetic medium with different substrates in absence or presence of cellulolytic enzymesa
| Substrate | No addition
|
With
Celluclast 1.5L
|
||
|---|---|---|---|---|
| Growth | Solvent concn (g/liter) | Growth | Solvent concn (g/liter) | |
| None | − | 0.6–0.8 | NDb | ND |
| Glucose | + | 11.1–11.2 | ND | ND |
| Cellobiose | + | 8.9–9.3 | ND | ND |
| Lichenan | + | 4.1–4.3 | + | 4.8–5.1 |
| CMC | − | 1.0–1.2 | − | 1.4–1.8 |
| CMCc | − | 1–1.1 | ||
| Avicel | − | 0.8–1.1 | + | 6.7–7.7 |
| Sigmacell type 50 | − | 0.3–0.7 | + | 5–5.4 |
Semisynthetic media (19) were inoculated (10% vol/vol) with an overnight culture grown on glucose, and the resulting cultures were grown for 96 h in the presence of 6% (wt/vol) glucose, 3% (wt/vol) cellobiose, 2% (wt/vol) lichenan, 2% (wt/vol) CMC, 6% (wt/vol) Sigmacell type 50, or 6% (wt/vol) Avicel. Celluclast 1.5L was added to the cultures at a concentration of 2% (wt/wt), unless indicated otherwise. Growth was monitored by measuring the optical density of a culture at 600 nm (+, optical density at 600 nm of more than 1.0 −, residual growth optical density at 600 nm of 0.01 or less). Normally, the acetone/butanol solvent ratio was 1:4; however, in the presence of lichenan the ratio was 1:3. All experiments were carried out in duplicate.
ND, not determined.
Celluclast 1.5L was added at a concentration of 10% (wt/wt) relative to the concentration of CMC.
In order to study the possibility of increasing solvent production from cellulose (Avicel, Sigmacell, or CMC) or lichenan, a commercial cellulase mixture, Celluclast 1.5L, was added to media along with these substrates at the same time as the inoculum. This enzyme mixture is obtained by fermentation from the fungus Trichoderma reesei and catalyzes the breakdown of cellulose into glucose, cellobiose, and higher glucose polymers since it has mainly cellulase activity and exhibits relatively low β-glucosidase activity (2, 4). When the cellulolytic enzymes were added to medium containing CMC as the sole carbon source, no growth was observed. This did not change even when the initial amount of cellulases was increased fivefold. However, in media containing microcrystalline cellulose supplemented with the cellulolytic enzymes, significant growth and solvent production were observed. The growth of C. beijerinckii on microcrystalline cellulose supplemented with cellulases was slower than the growth on glucose, since the optimal temperature of Celluclast 1.5L is higher (60°C) than the incubation temperature of the cultures (37°C) and growth depended on hydrolysis of cellulose. C. beijerinckii is able to utilize cellobiose, and addition of a purified preparation of fungal β-glucosidases (Novozyme N188) to medium containing microcrystalline cellulose supplemented with Celluclast 1.5L did not significantly affect solvent production (data not shown). Finally, addition of cellulolytic enzymes did not significantly improve the production of solvents on lichenan, and a solid residue was still present in the medium after fermentation.
These results indicate that C. beijerinckii NCIMB 8052 can utilize the products of degradation of crystalline cellulose by the T. reesei cellulases added (Celluclast 1.5L) for growth and solvent production. In addition, C. beijerinckii grew to some extent on lichenan, and solvent production on this polymer increased, although slightly, when the mixture of fungal cellulases was added to the medium. Therefore, we expected that sufficient production of appropriate glycoside hydrolase enzymes by C. beijerinckii should enable it to utilize cellulose or lichenan more efficiently without exogenous addition of enzymes.
Cloning and expression of the N. patriciarum celA and celD cDNAs using a clostridial secretion vector.
To allow extracellular production of fungal glycoside hydrolases by C. beijerinckii, a clostridial secretion vector was designed. This vector was based on the 6.4-kb shuttle vector pMTL500E and the well-characterized promoter and signal peptide coding sequences of the β-1,4-endoglucanase (eglA) gene from C. saccharobutylicum NCP262 (31).
In-frame fusions between the eglA signal peptide sequence and the N. patriciarum celA or celD coding sequence were created by adding adequate restriction sites at the 3′ end of the eglA sequence as well as at the 5′ end of the celA or celD coding sequence (Fig. 1). Transformants of E. coli harboring the resulting 6.8-kb plasmid pWUR3 (celA) or the 6.9-kb plasmid pWUR4 (celD) were readily obtained and were found to have the expected configuration upon sequence analysis of the fusion sites (Fig. 1). Subsequently, E. coli transformants harboring either pWUR3 or pWUR4 were analyzed for production of endoglucanase activity. Both strains were found to produce clear halos around colonies grown in agar plates supplemented with CMC upon staining with Congo red (results not shown). This confirmed that the N. patriciarum celA and celD genes were functionally expressed in E. coli (5, 15).
The expression plasmids pWUR3 and pWUR4 were subsequently introduced by electroporation into C. beijerinckii. While transformants with pWUR4 were obtained at a frequency similar to that observed with cloning vector pMTL500E (∼102 transformants/μg of DNA), the transformation frequency of pWUR3 was more than 10-fold lower. However, restriction analysis showed that the size and architecture of the expression plasmids isolated from C. beijerinckii transformants were as expected in all of the transformant colonies tested.
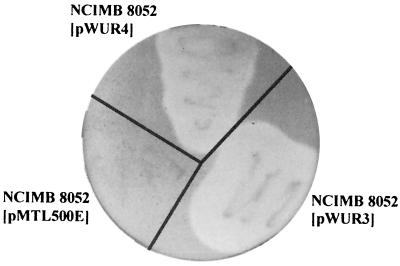
To prevent plasmid loss, erythromycin was added to the media at a concentration of 10 μg/ml when the transformants were cultivated. To detect production of the fungal glycoside hydrolase activity by the C. beijerinckii wild-type strain or the clones harboring pWUR3, pWUR4, or the cloning vector pMTL500E, colonies were grown on agar plates supplemented with 0.2% (wt/vol) CMC. Subsequently, CMCase activity was determined by staining CMC with Congo red (26). Around the colonies of C. beijerinckii harboring pWUR3 or pWUR4 clear halos were visible, indicating that degradation of CMC occurred. These halos were significantly larger than the halos around colonies of the untransformed strain or the transformant harboring pMTL500E (Fig. 2).
FIG. 2.
Endoglucanase activity plate assay with C. beijerinckii NCIMB 8052 harboring pMTL500E, pWUR3 (celA), or pWUR4 (celD). Cells were grown on agar plates supplemented with 0.2% (wt/vol) CMC for 48 h. CMC was stained with Congo red, and hydrolysis of CMC is indicated by clear halos around cells.
Enzymatic degradation of different polymeric substrates was determined in both cell extracts and concentrated extracellular medium from early-stationary-phase cultures of wild-type and recombinant C. beijerinckii grown on CBM with cellobiose or glucose as the carbon source. We determined these activities after 24 h of growth because at this time all of the sugar had been utilized and because in previous studies the highest levels of endoglucanase activity in supernatants of solvent-producing clostridial cultures were found at the end of the exponential growth phase or the early stationary phase (1, 13). No significant differences in glycoside hydrolase activity were found in cultures grown on cellobiose and cultures grown on glucose (results not shown). In the wild-type strain or transformant harboring pMTL500E, no hydrolytic activity with CMC was detectable, but the transformants harboring pWUR3 (celA) or pWUR4 (celD) showed significant endoglucanase activity (Table 2). The CMCase activity was predominantly (around 70% of the total activity) located in the culture supernatant (data not shown). This indicates that the fungal cellulases were produced in an active form and efficiently secreted into the medium. The recombinant enzymes were not detectable on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel, even after silver staining (results not shown).
TABLE 2.
Glucanase specific activities in C. beijerinckii culture supernatantsa
| Substrate | Sp act (U/mg of protein)
|
|||
|---|---|---|---|---|
| Wild type | pMTL500E | pWUR4 | pWUR3 | |
| CMC | <1 | <1 | 78 ± 3 | 123 ± 37 |
| Lichenan | 201 ± 17 | 181 ± 5 | 297 ± 34 | 447 ± 98 |
| Laminarin | 1,271 ± 122 | 1,144 ± 106 | 1,089 ± 57 | 1,395 ± 50 |
Untransformed wild-type and plasmid-harboring cultures were tested for enzymatic activity on CMC, lichenan, and laminarin. One unit of activity corresponded to the release of 1 nmol of reducing sugars per min. Cultures were grown in duplicate, and every sample was assayed in duplicate. The values are means ± standard deviations.
Lichenase activity was found in all cultures of C. beijerinckii, independent of the presence of the plasmids that were constructed. However, higher levels of activity were found in the cultures of C. beijerinckii harboring the fungal cellulase genes. CelA has higher lichenase activity than CelD (6, 29), and in agreement with this, C. beijerinckii harboring pWUR3 showed the highest lichenase activity (Table 2).
The activities of the cultures producing the fungal enzymes were also determined with laminarin, a polymer composed of β-1,3-linked glucose residues. We tested this substrate because we expected that the lichenase activity found in wild-type and recombinant cultures could be due to hydrolysis of β-1,3 linkages in lichenan by clostridial enzymes. We did not find laminarinase activity in cell extracts of E. coli harboring pWUR3 or pWUR4. There was not a significant difference in the laminarinase activities determined for the wild-type and recombinant cultures (Table 2).
Substrate utilization by C. beijerinckii producing fungal glycoside hydrolases.
Cultures of untransformed C. beijerinckii or cultures harboring pMTL500E, pWUR3, or pWUR4 were grown in CBM containing glucose, cellobiose, lichenan, Avicel, or CMC as the sole carbon source. In media containing glucose or cellobiose as the carbon source the transformants grew as well as the wild type. None of the strains grew in media containing CMC or Avicel. Also, cocultures of C. beijerinckii harboring pWUR3 and C. beijerinckii harboring pWUR4 did not grow on these substrates. Cultures were also grown in CBM containing 1% (wt/vol) CMC (high or low viscosity) or 6% (wt/vol) Avicel supplemented with cellobiose or glucose at a concentration of 1 or 0.1% (wt/vol). CMC and Avicel were not utilized by any of the strains, even after extended incubation (14 days). As expected, the growth and solvent production of these cultures were comparable to those of cultures containing only the sugars as carbon sources (results not shown).
In media containing lichenan, cultures of C. beijerinckii harboring the celA or celD gene produced significantly more solvents than the untransformed wild type or the transformant harboring the pMTL500E vector produced (Fig. 3). C. beijerinckii harboring pWUR3 grown on lichenan produced an amount of solvents similar to the amount produced by the wild-type strain grown on glucose at the same concentration in CBM when a 1% (vol/vol) inoculum was used (4.9 and 5.4 g of solvents per liter, respectively). At the concentration used in the media, lichenan was partially insoluble, and after fermentation residual undegraded polymer remained in all of the cultures except those of C. beijerinckii harboring pWUR3 (results not shown). This indicates that there was extensive degradation of this polymer by the fungal cellulase CelA, which can be explained by the high lichenase activity found in cultures of C. beijerinckii expressing the celA gene (Table 2).
FIG. 3.
Production of solvents by and growth of C. beijerinckii NCIMB 8052 carrying no plasmid (×), pMTL500E (□), pWUR4 (▵), or pWUR3 (○) on CBM containing 20 g of lichenan per liter as the sole carbon source. The 1% (vol/vol) inoculum was obtained from an overnight preculture grown in the presence of 1.5% (wt/vol) glucose. Because lichenan was insoluble and cultures were very turbid, growth was estimated by measuring the protein contents of cell extracts of culture samples. Solvents were produced at acetone/butanol ratios between 1:2 and 1:3. Averages based on two independent experiments are shown. The error bars indicate standard deviations based on at least triplicate samples.
DISCUSSION
C. beijerinckii NCIMB 8052 belongs to the group of well-known solventogenic clostridia, and its physiology and substrate utilization range have been the subjects of a number of studies (16). In this study we found that saccharification of cellulose, growth, and solvent production take place simultaneously when the medium is supplemented with fungal cellulolytic enzymes (Table 1). Remarkably, growth on CMC (either high or low viscosity) was not observed even when the same cellulases were added to the medium at high concentrations (Table 1). Since the bacteria can grow in media containing CMC and supplemented with glucose or cellobiose, the presence of toxic impurities in this substrate can be ruled out as an explanation for this observation. It seems likely that the carboxymethyl groups that are present in 7 of the 10 glucose residues in CMC confer a negative charge that could inhibit utilization of the oligosaccharides generated by the bacteria.
Addition of fungal cellulases (Celluclast 1.5L) to a medium with microcrystalline cellulose as the sole carbon source enabled C. beijerinckii to grow on this substrate (Table 1). Celluclast 1.5L contains a mixture of different cellulases, including exo- and endoglucanases, which act synergistically in degradation of sugar polymers (4, 15).
For cloning of the N. patriciarum glycoside hydrolase genes we used the promoter and signal sequences of the eglA gene from C. saccharobutylicum NCP262 (31), which appeared to be adequate to control expression of a cellulase gene. This system has been used recently for cloning of another heterologous gene in C. acetobutylicum (27); the only difference was that the signal peptide sequence used was one codon shorter. In this study we predicted a signal peptide with 38 amino acids (28) and showed that this is effective in directing the secretion of fungal cellulases. Although both CelD and CelA were successfully produced and excreted into the extracellular medium in an active form by C. beijerinckii NCIMB 8052 (Fig. 2 and Table 2), the transformant strains, alone or in combination, failed to utilize Avicel or Sigmacell as a carbon source under the conditions tested. While this finding could be attributed to a number of things, one of the possible explanations is that the levels of the fungal enzymes produced were too low to efficiently degrade cellulose. Moreover, the degradation of cellulose is a complex reaction in which many enzymes are involved, and it is likely that more than the two new fungal glycoside hydrolases is necessary for efficient hydrolysis of this substrate.
In contrast to the results obtained with other solventogenic clostridia (13), no CMCase activity was found in liquid cultures of C. beijerinckii NCIMB 8052 (Table 2) or in plate assays (Fig. 2). The experiment was repeated with a new culture of strain NCIMB 8052 received from the National Collections of Industrial, Food and Marine Bacteria, and the same results were obtained (data not shown). Since CMC is a soluble derivative of cellulose that is considered a typical substrate for endoglucanases, we concluded that this strain does not produce endo-β-1,4-glucanase activity, which contrasts with the conclusions of previous reports (12, 18). C. beijerinckii NCIMB 8052 showed extracellular hydrolytic activity on lichenan (β-1,3/β-1,4 glucan) and laminarin (β-1,3 glucan), indicating that it produces β-1,3-glucanases and lichenases. These enzymes, which have been placed in family 16 of the glycoside hydrolases, are widely distributed in bacteria (11). A number of lichenases (EC 3.2.1.73), endo-β-1,3-glucanases (EC 3.2.1.39), and 1,3(4)-β-glucanases (EC 3.2.1.6) have been characterized at the genetic level in several Bacillus spp. and Clostridium thermocellum. In solventogenic clostridia these activities have not been studied in detail yet, and this is the first report of lichenase and laminarinase activities in C. beijerinckii NCIMB 8052. During growth of wild-type or transformant strains on lichenan, the acetone/butanol ratio in the media varied between 1:2 and 1:3, in contrast to the ratio found during growth on glucose (1:4). The influence of substrate concentration on the solvent ratio has been described previously (25). Transformants harboring pWUR3 or pWUR4 showed increased extracellular lichenase activity as a result of expression of either the celA or celD gene. This increased activity resulted in increased solvent production when lichenan was the substrate (Fig. 3). Only the transformant carrying pWUR3 was able to utilize lichenan completely, which resulted in clear media after fermentation, whereas the turbidity did not disappear in cultures of the other strains. This observation is in agreement with the fact that in cell extracts of E. coli, fungal CelA exhibits approximately fivefold-higher lichenase activity than CelD (6, 29). Remarkably, the solvent production by the pWUR3-harboring transformant on lichenan was comparable to the solvent production by the wild-type strain on glucose.
This is the first example of cloning of cellulase genes from a eukaryotic organism into C. beijerinckii. The N. patriciarum celA and celD genes were functionally expressed in C. beijerinckii NCIMB 8052, and the resulting enzymes were exported to the medium. However, the recombinant strains individually or in cocultures did not grow on microcrystalline cellulose or CMC as a sole carbon source. It is likely that more proteins are needed for efficient degradation of cellulose to support growth. The C. beijerinckii strains producing the fungal enzymes showed increased utilization of lichenan, a polymer very similar to the mixed 1,3-/1,4-β-glucans that are part of the cell walls of cereals such as barley, rice, and sorghum. In conclusion, we show that cloning of fungal genes into Clostridium strains can produce strains with a substrate utilization range different from that of the wild-type strain. This may open possibilities for generating solventogenic strains that are able to grow on polymeric substrates suitable for economically viable ABE fermentation.
ACKNOWLEDGMENTS
We thank J. Springer for technical assistance.
This work was supported in part by an EU Madam Curie fellowship (grant FAIR-CT96-5047) to A. M. López-Contreras.
REFERENCES
- 1.Allcock E R, Woods D R. Carboxymethyl cellulase and cellobiase production by Clostridium acetobutylicumin an industrial fermentation medium. Appl Environ Microbiol. 1981;41:539–541. doi: 10.1128/aem.41.2.539-541.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breuil C, Chan M, Gilbert M, Saddler J N. Influence of β-glucosidase on the filter paper activity and hydrolysis of lignocellulosic substrates. Biores Technol. 1992;39:139–142. [Google Scholar]
- 3.Claassen P A M, van Lier J B, López-Contreras A M, van Niel E W J, Sijtsma L, Stams A J M, de Vries S S, Weusthuis R A. Utilisation of biomass for the supply of energy carriers. Appl Microbiol Biotechnol. 1999;52:741–755. [Google Scholar]
- 4.Claeyssens M, Aerts G. Characterisation of cellulolytic activities in commercial Trichoderma reeseipreparations: an approach using small chromogenic substrates. Biores Technol. 1992;39:143–146. [Google Scholar]
- 5.Coutinho P M, Henrissat B. Carbohydrate-active enzymes: an integrated database approach. In: Gilbert H J, Davies G, Henrissat B, Svensson B, editors. Recent advances in carbohydrate bioengineering. Cambridge, United Kingdom: The Royal Society of Chemistry; 1999. pp. 3–12. [Google Scholar]
- 6.Denman S, Xue G-P, Patel B. Characterization of a Neocallimastix patriciarum cellulase cDNA (celA) homologous to Trichoderma reeseicellobiohydrolase II. Appl Environ Microbiol. 1996;62:1889–1896. doi: 10.1128/aem.62.6.1889-1896.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dürre P. New insights and novel developments in clostridial acetone/butanol/isopropanol fermentation. Appl Microbiol Biotechnol. 1998;49:639–648. [Google Scholar]
- 8.Foong F, Hamamoto T, Osheyov O, Doi R H. Nucleotide sequence and characteristics of endoglucanase gene engB from Clostridium cellulovorans. J Gen Microbiol. 1991;137:1729–1736. doi: 10.1099/00221287-137-7-1729. [DOI] [PubMed] [Google Scholar]
- 9.Ghose T K. Measurement of cellulase activities. Pure Appl Chem. 1987;59:257–268. [Google Scholar]
- 10.Gosselink R J A, van Dam J E G, Zomers F H A. Combined HPLC analysis of organic acids and furans formed during organosolv pulping of fiber hemp. J Wood Chem Technol. 1995;15:1–25. [Google Scholar]
- 11.Gueguen Y, Voorhorst W, van der Oost J, de Vos W M. Molecular and biochemical characterization of an endo-β-1,3-glucanase of the hyperthermophilic archeon Pyrococcus furiosus. J Biol Chem. 1997;50:31258–31264. doi: 10.1074/jbc.272.50.31258. [DOI] [PubMed] [Google Scholar]
- 12.Kim A Y, Attwood G T, Holt S M, White B A, Blaschek H P. Heterologous expression of endo-β-1,4-d-glucanase from Clostridium cellulovorans in Clostridium acetobutylicum ATCC 824 following transformation of the engBgene. Appl Environ Microbiol. 1994;60:337–340. doi: 10.1128/aem.60.1.337-340.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S F, Forsberg C W, Gibbins L N. Cellulolytic activity of Clostridium acetobutylicum. Appl Environ Microbiol. 1985;50:220–228. doi: 10.1128/aem.50.2.220-228.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez-Contreras A M, Claassen P A M, Mooibroek A, de Vos W M. Utilisation of saccharides in extruded domestic organic waste by Clostridium acetobutylicumATCC 824 to produce acetone, butanol and ethanol. Appl Microbiol Biotechnol. 2000;54:162–167. doi: 10.1007/s002530000374. [DOI] [PubMed] [Google Scholar]
- 15.Medve J, Karlsson J, Lee D, Tjerneld F. Hydrolysis of microcrystalline cellulose by cellobiohydrolase I and endoglucanase II from Trichoderma reesei: adsorption, sugar production pattern, and synergism of the enzymes. Biotechnol Bioeng. 1998;59:621–634. [PubMed] [Google Scholar]
- 16.Mitchell W J. Physiology of carbohydrate to solvent conversion by clostridia. Adv Microb Physiol. 1998;39:31–130. doi: 10.1016/s0065-2911(08)60015-6. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell W J, Albasheri K A, Yazdanian M. Factors affecting utilization of carbohydrates by clostridia. FEMS Rev. 1995;17:317–319. [Google Scholar]
- 18.Montoya D, Espitia S, Silva E, Schwarz W H. Isolation of mesophilic solvent-producing clostridia from Colombian sources: physiological characterization, solvent production and polysaccharide hydrolysis. J Biotechnol. 2000;79:117–126. doi: 10.1016/s0168-1656(00)00218-2. [DOI] [PubMed] [Google Scholar]
- 19.Nimcevic D, Schuster M, Gapes J R. Solvent production by Clostridium beijerinckiiNRRL B592 growing on different potato media. Appl Microbiol Biotechnol. 1998;50:426–428. doi: 10.1007/s002530051315. [DOI] [PubMed] [Google Scholar]
- 20.O'Brien R W, Morris J G. Oxygen and the growth and metabolism of Clostridium acetobutylicum. J Gen Microbiol. 1971;68:307–318. doi: 10.1099/00221287-68-3-307. [DOI] [PubMed] [Google Scholar]
- 21.Oultram J D, Loughlin M, Swinfield T-J, Brehm J K, Thompson D E, Minton N P. Introduction of plasmids into whole cells of Clostridium acetobutylicumby electroporation. FEMS Microbiol Lett. 1988;56:83–88. [Google Scholar]
- 22.Pospiech A, Neumann B. A versatile quick-prep of genomic DNA from Gram-positive bacteria. Trends Genet. 1995;11:217–218. doi: 10.1016/s0168-9525(00)89052-6. [DOI] [PubMed] [Google Scholar]
- 23.Reysset G, Sebald M. Transformation/electrotransformation of clostridia. In: Woods D R, editor. The clostridia and bio/technology. Stoneham, Mass: Butterworth-Heinemann; 1993. pp. 151–156. [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 25.Shaheen R, Shirley M, Jones D T. Comparative fermentation studies of industrial strains belonging to four species of solvent-producing clostridia. J Mol Microbiol Biotechnol. 2000;2:115–124. [PubMed] [Google Scholar]
- 26.Teather R M, Wood P J. Use of congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol. 1982;43:777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Theys J, Nuyts S, Landuyt W, van Mellaert L, Dillen C, Böhringer M, Dürre P, Lambin P, Anné J. Stable Escherichia coli-Clostridium acetobutylicumshuttle vector for secretion of murine tumor necrosis factor alpha. Appl Environ Microbiol. 1999;65:4295–4300. doi: 10.1128/aem.65.10.4295-4300.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Von Heijne G. A new method for predicting signal peptide cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue G-P, Gobius K S, Orpin C G. A novel polysaccharide hydrolase cDNA (celD) from Neocallimastix patriciarumencoding three multi-functional catalytic domains with high endoglucanase, cellobiohydrolase and xylanase activities. J Gen Microbiol. 1992;138:2397–2403. doi: 10.1099/00221287-138-11-2397. [DOI] [PubMed] [Google Scholar]
- 30.Xue G-P, Orpin C G, Gobius K S, Aylward J H, Simpson G D. Cloning and expression of multiple cellulase cDNAs from the anaerobic rumen fungus Neocallimatix patriciarum in Escherichia coli. J Gen Microbiol. 1992;138:1413–1420. doi: 10.1099/00221287-138-7-1413. [DOI] [PubMed] [Google Scholar]
- 31.Zappe H, Jones W A, Jones D T, Woods D R. Structure of an endo-β-1,4-glucanase gene from Clostridium acetobutylicum P262 showing homology with endoglucanase genes from Bacillusspp. Appl Environ Microbiol. 1988;54:1289–1292. doi: 10.1128/aem.54.5.1289-1292.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]