Abstract
Tissue‐resident macrophages are present in all tissues where they perform homeostatic and immune surveillance functions. In many tissues, resident macrophages develop from embryonic progenitors, which mature into a self‐maintaining population through local proliferation. However, tissue‐resident macrophages can be supported by recruited monocyte‐derived macrophages during scenarios such as tissue growth, infection, or sterile inflammation. Circulating blood monocytes arise from hematopoietic stem cell progenitors and possess unique gene profiles that support additional functions within the tissue. Determining cell origins (ontogeny) and cellular turnover within tissues has become important to understanding monocyte and macrophage contributions to tissue homeostasis and disease. Fate mapping, or lineage tracing, is a promising approach to tracking cells based on unique gene expression driving reporter systems, often downstream of a Cre‐recombinase–mediated excision event, to express a fluorescent protein. This approach is typically deployed temporally with developmental stage, disease onset, or in association with key stages of inflammation resolution. Importantly, myeloid fate mapping can be combined with many emerging technologies, including single‐cell RNA‐sequencing and spatial imaging. The application of myeloid cell fate mapping approaches has allowed for impactful discoveries regarding myeloid ontogeny, tissue residency, and monocyte fate within disease models. This protocol outline will discuss a variety of myeloid fate mapping approaches, including constitutive and inducible labeling approaches in adult and embryo tissues. This article outlines basic approaches and models used in mice for fate mapping macrophages. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1: Adult Fate Mapping
Basic Protocol 2: Embryonic Fate Mapping
Keywords: fate mapping, macrophage origin, mononuclear phagocytes, reporter mice, tamoxifen
INTRODUCTION
Macrophages are mononuclear phagocytes that are present throughout all tissues of the body. Tissue‐resident macrophages are essential innate immune cells serving multiple functions to promote tissue homeostasis and immune surveillance (Epelman, Lavine, & Randolph, 2014; Varol, Mildner, & Jung, 2015). Importantly, tissue‐resident macrophages perform unique functions associated with each specific organ, and their phenotype is driven by local microenvironmental factors (Gautier et al., 2012; Gordon & Plüddemann, 2017; Williams, Giannarelli, Rahman, Randolph, & Kovacic, 2018). Initial work on macrophage origin and development by van Furth (van Furth & Cohn, 1968) suggested that bone marrow‐derived monocytes were the sole precursor for macrophages in tissue. This theory prevailed for 40 years until accumulating evidence revealed that tissue‐resident macrophages in the microglia of the brain develop prior to the formation of hematopoietic stem cells in the bone marrow (Ginhoux et al., 2010; Shepard & Zon, 2000). Pioneering work using fluorescent reporter mice and myeloid fate‐mapping strategies revealed the embryonic origins of many tissue‐resident macrophage lineages (Schulz et al., 2012). Indeed, in addition to bone marrow hematopoietic stem cells, the yolk sac and fetal liver are both sources for embryonically derived tissue‐resident macrophages. These key conclusions were addressed using various temporally induced myeloid fate‐mapping studies (Epelman et al., 2014; Perdiguero & Geissmann, 2016; Samokhvalov, Samokhvalova, & Nishikawa, 2007). Since these early studies, fate‐mapping tools have dramatically expanded and become a common approach for identifying and examining macrophage development and turnover in mouse models.
This unit will discuss a detailed protocol for monocyte and macrophage fate mapping in mouse models. We will discuss the application of fate‐mapping techniques, focusing on inducible fate‐mapping strategies for adult (Basic Protocol 1) and embryonic labeling (Basic Protocol 2). Both protocols utilize targeted genetic recombination in engineered animals to label cells of interest. The most common approach to achieving targeted genetic recombination is through the use of the Cre‐recombinase–LoxP (cre‐loxP) system, which employs the Cre recombinase enzyme to cleave DNA at target sequences called ‘loxP’ sites (Liu, Jin, & Zhou, 2020; Sauer & Henderson, 1988). Other site‐directed recombination approaches, such as Dre‐recombinase (Dre‐rox) (Anastassiadis et al., 2009), flippase (Flp‐FRT) (Schlake & Bode, 1994), and split‐Cre (or binary) (Kim et al., 2021) systems exist but are not widely used in myeloid research. Beyond genetic models, photoactivatable or photoconvertible protein labels are also commonly used to address myeloid cell dynamics (Cugurra et al., 2021; Genshaft et al., 2021). Details associated with these approaches are discussed in depth elsewhere (Lee, Rudd, & Smith, 2022). Cre activity is typically regulated downstream of a cell‐specific gene promoter that will restrict LoxP excision to cells of interest. Temporal regulation of Cre activity has recently become a common method that allows for the regulation of cell labeling in a discrete window of time (Hayashi & McMahon, 2002; Nakamura, Nguyen, & Mackem, 2006). This is achieved by creating Cre‐fusion proteins with estrogen receptor (CreER), CreERT2, or merCREmer variations (Indra et al., 1999; Sohal et al., 2001). Each of these approaches works by sequestering CreER in the cytoplasm until it is activated by drug administration, in this case, the estrogen analog tamoxifen, which will engage the ER and mediate translocation into the nucleus. Once in the cell nucleus, the Cre‐recombinase can target loxP sites to excise desired gene segments. Target sites are also important components of successful fate mapping approaches. The most common design involves a fluorescent reporter allele that is regulated by the presence of a stop codon flanked by LoxP‐sites. This stop codon will be excised by Cre, enabling the expression of the fluorescent reporter. Importantly, since this is genetic recombination, labeling is durable in the cell and passed on to any daughter cells that may arise by proliferation. Alternative and more complex reporter scenarios exist and are useful in various approaches. Examples include the use of stochastic reporter mice, such as confetti (Livet et al., 2007) or rainbow (Rinkevich, Lindau, Ueno, Longaker, & Weissman, 2011), induction of genetic bar codes (Pei et al., 2017), or expression of LacZ protein (Soriano, 1999).
To develop conditional and inducible fate mapping approaches, two mouse strains must be crossed (a Cre‐driver and a fluorescent reporter). First, appropriate promoters must be identified to track the cell type of interest and express the desired reporters. The ideal promoter will be cell‐restricted and highly expressed by the cells of interest to maximize specificity and efficiency when labeling. Fluorescent reporter mouse lines typically take advantage of highly and ubiquitously expressed genes to allow for versatility between Cre‐induction approaches. The most commonly used promoter is the Rosa26 locus because it is highly expressed in all immune cell types and has been used to drive fluorescent reporter proteins such as enhanced Green Fluorescent Protein (eGFP) or a variety of Red Fluorescent Proteins (RFP). An RFP variation, called tandem dimer Tomato (tdTomato), one of the brightest fluorescent reporter proteins, was used in the example datasets provided in this protocol. Other promoters, such as the CAG promoter, can be used to drive reporter alleles and have proven to be useful in this approach.
Fate mapping can be a powerful tool for determining cellular dynamics within steady‐state or disease models and has been used to obtain evidence for tissue residency or macrophage turnover. We will outline models using the inducible (temporal control) model for CreER induction. Basic Protocol 1 will describe conditional fate mapping approaches in adult animals. Basic Protocol 2 will describe embryonic fate mapping via in utero labeling, in which pregnant mothers are treated with tamoxifen at desired stages of embryonic development. This approach allows for the determination of the cellular origin of tissue‐resident macrophages. Fate mapping techniques largely depend on fluorescent reporter expression in genetically engineered mice. Thus, analyses of these data typically utilize flow cytometry to assess population representation or fluorescence microscopy to determine tissue localization. These approaches have allowed for insightful discoveries regarding the role of origin, tissue residency, and monocyte fates within disease models.
Basic Protocol 1. ADULT MOUSE – MYELOID FATE MAPPING WITH ORAL TAMOXIFEN GAVAGE
Most macrophage fate‐mapping models utilize fluorescent reporter activation following Cre‐recombinase–induced genetic recombination to label a population of interest. Therefore, precise and efficient induction of Cre becomes critical in fate‐mapping experiments. Mice expressing a Cre protein under the control of the estrogen‐nuclear receptor require exposure to tamoxifen (an estrogen analog) for translocation to the nucleus. The following Basic Protocol, adapted from the Jackson Laboratory, describes a tamoxifen preparation for adult animals (Heffner, 2011; Madisen et al., 2010; Sohal et al., 2001) that can be applied to any inducible fate mappers.
Tamoxifen is a chemically hazardous substance that must be handled appropriately. Administration procedures and mice receiving tamoxifen should be performed and housed in an approved facility under approved procedures. It is important to note that the tamoxifen should be prepared in light‐blocking vessels, as it is light sensitive. The solution should be kept at 4°C (short term) or −20°C (up to 6 months). In addition, tamoxifen is provided as a prohormone that is transformed in the liver into the bioactive form 4‐hydroxytamoxifen (4OH‐tamoxifen), which has significantly enhanced binding affinity for the estrogen receptor (Furr & Jordan, 1984). Tamoxifen has proven effective in adult mice or pregnant mothers, but OH‐tamoxifen may be preferred in some scenarios, such as targeting cells in vitro.
Materials
Experimental mice (expressing creER and fluorescent reporter constructs)
The described procedure was optimized using the CX3CR1creER/WT (strain #020940, Jackson Laboratory) R26tdTomato (strain #007909, Jackson Laboratory) mouse strain on a C57bl/6 background at 8 weeks of age. This procedure is effective in both male and female mice. Experimental cohort size is suggested to start at 5‐6 animals for each treatment group and adjust needed animal numbers based on observed variation and detectable differences within the experimental model. This is typically performed using a power calculation following initial pilot studies.
Tamoxifen (Sigma‐Aldrich #10540‐29‐1)
Corn oil (Sigma‐Aldrich #C8267)
Gavage feeding needle (or tube) with a round tip
1‐ml syringe (BD #309628)
Light‐blocking vessel/foil wrap
Incubating orbital shaker
Small animal scale
Laminar flow hood (Biosafety Class II (BSL2) or equivalent)
-
1
Working in a BSL2 biosafety cabinet, prepare 20 mg/ml tamoxifen in corn oil: dissolve tamoxifen in sterile corn oil by placing it on a shaking platform overnight at 37°C. This is done in a light‐blocking vessel (ex. Foil‐wrapped).
Storage at 4°C is for short‐term use for up to 2 months. Aliquots can be taken from the stock but should be replaced if contamination is observed.
-
2
Weigh animals on a scale prior to starting the procedure.
-
3
Determine the gavage dose by weight, 200 mg/kg for our example strain CX3CR1creER R26tdTomato
For new strains, we suggest starting with treatments in the range of 75‐250 mg tamoxifen/kg body weight. This has been effective for most mouse models that we have tested.
-
4
Remove the tamoxifen in corn oil solution from the refrigerator to a rocking plate 30 min prior to the gavage procedure to allow it to come to room temperature and become evenly mixed.
-
5Administer tamoxifen via oral gavage.
-
Use blunt‐tip gavage or feeding needles according to mouse size and connect to a 1‐ml syringe. Small animals will require a 22 gauge (FST #18061‐22), whereas adult animals can accommodate a larger 18‐gauge needle (FST #18061‐50). Fill the syringe with tamoxifen solution.Two groups of animals for control treatment are ideal for tamoxifen‐mediated fate‐mapping experiments. The first control group is treated with vehicle alone (corn oil) in the same volume as step 5a, using the same creER reporter mouse strain. This should act as a negative control, and treatment should not induce fluorescence labeling. The second control group comprises tamoxifen‐treated mice that do not carry the creER or fluorescent reporter, allowing confirmation of whether tamoxifen treatment leads to any defects.
- Restrain the animal by firmly grasping the neck skin over the shoulders so that the animal is stationary and the neck is vertical during the gavage.
- Insert the gavage needle in the diastema and advance the needle until the tip is fully inserted. If the needle is placed appropriately in the esophagus, the operation should exhibit minimal resistance. Do not force the needle if any resistance is experienced.
- Dispense the tamoxifen solution (∼200 µl; dose will depend upon the mouse strain being used).
- Once gavage is completed, gently retrieve the needle along the insertion angle.
- Release the animal back to the cage and observe for signs of distress or abnormal breathing. Monitor the animal for 5‐10 min before returning the cage to housing.
-
-
6
Animals should be closely monitored and weighed daily following the procedure to detect any adverse reactions to the gavage or tamoxifen exposure.
Observation is necessary to ensure that the gavage technique was performed effectively. Poor gavage technique may result in acute respiratory distress, which can be observed within 10 min of gavage and may result in the animal's death. Tamoxifen exposure has been reported to reduce animal activity immediately following treatment (Chen, Wu, Shi, & Xu, 2002a; Li et al., 2020). Weight loss in the days following gavage is unexpected and might indicate tamoxifen overdose in your animal strain. Higher tamoxifen dosing or prolonged treatment may result in toxicity that can lead to mortality, particularly when tamoxifen is given at doses above 250 mg/kg or if a strain is found to be highly sensitive. Tamoxifen toxicity has been reported in a number of studies to be associated with gastric dysfunction, liver damage, retinal epithelial cell death, nervous system dysfunction, and other phenotypes (Bersell et al., 2013; Chen, Wu, Shi, & Xu, 2002b; Hameyer et al., 2007; Huh et al., 2012). Thus, limiting tamoxifen exposure for the strain of interest and titering the dose is very important.
-
7
CreER activity and reporter expression can be monitored by flow cytometry or imaging analysis (outlined in Support Protocols). In some strains, recombination can be observed within hours of tamoxifen administration. If the efficiency in the target cell population is unsatisfactory, additional tamoxifen doses may be required and can be administered on consecutive days.
In our example strain, CX3CR1creER R26tdTomato, a single tamoxifen gavage will label ∼85% to 90% of circulating blood monocytes after 1 day (measured by flow cytometry), but 3 consecutive tamoxifen gavages over 3 days will label >98% of circulating blood monocytes. Thus, the treatment regime can be optimized for the desired labeling efficiency for your tissue and cell type of interest.
-
8
Animals can be sacrificed at any time after tamoxifen treatment to analyze the labeling of cells of interest. Experimental data are provided in Supporting Protocols 1 and 2 for examples of flow cytometry analyses of blood and cells isolated from brain tissue and cells imaged after cryosectioning of the adrenal glands and heart. Sacrifice time points can be performed as early as the day following tamoxifen labeling or can be extended even months after tamoxifen exposure for long‐term fate‐mapping experiments (Williams et al., 2020).
Alternate Protocol 1. INTRAPERITONEAL TAMOXIFEN INJECTION
Intraperitoneal (IP) injection is a common alternative to oral gavage. For IP injection, we chose tamoxifen dissolved in sterile corn oil (adapted from Jackson Labs (Heffner, 2011)). However, other oil options would be adequate substitutes, such as sunflower oil, which is a slightly better solvent for tamoxifen. One concern with the IP approach is that it will disrupt tissue‐resident macrophages in the peritoneal cavity, as the large bolus of oil will cause some level of cell death in the large peritoneal macrophage population.
Additional Materials
3/8 26‐G beveled needle (BD #305111)
70% Ethanol for disinfection
-
1
Weigh the animal prior to injection.
-
2
Draw tamoxifen solution (20 mg/ml tamoxifen in corn oil) into the syringe. We commonly use 100 µl for our CX3CR1creER strain, but the dose should be adjusted for each strain of interest.
-
3
Scruff the animal and position it head‐down.
-
4Administer tamoxifen via IP injection (adopted from UBC Animal Care Guidelines for IP injection (Andrews, 2014)).
- Sterilize the area of injection with 70% ethanol to prevent infection.
- Locate the injection site in the lower right quadrant of the abdomen.
- While holding the syringe parallel to the mouse tail, gently insert the needle with the bevel facing up. The action must be done with caution to avoid damage to the abdominal organs, including the urinary bladder and cecum.
- Depress the plunger to dispense the desired volume and gently extract the needle.
-
Return the animal to the cage and monitor for any adverse reactions.Acute responses will typically be observed within an hour of treatment and would likely be associated with poor injection technique. Other potential adverse reactions, such as weight loss, can be monitored over a series of days following treatment and would likely suggest the tamoxifen dosage was too high.
-
5
Repeat steps #2‐4 for consecutive or alternating days to label the desired cell populations. Throughout treatment, animals should be closely monitored daily and weighed to assess them for adverse reactions. For strains or experiments requiring more than 3 doses of tamoxifen, researchers should consider Alternative Protocol #2.
Alternate Protocol 2. TAMOXIFEN‐ENRICHED DIET
Overexposure to tamoxifen can cause CreER over‐activation, leading to DNA damage and higher mortality in the animals (Bersell et al., 2013). This is especially true when consecutive and acute tamoxifen pulsing is performed. Here, we introduce the tamoxifen‐enriched diet to avoid aggressive pulsing. This is a particularly favored method for continuous labeling. To induce consistent and effective Cre‐recombination, we suggest a minimum 2‐week feeding period (300‐500 mg tamoxifen per kg of food). Importantly, specialized diets can be designed with diet manufacturers, such as a tamoxifen‐enriched high‐fat diet. A potential concern with this method is that animals will resist the transition to the tamoxifen diet because it has a different texture and flavor than the normal diet. This may result in weight loss or even aggressive behaviors between animals. To improve mouse appetite, some tamoxifen‐diet can be softened with water and placed on the cage floor to make intake easier for animals undergoing diet transition. If the mice still refuse to eat, they can be rotated between the tamoxifen diet and normal chow for an alternating feeding cycle of 5 days on and 2 days off to maintain animal health.
Additional Materials
Tamoxifen‐supplemented diet (such as Harlan, Envigo Teklad Custom diet TD.130857)
-
1
Weigh mice prior to the diet switch.
-
2
Provide softened tamoxifen‐enriched food pellets on the cage floor to assist in diet transition.
-
3
House animals with the tamoxifen‐supplemented diet for a minimum period of 2 weeks.
In most animal strains we have worked with, we have observed that animals slowly transition to the new diet and will take up to 2 weeks of treatment to demonstrate significant recombination in target cell populations.
-
4
Animals should be weighed daily and monitored for any health concerns. Weight loss greater than 10% should be discussed with veterinary staff and may require nutrient supplementation.
We have consistently observed weight loss of up to 10% of total animal weight when transitioning to a tamoxifen‐enriched diet. Therefore, we suggest avoiding placing very young mice on the diet until at least 8 weeks of age, as they are more sensitive to weight fluctuations. Using a high‐fat, tamoxifen‐enriched diet appears to have a less dramatic effect and is eaten more readily by experimental mice. Additional alternative diet‐treatment options were discussed in the introduction to this section, including diet cycling, which can help to avoid undesired weight fluctuation.
-
5
If a tamoxifen‐enriched diet does not effectively induce Cre‐recombination in the target cells of interest after a 2‐week period, consider longer treatment or use an alternative approach such as gavage or IP administration.
Basic Protocol 2. EMBRYONIC LABELING
Embryonic fate‐mapping experiments allow for the tracking of macrophages that derive from distinct developmental stages, including yolk sac, fetal liver, or bone marrow origins. Macrophage ontogeny may influence the function of cells in some tissues. To investigate the origin of the macrophages that seed tissues and organs, an embryonic labeling technique is introduced (Perdiguero & Geissmann, 2016; Schulz et al., 2012). To perform this assay, timed breeder pairs need to be arranged so that tamoxifen can be administered at the appropriate developmental time points, such as yolk sac (E7.5‐8.5), fetal liver (E12.5‐14.5), or HSC (E18.5+). As outlined in Basic Protocol 1, tamoxifen can be administered to pregnant mothers by gavage or IP injection.
Since tamoxifen is an estrogen analog, it can disrupt the ability of some mothers to give natural birth and can lead to late‐term fetal abortion. Therefore, pregnant mothers should be given the lowest tamoxifen dose possible to induce the recombination to the desired efficiency. Additionally, it has been suggested that the addition of progesterone to embryonic tamoxifen treatment may help to avoid lost litters (Nakamura et al., 2006). Since these experiments are being performed in vivo, mice can receive unprocessed tamoxifen. However, an important consideration is if embryos are being pulsed ex vivo or cells are being labeled in vitro, the bioactive 4OH‐tamoxifen form will need to be used since tamoxifen requires processing in the liver to become active (Hayashi & McMahon, 2002). Since tamoxifen treatment, particularly when performed late in gestation, may lead to failure to give birth, mice may need to be sacrificed for cesarean section on embryonic day 20‐21 (Murphy, 1993), and recipient foster mothers will adopt the delivered pups. The following Basic Protocol section describes maternal tamoxifen gavage to induce CreER recombination and fluorescent protein expression in mouse embryos.
Materials
Full materials listed in Basic Protocol 1 for oral gavage
70% ethanol
Potential foster mother, with new litter same day or no more than 5 days old
Heating pad
Sterile Paper towel
Sterile surgical forceps
Sterile surgical scissors
Laminar flow hood (BSL2 or equivalent)
Timed breeding (adapted from Jackson Laboratories)
-
1
Studding the desired male mouse for 1‐2 weeks increases sperm counts and hormone levels. When possible, use males that have proven to be effective breeders. Males should be at minimum 8 weeks of age and can be used at an older age than females. Males can be used effectively out to 8 months of age if they have a history of successful breeding.
-
2
Select female breeders that are at least 8 weeks of age and not beyond 16 weeks of age to enhance the likelihood of successful mating. Mate timed breeders of interest together for one night, then separated the following morning.
Verifying the female is in the appropriate estrous state (proestrus or estrus) will help improve timed breeding success.
-
3
Visually check for a vaginal plug when separating the breeding pair to confirm if copulation occurred overnight.
A vaginal plug may be observed and can persist for 8‐24 hr after mating. However, the plugs from some strains, C57BL/6 in particular, may be thin and dissolve rapidly. Seeing a plug is a positive indication that mating occurred but does not imply successful pregnancy. Additionally, the lack of a plug does not imply that copulation did not happen.
-
4
Weigh the females each day to document weight gain and confirm successful pregnancy.
-
5
Perform labeling for the fate‐mapping procedure by oral tamoxifen gavage (as outlined in Basic Protocol 1) or IP tamoxifen injection of pregnant females at the time point of interest.
Treatment is typically performed at E7.5‐E8.5 for yok sac labeling, E11.5‐E14.5 for fetal liver labeling, and for late embryonic time points, E18.5 might be useful for identifying cells that developed prior to birth. Please see Adult Fate Mapping Basic Protocol 1 for tamoxifen administration procedures. For these experiments, a single dose is typically used to restrict the labeling window. For CX3CR1creER R26tdTomato labeling data provided in Figure 1A, pregnant dams received ∼200 mg/kg dosing by gavage at E14.5.
Figure 1.
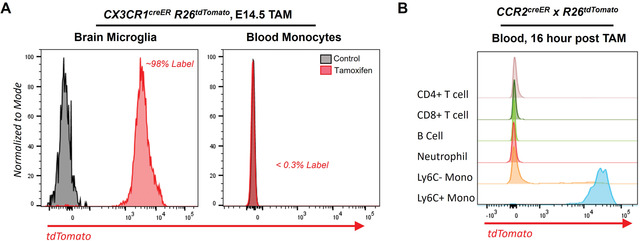
Flow cytometry analysis of fate‐mapped monocytes and macrophages.
A. CX3CR1creER R26tdTomato reporter mice were labeled in utero, embryonic day 14.5 (E14.5), by tamoxifen gavage of pregnant mothers. Labeled pups were weaned and then sacrificed at 8 weeks of age and assessed for labeling efficiency in brain microglia and blood. (Left panel) Brain microglia (CD45+ CD11b+ CD64+, Red line) were labeled with >98% efficiency from embryonic exposure to tamoxifen, whereas unlabeled age‐matched mice showed no labeling (control, Gray line). (Right panel) In the same mice, blood at 8 weeks of age showed no labeling of blood monocytes (CD45+ CD11b+ Ly6G‐ CD115+) in E14.5 labeled (Red) or control (Gray).
B. CCR2creER R26tdTomato reporter mice were gavaged with a single dose of tamoxifen, and labeling efficiency in the blood was analyzed 16 hr later. T cells (CD45+ TCRβ+), B Cells (CD45+ CD19+), Neutrophil (CD45+ CD11b+ Ly6G+), Ly6C‐ Monocytes (CD45+ CD11b+ Ly6G‐ CD115+ Ly6C‐), and Ly6C+ Monocytes (CD45+ CD11b+ Ly6G‐ CD115+ Ly6C+) were assessed for tdTomato labeling, where Ly6C monocytes were efficiently labeled.
Caesarian section of the pregnant female, adapted from Murphy, 1993
-
6
Continue to monitor and weigh the pregnant mice daily to detect any adverse reactions, such as lost litter.
-
7
Females reaching E21 without giving natural birth may require surgical recovery of the pups.
-
8
Euthanize the pregnant female via cervical dislocation.
Some female timed breeders may go into dystocia or fail to birth their entire litter, so animals should be monitored many times daily, and staff should be prepared for C‐section on or after E19.5.
-
9
Clean the abdomen with ethanol and cut open with sterile scissors. This will expose the uterus and allow access to the oviduct.
-
10
Remove the pups one at a time from membranes and cut the umbilical cord.
-
11
Wipe fluids from the pups’ face/mouth area using paper towels and check for breathing. Rhythmic chest pumping with forceps can be used to help assist breathing if pups do not immediately begin to take breaths.
-
12
Foster pups immediately in a cage with a fresh litter. This cage should be kept on a heating pad to maintain the temperature at approximately 32°C.
-
13
Donor mice should be mixed with foster pups to help facilitate acceptance by the foster mother. Rubbing new pups with nestlets or bedding from the new cage will enhance foster acceptance of new pups.
-
14
Embryonic labeled pups can then be assessed for labeling efficiency across tissues by flow cytometry or imaging analysis (discussed in Support Protocols 1 and 2).
For labeling efficiency of embryonically derived macrophages, the brain is often examined since microglia are exclusively derived from yolk sac progenitor cells (data provided in Fig. 1). Additional control data can be collected, such as assessing blood cells for labeling efficiency to address any potential for unwanted “off‐target” recombination, since blood monocytes are exclusively derived from bone marrow HSCs in the adult mice and should not be labeled. Embryonic labeled mice are often studied early, at 1 or 2 weeks after birth or later into early adulthood (8‐12 weeks of age).
Support Protocol 1. ANTIBODY STAINING AND FLOW CYTOMETRY
Flow cytometry remains a core technology that enables researchers to access the cellular composition within a tissue of interest. It is ideal for analyzing fate mapping results, as the targeted myeloid cells are labeled and become fluorescently active after tamoxifen‐induced CreER activation. However, to perform flow cytometry, the tissue samples need to be completely dissociated into a single‐cell suspension. This process negates the ability to assess any type of cell localization. Despite this disadvantage, flow cytometry still serves an essential role in fate‐mapping experiments due to its high throughput and ease of quantification across multiple samples. In addition, flow cytometry can be used as a cell sorting approach to allow for the isolation of labeled cells. Sorting strategies will follow this protocol, followed by additional techniques such as in vitro culture or lysis for gene expression analysis, like single‐cell RNA sequencing. The following protocol describes the materials and necessary steps for a successful flow cytometry experiment, but for the overarching purpose of this protocol, tissue dissociation and desired staining panels will need to be optimized for each organ being addressed. Please reference the recent protocol focused on assessing myeloid cells across tissues using flow cytometry for optimizing antibody staining panel design (Liu, Gu, Shin, Zhang, & Ginhoux, 2020).
Materials and Reagents
Mice to be sacrificed for tissue and blood harvest
CO2 source
Ice‐cold PBS
FACS Buffer: 0.1% EDTA, 10% FBS in HBSS (see recipe)
HBSS (ThermoFisher #88284)
0.5M EDTA (AccuGENE #51201)
Diluted EDTA: 10% EDTA in PBS
Distilled water
Red Blood Cell Lysis Buffer 1X (10X RBC, BioLegend #420301, 10% in distilled water) (see recipe)
RPMI‐1640 medium (Sigma‐Aldrich, R8758) with 2% Fetal Bovine Serum (FBS) (Sigma‐Aldrich, F2442)
Percoll (Sigma‐Aldrich, P1644)
Flow cytometry antibodies (Table 1)
Table 1.
Basic Flow Cytometry Panel for Myeloid Cells
| CD45 | All leukocytes |
|---|---|
| Ly6G | Neutrophils |
| CD11b | Myeloid cells |
| F4/80 | Macrophages |
| CD64 | Monocytes/macrophages |
| MerTK | Macrophages |
| CD115 | Monocytes/macrophages |
| Ly6C | Classical monocytes |
| Fluorescent Reporter Protein | Cre‐restricted cell expression |
An example of a flow cytometry panel that can be used in combination with a fluorescent fate‐mapping approach in a variety of tissues.
10‐ml Syringe (BD #309604)
Tuberculin syringe ½ ml (BD #305620)
Centrifuge
Dissection tools
BD LSRFortessa H1770 instrument or equivalent
FlowJo v.10
Tissue and Blood Harvesting
-
1
Euthanize the mouse in a CO2 smart‐box chamber.
Blood can also be collected by cheek bleeding (Golde, Gollobin, & Rodriguez, 2005). This alternative strategy could allow for monitoring of labeling efficiency in circulating blood cells without sacrificing the animal.
-
2
Open the animal and make an incision along the bottom of the rib cage to reveal the diaphragm.
-
3
Open the diaphragm and perform a cardiac puncture by injecting a heparin‐treated insulin syringe into the left ventricle to draw 200 µl of blood.
-
4
Store the blood sample in a 1.5‐ml tube containing 10 µl EDTA (10% in PBS) (or heparin/EDTA treated tube) to prevent clotting and keep it on ice to prevent cell death.
Blood samples are often important controls for experiments to show labeling specificity and can also be used for staining controls when establishing flow cytometer settings.
-
5
Perform perfusion by injecting 10 ml of ice‐cold PBS into the left heart ventricle. This will clear blood from tissues to allow for analysis of tissue‐associated macrophages rather than circulating blood cells.
-
6Harvest tissue(s) of interest for:
- Tissue digestion/dissociation, followed by flow cytometry. The brain (used in Fig. 1) processing protocol is outlined in a subsequent tissue dissociation section. Additional digestion protocols can be found in the literature for tissues of interest. Fresh tissue should be used for flow cytometry analysis, but single‐cell suspensions can be fixed and stored for delayed analysis once isolated and stained with antibodies. Regardless, it is suggested to run samples on a flow cytometer as quickly as possible to limit any potential issues with cell viability or decay of the fluorescence signal.
- Tissue fixation, then immunofluorescence imaging (supporting protocol #2). Fresh tissue can also be embedded directly in OCT for sectioning (fresh‐frozen), where samples should be kept at −20°C for storage. This approach may be desired for some epitopes that are lost during fixation protocols.
Single‐Stain Control and Sample Preparation
Blood, Red Blood Cell (RBC) Lysis Procedure
-
7
Add 1 ml RBC lysis buffer (10% RBC in distilled water) to each 1.5‐ml tube containing the blood sample, mix well, and lyse for 3 min.
-
8
While waiting, prepare a FACS tube filled with 1 ml FACS buffer for each sample and control tube.
-
9
Neutralize the lysis reaction by pipetting to move the blood samples to each FACS tube containing FACS buffer.
-
10
Centrifuge 5 min at 800 × g and 4°C.
-
11
Aspirate the supernatant.
-
12
Repeat the RBC lysis procedure (steps 7‐11) if the cell pellet remains pink (indicating the presence of RBCs).
-
13
For antibody labeling, stain at 1 µg/ml (or as titred) in a 50‐µl volume of FACS buffer, for 25 min in the dark, on ice.
-
14
Add 1 ml of FACS buffer to each tube to wash the samples.
-
15
Centrifuge as described in Step #10.
-
16
Head to the flow cytometer to collect data or cell sorter for cell isolation.
Tissue Dissociation for Single Cell Suspension
This protocol is used for processing brain samples, which are commonly assessed for fate‐mapping studies. However, it can be replaced with other dissociation or enzymatic digestion protocols for other tissues of interest.
-
17
Dissect and isolate the brain into a 1.5‐ml tube
-
18
Perform mechanical dissociation of the brain using scissors to liberate cells. The brain should turn to a mush‐like consistency with no visible chunks.
-
19
Add 1 ml RPMI medium with 2% FBS. Mix well with a 1‐ml pipette by pipetting up and down to further dissociate the cells.
-
20
Transfer brain mix to a 15‐ml conical tube and fill to 7 ml final volume with additional RPMI/2% FBS.
-
21
Make a 30/70 Percoll gradient by adding 3 ml of 100% Percoll to the sample solution. Underlay the cells with a 70% Percoll solution diluted in 1× PBS.
-
22
Centrifuge the Percoll gradient at 850 × g and 20°C for 20 min. Spin cells with the brake “off” or reduced to level 1 on the swing‐bucket centrifuge.
-
23
Carefully collect cells from the gradient interface using a pipette and transfer to a fresh conical tube.
-
24
Wash cells with 5 ml RPMI/2% FBS and centrifuge at 800 × g and 4°C for 5 min.
-
25
Aspirate supernatant, resuspend in 1 ml FACS buffer.
-
26
Count cells using a hemacytometer or counting instrument.
-
27
Add ∼1 million cells to each flow cytometry tube. This number can be adjusted for the tissue of interest, but antibody staining cocktail concentrations may need to be adjusted for increased cell numbers.
Antibody Labeling for Flow Cytometry or Cell Sorting
-
1
Sample tubes, unstained controls, and single‐stained controls should be organized. Compensation beads (such as ThermoFisher UltraComp Beads, cat. #01‐2222‐41) or cells can be used for single‐stained controls in most experimental setups.
-
2
Centrifuge cells and remove supernatant.
-
3
For antibody labeling, stain at 1 µg/ml (or as titered) in a 50‐µl volume of FACS buffer for 20 min in the dark, on ice. Antibody cocktails should be prepared as a master mix to help limit any potential pipetting errors.
An example staining panel is provided in Table 1 for a general monocyte/macrophage staining approach that can be useful for most tissues. These cells can then be assessed for labeling efficiency in fate‐mapping experiments.
-
4
Add 1 ml of FACS buffer to each tube to wash the samples.
-
5
Centrifuge for 5 min at 800 × g and 4°C.
-
6
Head to the flow cytometer to collect data or the cell sorter for cell isolation.
Table 1 outlines some common cell surface markers used in flow cytometry to identify monocytes and macrophages. A common gating strategy for exclusion of neutrophil contamination using Ly6G and identification of macrophages by MerTK and CD64 co‐expression is effective across most mouse tissues. For blood monocyte identification, we typically propose using a combination of CD45+ CD11b+ Ly6G‐ CD115+. This population can then be further separated using CCR2, Ly6C, or CX3CR1 into classical (CCR2+ Ly6C+ CX3CR1lo) or nonclassical (CCR2‐ Ly6C‐ CX3CR1hi) lineages. Once separated, cells can be assessed for expression of fluorescent reporter proteins such as tdTomato or GFP.
Figure 1 provides example flow cytometry data from fate‐mapping experiments. Figure 1A followed embryonic fate‐mapping strategies outlined in Basic Protocol 2. Pregnant CX3CR1creER R26tdTomato reporter mice were treated with a single dose of tamoxifen (200 mg/kg) by gavage when pups were at embryonic day 14.5 (E14.5). Labeled pups were born, then sacrificed at 8 weeks of age as adult mice to fate‐map the ability of embryonic cells to seed into adult macrophage lineages. Brain microglia (CD45+ CD11b+ CD64+) were labeled with >98% efficiency from embryonic exposure to tamoxifen. In the same mice, blood at 8 weeks of age showed no labeling of blood monocytes (CD45+ CD11b+ Ly6G‐ CD115+). Figure 1B illustrates the adult fate‐mapping experiment described in Basic Protocol 1, using CCR2creER R26tdTomato reporter mice. This strain will label CCR2‐expressing cells with tdTomato protein following tamoxifen exposure. Mice were gavaged with a single dose of tamoxifen (200 mg/kg). The mice were bled 16 hr later for tdTomato labeling analysis. T cells (CD45+ TCRβ+), B Cells (CD45+ CD19+), Neutrophils (CD45+ CD11b+ Ly6G+), Ly6C‐ Monocytes (CD45+ CD11b+ Ly6G‐ CD115+ Ly6C‐), and Ly6C+ Monocytes (CD45+ CD11b+ Ly6G‐ CD115+ Ly6C+) were assessed for tdTomato labeling. Only Ly6C+ monocytes were efficiently labeled.
Support Protocol 2. IMMUNOFLUORESCENCE IMAGING
To answer questions regarding cell population localization, an immunostaining method is introduced in this section. This protocol can often supplement flow cytometry to determine whether labeled cells of interest are localizing to features or regions of the organ of interest. Experiments can be performed in whole‐mount or following tissue sectioning. This approach is frequently paired with antibody labeling followed by fluorescence imaging. A detailed process for cryostat operation and immunohistochemistry staining is provided in the protocol unit 21.4 (Hofman & Taylor, 2013). This protocol will only provide a brief overview of the approach.
Materials
OCT (Sakura Tissue‐Tek O.C.T Compound #4583)
Dry ice
4% PFA fixation buffer (see recipe)
Triton X‐100 (tx‐100) (Sigma‐Aldrich, #T8787)
Antibody of choice
12‐well plate (Sigma Aldrich Corning Costar TC‐Treated Multiple Well Plates #CLS527)
Cryostat
Coverslips
Confocal microscope instrument
Leica software
Sample Fixation and Processing
Fixation in Paraformaldehyde (PFA)
-
1
Submerge freshly isolated samples in 4% PFA in PBS containing 30% sucrose. The addition of sucrose prevents fluorescence quenching. Samples can be kept at 4°C. The time required for appropriate fixation will vary for each tissue based on size and density. Samples like heart or kidney typically require overnight incubation.
Additional fixation strategies can be used, but over‐fixation may result in quenching fluorescent reporter proteins. These proteins can often be stained using antibodies (such as anti‐GFP) if endogenous fluorescence is lost.
-
2
For storage beyond 2 days, samples can be transferred to 1× PBS or 30% sucrose solution only for samples that will undergo cryosectioning.
Embedding in OCT and Cryosectioning
-
3
Trim sample to desired size. Sample can be previously fixed in PFA/sucrose or freshly isolated (fresh‐frozen). Fixed samples should be floated in 30% sucrose for 12 hr prior to embedding.
-
4
Place a metal plate or mount on dry ice (metal will allow for rapid energy transfer and faster freezing).
-
5
Drop a dab of OCT on the metal and, when partially frozen, place the tissue sample onto the OCT base with forceps. Cover the sample with more OCT and let the OCT freeze completely.
-
6
Trim the OCT‐tissue block to the desired size with a razor blade.
-
7
Store the sample embedded in OCT at −20°C until ready to section.
-
8
Section samples on a cryostat to the desired size (often 5‐20 µm) and adhere to positively charged glass slides.
Immunofluorescence Staining and Imaging
-
1
Cryosectioned samples should be warmed to room temperature on the bench.
-
2
Excess OCT should be washed away with two 5‐min incubations in 1× PBS.
-
3
Staining regions can be defined by a hydrophobic marker to restrict the amount of solution needed for immunostaining.
-
4
Block and permeabilize samples using 5% donkey serum diluted in 0.1% tx‐100. Blocking solution and permeabilization buffers can be adjusted for the tissue/staining approach. Stain for 30 min at room temperature.
Remember to cover your slides to shield them from light during the staining procedure to protect the fluorescent reporter proteins.
-
5
Wash with 1× PBS.
-
6
Stain with primary antibodies (diluted 1:500, or as titered) in 1× PBS solution for 1 hr at room temperature.
-
7
Wash three times with 1× PBS.
Staining times and treatment conditions may require optimization for the antigens being detected. For example, it may be desirable to stain at 4°C or extend the staining time to overnight.
-
8
Stain with secondary antibodies conjugated with fluorescent proteins (diluted 1:500 or as titered) in 1× PBS solution for 1 hr at room temperature.
-
9
Wash three times with 1× PBS.
-
10
Mount samples with a fluorescence‐mounting medium and place glass coverslip.
-
11
Collect images by epifluorescence or confocal microscopy.
Figure 2 shows example confocal microscope images of cryosectioned fate‐mapped samples. Figure 2A was obtained using Adult Fate‐Mapping Protocol 1 using a single gavage of tamoxifen using the CCR2creER R26tdTomato CX3CR1gfp dual reporter strain. Mice were gavaged, then sacrificed 48 hr later. Adrenal glands were assessed for tdTomato and GFP expression to determine the distribution of differentially expressing cells. Figure 2B was obtained using the continuous tamoxifen approach to fate mapping in the context of atherosclerosis progression. CX3CR1creER R26tdTomato Ldlr‐/‐ mice are susceptible to atherosclerosis following high‐fat diet feeding. To induce tdTomato expression on monocytes and macrophages and promote plaque formation in the arteries, reporter animals were fed a tamoxifen‐enriched high fat diet for 8 weeks (continuously). Mice were sacrificed and the hearts isolated and sectioned to 10 µm. Samples were stained for smooth muscle actin (SMA) and DAPI for nuclei. Samples were imaged by confocal microscopy for localization of tdTomato‐expressing macrophages within atherosclerotic lesions (Fig. 2B).
Figure 2.
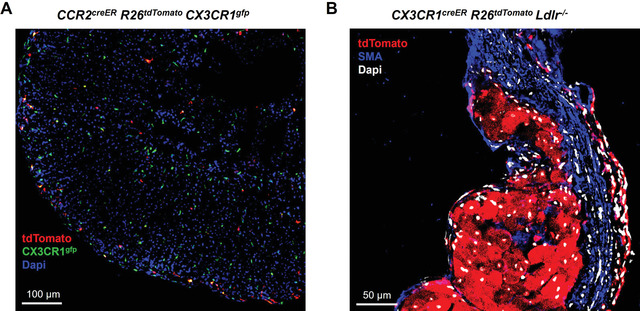
Fluorescent imaging analysis of fate‐mapped macrophages in tissues.
A. Dual reporter CCR2creER R26tdTomato CX3CR1gfp mice were given a single gavage of tamoxifen (200 mg/kg) and then sacrificed 48 hr later. Adrenal glands were cryosectioned and assessed by confocal microscopy for tdTomato (red) and GFP (green) expression to determine the distribution of differentially expressing cells. Nuclei are labeled with Dapi staining (blue).
B. CX3CR1creER R26tdTomato Ldlr‐/‐ mice were continuously fed a tamoxifen‐enriched high fat diet to induce atherosclerosis while labeling all CX3CR1‐expressing monocytes and macrophages throughout disease progression. After 8 weeks, hearts were collected, fixed in PFA/sucrose, and cryosectioned. Samples were immunostained for smooth muscle actin (SMA), then imaged by confocal microscopy for SMA (blue), tdTomato (red), and DAPI (white) for nuclei.
REAGENTS AND SOLUTIONS
PBS, 1×
Phosphate‐buffered saline 10× (PBS, Thermo Scientific Phosphate‐Buffered Saline 10×, pH 7.4 #J62036K7)
Distilled water
Store at room temperature for up to 2 years.
FACS Buffer
2% FBS, 2mM EDTA in HBSS
Fetal bovine serum (FBS, low‐endotoxin Sigma‐Aldrich #F2442)
0.5M EDTA (AccuGENE #51201)
Hank's Balanced Salt Solution (Ca‐ and Mg‐ free) (HBSS, ThermoFisher #88284)
Store at 4°C for up to 1 month.
1× PBS is also commonly used in FACS buffer but should be used without calcium/magnesium since these can promote enzyme activity that can lead to cleavage of surface molecules.
PFA Fixation Buffer
4% Formaldehyde (Sigma‐Aldrich #1004965000)
30% Sucrose (Sigma‐Aldrich #S9378)
Store for 1‐2 weeks at 4°C or at 20°C for up to 1 year.
RBC Lysis Buffer, 1×
10× Red Blood Cell Lysis Buffer (RBC, BioLegend #420301)
Distilled water
Store for up to 6 months at room temperature or 4°C.
COMMENTARY
Background Information
Fate‐mapping strategies have clarified the developmental origins of tissue‐resident macrophages (Epelman et al., 2014; Perdiguero & Geissmann, 2016; Williams et al., 2018). Initial studies used labeling systems where fluorescent reporter alleles were expressed by knock‐in or constitutive Cre‐lox systems. Using the CX3CR1‐gfp mouse model, Ginhoux, Merad, and colleagues observed the embryonic development of brain microglia (Ginhoux et al., 2010). These studies were followed with an application of inducible fate‐mapping approaches using the Csf1rMer‐iCre‐Mer to definitively track developmental time points for individual waves of embryonic macrophages and track their contribution into adulthood (Gomez Perdiguero et al., 2015; Schulz et al., 2012). Conclusions were supplemented with key work from other groups applying additional CX3CR1creER , Flt3cre , and RunxMerCreMer fate‐mapping systems (Bain et al., 2014; Bain et al., 2016; Epelman et al., 2014; Hoeffel et al., 2015; Yona et al., 2013). Since these initial studies, it has become increasingly clear that macrophages are derived from at least three sources: yolk sac, fetal liver, and bone marrow. During primitive hematopoiesis during embryonic days 6.5‐8.5, macrophage precursors in the yolk sac are observed and begin to differentiate into tissue‐resident macrophages (Ginhoux et al., 2010; Perdiguero & Geissmann, 2016; Samokhvalov, 2014; Schulz et al., 2012; Yona et al., 2013). Next, between E10.5‐E14.5, macrophage precursors emerge from the aorta‐gonad‐mesonephros (AGM) region to colonize the fetal liver and seed a second wave of tissue‐resident macrophages throughout the body (Frame, Mcgrath, & Palis, 2013; Mcgrath et al., 2015). After E14.5, hematopoietic stem cells begin to form in the bone marrow and give rise to blood monocytes that support tissue expansion during growth and seed macrophages in additional tissues (Orkin & Zon, 2008). To decipher this complex developmental network, research teams have turned to embryonic fate mapping strategies (outlined in Basic Protocol 2) to determine when macrophages are present in tissues and whether they can persist in the mouse.
Concurrent to the development of embryonic labeling approaches, macrophage research in the adult mouse defined a new ability for fully differentiated macrophages to self‐maintain in the tissue by proliferation (Hashimoto et al., 2013; Yona et al., 2013). Previous dogma held that fully differentiated macrophages were unable to proliferate in tissues. Thus, major efforts have focused on the maturation of monocytes and their contribution to tissue‐resident macrophages in the adult mouse in steady‐state and disease conditions. Multiple models to fate map monocytes have been developed recently. These are exciting tools, as they will allow researchers to uncover the fate of distinct monocyte lineages at unique stages of development and following injury responses. Two leading models are the Ms4a3creER (Liu et al., 2019) and CCR2creER (Chen et al., 2020; Croxford et al., 2015) models. CCR2creER fate mapping was recently used to track monocyte contributions to development (Dick et al., 2022), brain inflammation (Amorim et al., 2022; Chen et al., 2020), and thermogenesis (Gallerand et al., 2021). In addition to these new tools, fate‐mapping approaches can be applied to adult animals by labeling tissue‐resident macrophages by a ‘pulse‐chase’ type approach, such as those performed in recent studies of cardiac and arterial macrophages (Dick et al., 2019; Williams et al., 2020). These approaches all take advantage of Basic Protocol 1. Finally, to assist in future experimental design, a non‐exhaustive list of common fate‐mapping mouse models is provided in Table 2, highlighting key research articles that have developed or applied the approaches to advance macrophage biology.
Table 2.
Monocyte and Macrophage Fate Mapping Toolbox
| Promoter | Model | Target cell | Reference |
|---|---|---|---|
| Cx3cr1 | cre | Monocytes/macrophages | (Yona et al., 2013) |
| creER | Monocytes/macrophages | (Yona et al., 2013) | |
| GFP/+ | Monocytes/macrophages | (Bain et al., 2014; Ginhoux et al., 2010; Jung et al., 2000) | |
| Ccr2 | cre | Monocytes/macrophages /DCs | (Heung & Hohl, 2019) |
| creER | Monocytes/macrophages /DCs | (Amorim et al., 2022; Chen et al., 2020; Croxford et al., 2015; Dick et al., 2022; Gallerand et al., 2021; Xu et al., 2020) | |
| GFP/+ | Monocytes/macrophages /DCs | (Satpathy et al., 2013) | |
| RFP/+ | Monocytes/macrophages /DCs | (Saederup et al., 2010) | |
| Ms4a3 | cre | GMP/monocytes | (Amorim et al., 2022; Liu et al., 2019) |
| creER | GMP/monocytes | ||
| TdTomato | GMP/monocytes | ||
| Csf1r | cre | Monocytes/macrophages | (Deng et al., 2010; Schulz et al., 2012) |
| merCREmer | Monocytes/macrophages | (Epelman et al., 2014; Gomez Perdiguero et al., 2015; Qian et al., 2011; Schulz et al., 2012) | |
| EGFP (MacGreen) | Monocytes/macrophages | (Sasmono et al., 2003) | |
| Flt3 | cre | HSC‐derived cells | (Boyer, Schroeder, Smith‐Berdan, & Forsberg, 2011; Schulz et al., 2012) |
| Runx1 | merCREmer | HSC and embryonic progenitor cells | (Hoeffel et al., 2015; Hoeffel et al., 2012; Samokhvalov et al., 2007) |
| LysM | GFP/+ | Monocytes/neutrophil/macrophages | (Faust, Varas, Kelly, Heck, & Graf, 2000) |
| cre | Monocytes/neutrophil/macrophages | (Clausen, Burkhardt, Reith, Renkawitz, & Förster, 1999) | |
| Gata6 | (roxstoprox) icreER | Large peritoneal/serous cavity macrophages | (Jin et al., 2021) |
| F4/80 | cre | Macrophages | (Schaller et al., 2002) |
| cKIT | cre | Hematopoietic progenitor cells | (Hatzistergos et al., 2015) |
| creER | Hematopoietic progenitor cells | (Stremmel et al., 2018; Van Berlo et al., 2014) | |
| Lyve1 | cre | Lyve1+ macrophages | (Pham et al., 2010; Zhang et al., 2021) |
| creER | Lyve1+ macrophages | (Connor, Kelley, & Tempero, 2016) |
Common mouse models that are useful for applying macrophage fate‐mapping approaches to different macrophage subsets and tissues. Primary immune cell types labeled with these models are listed under “Target Cell”.
GMP (granulocyte‐monocyte progenitor), HSC (hematopoietic stem cell), DC (dendritic cells).
Overall, the fate‐mapping technique is an advanced tool that allows for tracking cells across multiple stages of development and performing kinetic studies on cell motility. The high specificity of the approach and efficiency of available animal models make it a prominent method for understanding macrophage biology. We suspect that application of this technique will continue to expand as other reporter systems are devised to incorporate this powerful tool with single‐cell RNA‐seq and spatial analysis approaches.
Critical Parameters
Fate‐mapping experiments are fairly simple to conduct, and treatment can be performed in a short period. The key is to select appropriate mouse models and to induce CreER activity efficiently with the minimum amount of tamoxifen required to limit potential adverse events. In addition, tamoxifen dissolves very slowly in oil. Researchers should allow significant time to completely dissolve tamoxifen before administration.
Basic Protocols 1 & 2 : Tamoxifen administration via oral gavage requires considerable experience with the gavage technique. As stated in the section, if resistance is experienced while advancing the gavage needle, the procedure must be halted, and the needle must be retrieved immediately because forcing it down could damage the trachea or esophagus. A second critical point of oral gavage is to avoid pushing the solution while the bent section of the tube is not fully extended in the esophagus. This is important because shallow embedding may cause corn oil to leak into the airway and choke the animal. After successful oral gavage, researchers should rest the animal before euthanizing to allow Cre recombination and fluorescence expression.
Alternative Protocol 1: IP injection can be a risk factor given the localization of the injection to many vital organs. This protocol is not suggested as a first choice for use on pregnant females. In addition, injecting corn oil can interfere with the mouse anatomy, especially if investigators are examining the peritoneal cavity.
Alternative Protocol 2: Mice will often avoid eating this diet when transitioning from typical chow. Mouse weight needs to be watched carefully and addressed if significant weight loss is observed. Feeding for 2 or more weeks is typically required to induce fluorescent reporter alleles. Due to these concerns, the tamoxifen‐supplemented diet is not an ideal approach for pregnant females.
Support Protocol 1 : To prevent cell death during flow cytometry preparation, samples should be kept on ice as much as possible during the experiment. Appropriate live/dead dyes may be required to help clean up samples with significant numbers of dead cells present (such as cells isolated from tissues requiring collagenase digestion).
Support Protocol 2 : Some epitopes for antibody staining can be lost by fixation. Therefore, some preparations may require samples to be “fresh” frozen, where they are directly (and rapidly) frozen in OCT and sections. This approach can help preserve epitopes for some hard‐to‐stain antibodies.
Troubleshooting
Basic Protocols 1 & 2 : Some tissues, such as the brain, are not sufficiently induced by a single gavage or IP tamoxifen injection in adult mice. For these difficult‐to‐induce locations, repeated dosing or switching to tamoxifen‐enriched diets may be required for efficient recombination. Before experimentation, all new mouse models should be assessed for optimal labeling efficiency and treatment protocols.
Basic Protocol 2 : For embryonic labeling approaches, a common issue is the loss of litters, especially when tamoxifen is given late‐term. A potentially helpful addition to the outlined procedure is the addition of progesterone to the tamoxifen treatment, which may help avoid abortion.
Support Protocol 1 : Depending on the tissue types being dissociated, researchers may experience cell sparsity. This could be due to the nature of the organ but most likely due to inappropriate sample preparation and storage. First, researchers have to ensure the samples sit on ice throughout the experiment to reduce the likelihood of cell death. Second, optimizing the enzyme cocktail concentrations and incubation time is vital if enzyme‐mediated tissue dissociation is utilized. Unnecessarily extending or shortening digestion time may trigger apoptosis or inadequate tissue breakdown.
Support Protocol 2 : During immunostaining of cryosections, researchers should ensure the slides are kept moisturized in PBS or supplemental reagent. Staining length varies for different tissue types, but a minimal 1 hr primary and secondary antibody staining is suggested for any 10‐µm sections. It should be extended accordingly if staining thicker sections or if staining yields poor‐quality images. Alternatively, detergents may be used to help permeabilize the tissue and increase antibody penetration.
Understanding Results
Basic and alternative protocols describe the same tamoxifen‐inducible CreER system to induce fluorescent reporter proteins. Either flow cytometry or immunofluorescence imaging are standard methods to access recombination efficiency. Positive experimental results should yield fluorescent reporter activation in desired cells. Positive and negative control samples should always be included when performing these types of experiments.
Time Considerations
Basic & Alternative Protocols: Preparing the tamoxifen solution can be time‐consuming and should be performed in advance of planned experimentation. Preparation of 20 mg/ml tamoxifen typically takes 3‐4 hr rotating at room temperature (and protected from light). It can be slightly warmed to accelerate solubility, if necessary. Tamoxifen delivery to pregnant mothers or adult mice requires less than 5 min per animal in handling time. The tamoxifen‐supplemented diet will typically take a minimum of 2 weeks to ensure effective CreER induction. Finally, it is generally true that following tamoxifen‐induced Cre‐recombination, the expression of fluorescent reporter alleles requires several hours (up to 24) to accumulate sufficient reporter protein within desired cells to be readily observable by conventional approaches.
Support Protocol 1 : Labeling tubes and setting up animals for the experiment will take 15 min. Euthanizing, cardiac puncture, and perfusion will generally take 15 min for each animal, although an experienced researcher may perform these techniques more quickly. Tissue dissociation may vary for different tissue types but will typically take about an hour from the beginning of tissue isolation to the completed single‐cell suspension. Staining should take approximately 45 min. Running samples on a flow cytometer and data processing can be completed within a day.
Support Protocol 2 : Cryosectioning is a time‐consuming process that may take up to 30 min per sample. Staining with primary and secondary antibodies will need a minimum of 2.5 hr to complete. Confocal imaging and analysis will often take 2 or more hours to complete.
Author Contributions
Yingzheng Xu: Original draft; Patricia R. Schrank: Review and editing; Jesse W. Williams: Conceptualization, Original draft, review, and editing.
Conflict of Interest
The authors declare no conflicts of interest relating to this manuscript.
Acknowledgments
The authors would like to thank Michael Patterson and Alisha Zhu (University of Minnesota) for their assistance in generating the data and figures presented in this article. JWW and YX were supported by grants from the National Institutes of Health HL138163 and American Heart Association (AHA) CDA855022.
Xu, Y. , Schrank, P. R. , & Williams, J. W. (2022). Macrophage fate mapping. Current Protocols, 2, e456. doi: 10.1002/cpz1.456
Published in the Immunology section
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Literature Cited
- Amorim, A. , De Feo, D. , Friebel, E. , Ingelfinger, F. , Anderfuhren, C. D. , Krishnarajah, S. , … Becher, B. (2022). IFNγ and GM‐CSF control complementary differentiation programs in the monocyte‐to‐phagocyte transition during neuroinflammation. Nature Immunology, 23, 217–228. doi: 10.1038/s41590-021-01117-7. [DOI] [PubMed] [Google Scholar]
- Anastassiadis, K. , Fu, J. , Patsch, C. , Hu, S. , Weidlich, S. , Duerschke, K. , … Stewart, A. F. (2009). Dre recombinase, like Cre, is a highly efficient site‐specific recombinase in E. coli, mammalian cells and mice. Disease Models & Mechanisms, 2, 508–515. doi: 10.1242/dmm.003087. [DOI] [PubMed] [Google Scholar]
- Andrews, K. (2014). UBC animal care guidelines: Intraperitoneal (IP) injection in rats and mice SOP. UBC Animal Care Guidelines, 1–4. [Google Scholar]
- Bain, C. C. , Bravo‐Blas, A. , Scott, C. L. , Perdiguero, E. G. , Geissmann, F. , Henri, S. , … Mowat, A. M. (2014). Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nature Immunology, 15, 929–937. doi: 10.1038/ni.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain, C. C. , Hawley, C. A. , Garner, H. , Scott, C. L. , Schridde, A. , Steers, N. J. , … Jenkins, S. J. (2016). Long‐lived self‐renewing bone marrow‐derived macrophages displace embryo‐derived cells to inhabit adult serous cavities. Nature Communication, 7, ncomms11852. doi: 10.1038/ncomms11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersell, K. , Choudhury, S. , Mollova, M. , Polizzotti, B. D. , Ganapathy, B. , Walsh, S. , … Kühn, B. (2013). Moderate and high amounts of tamoxifen in αmHC‐MerCreMer mice induce a DNA damage response, leading to heart failure and death. Disease Models & Mechanisms, 6, 1459–1569. doi: 10.1242/dmm.010447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer, S. W. , Schroeder, A. V. , Smith‐Berdan, S. , & Forsberg, E. C. (2011). All hematopoietic cells develop from hematopoietic stem cells through Flk2/Flt3‐positive progenitor cells. Cell Stem Cell, 9, 64–73. doi: 10.1016/j.stem.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D. , Wu, C‐F. , Shi, B. , & Xu, Y.‐M. (2002a). Tamoxifen and toremifene impair retrieval, but not acquisition, of spatial information processing in mice. Pharmacology Biochemistry and Behavior, 72, 417–421. doi: 10.1016/S0091-3057(01)00782-1. [DOI] [PubMed] [Google Scholar]
- Chen, D. , Wu, C. F. , Shi, B. , & Xu, Y. M. (2002b). Tamoxifen and toremifene cause impairment of learning and memory function in mice. Pharmacology Biochemistry and Behavior, 71, 269–276. doi: 10.1016/S0091-3057(01)00656-6. [DOI] [PubMed] [Google Scholar]
- Chen, H.‐R. , Sun, Y.‐Y. , Chen, C.‐W. , Kuo, Y.‐M. , Kuan, I. S. , Tiger Li, Z.‐R. , … Kuan, C.‐Y. (2020). Fate mapping via CCR2‐CreER mice reveals monocyte‐to‐microglia transition in development and neonatal stroke. Science Advances, 6, eabb2119. doi: 10.1126/sciadv.abb2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H.‐R. , Sun, Y.‐Y. , Chen, C.‐W. , Kuo, Y.‐M. , Kuan, I. S. , Tiger Li, Z.‐R. , … Kuan, C.‐Y. (2020). Fate mapping via CCR2‐CreER mice reveals monocyte‐to‐microglia transition in development and neonatal stroke. Science Advances, 6, 1–14. doi: 10.1126/sciadv.abb2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen, B. E. , Burkhardt, C. , Reith, W. , Renkawitz, R. , & Förster, I. (1999). Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Research, 8, 265–277. doi: 10.1023/A:1008942828960. [DOI] [PubMed] [Google Scholar]
- Connor, A. L. , Kelley, P. M. , & Tempero, R. M. (2016). Lymphatic endothelial lineage assemblage during corneal lymphangiogenesis. Laboratory Investigation, 96, 270–282. doi: 10.1038/labinvest.2015.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxford, A. L. , Lanzinger, M. , Hartmann, F. J. , Schreiner, B. , Mair, F. , Pelczar, P. , … Becher, B. (2015). The cytokine GM‐CSF drives the inflammatory signature of CCR2+ monocytes and licenses autoimmunity. Immunity, 43(3), 502–514. doi: 10.1016/j.immuni.2015.08.010. [DOI] [PubMed] [Google Scholar]
- Cugurra, A. , Mamuladze, T. , Rustenhoven, J. , Dykstra, T. , Beroshvili, G. , Greenberg, Z. J. , … Kipnis, J. (2021). Skull and vertebral bone marrow are myeloid cell reservoirs for the meninges and CNS parenchyma. Science (80‐.), 373, eabf7844. doi: 10.1126/science.abf7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, L. , Zhou, J.‐F. , Sellers, R. S. , Li, J.‐F. , Nguyen, A. V. , Wang, Y. , … Lin, E. Y. (2010). A novel mouse model of inflammatory bowel disease links mammalian target of rapamycin‐dependent hyperproliferation of colonic epithelium to inflammation‐associated tumorigenesis. American Journal of Pathology, 176, 952–967. doi: 10.2353/ajpath.2010.090622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick, S. A. , Macklin, J. A. , Nejat, S. , Momen, A. , Clemente‐Casares, X. , Althagafi, M. G. , … Epelman, S. (2019). Self‐renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nature Immunology, 20, 29–39. doi: 10.1038/s41590-018-0272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick, S. A. , Wong, A. , Hamidzada, H. , Nejat, S. , Nechanitzky, R. , Vohra, S. , … Epelman, S. (2022). Three tissue resident macrophage subsets coexist across organs with conserved origins and life cycles. Science Immunology, 7, 1–18. doi: 10.1126/sciimmunol.abf7777. [DOI] [PubMed] [Google Scholar]
- Epelman, S. , Lavine, K. J. , Beaudin, A. E. , Sojka, D. K. , Carrero, J. A. , Calderon, B. , … Mann, D. L. (2014). Embryonic and adult‐derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity, 40, 91–104. doi: 10.1016/j.immuni.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epelman, S. , Lavine, K. J. , & Randolph, G. J. (2014). Origin and Functions of Tissue Macrophages. Immunity, 41, 21–35. doi: 10.1016/j.immuni.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust, N. , Varas, F. , Kelly, L. M. , Heck, S. , & Graf, T. (2000). Insertion of enhanced green fluorescent protein into the lysozyme gene creates mice with green fluorescent granulocytes and macrophages. Blood, 96, 719–726. doi: 10.1182/blood.V96.2.719. [DOI] [PubMed] [Google Scholar]
- Frame, J. M. , Mcgrath, K. E. , & Palis, J. (2013). Erythro‐myeloid progenitors: ‘Definitive’ hematopoiesis in the conceptus prior to the emergence of hematopoietic stem cells. Blood Cells, Molecules, and Diseases, 51, 220–225. doi: 10.1016/j.bcmd.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furr, B. J. A. , & Jordan, V. C. (1984). The pharmacology and clinical uses of tamoxifen. Pharmacology & Therapeutics, 25, 127–205. doi: 10.1016/0163-7258(84)90043-3. [DOI] [PubMed] [Google Scholar]
- Gallerand, A. , Stunault, M. I. , Merlin, J. , Luehmann, H. P. , Sultan, D. H. , Firulyova, M. M. , … Ivanov, S. (2021). Brown adipose tissue monocytes support tissue expansion. Nature Communication, 12, 5255. doi: 10.1038/s41467-021-25616-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, E. L. , Shay, T. , Miller, J. , Greter, M. , Jakubzick, C. , Ivanov, S. , … and the Immunological Genome Consortium . (2012). Gene expression profiles and transcriptional regulatory pathways underlying mouse tissue macrophage identity and diversity. Nature Immunology, 13(11), 1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genshaft, A. S. , Ziegler, C. G. K. , Tzouanas, C. N. , Mead, B. E. , Jaeger, A. M. , Navia, A. W. , … Shalek, A. K. (2021). Live cell tagging tracking and isolation for spatial transcriptomics using photoactivatable cell dyes. Nature Communication, 12, 4995. doi: 10.1038/s41467-021-25279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux, F. , Greter, M. , Leboeuf, M. , Nandi, S. , See, P. , Gokhan, S. , … Merad, M. (2010). Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science (80‐.), 330, 841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golde, W. T. , Gollobin, P. , & Rodriguez, L. L. (2005). A rapid, simple, and humane method for submandibular bleeding of mice using a lancet. Lab Animal, 34, 39–43. doi: 10.1038/laban1005-39. [DOI] [PubMed] [Google Scholar]
- Gomez Perdiguero, E. , Klapproth, K. , Schulz, C. , Busch, K. , Azzoni, E. , Crozet, L. , … Rodewald, H.‐R. (2015). Tissue‐resident macrophages originate from yolk‐sac‐derived erythro‐myeloid progenitors. Nature, 518, 547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, S. , & Plüddemann, A. (2017). Tissue macrophages: Heterogeneity and functions. BMC Biology, 15(1), 53. doi: 10.1186/s12915-017-0392-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameyer, D. , Loonstra, A. , Eshkind, L. , Schmitt, S. , Antunes, C. , Groen, A. , … Bockamp, E. (2007). Toxicity of ligand‐dependent Cre recombinases and generation of a conditional Cre deleter mouse allowing mosaic recombination in peripheral tissues. Physiological Genomics, 31, 32–41. doi: 10.1152/physiolgenomics.00019.2007. [DOI] [PubMed] [Google Scholar]
- Hashimoto, D. , Chow, A. , Noizat, C. , Teo, P. , Beasley, M. B. , Leboeuf, M. , … Merad, M. (2013). Tissue‐resident macrophages self‐maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity, 38, 792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzistergos, K. E. , Takeuchi, L. M. , Saur, D. , Seidler, B. , Dymecki, S. M. , Mai, J. J. , … Hare, J. M. (2015). CKit+ cardiac progenitors of neural crest origin. Proceedings of the National Academy of Sciences of the United States of America, 112, 13051–13056. doi: 10.1073/pnas.1517201112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, S. , & Mcmahon, A. P. (2002). Efficient recombination in diverse tissues by a tamoxifen‐inducible form of Cre: A tool for temporally regulated gene activation/inactivation in the mouse. Developmental Biology, 244, 305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Heffner, C. (2011). Intraperitoneal Injection of Tamoxifen for Inducible Cre‐Driver Lines. Jackson Labs, 13. [Google Scholar]
- Heung, L. J. , & Hohl, T. M. (2019). Inflammatory monocytes are detrimental to the host immune response during acute infection with cryptococcus neoformans. Plos Pathogens, 15, e1007627. doi: 10.1371/journal.ppat.1007627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel, G. , Chen, J. , Lavin, Y. , Low, D. , Almeida, F. F. , See, P. , … Ginhoux, F. (2015). C‐Myb+ erythro‐myeloid progenitor‐derived fetal monocytes give rise to adult tissue‐resident macrophages. Immunity, 42, 665–678. doi: 10.1016/j.immuni.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel, G. , Wang, Y. , Greter, M. , See, P. , Teo, P. , Malleret, B. , … Ginhoux, F. (2012). Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac‐derived macrophages. Journal of Experimental Medicine, 209, 1167–1181. doi: 10.1084/jem.20120340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman, F. M. , & Taylor, C. R. (2013). Immunohistochemistry. Current Protocols in Immunology, 103, 21.4.1–21.4.26. doi: 10.1002/0471142735.im2104s103. [DOI] [PubMed] [Google Scholar]
- Huh, W. J. , Khurana, S. S. , Geahlen, J. H. , Kohli, K. , Waller, R. A. , & Mills, J. C. (2012). Tamoxifen induces rapid, reversible atrophy, and metaplasia in mouse stomach. Gastroenterology, 142, 21–24.e7. doi: 10.1053/j.gastro.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra, A. K. , Warot, X. , Brocard, J. , Bornert, J.‐ M. , Xiao, J.‐H. , Chambon, P. , & Metzger, D. (1999). Temporally‐controlled site‐specific mutagenesis in the basal layer of the epidermis: Comparison of the recombinase activity of the tamoxifen‐inducible Cre‐ER(T) and Cre‐ER(T2) recombinases. Nucleic Acids Research, 27, 4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H. , Liu, K. , Tang, J. , Huang, X. , Wang, H. , Zhang, Q. , … Zhou, B. (2021). Genetic fate‐mapping reveals surface accumulation but not deep organ invasion of pleural and peritoneal cavity macrophages following injury. Nature Communication, 12, 2863. doi: 10.1038/s41467-021-23197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, S. , Aliberti, J. , Graemmel, P. , Sunshine, M. J. , Kreutzberg, G. W. , Sher, A. , & Littman, D. R. (2000). Analysis of Fractalkine Receptor CX3CR1 Function by Targeted Deletion and Green Fluorescent Protein Reporter Gene Insertion. Molecular and Cellular Biology, 20, 4106–4114. doi: 10.1128/MCB.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.‐S. , Kolesnikov, M. , Peled‐Hajaj, S. , Scheyltjens, I. , Xia, Y. , Trzebanski, S. , … Jung, S. (2021). A binary cre transgenic approach dissects microglia and CNS border‐associated macrophages. Immunity, 54, 176–190.e7. doi: 10.1016/j.immuni.2020.11.007. [DOI] [PubMed] [Google Scholar]
- Lee, S. E. , Rudd, B. D. , & Smith, N. L. (2022). Fate‐mapping mice: New tools and technology for immune discovery. Trends in Immunology, xx, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Du, Z.‐J. , Chen, M.‐Q. , Chen, J.‐J. , Liang, Z.‐M. , Ding, X.‐T. , … Gao, T.‐M. (2020). The effects of tamoxifen on mouse behavior. Genes, Brain, and Behavior, 19, e12620. doi: 10.1111/gbb.12620. [DOI] [PubMed] [Google Scholar]
- Liu, K. , Jin, H. , & Zhou, B. (2020). Genetic lineage tracing with multiple DNA recombinases: A user's guide for conducting more precise cell fate mapping studies. Journal of Biological Chemistry, 295, 6413–6424. doi: 10.1074/jbc.REV120.011631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Gu, Y. , Chakarov, S. , Bleriot, C. , Kwok, I. , Chen, X. , … Ginhoux, F. (2019). Fate Mapping via Ms4a3‐expression history traces monocyte‐derived cells. Cell, 178(6), 1509–1525.e19. doi: 10.1016/j.cell.2019.08.009. [DOI] [PubMed] [Google Scholar]
- Liu, Z. , Gu, Y. , Shin, A. , Zhang, S. , & Ginhoux, F. (2020). Analysis of myeloid cells in mouse tissues with flow cytometry. STAR Protocols, 1, 100029. doi: 10.1016/j.xpro.2020.100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livet, J. , Weissman, T. A. , Kang, H. , Draft, R. W. , Lu, J. , Bennis, R. A. , … Lichtman, J. W. (2007). Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature, 450, 56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Madisen, L. , Zwingman, T. A. , Sunkin, S. M. , Oh, S. W. , Zariwala, H. A. , Gu, H. , … Zeng, H. (2010). A robust and high‐throughput Cre reporting and characterization system for the whole mouse brain. Nature Neuroscience, 13, 133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcgrath, K. E. , Frame, J. M. , Fegan, K. H. , Bowen, J. R. , Conway, S. J. , Catherman, S. C. , … Palis, J. (2015). Distinct sources of hematopoietic progenitors emerge before HSCs and provide functional blood cells in the mammalian embryo. Cell Reports, 11, 1892–1904. doi: 10.1016/j.celrep.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, D. (1993). Caesarean section and fostering. Methods in Molecular Biology, 18, 177–178. doi: 10.1385/0-89603-245-0:177. [DOI] [PubMed] [Google Scholar]
- Nakamura, E. , Nguyen, M.‐T. , & Mackem, S. (2006). Kinetics of tamoxifen‐regulated Cre activity in mice using a cartilage‐specific CreERT to assay temporal activity windows along the proximodistal limb skeleton. Developmental Dynamics, 235, 2603–2612. doi: 10.1002/dvdy.20892. [DOI] [PubMed] [Google Scholar]
- Orkin, S. H. , & Zon, L. I. (2008). Hematopoiesis: An evolving paradigm for stem cell biology. Cell, 132, 631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, W. , Feyerabend, T. B. , Rössler, J. , Wang, Xi , Postrach, D. , Busch, K. , … Rodewald, H.‐R. (2017). Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature, 548, 456–460. doi: 10.1038/nature23653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdiguero, E. G. , & Geissmann, F. (2016). The development and maintenance of resident macrophages. Nature Immunology, 17(1), 2–8. doi: 10.1038/ni.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, T. H. M. , Baluk, P. , Xu, Y. , Grigorova, I. , Bankovich, A. J. , Pappu, R. , … Cyster, J. G. (2010). Lymphatic endothelial cell sphingosine kinase activity is required for lymphocyte egress and lymphatic patterning. Journal of Experimental Medicine, 207, 17–27. doi: 10.1084/jem.20091619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, B.‐Z. , Li, J. , Zhang, H. , Kitamura, T. , Zhang, J. , Campion, L. R. , … Pollard, J. W. (2011). CCL2 recruits inflammatory monocytes to facilitate breast‐tumour metastasis. Nature, 475, 222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkevich, Y. , Lindau, P. , Ueno, H. , Longaker, M. T. , & Weissman, I. L. (2011). Germ‐layer and lineage‐restricted stem/progenitors regenerate the mouse digit tip. Nature, 476, 409–413. doi: 10.1038/nature10346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saederup, N. , Cardona, A. E. , Croft, K. , Mizutani, M. , Cotleur, A. C. , Tsou, C.‐L. , … Charo, I. F. (2010). Selective chemokine receptor usage by central nervous system myeloid cells in CCR2‐red fluorescent protein knock‐in mice. Plos One, 5, e13693. doi: 10.1371/journal.pone.0013693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samokhvalov, I. M. (2014). Deconvoluting the ontogeny of hematopoietic stem cells. Cellular and Molecular Life Sciences, 71, 957–978. doi: 10.1007/s00018-013-1364-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samokhvalov, I. M. , Samokhvalova, N. I. , & Nishikawa, S.‐I. (2007). Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature, 446, 1056–1061. doi: 10.1038/nature05725. [DOI] [PubMed] [Google Scholar]
- Sasmono, R. T. , Oceandy, D. , Pollard, J. W. , Tong, W. , Pavli, P. , Wainwright, B. J. , … Hume, D. A. (2003). A macrophage colony‐stimulating factor receptor‐green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood, 101, 1155–1163. doi: 10.1182/blood-2002-02-0569. [DOI] [PubMed] [Google Scholar]
- Satpathy, A. T. , Briseño, C. G. , Lee, J. S. , Ng, D. , Manieri, N. A. , Kc, W. , … Murphy, K. M. (2013). Notch2‐dependent classical dendritic cells orchestrate intestinal immunity to attaching‐and‐effacing bacterial pathogens. Nature Immunology, 14, 937–948. doi: 10.1038/ni.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, B. , & Henderson, N. (1988). Site‐specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proceedings of the National Academy of Sciences of the United States of America, 85, 5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, E. , Macfarlane, A. J. , Rupec, R. A. , Gordon, S. , Mcknight, A. J. , & Pfeffer, K. (2002). Inactivation of the F4/80 Glycoprotein in the Mouse Germ Line. Molecular and Cellular Biology, 22, 8035–8043. doi: 10.1128/MCB.22.22.8035-8043.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlake, T. , & Bode, J. (1994). Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry, 33, 12746–12751. doi: 10.1021/bi00209a003. [DOI] [PubMed] [Google Scholar]
- Schulz, C. , Perdiguero, E. G. , Chorro, L. , Szabo‐Rogers, H. , Cagnard, N. , Kierdorf, K. , … Geissmann, F. (2012). A lineage of myeloid cells independent of myb and hematopoietic stem cells. Science (80‐.), 336, 86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- Shepard, J. L. , & Zon, L. I. (2000). Developmental derivation of embryonic and adult macrophages. Current Opinion in Hematology, 7, 3–8. doi: 10.1097/00062752-200001000-00002. [DOI] [PubMed] [Google Scholar]
- Sohal, D. S. , Nghiem, M. , Crackower, M. A. , Witt, S. A. , Kimball, T. R. , Tymitz, K. M. , … Molkentin, J. D. (2001). Temporally regulated and tissue‐specific gene manipulations in the adult and embryonic heart using a tamoxifen‐inducible Cre protein. Circulation Research, 89, 20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- Soriano, P. (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genetics, 21, 70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stremmel, C. , Schuchert, R. , Wagner, F. , Thaler, R. , Weinberger, T. , Pick, R. , … Schulz, C. (2018). Yolk sac macrophage progenitors traffic to the embryo during defined stages of development. Nature Communication, 9, 75. doi: 10.1038/s41467-017-02492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Berlo, J. H. , Kanisicak, O. , Maillet, M. , Vagnozzi, R. J. , Karch, J. , Lin, S‐C. J. , … Molkentin, J. D. (2014). C‐kit+ cells minimally contribute cardiomyocytes to the heart. Nature, 509, 337–341. doi: 10.1038/nature13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Furth, R. , & Cohn, Z. A. (1968). The origin and kinetics of mononuclear phagocytes. Journal of Experimental Medicine, 128, 415–435. doi: 10.1084/jem.128.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varol, C. , Mildner, A. , & Jung, S. (2015). Macrophages: Development and tissue specialization. Annual Review of Immunology, 33, 643–675. doi: 10.1146/annurev-immunol-032414-112220. [DOI] [PubMed] [Google Scholar]
- Williams, J. W. , Zaitsev, K. , Kim, K.‐W. , Ivanov, S. , Saunders, B. T. , Schrank, P. R. , … Randolph, G. J. (2020). Limited proliferation capacity of aortic intima resident macrophages requires monocyte recruitment for atherosclerotic plaque progression. Nature Immunology, 21(10), 1194–1204. doi: 10.1038/s41590-020-0768-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, J. W. , Giannarelli, C. , Rahman, A. , Randolph, G. J. , & Kovacic, J. C. (2018). Macrophage biology, classification, and phenotype in cardiovascular disease: JACC macrophage in CVD series (Part 1). Journal of the American College of Cardiology, 72, 2166–2180. doi: 10.1016/j.jacc.2018.08.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z. , Rao, Y. , Huang, Y. , Zhou, T. , Feng, R. , Xiong, S. , … Peng, B. (2020). Efficient strategies for microglia replacement in the central nervous system. Cell Reports, 32, 108041. doi: 10.1016/j.celrep.2020.108041. [DOI] [PubMed] [Google Scholar]
- Yona, S. , Kim, Ki‐W. , Wolf, Y. , Mildner, A. , Varol, D. , Breker, M. , … Jung, S. (2013). Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity, 38, 79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, N. , Kim, S. H. , Gainullina, A. , Erlich, E. C. , Onufer, E. J. , Kim, J. , … Kim, K.‐W. (2021). Lyve1+ macrophages of murine peritoneal mesothelium promote omentum‐independent ovarian tumor growth. Journal of Experimental Medicine, 218, e20210924. doi: 10.1084/jem.20210924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


