Abstract
Aims
Recent studies suggest an association between cardiovascular disease (CVD) and cancer incidence/mortality, but the pathophysiological mechanisms underlying these associations are unclear. We aimed to examine biomarkers previously associated with CVD and study their association with incident cancer and cancer-related death in a prospective cohort study.
Methods and results
We used a proteomic platform to measure 71 cardiovascular biomarkers among 5032 participants in the Framingham Heart Study who were free of cancer at baseline. We used multivariable-adjusted Cox models to examine the association of circulating protein biomarkers with risk of cancer incidence and mortality. To account for multiple testing, we set a 2-sided false discovery rate <0.05. Growth differentiation factor-15 (also known as macrophage inhibitory cytokine-1) was associated with increased risk of incident cancer [hazards ratio (HR) per 1 standard deviation increment 1.31, 95% CI 1.17–1.47], incident gastrointestinal cancer (HR 1.85, 95% CI 1.37–2.50), incident colorectal cancer (HR 1.94, 95% CI 1.29–2.91), and cancer-related death (HR 2.15, 95% CI 1.72–2.70). Stromal cell-derived factor-1 showed an inverse association with cancer-related death (HR 0.75, 95% CI 0.65–0.86). Fibroblast growth factor-23 showed an association with colorectal cancer (HR 1.55, 95% CI 1.20–2.00), and granulin was associated with haematologic cancer (HR 1.61, 95% CI 1.30–1.99). Other circulating biomarkers of inflammation, immune activation, metabolism, and fibrosis showed suggestive associations with future cancer diagnosis.
Conclusion
We observed several significant associations between circulating CVD biomarkers and cancer, supporting the idea that shared biological pathways underlie both diseases. Further investigations of specific mechanisms that lead to both CVD and cancer are warranted.
Keywords: GDF15, cardio-oncology, cohort study, cancer, biomarker
Graphical Abstract
Graphical Abstract.
1. Introduction
Cancer and cardiovascular disease (CVD) are the two leading causes of death in the USA. While epidemiological studies focused on cause-specific death have traditionally considered CVD and cancer separately, recent studies link cancer and oncologic therapies with subsequent CVD, leading to the development of a new field of study, that of cardio-oncology.1–4 Conversely, some studies suggest that CVD itself may be a risk factor for cancer incidence/mortality, independent of shared risk factors like obesity, smoking, and diabetes.1,5,6 The exact mechanisms underlying the association of CVD and cancer are unclear, although recent investigations suggest possible common pathways.7
For example, clonal haematopoiesis of indeterminate potential, which increases the risk of haematologic malignancies, seems also associated with atherosclerotic CVD, probably by the mediation of aberrant inflammatory activity.8 In a recent experimental model a direct link between heart failure (HF) and increased tumorigenicity was hypothesized as several myocardial secreted proteins promoted tumour growth.2 Furthermore, baseline N-terminal pro-B-type natriuretic peptide was associated with cancer incidence in a population-based prospective cohort study during a 12-year follow-up.2 In light of these and similar studies, which demonstrate that circulating proteins may elucidate potential pathophysiological mechanisms linking CVD and cancer, we sought to examine a broad set of proteins previously linked to CVD and representing pathways, such as inflammation, neuro-hormonal activation, oxidative stress, and immune activation,1 and in turn examine these proteins in relation to development of future cancer diagnosis.
Specifically, we leveraged a panel of biomarkers through the Systems Approach to Biomarker Research in Cardiovascular Disease (SABRe CVD) Initiative. This is a targeted proteomic platform established by the National Heart, Lung, and Blood Institute with the aim of identifying biomarker signatures of atherosclerotic CVD and its risk factors. To that end, 85 candidate protein biomarkers for CVD were measured in participants in the Framingham Heart Study (FHS), based on their known or presumed association with CVD including HF, myocardial infarction (MI), and atherosclerotic cardiovascular disease. These circulating biomarkers reflected various pathways including inflammation, oxidative stress, adiposity, metabolism, and fibrosis and tissue remodelling. We previously demonstrated associations between many of these proteins and CVD.9 In this study, we hypothesized that these CVD-associated biomarkers may also be associated with cancer incidence and cancer-related death, and may elucidate potential biological pathways underlying the clinical/epidemiological observations that highlight greater cancer risk among individuals with CVD. If confirmed, these epidemiologic data could lead to mechanistic insights on the pathophysiological processes underlying both cancer and CVD.
2. Methods
2.1 Study sample
We studied participants from the FHS Offspring and Third Generation cohorts who attended Exam 7 (1998–2001) and Exam 1 (2002–05), respectively, were at least 40 years of age at their baseline examination, and were included in the SABRe CVD Initiative protein assay project.9 Since prevalent CVD and/or cancer may influence biomarker levels, individuals with baseline HF (n = 48), MI (n = 149), end-stage renal disease (n = 24), and history of cancer (n = 368) were excluded in order to reduce the risk of reverse causation. Furthermore, participants with missing key clinical covariates (n = 81) and follow-up time (n = 7) were also excluded. A final sample of 5032 participants was included in analyses. This study was approved by the appropriate Institutional Review Board and all participants provided written informed consent. This study abides by the principles outlined in the Declaration of Helsinki.
2.2 Study procedures
Participants underwent review of comprehensive medical history, physical examination, and anthropometrics at the baseline examination. Blood samples were obtained after an overnight fast and samples were processed immediately and stored at −80°C for further processing. As part of the SABRe CVD Initiative, we used a discovery-based proteomic platform (Luminex xMAP multiplex assay, Sigma-Aldrich, St. Louis, MO) to measure 85 circulating plasma biomarkers as previously described.9 Candidate biomarkers were selected based on previous associations with atherosclerotic CVD, gene expression profiling, published genome-wide association studies, and discovery proteomics. In brief, targets were assessed based on factors, such as dilution rate, cross-reactivity, and when the target was added to the assay list, to develop 17 unique multiplex panels. Previously described detailed protocols for multiplex assay development were followed.9 To account for low-abundance biomarkers, high-abundance biomarkers were depleted using an antibody-based resin designed to deplete 95% of total plasma protein (ProteoPrep 20, Sigma-Aldrich).9 Of 85 measured biomarkers, 14 had >25% of the samples with measurements below the detection limit. We therefore focused the current analysis on the remaining 71 biomarkers (details in Supplementary material online, Table S1).
2.3 Ascertainment of incident cancer outcomes
Participants were followed longitudinally after their baseline examination for the occurrence of incident cancer. Potential cancer cases were identified through surveillance of routine examinations, annual health updates, and medical records including outpatient visits, pathology reports, hospital admissions, or death records through 31 December 2016. We reviewed medical records and pathology reports. Cancer cases were adjudicated and coded based on topology, morphology, and graded by two independent physicians, with discrepancies resolved after discussion and re-review of cases with a third physician. All malignancies except non-melanoma skin cancers were included for analysis. Cause of death was ascertained by a 3-physician panel after review of death certificates, hospital admission records, and medical records. Cancer death was adjudicated based on a prior diagnosis of cancer and the identification of cancer as the primary cause of death.
2.4 Statistical analysis
Baseline characteristics were summarized using means ± standard deviation (SD) or medians and inter-quartile range (IQR) for continuous variables and percentages for dichotomous variables. We examined the association of CVD protein biomarkers with incident cancer using multivariable (MV) Cox regression models. Variables included in the model were chosen a priori based on clinical knowledge of factors known to be confounders for cancer incidence. Biomarkers were rank normalized with effect sizes expressed per 1-SD difference. Higher SD denotes higher absolute values. The proportional hazards assumption was tested for each biomarker and for each incident cancer analysis—there was no violation of model assumptions for any of the biomarkers. The primary outcome of interest was incident cancer (including fatal and non-fatal). Secondary outcomes included cancer-related death and incidence of specific cancer subtypes, including gastrointestinal (GI), lung, prostate, and breast cancers. We examined Cox models (i) adjusted for age and sex and (ii) further adjusted for body mass index, smoking status (current, former, and never), systolic blood pressure, hypertension treatment, diabetes mellitus, alcohol use, and aspirin use. For all analyses, follow-up was censored at time of event, death, or at 15-years of follow-up. Analyses accounted for cohort (Offspring vs. Third Generation) as a stratum variable.
For secondary analyses examining incident breast cancer, we restricted the sample to women given the low incidence of breast cancer among men and additionally adjusted for menopausal status. Similarly, prostate cancer analyses were restricted to men.
We performed two types of sensitivity analyses. First, we excluded cancer events occurring within 6 months of baseline examination, in order to minimize the inclusion of undiagnosed prevalent cancers prior to the baseline examination. Second, in the MV model, we adjusted for pack-years in addition to smoking status to further fine-tune this cancer risk factor. A 2-sided false discovery rate (FDR) Q-value <0.05 was considered statistically significant for primary analyses, with a P-value of <0.05 considered suggestive. Cox models met the proportionality hazards assumption. Statistical analyses were performed using SAS version 9.4 (SAS Institute Inc., Cary, NC).
3. Results
We studied 5032 FHS Offspring and Third Generation cohort participants who were without a cancer diagnosis at the baseline examination (mean age 55 ± 10, 54% women). CVD comorbidities included 24% with hypertension, 6% with diabetes mellitus, 29% with obesity (BMI ≥30 kg/m2), and 15% current smokers (Table 1 for all baseline characteristics). Over a median follow-up of 11.8 years (IQR 10.5–15.0), there were 841 incident cancer events, including 221 cancer deaths.
Table 1.
Baseline characteristics of participants by cohort (Offspring at Exam 7 and Gen 3 at Exam 1)
| Clinical characteristica | Offspring | Generation 3 | Overall |
|---|---|---|---|
| n = 2938 | n = 2094 | N = 5032 | |
| Age, years | 61 (9) | 47 (5) | 55 (10) |
| Women, n (%) | 1638 (56) | 1094 (52) | 2732 (54) |
| Body mass index, kg/m2 | 28.1 (5.4) | 27.6 (5.7) | 27.9 (5.5) |
| Waist circumference, cm | 100 (14) | 96 (15) | 98 (15) |
| Systolic blood pressure, mmHg | 127 (19) | 120 (15) | 124 (18) |
| Diastolic blood pressure, mmHg | 74 (10) | 77 (10) | 75 (10) |
| Hypertension treatment, n (%) | 940 (32) | 272 (13) | 1212 (24) |
| Diabetes mellitus, n (%) | 250 (9) | 67 (3) | 317 (6) |
| Total cholesterol, mmol/L | 5.23 (0.93) | 5.05 (0.91) | 5.15 (0.93) |
| HDL cholesterol, mmol/L | 1.40 (0.44) | 1.42 (0.44) | 1.42 (0.44) |
| LDL cholesterol, mmol/L | 3.13 (0.85) | 3.03 (0.83) | 3.08 (0.83) |
| Triglycerides, mmol/L | 1.52 (0.96) | 1.37 (1.03) | 1.46 (0.99) |
| eGFR, mL/min/1.73 m2 | 69 (14) | 99 (12) | 82 (20) |
| Physical activity index (PAI) | 38 (6) | 37 (8) | 38 (7) |
| Current smoking, n (%) | 400 (14) | 358 (17) | 758 (15) |
| Current aspirin users, n (%) | 934 (32) | 285 (14) | 1219 (24) |
| Modest-high alcohol intake, n (%) | 462 (16) | 173 (8) | 635 (13) |
| Post-menopausalb, n (%) | 1386 (85) | 324 (30) | 1710 (63) |
HDL, high-density lipoprotein; LDL, low-density lipoprotein; eGFR, estimated glomerular filtration rate.
Continuous variables presented as mean (SD), categorical variables presented as number (%).
Only among women.
3.1 Association of cardiovascular biomarkers with incident cancer and cancer death
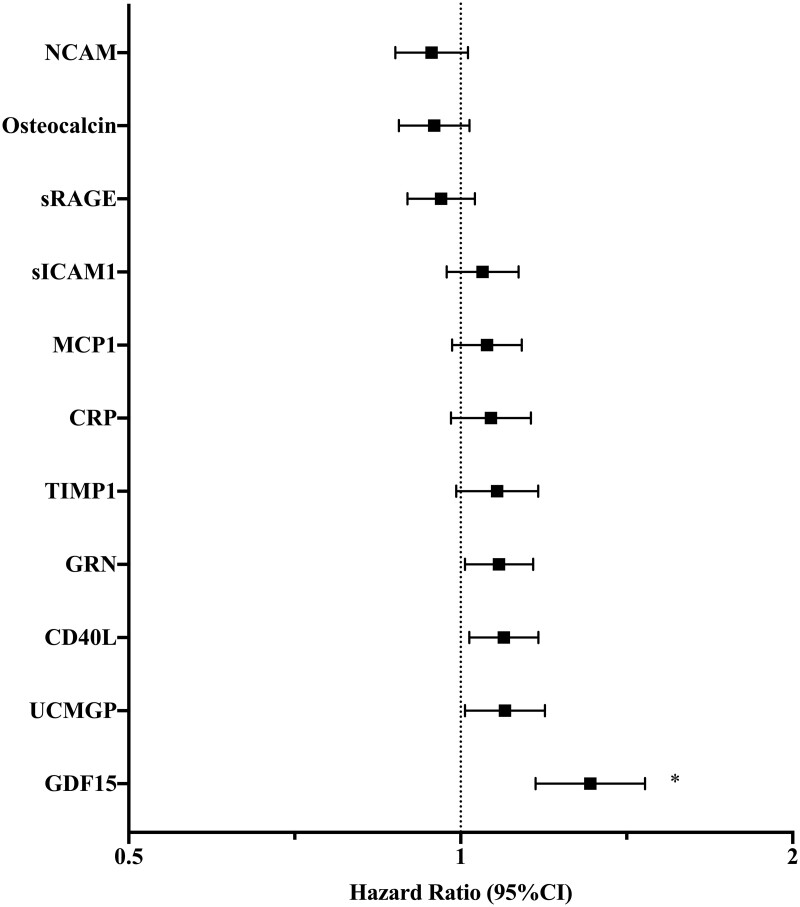
Of 71 biomarkers examined, 11 had suggestive associations with incident cancer in age- and sex-adjusted analyses at a P-value <0.05 (Table 2 and Figure 1). After MV-adjustment, four biomarkers remained associated with incident cancer at P < 0.05: growth differentiation factor-15 (GDF15), soluble CD40 ligand (CD40L), granulin (GRN), and uncarboxylated matrix gamma-carboxyglutamate protein (UCMGP). Of these biomarkers, GDF15 displayed the strongest association with incident cancer and met the pre-specified FDR threshold for significance. Specifically, a 1-SD increase in GDF15 was associated with a 31% increased risk of future cancer (HR 1.31, 95% CI 1.17–1.47, P < 0.0001, Q = 0.0002 in MV analyses). Among those with cancer incidence, the median GDF-15 value was 620 pg/mL (IQR 488–848), while among those without cancer it was 503 pg/mL (IQR 390–674; Supplementary material online, Table S6). The addition of GDF15 significantly improved the C-statistic of the MV model from 0.655 to 0.662 (P = 0.026).
Table 2.
Single biomarker associations with cancer incidence and cancer death
| Age- and sex-adjusted model | Multivariable model | ||||
|---|---|---|---|---|---|
| Biomarker | HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Cancer incidence (n = 841) | GDF15 | 1.38 (1.24–1.53) | <0.0001 * | 1.31 (1.17–1.47) | <0.0001 ** |
| CD40L | 1.11 (1.03–1.19) | 0.004 | 1.09 (1.02–1.18) | 0.01 | |
| NCAM | 0.90 (0.84–0.97) | 0.005 | 0.94 (0.87–1.02) | 0.12 | |
| GRN | 1.10 (1.02–1.18) | 0.01 | 1.08 (1.01–1.16) | 0.03 | |
| UCMGP | 1.11 (1.02–1.20) | 0.01 | 1.10 (1.01–1.19) | 0.03 | |
| CRP | 1.10 (1.02–1.19) | 0.01 | 1.07 (0.98–1.16) | 0.14 | |
| sICAM1 | 1.09 (1.02–1.17) | 0.02 | 1.05 (0.97–1.13) | 0.23 | |
| TIMP1 | 1.09 (1.01–1.19) | 0.04 | 1.08 (0.99–1.18) | 0.08 | |
| sRAGE | 0.93 (0.87–1.00) | 0.04 | 0.96 (0.89–1.03) | 0.26 | |
| Osteocalcin | 0.93 (0.86–1.00) | 0.04 | 0.95 (0.88–1.02) | 0.14 | |
| MCP1 | 1.08 (1.00–1.16) | 0.047 | 1.06 (0.98–1.14) | 0.14 | |
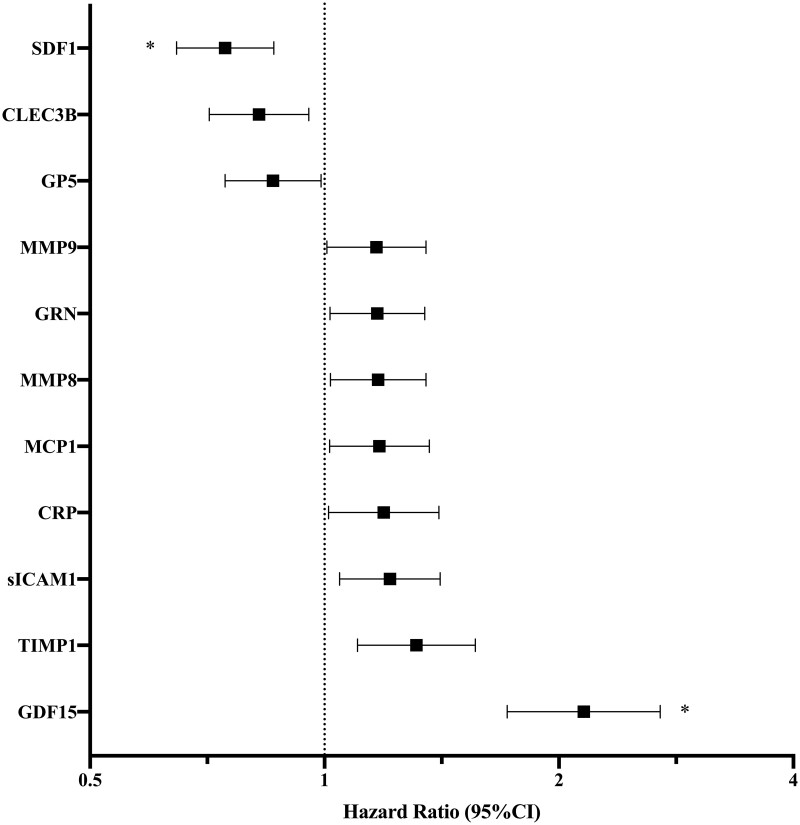
| Cancer death (n = 221) | GDF15 | 2.58 (2.10–3.15) | <0.0001* | 2.15 (1.72–2.70) | <0.0001 *** |
| SDF1 | 0.71 (0.62–0.82) | <0.0001* | 0.75 (0.65–0.86) | <0.0001 **** | |
| sICAM1 | 1.34 (1.16–1.55) | <0.0001* | 1.21 (1.05–1.41) | 0.01 | |
| MMP9 | 1.33 (1.14–1.54) | <0.0001* | 1.17 (1.01–1.35) | 0.04 | |
| TIMP1 | 1.39 (1.17–1.65) | 0.0002* | 1.31 (1.10–1.56) | 0.002 | |
| CRP | 1.28 (1.10–1.49) | 0.001* | 1.19 (1.01–1.40) | 0.04 | |
| MCP1 | 1.25 (1.08–1.44) | 0.003* | 1.18 (1.02–1.36) | 0.03 | |
| CLEC3B | 0.81 (0.70–0.93) | 0.003* | 0.82 (0.71–0.95) | 0.01 | |
| MMP8 | 1.23 (1.07–1.41) | 0.004* | 1.17 (1.02–1.35) | 0.03 | |
| GP5 | 0.81 (0.71–0.94) | 0.004* | 0.86 (0.75–0.99) | 0.04 | |
| GRN | 1.21 (1.06–1.39) | 0.006* | 1.17 (1.02–1.34) | 0.03 | |
| clusterin | 0.80 (0.68–0.95) | 0.01 | 0.82 (0.69–0.97) | 0.02 | |
| PAI1 | 1.21 (1.04–1.40) | 0.01 | 1.07 (0.91–1.26) | 0.04 | |
| MPO | 1.18 (1.03–1.35) | 0.01 | 1.13 (0.99–1.30) | 0.07 | |
| ADM | 1.23 (1.04–1.47) | 0.02 | 1.11 (0.92–1.34) | 0.28 | |
| LPA | 0.85 (0.74–0.97) | 0.02 | 0.87 (0.76–1.00) | 0.06 | |
| KLKB1 | 0.84 (0.73–0.98) | 0.02 | 0.84 (0.72–0.98) | 0.02 | |
| IGFBP2 | 1.18 (1.02–1.37) | 0.03 | 1.23 (1.04–1.44) | 0.01 | |
| IGF1 | 0.84 (0.72–0.98) | 0.03 | 0.90 (0.77–1.04) | 0.16 | |
| CD14 | 1.17 (1.02–1.35) | 0.03 | 1.08 (0.93–1.25) | 0.33 | |
| GMP_140 | 1.17 (1.01–1.34) | 0.03 | 1.08 (0.94–1.25 | 0.27 | |
| PMP2 | 0.86 (0.75–0.99) | 0.04 | 0.89 (0.77–1.02) | 0.09 | |
| sRAGE | 0.87 (0.76–0.99) | 0.04 | 0.93 (0.81–1.06) | 0.27 | |
| NCAM | 0.87 (0.75–1.00) | 0.05 | 0.96 (0.83–1.11) | 0.57 | |
Multivariable model adjusted for age, sex, BMI, systolic blood pressure, hypertension treatment, diabetes mellitus, smoking, alcohol use, and aspirin use. Displayed raw P-values <0.05 are suggestive of association between biomarker and cancer incidence. Bolded rows denote biomarkers with a significant false discovery rate Q-value <0.05 in age- and sex-adjusted models as well as multivariable models.
Denotes Q-value <0.05.
Q = 0.0002.
Q < 0.0001.
Q = 0.002.
Figure 1.
MV-adjusted associations of biomarkers with incident cancer. The MV Cox regression model was adjusted for age, sex, body mass index, smoking status (current, former, and never), systolic blood pressure, hypertension treatment, diabetes mellitus, alcohol use, and aspirin use. Displayed biomarkers meet P < 0.05 in age- and sex-adjusted Cox regression analyses. * denotes GDF15 P-value < 0.0001 and FDR Q-value = 0.0002. Remaining biomarkers do not meet statistical significance in MV analyses.
There were 24 biomarkers with at least suggestive associations for cancer death in age- and sex-adjusted analyses (P < 0.05), 11 of which also met an FDR Q-value <0.05 (Table 2). This included GDF15 in addition to other markers of inflammation [intercellular adhesion molecule-1 (sICAM1), C-reactive protein (CRP), monocyte chemotactic molecule-1 (MCP1)], and immune activation and fibrosis [matrix metallopeptidase-9 (MMP9), tissue inhibitor of metalloproteinases-1 (TIMP1), and matrix metallopeptidase-8 (MMP8)]. After adjustment for clinical covariates, GDF15 and stromal cell-derived factor-1 (SDF1) remained significantly associated with cancer death (Table 2 and Figure 2). Specifically, a 1-SD increase in GDF15 was associated with a greater than two-fold increased hazards of cancer death (HR 2.15, 95% CI 1.72–2.70, P < 0.0001, Q < 0.0001). By contrast, a 1-SD increase in SDF1 was associated with a 25% decreased hazards of cancer death (HR 0.75, 95% CI 0.65–0.86, P < 0.0001, Q = 0.002).
Figure 2.
MV-adjusted associations of biomarkers with cancer-related death. The MV Cox regression model was adjusted for age, sex, body mass index, smoking status (current, former, and never), systolic blood pressure, hypertension treatment, diabetes mellitus, alcohol use, and aspirin use. Displayed biomarkers meet P < 0.05 in age- and sex-adjusted Cox regression analyses. * denotes MV FDR Q-value <0.05, specifically SDF1 Q = 0.002 and GDF15 Q < 0.0001, P-value < 0.0001 for both. Remaining biomarkers do not meet statistical significance in MV analyses.
3.2 CVD biomarkers and cancer subtypes
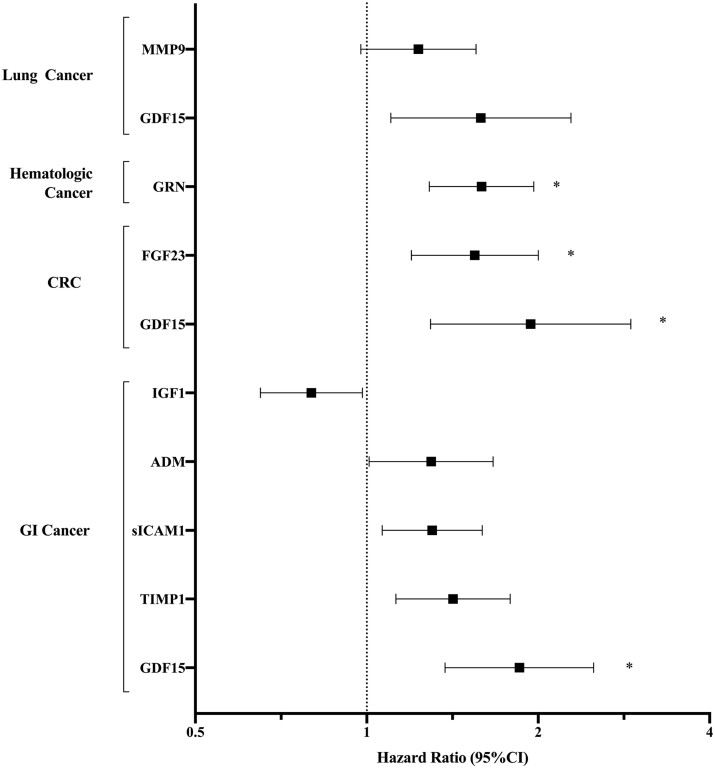
We examined biomarker associations with specific cancers [GI, colorectal cancer (CRC, a subset of GI cancers), haematologic, lung, melanoma, bladder, gynaecologic, breast, and prostate] in secondary analyses. We found that 15 biomarkers had suggestive associations with GI cancer (n = 118, P < 0.05 for all, Table 3) in age- and sex-adjusted analyses and 9 remained associated at P < 0.05 after further adjustment for clinical confounders, including GDF15, TIMP1, apolipoprotein B (APOB), and fibroblast growth factor-23 (FGF23) (Table 3 and Figure 3). GDF15 had the most robust association with GI cancer, with an 85% increase in risk of incident GI cancer per 1-SD change (HR 1.85, 95% CI 1.37–2.50, P < 0.0001, Q = 0.004). Notably, GDF15 was specifically associated with incident CRC (n = 65, HR 1.94, 95% CI 1.29–2.91, P = 0.001, Q = 0.047), as was FGF23 (HR 1.55, 95% CI 1.20–2.00, P = 0.0008, Q = 0.047) (Table 3 and Figure 3).
Table 3.
Single biomarker associations with site-specific cancer incidence
| Age- and sex-adjusted model | Multivariable model | ||||
|---|---|---|---|---|---|
| Biomarker | HR (95% CI) | P-value | HR (95% CI) | P-value | |
| GI cancer (n = 118) | GDF15 | 2.10 (1.59–2.76) | <0.0001 * | 1.85 (1.37–2.50) | <0.0001 ** |
| TIMP1 | 1.55 (1.23–1.95) | 0.00018* | 1.42 (1.13–1.79) | 0.003 | |
| sICAM1 | 1.38 (1.14–1.68) | 0.001* | 1.30 (1.06–1.60) | 0.01 | |
| ADM | 1.46 (1.16–1.85) | 0.002* | 1.30 (1.01–1.67) | 0.04 | |
| IGF1 | 0.73 (0.59–0.89) | 0.002* | 0.80 (0.65–0.98) | 0.03 | |
| sRAGE | 0.76 (0.64–0.92) | 0.004 | 0.83 (0.69–1.01) | 0.06 | |
| sGP130 | 1.32 (1.09–1.61) | 0.005 | 1.25 (1.04–1.54) | 0.02 | |
| APOB | 0.77 (0.63–0.94) | 0.009 | 0.74 (0.61–0.90) | 0.0025 | |
| FGF23 | 1.28 (1.06–1.55) | 0.009 | 1.29 (1.06–1.56) | 0.009 | |
| Osteocalcin | 0.79 (0.65–0.95) | 0.01 | 0.85 (0.69–1.03) | 0.10 | |
| UCMGP | 1.31 (1.05–1.62) | 0.02 | 1.22 (0.98–1.54) | 0.08 | |
| clusterin | 0.77 (0.61–0.96) | 0.02 | 0.76 (90.61–0.95) | 0.02 | |
| CLEC3B | 0.80 (0.66–0.97) | 0.03 | 0.83 (0.68–1.01) | 0.06 | |
| PAI1 | 1.24 (1.02–1.52) | 0.03 | 1.05 (0.84– 1.32) | 0.06 | |
| GRN | 1.21 (1.00–1.46) | 0.05 | 1.16 (0.96–1.40) | 0.12 | |
| CRC (n = 65) | GDF15 | 2.09 (1.45–3.02) | <0.0001 * | 1.94 (1.29–2.91) | 0.001 *** |
| FGF23 | 1.53 (1.19–1.97) | 0.0011 * | 1.55 (1.20–2.00) | 0.0008 *** | |
| ADM | 1.62 (1.19–2.21) | 0.002 | 1.51 (1.08–2.12) | 0.02 | |
| TIMP1 | 1.53 (1.13–2.07) | 0.007 | 1.45 (1.07–1.98) | 0.02 | |
| UCMGP | 1.47 (1.10–1.98) | 0.01 | 1.41 (1.04–1.92) | 0.03 | |
| PAI1 | 1.40 (1.07–1.82) | 0.01 | 1.27 (0.94–1.70) | 0.12 | |
| sRAGE | 0.74 (0.58–0.95) | 0.02 | 0.80 (0.62–1.03) | 0.08 | |
| DPP4 | 0.74 (0.58–0.95) | 0.02 | 0.75 (0.59–0.96) | 0.02 | |
| EFEMP1 | 1.45 (1.06–1.96) | 0.02 | 1.36 (0.99–1.86) | 0.05 | |
| sICAM1 | 1.34 (1.03–1.74) | 0.03 | 1.24 (0.95–1.63) | 0.12 | |
| APOB | 0.76 (0.58–0.99) | 0.04 | 0.72 (0.55–0.94) | 0.02 | |
| BCHE | 0.77 (0.60–1.00) | 0.05 | 0.75 (0.58–0.97) | 0.03 | |
| clusterin | 0.74 (0.55–1.00) | 0.05 | 0.74 (0.55–1.00) | 0.05 | |
| Haematologic cancer (n = 97) | GRN | 1.62 (1.30–1.99) | <0.0001 * | 1.61 (1.30–1.99) | <0.0001 **** |
| MMP9 | 1.37 (1.11–1.70) | 0.02 | 1.36 (1.09–1.69) | 0.02 | |
| B2M | 1.43 (1.10–1.86) | 0.02 | 1.44 (1.10–1.87) | 0.02 | |
| MCP1 | 1.30 (1.04–1.62) | 0.02 | 1.30 (1.04–1.62) | 0.02 | |
| COL18A1 | 1.27 (1.03–1.57) | 0.02 | 1.27 (1.02–1.57) | 0.02 | |
| SAA1 | 1.26 (1.01–1.57) | 0.02 | 1.31 (1.05–1.65) | 0.01 | |
| Lung cancer (n = 87) | GDF15 | 2.14 (1.55–2.97) | <0.0001* | 1.59 (1.10–2.28) | 0.01 |
| MMP9 | 1.52 (1.21–1.92) | 0.008* | 1.23 (0.98–1.55) | 0.08 | |
| sRAGE | 0.75 (0.61–0.93) | 0.008 | 0.77 (0.61–0.95) | 0.02 | |
| sICAM1 | 1.35 (1.07–1.70) | 0.01 | 1.15 (0.91–1.46) | 0.25 | |
| SDF1 | 0.75 (0.60–0.94) | 0.012 | 0.79 (0.63–0.99) | 0.04 | |
| DPP4 | 0.76 (0.62–0.94) | 0.01 | 0.77 (0.62–0.95) | 0.01 | |
| AGP1 | 1.30 (1.04–1.64) | 0.02 | 1.20 (0.95–1.52) | 0.12 | |
| Leptin | 0.76 (0.60–0.98) | 0.03 | 1.01 (0.73–1.40) | 0.95 | |
| PON1 | 0.78 (0.62–0.98) | 0.03 | 0.83 (0.67–1.03) | 0.09 | |
| NCAM | 0.78 (0.62–0.98) | 0.03 | 0.88 (0.70–1.10) | 0.26 | |
| ANGPTL3 | 0.78 (0.62–0.98) | 0.03 | 0.80 (0.63–1.02) | 0.07 | |
| CLEC3B | 0.79 (0.63–0.99) | 0.04 | 0.82 (0.65–1.04) | 0.10 | |
| PPBP | 1.25 (1.01–1.55) | 0.04 | 1.16 (0.94–1.44) | 0.18 | |
| IGFBP1 | 1.29 (1.01–1.65) | 0.04 | 1.02 (0.77–1.35) | 0.89 | |
| Notch1 | 0.80 (0.64–0.99) | 0.04 | 0.85 (0.67–1.06) | 0.15 | |
| CRP | 1.27 (1.00–1.62) | 0.05 | 1.29 (1.01–1.65) | 0.04 | |
| haemopexin | 0.79 (0.62–1.00) | 0.05 | 0.83 (0.65–1.06) | 0.14 | |
| Melanoma (n = 90) | MCP1 | 0.71 (0.58–0.88) | 0.0016 | 0.71 (0.57–0.88) | 0.002 |
| SERPINA10 | 1.30 (1.04–1.61) | 0.02 | 1.30 (1.05–1.61) | 0.02 | |
| CNTN1 | 1.32 (1.05–1.66) | 0.02 | 1.36 (1.07–1.72) | 0.01 | |
| MMP9 | 0.78 (0.63–0.97) | 0.02 | 0.78 (0.63–0.97) | 0.03 | |
| CXCL16 | 0.81 (0.65–1.00) | 0.05 | 0.80 (0.65–1.00) | 0.05 | |
| Bladder cancer (n = 58) | Osteocalcin | 0.71 (0.54–0.93) | 0.01 | 0.69 (0.52–0.93) | 0.01 |
| EFEMP1 | 0.72 (0.54–0.97) | 0.03 | 0.74 (0.55–1.01) | 0.06 | |
| PAI1 | 1.37 (1.01–1.84) | 0.04 | 1.59 (1.16–2.18) | 0.004 | |
| Gynaecologic cancer (n = 48) | Osteocalcin | 0.70 (0.52–0.94) | 0.02 | 0.73 (0.53–1.00) | 0.05 |
| IGFBP2 | 0.71 (0.52–0.96) | 0.03 | 0.79 (0.57–1.11) | 0.17 | |
| VEGF | 0.70 (0.51–0.97) | 0.03 | 0.67 (0.48–0.93) | 0.02 | |
| Breast cancer (n = 159) | ANGPTL3 | 1.20 (1.02–1.41) | 0.03 | 1.18 (0.99–1.40) | 0.07 |
| Prostate cancer (n = 149) | REG1A | 0.76 (0.64–0.91) | 0.002 | 0.76 (0.63–0.90) | 0.002 |
| Leptin-R | 1.26 (1.07–1.49) | 0.005 | 1.26 (1.07–1.49) | 0.005 | |
Multivariable model adjusted for age, sex, BMI, systolic blood pressure, hypertension treatment, diabetes mellitus, smoking, alcohol use, and aspirin use. Breast cancer and gynaecologic cancer analyses were performed only in women. The multivariable analysis was further adjusted for menopausal status. Prostate cancer analysis was performed only in men. Displayed raw P-values <0.05 are suggestive of association between biomarker and cancer incidence. Bolded rows denote biomarkers with a significant false discovery rate Q-value <0.05 in age- and sex-adjusted models as well as multivariable models.
Denotes Q-value <0.05.
Q = 0.004.
Q = 0.047.
Q = 0.0008.
Figure 3.
MV-adjusted associations of biomarkers with site-specific incident cancers. The MV Cox regression model was adjusted for age, sex, body mass index, smoking status (current, former, and never), systolic blood pressure, hypertension treatment, diabetes mellitus, alcohol use, and aspirin use. Breast cancer and gynaecologic cancer analyses were performed only in women and the MV model was further adjusted for menopausal status. Prostate cancer analysis was performed only in men. Displayed biomarkers meet P < 0.05 in age- and sex-adjusted Cox regression analyses. * denotes MV FDR Q-value <0.05. GRN P-value < 0.0001 and Q = 0.0008 for haematologic cancer; FGF23 P = 0.0008 and Q = 0.047, GDF15 P = 0.001 and Q = 0.047 for colorectal cancer (CRC); GDF15 P < 0.0001 and Q = 0.004 for GI cancer. Remaining biomarkers do not meet statistical significance in MV analyses.
GDF15 was also significantly associated with future risk of lung cancer in age- and sex-adjusted analyses (n = 87, HR 2.14, 95% CI 1.55–2.97, P < 0.0001), as was MMP9 (HR 1.52, 95% CI 1.21–1.92, P = 0.008), though both associations were attenuated after MV-adjustment (GDF15 HR 1.59, 95% CI 1.10–2.28, P = 0.01; MMP9 HR 1.23, 95% CI 0.98–1.55, P = 0.08). Other biomarkers with suggestive associations in MV analyses with lung cancer included SDF1, dipeptidyl dipeptidase (DPP4), and CRP (P < 0.05 for all, Table 3 and Figure 3).
We found that six biomarkers had suggestive associations with haematologic cancer (MV-adjusted P < 0.05). The strongest effect size was observed for GRN where a 1-SD higher GRN level was associated with a 61% increased risk of future haematologic cancer (HR 1.61, 95% CI 1.30–1.99, P < 0.0001, Q = 0.0008) (Table 3 and Figure 3). Suggestive associations for haematologic malignancies were also observed for MMP9, beta-2-microglobulin (B2M), MCP1, collagen type XVIII alpha 1 (COL18A1), and serum amyloid A1 (SAA1).
Other suggestive associations for cancer subtypes are summarized in Table 3 and include: five biomarkers for melanoma [MCP1, protein Z-dependent protease inhibitor (SERPINA10), contactin 1 (CNTN1), MMP9, and chemokine (C-X-C motif) ligand 16 (CXCL16)], three for bladder cancer [osteocalcin, EGF containing fibulin-like extracellular matrix protein 1 (EFEMP1), and plasminogen activator inhibitor 1 (PAI1)], three for gynaecologic cancers [osteocalcin, insulin-like growth factor binding protein 2 (IGFBP2), and vascular endothelial growth factor (VEGF)], one for breast cancer [angiopoietin-like 3 (ANGPTL3)], and two for prostate cancer [lithostathine-1-alpha (REG1A) and leptin receptor (leptin-R)]. None of these reached statistical significance at the specified FDR cut-point.
3.3 Sensitivity analyses
In sensitivity analyses, we excluded 38 cancer events that occurred within 6 months of baseline examination. We excluded these patients to reduce the risk of reverse causation under the assumption that cancers diagnosed within 6 months of baseline could have been present but undiagnosed at the baseline examination, thus influencing biomarker levels. Results (Supplementary material online, Table S2) were similar to those of our main analyses. GDF15 remained associated with incident cancer (HR 1.28, 95% CI 1.13–1.43, Q = 0.003), and both GDF15 and SDF1 remained associated with cancer death (GDF15 HR 2.06, 95% CI 1.64–2.60, Q < 0.0001; SDF1 HR 0.76, 95% CI 0.65–0.88, Q = 0.007). Site-specific cancer incidence sensitivity analyses showed similar suggestive associations as the main analysis (Supplementary material online, Table S3). In separate sensitivity analyses, we further adjusted for pack-years and results were similar to those of our main analyses (Supplementary material online, Tables S4 and S5). In some of these secondary analyses, the number of events was relatively small compared to the number of variables included in the MV model (Supplementary material online, Table S5).
4. Discussion
With the growth of cardio-oncology as a field and the recognition that cancer survivors may suffer cardiovascular complications, increasing attention has also been focused on the converse—the idea that CVD in and of itself may be linked to cancer development.1,6 This association may be driven by shared risk factors, though exact mechanisms remain unclear. We recently showed that traditional cardiovascular risk factors were associated with incident cancer among participants in the FHS.7 In this context, we leveraged a targeted proteomic approach to query CVD-associated biomarkers in relation to the development of new-onset cancer in an inception cohort. We found that there was overlap in proteins previously associated with CVD that also preceded the development of cancer. This included proteins representing distinct pathways including inflammation, immune activation, and fibrosis. We observed the most robust associations with cancer incidence and/or cancer-related mortality for GDF15, SDF1, GRN, and FGF23. Many of these proteins had also previously been linked to CVD,9 further strengthening the recently suggested clinical tie between these two diseases. Interestingly, it was recently reported that established tumour biomarkers are increased in patients with prevalent HF and are independently associated with poor outcomes,10 which provides additional support for the bi-directionality between cancer and CVD. Our findings further suggest that shared biological pathways may underlie or characterize both cancer and CVD and highlight the need for future work focused on the identification of at-risk individuals and potential therapeutic avenues.
Among the 71 circulating biomarkers, GDF15 was most consistently associated with risk of incident overall cancer, GI and CRC cancer, and overall cancer-related death. GDF15 is a pleiotropic cytokine with multiple functions, and is considered an indicator of cell injury, oxidative stress, and inflammation.11 GDF15 (also known as macrophage inhibitory cytokine-1) reduces the macrophage response immune surveillance, making it an important promoter of early cancer development, thus leading to increased cancer incidence in experimental models.12,13 In addition, GDF15 has substantial pro-neoplastic activity by stimulating tumour growth, migration, invasion, and immune escape,14,15 and plays a role in cancer-related cachexia, and thus cancer-related death.16 Prior epidemiological studies suggest that GDF15 was associated with increased all-cause and non-cardiovascular mortality.17–19 In prior prospective cohort studies, GDF15 was associated with an increased risk of CRC incidence20 and CRC-related mortality.21 Our findings confirm and expand on these studies. The recently observed association between CVD and cancer 1–4 further lends biological plausibility to our results. Several studies,22 including ours in the FHS,9 have found that GDF15 is associated with future CVD and CVD-related death, and GDF15 has consequently been incorporated in some risk-prediction scores for CVD-related death.23 Recent discovery-based proteomic analyses suggest that GDF15 is strongly correlated with chronological age,24,25 and therefore it is not surprising to observe its association with both CVD and cancer. In addition, a recent genome-wide DNA methylation study has identified 16 genes that influenced circulating GDF15 levels and which were differently methylated in patients with or without CVD.26 Five of these genes were linked to regulation of microRNA-21, one of the most frequently up-regulated miRNAs in solid tumours that post-transcriptionally down-regulates tumour suppressors and thus stimulates invasion, intravasation, and metastasis in cancer.26 While these studies offer some initial insight into the pathophysiological mechanisms that underlie the association between GDF15 and cancer/CVD at the molecular level, our knowledge is still quite limited. These results suggest that GDF15 may play a role in or serve as a biomarker of accelerated biological age, and thus constitute an early detection biomarker, a prognostic factor, or a potential therapeutic target for both cancer and CVD.14,27 The results from this prospective cohort study provide further strong clinical and epidemiological rationale for pursuing mechanistic studies of the role of GDF15 in relation to both CVD and cancer.
Of note, we found that SDF1 (widely known as CXCL12) was significantly associated with a reduced risk of cancer-related mortality though we did not find such an association with incident cancer. Some data suggest that SDF1 may lead to worse cancer outcomes due to its possible involvement in leukocyte trafficking, cross-talking between tumour cells and their microenvironment, and induction of angiogenesis, homing, and metastasis.28–30 Other experimental data, however, suggest that high levels of SDF1 may inhibit colon31–33 and pancreatic34 metastases, and may reduce the deleterious wasting associated with cancer.35 In cross-sectional human studies, high levels of SDF1 have been associated with better outcomes in smaller studies of patients with prevalent GI cancer.36–38 Separately, SDF1 has also been reported to be associated with atherogenic CVD, HF, and all-cause mortality,39,40 and it may be that an inverse association with cancer-related death is observed due to competing risk from CVD-related deaths. Further mechanistic studies to elucidate the association between SDF1 and cancer/CVD-related death are warranted.
Two additional results in our study deserve mention. First, we found that GRN was associated with an increased risk of future haematologic malignancies. This finding is notable because of biological plausibility based on prior smaller studies. Specifically, GRN is a secreted glycoprotein with pleiotropic functions, which include regulation of cell cycle progression, cell motility, and wound repair. It is overexpressed by many cancers and may contribute to their progression.41 In particular, progranulin, a GRN precursor, was found to be an independent predictor of disease progression and overall survival in chronic lymphocytic leukaemia,42 and experimental data suggest that GRN-expressing haematopoietic cells may predispose to other cancers as well.43 We now extend these findings and show that baseline circulating GRN levels portend the future development of haematologic malignancies in a large inception cohort. Additionally within the same sample, we have previously shown GRN to be associated with CVD and all-cause mortality.9 This finding is in line with other studies44,45 and supports the hypothesis that shared biologic pathways may underlie the development of both cancer and CVD.
Second, we found that FGF23 was associated with increased risk for CRC. FGF23, a member of the endocrine FGF subfamily, is normally expressed in osteocytes and has a critical role in phosphate homeostasis. Prior studies suggest that FGF23 is associated with the incidence46 and progression47 of prostate cancer, and with worse outcomes in patients with different types of cancers and bone metastases.48,49 While FGF23 has previously been associated with worse outcomes in CVD,50,51 our study now links FGF23 to CRC specifically, again confirming the possibility that similar processes underlie both conditions.
Finally, we found a number of other proteins representative of distinct pathways with suggestive associations with cancer incidence and/or cancer-related mortality. These include markers of inflammation [sICAM1, CRP, MCP1, interleukin-6 receptor beta (sGP130), butyrylcholine esterase (BCHE), B2M, SAA1, adrenomedullin (ADM)], immune activation [CD40L, GRN, plasma kallikrein (KLKB1), CXCL16, clusterin], metabolic and adipocyte homeostasis [IGFBP2, REG1A, insulin-like growth factor 1 (IGF1), APOB, DPP4, receptor for advanced glycation end-products (sRAGE), osteocalcin, leptin-R], fibrosis (MMP9, TIMP1, MMP8, EFEMP1, COL18A1), and markers of circulatory homeostasis [GP5, PAI1, SERPINA10, VEGF, tetranectin (CLEC3B)]. Many of these biomarkers have also been associated with CVD outcomes9, which suggests that shared biological mechanisms including inflammation and immune activation underlie both conditions. These hypothesis-generating results should be explored in future experimental and/or clinical studies.
There are several limitations to our study. First, our study was a discovery-based observational sample and causal inferences cannot be drawn. In addition, we did not confirm these findings in an independent population. For these reasons, our results need to be considered as exploratory and hypothesis-generating in nature. While the association between GDF15 and CRC has been observed in other prospective cohort studies similar to ours,20,21 no prospective cohort studies are available to confirm our results with regard to the other biomarkers mentioned above. This highlights the need for future studies to confirm our findings in other populations and to elucidate whether these proteins are mechanistically linked to cancer development. Second, although we observed a large number of overall cancers, analyses examining cancer subtype with modest event numbers and smaller effect sizes may not have been detectable. In addition, as an observational study, we cannot rule out the possibility of unmeasured residual confounding. However, we adjusted for many known potential confounding factors, and multivariate adjustment did not substantially alter our age/sex-adjusted effect estimates. Third, our sample was predominantly white, limiting potential generalizability. We also were not able to account for hereditary factors in this analysis. Fourth, a single measurement of biomarkers in relation to subsequent cancer diagnosis may underestimate associations due to regression dilution bias, potentially leading to false negative results. Furthermore, there is overlap in biomarker distributions among those with and without future cancer, and that discriminatory ability of a single biomarker cut-point will not be very useful, particularly in the absence of other clinical criteria. Fifth, we used the FDR method for accounting for multiple testing, which is not is not as robust as other stricter, and potentially over-conservative, methods (e.g. Bonferroni). However, considering the exploratory nature of our study, we were interested in minimizing the risk of false negatives, while at the same time accounting for multiple testing to avoid the risk of false positives. The FDR methods, widely used in epidemiologic research, seem to achieve both these goals. Sixth, considering that we excluded the relatively few patients with prevalent CVD, we cannot comment on the association of biomarkers with cancer incidence/mortality in this subgroup. Finally, limitations of the multiple reactions monitoring mass spectrometry platform include bias towards detection of more-abundant proteins. Nevertheless, our study’s strength includes a large, population-based, prospective study design with detailed follow-up that reduces the potential for recall or misclassification bias. In this context, we were able to leverage detailed CVD-related protein profiling to study associations with future cancer diagnosis.
In conclusion, we observed several significant associations between circulating CVD-related protein biomarkers and new-onset cancer. GDF15 was consistently associated with increased risk of overall cancer, GI and CRC cancer incidence, and overall cancer-related death, SDF1 was significantly associated with a reduced risk of cancer-related mortality, GRN was associated with an increased risk for haematological malignancies, and FGF23 with increased risk for CRC. Furthermore, proteins representing inflammation, immune activation, metabolism, and fibrosis had suggestive associations with future cancer diagnosis. These findings support the idea that shared biological pathways may underlie both CVD and cancer development, as many of the proteins share overlap in both future risk of CVD and cancer. Further investigations in the clinical settings of the role of these biomarkers, especially GDF15, as potential diagnostic and prognostic factors for cancer, particularly for GI cancer and CRC, are warranted. In addition, mechanistic studies on the molecular mechanisms underlying these associations could serve to inform a more precise, molecularly targeted approach to chemoprevention, early diagnosis, and future treatments.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
M.J. and J.E.H. contributed to study design. M.J., E.E.L., E.S.L., S.X.L., K.S.T., B.E.K., G.L.S, S.-J.H., C.Y., T.H., P.C., and J.E.H. contributed to data acquisition. M.J., E.E.L., S.M.P., R.A.d.B., A.D.J., M.G.L., D.L., A.T.C., and J.E.H. contributed to data analyses and interpretation. M.J., E.E.L., and J.E.H. contributed to drafting of the manuscript. All authors participated in critical review and revision of the final manuscript.
Funding
This work was supported by grants from the National Institutes of Health/National Heart, Lung, and Blood Institute [grant numbers N01-HC25195 and HHSN268201500001I (Framingham Heart Study), R01-CA137178 and U19 AG062682 to A.T.C., K24-HL153669, R01-HL134893 and R01-HL140224 to J.E.H.]. The Framingham Heart Study was funded by the National Institutes of Health [grant number N01-HC-25195], which funded the protein biomarker assays. Oversight of the laboratory work for this investigation was conducted by the Division of Intramural Research, National Heart, Lung, and Blood Institute, National Institutes of Health (D.L.).
Conflict of interest: A.T.C. has previously served as a consultant for Bayer Healthcare, Pfizer, Inc., and Janssen Pharmaceuticals. J.E.H. has previously received research grants from Bayer, AG and research supplies from EcoNugenics, Inc. Other authors: no conflicts of interest to declare.
Data availability
Data from Framingham Heart Study participants are publicly available at dbGap according to NIH data sharing policies (study accession phs000007.v29.p10).
Supplementary Material
Contributor Information
Manol Jovani, Division of Gastroenterology, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA; Clinical and Translational Epidemiology Unit, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA; Harvard Medical School, 25 Shattuck Street, Boston, MA 02115, USA; Division of Gastroenterology, University of Kentucky Albert B. Chandler Hospital, 800 Rose Street, Lexington, KY 40536, USA.
Elizabeth E Liu, Cardiovascular Research Center, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA.
Samantha M Paniagua, Cardiovascular Research Center, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA.
Emily S Lau, Corrigan Minehan Heart Center, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA; Cardiology Division, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA; Department of Medicine, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA.
Shawn X Li, Department of Medicine, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA.
Katherine S Takvorian, Department of Medicine, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA.
Bernard E Kreger, General Internal Medicine, Department of Medicine, Boston University School of Medicine, 72 E Concord Street, Boston, MA 02118, USA; The Framingham Heart Study, 73 Mt Wayte Avenue, Framingham, MA 01702, USA.
Greta Lee Splansky, The Framingham Heart Study, 73 Mt Wayte Avenue, Framingham, MA 01702, USA.
Rudolf A de Boer, Department of Cardiology, University Medical Centre Groningen, Hanzeplein 1, 9713 GZ, Groningen, The Netherlands.
Amit D Joshi, Division of Gastroenterology, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA; Clinical and Translational Epidemiology Unit, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA; Harvard Medical School, 25 Shattuck Street, Boston, MA 02115, USA.
Shih Jen Hwang, The Framingham Heart Study, 73 Mt Wayte Avenue, Framingham, MA 01702, USA; Population Sciences Branch, Division of Intramural Research, National Heart, Lung, and Blood Institute, National Institutes of Health, 9000 Rockville Pike, Bethesda, MD 20892, USA.
Chen Yao, The Framingham Heart Study, 73 Mt Wayte Avenue, Framingham, MA 01702, USA; Population Sciences Branch, Division of Intramural Research, National Heart, Lung, and Blood Institute, National Institutes of Health, 9000 Rockville Pike, Bethesda, MD 20892, USA.
Tianxiao Huan, The Framingham Heart Study, 73 Mt Wayte Avenue, Framingham, MA 01702, USA; Population Sciences Branch, Division of Intramural Research, National Heart, Lung, and Blood Institute, National Institutes of Health, 9000 Rockville Pike, Bethesda, MD 20892, USA.
Paul Courchesne, The Framingham Heart Study, 73 Mt Wayte Avenue, Framingham, MA 01702, USA; Population Sciences Branch, Division of Intramural Research, National Heart, Lung, and Blood Institute, National Institutes of Health, 9000 Rockville Pike, Bethesda, MD 20892, USA.
Martin G Larson, The Framingham Heart Study, 73 Mt Wayte Avenue, Framingham, MA 01702, USA; Department of Biostatistics, Boston University School of Public Health, 715 Albany Street, Boston, MA 02118, USA.
Daniel Levy, The Framingham Heart Study, 73 Mt Wayte Avenue, Framingham, MA 01702, USA; Population Sciences Branch, Division of Intramural Research, National Heart, Lung, and Blood Institute, National Institutes of Health, 9000 Rockville Pike, Bethesda, MD 20892, USA.
Andrew T Chan, Division of Gastroenterology, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA; Clinical and Translational Epidemiology Unit, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA; Harvard Medical School, 25 Shattuck Street, Boston, MA 02115, USA.
Jennifer E Ho, Clinical and Translational Epidemiology Unit, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA; Harvard Medical School, 25 Shattuck Street, Boston, MA 02115, USA; Cardiovascular Research Center, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA; Corrigan Minehan Heart Center, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA; Cardiology Division, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA; Department of Medicine, Massachusetts General Hospital, 55 Fruit Street, Boston, MA 02114, USA.
Translational Perspective.
In our prospective cohort study, baseline levels of biomarkers previously associated with CVD were found to be associated with future development of cancer. In particular, GDF15 was associated with increased risk of cancer incidence and mortality, including GI and colorectal cancers; SDF1 was inversely associated with cancer-related death; and FGF23 and GRN were associated with increased risk of colorectal and haematologic cancers, respectively. Other biomarkers of inflammation, immune activation, metabolism, and fibrosis showed suggestive associations. These results suggest potential shared biological pathways that underlie both development of cancer and CVD.
References
- 1. de Boer RA, Meijers WC, van der Meer P, van Veldhuisen DJ.. Cancer and heart disease: associations and relations. Eur J Heart Fail 2019;21:1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meijers WC, Maglione M, Bakker SJL, Oberhuber R, Kieneker LM, Jong S, de Haubner BJ, Nagengast WB, Lyon AR, Vegt B, van der Veldhuisen DJ, van Westenbrink BD, Meer P, van der Silljé HHW, Boer RD.. Heart failure stimulates tumor growth by circulating factors. Circulation 2018;138:678–691. [DOI] [PubMed] [Google Scholar]
- 3. Hasin T, Gerber Y, Weston SA, Jiang R, Killian JM, Manemann SM, Cerhan JR, Roger VL.. Heart failure after myocardial infarction is associated with increased risk of cancer. J Am Coll Cardiol 2016;68:265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Banke A, Schou M, Videbaek L, Møller JE, Torp-Pedersen C, Gustafsson F, Dahl JS, Køber L, Hildebrandt PR, Gislason GH.. Incidence of cancer in patients with chronic heart failure: a long-term follow-up study. Eur J Heart Fail 2016;18:260–266. [DOI] [PubMed] [Google Scholar]
- 5. Meijers WC, de Boer RA.. Common risk factors for heart failure and cancer. Cardiovasc Res 2019;115:844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aboumsallem JP, Moslehi J, de Boer RA.. Reverse cardio-oncology: cancer development in patients with cardiovascular disease. J Am Heart Assoc 2020;9:e013754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lau ES, Paniagua SM, Liu E, Jovani M, Li SX, Takvorian K, Suthahar N, Cheng S, Splansky GL, Januzzi JL, Wang TJ, Vasan RS, Kreger B, Larson MG, Levy D, de Boer RA, Ho JE.. Cardiovascular risk factors are associated with future cancer. JACC CardioOncol 2021;3:48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, McConkey M, Gupta N, Gabriel S, Ardissino D, Baber U, Mehran R, Fuster V, Danesh J, Frossard P, Saleheen D, Melander O, Sukhova GK, Neuberg D, Libby P, Kathiresan S, Ebert BL.. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med 2017;377:111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ho JE, Lyass A, Courchesne P, Chen G, Liu C, Yin X, Hwang S-J, Massaro JM, Larson MG, Levy D.. Protein biomarkers of cardiovascular disease and mortality in the community. J Am Heart Assoc 2018;7:e008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shi C, Wal HH, Silljé HHW, Dokter MM, den Berg F, Huizinga L, Vriesema M, Post J, Anker SD, Cleland JG, Ng LL, Samani NJ, Dickstein K, Zannad F, Lang CC, Haelst PL, Gietema JA, Metra M, Ameri P, Canepa M, Veldhuisen DJ, Voors AA, Boer RA.. Tumor biomarkers: association with heart failure outcomes. J Intern Med 2020;288:207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mimeault M, Batra SK.. Divergent molecular mechanisms underlying the pleiotropic functions of macrophage inhibitory cytokine-1 in cancer. J Cell Physiol 2010;224:626–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ratnam NM, Peterson JM, Talbert EE, Ladner KJ, Rajasekera PV, Schmidt CR, Dillhoff ME, Swanson BJ, Haverick E, Kladney RD, Williams TM, Leone GW, Wang DJ, Guttridge DC.. NF-κB regulates GDF-15 to suppress macrophage surveillance during early tumor development. J Clin Invest 2017;127:3796–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang X, Baek SJ, Eling TE.. The diverse roles of nonsteroidal anti-inflammatory drug activated gene (NAG-1/GDF15) in cancer. Biochem Pharmacol 2013;85:597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fang L, Li F, Gu C.. GDF-15: a multifunctional modulator and potential therapeutic target in cancer. Curr Pharm Des 2019;25:654–662. [DOI] [PubMed] [Google Scholar]
- 15. Corre J, Hébraud B, Bourin P.. Concise review: growth differentiation factor 15 in pathology: a clinical role? Stem Cells Transl Med 2013;2:946–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tsai VWW, Husaini Y, Sainsbury A, Brown DA, Breit SN.. The MIC-1/GDF15-GFRAL pathway in energy homeostasis: implications for obesity, cachexia, and other associated diseases. Cell Metab 2018;28:353–368. [DOI] [PubMed] [Google Scholar]
- 17. Wiklund FE, Bennet AM, Magnusson PKE, Eriksson UK, Lindmark F, Wu L, Yaghoutyfam N, Marquis CP, Stattin P, Pedersen NL, Adami H-O, Grönberg H, Breit SN, Brown DA.. Macrophage inhibitory cytokine-1 (MIC-1/GDF15): a new marker of all-cause mortality. Aging Cell 2010;9:1057–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eggers KM, Kempf T, Wallentin L, Wollert KC, Lind L.. Change in growth differentiation factor 15 concentrations over time independently predicts mortality in community-dwelling elderly individuals. Clin Chem 2013;59:1091–1098. [DOI] [PubMed] [Google Scholar]
- 19. Daniels LB, Clopton P, Laughlin GA, Maisel AS, Barrett-Connor E.. Growth-differentiation factor-15 is a robust, independent predictor of 11-year mortality risk in community-dwelling older adults: the Rancho Bernardo Study. Circulation 2011;123:2101–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mehta RS, Song M, Bezawada N, Wu K, Garcia-Albeniz X, Morikawa T, Fuchs CS, Ogino S, Giovannucci EL, Chan AT.. A prospective study of macrophage inhibitory cytokine-1 (MIC-1/GDF15) and risk of colorectal cancer. J Natl Cancer Inst 2014;106:dju016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mehta RS, Chong DQ, Song M, Meyerhardt JA, Ng K, Nishihara R, Qian Z, Morikawa T, Wu K, Giovannucci EL, Fuchs CS, Ogino S, Chan AT.. Association between plasma levels of macrophage inhibitory cytokine-1 before diagnosis of colorectal cancer and mortality. Gastroenterology 2015;149:614–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Y, Zhen C, Wang R, Wang G.. Growth-differentiation factor-15 predicts adverse cardiac events in patients with acute coronary syndrome: a meta-analysis. Am J Emerg Med 2019;37:1346–1352. [DOI] [PubMed] [Google Scholar]
- 23. Hijazi Z, Oldgren J, Lindbäck J, Alexander JH, Connolly SJ, Eikelboom JW, Ezekowitz MD, Held C, Hylek EM, Lopes RD, Yusuf S, Granger CB, Siegbahn A, Wallentin L, ARISTOTLE and RE-LY Investigators . A biomarker-based risk score to predict death in patients with atrial fibrillation: the ABC (age, biomarkers, clinical history) death risk score. Eur Heart J 2018;39:477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tanaka T, Biancotto A, Moaddel R, Moore AZ, Gonzalez-Freire M, Aon MA, Candia J, Zhang P, Cheung F, Fantoni G, Consortium CHI, Semba RD, Ferrucci L, CHI consortium . Plasma proteomic signature of age in healthy humans. Aging Cell 2018;17:e12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lehallier B, Gate D, Schaum N, Nanasi T, Lee SE, Yousef H, Moran Losada P, Berdnik D, Keller A, Verghese J, Sathyan S, Franceschi C, Milman S, Barzilai N, Wyss-Coray T.. Undulating changes in human plasma proteome profiles across the lifespan. Nat Med 2019;25:1843–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ek WE, Hedman ÅK, Enroth S, Morris AP, Lindgren CM, Mahajan A, Gustafsson S, Gyllensten U, Lind L, Johansson Å.. Genome-wide DNA methylation study identifies genes associated with the cardiovascular biomarker GDF-15. Hum Mol Genet 2016;25:817–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emmerson PJ, Duffin KL, Chintharlapalli S, Wu X.. GDF15 and growth control. Front Physiol 2018;9:1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Janssens R, Struyf S, Proost P.. Pathological roles of the homeostatic chemokine CXCL12. Cytokine Growth Factor Rev 2018;44:51–68. [DOI] [PubMed] [Google Scholar]
- 29. Meng W, Xue S, Chen Y.. The role of CXCL12 in tumor microenvironment. Gene 2018;641:105–110. [DOI] [PubMed] [Google Scholar]
- 30. Mousavi A. CXCL12/CXCR4 signal transduction in diseases and its molecular approaches in targeted-therapy. Immunol Lett 2020;217:91–115. [DOI] [PubMed] [Google Scholar]
- 31. Drury LJ, Ziarek JJ, Gravel S, Veldkamp CT, Takekoshi T, Hwang ST, Heveker N, Volkman BF, Dwinell MB.. Monomeric and dimeric CXCL12 inhibit metastasis through distinct CXCR4 interactions and signaling pathways. Proc Natl Acad Sci USA 2011;108:17655–17660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Drury LJ, Wendt MK, Dwinell MB.. CXCL12 chemokine expression and secretion regulates colorectal carcinoma cell anoikis through Bim-mediated intrinsic apoptosis. PLoS One 2010;5:e12895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wendt MK, Johanesen PA, Kang-Decker N, Binion DG, Shah V, Dwinell MB.. Silencing of epithelial CXCL12 expression by DNA hypermethylation promotes colonic carcinoma metastasis. Oncogene 2006;25:4986–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roy I, Zimmerman NP, Mackinnon AC, Tsai S, Evans DB, Dwinell MB.. CXCL12 chemokine expression suppresses human pancreatic cancer growth and metastasis. PLoS One 2014;9:e90400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martinelli GB, Olivari D, Re Cecconi AD, Talamini L, Ottoboni L, Lecker SH, Stretch C, Baracos VE, Bathe OF, Resovi A, Giavazzi R, Cervo L, Piccirillo R.. Activation of the SDF1/CXCR4 pathway retards muscle atrophy during cancer cachexia. Oncogene 2016;35:6212–6222. [DOI] [PubMed] [Google Scholar]
- 36. Goto M, Yoshida T, Yamamoto Y, Furukita Y, Inoue S, Fujiwara S, Kawakita N, Nishino T, Minato T, Yuasa Y, Yamai H, Takechi H, Seike J, Bando Y, Tangoku A.. CXCR4 expression is associated with poor prognosis in patients with esophageal squamous cell carcinoma. Ann Surg Oncol 2017;24:832–840. [DOI] [PubMed] [Google Scholar]
- 37. Stanisavljević L, Aßmus J, Storli KE, Leh SM, Dahl O, Myklebust MP.. CXCR4, CXCL12 and the relative CXCL12-CXCR4 expression as prognostic factors in colon cancer. Tumour Biol J Biol 2016;37:7441–7452. [DOI] [PubMed] [Google Scholar]
- 38. D'Alterio C, Avallone A, Tatangelo F, Delrio P, Pecori B, Cella L, Pelella A, D'Armiento FP, Carlomagno C, Bianco F, Silvestro L, Pacelli R, Napolitano M, Iaffaioli RV, Scala S.. A prognostic model comprising pT stage, N status, and the chemokine receptors CXCR4 and CXCR7 powerfully predicts outcome in neoadjuvant resistant rectal cancer patients. Int J Cancer 2014;135:379–390. [DOI] [PubMed] [Google Scholar]
- 39. Subramanian S, Liu C, Aviv A, Ho JE, Courchesne P, Muntendam P, Larson MG, Cheng S, Wang TJ, Mehta NN, Levy D.. Stromal cell-derived factor 1 as a biomarker of heart failure and mortality risk. Arterioscler Thromb Vasc Biol 2014;34:2100–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gao J-H, Yu X-H, Tang C-K.. CXC chemokine ligand 12 (CXCL12) in atherosclerosis: an underlying therapeutic target. Clin Chim Acta Int Acta 2019;495:538–544. [DOI] [PubMed] [Google Scholar]
- 41. Bateman A, Bennett HPJ.. The granulin gene family: from cancer to dementia. BioEssays 2009;31:1245–1254. [DOI] [PubMed] [Google Scholar]
- 42. Göbel M, Eisele L, Möllmann M, Hüttmann A, Johansson P, Scholtysik R, Bergmann M, Busch R, Döhner H, Hallek M, Seiler T, Stilgenbauer S, Klein-Hitpass L, Dührsen U, Dürig J.. Progranulin is a novel independent predictor of disease progression and overall survival in chronic lymphocytic leukemia. PLoS One 2013;8:e72107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Elkabets M, Gifford AM, Scheel C, Nilsson B, Reinhardt F, Bray M-A, Carpenter AE, Jirström K, Magnusson K, Ebert BL, Pontén F, Weinberg RA, McAllister SS.. Human tumors instigate granulin-expressing hematopoietic cells that promote malignancy by activating stromal fibroblasts in mice. J Clin Invest 2011;121:784–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kojima Y, Ono K, Inoue K, Takagi Y, Kikuta K, Nishimura M, Yoshida Y, Nakashima Y, Matsumae H, Furukawa Y, Mikuni N, Nobuyoshi M, Kimura T, Kita T, Tanaka M.. Progranulin expression in advanced human atherosclerotic plaque. Atherosclerosis 2009;206:102–108. [DOI] [PubMed] [Google Scholar]
- 45. Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, Pirruccello JP, Muchmore B, Prokunina-Olsson L, Hall JL, Schadt EE, Morales CR, Lund-Katz S, Phillips MC, Wong J, Cantley W, Racie T, Ejebe KG, Orho-Melander M, Melander O, Koteliansky V, Fitzgerald K, Krauss RM, Cowan CA, Kathiresan S, Rader DJ.. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature 2010;466:714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim HJ, Kim K-H, Lee J, Oh JJ, Cheong HS, Wong EL, Yang B-S, Byun SS, Myung SC.. Single nucleotide polymorphisms in fibroblast growth factor 23 gene, FGF23, are associated with prostate cancer risk. BJU Int 2014;114:303–310. [DOI] [PubMed] [Google Scholar]
- 47. Feng S, Wang J, Zhang Y, Creighton CJ, Ittmann M.. FGF23 promotes prostate cancer progression. Oncotarget 2015;6:17291–17301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mansinho A, Ferreira AR, Casimiro S, Alho I, Vendrell I, Costa AL, Sousa R, Abreu C, Pulido C, Macedo D, Pacheco TR, Correia L, Costa L.. Levels of circulating fibroblast growth factor 23 (FGF23) and prognosis in cancer patients with bone metastases. Int J Mol Sci 2019;20:695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Roskoski R. The role of fibroblast growth factor receptor (FGFR) protein-tyrosine kinase inhibitors in the treatment of cancers including those of the urinary bladder. Pharmacol Res 2020;151:104567. [DOI] [PubMed] [Google Scholar]
- 50. Parker BD, Schurgers LJ, Brandenburg VM, Christenson RH, Vermeer C, Ketteler M, Shlipak MG, Whooley MA, Ix JH.. The associations of fibroblast growth factor 23 and uncarboxylated matrix Gla protein with mortality in coronary artery disease: the Heart and Soul Study. Ann Intern Med 2010;152:640–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vogt I, Haffner D, Leifheit-Nestler M.. Leifheit-Nestler M. FGF23 and phosphate-cardiovascular toxins in CKD. Toxins 2019;11:647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from Framingham Heart Study participants are publicly available at dbGap according to NIH data sharing policies (study accession phs000007.v29.p10).