Abstract
Although there is significant interest in the potential interactions of microbes with gas hydrate, no direct physical association between them has been demonstrated. We examined several intact samples of naturally occurring gas hydrate from the Gulf of Mexico for evidence of microbes. All samples were collected from anaerobic hemipelagic mud within the gas hydrate stability zone, at water depths in the ca. 540- to 2,000-m range. The δ13C of hydrate-bound methane varied from −45.1‰ Peedee belemnite (PDB) to −74.7‰ PDB, reflecting different gas origins. Stable isotope composition data indicated microbial consumption of methane or propane in some of the samples. Evidence of the presence of microbes was initially determined by 4,6-diamidino 2-phenylindole dihydrochloride (DAPI) total direct counts of hydrate-associated sediments (mean = 1.5 × 109 cells g−1) and gas hydrate (mean = 1.0 × 106 cells ml−1). Small-subunit rRNA phylogenetic characterization was performed to assess the composition of the microbial community in one gas hydrate sample (AT425) that had no detectable associated sediment and showed evidence of microbial methane consumption. Bacteria were moderately diverse within AT425 and were dominated by gene sequences related to several groups of Proteobacteria, as well as Actinobacteria and low-G + C Firmicutes. In contrast, there was low diversity of Archaea, nearly all of which were related to methanogenic Archaea, with the majority specifically related to Methanosaeta spp. The results of this study suggest that there is a direct association between microbes and gas hydrate, a finding that may have significance for hydrocarbon flux into the Gulf of Mexico and for life in extreme environments.
Gas hydrate is an ice-like mineral that crystallizes under conditions of high pressure, low temperature, and high gas concentration (9). It is composed of hydrocarbon and nonhydrocarbon gases held in cages of water molecules. Marine gas hydrate is thought to comprise an extremely large reservoir of reduced carbon, with energy content exceeding that of all conventional subsurface reserves of oil, gas, and coal combined (25). There has been significant interest in gas hydrate as a future energy resource, as a positive feedback mechanism for global warming, and as an agent of catastrophic sediment failure (24). It has been implicated in transient greenhouse warming at the Paleocene/Eocene transition (19, 32) and in a Jurassic oceanic anoxic event (13). Gas hydrate is also found in permanently frozen soils and glacial ices at high latitudes on Earth and is thought to be a component of icy planets and satellites, comets, and the Mars polar ice caps (reviewed in reference 9).
Microbial communities physically associated with gas hydrates and related sediments are potentially critical for gas hydrate stability, composition, and crystal structure. Via methanogenesis, microbes are indirectly involved in the formation of the most common form of gas hydrate on Earth, biogenic methane hydrate (51). There are indications that microbes anaerobically oxidize methane in the seep environment (6, 30, 46, 48) and within gas hydrate after crystallization (39).
The Gulf of Mexico (GOM) is a natural laboratory for studying gas hydrate dynamics and microbiology for several reasons. Gas hydrate is often found in sediments associated with natural gas venting and at cold hydrocarbon seeps, both of which are abundant on the northern continental slope (8). In some cases, so much gas hydrate is present that massive gas hydrate mounds break through the sediment surface (29). The GOM is also one of the few sites globally where both thermogenic (i.e., composed primarily of hydrocarbon gases derived from thermal degradation of petroleum) and biogenic (i.e., composed primarily of methane from biological methanogenesis) gas hydrates have been recovered (43). Gas hydrate at seep sites hosts complex chemosynthetic communities, where primary production is based on microbial consumption of methane and hydrogen sulfide (40). Finally, authigenic carbonates with extremely light carbon isotope signatures, which have been linked to anaerobic biological oxidation of methane (36), as well as massive gas hydrates, have been recovered in sediment cores from this region.
Geochemical evidence has indirectly shown microbial consumption of methane within gas hydrate (39) and petroleum components within cold hydrocarbon seep regions (41) on the northern continental shelf of the GOM. Additionally, the microbial diversity of gas hydrate-containing sediments in other regions has been investigated in several previous studies (4, 15, 30, 33). However, no direct observation of microbes within massive gas hydrates has been reported.
This study is the first to characterize a microbial community directly associated with massive gas hydrate. We report geochemical, microscopy, and DNA-based data supporting such a direct physical association.
MATERIALS AND METHODS
Geologic setting.
The GOM continental slope is affected by large sheet-like salt thrusts that extend from the shelf edge across the continental slope to the Sigsbee Escarpment, near the edge of the abyssal plain (54). The geology is conducive to hydrocarbon seepage to the sea floor from a deeply buried petroleum system (53). Fracture zones associated with moving salt sheets and active faults provide conduits for fluid flow to the sea floor. Massive hydrocarbon seepage manifests itself at the Gulf sea floor as gas hydrate, oil-stained sediments, authigenic carbonate depleted in 13C, and chemosynthetic communities (1, 28, 36, 37). Seeps and gas hydrate are concentrated along salt-withdrawal basin margins, over salt ridges, and near the edge of the Sigsbee Escarpment (43).
Sample collection.
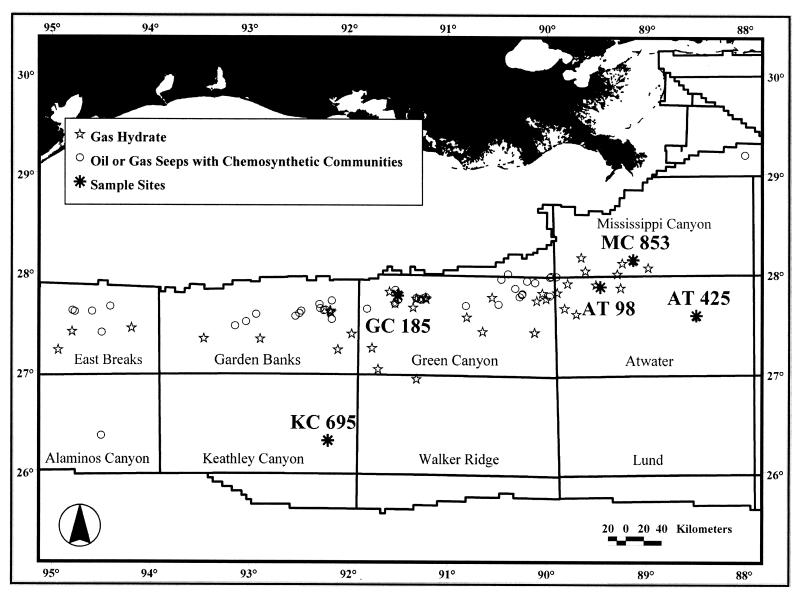
Samples were collected during 1998 to 2000 research cruises. The sites are described in Table 1 and Fig. 1 and were selected on the basis of seismic indications of hydrocarbon seepage within the gas hydrate stability zone. Samples were collected with a 6-m piston coring device (7-cm interior diameter). A high-speed winch facilitated rapid core recovery, before extensive gas hydrate decomposition could occur. Intact white-to-orange gas hydrate was observed in oil-stained sediments as vein fillings and as subspherical nodules with a radial pattern of crystallization. The gas hydrate was preserved by immersion in liquid nitrogen within minutes of core recovery. All samples had significant amounts of associated crude oil (ca. 30% [vol/vol]).
TABLE 1.
Description of sampled sites
| Samplea | Location | Water depth (m) | Gas hydrate | Associated features | Descriptors | Reference |
|---|---|---|---|---|---|---|
| AT425 | 27° 34.1′ N, 88° 29.7′ W | 1,920–1,930 | Structure II | Authigenic carbonate, H2S | Part of Mississippi Fan Foldbelt, salt ridge | 42 |
| KC695 | 26° 18′ N, 92° 12′ W | 2,000 | Structure I | H2S | Edge of Sigsbee escarpment | This study |
| MC853 | 28° 7.4′ N, 89° 8.2′ W | 1,060–1,070 | Structure II | Authigenic carbonate, H2S, free gas | Large sea floor mound (1.5 km across, ca. 30–40 m relief) | 43 |
| AT98 | 27° 51.1′ N, 89° 28.1′ W | 1,076 | Structure I | Free gas, authigenic carbonate, H2S | On the middle Gulf slope near a fault, shallow salt | This study |
| GC185 | 27° 45.7′ N, 91° 30.5′ W | 540 | NDb | Free gas, authigenic carbonate, H2S | Control sediment with no associated gas hydrate | 38, 39 |
AT, Atwater Canyon; KC, Keathley Canyon; MC, Mississippi Canyon; GC, Green Canyon.
ND, no data.
FIG. 1.
Map of sampling sites. Locations of known shallow gas hydrates, major gas and oil seeps, and chemosynthetic communities across the northen continental slope of the GOM are also indicated (38).
The MC853 and KC695 samples were massive gas hydrate with attached sediment. MC853 was located ca. 0.4 to 0.6 m, and KC695 was located ca. 1.4 m deep in the sediment column. Samples AT425 and AT98 were massive gas hydrate with no detectable attached sediment, located in the upper 20 cm of the sediment column. The GC185 sample was control sediment with no visible associated gas hydrate; however, we cannot exclude the possibility that small amounts of vein-filling hydrate were present in the GC185 sample, but decomposed prior to retrieval.
Analysis of gas hydrate samples.
Aliquots of intact gas hydrate were removed from liquid N2 storage and were picked to remove sediment when necessary. Cleaned samples were allowed to decompose under a water-filled bell jar to obtain large volumes of free gas. Aliquots of gas samples were immediately transferred to preevacuated metal vacutainers with a 60-ml gas-tight syringe and held at −20°C until analysis. Detailed analytical procedures for C1–C5 gas chromatography and measurement of isotopic properties of hydrocarbon gases have been described elsewhere (38). Concentrations of each hydrocarbon were expressed in parts per million by sediment volume and normalized as a percentage of total C1–C5 hydrocarbons. The δ13C values are reported as parts per thousand (‰) relative to the Peedee belemnite (PDB) standard (precision of ±0.2‰), and the δD values are reported as parts per thousand relative to standard mean ocean water (SMOW) (precision of ±5‰).
Direct microscopic counts of cells in hydrate.
Samples were removed from liquid N2 and allowed to melt (sediment) or decompose (gas hydrate) in sterile containers. The resulting liquid or liquid-sediment mix was centrifuged at approximately 400 × g to separate gas hydrate fluids, oil, and sediment. No sediment pellet was observed for samples AT425 and AT98. For decomposed gas hydrate with no attached sediment, 1 ml of the aqueous phase was transferred to a filtration tower with care taken to avoid the organic phase. The samples were stained with the DNA-staining dye 4,6-diamidino 2-phenylindole dihydrochloride (DAPI; Sigma), and cells were counted as previously described (22). Attached sediment from samples MC853 and KC695 and the control sample, GC185, was diluted 1,000-fold, stained with DAPI, and counted as previously described (7). Due to intrinsic autofluorescence, the hydrocarbons present in the samples led to high background fluorescence, therefore, the cell counts presented are a minimal estimate.
DNA isolation, PCR amplification, and cloning.
Remaining liquid from sample AT425 (ca. 50 ml) was filtered on to a 0.2-μm-pore-size Supor filter (Pall, Ann Arbor, Mich.). The filter was frozen in the presence of lysis buffer (20 mM Na-EDTA, 400 mM NaCl, 0.75 M sucrose, 50 mM Tris-HCl [pH 9.0]) and stored at −80°C. Total nucleic acids were extracted from the filters and purified as described elsewhere (12). Bacterial small subunit (SSU) rRNA genes were PCR amplified with primers S-D-Bact-0008-a-A-19 and S-D-Bact-1492-a-A-21 (14), and archaeal SSU rRNA genes were PCR amplified with primers A20F (11) and A958R (10). The PCR conditions used were 1 min of 95°C denaturation, 2 min of 55°C annealing, and 3 min of 72°C elongation for 35 cycles in an MJ Research thermal cycler. After a final 10-min incubation at 72°C, the product was purified with a gel extraction kit (Qiagen, Chatsworth, Calif.). Amplification products were cloned into the plasmid vector pCR2.1 by TA cloning (Invitrogen, Carlsbad, Calif.).
RFLP analysis and sequencing of clone libraries.
SSU rDNA inserts were PCR-amplified under the same conditions as above with M13R and T7 primers. The product was digested with HhaI restriction endonuclease (New England Biolabs, Beverly, Mass.) at 37°C for 2 h. The banding patterns were grouped according to similarity, and representative members of each pattern group were fully, bidirectionally sequenced with either an ABI 3700 (Applied Biosystems, Inc., Foster City, Calif.) or a Licor 4200 (Licor, Inc., Lincoln, Neb) automated DNA sequencer. Multiple representatives were sequenced for restriction fragment length polymorphism (RFLP) patterns that had more than five members.
Sequence analysis.
Sequences were initially aligned to their nearest neighbor by using the program ARB (Ludwig and Strunk, Technische Universität München, Munich, Germany [http://www.mpi-bremen.de/molecol/arb/]). The sequences were further manually aligned to sequences obtained from the GenBank database by using the Genetic Data Environment (GDE) version 2.0 sequence analysis software package (Smith, Millipore Corporation, Bedford, Mass.), as described elsewhere (35). Phylogenetic inference and evolutionary distance calculation were performed as described previously (35). Phylogenetic trees were constructed by the neighbor-joining method with the Kimura two-parameter model for nucleotide change (21).
Nucleotide sequence accession numbers.
The rDNA sequences were entered into the GenBank database and were assigned accession no. AY053466 to AY053496.
RESULTS
Gas hydrate molecular and isotopic composition.
The gas hydrate samples of the present study included examples of the two major types of gas hydrate found in the Gulf (Table 1). Samples KC695 and AT98 were representatives of biogenic gas hydrate (structure I). Methane was the main hydrocarbon component, with δ13C values of −70.1‰ PDB from KC695 and −74.7‰ PDB from AT98 (Table 2). The δD value of methane from AT98 was −155‰ SMOW. These δ13C and the δD values were consistent with a microbial methane source (38). Small percentages of ethane were present in both samples, indicating minor mixing with thermogenic hydrocarbon gases.
TABLE 2.
Normalized hydrocarbon concentrations and isotope properties of gasses from gas hydrate samples
| Sample | % Methane concn (δ13C)a | δD Methaneb | % Ethane concn (δ13C) | % Propane concn (δ13C) | % Isobutane concn (δ13C) | % n-Butane concn (δ13C) | % Isopentane concn (δ13C) | % n-Pentane concn (δ13C) | CO2 δ13C |
|---|---|---|---|---|---|---|---|---|---|
| MC853 | 75.3 (−45.1) | −166 | 6.8 (−28.4) | 11.3 (−24.5) | 3.4 (−27.0) | 0.7 (−23.4) | 0.3 (ND)c | <0.1 (ND) | +11.6 |
| AT425 | 77.3 (−49.3) | −148 | 5.0 (−38.2) | 12.5 (−32.0) | 3.0 (−31.6) | 1.2 (−28.3) | 0.9 (−28.8) | 0.2 (−28.4) | +12.4 |
| KC695 | 99.9 (−70.1) | ND | 0.1 (ND) | 0 (ND) | 0 (ND) | 0 (ND) | 0 (ND) | 0 (ND) | ND |
| AT98 | 99.9 (−74.7) | −155 | 0.1 (ND) | 0 (ND) | 0 (ND) | 0 (ND) | 0 (ND) | 0 (ND) | ND |
| GC185 | 84.1 (ND) | ND | 5.5 (ND) | 5.7 (ND) | 1.7 (ND) | 0.8 (ND) | 2.2 (ND) | 0 (ND) | ND |
δ13C values are reported in parts per thousand relative to the Peedee Belemnite standard.
δD values are reported in parts per thousand relative to the SMOW standard.
ND, no data.
Thermogenic gas hydrate (structure II) was represented by samples MC853 and AT425. Methane was the major hydrocarbon component, along with lesser percentages of thermogenic hydrocarbons in the order propane, ethane, isobutane, butane, and minor pentanes (Table 2). The δ13C value of methane was −45.1‰ PDB from MC853 and −49.3‰ PDB from AT425. These values were enriched in 13C relative to biogenic methane gas hydrate; therefore, the methane was likely thermogenic, from the deep subsurface hydrocarbon system of the Gulf (Table 2). However, in the seep environment, we cannot exclude the possibility that a minor fraction of the methane was microbial in origin (38).
The δD value of methane was −166‰ SMOW from MC853 and −148‰ SMOW from AT425, and the δ13C of propane was −24.5‰ PDB in MC853 (Table 2). These values are enriched in the heavy isotopes relative to unaltered vent gas (38, 39). Such enrichment is indicative of microbial consumption of these gaseous components from within the gas hydrate, an observation consistent with previous studies (38, 39). Isotope properties of other C1–C5 hydrocarbons and CO2 (Table 2) were similar to those observed in unaltered vent gases.
Cell counts.
Direct counts of DAPI-stained cells were ca. 106 cells ml−1 for the decomposed gas hydrate fluids and 109 cells g−1 (wet weight) for the sediments (Table 3). Cell counts for gas hydrate fluids with or without attached sediments were indistinguishable. The sediment with no intact associated gas hydrate (GC185) had similar cell counts to the sediments associated with gas hydrate. These direct counts are similar to those commonly obtained from standard marine systems (45). This result is unexpected, because other studies based on extractable lipid concentrations show up to 30-fold higher biomass, a value indirectly correlated to cellular abundance, in the Gulf seep system than in nearby marine sediments (C. Zhang, personal communication).
TABLE 3.
Direct counts of DAPI-stained cells from decomposed gas hydrate and sediment
| Sample | Direct count of cells froma:
|
|
|---|---|---|
| Hydrate (105 cells ml−1) | Sediment (108 cells/g−1 [wet wt]) | |
| AT425b | 9.99 ± 4.72 | |
| KC695 | 6.57 ± 1.68 | 11.7 ± 2.85 |
| MC853 | 13.2 ± 2.96 | 10.1 ± 3.37 |
| AT98b | 10.5 ± 4.13 | |
| GC185 | 8.18 ± 2.51 | |
Values are means ± standard deviations.
Gas hydrate with no attached sediment.
Microbial diversity.
We focused on one of the samples, AT425, to characterize the microbial diversity associated with GOM gas hydrate. This gas hydrate was chosen because it had no associated sediment; therefore, all microbes in this sample were physically attached to or included within the gas hydrate. AT425 was also chosen because it showed signs of microbial oxidation of methane within the gas hydrate (see above).
Bacteria.
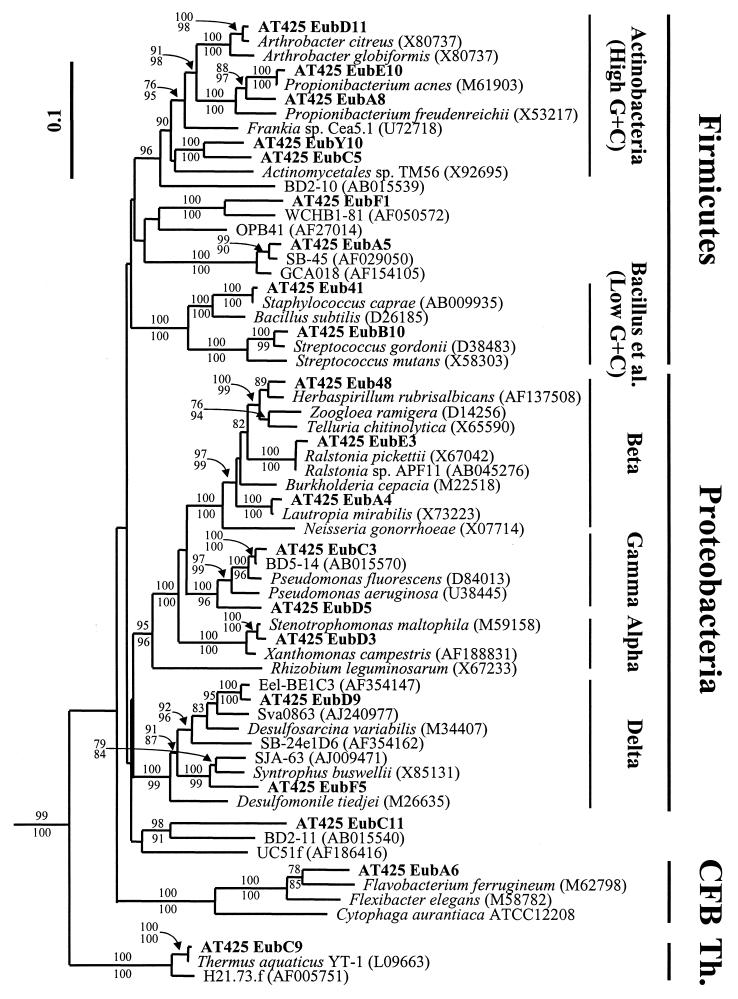
There was fairly high bacterial diversity associated with the AT425 hydrate (Fig. 2). Of 127 rRNA clones analyzed, there were 21 different HhaI RFLP patterns (Table 4). Sequencing of representatives of these RFLP patterns confirmed that they were phylogenetically distinct. Diverse phylotypes related to Actinobacteria and low G + C (Bacillus, etc.) Firmicutes; β-, γ-, α-, and δ-Proteobacteria; and a group without clear affiliation with broad phylogenetic clades (AT425 EubC11) were the most frequently recovered sequences (Fig. 2 and Table 4). Overall, of 127 16S rDNA clones, there were 42 Firmicutes-related and 63 Proteobacteria-related clones. Cytophaga/Flavobacterium/Bacteroides (CFB)- and Thermus-related sequences were also obtained, but only at low frequency (Fig. 2 and Table 4).
FIG. 2.
Phylogenetic analysis of bacterial SSU rRNA gene clones from sample AT425. This phylogenetic tree was rooted with Sulfolobus acidocaldarius. A mask of 989 nucleotides, including all nonambiguously aligned positions, spanning nearly the full length of the SSU rRNA gene, was included. Bootstrap values (100 replications) generated by the neighbor-joining method are shown above relevant nodes, and those generated by maximum-parsimony analysis are shown below. Only bootstrap values above 70 are shown. Sequences from isolates are in italics, sequences from environmental gene clones are in plain text, and sequences from the AT425 sample are in boldface. GenBank accession numbers of the sequences from other studies are included. CFB, Cytophaga/Bacteroides/Flavobacterium. Th, Thermus.
TABLE 4.
Grouping of microbes associated with sample AT425
| Representative sequence | No. of clones | Nearest relativea | Phylogenetic groupb | Similarityc |
|---|---|---|---|---|
| Bacteria (127 clones) | ||||
| AT425 EubE10 | 15 | Propionibacterium acnes | Actinobacteria | 0.99 |
| AT425 EubC5 | 11 | Actinomycetales sp. strain TM56 | Actinobacteria | 0.88 |
| AT425 EubA8 | 3 | Propionibacterium acnes | Actinobacteria | 0.95 |
| AT425 EubD11 | 3 | Arthrobacter citreus | Actinobacteria | 0.99 |
| AT425 EubY10 | 1 | Frankia sp. strain Cea5.1 | Actinobacteria | 0.89 |
| AT425 EubB10 | 8 | Streptococcus gordonii | Low G + C Firmicutes | 0.98 |
| AT425 Eub41 | 1 | Staphylococcus caprae | Low G + C Firmicutes | 0.99 |
| AT425 EubF1 | 2 | Hydrocarbon-contaminated aquifer clone WCHB1-81 | Firmicutes? | 0.91 |
| AT425 EubA5 | 1 | Benzene-mineralizing consortium clone SB-45 | Firmicutes? | 0.98 |
| AT425 EubD3 | 8 | Stenotrophomonas maltophila | α-Proteobacteria | 0.99 |
| AT425 EubE3 | 9 | Ralstonia pickettii | β-Proteobacteria | 0.99 |
| AT425 Eub48 | 1 | Herbaspirillum rubrisalbicans | β-Proteobacteria | 0.97 |
| AT425 EubA4 | 1 | Lautropia mirabilis | β-Proteobacteria | 0.99 |
| AT425 EubC3 | 36 | Pseudomonas fluorescens/deep sea clone BD5-14 | γ-Proteobacteria | 0.98 |
| AT425 EubD5 | 2 | Deep sea clone BD5-14 | γ-Proteobacteria | 0.94 |
| AT425 EubD9 | 5 | Hydrocarbon seep clone Eel-BE1C3 | δ-Proteobacteria | 0.98 |
| AT425 EubF5 | 1 | Syntrophus buswellii/trichlorobenzene-degrading consortium clone SJA-63 | δ-Proteobacteria | 0.93 |
| AT425 EubA6 | 1 | Flavobacterium ferrugineum | CFB | 0.92 |
| AT425 EubC9 | 4 | Thermus aquaticus YT-1 | Thermus and relatives | 1.00 |
| AT425 EubC11 | 14 | Deep sea clone BD2-11 | Unknown | 0.83 |
| Archaea (93 clones) | ||||
| AT425 ArC7 | 73 | Methanosaeta sp. strain clone A1 | Methanosarcinales | 0.95 |
| AT425 ArD2 | 2 | Hydrocarbon seep clone Eel-TA1a4 | ANME-2 | 0.95 |
| AT425 ArE12 | 1 | Hydrocarbon seep clone Eel-TA1a4 | ANME-2 | 0.95 |
| AT425 ArB7 | 8 | Hydrocarbon seep clone Eel-BA2e8 | ANME-1 | 0.99 |
| AT425 ArB9 | 8 | Hydrocarbon seep clone Eel-BA2e8 | ANME-1 | 0.93 |
| AT425 ArD10 | 1 | Salt marsh clone 2C84 | Salt marsh | 0.94 |
Two of the Firmicutes-related clone groups were only distantly related to their nearest neighbor and appeared to be a distinct branch of the Actinobacteria. These are a group of 11 clones, represented by AT425 EubC5, and a related group of one clone (AT425 EubY10). Two other groups, totaling three clones, may also be affiliated with the Firmicutes (i.e., AT425 EubF1 and AT425 EubA5), although there is poor statistical (bootstrap) support for that affiliation. However, the majority of the 16S rDNA sequences clearly affiliated with the Firmicutes were related at the species level (>97% 16S rDNA sequence similarity) (44) to previously cultured organisms (Table 4). The closest relatives of these clones have heterotrophic metabolism, and most are aliphatic or aromatic hydrocarbon utilizers (Table 4).
A high similarity to previously cultured organisms also held for most Proteobacteria in this system (Fig. 2 and Table 4). The most frequently recovered sequence group, represented by AT425 EubC3, was specifically related to the fluorescent pseudomonad subgroup of the γ-Proteobacteria. This group is physiologically diverse, widespread, and abundant. The other γ-proteobacterial group of two clones, represented by AT425 EubD5, are specifically related to BD5-14 (98% similarity), an environmental 16S rDNA clone from deep sea sediments (27). Six 16S rDNAs were relatives of δ-proteobacterial sulfate-reducing bacteria (SRB), specifically the Desulfosarcinales and Syntrophus spp. AT425 EubD9, representing five clones, is specifically related (98% similarity) to a group of environmental 16S rRNA gene sequences that were originally found in a variety of sediments associated with the anaerobic oxidation of methane (33). The remaining δ-proteobacterial clone, AT425 EubF5, is related to Syntrophus buswellii, an organism that requires syntrophic H2 shuttling for growth (50). The β- and α-Proteobacteria-affiliated 16S rDNA gene sequences, accounting for 19 clones, are related at the species level to previously cultured organisms with heterotrophic metabolism (Fig. 2 and Table 4).
AT425 EubC11, which represents 14 of 127 16S rDNA clones, is poorly affiliated with previously characterized 16S rDNA sequences in the public databases. This sequence showed a similarity of only 85% to its nearest neighbor, deep sea clone BD2-11 (Fig. 2 and Table 4). Other gene sequences that appear to affiliate with this group are primarily symbionts of marine sponges, such as UC51f (GenBank accession no. AF186416) and R11 (GenBank accession no. AF333520).
Only one clone related to the CFB group was recovered from this system. It is only distantly related to other members of the CFB group, being only 92% similar to its nearest neighbor (Table 4). The final group of four clones observed in this system, represented by AT425 EubA6, is related at the species level to Thermus aquaticus YT-1 (Fig. 2 and Table 4).
Archaea.
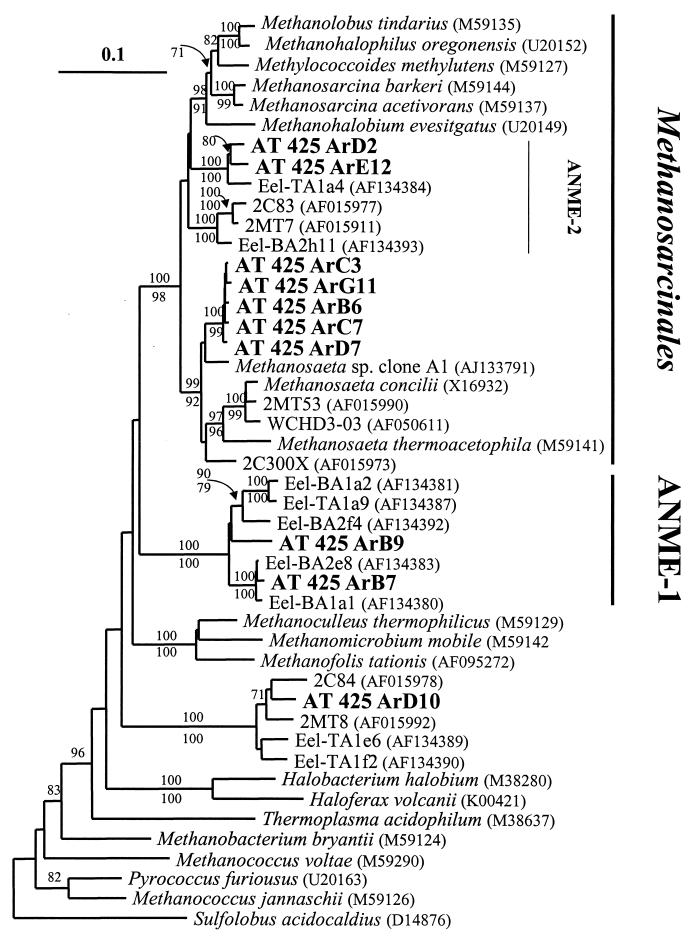
In contrast to the bacterial diversity, the archaeal diversity in the AT425 sample was quite low (Fig. 3). Of 93 rRNA clones analyzed, only eight distinct HhaI RFLP patterns were observed, representing five phylogenetically distinct groups. This level of archaeal diversity is much lower than that observed in studies of standard marine sediments (31, 49), but it is similar to that observed in other hydrocarbon seep systems (15, 33).
FIG. 3.
Phylogenetic analysis of archaeal SSU rRNA gene clones from sample AT425. This unrooted phylogenetic tree was obtained by methods described in the legend to Fig. 2. A mask of 695 nucleotides, including all nonambiguously aligned positions, spanning approximately 900 nucleotides of the 5′ end of the SSU rRNA gene, was included in the analysis. Sequences from isolates are in italics, sequences from environmental gene clones are in plain text, and sequences from the AT425 sample are in boldface. Sequences labeled “Eel” are from a study of the Eel River area off the coast of northern California (15), and sequences labeled “2C” or “2M” are from a study of a coastal salt marsh (31). GenBank accession numbers of the sequences from other studies are included.
Two of the archaeal groups, totaling 76 clones, were related to the methanogenic order Methanosarcinales. The most frequently recovered sequence group (73 clones) was specifically related to the genus Methanosaeta. The closest relative of these sequences, Methanosaeta sp. strain clone A1 (Fig. 3 and Table 4), is an rDNA gene sequence obtained from a consortium capable of degrading long-chain hydrocarbons (e.g., hexadecane) to methane, a process termed “microbial alkane cracking” (55). The remaining three clones within Methanosarcinales are specifically related (Fig. 3 and Table 4) to a group labeled ANME-2 (for anaerobic methane oxidizers 2) by Orphan and colleagues (33). ANME-2 has only been observed in sediments that exhibit anaerobic methane oxidation.
Two other groups totaled 16 clones and were related to the group ANME-1. This group has no previously cultured members and has only been described in sediments showing evidence of anaerobic methane oxidation (15, 33, 47). The final group, composed of one clone, was most closely related to a cluster of Euryarchaeota of unknown physiology found in marine sediments and also observed previously in anaerobic methane oxidation zones (15, 31, 33). No evidence of Crenarchaeota or other archaeal groups (e.g., Korarchaeaota) was observed.
One final group, comprised of a single clone (AT425 ArD10), was related to a group of environmental clones obtained from a salt marsh ecosystem (Fig. 3 and Table 4) (31). Related sequences were also obtained from the Eel River site and were associated with AOM (15). This group is composed entirely of environmental gene sequences, and no previously cultured organisms are associated.
DISCUSSION
Although of significant interest, little is known about gas hydrate crystallization or decomposition, or the role of gas hydrate in diagenetic processes. The presence of microbial cells directly associated with gas hydrate supports geochemical evidence that biology may have a significant effect on both the stability and composition of gas hydrate (38, 39). Because gas hydrate is estimated to be a larger reservoir of hydrocarbons than all oil, gas, and coal reserves combined (26), they could be an important, poorly understood carbon and/or energy source for microorganisms. Consumption of gas hydrate hydrocarbons by associated microbes could also play a significant role in global methane and carbon cycles and in diagenetic processes. This study was designed to find direct evidence of physical interactions between microbes and gas hydrates.
Molecular composition and isotope data.
In contrast to simple biogenic methane, thermogenic hydrocarbons preserve complex information on their origin and alteration. For example, unaltered thermogenic methane from subsurface reservoirs and from unaltered sea floor vent gas will not often show large variation in isotopic properties (38, 39). Because there is no isotopic fractionation as a consequence of gas hydrate crystallization, the gas hydrate isotopic properties are generally very similar to that of the vent gas. Differences are attributed to bacterial oxidation after crystallization (38, 39). The δD observed for methane from sample AT425 (Table 2) was similar to values previously observed for microbially altered gas hydrate from near the GC185 site and was enriched in D relative to unaltered vent gases (39, 43). Additionally, the propane from sample MC853 was enriched in 13C relative to unaltered vent gas. Therefore, although we lack vent gas samples from the MC853 and AT425 sites for direct reference and comparison, we can infer that some components of the Gulf thermogenic gas hydrate have been impacted by microbial alteration after crystallization. These results support previous data indicating that microbes are directly consuming hydrocarbons within gas hydrates in the Gulf (39, 43).
Fluorescence microscopy data.
We reasoned that microbes are likely to be physically associated with the gas hydrate, because direct consumption of gases would require a close physical interaction. Staining of hydrate-associated sediment and decomposed gas hydrate fluids with the DNA dye DAPI followed by fluorescence microscopy showed the presence of microbial cells within the gas hydrate samples. Three lines of evidence indicated that these cells were physically associated with the gas hydrate. First, all gas hydrate samples, including those that had no visible attached sediments, provided similar cell counts (Table 3). If the observed cells were due to contamination from sediment or seawater, cell counts lower than those normally observed for sediment or seawater would have been expected as a consequence of dilution. Second, the outer layers of the gas hydrate were lost upon retrieval by decomposition due to decreased pressure and increased temperature. Cells that were only peripherally associated with the gas hydrate would have been lost with the outer layers. Third, the isotopic evidence of methane and propane alteration within the gas hydrate requires the presence of microbes. Therefore, the microbes are most likely directly physically associated with the gas hydrates. At this time, we cannot determine the source of these microbes (i.e., seep sediments, oil associated with the gas hydrates, or an independent community specifically affiliated with the gas hydrate).
It is, perhaps, surprising that diverse (see below) groups of Bacteria and Archaea would be found in the highly crystalline environment of the gas hydrate. However, CH4, Ar, N2, and CO2 can all form highly porous (as high as 40% porosity), “sponge-like” gas hydrates (23). Typical pore sizes are 100 to 400 nm for CH4 hydrates, with occasional channels on the order of a few micrometers. Many Bacteria and Archaea, as well as their chemical substrates and waste products, would be able to freely move through pores of this size, allowing exchange with the external environment, which may be the primary source for microbes in this environment.
It is unclear why the gas hydrates had cell counts that were several orders of magnitude lower than for a similar amount of sediment. One possible explanation is that the gas hydrates have significantly higher cell counts than were observed here. Much of the surface of the gas hydrate samples was lost due to decomposition during retrieval. Additionally, due to differences in methodology for counting the sediment versus liquid samples, the concentration of autofluorescent hydrocarbons was much higher in the gas hydrate samples, which may have led to the underestimation of the number of cells in the gas hydrate samples. If this hypothesis is accurate, it might partially account for the difference between estimates of microbial biomass based on extractable lipids (C. Zhang, personal communication) and the direct microscopic counts reported here. However, this hypothesis must be tested further.
Despite these difficulties with the direct microscopic counting of microbial cells, this approach did show the presence of microbes within the gas hydrate structure. This is the first direct evidence of physical interactions between gas hydrates and microbes.
Phylogenetic analysis.
We used rRNA phylogenetic analysis to determine the identity of the microbes associated with one sample of gas hydrate, AT425, a massive thermogenic hydrate with no associated sediment. Fairly high bacterial diversity and low archaeal diversity were associated with this sample (Fig. 2).
Bacteria.
Several unusual features are apparent regarding the bacterial diversity in sample AT425. First, a large fraction (ca. 72%) of the recovered 16S rRNA gene sequences are related at the species level to previously cultured microbes (Fig. 2 and Table 4). A predominance of sequences that are nearly indistinguishable from previously cultured organisms is unusual in non-culture-based studies (17).
Second, the Firmicutes are more frequently recovered from this sample relative to other systems. Overall, 33 to 35% of the recovered gene sequences affiliated with the Actinobacteria or the low-G+C Firmicutes (Fig. 2 and Table 4). Although widespread in marine systems (17), to our knowledge, no other studies of marine systems, either planktonic or benthic, show such high recovery of Firmicutes-related gene sequences. None of the Firmicutes gene sequences in this sample appear to be affiliated with the so-called marine Actinobacteria (34) or other common, but previously uncultured, groups of Actinobacteria.
Third, a group of 11 clones, represented by AT425 EubC11, was recovered that is only poorly affiliated with 16S rRNA gene sequences in public databases (Fig. 2 and Table 4). Other gene sequences that may affiliate with this group, primarily symbionts of marine sponges such as UC51f (AF186416) and R11 (AF333520) (data not shown), were previously thought to be related to the Actinobacteria (52). However, our phylogenetic analysis does not support such a relationship (Fig. 2). Instead, this group appears to be a deep branch of the Bacteria. Further characterization is required to determine whether this group should be considered a candidate division of the Bacteria (See reference 17 for more information on candidate divisions.)
Fourth, four 16S rRNA gene clones, represented by AT425 EubC9, were recovered from this cold environment that are indistinguishable from Thermus aquaticus YT-1. It is unlikely that these sequences are contaminants from the Taq polymerase used for PCR amplification, because the brand used is recombinant and was purified from Escherichia coli. At this time, we are unable to provide an explanation for the recovery of multiple gene sequences nearly indistinguishable from a monophyletic group of obligately thermophilic and aerobic organisms in an anaerobic, cold environment. To our knowledge, there are no reports of Thermus spp. in nonthermophilic environments.
Archaea.
There were two major groups of Archaea present in sample AT425: those related to 16S rRNA gene sequences recovered from sediments with active anaerobic oxidation of methane (ANME-1 and ANME-2 [33] and salt marsh clones [31]) and those specifically related to the genus Methanosaeta (Fig. 3). The level of diversity observed here is extremely low compared to those in most other environmental, nonculture-based studies of Archaeal diversity (3), although it is similar to that observed in sediments that exhibit anaerobic methane oxidation (15, 33, 47).
It is intriguing that sequences specifically related to the genus Methanosaeta are so frequently recovered from sample AT425 (Fig. 3 and Table 4). This genus is a member of the Methanosarcinales, which are the only methanogenic Archaea capable of utilizing acetate (acetoclastic methanogenesis) or intermediate redox state C1 compounds, such as methylamines or methanol (methylotrophic methanogenesis [5]). In fact, the only known energy-generating metabolism for Methanosaeta spp. is acetoclastic methanogenesis (5).
ANME-1 and ANME-2, also well represented in sample AT425 (Fig. 3 and Table 4), have no previously cultivated members. ANME-2-related sequences form a distinct branch within the Methanosarcinales. ANME-1, while clearly related to the methanogenic Archaea, is a distinct branch and is not specifically affiliated with any previously cultured methanogenic Archaea. We assume that both groups have methanogenic enzymes, because they are clearly related to the methanogenic Archaea, which are monophyletic and all have similar physiology (18). However, their specific role in this system is not known.
Predicted roles for microbial communities in GOM gas hydrate.
We believe that it is likely that the microbial communities described here are active within the gas hydrates. High porosity of methane gas hydrate (pore sizes of 100 to 400 nm and pore volumes of approximately 25 to 40% [23]) allows potential substrates (e.g., sulfate) to enter and products of microbial metabolism (e.g., sulfide) to exit the gas hydrate structure without difficulty. Some pores may even be large enough for microbes to freely enter and leave the superstructure of the solid gas hydrate. Therefore, this microbial community may not be highly specialized and selected for life within a gas hydrate, but rather may freely exchange with the communities within the surrounding sediments.
Assuming that the microbial communities associated with the gas hydrates are active, two major metabolic activities are implied by the composition of the microbial community: anaerobic methane oxidation and nonmethane hydrocarbon oxidation.
AOM.
Biological oxidation of methane in solid gas hydrate from the GOM has been observed previously, as indicated by both the isotope composition of methane and the molecular composition of gases held in the gas hydrate structure relative to unaltered vent gas (39). Additionally, these isotope composition shifts imply that solid gas hydrate can act as a substrate for microbial metabolism and growth, a process that has not been observed directly. Of hydrocarbon gases, methane is least tightly held in the crystal structure of gas hydrate, especially structure II. Therefore, it is the most accessible target of microbial consumption. Such activity can potentially change the composition of the gases held in the gas hydrate, hydrate stability, gas hydrate geochemistry, and sediment diagenesis (39).
No organism capable of net anaerobic oxidation of methane (AOM) has been isolated, however, geochemical evidence has indicated that microbially driven net oxidization of methane can occur under anaerobic conditions (reviewed in reference 48). Hoehler and colleagues have proposed a model wherein a methanogen (working in reverse) coupled to sulfate-reducing Bacteria (SRB) anaerobically oxidizes methane (16). This model has received support from several recent studies of compound-specific stable isotopes (reviewed in reference 48). Some of the Archaea in sample AT425, specifically those related to ANME-1 and ANME-2 (Fig. 3 and Table 4), are closely related to those previously shown to be associated with AOM (15, 33, 47). Additionally, one group of Bacteria (AT425 EubD9) in AT425 is closely related to a group of δ-Proteobacteria that have been found in these same AOM systems and may be important in the process of AOM (33). Furthermore, gas hydrate samples from the nearby Green Canyon area of the Gulf have previously been shown to be affected by AOM activity (38, 39), and sample AT425 shows isotopic evidence of microbial oxidation of methane (Table 2). Each of these lines of evidence implies that gas hydrate-associated microbial communities in this region are involved in anaerobic oxidation of the methane in gas hydrate.
Nonmethane hydrocarbon oxidation.
We hypothesize that the large volume of hydrocarbons in the form of petroleum associated with the gas hydrate at this site may be a source of carbon and energy for many of the associated microbes. It has been noted that a significant amount of hydrocarbons enter the Gulf of Mexico through natural seeps, many of which also have associated gas hydrate (8, 20). Additionally, geochemical evidence based on comparisons of the isotopic composition of reservoir oils and those that enter the GOM indicates biological alteration of petroleum components (40). Aliphatic or aromatic hydrocarbon utilization is a widespread and common feature in the Bacteria (2). Therefore, it is possible that the bacterial community associated with natural gas hydrate may affect the flux of hydrocarbons into the GOM.
It is also possible that short-chain alkanes within thermogenic gas hydrate are a substrate for microbial activity. Propane δ13C in sample MC853 appears to be affected by microbial consumption (described above and as shown in Table 2). However, other studies in the GOM have indicated that methane is the primary gas hydrate component oxidized by microbes and that short-chain alkanes, up to C5, are relatively unaffected (39). Therefore, it is unclear whether consumption of short-chain alkanes in gas hydrates would be a significant carbon source for associated microbial communities.
Conclusions.
In this study, we have shown that microbes are physically associated with methane hydrate and characterized one of the communities. This is the first study to show direct physical interaction between microbes and gas hydrate, a finding with important implications for gas hydrate stability, composition, and geochemistry. Our results are consistent with the notion that the microbes in this system likely consume liquid and/or volatile methane and nonmethane hydrocarbons both from the seep system and directly within gas hydrate. We plan to examine more communities to determine whether the results reported here are widely applicable to all gas hydrate or are specific to this study system. Also, more detailed community characterization with gas hydrates as well as the rest of the seep system, including seeking similar physical interactions between SRB and methanogenic Archaea as those observed by Boetius and colleagues (6), is necessary for a complete understanding of the GOM seep system. Future studies will also focus on the mechanisms of microbe-gas hydrate interactions, anaerobic methane oxidation, and the significance of microbial consumption to the overall flux of hydrocarbons into the GOM.
ACKNOWLEDGMENTS
Support was provided to R.S. by the Applied Gas Hydrate Research Program at Texas A&M University and to K.H.N. by the NASA Astrobiology program.
We thank J. W. Ammerman for use of his fluorescent microscope, F. Carsey for providing travel support for B.D.L., D. A. DeFreitas for preparation of the GIS map, and the crew of the R. V. Powell for assistance with sample collection.
REFERENCES
- 1.Aharon P, Scharez H P, Roberts H H. Radiometric dating of hydrocarbon seeps in the Gulf of Mexico. GSA Bull. 1997;109:568–579. [Google Scholar]
- 2.Balows A, Truber H G, Dworkin M, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. [Google Scholar]
- 3.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bidle K A, Kastner M, Bartlett D H. A phylogenetic analysis of microbial communities associated with methane hydrate containing marine fluids and sediments in the Cascadia Margin (ODP Site 892B) FEMS Microbiol Ecol. 1999;177:101–108. doi: 10.1111/j.1574-6968.1999.tb13719.x. [DOI] [PubMed] [Google Scholar]
- 5.Blaut M. Metabolism of methanogens. Antonie Leeuwenhoek. 1994;66:187–208. doi: 10.1007/BF00871639. [DOI] [PubMed] [Google Scholar]
- 6.Boetius A, Ravenschlag K, Schubert C J, Rickert D, Widdel F, Gieske A, Amann R, Jorgensen B B, Witte U, Pfannkuche O. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–625. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- 7.Bottomley P J. Light microscopic methods for studying soil microorganisms. In: Weaver R W, Angle S, Bottomley P, Bezdicek D, Smith S, Tabatabai A, Wollum A, Mickelson S H, editors. Methods in soil analysis, part 2. Microbiological and biochemical properties. Madison, Wis: SSSA; 1994. pp. 81–105. [Google Scholar]
- 8.Brooks J M, Cox H B, Bryant W R, Kennicutt II M C, Mann R G, McDonald T J. Association of gas hydrates and oil seepage in the Gulf of Mexico. Org Geochem. 1986;10:221–234. [Google Scholar]
- 9.Buffett B A. Clathrate hydrates. Annu Rev Earth Planet Sci. 2000;28:477–507. [Google Scholar]
- 10.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giovannoni S J, DeLong E F, Olsen G J, Pace N R. Phylogenetic group-specific oligodeoxynucleotide probes identification of single microbial cells. J Bacteriol. 1988;170:720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giovannoni S J, Rappé M S, Vergin K L, Adair N L. 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the green non-sulfur bacteria. Proc Natl Acad Sci USA. 1996;93:7979–7984. doi: 10.1073/pnas.93.15.7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hesselbo S P, Gröcke D R, Jenkyns H C, Bjerrum C J, Farrimond P, Morgans Bell H S, Green O R. Massive dissociation of gas hydrate during a Jurassic oceanic anoxic event. Nature. 2000;406:392–395. doi: 10.1038/35019044. [DOI] [PubMed] [Google Scholar]
- 14.Hicks R E, Amann R I, Stahl D A. Dual staining of natural bacterioplankton with 4′,6-diamdino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992;58:2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinrichs K-U, Hayes J M, Sylva S P, Brewer P G, DeLong E F. Methane-consuming archaebacteria in marine sediments. Nature. 1999;398:802–805. doi: 10.1038/19751. [DOI] [PubMed] [Google Scholar]
- 16.Hoehler T M, Alperin M J, Albert D B, Martens C S. Field and laboratory studies of methane oxidation in an anoxic marine sediment: evidence for a methanogen-sulfate reducer consortium. Global Biogeochem Cycles. 1994;8:451–463. [Google Scholar]
- 17.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones W J, Nagle D P, Jr, Whitman W B. Methanogens and the diversity of archaebacteria. Microbiol Rev. 1987;51:135–177. doi: 10.1128/mr.51.1.135-177.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katz M E, Pak D K, Dickens G R, Miller K G. The source and fate of massive carbon input during the latest Palaeocene thermal maximum. Science. 1999;286:1531–5133. doi: 10.1126/science.286.5444.1531. [DOI] [PubMed] [Google Scholar]
- 20.Kennicutt M C, II, Brooks J M, Denoux G J. Leakage of deep, reservoired petroleum to the near surface of the Gulf of Mexico continental slope. Mar Chem. 1988;24:39–59. [Google Scholar]
- 21.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 22.Kogure K, Simidu U, Taga N. A tentative direct microscopic method for counting living marine bacteria. Can J Microbiol. 1979;25:415–420. doi: 10.1139/m79-063. [DOI] [PubMed] [Google Scholar]
- 23.Kuhs W F, Klapproth A, Gotthardt F, Techmer K, Heinrichs T. The formation of meso- and macroporous gas hydrates. Geophys Res Lett. 2000;27:2929–2932. [Google Scholar]
- 24.Kvenvolden K A. Potential effects of gas hydrate on human welfare. Proc Natl Acad Sci USA. 1999;96:3420–3426. doi: 10.1073/pnas.96.7.3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kvenvolden K A. A review of the geochemistry of methane in natural gas hydrate. Org Geochem. 1995;23:997–1008. [Google Scholar]
- 26.Kvenvolden K A, Ginsburg G D, Soloviev V A. World-wide distribution of subaquatic gas hydrates. Geo-Mar Lett. 1993;13:32–40. [Google Scholar]
- 27.Li L, Kato C, Horikoshi K. Bacterial diversity in deep-sea sediments from different depths. Biodivers Conserv. 1999;8:659–677. [Google Scholar]
- 28.MacDonald I R, Boland G S, Baker J S, Brooks J M, Kennicutt II M C, Bidigare R R. Gulf of Mexico hydrocarbon seep communities. II. Spatial distribution of seep organisms and hydrocarbons at Bush Hill. Mar Biol. 1989;101:235–247. [Google Scholar]
- 29.MacDonald I R, Guinasso N L, Jr, Sassen R, Brooks J M, Lee L, Scott K T. Gas hydrate that breaches the sea-floor on the continental slope of the Gulf of Mexico. Geology. 1994;22:699–702. [Google Scholar]
- 30.Marchesi J R, Weightman A J, Cragg B A, Parkes R J, Fry J C. Methanogen and bacterial diversity and distribution in deep gas hydrate sediments from the Cascadia Margin as revealed by 16S rRNA molecular analysis. FEMS Microbiol Ecol. 2001;34:221–228. doi: 10.1111/j.1574-6941.2001.tb00773.x. [DOI] [PubMed] [Google Scholar]
- 31.Munson M A, Nedwell D B, Embley T M. Phylogenetic diversity of Archaea in sediment samples from a coastal salt marsh. Appl Environ Microbiol. 1997;63:4729–4733. doi: 10.1128/aem.63.12.4729-4733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norris R D, Rohl U. Carbon cycling and chronology of climate warming during the Palaeocene/Eocene transition. Nature. 1999;401:775–778. [Google Scholar]
- 33.Orphan V J, Hinrichs K-U, Ussler III W, Paull C K, Taylor L T, Sylva S P, Hayes J M, DeLong E F. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl Environ Microbiol. 2001;67:1922–1934. doi: 10.1128/AEM.67.4.1922-1934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rappé M S, Gordon D A, Vergin K L, Giovannoni S J. Phylogeny of actinobacteria small subunit (SSU) rRNA gene clones recovered from marine bacterioplankton. Syst Appl Microbiol. 1999;22:106–112. [Google Scholar]
- 35.Rappé M S, Kemp P F, Giovannoni S J. Chromophyte plastid 16S ribosomal RNA genes found in a clone library from Atlantic ocean seawater. J Phycol. 1995;31:979–988. [Google Scholar]
- 36.Roberts H H, Aharon P. Hydrocarbon-derived carbonate buildups of the northern Gulf of Mexico slope: a review of submersible investigations. Geo-Mar Lett. 1994;14:135–148. [Google Scholar]
- 37.Roberts H H, Carney R S. Evidence of episodic fluid, gas, and sediment venting on the northern Gulf of Mexico continental slope. Econ Geol. 1997;92:863–879. [Google Scholar]
- 38.Sassen R, Joye S, Sweet S T, DeFrietas D A, Milkov A V, MacDonald I R. Thermogenic gas hydrates and hydrocarbon gases in complex chemosynthetic communities, Gulf of Mexico continental slope. Org Geochem. 1999;30:485–497. [Google Scholar]
- 39.Sassen R, MacDonald I R, Guinasso N L J, Joye S, Requejo A G, Sweet S T, Alcala-Herrera J, DeFreitas D A, Schink D R. Bacterial methane oxidation in sea-floor gas hydrate: significance to life in extreme environments. Geology. 1998;26:851–854. [Google Scholar]
- 40.Sassen R, MacDonald I R, Requejo A G, Guinasso N L, Jr, Kennicutt II M C, Sweet S T, Brooks J M. Organic geochemistry of sediments from chemosynthetic communities, Gulf of Mexico slope. Geo-Mar Lett. 1994;14:110–119. [Google Scholar]
- 41.Sassen R, Roberts H H, Aharon P, Larkin J, Carney R. Chemosynthetic bacterial mats at cold hydrocarbon seeps, Gulf of Mexico continental slope. Org Geochem. 1993;20:77–89. [Google Scholar]
- 42.Sassen R, Sweet S T, DeFreitas D A, Milkov A V. Exclusion of 2-methylbutane (isopentane) during crystallization of structure II gas hydrate in sea-floor sediment, Gulf of Mexico. Org Geochem. 2000;31:1257–1262. [Google Scholar]
- 43.Sassen R, Sweet S T, Milkov A V, DeFreitas D A, Salata G G, McDade E C. Geology and geochemistry of gas hydrates, central Gulf of Mexico continental slope. Trans Gulf Coast Assoc Geol Soc. 1999;49:462–468. [Google Scholar]
- 44.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 45.Staley J T, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 46.Thiel V, Peckmann J, Seifert R, Wehrung P, Reitner J, Michaelis W. Highly isotopically depleted isoprenoids: molecular markers for ancient methane venting. Geochim Cosmochim Acta. 1999;63:3959–3966. [Google Scholar]
- 47.Thomsen T R, Finster K, Ramsing N B. Biogeochemical and molecular signatures of anaerobic methane oxidation in a marine sediment. Appl Environ Microbiol. 2001;67:1646–1656. doi: 10.1128/AEM.67.4.1646-1656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valentine D L, Reeburgh W S. New perspectives on anaerobic methane oxidation. Environ Microbiol. 2000;2:477–484. doi: 10.1046/j.1462-2920.2000.00135.x. [DOI] [PubMed] [Google Scholar]
- 49.Vetriani C, Jannasch H W, MacGregor B J, Stahl D A, Reyesenbach A-L. Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl Environ Microbiol. 1999;65:4375–4384. doi: 10.1128/aem.65.10.4375-4384.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallrabenstein C, Gorny N, Springer N, Ludwig W, Schink B. Pure culture of Syntrophus buswellii, definition of its phylogenetic status, and description of Syntrophus gentianae sp. nov. Syst Appl Microbiol. 1995;18:62–66. [Google Scholar]
- 51.Waseda A. Organic carbon content, bacterial methanogenesis, and accumulation processes of gas hydrates in marine sediments. Geochem J. 1998;32:143–157. [Google Scholar]
- 52.Webster N C, Wilson K J, Blackall L L, Hill R T. Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odorabile. Appl Environ Microbiol. 2001;67:434–444. doi: 10.1128/AEM.67.1.434-444.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wenger L M, Goodoff L R, Gross O P, Harrison S C, Hood K C. Geological aspects of petroleum systems. Proceedings of the First Joint American Association of Petroleum Geologists/Asociasion Mexicana de Geologos Petroleos Research Conference, Mexico City, Mexico. 1994. Northern Gulf of Mexico: an integrated approach to source, maturation, and migration; p. 6. [Google Scholar]
- 54.Worrall D M, Snelson S. Evolution of the northern Gulf of Mexico with emphasis on Cenozoic growth faulting and the role of salt. In: Bally A W, Palmer E R, editors. The geology of north america—an overview. Boulder, Colo: Geological Society of America; 1989. pp. A97–A138. [Google Scholar]
- 55.Zengler K, Richnow H H, Rossello-Mora R, Michaelis W, Widdel F. Methane formation from long-chain alkanes by anaerobic microorganisms. Nature. 1999;401:266–269. doi: 10.1038/45777. [DOI] [PubMed] [Google Scholar]