Abstract
Background and Purpose
Currently, there are no licensed vaccines and limited antivirals for the treatment of COVID‐19. Heparin (delivered systemically) is currently used to treat anticoagulant anomalies in COVID‐19 patients. Additionally, in the United Kingdom, Brazil and Australia, nebulised unfractionated heparin (UFH) is being trialled in COVID‐19 patients as a potential treatment. A systematic comparison of the potential antiviral effect of various heparin preparations on live wild type SARS‐CoV‐2, in vitro, is needed.
Experimental Approach
Seven different heparin preparations including UFH and low MW heparins (LMWH) of porcine or bovine origin were screened for antiviral activity against live SARS‐CoV‐2 (Australia/VIC01/2020) using a plaque inhibition assay with Vero E6 cells. Interaction of heparin with spike protein RBD was studied using differential scanning fluorimetry and the inhibition of RBD binding to human ACE2 protein using elisa assays was examined.
Key Results
All the UFH preparations had potent antiviral effects, with IC50 values ranging between 25 and 41 μg·ml−1, whereas LMWHs were less inhibitory by ~150‐fold (IC50 range 3.4–7.8 mg·ml−1). Mechanistically, we observed that heparin binds and destabilizes the RBD protein and furthermore, we show heparin directly inhibits the binding of RBD to the human ACE2 protein receptor.
Conclusion and Implications
This comparison of clinically relevant heparins shows that UFH has significantly stronger SARS‐CoV‐2 antiviral activity compared to LMWHs. UFH acts to directly inhibit binding of spike protein to the human ACE2 protein receptor. Overall, the data strongly support further clinical investigation of UFH as a potential treatment for patients with COVID‐19.
Keywords: COVID‐19, heparin, LMWH, nebulised, SARS‐CoV‐2, UFH
Abbreviations
- BCA
bicinchoninic acid
- COVID‐19
coronavirus disease 2019
- DSF
differential scanning fluorimetry
- ECACC
European Collection of Authenticated Cell Cultures
- hCoV
human coronavirus
- ICU
intensive care unit
- LMWH
low MW heparin
- NIBSC
National Institute for Biological Standards and Control
- PEI
polyethyleneimine
- RBD
receptor binding site
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- SD1
spike protein subdomain 1
- UFU
unfractionated heparin
- VIC01
Victoria isolate 01
What is already known
Systemically administered heparin is used to help treat COVID‐19. Evidence suggests heparin has antiviral activity. What this study adds
What this study adds
Systematic comparison of the antiviral activity of a range of clinically relevant UFH and LMWH.
What is the clinical significance
UFH has potent antiviral properties at clinically relevant doses and is superior compared to LMWH.
1. INTRODUCTION
Currently, there are no licensed vaccines and limited antivirals for the treatment of COVID‐19. Repurposing existing clinical drugs with proven safety profiles provides a rapid approach to address this gap in treatment. Throughout the COVID‐19 pandemic, anticoagulant drugs such as unfractionated heparin (UFH) and low MW heparin (LMWH) delivered systemically have been widely used across the world as part of standard treatment for patients in intensive care units (ICU). The use of these drugs has been shown to be effective for dealing with coagulopathies seen during the late stage of the disease (Kollias et al., 2020; Tang, Li, Wang, & Sun, 2020). Furthermore, heparin exhibits a wide range of anti‐inflammatory properties (Mulloy, Hogwood, Gray, Lever, & Page, 2016), thus providing an additional rationale for its clinical use to treat the hyperinflammatory response observed in patients with COVID‐19.
There is a recent and growing body of evidence suggesting that UFH has antiviral properties against SARS‐CoV‐2, the causative agent of COVID‐19. UFH has been shown to bind to the receptor binding domain (RBD) of the SARS‐CoV‐2 spike protein, which induces a conformational change (Mycroft‐West et al., 2020) and also inhibits binding of spike protein to cells (Mycroft‐West et al., 2020; Partridge, Green, & Monk, 2020). Additional work with a pseudotyped assay indicated that UFH has a half inhibitory maximal concentration (IC50) of 0.6 μg·ml−1 (Tandon et al., 2020). Recent studies, performed with live SARS‐CoV‐2 and Vero E6 cells, were limited to one heparin brand (Celsus, USA) and showed SARS‐CoV‐2 virus was inhibited by 44–80% with 6.25–200 μg·ml−1 of UFH in vitro (Mycroft‐West, Su, Elli, et al., 2020). Conzelmann et al. also report that heparin, sourced from the company Sigma‐Aldrich, suppressed viral replication of SARS‐CoV‐2 in Vero E6 cells by 60% using 125–250 μg·ml−1 (Conzelmann et al., 2020) and most recently, Clausen et al. also show a concentration‐dependent inhibitory effect of UFH across the range of 1–100 μg·ml−1 in Vero E6 cells. They reported a fivefold reduction in virus infection in primary human bronchial epithelial cells (Clausen et al., 2020).
The accurate determination of the amount of heparin (μg·ml−1) required to inhibit 50% of wild type, authentic, live SARS‐CoV‐2 virus has yet to be reported in an in vitro system and a systematic assessment of the antiviral activity of different commercially available heparins, already in clinical use, has yet to be performed. Once determined, though, such an analysis should inform the selection of the most appropriate heparin preparation having the most antiviral activity. The determination of the IC50 of different heparin preparations will also enable a quantitative comparison between different heparin sources, for example bovine and porcine, and heparins with different MWs, for example UF and LMWH. Such a systematic analysis should also enable a quantitative comparison with other drugs under investigation as treatments for COVID‐19, for example remdesivir.
Independent from the systemic use of heparin in COVID‐19 patients, nebulised heparin has been proposed as a unique and potentially effective treatment for different stages of COVID‐19 disease (van Haren et al., 2020). In the United Kingdom, a clinical trial evaluating the effectiveness of nebulised UFH in hospitalised COVID‐19 patients is currently underway (https://accord-trial.org). The strategy underpinning this treatment stems from positive observations made, based on anticoagulant and anti‐inflammatory activity, when running trials of nebulised heparin in patients with acute lung injury and other respiratory conditions (van Haren et al., 2020). The delivery of an aerosolised antiviral agent directly to the lungs where SARS‐CoV‐2 virus is known to be present (both upper and lower respiratory tract) (Schaefer et al., 2020) may help to treat the alveolar coagulopathy that is often a feature of COVID‐19, reduce the hyperinflammatory response and prevent patients from developing acute respiratory distress syndrome (ARDS) and pulmonary fibrosis (PF), the major complications of infection with SARS‐CoV‐2.
Here, we report for the first time a systematic assessment of the antiviral activity of a range of heparins (UFH and LMWH) in vitro against live wild type SARS‐CoV‐2, thereby shedding light on the antiviral potency of a range of different heparin preparations, including the nebulised UFH already undergoing clinical trials in COVID‐19 patients. We used a 24‐well, 5‐day plaque inhibition assay to assess the antiviral activity of heparin, in a biosafety level 3 containment laboratory. This classical, virological technique has been used previously to assess the antiviral activity of compounds against other viruses, for example influenza virus (Hayden, Cote, & Douglas, 1980). We also report evidence for a direct mechanism of action via inhibition of binding of the spike protein receptor binding domain (RBD) on the angiotensin‐converting enzyme (ACE2).
2. METHODS
2.1. Heparin preparations
Clinically used LMWH preparations investigated were Innohep (tinzaparin sodium, LEO Pharma), Clexane (enoxaparin sodium, Sanofi), and Fragmin (dalteparin sodium, Pfizer), along with a porcine mucosal UFH (heparin sodium, Wockhardt, UK) [currently being investigated by nebulisation in a UK human clinical trial]. Further heparin preparations investigated were a porcine mucosal UFH preparation (Celsus, USA) and both a bovine lung heparin (Calbiochem) and a bovine mucosal heparin (15/110, NIBSC, UK).
Specific anticoagulant activity for the UFH preparations was measured as described in the United States Pharmacopeia (USP), n.d, a general monograph for the assay of heparin. The MWs for the UFH and LMWH samples were measured as previously described (Mulloy & Hogwood, 2015). The specific activity of the clinical LMWHs was taken from the clinical product information (Clexane, 100 IU·mg−1; Innohep, 100 IU·mg−1; Fragmin 130 IU·mg−1).
2.2. Plaque inhibition assay
SARS‐CoV‐2 (hCoV‐19/Australia/VIC01/2020) (Caly et al., 2020) was generously provided by The Doherty Institute, Melbourne, Australia, at P1 and passaged twice in Vero/hSLAM cells [ECACC 04091501]. Whole genome sequencing was performed, on the working stock at Passage 3, using both Nanopore and Illumina as described previously (Lewandowski et al., 2019) and no significant changes in the viral sequence were observed. Virus titre was determined by a plaque assay on Vero E6 cells [ECACC 85020206]. Cell lines were obtained from the European Collection of Authenticated Cell Cultures (ECACC) PHE, Porton Down, UK. Cell cultures were maintained at 37°C in minimal essential media (MEM) (Life Technologies, California, USA) supplemented with 10% FBS (Sigma, Dorset, UK) and 25 mM HEPES (Life Technologies).
All heparin preparations were diluted, fivefold in MEM (Life Technologies) containing 1% (v/v) FBS (Life Technologies), 1× antibiotic/antimycotic (Life Technologies) and 25 mM HEPES buffer (Sigma) by diluting down in a 96‐well plate. Dilutions were made fresh on the day of the assay. SARS‐CoV‐2 was diluted to a concentration of 933 pfu·ml−1 (70 pfu/75 μl) in the same media and mixed 50:50 with heparin dilutions, in a 96‐well V‐bottomed plate. The plate was incubated at 37°C in a humidified box for 1 h to allow the virus to be exposed to heparin. The neutralised virus was transferred onto the wells of a washed 24‐well plate that had been seeded with Vero E6 cells the previous day at 1.5 × 105 cells per well. The virus/heparin mixture was left to adsorb for an hour at 37°C and then plaque assay overlay media was applied (MEM containing 1.5% carboxymethylcellulose [Sigma], 1× antibiotic/antimycotic [Life Technologies], 4% (v/v) FBS and 25 mM HEPES buffer). After incubation at 37°C in a humidified box, for 5 days, plates were fixed overnight with 20% (v/v) formalin/PBS, washed with tap water and then stained with methyl crystal violet solution (0.2% v/v) (Sigma),and plaques were counted. A series of eight dilutions (fivefold) were prepared for each heparin preparation, in triplicate (technical replicates) to ensure that the most accurate prediction of the IC50 could be obtained with the narrowest 95% confidence interval. Repeat experiments (n = 5) were performed on different days (independent experiments). Heparin dilutions with cells only were also run in duplicate, to determine if there was any cell cytotoxicity.
An internal positive control for the plaque inhibition assay was run in duplicate using a sample of heat‐inactivated human MERS convalescent serum known to neutralise SARS‐CoV‐2 (National Institute for Biological Standards and Control [NIBSC], UK).
2.3. Recombinant RBD‐SD1 expression and purification
Secreted Spike protein RBD‐SD1 was transiently produced in suspension HEK293‐6E cells. A plasmid encoding RBD‐SD1, residues 319–591 of 2019‐nCoV S were cloned upstream of a C‐terminal HRV3C protease cleavage site; a monomeric Fc tag and an His8x Tag were a gift from Jason S. McLellan, University of Texas at Austin, USA. Briefly, 100 ml of HEK293‐6E cells were seeded at a cell density of 0.5 × 106 cells·ml−1 24 h before transfection with polyethyleneimine (PEI). For transfection, 100 μg of the ACE2 plasmid and 300 μg of PEI (1:3 ratio) were incubated for 15 min at room temperature. Transfected cells were cultured for 48 h and fed with 100 ml fresh media for additional 48 h before harvest. RBD‐SD1 was purified by HiTrap Protein G HP column (GE Healthcare, USA) pre‐equilibrated in PBS and eluted with 0.1 M glycine (pH 2.7). Purity of proteins was evaluated by Coomassie staining of SDS‐PAGE gels and proteins were quantified by BCA Protein Assay Kit (Thermo Scientific).
2.4. Differential scanning fluorimetry
Differential scanning fluorimetry (DSF) (Katarzna et al., 2010) was performed employing the hydrophobic dye Sypro orange (1.25 X; Invitrogen) to monitor the thermal denaturation of RBD‐SD1 (1 μg per well) alone or in the presence of 200 μg heparin (Celsus or Wockhardt) in PBS pH 7.6 with a total well volume of 40 μl. To ensure that a change in melting temperature was not a result of interactions between Sypro orange and heparin (Celsus or Wockhardt), control wells containing H2O or heparin (Celsus or Wockhardt) without mS1‐RBD were also screened. Melt curve experiments were performed in 96‐well qPCR plates (AB Biosystems) using an AB biosystems StepOne plus qPCR machine with the TAMRA filter setting enabled. An initial incubation of 2 min at 25°C was set, increasing sequentially by 0.5°C increments every 30 s up to 90°C. Melt curves were smoothed (nine neighbours, second‐order polynomial, Savitxky‐Golay) and first differential plots were constructed utilising Prism 8 (GraphPad). The peak of the first differential plots was used to calculate Tm values using MatLab software (R20018a, MathWorks) and the difference between mean values (n = 6 independent experiments) was calculated using a Student's t‐test in Prism 8 (GraphPad). Studies on Wockhardt heparin were exploratory (n = 3 independent experiments) due to limited availability of the RBD protein.
2.5. elisa assay for measuring inhibition of RBD‐ACE2 binding
Streptavidin (3 μg·ml−1; Fisher) in 50 mM sodium carbonate buffer pH 9.6 (50 μl per well) was incubated for 1 h at 37°C in high binding 96‐well plates (Greiner). Plates were blocked with PBS, 0.2% Brij35 (w/v) + 1% casein (w/v) for 1 h at 37°C, after washing thrice with PBS, 0.2% Brij35 (w/v) (PBSB). Plates were washed again with PBSB and then incubated with biotinylated‐ACE2 (Sino Biological) in PBSB + 1% (w/v) casein for 1 h at 37°C. mS1‐RBD in PBSB + 1% casein (w/v) was preincubated separately for 30 min at r.t. with or without heparin at distinct concentrations (100–0.7 μg·ml−1), before addition to the prewashed plates containing immobilised ACE2. Plates were subsequently incubated for 1 h at 37°C and washed with PBSB. Bound mS1‐RBD was detected by incubation with 0.5 μg·ml−1 Rabbit‐SARS‐CoV‐2 (2019‐nCoV) Spike RBD Antibody (Stratech) in PBSB + 1% (w/v) casein (50 μl per well) for 1 h at 37°C. Plates were washed thrice with PBSB before being incubated for 30 min at 37°C with HRP‐conjugated Donkey anti‐Rabbit IgG, diluted 1:1000 (v/v) in PBSB + 1% casein (w/v) (Biolegend). Plates were washed thoroughly with PBSB before being developed with 3,3′,5,5′‐tetramethylbenzidine prepared according to the manufacturer's instructions (Sigma). After 10 min colour development, the reaction was stopped through the addition of 20 μl, 2 M HsSO4 and well‐absorbances were determined at λ = 450 nm using a Tecan Infinate M200 Pro multiwell plate reader (Tecan Group). Control wells containing no biotinylated ACE2 were employed to ensure binding was specific. These elisa studies were exploratory (n = 3 independent experiments) due to limited availability of the key reagents (RBD, biotinylated ACE2 and Spike antibody).
2.6. Data and statistical analysis
The data and statistical analysis comply with the recommendations of the British Journal of Pharmacology on experimental design and analysis in pharmacology (Curtis et al., 2018). A series of eight dilutions (fivefold) were prepared for each heparin preparation, in triplicate (technical replicates). Repeat plaque inhibition assays were run on different days, at random, to ensure true independence and unbias treatment. The samples were run in a partly blinded style as the operator conducting the assays did not see the results. The repeat of assays was limited to five occasions because these biological experiments were lengthy (5 days) and run at biosafety level 3 containment. Competing demands for the high containment facilities during the current pandemic meant 1 heparin preparation (Fragmin LMWH) was repeated only four times. A midpoint probit analysis (written in R programming language for statistical computing and graphics) was used to determine the amount of heparin (μg·ml−1) required to inhibit SARS‐CoV‐2 viral plaques by 50% (IC50) compared with the virus only control (n = 10). All outliers were included in the midpoint probit analysis. The analysis was conducted in R (R Project, 2019) and the script was based on a source script from Johnson, Dahlgren, Siegfried, & Ellis, 2013 (Johnson et al., 2013). Data are presented as geometric mean ± SEM (n = 5) and were tested for normality via Shapiro–Wilk test. When a statistical comparison between two groups was performed, a Welch's two‐sample t‐test was used in R. This test was only performed when group sizes were n ≥ 5. P < 0.05 value was considered statistically significant. The declared group size is the number of independent values and the statistical analysis was done using these independent values.
2.7. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in the IUPHAR/BPS Guide to PHARMACOLOGY http://www.guidetopharmacology.org and are permanently archived in the Concise Guide to PHARMACOLOGY 2017/1820 (Alexander et al., 2017).
3. RESULTS
The characteristics of the different unfractionated heparins (UFHs) and low MW heparins (LMWHs) are detailed in Table 1. A concentration‐dependent relationship between the amount of heparin (μg·ml−1) and the antiviral activity against SARS‐CoV‐2 was observed for all heparin preparations tested. Increasing concentrations of heparin caused a reduction in the number of viral plaques compared to the control. Five independent plaque inhibition experiments were run for each heparin preparation, unless otherwise stated. The geometric mean IC50 and the pIC50 values for each heparin preparation are stated in Table 1. Representative data (midpoint probit curve ± 95% confidence intervals) from one independent plaque inhibition experiment for each UFH is presented graphically in Figure 1a–d, a summary of all the experimental data is presented in Table 1. Similarly, representative data from one independent plaque inhibition experiment for each LMWH preparation are presented in Figure 2a–c and a summary is presented in Table 1. The midpoint probit plots show the spread of data across different dose ranges.
TABLE 1.
Properties of different heparin preparations and the geometric mean midpoint probit IC50/pIC50 values
| Product | Brand | Origin | Weight average molecular weight Da ± 1 SD a | Specific anticoagulant activity IU·mg−1 (95% confidence interval) | Geometric mean probit midpoint (IC50) μg·ml−1 ± 1 SE | pIC50 | No. independent plaque inhibition experiments |
|---|---|---|---|---|---|---|---|
| Unfractionated heparins (UFs) | |||||||
| Heparin sodium | Wockhardt (clinical batch b ) | Porcine mucosa | 16,300 ± 100 | 192 (187–197) | 41 ± 26 | 5.6 | 5 |
| Heparin sodium | Celsus | Porcine mucosa | 15,650 ± 550 | 192 (186–198) | 31 ± 8 | 5.7 | 5 |
| Heparin sodium | Calbiochem | Bovine lung | 15,950 ± 550 | 108 (104–112) | 25 ± 8 | 5.8 | 5 |
| Heparin sodium | NIBSC | Bovine mucosa | 19,100 ± 200 | 146 (142–152) | 34 ± 15 | 5.7 | 5 |
| Low MW heparins (LMWHs) | |||||||
| Tinzaparin sodium | Innohep | Porcine mucosa | 6,650 ± 100 | 100 c | 3,721 ± 4,402 | 3.3 | 5 |
| Dalteparin sodium | Fragmin | Porcine mucosa | 6,110 ± 50 | 130 c | 3,428 ± 1,337 | 3.3 | 4 |
| Enoxaparin sodium | Clexane | Porcine mucosa | 4,200 ± 250 | 100 c | 7,809 ± 2,237 | 2.7 | 5 |
Rounded to the nearest 50 Da, n = 2 independent measurements.
This is the same batch of Wockhardt heparin used to treat patients with COVID‐19, UK.
These values are based on specific activities quoted by the manufacturers.
FIGURE 1.
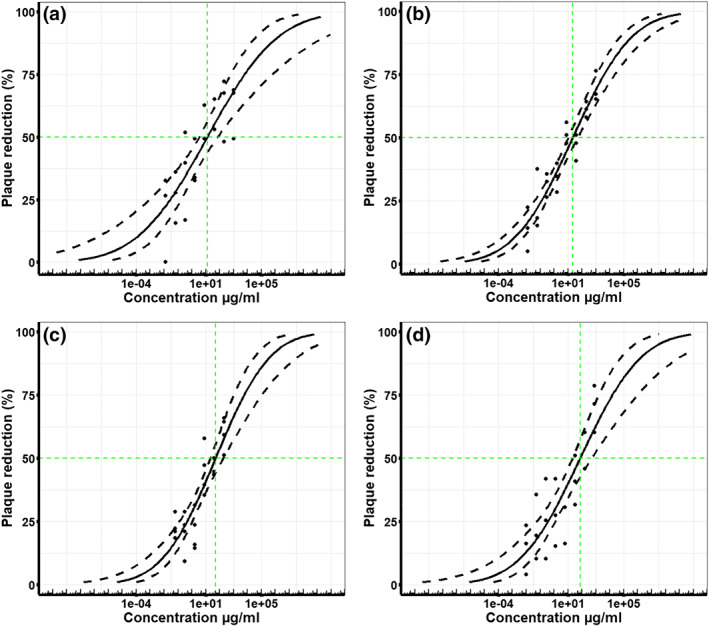
Unfractionated heparins (UFHs) inhibit live wild type SARS‐CoV‐2 infectivity in Vero E6 cells. The % viral plaque reduction, compared to virus only control, caused by increasing concentrations (μg·ml−1) of different unfractionated heparins in vitro with Vero E6 cells. Five independent experiments were run for Wockhardt UFH (n = 5), Celsus UFH (n = 5), bovine lung UFH (n = 5),and bovine mucosa UFH (n = 5). Representative data presented here graphically are taken from one independent experiment that had three technical replicates and a summary of all the data is presented in Table 1. Midpoint probit curves are plotted with 95% confidence intervals (dashed lines): (a) Wockhardt UFH (clinical batch), (b) Celsus UFH, (c) bovine UFH (lung), (d) bovine UFH (mucosa)
FIGURE 2.
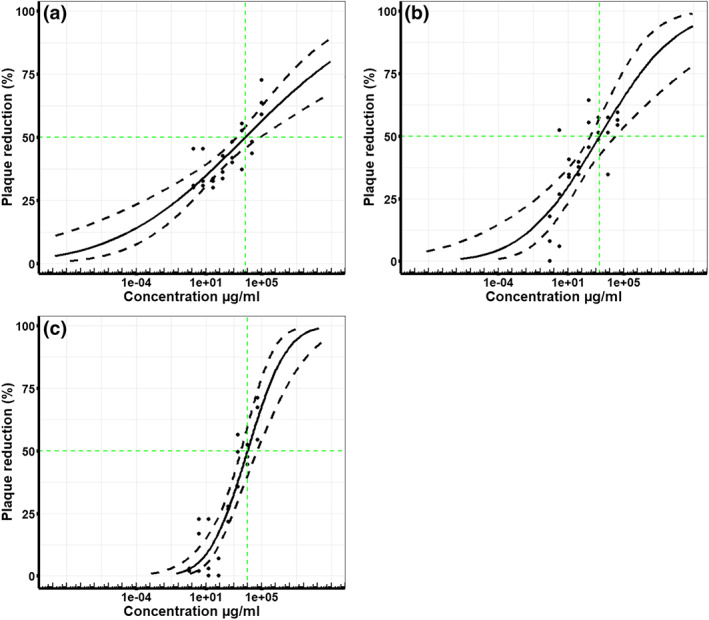
Low MW heparins (LMWHs) inhibit live wild type SARS‐CoV‐2 infectivity in Vero E6 cells. The % viral plaque reduction, compared to the virus only control, caused by increasing concentrations (μg·ml−1) of different LMWH preparations, in vitro with Vero E6 cells. Five independent experiments were run for Clexane LMWH (n = 5) and Innohep LMWH (n = 5); only four independent experiments were run for Fragmin LMWH (n = 4), so these data are preliminary. Representative data presented graphically here are taken from one independent experiment that had three technical replicates and a summary of all the data is presented in Table 1. Midpoint probit curves are plotted with 95% confidence intervals (dashed lines): (a) Innohep LMWH, (b) Fragmin LMWH and (c) Clexane LMWH
3.1. Unfractionated heparins (UFHs)
The antiviral activity of different UFHs fell within the range of 25–41 μg·ml−1 (Table 1). The two porcine unfractionated heparins obtained from Wockhardt (clinical batch) and Celsus had geometric mean IC50 values of 41 and 31 μg·ml−1, respectively (Table 1). There was no significant difference between the IC50 values for Celsus and Wockhardt heparin (Welch's two‐sample t‐test, n = 5, P > 0.05). Similarly, there was no significant difference between the IC50 values for heparin sourced from bovine lung (geometric mean IC50 25 μg·ml−1) or bovine mucosa (geometric mean IC50 34 μg·ml−1) (Welch's two‐sample t‐test, n = 5, P > 0.05) (Table 1). To determine if there was an effect of species on the antiviral activity of UFHs, the bovine mucosa and bovine lung data were grouped (n = 10) and the two porcine UFHs were grouped (n = 10). When compared using a Welch's t‐test, there was no significant difference (P > 0.05) between the species.
3.2. Low MW heparins (LMWHs)
The LMW heparins Innohep and Clexane had an inhibitory effect on the growth of SARS‐CoV‐2, with geometric mean IC50 values of 3.7 and 7.8 mg·ml−1, respectively. There was no significant difference between the IC50 values for Clexane and Innohep heparin preparations (Welch's two‐sample t‐test, n = 5, P > 0.05) Mycroft‐West et al. (2020). Exploratory experiments were performed with the LMW heparin Fragmin, this clinical preparation also had antiviral activity with an IC50 value of 3.4 mg·ml−1.
3.3. Comparison unfractionated heparins (UFHs) and low MW heparins (LMWHs)
Overall, the antiviral activity of LMWH was ~150‐fold lower than the activity of UFH (Table 1); thus, more LMW heparin (μg·ml−1) was required to reduce the same amount of SARS‐CoV‐2 virus. Since there was no significant difference between the antiviral activity of Celsus and Wockhardt the data was grouped (n = 10). Similarly, the data were grouped for the LMWHs Clexane and Innohep (n = 10). When the grouped UFH and the grouped LMWH data were compared using a Welch's two‐sample t‐test there was a significant difference (P < 0.05) between the IC50 values of UFH and LMWH, indicating the antiviral activity of porcine mucosa UFH is significantly greater than porcine mucosa LMWH.
3.4. Effect of molecular weight
The antiviral activity shown by the different heparins appeared to correlate broadly with their MW. Overall, UFHs with the highest MWs ranging between 15,650 and 19,100 Da were more potent inhibitors of SARS‐CoV‐2 than low MW heparins ranging between 4,200 and 6,650 Da (Table 1).
3.5. Mechanisms of action of heparin against SARS‐CoV‐2—Destabilisation of spike protein RBD and inhibition of RBD binding to human ACE2
Since previous work has indicated interactions of heparin with spike protein RBD, we first confirmed a direct interaction of UF heparins with soluble S1 RBD domain (RBD‐SD1) expressed in HEK239 cells by exploring their effects on protein thermal stability using differential scanning fluorimetry (DSF). DSF exploits conventional real time PCR machines to monitor the thermal denaturation of a protein through a range of temperatures, in the presence of a hydrophobic fluorescent dye. Upon heating, the hydrophobic regions of proteins become exposed upon unfolding, resulting in increased fluorescence. The midpoint of unfolding (Tm) can be determined through the first differential of the resulting melt curve and changes in Tm values in the presence of ligands can be used to determine binding affinity (Niesen, Berglund, & Vedadi, 2007). This method has previously been exploited to characterise the structural requirements of heparin for binding to FGFs (Uniewicz et al., 2010). Celsus heparin with RBD‐SD1 elicited a significant reduction in the melting temperature (ΔTm) of 3.25°C (Figure 3a; P < 0.05, n = 6 independent experiments). Exploratory data on Wockhardt heparin indicated that it induces a similar reduction in Tm of 2.4°C (Figure 3b). This confirmed a direct interaction of heparin with RBD and furthermore suggests a significant destabilisation of the RBD protein structure which could alter its ability to interact with the ACE2 cell receptor.
FIGURE 3.
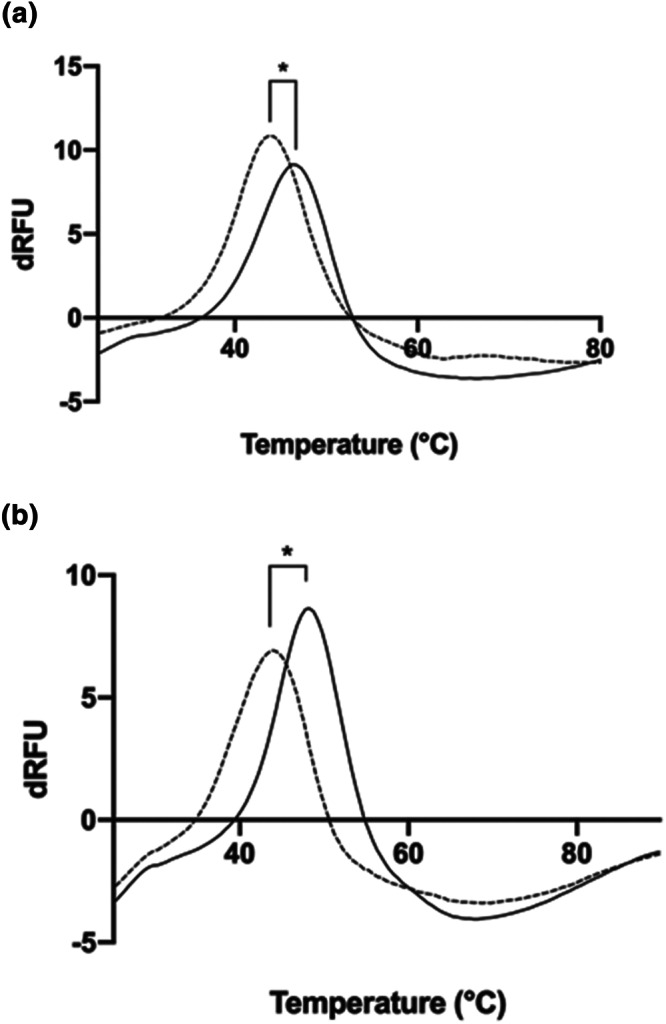
Unfractionated heparins (UFs) interact directly with spike protein RBD domain. Differential scanning fluorimetry was employed to measure the thermal stability curve for recombinant RBD in the absence of presence of UF heparin. First differential of the thermal stability of 1 μg RBD alone (solid line) or with 200 μg UF heparin (a, Celsus UF, dashed line; b, Wockhardt UF, dashed line). (Figure 3a; P < 0.05, n = 6 independent experiments). *Significant difference between the Tm value of RBD (46.4°C ± 0.25 (±SD); n = 6 independent experiments) compared to RBD + Celsus UF (43.1°C ± 0.8; n = 6), t(10) = 8.854, P < 0.05. ΔTm = 3.25°C. In exploratory experiments (n = 3 independent experiments) Wockhardt UF displayed a Tm value of 44.0°C ± 0.6 (n = 3), indicating a ΔTm of 2.4°C
In order to both validate the human relevance of our data and also evaluate the potential mechanism by which heparin might inhibit SARS‐CoV‐2 binding to human cells, we utilized an elisa assay with surface immobilised recombinant human ACE2 to measure the binding of RBD‐SD1. Binding of RBD‐SD1 to human ACE2 protein was confirmed by detection with an anti‐RBD antibody (Figure 4). In exploratory experiments, a concentration response assay using Celsus UFH demonstrated potent inhibition of binding, with an IC50 geometric mean of ~16.2 ± 9.3 μg·ml−1 (n = 3 independent experiments, ±SEM) (Figure 4). These data are in very close agreement with the heparin inhibition data for live SARS‐CoV‐2 infectivity of Vero E6 cells (Table 1). Notably, these data support the human relevance of our data on the inhibition of SARS‐CoV‐2 infection of Vero E6 cells by heparins and provides strong evidence for a direct mechanism in which heparin destabilises the spike protein RBD and inhibits its binding to the ACE2 cell receptor.
FIGURE 4.
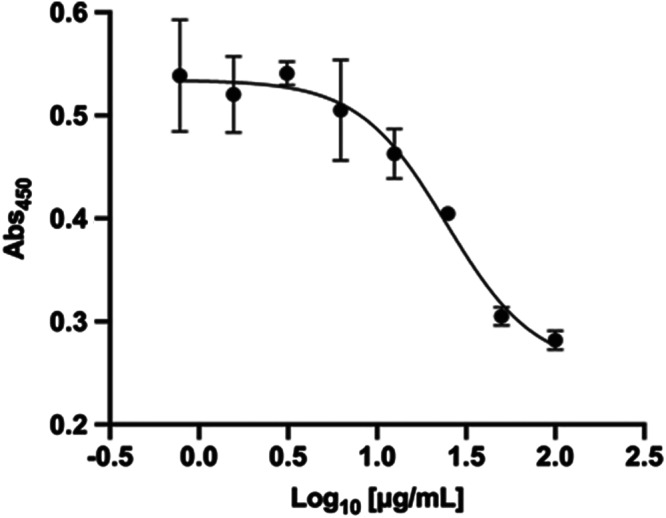
Unfractionated heparin (UF) hdirectly inhibits binding of spike protein RBD to ACE2 protein receptor. Biotinylated ACE2 was captured onto a high binding microplate coated with streptavidin prior to the addition of RBD preincubated with or without varying concentrations of Celsus UF heparin. Detection of bound RBD was achieved with an anti‐RBD antibody. In exploratory studies (n = 3 independent experiments), the IC50 observed was 16.2 ± 9.3 μg·ml−1 (±SEM). Representative example shown (IC50, 24.6 μg·ml−1)
4. DISCUSSION
Some studies have already reported the association between treatment with systemically administered heparin and lower mortality in patients admitted to hospital with COVID‐19 (Ayerbe, Risco, & Ayis, 2020; Tang et al., 2020), assumed to be a consequence of the known anticoagulant effect. Coagulopathies have caused major problems in late stage COVID‐19 disease (Kollias et al., 2020) and the use of heparin in the treatment of COVID‐19 has become part of the standard care for many patients in ICU. However, the dose and type of heparin (UFH or LMWH) delivered either by the subcutaneous or intravenous route varies across the world. Regarding standard systemic usage, the therapeutic range for UFH is typically 0.3–0.7 IU·ml−1 (~2–4 μg·ml−1) (Hirsh, Anand, Halperin, & Fuster, 2001), with peak dosing concentrations reaching ~10–20 μg·ml−1. It is reassuring to see that the IC50 values for porcine UFH is 31–41 μg·ml−1, which is close to the peak dosing range suggesting that at least partial antiviral effects could be expected in this setting. It is also feasible that heparin could be used in combination with alternative anti‐viral agents acting via different mechanisms, to gain therapeutic advantage.
In the United Kingdom, Brazil and Australia, in addition to systemic heparin use, the novel approach of delivering UFH via nebulisation directly to lungs of COVID‐19 patients is undergoing evaluation in a clinical trial by the ACCORD clinical trials platform (https://accord-trial.org/) (van Haren et al., 2020). This is to determine if there is a benefit from the additional known anti‐inflammatory activity of heparin and to provide high local anti‐coagulant effects on the alveolar region of patients with severe COVID‐19. Nebulisation allows for the targeting of lung tissue directly and therefore impact upon the local hyperinflammatory response and alveolar coagulation resulting from SARS‐CoV‐2 viral load in the lung. During the UK trial, UFH (Wockhardt) will be administered at 25,000 IU (130 mg) every 6 h to patients. The efficiency of nebulising UFH through a high efficiency mesh nebuliser is estimated to be about 20%; thus, the delivered dose to the lung is ~26 mg. Assuming the normal human airway surface fluids are in the range 10–60 ml (Frohlich, Mercuri, Wu, & Salar‐Behzadi, 2016), the peak amount of UFH delivered to the lung should be ~400–2,600 μg·ml−1, though these values could be lower if diluted by increased fluid volumes as a result of pulmonary oedema. Even allowing for this, these values greatly exceed the IC50 of 41 μg·ml−1 reported here for the same batch of Wockhardt UFH as used in the current UK clinical trial. Thus, nebulisation of UFH should provide strong antiviral effects, in vivo. Importantly, inhaled UFH does not enter the systemic coagulation in any meaningful way so that nebulised UFH can be used safely on top of standard of care involving the systemic use of UFH or LMWH as an anticoagulant.
Previous work has demonstrated that LMWHs also bind to the SARS‐CoV‐2 (Mycroft‐West, Su, Li, et al., 2020). However, in the present study, we observed that LMWHs were markedly less potent in live SARS‐CoV‐2 virus assays (IC50 values of 3.4–7.8 mg·ml−1) than UFH. Using the results reported here LMWH would appear unlikely to reach sufficient concentration to achieve significant antiviral activity for either systemic or nebulisation delivery. The typical therapeutic range for LMWH for anticoagulant therapy is ~5–8 μg·ml−1 (0.6–1 IU·ml−1). Consistent with our data, the relationship (fold difference, ~170‐fold) between UFH and enoxaparin (Clexane) seen here was close to that observed by Tandon et al. (2020) using a pseudotyped lentivirus inhibition assay where 180‐fold difference was seen (Tandon et al., 2020). In addition, LMWHs were also observed to be less potent than UFH for inhibition of cell binding by spike protein (Partridge et al., 2020). However, an important caveat is that the potency of UFH and LMWHs remains to be determined in a suitable range of human cells relevant to those affected in individuals infected with SARS‐CoV‐2, perhaps especially from respiratory tract tissues.
The results of the present study also suggest a dependency on MW for different UFH and LMWH preparations. A positive correlation between MW and antiviral activity was noted for the various porcine LMWHs (4,200–6,650 Da) and UFH preparations tested which supports the hypothesis that UFH is more active due to its higher MW (15,650–19,100 Da). Consistent with these live virus data, MW dependency for binding of heparin and heparan sulfate saccharides to spike protein has been observed (Liu et al., 2020; Mycroft‐West, Su, Pagani, et al., 2020). In terms of a molecular explanation for this effect, data from molecular modelling have indicated multiple binding sites on RBD for heparin, including some near the RBD‐ACE2 interface (Clausen et al., 2020; Mycroft‐West, Su, Elli, et al., 2020). Based on these data, we anticipate that longer heparin chains may be able to bind more efficiently to multiple sites and also potentially extend beyond RBD protein to interfere directly with the ACE2 binding of the spike protein and this would be consistent with our elisa data demonstrating inhibition of RBD‐ACE2 interaction by heparin.
Recent work has demonstrated that cell surface heparan sulfate is a necessary coreceptor with ACE2 that mediates SARS‐CoV‐2 virus infection of cells (Bermejo‐Jambrina et al., 2020; Clausen et al., 2020). Regarding the underlying mechanisms, we provided additional evidence here for the action of heparin to destabilise the RBD protein structure (Figure 3) and notably the first evidence for the ability of heparin to directly inhibit binding of the RBD to its protein receptor human ACE2 (Figure 4). Both these pieces of data are supported by recent preliminary observations using native MS (Yang, Du, & Kaltashov, 2020). Thus, there is a clear mechanistic basis supporting the notion of targeting these interactions with heparin and related compounds. It is also notable that binding of RBD to human lung cancer cell lines can be potently inhibited by heparin (Clausen et al., 2020), strongly supporting the view that our data showing inhibition of live SARS‐CoV‐2 infectivity of monkey Vero E6 cells, a widely used experimental model, will also be relevant in vivo for inhibiting infection of human cells.
In the present study, bovine UFH (IC50 25–34 μg·ml−1) had a similar level of potency when compared to porcine UFH (IC50 31–41 μg·ml−1). The potent antiviral activity seen for both porcine and bovine heparins suggests that this property is not species dependent. Bovine UFH may provide an additional source of heparin to use during the coronavirus pandemic. Currently, therapeutic UFH available in Europe and United States is of porcine mucosal origin. However, owing to supply issues there is now interest, specifically in the United States, employing bovine mucosal UFH as an additional source to improve the robustness of supply chains (Hogwood, Mulloy, & Gray, 2017; Szajek et al., 2015).
The antiviral (IC50) data for the different UFH and LMWHs display no obvious correlation with anticoagulant activities (IU·mg−1), indicating that different structure–activity relationships exist for antiviral activity. Importantly, this suggests that further investigation of non‐anticoagulant heparins (Cassinelli, Torri, & Naggi, 2020; Lindahl & Li, 2020) and heparin mimetics (Guimond et al., 2020; Lindahl & Li, 2020) is warranted. Mimetics have significant potential to target similar antiviral mechanisms and could be delivered systemically at higher doses to improve efficacy without potential side effects such as bleeding. This would apply to a broad range of patients, not only those requiring anticoagulation. Moreover, mimetics would also provide a fully synthetic route to bypass limitations of heparin supply.
Here, we provide evidence for the first time that various types of commercially available and clinically used UFH preparations exhibit potent antiviral efficacy against live wild type SARS‐CoV‐2 in vitro. This activity was seen across different preparations of UFH and was also observed with both porcine and bovine heparins and is supported by data indicating a mechanism of action via inhibition of RBD binding to human ACE2. These data indicate that current clinical use of systemic UFH in the treatment of COVID‐19 patients in an ICU setting may provide useful antiviral benefits. Moreover, we predict that the delivery of UFH to the lung (via nebulisation) should provide a strong direct antiviral therapy in addition to other documented beneficial effects of heparins.
AUTHOR CONTRIBUTIONS
J.A.T., J.E.T., M.W.C., E.G., T.W., D.S. and C.P.P. conceived the study. J.A.T., J.E.T., M.W.C. and C.P.P. wrote the first draft of the manuscript. K.J.S., C.M.S., T.W. and D.S. provided critical review. K.R.B. and N.C. performed the virology. J.H. and E.Y. determined the molecular mass and heparin anti‐coagulant activity. E.Y., C.J., M.‐W., M.A.L. and M.A.K. expressed RBD in E. coli and performed the DSF and ACE2 binding assays. R.K., Y.‐H.C. and Y.Z. expressed RBD protein in mammalian cells. M.J.E. performed the statistical analysis. All the authors contributed to the preparation of the manuscript.
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
DECLARATION OF TRANSPARENCY AND SCIENTIFIC RIGOUR
This Declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research as stated in the BJP guidelines for Design and Analysis and as recommended by funding agencies, publishers and other organisations engaged with supporting research.
ACKNOWLEDGEMENTS
We thank Professor Jan Shute (University of Portsmouth) for helpful comments on nebulised heparin. We add thanks to Dr Julian Druce, Doherty Institute, Melbourne, Australia, for donating the virus used in this study. We thank Dr Kevin Bewley, Public Health England, for designing and optimising the PRNT assay used in this study. We thank Mrs Elizabeth Penn for preparing the cell cultures, Mrs Harriet Garlant and Mrs Joanna McGlashan for technical assistance. We are grateful for receiving MERS convalescent sera from Dr Mark Page and Dr Giada Mattiuzzo, NIBSC, UK. D.S. is supported by the National Institute for Health Research (NIHR) Manchester Biomedical Research Centre (BRC). J.E.T. acknowledges the support of University of Liverpool, and M.A.S. the support of Keele University. Z.Y. acknowledges the Danish National Research Foundation (DNRF107) and the Lundbeck Foundation, Y.‐H.C. the Innovation Fund Denmark and R.K. the European Commission (GlycoImaging H2020‐MSCA‐ITN‐721297). This work was funded by Public Health England, the University of Liverpool and the University of Keele.
Tree JA, Turnbull JE, Buttigieg KR, et al. Unfractionated heparin inhibits live wild type SARS‐CoV‐2 cell infectivity at therapeutically relevant concentrations. Br J Pharmacol. 2021;178:626–635. 10.1111/bph.15304
Julia A. Tree and Jeremy E. Turnbull are joint first authors.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request. Some data may not be made available because of privacy or ethical restrictions.
REFERENCES
- Alexander, S. P. H. , Kelly, E. , Marrion, N. V. , Peters, J. A. , Faccenda, E. , Harding, S. D. , … CGTP Collaborators . (2017). S7 Blood coagulation components in the concise guide to Pharmacology 2017/18, Overview S1‐S16. British Journal of Pharmacology, 174, S1–S16. 10.1111/bph.13882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayerbe, L. , Risco, C. , & Ayis, S. (2020). The association between treatment with heparin and survival in patients with Covid‐19. Journal of Thrombosis and Thrombolysis, 50, 298–301. 10.1007/s11239-020-02162-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo‐Jambrina, M. , Eder, J. , Kaptein, T. M. , Helgers, L. C. , Brouwer, J. M. , van Hamme, J. L. , … Geijtenbeek, T. B. H. (2020). SARS‐CoV‐2 infection and transmission depends on heparan sulfates and is blocked by low molecular weight heparins. BioRxiv, 1–27. 10.1101/2020.08.18.255810 [DOI] [Google Scholar]
- Caly, L. , Druce, J. , Roberts, J. , Bond, K. , Tran, T. , Kostecki, R. , … Catton, M. G. (2020). Isolation and rapid sharing of the 2019 novel coronavirus (SARS‐CoV‐2) from the first patient diagnosed with COVID‐19 in Australia. The Medical Journal of Australia, 212(10), 459–462. 10.5694/mja2.50569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassinelli, G. , Torri, G. , & Naggi, A. (2020). Non‐anticoagulant heparins as heparanase inhibitors. Advances in Experimental Medicine and Biology, 1221, 493–522. 10.1007/978-3-030-34521-1_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen, T. M. , Sandoval, D. R. , Spliid, C. B. , Pihl, J. , Painter, C. D. , Narayanan, A. , … Esko, J. D. (2020). SARS‐CoV‐2 infection depends on cellular heparan sulfate and ACE2. Cell, 183(4), 1043–1057. 10.1016/j.cell.2020.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelmann, C. , Müller, J. A. , Perkhofer, L. , Sparrer, K. M. J. , Zelikin, A. N. , Münch, J. , & Kleger, A. (2020). Inhaled and systemic heparin as a repurposed direct antiviral drug for prevention and treatment of COVID‐19. Clinical Medicine, 20, clinmed.2020–clinmed.0351. 10.7861/clinmed.2020-0351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M. J. , Alexander, S. , Cirino, G. , Docherty, J. R. , George, C. H. , Giembycz, C. G. , … Ahluwalia, A. (2018). Experimental design and analysis and their reporting II: Updated and simplified guidance for authors and peer reviewers. Brit J Pharm, 175, 987–993. 10.1111/bph.14153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich, E. , Mercuri, A. , Wu, S. , & Salar‐Behzadi, S. (2016). Measurements of deposition, lung surface area and lung fluid for simulation of inhaled compounds. Frontiers in Pharmacology, 7, 1–10. 10.3389/fphar.2016.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimond, S. , Mycroft‐West, C. , Gandhi, N. , Tree, J. , Buttigieg, K. , Coombes, N. , … Turnbull, J. (2020). Pixatimod (PG545), a clinical‐stage heparan sulfate mimetic, is a potent inhibitor of the SARS‐CoV‐2 virus. bioRxiv, 1–24. 10.1101/2020.06.24.169334 [DOI] [Google Scholar]
- Hayden, F. G. , Cote, K. M. , & Douglas, R. G. (1980). Plaque inhibition assay for drug susceptibility testing of influenza viruses. Antimicrobial Agents and Chemotherapy, 17, 865–870. 10.1128/AAC.17.5.865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh, J. , Anand, S. S. , Halperin, J. L. , & Fuster, V. (2001). Guide to anticoagulant therapy: Heparin: A statement for healthcare professionals from the American Heart Association. Circulation, 103(24), 2994–3018. 10.1161/01.cir.103.24.2994 [DOI] [PubMed] [Google Scholar]
- Hogwood, J. , Mulloy, B. , & Gray, E. (2017). Precipitation and neutralization of heparin from different sources by protamine sulfate. Pharmaceuticals (Basel), 10(59), 1–12. 10.3390/ph10030059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R. M. , Dahlgren, L. , Siegfried, B. D. , & Ellis, M. D. (2013). Acaricide, fungicide and drug interactions in honey bees (Apis mellifera). PLoS ONE, 8(1), e54092. 10.1371/journal.pone.0054092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katarzna, A. U. , Orio, A. , R, X. , Ahmed, Y. , Wilkinson, M. , Fernig, G. D. , & Yates, A. E. (2010). Differential scanning fluorimetry measurements of protein stability changes upon binding to glycosaminoglycans: A screening test for binding specificity. Analytical Chemistry, 82(9), 3796–3802. [DOI] [PubMed] [Google Scholar]
- Kollias, A. , Kyriakoulis, K. G. , Dimakakos, E. , Poulakou, G. , Stergiou, G. S. , & Syrigos, K. (2020). Thromboembolic risk and anticoagulant therapy in COVID‐19 patients: Emerging evidence and call for action. British Journal of Haematology, 189(5), 846–847. 10.1111/bjh.16727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski, K. , Xu, Y. , Pullan, S. T. , Lumley, S. F. , Foster, D. , Sanderson, N. , … Matthews, P. C. (2019). Metagenomic nanopore sequencing of influenza virus direct from clinical respiratory samples. Journal of Clinical Microbiology, 58(1), 1–15. 10.1128/JCM.00963-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl, U. , & Li, J. P. (2020). Heparin—An old drug with multiple potential targets in Covid‐19 therapy. Journal of Thrombosis and Haemostasis, 18, 2422–2424. 10.1111/jth.14898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L. , Chopra, P. , Li, X. , Wolfert, M. A. , Tompkins, S. M. , & Boons, G.‐J. (2020). SARS‐CoV‐2 spike protein binds heparan sulfate in a length‐ and sequence‐dependent manner. bioRxiv, 1–15. 10.1101/2020.05.10.087288 [DOI] [Google Scholar]
- Mulloy, B. , & Hogwood, J. (2015). Chromatographic molecular weight measurements for heparin, its fragments and fractions, and other glycosaminoglycans. Methods in Molecular Biology, 1229, 105–118. 10.1007/978-1-4939-1714-3_11 [DOI] [PubMed] [Google Scholar]
- Mulloy, B. , Hogwood, J. , Gray, E. , Lever, R. , & Page, C. P. (2016). Pharmacology of heparin and related drugs. Pharmacological Reviews, 68(1), 76–141. 10.1124/pr.115.011247 [DOI] [PubMed] [Google Scholar]
- Mycroft‐West, C. , Su, D. , Elli, S. , Li, Y. , Guimond, S. , Miller, G. , … Skidmore, M. (2020). The 2019 coronavirus (SARS‐CoV‐2) surface protein (spike) S1 receptor binding domain undergoes conformational change upon heparin binding. bioRxiv, 1–9. 10.1101/2020.02.29.971093 [DOI] [Google Scholar]
- Mycroft‐West, C. J. , Su, D. , Li, Y. , Guimond, S. E. , Rudd, T. R. , Elli, S. , & Skidmore, M. A. (2020). SARS‐CoV‐2 Spike S1 Receptor Binding Domain undergoes Conformational Change upon Interaction with Low Molecular Weight Heparins. bioRxiv, 1–15. 10.1101/2020.04.29.068486 [DOI] [Google Scholar]
- Mycroft‐West, C. J. , Su, D. , Pagani, I. , Rudd, T. R. , Elli, S. , Guimond, S. E. , … Skidmore, M. A. (2020). Heparin inhibits cellular invasion by SARS‐CoV‐2: Structural dependence of the interaction of the surface protein (spike) S1 receptor binding domain with heparin. J bioRxiv, 1–22. 10.1101/2020.04.28.066761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesen, H. F. , Berglund, H. , & Vedadi, M. (2007). The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nature Protocols, 2(9), 2212–2221. 10.1038/nprot.2007.321 [DOI] [PubMed] [Google Scholar]
- Partridge, L. J. , Urwin, L. , Nicklin, M. J. H. , James, D. C. , Green, V. L. R. , & Monk, P. N. (2020). Unfractionated heparin potently inhibits the binding of SARS‐CoV‐2 spike protein to a human cell line. bioRxiv, 1‐31 10.1101/2020.05.21.107870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Project, R. (2019). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. https://www.R-project.org/ [Google Scholar]
- Schaefer, I. M. , Padera, R. F. , Solomon, I. H. , Kanjilal, S. , Hammer, M. M. , Hornick, J. L. , & Sholl, L. M. (2020). In situ detection of SARS‐CoV‐2 in lungs and airways of patients with COVID‐19. Modern Pathology, 33, 2104–2114. 10.1038/s41379-020-0595-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajek, A. , Gray, E. , Keire, D. , Mulloy, B. , Al‐Hakim, A. , Chase, C. , Soares, M. , Cairatti, D. , Hogwood, J. , Mourão, P. , & Morris, T. S. ; (2015). Diversifying the global heparin supply chain reintroduction of bovine heparin in the United States? Pharmaceutical Technology, 39(11), 28–35. [Google Scholar]
- Tandon, R. , Sharp, J. S. , Zhang, F. , Pomin, V. H. , Ashpole, N. M. , Mitra, D. , … Linhardt, R. J. (2020). Effective inhibition of SARS‐CoV‐2 entry by heparin and enoxaparin derivatives. bioRxiv. 10.1101/2020.06.08.140236 Epub ahead of print. PMID: 33173010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, N. , Bai, H. , Chen, X. , Gong, J. , Li, D. , & Sun, Z. (2020). Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. Journal of Thrombosis and Haemostasis, 18(5), 1094–1099. 10.1111/jth.14817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, N. , Li, D. , Wang, X. , & Sun, Z. (2020). Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. Journal of Thrombosis and Haemostasis, 18(4), 844–847. 10.1111/jth.14768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S.P. United States Pharmacopeia . (n.d.) https://www.usp.org/, General Chapter 208.
- Uniewicz, K. A. , Ori, A. , Xu, R. , Ahmed, Y. , Wilkinson, M. C. , Fernig, D. G. , & Yates, E. A. (2010). Differential scanning fluorimetry measurement of protein stability changes upon binding to glycosaminoglycans: A screening test for binding specificity. Anal. Chem. May 1, 82(9), 3796–3802. 10.1021/ac100188x PMID: 20353159 [DOI] [PubMed] [Google Scholar]
- van Haren, F. , Page, C. P. , Laffey, M. D. , Artigas, A. , Camprubi‐Rimblas, M. , Nunes, Q. , … Dixon, B. (2020). Nebulised heparin as a treatment for COVID‐19: Scientific rationale and a global call for randomised evidence. Critical Care in press, 24, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Du, Y. , & Kaltashov, I. A. (2020). The utility of native MS for understanding the mechanism of action of repurposed therapeutics in COVID‐19: Heparin as disruptor of the SARS‐CoV‐2 interaction with its host cell receptor. bioRxiv, 1–18. 10.1101/2020.06.09.142794 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Some data may not be made available because of privacy or ethical restrictions.


