Abstract
Background and Purpose
The enzyme α/β‐hydrolase domain containing 6 (ABHD6), a new member of the endocannabinoid system, is a promising therapeutic target against neuronal‐related diseases. However, how ABHD6 activity is regulated is not known. ABHD6 coexists in protein complexes with the brain‐specific carnitine palmitoyltransferase 1C (CPT1C). CPT1C is involved in neuro‐metabolic functions, depending on brain malonyl–CoA levels. Our aim was to study CPT1C–ABHD6 interaction and determine whether CPT1C is a key regulator of ABHD6 activity depending on nutritional status.
Experimental Approach
Co‐immunoprecipitation and FRET assays were used to explore ABHD6 interaction with CPT1C or modified malonyl–CoA‐insensitive or C‐terminal truncated CPT1C forms. Cannabinoid CB1 receptor‐mediated signalling was investigated by determining cAMP levels. A novel highly sensitive fluorescent method was optimized to measure ABHD6 activity in non‐neuronal and neuronal cells and in brain tissues from wild‐type (WT) and CPT1C–KO mice.
Key Results
CPT1C interacted with ABHD6 and negatively regulated its hydrolase activity, thereby regulating 2‐AG downstream signalling. Accordingly, brain tissues of CPT1C–KO mice showed increased ABHD6 activity. CPT1C malonyl–CoA sensing was key to the regulatory role on ABHD6 activity and CB1 receptor signalling. Fasting, which attenuates brain malonyl–CoA, significantly increased ABHD6 activity in hypothalamus from WT, but not CPT1C–KO, mice.
Conclusions and Implications
Our finding that negative regulation of ABHD6 activity, particularly in the hypothalamus, is sensitive to nutritional status throws new light on the characterization and the importance of the proteins involved as potential targets against diseases affecting the CNS.
Keywords: ABHD6, CPT1C, endocannabinoid signalling, fasting, hippocampus, hypothalamus, malonyl–CoA
Abbreviations
- 2‐AG
2‐arachidonoylglycerol
- 4‐MUH
4‐methylumbelliferyl‐heptanoate
- ABHD6
α/β‐hydrolase domain containing 6
- ACEA
arachidonyl‐2′‐chloroethylamide
- CPT1C
carnitine palmitoyltransferase 1C
- ER
endoplasmic reticulum
- MAGL
monoacylglycerol lipase
- OEA‐d2
N‐Oleylethanolamine‐d2
What is already known
ABHD6, a new member of the endocannabinoid system, is a promising target against neuron‐related diseases.
Proteomic data indicates that the neuron‐specific protein CPT1C interacts with ABHD6.
What this study adds
Identification of the neuron‐specific protein CPT1C as a functional interactor in ABHD6 activity.
In vivo, negative regulation of ABHD6 activity by CPT1C is sensitive to nutritional status.
What is the clinical significance
New insights regarding the characterization of CPT1C and ABHD6 as therapeutic targets against neuron‐related diseases.
1. INTRODUCTION
The enzyme α/β‐hydrolase domain containing 6 (ABHD6) is one of the most recently recognized members of the endocannabinoid system that regulates levels of the endocannabinoid, 2‐arachidonoylglycerol (2‐AG), by hydrolyzing 2‐AG to arachidonic acid and glycerol (Cao et al., 2019). 2‐AG, a key signalling lipid in the CNS, acts as a retrograde messenger in activating presynaptic cannabinoid CB1 receptors to regulate neurotransmitter release and, consequently, a wide range of physiological processes. From its main localization at the postsynaptic neurons, ABHD6 is responsible for ~4% of 2‐AG hydrolysis in mouse brain homogenates (Blankman et al., 2007). Although monoacylglycerol lipase (MAGL) is the major enzyme in 2‐AG degradation, the subcellular distribution of ABHD6 is such that it regulates a different 2‐AG pool. This would suggest that ABHD6 and MAGL affect different physiological functions (Marrs et al., 2010; van Esbroeck et al., 2019).
ABHD6 has emerged as a promising therapeutic target against neuron‐related diseases (Cao et al., 2019), in particular, in combatting traumatic brain injury (Tchantchou & Zhang, 2013), multiple sclerosis (Manterola et al., 2018; Wen et al., 2015), GABAA receptor activity in epilepsy (Naydenov et al., 2014; Sigel et al., 2011), and neuronal differentiation (van Esbroeck et al., 2019). ABHD6 is also physically associated with AMPA receptors in the CNS (Erlenhardt et al., 2016), in negatively regulating their delivery to the cell surface and synaptic functions in a hydrolase activity‐independent fashion (Wei et al., 2016, 2017). In obesity, ABHD6 regulates metabolic flexibility by controlling 2‐AG levels in the ventromedial nucleus of the hypothalamus (Fisette et al., 2016). In addition to 2‐AG as a substrate, ABHD6 is able to hydrolyse in brain and liver non‐cannabinoid lipids such as bis (monoacylglycero)phosphate (Pribasnig et al., 2015), a lipid enriched in late endosomal/lysosomes. This would suggest its participation in lipid sorting machinery (Grabner et al., 2019; Pribasnig et al., 2015). Although many pathophysiological functions have been related to ABHD6, how its enzymic activity is regulated remains unknown.
High‐resolution proteomics has recently revealed that ABHD6 and the brain‐specific carnitine palmitoyltransferase 1C (CPT1C) are part of the AMPA receptor macromolecular complexes (Brechet et al., 2017; Schwenk et al., 2019, 2012), with CPT1C acting as a potential interactor with ABHD6. CPT1C is the most enigmatic of the CPT1 isoforms as, in contrast with the canonical mitochondrial isoforms, CPT1A and CPT1B, it is localized in the endoplasmic reticulum (ER) (Sierra et al., 2008). Furthermore, although it has minimal catalytic activity, it still binds malonyl–CoA, a metabolic intermediate that acts as an energy sensor (Price et al., 2002; Wolfgang et al., 2006). CPT1C plays a critical role in energy homeostasis, cognition, and motor function (Casals et al., 2016). We have previously demonstrated that CPT1C is involved in synaptic plasticity and learning via AMPA receptor trafficking (Carrasco et al., 2012; Fadó et al., 2015; Gratacòs‐Batlle et al., 2018) and in axonal growth regulation (Palomo‐Guerrero et al., 2019). In addition, CPT1C within the hypothalamus is crucial in fasting conditions for fuel selection (Pozo et al., 2017), food preference determination (Okamoto et al., 2018), and activation of brown fat thermogenesis (Rodríguez‐Rodríguez et al., 2019). To perform those neurometabolic functions, CPT1C seems to interact with other proteins (Casas et al., 2020; Palomo‐Guerrero et al., 2019; Schwenk et al., 2019) and to be sensitive to malonyl–CoA levels (Casals et al., 2016; Casas et al., 2020; Fadó et al., 2021; Palomo‐Guerrero et al., 2019; Rodríguez‐Rodríguez et al., 2019). Levels of malonyl–CoA, a precursor of fatty acid synthesis, fluctuate greatly depending on cell energy status, increasing after feeding and falling during fasting (Tokutake et al., 2012, 2010).
In this study, we have investigated the role of CPT1C as a regulator of ABHD6 hydrolase activity in cells and the brain and explore whether this regulation is subject to a metabolic challenge that involves brain malonyl–CoA fluctuations. Results show that CPT1C negatively regulates ABHD6 and that this regulation depends on nutritional status. Our investigation, in underscoring a novel link between CPT1C and the endocannabinoid system, sheds light on the therapeutic potential of both CPT1C and ABHD6 against diseases affecting the CNS.
2. METHODS
2.1. Cell culture and expressing vectors
Primary cortical mouse neurons were prepared from E16 wild‐type (WT) or CPT1C–KO embryos and cultured and maintained as described previously (Fadó et al., 2015) in Neurobasal supplemented with B27, glutaMAX (Gibco, Thermo Fisher, Massachusetts, USA) and antibiotics. HEK‐293T and hypothalamic neuronal GT1‐7 cell lines were grown according to the American Type Culture Collection (ATCC). HEK‐293T cells (RRID:CVCL_0063) were maintained in high‐glucose DMEM (Sigma‐Aldrich, Madrid, Spain) containing 10% FBS (Sigma‐Aldrich, Madrid, Spain), 1% antibiotics (penicillin/streptomycin) and 1% glutamine. GT1‐7 cells (RRID:CVCL_0281) were maintained in pyruvate‐ and glutamine‐enriched DMEM containing 10% FBS and 1% antibiotics. Cell cultures were incubated at 37°C in a humidified atmosphere of 5% CO2/95% air. Cell lines were authenticated by genotyping (ATCC Cell Line Authentication, STR Profiling Results, Manassas, VA, USA) and were regularly tested for mycoplasma contamination.
Mouse CPT1C was cloned in pWPI–IRES–GFP (empty vector, EV), generating pWPI–CPT1C–IRES–GFP. To assess the role of malonyl–CoA, we generated a CPT1C mutated form insensitive to malonyl–CoA by side‐directed mutagenesis, pWPI–CPT1CM589S–IRES–GFP (Rodríguez‐Rodríguez et al., 2019), referred to as M589S in this study. The role of the CPT1C C‐terminal motif was analysed using a form that substitutes its last 39 amino acids (772–810) for the FLAG epitope (DYKDDDDK), pWPI–CPT1CΔCter–FLAG–IRES–GFP, referred to as ΔCter in this study. Human CPT1C–SYFP2 was kindly supplied by Dr. Dolors Serra (University of Barcelona, Spain). mTurquoise‐ER‐5 (RRID:Addgene_55550, Addgene, Teddington, UK) was used to express the turquoise protein fused to KDEL (lysine [K], aspartic acid [D], glutamic acid [E], and leucine [L]), an ER retaining sequence. mTurquoise‐Calnexin‐N‐14 was purchased from Addgene (RRID:Adggene_55539). The mTurquoise2‐N1 and pSYFP2‐C1 vectors (RRID:Adggene_54843 and 22878, respectively) were used as EVs and cloned at BamHI (Table 1) to obtain the following fusion proteins: GluA1–mTurq2, ABHD6–mTurq2, ABHD6–SYFP2, M589S–SYFP2, and ΔCter–SYFP2. cDNA for the human version of CB1R was cloned in pRluc–N1 (PerkinElmer, Wellesley, MA) plasmid, as previously described (Reyes‐Resina et al., 2018), obtaining CB1R–Rluc. These constructs were used for FRET (FRET) assays, co‐immunoprecipitation, the ABHD6 activity assay and cAMP determination.
TABLE 1.
Primers obtained for recombination of plasmids
| Plasmid | Primers |
|---|---|
| GluA1‐CFP |
F: 5′ GCGACCGGTGGATCCAACAATCCTGTGGCTCCCAAGGGCATCC 3′ R: 5′ CGCGGGCCCGGGATCCATGCCGTACATCTTTGCCTTTTTCTGC 3′ |
| ABHD6‐YFP |
F: 5′ CGCGGGCCCGGGATCCATGGATCTCGATGTGGTTAACATGT 3′ R: 5′ GCGACCGGTGGATCCAAGTTCAGCTTCTTGTTGTCTGTGTT 3′ |
| ABHD6‐CFP |
F: 5′ CGCGGGCCCGGGATCCATGGATCTCGATGTGGTTAACATGT 3′ R: 5′ GCGACCGGTGGATCCAAGTTCAGCTTCTTGTTGTCTGTGTT 3′ |
| M589S‐YFP |
F: 5′ CGCGGGCCCGGGATCCATGGCTGAGGCACACCAGGCCTCGA 3′ R: 5′ GCGACCGGTGGATCCAACAAGTTGGTGGAGGATGTAGGGGT 3′ |
| ΔCter‐YFP |
F: 5′ CGCGGCCCGGGATCCATGGCTGAGGCACACCAGGC 3′ R: 5′ GCGACCGGTGGATCCAAAACCCGGAACAGGGAGGCTACATCC 3′ |
2.2. Animals and sample collection
All animal care and experimental procedures were carried out in strict accordance with European directive 2010/63/EU and Spanish legislation (BOE 252/34367‐91, 2005) regulating animal research and were approved by the Clinical Research Ethics Committee of the University of Barcelona (Procedure Ref. 9659, Generalitat de Catalunya). All efforts were made to minimize animal suffering and to minimize the number of animals used. Animal studies are reported in compliance with the ARRIVE guidelines (Percie du Sert et al., 2020) and with the recommendations made by the British Journal of Pharmacology (Lilley et al., 2020).
Male or female (8–10 weeks old) CPT1C–KO mice and their WT littermates with the same genetic background (C57BL/6J) were used for the experiments. All animals were housed according to a 12‐h/12‐h light/dark cycle (from 8 a.m. to 8 p.m.) in a temperature‐ and humidity‐controlled room and were allowed free access to water and standard laboratory chow diet. For the fasting experiments, animals were randomly assigned to a food‐deprived overnight group or an ad libitum feeding group (fasted and fed groups, respectively). Animals were killed by cervical dislocation under isoflurane anaesthesia during the light phase, and the hypothalamus and hippocampus were quickly removed, weighed, and stored at −80°C.
2.3. Co‐immunoprecipitation
HEK‐293T cells were transfected according to a calcium‐phosphate standard protocol with 4.5 μg of cDNA on a 100‐mm plate for 48 h. Two different co‐immunoprecipitation protocols were used, as described below.
G‐Sephadex beads. Cells were transfected with pWPI–CPT1C–IRES–GFP and pWPI–CPT1CM589S–IRES–GFP in combination with ABHD6–mTurq2 and scraped using 500 μl for 60‐mm plates of ComplexioLyte‐47 buffer (CL‐47, Logopharm, Freiburg, Germany). CL‐47 was diluted 1/4 with its buffer solution, sonicated for 5 min, supplemented with protease and phosphatase inhibitor cocktails and homogenates (1 mg of total protein) and kept for 30 min at 4°C in an orbital shaker for solubilization of the membrane proteins. Samples were processed as previously described (Casas et al., 2020; Fadó et al., 2015; Palomo‐Guerrero et al., 2019). CPT1C antibody (Sierra et al., 2008) was coupled to the sepharose beads (G‐Sephadex, GE Healthcare Life Science, Barcelona, Spain) for 1 h at 4°C in an orbital shaker. ABHD6 fraction bound to CPT1C or M589S was co‐immunoprecipitated. The amount of antibody used was 2 μg.
GFP‐Trap assay. Cells were collected after transfection with pWPI–CPT1C–IRES–GFP and pWPI–CPT1CΔCter–FLAG–IRES–GFP in combination with ABHD6–mTurq2. ABHD6 was immunoprecipitated from the supernatant of the cellular lysates using the GFP–Trap assay following the manufacturer's protocol (GTA‐100, Chromotek, Planegg, Germany). An additional ultra‐centrifugation of 100,000 × g was performed, before the immunoprecipitation, to eliminate non‐solubilized proteins from the supernatant of the cellular lysates. CPT1C or ΔCter fraction bound to ABHD6 was co‐immunoprecipitated.
Finally, immunoprecipitated proteins and their interactors were detected by western blot in whole lysates (input) and the immunoprecipitated samples.
2.4. Western blotting
The antibody‐based procedures used in this study comply with the recommendations made by the British Journal of Pharmacology (Alexander et al., 2018). Western blots was carried out as previously described (Pozo et al., 2017). For the co‐immunoprecipitation, precipitated complexes were washed three times in solubilization buffer and eluted with 2 x SDS/DTT sample buffer. Whole cell lysates were eluted with 1 x SDS sample buffer. Protein extracts were heated for 5 min at 95°C, separated on 10% SDS–PAGE, and transferred into PVDF membranes. Proteins were probed with antibodies against rabbit anti‐CPT1C produced in our laboratory (Sierra et al., 2008), rabbit anti‐GFP (1/500; Cell Signaling Cat#2956, RRID:AB_1196615), anti‐FLAG (Sigma‐Aldrich Cat# F3165, RRID:AB_259529), rabbit anti‐ABHD6 (1/1000; Abcam Cat#ab87532, RRID:AB_1951400), and mouse anti‐GAPDH (1/10.000; Abcam Cat#ab36845, RRID:AB_732650). Each membrane was then incubated with the corresponding secondary antibody and developed using Luminata Forte Western HRP substrate (Merck Millipore, Madrid, Spain). Images were captured using Gene Tools software (Syngene, Cambridge, UK) and quantified by densitometry using ImageJ‐1.33 software (NIH, Bethesda, MD, USA). The results were expressed as the target protein/housekeeping protein ratio and normalized to the control group, if applicable (with the Y axis representing “fold mean of the control values”).
2.5. FRET assays
For FRET sensitized emission saturation curves, HEK‐293T cells were transiently co‐transfected with the plasmid cDNA corresponding to ABHD6–mTurq2 or mTurq2–KDEL (donor proteins) and CPT1C–SYFP2 (acceptor protein) using the donor to acceptor ratio specified in Figure 1e. Cell suspension (20 μg of protein), distributed between 96‐well black microplates, was read in a fluorescence microplate reader (BioTek Synergy H1, Bad Friedrichshall, Germany), using an excitation filter at 420/50 or 485/20 and emission filters corresponding to 485/20‐nm filter (Ch 1) and 530/25‐nm filter (Ch 2). Gain settings were identical for all experiments to keep constant the relative contribution of the fluorophores to the detection channels for spectral unmixing. The contribution to the two detection channels (spectral signature) of the mTurq2 and SYFP2 proteins alone was measured in experiments with cells expressing only one of those proteins and was normalized to the sum of the signals obtained for the two detection channels. FRET quantification was performed as described elsewhere (Navarro et al., 2010).
FIGURE 1.
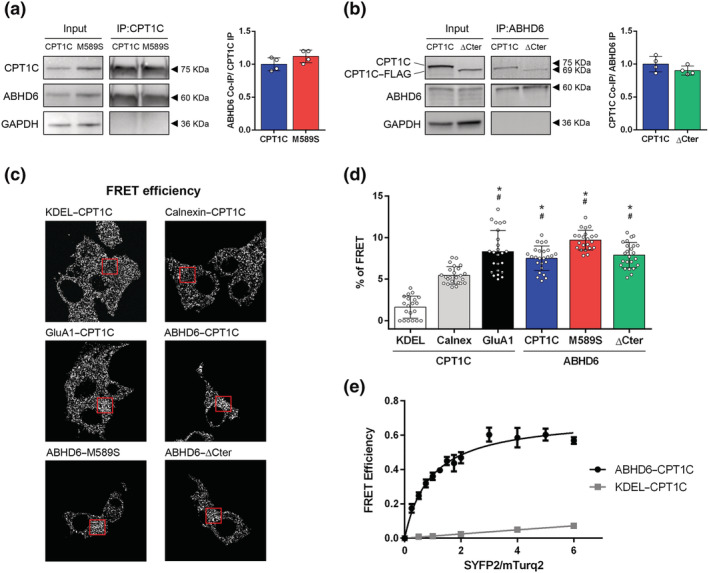
ABHD6 interacts with CPT1C. (a) Co‐immunoprecipitation of ABHD6‐SYFP2 in HEK‐293T cells using CPT1C antibody for CPT1C and M589S (n = 4). (b) Co‐immunoprecipitation of CPT1C and ΔCter in HEK‐293T cells using GFP–Trap for ABHD6–mTurq2 (n = 4). Proteins were detected in whole lysate (input) and immunoprecipitated (IP) samples. (c) Representative pictures of FRET efficiency. (d) FRET efficiency of CPT1C–SYFP2 interaction with mTurq2–KDEL and mTurq2–Calnexin as negative FRET interactions, and CPT1C–SYFP2 with GluA1–mTurq2 as a positive FRET interaction. ABHD6–mTurq2 interaction with CPT1C–SYFP2, M589S–SYFP2, and ΔCter‐SYFP2. Percentage of FRET was measured by the increase in donor intensity after photobleaching (n = 25). (e) FRET sensitized emission of CPT1C with either ABHD6 or KDEL in living cells. Assays were performed 48 h post transfection in HEK‐293T cells expressing ABHD6–mTurq2 or mTurq2–KDEL and with increasing amounts of the cDNA for CPT1C–SYFP2. FRET saturation curves for both protein pairs were obtained by monitoring SYFP2 fluorescence emission at 530 nm after excitation of mTurq2 at 420 nm and subtracting values obtained with cells expressing the same amount of donor protein. Means ± SD (n = 5). *P < 0.05, significantly different from KDEL‐CPT1C; # P < 0.05, significantly different from Calnexin‐CPT1C; one‐way ANOVA followed by Bonferroni's post hoc correction
The saturating interaction CPT1C–ABHD6 was also analysed by FRET sensitized emission after cell treatment with an inhibitor of malonyl–CoA synthesis, TOFA (20 μg·ml−1, incubation 2 h) (ab141578; Abcam) or vehicle DMSO.
For FRET imaging experiments, HEK‐293T cells were cultured on 24‐well plates on coverslips pre‐coated with poly‐L‐lysine for 1 h at 37°C. The day after seeding, cells were transiently transfected, applying a calcium‐phosphate standard protocol, with 0.5 μg per well of donor plasmids containing mTurquoise (mTurq) or 0.75 μg per well of acceptor plasmids containing SYFP2. cDNAs encoding for EV, mTurq–KDEL, mTurq–Calnexin, GluA1–mTurq2, CPT1C–SYFP2, M589S–SYFP2, ΔCter–SYFP2 and ABHD6–mTurq2 were used. Twenty‐four hours after transfection, cells were fixed with 4% paraformaldehyde. Coverslips were mounted with Fluoromount Mounting Medium (Thermo Fisher Scientific, Madrid, Spain). All FRET imaging experiments were performed using a Leica TCS SP2 microscope following a previously described protocol (Palomo‐Guerrero et al., 2019).
2.6. 4‐MUH‐based activity assay
The ABHD6 activity assay is based on its natural capacity to hydrolyse 4‐methylumbelliferyl‐heptanoate (4‐MUH), a fluorogenic substrate whose hydrolysis produces the fluorescent compound 4‐methylumbelliferone (4‐MU) (λ ex = 355 nm and λ em = 460 nm; Figure 2a). 4‐MUH was dissolved in ethanol at 20 times and kept at −20°C. For the 4‐MUH assay carried out in cell homogenates, HEK‐293T cells were seeded on six‐well plates and transiently transfected with 1.2 μg per well of cDNAs encoding EV–SYFP2, ABHD6–SYFP2, CPT1C–SYFP2, M589S–SYFP2, or ΔCter–SYFP2, alone or in combination, using FuGENE HD (Promega, Madrid, Spain). To maintain the same ratio of DNA in co‐transfections, we used EV–SYFP2 to equilibrate the amount of total cDNA transfected. After 48 h, cells were washed twice with ice‐cold PBS, scraped, and centrifuged for collection. Pellets were then homogenized with 100 μl of Tris‐EDTA 50:1‐mM buffer at pH 7.4 and briefly sonicated. The same protocol was applied to cells endogenously expressing ABHD6 and CPT1C, GT1‐7 cells and cortical primary neurons. In both neuronal and non‐neuronal cell cultures, ABHD6 activity was calculated after subtracting the curve in the presence of the ABHD6 inhibitor WWL70 (concentration ranges are specified in the figure legends) (Blankman et al., 2007; Cao et al., 2019). Some of these experiments were performed after cell treatment with an inhibitor of malonyl–CoA synthesis, TOFA (20 μg·ml−1, 1 or 2 h of incubation) or vehicle DMSO.
FIGURE 2.
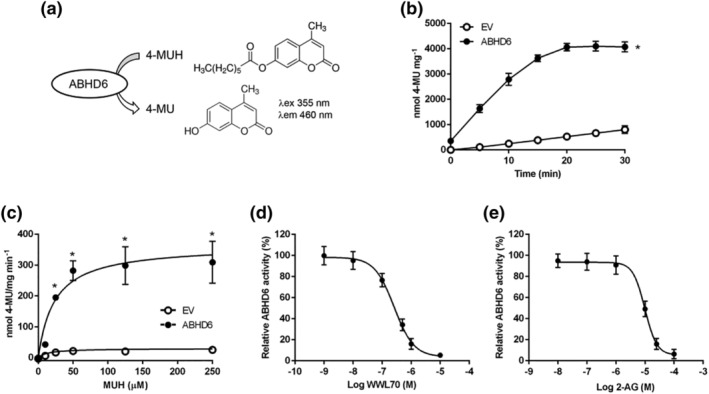
ABHD6 assay based on 4‐MUH hydrolysis in HEK‐293T cells overexpressing ABHD6. (a) ABHD6 hydrolyses 4‐MUH producing 4‐MU, a highly florescent compound (excitation 355 nm; emission 460 nm). (b) 4‐MUH hydrolysis by cells expressing empty vector (EV) or ABHD6 (n = 7). (c) ABHD6 activity dependent on 4‐MUH concentration at t = 10 min (n = 6). (d) ABHD6 activity inhibition by WWL70 at t = 10 min (n = 8). (e) 2‐AG competition with 4‐MUH in cells expressing ABHD6 (n = 6). Data shown are means ± SD. *P < 0.05, significantly different from EV; two‐way ANOVA followed by Bonferroni's post hoc correction
For the 4‐MUH assay carried out in tissue homogenates, tissue from hypothalamus (6–8 mg) and hippocampus (11–12 mg) was quickly removed and dounce‐homogenized in 200 μl of Tris‐EDTA 50:1‐mM buffer at pH 7.4 and briefly sonicated. The assay was performed in black 96‐well plates using 1 μg per well of protein in a total volume of 100 μl per well. Cell homogenates were incubated with either 5 μl of WWL70 (Blankman et al., 2007; Cao et al., 2019), in a final concentration range from 1 nM to 50 μM, or 2‐AG in a final concentration range from 25 to 100 μM for 20 min at 37°C. In hypothalamic homogenates, dose–response inhibition of either WWL70 or KT‐182 (another specific ABHD6 inhibitor) was also assessed (Blankman et al., 2007; Cao et al., 2019) (Figure S4). Subsequently, 5 μl of 4‐MUH were added (50‐μM final concentration) and fluorescence was immediately measured in 5‐min intervals for 60 min or 120 min using a Synergy HT Reader (BioTek, Bad Friedrichshall, Germany). In brain tissue homogenates, ABHD6 activity was calculated after subtracting the curve in the presence of 10‐μM WWL70. 4‐MU was used to conduct a standard curve to express ABHD6 activity as nmol·mg−1.
2.7. cAMP determination
HEK‐293T cells were seeded on six‐well plates and transfected for 48 h using polyethyleneimine (Sigma, Madrid, Spain), as previously described (Carriba et al., 2008), with 1.5 μg per well of plasmids encoding for CB1R–Rluc, ABHD6–SYFP2, CPT1C–SYFP2, M589S–SYFP2, and ΔCter–SYFP2, alone or in combination. To maintain the same ratio of DNA in co‐transfections, we used EV–SYFP2 to equilibrate the amount of total cDNA transfected. Two hours before adding reagents, HEK‐293T cells were placed in serum‐free medium. Cells were then detached by suspension in a medium containing 50 μM of zardaverine and were placed on white 384‐well plates (2,500 cells per well), treated for 15 min with 1 μM of rimonabant (Sigma, Madrid, Spain)—a CB1 receptor antagonist—and stimulated, for 15 min, with 100 nM of arachidonyl‐2′‐chloroethylamide (ACEA)—a highly selective CB1 receptor agonist —or increasing concentrations of 2‐AG (from 100 nM to 1 μM). Cells transfected only with CB1 receptors were next treated with 0.5‐μM forskolin for 15 min to increase intracellular cAMP levels. Cells transfected with CB1R–ABHD6, CB1R–ABHD6–CPT1C, CB1R–ABHD6–M589S, and CB1R–ABHD6–ΔCter were not incubated with forskolin. The non‐treated condition in these combinations of plasmids is referred as basal. Homogeneous time‐resolved fluorescence energy transfer (HTRF) measurements were made, as previously described (Reyes‐Resina et al., 2018), using the Lancer Ultra cAMP Kit (PerkinElmer, Waltham, MA, USA), after 1 h of plate incubation at room temperature, protected from light. Fluorescence at 665 nm was analysed using a PHERAstar Flagship plate reader equipped with an HTRF optical module (BMG Lab technologies, Offenburg, Germany).
2.8. Endocannabinoid extraction and analysis
HEK‐293T cell endocannabinoids were extracted and measured by HPLC–MS/MS, as previously described (Miralpeix et al., 2019), but with some modifications. Briefly, cells were seeded on a six‐well plate and transfected with EV–SYFP2, ABHD6–SYFP2, and CPT1C–SYFP2 using FuGENE HD (Promega, Madrid, Spain). Forty‐eight hours after transfection, cells were washed twice with ice‐cold PBS and centrifuged (10,000 × g, 5 min, 4°C). Cells were then immediately suspended in 200 μl of ice‐cooled deionized water containing a final concentration of 0.362 μM of N‐oleylethanolamine‐d2 (OEA‐d2) as internal standard, 100 μM of PMSF and 0.01% of butylated hydroxytoluene and were then homogenized in a brief sonication. Next, 50 μl of the homogenized sample were kept at −20°C for protein quantification, and the remaining 150 μl were mixed with 600 μl ethyl acetate/n‐hexane (9:1 v/v) and vortexed for 5 min. After centrifugation (14,000 × g, 5 min, 4°C), the upper layer was collected, evaporated in a nitrogen stream and analysed by HPLC–MS/MS, as described previously (Miralpeix et al., 2019).
2.9. Data and statistical analysis
The data and statistical analysis comply with British Journal of Pharmacology recommendations on experimental design and analysis (Curtis et al., 2018). Mice were randomly assigned to the treatment and experimental groups and data was analysed in a manner completely blind to experimental groups. All results are expressed as mean ± SD. Student's t test was used to compare two groups and multiple comparisons were analysed by one‐ or two‐way ANOVA followed by Bonferroni's post hoc correction. The normality of populations and homogeneity of variances were tested prior to the ANOVA. Post hoc tests were run only if F achieved P < 0.05, and there was no significant variance in homogeneity. Kinetics parameters were analysed using nonlinear regression with variable slope and the Michaelis–Menten model. Sample size was determined via an a priori calculation based on the assumption that SD was less than 20% of variance. Statistical analysis was undertaken only when each group size was at least n = 5 independent variables (not including technical replicates). Used exceptionally, even though the number of experiments was less than 5, were inclusion data obtained in immunoprecipitation assays, as only experiments in which the bands were appropriately observed in the studied fractions were considered; the results were then considered to be preliminary until confirmed by other experimental approaches.
While the experiments were designed to generate groups of equal size, unequal group size in the experimental approach with transgenic mice under fasting or feeding conditions were due to the limited availability of CPT1C–KO mice.
The threshold for statistical significance was P < 0.05 throughout. Statistical analysis was performed using GraphPad Prism 6 Software (RRID:SCR_002798; GraphPad Software, La Jolla, CA, USA). Outlier tests were not used.
2.10. Materials
AG, KT‐182, N‐oleylethanolamine‐d2 (OEA‐d2) and WWL70 were supplied by Cayman Chemicals (Ann Arbor, MI, USA) and 4‐MUH, 4‐MU, ACEA, butylated hydroxytoluene and zardaverine by Sigma‐Aldrich (Madrid, Spain). TOFA was supplied by Abcam (Cambridge, UK).
2.11. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in the IUPHAR/BPS Guide to PHARMACOLOGY (http://www.guidetopharmacology.org) and are permanently archived in the Concise Guide to PHARMACOLOGY 2019/20 (Alexander, Christopoulos, et al., 2019; Alexander, Fabbro, et al., 2019; Alexander, Mathie et al., 2019).
3. RESULTS
3.1. CPT1C interacts with ABHD6 and the interaction is maintained with modified forms of CPT1C (malonyl–CoA‐insensitive or without the C‐terminal region)
Both CPT1C and ABHD6 were recently described, using a proteomic approach, as constituents of the AMPA receptor proteome and ABHD6 as an interactor with CPT1C in brain homogenates (Brechet et al., 2017; Schwenk et al., 2019). We aimed to demonstrate that CPT1C and ABHD6 may interact outside the AMPA receptor complex and to explore the importance of malonyl–CoA sensing and the C‐terminal region of CPT1C in this interaction. These two features of CPT1C are interesting as malonyl–CoA is a within‐cell energy sensor important for CPT1C function and also because the last 39 residues of CPT1C (the C‐terminal region) are not present in the canonical CPT1 isoforms (CPT1A and CPT1B). We therefore used a heterologous system consisting of HEK‐293T cells co‐expressing ABHD6 and either CPT1C—the mutated form M589S, insensitive to malonyl–CoA (Morillas et al., 2003; Rodríguez‐Rodríguez et al., 2019)—or the truncated form ΔCter—in which the last 39 amino acids were replaced by the FLAG tag. We first performed a co‐immunoprecipitation assay using CPT1C antibodies, observing that ABHD6 interacted with CPT1C and that this interaction was still maintained with the mutated M589S form (Figure 1a). We then immunoprecipitated ABHD6 using the GFP‐Trap system, observing that this too was able to interact with ΔCter (Figure 1b).
The preliminary co‐immunoprecipitation results were confirmed by FRET assays, performed in HEK‐293T cells (Figures 1c–e, S1, and S2). Plasmids encoding for the mTurquoise‐ER, KDEL, and calnexin were used as negative interaction controls and, as a positive interaction control, we used GluA1, a subunit of AMPA receptors that is known to interact with CPT1C (Brechet et al., 2017; Fadó et al., 2015; Schwenk et al., 2019). Results showed that, as expected, the FRET efficiency of the positive control pair CPT1C/GluA1 was significantly different from that for the negative control pairs CPT1C/KDEL and CPT1C/calnexin (Figure 1c,d). Moreover, FRET efficiency for CPT1C/ABHD6 was similar to that found for CPT1C/GluA1, indicating a positive interaction (Figures 1c,d and S1). Similar percentages of FRET efficiency were observed for the pairs M589S/ABHD6 and ΔCter/ABHD6 compared to CPT1C/ABHD6 (Figures 1c,d and S1), suggesting that both malonyl–CoA sensing and the presence of the C‐terminal region of CPT1C are not critical to the interaction with ABHD6, corroborating Figure 1a,b.
These results were supported by FRET sensitized emission assays of CPT1C, with either ABHD6 or KDEL, in living cells expressing ABHD6–mTurq2 or mTurq–KDEL and with increasing amounts of the cDNA for the CPT1C–SYFP2 acceptor. The positive interaction was confirmed by a FRET curve for the ABHD6–CPT1C protein pair that was saturating—unlike the curve for the KDEL–CPT1C protein pair, which was non‐saturating (Figure 1e). Under the same experimental conditions, the positive and saturating curve of the ABHD6–CPT1C pair was not altered after TOFA treatment (Figure S2), supporting the finding that the interaction of both proteins does not depend on malonyl–CoA levels.
In summary, co‐immunoprecipitation and FRET approaches confirm that CPT1C interacts with ABHD6. Neither the CPT1C mutation affecting malonyl–CoA sensing or the C‐terminus region is essential for this interaction.
3.2. Optimization and validation of a 4‐MUH‐based ABHD6 activity assay in non‐neuronal and neuronal cells and mouse brain tissue homogenates
The CPT1C–ABHD6 interaction raised the question of whether CPT1C was regulating ABHD6 activity. To answer this question, we optimized a rapid, direct, and highly sensitive fluorescent activity assay for ABHD6, based on hydrolysis of a 4‐MUH fluorogenic substrate (Figure 2a). HEK‐293T cell lysates expressing ABHD6 showed a higher accumulation of fluorescence over time than cells expressing EV when 50 μM of MUH was added as substrate (Figure 2b). The fluorescence progression curve for ABHD6 displayed linear kinetics from 5 up to 20 min, after which the fluorescence signal plateaued, while the background signal continued to increase (Figure 2b). Unless otherwise stated, further experiments described below use the 10‐min point within the linear increase of fluorescence.
ABHD6 activity depending on 4‐MUH concentration also showed that, up to 50 μM of 4‐MUH, the formation of 4‐MU was constant, whereas a plateau was reached at higher 4‐MUH concentrations (Figure 2c). Nonlinear regression analysis was used to calculate the apparent kinetics (V max and K m) of ABHD6 for 4‐MUH as substrate (Table 2). To validate the ABHD6 activity assay, we tested the effectiveness of a specific ABHD6 inhibitor, WWL70 (Blankman et al., 2007; Cao et al., 2019). We observed that WWL70 dose‐dependently inhibited ABHD6 activity with an IC50 of 274 nM (Figure 2d). Moreover, to corroborate the specificity of 4‐MUH as a substrate for ABHD6, we performed a competitive substrate assay by adding increasing concentrations of the natural substrate 2‐AG (Figure 2e). We found that 2‐AG decreased 4‐MUH hydrolysis with an IC50 value of 10 μM (Figure 2e).
TABLE 2.
V max and K m values of the time course of 4‐MUH hydrolysis by cells overexpressing EV and ABHD6 alone or in combination with CPT1C, M589S, and Δcter
| V max (nmol·mg−1·min−1) | K m (μM) | |
|---|---|---|
| ABHD6 | 322.3 ± 15.3 | 18.8 ± 3.5 |
| ABHD6–CPT1C | 111.8 ± 4.6 * | 4.6 ± 1.4 * |
| ABHD6–M589S | 230.3 ± 11.2 * , # | 13.6 ± 2.9 * , # |
| ABHD6–ΔCter | 224.3 ± 15.8 * , # | 7.3 ± 3.0 * , † |
These values were obtained by Michaelis–Menten analysis and are means ± SD (n = 9).
P < 0.05, significantly different from ABHD6;
P < 0.05, significantly different from ABHD6‐CPT1C;
P < 0.05, significantly different from ABHD6‐M589S.
Previous ABHD6 activity assays were unable to detect that activity in systems endogenously expressing ABHD6 (Navia‐Paldanius et al., 2012; van der Wel et al., 2015). For this reason, we validated our new fluorescent assay in mouse hypothalamic neuronal cell line GT1–7 (Figure 3a,b) and in primary cortical neurons (Figure 3c,d). In both models, we were able to measure and to inhibit ABHD6 activity by treating the cells with 1–10 μM of WWL70 and 50 μM of WWL70, which inhibited around 25–30% and 50% of hydrolase activity, respectively (Figure 3).
FIGURE 3.
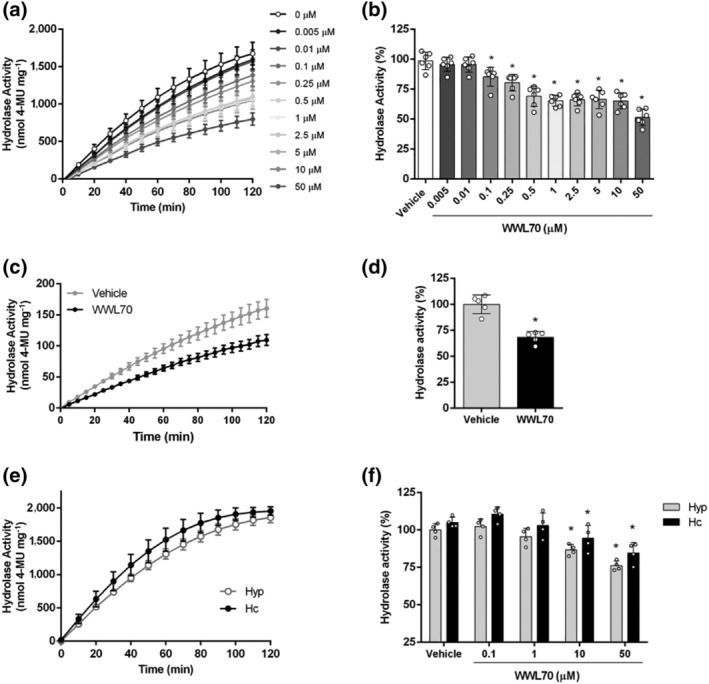
ABHD6 activity assay based on 4‐MUH hydrolysis in cells endogenously expressing ABHD6 and in brain tissue homogenates. (a) ABHD6 activity inhibition by WWL70 (0.005–50 μM) in GT1‐7 mouse hypothalamic neuronal cell line; (b) at t = 120 min (n = 6). One‐way ANOVA followed by Bonferroni's post hoc correction was used for statistical analysis. (c) ABHD6 activity inhibition by WWL70 (1 μM) in cortical neurons of WT mice; (d) at t = 120 min (related to the WT vehicle) (n = 5). Student's t test was used for statistical analysis. (e) 4‐MUH hydrolysis in hypothalamus (Hyp) and hippocampus (Hc) of WT mice (n = 5). (f) ABHD6 activity inhibition by WWL70 (0.1–50 μM) in hypothalamus and hippocampus homogenates at t = 120 min (n = 5). Data shown are means ± SD. *P < 0.05, significantly different from vehicle; two‐way ANOVA followed by Bonferroni's post hoc correction
We also confirmed that the 4‐MUH‐based assay was useful in determining enzyme activity in murine hypothalamus and hippocampus homogenates (Figures 3e,f and S3). The fluorescent progression curve displayed linear kinetics up to 60 min in tissue homogenates (Figure 3e). Dose–response inhibition of ABHD6 activity by WWL70 showed that ABHD6 achieved up to 20–25% of 4‐MUH hydrolysis by brain tissue homogenates (Figure 3f). WWL70 is a selective early‐generation carbamate‐based ABHD6 inhibitor that has been widely used to study this hydrolase (Bottemanne et al., 2019; Cao et al., 2019). Our use of this inhibitor is therefore supported by the literature ruling out the activity of other hydrolases. Since next‐generation carbamate‐based ABHD6 inhibitors have been generated with a better in vivo profile and capacity to reach the brain, for example, KT‐182 (Hsu et al., 2013; Manterola et al., 2018), we also inhibited ABHD6 activity in hypothalamic homogenates using KT‐182 (Figure S4), finding that the percentage inhibition, using the range of concentrations as recommended in previous studies, was similar to that observed for WWL70 and even higher at the maximum tested dosage. The use of both inhibitors in brain tissue sustains our findings attributed to ABHD6 activity.
In summary, the 4‐MUH‐based activity assay is a suitable and reliable method to study ABHD6 activity in both heterologous and natural systems.
3.3. CPT1C regulates ABHD6 activity depending on malonyl–CoA sensing
Once we set up the ABHD6 activity assay, we were interested in evaluating whether CPT1C was able to regulate ABHD6 activity. ABHD6 activity was assessed in HEK‐293T cells co‐expressing ABHD6 and CPT1C, M589S, or ΔCter. We observed that CPT1C significantly depressed ABHD6 activity (Figure 4a). When ABHD6 was expressed alone, the activity curve displayed linear kinetics from 5 to 20 min, after which a plateau was reached (Figure 4a). In contrast, when CPT1C was present, ABHD6 activity plateaued much later, after 45 min of reaction (Figure 4a). Normalization of ABHD6 activity by its protein expressing levels (Figure S5A) showed that CPT1C reduced ABHD6 activity by almost 60% (Figure 4b). Furthermore, nonlinear regression analysis of ABHD6 activity as a function of 4‐MUH concentration demonstrated that CPT1C decreased ABHD6 maximal activity almost threefold and substrate affinity fourfold (Figure 4c and Table 2).
FIGURE 4.
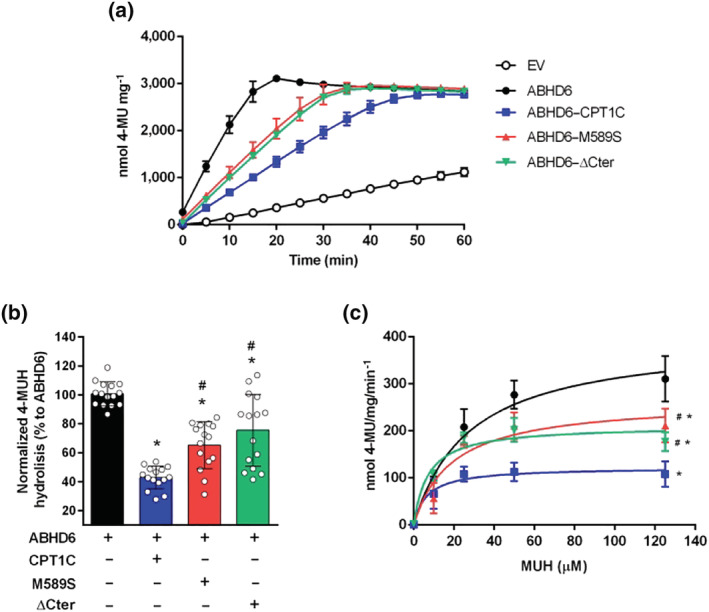
CPT1C regulates ABHD6 activity depending on malonyl–CoA sensing and CPT1C C‐terminus. (a) Time course of 4‐MUH hydrolysis by cells overexpressing empty vector (EV) and ABHD6, alone or in combination with CPT1C, M589S or ΔCter (n = 5). (b) Normalization of ABHD6 activity with ABHD6 protein expression levels at t = 10 min (n = 15). (c) ABHD6 activity dependent on 4‐MUH concentration at t = 10 min (n = 9). Data shown are means ± SD. *P < 0.05, significantly different from ABHD6; # P < 0.05, significantly different from ABHD6–CPT1C; one‐way ANOVA followed by Bonferroni's post hoc correction
Interestingly, cells co‐expressing ABHD6 and the CPT1C mutant insensitive to malonyl–CoA, M589S, showed a lower reduction of ABHD6 activity (35%, Figure 4a,b) compared to CPT1C, while both maximal velocity and substrate affinity were partly restored (Figure 4c and Table 2). In line with these results, ABHD6 activity in the hypothalamic cell line GT1‐7 was significantly increased after inhibition of malonyl–CoA synthesis with TOFA (Figure S6). The expression of ΔCter in HEK‐293T cells also attenuated ABHD6 activity (27%, Figure 4), but to a lesser extent than CPT1C. Cells expressing CPT1C, M589S, or ΔCter alone showed similar fluorescent accumulation as EV transfected cells, demonstrating that CPT1C and its modified forms had no 4‐MUH hydrolysis activity per se (Figure S5B). In summary, our results demonstrate that CPT1C negatively regulates ABHD6 activity, that malonyl–CoA sensing by CPT1C is required for this regulation and that the presence of the C‐terminal region of CPT1C is important for its inhibitory effect.
Levels were also measured of endogenous 2‐AG (a substrate of ABHD6) and AEA (anandamide; an endocannabinoid not metabolized by ABHD6) in HEK‐293T expressing ABHD6 alone or in combination with CPT1C. Under these conditions, no changes in 2‐AG and AEA levels were found (Figure S5C), indicating that ABHD6 expression in HEK‐293T cells is not enough to produce changes in endogenous 2‐AG levels.
It is known that 2‐AG is the main ligand of CB1 receptors, GPCRs that, upon Gi coupling, inhibit adenylyl cyclase (AC) and thus modulates the cAMP/PKA signalling pathway (Busquets‐Garcia et al., 2018). To elucidate whether CPT1C regulation of ABHD6 could modulate a physiological response in living cells—such as intracellular cAMP levels depending on 2‐AG availability—we measured this regulation in HEK‐293T cells transiently expressing CB1 receptors in combination with ABHD6 and CPT1C or M589S or ΔCter.
We first set up cAMP detection in cells expressing CB1 receptors alone (Figure S7A) and incubated with forskolin, an activator of AC. As expected, ACEA, a CB1 receptor agonist, along with increasing concentrations of 2‐AG decreased forskolin‐induced cAMP levels (Figure S7A). In contrast, rimonabant, a CB1 receptor antagonist, was able to block the 2‐AG‐induced decrease in cAMP levels (Figure S7A). We also tested the effects of products derived from 2‐AG hydrolysis, namely, arachidonic acid and glycerol. Surprisingly, arachidonic acid was able to increase cAMP levels in the cells, while no effect was evident for glycerol (Figure S7A). Moreover, when CB1 receptors and ABHD6 were co‐expressed, the addition of 2‐AG significantly increased forskolin‐induced cAMP levels above the forskolin levels (data not shown), probably due to the accumulation of arachidonic acid derived from ABHD6‐mediated hydrolysis of 2‐AG. That fact led us to postulate that treating cells expressing ABHD6 with forskolin could influence the increase in cAMP levels. Subsequent experiments were therefore performed without forskolin (Figures 5 and S7B), assessing the effects of increasing concentrations of 2‐AG, arachidonic acid, and glycerol (Figure S7B).
FIGURE 5.
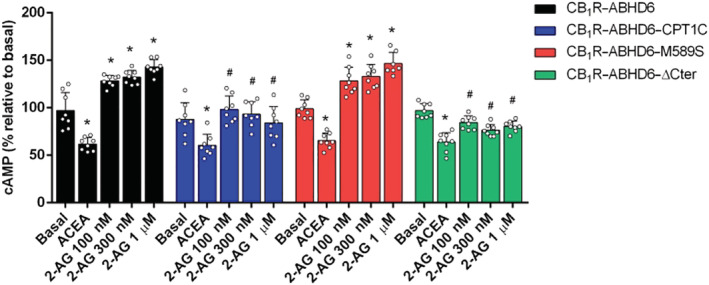
Effect of ABHD6 and CPT1C on the CB1R/cAMP signalling pathway in HEK‐293T cells. cAMP baseline levels in cells expressing CB1R and ABHD6 in combination with CPT1C, M589S, or ΔCter and treated with ACEA and with increasing concentrations of 2‐AG (n = 8). Data shown are means ± SD. *P < 0.05, significantly different from baseline; # P < 0.05, significantly different from corresponding treatment in CB1R‐ABHD6; two‐way ANOVA followed by Bonferroni's post hoc correction
Under these conditions, results showed that 2‐AG hydrolysis by ABHD6 (CB1R–ABHD6) increased intracellular cAMP levels, whereas CB1 receptor activation via ACEA reduced those levels (Figure 5). Interestingly, expression of CPT1C in combination with ABHD6 (CB1R–ABHD6–CPT1C) restored cAMP levels to baseline and was significantly different from that for CB1R–ABHD6 (Figure 5)—in line with the fact that CPT1C is an inhibitor of ABHD6 activity. When M589S was expressed (CB1R–ABHD6–M589S), cAMP levels were similar to those for the CB1R–ABHD6 condition (Figure 5), indicating that CPT1C binding of malonyl–CoA is necessary to blunt ABHD6 activity. The C‐terminal region of CPT1C, furthermore, seems not to be essential for the regulatory role of CPT1C in ABHD6‐mediated cAMP levels, since expression of ΔCter (CB1R–ABHD6–ΔCter) did not alter cAMP levels, compared to those for CB1R–ABHD6–CPT1C (Figure 5).
3.4. Brain tissue homogenates of CPT1C–KO mice show higher ABHD6 activity than control mice
To study the ability of CPT1C to regulate ABHD6 activity in a physiological system, we measured 4‐MUH hydrolysis in both hypothalamic and hippocampal homogenates of WT and CPT1C–KO mice. Hydrolase activity was measured in the absence or presence of the ABHD6 inhibitor, WWL70, in order to determine ABHD6‐dependent hydrolase activity (Figure S8).
Corroborating the results for neuronal and non‐neuronal cells, the hypothalamus from CPT1C‐deficient male mice showed higher ABHD6 activity than that of the hypothalamus from WT mice (Figure 6a,b). Hippocampal homogenates of CPT1C–KO mice only exhibited increases in ABHD6 activity, compared to WT mice, at the last time of the activity assay (Figure 6c); however, the AUCs for both KO‐ and WT‐derived hippocampus were not significantly different (Figure 6d). These results suggest a more prominent role for CPT1C in the regulation of ABHD6 activity in the hypothalamus than in the hippocampus.
FIGURE 6.
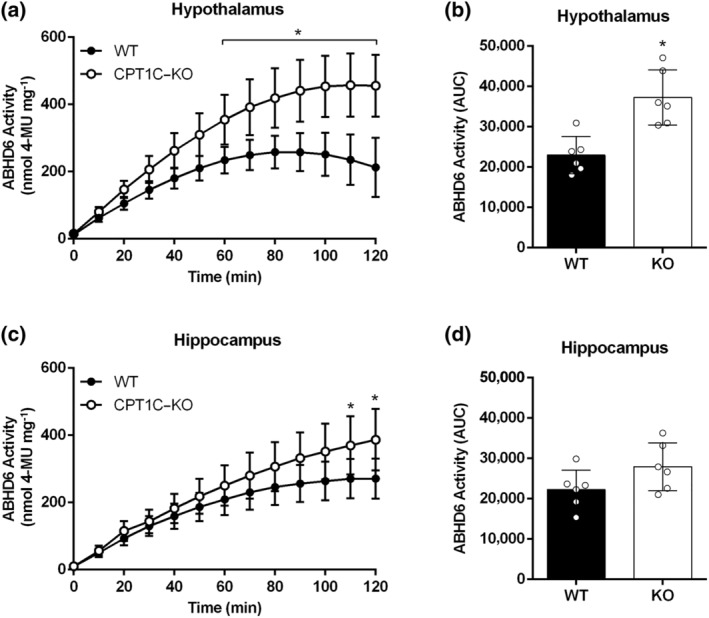
Brain tissues from CPT1C–KO mice compared to WT mice show increased ABHD6 activity. (a) ABHD6 activity in hypothalamic homogenates (n = 6) for CPT1C–KO compared to WT mice; (b) AUCs for hypothalamic activity. (c) ABHD6 activity in hippocampal homogenates (n = 6) for CPT1C–KO compared to WT mice; (d) AUCs for hippocampal activity. Data shown are means ± SD. *P < 0.05, significantly different from WT; Two‐way ANOVA followed by Bonferroni's post hoc correction
3.5. ABHD6 activity is increased in brain homogenates after fasting conditions
ABHD6 activity was analysed in fasted mice, because fasting provides a metabolic challenge in which brain malonyl–CoA levels are significantly decreased (Tokutake et al., 2012, 2010) and, in such conditions, the CPT1C effect on ABHD6 activity may be compromised. Fasting also increases endocannabinoid levels (Kirkham et al., 2002). Overnight deprivation of food induced a significant increase in ABHD6 activity in both hypothalamic (Figure 7a–c) and hippocampal (Figure 7d–f) homogenates for the control male mice, whereas no differences were apparent in brain tissues from the CPT1C–KO mice, indicating that CPT1C inhibition of ABHD6 is blunted when malonyl–CoA levels drop. The effect of fasting on ABHD6 activity is more evident in the hypothalamus than in the hippocampus (Figure 7). Noteworthy is the fact that no significant differences were observed between fed and fasted conditions for CPT1C or ABHD6 protein expression levels in hypothalamus or hippocampus from control mice (Figure S9).
FIGURE 7.
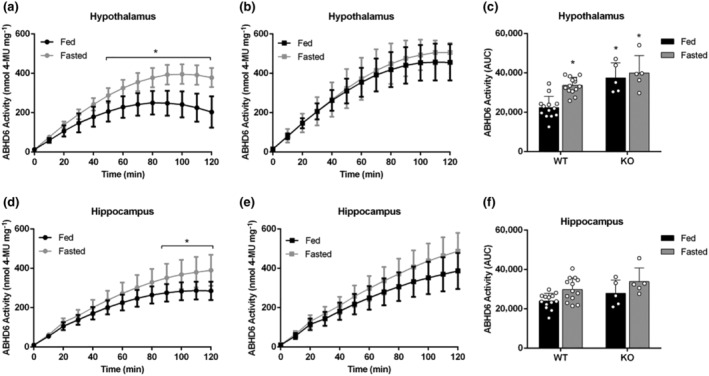
Effect of fasting on ABHD6 activity of male WT or CPT1C–KO mouse brain tissues. (a–c) ABHD6 activity in hypothalamic homogenates of (a) WT mice (n = 13) and (b) CPT1C–KO mice (n = 5) and (c) AUCs for hypothalamic activity in fed and fasted mice. (d–f) ABHD6 activity in hippocampal homogenates of (d) WT mice (n = 13) and (e) CPT1C–KO mice (n = 5) and (f) AUCs for hippocampal activity in fed and fasted mice. Data shown are means ± SD. *P < 0.05, significantly different from WT‐fed; two‐way ANOVA followed by Bonferroni's post hoc correction
Tissue samples from female mice were also assayed, with results similar to those for male mice, that is, the fasting‐induced increase in ABHD6 activity was only apparent in hypothalamic but not in hippocampal homogenates (Figure S10).
4. DISCUSSION
ABHD6 is one of the newest 2‐AG hydrolases to be incorporated into the so‐called endocannabinoid system, which includes endocannabinoids, cannabinoid receptors, and the enzymes that produce and degrade endocannabinoids. Since its discovery, ABHD6 has been linked to important neural functions (Cao et al., 2019), but it is not known how its activity is regulated. We identified CPT1C as the first protein to interact with ABHD6, also observing that it acts as a negative regulator of activity depending on the nutritional state (fed or fasted). These results are relevant as inhibiting ABHD6 activity has emerged as an important therapeutic approach in preclinical models of neuron‐related, devastating diseases (Cao et al., 2019). Our study also throws light on the interplay between those two proteins and the physiological relevance of their interaction.
Our finding that CPT1C directly interacts with ABHD6 in transfected HEK‐293T cells is in line with previous proteomic studies that identified both CPT1C and ABHD6 as interacting components in AMPA receptor complexes (Brechet et al., 2017; Casas et al., 2020; Schwenk et al., 2019). Within these complexes, ABHD6 stabilizes pore‐forming GluA monomers, while the FRRS1I/CPT1C proteins promote GluA oligomerization and trafficking to the cell surface (Schwenk et al., 2019). Moreover, ABHD6 is able to interact with GluA1‐3 and to negatively regulate its surface delivery, independently of its hydrolase activity (Wei et al., 2017, 2016). We have shown that CPT1C and ABHD6 are able to interact in cells, such as HEK‐293T, that do not express AMPA receptors, using immunoprecipitation and FRET‐based approaches. This finding may be of relevance to cancer research as both proteins are involved in tumour proliferation and in metastasis (Casals et al., 2016; Tang et al., 2020; Zaugg et al., 2011). CPT1C has attracted interest because it can bind malonyl–CoA, an important nutritional sensor in the brain, and has a longer tail at the C‐terminus, compared to other CPT1 forms. Our group recently demonstrated that both characteristics of CPT1C are important to regulate activity or interaction for other proteins (Casas et al., 2020; Palomo‐Guerrero et al., 2019). We consequently used two versions of CPT1C: the M589S mutant that is insensitive to malonyl–CoA and ΔCter, which lacks the C‐terminal region. FRET and co‐immunoprecipitation assays revealed that CPT1C and ABHD6 interaction is not modified when using the malonyl–CoA insensitive or the C‐terminal truncated form of CPT1C. The effects of malonyl–CoA on CPT1C–ABHD6 interactions were further underpinned by FRET experiments in the presence of TOFA.
The activity of endocannabinoid system enzymes has been commonly studied using laborious or expensive assays, specifically, radiolabelled substrates (Dinh et al., 2002; Gopapaju et al., 1999; Karlsson et al., 2000; Sakurada & Noma, 1981; Somma‐Delpéro et al., 1995; Tornqvist et al., 1974) or mass spectrometry‐based assays (Blankman et al., 2007) and HPLC detection of arachidonic acid (Saario et al., 2005). Assays have also been developed for the study of different hydrolases using chromogenic (Imamura & Kitaura, 2000) and fluorogenic (Wang et al., 2008; Zvonok et al., 2008) substrates, including ABHD6 (Navia‐Paldanius et al., 2012; Shields et al., 2019). However, those assays require multiple‐enzymic steps or long incubation times. To address these problems, we have optimized an easy, rapid, inexpensive, and highly sensitive fluorescent ABHD6 activity assay using 4‐MUH as the substrate. 4‐MUH has been used to study lipase activity for decades. As it is hydrosoluble, 4‐MUH is not easily hydrolysed by non‐lipolytic esterases, and its hydrolysis produces a highly fluorescent compound, 4‐MU (Dolinsky et al., 2004; Gilham et al., 2005). The apparent kinetic parameters, V max ~ 322 nmol·mg−1·min−1 and K m ~ 19 μM, demonstrate that 4‐MUH is a high‐affinity substrate that is rapidly hydrolysed by ABHD6. In fact, those parameters were better than those previously reported for 2‐AG (V max ~ 45 nmol·mg−1·min−1 and K m ~ 159 μM; Navia‐Paldanius et al., 2012) and for another fluorogenic substrate, the arachidonoyl coumarin ester AHMMCE (V max ~ 67 pmol·mg−1·min−1 and K m ~ 26 μM; Shields et al., 2019). To validate our method, we inhibited ABHD6 activity using WWL70 (IC50 = 274 nM) and performed substrate competition with 2‐AG (IC50 = 10 μM). The constant of inhibition obtained for WWL70 was higher than in previous assays using activity‐based protein profiling (IC50 = 70 nM; Li et al., 2007) or 2‐AG as substrate (IC50 = 85 nM; Navia‐Paldanius et al., 2012). However, assays using the other fluorogenic substrate obtained an inhibition constant (IC50 = 285 nM; Shields et al., 2019) that was similar to that for our method. Using this enzymatic assay, we demonstrated that CPT1C acts as an inhibitor of ABHD6 activity by reducing ABHD6 activity in cells simultaneously expressing both proteins by more than 50%. In a more physiological environment, ABHD6 activity was assessed in cultured cells endogenously expressing both ABHD6 and CPT1C (primary neurons and a hypothalamic neuronal cell line GT1‐7) and, for the first time, in brain tissue homogenates of WT and CPT1C–KO mice in the presence of WWL70 or KT‐182. The results regarding ABHD6 activity in brain homogenates of WT and CPT1C–KO mice demonstrate that brain tissues, and particularly hypothalamic tissue, for CPT1C‐deficient mice show more ABHD6 activity than WT mice, in accordance with the results obtained for heterologous cells.
Although ABHD6 activity was successfully measured in neuronal and brain tissue models, the ability of WWL70 to inhibit ABHD6 activity was less (by 25–30%) than that shown in cell models overexpressing ABHD6. This finding might be related to the fact that 4‐MUH is not a specific substrate for ABHD6 and can be hydrolysed by other enzymes with lipase activity. Therefore, 4‐MUH in HEK‐293T cells overexpressing ABHD6 is mainly hydrolysed by ABHD6, as observed for the low 4‐MUH activity reported for cells transfected with EV. However, in brain tissue homogenates and cells endogenously expressing ABHD6, other enzymes can hydrolyse 4‐MUH. Therefore, the inhibitory effect of WWL70 in brain homogenates might be partly masked for the 4‐MUH‐hydrolysing activity of other lipases in the sample. Note that this finding is in line with previous literature pointing to low hydrolase activity of ABHD6 in neurons compared to other hydrolases such as MAGL (Blankman et al., 2007; Cao et al., 2019).
In recent years, it has been postulated that the neuron‐specific CPT1C acts as a regulator of other interacting proteins in response to changes in malonyl–CoA levels (Casals et al., 2016; Casas et al., 2020; Palomo‐Guerrero et al., 2019; Wolfgang & Lane, 2011)and such effects have also been demonstrated for two interacting proteins, protrudin and SAC1 (Casas et al., 2020; Palomo‐Guerrero et al., 2019). Our results show that CPT1C sensing of malonyl–CoA is necessary for regulation of ABHD6 activity, which is inhibited only when CPT1C detects malonyl–CoA. These results are relevant since malonyl–CoA acts as a nutritional‐sensing mechanism in the brain and is crucial for CPT1C regulation of energy expenditure (Rodríguez‐Rodríguez et al., 2019; Wolfgang & Lane, 2011) and food intake (Gao et al., 2011; Ramírez et al., 2013). We have also demonstrated that CPT1C in the hypothalamus is crucial for the proper adaptation to a fasting situation, when malonyl–CoA levels drop (Pozo et al., 2017). Interestingly, brain tissue homogenates from fasting control mice showed a significant increase in ABHD6 activity in both male and female mice, especially in the hypothalamus. Thus, when malonyl–CoA levels are low, the inhibitory effect of CPT1C on ABHD6 activity is blunted—a hypothesis that fits with the lack of any effect on fasting in CPT1C–KO mice. Note that under fasting conditions, not only malonyl–CoA levels, but also those of 2‐AG, increase in brain tissues (Kirkham et al., 2002). This increase may also contribute to increased ABHD6 activity, as there are more 2‐AG levels to hydrolase. Fasting not only decreases intracellular malonyl–CoA levels but also produces a profound rewiring of brain metabolism and function. As this consequence suggested the effects we observed could be a non‐specific response, to partly address this limitation of our study, we demonstrated that fasting did not induce differences in CPT1C and ABHD6 expression levels in either hypothalamic or hippocampal homogenates. The changes in ABHD6 activity observed under fasting conditions are therefore not a consequence of alterations in ABHD6 expression level, but possibly a consequence of the negative regulation of CPT1C in ABHD6. Our findings are coherent with ABHD6 activity being under the control of sensing malonyl–CoA by CPT1C; that is, cell energy status can regulate ABHD6 activity through the malonyl–CoA/CPT1C axis. Further in vivo experiments are needed, however, to demonstrate that malonyl–CoA is the only metabolic intermediary regulating ABHD6 activity in fasting conditions.
We observed no significant change in endogenous endocannabinoid levels in cells expressing ABHD6 or ABHD6‐CPT1C, a finding in line with the fact that ABHD6 only hydrolyses 2‐AG when it is produced on demand (Marrs et al., 2010). Levels of 2‐AG and AEA in HEK‐293T cells expressing ABHD6 after stimulation of 2‐AG production need to be measured, as the specific effects of ABHD6 on 2‐AG levels depend on the cell type and the experimental setting. Recent data suggest that ABHD6 is a dual DAG/MAG‐lipase that produces 2‐AG in Neuro‐2a cells during differentiation and neurite outgrowth (van Esbroeck et al., 2019).
Although we did not observe changes in endogenous levels of 2‐AG in HEK‐293T cells, we explored whether CPT1C regulation of ABHD6 activity could influence intracellular response in the form of cAMP levels. The canonical intracellular pathway for 2‐AG production starts by activating CB1 receptors, which leads to AC inhibition and, consequently, to reduced intracellular cAMP levels (Busquets‐Garcia et al., 2018). Our results show that ABHD6 hydrolysis of 2‐AG raised cAMP intracellular levels due to accumulation of the 2‐AG hydrolysis product arachidonic acid, which increases cAMP in a CB1 receptor‐independent manner. In contrast, CPT1C nullifies that increase, in line with its inhibitory effect on ABHD6 activity. Moreover, the effect of CPT1C on ABHD6‐dependent cAMP levels requires malonyl–CoA sensing, but not the presence of the C‐terminal region. This is undoubtedly an intriguing result as the C‐terminal region in our study was important for its regulatory effect of CPT1C on ABHD6‐mediated 4‐MUH hydrolysis, but not on cAMP response. This finding may be related to the fact that the cAMP determination assay is a more sensitive and more physiological assay than the 4‐MUH‐based assay and also that malonyl–CoA sensing is much more crucial to CPT1C regulation of other enzyme activities, than the presence of the C‐terminal region (Casas et al., 2020). Therefore, CPT1C control of ABHD6 modifies cAMP levels in a malonyl–CoA‐dependent manner, confirming ABHD6 activity assay results for both cells and mouse brain homogenates.
CPT1C regulation of ABHD6 activity may be an important player in CNS‐related ABHD6 functions. For instance, both proteins have been reported to play an important role in regulating energy homeostasis in the hypothalamus under fasting and high‐fat diet conditions (Fisette et al., 2016; Pozo et al., 2017; Rodríguez‐Rodríguez et al., 2019). Interestingly, ABHD6 can hydrolyse other monoacylglycerols in the brain, for example, bis (monoacylglycero)phosphate, the amount and composition of which is sensitive to a high‐fat diet (Grabner et al., 2019; Pribasnig et al., 2015). We suggest that it might be interesting to study the role of the CPT1C–ABHD6 complex in regulating levels of bis (monoacylglycero)phosphate in response to metabolic challenges. CPT1C regulation of ABHD6 could also be of interest for epilepsy, since inhibition of ABHD6 activity has been reported to attenuate seizure development (Naydenov et al., 2014; Sigel et al., 2011). Finally, although our study strongly demonstrates that CPT1C is a physiological inhibitor of ABHD6 depending on malonyl–CoA sensing, the exact molecular mechanisms mediating this inhibition remain unclear. Bearing in mind that CPT1C can modulate palmitoylation in other proteins such as GluA1 (Gratacòs‐Batlle et al., 2018), potential palmitoylation of ABHD6 could not be ruled out and, consequently, the localization and activity of ABHD6.
As far as we are aware, our study describes, for the first time, a link between CPT1C and the endocannabinoid system, thereby yielding valuable information on the characterization of both CPT1C and ABHD6. We report CPT1C as the first physiological inhibitor of ABHD6 activity, with the sensing of cell nutritional status playing a key role in this regulatory role of CPT1C. These findings are notable, as blocking ABHD6 activity may have therapeutic benefits for epilepsy, traumatic brain injury, multiple sclerosis and obesity (Cao et al., 2019). CPT1C, therefore, as a negative regulator of ABHD6, might be an interesting target in combating these pathologies.
AUTHOR CONTRIBUTIONS
C.M., A.C.R., A.F., M.C., J.L., G.N., and J.C. performed experiments. C.M., N.C., and R.R.‐R. designed the research study. C.M., A.C.R. and R.R.‐R. analysed the data. C.M, R.R.‐R., and N.C. wrote the manuscript. R.F. and S.P.H.A. actively participated in data interpretation, writing, and editing.
CONFLICT OF INTEREST
The authors declared no conflict of interest.
DECLARATION OF TRANSPARENCY AND SCIENTIFIC RIGOUR
This Declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research as stated in the BJP guidelines for Design & Analysis, Immunoblotting and Immunochemistry, and Animal Experimentation, and as recommended by funding agencies, publishers and other organizations engaged with supporting research.
Supporting information
Figure S1. Representative FRET images of transfected HEK‐293 T cells with proteins fused to mTurquoise2 (donor; blue) or SYFP2 (acceptor; yellow) before and after bleaching.
Figure S2. FRET sensitized emission of CPT1C with ABHD6 in the presence of an ACC inhibitor in living cells. Assays were performed 48 h post‐transfection in HEK‐293 T cells expressing ABHD6‐mTurq2 and increasing amounts of the cDNA for CPT1C‐SYFP2, in the presence or absence of the ACC inhibitor, TOFA (20 μg/ml, 2 h). FRET saturation curves were obtained by monitoring the SYFP2 fluorescence emission at 530 nm after excitation of mTurq2 at 420 nm, with subtraction of the values obtained with cells expressing the same amount of donor protein. Mean±SD (n = 4).
Figure S3. ABHD6 assay based on 4‐MUH hydrolysis in hypothalamus of WT and CPT1C‐KO mice. ABHD6 activity dependent on 4‐MUH concentration at t = 30 min in the absence (A) or presence of 10 μM WWL70 (B). Two‐way ANOVA followed by Bonferroni's post‐hoc correction was used for statistical analysis. Mean±SD (n = 6). *P < 0.05 versus WT.
Figure S4. Kinetic hydrolase activity (A) and areas under the curve (B) in the absence or presence of the ABHD6 inhibitors WWL70 and KT182. Mean±SD (n = 7). *P < 0.05 versus Vehicle.
Figure S5. ABHD6 activity assay based on 4‐MUH hydrolysis. (A) Blotting and protein expression levels of ABHD6 in combination with CPT1C, M589S or ΔCter in HEK‐293 T for ABHD6 activity assays (n = 15). (B) 4‐MUH hydrolysis of HEK‐293 T cells expressing EV, CPT1C, M589S and Δcter (n = 4). (C) HEK‐293 T endogenous endocannabinoids (eCBs) levels, 2‐AG and AEA, when transfected with EV, ABHD6 or ABHD6‐CPT1C (n = 15). Mean±SD.
Figure S6. ABHD6 activity assay based on 4‐MUH hydrolysis in a neuronal cell line, GT1–7, endogenously expressing ABHD6 and CPT1C. Time course ABHD6 activity assay (A) and areas under the curve (B) in GT1–7 cells with or without TOFA treatment (20 μg/mL) for 1 h and 2 h. Mean±SD (n = 6). *P < 0.05 versus vehicle.
Figure S7. Intracellular cAMP assay was set up in HEK‐293 T. (A) Forskolin (FK)‐induced cAMP levels of HEK‐293 T expressing CB1R incubated with an agonist of CB1R, ACEA, an antagonist, rimonabant (Rimo), Rimo + 2‐AG and increasing concentrations of 2‐AG, arachidonic acid (ARA) and glycerol. (B) Effect of ACEA, 2‐AG, ARA and glycerol in basal cAMP levels of HEK‐293 T cells alone. Mean±SD (n = 8). + P < 0.05 versus FK and *P < 0.05 versus basal.
Figure S8. ABHD6 assay based on 4‐MUH hydrolysis in brain tissue homogenates of WT or CPT1C‐KO male mice in the absence or presence of the ABHD6 inhibitor, WWL70 (10 μM). (A) 4‐MUH hydrolysis in hypothalamic homogenates. (B) 4‐MUH hydrolysis in hippocampal homogenates. Mean±SD (n = 6).
Figure S9. CPT1C and ABHD6 protein expression on brain tissue homogenates of fed or fasted mice. Representative blots (A) and quantification (% relative to WT‐fed) of CPT1C (B) and ABHD6 (C) protein levels in either hypothalamus (Hyp) or hippocampus (Hc) of WT mice under fed (n = 6) or fasting conditions (n = 5). Mean±SD.
Figure S10. Effect of fasting on ABHD6 activity of female WT brain tissues. (A, B) ABHD6 activity in hypothalamic homogenates and area under the curves of mice after fed or fasted state. (C, D) ABHD6 activity in hippocampal homogenates and area under the curves of mice after fed or fasted state. Mean±SD (n = 5). *P < 0.05 versus WT‐fed.
ACKNOWLEDGEMENTS
We thank Dr. Gemma Fabriàs and Alexandre García from the Research Unit of BioActive Molecules (Institut de Química Avançada de Catalunya, IQAC) for technical assistance in the analysis of endocannabinoid levels. We also thank Prof. Kenneth Mackie from the Department of Psychological and Brain Sciences, Indiana University Bloomington (USA), for providing ABHD6 antibody for western blotting assays in brain tissue. This work was supported by the Ministerio de Economía, Industria y Competitividad (MINECO), Agencia Estatal de Investigación (AEI) and Fondo Europeo de Desarrollo Regional (FEDER) (SAF2017‐83813‐C3‐3‐R to N.C. and R.R.‐R.), Joint Bilateral Project Japan‐Spain/AEI (PCI2018‐092997/AEI to R.R.‐R.) and Fundació La Marató de TV3 (Grant 87/C/2016 to N.C.). A.F. and C.R. are recipients of a fellowship from the Agència de Gestió d'Ajuts Universitaris i de la Recerca (AGAUR) in Catalonia.
Miralpeix C, Reguera AC, Fosch A, et al. Carnitine palmitoyltransferase 1C negatively regulates the endocannabinoid hydrolase ABHD6 in mice, depending on nutritional status. Br J Pharmacol. 2021;178:1507–1523. 10.1111/bph.15377
Cristina Miralpeix and Ana Cristina Reguera contributed equally to this work.
In this article, we demonstrate how CPT1C, a neuron‐specific protein, interacts and regulates the activity of ABHD6, a new member of the endocannabinoid system. The interplay between these two proteins is key to the treatment of neuron‐related diseases.
Contributor Information
Núria Casals, Email: ncasals@uic.es.
Rosalía Rodríguez‐Rodríguez, Email: rrodriguez@uic.es.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request. Some data may not be made available because of privacy or ethical restrictions.
REFERENCES
- Alexander, S. P. H. , Christopoulos, A. , Davenport, A. P. , Kelly, E. , Mathie, A. , Peters, J. A. , … CGTP Collaborators . (2019). THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: G protein‐coupled receptors. British Journal of Pharmacology, 176, S21–S141. 10.1111/bph.14748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Fabbro, D. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , … CGTP Collaborators . (2019). THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Enzymes. British Journal of Pharmacology, 176, S297–S396. 10.1111/bph.14752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Mathie, A. , Peters, J. A. , Veale, E. L. , Striessnig, J. , Kelly, E. , … CGTP Collaborators . (2019). THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Ion channels. British Journal of Pharmacology, 176, S142–S228. 10.1111/bph.14749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Roberts, R. E. , Broughton, B. R. S. , Sobey, C. G. , George, C. H. , Stanford, S. C. , … Ahluwalia, A. (2018). Goals and practicalities of immunoblotting and immunohistochemistry: A guide for submission to the British Journal of Pharmacology. British Journal of Pharmacology, 175, 407–411. 10.1111/bph.14112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankman, J. L. , Simon, G. M. , & Cravatt, B. F. (2007). A comprehensive profile of brain enzymes that hydrolyze the endocannabinoid 2‐arachidonoylglycerol. Chemistry & Biology, 14, 1347–1356. 10.1016/j.chembiol.2007.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottemanne, P. , Paquot, A. , Ameraoui, H. , Alhouayek, M. , & Muccioli, G. G. (2019). The α/β–hydrolase domain 6 inhibitor WWL70 decreases endotoxin‐induced lung inflammation in mice, potential contribution of 2‐arachidonoylglycerol, and lysoglycerophospholipids. The FASEB Journal, 33, 7635–7646. 10.1096/fj.201802259R [DOI] [PubMed] [Google Scholar]
- Brechet, A. , Buchert, R. , Schwenk, J. , Boudkkazi, S. , Zolles, G. , Siquier‐Pernet, K. , … Fakler, B. (2017). AMPA‐receptor specific biogenesis complexes control synaptic transmission and intellectual ability. Nature Communications, 8, 15910. 10.1038/ncomms15910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busquets‐Garcia, A. , Bains, J. , & Marsicano, G. (2018). CB 1 receptor signaling in the brain: Extracting specificity from ubiquity. Neuropsychopharmacology, 43, 4–20. 10.1038/npp.2017.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, J. K. , Kaplan, J. , & Stella, N. (2019). ABHD6: Its place in endocannabinoid signaling and beyond. Trends in Pharmacological Sciences, 40, 267–277. 10.1016/j.tips.2019.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco, P. , Sahún, I. , McDonald, J. , Ramírez, S. , Jacas, J. , Gratacós, E. , … Casals, N. (2012). Ceramide levels regulated by carnitine palmitoyltransferase 1C control dendritic spine maturation and cognition. The Journal of Biological Chemistry, 287, 21224–21232. 10.1074/jbc.M111.337493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carriba, P. , Navarro, G. , Ciruela, F. , Ferré, S. , Casadó, V. , Agnati, L. , … Franco, R. (2008). Detection of heteromerization of more than two proteins by sequential BRET‐FRET. Nature Methods, 5, 727–733. 10.1038/nmeth.1229 [DOI] [PubMed] [Google Scholar]
- Casals, N. , Zammit, V. , Herrero, L. , Fadó, R. , Rodríguez‐Rodríguez, R. , & Serra, D. (2016). Carnitine palmitoyltransferase 1C: From cognition to cancer. Progress in Lipid Research, 61, 134–148. 10.1016/j.plipres.2015.11.004 [DOI] [PubMed] [Google Scholar]
- Casas, M. , Fadó, R. , Domínguez, J. L. , Roig, A. , Kaku, M. , Chohnan, S. , … Casals, N. (2020). Sensing of nutrients by CPT1C controls SAC1 activity to regulate AMPA receptor trafficking. The Journal of Cell Biology, 219. 10.1083/jcb.201912045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, M. J. , Alexander, S. , Cirino, G. , Docherty, J. R. , George, C. H. , Giembycz, M. A. , … Ahluwalia, A. (2018). Experimental design and analysis and their reporting II: Updated and simplified guidance for authors and peer reviewers. British Journal of Pharmacology, 175, 987–993. 10.1111/bph.14153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh, T. P. , Carpenter, D. , Leslie, F. M. , Freund, T. F. , Katona, I. , Sensi, S. L. , … Piomelli, D. (2002). Brain monoglyceride lipase participating in endocannabinoid inactivation. Proceedings of the National Academy of Sciences, 99, 10819–10824. 10.1073/pnas.152334899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky, V. W. , Douglas, D. N. , Lehner, R. , & Vance, D. E. (2004). Regulation of the enzymes of hepatic microsomal triacylglycerol lipolysis and re‐esterification by the glucocorticoid dexamethasone. The Biochemical Journal, 378, 967–974. 10.1042/bj20031320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlenhardt, N. , Yu, H. , Abiraman, K. , Yamasaki, T. , Wadiche, J. I. , Tomita, S. , & Bredt, D. S. (2016). Porcupine controls hippocampal AMPAR levels, composition, and synaptic transmission. Cell Reports, 14, 782–794. 10.1016/j.celrep.2015.12.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadó, R. , Rodríguez‐Rodríguez, R. , & Casals, N. (2021). The return of malonyl–CoA to the brain: Cognition and other stories. Progress in Lipid Research, 81, 101071. 10.1016/j.plipres.2020.101071 [DOI] [PubMed] [Google Scholar]
- Fadó, R. , Soto, D. , Miñano‐Molina, A. J. , Pozo, M. , Carrasco, P. , Yefimenko, N. , … Casals, N. (2015). Novel regulation of the synthesis of α‐Amino‐3‐hydroxy‐5‐methyl‐4‐isoxazolepropionic acid (AMPA) receptor subunit GluA1 by carnitine palmitoyltransferase 1C (CPT1C) in the hippocampus. The Journal of Biological Chemistry, 290, 25548–25560. 10.1074/jbc.M115.681064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisette, A. , Tobin, S. , Décarie‐Spain, L. , Bouyakdan, K. , Peyot, M.‐L. , Madiraju, S. R. M. , … Alquier, T. (2016). α/β‐hydrolase domain 6 in the ventromedial hypothalamus controls energy metabolism flexibility. Cell Reports, 17, 1217–1226. 10.1016/j.celrep.2016.10.004 [DOI] [PubMed] [Google Scholar]
- Gao, S. , Zhu, G. , Gao, X. , Wu, D. , Carrasco, P. , Casals, N. , … Lopaschuk, G. D. (2011). Important roles of brain‐specific carnitine palmitoyltransferase and ceramide metabolism in leptin hypothalamic control of feeding. Proceedings of the National Academy of Sciences of the United States of America, 108, 9691–9696. 10.1073/pnas.1103267108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilham, D. , Alam, M. , Gao, W. , Vance, D. E. , & Lehner, R. (2005). Triacylglycerol hydrolase is localized to the endoplasmic reticulum by an unusual retrieval sequence where it participates in VLDL assembly without utilizing VLDL lipids as substrates. Molecular Biology of the Cell, 16, 984–996. 10.1091/mbc.e04-03-0224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopapaju, S. K. , Ueda, N. , Taniguchi, K. , & Yamamoto, S. (1999). Different enzymes of porcine brain hydrolyzing 2‐Arachidonoylglycerol, an endogenous ligand for cannabinoid receptors. Prostaglandins & Other Lipid Mediators, 59, 208. [DOI] [PubMed] [Google Scholar]
- Grabner, G. F. , Fawzy, N. , Pribasnig, M. A. , Trieb, M. , Taschler, U. , Holzer, M. , … Zimmermann, R. (2019). Metabolic disease and ABHD6 alter the circulating bis (monoacylglycerol)phosphate profile in mice and humans. Journal of Lipid Research, 60, 1020–1031. 10.1194/jlr.M093351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratacòs‐Batlle, E. , Olivella, M. , Sánchez‐Fernández, N. , Yefimenko, N. , Miguez‐Cabello, F. , Fadó, R. , … Soto, D. (2018). Mechanisms of CPT1C‐dependent AMPAR trafficking enhancement. Frontiers in Molecular Neuroscience, 11, 275. 10.3389/fnmol.2018.00275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, K. L. , Tsuboi, K. , Chang, J. W. , Whitby, L. R. , Speers, A. E. , Pugh, H. , & Cravatt, B. F. (2013). Discovery and optimization of piperidyl‐1,2,3‐triazole ureas as potent, selective, and in vivo‐active inhibitors of α/β‐hydrolase domain containing 6 (ABHD6). Journal of Medicinal Chemistry, 56, 8270–8279. 10.1021/jm400899c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura, S. , & Kitaura, S. (2000). Purification the moderately characterization thermophilic of a monoacylglycerol bacillus lipase from. 127, 419–425. [DOI] [PubMed] [Google Scholar]
- Karlsson, M. , Tornqvist, H. , & Holm, C. (2000). Expression, purification, and characterization of histidine‐tagged mouse monoglyceride lipase from baculovirus‐infected insect cells. Protein Expression and Purification, 18, 286–292. 10.1006/prep.1999.1194 [DOI] [PubMed] [Google Scholar]
- Kirkham, T. C. , Williams, C. M. , Fezza, F. , & Di Marzo, V. (2002). Endocannabinoid levels in rat limbic forebrain and hypothalamus in relation to fasting, feeding and satiation: Stimulation of eating by 2‐arachidonoyl glycerol. British Journal of Pharmacology, 136, 550–557. 10.1038/sj.bjp.0704767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Blankman, J. L. , & Cravatt, B. F. (2007). A functional proteomic strategy to discover inhibitors for uncharacterized hydrolases. Journal of the American Chemical Society, 129, 9594–9595. 10.1021/ja073650c [DOI] [PubMed] [Google Scholar]
- Lilley, E. , Stanford, S. C. , Kendall, D. E. , Alexander, S. P. , Cirino, G. , Docherty, J. R. , … Ahluwalia, A. (2020). ARRIVE 2.0 and the British Journal of Pharmacology: Updated guidance for 2020. British Journal of Pharmacology, 177, 3611–3616. https://bpspubs.onlinelibrary.wiley.com/doi/full/10.1111/bph.15178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manterola, A. , Bernal‐Chico, A. , Cipriani, R. , Ruiz, A. , Pérez‐Samartín, A. , Moreno‐Rodríguez, M. , … Mato, S. (2018). Re‐examining the potential of targeting ABHD6 in multiple sclerosis: Efficacy of systemic and peripherally restricted inhibitors in experimental autoimmune encephalomyelitis. Neuropharmacology, 141, 181–191. 10.1016/j.neuropharm.2018.08.038 [DOI] [PubMed] [Google Scholar]
- Marrs, W. R. , Blankman, J. L. , Horne, E. A. , Thomazeau, A. , Lin, Y. H. , Coy, J. , … Stella, N. (2010). The serine hydrolase ABHD6 controls the accumulation and efficacy of 2‐AG at cannabinoid receptors. Nature Neuroscience, 13, 951–957. 10.1038/nn.2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralpeix, C. , Fosch, A. , Casas, J. , Baena, M. , Herrero, L. , Serra, D. , … Casals, N. (2019). Hypothalamic endocannabinoids inversely correlate with the development of diet‐induced obesity in male and female mice. Journal of Lipid Research, 60, 1260–1269. 10.1194/jlr.M092742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillas, M. , Gómez‐Puertas, P. , Bentebibel, A. , Sellés, E. , Casals, N. , Valencia, A. , … Serra, D. (2003). Identification of conserved amino acid residues in rat liver carnitine palmitoyltransferase I critical for malonyl–CoA inhibition: Mutation of methionine 593 abolishes malonyl–CoA inhibition. The Journal of Biological Chemistry, 278, 9058–9063. 10.1074/jbc.M209999200 [DOI] [PubMed] [Google Scholar]
- Navarro, G. , Ferré, S. , Cordomi, A. , Moreno, E. , Mallol, J. , Casadó, V. , … Woods, A. S. (2010). Interactions between intracellular domains as key determinants of the quaternary structure and function of receptor heteromers. The Journal of Biological Chemistry, 285, 27346–27359. 10.1074/jbc.M110.115634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navia‐Paldanius, D. , Savinainen, J. R. , & Laitinen, J. T. (2012). Biochemical and pharmacological characterization of human/ ‐hydrolase domain containing 6 (ABHD6) and 12 (ABHD12). Journal of Lipid Research, 53, 2413–2424. 10.1194/jlr.M030411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naydenov, A. A. V. , Horne, E. A. , Cheah, C. S. , Swinney, K. , Hsu, K.‐L. L. , Cao, J. K. , … Stella, N. (2014). ABHD6 blockade exerts antiepileptic activity in PTZ‐induced seizures and in spontaneous seizures in R6/2 mice. Neuron, 83, 361–371. 10.1016/j.neuron.2014.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, S. , Sato, T. , Tateyama, M. , Kageyama, H. , Maejima, Y. , Nakata, M. , … Minokoshi, Y. (2018). Activation of AMPK‐regulated CRH neurons in the PVH is sufficient and necessary to induce dietary preference for carbohydrate over fat. Cell Reports, 22, 706–721. 10.1016/j.celrep.2017.11.102 [DOI] [PubMed] [Google Scholar]
- Palomo‐Guerrero, M. , Fadó, R. , Casas, M. , Pérez‐Montero, M. , Baena, M. , Helmer, P. O. , … Casals, N. (2019). Sensing of nutrients by CPT1C regulates late endosome/lysosome anterograde transport and axon growth. eLife, 8, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percie du Sert, N. , Hurst, V. , Ahluwalia, A. , Alam, S. , Avey, M. T. , Baker, M. , … Würbel, H. (2020). The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. PLoS Biology, 18(7), e3000410. 10.1371/journal.pbio.3000410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo, M. , Rodriguez‐Rodriguez, R. , Raḿirez, S. , Seoane‐Collazo, P. , López, M. , Serra, D. , … Casals, N. (2017). Hypothalamic regulation of liver and muscle nutrient partitioning by brain‐specific carnitine palmitoyltransferase 1c in male mice. Endocrinology, 158, 2226–2238. 10.1210/en.2017-00151 [DOI] [PubMed] [Google Scholar]
- Pribasnig, M. A. , Mrak, I. , Grabner, G. F. , Taschler, U. , Knittelfelder, O. , Scherz, B. , … Zimmermann, R. (2015). α/β hydrolase domain‐containing 6 (ABHD6) degrades the late endosomal/lysosomal lipid Bis (Monoacylglycero)phosphate. The Journal of Biological Chemistry, 290, 29869–29881. 10.1074/jbc.M115.669168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, N. , van der Leij, F. , Jackson, V. , Corstorphine, C. , Thomson, R. , Sorensen, A. , & Zammit, V. A. (2002). A novel brain‐expressed protein related to carnitine palmitoyltransferase I. Genomics, 80, 433–442. 10.1006/geno.2002.6845 [DOI] [PubMed] [Google Scholar]
- Ramírez, S. , Martins, L. , Jacas, J. , Carrasco, P. , Pozo, M. , Clotet, J. , … Casals, N. (2013). Hypothalamic ceramide levels regulated by CPT1C mediate the orexigenic effect of ghrelin. Diabetes, 62, 2329–2337. 10.2337/db12-1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes‐Resina, I. , Navarro, G. , Aguinaga, D. , Canela, E. I. , Schoeder, C. T. , Załuski, M. , … Franco, R. (2018). Molecular and functional interaction between GPR18 and cannabinoid CB2 G‐protein‐coupled receptors. Relevance in neurodegenerative diseases. Biochemical Pharmacology, 157, 169–179. 10.1016/j.bcp.2018.06.001 [DOI] [PubMed] [Google Scholar]
- Rodríguez‐Rodríguez, R. , Miralpeix, C. , Fosch, A. , Pozo, M. , Calderón‐Domínguez, M. , Perpinyà, X. , … Casals, N. (2019). CPT1C in the ventromedial nucleus of the hypothalamus is necessary for brown fat thermogenesis activation in obesity. Molecular Metabolism, 19, 75–85. 10.1016/j.molmet.2018.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saario, S. M. , Salo, O. M. H. , Nevalainen, T. , Poso, A. , Laitinen, J. T. , Järvinen, T. , & Niemi, R. (2005). Characterization of the sulfhydryl‐sensitive site in the enzyme responsible for hydrolysis of 2‐arachidonoyl‐glycerol in rat cerebellar membranes. Chemistry & Biology, 12, 649–656. 10.1016/j.chembiol.2005.04.013 [DOI] [PubMed] [Google Scholar]
- Sakurada, T. , & Noma, A. (1981). Subcellular localization and some properties of monoacylglycerol lipase in rat adipocytes. Journal of Biochemistry, 90, 1413–1419. 10.1093/oxfordjournals.jbchem.a133607 [DOI] [PubMed] [Google Scholar]
- Schwenk, J. , Boudkkazi, S. , Kocylowski, M. K. , Brechet, A. , Zolles, G. , Bus, T. , … Fakler, B. (2019). An ER assembly line of AMPA‐receptors controls excitatory neurotransmission and its plasticity. Neuron, 104, 680–692.e9. [DOI] [PubMed] [Google Scholar]
- Schwenk, J. , Harmel, N. , Brechet, A. , Zolles, G. , Berkefeld, H. , Müller, C. S. , … Fakler, B. (2012). High‐resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron, 74, 621–633. 10.1016/j.neuron.2012.03.034 [DOI] [PubMed] [Google Scholar]
- Shields, C. M. , Zvonok, N. , Zvonok, A. , & Makriyannis, A. (2019). Biochemical and proteomic characterization of recombinant human α/β hydrolase domain 6. Scientific Reports, 9, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra, A. Y. , Gratacós, E. , Carrasco, P. , Clotet, J. , Ureña, J. , Serra, D. , … Casals, N. (2008). CPT1c is localized in endoplasmic reticulum of neurons and has carnitine palmitoyltransferase activity. The Journal of Biological Chemistry, 283, 6878–6885. 10.1074/jbc.M707965200 [DOI] [PubMed] [Google Scholar]
- Sigel, E. , Baur, R. , Rácz, I. , Marazzi, J. , Smart, T. G. , Zimmer, A. , & Gertsch, J. (2011). The major central endocannabinoid directly acts at GABAA receptors. Proceedings of the National Academy of Sciences of the United States of America, 108, 18150–18155. 10.1073/pnas.1113444108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somma‐Delpéro, C. , Valette, A. , Lepetit‐Thévenin, J. , Nobili, O. , Boyer, J. , & Vérine, A. (1995). Purification and properties of a monoacylglycerol lipase in human erythrocytes. The Biochemical Journal, 312, 519–525. 10.1042/bj3120519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Z. , Xie, H. , Heier, C. , Huang, J. , Zheng, Q. , Eichmann, T. O. , … Hao, H. (2020). Enhanced monoacylglycerol lipolysis by ABHD6 promotes NSCLC pathogenesis. eBioMedicine, 53, 102696. 10.1016/j.ebiom.2020.102696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchantchou, F. , & Zhang, Y. (2013). Selective inhibition of alpha/beta‐hydrolase domain 6 attenuates neurodegeneration, alleviates blood brain barrier breakdown, and improves functional recovery in a mouse model of traumatic brain injury. Journal of Neurotrauma, 30, 565–579. 10.1089/neu.2012.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokutake, Y. , Iio, W. , Onizawa, N. , Ogata, Y. , Kohari, D. , Toyoda, A. , & Chohnan, S. (2012). Effect of diet composition on coenzyme A and its thioester pools in various rat tissues. Biochemical and Biophysical Research Communications, 423, 781–784. 10.1016/j.bbrc.2012.06.037 [DOI] [PubMed] [Google Scholar]
- Tokutake, Y. , Onizawa, N. , Katoh, H. , Toyoda, A. , & Chohnan, S. (2010). Coenzyme A and its thioester pools in fasted and fed rat tissues. Biochemical and Biophysical Research Communications, 402, 158–162. 10.1016/j.bbrc.2010.10.009 [DOI] [PubMed] [Google Scholar]
- Tornqvist, H. , Krabisch, L. , & Belfrage, P. (1974). Simple assay for monoacylglycerol hydrolase activity of rat adipose tissue. Journal of Lipid Research, 15, 291–294. 10.1016/S0022-2275(20)36810-3 [DOI] [PubMed] [Google Scholar]
- van der Wel, T. , Janssen, F. J. , Baggelaar, M. P. , Deng, H. , den Dulk, H. , Overkleeft, H. S. , & van der Stelt, M. (2015). A natural substrate‐based fluorescence assay for inhibitor screening on diacylglycerol lipase α. Journal of Lipid Research, 56, 927–935. 10.1194/jlr.D056390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esbroeck, A. C. M. , Kantae, V. , Di, X. , van der Wel, T. , den Dulk, H. , Stevens, A. F. , … van der Stelt, M. (2019). Identification of α,β‐hydrolase domain containing protein 6 as a diacylglycerol lipase in neuro‐2a cells. Frontiers in Molecular Neuroscience, 12, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Chanda, P. , Jones, P. G. , & Kennedy, J. D. (2008). A fluorescence‐based assay for Monoacylglycerol lipase compatible with inhibitor screening. Assay and Drug Development Technologies, 6, 387–393. 10.1089/adt.2007.122 [DOI] [PubMed] [Google Scholar]
- Wei, M. , Jia, M. , Zhang, J. , Yu, L. , Zhao, Y. , Chen, Y. , … Zhang, C. (2017). The inhibitory effect of α/β‐hydrolase domain‐containing 6 (ABHD6) on the surface targeting of GluA2‐ and GluA3‐containing AMPA receptors. Frontiers in Molecular Neuroscience, 10, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, M. , Zhang, J. , Jia, M. , Yang, C. , Pan, Y. , Li, S. , … Zhang, C. (2016). α/β‐hydrolase domain‐containing 6 (ABHD6) negatively regulates the surface delivery and synaptic function of AMPA receptors. Proceedings of the National Academy of Sciences, 113, E2695–E2704. 10.1073/pnas.1524589113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, J. , Ribeiro, R. , Tanaka, M. , & Zhang, Y. (2015). Activation of CB2 receptor is required for the therapeutic effect of ABHD6 inhibition in experimental autoimmune encephalomyelitis. Neuropharmacology, 99, 196–209. 10.1016/j.neuropharm.2015.07.010 [DOI] [PubMed] [Google Scholar]
- Wolfgang, M. J. , Kurama, T. , Dai, Y. , Suwa, A. , Asaumi, M. , Matsumoto, S.‐I. , … Lane, M. D. (2006). The brain‐specific carnitine palmitoyltransferase‐1c regulates energy homeostasis. Proceedings of the National Academy of Sciences of the United States of America, 103, 7282–7287. 10.1073/pnas.0602205103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang, M. J. , & Lane, M. D. (2011). Hypothalamic malonyl–CoA and CPT1c in the treatment of obesity. The FEBS Journal, 278, 552–558. 10.1111/j.1742-4658.2010.07978.x [DOI] [PubMed] [Google Scholar]
- Zaugg, K. , Yao, Y. , Reilly, P. T. , Kannan, K. , Kiarash, R. , Mason, J. , … Mak, T. W. (2011). Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes & Development, 25, 1041–1051. 10.1101/gad.1987211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvonok, N. , Pandarinathan, L. , Williams, J. , Johnston, M. , Karageorgos, I. , Janero, D. R. , … Makriyannis, A. (2008). Covalent inhibitors of human Monoacylglycerol lipase: Ligand‐assisted characterization of the catalytic site by mass spectrometry and mutational analysis. Chemistry & Biology, 15, 854–862. 10.1016/j.chembiol.2008.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Representative FRET images of transfected HEK‐293 T cells with proteins fused to mTurquoise2 (donor; blue) or SYFP2 (acceptor; yellow) before and after bleaching.
Figure S2. FRET sensitized emission of CPT1C with ABHD6 in the presence of an ACC inhibitor in living cells. Assays were performed 48 h post‐transfection in HEK‐293 T cells expressing ABHD6‐mTurq2 and increasing amounts of the cDNA for CPT1C‐SYFP2, in the presence or absence of the ACC inhibitor, TOFA (20 μg/ml, 2 h). FRET saturation curves were obtained by monitoring the SYFP2 fluorescence emission at 530 nm after excitation of mTurq2 at 420 nm, with subtraction of the values obtained with cells expressing the same amount of donor protein. Mean±SD (n = 4).
Figure S3. ABHD6 assay based on 4‐MUH hydrolysis in hypothalamus of WT and CPT1C‐KO mice. ABHD6 activity dependent on 4‐MUH concentration at t = 30 min in the absence (A) or presence of 10 μM WWL70 (B). Two‐way ANOVA followed by Bonferroni's post‐hoc correction was used for statistical analysis. Mean±SD (n = 6). *P < 0.05 versus WT.
Figure S4. Kinetic hydrolase activity (A) and areas under the curve (B) in the absence or presence of the ABHD6 inhibitors WWL70 and KT182. Mean±SD (n = 7). *P < 0.05 versus Vehicle.
Figure S5. ABHD6 activity assay based on 4‐MUH hydrolysis. (A) Blotting and protein expression levels of ABHD6 in combination with CPT1C, M589S or ΔCter in HEK‐293 T for ABHD6 activity assays (n = 15). (B) 4‐MUH hydrolysis of HEK‐293 T cells expressing EV, CPT1C, M589S and Δcter (n = 4). (C) HEK‐293 T endogenous endocannabinoids (eCBs) levels, 2‐AG and AEA, when transfected with EV, ABHD6 or ABHD6‐CPT1C (n = 15). Mean±SD.
Figure S6. ABHD6 activity assay based on 4‐MUH hydrolysis in a neuronal cell line, GT1–7, endogenously expressing ABHD6 and CPT1C. Time course ABHD6 activity assay (A) and areas under the curve (B) in GT1–7 cells with or without TOFA treatment (20 μg/mL) for 1 h and 2 h. Mean±SD (n = 6). *P < 0.05 versus vehicle.
Figure S7. Intracellular cAMP assay was set up in HEK‐293 T. (A) Forskolin (FK)‐induced cAMP levels of HEK‐293 T expressing CB1R incubated with an agonist of CB1R, ACEA, an antagonist, rimonabant (Rimo), Rimo + 2‐AG and increasing concentrations of 2‐AG, arachidonic acid (ARA) and glycerol. (B) Effect of ACEA, 2‐AG, ARA and glycerol in basal cAMP levels of HEK‐293 T cells alone. Mean±SD (n = 8). + P < 0.05 versus FK and *P < 0.05 versus basal.
Figure S8. ABHD6 assay based on 4‐MUH hydrolysis in brain tissue homogenates of WT or CPT1C‐KO male mice in the absence or presence of the ABHD6 inhibitor, WWL70 (10 μM). (A) 4‐MUH hydrolysis in hypothalamic homogenates. (B) 4‐MUH hydrolysis in hippocampal homogenates. Mean±SD (n = 6).
Figure S9. CPT1C and ABHD6 protein expression on brain tissue homogenates of fed or fasted mice. Representative blots (A) and quantification (% relative to WT‐fed) of CPT1C (B) and ABHD6 (C) protein levels in either hypothalamus (Hyp) or hippocampus (Hc) of WT mice under fed (n = 6) or fasting conditions (n = 5). Mean±SD.
Figure S10. Effect of fasting on ABHD6 activity of female WT brain tissues. (A, B) ABHD6 activity in hypothalamic homogenates and area under the curves of mice after fed or fasted state. (C, D) ABHD6 activity in hippocampal homogenates and area under the curves of mice after fed or fasted state. Mean±SD (n = 5). *P < 0.05 versus WT‐fed.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Some data may not be made available because of privacy or ethical restrictions.


