Abstract
pH controls a large repertoire of chemical and biochemical processes in water. Densely arrayed pH microenvironments would parallelize these processes, enabling their high-throughput studies and applications. However, pH localization, let alone its arrayed realization, remains challenging because of fast diffusion of protons in water. Here, we demonstrate arrayed localizations of picoliter-scale aqueous acids, using a 256-electrochemical cell array defined on and operated by a complementary metal oxide semiconductor (CMOS)–integrated circuit. Each cell, comprising a concentric pair of cathode and anode with their current injections controlled with a sub-nanoampere resolution by the CMOS electronics, creates a local pH environment, or a pH “voxel,” via confined electrochemistry. The system also monitors the spatiotemporal pH profile across the array in real time for precision pH control. We highlight the utility of this CMOS pH localizer-imager for high-throughput tasks by parallelizing pH-gated molecular state encoding and pH-regulated enzymatic DNA elongation at any selected set of cells.
A CMOS electrochemical cell array achieves picoliter-scale pH localization for high-throughput pH-regulated chemistry.
INTRODUCTION
pH, the proton concentration in an aqueous solution, regulates a large number of natural and engineered biochemical processes, such as protein activities (1, 2), cell signaling (3), viral infection (4, 5), tumor growth (6, 7), and DNA deprotection (8–10), to name a few. A platform that can localize target pH values in a densely arrayed manner can parallelize such pH-dependent processes in aqueous solutions for high-throughput experiments and applications. A timely example is a high-throughput enzymatic DNA synthesis. Whereas phosphoramidite DNA synthesis in a nonaqueous solution has been the dominant synthesis method (11–13), enzymatic synthesis in an aqueous solution is being actively pursued, for it may reduce cost, molecular damages, and hazardous wastes (14–16). As pH can gate each nucleotide incorporation step in enzymatic synthesis (9, 10), densely arrayed pH localization could drive it in high throughput.
A microelectrode array, which can electrochemically produce protons at any number of electrodes used as anodes, is a natural platform with which arrayed pH localization can be pursued. Previous works (17–23) used microelectrode arrays defined on complementary metal oxide semiconductor (CMOS) chips (17–22) or glass substrates (23) to electrochemically produce protons, with all array electrodes used as anodes. They, however, used pH buffers to chemically––as opposed to electrochemically––confine the locally generated protons. The resulting arrayed pH localization was thus not via a fully electrochemical means. Generally, chemical confinement of protons becomes increasingly less effective with higher electrode densities (24, 25), an issue that would be particularly pronounced in water, where protons diffuse notoriously fast (26–28). Aside this issue with the chemical confinement, none of these previous works (17–23) implemented on-chip pH sensors that can measure pH across the array in real time; in one of the works (23), pH was quantified in real time, but it was based on fluorescence pH indicators with off-chip optics.
Here, we fully electrochemically realize densely arrayed pH localization in an aqueous solution. We use an array of 256 electrochemical cells, or “pixels,” constructed on a CMOS microelectrode array (Fig. 1A). Each pixel, consisting of two concentric metallic rings, cannot only electrochemically generate protons with a positive current through the inner anodic ring but also electrochemically create a basic wall with a negative current through the outer cathodic ring. The basic wall prevents the diffusion of the protons, confining them within a picoliter-scale volume, thus creating an acidic pH “voxel” above the CMOS surface pixel. The positive and negative currents are defined by the underlying CMOS electronics with a sub-nanoampere resolution, and an optimal electrochemical balance to create and maintain the acidic pH voxel is found when the anodic and cathodic currents are of the same magnitude. By altering this magnitude, the local acidic pH value can be tuned electrochemically with a high precision and in a repeatable manner. Last, each pixel is randomly accessible, and thus, any set of pixels can be chosen to create pH voxels, allowing for spatioselective pH programming.
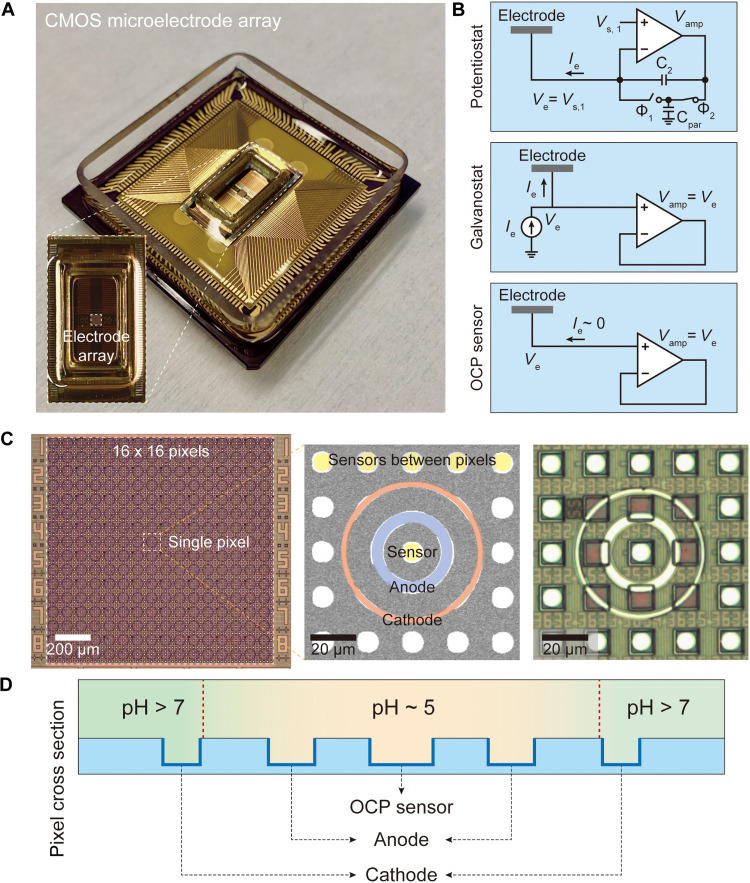
Fig. 1. CMOS electrochemical cell array.
(A) Image of the packaged CMOS IC featuring a predefined array of 64 × 64 = 4096 Al pads. (B) The CMOS circuit connected to each Al pad can be configured into one of the three modes: potentiostat, galvanostat, and OCP sensor. (C) Pt electrodes are postfabricated on top of the predefined Al pad array, resulting in an array of 16 × 16 = 256 electrochemical cells or pixels. Each pixel consists of an inner anodic Pt ring electrode connected to a set of four underlying Al pads, an outer cathodic Pt ring electrode connected to another set of four underlying Al pads, and a center OCP sensor with a circular Pt electrode connected to one underlying Al pad. In addition, circular Pt electrodes (with each connected to one underlying Al pad) are postfabricated in between pixels to be used as additional OCP sensors. A scanning electron microscope image and an optical image of a pixel and its surrounding are shown in the middle and at the right, respectively. (D) In any given pixel, if both of its two concentric rings are operated in the galvanostat mode with the outer cathodic ring with a negative current injection and the inner anodic ring with a positive current injection, the base generated by the former serves as an electrochemical wall that confines the protons electrochemically generated by the latter.
Our system also electrochemically measures pH in real time with open circuit potential (OCP) sensors integrated across the CMOS pixel array. This monitoring of the spatiotemporal pH profile enables the precision pH tuning. A variety of CMOS pH sensors and their arrays have already been developed over the past decades, with a notable example being CMOS ion-sensitive field-effect transistor (ISFET) pH sensors (29–39). Our OCP sensors may be viewed as a variant of such CMOS pH sensors. The key point, however, is the cointegration of the pH sensing and pH localization capabilities to enable high-precision control of pH across the array.
Our fully electrochemical aqueous pH localizer-imager can be compared also to microelectrode arrays for nonaqueous acid localization (24, 25, 40), which were used for acid-controlled phosphoramidite DNA synthesis to build DNA microarrays (24, 25) or pursue DNA data storage (40). A previous work (24, 25) fabricated two types of electrode arrays on CMOS chips. In one, a concentric pair of anodic disk and cathodic ring reminiscent of our structure is arrayed, but with no experimental details reported; its ability for fully electrochemical acid localization is unknown. In the other, while each anodic electrode is surrounded by four nearest cathodic electrodes, the anode-generated acids were confined not only by a basic wall electrochemically produced with the cathodic electrodes but also with a chemical aid using organic base molecules. Another work (40) reported a fully electrochemical acid localization with a local anodic electrode surrounded by nearby common cathodes. However, as each electrochemical cell was not randomly accessible, only a very limited spatial pattern of acid localization was possible. Moreover, as none of these works (24, 25, 40) implemented on-chip acid sensors or used fluorescence acid measurements, no real-time acid monitoring was done, rendering the accuracy of acid localization and control unknown. In summary, even setting aside whether the localization is done in aqueous or nonaqueous solutions, our work is distinctive for its explicit demonstration of fully electrochemical localization, random access to each electrochemical cell, and real-time spatiotemporal quantification for precision control, while each of these works (24, 25, 40) achieves only some, but not all, of these traits.
To highlight the utility of our CMOS pH localizer-imager, we performed pH-regulated enzymatic incorporation of nucleotides to single-stranded DNA molecules in parallel at any selected locations in the 256-pixel array, exemplifying a high-throughput control of aqueous biochemical processes enabled by the densely arrayed and spatioselective pH localization. This example, the pH-regulated enzymatic DNA elongation, is an aqueous counterpart to the acid-regulated phosphoramidite DNA elongation in a nonaqueous solution (24, 25, 40) and can have a powerful application in enzymatic DNA synthesis for such applications as molecular data storage.
RESULTS
CMOS electrochemical cell array
The CMOS integrated circuit (IC) (41, 42), on which the array of 16 × 16 = 256 electrochemical cells is postfabricated, features on its surface an array of 64 × 64 = 4,096 aluminum (Al) pad electrodes in its foundry fabricated form, with an individual pad dimension of 10.5 × 10.5 μm and a pad-to-pad (center-to-center) pitch of 20 μm (see Materials and Methods). Each pad is connected to its own electronic circuit integrated in the underlying CMOS chip, which is individually addressable and highly configurable (Fig. 1B). The pad electronics can be configured as a galvanostat that can inject a current and simultaneously measure the resulting voltage of the pad electrode, a potentiostat that can apply a voltage and concurrently measure the resulting current through the pad electrode or an OCP sensor that can measure the voltage of the pad electrode while blocking a current flow. This CMOS IC has previously been designed and used with vertical nanoelectrodes postfabricated on the pads for highly sensitive intracellular recording of neuronal membrane potentials (a varied version of the galvanostat configuration was used there for current clamp recording) for fundamental and applied neuroscience (41–43). In the present work, its application is changed to electrochemical pH control and imaging by transforming, via postfabrication, its surface into the new 256-electrochemical-cell array structure.
Each cell, or pixel, consists of an inner anodic platinum (Pt) ring and an outer cathodic Pt ring arranged in a concentric geometry (Fig. 1, C and D). Each ring is postfabricated to make contacts with four Al pads, and the four corresponding CMOS circuits are used together as one effective circuit. pH manipulation is enabled by interfacing the chip surface with an aqueous solution of the quinone redox couple––2,5-dimethyl-1,4-hydroquinone (DMHQ) and 2,5-dimethyl-1,4-benzoquinone (DMBQ) (44–46)––and by making the concentric electrochemical cell induce redox reactions (see Materials and Methods for the solution composition). To create the local pH voxel above a pixel, both anodic and cathodic rings of the pixel are configured in the galvanostat mode, where the anodic ring injects a positive current to oxidize DMHQ into DMBQ for the generation of protons and the cathodic ring injects a negative current to reduce DMBQ for the generation of base molecules (Fig. 2A; see also fig. S1 for a full description of the relevant chemical reactions). The base molecules generated by the outer cathodic ring serves as an electrochemical wall that confines the protons generated from the inner anodic ring (Fig. 1D), thus creating an acidic pH voxel in the local volume above around the pixel center.
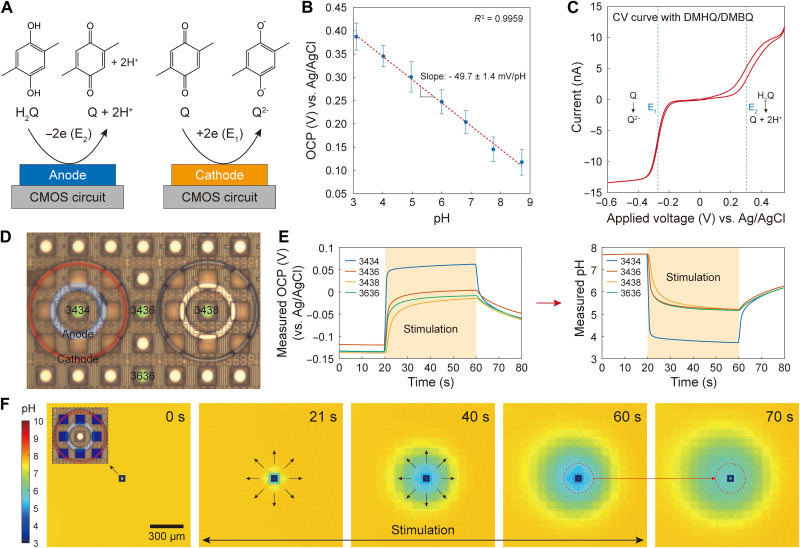
Fig. 2. pH manipulation with quinone chemistry and proton diffusion.
(A) Illustration of redox reactions of DMHQ or H2Q for brevity and DMBQ (or Q). The oxidation reaction generates protons, while the reduction reaction generates quinone dianions (Q2−), which serve as base. (B) OCP versus pH calibration. The OCP measured by our OCP sensors with the circular Pt electrodes has a linear relationship to pH with an experimentally determined sensitivity of −49.7 ± 1.4 mV/pH. This calibration was conducted by measuring OCP of various pH buffer solutions. Only the slope of this calibration line is used in converting OCP to pH (for details, see Materials and Methods). (C) Cyclic voltammogram at a single circular Pt electrode (which in this case is used not in the OCP sensing mode but in the potentiostat mode) with a scan range from −0.6 to 0.5 V and a scan rate of 20 mV/s. (D) A positive current is injected into the anodic ring around pad “3434” (i.e., the Al pad located at row 34 and column 34), with the results presented in parts (E) and (F). (E) The OCP measured with the OCP sensor at pad 3434 (left) and its conversion to pH (right) using the calibration result of part (B) (see Materials and Methods). (F) Spatiotemporal pH imaging during the stimulation of the single anodic ring around pad 3434. The temporal evolution of the pH heatmap shows a radial diffusion of protons generated by the anodic stimulation. The ceiling height is about 39 μm.
In this electrochemical approach to pH localization, the anodic and cathodic rings in a pixel function independently as two working electrodes (WEs). In the WE/WE configuration, positive and negative current injections can have different magnitudes, with the charge balancing completed by an external large-scale Ag/AgCl pseudo–reference electrode located outside the electrode array (see fig. S2 for a detailed description of the pseudo–reference electrode). This enables an independent tuning of each of the anodic and cathodic currents to achieve an optimal acidic pH confinement. The optimal current ratio is found to be 1:−1 for the concentric ring pair; pH is successfully localized when anodic and cathodic currents are equal and opposite.
The CMOS electrochemical cell array can also measure the spatiotemporal profile of pH across the array in real time using OCP sensors. This is achieved by implementing OCP sensors both at pixel centers and in between pixels (Fig. 1C): overall, the array features a total of 2048 OCP sensors. Each OCP sensor is a circular Pt electrode postfabricated on an Al pad, whose corresponding CMOS circuit is configured into the OCP sensing mode. Here, Pt was chosen instead of commonly used and more pH-sensitive metal oxides, such as iridium oxide and tantalum oxide (47, 48), because of the limitation in our in-house fabrication facility. Nonetheless, the OCP sensor with the Pt electrode exhibits a sufficient pH sensitivity of −49.7 ± 1.4 mV/pH, which is determined on the basis of the principle of potentiometric pH measurement (Fig. 2B; see also Materials and Methods for a detailed description of the pH calculation from the measured OCP) (47, 48). The OCP sensor at each pixel center examines whether a target acidic pH has been reached within the pixel. The localization of this acidic pH within the pixel is verified by the OCP sensors in between pixels, which would measure pH values higher than the in-pixel acidic pH. Notice that this on-chip electronic pH measurement based on the linear pH-OCP relationship has a broader pH monitoring range than the off-chip optical measurement based on the nonlinear pH-fluorescence relation of pH-sensitive fluorescent molecules. Last, we note that while the concentric rings of a pixel used for pH localization are operated in the galvanostat mode, the concentric rings in pixels not engaged in pH localization are operated in the OCP sensing mode for a denser monitoring of the spatial pH profile.
Quinone chemistry
Because quinone chemistry (44–46) is used for pH manipulation, electrochemical reactions of quinone were first characterized in the aqueous solution (see Materials and Methods) using cyclic voltammetry (CV). For this particular experiment, only one circular Pt electrode was activated and was operated in the potentiostat mode as opposed to its usual OCP sensing mode. The CV curve (Fig. 2C) shows that DMHQ and DMBQ are the main redox species. DMBQ (labeled as Q) is reduced to a dimethyl quinone dianion (labeled as Q2−), as the voltage is decreased across a half-wave potential of E1 ~ −0.3 V versus Ag/AgCl. The Q2− dianions then undergo a series of chemical equilibria, generating HQ− and OH− base molecules (fig. S1). On the other hand, as the voltage is increased across a half-wave potential of E2 ~ 0.3 V versus Ag/AgCl, DMHQ (labeled as H2Q) is oxidized into DMBQ, generating protons. For pH localization in a pixel where its concentric rings cooperate in the WE/WE galvanostat configuration, the protons generated from the inner anodic ring are consumed by a volume of the base molecules (i.e., Q2−, HQ−, and OH−) generated from the cathodic ring. That is, the base molecules set up an electrochemical barrier that fastens protons within the pixel, thus achieving pH localization.
Control: Spatiotemporal imaging of proton diffusion
Before demonstrating the pH localization, a control experiment is first presented, where protons are allowed to diffuse away with no electrochemical walls set up. For this control experiment, a positive current was injected from a single anodic ring in one chosen pixel (“stimulation pixel”), and all the rest electrodes––the cathodic ring of the stimulation pixel, all concentric rings of the remaining pixels, and all circular electrodes––were engaged in the OCP sensing mode to perform a spatiotemporal imaging of pH diffusion across the array.
When the anodic ring of the stimulation pixel injected a positive current of 57 nA for 40 s to oxidize DMHQ for proton generation, the pH measured at the OCP sensor inside the stimulation pixel dropped as expected (Fig. 2, D and E). At the same time, the array-wide pH measurement showed that the electrochemically generated acids spread out radially during the current injection (Fig. 2F; see also movie S1; fig. S3 shows another control: when a negative current was injected to the single cathodic ring of the stimulation pixel, electrochemically generated base spread out radially). This demonstrates that the acidic pH created by the anodic ring cannot be confined without an activation of the cathodic ring.
One solution to stopping the proton diffusion is a chemical approach, where one activates the anodic ring only and localizes pH by using a strong pH buffer (17–23). As mentioned earlier, this chemical confinement does not scale well with the electrode density, especially in an aqueous solution. Instead, we use the fully electrochemical approach using both anodic and cathodic rings to obtain pH localization. This full-fledged electrochemical approach for pH confinement is what shall be described now.
Array-wide localized control of pH
Here, a densely arrayed pH localization, i.e., the construction of an array of pH voxels, is demonstrated in an aqueous solution using the concentric pixel array. To start, in a single concentric pixel, 57 nA of current was injected to the anodic ring to produce protons, and − 57 nA of current was injected to the cathodic ring to create an electrochemical wall. Spatiotemporal pH imaging shows that an acidic pH of 5.26 was obtained and maintained within the pixel throughout 40 s of stimulation, during which pH measured anywhere outside this single activated pixel is larger than 7, showing that the acidic pH inside the pixel remains localized, instead of breaking out (Fig. 3, A and B; see also movie S2, left). Figure 3A also shows that the confined acid vanished as soon as the current injections were terminated. The localized pH value at the pixel center can be tuned by varying the anodic and cathodic currents with their ratio kept about 1:−1, where this optimal ratio was found experimentally by leveraging the independent current control of the WE/WE configuration via trial and error (Fig. 3C; also see figs. S4 and S5).
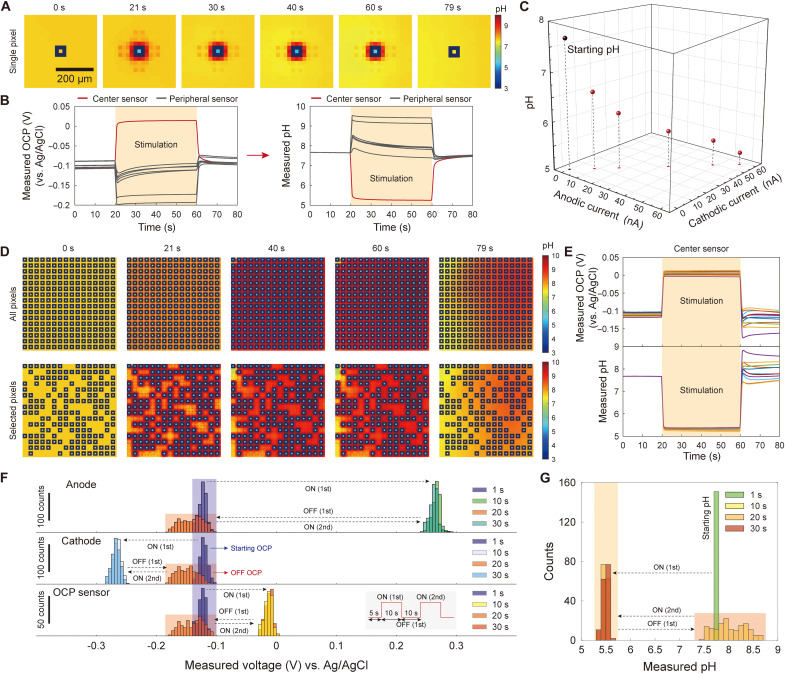
Fig. 3. Array-wide pH localization.
(A) pH localization at a single concentric pixel. (B) OCP and pH measured with the OCP sensor at the center of the pixel of part (A) (red line) and peripheral OCP sensors (gray lines). (C) By altering the magnitude of anodic and cathodic currents in optimal ratio 1:−1, the localized pH value can be tuned. (D) Array-wide pH localization for all 256 pixels (top) and selected pixels (bottom). (E) OCP and corresponding pH measured at centers of randomly sampled pixels in the experiment of (D), top. (F) Distributions of measured potentials at anodes, cathodes, and pixel center OCP sensors during application of two sets of anodic and cathodic current pulses at all 256 pixels. Bin size = 5 mV. The distribution of the measured potentials for each electrode type remains almost identical from the first pulse to the second pulse, indicating that concentration overpotentials do not develop during the stimulation. The absence of concentration overpotentials indicates that the redox species (i.e., quinones) are not depleted, enabling a repeatable pH control. (G) pH converted version of the bottom of (F), i.e., distribution of localized pH values measured at pixel center OCP sensors during the current pulse stimulation. Bin size = 0.1. The distribution of localized pH values remains almost identical from the first pulse to the second pulse. The ceiling height is about 39 μm throughout the figure.
The on-chip OCP sensors measure pH only on the chip surface and provide a limited spatial resolution even on the surface as they are not at every surface position. We thus performed COMSOL simulations to understand the full three-dimensional (3D) pH profile with a single pixel activated (figs. S6 and S7). These simulations confirm the pH localization, or the acidic pH voxel formation, with a steep pH gradient from within to outside the voxel, and also show that the cross-sectional area of the acidic pH voxel shrinks as the distance from the chip surface is increased. This specific 3D pH profile––and the associated 3D proton concentration gradient profile––is a direct consequence of the concentric ring geometry (different electrochemical cell structures would lead to different concentration gradient profiles) (49–53).
pH can also be localized in any number of chosen pixels, where each chosen pixel is still activated with the anodic and cathodic current ratio of 1:−1 optimized for the single pixel pH localization. Fig. 3D and corresponding movie S2 (middle and right) show two pH localization patterns, and fig. S8 shows yet two additional pH localization patterns (see also movie S3 that juxtaposes the cases where all-pixel pH localization succeeds and fails, with the latter obtained by detuning the anodic and cathodic current ratio from 1:−1; see also fig. S9 for all-pixel pH localization with no ceiling; last, see movie S4 for comparison with a different electrode structure that is more susceptible to pixel-to-pixel cross-talk). The pH values within all chosen pixels were close to one another (Fig. 3E). For example, when activating every concentric pixel on the array (Fig. 3D, top), the distribution of pixel center pH values had a median of 5.33 and an SD of 0.03 after 20 s (half point) of stimulation. This precision pH control with the array-wide localization could be achieved for various solution compositions, even with no pH buffering at all (fig. S10). The pH can be controlled with such accuracy because the magnitude of the optimal 1:−1 ratio of cathodic and anodic currents of the concentric rings can be tuned with a sub-nanoampere resolution because of the particular CMOS design of the current injector in our galvanostat configuration (42). (In the galvanostat mode here, voltages can be measured at the anodic and cathodic rings during the current injection. As seen in fig. S11, these voltages are constant during the current injection, which shows that the injected currents are Faradaic.) This precision current control is critical, given the high dependence of the localized pH value on the current magnitudes, i.e., the rates of redox reactions (fig. S5) (54, 55). [It would be interesting to explore in the future the use of the potentiostat configuration for the activation of the concentric rings for the high-precision pH localization and control, as opposed to the galvanostat configuration as used here (56, 57).]
The local pH control can be repeated multiple times. To test, two successive sets of square pulses of anodic and cathodic currents were applied to all 256 pixels (57 nA in anodic rings and − 57 nA in cathodic rings), with each set of pulses having a duration of 10 s and the time lapse between the two sets of pulses being 10 s. At the same time, potentials at all anodic and cathodic rings were monitored (Fig. 3F, top and middle), which was possible as they operated in the galvanostat mode (these potentials measured with current injections are not OCPs and are difficult to convert to pH). The distribution of the measured potentials at anodes and cathodes remained almost identical between the two sets of pulses (Fig. 3F, top and middle) and did not show any hint of concentration overpotentials (58, 59). This shows that redox species (i.e., quinones) were not significantly depleted to the extent of rendering the control of the reaction dynamics unrepeatable. Owing to this repeatable control, the distribution of OCPs (Fig. 3F, bottom) or the corresponding distribution of pH values (Fig. 3G), measured at all 256 pixel center OCP sensors, also remained almost identical between the two sets of pulses, despite the wide variation of starting pH values before the second pulse (see also fig. S12 for repeated pH localization at a single pixel in different solution compositions) (Fig. 3G).
Parallelizing pH-regulated processes
The utility of this repeatable, arrayed creation of pH voxels in parallelizing pH-regulated chemical processes in aqueous solutions is now demonstrated with two examples. First, a parallelized pH-gated molecular state encoding is demonstrated. Fluorescein, a widely used pH indicator (60, 61), is chosen as the demonstration molecule because its pH-gated state change can be cross-checked with fluorescence. An individual fluorescein molecule can be either in the unprotonated anionic state “0,” which fluoresces, with a probability P0, or in the fully protonated neutral state “1,” which does not fluoresce, with a probability P1 = 1 – P0 (fig. S13). The probability P0 increases with pH and saturates to 1 as pH exceeds 6.65 in the 10 μM aqueous fluorescein solution (see Materials and Methods). Thus, the collective fluorescence intensity, which is proportional to P0, increases with pH and saturates to the maximum as pH exceeds 6.65, creating a plateau where all fluorescein molecules are in 0 with P0 = 1 (Fig. 4A). Conversely, as pH is lowered from 6.65, P0 decreases, reducing the collective fluorescence intensity. Therefore, when acidic pH voxels are formed above a select set of pixels (fig. S14), the fluorescence intensity from each voxel will be reduced, as an appreciable fraction of fluorescein molecules there will be encoded into state 1. In contrast, the fluorescein molecules in the remaining inactivated pixels will preferentially remain in the state 0, exhibiting stronger fluorescence intensity.
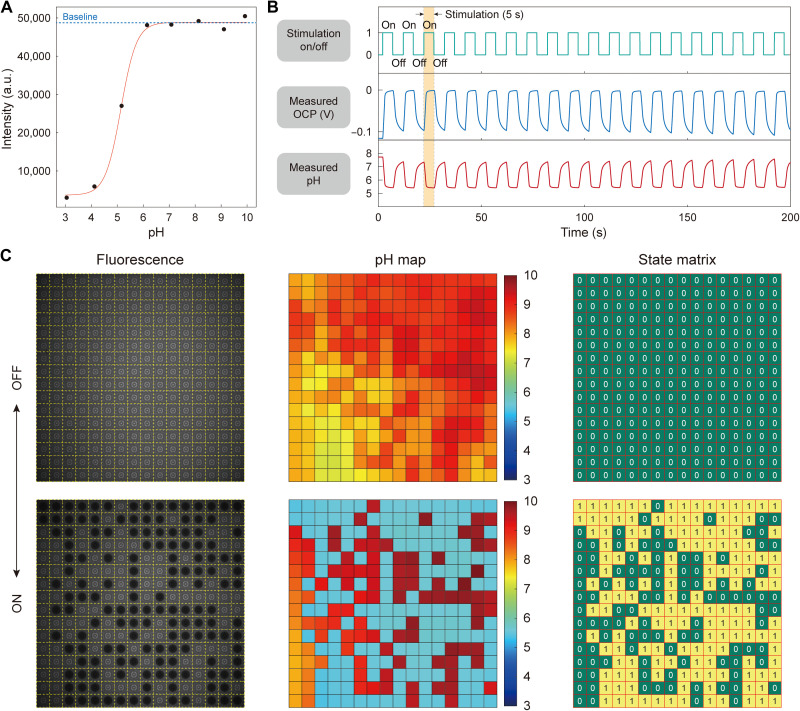
Fig. 4. Parallelizing pH-gated molecular state encoding.
(A) Epifluorescence intensity versus pH for an aqueous fluorescein solution (10 μM). (B) On-chip measured pixel-center OCP and pH as a function of time in an example stimulated pixel. (C) Epifluorescence imaging and on-chip pH imaging juxtaposed side by side (left and middle). In each of the pixels selected for stimulation, an appreciable fraction of fluorescein molecules enter state “1” every time acidic pH voxels are formed (e.g., pH 5.54 converts approximately 20% to state 1), while nearly all molecules are left in state 0 in each of the unstimulated pixels. In the state matrix (right), matrix element 1 refers to a pixel’s collective status where an appreciable fraction of fluorescein molecules are in state 1, whereas matrix element 0 refers to the status where nearly all molecules are in state 0. The ceiling height is about 14 μm. a.u., arbitrary units.
To demonstrate, a sequence of 20 sets of anodic and cathodic current pulses (48 and −48 nA; each pulse was 5 s long, and there was a 5-s time lapse between two adjacent pulses) was applied to an arbitrarily selected group of pixels in parallel. At the same time, pH values at all pixels were monitored using the pixel center OCP sensors. Figure 4B shows the measured OCP and pH versus time in an example stimulated pixel (see also fig. S15 for a similar experiment with 160-s duration current pulses; see also movie S5). The acidic pH values reached within all select pixels were close to one another at any given time, with their distribution nearly identical from pulse to pulse: the median/SD at t = 6 s (1st pulse), 95 s (10th pulse), and 196 s (20th pulse) are 5.54/0.06, 5.58/0.07, and 5.65/0.10, respectively. By concurrently performing epifluorescence measurement of pH, which was consistent with the on-chip electronic pH measurement, we confirmed that in each of these selected pixels, an appreciable fraction of fluorescein molecules converted to state 1 every time acidic pH voxels formed (e.g., approximately 20% of fluorescein molecules were in state 1 for pH 5.54), whereas in each of the unstimulated pixels, nearly all fluorescein molecules remained in state 0 (Fig. 4C). The key point of this first example is the parallel execution of the pH-gated molecular state encoding at any selected pixels.
We remark that for Fig. 4C, a pH localization pattern different from the one shown in Fig. 3D was chosen to emphasize the programmability for random pH localization patterns. Also note that while our wide-field measurement system resolves the lateral fluorescence variation, it does not resolve, but aggregates, the fluorescence variation along the direction vertical to the chip surface. Therefore, the consistency of the array-wide fluorescence pattern with the programmed distribution of the acidic pH voxels confirms that the acidic pH voxels indeed achieve low pH in the vertically averaged sense.
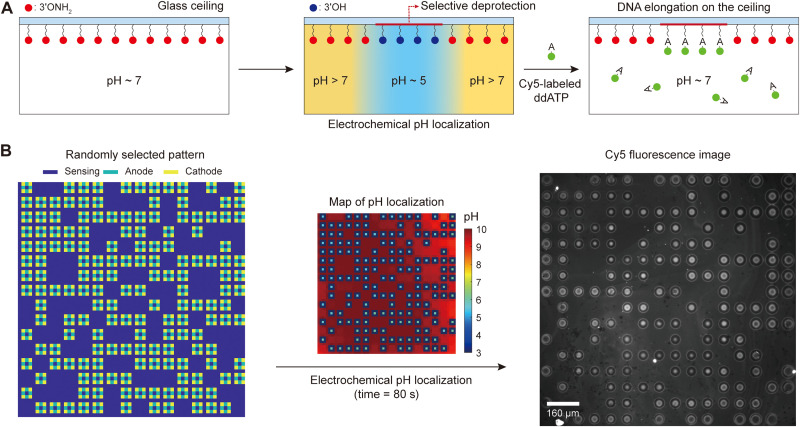
The second example uses the CMOS pH localizer-imager to perform pH-regulated enzymatic incorporation of chain-terminating nucleotides, dideoxyadenosine triphosphate (ddATP), onto substrate single-stranded DNA molecules at any prescribed locations in parallel. The substrate DNA molecules are anchored on a glass ceiling (62, 63) facing the CMOS chip surface at a distance of 14 μm. The acidic pH localization is performed in parallel in a select set of sites to deprotect the DNA strands only on those sites, making them available for the binding of nucleotides. Then, the enzyme, terminal deoxynucleotidyl transferase (TdT), is used as a template-free and template-independent polymerase that catalyzes the incorporation of nucleotides to the deprotected 3′OH ends of the substrate DNA molecules. As described earlier, this pH-regulated enzymatic DNA elongation is an aqueous counterpart to the acid-regulated phosphoramidite DNA elongation in a nonaqueous solution. Our platform parallelizes this aqueous pH-regulated enzymatic DNA elongation.
To start, single-stranded DNA molecules as substrates were anchored to a removable glass ceiling coated with silane-free acrylamide (SFA) (see Materials and Methods). To spatioselectively incorporate nucleotides to these DNA strands, their 3′ONH2-protecting group should be converted to the 3′OH-deprotected group at the selected sites. A sodium nitrite solution buffered to a pH of 5.5 can perform this deprotection step within minutes (8). The quinone solution contained sodium nitrite exactly for this reason (see Materials and Methods), and the deprotection was done by creating locally acidic microenvironments (Fig. 5A) by applying 48 nA of anodic current and −48 nA of cathodic current to an arbitrarily selected group of pixels in parallel for 80 s: the localized pH distribution had a median of 5.59 and an SD of 0.06 after 40 s of stimulation. Subsequently, Cy5-labeled ddATP was enzymatically incorporated into the spatioselectively deprotected strands (see Materials and Methods). The measured Cy5 fluorescence pattern was identical to the pixel activation pattern (Fig. 5B), which attests to the enzymatic nucleotide incorporation at the selected sites. To emphasize the programmability for random pH localization patterns once again, for Fig. 5B, a localization pattern different from those of Figs. 3D and 4C was chosen. The Cy5 fluorescence signals were localized at each selected site, indicating successful pH localization at the ceiling. On the other hand, the variation of the fluorescence intensity and shape at the Cy5-labeled sites might have been caused by the nonuniform surface coating and, thus, nonuniform distribution of substrate DNA strands on the glass, and/or by the site-to-site pH variation, albeit small. This pH variation might have arisen, for example, from the postfabricated process variations of electrodes and/or from the pixel-to-pixel variations of the CMOS control electronics, with which the 3D pH profiles of acidic pH voxels created would not be exactly identical to one another.
Fig. 5. Parallelizing pH-regulated enzymatic incorporation of nucleotides (ddATP) onto single-stranded DNA molecules.
(A) Schematic illustration of pH-regulated enzymatic incorporation of nucleotides to substrate DNA molecules on a glass ceiling with spatioselectivity. Localized pH control induces a spatioselective deprotection of DNA strands. These deprotected strands can then be enzymatically elongated with Cy5-labeled ddATP nucleotides in parallel. The ceiling height is about 14 μm. (B) After parallelized enzymatic incorporation of Cy5-labeled ddATP to spatioselectively deprotected sites, epifluorescence imaging shows that the Cy5 fluorescence pattern (right) matches the randomly selected current stimulation pattern (left) exactly, thus confirming the enzymatic incorporation of nucleotides at the spatioselectively deprotected sites. The pH map at the midpoint of the 80-s-long stimulation is shown in the middle.
DISCUSSION
We have reported an advance in scalable pH microelectrochemistry in an aqueous solution. An array of 256 electrochemical cells with concentric geometry postfabricated on and operated by a CMOS IC can fully electrochemically localize pH at any arbitrarily selected subset of the 256 sites in a parallel fashion. This electrochemical pH localizer also performs a real-time measurement of the spatiotemporal pH profile, which allows for precise tuning of localized pH values. We demonstrated the utility of this fully electrochemical pH localizer-imager first by parallelizing pH-gated molecular state encoding and then by parallelizing pH-regulated enzymatic DNA elongation with the spatioselection programmability. This parallel execution of pH-regulated enzymatic DNA elongation can now be substantially expanded to densely arrayed, high-throughput enzymatic synthesis of DNA in a mild aqueous medium, which can provide access to a range of molecular biology tools (14–16) and also can be applied for DNA data storage and archiving (15, 16, 40, 64). In addition, because pH is a fundamental parameter in manipulating many other processes in chemistry and biology, the highly densified and parallelized pH control may enable high-throughput work (14–25, 62–68) for a broad variety of pH-regulated chemical and biochemical processes beyond enzymatic DNA synthesis. The number and density of electrochemical cells can be readily increased, for scalability is at the heart of CMOS technology. Scaling the electrochemistry itself along with such CMOS scaling will stimulate new inquiries at the research frontier. Last, from a broader point of view, CMOS electronics has been increasingly used for biological and biotechnological applications in direct interface with electrolytes, molecules, and cells (29–39, 41–43, 69–77), and this work adds an advance in this direction of applying CMOS technology to biology and biotechnology.
MATERIALS AND METHODS
Postfabrication and packaging of the CMOS electrochemical cell array
The custom-designed CMOS IC was outsourced to United Microelectronics Corporation for fabrication in 0.18-μm technology. Each chip from the foundry has an array of 64 × 64 = 4096 Al pads (10.5 × 10.5 μm square) with a pitch of 20 μm and a passivation layer. An array of 16 × 16 = 256 concentric pixels was postfabricated onto this predefined array with the following steps. First, photolithography was used to define a desired pattern for circle electrodes (diameter: 8 μm), anodic rings (inner and outer diameters: 26 and 36 μm), and cathodic rings (inner and outer diameters: 58 and 62 μm). Then, the foundry passivation layer was removed to expose the Al pads via reactive-ion etching. Last, a thick metal layer (15-nm Ti and 200-nm Pt) was sequentially deposited via sputtering. After the electrode fabrication, a 14-μm-thick SU-8 spacer layer was fabricated above and below the array with 1-mm spacings via photolithography. After the postfabrication, the chip was wire bonded to a chip carrier (Spectrum Semiconductor Materials, San Jose, CA) via a custom-designed interposer printed circuit board (PCBWay, Shenzhen, China). A glass outer ring (Friedrich & Dimmock, Millville, NJ) and a laser-cut acrylic inner ring were glued to the chip carrier and the chip, respectively, with polydimethylsiloxane (PDMS). PDMS was then poured in between the rings to encapsulate the wire bonds and metal interconnect lines, as shown in fig. S2.
Array-wide pH localization
The electrochemical setup (fig. S2) was first filled with a quinone solution, consisting of 10 mM DMHQ (Alfa Chemistry, Ronkonkoma, NY), 5 mM DMBQ (Sigma-Aldrich, Atlanta, GA), 1 M NaCl (aq), 1.1 M NaNO2 (aq), and 5% (v/v) dimethyl sulfoxide (DMSO). Here, NaNO2 (aq) is a weak pH buffer and was added for the demonstration of pH-regulated DNA deprotection (Fig. 5B). In the case of an epifluorescence measurement with fluorescein molecules, 10 μM fluorescein disodium (VWR International, Pittsburgh, PA) was additionally added to the quinone solution. Then, a glass ceiling was placed on top of the electrode array at a fixed ceiling height, which was determined by the thickness of the spacer layer. For a ceiling height of 39 μm, a 25-μm-thick Kapton tape (McMaster Carr, Princeton, NJ) was used together with a 14-μm-thick SU-8 spacer layer. For a ceiling height of 14 μm, only the SU-8 layer was used. An Ag/AgCl electrode was used as a pseudo–reference electrode, serving the roles of both a reference electrode and a counter electrode. Given this setup, a desired set of stimulation currents was applied to the anodic and cathodic rings at the selected pixels for array-wide pH localization, while OCP sensors monitored the spatiotemporal pH profile concurrently.
To convert the measured OCP to pH, pH of the quinone solution was first measured with an external pH meter. Then, the starting OCP values were measured across the array. The starting OCP values might vary as a result of surface inhomogeneity, resulting from nonuniform adsorption of impurities on the electrodes for instance. We assumed that despite this variation, all starting OCP values corresponded to the same pH (i.e., the solution pH measured with an external pH meter), because no reaction had occurred yet to cause any pH change. Once the current stimulation began, the pH change was calculated by dividing the measured OCP change by the experimentally determined pH sensitivity (−49.7 mV/pH) of the Pt electrodes, and then added to the solution pH to obtain the resulting pH at each OCP sensor. Note that we use only the slope (i.e., pH sensitivity) of the calibration curve (Fig. 2B); unlike the universal pH buffer solutions used for calibration, the pH modulation solution contains quinones as well as sodium nitrite, which add an offset to the measured OCP while the slope remains the same.
Enzymatic incorporation of nucleotides to substrate DNAs on a removable glass ceiling
SFA coating of glass ceilings
A glass ceiling was cleaned sequentially in 1 M NaOH (aq), 0.1 M HCl (aq), and ethanol. In between the solutions, the ceiling was rinsed with deionized water. Then, it was treated with vacuum plasma. A 2% (v/v) acrylamide (VWR International, Pittsburgh, PA) solution was made by dissolving 1.3 g of acrylamide in 65 ml of deionized water and was purged with nitrogen gas for 15 min. Then, 103 mg of N-(5-bromoacetamidylpentyl) acrylamide (Ark Pharma Scientific Ltd., Cambridge, UK) was dissolved in 1.07 ml of N,N-dimethyl formamide and then added to the acrylamide solution. Subsequently, 75 μl of PlusOne tetramethylethylenediamine catalyst (VWR International, Pittsburgh, PA) and 0.65 ml of potassium persulfate (0.05 g/ml) were sequentially added to the acrylamide solution. Cleaned glass ceilings were placed in the final acrylamide solution for 90 min. After 90 min of coating, the slides were rinsed with deionized water and dried with nitrogen gas.
Coupling DNA to SFA-coated glass ceilings
Single-stranded DNA molecules (20 μM; 5′thiophosphate-TTT-TTT-TTT-TrUT-TTT-FAMdTTG-TGA-GAG-TGA-AAT-GAG-G, Eurogentec, Seraing, Belgium) in 10 mM phosphate-buffered saline (PBS) solution was spotted on an SFA-coated glass ceiling for an hour. Then, the glass ceiling was sequentially rinsed with 0.1 M PBS buffer, 10 mM tris/10 mM EDTA buffer at pH 8.0, and deionized water. Last, it was dried with nitrogen gas.
Spatioselective DNA deprotection
To change the 3′ end of the coupled DNA strands from 3′OH to 3′ONH2, a DNA-coupled glass ceiling was spotted for 5 min with an enzymatic solution, consisting of 500 μM dTTP-3′ONH2 (DNA Script, Paris, France), 8 μM terminal deoxynucleotidyl transferase (TdT) enzyme (Mutant “F6,” DNA Script, Paris, France), 1 mM CoCl2, and 1× TdT buffer (DNA Script, Paris, France). Here, the TdT enzyme serves as a template-independent polymerase that catalyzes the addition of deoxynucleotides to the 3′OH end of DNA molecules. Then, the glass ceiling was washed with a quinone solution, consisting of 10 mM DMHQ (Alfa Chemistry, Ronkonkoma, NY), 5 mM DMBQ (Sigma-Aldrich, Atlanta, GA), 1 M NaCl (aq), 0.7 M NaNO2 (aq), and 5% (v/v) DMSO. The same quinone solution was used for pH control. After placing the glass ceiling on top of the electrode array, 48 nA of anodic current and −48 nA of cathodic current were applied to a group of randomly selected pixels for 80 s to spatioselectively deprotect the DNA strands’ 3’ONH2 protecting group. After deprotection, the glass ceiling was washed with deionized water and dried with nitrogen gas.
Enzymatic elongation of the deprotected DNA strands
For elongation of the deprotected strands with Cy5-labeled ddATP, the glass ceiling was spotted for 5 min with a Cy5-labeling enzymatic solution, consisting of 50 μM ddATP-Cy5 (Jena Bioscience, Jena, Germany), 8 μM TdT enzyme (Mutant F6, DNA Script, Paris, France), 1 mM CoCl2, and 1× TdT buffer (DNA Script, Paris, France). After the elongation, the glass ceiling was washed with deionized water and dried with nitrogen gas. Last, the slide was examined under an epifluorescence microscope to read the Cy5 signals.
Acknowledgments
Funding: This research is based upon work supported in part by the Office of the Director of National Intelligence (ODNI), Intelligence Advanced Research Projects Activity (IARPA), via 2019-19081900002. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of ODNI, IARPA, or the U.S. government. The U.S. government is authorized to reproduce and distribute reprints for governmental purposes notwithstanding any copyright annotation therein. Postfabrication and characterization were performed, in part, at the Center for Nanoscale Systems at Harvard University.
Author contributions: H.S.J., W.-B.J., J.A., A.H., X.G., R.N., H.P., and D.H. conceived the project. H.S.J. and W.-B.J. designed the experiments. H.S.J. and W.-B.J. performed the device postfabrication, packaging, and experiments. J.A., who had designed the CMOS IC, performed initial electrochemistry measurements. J.W., J.A., and H.H. developed the electrochemical control software for the CMOS IC. J.W. assembled the electrochemical control system based upon the CMOS IC. H.S.J., W.-B.J., J.W., A.H., M.F., H.H., and Y.-H.H. analyzed the data. A.H., M.F., and X.G. developed the enzymatic DNA synthesis protocol. D.H., H.P., R.N., and X.G. supervised the project. H.S.J., W.-B.J., H.P., and D.H. wrote the manuscript, and all authors read and discussed it.
Competing interests: H.S.J., W.-B.J., J.W., J.A., H.H., Y.-H.H., H.P., and D.H. are inventors on a provisional patent application related to this work filed by Harvard University (no. 63/210,430, filed 14 June 2021). D.H. is also a Fellow of Samsung Electronics, but there is no competing interest for this work D.H. performed exclusively at Harvard University. A.H., R.N., and X.G are inventors on a provisional patent application related to this work filed by DNA Script (no. WO2020020608A1, filed 5 July 2019). A.H., M.F., and X.G have been employed by DNA Script, including the duration of this work. R.N. consulted for DNA Script for the duration of this work. The other authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The DNA synthesis reagents can be provided by DNA Script pending scientific review and a completed material transfer agreement. Requests for the DNA synthesis reagents should be submitted to X.G. (xg@dnascript.co).
Supplementary Materials
This PDF file includes:
Figs. S1 to S15
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S5
REFERENCES AND NOTES
- 1.Thomas P. G., Russell A. J., Fersht A. R., Tailoring the pH dependence of enzyme catalysis using protein engineering. Nature 318, 375–376 (1985). [Google Scholar]
- 2.Liu Q., Chen L., Zhang Z., Du B., Xiao Y., Yang K., Gong L., Wu L., Li X., He Y., pH-dependent enantioselectivity of d-amino acid oxidase in aqueous solution. Sci. Rep. 7, 2994 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ludwig M.-G., Vanek M., Guerini D., Gasser J. A., Jones C. E., Junker U., Hofstetter H., Wolf R. M., Seuwen K., Proton-sensing G-protein-coupled receptors. Nature 425, 93–98 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Bullough P. A., Hughson F. M., Skehel J. J., Wiley D. C., Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371, 37–43 (1994). [DOI] [PubMed] [Google Scholar]
- 5.Stegmann T., Booy F. P., Wilschut J., Effects of low pH on influenza virus. Activation and inactivation of the membrane fusion capacity of the hemagglutinin. J. Biol. Chem. 262, 17744–17749 (1987). [PubMed] [Google Scholar]
- 6.DiGiammarino E. L., Lee A. S., Cadwell C., Zhang W., Bothner B., Ribeiro R. C., Zambetti G., Kriwacki R. W., A novel mechanism of tumorigenesis involving pH-dependent destabilization of a mutant p53 tetramer. Nat. Struct. Biol. 9, 12–16 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Webb B. A., Chimenti M., Jacobson M. P., Barber D. L., Dysregulated pH: A perfect storm for cancer progression. Nat. Rev. Cancer 11, 671–677 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Hutter D., Kim M.-J., Karalkar N., Leal N. A., Chen F., Guggenheim E., Visalakshi V., Olejnik J., Gordon S., Benner S. A., Labeled nucleoside triphosphates with reversibly terminating aminoalkoxyl groups. Nucleosides Nucleotides Nucleic Acids 29, 879–895 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.A. C. Hiatt, F. Rose, Compositions for enzyme catalyzed template-independent formation of phosphodiester bonds using protected nucleotides, U.S. Patent 6214987B1 (2001).
- 10.J. W. Efcavitch, J. E. Sylvester, Modified template-independent enzymes for polydeoxynucleotide synthesis, U.S. Patent 2016/0108382A1 (2016).
- 11.Beaucage S. L., Caruthers M. H., Deoxynucleoside phosphoramidites-a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 22, 1859–1862 (1981). [Google Scholar]
- 12.McBride L. J., Caruthers M. H., An investigation of several deoxynucleoside phosphoramidites useful for synthesizing deoxyoligonucleotides. Tetrahedron Lett. 24, 245–248 (1983). [Google Scholar]
- 13.Matteucci M. D., Caruthers M. H., Synthesis of deoxyoligonucleotides on a polymer support. J. Am. Chem. Soc. 103, 3185–3191 (1981). [PubMed] [Google Scholar]
- 14.Palluk S., Arlow D. H., Rond T., Barthel S., Kang J. S., Bector R., Baghdassarian H. M., Truong A. N., Kim P. W., Singh A. K., Hillson N. J., Keasling J. D., De novo DNA synthesis using polymerase-nucleotide conjugates. Nat. Biotechnol. 36, 645–650 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Lee H. H., Kalhor R., Goela N., Bolot J., Church G. M., Terminator-free template-independent enzymatic DNA synthesis for digital information storage. Nat. Commun. 10, 2383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee H., Wiegand D. J., Griswold K., Punthambaker S., Chun H., Kohman R. E., Church G. M., Photon-directed multiplexed enzymatic DNA synthesis for molecular digital data storage. Nat. Commun. 11, 5246 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D. D. Montgomery, Electrochemical solid phase synthesis, U.S. Patent 6093302 (2000).
- 18.D. D. Montgomery, Electrochemical solid phase synthesis, U.S. Patent 6280595B1 (2001).
- 19.D. D. Montgomery, Electrochemical solid phase synthesis of polymers, U.S. Patent 6444111B1 (2002).
- 20.K. Maurer, J. Cooper, M. Strathmann, A. Gindilis, Electrode array device having an adsorbed porous reaction layer, U.S. Patent 2006/0102471A1 (2006).
- 21.A. Gindilis, Electrode array device having an adsorbed porous reaction layer, U.S. Patent 9339782B1 (2016).
- 22.Maurer K., Cooper J., Caraballo M., Crye J., Suciu D., Ghindilis A., Leonetti J. A., Wang W., Rossi F. M., Stöver A. G., Larson C., Gao H., Dill K., McShea A., Electrochemically generated acid and its containment to 100 micron reaction areas for the production of DNA microarrays. PLOS ONE 1, e34 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fomina N., Johnson C. A., Maruniak A., Bahrampour S., Lang C., Davis R. W., Kavusi S., Ahmad H., An electrochemical platform for localized pH control on demand. Lab Chip 16, 2236–2244 (2016). [DOI] [PubMed] [Google Scholar]
- 24.K. Maurer, J. J. Cooper, H. S. Fujii, J. Leonetti, Neutralization and containment of redox species produced by circumferential electrodes, U.S. Patent 2016/0354751A1 (2016).
- 25.K. Maurer, J. J. Cooper, H. S. Fujii, J. Leonetti, Neutralization and containment of redox species produced by circumferential electrodes, U.S. Patent 10525436B2 (2020).
- 26.Bernal J. D., Fowler R. H., A theory of water and ionic solution, with particular reference to hydrogen and hydroxyl ions. J. Chem. Phys. 1, 515–548 (1933). [Google Scholar]
- 27.Agmon N., Bakker H. J., Campen R. K., Henchman R. H., Pohl P., Roke S., Thämer M., Hassanali A., Protons and hydroxide ions in aqueous systems. Chem. Rev. 116, 7642–7672 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen M., Zheng L., Santra B., Ko H.-Y., DiStasio R. A. Jr., Klein M. L., Car R., Wu X., Hydroxide diffuses slower than hydronium in water because its solvated structure inhibits correlated proton transfer. Nat. Chem. 10, 413–419 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Milgrew M. J., Hammond P. A., Cumming D. R. S., The development of scalable sensor arrays using standard CMOS technology. Sens. Actuators B Chem. 103, 37–42 (2004). [Google Scholar]
- 30.Milgrew M. J., Riehle M. O., Cumming D. R. S., A large transistor-based sensor array chip for direct extracellular imaging. Sens. Actuators B Chem. 111-112, 347–353 (2005). [Google Scholar]
- 31.Rothberg J. M., Hinz W., Rearick T. M., Schultz J., Mileski W., Davey M., Leamon J. H., Johnson K., Milgrew M. J., Edwards M., Hoon J., Simons J. F., Marran D., Myers J. W., Davidson J. F., Branting A., Nobile J. R., Puc B. P., Light D., Clark T. A., Huber M., Branciforte J. T., Stoner I. B., Cawley S. E., Lyons M., Fu Y., Homer N., Sedova M., Miao X., Reed B., Sabina J., Feierstein E., Schorn M., Alanjary M., Dimalanta E., Dressman D., Kasinskas R., Sokolsky T., Fidanza J. A., Namsaraev E., McKernan K. J., Williams A., Roth G. T., Bustillo J., An integrated semiconductor device enabling non-optical genome sequencing. Nature 475, 348–352 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Nemeth B., Symes M. D., Boulay A. G., Busche C., Cooper G. J. T., Cumming D. R. S., Cronin L., Real-time ion-flux imaging in the growth of micrometer-scale structures and membranes. Adv. Mater. 24, 1238–1242 (2012). [DOI] [PubMed] [Google Scholar]
- 33.C. Giagkoulovits, M. A. Al-Rawhani, B. C. Cheah, C. Martin, C. Busche, L. Cronin, D. R. S. Cumming, Hybrid Amperometric and Potentiometric Sensing Based on a CMOS ISFET Array, paper presented at the 2017 IEEE Sensors, Glasgow, UK, 25 December 2017. [Google Scholar]
- 34.Xu G., Abbott J., Ham D., Optimization of CMOS-ISFET-based biomolecular sensing: Analysis and demonstration in DNA detection. IEEE Trans. Electron Devices 63, 3249–3256 (2016). [Google Scholar]
- 35.Nemeth B., Piechocinski M. S., Cumming D. R. S., High-resolution real-time ion-camera system using a CMOS-based chemical sensor array for proton imaging. Sens. Actuators B Chem. 171–172, 747–752 (2012). [Google Scholar]
- 36.Hammond P. A., Ali D., Cumming D. R. S., Design of a single-chip pH sensor using a conventional 0.6-μm CMOS process. IEEE Sensors J. 4, 706–712 (2004). [Google Scholar]
- 37.Shields P., Nemeth B., Green R. B., Riehle M. O., Cumming D. R. S., High-speed imaging of 2D ionic diffusion using a 16 × 16 pixel CMOS ISFET array on the microfluidic scale. IEEE Sensors J. 12, 2744–2749 (2012). [Google Scholar]
- 38.B. Nemeth, Ion camera development for real-time acquisition of localised pH responses using the CMOS based 64×64-pixel ISFET sensor array technology, thesis, University of Glasgow, Glasgow, Scotland (2012). [Google Scholar]
- 39.M. J. Milgrew, M. O. Riehle, D. R. S. Cumming, paper presented at the 2008 IEEE International Solid-State Circuits Conference-Digest of Technical Papers, San Francisco, CA, 4 March 2009. [Google Scholar]
- 40.Nguyen B. H., Takahashi C. N., Gupta G., Smith J. A., Rouse R., Berndt P., Yekhanin S., Ward D. P., Ang S. D., Garvan P., Parker H.-Y., Carlson R., Carmean D., Ceze L., Strauss K., Scaling DNA data storage with nanoscale electrode wells. Sci. Adv. 7, eabi6714 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abbott J., Ye T., Krenek K., Gertner R. S., Ban S., Kim Y., Qin L., Wu W., Park H., Ham D., A nanoelectrode array for obtaining intracellular recordings from thousands of connected neurons. Nat. Biomed. Eng. 4, 232–241 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abbott J., Ye T., Krenek K., Qin L., Kim Y., Wu W., Gertner R. S., Park H., Ham D., The design of a cmos nanoelectrode array with 4096 current-clamp/voltage-clamp amplifiers for intracellular recording/stimulation of mammalian neurons. IEEE J. Solid State Circuits 55, 2567–2582 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ham D., Park H., Hwang S., Kim K., Neuromorphic electronics based on copying and pasting the brain. Nat. Electron. 4, 635–644 (2021). [Google Scholar]
- 44.Huynh M. T., Anson C. W., Cavell A. C., Stahl S. S., Hammes-Schiffer S., Quinone 1 e− and 2 e−/2 H+ reduction potentials: Identification and analysis of deviations from systematic scaling relationships. J. Am. Chem. Soc. 138, 15903–15910 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quan M., Sanchez D., Wasylkiw M. F., Smith D. K., Voltammetry of quinones in unbuffered aqueous solution: Reassessing the roles of proton transfer and hydrogen bonding in the aqueous electrochemistry of quinones. J. Am. Chem. Soc. 129, 12847–12856 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Tang Y., Wu Y., Wang Z., Spectroelectrochemistry for electroreduction of p-benzoquinone in unbuffered aqueous solution. J. Electrochem. Soc. 148, E133 (2001). [Google Scholar]
- 47.Ghoneim M. T., Nguyen A., Dereje N., Huang J., Moore G. C., Murzynowski P. J., Dagdeviren C., Recent progress in electrochemical pH-sensing materials and configurations for biomedical applications. Chem. Rev. 119, 5248–5297 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Kreider K. G., Tarlov M. J., Cline J. P., Sputtered thin-film pH electrodes of platinum, palladium, ruthenium, and iridium oxides. Sens. Actuators B Chem. 28, 167–172 (1995). [Google Scholar]
- 49.A. J. Bard, L. R. Faulkner, Electrochemical Methods: Fundamentals and Applications (Wiley, ed. 2, 2001). [Google Scholar]
- 50.Hasegawa J., Uno S., Nakazato K., Amperometric electrochemical sensor array for on-chip simultaneous imaging: Circuit and microelectrode design considerations. Jpn. J. Appl. Phys. 50, 04DL03 (2011). [Google Scholar]
- 51.Kuno T., Niitsu K., Nakazato K., Amperometric electrochemical sensor array for on-chip simultaneous imaging. Jpn. J. Appl. Phys. 53, 04EL01 (2014). [Google Scholar]
- 52.Guo J., Lindner E., Cyclic voltammograms at coplanar and shallow recessed microdisk electrode arrays: Guidelines for design and experiment. Anal. Chem. 81, 130–138 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hurth C., Li C., Bard A. J., Direct probing of electrical double layers by scanning electrochemical potential microscopy. J. Phys. Chem. C 111, 4620–4627 (2007). [Google Scholar]
- 54.Sandford C., Edwards M. A., Klunder K. J., Hickey D. P., Li M., Barman K., Sigman M. S., White H. S., Minteer S. D., A synthetic chemist’s guide to electroanalytical tools for studying reaction mechanisms. Chem. Sci. 10, 6404–6422 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elgrishi N., Rountree K. J., McCarthy B. D., Rountree E. S., Eisenhart T. T., Dempsey J. L., A practical beginner’s guide to cyclic voltammetry. J. Chem. Educ. 95, 197–206 (2017). [Google Scholar]
- 56.Rothe J., Frey O., Stettler A., Chen Y., Hierlemann A., Fully integrated CMOS microsystem for electrochemical measurements on 32 × 32 working electrodes at 90 frames per second. Anal. Chem. 86, 6425–6432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giagkoulovits C., Cheah B. C., Al-Rawhani M. A., Accarino C., Busche C., Grant J. P., Cumming D. R. S., A 16 × 16 CMOS amperometric microelectrode array for simultaneous electrochemical measurements. IEEE Trans. Circuits Syst. I, Reg. Papers 65, 2821–2831 (2018). [Google Scholar]
- 58.Short G. D., Bishop E., Concentration overpotentials on antimony electrodes in differential electrolytic potentiometry. Anal. Chem. 37, 962–967 (1965). [Google Scholar]
- 59.Xiang C., Papadantonakis K. M., Lewis N. S., Principles and implementations of electrolysis systems for water splitting. Mater. Horiz. 3, 169–173 (2016). [Google Scholar]
- 60.Martin M. M., Lindqvist L., The pH dependence of fluorescein fluorescence. JOL 10, 381–390 (1975). [Google Scholar]
- 61.Sjoback R., Nygren J., Kubista M., Absorption and fluorescence properties of fluorescein. Spectrochim. Acta Part A 51, L7–L21 (1995). [Google Scholar]
- 62.Egeland R. D., Marken F., Southern E. M., An electrochemical redox couple activitated by microelectrodes for confined chemical patterning of surfaces. Anal. Chem. 74, 1590–1596 (2002). [DOI] [PubMed] [Google Scholar]
- 63.Egeland R. D., Southern E. M., Electrochemically directed synthesis of oligonucleotides for DNA microarray fabrication. Nucleic Acids Res. 33, e125 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kosuri S., Church G. M., Large-scale de novo DNA synthesis: Technologies and applications. Nat. Methods 11, 499–507 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao X., LeProust E., Zhang H., Srivannavit O., Gulari E., Yu P., Nishiguchi C., Xiang Q., Zhou X., A flexible light-directed DNA chip synthesis gated by deprotection using solution photogenerated acids. Nucleic Acids Res. 29, 4744–4750 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pellois J. P., Zhou X., Srivannavit O., Zhou T., Gulari E., Gao X., Individually addressable parallel peptide synthesis on microchips. Nat. Biotechnol. 20, 922–926 (2002). [DOI] [PubMed] [Google Scholar]
- 67.Pellois J. P., Wang W., Gao X., Peptide synthesis based on t-Boc chemistry and solution photogenerated acids. J. Am. Chem. Soc. 2, 355–360 (2000). [DOI] [PubMed] [Google Scholar]
- 68.Emili A. Q., Cagney G., Large-scale functional analysis using peptide or protein arrays. Nat. Biotechnol. 18, 393–397 (2000). [DOI] [PubMed] [Google Scholar]
- 69.Abbott J., Ye T., Krenek K., Gertner R. S., Wu W., Jung H. S., Ham D., Park H., Extracellular recording of direct synaptic signals with a CMOS-nanoelectrode array. Lab Chip 20, 3239–3248 (2020). [DOI] [PubMed] [Google Scholar]
- 70.Abbott J., Mukherjee A., Wu W., Ye T., Jung H. S., Cheung K. M., Gertner R. S., Basan M., Ham D., Park H., Multi-parametric functional imaging of cell cultures and tissues with a CMOS microelectrode array. Lab Chip 22, 1286–1296 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Müller J., Ballini M., Livi P., Chen Y., Radivojevic M., Shadmani A., Viswam V., Jones I. L., Fiscella M., Diggelmann R., Stettler A., Frey U., Bakkuma D. J., Hierlemann A., High-resolution CMOS MEA platform to study neurons at subcellular, cellular, and network levels. Lab Chip 15, 2767–2780 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Franks W., Schenker I., Schmutz P., Hierlemann A., Impedance characterization and modeling of electrodes for biomedical applications. I.E.E.E. Trans. Biomed. Eng. 52, 1295–1302 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Hafizovic S., Heer F., Ugniwenko T., Frey U., Blau A., Ziegler C., Hierlemann A., A CMOS-based microelectrode array for interaction with neuronal cultures. J. Neurosci. Methods 164, 93–106 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Heer F., Hafizovic S., Franks W., Blau A., Ziegler C., Hierlemann A., CMOS microelectrode array for bidirectional interaction with neuronal networks. IEEE J. Solid State Circuits 41, 1620–1629 (2006). [Google Scholar]
- 75.Ballini M., Müller J., Livi P., Chen Y., Frey U., Stettler A., Shadmani A., Viswam V., Jones I. L., Jäckel D., Radivojevic M., Lewandowska M. K., Gong W., Fiscella M., Bakkum D. J., Heer F., Hierlemann A., A 1024-channel CMOS microelectrode array with 26,400 electrodes for recording and stimulation of electrogenic cells in vitro. IEEE J. Solid State Circuits 49, 2705–2719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frey U., Sedivy J., Heer F., Pedron R., Ballini M., Muellwe J., Bakkum D., Hafizovic S., Faraci F. D., Greve F., Kirstein K.-U., Hierlemann A., Switch-matrix-based high-density microelectrode array in CMOS technology. IEEE J. Solid State Circuits 45, 467–482 (2010). [Google Scholar]
- 77.Tsai D., Sawyer D., Bradd A., Yuste R., Shepard K. L., A very large-scale microelectrode array for cellular-resolution electrophysiology. Nat. Commun. 8, 1802 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S15
Movies S1 to S5