Abstract
Transforming growth factor–β is well known to restrain cytotoxic T cell responses to maintain self-tolerance and to promote tumor immune evasion. In this study, we have investigated the role of SMAD4, a core component in the TGF-β signaling pathway, in CD8+ T cells. Unexpectedly, we found that SMAD4 was critical in promoting CD8+ T cell function in both tumor and infection models. SMAD4-mediated transcriptional regulation of CD8+ T cell activation and cytotoxicity was dependent on the T cell receptor (TCR) but not TGF-β signaling pathway. Following TCR activation, SMAD4 translocated into the nucleus, up-regulated genes encoding TCR signaling components and cytotoxic molecules in CD8+ T cells and thus reinforced T cell function. Biochemically, SMAD4 was directly phosphorylated by ERK at Ser367 residue following TCR activation. Our study thus demonstrates a critical yet unexpected role of SMAD4 in promoting CD8+ T cell–mediated cytotoxic immunity.
SMAD4, which is phosphorylated by ERK following TCR activation, plays a critical role in promoting CD8+ T cell cytotoxic function.
INTRODUCTION
Transforming growth factor–β (TGF-β) has pleiotropic roles in lymphocyte regulation (1). TGF-β is important in T cells by controlling cell differentiation into specific lineages and regulating cell survival, proliferation, and effector functions. In contrast to its role in supporting the maintenance of naive T cell pools, TGF-β markedly restrains the activity of effector T cells. In particular, TGF-β inhibits CD8+ T cell proliferation; in the absence of TGF-β signaling, CD8+ T cells mount a potent antitumor cytotoxic response in tumor eradication (2). In these cells, TGF-β signaling was shown to repress the expression of interferon-γ (IFN-γ), genes encoding cytolytic machinery components and transcription mediators that orchestrate their expression, thus inhibiting the antitumor activity (3). TGF-β is thus an attractive target in cancer immunotherapy.
SMAD (mothers against decapentaplegic homologs) molecules are the core components in TGF-β signaling pathway. TGF-β binding to its receptor induces phosphorylation and activation of receptor-regulated SMADs (R-SMADs), i.e., SMAD2 and SMAD3, which subsequently associate with their partner SMAD4 and translocate from cytoplasm to nucleus (4). Formation of R-SMAD–SMAD4 complexes is essential in signaling of most TGF-β family members (5). TGF-β–SMAD signaling also has a key role in CD8+ T cell responses. Reduced SMAD2 phosphorylation in the dominant-negative TGFβRII T cells was associated with increased survival and expansion of virus-specific CD8+ T cells during chronic lymphocytic choriomeningitis virus (LCMV) infection (6). As a result, CD8+ T cells exhibited enhanced cytotoxicity, increased production of antiviral cytokines, and down-regulation of the inhibitory molecules, suggesting that TGF-β–SMAD2 signaling is important for T cell exhaustion. However, Cd4CreSmad4fl/fl or LckCreSmad4fl/fl mice were reported to spontaneously develop cancer but not autoimmunity (7, 8). In CD8+ T cells, the role of SMAD4 is still controversial. A study showed that SMAD4 deficiency reduced memory CD8+ T cell responses due to aberrant transcriptional programs (9). On the contrary, SMAD4 was indicated by another study to be essential in the development of central memory CD8+ T cells in Listeria monocytogenes infection model; reduced short-lived effector T cells but increased memory precursor effector T cells were generated in LckCreSmad4fl/fl mice, where SMAD4 was deficient in both CD4+ and CD8+ T cells (10). However, another study using Cd4CreTgfbr2fl/flSmad4fl/fl mice reported that SMAD4 was dispensable for T cell generation, homeostasis, and effector function, but it was essential for both CD4+ and CD8+ T cell proliferation after activation in vitro and in vivo through regulating Myc expression (11). Hence, until now, whether and how SMAD4 regulates CD8+ T cell effector function are still not clear.
In this study, we unexpectedly found that SMAD4 was critical in promoting CD8+ T cell cytotoxic function. SMAD4-mediated transcriptional regulation of CD8+ T cell activation and cytotoxicity is regulated by T cell receptor (TCR) but not TGF-β signaling pathway. Mechanistically, SMAD4 was directly phosphorylated by extracellular signal–regulated kinase (ERK) at Ser367 residue following TCR activation. Therefore, our work has offered previously unidentified, important insights into the function of SMAD4 in promoting CD8+ T cell–mediated cytotoxic immune responses.
RESULTS
SMAD4 expression in CD8+ T cells is required for their cytotoxic function
To understand the role of SMAD4 in T cell–mediated tumor immunity, we first subcutaneously inoculated E.G7 cells (106 cells per mouse), a chicken ovalbumin (OVA)–expressing mouse T cell lymphoma derived from EL4 cells, into Cd4CreSmad4fl/fl mice and Smad4fl/fl mice [wild type (WT)], and compared tumor growth between these two groups. Cd4CreSmad4fl/fl group developed much larger tumors than WT control mice (fig. S1A). In tumor-infiltrating immune cells, cells expressing CD11b, Gr-1, and NK1.1 showed no significant difference between the two groups (fig. S1B). SMAD4 deletion did not alter the numbers of total tumor-infiltrating lymphocytes (TILs) or CD3+ TILs, but cell numbers per cubic millimeter of tumor mass were greatly reduced in Smad4-deficient group (fig. S1C). Further flow cytometric analysis showed that SMAD4 deletion decreased the percentage of CD8+, but not CD4+ TILs (fig. S1D). Consistent with the reduced frequencies, CD8+ TILs from Cd4CreSmad4fl/fl mice expressed greatly reduced levels of granzyme B (GZMB), the major mediator of CD8+ T cell cytotoxicity, compared with those from WT mice (fig. S1E). The expression levels of programmed cell death protein 1 (PD-1) and IFN-γ were also slightly decreased in Smad4-deficient CD8+ TILs, although the expression of tumor necrosis factor–α (TNFα) showed no difference between these two groups (fig. S1E). In CD4+ TILs, the expression of TNFα, IFN-γ, and forkhead box P3 (Foxp3) displayed no significant difference between WT and Cd4CreSmad4fl/fl groups (fig. S1F). Similar to E.G7 tumor model, experiments using subcutaneous injection of mouse colon carcinoma MC38 cells also showed increased tumor growth in Cd4CreSmad4fl/fl mice compared with WT mice (fig. S1G).
The above studies suggest that SMAD4 deletion unexpectedly inhibited CD8+ T cell antitumor response, without affecting the expression of TNFα, IFN-γ, and Foxp3 in CD4+ T cells. Then, to determine whether SMAD4 expression in CD8+ T cells was intrinsically required for their function, we generated Cd8CreSmad4fl/fl mice, which deleted Smad4 specifically in peripheral CD8+ T cells. We subcutaneously injected E.G7 cells (106 cells per mouse) into WT and Cd8CreSmad4fl/fl mice. As above in Cd4CreSmad4fl/fl mice, E.G7 tumors in Cd8CreSmad4fl/fl mice exhibited increased growth compared with those in WT control mice (Fig. 1A). Then, we studied tumor antigen-specific immune responses. The percentages of OVA-specific CD8+ TILs stained with a SIINFEKL H-2Kb tetramer were greatly reduced in Smad4-defecient group compared with WT group (Fig. 1B). GZMB and IFN-γ expression was also much lower in Smad4-deficient CD8+ TILs than WT CD8+ TILs when gating total CD8+ TILs (Fig. 1B). As expected, consistent with reduced effector molecule production, expression of the exhaustion markers PD-1 and TIM-3 (T cell membrane protein 3) was also lower in Smad4-deficient CD8+ TILs (fig. S2A). Similar levels of Ly108 and Ki-67 expression indicated that the stemness and proliferation of CD8+ T cells, respectively, were unaffected in live cells in the absence of SMAD4 (fig. S2A). Moreover, we performed recall response experiments to analyze CD8+ TILs following restimulation by OVA peptides in vitro. Both the percentages of CD8+ TILs and the expression levels of effector molecules GZMB and KLRG1 (killer cell lectin-like receptor G1) were reduced in SMAD4-deficient T cells (fig. S2, B and C). In addition, we compared E.G7 tumor growth side by side among Cd8CreSmad4fl/fl mice, Cd4CreSmad4fl/fl mice, and WT mice. Similar trends of tumor growth were seen in Cd8CreSmad4fl/fl mice and Cd4CreSmad4fl/fl mice, both of which generated much larger tumors than WT control mice (fig. S3A). GZMB and IFN-γ expression in OVA-specific CD8+ T cells from both Cd8CreSmad4fl/fl mice and Cd4CreSmad4fl/fl mice was significantly reduced compared with WT mice (fig. S3B).
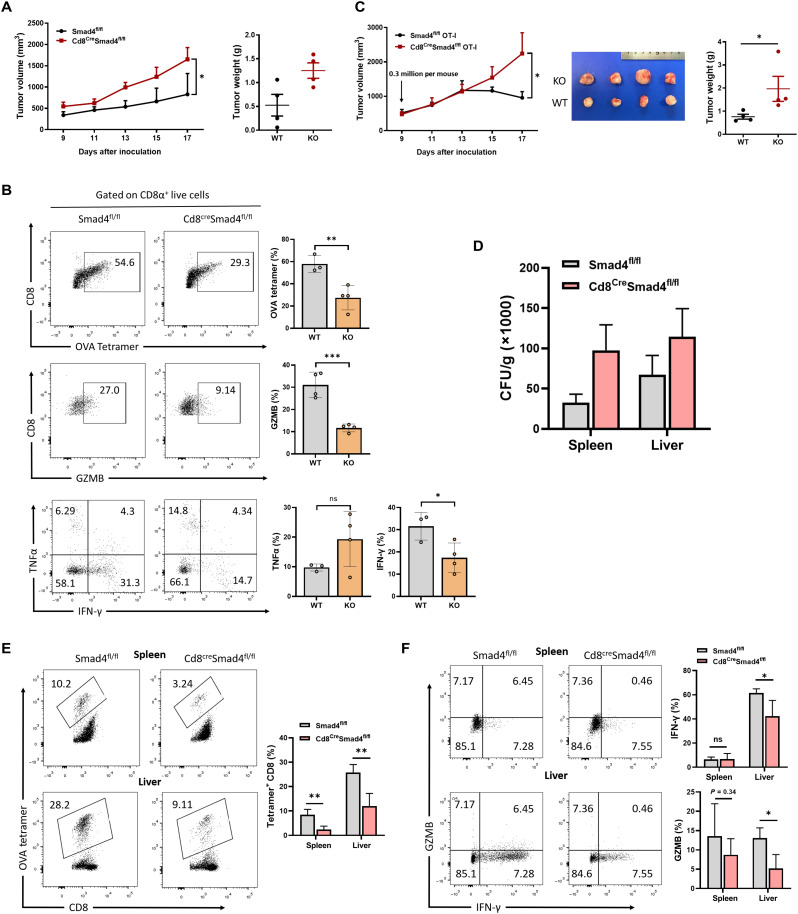
Fig. 1. SMAD4-deficient CD8+ T cells exhibit impaired cytotoxic function.
(A) E.G7 cells (1 × 106 cells per mouse) were inoculated subcutaneously into WT (n = 5) and Cd8CreSmad4fl/fl mice (n = 5). Tumor growth was monitored from day 9 after inoculation. (B) TILs were isolated at day 17 from E.G7-bearing mice. OVA-specific CD8+ T cell percentages and GZMB, TNFα, and IFN-γ expression in CD8+ TILs were analyzed by flow cytometry. (C) Left: Growth of E.G7 tumor in mice receiving WT or Smad4-deficient OT-I cells (3 × 105 cells per mouse). Right: Representative images of tumors, 17 days after transplant. KO, knockout. (D) OVA-modified L. monocytogenes (LM-OVA) colony-forming units (CFUs) from spleens and livers were calculated at day 8 after infection. (E) The percentages of OVA-specific CD8+ T cells in spleens and livers from LM-OVA–infected WT (n = 5) or Cd8CreSmad4fl/fl mice (n = 5). Measured by flow cytometry. (F) Expression levels of GZMB and IFN-γ in CD8+ T cells from LM-OVA–infected WT or Cd8CreSmad4fl/fl mice. These experiments were repeated three times. Data are represented as means ± SEM. ns, not significant. *P < 0.05, **P < 0.01, and ***P < 0.001.
To validate the above results further, we sorted CD8+ T cells recognizing OVA257-264 peptide from Cd8CreSmad4fl/fl OT-I mice and Smad4fl/fl OT-I (WT OT-I) mice and adoptively transferred them (3 × 105 cells per mouse) into E.G7 tumor–bearing TCRbd−/− mice at day 9 after inoculation. WT OT-I cells inhibited tumor development since day 13, but Smad4-defecient OT-I cells did not at all (Fig. 1C). The percentages of OT-I cells were significantly reduced in Smad4-defecient group (fig. S4A). Similar as above, expression of GZMB was strongly suppressed in Smad4-defecient OT-I cells in contrast with WT OT-I cells. Although WT and Smad4-defecient OT-I cells did not show much difference in their expression of IFN-γ and TNFα (fig. S4B), surface PD-1 and TIM-3 expression was significantly decreased in the absence of SMAD4 (fig. S4C). These results revealed that Smad4-defecient CD8+ TILs exhibited defective maintenance and impaired effector function.
The above results indicate a critical role of SMAD4 in CD8+ T cell response in tumors. To confirm this using an infection model, we challenged Cd8CreSmad4fl/fl mice and their littermate controls with OVA-modified L. monocytogenes (LM-OVA). LM-OVA colony-forming units (CFUs) and CD8+ T cells from both spleens and livers were analyzed at day 8 after infection. Cd8CreSmad4fl/fl mice had much higher levels of LM-OVA CFUs than their littermate controls (Fig. 1D). Percentages of OVA-specific CD8+ T cells from both spleen and liver were much lower in the Cd8CreSmad4fl/fl group than the WT group (Fig. 1E). GZMB was notably reduced in Smad4-deficient CD8+ splenic and hepatic T cells upon restimulation with the SIINFEKL peptide (Fig. 1F). IFN-γ expression also decreased in Smad4-deficient hepatic CD8+ T cells but showed no difference in spleen (Fig. 1F). Thus, we conclude that SMAD4 is intrinsically required for the cytotoxic function of CD8+ T cells in vivo.
SMAD4 regulates the expression of genes related to CD8+ T cell activation and effector function
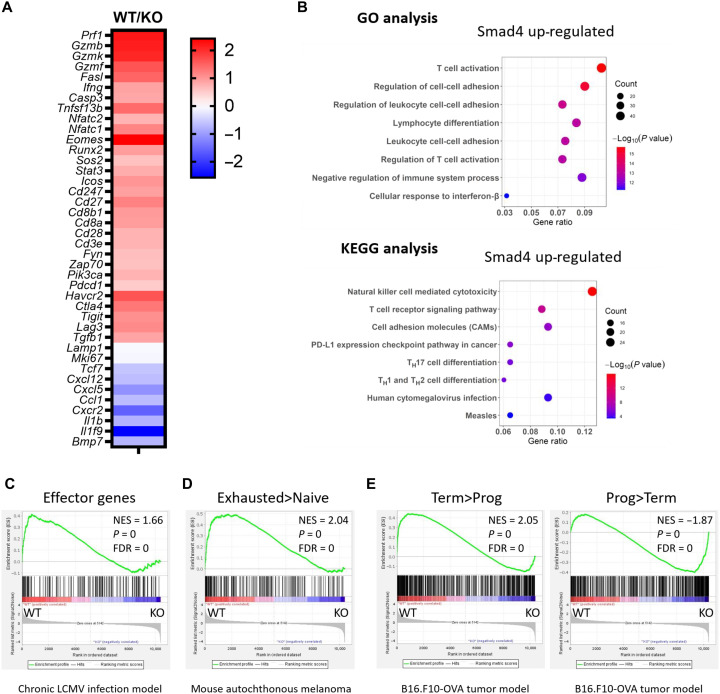
To understand the mechanisms whereby SMAD4 regulates CD8+ T cell function, we conducted transcriptional profile analysis of OVA-specific CD8+ TILs from E.G7-bearing WT and Cd8CreSmad4fl/fl mice (day 19). Overall, SMAD4 deficiency led to down-regulation of 508 genes (P < 0.01, fold change ≥ 1.5). Gene Ontology pathway analysis of these genes revealed that the top affected pathway involved T cell activation, including genes encoding TCR and costimulatory receptors (e.g., Cd3e, Cd28, Cd8a, and Icos), TCR signaling–related tyrosine kinase (e.g., Zap70 and Fyn), activation or effector markers (e.g., Cd27, Ifng, and Tnfsf13b), and coinhibitory receptors (e.g., Lag3, Tigit, and Ctla4) (Fig. 2, A and B). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis also revealed that the top two affected pathways were nature killer cell–mediated cytotoxicity, also expressed during CD8+ T cell–mediated cytotoxicity, such as Prf1, Gzmb, Fasl, Ifng, and Klrc1, and TCR signaling pathway, such as Cd247, Pik3r1, Sos2, Nfatc1, and Nfatc2 (Fig. 2, A and B). The gene Lamp1 (encoding CD107a) showed no difference (Fig. 2A), indicating that SMAD4 abrogation did not affect CD8+ T cell degranulation. In addition, Mki67 (encoding Ki-67) exhibited no difference (Fig. 2A), accordant with our flow cytometric results, demonstrating that SMAD4 abrogation did not affect CD8+ T cell proliferation. Genes up-regulated as a result of SMAD4 abrogation were mainly involved in T cell chemotaxis and migration (Fig. 2A and fig. S5). Thus, the pathway analysis reveals that SMAD4 contributes to CD8+ T cell effector function possible via TCR-mediated signaling pathway.
Fig. 2. SMAD4 regulates CD8+ T cell activation and effector function.
(A) Heatmap of selected differentially expressed genes showing OVA-specific CD8+ TILs from WT mice versus that from Cd8CreSmad4fl/fl mice (WT versus KO). (B) Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of SMAD4 up-regulated (SMAD4 abrogation down-regulated) genes between OVA-specific CD8+ TILs from WT and Cd8CreSmad4fl/fl mice. TH, T helper cell. (C) Gene set enrichment analysis of effector genes in chronic LCMV infection (12). Genes from the left to right of the rank-ordered list are enriched in WT and Smad4-deficient (KO) groups, respectively. (D) Gene set enrichment of genes describing for exhausted CD8+ T cells in mouse autochthonous melanoma, relative to naive CD8+ T cells (13). (E) Gene set enrichment of genes describing for terminally exhausted CD8+ T cells in B16-OVA tumor model, relative to progenitor exhausted CD8+ T cells (14). NES, normalized enrichment score.
Gene set enrichment analysis was also conducted, which revealed that gene sets related to effector T cells in chronic LCMV infection (12) were enriched in OVA-specific CD8+ TILs from WT mice, but not Smad4-deficient CD8+ TILs (Fig. 2C). On the other hand, the gene sets defined as up-regulated in exhausted T cells when comparing with naive T cells in autochthonous melanoma (13) were enriched in OVA-specific CD8+ TILs from WT mice, but not from Cd8CreSmad4fl/fl mice (Fig. 2D). In addition, gene sets defined as up-regulated in terminally exhausted CD8+ T cells in B16-OVA tumor model (14) were biased toward the WT group; those defined as up-regulated in progenitor exhausted CD8+ T cells were biased toward the Smad4-deficient group (Fig. 2E).
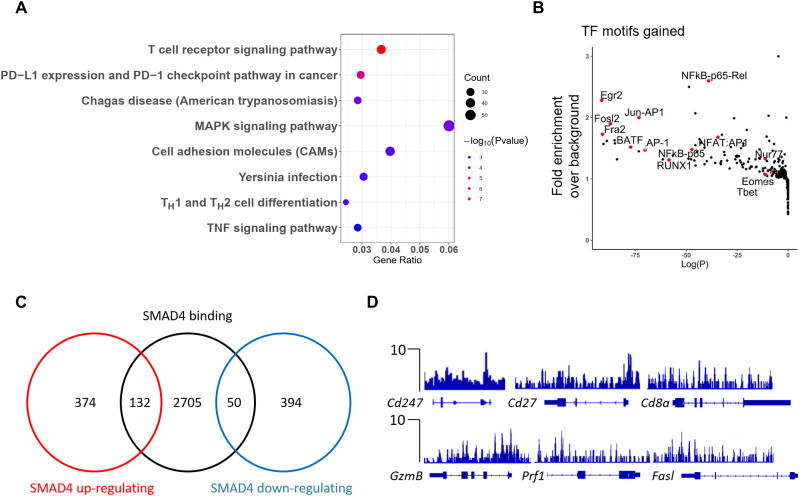
To characterize the direct targets of SMAD4 in CD8+ T cells, genome-wide SMAD4 chromatin immunoprecipitation sequencing (ChIP-seq) assay was conducted in CD8+ T cells activated for 4 days in vitro. In total, SMAD4 bound 2800 to 3000 gene loci in activated CD8+ T cells. Notably, KEGG pathway analysis revealed that the top one listed pathway with SMAD4-bound genes is involved in TCR signaling pathway (Fig. 3A). SMAD4 occupancy sites were enriched with binding motifs for the central transcription factors in TCR-mediated signaling pathway, such as activating protein 1 (AP1), Jun-AP1, nuclear factor of activated T cell:AP1, nuclear factor κB (NFκB)–p65, and NFκB-Rel (Fig. 3B). Combinational analysis of ChIP-seq and transcriptome data revealed that 132 genes were directly bound and up-regulated by SMAD4 (Fig. 3C), such as those encoding TCR complex and costimulatory receptors (e.g., Cd247, Cd28, Cd8a, and Icos) and activation or cytotoxic markers (e.g., Cd27, Gzmb, Prf1, and Fasl) (Fig. 3D). Therefore, SMAD4 directly promotes CD8+ T cell activation and effector function.
Fig. 3. SMAD4-mediated transcriptional regulation of CD8+ T cell activation and cytotoxicity.
(A) KEGG pathway analysis of SMAD4-bound gene loci in activated CD8+ T cells. (B) Transcription factor motif analysis following SMAD4 ChIP-seq experiment on activated CD8+ T cells. (C) Venn diagram of SMAD4-regulated genes and SMAD4-bound genes. (D) SMAD4 peaks at the Cd247, Cd27, Cd8a, Gzmb, Prf1, and Fasl gene loci in activated CD8+ T cells (versus control input DNA).
SMAD4 regulation of CD8+ T cell function is independent of TGF-β signaling
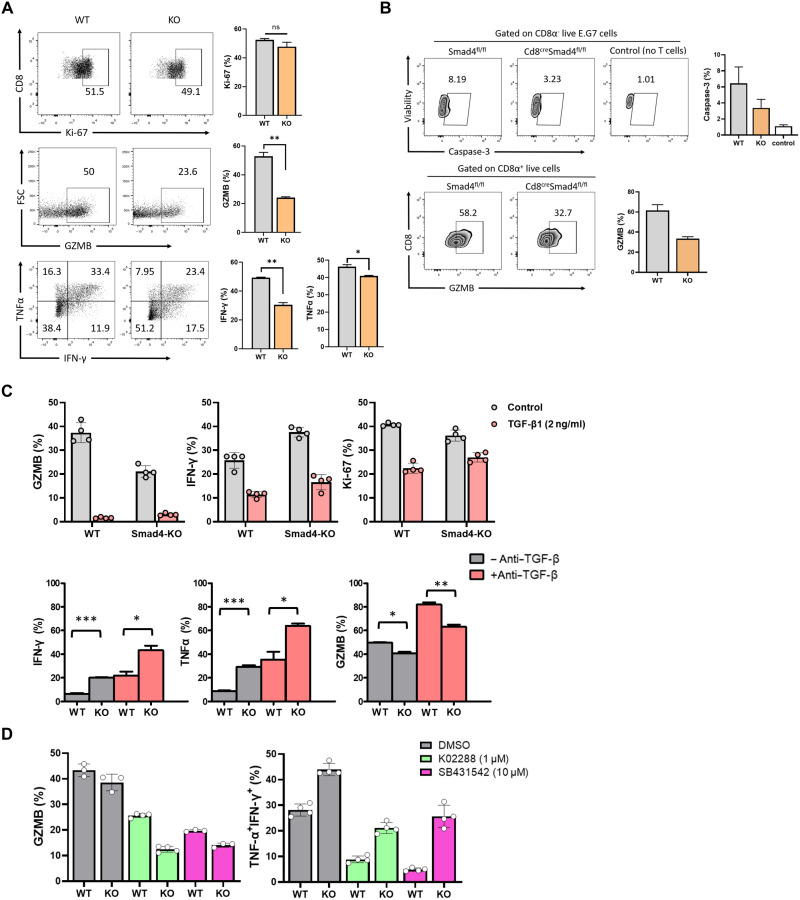
To further investigate the mechanisms underlying SMAD4-mediated CD8+ T cell activation and effector function, naive CD8+ T cells from WT and Cd8CreSmad4fl/fl mice were cultured in vitro with plate-bound anti-CD3 (5 μg/ml) plus anti-CD28 (5 μg/ml) for 3 to 4 days. We found that similar to in vivo results, GZMB, TNFα, and IFN-γ production in Smad4-deficient CD8+ T cells was significantly decreased in contrast with WT CD8+ T cells (Fig. 4A). Expression of Ki-67 showed no difference. Next, we compared the cytolytic activity between WT and Smad4-deficient CD8+ T cells using in vitro cytolytic assay. WT or Smad4-deficient OT-I cells were activated in vitro for 3 days and cocultured with E.G7 cells. Intracellular cleaved caspase-3 was measured at 5 hours in E.G7 cells and GZMB at 24 hours in T cells, after coculture. WT group displayed both significantly elevated caspase-3 levels in E.G7 cells and increased GZMB levels in CD8+ T cells compared with those from the knockout (KO) group (Fig. 4B), suggesting that Smad4-deficient CD8+ T cells had defective cytolytic activity to target cells in vitro.
Fig. 4. SMAD4 regulation of CD8+ T cell cytotoxicity is independent of TGF-β signaling.
(A) Expression levels of GZMB, IFN-γ, Ki-67, and TNFα in WT and Smad4-deficient CD8+ T cells. Cells were activated with plate-bound anti-CD3 (5 μg/ml) plus anti-CD28 (5 μg/ml) and cultured in the presence of IL-2 (1 ng/ml) in vitro for 3 to 4 days. Measured by flow cytometry. FSC, forward scatter. (B) WT or Smad4-deficient OT-I cells were activated in vitro for 3 days and mixed with E.G7 cells at 1:1 ratio. Intracellular cleaved caspase-3 was measured by anti–cleaved caspase 3 (1:100) at 5 hours after coculture. GZMB was measured by anti-GZMB (1:400) at 24 hours after coculture. (C) Top: Expression levels of GZMB, IFN-γ, and Ki-67 in WT and Smad4-deficient CD8+ T cells in the presence of TGF-β1 (2 ng/ml). Bottom: Expression of GZMB, IFN-γ, and TNFα in WT and Smad4-deficient CD8+ T cells in the presence of anti–TGF-β (1 μg/ml). Measured by flow cytometry after 3 days of culture. (D) Expression levels of effector molecules in WT and Smad4-deficient CD8+ T cells in the presence of TGF-β receptor inhibitor (SB431542; 1 μM) or BMP receptor inhibitor (K02288; 10 μM). Measured by flow cytometry after 3 to 4 days of culture. These experiments were repeated three times. Data are represented as means ± SEM. *P < 0.05 and ***P < 0.001. DMSO, dimethyl sulfoxide.
Because SMAD4 is a central cofactor involved in TGF-β signaling, we analyzed CD8+ T cell function in the presence of TGF-β1 (2 ng/ml) in vitro. Expression of GZMB, IFN-γ, and Ki-67 in CD8+ T cells from WT and Cd8CreSmad4fl/fl mice was analyzed after 3 days of culture. We found that TGF-β1 reduced the expression levels of these molecules not only in WT CD8+ T cells, consistent to its inhibitory roles in CD8+ T cells, but also in Smad4-deficient CD8+ T cells (Fig. 4C). Anti–TGF-β, which was used to neutralize endogenous TGF-β1, elevated the expression levels of GZMB, IFN-γ, and TNFα but still did not change the differences between WT and Smad4-deficient CD8+ T cells (Fig. 4C). These data suggested that TGF-β suppression of CD8+ T cell function was independent of SMAD4. Moreover, we compared GZMB, TNFα, and IFN-γ expression between WT and Smad4-deficient CD8+ T cells in the presence of TGF-β receptor inhibitor (SB431542; 1 μM) or bone morphogenetic protein (BMP) receptor inhibitor (K02288; 10 μM). These two inhibitors reduced the expression of effector molecules in both WT and KO groups (Fig. 4D), suggesting that other TGF-β superfamily members, such as BMPs, Activins, and Nodal, maybe had an active role in maintaining CD8+ T cell function. However, neither inhibitor narrowed the differences between WT and Smad4-deficient CD8+ T cells. In addition, we subcutaneously injected E.G7 cells (106 cells per mouse) into Cd4CreSmad2fl/fl mice and Cd4CreTrim33fl/fl mice to monitor the tumor growth. TRIM33 (tripartite motif containing 33) in TGF-β signaling can compete with SMAD4 for binding to phosphorylated SMAD2 or SMAD3 and mediate SMAD4-independent transcription of a distinct set of genes (15). We found that E.G7 tumor growth showed no differences among Cd4CreSmad2fl/fl mice, Cd4CreTrim33fl/fl mice, and WT control mice (fig. S6, A and B). In addition, similar to Cd4CreSmad4fl/fl mice, Cd4CreSmad4fl/flTrim33fl/fl mice formed much larger tumors than WT control mice (fig. S6C). Accordingly, these data suggest that the role of SMAD4 in regulation of CD8+ T cell cytotoxicity is independent of TGF-β signaling.
SMAD4 nuclear translocation in CD8+ T cells is regulated by TCR activation
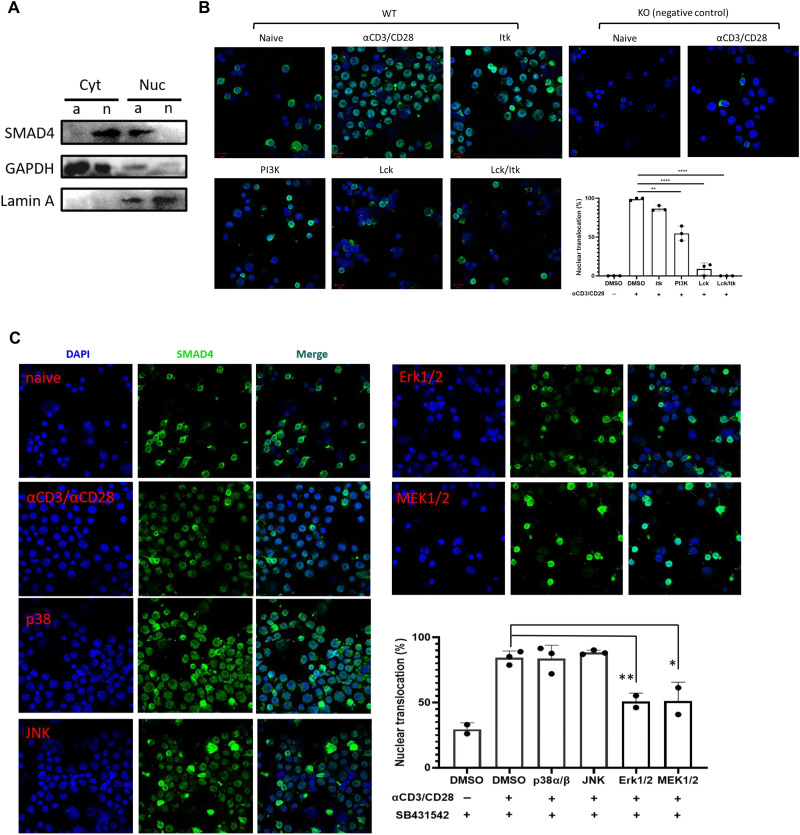
According to the above results, SMAD4 regulation of CD8+ T cell effector function appears independent of TGF-β signaling but closely related with TCR activation. Moreover, previous studies have reported that posttranslational modifications defining SMAD4 interactome, subcellular localization, and stability could be regulated by some receptor kinases (16, 17). Thus, we examined the subcellular localization of SMAD4 in CD8+ T cells by immunofluorescence microscopy to investigate whether TCR signaling could regulate SMAD4 activity. First, we found that SMAD4 translocated into nucleus, at 24 hours, but not at 4 or 8 hours, after TCR stimulation in vitro by anti-CD3/CD28 (fig. S7A). In contrast, when we examined the subcellular localization of SMAD4 in immortalized T cell lines, such as Jurkat and E.G7, SMAD4 was totally located in the nucleus regardless of whether these cells were activated by anti-CD3/CD28 or not (fig. S7, B and C). In addition, we compared expression levels of SMAD4 in cytoplasm and nucleus between naive and activated CD8+ T cells by Western blotting. In naive CD8+ T cells, SMAD4 was mainly found in cytoplasm, but in activated CD8+ T cells, it was mainly in the nucleus (Fig. 5A).
Fig. 5. SMAD4 nuclear translocation in CD8+ T cells is triggered by TCR signaling.
(A) Expression levels of SMAD4 in cytoplasm and nucleus between naive and activated CD8+ T cells by Western blotting. Cytoplasmic and nuclear protein was extracted from naive and in vitro activated CD8+ T cells. The cells were activated by αCD3/CD28 for 24 to 48 hours. Cyt, cytoplasm; Nuc, nucleus; n, naive; a, activated; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (B) Subcellular localization of SMAD4 in naive and αCD3/CD28 activated CD8+ T cells (in the presence of DMSO control, Itk inhibitor, PI3K inhibitor, Lck inhibitor, and combination of Lck/Itk inhibitors, respectively). The cells were collected, spun down to a cytospin microscope slide, and fixed and stained with αSMAD4 followed by staining with Alexa Fluor 488–conjugated secondary antibody. The results shown here represent the merged photos of SMAD4 (green) and 4′ (green) andcondary antibod (DAPI) (blue; indicated for nuclear location) staining. Scale bars, 10 μm. Right: Statistic data of SMAD4 nuclear translocation ratio, which was determined by manually counting the percentage of cells containing higher SMAD4 staining intensity in the nucleus versus cytoplasm in three representative fields revealed by Image-Pro Plus software. (C) Subcellular localization of SMAD4 in αCD3/CD28 activated CD8+ T cells in the presence of DMSO control, p38α/β inhibitor, c-Jun N-terminal kinase 1/2 inhibitor, MEK1/2 inhibitor, and ERK1/2 inhibitor, respectively. All the groups were added TGF-β receptor inhibitor. These experiments were repeated two or three times with consistent results. The statistics were performed by Student’s t test. *P < 0.05, **P < 0.01, and ****P < 0.0001.
Then, we examined SMAD4 translocation in the presence of Lck (lymphocyte-specific protein tyrosine kinase) inhibitor (10 μM), Itk (IL2 inducible T cell kinase) inhibitor (BMS-509744; 0.1 μM), or phosphatidylinositol 3-kinase (PI3K) inhibitor (PI-103; 0.5 μM). Earlier published studies have showed that Lck is a critical component of TCR signaling (18–20). Inhibition of Lck nearly completely suppressed SMAD4 nuclear localization whether in the presence of TGF-β receptor inhibitor (SB431542; 1 μM) and BMP receptor inhibitor (K02288; 10 μM) or not (fig. S7D and Fig. 5B). Treatment with Itk inhibitor, which inhibits the generation of diacylglycerol and inositol trisphosphate, suppressed SMAD4 nuclear localization only in a small part of cells, and inhibition of PI3K suppressed SMAD4 nuclear localization in about 50% cells (Fig. 5B). We then continued to test some inhibitors for mitogen-activated protein kinases (MAPKs). c-Jun N-terminal kinase 1/2 inhibitor (SP600125; 20 μM) and p38α/β inhibitor (doramapimod; 5 μM) exhibited no effect on nuclear translocation of SMAD4 (Fig. 5C). MAPK kinase 1/2 (MEK1/2) inhibitor (trametinib; 0.1 μM) and ERK1/2 inhibitor (SCH772984; 0.4 μM) significantly reduced SMAD4 nuclear localization (Fig. 5C), suggesting that nuclear translocation of SMAD4 is regulated by TCR-triggered MEK/ERK signaling.
ERK phosphorylation of SMAD4 at Ser367 regulates SMAD4 nuclear translocation
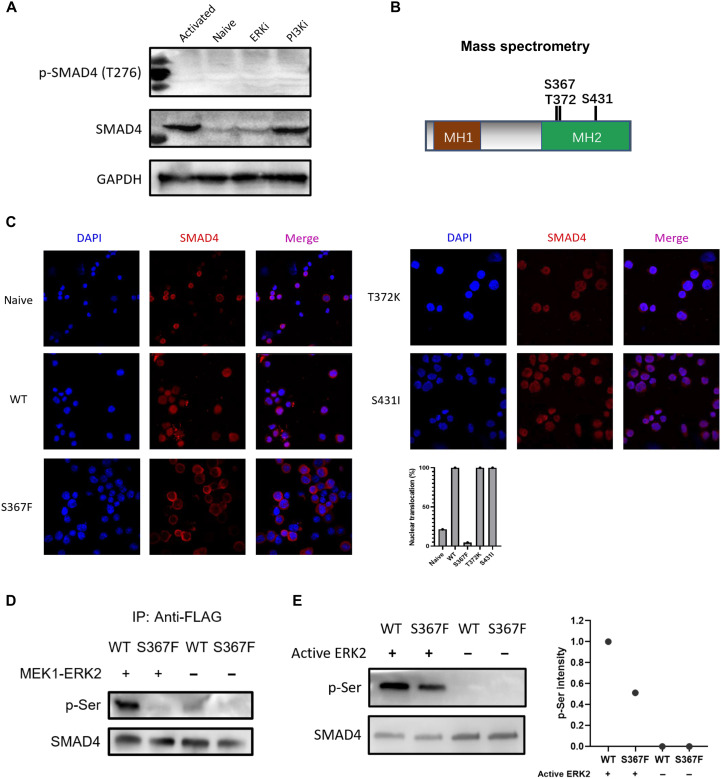
The above studies suggest that SMAD4 nuclear translocation is regulated by MEK/ERK signaling. It has been reported that SMAD4 could be directly phosphorylated by ERK at Thr276 (21), so we tested whether phosphorylation of Thr276 could be detected on SMAD4 in CD8+ T cells. High expression levels of total SMAD4 were detected in anti-CD3/CD28–activated CD8+ T cells and anti-CD3/CD28 plus PI3K inhibitor (PI-103; 0.5 μM)–treated CD8+ T cells. Naive and CD8+ T cells treated with anti-CD3/CD28 plus ERK inhibitor (SCH772984; 0.4 μM) exhibited reduced expression of total SMAD4. However, in all these four groups, we did not detect phosphorylation of Thr276 on SMAD4 by Thr276 antibody (Fig. 6A). Thus, we further performed phosphorylation site screening in activated CD8+ T cells. T cells were subjected to anti-SMAD4 immunoprecipitation followed by mass spectrometry analysis. In activated CD8+ T cells, three candidate serine/threonine phosphorylation residues (Ser367, Thr372, and Ser431) were detected on SMAD4, which were not detected in naive or ERK inhibitor–treated CD8+ T cells (Fig. 6B). To validate the three phosphorylation sites, we generated the phosphorylation defective S367F, T372K, and S431I mutants of SMAD4 and overexpressed SMAD4-WT (WT SMAD4) or these three SMAD4 mutants in Smad4−/− CD8+ T cells. SMAD4 nuclear translocation was measured at 24 hours after infection. Figure 6C showed that SMAD4-S367F mutant did not respond to TCR signaling in their nuclear localization and mostly retained in the cytoplasm. SMAD4-T372K and SMAD4-S431I mutants responded well to TCR signaling and translocated into nuclei in all cells, just like SMAD4-WT protein.
Fig. 6. SMAD4 phosphorylation at Ser367 by ERK regulates SMAD4 nuclear translocation.
(A) Expression levels of total SMAD4 and phosphorylated SMAD4 at Thr276 in naive and αCD3/CD28 activated CD8+ T cells (in the presence of DMSO control, PI3K inhibitor, ERK inhibitor, respectively). ERKi, ERK1/2 inhibitor; PI3Ki, PI3K inhibitor. (B) Three candidate serine/threonine phosphorylation sites (Ser367, Thr372, and Ser431) on SMAD4. Cell lysates from αCD3/CD28 activated CD8+ T cells were subjected to anti-SMAD4 purification via immunoprecipitation followed by mass spectrometry analysis. (C) Immunofluorescence staining data of SMAD4 (red; Alexa Fluor 594) in naive or αCD3/CD28 activated CD8+ T cells infected with retrovirus harboring WT Smad4 or the three mutant Smad4 gene (24 hours after retrovirus infection). Statistic data of SMAD4 nuclear translocation ratio were determined by manually counting the percentage of cells containing higher SMAD4 staining intensity in the nucleus versus cytoplasm in three representative fields. (D) Co-overexpressed MEK1 and ERK2 phosphorylated SMAD4 at Ser367 site. FLAG-SMAD4-WT or FLAG-SMAD4-S367F and MEK1-ERK2 were cotransfected into HEK-293T cells. Immunoprecipitation by anti-FLAG and Western blotting were carried out. (E) Western blotting results of in vitro phosphorylation assay by using recombinant active ERK2. FLAG-SMAD4-WT and FLAG-SMAD4-S367F were immunoprecipitation-purified with anti-FLAG affinity gel, washed by 3× FLAG peptide. Phospho-serine (p-Ser) levels (relative to total SMAD4) were quantified by ImageJ software. These experiments were repeated two or three times with consistent results.
To validate whether S367 on SMAD4 is phosphorylated by ERK, we performed kinase assays in vivo and in vitro, using a phospho-serine (p-Ser) antibody. In transfected human embryonic kidney (HEK) 293T cells, co-overexpressed MEK1 and ERK2 apparently phosphorylated SMAD4-WT. In contrast, SMAD4-S367F mutant failed to be phosphorylated by co-overexpressed MEK1 and ERK2 (Fig. 6D). In vitro phosphorylation assay showed that recombinant active ERK2 phosphorylated SMAD4-WT more efficiently than SMAD4-S367F mutant, both of which were expressed in HEK-293T cells and purified via immunoprecipitation (Fig. 6E). All of these results demonstrate that SMAD4 can be phosphorylated by ERK at Ser367 residue.
CD8+ T cells with SMAD4-S367F mutation exhibit impaired cytotoxicity
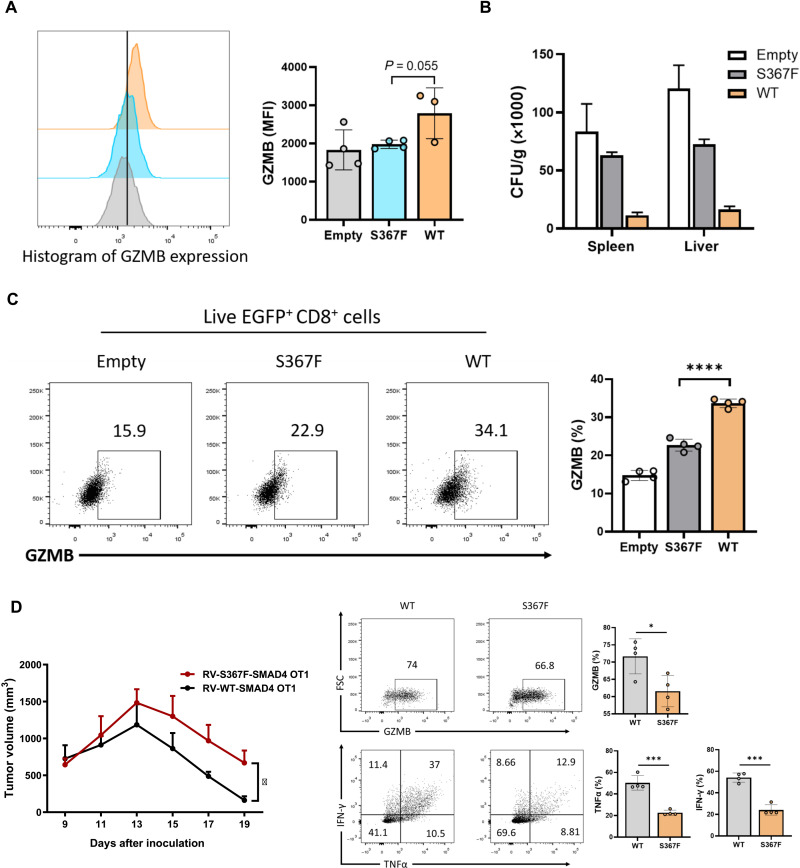
To further validate the role of S367 phosphorylation in T cells, we tested the cytotoxic function of CD8+ T cells with SMAD4-S367F mutation in vitro and in vivo. We infected Smad4−/− CD8+ T cells with empty control retrovirus or retrovirus containing either WT or S367F mutant Smad4. Forty-eight hours after infection, GZMB expression level in S367F-infected CD8+ T cells was as low as that in Smad4−/− CD8+ T cells (empty control group) and was apparently lower than that in CD8+ T cells reconstituted with SMAD4-WT molecule (Fig. 7A). In addition, we infected Smad4−/− OT-I cells with empty control retrovirus or retrovirus containing either WT or S367F mutant Smad4, and the infected OT-I cells (EGFPhigh cells) were sorted and introduced into C57BL/6 mice, followed by LM-OVA infection. LM-OVA CFUs from spleens and livers and splenic OT-I cells were analyzed at day 7 after infection. Consistent with the LM-OVA model performed with WT and Cd8creSmad4fl/fl mice, SMAD4-S367F group displayed much higher levels of LM-OVA CFUs in both spleens and livers (Fig. 7B), with reduced GZMB production in OT-I cells (Fig. 7C), compared with SMAD4-WT group. We also tested the antitumor activity of OT-I cells with SMAD4-S367F mutation in E.G7-bearing TCRbd−/− mice. SMAD4-S367F OT-I cells showed reduced antitumor activity compared with SMAD4-WT OT-I cells (Fig. 7D). In addition, the expression levels of GZMB, TNFα, and IFN-γ were significantly reduced in SMAD4-S367F OT-I cells when compared with SMAD4-WT OT-I cells (Fig. 7E). Together, our results demonstrate a key role for SMAD4, activated by the TCR-ERK pathway, in regulation of CD8+ T cell function.
Fig. 7. SMAD4-S367F mutant CD8+ T cells exhibit impaired cytotoxicity.
(A) Expression levels of GZMB in empty control, SMAD4-WT, or SMAD4-S367F retrovirus–infected CD8+ T cells. CD8+ T cells from Cd8CreSmad4fl/fl mice were isolated and infected with empty control retrovirus or retrovirus containing either WT or S367F mutant Smad4. Forty-eighty hours after infection, GZMB expression was measured by flow cytometry. (B) LM-OVA CFUs from spleens and livers were calculated at day 7 after infection. OT-I cells isolated from Cd8CreSmad4fl/fl OT-I mice were infected with empty control retrovirus or retrovirus containing either WT or S367F mutant Smad4. The infected OT-I cells (EGFP+ cells) were sorted and intravenously transferred into C57BL/6 mice. Twelve hours later, LM-OVA (2 × 104 CFUs per mouse) was also intravenously injected into the recipient mice. (C) Expression of GZMB in splenic OT-I cells. Measured by flow cytometry. (D) E.G7 cells (1 × 106 cells per mouse) were inoculated subcutaneously into TCRbd−/− mice. Tumor growth were monitored from day 9 after inoculation. OT-I cells isolated from Cd8CreSmad4fl/fl OT-I mice were infected with retrovirus containing either WT or S367F mutant Smad4. The infected OT-I cells (EGFP+ cells) were sorted and intravenously transferred into E.G7-bearing TCRbd−/− mice at day 9 after E.G7 cells inoculation. (E) Expression of GZMB, TNFα, and IFN-γ in CD8+ TILs from E.G7-bearing TCRbd−/− mice was measured by flow cytometry. Data are represented as means ± SEM. The statistics were performed by Student’s t test. *P < 0.05, ***P < 0.001, and ****P < 0.0001.
DISCUSSION
In our current study, we unexpectedly discovered SMAD4 as a critical factor in TCR-mediated intracellular signaling in CD8+ T cells. It functions to directly up-regulate the expression of the genes encoding TCR complex subunits and cytotoxic molecules in CD8+ T cells. SMAD4 is activated after phosphorylation by ERK at the Ser367 residue.
Structural and functional differences separate the 32 members of the TGF-β superfamily cytokines into TGF-β and BMP subfamilies (22). Our results indicate that SMAD4 plays a critical role in CD8+ T cell activation and effector function via TCR-mediated rather than TGF-β–mediated signaling pathway by using TGF-β receptor inhibitor and BMP receptor inhibitor. Moreover, we found that SMAD4 contributes to T cell activation and effector function specifically in CD8+ T cells, which was not found previously in CD4+ T cells. The basis for the differential functions of SMAD4 in CD4+ and CD8+ T cells is not understood, maybe because of different kinase-substrate interaction, complex formation, or different phosphorylation site, which still needs further investigation. We have not formally ruled out a potential role of SMAD4 in TGF-β signaling in CD8+ T cells, although it has not been revealed in our genetic analysis.
Multiple phosphorylation sites have been reported in SMAD4, including constitutive phosphorylation at Thr276 in the linker region by ERK (21), Ser343 phosphorylation in the MH2 domain by MPK38 (23), Thr77 phosphorylation in the amino-terminal MH1 domain by liver kinase B1 (24), and Tyr95 phosphorylation in the MH1 domain by anaplastic lymphoma kinase (17), although not all the phosphorylation sites have been identified. Our results do not exclude the possibility that SMAD4 is phosphorylated on other residues in CD8+ T cells. However, Ser367 is certainly the most critical phosphorylation site of SMAD4 in TCR-triggered MEK/ERK signaling in CD8+ T cells. Functional and structural analyses of the MH1 and MH2 domains have provided critical insights into the principal interactions of SMAD4 with other proteins and with DNA. The MH1 domain is a DNA binding module, while the highly conserved MH2 domain is one of the most versatile protein-interacting modules in signal transduction (25). Therefore, phosphorylation of Ser367 residue in the MH2 domain by ERK, which regulates SMAD4 nuclear accumulation, is consistent with the function of SMAD4 MH2 domain.
Our results revealed that SMAD4 expression levels were elevated after TCR signaling in CD8+ T cells (Fig. 6A). Quantitative analysis from a published study also showed that SMAD4 protein expression levels were significantly elevated in activated CD4+ and CD8+ T cells, compared with naive T cells by mass spectrometry (fig. S8) (26). Thus, during CD8+ T cell activation and effector differentiation, the expression levels of SMAD4 increase. Meanwhile, SMAD4 translocates into the nucleus and up-regulates the genes encoding TCR complex subunits and costimulatory receptors (e.g., Cd3e, Cd28, Cd8a, and Icos) and TCR signaling–related tyrosine kinase (e.g., Zap70 and Fyn). These findings indicate that SMAD4 reinforces the TCR activation signals through a feedforward loop. Reduced TCR activation and insufficient TCR signaling strength led to defective maintenance and survival of Smad4-deficient T cells. This is likely the reason why Smad4-deficient T cells were reduced in cell numbers and percentages in tumor and infection models, although the expression levels of Ki-67 between live WT and SMAD4-deficient CD8+ T cells show no differences in both in vivo (fig. S2A) and in vitro (Fig. 4A) results. In addition, increased proportions of CD127+KLRG1− subsets were found in SMAD4-deficient CD8+ TILs (fig. S2C), suggesting that SMAD4 deficiency led to memory T cell precursor generation.
Moreover, our results suggest that SMAD4 regulates the expression of many genes involved in TCR-mediated MAPK signaling pathway (Fig. 3A), such as Map2k3, Map3k4, and Map4k2. The results indicate complex interactions between SMAD4 and the MAPK pathway (containing MEK and ERK), which is still not fully understood. Future analysis of the dynamic interaction of SMAD4 and MAPK will be needed, which may also exist in other types of cells, in addition to CD8+ T cells.
In light of the critical role of TGF-β signaling on cancer progression and immune evasion, a wide range of anticancer therapeutic modalities have been developed targeting TGF-β. TGF-β is thought to suppress CD8+ T cell function through canonical (SMAD-dependent) or non-canonical (SMAD-independent) pathway (27), the signaling mechanisms of which still need further investigation. However, our study found that SMAD4 was critical in promoting CD8+ T cell cytotoxicity, which was dependent on TCR but not on TGF-β signaling. Our results suggest that SMAD4, dispensable in TGF-β signaling, is important in TCR-triggered CD8+ T cell cytotoxic function.
Natural killer (NK) cells and CD8+ cytotoxic T cells share functions in cytolytic immune response. In mice, SMAD4-deficient NK cells were reported to have a gene signature that was ILC1 (innate lymphoid cell 1)-like and was indicative of imprinting by TGF-β. NK cell effector functions require SMAD4 (28). Similar with our results in CD8+ T cells, SMAD4-deficient NK cells were unable to control tumor metastasis or viral infection. However, the mechanisms underlying SMAD4 activation and function in NK cells will need to be further studied, where perhaps our results in CD8+ T cells will be applicable.
In summary, we discovered an unexpected role of SMAD4 in TCR-triggered CD8+ T cell activation and cytotoxic function. Our study thus offers important insights into the molecular mechanisms underlying cytotoxic CD8+ T cell function in pathogen elimination and tumor surveillance.
MATERIALS AND METHODS
Mice
Trim33fl/fl (29) and Smad2fl/fl mice (30) were crossed with Cd4Cre mice (31). Smad4fl/fl mice (32) were crossed with Cd4Cre (31) and Cd8cre mice to generate conditional Smad4 KO mice. The Cd8cre, Tcrbd−/−, and OT-I TCR transgenic mice were purchased from the Jackson Laboratory. All the mice were housed in the specific pathogen–free animal facility at Tsinghua University. All the animal experiments were performed according to the protocols approved by the Institutional Animal Care and Use Committee.
Murine tumor models
E.G7 cell line was purchased from American Type Culture Collection. MC38 cell line was generously provided by Y. Zhang (Tsinghua University, China). E.G7 and MC38 cells were cultured in RPMI 1640 medium plus 10% fetal bovine serum and penicillin-streptomycin (100 U/ml). E.G7 or MC38 cells (1 × 106) resuspended in 100 μl of phosphate-buffered saline (PBS) was injected subcutaneously into the right flank of 6- to 10-week-old mice.
For adoptive transfer experiments, E.G7 cells (1 × 106 cells per mouse) were implanted subcutaneously into the right flank of Tcrbd−/− recipient mice. At 9 days after tumor transplant, naive (CD8+CD62LhiCD44lo) OT-I cells (3 × 105 cells per mouse) from Smad4fl/fl OT-I mice or Cd8CreSmad4fl/fl OT-I mice were sorted and adoptively transferred into tumor-bearing mice intravenously.
To test the cytotoxic function of CD8+ T cells with SMAD4-S367F mutation, E.G7 cells (1 × 106 cells per mouse) were inoculated subcutaneously into TCRbd−/− mice. Tumor growth was monitored from day 9 after inoculation. OT-I cells isolated from Cd8CreSmad4fl/fl OT-I mice were infected with retrovirus containing either WT or S367F mutant Smad4. The infected OT-I cells [enhanced green fluorescent protein–positive (EGFP+) cells] were sorted and intravenously transferred into E.G7-bearing TCRbd−/− mice at day 9 after E.G7 cells inoculation.
For all the tumor models, tumor growth was monitored every 2 days. Tumor volume was calculated by the following formula: tumor volume = (π/6) × (L × W2), where L and W are the longer and shorter dimensions of the tumor, respectively. Mice were sacrificed after 18 to 22 days for phenotypic analyses.
L. monocytogenes infection model
To test the cytotoxic function of Smad4-deficient CD8+ T cells, Cd8CreSmad4fl/fl or Smad4fl/f mice were intravenously infected with 1 × 104 CFUs of LM-OVA. To test the cytotoxic function of CD8+ T cells with SMAD4-S367F mutation, C57BL/6 recipient mice were intravenously infected with 2 × 104 CFUs of LM-OVA. Twelve hours later, mice were adoptively transferred with empty control, WT-Smad4, or S367F-Smad4 retrovirus plasmid–infected OT-I cells. To determine the organ Listeria burden, spleens and livers from infected mice were homogenized separately in PBS and then 1:1 mixed with 0.1% Triton X-100. Serial dilutions of homogenate were plated on brain heart infusion agar plates, and bacterial CFUs were assessed after 24 to 48 hours growth at 37°C. Spleens and livers were minced using razor blades in ice-cold PBS followed by passing through a 70-mm cell strainer to achieve a single-cell suspension. After lysis of red blood cells and washing with cold PBS, cells were stimulated with the SIINFEKL peptide (10 ng/ml) or phorbol 12-myristate 13-acetate (PMA; 50 ng/ml; Sigma-Aldrich, MO) and ionomycin (500 ng/ml; Sigma-Aldrich, MO) in the presence of brefeldin A (GolgiPlug, BD Biosciences) for 4 hours before staining with fluorescence-conjugated antibodies for flow cytometric analysis.
Plasmid construction and retroviral transduction
The genes of MEK1, ERK2, and SMAD4 were polymerase chain reaction (PCR)–amplified, cloned into the pRVKM retroviral vector, and then used for constructing the MEK1-ERK2 co-overexpression or SMAD4-S367F/T372K/S431I mutant plasmid by site-direct mutagenesis. The plasmids were transfected together with pcl-ECO into HEK-293T cells for preparing retrovirus. Naive CD8+ T cells [isolated using a magnetic-activated cell sorting (MACS) mouse CD8+ T cell isolation kit (Invitrogen) and CD8+CD25−CD44lowCD62Lhigh naive CD8+ T cells were sorted by FACSAria III cell sorter] were activated with plate-bound anti-CD3 plus anti-CD28 for 24 hours under neutral condition and were infected with virus harboring the WT or SMAD4-mutant genes by spinning.
Isolation of TILs
E.G7 or MC38 tumors were digested with collagenase D (1 mg/ml) supplemented with deoxyribonuclease I (10 U/ml) for 40 min at 37°C before centrifugation on a discontinuous Percoll gradient (GE Healthcare).
Flow cytometry
Single-cell suspensions were stained with antibodies against surface molecules. For intracellular cytokine staining, cells were stimulated with PMA (50 ng/ml; Sigma-Aldrich, MO) and ionomycin (500 ng/ml; Sigma-Aldrich, MO) in the presence of brefeldin A (GolgiPlug, BD Biosciences) for 4 hours before staining with antibodies against surface proteins followed by fixation and permeabilization by using the eBioscience Fix/Perm or BD Fix/Perm buffer kit and staining with antibodies against intracellular antigens. For recall response experiment, cells were stimulated with OVA257–264 peptide (10 ng/ml) overnight and then added GolgiPlug for 2 hours. Cells were analyzed on an LSRFortessa (33) flow cytometer, and data were analyzed using FlowJo X. Dead cells were excluded on the basis of viability dye staining (Fixable Viability Dye eF506, eBioscience).
In vitro cytolytic assay
Naive OT-I cells were activated in vitro for 3 days and mixed with E.G7 cells at 1:1 ratio. Intracellular cleaved caspase-3 was measured by anti–cleaved caspase 3 (1:100) at 5 hours after coculture. GZMB was measured by anti-GZMB (1:400) at 24 hours after coculture.
Cytospin and immunofluorescence staining
Naive T cells were activated for 24 hours and then resuspended in culture medium at 1 million/ml. One hundred microliters of each cell suspension was added to a slide chamber and spun down onto the slide using a cytocentrifuge (800 rpm/3 min). The cells were fixed on slide with 4% paraformaldehyde, permeabilized with 0.01% Trion X-100, and blocked with goat serum and stained with SMAD4 antibody (Santa Cruz Biotechnology, catalog no. sc-7966) overnight. The slides were washed and then incubated with Alexa Fluor 488– or Alexa Fluor 594–conjugated goat anti-mouse immunoglobulin G (IgG) (BioLegend) secondary antibody for 1 hour at room temperature and lastly mounted with mounting medium containing 4′,6-diamidino-2-phenylindole. The images were obtained by LSM780 fluorescence microscope (Zeiss).
Chromatin immunoprecipitation sequencing
The ChIP assay was performed using an Active Motif’s ChIP assay kit (53035) according to the manufacturer’s instructions with slight modifications (34). Briefly, CD8+ T cells were harvested and then cross-linked with 1% paraformaldehyde for 10 min and stopped with 125 mM glycine for 5 min at room temperature. The cells were lysed and digested with shearing enzyme followed by 10 cycles’ sonication. The cell lysate was then used for immunoprecipitation with antibodies anti-SMAD4 (Abcam, catalog no. ab40759) or control IgG (Abcam, catalog no. ab46540) followed by Dynabeads Protein A (Life Technologies, catalog no. 10002D) pull-down. The precipitated DNA was then washed, eluted, de–cross-linked and purified for real-time PCR analysis or for deep sequencing carried by BGI Genomics. ChIP-qPCR primers included Eomes [(positive control) forward, AGATGGAAATTTGGGAATGAA; reverse, GGCTACTACGGCCTGAAACT] and Fp3-CNS2 [(negative control) forward, CCCAACAGACAGTGCAGGAA; reverse, TGGTGTGACTGTGTGATGCA].
Clean reads after filtering were aligned to the reference sequence mm10 genome by using bowtie2 (35). PCR duplicates were removed using picard MarkDuplicates. The uniquely mapped reads were used to call peak with MACS2 (36) using a P value of 0.01 as a cutoff. ChIPseeker was used for peak annotation (37). deepTools was used to generate coverage track file (bigWig), which can be visualized in Integrative Genomics Viewer.
RNA sequencing
OVA-specific CD8+ TILs from E.G7-bearing WT and Cd8CreSmad4fl/fl mice were sorted into lysis buffer using BD FACSAria Cell Sorter. cDNA was extracted, and library was constructed using the SMARTer Ultra Low Input RNA for Illumina Sequencing-HV kit and the Creator SMART cDNA Library Construction Kit (Clontech) according to the manufacturer’s instructions. The library products were sequenced via Illumina HiSeq2500 or Hiseq4000 by BerryGenomics or BGI Genomics. Low-quality reads and adaptor sequences were removed by Trim Galore v0.4.4. The clean reads were aligned to mm10 by Bowtie2 with default parameter, and uniquely mapping reads were summarized by FeatureCounts (from Subread package). Differentially expressed genes are identified by DESeq2 using at least 1.5 fold change and false discovery rate (FDR) adjusted P value 0.05. clusterProfiler (R package) was used for pathway analysis.
Immunoprecipitation and Western blotting analysis
Cells were collected in lysis buffer [25 mM tris-HCl (pH 7.5), 150 mM NaCl, 0.5% NP-40; protease and phosphatase inhibitors added before use]. The lysate was centrifuged and immunoprecipitated with primary antibody at 4°C overnight. Target protein was separated by protein A/G agarose beads (Invitrogen). Precipitated proteins and initial whole-cell lysates were boiled with SDS loading buffer, separated by SDS–polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membrane, incubated with the primary and secondary antibodies, and detected with an enhanced chemiluminescence staining kit.
Mass spectrometry analysis
Cell lysates from anti-CD3/CD28 activated CD8+ T cells, naïve CD8+ T cells, or ERK inhibitor–treated CD8+ T cells were subjected to anti-SMAD4 purification via immunoprecipitation followed by mass spectrometry analysis. For liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis, the peptides were separated by an 85-min gradient elution at a flow rate 0.30 μl/min with a Thermo-Dionex UltiMate 3000 HPLC system, which was directly interfaced with a Thermo Scientific Q Exactive mass spectrometer. The analytical column was a homemade fused silica capillary column (75 μm inside diameter, 150 mm length; Upchurch, Oak Harbor, WA) packed with C-18 resin (300 Å, 5 μm, Varian, Lexington, MA). Mobile phase consisted of 0.1% formic acid, while mobile phase B consisted of 80% acetonitrile and 0.1% formic acid. The Q Exactive mass spectrometer was operated in the data-dependent acquisition mode using Xcalibur 2.2 software, and there was a single full-scan mass spectrum in the orbitrap (300 to 1800 mass/charge ratio, 70,000 resolution) followed by 20 data-dependent MS/MS scans at 27% normalized collision energy [higher-energy collisional dissociation (HCD)].
The MS/MS spectra from each LC-MS/MS run were searched against the mouse.fasta from UniProt using an in-house Proteome Discoverer (version PD1.4, Thermo Fisher Scientific, USA). The search criteria were as follows: full tryptic specificity was required; two missed cleavage was allowed; carbamidomethyl (C) was set as the fixed modifications; the oxidation (M) and phosphorylation (P) were set as the variable modification; precursor ion mass tolerances were set at 20 ppm for all MS acquired in an orbitrap mass analyzer; and the fragment ion mass tolerance was set at 0.02 Da for all MS2 spectra acquired. The peptide FDR was calculated using Percolator provided by Proteome Discoverer. When the q value was smaller than 1%, the peptide spectrum match (PSM) was considered to be correct. FDR was determined on the basis of PSMs when searched against the reverse, decoy database. Peptides only assigned to a given protein group were considered as unique. The FDR was also set to 0.01 for protein identifications.
In vitro phosphorylation assay
Recombinant proteins of FLAG–SMAD4-WT and FLAG–SMAD4-S367F were first prepared and purified from HEK-293T cells. Anti-FLAG affinity gel was used to immunoprecipitate the proteins followed by washing with 3× FLAG peptide. In a typical phosphorylation reaction, 2 μg of FLAG–SMAD4-WT or FLAG–SMAD4-S367F protein was incubated with 2 μg of recombinant active ERK2 in 80 μl of kinase reaction buffer [25 mM tris-HCl (pH 7.5), 10 mM MgCl2, 5 mM β-glycerophosphate, 2 mM dithiothreitol, 0.1 mM Na3VO4, 200 μM adenosine 5′-triphosphate] at 37°C for 0.5 hours. Phosphorylation of SMAD4 was analyzed by Western blotting with SMAD4 antibody or p-Ser antibody.
Statistical analysis
Statistical comparisons were performed using ordinary one-way analysis of variance (ANOVA) analysis followed by multiple comparisons or Student’s t test. P < 0.05 was considered to indicate a statistically significant difference.
Acknowledgments
We thank the Protein Research Technology Center and Protein Chemistry and Omics Platform at Tsinghua University for help with MS analysis, the Immunology Core Facility at the Institute for Immunology at Tsinghua University for assistance on FACS, E. J. Robertson for providing Smad4fl/fl and Smad2fl/fl mice, and all Dong laboratory members for help.
Funding: This work was supported by grants from the Natural Science Foundation of China (31991173 and 31991170), the Tsinghua University Spring Breeze Fund (2020Z99CFG008), and the Tsinghua University-Xiamen Chang Gung Hospital Joint Research Center for Anaphylactic Disease.
Author contributions: X.L. and C.D. designed the project and analyzed the data. X.L. performed the experiments. J.H., P.W., Q.L., Lu Ni, Y.C., and X.B. helped with the experiments. X.Z. analyzed the bioinformatics data. X.L., Ling Ni, and C.D. prepared the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The accession number for RNA sequencing data reported in this paper is GenBank: GSE192621. The accession number for ChIP-seq data reported in this paper is GenBank: GSE192620.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Other Supplementary Material for this manuscript includes the following:
Supplemental excel files S1 to S3
REFERENCES AND NOTES
- 1.Sanjabi S., Oh S. A., Li M. O., Regulation of the immune response by TGF-β: From conception to autoimmunity and infection. Cold Spring Harb. Perspect. Biol. 9, a022236 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorelik L., Flavell R. A., Immune-mediated eradication of tumors through the blockade of transforming growth factor-β signaling in T cells. Nat. Med. 7, 1118–1122 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Thomas D. A., Massagué J., TGF-β directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell 8, 369–380 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Martin-Malpartida P., Batet M., Kaczmarska Z., Freier R., Gomes T., Aragón E., Zou Y., Wang Q., Xi Q., Ruiz L., Vea A., Márquez J. A., Massagué J., Macias M. J., Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors. Nat. Commun. 8, 2070 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Y., Massagué J., Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113, 685–700 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Tinoco R., Alcalde V., Yang Y., Sauer K., Zuniga E. I., Cell-intrinsic transforming growth factor-β signaling mediates virus-specific CD8+ T cell deletion and viral persistence in vivo. Immunity 31, 145–157 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hahn J. N., Falck V. G., Jirik F. R., Smad4 deficiency in T cells leads to the Th17-associated development of premalignant gastroduodenal lesions in mice. J. Clin. Invest. 121, 4030–4042 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim B.-G., Li C., Qiao W., Mamura M., Kasprzak B., Anver M., Wolfraim L., Hong S., Mushinski E., Potter M., Kim S.-J., Fu X.-Y., Deng C., Letterio J. J., Smad4 signalling in T cells is required for suppression of gastrointestinal cancer. Nature 441, 1015–1019 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Cao J., Zhang X., Wang Q., Qiu G., Hou C., Wang J., Cheng Q., Lan Y., Han H., Shen H., Zhang Y., Yang X., Shen B., Zhang J., Smad4 represses the generation of memory-precursor effector T cells but is required for the differentiation of central memory T cells. Cell Death Dis. 6, e1984 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Y., Lee Y. T., Kaech S. M., Garvy B., Cauley L. S., Smad4 promotes differentiation of effector and circulating memory CD8 T cells but is dispensable for tissue-resident memory CD8 T cells. J. Immunol. 194, 2407–2414 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu A.-D., Zhang S., Wang Y., Xiong H., Curtis T. A., Wan Y. Y., A critical role for transcription factor Smad4 in T cell function that is independent of transforming growth factor β receptor signaling. Immunity 42, 68–79 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pauken K. E., Sammons M. A., Odorizzi P. M., Manne S., Godec J., Khan O., Drake A. M., Chen Z., Sen D. R., Kurachi M., Barnitz R. A., Bartman C., Bengsch B., Huang A. C., Schenkel J. M., Vahedi G., Haining W. N., Berger S. L., Wherry E. J., Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giordano M., Henin C., Maurizio J., Imbratta C., Bourdely P., Buferne M., Baitsch L., Vanhille L., Sieweke M. H., Speiser D. E., Auphan-Anezin N., Schmitt-Verhulst A.-M., Verdeil G., Molecular profiling of CD8 T cells in autochthonous melanoma identifies Maf as driver of exhaustion. EMBO J. 34, 2042–2058 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller B. C., Sen D. R., Abosy R. A., Bi K., Virkud Y. V., La Fleur M. W., Yates K. B., Lako A., Felt K., Naik G. S., Manos M., Gjini E., Kuchroo J. R., Ishizuka J. J., Collier J. L., Griffin G. K., Maleri S., Comstock D. E., Weiss S. A., Brown F. D., Panda A., Zimmer M. D., Manguso R. T., Hodi F. S., Rodig S. J., Sharpe A. H., Haining W. N., Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He W., Dorn D. C., Erdjument-Bromage H., Tempst P., Moore M. A. S., Massagué J., Hematopoiesis controlled by distinct TIF1γ and Smad4 branches of the TGFβ pathway. Cell 125, 929–941 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Xu P., Lin X., Feng X. H., Posttranslational regulation of smads. Cold Spring Harb. Perspect. Biol. 8, a022087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q., Xiao M., Gu S., Xu Y., Liu T., Li H., Yu Y., Qin L., Zhu Y., Chen F., Wang Y., Ding C., Wu H., Ji H., Chen Z., Zu Y., Malkoski S., Li Y., Liang T., Ji J., Qin J., Xu P., Zhao B., Shen L., Lin X., Feng X.-H., ALK phosphorylates SMAD4 on tyrosine to disable TGF-β tumour suppressor functions. Nat. Cell Biol. 21, 179–189 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Gaud G., Lesourne R., Love P. E., Regulatory mechanisms in T cell receptor signalling. Nat. Rev. Immunol. 18, 485–497 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Li M., Ong S. S., Rajwa B., Thieu V. T., Geahlen R. L., Harrison M. L., The SH3 domain of Lck modulates T-cell receptor-dependent activation of extracellular signal-regulated kinase through activation of Raf-1. Mol. Cell. Biol. 28, 630–641 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Opavsky M. A., Martino T., Rabinovitch M., Penninger J., Richardson C., Petric M., Trinidad C., Butcher L., Chan J., Liu P. P., Enhanced ERK-1/2 activation in mice susceptible to coxsackievirus-induced myocarditis. J. Clin. Invest. 109, 1561–1569 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roelen B. A. J., Cohen O. S., Raychowdhury M. K., Chadee D. N., Zhang Y., Kyriakis J. M., Alessandrini A. A., Lin H. Y., Phosphorylation of threonine 276 in Smad4 is involved in transforming growth factor-β-induced nuclear accumulation. Am. J. Physiol. Cell Physiol. 285, C823–C830 (2003). [DOI] [PubMed] [Google Scholar]
- 22.David C. J., Massagué J., Contextual determinants of TGFβ action in development, immunity and cancer. Nat. Rev. Mol. Cell Biol. 19, 419–435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seong H.-A., Jung H., Ha H., Murine protein serine/threonine kinase 38 stimulates TGF-β signaling in a kinase-dependent manner via direct phosphorylation of Smad proteins. J. Biol. Chem. 285, 30959–30970 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moren A., Raja E., Heldin C. H., Moustakas A., Negative regulation of TGFβ signaling by the kinase LKB1 and the scaffolding protein LIP1. J. Biol. Chem. 286, 341–353 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massague J., Seoane J., Wotton D., Smad transcription factors. Genes Dev. 19, 2783–2810 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Howden A. J. M., Hukelmann J. L., Brenes A., Spinelli L., Sinclair L. V., Lamond A. I., Cantrell D. A., Quantitative analysis of T cell proteomes and environmental sensors during T cell differentiation. Nat. Immunol. 20, 1542–1554 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tauriello D. V. F., Sancho E., Batlle E., Overcoming TGFβ-mediated immune evasion in cancer. Nat. Rev. Cancer 22, 25–44 (2022). [DOI] [PubMed] [Google Scholar]
- 28.Cortez V. S., Ulland T. K., Cervantes-Barragan L., Bando J. K., Robinette M. L., Wang Q., White A. J., Gilfillan S., Cella M., Colonna M., SMAD4 impedes the conversion of NK cells into ILC1-like cells by curtailing non-canonical TGF-β signaling. Nat. Immunol. 18, 995–1003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J., Kaartinen V., Generation of mice with a conditional allele for Trim33. Genesis 46, 329–333 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vincent S. D., Dunn N. R., Hayashi S., Norris D. P., Robertson E. J., Cell fate decisions within the mouse organizer are governed by graded Nodal signals. Genes Dev. 17, 1646–1662 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee P. P., Fitzpatrick D. R., Beard C., Jessup H. K., Lehar S., Makar K. W., Pérez-Melgosa M., Sweetser M. T., Schlissel M. S., Nguyen S., Cherry S. R., Tsai J. H., Tucker S. M., Weaver W. M., Kelso A., Jaenisch R., Wilson C. B., A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15, 763–774 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Chu G. C., Dunn N. R., Anderson D. C., Oxburgh L., Robertson E. J., Differential requirements for Smad4 in TGFβ-dependent patterning of the early mouse embryo. Development 131, 3501–3512 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Papageorgis P., Cheng K., Ozturk S., Gong Y., Lambert A. W., Abdolmaleky H. M., Zhou J.-R., Thiagalingam S., Smad4 inactivation promotes malignancy and drug resistance of colon cancer. Cancer Res. 71, 998–1008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang Y., Liu Y., Lu H., Sun S.-C., Jin W., Wang X., Dong C., Epigenetic activation during T helper 17 cell differentiation is mediated by Tripartite motif containing 28. Nat. Commun. 9, 1424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu G., Wang L. G., He Q. Y., ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Supplemental excel files S1 to S3