Abstract
Mitochondrial quality in skeletal muscle is crucial for maintaining energy homeostasis during metabolic stresses. However, how muscle mitochondrial quality is controlled and its physiological impacts remain unclear. Here, we demonstrate that mitoprotease LONP1 is essential for preserving muscle mitochondrial proteostasis and systemic metabolic homeostasis. Skeletal muscle–specific deletion of Lon protease homolog, mitochondrial (LONP1) impaired mitochondrial protein turnover, leading to muscle mitochondrial proteostasis stress. A benefit of this adaptive response was the complete resistance to diet-induced obesity. These favorable metabolic phenotypes were recapitulated in mice overexpressing LONP1 substrate ΔOTC in muscle mitochondria. Mechanistically, mitochondrial proteostasis imbalance elicits an unfolded protein response (UPRmt) in muscle that acts distally to modulate adipose tissue and liver metabolism. Unexpectedly, contrary to its previously proposed role, ATF4 is dispensable for the long-range protective response of skeletal muscle. Thus, these findings reveal a pivotal role of LONP1-dependent mitochondrial proteostasis in directing muscle UPRmt to regulate systemic metabolism.
Mitochondrial proteostasis directs ATF4-independent muscle mitochondrial unfolded protein response to alleviate dietary obesity.
INTRODUCTION
Mitochondria are multifunctional and essential organelles that require accurate surveillance to maintain their functional integrity. Mitochondrial proteases serve as the frontline quality control mechanism by selectively degrading damaged or dysfunctional mitochondrial proteins to ensure proper mitochondrial function (1–4). Imbalanced mitochondrial proteostasis occurs in response to cellular stresses and is observed in many chronic illnesses, including metabolic disorders, cancer, and neurodegenerative diseases (1, 3–6). However, the in vivo physiological relevance and control mechanisms of mitochondrial proteolysis remain unclear.
Preserving the integrity of the mitochondrial network is particularly important in skeletal muscle, the largest metabolically active and endocrine organ that depends critically on mitochondrial quality. Numerous studies have suggested that mitochondrial quality control mechanisms in skeletal muscle are crucial for maintaining systemic energy homeostasis during the adaptive stress response (3, 7–11). Many effects of skeletal muscle on systemic metabolism have been linked to non–cell-autonomous mitochondrial communications (7–12). Substantial evidence suggests that mitochondrial stress in skeletal muscle not only induces local adaptive responses but also sends long-range stress signals to nonaffected tissues. Many “mitokines” (soluble factors that can be released from cells with stressed mitochondria) released from skeletal muscle are known messengers to mediate interorgan metabolic communications. For instance, the mitokines FGF21 and GDF15, known to have favorable effects on systemic energy metabolism (13–17), have been shown to be induced and released from skeletal muscle upon mitochondrial stress in both mice and humans (7, 8, 10, 18–23). Evidence is emerging that mitochondrial proteostasis imbalance can trigger mitochondrial-to-nuclear communications [mitochondrial unfolded protein response (UPRmt)] and mediate adaptive genomic reprogramming (9, 11, 18). However, most of the UPRmt mechanistic studies were delineated in Caenorhabditis elegans (9, 11, 18, 24, 25), and how the UPRmt is activated and executed in mammals remains largely unknown. Thus, it is of great interest to understand the fundamental principles of the regulation of muscle mitochondrial proteostasis and the tailored responses to them.
Approximately two-thirds of the 1200 mitochondrial proteins reside in the matrix, and the regulation of the complex protein-folding environment within the matrix is therefore essential for maintaining mitochondrial proteostasis. Lon protease homolog, mitochondrial (LONP1), an evolutionarily conserved serine peptidase from yeast to human, is the major protease stationed in the mitochondrial matrix. Underscoring the importance of LONP1 in regulating a wide range of biological processes, mutations in LONP1 are associated with cerebral, ocular, dental, auricular, and skeletal anomalies (CODAS) syndrome, a human genetic disease that causes multisystem malfunctions (4, 26). Several mitochondrial regulatory proteins have been identified as LONP1 substrates in cells under basal or stress conditions (3, 4, 27, 28). Studies in mouse models further demonstrated the essential role of LONP in survival (29). Moreover, a growing body of evidence has connected LONP1 to the UPRmt, and the levels of LONP1 are also induced in response to mitochondrial stresses (3, 4, 30). However, it has yet to be unraveled whether LONP1-mediated proteolysis is implicated in controlling the muscle UPRmt and systemic metabolism.
In this study, we investigated the metabolic role of LONP1-dependent mitochondrial protein quality control in skeletal muscle, a metabolically active organ sensitive to stressed mitochondria. We generated skeletal muscle–specific LONP1 knockout mice and uncovered an essential role of the protease LONP1 in controlling muscle mitochondrial proteostasis as well as systemic metabolic homeostasis. Mice lacking LONP1 in muscle showed impaired mitochondrial protein turnover, prevention of high-fat diet (HFD)–induced obesity and insulin resistance, and reduced liver steatosis. Many of these metabolic phenotypes were recapitulated in mice overexpressing a mitochondrial-retained ΔOTC, a known unfolded protein degraded by LONP1, in skeletal muscle. Mechanistically, we demonstrated that mitochondrial proteostasis imbalance elicits an UPRmt in skeletal muscle to induce favorable metabolic remodeling in adipose tissue and liver. Muscle mitochondrial proteostasis stress directs the long-range metabolic response independent of ATF4, highlighting the ATF4-independent UPRmt in skeletal muscle.
RESULTS
Skeletal muscle–specific deletion of LONP1 confers resistance to diet-induced obesity and improves insulin sensitivity
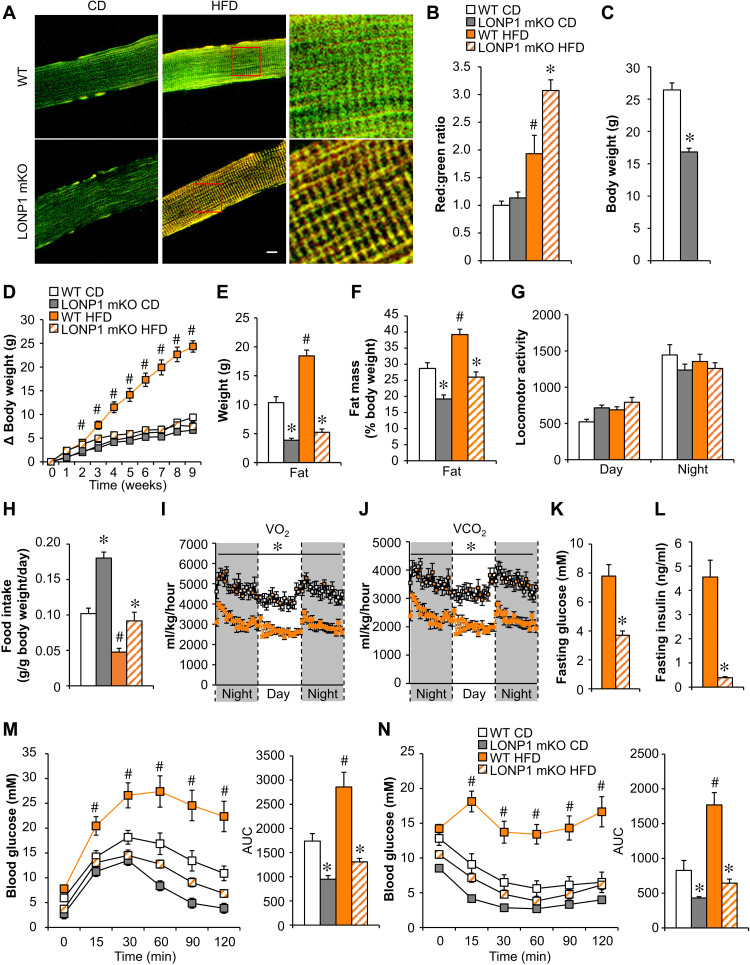
As an initial step, to explore the metabolic role of mitochondrial proteostasis in skeletal muscle in mice, we used a mouse model that allowed us to monitor the overall protein turnover status of the mitochondrial reticulum in skeletal muscles in vivo upon HFD metabolic stress. Briefly, transgenic mice express the fluorescent reporter MitoTimer; therefore, mitochondrial protein turnover in skeletal muscle can be quantified by confocal microscopy imaging. Consistent with previously published data (31), we found that the red:green ratio of MitoTimer was shifted toward red fluorescence, an indicator of slow mitochondrial protein turnover, in the soleus muscles of wild-type (WT) mice fed an HFD (60% kcal from fat) for 3 weeks (Fig. 1, A and B). LONP1 resides in the mitochondrial matrix and is the most conserved mitochondrial protease from yeast to humans (1, 4). We recently generated skeletal muscle–specific Lonp1-knockout (LONP1 mKO) mice (32), and we examined the consequences of LONP1 loss of function on muscle mitochondrial proteostasis by breeding LONP1 mKO mice with MitoTimer reporter mice. As shown in Fig. 1A, 3 weeks of HFD feeding caused a much greater increase in MitoTimer red fluorescence in skeletal muscles from LONP1 mKO mice than in WT controls (Fig. 1, A and B). These data demonstrate the crucial role of LONP1 in preserving mitochondrial proteostasis in skeletal muscle upon HFD metabolic stress.
Fig. 1. Skeletal muscle–specific deletion of LONP1 confers resistance to diet-induced obesity and improves insulin sensitivity.
(A) Confocal images of soleus muscle fibers from LONP1 mKO/MitoTimer and Lonp1f/f/MitoTimer mice following chow diet (CD) or HFD feeding. Scale bar, 10 μm. (B) Quantification of MitoTimer red:green ratio normalized (= 1.0) is shown. n = 3 to 6. (C) Body weight at the age of 6 weeks. n = 9 to 13. (D) Increase in body weight following HFD feeding. n = 8 to 13. (E) Body composition measured to determine fat mass. n = 8 to 10. (F) Fat mass plotted as a percentage of body weight. (G) Locomotor activity. n = 7 to 8. (H) Food consumption. n = 7 to 8. Data are expressed as gram consumed per gram of body weight per day. (I and J) Oxygen consumption and carbon dioxide production normalized to body weight. n = 8. (K and L) Fasting glucose and insulin levels. n = 8 to 10. (M and N) Left: Glucose and insulin tolerance test (GTT and ITT). Right: Area under the curve for GTT or ITT is shown. n = 8 to 10. Color legend for the panel: white, WT CD; gray, LONP1 mKO CD; orange, WT HFD; diagonal hatch, LONP1 mKO HFD. Values represent means ± SEM. *P < 0.05 versus corresponding WT controls; #P < 0.05 versus corresponding CD controls. Two-tailed unpaired Student’s t test (C and I to L) or one-way analysis of variance (ANOVA) (B, D to H, M, and N) was performed.
We speculated that LONP1-dependent mitochondrial proteolysis might contribute to the metabolic adaptation of skeletal muscle under excess nutrient stress. We next evaluated how LONP1 mKO mice could adapt to HFD metabolic stress. Although LONP1 mKO mice were smaller and showed reduced muscle mass (32), we followed a cohort of LONP1 mKO mice that show survival similar to that of respective WT littermates at least up to 1.5 years of age. LONP1 mKO mice were also leaner when fed standard chow diet (CD) and were resistant to obesity on HFD, gaining body weight similarly to WT mice on CD (Fig. 1, C to F, and fig. S1, A and B). Total lean mass was equivalently decreased because of reduced muscle fiber size in LONP1 mKO mice fed either CD or HFD (fig. S1, C to E). Fat mass but not lean mass, when normalized to body weight, was significantly lower in LONP1 mKO mice than in WT littermates regardless of the diet (Fig. 1F and fig. S1D). LONP1 mKO mice fed either CD or HFD have approximately similar patterns of activity during both the light and dark cycles in comparison with WT controls (Fig. 1G). However, food intake, when normalized to body weight, was significantly higher in LONP1 mKO mice than in WT littermates fed either CD or HFD (Fig. 1H and fig. S1F), indicating that LONP1 mKO mice have a decreased efficiency in converting food into body mass. In addition, LONP1 mKO mice fed either CD or HFD have a marked increase in oxygen consumption (VO2) and carbon dioxide (VCO2) production during both dark and light cycles in comparison with WT controls (Fig. 1, I and J, and fig. S1, G and H). We further determined glucose homeostasis in WT and LONP1 mKO mice. The fasting glucose and insulin levels were significantly reduced in HFD-fed LONP1 mKO mice compared to HFD-fed WT controls (Fig. 1, K and L), indicating that HFD-fed LONP1 mKO mice have an improved sensitivity to insulin. Glucose tolerance tests (GTTs) showed that LONP1 mKO mice were more tolerant to glucose challenge compared to the WT littermates or human α-skeletal actin (HSA)–Cre controls after HFD feeding (Fig. 1M and fig. S1I). Similarly, insulin tolerance tests (ITTs) demonstrated that HFD-fed LONP1 mKO mice were more sensitive to insulin stimulation than HFD-fed WT controls (Fig. 1N). Collectively, these results suggest that muscle LONP1 deficiency confers resistance to HFD-induced obesity and improves insulin sensitivity. Together, these data established a crucial role of muscle LONP1 for mitochondrial proteostasis and systemic metabolic homeostasis.
Muscle LONP1 regulates the response of adipose tissue and liver to HFD
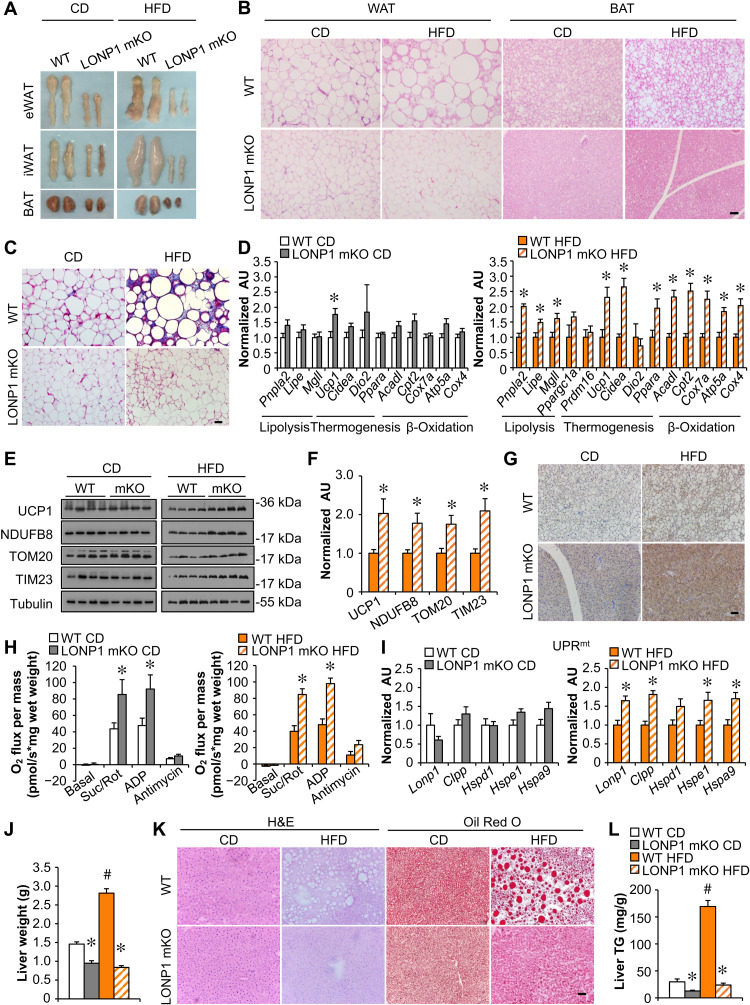
Evidence has emerged that skeletal muscle metabolic reprogramming could be a modifier of adipose and liver metabolism. We next examined lipid metabolism in adipose tissue of LONP1 mKO mice. Both the white and brown adipose tissue (WAT and BAT) pads were smaller with less lipid accumulation in LONP1 mKO mice under CD and particularly HFD conditions as compared to their WT controls (Fig. 2A and fig. S1J). Histological analysis also showed smaller adipocytes in adipose tissue of LONP1 mKO mice, in particular under HFD conditions (Fig. 2B). Increased macrophage infiltration and fibrosis in adipose tissue have been linked to obesity. We found that the expression of macrophage (e.g., F4/80, Cd68, and Cd11c)– and profibrosis (e.g., Hif1α, Lox, and Tgfβ)–related genes were markedly reduced in WAT from HFD-fed LONP1 mKO mice compared with WT controls (fig. S2A). Masson’s trichrome staining confirmed a marked decrease in HFD-induced fibrosis in WAT from LONP1 mKO mice (Fig. 2C). On a CD, LONP1 mKO mice showed minimal induction of thermogenic gene expression in BAT (Fig. 2D). However, HFD-fed LONP1 mKO mice had a significant increase in the mRNA expression of genes involved in lipolysis (e.g., Pnpla2, Lige, and Mgll), thermogenesis (e.g., Ucp1 and Cidea), and mitochondrial oxidation (e.g., Cpt2, Cox7a, and Atp5a) in BAT in comparison with WT control mice fed HFD (Fig. 2D). An increase in the mRNA expression of genes associated with lipolysis was also observed in WAT from HFD-fed LONP1 mKO mice (fig. S2B). Moreover, HFD feeding led to a greater induction of mitochondrial protein expression in BAT from LONP1 mKO mice (Fig. 2, E and F, and fig. S2C). In LONP1 mKO mice, HFD appears to more markedly induce uncoupling protein 1 (UCP1) protein expression in BAT as evidenced by both Western blot and immunohistochemistry (IHC) (Fig. 2, E to G). Mitochondrial respiration rates were significantly higher in the BAT of LONP1 mKO mice than that of WT littermates fed either CD or HFD (Fig. 2H). Notably, LONP1 mKO mice fed CD showed no differences in the mRNA expression of genes associated with UPRmt in BAT (Fig. 2I). However, we observed an induction of UPRmt in BAT of LONP1 mKO mice. The expression levels of mitochondrial proteases (e.g., Lonp1 and Clpp) and chaperones (e.g., Hspe1 and Hspa9) were significantly elevated in the BAT of HFD-fed LONP1 mKO mice (Fig. 2I). Notably, the mRNA levels of genes encoding amino acid metabolism (e.g., Asns, Aldh18a1, and Slc7a11), one-carbon metabolism (e.g., Psat1 and Mthfd2) and Fgf21 were not increased in HFD-fed LONP1 mKO BAT (fig. S2D). These data suggest that LONP1-deficient muscles send stress signals to nonaffected tissues. Together, these data indicate that mitochondrial proteostasis stress in skeletal muscle upon loss of LONP1 triggers thermogenic remodeling and UPRmt in distal adipose tissue.
Fig. 2. Muscle LONP1 regulates the response of adipose tissue and liver to HFD.
(A) Pictures of adipose pads from indicated mice fed CD or HFD. (B) Hematoxylin and eosin (H&E) staining of epididymal WAT (eWAT) and BAT. Scale bar, 50 μm. n = 5 to 9. (C) Masson’s trichrome staining in eWAT. Scale bar, 50 μm. n = 5 to 9. (D) Expression of genes [quantitative reverse transcription polymerase chain reaction (qRT-PCR)] in BAT from indicated mice fed CD or HFD. n = 6. (E) Western blot analysis of BAT from indicated mice. n = 4. (F) Quantification of UCP1/tubulin, NDUFB8/tubulin, TOM20/tubulin, and TIM23/tubulin in BAT from indicated mice fed HFD. n = 4. (G) UCP1 immunohistochemistry (IHC) staining in BAT from indicated mice fed CD or HFD. Scale bar, 50 μm. n = 4 to 5. (H) Mitochondrial respiration rates were determined from BAT of indicated genotypes. Succinate/rotenone (Suc/Rot)–stimulated, adenosine 5′-diphosphate (ADP)–dependent, and antimycin-induced respiration is shown. n = 5 to 8. (I) Expression of genes involved in UPRmt in BAT from indicated mice fed CD or HFD. n = 6. (J) Liver weight. n = 8 to 10. (K) H&E and Oil Red O staining of livers. Scale bar, 50 μm. n = 5 to 6. (L) Liver triglyceride levels. n = 6 to 8. Color legend for the panel: white, WT CD; gray, LONP1 mKO CD; orange, WT HFD; diagonal hatch, LONP1 mKO HFD. Values represent means ± SEM. *P < 0.05 versus corresponding WT controls; #P < 0.05 versus corresponding CD controls. Two-tailed unpaired Student’s t test (D, F, H, and I) or one-way ANOVA (J and L) was performed. AU, arbitrary units.
Lonp1 was not disrupted in the livers from LONP1 mKO mice (fig. S2E). Gross anatomical analysis of livers revealed markedly reduced hepatic steatosis in HFD-fed LONP1 mKO mice compared to WT controls (fig. S2F). This was supported by reduced liver weight in HFD-fed LONP1 mKO mice (Fig. 2J). Histological examination of liver sections with hematoxylin and eosin (H&E) and Oil Red O staining provided further evidence of markedly reduced liver fat deposition in HFD-fed LONP1 mKO mice (Fig. 2K). Biochemical measurements confirmed that the increase in triglyceride (TG) levels observed in WT livers after HFD feeding was completely blunted in LONP1 mKO livers (Fig. 2L). To further determine liver metabolic reprogramming in LONP1 mKO mice, we analyzed the expression profile of metabolic genes in the livers. HFD-fed LONP1 mKO mice had markedly reduced expression of genes involved in fatty acid import (e.g., Cd36) and de novo lipogenesis (e.g., Fasn and Acaca) in the livers (fig. S2G), whereas we did not observe a change in the mRNA expression of genes associated with fatty acid oxidation and gluconeogenesis in the livers from HFD-fed LONP1 mKO mice (fig. S2G). The markedly decreased fatty acid synthase (FASN) protein levels in the livers of HFD-fed LONP1 mKO mice were also confirmed by immunoblotting (fig. S2H). We did not detect induction of UPRmt-related gene expression in livers from LONP1 mKO mice on HFD (fig. S2I). These findings implicate that LONP1 deletion in skeletal muscle decreases free fatty acid uptake and de novo lipogenesis in the liver, leading to protection from diet-induced hepatic steatosis. Together, these data suggest that LONP1-dependent mitochondrial signaling in skeletal muscle mediates the interorgan metabolic communications.
Loss of LONP1 elicits the mitochondrial UPR in skeletal muscle
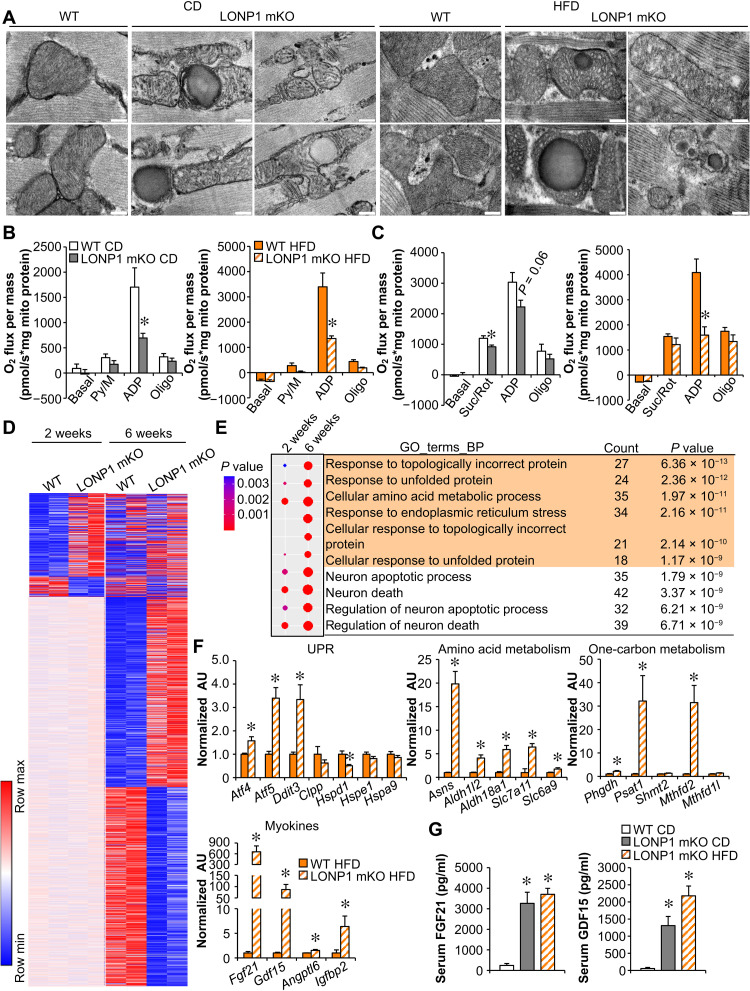
To understand the molecular basis of the lean phenotype in LONP1 mKO mice, we first examined mitochondrial remodeling in skeletal muscle from LONP1 mKO mice. Mitochondrial quality was assessed by electron microscopy (EM) in the soleus muscles of LONP1 mKO and WT control mice on CD or HFD (Fig. 3A). The quantitative EM analysis revealed that the relative mitochondrial area per muscle fiber area was not different in LONP1 mKO muscles compared with WT controls regardless of the diet (fig. S3A). In addition, we did not observe a change in mtDNA content or mitochondrial ETC protein levels in LONP1 mKO muscles relative to WT controls under both CD and HFD conditions (fig. S3, B and C). However, LONP1 deficiency led to disturbed mitochondrial ultrastructure, characterized by loss of cristae content, abnormal vacuolated structure, and electrodense aggregates under both CD and HFD conditions (Fig. 3A). Consistent with the structural derangements, both pyruvate- and succinate-driven mitochondrial respiration rates were markedly reduced in LONP1 mKO muscles compared to the WT controls under both CD and HFD conditions (Fig. 3, B and C). Together, these data demonstrate that LONP1 preserves mitochondrial quality in skeletal muscle.
Fig. 3. Loss of LONP1 elicits the mitochondrial UPR in skeletal muscle.
(A) Electron micrographs of soleus muscle showing intermyofibrillar mitochondria in sections from indicated mice fed CD or HFD. Scale bars, 200 nm. n = 3 to 4. (B and C) Mitochondrial respiration rates were determined on mitochondria isolated from muscles of indicated mice using pyruvate (B) or succinate (C) as substrates. Pyruvate/malate (Py/M)- or Suc/Rot-stimulated respiration is shown. n = 3 to 7. (D) Heatmap analysis of genes differentially regulated in LONP1 mKO muscles. Each group is represented by RNA sequencing (RNA-seq) data from two independent samples generated from muscles of 2- and 6-week-old mice. Red, relative increase in abundance; blue, relative decrease. (E) Gene Ontology (GO) enrichment analysis of genes that were induced early at 2 weeks old and were further enhanced by LONP1 abrogation at 6 weeks old, with top 10 terms shown. The dot size reflects the gene count. (F) Expression of genes (qRT-PCR) related to UPR, amino acid metabolism, one-carbon metabolism, and myokines in muscles from HFD-fed LONP1 mKO mice. n = 6. (G) Serum FGF21 and GDF15 levels. n = 6 to 9. Color legend for the panel: white, WT CD; gray, LONP1 mKO CD; orange, WT HFD; diagonal hatch, LONP1 mKO HFD. Values represent means ± SEM. *P < 0.05 versus corresponding WT controls. Two-tailed unpaired Student’s t test (B, C, and F) or one-way ANOVA (G) was performed.
Substantial evidence suggests that stressed mitochondria can communicate with the nucleus and mediate adaptive genomic reprogramming. To thoroughly analyze the gene expression changes that result from loss of muscle LONP1, we performed RNA sequencing (RNA-seq) transcriptome analysis of skeletal muscles from LONP1 mKO and WT mice at 2 or 6 weeks of age. This transcriptome analysis revealed remarkable muscle genomic reprogramming early at 2 weeks of age that became more marked at 6 weeks of age in LONP1 mKO mice (Fig. 3D). Comparison of gene expression profiles between LONP1 mKO and WT muscles showed 457 and 1922 genes that were regulated (with a cutoff of 1.5-fold change and P < 0.05) by LONP1 abrogation at 2 and 6 weeks of age, respectively (Fig. 3D). As LONP1 abrogation appeared to affect genes involved in a great variety of biological processes (Fig. 3E and fig. S3, D to G), we focused on the genes that were induced early at 2 weeks of age and were further enhanced by LONP1 abrogation at 6 weeks of age (Fig. 3E). Gene Ontology (GO) pathway analysis revealed that muscle LONP1 deficiency triggers a progressive mitochondrial UPR. We then confirmed the mitochondrial UPR in HFD-fed LONP1 mKO muscles by quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis. The mRNA expression of Atf4, Atf5, and Ddit3, known markers of the UPR, was up-regulated in HFD-fed LONP1 mKO muscles (Fig. 3F). The mRNA levels of genes encoding amino acid metabolism (e.g., Asns, Slc7a11, and Slc6a9) and one-carbon metabolism (e.g., Phgdh, Psat1, and Mthfd2) were also up-regulated, as well as a broad array of genes encoding myokines (e.g., Fgf21 and Gdf15) (Fig. 3F and fig. S3H). The marked induction of FGF21 and GDF15 in LONP1 mKO muscles is of interest given their well-known favorable effect on systemic metabolism (13–17). Numerous studies in mice, and in humans, have shown that stressed muscle secretes FGF21 and GDF15 into circulation to exert an endocrine effect (7, 8, 10, 18–23). We also detected significantly increased abundance of circulating FGF21 and GDF15 protein in LONP1 mKO mice relative to their WT littermates (Fig. 3G). Therefore, our results suggest that muscle mitochondrial proteostasis stress upon loss of LONP1 stimulates the expression of mitokines FGF21 and GDF15 to mediate interorgan metabolic communications. Thus, these results link the LONP1-dependent muscle mitochondrial UPR to systemic metabolic benefits.
Mitochondria overloaded by unfolded proteins beyond the LONP1 capacity induce the UPRmt in skeletal muscle
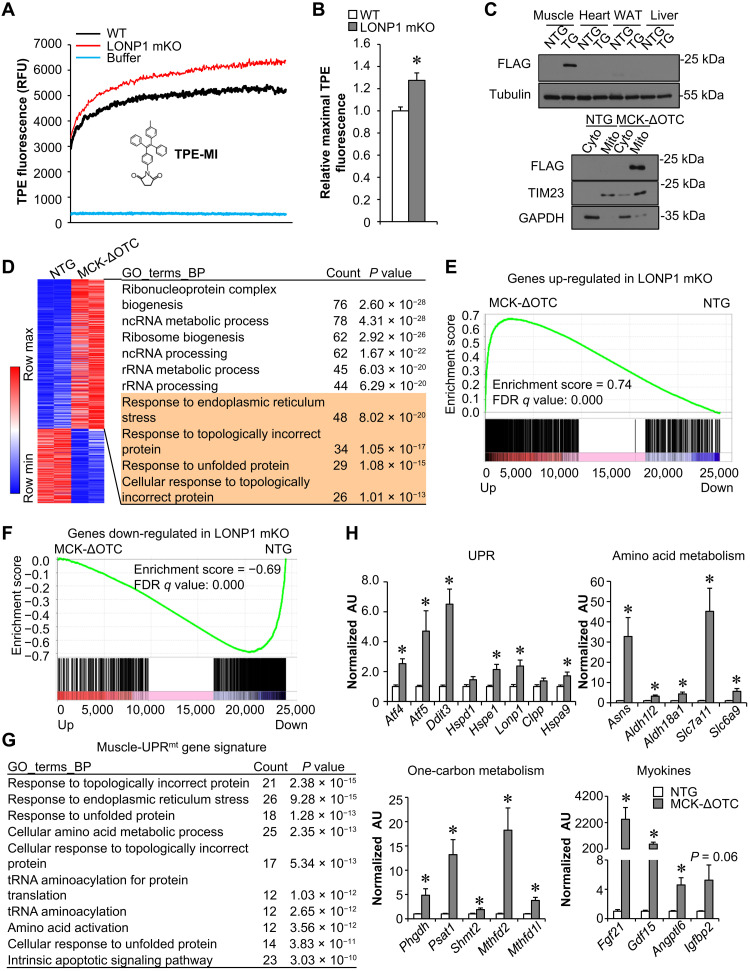
LONP1 is well known as a quality control protease responsible for the degradation of misfolded or damaged proteins in the mitochondrial matrix (1, 2, 4). We speculated that the activation of the muscle mitochondrial stress response can be due to the accumulation of mitochondrial-retained protein in muscle mitochondria upon loss of LONP1. To provide direct support for this notion, we measured mitochondrial unfolded protein levels in LONP1 mKO mitochondria. Tetraphenylethene maleimide (TPE-MI) is a turn-on fluorogenic dye that could be used to probe unfolded proteins, thus reporting proteostasis imbalance. We observed an increased level of TPE-MI fluorescence, an indicator of higher unfolded protein abundance, in the mitochondria isolated from LONP1 mKO muscles compared to WT controls (Fig. 4, A and B). We next asked whether mitochondria overloaded by unfolded proteins beyond the LONP1 capacity could also lead to similar genomic reprogramming effects in skeletal muscle. Mitochondrial-retained mutant ornithine transcarbamylase (ΔOTC) is a known unfolded protein degraded by LONP1 and an established model for studying mitochondrial proteostasis imbalance (33–35). We generated transgenic mice overexpressing ΔOTC specifically in skeletal muscle using the muscle creatine kinase promoter (MCK-ΔOTC). The ΔOTC protein was expressed specifically in skeletal muscle mitochondria, and we observed no overexpression of ΔOTC protein in other tissues such as the heart, liver, and fat (Fig. 4C). We thus analyzed the gene expression changes that result from mitochondria overloaded by ΔOTC-unfolded proteins in skeletal muscles. Transcriptome analysis was performed by whole-genome gene expression profiling experiments in gastrocnemius (GC) muscles from MCK-ΔOTC mice and nontransgenic (NTG) littermate controls. Consistent with the observations in LONP1 mKO muscles, the comparative mRNA profiling strategy revealed extensive genomic reprogramming in MCK-ΔOTC muscles, with 1051 up-regulated and 519 down-regulated genes (1.5-fold change and P < 0.05), respectively (Fig. 4D). GO analysis of the regulated genes in MCK-ΔOTC muscles also revealed significant enrichment in UPR as well as RNA processing (Fig. 4D and fig. S4, A to C). Gene set enrichment analysis (GSEA) further demonstrated that ΔOTC overexpression significantly influenced the expression of LONP1-regulated genes in muscles (Fig. 4, E and F). These data suggest that mitochondria overloaded by ΔOTC-unfolded proteins recapitulate the genomic reprogramming effects in LONP1 mKO muscles.
Fig. 4. Mitochondria overloaded by unfolded proteins beyond the LONP1 capacity induce the UPRmt in skeletal muscle.
(A) Mitochondria were isolated from indicated mice and incubated with TPE-MI dye, followed by measurement of fluorescence intensity for 2 hours. n = 6 to 8. (B) Quantification of the maximal fluorescence ratios normalized (= 1.0) to WT controls. n = 6 to 8. (C) Top: Western blot analysis of ΔOTC expression in tissues from indicated mice. Bottom: Western blot analysis performed with mitochondrial and cytosolic fractions isolated from muscles of the indicated mice. (D) Left: Heatmap of differentially expressed genes. RNA-seq data (n = 2 independent) were generated from muscles of 2-week-old indicated mice. Red, relative increase in abundance; blue, relative decrease. Right: GO enrichment analysis of gene transcripts up-regulated in MCK-ΔOTC muscles, with top 10 terms shown. (E and F) GSEA of genes regulated in LONP1-deficient muscles in relation to genes altered in MCK-ΔOTC muscles. Genes regulated by ΔOTC overexpression were ranked by fold difference and expressed on the x axis. This dataset was compared with regulated genes identified in LONP1 mKO muscles. (G) GO enrichment analysis of the muscle-UPRmt gene signature, the core set of genes commonly up-regulated by both ΔOTC overexpression and LONP1 mKO in muscle, with top 10 terms shown. (H) Expression of genes (qRT-PCR) related to UPR, amino acid metabolism, one-carbon metabolism, and myokines in muscles from indicated mice. n = 6. Values represent means ± SEM. *P < 0.05 versus corresponding nontransgenic (NTG) controls. Two-tailed unpaired Student’s t tests were performed.
We then focused on the core set of genes commonly up-regulated by both ΔOTC overexpression and LONP1 mKO in skeletal muscle, hereafter referred to as the muscle-UPRmt gene signature (table S1). GO analysis revealed that the primary muscle-UPRmt gene sets were the response to unfolded protein and amino acid metabolism (Fig. 4G). Gene expression validation studies demonstrated that a broad array of genes involved in UPR (e.g., Atf4, Atf5, and Ddit3), amino acid metabolism (e.g., Asns, Slc7a11, and Slc6a9), one-carbon metabolism (e.g., Phgdh, Psat1, and Mthfd2), and myokines (e.g., Fgf21 and Gdf15) were induced in MCK-ΔOTC muscles (Fig. 4H and fig. S4D). This observation points to the possibility that muscle-UPRmt signals may mediate the long-range protective response that alleviates dietary obesity.
Mitochondrial proteostasis stress in skeletal muscle protects against HFD-induced obesity and improves glucose homeostasis
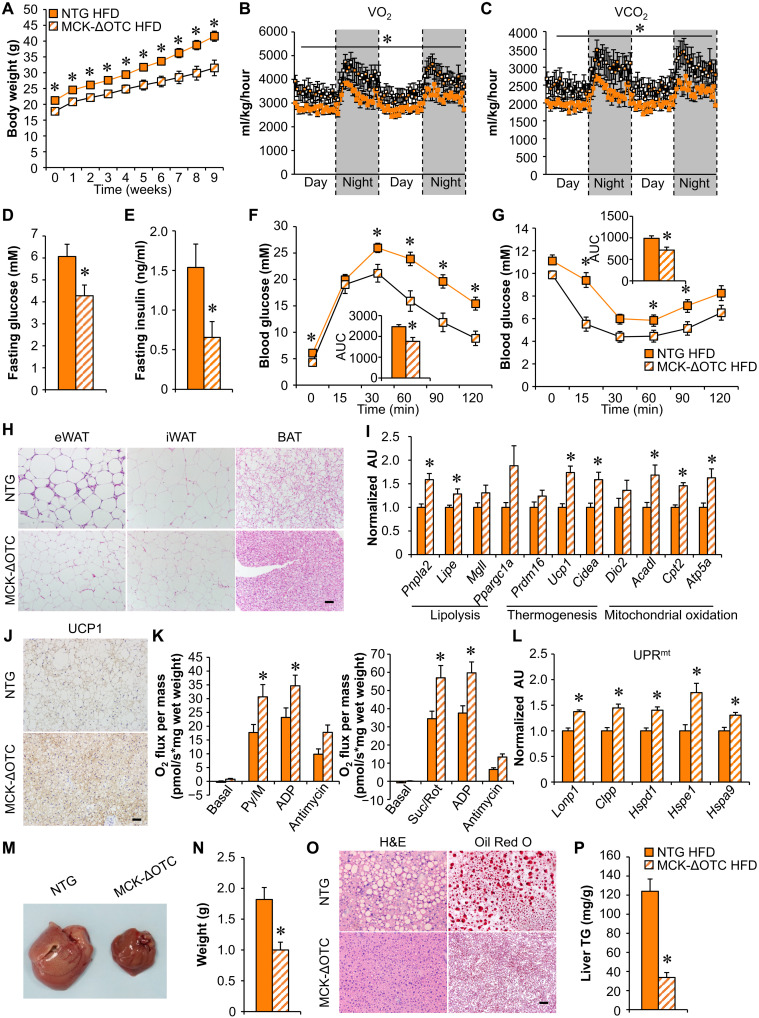
To further confirm the importance of the skeletal muscle mitochondrial proteostasis stress response in regulating systemic metabolism, MCK-ΔOTC mice and NTG littermates were fed either a normal CD or HFD. Similar to the LONP1 mKO mice, the body weights of MCK-ΔOTC mice were lower than those of NTG littermate controls fed CD (fig. S5A). A decrease in muscle mitochondrial respiration rates was also observed in CD-fed MCK-ΔOTC mice (fig. S5B), but unlike LONP1 mKO mice, mitochondrial respiration rates were not different between MCK-ΔOTC and NTG littermates following HFD feeding (fig. S5B). MCK-ΔOTC mice were protected against obesity when challenged with an HFD for 3 months (Fig. 5A and fig. S5, C and D), showing significantly less weight gain following HFD feeding than NTG littermates despite higher food consumption (fig. S5, C and E). HFD-fed MCK-ΔOTC mice exhibited an increase in oxygen consumption (VO2) and carbon dioxide (VCO2) production during both dark and light cycles (Fig. 5, B and C). Notably, HFD-fed MCK-ΔOTC mice had increased locomotor activity at night (fig. S5F). In addition, MCK-ΔOTC mice showed improved whole-body glucose and insulin sensitivity after 12 weeks of HFD feeding (Fig. 5, D to G). Both fasting glucose and insulin levels were significantly decreased in HFD-fed MCK-ΔOTC mice compared to HFD-fed WT controls (Fig. 5, D and E). GTT and ITT further demonstrated that HFD-fed MCK-ΔOTC mice were more tolerant to glucose challenge and more sensitive to insulin stimulation than HFD-fed WT controls (Fig. 5, F and G). Thus, this metabolic phenotype exhibited by MCK-ΔOTC mice appears to resemble that of LONP1 mKO mice.
Fig. 5. Mitochondrial proteostasis stress in skeletal muscle protects against HFD-induced obesity and improves glucose homeostasis.
(A) Growth curves following HFD feeding. n = 12 to 14. (B and C) Oxygen consumption and carbon dioxide production normalized to body weight. n = 7 to 8. (D and E) Fasting glucose and insulin levels. n = 12 to 14. (F and G) GTT and ITT. Inset: Area under the curve for GTT or ITT is shown. n = 12 to 14. (H) H&E staining of eWAT, inguinal WAT (iWAT), and BAT of indicated mice following HFD feeding. Scale bar, 50 μm. n = 7 to 9. (I) Expression of genes (qRT-PCR) in BAT from indicated mice. n = 6. (J) UCP1 IHC staining in BAT. Scale bar, 50 μm. n = 4 to 5. (K) Mitochondrial respiration rates were determined from the BAT of indicated genotypes using pyruvate or succinate as substrates. n = 9 to 11. (L) Expression of genes (qRT-PCR) involved in UPRmt in BAT from indicated mice. n = 6. (M) Pictures of livers from the indicated mice fed HFD. (N) Liver weight. n = 12. (O) H&E and Oil Red O staining of livers. Scale bar, 50 μm. n = 6. (P) Liver triglyceride levels. n = 6 to 9. Color legend for the panel: orange, NTG HFD; diagonal hatch, MCK-ΔOTC HFD. Values represent means ± SEM. *P < 0.05 versus corresponding NTG controls. Two-tailed unpaired Student’s t tests were performed.
We next examined the interorgan metabolic communications in HFD-fed MCK-ΔOTC mice. Similar to the LONP1 mKO mice, HFD-fed MCK-ΔOTC mice also showed significantly less fat mass and smaller adipocytes than NTG littermates (Fig. 5H and fig. S5, G and H). HFD-fed MCK-ΔOTC mice had decreased macrophage infiltration and fibrosis in adipose tissue (fig. S5, I and J). In addition, HFD-fed MCK-ΔOTC mice also had increased expression of genes encoding lipolysis, thermogenesis, and mitochondrial oxidation in BAT (Fig. 5I). The number of UCP1-positive adipocytes and mitochondrial respiration rates were increased in the BAT of HFD-fed MCK-ΔOTC mice (Fig. 5, J and K). Moreover, we further confirmed non–cell-autonomous UPRmt activation in BAT from HFD-fed MCK-ΔOTC mice (Fig. 5L). Furthermore, HFD-fed MCK-ΔOTC mice also had reduced hepatic steatosis compared to NTG mice (Fig. 5, M to P). Together, the similarity of the phenotypes of MCK-ΔOTC and LONP1 mKO mice strongly demonstrates that the mitochondrial proteostasis stress response in skeletal muscle confers resistance to HFD-induced obesity and improves insulin sensitivity.
Mitochondrial proteostasis stress programs local amino acid and one-carbon metabolism in skeletal muscle via the ATF4 transcription factor
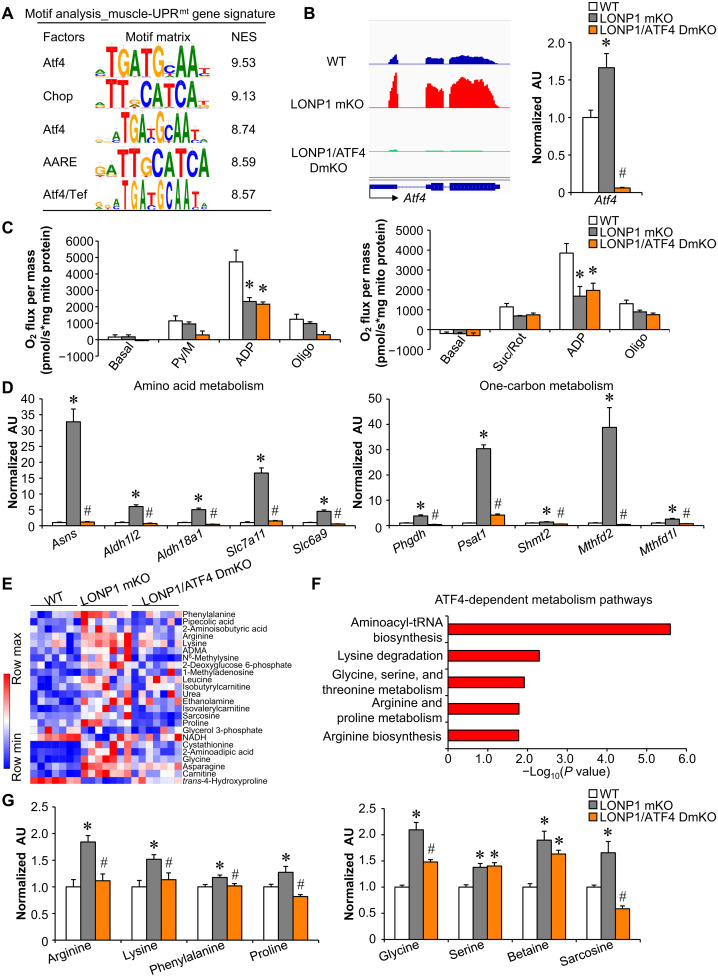
What might be the molecular interface between mitochondrial proteostasis imbalance and genomic reprogramming in skeletal muscle? The search for enriched motifs within the muscle-UPRmt gene sets using i-cisTarget revealed an ATF4 sequence element, 5′-ATGATGCAAT-3′, as the highest-score motif with a normalized enrichment scores (NES) of 9.53 (Fig. 6A). C/EBP homologous protein (CHOP) and amino acid response element (AARE) motifs were also enriched within the muscle-UPRmt gene sets (Fig. 6A). Previous studies have suggested an important role of ATF4 in regulating the mitochondrial stress response in mammals (9, 11, 30, 36, 37). We confirmed that Atf4 mRNA expression levels were up-regulated by both ΔOTC overexpression and LONP1 mKO in skeletal muscles (Fig. 6B and fig. S6A). We next asked whether knocking out ATF4 would be sufficient to ablate genomic and metabolic reprogramming in muscles from LONP1 mKO mice. We bred LONP1 mKO mice with muscle-specific ATF4 KO mice to generate LONP1/ATF4f/f/HSA-Cre (LONP1/ATF4 DmKO) mice, in which Atf4 gene was disrupted in muscle in the LONP1 mKO background (Fig. 6B). We confirmed that the induced expression of Atf4 was completely abolished in the LONP1/ATF4 DmKO muscles (Fig. 6B). Deletion of ATF4 failed to affect mitochondrial respiration defects in skeletal muscles lacking LONP1 (Fig. 6C). However, LONP1 deficiency–mediated induction of amino acid metabolism gene expression (e.g., Asns, Slc7a11, and Slc6a9) was significantly reduced in the absence of ATF4 (Fig. 6D). Moreover, muscle-specific disruption of ATF4 also abolished the marked induction of one-carbon metabolism gene expression (e.g., Phgdh, Psat1, and Mthfd2) in LONP1 mKO muscles (Fig. 6D). These data suggest that ATF4 is required for the adaptive increase in amino acid and one-carbon metabolism gene expression in LONP1 mKO muscles. We also conducted metabolomic analysis on skeletal muscles using capillary electrophoresis–mass spectrometry (CE-MS). We measured a large panel (166 intermediates) of organic acids and amino acids in seven replicates of skeletal muscle from WT, LONP1 mKO, and LONP1/ATF4 DmKO mice (Fig. 6E and fig. S6C). Principal components analysis (PCA) showed a clear global metabolite profile shift in LONP1 mKO muscles, suggesting that LONP1 ablation induces divergent skeletal muscle metabolic changes (fig. S6B). LONP1 deficiency resulted in elevated levels of a broad array of amino acid intermediates in skeletal muscle (Fig. 6E and fig. S6C). Many of these effects were blocked upon ATF4 ablation (Fig. 6, E to G), which is consistent with the gene expression results. Notably, glycine and sarcosine are known cellular donors of one-carbon units (Fig. 6G). LONP1 deficiency–mediated induction of glycine and sarcosine was also significantly reduced in the absence of ATF4 (Fig. 6G and fig. S6D). Together, these results demonstrate that ATF4 is necessary for the mitochondrial proteostasis stress–mediated activation of local amino acids and one-carbon metabolism in skeletal muscle.
Fig. 6. Mitochondrial proteostasis stress programs local amino acid and one-carbon metabolism in skeletal muscle via the ATF4 transcription factor.
(A) Motif enrichment analysis of the muscle-UPRmt gene signature using i-CisTarget. The table includes the transcription factor gene symbols, consensus binding sites, and their normalized enrichment scores (NES). (B) Left: Genome browser tracks of RNA-seq data were visualized in Integrative Genomics Viewer. Right: Expression of the Atf4 (qRT-PCR) in GC muscle from indicated genotypes. n = 6. (C) Mitochondrial respiration rates were determined on mitochondria isolated from muscles of the indicated mice using pyruvate or succinate as substrates. n = 3 to 5. (D) Expression of genes (qRT-PCR) involved in amino acid and one-carbon metabolism in GC muscles from indicated mice. n = 6. (E) Heatmap analysis of muscle metabolites. Capillary electrophoresis–mass spectrometry–based metabolite analysis was performed with GC muscles from indicated mice. n = 7. Red, relative increase in abundance; blue, relative decrease. ADMA, asymmetric dimethylarginine; NADH, reduced form of nicotinamide adenine dinucleotide. (F) Pathway analysis of ATF4-dependent metabolites using online software MetaboAnalyst 5.0. (G) Amino acid levels in WT, LONP1 mKO, and LONP1/ATF4 DmKO muscles. n = 7. Color legend for the panel: white, WT; gray, LONP1 mKO; orange, LONP1/ATF4 DmKO. Values represent means ± SEM. *P < 0.05 versus WT controls; #P < 0.05 versus LONP1 mKO. One-way ANOVA tests were performed.
Mitochondrial proteostasis stress directs a long-range metabolic response independent of ATF4 in skeletal muscle
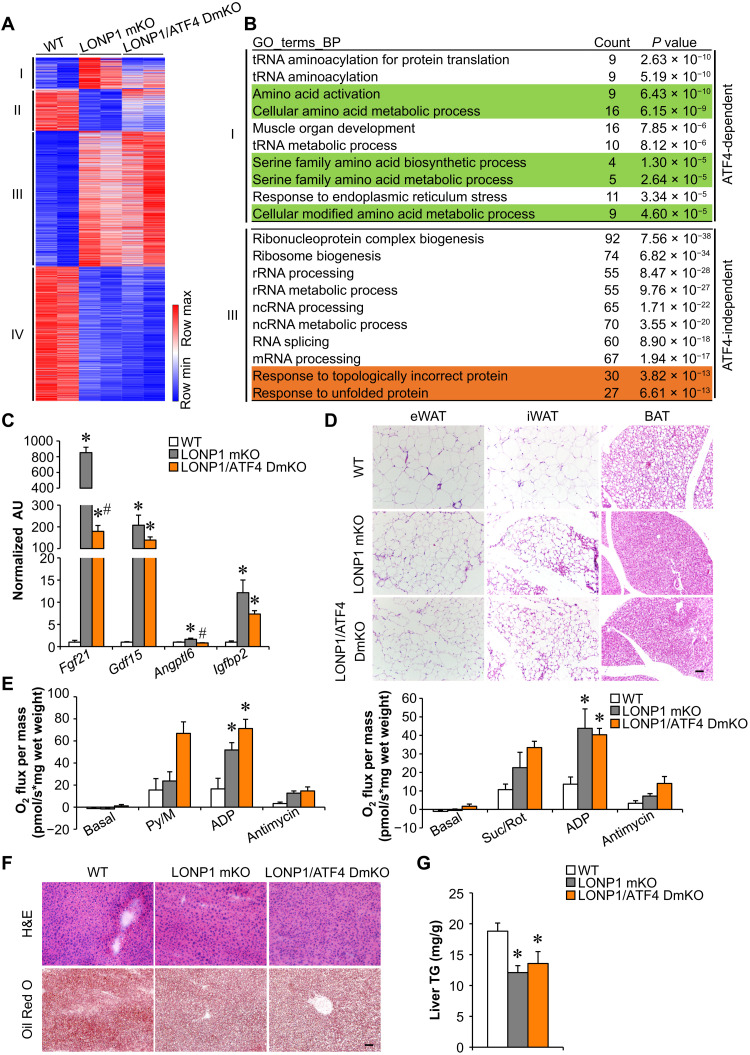
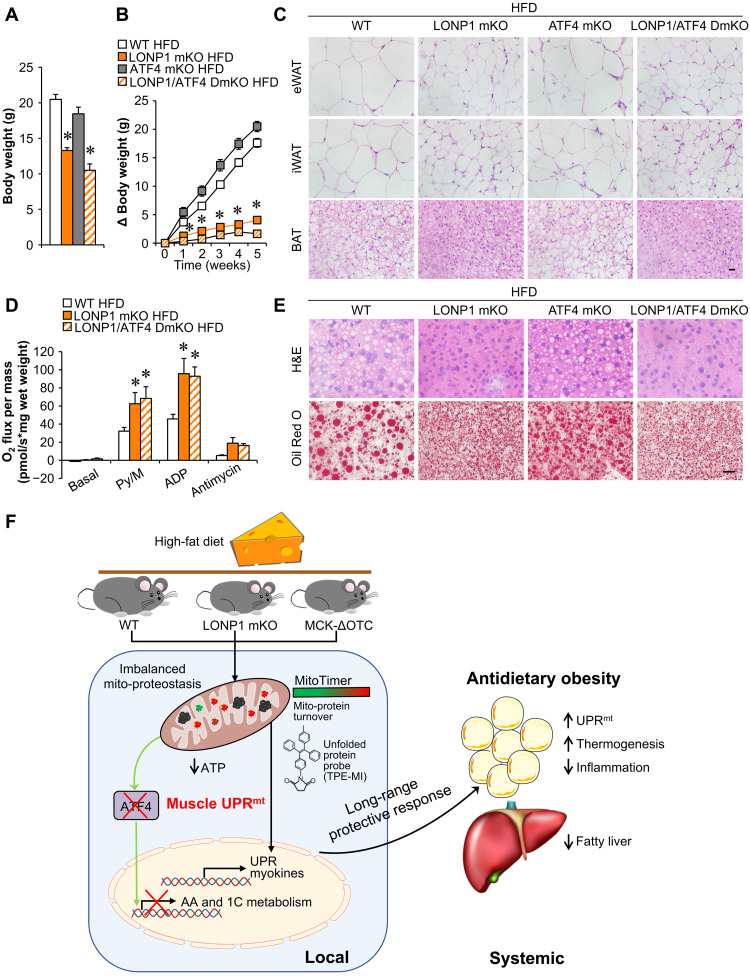
To thoroughly analyze pathways that are affected by ATF4 ablation in the context of LONP1 mKO, we performed RNA-seq transcriptome analysis of skeletal muscles from WT, LONP1 mKO, and LONP1/ATF4 DmKO mice. We identified a total of 3017 differentially expressed genes with a cutoff of 1.5-fold and P < 0.05. Unexpectedly, whereas we found that ATF4 loss blunted a subset of regulated genes in LONP1 mKO muscles (Fig. 7A), the majority of differentially regulated genes in LONP1 mKO muscles were not affected by ATF4 ablation (Fig. 7A). We further clustered the differentially regulated genes into four groups (Fig. 7A). The numbers of genes were 276 (cluster I), 367 (cluster II), 1192 (cluster III), and 1182 (cluster IV) (Fig. 7A). Clusters I and II showed ATF4-dependent genes regulated by LONP1 ablation, whereas clusters III and IV showed ATF4-independent genes affected by LONP1 deficiency (Fig. 7A). We also performed GO analysis of differentially expressed genes in different clusters (Fig. 7B and fig. S7A). Genes in cluster I revealed significant enrichment in amino acid metabolic processes, which is consistent with ATF4-dependent amino acid metabolism in LONP1 mKO muscles described above (Fig. 7B and fig. S7B). However, LONP1 deficiency–induced activation of muscle UPR and UPR-related RNA processing as well as myokine pathways were not affected by ATF4 ablation (Fig. 7B and fig. S7C). Moreover, gene expression validation studies demonstrated that many myokine genes (e.g., Fgf21 and Gdf15) were induced in both LONP1 mKO and LONP1/ATF4 DmKO muscles (Fig. 7C). As expected, histological analysis also showed smaller adipocytes in adipose tissue of LONP1/ATF4 DmKO mice than in adipose tissue of WT mice (Fig. 7D). Consistently, BAT mitochondrial respiration rates were increased in LONP1 mKO mice but not affected by disruption of ATF4 in muscle (Fig. 7E). Moreover, LONP1/ATF4 DmKO mice also had reduced liver lipid accumulation compared to WT mice, as evidenced by H&E and Oil Red O staining (Fig. 7F). Biochemical measurements confirmed reduced TG levels in livers from LONP1 mKO and LONP1/ATF4 DmKO mice (Fig. 7G). LONP1/ATF4 DmKO mice also showed lower body weight than those of ATF4 mKO or WT mice on CD (Fig. 8A). We further challenged LONP1/ATF4 DmKO mice with HFD. Similar to the LONP1 mKO mice, LONP1/ATF4 DmKO mice exhibited complete resistance to dietary obesity. We observed significantly lower weight gain in LONP1/ATF4 DmKO mice than in ATF4 mKO or WT control mice during HFD feeding (Fig. 8B). Moreover, no significant changes in adipocyte size and mitochondrial respiration rates were observed in the adipose tissue of LONP1/ATF4 DmKO mice compared to LONP1 mKO mice following HFD feeding (Fig. 8, C and D, and fig. S8, A and B). Gene expression studies showed that the expression of the genes involved in UPRmt was not increased in BAT of LONP1/ATF4 DmKO mice compared to the WT control or ATF4 mKO mice after HFD feeding for 5 weeks (fig. S8C). Although HFD induced hepatic steatosis in WT and ATF4 mKO mice, very little hepatic lipid and TG accumulation was observed in both LONP1 mKO and LONP1/ATF4 DmKO mice on HFD (Fig. 8E and fig. S8, D and E). Together, the similarity of the adipose and liver metabolic phenotypes of LONP1 mKO and LONP1/ATF4 DmKO mice under both CD and HFD conditions suggests that ATF4 is dispensable for the muscle mitochondrial proteostasis stress–mediated long-range metabolic response, highlighting the ATF4-independent UPRmt in skeletal muscle.
Fig. 7. Mitochondrial proteostasis stress directs a long-range metabolic response independent of ATF4 in skeletal muscle.
(A) Heatmap analysis of genes differentially regulated in muscles from indicated mice. Each group is represented by RNA-seq data from two independent samples generated from muscles from indicated mice. The differentially regulated genes were clustered into four groups, and a color scheme for fold change is provided. Red, relative increase in abundance; blue, relative decrease. (B) GO enrichment analysis of clusters I (ATF4-dependent) and III (ATF4-independent) genes, with top 10 terms shown. (C) Expression of myokines genes (qRT-PCR) in GC muscles from the indicated mice. n = 6. (D) H&E staining of eWAT, iWAT, and BAT of indicated mice. Scale bar, 50 μm. n = 4 to 8. (E) Mitochondrial respiration rates were determined from the BAT of indicated genotypes using pyruvate or succinate as substrates. n = 4 to 10. (F) H&E and Oil Red O staining of livers from indicated mice. Scale bar, 50 μm. n = 4 to 6. (G) Liver triglyceride levels from indicated mice. n = 5 to 7. Color legend for the panel: white, WT; gray, LONP1 mKO; orange, LONP1/ATF4 DmKO. Values represent means ± SEM. *P < 0.05 versus WT controls; #P < 0.05 versus LONP1 mKO. One-way ANOVA tests were performed.
Fig. 8. Mitochondrial proteostasis stress in skeletal muscle directs a long-range metabolic response to alleviate dietary obesity independent of ATF4.
(A) Body weight of indicated mice at the age of 6 weeks. n = 7 to 14. (B) Increase in body weight following HFD feeding. n = 7 to 14. (C) H&E staining of eWAT, iWAT, and BAT of indicated mice. Scale bar, 20 μm. n = 4 to 7. (D) Mitochondrial respiration rates were determined from the BAT of indicated genotypes using pyruvate as substrate. n = 5 to 7. (E) H&E and Oil Red O staining of livers from indicated mice. Scale bar, 20 μm. n = 5 to 10. (F) The schematic depicts the proposed model for the mitochondrial proteostasis stress in skeletal muscle that drives a long-range protective response to alleviate dietary obesity. Color legend for the panel: white, WT HFD; gray, ATF4 mKO HFD; orange, LONP1 mKO HFD; diagonal hatch, LONP1/ATF4 DmKO HFD. Values represent means ± SEM. *P < 0.05 versus WT controls. One-way ANOVA tests were performed.
DISCUSSION
The quality of mitochondria is of fundamental importance in skeletal muscle, the largest metabolically active and endocrine organ of the human body that depends critically on mitochondrial function. Proteolytic rewiring of mitochondria is emerging as a critical regulatory step to safeguard mitochondrial function (1–4), but our understanding of the in vivo physiological relevance and control mechanism of mitochondrial proteostasis stress remains largely convoluted. In this study, we uncovered a crucial role for the mitochondrial protease LONP1 in controlling the muscle mitochondrial proteostasis stress response that governs systemic metabolic homeostasis (Fig. 8F). Skeletal muscle–specific ablation of LONP1 resulted in impaired mitochondrial proteostasis. Mice lacking muscle LONP1 were markedly resistant to HFD-induced obesity with improved systemic insulin sensitivity and prevented liver steatosis. Many of these metabolic phenotypes were recapitulated in mice overexpressing a ΔOTC-unfolded protein in skeletal muscle. Mechanistically, we demonstrated that mitochondrial proteostasis imbalance triggers an UPRmt in skeletal muscle that communicates with adipose tissue and liver, thereby promoting favorable metabolic remodeling in adipose tissue and liver. Unexpectedly, mitochondrial proteostasis stress directs a long-range metabolic response independent of ATF4 in skeletal muscle. Together, our study establishes a pivotal role of LONP1 in preserving muscle mitochondrial proteostasis, functionally linking muscle mitochondrial proteostasis stress to systemic metabolic homeostasis. Our current study also provided previously unknown mechanistic insights into the UPRmt in mammals.
Mitochondrial proteases have been considered as the first-line mitochondrial quality control mechanism in mammalian cells (1–4). Both ATP-dependent proteases LONP1 and caseinolytic peptidase P (CLPP) reside in the mitochondrial matrix and regulate mitochondrial protein degradation. In this study, we clearly showed that LONP1-mediated proteolysis regulates mitochondrial protein turnover in skeletal muscle in vivo. We detected a marked increase in MitoTimer red fluorescence and significantly higher mitochondrial unfolded protein abundance in LONP1 mKO muscles. Moreover, we show that functionally, loss of muscle LONP1 triggers extensive metabolic reprogramming in skeletal muscle and protects the mice from diet-induced obesity. Notably, we have recently demonstrated that LONP1 deficiency activates autophagy-lysosome degradation program of muscle loss, resulting in reduced skeletal muscle mass and strength in particular during muscle disuse (32). Total lean mass was equivalently reduced in LONP1 mKO mice regardless of the diet; thus, the reduced body weight in HFD-fed LONP1 mKO mice can be attributed to both reduced fat mass and lean mass. The metabolic phenotype of LONP1 mKO mice were recapitulated in a mouse model that transgenic overexpression of ΔOTC in muscle mitochondria, including increased energy expenditure, improved insulin sensitivity, and reversed hepatic steatosis. Muscle-specific deletion of CLPP using muscle creatine kinase promoter–driven Cre does not lead to a metabolic beneficial phenotype compared to WT controls (38). It is possible that LONP1 could compensate for the loss of CLPP in skeletal muscles. Thus, our findings highlight the essential role of LONP1-dependent proteolysis for preserving muscle mitochondrial proteostasis and controlling systemic metabolic homeostasis.
Although UPRmt was first reported in mammalian cells, most of the subsequent mechanisms were studied in C. elegans (5, 18, 30, 35). Many aspects of mammalian UPRmt signaling are largely unknown. Previous studies have proposed an important role of the transcription factor ATF4 in regulating mammalian UPRmt (9, 11, 18, 30, 36, 37). However, here we provided clear genetic data that demonstrate the transcriptional response of the muscle UPRmt can only be explained partially by ATF4, that is, only ~20% of genes depend on ATF4. Although we found that LONP1 exerts control upon the local skeletal muscle amino acid and one-carbon metabolic program through ATF4 signaling, the many effects of LONP1 on muscle genomic reprogramming are independent of ATF4. Therefore, our results highlight an ATF4-independent mechanism in mediating the UPRmt in mammals. Studies in C. elegans have also demonstrated that UPRmt can be transmitted nonautonomously to tissues unaffected by the initial insult (11, 18, 30). It is not clear whether this mechanism is conserved in mammals. We found that loss of muscle LONP1 resulted in UPRmt activation not only within the skeletal muscle but also within the adipose tissues upon HFD feeding, suggesting the presence of similar mechanism to coordinate UPRmt activation between distal tissues in mammals. Our data also suggest a possible involvement of muscle ATF4 in mediating the UPRmt activation in BAT of HFD-fed LONP1 mKO mice. Whereas our results provide substantial evidence to suggest an ATF4-independent UPRmt mechanism in skeletal muscle, the precise mechanisms involved in the activation of muscle UPRmt program were not fully determined in this study. Previous studies have implicated multiple other transcriptional signaling, such as ATF5 and DDIT3/CHOP, in the regulation of mammalian UPRmt (11, 36, 39). Notably, our data also showed that expression levels of Atf5 and Ddit3 genes were up-regulated in both LONP1 mKO and MCK-ΔOTC muscles, thereby suggesting alternative ATF4-independent mechanisms. In addition, evidence has emerged that intermediary metabolites derived from mitochondrial metabolism are critical for genomic reprogramming (40, 41). Given that LONP1 deficiency also caused the divergent mitochondrial metabolic changes in muscle, it is also possible that the altered mitochondrial metabolism leads to activation of UPRmt gene programs in LONP1 mKO muscles. Future studies will be necessary to further delineate the muscle-UPRmt signaling.
A large number of studies have suggested that impaired activity of mitochondrial oxidative phosphorylation could lead to obesity and insulin resistance (42, 43), whereas increasing evidence also suggests that mice with skeletal muscle mitochondrial energetics deficiency could be more insulin sensitive and protect against diet-induced obesity (8, 10, 44, 45), and this phenomenon may be related to a non–cell-autonomous role of the muscle mitochondrial stress response (11). LONP1 mKO mice fed either CD or HFD had impaired mitochondrial structure and respiration function in skeletal muscles. HFD feeding appears to induce more marked metabolic remodeling in LONP1 mKO mice, which is consistent with the metabolically favorable synergy between HFD and mitochondrial stress (46). The concept of mitokines, induced by the stressed mitochondria, has been put forth to explain the systemic metabolic benefits in the context of mitochondrial stress in skeletal muscles (7, 8, 10, 11, 19, 21). The marked induction of mitokines FGF21 and GDF15 in LONP1 mKO and MCK-ΔOTC muscles is of interest given their well-known favorable effects on systemic energy metabolism (13–17). Our data suggest that LONP1 mKO and MCK-ΔOTC muscles secrete FGF21 and GDF15 into circulation to exert endocrine effects, leading to diet-induced obesity resistance and improved whole-body metabolism. Given that a broad array of myokine genes were induced in LONP1 mKO and MCK-ΔOTC muscles, it is certainly possible, however, that other unknown mitokine factors may also contribute to the improved metabolic profile in LONP1 mKO and MCK-ΔOTC mice.
In summary, we have uncovered an essential role of LONP1-dependent mitochondrial proteolysis in controlling muscle mitochondrial proteostasis stress response as well as systemic metabolic homeostasis. Given the remarkable benefits of muscle UPRmt on metabolic health, these findings highlight the potential therapeutic opportunities for targeting muscle mitochondrial protein quality control pathways to counteract obesity and metabolic diseases.
MATERIALS AND METHODS
Animal studies
All animal studies were conducted in strict accordance with the institutional guidelines for the humane treatment of animals and were approved by the Institutional Animal Care and Use Committee at the Model Animal Research Center of Nanjing University (approval no. GZJ07). Male C57BL/6J WT mice were obtained from GemPharmatech Co. Ltd. (Jiangsu, China). The generation of Lonp1f/f mice has been described elsewhere (47). Mice were back-crossed to the C57BL/6J background for more than six generations. To generate mice with muscle-specific disruption of the Lonp1 allele, Lonp1f/f mice were crossed with mice expressing Cre recombinase under the control of an HSA promoter (the Jackson Laboratory, stock no. 006139) to achieve muscle-specific deletion of Lonp1 (LONP1 mKO). To generate mice with muscle-specific overexpression of a mitochondrial-localized mutant OTC (MCK-ΔOTC), cDNA encoding the rat mutant Otc gene was cloned into the Eco RV site downstream of the mouse Mck gene promoter (gift of E. N. Olson, University of Texas Southwestern). The transgene was linearized with Xho I and Sac II digestion and microinjected into C57BL/6J embryos by the transgenic mouse facility at the Model Animal Research Center of Nanjing University. Transgenic mice were identified by PCR amplification of a 552–base pair product using primers specific for the component of the Mck gene promoter (5′-GGAACCAGTGAGCAAGTCAG) and ΔOtc (5′-GTGAGACCTTTGAGAGAGCC). The CAG-CAT-MitoTimer reporter mice were provided by Z. Yan (University of Virginia). CAG-CAT-MitoTimer mice were crossed with HSA-Cre or LONP1 mKO mice to obtain HSA-Cre/MitoTimer or LONP1 mKO/MitoTimer mice to monitor the overall protein turnover status of skeletal muscle mitochondria. To generate mice with a muscle-specific disruption of the Atf4 allele in LONP1 mKO mice, Atf4f/f mice (GemPharmatech Co. Ltd.) were crossed with LONP1 mKO mice to obtain Lonp1f/f, Atf4f/f/HSA-Cre mice (herein named LONP1/ATF4 DmKO mice). Male offspring were genotyped, and mice aged 2 to 18 weeks were used. Mice were allowed ad libitum access to standard laboratory rodent CD or an HFD (60% of calories from fat, research diets no. D12492) as indicated. Mice were randomly assigned to various analyses. Littermate controls (Lonp1f/f mice) were used in all cases. Investigators involved in the fluorescence imaging, RNA-seq, and histological analysis were blinded. Investigators performing animal handling, sampling, and raw data collection were not blinded.
Measuring in vivo mitochondrial protein turnover
In vivo MitoTimer measurements in skeletal muscle were conducted as previously described (31). Briefly, fresh muscles were fixed directly with 4% paraformaldehyde for 2 hours at 4°C. Single-muscle fibers were separated with microforceps under a stereomicroscope. Fluorescence of MitoTimer was imaged in two channels via two sequential excitations (488 nm, green; 561 nm, red) using a Zeiss LSM880 confocal microscope (Carl Zeiss MicroImaging). Identical acquisition parameters were set for all samples under the same experimental conditions. The mean intensity of the MitoTimer signal was calculated using ImageJ software.
Measurement of mitochondrial unfolded proteins
Measurements of mitochondrial unfolded proteins were performed as previously described (48). Briefly, isolated muscle mitochondria were resuspended in phosphate-buffered saline with 1% Triton X-100. A mixture of 40 μg of mitochondrial protein and 50 μM TPE-MI dye (AIEgen Biotech Co. Ltd.) in a final volume of 50 μl was added to a 96-well plate for fluorescence reading up to 2 hours. The load of mitochondrial unfolded protein was represented as the highest relative fluorescence unit in each group.
Mitochondrial isolation and respiration studies
Mitochondria were isolated from mouse skeletal muscle as previously described (10, 49). Briefly, minced muscle was homogenized with a glass dounce on ice, and then centrifuged twice at 600g for 15 min at 4°C. The resulting supernatants were carefully transferred to a new tube and centrifuged at 8000g for 15 min at 4°C, and the resulting supernatants were discarded. The pellet containing the mitochondria was washed twice and centrifuged at 8000g for 15 min at 4°C before resuspension.
Mitochondrial respiration rates were measured in muscle mitochondria with pyruvate or succinate as substrates as described previously (10, 49). Briefly, muscle mitochondria were resuspended in buffer Z [105 mM potassium 2-[N-morpholino]-ethanesulfonic acid, 30 mM KCl, 10 mM KH2PO4, 5 mM MgCl2, bovine serum albumin (5 mg/ml), and 1 mM EGTA (pH 7.4)]. Measurements of oxygen consumption in muscle mitochondria were performed in buffer Z at 37°C and in the oxygen concentration range 220 to 150 nmol O2/ml in the respiration chambers of an Oxygraph-2K (Oroboros Inc., Innsbruck, Austria). Following the measurement of basal, pyruvate (10 mM)/malate (5 mM), or succinate (5 mM)/rotenone (10 μM) respiration, maximal [adenosine 5′-diphosphate (ADP)–stimulated] respiration was determined by exposing the mitochondria to 4 mM ADP. Uncoupled respiration was evaluated following the addition of oligomycin (1 μg/ml). Respiration rates were determined and normalized to mitochondrial protein contents using DatLab 5 software (Oroboros Inc., Innsbruck, Austria), and the data were expressed as “pmol O2 s−1 mg mito protein−1.”
For mitochondrial respiration rate measurement in BAT, approximately 2 mg of BAT was dissected and permeabilized with saponin (5 mg/ml). Following measurement of basal, pyruvate (10 mM)/malate (5 mM), and succinate (5 mM)/rotenone (10 μM) respiration, maximal (ADP stimulated) respiration was determined by exposing the mitochondria to 4 mM ADP. Uncoupled respiration was evaluated following the addition of antimycin (1.5 μg/ml). Respiration rates were determined and normalized to tissue wet weight using DatLab 5 software (Oroboros Inc., Innsbruck, Austria), and the data were expressed as “pmol O2 s−1 mg wet weight−1.”
RNA-seq studies
Transcriptomics analyses were performed using RNA-seq as described previously (50, 51). Total RNA was isolated from the entire GC muscle of male LONP1 mKO, MCK-ΔOTC, or LONP1/ATF4 DmKO mice using RNAiso Plus (Takara Bio). RNA-seq using an Illumina HiSeq 4000 was performed by Beijing Novogene Bioinformatics Technology Co. Ltd. Two independent samples per group were analyzed. Paired-end, 150-nucleotide reads were obtained from the same sequencing lane. The sequencing reads were then aligned to the UCSC mm10 genome assembly using TopHat 2.0.14 with the default parameters. Fragments per kilobase of exon per million mapped reads were calculated using Cufflinks 2.2.1. The criteria for a regulated gene were a fold change greater than 1.5 (either direction) and a significant P value (<0.05) versus WT. GO enrichment analysis of differentially regulated genes was performed using the R package “clusterProfiler” (52) with fun = “enrichGO,” OrgDb = “org.Mm.eg.db,” ont = “ALL,” pAdjustMethod = “BH,” pvalueCutoff = 0.01, qvalueCutoff = 0.05, readable = TRUE” arguments. The compare cluster function was used for comparisons between groups. The regulated GO terms were ranked by P value and visualized using the packages of clusterProfiler and ggplot2 in R. GSEA was carried out by comparing a set of differentially regulated genes in LONP1 mKO muscles with the gene list of regulated genes identified in MCK-ΔOTC muscles. To search for enriched motifs within the muscle-UPRmt gene sets, motif analysis was performed using i-cisTarget. Genome browser tracks of RNA-seq data were visualized in Integrative Genomics Viewer, version 2.8.10. The volcano plot of regulated genes was generated by using R software, version 4.0.2, and the ggplot2 package. Heatmaps were generated using Morpheus (https://software.broadinstitute.org/morpheus). The RNA-seq data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO series accession numbers GSE166071 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE166071), GSE192990, and GSE192991.
RNA analyses
Quantitative RT-PCR was performed as described previously (49, 53). Briefly, total RNA was extracted from mouse muscle using RNAiso Plus (Takara Bio). Isolated total RNA integrity was electrophoretically verified by ethidium bromide staining. One-microgram total RNA samples were then reverse transcribed using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara Bio). Real-time qRT-PCR was performed using the ABI Prism Step-One system with Reagent Kit from Takara Bio. Specific oligonucleotide primers for target gene sequences are listed in table S2. Arbitrary units of target mRNA were corrected to the expression of 36b4.
Transmission electron microscopy
Mice were euthanized and perfused with sodium phosphate buffer [PB; 100 mM (pH 7.4)] and prefixed solution [2.5% (v/v) glutaraldehyde and 1% paraformaldehyde in PB]. Soleus muscle was dissected, cut into small pieces, and fixed in the same prefixed solution overnight at 4°C. After rinsing with PB, tissues were immersed in 0.2 M imidazole in PB for 15 min, and then postfixed with 1% osmium tetraoxide in PB. After rinsing with high-purity water, the samples were stained with 1% aqueous lead at 4°C overnight. Gradient dehydration was accomplished by incrementing the concentration of acetone and embedded in epoxy resin (60°C for 24 hours). Samples were sectioned using a Leica EM UC7 and placed on copper grids. Images were taken on an FEI Tecnai G2 20 Twin electron microscope equipped with an Eagle 4k charge-coupled device digital camera (FEI; USA) in a double-blind manner (32).
Mitochondrial DNA analyses
Genomic/mitochondrial DNA was measured as described previously (10, 49). Mitochondrial DNA content was determined by SYBR Green analysis (Takara Bio). The levels of NADH (reduced form of nicotinamide adenine dinucleotide) dehydrogenase subunit 1 (mitochondrial DNA) were normalized to the levels of lipoprotein lipase (genomic DNA). The primer sequences are noted in Table S2.
Metabolic measurements in vivo
Mice were housed individually in metabolic cages under a 12-hour light and dark cycle with free access to food and water using CLAMS (Columbus Instruments). Mice were acclimated to the metabolic cage for 1 day before recording according to the instructions of the manufacturer. Food, energy expenditure, physical activity, VO2, and VCO2 were assessed simultaneously (10).
Body composition analyses
Mouse body composition parameters including fat mass and lean tissue mass were determined via dual-energy x-ray absorptiometry using a Lunar PIXImus II densitometer (GE Healthcare) according to the instructions of the manufacturer (10).
Glucose and insulin tolerance testing
Before studies, mice were fasted overnight (GTT) or for 4 hours (ITT). For GTT studies, mice were injected with d-glucose (1.5 g/kg). For ITT, mice received an intraperitoneal injection of human regular insulin (Sigma-Aldrich) at a dose of 0.75 U/kg body weight. Blood glucose levels were determined at 0, 15, 30, 60, 90, and 120 min after challenge using a OneTouch UltraMini glucose meter (OneTouch). The area under the curve (AUC) was defined as the difference between baseline glucose levels and the deflection caused by the glucose or insulin challenge. Total AUC was calculated using the trapezoidal rule (10).
Blood and tissue chemistry
After a 12-week HFD regimen, mice were fasted overnight (16 hours) beginning at 5 p.m. Blood samples were obtained for glucose and insulin measurements. Plasma insulin levels were determined using an insulin ELISA kit (Millipore). Nonfasting serum FGF21 levels were measured by an FGF21 ELISA kit (Proteintech, KE10042) according to the manufacturer’s instructions. Nonfasting serum GDF15 levels were determined using a GDF15 ELISA kit (ImmunoDiagnostics, 32980). Liver tissue (50 mg) was homogenized, and centrifuged supernatants were harvested. The TG levels were determined with the Free Glycerol Reagent (Sigma-Aldrich, F6428) using glycerol (Sigma-Aldrich, G7793) as the standard for calculation (10).
Histological analyses
Mouse muscle tissues were frozen in isopentane that had been cooled in liquid nitrogen. Wheat germ agglutinin (WGA) staining was performed using fluorescein isothiocyanate (FITC)–conjugated WGA (Sigma-Aldrich, #L4859). Quantification of cross-sectional area of the myofibers was performed with National Institutes of Health ImageJ software. Adipose tissues were fixed in 4% formaldehyde overnight at 4°C immediately after euthanasia, embedded in paraffin, and cut to 7-μm sections on slides using a Leica RM2016 microtome. H&E and Masson’s trichrome staining were conducted according to the standard protocol. For UCP1 IHC, brown adipose tissue sections were rehydrated and then boiled in 10 mM citric acid buffer, pH 6.0, at 95°C for antigen retrieval. IHC was performed using the UltraSensitive SP (Rabbit) IHC Kit (Maxim) according to the manufacturer’s instructions using rabbit UCP1 antibodies (1:500; ab10983, Abcam). DAB Staining Kit (DAB-0031, Maxim) was used according to the manufacturer’s instructions. Livers of mice were embedded in Tissue-Tek OCT cryostat molds (Leica) and frozen at −80°C. Ten-micrometer-thick sections of livers were generated in a cryostat. Tissue sections were stained in 0.5% Oil Red O and counterstained with H&E.
Metabolite analysis by CE-MS
Analysis of skeletal muscle metabolites was performed as previously described (54). Briefly, we conducted CE-MS–based metabolite analysis on CE (G7100A, Agilent) coupled to time-of-flight (TOF) MS (G6224A, Agilent). Whole pieces of GC muscle were weighed and ground in 1 ml of methanol with internal standards [grinding apparatus (Scientz-48), homogenize for 2 min at 40 Hz]. The lysate was then mixed with 1 ml of chloroform and 400 μl of water by 30 s of vortexing. After centrifugation at 16,000g for 15 min at 4°C, 400 μl of aqueous phase was collected and then filtered through a 5-kDa cutoff filter (Millipore, cat. UFC3LCCNB-HMT) by centrifuging at 13,000g for 4 hours at 4°C. The filtered aqueous phase was then freeze dried in a vacuum concentrator and dissolved in water containing IS3 (1:200; Human Metabolome Technologies, H3304-1104) to adjust the migration time. Ten microliters of redissolved solution was then loaded into an injection vial with a conical insert for CE–TOF MS analysis. The fused silica capillary [50 μm internal diameter (i.d.) × 80 cm, Human Metabolome Technologies (HMT), Tsuruoka, Japan] was used for sample separation. The qualitative analysis was preceded on the basis of the preanalyzed metabolite standard library (HMT). Peak extraction and identification were carried out with Quantitative Analysis Software (Agilent). PCA analysis was generated by using R software, version 4.0.2 with the factoextra/FactoMineR/ggpubr and gplots packages. Heatmaps were generated using Morpheus (https://software.broadinstitute.org/morpheus). Pathway analysis was conducted by using online software MetaboAnalyst 5.0.
Antibodies and immunoblotting studies
The antibodies with the indicated dilutions were as follows: anti-UCP1 (1:1000; Abcam, ab10983), anti-LONP1 (1:2500; Proteintech, 15440-1-AP), anti-CLPP (1:2,500; Proteintech,15698-1-AP), anti-NDUFB8 (1:1000; Proteintech, 14794-1-AP), anti-SDHA (1:1000; Proteintech, 14865-1-AP), anti-UQCRC2 (1:1000; Proteintech, 14742-1-AP), anti-COX4 (1:1000; Proteintech, 11242-1-AP), anti-ATP5A (1:1000; Proteintech, 14676-1-AP), anti-GAPDH (1:1000; Proteintech, 60004-1-Ig), anti-TIM23 (1:1000; BD Biosciences, #611222), anti-TOM20 (1:1000 dilution; Abclonal, #A6774), anti–α-tubulin (1:5000; Bioworld, bs1699), anti-FLAG (1:1000; Sigma-Aldrich, #F1804), and anti-FASN (1:1000; Cell Signaling Technology, #3180). Western blotting studies were performed as previously described (49, 50, 53). Blots were normalized to α-tubulin. The total protein concentration was measured by bicinchoninic acid (BCA) assay using the Pierce BCA Assay Kit Protocol (Thermo Fischer Scientific). Equal total protein was loaded to each lane.
Statistical analyses
All studies were analyzed by Student’s t test or one-way analysis of variance (ANOVA) coupled to a Fisher’s least significant difference post hoc test when more than two groups were compared. No statistical methods were used to predetermine sample sizes, and sample size (range from n = 3 to n = 14) is explicitly stated in the figure legends. All data points were used in statistical analyses. Data represent the means ± SEM, with a statistically significant difference defined as a value of P < 0.05.
Acknowledgments
We thank Z. Yan (University of Virginia) for providing the MitoTimer reporter mice.
Funding: This work was supported by grants from the National Natural Science Foundation of China (nos. 91857105, 31771291, and 31922033 to Z.G.; 32071136 and 32100922 to T.F.; and 32125021 to X.-W.C.), the Ministry of Science and Technology of China (National Key R&D Program of China 2018YFA0800700) and Natural Science Foundation of Jiangsu Province (BK20170014 and SWYY-002) (to Z.G.), and Fundamental Research Funds for the Central Universities 090314380036 and 021414380517 (to T.F.), as well as 021414380524 and 021414380511 (to Z.G.).
Author contributions: Z.G. conceived the ideas for experimental designs. Q.G. and Z.G. designed experiments, discussed the data, and wrote the manuscript. Q.G. and Z.X. conducted most of the experiments, analyzed the data, and prepared and revised the manuscript. D.Z. conducted mitochondrial respiration analysis. W.W. carried out metabolomics analysis and interpreted the data. T.F., W.S., L.X., L.L., C.D., Y.Y., Z.Z., Z.S., Y.J., and J.X. conducted sample collection and helped compose the experiments. Y.Z. and W.Z. performed EM analysis. Y.C., X.-W.C., H.-L.P., and B.L. contributed reagents and provided scientific insight and discussion. Z.G. developed the concept and supervised the work. All authors reviewed and contributed to the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The RNA-seq data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO series accession numbers GSE166071 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE166071), GSE192990, and GSE192991.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 and S2
REFERENCES AND NOTES
- 1.Deshwal S., Fiedler K. U., Langer T., Mitochondrial proteases: Multifaceted regulators of mitochondrial plasticity. Annu. Rev. Biochem. 89, 501–528 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Song J., Herrmann J. M., Becker T., Quality control of the mitochondrial proteome. Nat. Rev. Mol. Cell Biol. 22, 54–70 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Gan Z., Fu T., Kelly D. P., Vega R. B., Skeletal muscle mitochondrial remodeling in exercise and diseases. Cell Res. 28, 969–980 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quiros P. M., Langer T., Lopez-Otin C., New roles for mitochondrial proteases in health, ageing and disease. Nat. Rev. Mol. Cell Biol. 16, 345–359 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Mottis A., Herzig S., Auwerx J., Mitocellular communication: Shaping health and disease. Science 366, 827–832 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Sorrentino V., Menzies K. J., Auwerx J., Repairing mitochondrial dysfunction in disease. Annu. Rev. Pharmacol. Toxicol. 58, 353–389 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Tezze C., Romanello V., Desbats M. A., Fadini G. P., Albiero M., Favaro G., Ciciliot S., Soriano M. E., Morbidoni V., Cerqua C., Loefler S., Kern H., Franceschi C., Salvioli S., Conte M., Blaauw B., Zampieri S., Salviati L., Scorrano L., Sandri M., Age-associated loss of opa1 in muscle impacts muscle mass, metabolic homeostasis, systemic inflammation, and epithelial senescence. Cell Metab. 25, 1374–1389.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pereira R. O., Tadinada S. M., Zasadny F. M., Oliveira K. J., Pires K. M. P., Olvera A., Jeffers J., Souvenir R., McGlauflin R., Seei A., Funari T., Sesaki H., Potthoff M. J., Adams C. M., Anderson E. J., Abel E. D., OPA1 deficiency promotes secretion of FGF21 from muscle that prevents obesity and insulin resistance. EMBO J. 36, 2126–2145 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quiros P. M., Mottis A., Auwerx J., Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 17, 213–226 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Fu T., Xu Z., Liu L., Guo Q., Wu H., Liang X., Zhou D., Xiao L., Liu L., Liu Y., Zhu M. S., Chen Q., Gan Z., Mitophagy directs muscle-adipose crosstalk to alleviate dietary obesity. Cell Rep. 23, 1357–1372 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Bar-Ziv R., Bolas T., Dillin A., Systemic effects of mitochondrial stress. EMBO Rep. 21, e50094 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy R. M., Watt M. J., Febbraio M. A., Metabolic communication during exercise. Nat. Metab. 2, 805–816 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Wang D., Day E. A., Townsend L. K., Djordjevic D., Jorgensen S. B., Steinberg G. R., GDF15: Emerging biology and therapeutic applications for obesity and cardiometabolic disease. Nat. Rev. Endocrinol. 17, 592–607 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Keipert S., Ost M., Stress-induced FGF21 and GDF15 in obesity and obesity resistance. Trends Endocrinol. Metab. 32, 904–915 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Kharitonenkov A., Shiyanova T. L., Koester A., Ford A. M., Micanovic R., Galbreath E. J., Sandusky G. E., Hammond L. J., Moyers J. S., Owens R. A., Gromada J., Brozinick J. T., Hawkins E. D., Wroblewski V. J., Li D. S., Mehrbod F., Jaskunas S. R., Shanafelt A. B., FGF-21 as a novel metabolic regulator. J. Clin. Investig. 115, 1627–1635 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inagaki T., Dutchak P., Zhao G., Ding X., Gautron L., Parameswara V., Li Y., Goetz R., Mohammadi M., Esser V., Elmquist J. K., Gerard R. D., Burgess S. C., Hammer R. E., Mangelsdorf D. J., Kliewer S. A., Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab. 5, 415–425 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Owen B. M., Ding X. S., Morgan D. A., Coate K. C., Bookout A. L., Rahmouni K., Kliewer S. A., Mangelsdorf D. J., FGF21 acts centrally to induce sympathetic nerve activity, energy expenditure, and weight loss. Cell Metab. 20, 670–677 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shpilka T., Haynes C. M., The mitochondrial UPR: Mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol. 19, 109–120 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Forsstrom S., Jackson C. B., Carroll C. J., Kuronen M., Pirinen E., Pradhan S., Marmyleva A., Auranen M., Kleine I. M., Khan N. A., Roivainen A., Marjamaki P., Liljenback H., Wang L., Battersby B. J., Richter U., Velagapudi V., Nikkanen J., Euro L., Suomalainen A., Fibroblast growth factor 21 drives dynamics of local and systemic stress responses in mitochondrial myopathy with mtDNA deletions. Cell Metab. 30, 1040–1054.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Khan N. A., Nikkanen J., Yatsuga S., Jackson C., Wang L., Pradhan S., Kivela R., Pessia A., Velagapudi V., Suomalainen A., mTORC1 regulates mitochondrial integrated stress response and mitochondrial myopathy progression. Cell Metab. 26, 419–428.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Chung H. K., Ryu D., Kim K. S., Chang J. Y., Kim Y. K., Yi H. S., Kang S. G., Choi M. J., Lee S. E., Jung S. B., Ryu M. J., Kim S. J., Kweon G. R., Kim H., Hwang J. H., Lee C. H., Lee S. J., Wall C. E., Downes M., Evans R. M., Auwerx J., Shong M., Growth differentiation factor 15 is a myomitokine governing systemic energy homeostasis. J. Cell Biol. 216, 149–165 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suomalainen A., Elo J. M., Pietilainen K. H., Hakonen A. H., Sevastianova K., Korpela M., Isohanni P., Marjavaara S. K., Tyni T., Kiuru-Enari S., Pihko H., Darin N., Ounap K., Kluijtmans L. A. J., Paetau A., Buzkova J., Bindoff L. A., Annunen-Rasila J., Uusimaa J., Rissanen A., Yki-Jarvinen H., Hirano M., Tulinius M., Smeitink J., Tyynismaa H., FGF-21 as a biomarker for muscle-manifesting mitochondrial respiratory chain deficiencies: A diagnostic study. Lancet Neurol. 10, 806–818 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tyynismaa H., Carroll C. J., Raimundo N., Ahola-Erkkila S., Wenz T., Ruhanen H., Guse K., Hemminki A., Peltola-Mjosund K. E., Tulkki V., Oresic M., Moraes C. T., Pietilainen K., Hovatta I., Suomalainen A., Mitochondrial myopathy induces a starvation-like response. Hum. Mol. Genet. 19, 3948–3958 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Nargund A. M., Pellegrino M. W., Fiorese C. J., Baker B. M., Haynes C. M., Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337, 587–590 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian Y., Garcia G., Bian Q., Steffen K. K., Joe L., Wolff S., Meyer B. J., Dillin A., Mitochondrial stress induces chromatin reorganization to promote longevity and UPR(mt). Cell 165, 1197–1208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strauss K. A., Jinks R. N., Puffenberger E. G., Venkatesh S., Singh K., Cheng I., Mikita N., Thilagavathi J., Lee J., Sarafianos S., Benkert A., Koehler A., Zhu A., Trovillion V., McGlincy M., Morlet T., Deardorff M., Innes A. M., Prasad C., Chudley A. E., Lee I. N., Suzuki C. K., CODAS syndrome is associated with mutations of LONP1, encoding mitochondrial AAA+ Lon protease. Am. J. Hum. Genet. 96, 121–135 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu B., Lee J., Nie X., Li M., Morozov Y. I., Venkatesh S., Bogenhagen D. F., Temiakov D., Suzuki C. K., Phosphorylation of human TFAM in mitochondria impairs DNA binding and promotes degradation by the AAA+ Lon protease. Mol. Cell 49, 121–132 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bota D. A., Davies K. J., Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat. Cell Biol. 4, 674–680 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Quiros P. M., Espanol Y., Acin-Perez R., Rodriguez F., Barcena C., Watanabe K., Calvo E., Loureiro M., Fernandez-Garcia M. S., Fueyo A., Vazquez J., Enriquez J. A., Lopez-Otin C., ATP-dependent Lon protease controls tumor bioenergetics by reprogramming mitochondrial activity. Cell Rep. 8, 542–556 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Anderson N. S., Haynes C. M., Folding the mitochondrial UPR into the integrated stress response. Trends Cell Biol. 30, 428–439 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laker R. C., Xu P., Ryall K. A., Sujkowski A., Kenwood B. M., Chain K. H., Zhang M., Royal M. A., Hoehn K. L., Driscoll M., Adler P. N., Wessells R. J., Saucerman J. J., Yan Z., A novel mitotimer reporter gene for mitochondrial content, structure, stress, and damage in vivo. J. Biol. Chem. 289, 12005–12015 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Z., Fu T., Guo Q., Zhou D., Sun W., Zhou Z., Chen X., Zhang J., Liu L., Xiao L., Yin Y., Jia Y., Pang E., Chen Y., Pan X., Fang L., Zhu M. S., Fei W., Lu B., Gan Z., Disuse-associated loss of the protease LONP1 in muscle impairs mitochondrial function and causes reduced skeletal muscle mass and strength. Nat. Commun. 13, 894 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bezawork-Geleta A., Brodie E. J., Dougan D. A., Truscott K. N., LON is the master protease that protects against protein aggregation in human mitochondria through direct degradation of misfolded proteins. Sci. Rep. 5, 17397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin S. M., Youle R. J., The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy 9, 1750–1757 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Q., Wang J. H., Levichkin I. V., Stasinopoulos S., Ryan M. T., Hoogenraad N. J., A mitochondrial specific stress response in mammalian cells. EMBO J. 21, 4411–4419 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaspar S., Oertlin C., Szczepanowska K., Kukat A., Senft K., Lucas C., Brodesser S., Hatzoglou M., Larsson O., Topisirovic I., Trifunovic A., Adaptation to mitochondrial stress requires CHOP-directed tuning of ISR. Sci. Adv. 7, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quiros P. M., Prado M. A., Zamboni N., D’Amico D., Williams R. W., Finley D., Gygi S. P., Auwerx J., Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 216, 2027–2045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Becker C., Kukat A., Szczepanowska K., Hermans S., Senft K., Brandscheid C. P., Maiti P., Trifunovic A., CLPP deficiency protects against metabolic syndrome but hinders adaptive thermogenesis. EMBO Rep. 19, e45126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiorese C. J., Schulz A. M., Lin Y. F., Rosin N., Pellegrino M. W., Haynes C. M., The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr. Biol. 26, 2037–2043 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L. P., Chen K. S., Wang T. Y., Wu Y., Xing G. S., Chen M. Q., Hao Z. H., Zhang C., Zhang J. Y., Ma B. C., Liu Z. H., Yuan H., Liu Z. J., Long Q., Zhou Y. S., Qi J. T., Zhao D. Y., Gao M., Pei D. Q., Nie J. F., Ye D., Pan G. J., Liu X. G., Glis1 facilitates induction of pluripotency via an epigenome-metabolome-epigenome signalling cascade. Nat. Metab. 2, 882–892 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Matilainen O., Quiros P. M., Auwerx J., Mitochondria and epigenetics - crosstalk in homeostasis and stress. Trends Cell Biol. 27, 453–463 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Muoio D. M., Neufer P. D., Lipid-induced mitochondrial stress and insulin action in muscle. Cell Metab. 15, 595–605 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szendroedi J., Phielix E., Roden M., The role of mitochondria in insulin resistance and type 2 diabetes mellitus. Nat. Rev. Endocrinol. 8, 92–103 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Kim K. H., Jeong Y. T., Oh H., Kim S. H., Cho J. M., Kim Y. N., Kim S. S., Kim D. H., Hur K. Y., Kim H. K., Ko T., Han J., Kim H. L., Kim J., Back S. H., Komatsu M., Chen H. C., Chan D. C., Konishi M., Itoh N., Choi C. S., Lee M. S., Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat. Med. 19, 83–92 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Pospisilik J. A., Knauf C., Joza N., Benit P., Orthofer M., Cani P. D., Ebersberger I., Nakashima T., Sarao R., Neely G., Esterbauer H., Kozlov A., Kahn C. R., Kroemer G., Rustin P., Burcelin R., Penninger J. M., Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell 131, 476–491 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Wall C. E., Whyte J., Suh J. M., Fan W., Collins B., Liddle C., Yu R. T., Atkins A. R., Naviaux J. C., Li K., Bright A. T., Alaynick W. A., Downes M., Naviaux R. K., Evans R. M., High-fat diet and FGF21 cooperatively promote aerobic thermogenesis in mtDNA mutator mice. Proc. Natl. Acad. Sci. U.S.A. 112, 8714–8719 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.B. Lu, F. Shangguan, D. Huang, S. Gong, S. Yingchao, Z. Song, L. Jia, J. Xu, C. Yan, T. Chen, M. Xu, Y. Li, S. Han, N. Song, P. Chen, L. Wang, Y. Liu, X. Huang, C. Suzuki, G. Yang, LonP1 Orchestrates UPRmt and UPRER and Mitochondrial Dynamics to Regulate Heart Function, bioRxiv:10.1101/564492 (2019).
- 48.Chen M. Z., Moily N. S., Bridgford J. L., Wood R. J., Radwan M., Smith T. A., Song Z., Tang B. Z., Tilley L., Xu X., Reid G. E., Pouladi M. A., Hong Y., Hatters D. M., A thiol probe for measuring unfolded protein load and proteostasis in cells. Nat. Commun. 8, 474 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao L., Liu J., Sun Z., Yin Y., Mao Y., Xu D., Liu L., Xu Z., Guo Q., Ding C., Sun W., Yang L., Zhou Z., Zhou D., Fu T., Zhou W., Zhu Y., Chen X. W., Li J. Z., Chen S., Xie X., Gan Z., AMPK-dependent and -independent coordination of mitochondrial function and muscle fiber type by FNIP1. PLOS Genet. 17, e1009488 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu L., Ding C., Fu T., Feng Z., Lee J. E., Xiao L., Xu Z., Yin Y., Guo Q., Sun Z., Sun W., Mao Y., Yang L., Zhou Z., Zhou D., Xu L., Zhu Z., Qiu Y., Ge K., Gan Z., Histone methyltransferase MLL4 controls myofiber identity and muscle performance through MEF2 interaction. J. Clin. Invest. 130, 4710–4725 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu L., Cai J., Wang H., Liang X., Zhou Q., Ding C., Zhu Y., Fu T., Guo Q., Xu Z., Xiao L., Liu J., Yin Y., Fang L., Xue B., Wang Y., Meng Z. X., He A., Li J. L., Liu Y., Chen X. W., Gan Z., Coupling of COPII vesicle trafficking to nutrient availability by the IRE1α-XBP1s axis. Proc. Natl. Acad. Sci. U.S.A. 116, 11776–11785 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu G., Wang L. G., Han Y., He Q. Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J., Liang X., Zhou D., Lai L., Xiao L., Liu L., Fu T., Kong Y., Zhou Q., Vega R. B., Zhu M. S., Kelly D. P., Gao X., Gan Z., Coupling of mitochondrial function and skeletal muscle fiber type by a miR-499/Fnip1/AMPK circuit. EMBO Mol. Med. 8, 1212–1228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan M., Qi H., Xia T., Zhao X., Wang W., Wang Z., Lu C., Ning Z., Chen H., Li T., Tekcham D. S., Liu X., Liu J., Chen D., Liu X., Xu G., Piao H. L., Metabolomics profiling of metformin-mediated metabolic reprogramming bypassing AMPKα. Metabolism 91, 18–29 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 and S2