Abstract
Members of the group of 7-kDa cold-shock proteins (CSPs) are the proteins with the highest level of induction upon cold shock in the lactic acid bacterium Lactococcus lactis MG1363. By using double-crossover recombination, two L. lactis strains were generated in which genes encoding CSPs are disrupted: L. lactis NZ9000ΔAB lacks the tandemly orientated cspA and cspB genes, and NZ9000ΔABE lacks cspA, cspB, and cspE. Both strains showed no differences in growth at normal and at low temperatures compared to that of the wild-type strain, L. lactis NZ9000. Two-dimensional gel electrophoresis showed that upon disruption of the cspAB genes, the production of remaining CspE at low temperature increased, and upon disruption of cspA, cspB, and cspE, the production of CspD at normal growth temperatures increased. Northern blot analysis showed that control is most likely at the transcriptional level. Furthermore, it was established by a proteomics approach that some (non-7-kDa) cold-induced proteins (CIPs) are not cold induced in the csp-lacking strains, among others the histon-like protein HslA and the signal transduction protein LlrC. This supports earlier observations (J. A. Wouters, M. Mailhes, F. M. Rombouts, W. M. De Vos, O. P. Kuipers, and T. Abee, Appl. Environ. Microbiol. 66:3756–3763, 2000). that the CSPs of L. lactis might be directly involved in the production of some CIPs upon low-temperature exposure. Remarkably, the adaptive response to freezing by prior exposure to 10°C was significantly reduced in strain NZ9000ΔABE but not in strain NZ9000ΔAB compared to results with wild-type strain NZ9000, indicating a notable involvement of CspE in cryoprotection.
Lactic acid bacteria (LAB) are frequently used to start industrial food fermentations. During production of these starter cultures and during manufacture and storage of the fermented products, LAB experience a variety of temperature changes. For these reasons, the cold-adaptive response of LAB attracts increasing interest. Lactococcus lactis is a mesophilic LAB that is widely used in the manufacturing of cheese. In recent years, this bacterium has become the model organism for LAB because of its relevance in the food industry and the development of a wide variety of genetic systems.
Several bacteria react to a sudden downshift in temperature by the production of a set of proteins, together forming the cold-shock stimulon, that includes small (7-kDa) cold-shock proteins (CSPs). In a variety of bacteria, families of CSPs, consisting of three to nine members, have been described of which CspA in Escherichia coli (CspAE) and CspB in Bacillus subtilis (CspBB) are the best characterized (see reviews in references 15, 37, and 39). CspAE and CspBB are capable of binding to single-stranded DNA and RNA, and based on these characteristics, several functions for CSPs have been suggested, such as transcriptional activators (7, 19, 25), RNA chaperones that facilitate the initiation of translation (16, 18), and freeze-protective proteins (34). Recently it has been shown that CSPs might regulate the expression of cold-induced genes as antiterminators (3). Regulation of csp genes takes place at several levels, and for CspAE it was shown that cold-shock induction is achieved at the transcriptional level (13, 29) as well as at the level of mRNA and protein stability (6, 10, 12). For L. lactis MG1363, a family of five csp genes has been identified. The L. lactis chromosome was found to contain two sets of two tandemly located and cold-inducible csp genes (cspA/cspB and cspC/cspD) and a single, constitutively expressed cspE gene (38). By using L. lactis strains specifically overproducing the respective CSPs, it was found that these proteins protect against freezing and might be involved in the regulation of (non-7-kDa) cold-induced proteins (CIPs) (36).
Because of the implications of CSPs in freeze protection and their presumed central role in cold adaptation, it is of great interest to further investigate the role of these proteins for L. lactis, especially in relation to food or dairy production processes. In this work, we characterized the effects of multiple csp gene disruptions on adaptation to cold and gene regulation of L. lactis. Deletion of csp genes affects freeze survival of L. lactis, the production of the remaining counterparts of the lactococcal CSP family as well as the production of several CIPs.
MATERIALS AND METHODS
Strains and culturing conditions.
L. lactis NZ9000 (24) (Table 1) was used as the wild-type strain for the generation of csp-lacking strains and has been generated from strain MG1614, which is a rifampin- and streptomycin-resistant derivative of strain MG1363 (11). L. lactis was cultured at 30°C or at lower temperatures (as indicated) on M17 medium containing 0.5% glucose (GM17). Growth was monitored by measuring the optical density at 600 nm. E. coli MC1061 (8) was used as a host strain in cloning experiments and was grown in tryptone yeast extract medium with aeration at 37°C (28). Antibiotics were used in the following concentrations: ampicillin, 50 μg ml−1; erythromycin, 2.5 μg ml−1.
Generation of csp deletions.
L. lactis strains carrying deletions in their csp genes were constructed using a double-crossover replacement strategy (26). For the generation of L. lactis strains with deletions in their csp genes, the regions flanking (approximately 800 bp) the respective genes were amplified using PCR as described by Kuipers et al. (23). The oligonucleotides used for the PCR (Table 1) contain various restriction sites: the forward fragment 1 primers contain an EcoRI site, the reverse fragment 1 primers contain a KpnI site, the forward fragment 2 primers contain an XbaI site, and the reverse fragment 2 primers contain a SalI site. The cloning of the amplified fragments in pUC18ERY (30) resulted in the plasmids pUCEryΔAB12 and pUCEryΔE12 (Ampr Eryr) (Table 1). The subsequent replacement strategy results in deletion of the cspA-cspB tandem repeat starting from the codon for the 12th amino acid (Asp) of CspA to 8 bp downstream of the coding region of cspB. For the deletion of cspE the region from the 8th amino acid residue (Trp) to 18 bp downstream of the coding region was removed. The plasmids obtained were transformed into L. lactis NZ9000 by electroporation (33) and selected for erythromycin resistance in which the first integration took place. The erythromycin-resistant strains were cultured without selective pressure of erythromycin for 200 generations and were analyzed for the occurrence of the second crossover event. Candidates were verified using PCR and ultimately in a Southern blotting experiment using PCR fragment 1 as a probe as described previously (38). Using this strategy, an L. lactis strain lacking cspA and cspB (NZ9000ΔAB) was created and, sequentially, also a L. lactis strain additionally lacking cspE (NZ9000ΔABE) was obtained. Attempts to generate L. lactis strains lacking cspE only or lacking the cspA, cspB, cspC, cspD, and cspE genes using a similar strategy failed at the second crossover event. All manipulations with recombinant DNA were carried out following standard procedures (28) and according to the specifications of the manufacturer (GIBCO/BRL Life technologies, Breda, The Netherlands).
Protein analysis.
The protein composition of cell extracts was determined using two-dimensional gel electrophoresis (2D-E) as described previously (35). Total protein was extracted from cultures growing at mid-exponential phase at the optimal temperature (30°C) and from cultures exposed to a cold shock to 10°C for several h using a cell MSK cell homogenizer (Braun Biotech International, Melsungen, Germany) and zirconium beads (Biospec Products, Bartlesville, Okla.). Equal amounts of protein were analyzed using the Multiphor 2D electrophoresis system (Pharmacia Biotech, Uppsala, Sweden), and protein spots were visualized using silver staining (4). For the 2-DE, a representative gel for two independent samples is shown. The spots on the 2D-E gels were compared, calibrated, and calculated using the GEMINI program (Applied Imaging, Sunderland, England). N-terminal sequences of specific spots in 2-DE were determined as described previously (35), and by using the BlastP database and the L. lactis IL1403 genome database (5) (http://spock.jouy.inra.fr/cgi-bin/blast.cgi) the derived N-terminal sequences were screened for sequence similarities.
mRNA analysis.
RNA isolation, Northern blotting, and subsequent hybridization with radiolabeled probes was performed as described previously (22, 32). For the specific detection of the mRNAs of the csp genes, previously described primers were used (38). Quantification of the csp transcripts in Northern blotting was performed using the Dynamics Phosphor Imaging System (Dynamics, Rochester, N.Y.). Equal amounts of total RNA were applied on the gel as was shown using a probe specific for lactococcal 16S rRNA (5′-ATCTACGCATTTCACCGCTAC-3′) (21).
Freeze-thaw challenge.
The generated mutant strains and the wild-type strain were tested for their susceptibility towards freezing in a previously described freeze-thaw challenge (35). In short, cells were cultured in GM17 medium until mid-exponential phase (optical density at 600 nm, 0.5) at 30°C and subsequently were rapidly downshifted in growth temperature from 30 to 10°C. The cultures were exposed to 10°C for different time periods (0, 2, and 4 h), and subsequently, 1 ml of these cultures was frozen at −20°C. After a 24-h freezing period the number of remaining viable cells (CFU) was determined using plate counting following incubating for 48 h at 30°C. This freeze-thaw cycle was performed four times in total.
RESULTS
Generation and growth characteristics of strains lacking csp.
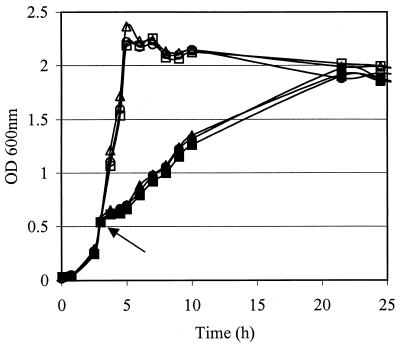
Using a double-crossover replacement strategy, L. lactis strains with deletions in the tandem cspA and cspB genes (NZ9000ΔAB) and carrying an additional deletion in cspE (NZ9000ΔABE) were obtained. The chromosomal configuration was confirmed by PCR and Southern hybridization (data not shown). Deletion of cspAB or cspABE did not affect the growth rate at optimal growth temperature (30°C) or at other temperatures (4, 7, 10, 15, 20, or 42°C). Upon cold-shock treatment at 10°C, identical adaptation times and growth rates were observed for the wild-type strain and the mutants (Fig. 1). Moreover, no differences in the number of CFU, the estimated lag time, and the appearance of the colonies were observed upon incubation of the csp mutants on GM17 plates at different temperatures for a 14-day period. Furthermore, the csp mutants and the wild-type cells showed similar sizes and chain lengths (data not shown).
FIG. 1.
Growth of NZ9000 (triangles), NZ9000ΔAB (circles), and NZ9000ΔABE (squares) at 30°C (open symbols) and after cold shock to 10°C (closed symbols). The arrow indicates the time point of cold shock.
Deletion of cspAB or cspABE affects the production of the remaining CSPs.
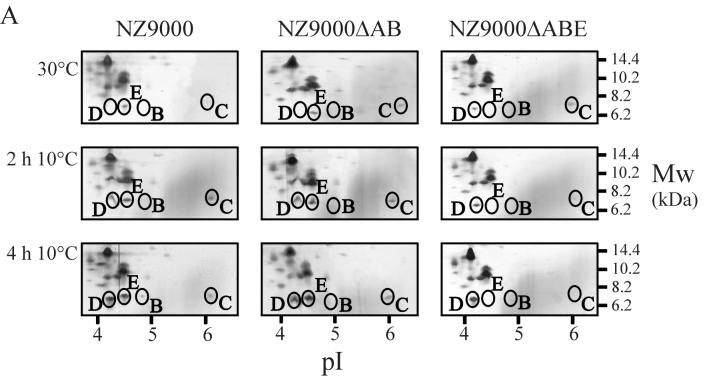
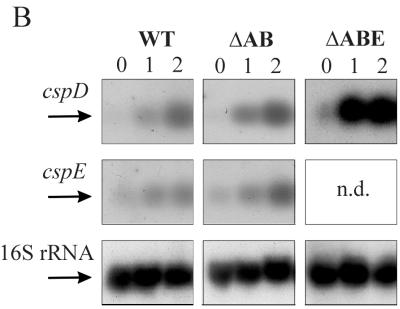
Since no changes in the growth of L. lactis were observed upon deletion of csp genes, the production levels of the remaining CSPs were analyzed by 2D-E with L. lactis NZ9000ΔAB and L. lactis NZ9000ΔABE (Fig. 2A). The positions of the CSPs on the 2D-E gels have been determined previously using overproduction of the individual CSPs and analysis of their N-terminal sequences (35, 36). For wild-type strain NZ9000, the only observed CSP at mid-exponential phase at 30°C was CspE. Upon exposure to 10°C, induction of CspD and, to a lesser extent, of CspA (Fig. 3), CspB, CspC, and CspE was observed (Fig. 2A). For strain L. lactis NZ9000ΔAB, identical production levels of CspD were found at high and low temperatures as observed for the wild-type strain, while CspE production increased slightly (twofold) at cold-shock conditions. At 30°C the level of CspC was higher for NZ9000ΔAB than for NZ9000, but it reached similar production levels at cold-shock conditions for NZ9000ΔAB and NZ9000. Strikingly, in strain NZ9000ΔABE, CspD was present at mid-exponential phase at 30°C, whereas it is not detectable at these conditions in wild-type cells. At low temperature the level of CspD increased but did not exceed the level found in wild-type cells. Remarkably, the CspC level slightly decreased upon low-temperature incubation with strain NZ9000ΔABE compared to results with wild-type cells (Fig. 2A). Thus, the loss of CspA and CspB is compensated for by an increased production of CspE at low temperature, and the additional deletion of cspE is compensated for by increased production of CspD at 30°C. At 10°C the production of the remaining CSPs, CspC, and CspD, in NZ9000ΔABE was not greater than in wild-type cells. To further assess the changes in the production levels of CspD and CspE, the mRNA levels of these genes also were analyzed in wild-type cells and in the strains with csp deleted (Fig. 2B). Indeed, an increased amount of cspE transcript (twofold induction) was observed for NZ9000ΔAB compared to its level in wild-type cells. By analogy, also for the cspD transcript an increased amount was found at 30°C compared to results for both wild-type cells and NZ9000ΔAB cells (fivefold induction). The cspD mRNA level also increased (similar fivefold induction) upon low-temperature incubation. However, at these conditions no significant increase in CspD production was noted (Fig. 2A). As a control, the 16S rRNA mRNA level was determined, and the levels showed maximally 9% variance (Fig. 2B).
FIG. 2.
CSP production and cspD and cspE mRNA levels at different time points after cold shock in strains NZ9000 (wild type), NZ9000ΔAB, and NZ9000ΔABE. (A) Proteins were extracted at mid-exponential phase (30°) and at 2 and 4 h after cold shock to 10°C. Only fragments of the 2D-E gels containing the region with the CSPs of L. lactis are shown, as was determined previously (35, 36). Equal amounts of the cell extracts were separated on a pI range of 3 to 10, and the proteins were visualized using silver staining. Molecular size markers are indicated on the right, and a pI scale is given at the bottom. CspA levels are not depicted on these gels and can be seen in Fig. 3. (B) mRNA levels of cspD, cspE, and 16S rRNA in strains NZ9000, NZ9000ΔAB, and NZ9000ΔABE. Northern blots of RNA extracted at 0, 1, and 2 h after cold shock from 30 to 10°C of strains NZ9000 (left), NZ9000ΔAB (center), and NZ9000ΔABE (right) are shown. Transcript sizes are about 300 nt for all csp genes and approximately 1,500 nt for 16S rRNA. N.d., not determined.
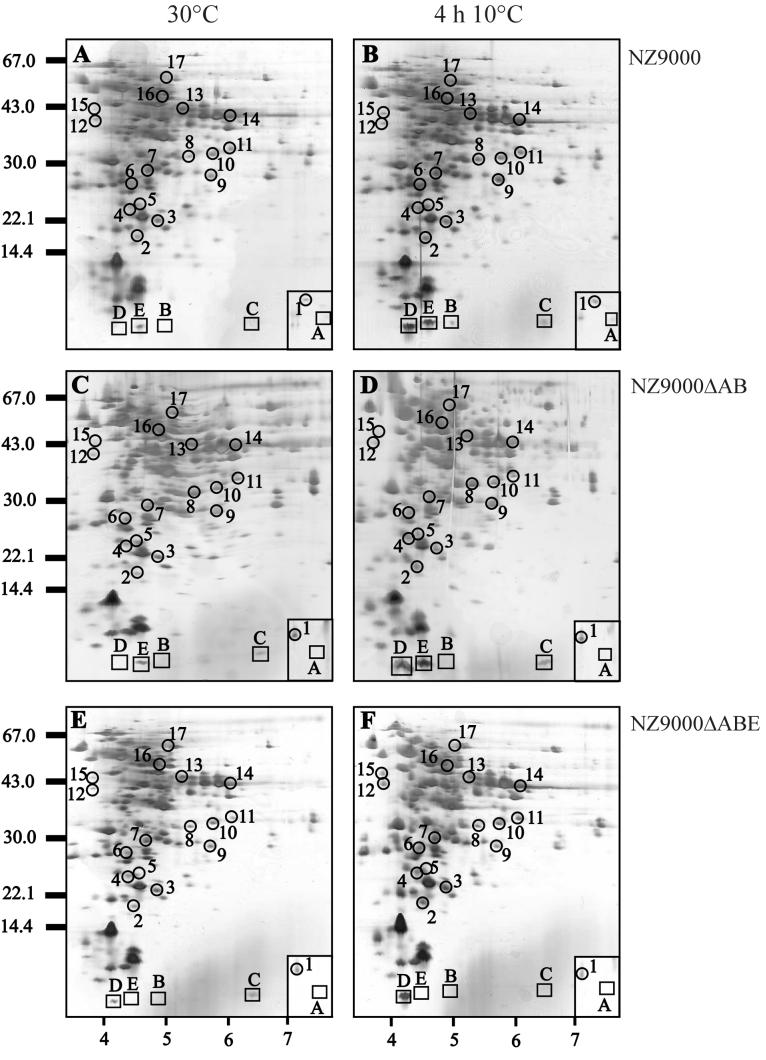
FIG. 3.
Protein analysis of cell extracts of strains NZ9000 (A and B), NZ9000ΔAB (C and D), and NZ9000ΔABE (E and F) by 2D-E. Cell extracts were isolated from these strains prior to cold shock (30°C mid-exponential phase) (A, C, and E) and at 4 h after cold shock to 10°C (B, D, and F) and were separated on a pI range of 3 to 10. Equal amounts of protein were loaded on the gel, and the proteins were visualized using silver staining. Molecular size markers are indicated on the left, and a pI scale is given at the bottom. The CSPs of L. lactis are boxed and have been identified previously (35). CIPs are circled and numbered as described previously (35). The box in the lower right corner of each gel indicates the region between pI values of approximately 9 and 10 of the same molecular weight. The N-terminal sequences of a number of spots are given in Table 2.
Disruption of csp genes affects the production of CIPs.
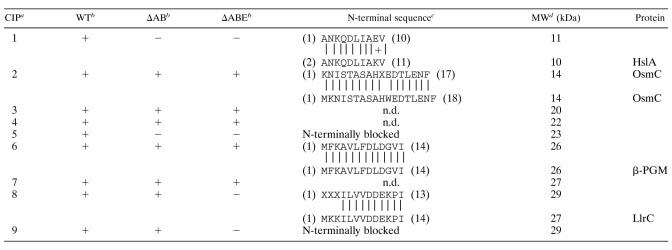
In a previous study we identified a group of 17 CIPs in L. lactis (35). We also showed that the levels of some CIPs increased upon specific overproduction of CSPs, which might point to the involvement of CSPs in the regulation of CIPs (36). To further study this phenomenon, we analyzed the levels of the lactococcal CIPs in the mutants with csp deleted. Separation of cell extracts by 2D-E revealed that the production level of several CIPs was affected in the strains NZ9000ΔAB and NZ9000ΔABE compared to results with the wild-type strain (Fig. 3). For a subset of proteins belonging to this group, the N-terminal amino acid sequence was determined (Table 2). CIP1 (histon-like protein HslA) and CIP5 (unidentified) were cold induced in wild-type cells but were no longer induced at low temperature for strains NZ9000ΔAB and NZ9000ΔABE. For HslA (CIP1), a higher production level was observed for strain NZ9000ΔAB than for wild-type cells at 30°C, which might point to derepression of this protein upon deletion of CspA and/or CspB. CIP8 (signal transduction protein LlrC) and CIP9 (unidentified) were not cold induced in strain NZ9000ΔABE. For the other CIPs, including OsmC (CIP2) and β-PGM (CIP6), no differences were observed in their production levels between the wild-type strain and the strains in which csp was deleted (Table 2).
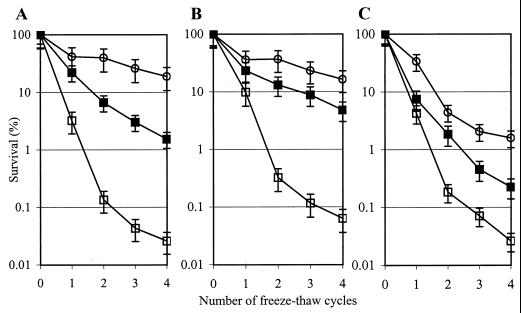
Deletion of cspABE decreases the freeze survival of L. lactis.
Since LAB starter cultures are frequently stored frozen, it is of practical relevance to monitor the abilities of these bacteria to survive freezing temperatures and to determine the critical factors for survival. L. lactis is able to adapt to freezing conditions by preexposure to 10°C, yielding increased survival in freezing conditions (20, 35). For strain NZ9000, preexposure to 10°C for 2 and 4 h increased the survival of freezing approximately 50- and nearly 1,000-fold, respectively, after four repetitive freeze-thaw cycles (Fig. 4A). Strain NZ9000ΔAB showed a response to freezing identical to the wild-type strain's (Fig. 4B). However, the capacity of strain NZ9000ΔABE to survive a freeze increased only approximately 10- and 100-fold after exposure to 10°C for 2 and 4 h, respectively (Fig. 4C). This indicates that the strain NZ9000ΔABE is less well able to adapt to freezing conditions at 10°C, possibly explained by the combined effect of the absence of CspE, the lower total amount of CSPs present, and/or the decreased production of certain CIPs.
FIG. 4.
Abilities of NZ9000 (A), NZ9000ΔAB (B), and NZ9000ΔABE (C) to survive a freeze after pretreatment at 10°C for several hours. Survival of freezing (number of viable cells prior to freezing is set at 100%) of L. lactis after preexposure to 10°C for 0 (open squares), 2 (closed squares), and 4 h (open circles) is shown. The data are shown as an average from two independent experiments, and the error bars indicate the variation for each sample point.
DISCUSSION
In this report we describe the effect of the disruption of two or three genes encoding CSPs in L. lactis on the cold-adaptive response of this bacterium. An L. lactis strain lacking the tandem cspA-cspB genes and a strain additionally lacking cspE were generated. In the absence of the encoded CSPs, the growth characteristics (lag time and growth rate) of L. lactis were not affected. By analogy, deletions of cspAE and cspBB show no effect on the growth characteristics of E. coli and B. subtilis, respectively, which can possibly be explained by increased production of the remaining CSPs (1, 16). We suggest that the presence of the remaining CSPs is sufficient to allow growth of L. lactis NZ9000ΔABE at both low and high temperatures. Indeed, deletion of cspA-cspB is compensated for by increased production of CspE and CspC, while deletion of cspA-cspB-cspE is compensated for by increased CspC and CspD production, mainly at 30°C. For cspD and for cspE, an increase was seen in the mRNA level at these conditions, indicating that the increased production is regulated at the transcriptional level. The increased production of CSPs upon deletion of their counterparts would point to an overlap in their functioning and to the necessity for a minimal CSP level in a cell. The latter aspect might also explain the inability to further inactivate the remaining cspC and cspD genes in strain L. lactis NZ9000ΔABE, similar to observations for B. subtilis (16). Multiple deletion of csp genes in B. subtilis revealed that the presence of at least one CSP is essential for growth at optimal and low temperatures (16). Next, the increased production of the remaining CSPs upon deletion of csp genes also suggests that CSPs directly or indirectly down-regulate the production of their family members. Based on the 2D-E observations for L. lactis NZ9000ΔABE, we speculate that CspE negatively regulates the production of at least CspC and CspD at 30°C. Also, for E. coli it has been found that CSPs are involved in the regulation of their family members: CspE functions as a negative regulator of cspA expression (2).
In a variety of bacteria, non-7-kDa cold-induced proteins have been identified which play a role in a variety of cellular processes, such as chromosome structuring, transcription, translation, general metabolism, sugar metabolism, and stress response (see reviews in references 15, 37, and 39). In this study, the amino-terminal sequences of a selection of these CIPs were determined, and in L. lactis also these proteins seem to be involved in a variety of cellular processes. CIP1 was identified as the histon-like protein HslA of L. lactis. Remarkably, H-NS of E. coli was also found to be cold induced, and a role for this protein in optimizing DNA supercoiling at low temperature has been suggested (7, 25). CIP2 was identical to OsmC in the L. lactis IL1403 genome (5), which is 49% identical to the osmotically inducible protein OsmC of E. coli (17). OsmC of E. coli and its ortholog, YkzA of B. subtilis, belong to the RpoS and SigB stress regulons, respectively (14, 31), which indicates that this type of protein is involved in stress adaptation. Remarkably, in the L. lactis genome no stress sigma factor is found (5), which necessarily points to an alternative regulatory mechanism for lactococcal OsmC. The precise function of OsmC and its orthologs remains to be established. Furthermore, CIP6 was identified as β-phosphoglucomutase of L. lactis (β-PGM) (27), and CIP8 was identified as LlrC in the L. lactis IL1403 genome (5). LlrC was found to be homologous to YycF, a member of a two-component signal transduction system of B. subtilis, which is probably involved in temperature sensing and is essential for growth of B. subtilis (9). It is tempting to speculate that the LlrC two-component signal transduction is involved in a temperature-sensing pathway of L. lactis. These data indicate that the low-temperature response of L. lactis includes adaptation at several levels and support the observation of multilevel cold adaptation for other organisms (15, 39).
In this work we show that the cold-induced production of HslA (CIP1) and CIP5 and of LlrC (CIP8) and CIP9 was affected upon disruption of cspAB or cspABE, respectively, indicating that CSPs regulate proteins most likely involved in cold adaptation. Previously we observed that overproduction of CSPs in L. lactis induces a variety of proteins, among which also are a number of CIPs: CIP2 (OsmC), CIP4, CIP5, and CIP9 (36). Collectively, these data indicate that the production of CIP5 and CIP9 is reduced upon deletion of CspABE, and production of these CIPs is increased upon overproduction of CspA and CspE (36). This would strongly suggest a regulatory role of CspA or CspE in the production of CIP5 and CIP9. For E. coli, it has been reported that CspAE functions as a transcriptional activator of several cold-induced genes, possibly by interacting with Y-boxes located in their promoter regions (7, 25). A similar regulation may be operating for the CSPs of L. lactis; however, no Y-boxes (ATTGG or the complementary CCAAT) were observed in the upstream regions of the genes encoding the identified proteins HslA (CIP1), OsmC (CIP2), β-PGM (CIP6), or LlrC (CIP8) of L. lactis.
In view of practical applications, it is an important finding that L. lactis adapts to freezing conditions during prior exposure to 10°C for several hours, resulting in increased survival rates (20, 35). More detailed analysis revealed that cells specifically overproducing CspB, CspD, or CspE show improved survival under freezing conditions (35, 36). In this work, we show that the level of freeze protection of strain NZ9000ΔABE is lower than that of strains NZ9000 and NZ9000ΔAB upon exposure to 10°C for 2 and 4 h. We suggest that deletion of cspA and cspB in NZ9000ΔAB is compensated for by the observed increased production of CspE. For strain NZ9000ΔABE, the decreased freeze survival can be explained by the absence of CspE, the lower total amount of CSPs present, and/or the decreased production of certain CIPs. These data suggest that CSPs are important for the survival of freezing for L. lactis. CSPs may either have a direct effect during freezing, e.g., by stabilizing RNA and/or DNA, or may regulate the expression of other factors involved in the cryoprotective response (i.e., CIP8 and CIP9). Further elucidation of gene regulation by CSPs and identification of the CIPs of L. lactis will undoubtedly result in an improved understanding of low-temperature adaptation of this organism.
TABLE 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Relevant properties or sequence | Reference |
|---|---|---|
| Strains | ||
| L. lactis NZ9000 | 24 | |
| L. lactis NZ9000ΔAB | NZ9000 derivative; ΔcspA ΔcspB | This study |
| L. lactis NZ9000ΔABE | NZ9000 derivative; ΔcspA ΔcspB ΔcspE | This study |
| Plasmids | ||
| pUC18ERY | Ampr Eryr | 30 |
| pUC18ERYΔAB12 | Ampr Eryr | This study |
| pUC18ERYΔE12 | Ampr Eryr | This study |
| Oligonucleotides | ||
| CspAB1For | 5′-GCAGG AATTC ATGCT CGCTC AGGCT CTTTT-3′ | |
| CspAB1Rev | 5′-AGCCG GTACC CATAT TGAAC CATTT TACTG-3′ | |
| CspAB2For | 5′-AGCGT CTAGA TAATT AATGA AAAAC GGAGC-3′ | |
| CspAB2Rev | 5′-CACGG TCGAC AAATC ACCAA ATTGA GGCAA-3′ | |
| CspE1For | 5′-CGCCG AATTC ATATG GATAA AGTTC GCCGT-3′ | |
| CspE1Rev | 5′-CGTCG GTACC ATTAA CAGTT CCTTG TGCCA-3′ | |
| CspE2For | 5′-GCTGT CTAGA CACTG ACAAA ATTGT CAGTG-3′ | |
| CspE2Rev | 5′-TCCCG TCGAC ATTCC GAGTC TAGCA TGGTG-3′ |
TABLE 2.
List of CIPs of strain NZ9000 (WT) and their production in strains NZ9000ΔAB (ΔAB) and NZ9000ΔABE (ΔABE) 4 h after cold shock to 10°C
For CIP10 to CIP17, no differences in production were observed for the strains NZ9000ΔAB or NZ9000ΔABE, and no N-terminal sequences were determined.
+ indicates induction (at least twofold) of spot after cold shock from 30 to 10°C for 4 h. − indicates no induction of spot after cold shock from 30 to 10°C for 4 h.
The upper sequence is the N-terminal derived sequence, and the lower sequence represents the N-terminal sequence for the respective proteins in the L. lactis IL1403 genome sequence (X indicates unidentified residues in N-terminal sequences).
MW, molecular mass. The upper molecular mass indicates the molecular mass based on the migration on a 2D-E gel, and the lower molecular mass indicates the calculated molecular mass based on the encoded gene sequence. n.d., N-terminal sequence not determined.
ACKNOWLEDGMENTS
J. A. W. and H. F. contributed equally to this report.
REFERENCES
- 1.Bae W, Jones P G, Inouye M. CspA, the major cold shock protein of Escherichia coli, negatively regulates its own expression. J Bacteriol. 1997;176:7081–7088. doi: 10.1128/jb.179.22.7081-7088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae W, Phadtare S, Severinov K, Inouye M. Characterization of Escherichia coli cspE, whose product negatively regulates transcription of cspA, the gene for the major cold shock protein. Mol Microbiol. 1999;31:1429–1441. doi: 10.1046/j.1365-2958.1999.01284.x. [DOI] [PubMed] [Google Scholar]
- 3.Bae W, Xia B, Inouye M, Severinov K. Escherichia coli CspA-family RNA chaperones are transcription terminators. Proc Natl Acad Sci USA. 2000;97:7784–7789. doi: 10.1073/pnas.97.14.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blum H, Beier H, Gross H J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 5.Bolotin A, Mauger S, Malarme K, Ehrlich S D, Sorokin A. Low-redundancy sequencing of the entire Lactococcus lactis IL1403 genome. Antonie Leeuwenhoek. 1999;76:27–76. [PubMed] [Google Scholar]
- 6.Brandi A, Pietroni P, Gualerzi C O, Pon C L. Post-transcriptional regulation of CspA expression in Escherichia coli. Mol Microbiol. 1996;19:231–240. doi: 10.1046/j.1365-2958.1996.362897.x. [DOI] [PubMed] [Google Scholar]
- 7.Brandi A, Pon C L, Gualerzi C O. Interaction of the main cold shock protein CS7.4 (CspA) of Escherichia coli with the promoter region of hns. Biochimie. 1994;76:1090–1098. doi: 10.1016/0300-9084(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 8.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 9.Fabret C, Hoch J A. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J Bacteriol. 1998;180:6375–6383. doi: 10.1128/jb.180.23.6375-6383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang L, Jiang W, Bae W, Inouye M. Promoter-independent cold-shock induction of cspA and its derepression at 37°C by mRNA stabilization. Mol Microbiol. 1997;23:355–364. doi: 10.1046/j.1365-2958.1997.2351592.x. [DOI] [PubMed] [Google Scholar]
- 11.Gasson M J. Plasmid components of Streptococcus lactis NCDO712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldenberg D, Azar I, Oppenheim A B. Differential mRNA stability of the cspA gene in the cold-shock response of Escherichia coli. Mol Microbiol. 1996;19:241–248. doi: 10.1046/j.1365-2958.1996.363898.x. [DOI] [PubMed] [Google Scholar]
- 13.Goldenberg D, Azar I, Oppenheim A B, Brandi A, Gualerzi C O, Pon C L. Role of Escherichia coli cspA promoter sequences and adaptation of translational apparatus in the cold shock response. Mol Gen Genet. 1997;256:282–290. doi: 10.1007/s004380050571. [DOI] [PubMed] [Google Scholar]
- 14.Gordia S, Guiterrez C. Growth-phase dependent expression of the osmotically inducible gene osmC of Escherichia coli K-12. Mol Microbiol. 1996;19:729–736. doi: 10.1046/j.1365-2958.1996.418945.x. [DOI] [PubMed] [Google Scholar]
- 15.Graumann P, Marahiel M A. A superfamily of proteins that contain the cold-shock domain. Trends Biochem Sci. 1998;23:286–290. doi: 10.1016/s0968-0004(98)01255-9. [DOI] [PubMed] [Google Scholar]
- 16.Graumann P, Wendrich T M, Weber M H W, Schröder K, Marahiel M A. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol Microbiol. 1997;25:741–756. doi: 10.1046/j.1365-2958.1997.5121878.x. [DOI] [PubMed] [Google Scholar]
- 17.Guiterrez C, Devedjian J C. Osmotic induction of gene osmC expression in Escherichia coli K12. J Mol Biol. 1991;220:959–973. doi: 10.1016/0022-2836(91)90366-e. [DOI] [PubMed] [Google Scholar]
- 18.Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 19.Jones P G, Krah R, Tafuri S R, Wolffe A P. DNA gyrase, CS7.4, and the cold shock response in Escherichia coli. J Bacteriol. 1992;174:5798–5802. doi: 10.1128/jb.174.18.5798-5802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim W S, Dunn N W. Identification of a cold shock gene in lactic acid bacteria and the effect of cold shock on cryotolerance. Curr Microbiol. 1997;35:59–63. doi: 10.1007/s002849900212. [DOI] [PubMed] [Google Scholar]
- 21.Klijn N, Weerkamp A H, De Vos W M. Identification of mesophilic lactic acid bacteria using PCR-amplified variable regions of 16S rRNA and specific DNA probes. Appl Environ Microbiol. 1991;57:3390–3393. doi: 10.1128/aem.57.11.3390-3393.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuipers O P, Beerthuyzen M M, Siezen R J, De Vos W M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis: requirement of expression of the nisA and nisI genes for development of immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 23.Kuipers O P, Boot H J, De Vos W M. Improved site-directed mutagenesis method using PCR. Nucleic Acids Res. 1991;19:4558. doi: 10.1093/nar/19.16.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuipers O P, De Ruyter P G G A, Kleerebezem M, De Vos W M. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol. 1998;64:15–21. [Google Scholar]
- 25.LaTeana A, Brandi A, Falconi M, Spurio R, Pon C L, Gualerzi C O. Identification of a cold shock transcriptional enhancer of the Escherichia coli major cold shock gene encoding nucleoid protein H-NS. Proc Natl Acad Sci USA. 1991;88:10907–10911. doi: 10.1073/pnas.88.23.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leenhouts K J, Venema G. Lactococcal plasmid vectors. In: Hardy K G, editor; Hardy K G, editor. Plasmids. A practical approach. New York, N.Y: Oxford University Press; 1993. pp. 65–94. [Google Scholar]
- 27.Qian N, Stanley G A, Bunte A, R°dström P. Product formation and phosphoglucomutase activities in Lactococcus lactis: cloning and characterization of a novel phosphoglucomutase gene. Microbiology. 1997;143:855–865. doi: 10.1099/00221287-143-3-855. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 29.Tanabe H, Goldstein J, Yang M, Inouye M. Identification of the promoter region of the Escherichia coli major cold shock gene, cspA. J Bacteriol. 1992;174:3867–3873. doi: 10.1128/jb.174.12.3867-3873.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Kranenburg R, Marugg J D, Van Swam I I, Willem N J, De Vos W M. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol Microbiol. 1997;24:387–397. doi: 10.1046/j.1365-2958.1997.3521720.x. [DOI] [PubMed] [Google Scholar]
- 31.Völker U, Andersen K K, Antelmann H, Devine K M, Hecker M. One of two OsmC homologs in Bacillus subtilis is part of the ςB-dependent general stress regulon. J Bacteriol. 1998;180:4212–4218. doi: 10.1128/jb.180.16.4212-4218.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vos P, Van Asseldonk M, Van Jeveren F, Siezen R J, Simons G, De Vos W M. A maturation protein is essential for the production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J Bacteriol. 1989;171:2795–2802. doi: 10.1128/jb.171.5.2795-2802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wells J M, Wilson P W, Le Page R W F. Improved cloning vectors and transformation procedure for Lactococcus lactis. J Appl Bacteriol. 1993;74:629–636. doi: 10.1111/j.1365-2672.1993.tb05195.x. [DOI] [PubMed] [Google Scholar]
- 34.Willimsky G, Bang H, Fischer G, Marahiel M A. Characterization of cspB, a Bacillus subtilis inducible cold shock gene affecting cell viability at low temperatures. J Bacteriol. 1992;174:6326–6335. doi: 10.1128/jb.174.20.6326-6335.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wouters J A, Jeynov B, Rombouts F M, De Vos W M, Kuipers O P, Abee T. Analysis of the role of 7 kDa cold-shock proteins of Lactococcus lactis MG1363 in cryoprotection. Microbiology. 1999;145:3185–3194. doi: 10.1099/00221287-145-11-3185. [DOI] [PubMed] [Google Scholar]
- 36.Wouters J A, Mailhes M, Rombouts F M, De Vos W M, Kuipers O P, Abee T. Physiological and regulatory effects of controlled overproduction of five cold-shock proteins of Lactococcus lactis MG1363. Appl Environ Microbiol. 2000;66:3756–3763. doi: 10.1128/aem.66.9.3756-3763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wouters J A, Rombouts F M, Kuipers O P, de Vos W M, Abee T. The role of cold-shock proteins in low-temperature adaptation of food-related bacteria. Syst Appl Microbiol. 2000;23:165–173. doi: 10.1016/S0723-2020(00)80001-6. [DOI] [PubMed] [Google Scholar]
- 38.Wouters J A, Sanders J-W, Kok J, De Vos W M, Kuipers O P, Abee T. Clustered organization and transcriptional analysis of a family of five csp genes of Lactococcus lactis MG1363. Microbiology. 1998;144:2885–2893. doi: 10.1099/00221287-144-10-2885. [DOI] [PubMed] [Google Scholar]
- 39.Yamanaka K, Fang L, Inouye M. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol Microbiol. 1998;27:247–255. doi: 10.1046/j.1365-2958.1998.00683.x. [DOI] [PubMed] [Google Scholar]