Summary
Mevalonate (MVA) pathway is the core for terpene and sterol biosynthesis, whose metabolic flux influences the synthesis efficiency of such compounds. Saccharomyces cerevisiae is an attractive chassis for the native active MVA pathway. Here, the truncated form of Enterococcus faecalis MvaE with only 3‐Hydroxy‐3‐methylglutaryl coenzyme A reductase (HMGR) activity was found to be the most effective enzyme for MVA pathway flux using squalene as the metabolic marker, resulting in 431‐fold and 9‐fold increases of squalene content in haploid and industrial yeast strains respectively. Furthermore, a positive correlation between MVA metabolic flux and β‐alanine metabolic activity was found based on a metabolomic analysis. An industrial strain SQ3‐4 with high MVA metabolic flux was constructed by combined engineering HMGR activity, NADPH regeneration, cytosolic acetyl‐CoA supply and β‐alanine metabolism. The strain was further evaluated as the chassis for terpenoids production. Strain SQ3‐4‐CPS generated from expressing β‐caryophyllene synthase in SQ3‐4 produced 11.86 ± 0.09 mg l−1 β‐caryophyllene, while strain SQ3‐5 resulted from down‐regulation of ERG1 in SQ3‐4 produced 408.88 ± 0.09 mg l−1 squalene in shake flask cultivations. Strain SQ3‐5 produced 4.94 g l−1 squalene in fed‐batch fermentation in cane molasses medium, indicating the promising potential for cost‐effective production of squalene.
The truncated form of Enterococcus faecalis MvaE with only 3‐Hydroxy‐3‐methylglutaryl coenzyme A reductase (HMGR) activity was found the most effective enzyme for MVA pathway flux using squalene as the metabolic marker. A positive correlation between MVA metabolic flux and alanine metabolic activity was found based on a metabolomic analysis. 4.94 g l−1 of squalene was obtained by the engineered yeast strain in fed‐batch fermentation in cane molasses medium, indicating the promising potential for cost‐effective production of squalene.
Introduction
Terpenes and sterols are value‐added compounds with extensive applications in cosmetics, food, healthcare, pharmaceutical and bioenergy industries due to their particular physiological activities such as anti‐oxidation, anti‐inflammatory and anti‐tumour (Huang et al., 2009; Bhilwade et al., 2010; Andre et al., 2012; Meadows et al., 2016; Bian et al., 2017). Microbial sources for these valuable compounds have become more attractive recently and have been proposed as the much competitive production routes because of the advantages in cost control, sustainability and environmental benefits when compared to chemical synthesis and extraction from plant or animal tissues (Ye et al., 2016; Paramasivan and Mutturi, 2017a; Yao et al., 2018; Moser and Pichler, 2019; Muhammad et al., 2020). Among the microorganisms, yeasts are regarded as attractive cell factories for production of terpenes and sterols owing to ease of genetic manipulation and the native supply of precursors (Meadows et al., 2016; Paramasivan and Mutturi, 2017a; Muhammad et al., 2020).
Isopentenyl pyrophosphate and its isomer dimethylallyl pyrophosphate are the common precursors for most terpenes and sterols, which are synthesized through the mevalonate (MVA) pathway in yeast species. Thus, the metabolic flux of MVA pathway has a significant influence on the synthesis efficiency of such compounds. 3‐Hydroxy‐3‐methylglutaryl coenzyme A (HMG‐CoA) reductase (HMGR) is the key rate‐limiting enzyme of the MVA pathway (Donald et al., 1997). Overexpression of the truncated form of Saccharomyces cerevisiae HMG1 (tHMG1) has been the general and essential strategy for enhancement of terpene and sterol production (Polakowski et al., 1998; Ro et al., 2006; Paramasivan and Mutturi, 2017b; Yao et al., 2018; Liu et al., 2020). However, HMGRs from diverse sources should be further explored to provide more options for optimal level of HMGR activity.
Acetyl‐CoA is the direct precursor of MVA pathway, thus increasing the synthesis of acetyl‐CoA in the cytoplasm and the flux to MVA pathway is particularly important for improving the synthesis of terpenes and sterols. The cytosolic acetyl‐CoA pool has been enlarged by rewriting central carbon metabolism in yeast cells through modulating native enzyme activities and introducing heterologous enzymes, which contributed to the synthesis of terpenes (Shiba et al., 2007; Lv et al., 2016; Meadows et al., 2016). As the most important intermediate, metabolism of acetyl‐CoA is quite complex in yeast, which not only serves as the precursor of various metabolic pathways, but involves histone acetylation as well (Nielsen, 2014). Hence, the metabolic flux of acetyl‐CoA should be tuned finely in yeast cells to balance the cell growth and metabolite synthesis. Furthermore, apart from engineering the central carbon metabolism and overexpression of key genes in MVA pathway, other metabolic pathways in host cells might also affect the metabolic flux of MVA pathway due to the complex mechanism, which is required to be explored furtherly.
Squalene and β‐caryophylene are two important terpenes. Squalene can be synthesized naturally in yeast as an intermediate of the sterol pathway, while β‐caryophylene is only found naturally in plants. Squalene is widely used in food, medicine and cosmetics owing to its biological activities such as anti‐inflammatory, anti‐tumour, antioxidant and moisturizing activities (Huang et al., 2009; Bhilwade et al., 2010). Microbial production of squalene has become increasingly attractive since the regulations of European Union forbids squalene from shark liver in 2019. Many microorganisms, such as Aurantiochytrium sp., Pseudozyma sp., Rhodoseudomonas palustris and yeast of different species, have been investigated for squalene synthesis (Chang et al., 2008; Nakazawa et al., 2012; Drozdíková et al., 2015; Xu et al., 2016; Ye et al., 2016; Paramasivan and Mutturi, 2017a; Muhammad et al., 2020). However, the squalene accumulation in native strains is very limited (less than 1 mg g−1 cell dry weight generally) due to its rapid conversion to the downstream metabolites (Paramasivan and Mutturi, 2017a). Thus, manipulation of squalene synthetic and its downstream pathways is required for enhancement of squalene production. Beta‐caryophyllene not only possesses antimicrobial, anti‐inflammatory and antioxidant activities but also contributes largely to anti‐depression and delaying Alzheimer's disease (Basha and Sankaranarayanan, 2016; Zhang et al., 2017; Machado et al., 2018). Moreover, it can be used as an environment‐friendly material for ozone removal (Parshintsev et al., 2008), or as the direct precursor of other important terpenoids such as β‐caryophyllene oxide‐natural compounds (Fidyt et al., 2016). Microbial synthetic β‐caryophyllene has great importance for its wide applications (Godara and Kao, 2015; Yang and Nie, 2016).
The availability of precursors and the activity of synthases or modifying enzymes are two bottlenecks determining the yield of terpenes and sterols (Yao et al., 2018). Herein, contribution of precursor supply was mainly considered to improve terpenes synthesis. Although several strategies have been employed to enhance the precursor supply, such as overexpression of key native genes in the MVA pathway and expansion of cytosolic acetyl‐CoA pool as stated above, further exploring alternative rate‐limiting enzymes and novel targets for enhancing the metabolic flux of MVA pathway remains due to the involving complex regulatory mechanism. In this study, a metabonomic strategy was applied to explore novel genomic targets related to metabolic flux of MVA pathway. S. cerevisiae chassis with enhanced MVA pathway flux was developed by concurrent engineering HMGR activity, cofactors balance, flux distributions in acetyl‐CoA synthesis and β‐alanine metabolism (Fig. 1). The potential of the engineered yeast strain for efficient production of terpenes was characterized via squalene and β‐caryophylene as the terpenes of interest, which suggests the availability of an efficient chassis for the production of valuable terpenes and sterols.
Fig. 1.
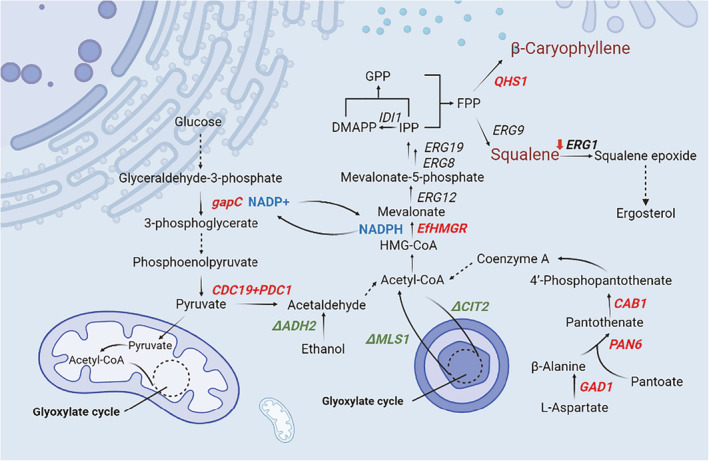
Strategies employed to enhance MVA pathway in Saccharomyces cerevisiae. Solid lines indicate each reaction step. Dashed lines represent the omission of more than one reaction step. The red bold fonts indicate overexpression of genes. The green bold fonts indicate knockout of genes. The blue bold fonts indicate NADPH regeneration. The red downward arrow indicates down‐regulation of ERG1 transcription.
Results and discussion
Screening of HMGR with high activity
HMGR catalyses the rate‐limiting step in MVA pathway, thus HMGR activity levels have significant effects on the metabolic flux of MVA pathway. To obtain HMGR with higher activity, the HMGRs from Hevea brasiliensis, Catharanthus roseus, Yarrowia lipolytica and Enterococcus faecalis were compared first in a S. cerevisiae haploid laboratory strain YS58 with the S. cerevisiae tHmg1 as the positive control using squalene as the index. Among the yeast strains, strain YS58‐EfmvaE expressing E. faecalis mvaE produced the highest level of squalene (28.02 ± 0.09 mg g−1 DCW), which was 59‐fold higher than that of strain YS58‐tHMGR expressing S. cerevisiae tHMG1 (Fig. 2A). Transcriptional analysis indicated that mRNA levels of HMGR genes from H. brasiliensis and C. roseus were much higher than those from S. cerevisiae and Y. lipolytica, while the mRNA level of E. faecalis mvaE was the lowest (Fig. 2B). These results suggest that E. faecalis MvaE has the highest specific activity among the tested enzymes. Transcription efficiency was influenced largely by the nucleotide sequences of target genes (Maertens et al., 2010). Thus, the divergence in mRNA levels might result from the specific sequences of different HMGR genes. E. faecalis mvaE encodes a bifunctional enzyme with both acetyl‐CoA C‐acetyltransferase activity and HMGR activity (Hedl et al., 2002), indicating much more complex sequence structure. To investigate whether the sequence complexity affects the transcriptional efficiency and to confirm the importance of HMGR activity in MvaE to MVA pathway, the mvaE was truncated by removing the region encoding acetyl‐CoA C‐acetyltransferase to generate EfHMGR. The transcription of EfHMGR in strain YS58‐EfHMGR was enhanced to be 288‐fold of the mvaE transcription in strain YS58‐EfMvaE, which was 39.1% higher compared to S. cerevisiae HMG1 in YS58 (Fig. 2B), indicating the negative influence of complex long sequence in mvaE on transcription efficiency. Importantly, the truncated EfHMGR improved the metabolic flux of the MVA pathway further, exhibiting a 54.5% increase of squalene synthesis when compared with MvaE (Fig. 2A).
Fig. 2.
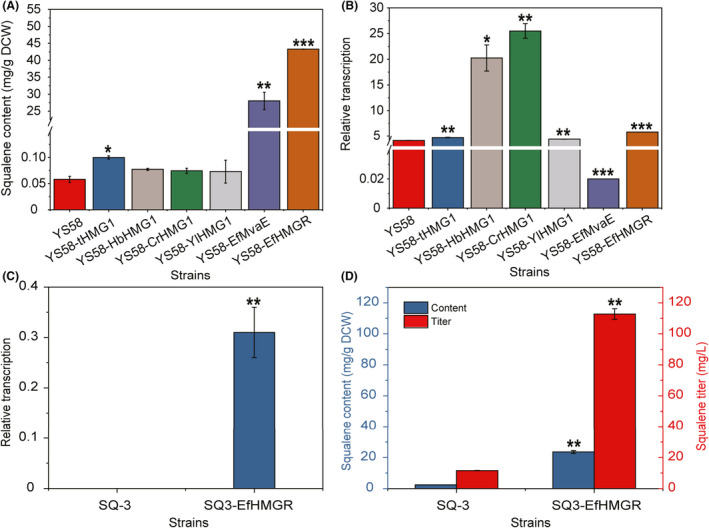
Screening of HMGR with high activity. (A) Squalene content. (B) Relative transcription levels of HMGR genes from different sources. (C) Relative transcription level of the truncated EfHMGR in industrial strains. (D) Squalene production of industrial yeast strains. Yeast cells were cultivated in YPD at 30°C for 24 h. Data are presented as the means of three biological replicates. Error bars represent standard deviations (n = 3). For significance analysis, results of strains YS58 (A, B) and SQ‐3 (C, D) were used as the controls. Student’s t‐test was used for statistical analysis (*P < 0.05; * *P < 0.01; ***P < 0.001).
To evaluate the performance of truncated EfHMGR in the context of industrial yeast strain, the HMG1 in S. cerevisiae SQ‐3 was replaced by EfHMGR expression cassette P TEF1 ‐EfHMGR‐T HMG1 (Fig. 1). The expression of EfHMGR was confirmed by transcriptional analysis of the resulting strain SQ3‐EfHMGR (Fig. 2C). After 24 h shake flasks cultivation, strain SQ3‐EfHMGR produced 112.76 ± 3.39 mg l−1 squalene with intracellular squalene content of 23.57 ± 0.95 mg g−1 DCW, which was 8.7‐fold higher in squalene titer than strain SQ‐3 (Fig. 2D). Structure analysis using SMART tool (http://smart.embl‐heidelberg.de/) indicated that HMGRs from plant and fungal sources have complex structures including positioning sequences and transmembrane domains, while structures of most HMGRs from bacteria are relatively simple. Analysis of the adaptability of HMGRs from various sources in S. cerevisiae showed that HMGRs with a simple protein structure were more effective for increasing the metabolic flux of MVA pathway. E. faecalis mvaE has been expressed in S. cerevisiae for production of lathyrane diterpenoids (Wong et al., 2018). Here, results in both haploid and industrial strains proved that the truncated form of MvaE with only HMGR activity rather than the full MvaE was much more effective due to the efficient transcription.
Cofactor cycle and enhancement of acetyl‐CoA supply for MVA pathway
As the direct precursor of MVA pathway, cytosolic acetyl‐CoA pool has a significant effect on the metabolic flux of MVA pathway. Deletion of peroxisomal citrate synthase gene CIT2 and cytosolic malate synthase gene MLS1 prevents acetyl‐CoA from entering the glyoxylate cycle (Chen et al., 2013; Nielsen, 2014), thus CIT2 and MLS1 loci were chosen as the integration sites. The integration of EfHMGR expression cassette into CIT2 locus of strain SQ‐3 resulted in 25.18 ± 0.02 mg g−1 DCW of squalene in strain SQ3‐1 (Fig. 3A).
Fig. 3.
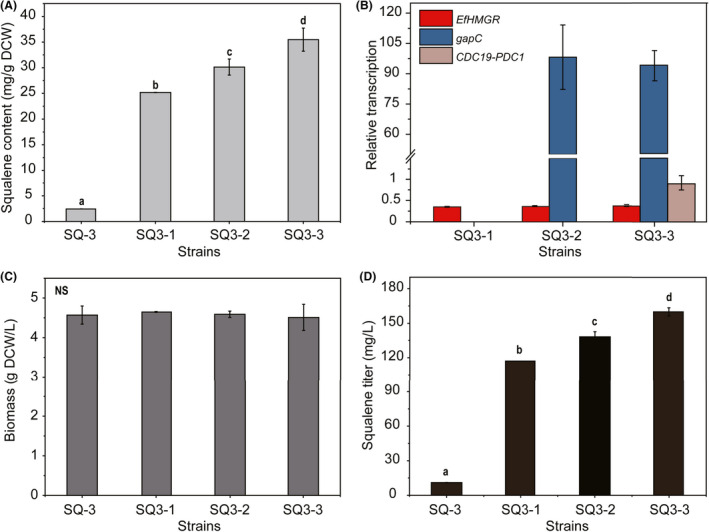
Characterization of the metabolic flux of MVA pathway of industrial strains using squalene as the metabolic marker. (A) Increase in squalene content in yeast strains via engineering HMGR activity, NADPH regeneration and cytosolic acetyl‐CoA supply. (B) Verification of the transcription of EfHMGR, gapC and CDC19‐PDC1 in different strains. (C) Cell growth. (D) Squalene production. Yeast cells were cultivated in YPD at 30°C for 24 h. Data are presented as the means of three biological replicates. Error bars represent standard deviations (n = 3). Student’s t‐test was used for statistical analysis. The lower‐case letters a, b, c and d in (A) and (D) indicate significant differences among different strains (P < 0.05). NS indicates that there are no significant differences among the different strains.
Reaction catalysed by S. cerevisiae HMGR or EfHMGR utilizes NADPH as the cofactor. In view of the limitation of insufficient NADPH on HMGR function, combined engineering of HMGR activity and NADPH regeneration should be conducted. Clostridium acetobutylicum glyceraldehyde 3‐phosphate dehydrogenase encoded by gapC is a NADP‐dependent enzyme (Martínez et al., 2008). Hence, codon optimized gapC expression cassette P IRA1 ‐gapC‐T MLS1 was integrated into MLS1 locus of strain SQ3‐1 to generate strain SQ3‐2, which not only promoted cytosolic NADPH regeneration but also improved the metabolic efficiency of glycolytic pathway and reduced the consumption of acetyl‐CoA by glyoxylate cycle (Chen et al., 2013). The intracellular squalene content was further increased to 30.13 ± 1.55 mg g−1 DCW in strain SQ3‐2 (Fig. 3A).
Conversion of pyruvate into acetyl‐CoA by pyruvate dehydrogenase (PDH) complex in mitochondria is the main competitive shunt of pyruvate into cytosolic acetyl‐CoA catalyzed first by pyruvate decarboxylase (PDC). To enhance the cytosolic route to acetyl‐CoA, expression of PDH complex in cytosol was attempted by several groups (Nielsen, 2014). However, only the E. faecalis PDH complex was expressed functionally in yeast cytosol, which activity required the presence of lipoic acid in the medium (Kozak et al., 2014; Zhang et al., 2020). Herein, pyruvate kinase encoded by CDC19 involving the formation of pyruvate from phosphoenolpyruvate and pyruvate decarboxylase encoded by PDC1 were coexpressed as a fusion protein in strain SQ3‐2 to generate strain SQ3‐3, which produced 35.47 ± 2.25 mg g−1 DCW of squalene (Fig. 3A). Pyruvate decarboxylase plays an important role in the synthesis of cytoplasmic acetyl‐CoA. Overexpression of PDC1 increased the titre of 3‐hydroxypropionic acid largely by enhancing the supply of acetyl‐CoA in S. cerevisiae (Kildegaard et al., 2016). In pyruvate decarboxylase minus (Pdc−) strains, downregulating PYK1 (a.k.a. CDC19) relieved the glucose repression, while mutation in CDC19 significantly increased the growth rate (Yu et al., 2018; Gambacorta et al., 2020), implying the importance of the coupling of pyruvate kinase and pyruvate decarboxylase in rewiring yeast carbon metabolism. Fusion of two enzymes catalyzing the consecutive reactions improved substrate accessibility (Jiang et al., 2017; Chen et al., 2019). All of these previous reports support the contribution of fusion protein of Cdc19‐Pdc1 to more conversion of pyruvate into cytoplasmic acetyl‐CoA, which then increases the metabolic flux through the MVA pathway.
The metabolic flux of MVA pathway was enhanced by combined engineering HMGR activity, NADPH regeneration and cytosolic acetyl‐CoA supply (Fig. 3A and B). No obvious influence on cell growth of these manipulations was observed (Fig. 3C). The combined engineering resulted in 159.97 ± 3.54 mg l−1 of squalene for strain SQ3‐3, 13.4‐fold higher squalene production than the original strain SQ‐3 after 24 h cultivation in YPD (Fig. 3D). Among the engineering strategies, introduction of the truncated E. faecalis HMGR contributed largely to enhance intracellular squalene accumulation, confirming the importance of high enzyme activity of HMGR in metabolic flux of MVA pathway. Previously, the truncated form of S. cerevisiae HMGR (tHmg1) was generally used to enhance the flux of MVA pathway (Polakowski et al., 1998; Ro et al., 2006; Paramasivan and Mutturi, 2017b; Yao et al., 2018; Liu et al., 2020), which was 19‐fold higher than the complete HMGR in specific activity (Donald et al., 1997). Here, the E. faecalis HMGR exhibited a much higher activity than the tHmg1 (Fig. 2A and B), indicating its wide application potentials for terpene and sterol production.
Metabolomics analysis of strain SQ3‐3 grown in YPD with two kinds of yeast extract
Fortuitously, significant impacts of yeast extract from different sources on squalene production were observed during the analysis of fermentation performance of strain SQ3‐3. Yeast extract from N company (designated as YE‐N) remarkably improved the squalene synthesis, which was 1.6‐fold of that in YPD containing the original yeast extract (designated as YE‐O) after 24 h of cultivation (Fig. 4A). In view of the complex composition of yeast extract, it is difficult to find the key factors directly related to the change in MVA pathway flux. To reveal the influence of different yeast extract on endo‐metabolism and explore the potential targets related to MVA pathway, yeast cells of SQ3‐3 grown in YPD with yeast extract YE‐N or YE‐O for 18 h were collected for metabolomics analysis. As depicted in Fig. 4B, significant differences occurred in amino acid metabolism. Higher amino acid metabolism flux was associated with higher MVA pathway activity, among which the β‐alanine metabolism was influenced by yeast extract significantly. Therefore, β‐alanine metabolism was speculated as the key potential target for improving MVA pathway flux.
Fig. 4.
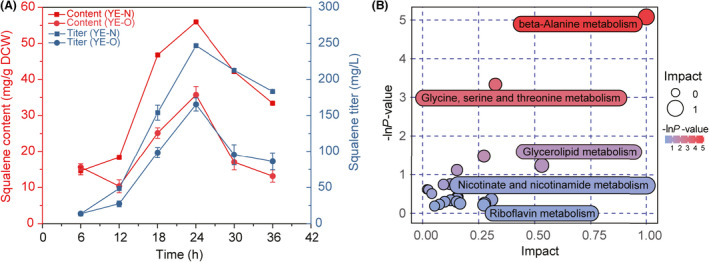
Metabolomics analysis of yeast strain SQ3‐3 grown in YPD with yeast extract from different sources. (A) Time course of squalene production in shake flask cultivation with yeast extract from different sources (YE‐O and YE‐N). (B) Differential pathway analysis based on metabolomics. Each bubble in the bubble chart represents a metabolic pathway. Impact values on the abscissa represent the influence degree of the pathway in the topology analysis. The ‐lnP‐value on ordinate is the negative natural logarithm of P‐value, which represents the significant differences of the enrichment analysis. Data are presented as the means of four biological replicates. Error bars represent standard deviations (n = 4).
Up‐regulation of β‐alanine metabolism enhances metabolic flux of MVA pathway
Beta‐alanine is a platform compound, which can be condensed with pantoate catalysed by pantothenate synthase to form pantothenic acid, the precursor of coenzyme A (CoA) and acyl carrier protein (White et al., 2001). Herein, to explore the effect of up‐regulating β‐alanine metabolic pathway on the metabolic flux of MVA pathway, the expression of GAD1 encoding glutamate decarboxylase, PAN6 encoding pantothenate synthase and CAB1 encoding pantothenate kinase was enhanced in haploid yeast strain YS58 firstly by integrating expression cassette P TEF2 ‐GAD1‐T SUP4 , P TEF2 ‐PAN6‐T SUP4 or P TEF2 ‐CAB1‐T ADH2 into the ADH2 locus respectively (Fig. 5A). The intracellular squalene content in the resulted strain YS58‐GAD1, YS58‐PAN6 or YS58‐CAB1 was increased compared to strain YS58 cultivated in YPD medium containing either yeast extract YE‐O or YE‐N, which was about 1.3‐fold, 1.7‐fold or 1.4‐fold of that in strain YS58 (Fig. 5B). The cell growth was influenced by overexpression of GAD1, PAN6 or CAB1 at different degrees (Fig. S1). Improving acetyl‐CoA biosynthesis was reported via enhancing coenzyme A synthesis in S. cerevisiae (Liu et al., 2017; Olzhausen et al., 2021). Hence, the positive effect endowed by overexpression of GAD1, PAN6 and CAB1 on the metabolic flux of MVA pathway might be due to the increase in acetyl‐CoA supply. The reaction catalyzed by pantothenate kinase (Cab1p) is considered as the rate‐limiting step in CoA synthetic pathway (Olzhausen et al., 2009). However, the enhanced expression of PAN6 caused the highest increase of squalene synthesis, perhaps because of the highest transcriptional levels of PAN6 compared to GAD1 and CAB1 (Fig. 5A). In Escherichia coli, β‐alanine is produced from the decarboxylation of L‐aspartate catalyzed by aspartate decarboxylase PanD (Cronan et al., 1982). In view of the absence of a homolog of PanD in yeast (White et al., 2001), GAD1 encoding glutamate decarboxylase was chosen to perform the decarboxylation of L‐aspartate, which increased the metabolic flux of MVA pathway (Fig. 5B).
Fig. 5.
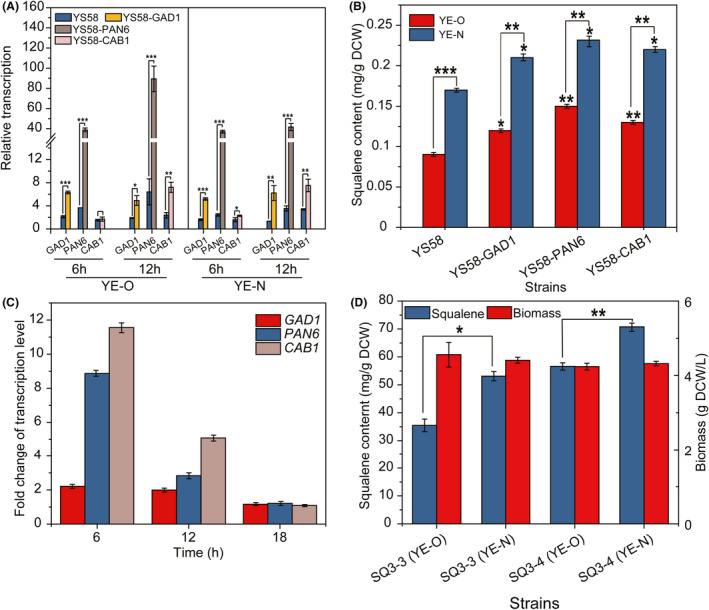
Influence of up‐regulation of β‐alanine metabolism on the metabolic flux of MVA pathway. (A) Relative transcription levels of GAD1, PAN6, and CAB1 in YS58‐derived strains. (B) Squalene content of YS58‐derived strains. (C) Fold change of transcription levels of GAD1, PAN6 and CAB1 in strain SQ3‐4 vs SQ3‐3. (D) Cell growth and squalene content of strains SQ3‐3 and SQ3‐4. Yeast cells were cultivated in YPD with yeast extract YE‐O or YE‐N, respectively, at 30°C and 200 rpm for 24 h. Data are presented as the means of three biological replicates. Error bars represent standard deviations (n = 3). Student’s t‐test was used for statistical analysis (*P < 0.05; * *P < 0.01; ***P < 0.001).
The expression cassettes P TEF2 ‐GAD1‐T SUP4 , P IRA1 ‐PAN6‐T SUP4 and P PGK1 ‐CAB1‐T ADH2 were further integrated into the ADH2 locus of chromosome XIII of strain SQ3‐3 to generate strain SQ3‐4. The transcription levels of GAD1, PAN6 and CAB1 in strain SQ3‐4 were enhanced, which were 2.2‐fold, 8.9‐fold and 11.6‐fold of those in strain SQ3‐3, respectively, at 6 h of cultivation (Fig. 5C). The intracellular accumulation of squalene in strain SQ3‐4 was 70.76 ± 2.72 mg g−1 DCW or 56.59 ± 1.29 mg g−1 DCW in YPD medium containing yeast extract YE‐N or YE‐O, respectively, indicating 33.2% or 59.5% increase when compared to strain SQ3‐3 (Fig. 5D). The squalene titre in strain SQ3‐4 reached 305.68 ± 2.72 mg l−1 in YE‐N medium, 1.3‐fold of that in strain SQ3‐3. The squalene production of strain SQ3‐4 in a medium containing yeast extract YE‐O matched that of strain SQ3‐3 in a medium containing yeast extract YE‐N, confirming the role of β‐alanine metabolism in the effect of yeast extract on metabolic flux of MVA pathway. However, for strain SQ3‐4, a 27.5% increase in squalene production in a medium with yeast extract YE‐N was still observed compared to a medium with yeast extract YE‐O, suggesting that the specific constituents in YE‐N might stimulate the Gad1p, Pan6p and/or Cab1p activity to further increase the β‐alanine metabolism, and then to promote the flux of MVA pathway. However, other factors, such as other amino acid metabolisms, might also contribute to the effect of yeast extract on MVA pathway, which are required to be investigated further. Moreover, in S. cerevisiae, an amine oxidase encoded by FMS1 was reported to catalyze the rate‐limiting step for β‐alanine and pantothenic acid biosynthesis (White et al., 2001; Schadeweg and Boles, 2016). Thus, the effect of enhancing FMS1 expression on MVA pathway flux should also be investigated in the future.
Efficient production of squalene using SQ3‐4 as the chassis
Squalene is an intermediate of the sterol biosynthetic pathway, which can be converted to squalene epoxide by squalene epoxidase (Erg1p) and then sterols through a serial of enzymatic reactions to reduce the intracellular accumulation of squalene. To evaluate the potential of strain SQ3‐4 for high yield production of squalene, a strategy for down‐regulation of ERG1 transcription via heterologous ERG1 promoter (P ERG1 ) was explored.
To obtain heterologous ERG1 promoters with lower activity, P ERG1 of Pichia pastoris, Hansenula polymorpha and Phaffia rhodozyma were chosen due to the lower metabolic flux of native sterol biosynthetic pathway compared to S. cerevisiae, and then were characterized in S. cerevisiae using GFP as the reporter. As shown in Fig. S2, the GFP fluorescence of strain YS58‐HpE1G or YS58‐PrE1G harbouring H. polymorpha P ERG1 (P HpERG1 ) or P. rhodozyma P ERG1 (P PrERG1 ) was much lower than that of other strains, which was reduced by 92.5% in strain YS58‐PrE1G after 12 h of cultivation when compared to strain YS58‐ScE1G with native ERG1 promoter. The replacement of native promoter P ERG1 in strain YS58 with P. rhodozyma promoter P PrERG1 significantly increased the intracellular squalene accumulation in strain YS58‐PrE1 (13.95 ± 1.53 mg g−1 DCW), which was 73.4‐fold of strain YS58 (Fig. S2). Hence, P. rhodozyma promoter P PrERG1 was used to replace the native promoter P ERG1 in SQ3‐4 genome to generate strain SQ3‐5. The transcription of ERG1 in SQ3‐5 was down‐regulated significantly via using promoter P PrERG1 (Fig. 6A). Although the growth of strain SQ3‐5 was affected by down‐regulation of ERG1, the intracellular squalene content in strain SQ3‐5 reached 152.01 ± 4.52 mg g−1 DCW after 24 h of cultivation, which was 1.2‐fold higher compared to SQ3‐4 (Fig. 6B). Strain SQ3‐5 produced 408.88 ± 8.30 mg l−1 squalene, indicating 37.2% increase in squalene titre compared to SQ3‐4. P. rhodozyma is a carotenoid‐producing strain, which diverts the metabolic flux from sterol synthesis to pigment synthesis largely (Johnson, 2003). Thus, we speculated that the P. rhodozyma ERG1 promoter should be weaker in transcriptional activity than the S. cerevisiae native ERG1 promoter. Significant decreases in GFP fluorescence and ERG1 transcriptional level were observed when promoter P PrERG1 was used, in line with our prediction. The down‐regulation of ERG1 using P PrERG1 caused large reduction (35.1%) in cell growth of SQ3‐5, perhaps due to the limitation on flux toward ergosterol synthesis or accumulation of toxic intermediates (Martin et al., 2003; Yuan and Ching, 2015). The glucose‐sensing HXT1 promoter, methionine‐repressible MET3 promoter and tetO7‐CYC1 promoter were used for the down‐regulation of gene expression in S. cerevisiae (Paddon et al., 2013; Yuan and Ching, 2015; Liu et al., 2020). However, the function of these promoters depends heavily upon the components of fermentation broth. Promoter P ERG1(M5) , an engineered ERG1 promoter of S. cerevisiae was developed to minimize the ERG1 transcription in our previous study (Zhou et al., 2021). This study provides an alternative for down‐regulation of gene expression to minimize the downstream metabolism. About 350 mg l−1 of squalene titre (about 80 mg g−1 DCW) was yielded by overexpressing genes involved in squalene synthesis from acetyl‐CoA and gene encoding NADH‐HMGR in S. cerevisiae after 24 h of cultivation in YPD medium, which was increased to about 400 mg l−1 by further peroxisomal engineering for synthesis and storage of squalene (Liu et al., 2020). In our previous study, 100.31 ± 9.90 mg g−1 CDW of intracellular squalene was obtained after 24 h of cultivation in YPD medium, which was the highest specific synthesis rate reported to date at the shake flask stage (Zhou et al., 2021). Here, squalene content in strain SQ3‐5 reached 152.01 ± 4.52 mg g−1 DCW, indicating the effectiveness of the used engineering strategies.
Fig. 6.
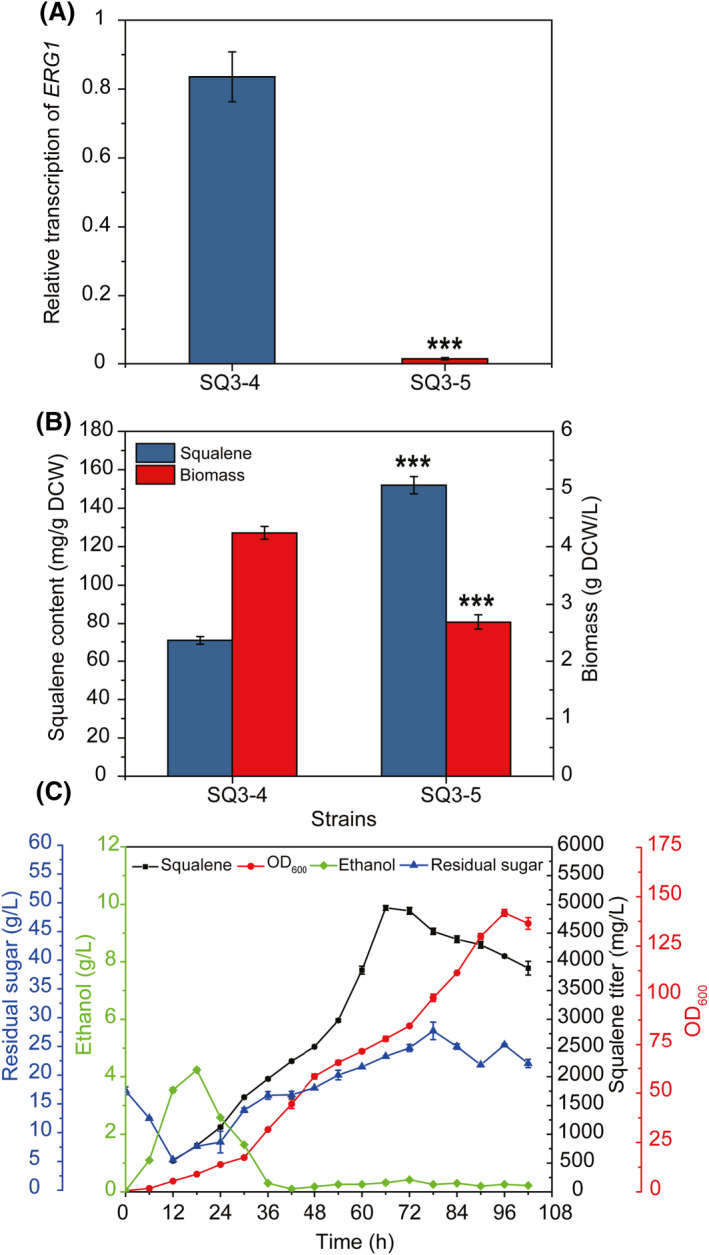
Squalene production by down‐regulation of ERG1 transcription in the chassis SQ3‐4. (A) Down‐regulation of ERG1 transcription of industrial strain by P. rhodozyma ERG1 promoter. (B) Influence of down‐regulating ERG1 transcription on cell growth and squalene content of industrial strain. Yeast cells were cultivated in YPD containing yeast extract YE‐N at 30°C and 200 rpm for 24 h. (C) Squalene production by the engineered strain SQ3‐5 in fed‐batch fermentation in cane molasses medium. Fermentation was conducted in 1‐L bioreactor with cane molasses and ammonium sulphate as the main feedstock. Data are presented as the means of three biological replicates. Error bars represent standard deviations (n = 3). For significance analysis, results of strain SQ3‐4 were used as the controls. Student’s t‐test was used for statistical analysis (***P < 0.001).
To evaluate the potential of the engineered yeast strain SQ3‐5 for cost‐effective production of squalene, fed‐batch fermentation was conducted using cane molasses and ammonium sulphate as the main substrates. The highest squalene titre was 4940.31 ± 41.15 mg l−1 after 66 h of cultivation with a specific synthesis rate of 74.85 mg l−1 h−1 (Fig. 6C). Molasses is a major by‐product of sugar industry, which is regarded as inexpensive and abundant feedstock for industrial production of value‐added bioproducts (Zhang et al., 2021). In this study, it was found that the usage of molasses medium to replace medium containing glucose, peptone, yeast extract and other substances could significantly reduce the cost of squalene production.
Evaluation of strain SQ3‐4 as the chassis for other terpene synthesis
The potential of strain SQ3‐4 as the chassis for biosynthesis of terpenes was further investigated using β‐caryophylene as the compound of interest. Beta‐caryophyllene synthase (CPS) catalyzes the conversion from farnesyl diphosphate (FPP) to β‐caryophyllene (Cai et al., 2002). Thus, a codon optimized coding sequence of Artemisia annua CPS designated QHS1 was overexpressed on the plasmid pYE‐CPS in strain SQ3‐4 and the original strain SQ‐3 to obtain strains SQ3‐4‐CPS and SQ‐3‐CPS respectively (Fig. 7A). Strain SQ3‐4‐CPS produced 11.86 ± 0.09 mg l−1 β‐caryophyllene, while no β‐caryophyllene was detected in both cells and extracellular broth of strain SQ‐3‐CPS (Fig. 7B). Reduction in squalene production in both SQ3‐4‐CPS and SQ‐3‐CPS was observed (Fig. 7B), indicating the shunt of metabolic flux from squalene production to β‐caryophyllene synthesis. These results demonstrated that strain SQ3‐4 is a potential chassis for synthesis of endogenous or heterologous terpenes. Further investigation will focus on increasing the activity of CPS and engineering the transporter of β‐caryophyllene to promote the yield of such terpenes.
Fig. 7.
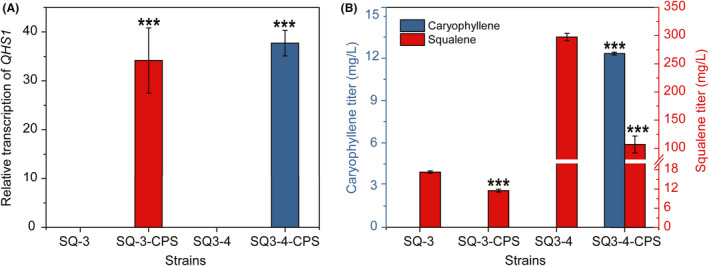
Evaluation of strain SQ3‐4 as the chassis for other terpene synthesis. (A) Relative transcription level of β‐caryophyllene synthase gene QHS1 (12 h cultures). (B) Comparison of β‐caryophyllene titre and squalene titre among various strains. Yeast cells were cultivated in YPD containing yeast extract YE‐N at 30°C and 200 rpm for 48 h. Data are presented as the means of three biological replicates. Error bars represent standard deviations (n = 3). For significance analysis, strains SQ‐3 and SQ3‐4 were used as the controls respectively. Student’s t‐test was used for statistical analysis (***P < 0.001).
Conclusion
For the synthesis of terpenes and sterols by engineered yeast strains, the truncated form of S. cerevisiae HMGR (tHmg1) was generally used to enhance the flux of MVA pathway. Here, we found that the E. faecalis HMGR (EfHMGR) exhibited much higher activity than the tHmg1, which is the most effective HMGR for enhancement of the MVA pathway flux so far. Furthermore, a positive effect of β‐alanine metabolism on the flux of MVA pathway was revealed by a metabonomic analysis and verified via its stimulation of squalene synthesis. These results provide novel function modules for enhancing the production of terpenes and sterols. Then an industrial S. cerevisiae chassis with enhanced metabolic flux of MVA pathway was developed by combined engineering HMGR activity, cofactor balance, β‐alanine metabolism and cytosolic acetyl‐CoA synthesis. The potential of this chassis for bioproduction of terpenoids was verified by using β‐caryophyllene and squalene as the terpene compounds of interest. The squalene production reached 4.94 g l−1 in cane molasses medium, indicating the promising potential for cost‐effective production of squalene.
Experimental procedures
Strains and culture conditions
All of the yeast strains used in this study are listed in Table 1. E. coli DH5α (supE44 ∆lacU169 (φ80lacZ∆M15) hsdR17 recA1 endA1 gyrA96 thi‐1 relA1) was used as a general host for plasmid construction and propagation. Haploid S. cerevisiae YS58 (Teunissen et al., 1993) was used to investigate the roles of target genes. Industrial S. cerevisiae strain SQ‐3 (Zhou et al., 2021) was used as the chassis for engineering metabolic flux of MVA pathway. E. coli cells were grown at 37°C in Luria–Bertani (LB) medium with 100 μg ml−1 of ampicillin when necessary. Yeast cells were cultivated generally at 30°C in YPD (10 g l−1 yeast extract, 20 g l−1 peptone, 20 g l−1 glucose) with 100 μg ml−1 G418 or 100 μg ml−1 zeocin when required. SCG medium containing 6.7 g l−1 yeast nitrogen base, 20 g l−1 galactose, 40 mg l−1 histidine, 40 mg l−1 leucine and 40 mg l−1 tryptophan was used for counter‐selection of PGAL1‐mazF‐zeoR (GMZ) knockout strains. For fed‐batch fermentation, cane molasses medium containing molasses with sugar amounting to 20 g l−1 of reducing sugar was used (Zhou et al., 2021).
Table 1.
Yeast strains used in this study.
| Strains | Description | Source |
|---|---|---|
| YS58 | MATα flo1 ura3‐52 leu2‐3,112 his4‐519 trp1‐789 | Teunissen et al. (1993) |
| YS58‐tHMGR | YS58, Δhmg1::P TEF1 ‐tHMG1‐T CYC1 | This work |
| YS58‐HbHMGR | YS58, Δhmg1::P TEF1 ‐HbHMG1‐T CYC1 | This work |
| YS58‐CrHMGR | YS58, Δhmg1::P TEF1 ‐CrHMG1‐T CYC1 | This work |
| YS58‐YlHMGR | YS58, Δhmg1::P TEF1 ‐YlHMG1‐T CYC1 | This work |
| YS58‐EfMvaE | YS58, Δhmg1::P TEF1 ‐EfmvaE‐T CYC1 | This work |
| YS58‐EfHMGR | YS58, Δhmg1::P TEF1 ‐EfHMGR‐T CYC1 | This work |
| YS58‐GAD1 | YS58, Δadh2::P TEF2 ‐GAD1‐T SUP4 | This work |
| YS58‐PAN6 | YS58, Δadh2::P TEF2 ‐PAN6‐T SUP4 | This work |
| YS58‐CAB1 | YS58, Δadh2::P TEF2 ‐CAB1‐T SUP4 | This work |
| YS58‐GPC | YS58, Δadh2::P TEF2 ‐GAD1‐T SUP4 ‐P IRA1 ‐PAN6‐T SUP4 ‐ P PGK1 ‐CAB1‐T SUP4 | This work |
| YS58‐ScE1G | YS58 containing plasmid pYC‐SEGA | This work |
| YS58‐PpE1G | YS58 containing plasmid pYC‐PEGA | This work |
| YS58‐HpE1G | YS58 containing plasmid pYC‐HEGA | This work |
| YS58‐PrE1G | YS58 containing plasmid pYC‐XEGA | This work |
| YS58‐PrE1 | YS58 with replacement of wild‐type ERG1 promoter by P. rhodozyma ERG1 promoter | This work |
| YEH‐56 | S. cerevisiae with high sterol pathway activity | He et al. (2000) |
| SQ‐3 | YEH‐56 with overexpression of ERG10, ERG13, tHMG1, ERG12, ERG8, ERG19, IDI1, ERG20 and ERG9 | Zhou et al. (2021) |
| SQ3‐EfHMGR | SQ‐3, Δhmg1::P TEF1 ‐EfHMGR‐T HMG1 | This work |
| SQ3‐1 | SQ‐3, Δcit2::P IRA1 ‐EfHMGR‐T CIT2 | This work |
| SQ3‐2 | SQ3‐1, Δmls1::P IRA1 ‐gapC‐T MLS1 | This work |
| SQ3‐3 | SQ3‐2, Δpdc1::P TEF2 ‐CDC19‐linker‐PDC1‐T PDC1 | This work |
| SQ3‐4 | SQ3‐3, Δadh2::P TEF2 ‐GAD1‐T SUP4 ‐P IRA1 ‐PAN6‐T SUP4 ‐P PGK1 ‐CAB1‐T ADH2 | This work |
| SQ3‐5 | SQ3‐4 with replacement of wild‐type ERG1 promoter by P. rhodozyma ERG1 promoter | This work |
| SQ‐3‐CPS | SQ‐3 containing plasmid pYE‐CPS | This work |
| SQ3‐4‐CPS | SQ3‐4 containing plasmid pYE‐CPS | This work |
For shake flask cultivation, a single colony of S. cerevisiae strain was grown in 2 ml YPD for 16 h at 30°C and 200 rpm. The seed culture was prepared by transferring 0.5 ml of cell culture into 5 mL YPD and cultivated at 30°C and 200 rpm for 18 h, which was then inoculated into 45 ml of YPD in 250‐mL shake flasks. In general, cultivation was conducted at 30°C and 200 rpm for 24 h. To evaluate the potential of the engineered yeast strain for squalene production, fed‐batch fermentation in cane molasses medium was conducted in the 1‐L stirred‐tank bioreactor (Infors HT, Switzerland) as previously reported (Zhou et al., 2021).
Plasmid construction
All the plasmids and primers used in this study are described in Table S1 and Table S2. Plasmid pFA6a‐kanMX4 was used as the template for amplification of G418‐resistant cassette KanMX. Plasmid pGMZC was used to amplify selective marker P GAL1 ‐mazF‐zeoR (GMZ) including zeocin‐resistant cassette P TEF1 ‐zeoR‐T CYC1 and mazF expression cassette P GAL1 ‐mazF‐T AOX1 (Zhou et al., 2021). To compare promoter strength, ERG1 promoter was amplified from S. cerevisiae, H. polymorpha, P. pastoris and P. rhodozyma genomes, respectively, and then was ligated into plasmid pYC‐GA (Zhou et al., 2021) to generate plasmids pYC‐SEGA, pYC‐HEGA, pYC‐PEGA and pYC‐XEGA with green fluorescence protein (GFP) as the reporter. For expression of β‐caryophyllene synthase (CPS), plasmid pYE‐CPS was constructed with the codon‐optimized coding sequence of β‐caryophyllene synthase gene QHS1 regulated by promoter P IRA1 and terminator T SUP4 .
Yeast strain construction
For genomic integration of the expression cassettes of target genes, the homologous arms, promoter sequences, coding sequences of target genes, terminator sequences and selective markers were amplified from genome of S. cerevisiae or the relative plasmids using the primers listed in Table S2. For heterologous expression, the codon‐optimized coding sequences of truncated HMGRs from H. brasiliensis, C. roseus, Y. lipolytica, mvaE from E. faecalis, gapC from C. acetobutylicum, as well as β‐caryophyllene synthase gene QHS1, were synthesized by Nanjing GenScript Biotechnology Co., Ltd (China). DNA fragments covering the target sequences were created by overlap extension PCR and transformed into S. cerevisiae strains by electroporation (Bio‐Rad Gene‐Pulser Apparatus). To compare the activity of HMGR from various sources, yeast strains YS58‐tHMGR, YS58‐HbHMGR, YS58‐CrHMGR, YS58‐YlHMGR, YS58‐EfMvaE and YS58‐EfHMGR with replacement of the native HMG1 by tHMG1 or coding sequence of HMGR from H. brasiliensis, C. roseus, Y. lipolytica, and E. faecalis were constructed. For engineering the metabolic flux of MVA pathway, the expression cassettes P IRA1 ‐EfHMGR‐TCIT2 , P IRA1 ‐gapC‐T MLS1 and P TEF2 ‐CDC19‐linker‐PDC1‐T PDC1 were integrated into the CIT2, MLS1 and PDC1 sites of SQ‐3 in turn to generate yeast strain SQ3‐1, SQ3‐2 and SQ3‐3 respectively. To explore the influence of β‐alanine metabolism, expression cassettes of GAD1, PAN6 and CAB1 were integrated into the ADH2 site of YS58 or SQ3‐3 by endogenous homologous recombination to generate strains YS58‐GAD1, YS58‐PAN6, YS58‐CAB1, YS58‐GPC and SQ3‐4. To compare the activity of ERG1 promoter from different sources, plasmids pYC‐SEGA, pYC‐HEGA, pYC‐PEGA and pYC‐XEGA were transformed into YS58 to generate strains YS58‐ScE1G, YS58‐PpE1G, YS58‐HpE1G and YS58‐PrE1G. For down‐regulation of ERG1, the native ERG1 promoter in strain YS58 and SQ3‐4 was replaced by P. rhodozyma ERG1 promoter to generate strains YS58‐PrE1 and SQ3‐5. For synthesis of β‐caryophyllene, plasmid pYE‐CPS was transformed into SQ‐3 and SQ3‐4, resulting in strains SQ3‐CPS and SQ3‐4‐CPS.
Quantification of transcriptional levels of the target genes by qRT‐PCR
Yeast cells were cultured in YPD at 30°C and 200 rpm for 6–24 h. RNA was extracted using the RNAprep pure Cell/Bacteria Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. The FastQuant RT kit (Tiangen, Beijing, China) was used for reverse transcription. Quantitative PCR was performed using RealMasterMix (SYBR Green I) (Tiangen, Beijing, China) in LightCycler 96 System (Roche, Switzerland) with primers in Table S2. Data were collected and processed by the second‐derivative maximum method of LightCycler 96 software SW1.1 with housekeeping gene ACT1 as the control to normalize different samples. Relative transcription is defined as the ratio of transcription of each target gene to that of ACT1 in each sample.
Metabonomic analysis
Yeast cells were collected after 18 h cultivation in 50 ml YPD by centrifugation at 5000 rpm for 5 min and washed twice with distilled water. Cell sample (50 mg) was resuspended in 1.0 ml extract solution (acetonitrile : methanol : water = 2 : 2 : 1) with isotopically labelled internal standard mixture. After 30 sec of vortex, the samples were homogenized at 35 Hz for 4 min and sonicated for 5 min in ice‐water bath. The cycle of homogenization and sonication was repeated 3 times. Then the samples were incubated for 1 h at −40°C and centrifuged at 12 000 rpm for 15 min at 4°C to collect the supernatant. Two groups of sample (YE‐N and YE‐O) were collected for metabonomic analysis with four biological replicates for each group.
LC‐MS/MS analyses were performed using an UHPLC system (Vanquish, Thermo Fisher Scientific) with a UPLC BEH Amide column (100 mm × 2.1 mm × 1.7 μm) coupled to Q Exactive HFX mass spectrometer (Orbitrap MS, Thermo). The mobile phase consisted of 25 mmol l−1 ammonium acetate and 25 mmol l−1 ammonia hydroxide in water (pH 9.75) (A) and acetonitrile (B). The auto‐sampler temperature was 4°C, and the injection volume was 3 μl.
The QE HFX mass spectrometer was used for its ability to acquire MS/MS spectra on information‐dependent acquisition (IDA) mode in the control of the acquisition software (Xcalibur, Thermo). In this mode, the acquisition software continuously evaluates the full scan MS spectrum. The ESI source conditions were set as follows: sheath gas flow rate as 30 Arb, aux gas flow rate as 25 Arb, capillary temperature 350°C, full MS resolution as 60000, MS/MS resolution as 7500, collision energy as 10/30/60 in NCE mode, spray Voltage as 3.6 kV (positive) or −3.2 kV (negative) respectively.
The raw data were converted to the mzXML format using ProteoWizard and processed with an in‐house program, which was developed using R and based on XCMS, for peak detection, extraction, alignment and integration. Then an in‐house MS2 database (BiotreeDB) was applied in metabolite annotation. The cut‐off for annotation was set at 0.3. Univariate statistical analysis and multivariate statistical analysis of the qualitative and quantitative results of the metabolome were performed to screen metabolites with significant differences, while optional data analysis is based on basic data analysis. A series of bioinformatics analyses of significantly different metabolites, such as KEGG analysis, pathway analysis and network analysis, were performed.
Quantitative analysis of cell growth, metabolites and GFP fluorescence
Cell growth was characterized by the optical density at 600 nm (OD 600) or the biomass. The value of OD 600 was measured with spectrophotometer (UV1800; Shimadzu, Tokyo, Japan), while the biomass was determined as dry cell weight (DCW) per litre of fermentation broth (g DCW l−1). Intracellular squalene content was quantified by HPLC using Agilent 1260 infinity equipped with a Plus‐C18 column (100 mm × 4.6 mm × 3.5 μm) and diode array detector (Agilent G1315D) (Zhou et al., 2021). Squalene production was determined as intracellular squalene content designated as the milligram of squalene per gram DCW (mg g−1 DCW) or squalene titre defined as the milligram of squalene per litre of fermentation broth (mg l−1). The fluorescence intensity of GFP in yeast cells was assayed and normalized to OD 600 (Zhou et al., 2021). Residual sugar content and ethanol concentration were detected by SBA‐40C biosensor (Institute of Biology, Shandong Academy of Sciences, China).
For quantitative analysis of β‐caryophyllene, cell pellet and the supernatant were collected respectively. An equal volume of ethyl acetate was used to extract β‐caryophyllene from the supernatant by vortexing for 30 min. The ethyl acetate phase was collected for further analysis. The cell pellet (about 0.2 g) was resuspended in 4 ml of 3 mol l−1 HCl, heated at 95°C for 7 min and cooled immediately in an ice bath for 10 min. After centrifugation at 12 000 rpm for 5 min, the precipitate was washed twice with sterile water and resuspended in 1 ml of ethyl acetate. The mixture was vortexed at room temperature for 30 min and then centrifuged to collect the ethyl acetate phase. The β‐caryophyllene content in ethyl acetate phase was measured via GC‐MS using SHIMADZU GCMS‐QP2010 Ultra infinity equipped with a RTX‐1701 column (30 mm × 0.32 mm × 0.25 μm) according to the following separation conditions and temperature programmes: injector port 240°C, FID 240°C, split ratio 50 : 1, helium flow 3.0 ml min−1, 150°C held for 1 min, ramped at rate 20°C min−1 to 280°C, held for 6 min. The sample injection volume was 1.0 μl.
Conflict of interest
The authors declare that they have no conflict of interest.
Author contributions
X. He and S. Lu conceived and designed the experiments. S. Lu and C. Zhou performed the experiments. X. Guo and Y. Cheng analysed the data. S. Lu and Z. Du performed the bioinformatics analyses. S. Lu, Z. Wang and X. He wrote and revised the manuscript. All authors read and approved the final manuscript.
Supporting information
Table S1. Plasmids used in this study.
Table S2. Primers used in this study.
Fig. S1. Cell growth of haploid YS58 derived strains in YPD with yeast extract YE‐O and YE‐N. Yeast cells were cultivated at 30°C and 200 rpm for 24 h. Data are presented as the means of three biological replicates. Error bars represent standard deviations (n=3). For significance analysis, results of strains YS58 were used as the controls. Student’s t‐test was used for statistical analysis (*P < 0.05; * *P < 0.01; ***P < 0.001).
Fig. S2. Characterization of ERG1 promoters from various sources and influence on squalene production in haploid strain YS58. (A) Promoter activity using GFP as the reporter. (B) Influence of down‐regulating ERG1 transcription on cell growth and squalene content. Data are presented as the means of three biological replicates. Error bars represent standard deviations (n=3). For significance analysis, results of strains YS58 were used as the controls. Student’s t‐test was used for statistical analysis (***P < 0.001).
Acknowledgements
This work was supported by the Science and Technology Service Network Initiative (STS) of Chinese Academy of Sciences (Grant No. KFJ‐STS‐QYZD‐201).
Microbial Biotechnology (2022) 15(8), 2292–2306
Funding information
The Science and Technology Service Network Initiative (STS) of Chinese Academy of Sciences (Grant/Award Number: KFJ‐STS‐QYZD‐201).
References
- Andre, C.M. , Greenwood, J.M. , Walker, E.G. , Rassam, M. , Sullivan, M. , Evers, D. , et al. (2012) Anti‐inflammatory procyanidins and triterpenes in 109 apple varieties. J Agric Food Chem 60: 10546–10554. [DOI] [PubMed] [Google Scholar]
- Basha, R.H. , and Sankaranarayanan, C. (2016) β‐Caryophyllene, a natural sesquiterpene lactone attenuates hyperglycemia mediated oxidative and inflammatory stress in experimental diabetic rats. Chem‐Biol Interact 245: 50–58. [DOI] [PubMed] [Google Scholar]
- Bhilwade, H.N. , Tatewaki, N. , Nishida, H. , and Konishi, T. (2010) Squalene as novel food factor. Curr Pharm Biotechnol 11: 875–880. [DOI] [PubMed] [Google Scholar]
- Bian, G. , Deng, Z. , and Liu, T. (2017) Strategies for terpenoid overproduction and new terpenoid discovery. Curr Opin Biotechnol 48: 234–241. [DOI] [PubMed] [Google Scholar]
- Cai, Y. , Jia, J.W. , Crock, J. , Lin, Z.X. , Chen, X.Y. , and Croteau, R. (2002) A cDNA clone for β‐caryophyllene synthase from artemisia annua . Phytochemistry 61: 523–529. [DOI] [PubMed] [Google Scholar]
- Chang, M.H. , Kim, H.J. , Jahng, K.Y. , and Hong, S.C. (2008) The isolation and characterization of Pseudozyma sp. JCC 207, a novel producer of squalene. Appl Microbiol Biotechnol 78: 963–972. [DOI] [PubMed] [Google Scholar]
- Chen, X. , Shukal, S. , and Zhang, C. (2019) Integrating enzyme and metabolic engineering tools for enhanced α‐ionone production. J Agric Food Chem 67: 13451–13459. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Davier, L. , Schalk, M. , Siewers, V. , and Nielsen, J. (2013) Establishing a platform cell factory through engineering of yeast acetyl‐CoA metabolism. Metab Eng 15: 48–54. [DOI] [PubMed] [Google Scholar]
- Cronan, J.E. , Little, K.J. , and Jackowski, S. (1982) Genetic and biochemical analyses of pantothenate biosynthesis in Escherichia coli and Salmonella typhimurium . J Bacteriol 149: 916–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald, K.A. , Hampton, R.Y. , and Fritz, I.B. (1997) Effects of overproduction of the catalytic domain of 3‐hydroxy‐3‐methylglutaryl coenzyme a reductase on squalene synthesis in Saccharomyces cerevisiae . Appl Environ Microbiol 63: 3341–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdíková, E. , Garaiová, M. , Csáky, Z. , Obernauerová, M. , and Hapala, I. (2015) Production of squalene by lactose ‐ fermenting yeast Kluyveromyces lactis with reduced squalene epoxidase activity. Lett Appl Microbiol 61: 77–84. [DOI] [PubMed] [Google Scholar]
- Fidyt, K. , Fiedorowicz, A. , Strzdaa, L. , and Szumny, A. (2016) β‐aryophyllene and β‐aryophyllene oxide‐natural compounds of anticancer and analgesic properties. Cancer Med 5: 3007–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambacorta, F.V. , Dietrich, J.J. , Yan, Q. , and Pfleger, B.F. (2020) Rewiring yeast metabolism to synthesize products beyond ethanol. Curr Opin Chem Biol 59: 182–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godara, A. , and Kao, K.C. (2015) Adaptive laboratory evolution of β‐caryophyllene producing Saccharomyces cerevisiae . Microb Cell Fact 20: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X.P. , Huai, W.H. , Tie, C.J. , Liu, Y.F. , and Zhang, B.R. (2000) Breeding of high ergosterol‐producing yeast strains. J Ind Microbiol Biotechnol 25: 39–44. [Google Scholar]
- Hedl, M. , Sutherlin, A. , Wilding, E.I. , Mazzulla, M. , Mcdevitt, D. , Lane, P. , et al. (2002) Enterococcus faecalis Acetoacetyl‐coenzyme A thiolase/3‐hydroxy ‐3‐methylglutaryl‐coenzyme A reductase, a dual‐function protein of isopentenyl diphosphate biosynthesis. J Bacteriol 184: 2116–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Z.R. , Lin, Y.K. , and Fang, J.Y. (2009) Biological and pharmacological activities of squalene and related compounds: potential uses in cosmetic dermatology. Molecules 14: 540–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, G.‐Z. , Yao, M.‐D. , Wang, Y. , Zhou, L. , Song, T.‐Q. , Liu, H. , et al. (2017) Manipulation of GES and ERG20 for geraniol overproduction in Saccharomyces cerevisiae . Metab Eng 41: 57–66. [DOI] [PubMed] [Google Scholar]
- Johnson, E.A. (2003) Phaffia rhodozyma: colorful odyssey. Int Microbiol 6: 169–174. [DOI] [PubMed] [Google Scholar]
- Kildegaard, K.R. , Jensen, N.B. , Schneider, K. , Czarnotta, E. , Özdemir, E. , Klein, T. , et al. (2016) Engineering and systems‐level analysis of Saccharomyces cerevisiae for production of 3‐hydroxypropionic acid via malonyl‐CoA reductase‐dependent pathway. Microb Cell Fact 15: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, B.U. , van Rossum, H.M. , Luttik, M.A.H. , Akeroyd, M. , Benjamin, K.R. , Wu, L. , et al. (2014) Engineering acetyl coenzyme A supply: functional expression of a bacterial pyruvate dehydrogenase complex in the cytosol of Saccharomyces cerevisiae . MBio 5: e01696‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G.‐S. , Li, T. , Zhou, W. , Jiang, M. , Tao, X.‐Y. , Liu, M. , et al. (2020) The yeast peroxisome: a dynamic storage depot and subcellular factory for squalene overproduction. Metab Eng 57: 151–161. [DOI] [PubMed] [Google Scholar]
- Liu, W. , Zhang, B. , and Jiang, R. (2017) Improving acetyl‐CoA biosynthesis in Saccharomyces cerevisiae via the overexpression of pantothenate kinase and PDH bypass. Biotechnol Biofuels 10: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, X. , Wang, F. , Zhou, P. , Ye, L. , Xie, W. , Xu, H. , and Yu, H. (2016) Dual regulation of cytoplasmic and mitochondrial acetyl‐CoA utilization for improved isoprene production in Saccharomyces cerevisiae . Nat Commun 7: 12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado, K.D.C. , Islam, M.T. , Ali, E.S. , Rouf, R. , Uddin, S.J. , Dev, S. , et al. (2018) A systematic review on the neuroprotective perspectives of beta‐caryophyllene. Phytother Res 32: 2376–2388. [DOI] [PubMed] [Google Scholar]
- Maertens, B. , Spriestersbach, A. , von Groll, U. , Roth, U. , Kubicek, J. , Gerrits, M. , et al. (2010) Gene optimization mechanisms: a multi‐gene study reveals a high success rate of full‐length human proteins expressed in Escherichia coli . Protein Sci 19: 1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, V.J. , Pitera, D.J. , Withers, S.T. , Newman, J.D. , and Keasling, J.D. (2003) Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol 21: 796–802. [DOI] [PubMed] [Google Scholar]
- Martínez, I. , Zhu, J. , Lin, H. , Bennett, G.N. , and San, K.‐Y. (2008) Replacing Escherichia coli NAD‐dependent glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) with a NADP‐dependent enzyme from Clostridium acetobutylicum facilitates NADPH dependent pathways. Metab Eng 10: 352–359. [DOI] [PubMed] [Google Scholar]
- Meadows, A.L. , Hawkins, K.M. , Tsegaye, Y. , Antipov, E. , Kim, Y. , Raetz, L. , et al. (2016) Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nature 537: 694–697. [DOI] [PubMed] [Google Scholar]
- Moser, S. , and Pichler, H. (2019) Identifying and engineering the ideal microbial terpenoid production host. Appl Microbiol Biotechnol 103: 5501–5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad, A. , Feng, X. , Rasool, A. , Sun, W. , and Li, C. (2020) Production of plant natural products through engineered Yarrowia lipolytica . Biotechnol Adv 43: 107555. [DOI] [PubMed] [Google Scholar]
- Nakazawa, A. , Matsuura, H. , Kose, R. , Kato, S. , Honda, D. , Inouye, I. , et al. (2012) Optimization of culture conditions of the thraustochytrid Aurantiochytrium sp. strain 18W–13a for squalene production. Bioresour Technol 109: 287–291. [DOI] [PubMed] [Google Scholar]
- Nielsen, J. (2014) Synthetic biology for engineering acetyl coenzyme A metabolism in yeast. MBio 5: e02153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzhausen, J. , Grigat, M. , Seifert, L. , Ulbricht, T. , and Schüller, H.J. (2021) Increased biosynthesis of acetyl‐CoA in the yeast Saccharomyces cerevisiae by overexpression of a deregulated pantothenate kinase gene and engineering of the coenzyme A biosynthetic pathway. Appl Microbiol Biotechnol 105: 7321–7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzhausen, J. , Schübbe, S. , and Schüller, H.J. (2009) Genetic analysis of coenzyme A biosynthesis in the yeast Saccharomyces cerevisiae: identification of a conditional mutation in the pantothenate kinase gene CAB1 . Curr Genet 55: 163–173. [DOI] [PubMed] [Google Scholar]
- Paddon, C.J. , Westfall, P.J. , Pitera, D.J. , Benjamin, K. , Fisher, K. , McPhee, D. , et al. (2013) High‐level semi‐synthetic production of the potent antimalarial artemisinin. Nature 496: 528–532. [DOI] [PubMed] [Google Scholar]
- Paramasivan, K. , and Mutturi, S. (2017a) Progress in terpene synthesis strategies through engineering of Saccharomyces cerevisiae . Crit Rev Biotechnol 37: 974–989. [DOI] [PubMed] [Google Scholar]
- Paramasivan, K. , and Mutturi, S. (2017b) Regeneration of NADPH coupled with HMG‐CoA reductase activity increases squalene synthesis in Saccharomyces cerevisiae . J Agric Food Chem 65: 8162–8170. [DOI] [PubMed] [Google Scholar]
- Parshintsev, J. , Nurmi, J. , Kilpeläinen, I. , Hartonen, K. , Kulmala, M. , and Riekkola, M.L. (2008) Preparation of beta‐caryophyllene oxidation products and their determination in ambient aerosol samples. Anal Bioanal Chem 390: 913–919. [DOI] [PubMed] [Google Scholar]
- Polakowski, T. , Stahl, U. , and Lang, C. (1998) Overexpression of a cytosolic hydroxyl methylglutaryl‐CoA reductase leads to squalene accumulation in yeast. Appl Environ Microbiol 49: 66–71. [DOI] [PubMed] [Google Scholar]
- Ro, D.‐K. , Paradise, E.M. , Ouellet, M. , Fisher, K.J. , Newman, K.L. , Ndungu, J.M. , et al. (2006) Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440: 940–943. [DOI] [PubMed] [Google Scholar]
- Schadeweg, V. , and Boles, E. (2016) Increasing n‐butanol production with Saccharomyces cerevisiae by optimizing acetyl‐CoA synthesis, NADH levels and trans‐2‐enoyl‐CoA reductase expression. Biotechnol Biofuels 9: 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba, Y. , Paradise, E.M. , Kirby, J. , Ro, D.K. , and Keasling, J.D. (2007) Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae for high‐level production of isoprenoids. Metab Eng 9: 160–168. [DOI] [PubMed] [Google Scholar]
- Teunissen, A. , Berg, J. , and Steensma, H.Y. (1993) Physical localization of the flocculation gene FLO1 on chromosomeⅠof saccharomyces cerevisiae . Yeast 9: 1–10. [DOI] [PubMed] [Google Scholar]
- White, W.H. , Gunyuzlu, P.L. , and Toyn, J.H. (2001) Saccharomyces cerevisiae is capable of de novo pantothenic acid biosynthesis involving a novel pathway of β‐alanine production from spermine. J Biol Chem 276: 10794–10800. [DOI] [PubMed] [Google Scholar]
- Wong, J. , de Rond, T. , d’Espaux, L. , van der Horst, C. , Dev, I. , Rios‐Solis, L. , et al. (2018) High‐titer production of lathyrane diterpenoids from sugar by engineered Saccharomyces cerevisiae . Metab Eng 45: 142–148. [DOI] [PubMed] [Google Scholar]
- Xu, W. , Chai, C. , Shao, L. , Yao, J. , and Wang, Y. (2016) Metabolic engineering of Rhodopseudomonas palustris for squalene production. J Ind Microbiol Biotechnol 43: 1–7. [DOI] [PubMed] [Google Scholar]
- Yang, J. , and Nie, Q. (2016) Engineering Escherichia coli to convert acetic acid to β‐caryophyllene. Microb Cell Fact 15: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, Z. , Zhou, P. , Su, B. , Su, S. , Ye, L. , and Yu, H. (2018) Enhanced isoprene production by reconstruction of metabolic balance between strengthened precursor supply and improved isoprene synthase in Saccharomyces cerevisiae . ACS Synth Biol 7: 2308–2316. [DOI] [PubMed] [Google Scholar]
- Ye, L. , Lv, X. , and Yu, H. (2016) Engineering microbes for isoprene production. Metab Eng 38: 125–138. [DOI] [PubMed] [Google Scholar]
- Yu, T. , Zhou, Y.J. , Huang, M. , Liu, Q. , Pereira, R. , David, F. , and Nielsen, J. (2018) Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell 174: 1549–1558. [DOI] [PubMed] [Google Scholar]
- Yuan, J. , and Ching, C.B. (2015) Dynamic control of ERG9 expression for improved amorpha‐4,11‐diene production in Saccharomyces cerevisiae . Microb Cell Fact 14: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , An, R. , Tian, X. , Yang, M. , Li, M. , Lou, J. , et al. (2017) β‐Caryophyllene pretreatment alleviates focal cerebral ischemia‐reperfusion injury by activating PI3K/Akt signaling pathway. Neurochem Res 42: 1–11. [DOI] [PubMed] [Google Scholar]
- Zhang, S.H. , Wang, J.J. , and Jiang, H. (2021) Microbial production of value added bioproducts and enzymes from molasses, a by‐product of sugar industry. Food Chem 346: 128860. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Su, M. , Qin, N. , Nielsen, J. , and Liu, Z. (2020) Expressing a cytosolic pyruvate dehydrogenase complex to increase free fatty acid production in Saccharomyces cerevisiae . Microb Cell Fact 19: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, C. , Li, M. , Lu, S. , Cheng, Y. , Guo, X. , He, X. , et al. (2021) Engineering of cis‐element in Saccharomyces cerevisiae for efficient accumulation of value‐added compound squalene via downregulation of the downstream metabolic flux. J Agric Food Chem 69: 12474–12484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Plasmids used in this study.
Table S2. Primers used in this study.
Fig. S1. Cell growth of haploid YS58 derived strains in YPD with yeast extract YE‐O and YE‐N. Yeast cells were cultivated at 30°C and 200 rpm for 24 h. Data are presented as the means of three biological replicates. Error bars represent standard deviations (n=3). For significance analysis, results of strains YS58 were used as the controls. Student’s t‐test was used for statistical analysis (*P < 0.05; * *P < 0.01; ***P < 0.001).
Fig. S2. Characterization of ERG1 promoters from various sources and influence on squalene production in haploid strain YS58. (A) Promoter activity using GFP as the reporter. (B) Influence of down‐regulating ERG1 transcription on cell growth and squalene content. Data are presented as the means of three biological replicates. Error bars represent standard deviations (n=3). For significance analysis, results of strains YS58 were used as the controls. Student’s t‐test was used for statistical analysis (***P < 0.001).


